Summary
Background
Some observational studies found that dyslipidaemia is a risk factor for non-alcoholic fatty liver disease (NAFLD), and lipid-lowering drugs may lower NAFLD risk. However, it remains unclear whether dyslipidaemia is causative for NAFLD. This Mendelian randomisation (MR) study aimed to explore the causal role of lipid traits in NAFLD and evaluate the potential effect of lipid-lowering drug targets on NAFLD.
Methods
Genetic variants associated with lipid traits and variants of genes encoding lipid-lowering drug targets were extracted from the Global Lipids Genetics Consortium genome-wide association study (GWAS). Summary statistics for NAFLD were obtained from two independent GWAS datasets. Lipid-lowering drug targets that reached significance were further tested using expression quantitative trait loci data in relevant tissues. Colocalisation and mediation analyses were performed to validate the robustness of the results and explore potential mediators.
Findings
No significant effect of lipid traits and eight lipid-lowering drug targets on NAFLD risk was found. Genetic mimicry of lipoprotein lipase (LPL) enhancement was associated with lower NAFLD risks in two independent datasets (OR1 = 0.60 [95% CI 0.50–0.72], p1 = 2.07 × 10−8; OR2 = 0.57 [95% CI 0.39–0.82], p2 = 3.00 × 10−3). A significant MR association (OR = 0.71 [95% CI, 0.58–0.87], p = 1.20 × 10−3) and strong colocalisation association (PP.H4 = 0.85) with NAFLD were observed for LPL expression in subcutaneous adipose tissue. Fasting insulin and type 2 diabetes mediated 7.40% and 9.15%, respectively, of the total effect of LPL on NAFLD risk.
Interpretation
Our findings do not support dyslipidaemia as a causal factor for NAFLD. Among nine lipid-lowering drug targets, LPL is a promising candidate drug target in NAFLD. The mechanism of action of LPL in NAFLD may be independent of its lipid-lowering effects.
Funding
Capital’s Funds for Health Improvement and Research (2022-4-4037). CAMS Innovation Fund for Medical Sciences (CIFMS, grant number: 2021-I2M-C&T-A-010).
Keywords: Mendelian randomization, Statins, TG, LDL-C, eQTL
Research in context.
Evidence before this study
Some observational studies found that dyslipidaemia was independently associated with incident non-alcoholic fatty liver disease (NAFLD), and lipid-lowering treatments (e.g. statin) may reduce the risk of NAFLD. However, epidemiological studies have methodological constraints and are subject to residual confounding; the causal relationship between dyslipidaemia and the risk of NAFLD has not been fully determined. With the growing availability of large-scale genome-wide association studies, Mendelian randomisation (MR) can efficiently explore the causal factors for diseases and can predict drug effectiveness by mimicking randomised controlled trials.
Added value of this study
Integrating genomics with lipid traits, lipid-lowering drug targets, and NAFLD traits, this study aimed to assess the causal roles of lipid traits in NAFLD and to explore the potential effects of drug targets on NAFLD. The association of lipid traits (i.e. low-density lipoprotein cholesterol, triglyceride, and total cholesterol) and lipid-lowering drug targets for statins, ezetimibe, proprotein convertase subtilisin/kexin type 9 inhibitors, bile acid sequestrants, angiopoietin-like 3 inhibitors, and antisense oligonucleotide targeting apolipoprotein C-III mRNA with NAFLD risk was not found in this study. Only one drug target, lipoprotein lipase (LPL), was found to have the ability to significantly lower the risk of NAFLD. This finding was validated in two independent NAFLD datasets using different approaches to mimic LPL activation. The part of the mechanism of action of LPL in lowering the NAFLD risk may be through the regulation of insulin levels other than lipid levels.
