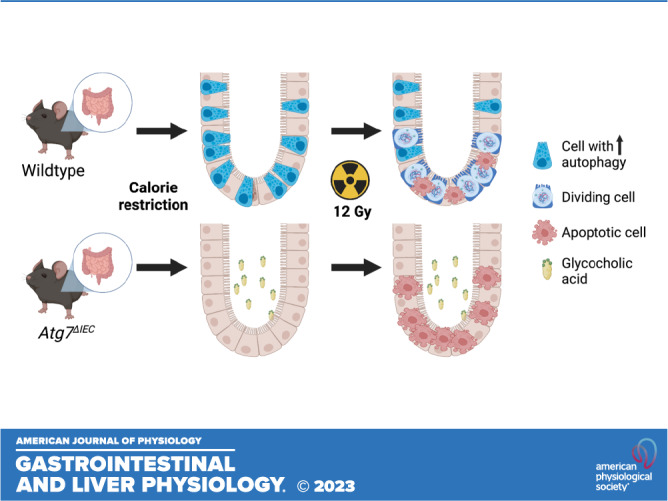
Keywords: autophagy, calorie restriction, glycocholic acid, intestinal epithelium, regeneration
Abstract
Calorie restriction can enhance the regenerative capacity of the injured intestinal epithelium. Among other metabolic changes, calorie restriction can activate the autophagy pathway. Although independent studies have attributed the regenerative benefit of calorie restriction to downregulation of mTORC1, it is not known whether autophagy itself is required for the regenerative benefit of calorie restriction. We used mouse and organoid models with autophagy gene deletion to evaluate the contribution of autophagy to intestinal epithelial regeneration following calorie restriction. In the absence of injury, mice with intestinal epithelial-specific deletion of autophagy gene Atg7 (Atg7ΔIEC) exhibit weight loss and histological changes similar to wild-type mice following calorie restriction. Conversely, calorie-restricted Atg7ΔIEC mice displayed a significant reduction in regenerative crypt foci after irradiation compared with calorie-restricted wild-type mice. Targeted analyses of tissue metabolites in calorie-restricted mice revealed an association between calorie restriction and reduced glycocholic acid (GCA) in wild-type mice but not in Atg7ΔIEC mice. To evaluate whether GCA can directly modulate epithelial stem cell self-renewal, we performed enteroid formation assays with or without GCA. Wild-type enteroids exhibited reduced enteroid formation efficiency in response to GCA treatment, suggesting that reduced availability of GCA during calorie restriction may be one mechanism by which calorie restriction favors epithelial regeneration in a manner dependent upon epithelial autophagy. Taken together, our data support the premise that intestinal epithelial Atg7 is required for the regenerative benefit of calorie restriction, due in part to its role in modulating luminal GCA with direct effects on epithelial stem cell self-renewal.
NEW & NOTEWORTHY Calorie restriction is associated with enhanced intestinal regeneration after irradiation, but the requirement of autophagy for this process is not known. Our data support the premise that intestinal epithelial autophagy is required for the regenerative benefit of calorie restriction. We also report that luminal levels of primary bile acid glycocholic acid are modulated by epithelial cell autophagy during calorie restriction with direct effects on epithelial stem cell function.
INTRODUCTION
Calorie restriction is a dietary regiment that reduces caloric intake while maintaining appropriate micronutrient levels. The benefits of a reduced calorie diet have been studied for decades in multiple organisms as a nutritional intervention strategy for extension of both health span and lifespan (1, 2). Previous studies demonstrated that calorie restriction increased the number of intestinal stem cells (ISCs), which primed the epithelium for regeneration in the face of damage (3–5). For example, in one study, mice on a calorie-restricted diet demonstrated increased numbers of Bmi1+ “quiescent” stem cells that are relatively radioresistant. A different study showed an increase in active cycling, Lgr5+ ISCs (6). Stem cells from calorie-restricted mice have a reduced number of genetic mutations present over time (5). Finally, although calorie restriction increases the number of ISCs, it is associated with a reduction of differentiated cells suggesting that calorie restriction may enhance stem cell proliferation but not differentiation (6). Paneth cells are long-lived, differentiated cells located in the intestinal crypt in close proximity to ISCs. In experiments where Paneth cells are cocultured with ISCs, it was observed that Paneth cells from calorie-restricted mice promote ISC proliferation compared with Paneth cells from ad libitum fed mice, suggesting a direct contribution of Paneth cells to epithelial regeneration following calorie restriction (6). Paneth cells are also thought to be a central modulator of the intestinal response to inflammation via regulation of both the unfolded protein response and autophagy (7).
Autophagy is an evolutionarily conserved process important for cellular responses to nutrient stress (8) and involves the breakdown of aberrant cellular content and organelles. In addition, Paneth cells utilize secretory autophagy to secrete lysozyme, which is important to protect intestinal epithelial cells from luminal bacteria (9). Genome-wide association studies of patients with chronic intestinal disorders such as inflammatory bowel disease identified mutations in key autophagy genes ATG16Ll1, NOD2, and LRRK2, linking aberrant autophagy directly to human disease (10–12). More recently, autophagy has been specifically implicated in ISC function and response to tissue damage, where deletion of Atg7 or Atg5 sensitizes mice to intestinal injury (13–15). Finally, we reported recently that crypt cells (including differentiated Paneth and enteroendocrine cells) with high autophagic vesicle content exhibit higher organoid formation potential compared with cells with low levels of autophagy (16). Taken together, prior studies suggest that contexts where autophagy is increased may favor enhanced regeneration by increasing the pool of crypt cells that can behave as stem cells.
In the current study, our goal was to evaluate the relative contribution and mechanisms of autophagy to the regenerative benefit of calorie restriction. As such, we characterized the consequence of autophagy deletion in vivo and used enteroids to further understand mechanisms by which calorie-restriction-induced changes may augment epithelial cell biology in the presence or absence of autophagy.
METHODS
Mouse Models
Mice carrying floxed alleles for the Atg7 gene [kindly provided by RIKEN BRC through National Bio-Resource Project of MEXT, Japan (17)] were crossed with VillinCre (The Jackson Laboratory No. 021504) mice. Enteroids for bile acid experiments with Atg7 deletion were generated from Atg7flox/flox mice crossed with VillinCreERT2 mice (The Jackson Laboratory No. 020282). Both male and female mice were used. Mice without VillinCre were used as controls for in vivo experiments and enteroids without 2% 4-hydroxytamoxifen (4OHT) were used for Atg7 deletion enteroid experiments. Deletion of Atg7 was confirmed by RT-qPCR with Taqman primers (Thermo Fisher) listed in Table 1. RNA isolations were performed with Quick-RNA MicroPrep kits (Zymo Research, R1051), followed by cDNA generation (Applied Biosystems Reverse Transcription Reagents, N8080234), and qPCR with Taqman Fast Universal PCR Master Mix (Applied Biosystems, 4444556). All mouse experiments and handling were approved under IACUC Protocol 001278 at the Children’s Hospital of Philadelphia. Mice were housed individually at the onset of diet studies in a temperature-controlled room with 12-h light and dark cycles and continuous access to water.
Table 1.
Antibodies and primers
| Antibody | Company | Ref. Number | Dilution (Diluent) | Source |
|---|---|---|---|---|
| Atg7 | Cell Signaling | D12B11 | 1 to 1,000 (milk) | Rabbit |
| Atg5 | Cell Signaling | D5F5U | 1 to 1,000 (milk) | Rabbit |
| Lc3 | Cell Signaling | D3U4C | 1 to 1,000 (milk) | Rabbit |
| p62 | AbCam | ab56416 | 1 to 1,000 (milk) | Mouse |
| Gapdh | Santa Cruz | sc-32233 | 1 to 1,000 (milk) | Mouse |
| Mouse HRP | AbCam | Ab6728 | 1 to 1,000 (milk) | Rabbit |
| Rabbit HRP | Cell Signaling | 7074 | 1 to 1,000 (milk) | Goat |
| E-Cadherin | BD Biosciences | 610181 | 1 to 200 (PBS) | Mouse |
| Lysozyme | Agilent/Dako | A009902-2 | 1 to 200 (PBS) | Rabbit |
| Cy2 Anti-rabbit | Jackson | 711-225-152 | 1 to 600 (PBS) | Donkey |
| Cy3 Anti-mouse | Jackson | 715-165-151 | 1 to 600 (PBS) | Donkey |
| Target Gene | Company | Assay No. | ||
|---|---|---|---|---|
| Atg7 | Thermo Fisher | Mm00512209_m1 | ||
| Gapdh | Thermo Fisher | Mm99999915_g1 |
Calorie Restriction Diet
Atg7ΔIEC and control mice between the age of 10 and 14 wk were randomly divided into ad libitum or calorie restriction groups. All sample sizes are listed in individual figures and were determined based on prior published studies (3). Ad libitum intake was calculated for each mouse by monitoring and weighing daily food intake for 3 wk before initiation of calorie restriction. The calorie restriction group was then placed on a 60% calorie-restricted diet relative to their ad libitum counterparts. A 40%-increased micronutrient diet matched to the standard mouse facility chow was given to mice on calorie restriction to control for the decrease in food provided (Lab Diet 5015, Animal Specialties and Provisions). Calorie-restricted mice were administered the same amount of weighed food at the same time (±1 h) each day for 5 wk and mice were weighed daily. At the end of week 5, a subset from each group was subjected to a single dose of 12 Gy whole body irradiation (X-RAD 320 Cabinet X-ray System). All mice were euthanized 3.5 days postirradiation. No mice were excluded from the experiment. On the day of euthanasia, each mouse received a single intraperitoneal injection of 5-ethynyl-2′-deoxyuridine (EdU) 90 min before euthanasia. A subset of calorie restriction and ad libitum fed mice were euthanized at the end of week 5 before irradiation, and small intestine flushes and stool were collected for microbial metabolite analyses.
Targeted Microbial Metabolite Analyses via Liquid Chromatography/Mass Spectrometry
Bile acids were quantified using a Waters Acquity uPLC System with a Cortecs UPLC C-18 + 1.6 μm 2.1 × 50 mm column and a QDa single quadrupole mass detector (18, 19). Amino acids were quantified using a Waters Acquity uPLC System with an AccQ-Tag Ultra C18 1.7 μm 2.1 × 100 mm column and a Photodiode Detector Array using the Waters AccQ-Tag Ultra Amino Acid Derivatization Kit (Waters Corporation, Milford, MA) and UPLC AAA H-Class Application Kit (Waters Corporation, Milford, MA) as previously described (18, 20).