Implications of all the available evidence
Available evidence does not support dyslipidaemia as a causal factor for NAFLD. Among nine lipid-lowering drug targets, LPL is the only drug target associated with a lower NAFLD risk. The pharmacological properties of LPL modulation can be observed in various medications, such as fibrates, thiazolidinediones, and omega-3 fatty acid; however, it is not the primary mechanism of action of these drugs. Given the lack of pharmacological therapies for NAFLD at present, there is a need to develop new drugs targeting LPL activation as their central mechanistic effect. Further investigation is required to understand whether LPL activators will show similar effects in basic and even clinical trials, and to elucidate the underlying biological mechanisms.
Introduction
Non-alcoholic fatty liver disease (NAFLD), characterised by excessive liver triglyceride (TG) accumulation due to metabolic dysfunction, is now the fastest-growing contributor to liver-related mortality and will affect one-third of the population worldwide in 2030.1,2 Considering the lack of pharmacological therapies for NAFLD, early identification and modification of risk factors for NAFLD in at-risk populations are warranted to lower the societal burden of this disease.2
Some observational studies supported dyslipidaemia as a risk factor for NAFLD, as dyslipidaemia was independently associated with severe NAFLD-related liver disease (i.e. liver cirrhosis, complications of cirrhosis or liver-related death).3 Moreover, dyslipidaemia often occurs in NAFLD: 72% of patients with non-alcoholic steatohepatitis (NASH), a more progressive type of NAFLD, have comorbid dyslipidaemia.4 Despite these observations, epidemiological studies have methodological constraints and are subject to residual confounding; the causal relationship between dyslipidaemia and the risk of NAFLD has not been fully determined.
Given that dyslipidaemia is closely associated with NAFLD development and severity, lipid-lowering drugs have been proposed as NAFLD repurposing candidates (expanding the indication of approved drugs into other indications). Statins reduced the risk of NAFLD and improved hepatic fibrosis in a recent observational study involving 11,593,409 Korean subjects.5 A randomized controlled trial (RCT) is the standard method for determining an agent's efficacy. However, few large-scale RCTs are present and several challenges remain in conducting this process: because invasive liver biopsy remains the gold standard in clinical trials, monitoring drug effect dynamically for individuals is difficult.6,7 Decades-long disease course of NAFLD results in difficulties in defining clinical trial endpoints, and drug testing is costly and time-consuming.8 Moreover, concerns remain about potential adverse effects of lipid-lowering drugs on liver function. Therefore, whether lipid-lowering drugs are effective for NAFLD remains unclear.
With the growing availability of genome-wide association studies (GWASs), Mendelian randomisation (MR) could be an efficient way to overcome the aforementioned issues.9 Because genetic variants (alleles) are randomly assigned during meiosis, participants in an MR study are ‘randomised’ according to the presence of alleles. This is similar to RCTs where participants are randomly assigned to an experimental treatment or a control group. Therefore, MR uses a ’randomised’ way to examine whether carriers of risk factors' alleles have high or low disease risk compared with noncarriers (Figure S1).10 It can explore the causal relationship between risk factors (referring to ‘Biomarker MR’) or therapeutic drug targets (referring to ‘Drug target MR’) and the outcome.11 For ‘Drug target MR’, genetic variants within the genes encoding protein targets can affect the expression or functions of target genes similar to the mechanisms of actions of drugs. It can foreshadow the results of RCTs.12,13 A classic example is that individuals carrying genetic variants associated with lower low-density lipoprotein cholesterol (LDL-C) levels in the PCSK9 gene had a lower incidence of coronary heart disease.14 This finding laid the foundation for developing proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors, and the efficacy of PCSK9 inhibitors was confirmed in subsequent RCTs.15 Now, MR analyses have been widely used to predict the potential effects of drug targets on cardiovascular diseases, neurological diseases and neoplasms.16, 17, 18
Therefore, in this study, we conducted MR analyses to determine the effects of lipid traits on NAFLD and to explore the potential effects of lipid-lowering drug targets on NAFLD and liver function traits.
Methods
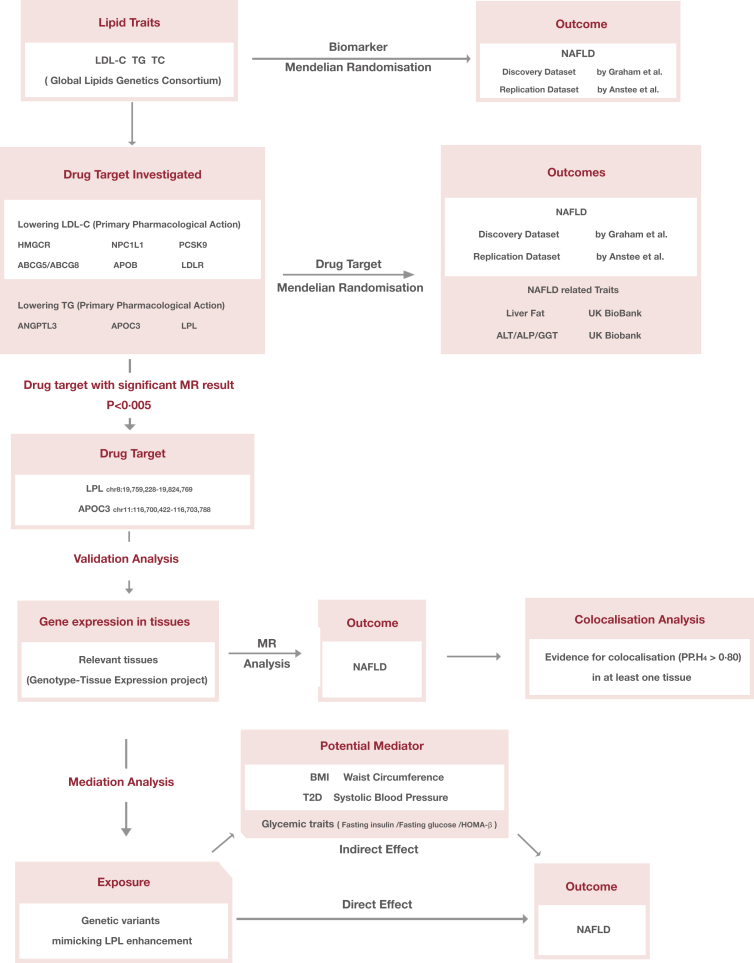
This study followed the Strengthening the Reporting of Observational Studies in Epidemiology-Mendelian Randomisation reporting guidelines (Table S1).19 An outline of the study design is shown in Fig. 1. Data sources were based on publicly available summary-level data from GWAS studies and expression quantitative trait loci (eQTL) study. Detailed information on these datasets is summarised in Table S2.
Fig. 1.
Outline of the study design. Abbreviations: LDL-C, low-density lipoprotein cholesterol; TG, triglyceride; TC, total cholesterol; NAFLD, non-alcoholic fatty liver disease; GWAS, genome-wide association study; HMGCR, HMG-CoA reductase; NPC1L1, Niemann-Pick C1-like protein 1; PCSK9, proprotein convertase subtilisin/kexin type 9; ABCG5, ATP Binding Cassette Subfamily G Member 5; ABCG8, ATP Binding Cassette Subfamily G Member 8; APOB, Apolipoprotein B-100; LDLR, LDL Receptor; ANGPTL3, angiopoietin-like 3; APOC3, Apolipoprotein C-III; LPL, lipoprotein lipase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; GGT, γ-glutamyl transferase; T2D, type 2 diabetes; BMI, body mass index; HOMA-beta, homeostasis model assessment of β-cell function.