Statistical Analyses of Metabolites
Metabolites that were detected in more than 10% of all the samples were kept for analysis. The concentration of the metabolites was log transformed and fit into a mixed effects model. In the model, the gene type and diet were considered as independent variables and participates as random effects. P values were corrected to reduce false discovery rate using Benjamini–Hochberg procedure. The bile acids were grouped into primary conjugated [glycochenodeoxycholic acid (GCDCA), glycocholic acid (GCA), taurochenodeoxycholic acid (TCDCA), taurine-conjugated cholic acid (TCA)], primary unconjugated [chenodeoxycholic acid (CDCA), cholic acid (CA)], secondary conjugated [glycodeoxycholic acid (GDCA), glycolithocholic acid (GLCA), taurodeoxycholic acid (TDCA), taurolithocholic acid (TLCA)], and secondary unconjugated [deoxycholic acid (DCA), lithocholic acid (LCA)] for analysis.
Generation of Cell Lysates and Western Blot
Epithelial cells were purified via EDTA dissociation as described later for enteroid culture. Cells were then lysed with lysis buffer (Cell Lysis reagent, Cell Signaling 9803S), Halt Protease Inhibitor Cocktail (Life technologies, 78430), 1 mM sodium orthovanadate, 10 μM sodium fluoride on ice and spun at 9391G at 4°C for 10 min. The supernatant was collected and combined with homemade running buffer (final concentration 62.5 mM Tris·HCl pH 6.8, 2.5% SDS, 0.002% bromophenol blue, 5% B-mercaptoethanol, and 10% glycerol) before it was boiled for 6 min. Samples were then run on an SDS-PAGE gel. Samples were then transferred onto PVDF membrane using the Bio-Rad Trans-blot Turbo system and blocked in 5% Milk in Tris-buffered saline-Tween 20 (TBS-T) (20.7 mM Tris Base, 150.7 NaCl, 0.1% Tween-20, pH 7.6) for 1 h at room temperature. Primary antibodies were diluted in 5% milk in TBS-T and incubated with gentle rocking at 4°C for 24 h. Blots were then washed with TBS-T and incubated in secondary antibody while rocking at room temperature for 1 h. Blots were finally incubated with luminol reagent (Santa Cruz Biotechnology, sc-2048) per manufacturer protocol and exposed to autoradiography film. Lc3-II bands were quantified using ImageJ v.2.1.0. All antibodies are listed in Table 1. Validation of Western blot antibody specificity (where available) is as follows: Atg7: validated via Western blot with Atg7 siRNA transfected cells per manufacturer’s website; Atg5: validated via Western blot with mouse embryonic fibroblasts (MEFs) from Atg5 knockout mice per manufacturer’s website; Lc3: validated via Western blot with rhabdomyosarcoma cells with or without Torin treatment to induce Lc3 per manufacturer’s website; p62: validated via Western blot in HEK293T knockout cell line per manufacturer’s website.
Immunofluorescent Staining
Proximal jejunum (5–10 cm) were cut open lengthwise, Swiss-rolled, fixed overnight in Zn formalin, and then processed for paraffin embedding. Five-micron sections from paraffin blocks were used for immunofluorescence staining with the antibodies listed in Table 1. Primary antibodies were used at 1:200 and incubated overnight at 4°C. All secondary antibodies were used at a 1:600 dilution at room temperature for 1 h. EdU staining was performed using the Click-iT Plus EdU kit (Fisher; C10639). After the EdU staining incubation period, sections were incubated with DAPI solutions and mounted for imaging. Z-stack images were captured and stitched with a ×40 objective. Imaging was performed using Keyence BZ-X100. At least eight well-oriented (clear crypt-basement membrane orientation), independent images per mouse were required to include mice for EdU quantification. The number of EdU+ foci were counted by two independent, genotype-blind investigators. For Paneth cell scoring, lysozyme and e-cadherin-stained sections were evaluated by a genotype blinded investigator based on published scoring criteria (21).
Enteroid Culture
After mouse euthanasia, 8 cm of distal jejunum were dissected and washed briefly in fresh PBS. Tissue was splayed open and transferred to a tube with 10 mL of 1× HBSS and 1 mM N-acetyl-l-cysteine (NAC, Sigma Aldrich, A9165). The tissue was vortexed for 15 s followed by 15 s rest on ice for a 2-min period before it was transferred to a second tube containing 10 mL of 1× HBSS with 1 mM NAC and 10 mM EDTA and placed on a rotator for 45 min at 4°C. After this incubation, the tissue was vortexed for 30 s followed by a 30-s rest on ice for a total of 3 min. The resulting tissue digestion was filtered through a 70-μm filter, and the flow through was centrifuged for 3 min at 300 g to generate a pellet of intestinal crypts. Intestinal crypts were plated in Matrigel droplets at a density of 100 crypts per 20 μL droplet on a 24-well plate. Each Matrigel droplet was overlaid with 450 mL of a specialized medium: advanced DMEM/F12 media containing 1× GlutaMax (Thermo Fisher, 35050061), 10 mM HEPES buffer (Thermo Fisher, 15-630-080), 1× Antibiotic-Antimycotic (Anti-Anti, Thermo Fisher 15240062), titrated R-spo1 and Noggin-containing conditioned media (22), 1× N2 supplement (Thermo Fisher, 17502048) HEPES, 1× B-27 supplement (Thermo Fisher, 0080085SA), 5 μM CHIR99021 (Cayman Chemical Company, 13122), 1 mM NAC, 50 ng/mL mEGF (PeproTech, 315-09), 5% Noggin/R-spondin conditioned medium, and 1× Anti-Anti. Media was replaced every other day after plating.
Bile Acid Treatment
GCA was made from glycocholic acid hydrate (G2878; Sigma, St. Louis, MO) dissolved in DMSO at the indicated concentrations. A pilot study was conducted based on previously published studies (23), where a range of 10–100 µM was used. We found that treatment with 100 µM GCA gave the most consistent effect on enteroid formation, although we saw similar effects at 50 µM. For organoid formation rate experiments, enteroids were treated with 100 µM GCA or DMSO for 3 days before single-cell passaging at 4,000 cells per 20 µL droplet, then treated for an additional 4 days. Each day, enteroid growth was imaged on the Keyence Microscope. For enteroid area experiments, enteroids were treated with GCA for two days after plating as crypts, and the media was switched every two days. After 6 days of treatment, enteroids were imaged with the Keyence Microscope and harvested for RT-PCR analysis. The average area of enteroids was determined using the analyzed particles function on ImageJ. The area of each enteroid per dome was calculated and the average area per organoid dome was calculated. Any overlapping enteroids that created one area measurement were manually removed from the average.
Statistical Analyses
Data were analyzed using paired and unpaired t tests, one-way or two-way ANOVA with Turkey’s pairwise comparisons or Sidak’s multiple comparisons, and Kruskal–Wallis test with Dunn’s multiple comparisons, with specific tests indicated in all figure legends. Weight changes over time were analyzed using repeated-measures ANOVA. All analyses were performed on data with at least three independent experiments unless noted otherwise, with each experiment containing technical replicates. Data are presented as means ± SE. All P values are indicated in the figures and legends. Statistical analyses were performed using Prism and SAS.
RESULTS
Intestinal Atg7 is Dispensable for Normal Homeostasis in Calorie-Restricted Mice
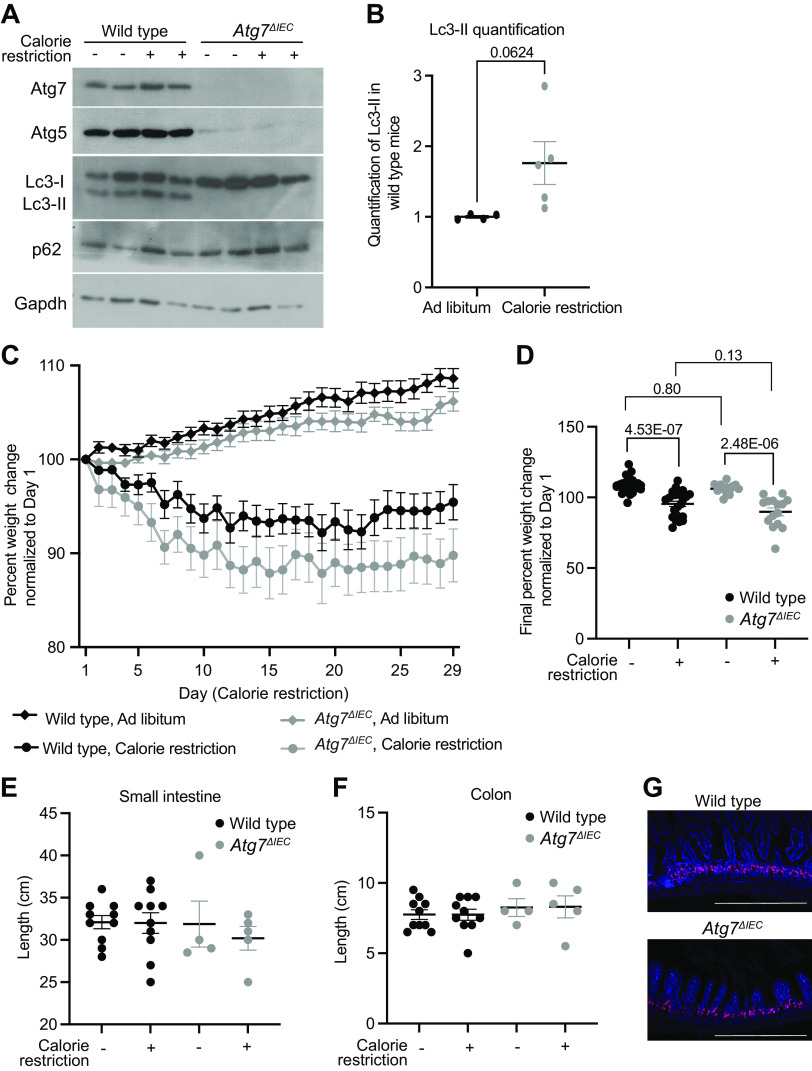
Nutrient deprivation is a classical inducer of autophagy in mammalian cells that may directly impart tissue regenerative potential. We therefore evaluated mice with intestinal epithelial-specific deletion of essential autophagy gene Atg7 (Atg7ΔIEC) in the context of calorie restriction. Atg7ΔIEC and control mice were subjected to 5 wk of either ad libitum diet or calorie restriction diet, which consisted of a 40% reduction of normal daily food intake with chow fortified in vitamins and minerals to control for reduction in essential micronutrients with reduced food (3). Western blot of lysates from small intestinal epithelium was used to evaluate autophagy proteins Atg7, Atg5, and Lc3-I/Lc3-II. Atg7 and Atg5 proteins were absent or reduced, respectively, in Atg7ΔIEC mice compared with wild-type mice, consistent with prior reports (7, 24). We observed an increase in Lc3-II in calorie restriction compared with ad libitum wild-type mice (1.00 ± 0.04 vs. 1.76 ± 0.68; Fig. 1B), supporting the conclusion that calorie restriction induces autophagy as previously reported (25, 26), and that Atg7 deletion inhibits autophagy during calorie restriction (Fig. 1A).