Genetic variant selection
Independent genetic variants (linkage disequilibrium [LD] clumping threshold of r2 < 0.001 with physical distance threshold 10,000 kb) associated with LDL-C, TG and TC at genome-wide significance (p < 5 × 10−8) were identified in a GWAS meta-analysis from the Global Lipids Genetics Consortium.20
Common lipid-lowering drugs and novel therapeutics were selected based on recent guidelines for the management of dyslipidaemia, such as statins, ezetimibe, PCSK9 inhibitors, bile acid sequestrants, mipomersen, fibrates, angiopoietin-like 3 (ANGPTL3) inhibitors and antisense oligonucleotide targeting apolipoprotein C-III (APOC3) mRNA (Table 1).21,22 Genes encoding pharmacologic targets of these drugs were identified using the DrugBank database(https://go.drugbank.com/) and relevant reviews.23, 24, 25 According to primary pharmacological action, these target genes were further classified as LDL-C-lowering target genes (i.e. LDLR, HMGCR, NPC1L1, PCSK9, APOB, ABCG5 and ABCG8) and TG-lowering target genes (i.e. LPL, PPARA, ANGPTL3 and APOC3) (Table 1).
Table 1.
Lipid-lowering drug classes, substances, and target genes.
| Primary pharmacological action | Drug class | Substance | Drug targets | Target genes | Gene region (GRCh37/hg19 by Ensembl) | Genetic instruments |
|---|---|---|---|---|---|---|
| Reduced LDL-C | Key regulator | LDL Receptora | LDLR | chr19:11,200,038-11,244,492 | 12 SNPs | |
| HMG-CoA reductase inhibitors | Pravastatin Simvastatin Lovastatin Fluvastatin Atorvastatin Rosuvastatin |
HMG-CoA reductase | HMGCR | chr5:74,632,154-74,657,929 | 5 SNPs | |
| Cholesterol absorption inhibitors | Ezetimibe | Niemann-Pick C1-like protein 1 | NPC1L1 | chr7:44,552,134-44,580,914 | 3 SNPs | |
| Proprotein convertase subtilisin/kexin type 9 inhibitors | Alirocumab Evolocumab |
Proprotein convertase subtilisin/kexin type 9 | PCSK9 | chr1:55,505,221-55,530,525 | 11 SNPs | |
| Antisense oligonucleotide targeting ApoB-100 mRNA | Mipomersen | Apolipoprotein B-100 | APOB | chr2:21,224,301-21,266,945 | 15 SNPs | |
| Bile acid sequestrants | Colesevelam Colestipol Cholestyramine |
ATP Binding Cassette Subfamily G Member 5/ATP Binding Cassette Subfamily G Member 8 | ABCG5/ABCG8b | chr2:44,039,611-44,066,004/chr2:44,066,103-44,105,605 | 7 SNPs | |
| Reduced TG | Key regulator | Lipoprotein Lipasea | LPL | chr8:19,759,228-19,824,769 | 15 SNPS | |
| Fibrates | Fenofibrate Gemfibrozil |
Peroxisome proliferator-activated receptor-a | PPARA | chr22:46,546,424-46,639,653 | 0 SNPsc | |
| Angiopoietin-like 3 Inhibitor | Evinacumab | Angiopoietin-related protein 3 | ANGPTL3 | chr1:63,063,158-63,071,830 | 3 SNPs | |
| Antisense oligonucleotide targeting ApoC-III mRNA | Volanesorsen | Apolipoprotein C-III | APOC3 | chr11:116,700,422-116,703,788 | 12 SNPs |
Abbreviation: SNPs, single-nucleotide polymorphisms; chr, chromosome; mRNA, messenger ribonucleic acid; LDL-C, low-density lipoprotein cholesterol; TG, triglyceride.
LDL receptor and lipoprotein lipase are central players in LDL-C and TG metabolisms and are extensively involved in the lipid-lowering action.
Drug targets of bile acid sequestrants were not specified in the DrugBank. They were identified from a previous study.25
Since none of the fibrate variants survived through instrument construction, they were excluded from further evaluation.
Following a similar methodology as in previous studies for selection of genetic variants,17,26 single-nucleotide polymorphisms (SNPs) were identified within the corresponding genes (±100 kb of the gene location) and were robustly associated with LDL-C or TG levels at genome-wide significance (p < 5 × 10−8) in a GWAS meta-analysis from the Global Lipids Genetics Consortium.20 They were further clumped to a LD threshold of r2 < 0.20 with a physical distance threshold of 250 kb and were selected as proxies for lipid-lowering drug targets. Since none of the genetic variants of PPARA were found in the variant selection process, it was excluded from further evaluation. Due to the proximity of the genes encoding ATP Binding Cassette Subfamily G Member 5 (ABCG5) and ATP Binding Cassette Subfamily G Member 8 (ABCG8) (ABCG5: chr2:44,039,611-44,066,004 [Ensembl]; ABCG8: chr2:44,066,103-44,105,605 [Ensembl]), variants in the vicinity of these genes were combined in our analyses. Finally, nine drug targets were included in the study, that is: HMG-CoA reductase (HMGCR), Niemann-Pick C1-like protein 1 (NPC1L1), PCSK9, Apolipoprotein B-100 (APOB), ABCG5/ABCG8, LDL Receptor (LDLR), ANGPTL3, APOC3 and lipoprotein lipase (LPL).
For drug targets that reached significance for the risk of NAFLD in the MR analysis, we used publicly available eQTLs data for relevant tissues in which target genes were highly expressed from the Genotype-Tissue Expression project (GTEx-V8).27 A total of 84.6% of participants in the GTEx project were white. eQTLs are genetic variants associated with expression levels of genes. SNPs, as genetic variants with a false discovery rate-corrected p value < 0.05, were selected and further clumped to an LD r2 threshold of 0.20.
Outcome
The primary outcome was NAFLD, and the secondary outcomes were liver function traits, including liver fat percentage and three liver enzymes (i.e. alanine aminotrans-ferase [ALT], alkaline phosphatase [ALP], and γ-glutamyl transferase [GGT]).
For the primary outcome, summary genetic association data were extracted from a GWAS meta-analysis of four cohorts of electronic health record-documented NAFLD with participants of European ancestry including 8434 cases with hepatic steatosis, NASH, or liver fibrosis and 770,180 controls.28 It is the largest GWAS dataset for analyses of clinical diagnosis of NAFLD. For replication analyses, another NAFLD GWAS dataset was obtained from 11 leading European tertiary liver centres, comprising 1483 biopsied NAFLD cases and 17,781 controls, among which 56% of the patients had NASH and 26% had advanced fibrosis (fibrosis stage 3 or 4).29
As for secondary outcomes, data pertaining to genetic associations with liver fat percentage, including 36,703 participants of European ancestry, were obtained from a GWAS carried out in UK-Biobank (UKB).30 The liver fat percentage was quantified via machine learning of abdominal magnetic resonance images. Excess liver fat (defined as liver fat content >5.5%) was found in 17% of the imaged participants; Liver fat content >20% was found in 1.6% of the participants. Moreover, summary statistics for three liver enzymes were obtained from a GWAS by Pazoki et al. that enrolled 437,438 individuals aged 40–69 years.31 Information on statistical analysis, imputation and quality control measures can be found in the original publications.20,27, 28, 29, 30, 31
Statistical analysis
The inverse-variance weighted method (fixed/random effects) was used to generate an overall estimate of the causal effect of genetically proxied circulating lipid traits on NAFLD and genetically proxied lipid-lowering treatment on NAFLD and liver function. All estimates (odds ratios [ORs] for NAFLD risk and effect estimates [βs] for liver fat and liver enzymes) were scaled up from the individual SNP-level effects on lipid levels to reflect the equivalent of a 1-mmol/L (i.e. LDL-C, 38.7 mg/dL; TG, 88.9 mg/dL; TC, 41.8 mg/dl) change in lipid levels. eQTL data were based on 1-SD changes in gene expression levels for each additional effect allele.