Figure 1.
Intestinal Atg7 is dispensable for normal homeostasis in calorie-restricted mice. A: representative Western blot on intestinal epithelial cells from indicated genotypes on either ad libitum or calorie restriction diets probed for indicated proteins. B: quantification of Lc3-II in wild-type mice for n = 4 ad libitum and five calorie-restricted mice normalized to loading control analyzed via unpaired, two-tailed t test with indicated P value. C: average weights normalized to day 1 of mice of indicated genotype and diet over time. Repeated-measures ANOVA showed significant effects of diet (P < 0.001) and genotype (P < 0.0194) on normalized weights over time, but no significant effect due to the interaction of diet and genotype (P = 0.4053). Data are presented as means ± SE. D: average final weights on day 29 normalized to the starting weight of each individual mouse by two-way ANOVA demonstrates that weight differences are not observed between genotypes for specific diets. Adjusted P values are listed on the graph for indicated comparisons. n = 26 wild-type, ad libitum; n = 24 wild-type, calorie restriction, n = 13 Atg7ΔIEC, ad libitum, n = 15 Atg7ΔIEC, calorie restriction. Length of small intestine (E) and colon (F) from indicated genotypes and diets. No statistical significance was observed. n = 10 wild-type, ad libitum; n = 10 wild-type, calorie restriction, n = 4 Atg7ΔIEC, ad libitum, n = 5 Atg7ΔIEC, calorie restriction. G: representative images of 5-ethynyl-2′-deoxyuridine (EdU) staining in small intestines of calorie-restricted wild-type and Atg7ΔIEC mice. n = 3 mice per genotype were stained for EdU in calorie-restricted, nonirradiated mice. Scale bar = 200 μm.
Calorie restriction was associated with a 5% to 10% weight decrease compared with initial weight, whereas ad libitum fed mice exhibited a 6% to 9% weight increase from initial weight. Repeated-measures ANOVA showed significant effects of diet and genotype on normalized weights over time, but no significant effect due to interaction between diet and genotype (Fig. 1C). At the end of the calorie restriction period, wild-type mice exhibited a mean weight of 95.4% of their initial weight and Atg7ΔIEC mice exhibited a mean weight of 89.8% of their initial weight; however, this was not statistically significant (Fig. 1D). In addition, there was no difference in small intestine or colon lengths between genotypes (Fig. 1, E and F) or in proliferative cells (Fig. 1G) with calorie restriction. These data together indicate an absence of statistically significant changes in gross morphology between wild-type and Atg7ΔIEC mice following calorie restriction.
Paneth Cell Granule Morphology is Unaltered with Calorie Restriction
Paneth cells are intestinal epithelial cells found in the crypt base adjacent to ISCs that secrete antimicrobial proteins including lysozyme. In addition, Paneth cells have been implicated as key ISC support cells, including during calorie restriction. Mice on a fasted diet have an abundance of lysozyme-labeled autophagosomes within their Paneth cells, confirming that autophagy is active in these cells (27). Finally, prior studies show that disrupting autophagy in the intestinal epithelium inhibits the release of lysozyme and increases the risk of inflammation (9). Because of the multiple protective roles for Paneth cells, we wanted to evaluate Paneth cell morphology in the context of calorie restriction using previously described Paneth cell scoring methods (21). Immunofluorescent staining of lysozyme from jejunal sections revealed the expected and previously reported diffuse staining of Paneth cells from Atg7ΔIEC mice compared with wild-type mice under ad libitum conditions (Fig. 2A). There was no difference in lysozyme granule morphology between wild-type and Atg7ΔIEC mice in ad libitum or calorie restriction conditions (Fig. 2, B and C). Taken together, these data suggest that Paneth cell morphology is unaffected by calorie restriction and that changes due to autophagy gene deletion persist regardless of nutrient status.
Figure 2.
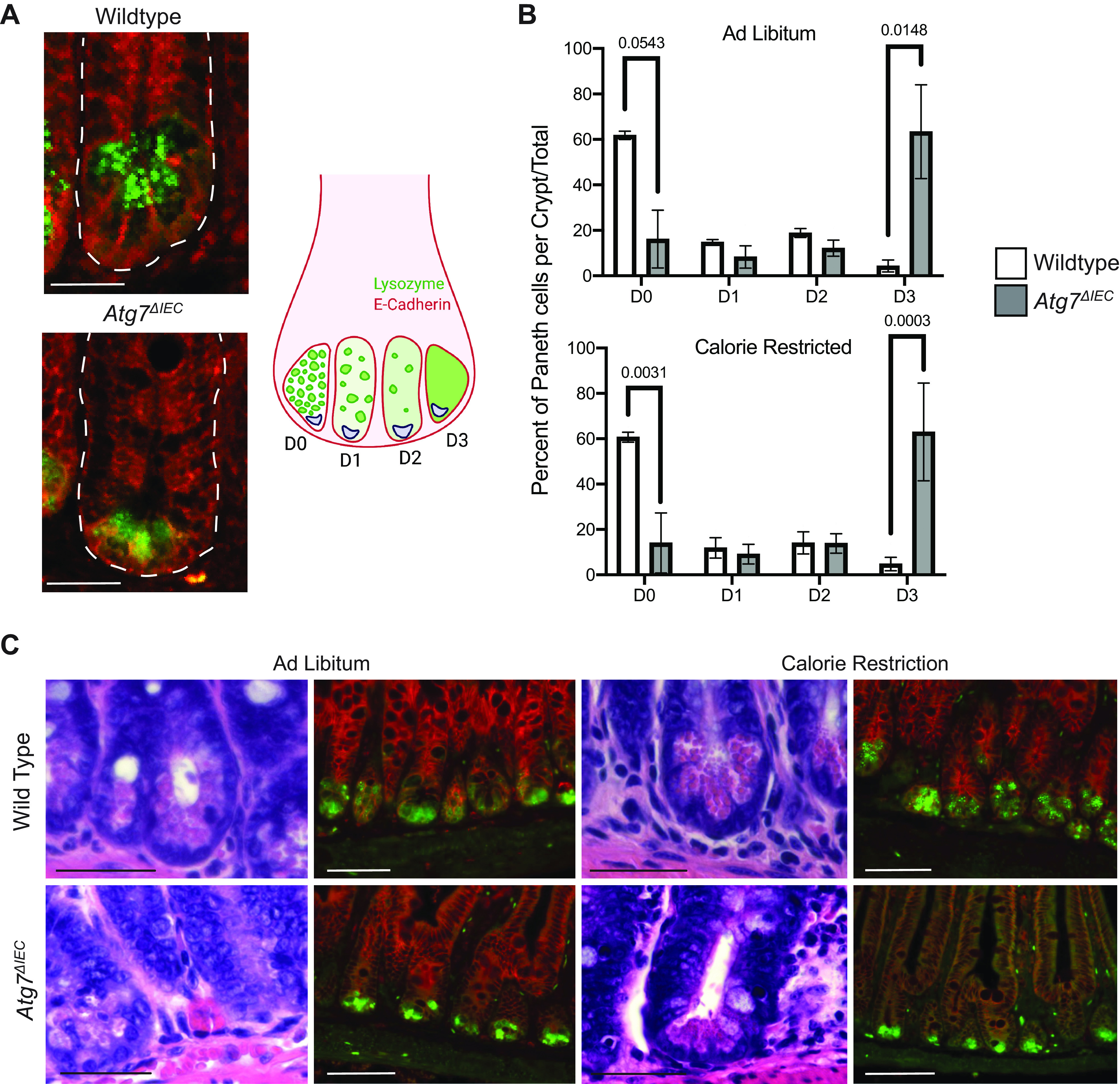
Paneth cells morphology is unaffected by calorie restriction. A: representative lysozyme staining of wild-type and Atg7ΔIEC crypts in ad libitum-fed mice with scoring scheme represented. Scale bar = 100 μm. B: percentage of total Paneth cells with each of the four granule morphology phenotypes (D0 = normal, D1 = disordered, D2 = depleted, D3 = diffuse) counted within each diet condition. n = 3 mice/condition. A two-way ANOVA with Sidak’s multiple comparisons test was done for each diet regiment. C: representative hematoxylin-eosin (H&E) and immunofluorescence staining of distal small intestinal sections of indicated control mice. Immunofluorescence shows E-cadherin (red) and lysozyme (green). Scale bar = 200 μm for H&E staining and 250 μm for immunofluorescence staining.
Intestinal Atg7 Deletion Mitigates the Regenerative Benefits of Calorie Restriction
Prior studies demonstrate that calorie restriction favors intestinal regeneration (3, 6). In addition, calorie restriction can induce autophagy, as demonstrated by others, and confirmed in Fig. 1. We therefore sought to determine if autophagy is required for this regenerative benefit. Calorie-restricted wild-type and Atg7ΔIEC mice were irradiated with a dose of 12 Gy followed by euthansia 3.5 days later to evaluate regenerative crypt foci (Fig. 3A). Comparing body weight loss between irradiation and day 3 after recovery, mice on ad libitum diet lost 10% more weight compared with those on a calorie restriction diet, and although there was slightly more weight loss in Atg7ΔIEC mice compared with wild type, this was not statistically significant (Fig. 3B).
Figure 3.