Three fundamental assumptions were behind the MR approach32: 1) Genetic variants and exposure are strongly correlated (“relevance”); 2) Genetic variants are independent of the confounders influencing the relationship between exposure and outcome (“independence”); 3) Genetic variants affect the outcome only through the exposure of interest (“exclusion restriction”) (Figure S1).
To test the relevance assumption, the strength of each genetic variant was assessed with F statistics analysis. Typically, an F statistic of at least 10 indicates no weak instrument bias. Statistical power was calculated using the online tool mRnd to ensure sufficient statistical power (http://cnsgenomics.com/shiny/mRnd/). To validate our selection of drug target genetic variants, positive control analyses were performed with coronary artery disease (CAD) and type 2 diabetes (T2D) as the outcomes. Summary statistics for CAD and T2D were obtained from the Coronary Artery Disease Genome-wide Replication and Meta-analysis plus the Coronary Artery Disease Genetics Consortium (CARDIo-GRAMplusC4D) and a GWAS meta-analysis combining three diabetes datasets (DIAbetes Genetics Replication and Meta-analysis [DIAGRAM], Genetic Epidemiology Research on Aging [GERA], and UKB).33,34
For a drug target that reached significance for risk of NAFLD, colocalisation analysis35 was performed to test the exclusion restriction assumption. It assessed the probability (PP.H4) that SNPs associated with the drug target and NAFLD are shared by the same causal variant at a given locus and the probability (PP.H3) that drug targets and NAFLD are affected by distinct causal variants that are in LD with each other. A posterior probability greater than 0.80 supported a tested configuration.35 Drug targets that strongly colocalised with NAFLD (PP.H4 > 0.80) were considered to be potential target genes.
To determine whether the observed association between drug targets and NAFLD was a direct association, we assessed the relationship between genetically proxied lipid-lowering therapies and previously established risk factors for NAFLD (i.e. body mass index[BMI], waist circumference, T2D and systolic blood pressure) in MR analyses.36 For significant associations, potential mediation effects (the exposure-mediator-outcome pathway) may exist. To assess the direct effect of genetically proxied lipid-lowering therapies on NAFLD risks after adjusting for mediator variables, the “Two-Step Cis-MR” method37 was used. Compared with multivariable MR method, “Two-Step Cis-MR” can attenuate the bias of high LD correlation among genetic variants in cis-MR analysis.37 Indirect effects, the effect of genetically proxied lipid-lowering therapies on NAFLD risk via each potential mediator, and mediated proportions were assessed with the “Product of coefficients” method. Standard errors for the indirect effects were derived with the delta method.38
The heterogeneity and pleiotropy between SNPs were evaluated by Cochran's Q test and the MR-Egger intercept test. For MR sensitivity analysis, Pleiotropy Residual Sum and Outlier (MR-PRESSO) was used to reduce bias caused by correlated horizontal pleiotropy; For uncorrelated horizontal pleiotropy, we applied weighted median and weighted mode methods. To assess whether the removal of a single influential SNP in drug-target proxies influenced the overall estimates of a causal effect, leave-one-out analyses were performed. As the genetic instruments selected as proxies for the drug target were in weak LD (r2 < 0.2), we calculated the LD correlation between genetic variants using LDmatrix Tool (https://ldlink.nci.nih.gov/) and adjusted LD structure in the sensitivity analyses. For significant drug-target MR associations, more stringent LD thresholds (r2 < 0.1, r2 < 0.01 and r2 < 0.001, respectively) were utilised to test the robustness of our observations.
Bonferroni-corrected significance levels of p-value < 0.016 (0.05/3) and p-value < 0.005 (0.05/9) were used to adjust for multiple testing of three lipid traits and nine drug targets, respectively. For other analyses, an observed 2-sided p < 0.05 was considered statistically significant. All aforementioned statistical analyses were conducted using ‘TwoSampleMR’, ‘MendelianRandomization’, ‘coloc’, and ‘TwoStepCisMR’ in R (version 4.1.2).