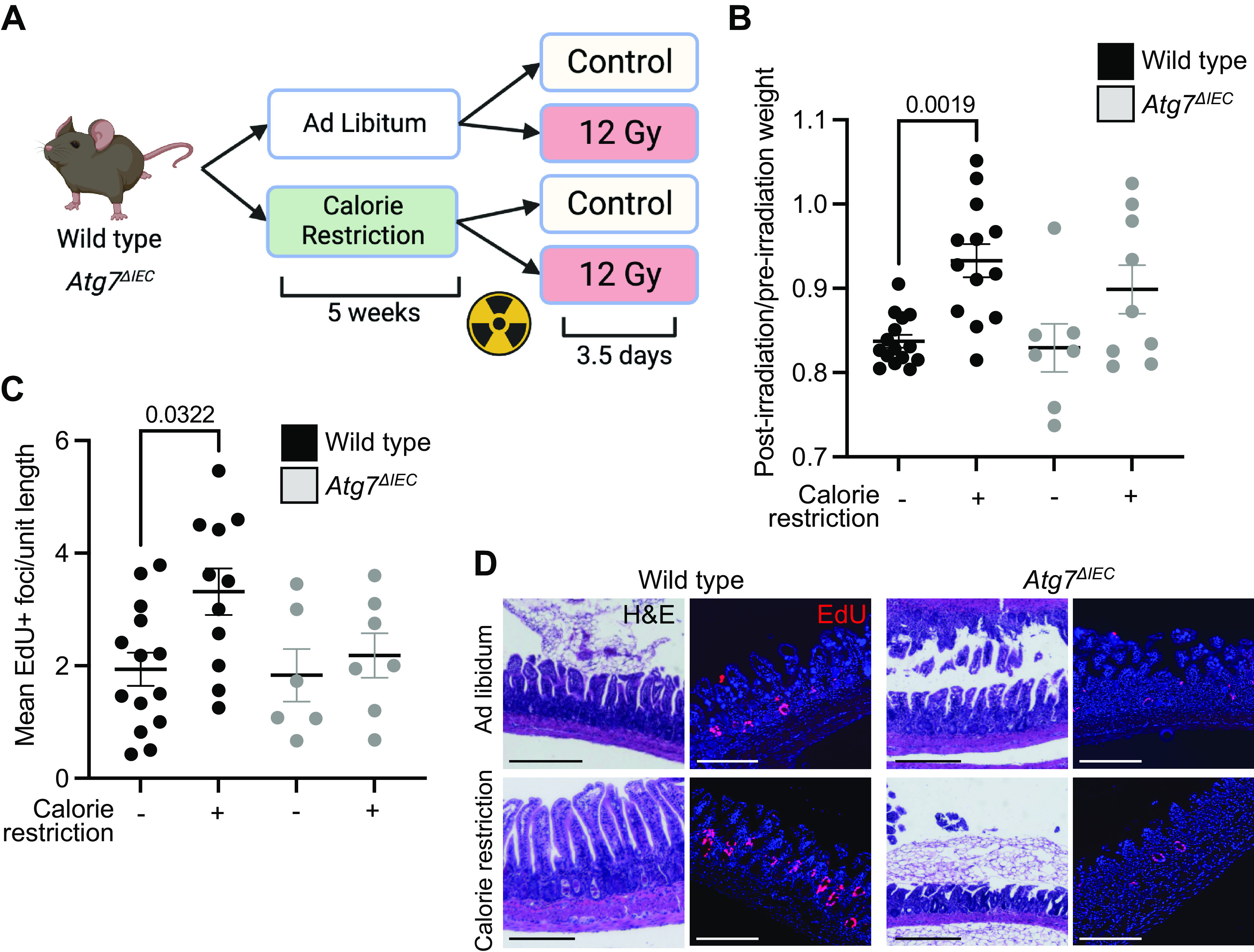
Atg7ΔIEC mice exhibit reduced intestinal regeneration following calorie restriction. A: schematic depicting experimental time course for calorie restriction and irradiation. B: postirradiation weight change calculated by dividing postirradiation weight by preirradiation weight for each mouse with significant P values indicated. n = 15 wild-type, ad libitum; n = 13 wild-type, calorie restriction, n = 7 Atg7ΔIEC, ad libitum, n = 7 Atg7ΔIEC, calorie restriction. C: quantification of EdU+ foci per unit length of distal small intestine from indicated groups, with each data point representing the average for each mouse. n = 14 wild-type, ad libitum; n = 11 wild-type, calorie restriction, n = 6 Atg7ΔIEC, ad libitum, n = 7 Atg7ΔIEC, calorie restriction. D: representative images of hematoxylin-eosin (H&E) and EdU staining of distal small intestine from each group. Scale bars = 150 μm. All data are displayed with standard error and indicated P values for all comparisons with P < 0.05. Data analyzed by ordinary one-way ANOVA with Tukey’s multiple comparison test. Figure was created with BioRender.
To measure the regenerative response of mice on calorie restriction, mice were injected with the nucleoside analogue EdU, which is incorporated into replicating DNA and is used to mark regenerative crypt foci (28). As reported previously, wild-type mice on a calorie restriction diet exhibited more regenerative crypt foci compared with wild-type mice on an ad libitum diet. By contrast, Atg7ΔIEC mice on calorie restriction diet did not exhibit more regenerative crypt foci compared with calorie-restricted wild-type or Atg7ΔIEC mice (Fig. 3, C and D). Taken together, these data demonstrate that Atg7 is required for the regenerative benefit of calorie restriction.
Calorie Restriction is Associated with a Shift in Luminal Glycocholic Acid in Wild-Type Mice
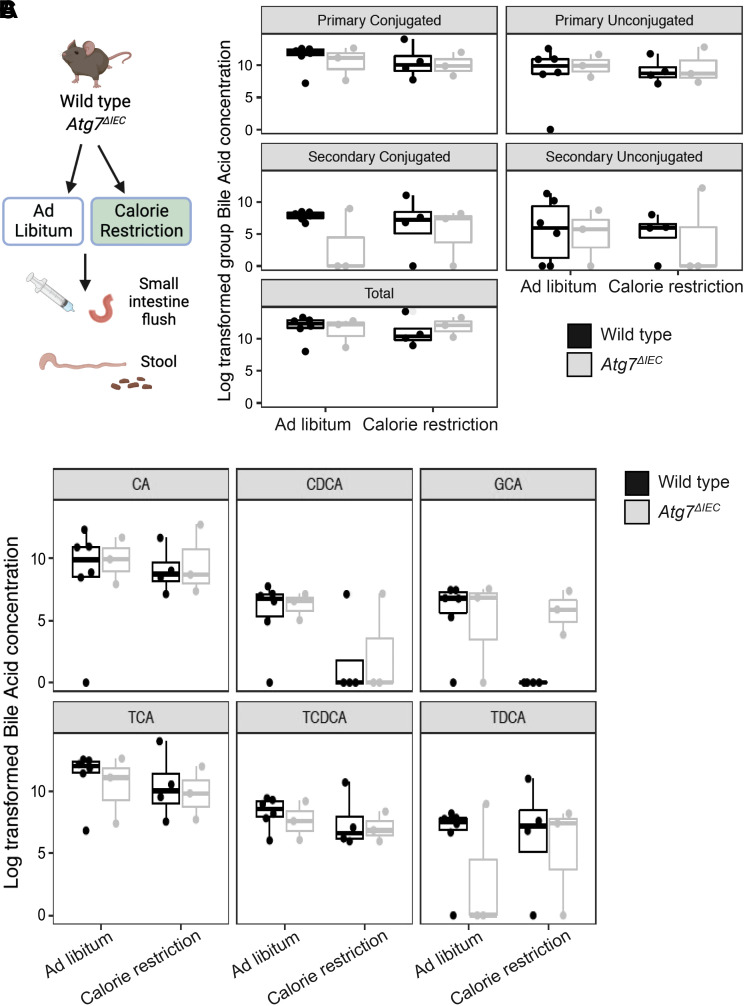
The microbiota and related metabolites can be directly affected by calorie restriction in humans (29) and have been implicated in intestinal health (23, 30, 31). We therefore performed targeted metabolomics on a small subset of mice to evaluate luminal and stool amino acid and bile acid composition following calorie restriction (Fig. 4A). There were no clear shifts in groups of bile acids from small intestine flushes or stool between genotypes, although we cannot exclude the possibility that this is due to small sample size [Fig. 4B, Supplemental Fig. S1A; Tables 2 and 3]. We next evaluated small intestine flushes for changes in specific bile acids. Variability within groups and sample size precluded us from making definitive conclusions about most individual analytes, however, we did observe a reduction in the conjugated bile acid glycocholic acid (GCA) in calorie-restricted wild-type but not Atg7ΔIEC mice (Fig. 4C). These data prompted us to pursue a potential role for GCA in modulating intestinal epithelial cell growth in subsequent studies using small intestinal enteroids.
Figure 4.
Glycocholic acid (GCA) is reduced in calorie-restricted wild-type but not Atg7ΔIEC mice. A: schematic of small intestine flush and stool collection from ad libitum fed and calorie-restricted mice. B: box plot of grouped bile acid concentrations in small intestinal flushes. C: boxplot of individual bile acid concentrations in small intestinal flushes. Mixed effects linear models were used to test for differences between groups. Each dot represents an individual mouse. n = 6 wild-type, ad libitum; n = 4 wild-type, calorie restriction, n = 3 Atg7ΔIEC, ad libitum, n = 3 Atg7ΔIEC, calorie restriction. CA, cholic acid; CDCA, chenodeoxycholic acid; TCA, taurine-conjugated cholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid. Figure was created with BioRender.
Table 2.
Linear models to test grouped bile acids from small intestine flushes
| Bile Acid Category | Comparison | Value | Std. Error | FDR |
|---|---|---|---|---|
| Primary conjugated | WT-Knockout | −0.82354 | 1.611765 | 0.841407 |
| Primary conjugated | Ad Lib-Calorie Restricted | −0.82596 | 1.471333 | 0.841407 |
| Primary conjugated | WT-Knockout:Ad Lib-Calorie Restricted | 0.421481 | 2.372453 | 0.875358 |
| Primary unconjugated | WT-Knockout | 1.220418 | 2.41429 | 0.905053 |
| Primary unconjugated | Ad Lib-Calorie Restricted | 0.298603 | 2.21377 | 0.905053 |
| Primary unconjugated | WT-Knockout:Ad Lib-Calorie Restricted | −0.71798 | 3.516938 | 0.905053 |
| Secondary conjugated | WT-Knockout | −4.82857 | 2.59051 | 0.183836 |
| Secondary conjugated | Ad Lib-Calorie Restricted | −1.45771 | 2.364801 | 0.600433 |
| Secondary conjugated | WT-Knockout:Ad Lib-Calorie Restricted | 3.671636 | 3.813127 | 0.582932 |
| Secondary unconjugated | WT-Knockout | −0.73448 | 3.489608 | 0.969616 |
| Secondary unconjugated | Ad Lib-Calorie Restricted | −0.54958 | 3.185562 | 0.969616 |
| Secondary unconjugated | WT- Knockout:Ad Lib-Calorie Restricted | −0.22082 | 5.136564 | 0.969616 |
| Total | WT-Knockout | −0.53583 | 1.427472 | 0.715225 |
| Total | Ad Lib-Calorie Restricted | −0.75594 | 1.303097 | 0.715225 |
| Total | WT-Knockout:Ad Lib-Calorie Restricted | 1.407466 | 2.101181 | 0.715225 |
The gene type and diet were considered as independent variables and participates as random effects. P values were corrected to reduce false discovery rate using Benjamini–Hochberg procedure.
Table 3.
Linear models to test grouped bile acids from stool
| Bile Acid Category | Comparison | Value | Std. Error | FDR |
|---|---|---|---|---|
| Primary conjugated | WT-Knockout | −1.43483 | 1.029816 | 0.260414 |
| Primary conjugated | Ad Lib-Calorie Restricted | −0.51052 | 0.882966 | 0.568307 |
| Primary conjugated | WT-Knockout:Ad Lib-Calorie Restricted | 1.462726 | 1.099246 | 0.260414 |
| Primary unconjugated | WT-Knockout | 0.228894 | 0.519753 | 0.667483 |
| Primary unconjugated | Ad Lib-Calorie Restricted | 1.026799 | 0.443824 | 0.058408 |
| Primary unconjugated | WT-Knockout:Ad Lib-Calorie Restricted | −1.13664 | 0.713544 | 0.164984 |
| Secondary conjugated | WT-Knockout | 0.000176 | 0.350649 | 0.999609 |
| Secondary conjugated | Ad Lib-Calorie Restricted | 0.136256 | 0.299423 | 0.999609 |
| Secondary conjugated | WT-Knockout:Ad Lib-Calorie Restricted | −0.03473 | 0.481389 | 0.999609 |
| Secondary unconjugated | WT-Knockout | 0.401038 | 0.446426 | 0.515572 |
| Secondary unconjugated | Ad Lib- Calorie Restricted | 0.39249 | 0.365533 | 0.515572 |
| Secondary unconjugated | WT-Knockout:Ad Lib-Calorie Restricted | −0.18051 | 0.582145 | 0.759076 |
| Total | WT-Knockout | 0.243458 | 0.324485 | 0.467537 |
| Total | Ad Lib-Calorie Restricted | 0.640345 | 0.277082 | 0.058725 |
| Total | WT-Knockout:Ad Lib-Calorie Restricted | −0.42255 | 0.44547 | 0.467537 |
The gene type and diet were considered as independent variables and participates as random effects. P values were corrected to reduce false discovery rate using Benjamini–Hochberg procedure.
Analyses of amino acids from both small intestinal flushes and stool (Supplemental Fig. S1, B and C, respectively) revealed minimal changes between groups, except for a significant decrease in stool arginine and serine in both genotypes with calorie restriction compared with ad libitum diet (Tables 4 and 5). In summary, calorie restriction was associated with variable, nonsignificant changes in most of the analyzed metabolites in wild-type and Atg7ΔIEC mice, though these studies likely require larger numbers of mice to draw definitive conclusions. However, we did observe a decrease in GCA in wild-type mice that was not observed in Atg7ΔIEC mice, supporting the hypothesis that GCA may have direct effects on intestinal epithelial cells.
Table 4.
Linear models to test individual amino acids from small intestine flushes
| Amino Acid | Comparison | Value | Std. Error | FDR |
|---|---|---|---|---|
| Alanine | WT-Knockout | 0.221651 | 0.90938 | 0.998073 |
| Alanine | Ad Lib-Calorie Restricted | −0.18464 | 0.833089 | 0.998073 |
| Alanine | WT-Knockout:Ad Lib-Calorie Restricted | 0.003618 | 1.327401 | 0.998073 |
| Arginine | WT-Knockout | 0.384864 | 1.025699 | 0.919277 |
| Arginine | Ad Lib-Calorie Restricted | −0.10738 | 0.937585 | 0.919277 |
| Arginine | WT-Knockout:Ad Lib-Calorie Restricted | −0.20738 | 1.504726 | 0.919277 |
| Asparagine | WT-Knockout | −0.04459 | 0.945696 | 0.96332 |
| Asparagine | Ad Lib-Calorie Restricted | −0.58023 | 0.876356 | 0.96332 |
| Asparagine | WT-Knockout:Ad Lib-Calorie Restricted | 0.511835 | 1.344607 | 0.96332 |
| Cysteine | WT-Knockout | 0.065875 | 0.633123 | 0.919188 |
| Cysteine | Ad Lib-Calorie Restricted | 0.129681 | 0.57796 | 0.919188 |
| Cysteine | WT-Knockout:Ad Lib-Calorie Restricted | −0.27025 | 0.931932 | 0.919188 |
| Glutamine | WT-Knockout | 0.192262 | 0.813508 | 0.9874 |
| Glutamine | Ad Lib-Calorie Restricted | −0.29716 | 0.754948 | 0.9874 |
| Glutamine | WT-Knockout:Ad Lib-Calorie Restricted | 0.020539 | 1.152532 | 0.9874 |
| Glycine | WT-Knockout | 0.166838 | 0.828649 | 0.936347 |
| Glycine | Ad Lib-Calorie Restricted | −0.15839 | 0.758552 | 0.936347 |
| Glycine | WT-Knockout:Ad Lib-Calorie Restricted | −0.10929 | 1.211625 | 0.936347 |
| Histidine | WT-Knockout | −0.15996 | 0.885239 | 0.872314 |
| Histidine | Ad Lib-Calorie Restricted | 0.150119 | 0.824532 | 0.872314 |
| Histidine | WT-Knockout:Ad Lib-Calorie Restricted | −0.58992 | 1.242363 | 0.872314 |
| Isoleucine | WT-Knockout | 0.070691 | 0.968019 | 0.990758 |
| Isoleucine | Ad Lib-Calorie Restricted | −0.1866 | 0.883676 | 0.990758 |
| Isoleucine | WT-Knockout:Ad Lib-Calorie Restricted | −0.01862 | 1.424885 | 0.990758 |
| Leucine | WT-Knockout | 0.086911 | 1.08759 | 0.940835 |
| Leucine | Ad Lib-Calorie Restricted | −0.32032 | 0.992829 | 0.940835 |
| Leucine | WT-Knockout:Ad Lib-Calorie Restricted | 0.134185 | 1.600889 | 0.940835 |
| Lysine | WT-Knockout | 0.072307 | 0.978379 | 0.971749 |
| Lysine | Ad Lib-Calorie Restricted | −0.34161 | 0.893134 | 0.971749 |
| Lysine | WT-Knockout:Ad Lib-Calorie Restricted | 0.057561 | 1.440136 | 0.971749 |
| Methionine | WT-Knockout | −0.11741 | 0.946531 | 0.903741 |
| Methionine | Ad Lib-Calorie Restricted | −0.36316 | 0.864061 | 0.903741 |
| Methionine | WT-Knockout:Ad Lib-Calorie Restricted | 0.209516 | 1.393256 | 0.903741 |
| Phenylalanine | WT-Knockout | −0.05419 | 0.973432 | 0.956699 |
| Phenylalanine | Ad Lib-Calorie Restricted | −0.32061 | 0.890761 | 0.956699 |
| Phenylalanine | WT-Knockout:Ad Lib-Calorie Restricted | 0.264183 | 1.424508 | 0.956699 |
| Proline | WT-Knockout | 0.066756 | 1.040858 | 0.950126 |
| Proline | Ad Lib-Calorie Restricted | −0.69238 | 0.959398 | 0.950126 |
| Proline | WT-Knockout:Ad Lib-Calorie Restricted | 0.578246 | 1.49871 | 0.950126 |
| Serine | WT-Knockout | 0.148125 | 1.13969 | 0.953477 |
| Serine | Ad Lib-Calorie Restricted | −0.55864 | 1.040389 | 0.953477 |
| Serine | WT-Knockout:Ad Lib-Calorie Restricted | 0.110493 | 1.677578 | 0.953477 |
| Threonine | WT-Knockout | 0.115375 | 0.94731 | 0.931243 |
| Threonine | Ad Lib-Calorie Restricted | −0.29989 | 0.864792 | 0.931243 |
| Threonine | WT-Knockout:Ad Lib-Calorie Restricted | 0.1359 | 1.394313 | 0.931243 |
| Tyrosine | WT-Knockout | −0.02646 | 0.952514 | 0.978384 |
| Tyrosine | Ad Lib-Calorie Restricted | −0.40503 | 0.869523 | 0.978384 |
| Tyrosine | WT-Knockout:Ad Lib-Calorie Restricted | 0.263054 | 1.402063 | 0.978384 |
| Valine | WT-Knockout | 0.086103 | 0.972137 | 0.976815 |
| Valine | Ad Lib-Calorie Restricted | −0.23157 | 0.887435 | 0.976815 |
| Valine | WT-Knockout:Ad Lib-Calorie Restricted | 0.046931 | 1.430946 | 0.976815 |
The gene type and diet were considered as independent variables and participates as random effects. P values were corrected to reduce false discovery rate using Benjamini-Hochberg procedure.
Table 5.
Linear models to test individual amino acids from stool
| Amino Acid | Comparison | Value | Std. Error | FDR | Sig Label |
|---|---|---|---|---|---|
| Alanine | WT-Knockout | −0.1073 | 0.615342 | 0.956471 | |
| Alanine | Ad Lib-Calorie Restricted | −0.68761 | 0.511469 | 0.509066 | |
| Alanine | WT-Knockout:Ad Lib-Calorie Restricted | −0.24471 | 0.695283 | 0.943088 | |
| Arginine | WT-Knockout | −0.10765 | 0.936506 | 0.956471 | |
| Arginine | Ad Lib-Calorie Restricted | −2.18275 | 0.761837 | 0.033315 | * |
| Arginine | WT-Knockout:Ad Lib-Calorie Restricted | −0.02785 | 1.104215 | 0.985223 | |
| Asparagine | WT-Knockout | 0.094295 | 0.686341 | 0.956471 | |
| Asparagine | Ad Lib-Calorie Restricted | −0.56615 | 0.553948 | 0.667634 | |
| Asparagine | WT-Knockout:Ad Lib-Calorie Restricted | −0.39857 | 0.866269 | 0.943088 | |
| Glutamine | WT-Knockout | −0.62699 | 0.739647 | 0.792071 | |
| Glutamine | Ad Lib-Calorie Restricted | −0.6151 | 0.610566 | 0.667634 | |
| Glutamine | WT-Knockout:Ad Lib-Calorie Restricted | 0.127152 | 0.846355 | 0.956471 | |
| Glycine | WT-Knockout | −0.63792 | 0.965301 | 0.890945 | |
| Glycine | Ad Lib-Calorie Restricted | −1.61922 | 0.795852 | 0.160397 | |
| Glycine | WT-Knockout:Ad Lib-Calorie Restricted | 0.605794 | 1.107135 | 0.919589 | |
| Histidine | WT-Knockout | 0.156234 | 0.553065 | 0.956471 | |
| Histidine | Ad Lib-Calorie Restricted | −0.44839 | 0.472269 | 0.702966 | |
| Histidine | WT-Knockout:Ad Lib-Calorie Restricted | −0.08448 | 0.759276 | 0.956471 | |
| Isoleucine | WT-Knockout | −0.28877 | 0.725687 | 0.943088 | |
| Isoleucine | Ad Lib-Calorie Restricted | −0.63274 | 0.601044 | 0.667634 | |
| Isoleucine | WT-Knockout:Ad Lib-Calorie Restricted | −0.3511 | 0.825287 | 0.943088 | |
| Leucine | WT-Knockout | −0.24831 | 0.738022 | 0.943088 | |
| Leucine | Ad Lib-Calorie Restricted | −0.64633 | 0.608206 | 0.667634 | |
| Leucine | WT-Knockout:Ad Lib-Calorie Restricted | −0.34648 | 0.847153 | 0.943088 | |
| Lysine | WT-Knockout | −0.54022 | 0.746722 | 0.859137 | |
| Lysine | Ad Lib Calorie Restricted | −1.49522 | 0.636885 | 0.096383 | |
| Lysine | WT-Knockout:Ad Lib-Calorie Restricted | 0.099118 | 0.805104 | 0.956471 | |
| Methionine | WT-Knockout | −0.56342 | 0.69592 | 0.793471 | |
| Methionine | Ad Lib-Calorie Restricted | −0.88884 | 0.580019 | 0.383931 | |
| Methionine | WT-Knockout:Ad Lib-Calorie Restricted | 0.191979 | 0.782495 | 0.956471 | |
| Phenylalanine | WT-Knockout | −0.46747 | 0.773454 | 0.890945 | |
| Phenylalanine | Ad Lib-Calorie Restricted | −0.82785 | 0.643093 | 0.516385 | |
| Phenylalanine | WT-Knockout:Ad Lib-Calorie Restricted | −0.01634 | 0.873442 | 0.985223 | |
| Proline | WT-Knockout | −0.17087 | 0.471709 | 0.943088 | |
| Proline | Ad Lib-Calorie Restricted | −0.73485 | 0.381019 | 0.189684 | |
| Proline | WT-Knockout:Ad Lib-Calorie Restricted | 0.05283 | 0.567567 | 0.956471 | |
| Serine | WT-Knockout | −0.61613 | 0.739111 | 0.792071 | |
| Serine | Ad Lib-Calorie Restricted | −1.69246 | 0.631136 | 0.048153 | * |
| Serine | WT-Knockout:Ad Lib-Calorie Restricted | 0.368377 | 1.014689 | 0.943088 | |
| Threonine | WT-Knockout | −0.4354 | 0.697263 | 0.890945 | |
| Threonine | Ad Lib-Calorie Restricted | −1.19757 | 0.574495 | 0.15194 | |
| Threonine | WT-Knockout:Ad Lib-Calorie Restricted | 0.25633 | 0.800686 | 0.943088 | |
| Tyrosine | WT-Knockout | −0.36453 | 0.700451 | 0.932937 | |
| Tyrosine | Ad Lib-Calorie Restricted | −0.75272 | 0.581825 | 0.516385 | |
| Tyrosine | WT-Knockout:Ad Lib-Calorie Restricted | −0.15028 | 0.792401 | 0.956471 | |
| Valine | WT-Knockout | −0.11812 | 0.682888 | 0.956471 | |
| Valine | Ad Lib-Calorie Restricted | −0.67426 | 0.565532 | 0.579218 | |
| Valine | WT-Knockout:Ad Lib-Calorie Restricted | −0.47436 | 0.776773 | 0.890945 |
The gene type and diet were considered as independent variables and participates as random effects. P values were corrected to reduce false discovery rate using Benjamini–Hochberg procedure. *FDR adjusted significant P value.
Glycocholic Acid Treatment Decreases Enteroid Formation Efficiency in Wild-Type Mice
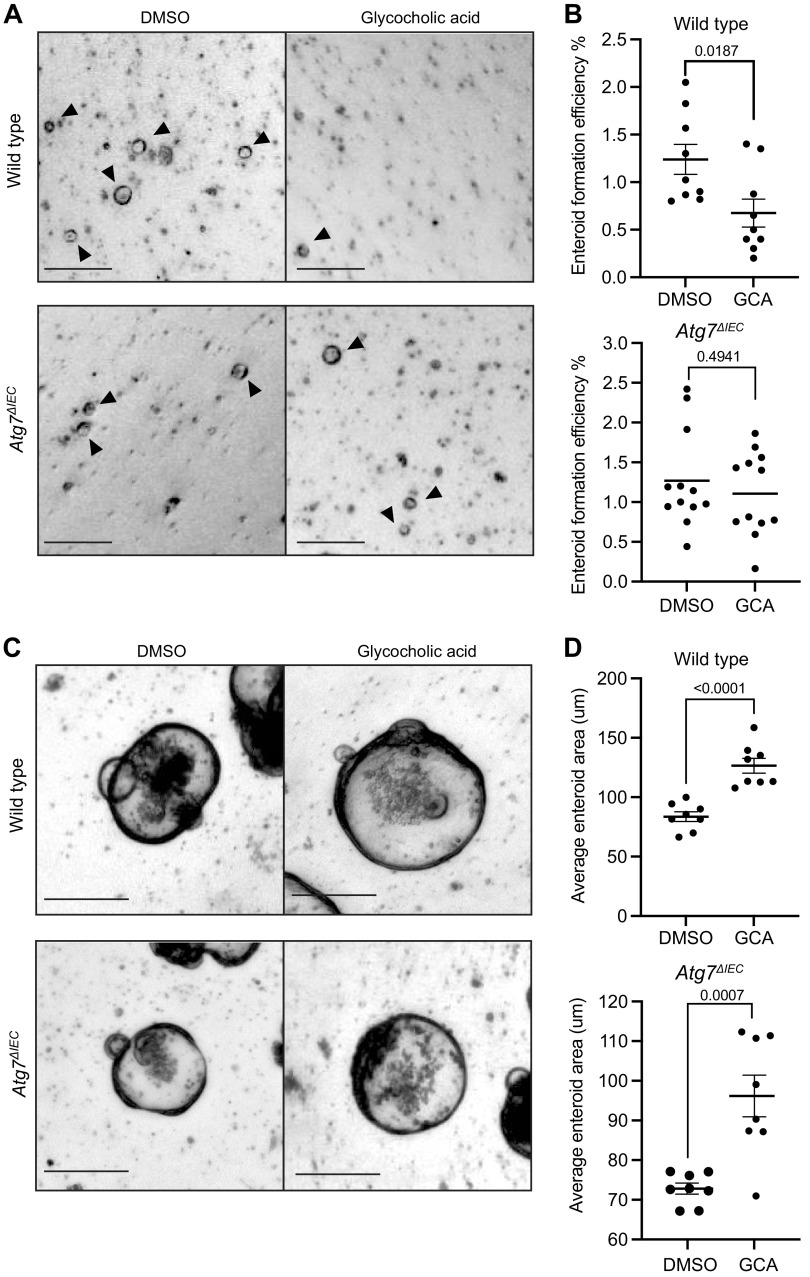
We next used intestinal crypt enteroids to dissect whether GCA has a direct contribution to epithelial cell self-renewal or growth. We hypothesized that since GCA is present in calorie-restricted Atg7ΔIEC mice but not wild-type mice, the presence of GCA may be detrimental to intestinal epithelial cells. We cultured enteroids in the presence or absence of GCA for 3 days before dissociation into single cells to mimic epithelial disruption in vivo followed by re-plating at equal density with continued treatment of GCA for two days to evaluate enteroid formation efficiency. We found that GCA treatment decreased enteroid formation compared with vehicle (DMSO) controls (1.24 ± 0.47 vs. 0.68 ± 0.44, Fig. 5, A and B), suggesting that the presence of GCA decreases stem cell self-renewal capacity following epithelial disruption. We next performed the same dissociation experiments in enteroids from mice with Atg7 deletion (VillinCreERT2;Atg7flox/flox with or without 4OHT, Supplemental Fig. 2). Although experiments in wild-type and VillinCreERT2;Atg7ΔIEC enteroids were performed sequentially rather than simultaneously, we found similar mean enteroid formation efficiencies in both untreated groups (1.24 ± 0.47 in wild-type vs. 1.23 ± 0.62 in VillinCreERT2;Atg7ΔIEC). We did not observe a significant change in enteroid formation with GCA treatment in VillinCreERT2;Atg7ΔIEC (1.23 ± 0.62 vs. 1.11 ± 0.53, Fig. 5, A and B). This is intriguing, as it suggests that epithelial Atg7 may directly mediate the effects of GCA on stem cell self-renewal at homeostasis.
Figure 5.
Glycocholic acid (GCA) inhibits enteroid formation in wild-type mice. A: representative images of enteroids grown from single cells from wild-type or VillinCreERT2;Atg7ΔIEC mice. Enteroids were treated with vehicle (DMSO) or GCA (100 µm) for 3 days before dissociation into single cells followed by replating at equal density with continued treatment of GCA for 4 days. Arrows point to live enteroids. Scale bar = 200 μm. B: enteroid formation rates of wild-type or VillinCreERT2;Atg7 ΔIEC enteroids treated with GCA. C: representative images of DMSO or GCA-treated enteroids on day 6 post-treatment of plated crypts. Scale bar = 250 μm. D: average enteroid area. Unpaired, two-tailed t tests were performed with P values indicated. n = 3 independent passages per genotype with at least 3 wells per passage for enteroid formation efficiencies, and n = 3 independent passages per genotype with at least 2 wells of approximately 36 enteroids per well for enteroid area measurements.
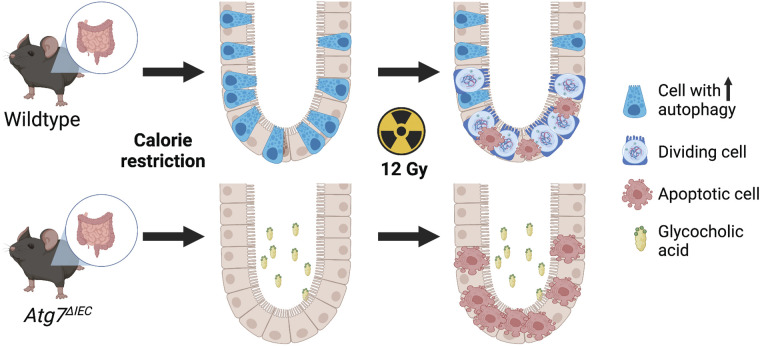
We also evaluated the effects of GCA on enteroid proliferation by measuring enteroid area 6 days after treating them with GCA. We observed a significant increase in enteroid area with GCA treatment in both wild-type and VillinCreERT2;Atg7ΔIEC enteroids compared with their untreated counterparts (Fig. 5, C and D). We do note, however, that the average enteroid area of untreated wild-type enteroids was larger than untreated VillinCreERT2;Atg7ΔIEC enteroids (83.65 ± 11.49 vs. 72.82 ± 5.81). As such, these data suggest that GCA has similar effects on proliferation regardless of epithelial Atg7 status. More globally, these data support the hypothesis that in wild-type mice, decreased GCA during calorie restriction may contribute to the increased regenerative response postirradiation compared with calorie-restricted Atg7ΔIEC mice (Fig. 6). These data also suggest that Atg7 contributes to stem cell responses to GCA in an epithelial-intrinsic manner, since VillinCreERT2;Atg7ΔIEC enteroids do not exhibit decreased self-renewal in response to GCA.
Figure 6.
Working model. In wild-type mice, decreased glycocholic acid (GCA) during calorie restriction may contribute to the increased regenerative response postirradiation compared with calorie-restricted Atg7ΔIEC mice, where GCA is present at noncalorie restriction levels. Figure was created with BioRender.
DISCUSSION
Previous research suggests that calorie restriction preserves stem cell viability (32) and expands the pool of radioresistant ISCs by shifting the equilibrium toward stem cell self-renewal instead of differentiation (3, 6), with a mechanistic focus on mTORC1. Similarly, a fasting-mimicking diet (1 day at 50% of normal caloric intake and 3 days of 10% of normal caloric intake) was shown to mitigate colon shortening in response to the dextran sodium sulfate (DSS) model of colonic damage. Protection from DSS-mediated damage was associated with increased Lgr5+ ISCs and increased proliferative index in both the small intestine and colon in mice on the fasting-mimicking diet (33). However, the contribution of autophagy to the regenerative benefit of calorie restriction has not been reported in vivo. In the current study, we first confirmed that calorie restriction can enhance autophagy via Western blot for Lc3-I/Lc3-II. Although we observed that calorie restriction was associated with increased Lc3-II, the effect was variable and not statistically significant. Furthermore, there were no apparent histological differences between genotypes associated with calorie restriction. The lack of robust differences between genotypes at homeostasis could reflect tissue adaptation in response to long-term calorie restriction and suggests that further challenge is required to reveal the requirement for autophagy. Accordingly, our prior studies showed that autophagy is markedly upregulated in the intestine following irradiation (34), which highlights putative differences between homeostatic increases as shown in the present study with calorie restriction versus damage-induced increases as with irradiation. Although autophagy upregulation was modest with calorie restriction in unchallenged wild-type mice, Atg7ΔIEC mice on a calorie restriction diet had fewer regenerative crypt foci than calorie-restricted wild-type mice following irradiation-induced intestinal damage. We recently demonstrated that relatively high levels of autophagic vesicles can prospectively identify cells with facultative ISC capacity that replenish crypt base columnar ISCs after injury (16). The activation of autophagy during calorie restriction likely increases the facultative ISC pool, priming it for regeneration after irradiation. Similarly, due to autophagy’s role in promoting the degradation of damaged organelles and abnormal protein aggregates (35), elevated autophagy before irradiation may prepare the cell to efficiently deal with damage caused by irradiation. Taken together, the present study and prior work support a working model whereby intestinal epithelial autophagy may both protect from damage and facilitate tissue regeneration.
Prior studies suggest that mice on a fasted diet have reduced intestinal inflammation and increased stem cell numbers due in part to enrichment of microbes and their derived metabolites that protect the intestinal barrier (33). We therefore evaluated microbial metabolites in a small subset of mice to identify differences that may contribute to regenerative phenotypes we observed in wild-type mice versus Atg7ΔIEC mice following calorie restriction. We found that wild-type mice on a calorie restriction diet exhibited lower luminal levels of primary bile acid glycocholic acid (GCA) compared with their Atg7ΔIEC counterparts, suggesting that epithelial autophagy may contribute to enterohepatic signaling during calorie restriction. Bile acids can play roles in cell responses to DNA damage, reactive oxygen species production, and activation of programmed cell death pathways in epithelial cells (36, 37). Increased bile acids are associated with high-fat diet, and recent studies show that Paneth cells upregulate the G protein-coupled bile acid receptor (Gpbar1, also known as Tgr5) in response to high-fat diet or normal chow with cholic acid supplementation. The same study suggested that increased bile acids in response to high-fat diet leads to Paneth cell toxicity, as there was an overall decrease in Paneth cells in this model (38). Conversely, in response to nutrient deprivation, Paneth cells are suggested to support the proliferative capacity of their neighboring ISCs (6). Although we did not observe changes in Paneth cells in Atg7ΔIEC mice in response to calorie restriction, it is possible that autophagy deletion in epithelial cells can alter their ability to sense and appropriately respond to luminal changes in bile acids. It is also possible that altered microbial communities in response to epithelial cell autophagy deletion may have a global effect on bile acid production or composition, shifting the proportion of bile acids toward those that are less favorable to ISC survival or regeneration.
As central mediators of vitamins and dietary fat absorption in the intestine, bile acids ultimately influence lipid and glucose metabolism (39), inflammatory pathways (40), intestinal barrier function (30), and stem cell proliferation (23). Previous literature suggested the bile acid lithocholic acid (LCA), which accumulates in stool with high-fat diet, can increase Lgr5+ stem cells via the direct activation of Tgr5 (23). Intriguingly, treatment with 10 µM LCA was associated with increased enteroid budding, but decreased enteroid size, and doses of LCA above 50 µM decreased enteroid formation. We did not observe a difference in fecal accumulation of LCA between genotypes in our study (not shown). Of note, LCA is a secondary bile acid generated in the colon and so its direct physiological relevance to small intestinal epithelial cells is not clear (41). In other studies, different bile acids have opposing effects on intestinal epithelial cell proliferation. For example, taurine-conjugated cholic acid (TCA) was associated with enhanced proliferation, whereas LCA and deoxycholic acid (DCA) were associated with decreased proliferation in both rat small intestinal cells (IEC-6) and in immortalized young adult mouse colon (YAMC) cells (41). Direct effects of bile acids on epithelial cell proliferation were shown to converge upon modulation of EGFR/Src/ERK in part through the nuclear farnesoid X receptor (FXR) (41).
Because GCA was the target most effected in our metabolite analyses, we examined the direct association of GCA with enteroid self-renewal and proliferation. We treated enteroids with GCA for 3 days and then disrupted them and plating single cells in the presence of GCA for four additional days to evaluate enteroid formation. We observed that enteroid formation efficiency was decreased with GCA treatment. Conversely, treatment of established enteroids with GCA was associated with increased enteroid size, which may suggest that GCA can enhance proliferation at homeostasis. We also performed the same experiments in inducible Atg7ΔIEC enteroids, where we did not observe decreased enteroid formation. Since we observe similar plating efficiencies between untreated wildtype and Atg7ΔIEC enteroids, our data support the conclusion that GCA appears to decrease self-renewal in wildtype enteroids only. Taken together, these data suggest that the presence of GCA is associated with reduced enteroid self-renewal in a manner dependent upon intact epithelial Atg7. More globally, our data support the conclusion that epithelial Atg7 may: 1) contribute to modulation of luminal levels of GCA during calorie restriction and 2) exhibit epithelial-intrinsic roles in modulating how ISCs respond to GCA at homeostasis. Although the direct mechanisms underlying these findings are not known, prior studies demonstrate an interplay between the autophagy pathway and FXR in other tissues. In liver, FXR binds directly near transcription start sites of multiple autophagy-related genes, including Atg7, and activation of FXR via agonist GW4064 has repressive effects on autophagy gene expression (42). In a separate study, bile acid activation of FXR in liver can induce Rubicon to inhibit autophagy (43). Although these studies place bile acid and FXR signaling upstream of autophagy, our future studies will focus on whether there is dynamic signaling or feedback between intestinal epithelial autophagy and bile acid or FXR signaling.
Of note, one limitation of our study is the relatively small sample size and variability within groups for our metabolite analyses. As such, while we do demonstrate a contribution of GCA to enteroid self-renewal and growth, it is likely that other luminal factors, including other bile acids, may contribute to modulating epithelial cell dynamics during calorie restriction. GCA represents a small proportion of total small intestinal bile acids (44), which is likely why the change in GCA is not reflected in the pooled bile acid data. Therefore, changes in GCA alone are unlikely to be the sole mechanism underlying differences observed between genotypes following irradiation. Nevertheless, we do find that GCA treatment can both reduce enteroid formation while at the same time increase enteroid size, demonstrating that there are direct effects if GCA on epithelial cells. An additional limitation to the study is that while luminal amounts of GCA are in the 0.5–2 µM range, we used supraphysiological doses of GCA (100 µM) in enteroid culture to ensure that it was able to penetrate the three-dimensional (3-D) matrix.
The role of GCA in intestinal epithelial cell function may have broad implications beyond this study, as bile acid composition is associated with microbial community structure and a myriad of microbiota-associated diseases. Future studies in Atg7ΔIEC mice will evaluate whether perturbations to the microbiota have unappreciated consequences on epithelial cell regeneration via host-bile acid cross talk (45–47). Ultimately, our findings demonstrate a previously unknown requirement for intestinal epithelial autophagy for the regenerative benefit of calorie restriction and provides additional evidence that associated shifts in bile acids may have direct effects on ISC self-renewal or proliferation.
DATA AVAILABILITY
Data will be made available upon reasonable request.
SUPPLEMENTAL DATA
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.21382275.
Supplemental Fig. S2: https://doi.org/10.6084/m9.figshare.21971096.
GRANTS
This work was supported by National Institutes of Health Grants F31-DK124956 (to L. R. Parham), NIH R01-DK124369 (to K. E. Hamilton), Children’s Hospital of Philadelphia Institutional Development Funds and Gastrointestinal Epithelium Modeling Program (to K. E. Hamilton), and the following cores via the Penn Center for Molecular Studies in Digestive and Liver Diseases NIH P30-DK050306: the Microbial Culture & Metabolomics and the High-Throughput Sequencing and Analytical Cores of the Penn CHOP Microbiome Program and the Host-Microbial Analytic and Repository Core, and the Molecular Pathology and Imaging Core.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
P.A.W., K.E.N., and K.E.H. conceived and designed research; P.A.W., K.E.N., L.A.S., L.R.P., E.S.F. H.M., and K.E.H., performed experiments; P.A.W., K.E.N., L.A.S., G.E.S., L.R.P., X.M., C.H.D., W.H., E.S.F., H.M., M.A.S., J.S.O., K.B., and K.E.H. analyzed data; P.A.W., K.E.N., G.E.S., X.M., C.H.D., W.H., E.S.F., E.A.M., J.S.O., J.Z., K.B., K.A.W., T.A.K., and K.E.H. interpreted results of experiments; P.A.W., K.E.N., G.E.S., W.H., E.S.F., E.A.M., K.B., and K.E.H. prepared figures; P.A.W., K.E.N., and K.E.H. drafted manuscript; P.A.W., K.E.N., W.H., E.S.F., J.S.O., J.Z., K.B., T.A.K., and K.E.H., edited and revised manuscript; P.A.W., K.E.N., L.A.S., G.E.S., L.R.P., X.M., C.H.D., W.H., E.S.F., E.A.M., H.M., M.A.S., J.S.O., J.Z., K.B., K.A.W., T.A.K., and K.E.H. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Drs. Gary Wu, Victoria Gershuni, Christopher Lengner, and Maryam Yousefi for guidance on mouse diet and calorie restriction protocols. We thank Dr. Walter Faig of the CHOP Biostatistics and Data Management Core for biostatistical support.
REFERENCES
- 1. Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418: 344–348, 2002. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 2. Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr 116: 641–654, 1986. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 3. Yousefi M, Nakauka-Ddamba A, Berry CT, Li N, Schoenberger J, Simeonov KP, Cedeno RJ, Yu Z, Lengner CJ. Calorie restriction governs intestinal epithelial regeneration through cell-autonomous regulation of mTORC1 in reserve stem cells. Stem Cell Rep 10: 703–711, 2018. doi: 10.1016/j.stemcr.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Igarashi M, Guarente L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell 166: 436–450, 2016. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 5. Bruens L, Ellenbroek SIJ, Suijkerbuijk SJE, Azkanaz M, Hale AJ, Toonen P, Flanagan DJ, Sansom OJ, Snippert HJ, van Rheenen J. Calorie restriction increases the number of competing stem cells and decreases mutation retention in the intestine. Cell Rep 32: 107937, 2020. doi: 10.1016/j.celrep.2020.107937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, Nielsen GP, Mino-Kenudson M, Zukerberg LR, Bhan AK, Deshpande V, Sabatini DM. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486: 490–495, 2012. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, Flak MB, Cusick JL, Kohno K, Iwawaki T, Billmann-Born S, Raine T, Bharti R, Lucius R, Kweon MN, Marciniak SJ, Choi A, Hagen SJ, Schreiber S, Rosenstiel P, Kaser A, Blumberg RS. Paneth cells as a site of origin for intestinal inflammation. Nature 503: 272–276, 2013. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25: 1025–1040, 2005. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, Hooper LV. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357: 1047–1052, 2017. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 47: 979–986, 2015. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Z, Lenardo MJ. The role of LRRK2 in inflammatory bowel disease. Cell Res 22: 1092–1094, 2012. doi: 10.1038/cr.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iida T, Onodera K, Nakase H. Role of autophagy in the pathogenesis of inflammatory bowel disease. World J Gastroenterol 23: 1944–1953, 2017. doi: 10.3748/wjg.v23.i11.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagy P, Sandor GO, Juhasz G. Autophagy maintains stem cells and intestinal homeostasis in Drosophila. Sci Rep 8: 4644, 2018. doi: 10.1038/s41598-018-23065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Asano J, Sato T, Ichinose S, Kajita M, Onai N, Shimizu S, Ohteki T. Intrinsic autophagy is required for the maintenance of intestinal stem cells and for irradiation-induced intestinal regeneration. Cell Rep 20: 1050–1060, 2017. doi: 10.1016/j.celrep.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 15. Trentesaux C, Fraudeau M, Pitasi CL, Lemarchand J, Jacques S, Duche A, Letourneur F, Naser E, Bailly K, Schmitt A, Perret C, Romagnolo B. Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proc Natl Acad Sci USA 117: 11136–11146, 2020. doi: 10.1073/pnas.1917174117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson NM, Parham LR, Na J, Monaghan KE, Kolev HM, Klochkova A, Kim MS, Danan CH, Cramer Z, Simon LA, Naughton KE, Adams-Tzivelekidis S, Tian Y, Williams PA, Leu NA, Sidoli S, Whelan KA, Li N, Lengner CJ, Hamilton KE. Autophagic state prospectively identifies facultative stem cells in the intestinal epithelium. EMBO Rep 23, e55209, 2022. doi: 10.15252/embr.202255209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434, 2005. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramsteijn AS, Jasarevic E, Houwing DJ, Bale TL, Olivier JD. Antidepressant treatment with fluoxetine during pregnancy and lactation modulates the gut microbiome and metabolome in a rat model relevant to depression. Gut Microbes 11: 735–753, 2020. doi: 10.1080/19490976.2019.1705728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedman ES, Li Y, Shen TD, Jiang J, Chau L, Adorini L, Babakhani F, Edwards J, Shapiro D, Zhao C, Carr RM, Bittinger K, Li H, Wu GD. FXR-dependent modulation of the human small intestinal microbiome by the bile acid derivative obeticholic acid. Gastroenterology 155: 1741–1752e.5, 2018. doi: 10.1053/j.gastro.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ni J, Shen TD, Chen EZ, Bittinger K, Bailey A, Roggiani M, Sirota-Madi A, Friedman ES, Chau L, Lin A, Nissim I, Scott J, Lauder A, Hoffmann C, Rivas G, Albenberg L, Baldassano RN, Braun J, Xavier RJ, Clish CB, Yudkoff M, Li H, Goulian M, Bushman FD, Lewis JD, Wu GD. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med 9: eaah6888, 2017. doi: 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HWt. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456: 259–263, 2008. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 23. Sorrentino G, Perino A, Yildiz E, El Alam G, Bou Sleiman M, Gioiello A, Pellicciari R, Schoonjans K. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 159: 956–968e.8, 2020. doi: 10.1053/j.gastro.2020.05.067. [DOI] [PubMed] [Google Scholar]
- 24. Cadwell K, Patel KK, Komatsu M, Virgin HW IV, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 5: 250–252, 2009. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bagherniya M, Butler AE, Barreto GE, Sahebkar A. The effect of fasting or calorie restriction on autophagy induction: a review of the literature. Ageing Res Rev 47: 183–197, 2018. doi: 10.1016/j.arr.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 26. Chung KW, Chung HY. The effects of calorie restriction on autophagy: role on aging intervention. Nutrients 11: 2923, 2019. doi: 10.3390/nu11122923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 140: 313–326, 2010. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2'-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2'-deoxyuridine antibodies. Biotechniques 44: 927–929, 2008. doi: 10.2144/000112812. [DOI] [PubMed] [Google Scholar]
- 29. von Schwartzenberg RJ, Bisanz JE, Lyalina S, Spanogiannopoulos P, Ang QY, Cai J, Dickmann S, Friedrich M, Liu SY, Collins SL, Ingebrigtsen D, Miller S, Turnbaugh JA, Patterson AD, Pollard KS, Mai K, Spranger J, Turnbaugh PJ. Caloric restriction disrupts the microbiota and colonization resistance. Nature 595: 272–277, 2021. doi: 10.1038/s41586-021-03663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raimondi F, Santoro P, Barone MV, Pappacoda S, Barretta ML, Nanayakkara M, Apicella C, Capasso L, Paludetto R. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am J Physiol Gastrointest Liver Physiol 294: G906–G913, 2008. doi: 10.1152/ajpgi.00043.2007. [DOI] [PubMed] [Google Scholar]
- 31. Kim DH, Park JS, Choi HI, Kim CS, Bae EH, Ma SK, Kim SW. The critical role of FXR is associated with the regulation of autophagy and apoptosis in the progression of AKI to CKD. Cell Death Dis 12: 320, 2021. doi: 10.1038/s41419-021-03620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tinkum KL, Stemler KM, White LS, Loza AJ, Jeter-Jones S, Michalski BM, Kuzmicki C, Pless R, Stappenbeck TS, Piwnica-Worms D, Piwnica-Worms H. Fasting protects mice from lethal DNA damage by promoting small intestinal epithelial stem cell survival. Proc Natl Acad Sci USA 112: E7148–E7154, 2015. doi: 10.1073/pnas.1509249112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rangan P, Choi I, Wei M, Navarrete G, Guen E, Brandhorst S, Enyati N, Pasia G, Maesincee D, Ocon V, Abdulridha M, Vd L. Fasting-mimicking diet modulates microbiota and promotes intestinal regeneration to reduce inflammatory bowel disease pathology. Cell Rep 26: 2704–2719e.6, 2019. doi: 10.1016/j.celrep.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chatterji P, Williams PA, Whelan KA, Samper FC, Andres SF, Simon LA, Parham LR, Mizuno R, Lundsmith ET, Lee DS, Liang S, Wijeratne HS, Marti S, Chau L, Giroux V, Wilkins BJ, Wu GD, Shah P, Tartaglia GG, Hamilton KE. Posttranscriptional regulation of colonic epithelial repair by RNA binding protein IMP1/IGF2BP1. EMBO Rep 20: e47074, 2019. doi: 10.15252/embr.201847074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell 176: 11–42, 2019. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Powolny A, Xu J, Loo G. Deoxycholate induces DNA damage and apoptosis in human colon epithelial cells expressing either mutant or wild-type p53. Int J Biochem Cell Biol 33: 193–203, 2001. doi: 10.1016/s1357-2725(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 37. Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol 15: 1677–1689, 2009. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou H, Zhou SY, Gillilland M III, Li JY, Lee A, Gao J, Zhang G, Xu X, Owyang C. Bile acid toxicity in Paneth cells contributes to gut dysbiosis induced by high-fat feeding. JCI Insight 5: e138881, 2020. doi: 10.1172/jci.insight.138881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol 25: 2020–2030, 2005. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 40. Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 103: 3920–3925, 2006. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dossa AY, Escobar O, Golden J, Frey MR, Ford HR, Gayer CP. Bile acids regulate intestinal cell proliferation by modulating EGFR and FXR signaling. Am J Physiol Gastrointest Liver Physiol 310: G81–G92, 2016. doi: 10.1152/ajpgi.00065.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 516: 108–111, 2014. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Panzitt K, Jungwirth E, Krones E, Lee JM, Pollheimer M, Thallinger GG, Kolb-Lenz D, Xiao R, Thorell A, Trauner M, Fickert P, Marschall HU, Moore DD, Wagner M. FXR-dependent Rubicon induction impairs autophagy in models of human cholestasis. J Hepatol 72: 1122–1131, 2020. doi: 10.1016/j.jhep.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 44. Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, Chen H, Zhao X, Zhang S, Zhou Z, Zeisel SH, Jia W. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J 27: 3583–3593, 2013. doi: 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1: e00045-15, 2016. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome 9: 140, 2021. doi: 10.1186/s40168-021-01101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 15: 111–128, 2018. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.21382275.
Supplemental Fig. S2: https://doi.org/10.6084/m9.figshare.21971096.
Data Availability Statement
Data will be made available upon reasonable request.