Ethics
No ethical approval was required for the present study, for all data sources were based on publicly available summary-level data. All these studies were approved by the relevant institutional review committees.
Role of funders
The funders had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Results
Lipid traits and NAFLD risk
Seventy-seven independent SNPs associated with LDL-C, 50 SNPs associated with TG and 80 SNPs associated with TC were identified as instrumental variables for lipid traits (Table S3–S5). Increases in genetically proxied TG levels were nominally associated with an increased risk of NAFLD in the discovery dataset (OR = 1.13 [95% confidence interval (CI), 1.02–1.26]; p = 0.02) (Tables 2 and S6). This finding was close to null in the replication dataset (OR = 1.01 [95% CI, 0.92–1.10]; p = 0.87), in the pooled dataset (OR = 1.06 [95% CI, 0.99–1.13]; p = 0.11) and in the multivariate MR analysis, including TG, LDL-C and TC (OR = 1.18 [95% CI, 0.97–1.44]; p = 0.09). No association was found between either LDL-C or TC and NAFLD.
Table 2.
Mendelian randomisation results of lipid traits with risk of non-alcoholic fatty liver disease.
| Lipid trait | Methods | NAFLD (discovery dataset) |
NAFLD (replication dataset) |
||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| TG | Inverse variance weighted | 1.131 (1.018, 1.256) | 0.02 | 1.007 (0.921, 1.102) | 0.87 |
| Weighted median method | 1.141 (0.959, 1.357) | 0.14 | 0.962 (0.851, 1.087) | 0.54 | |
| Weighted mode method | 1.170 (0.949, 1.443) | 0.15 | 0.965 (0.263, 3.543) | 0.96 | |
| MR-PRESSO | 1.167 (1.016, 1.341) | 0.03 | 0.981(0.876, 1.098) | 0.74 | |
| LDL-C | Inverse variance weighted | 0.967 (0.901, 1.036) | 0.33 | 0.998 (0.908, 1.096) | 0.97 |
| Weighted median method | 1.003 (0.905, 1.112) | 0.95 | 1.006 (0.877, 1.154) | 0.93 | |
| Weighted mode method | 0.996 (0.907, 1.094) | 0.94 | 1.026 (0.779, 1.353) | 0.85 | |
| MR-PRESSO | 0.980 (0.898, 1.069) | 0.65 | 1.125 (0.909, 1.011) | 0.84 | |
| TC | Inverse variance weighted | 0.944 (0.875, 1.018) | 0.13 | 1.016 (0.913, 1.129) | 0.78 |
| Weighted median method | 0.974 (0.853, 1.111) | 0.69 | 1.007 (0.865, 1.173) | 0.93 | |
| Weighted mode method | 0.973 (0.877, 1.079) | 0.60 | 1.008 (0.798, 1.275) | 0.94 | |
| MR-PRESSO | 0.960 (0.868, 1.061) | 0.42 | 1.012 (0.900, 1.138) | 0.84 | |
Abbreviations: LDL-C, low density lipoprotein cholesterol; TG, triglyceride; TC, total cholesterol; SNP, single-nucleotide polymorphisms; CI, confidence interval.
Lipid-lowering drug targets and NAFLD risk
We identified five SNPs as genetic instruments in HMGCR, three SNPs in NPC1L1, 11 SNPs in PCSK9, 15 SNPs in APOB, seven SNPs in ABCG5 and ABCG8, 12 SNPs in LDLR, three SNPs in ANGPTL3, 12 SNPs in APOC3 and 15 SNPs in LPL (Table S7). Except for ANGPTL3, significant associations between genetically proxied drug targets and a decreased risk of CAD were identified in the positive control analyses, ensuring the efficacy of the genetic instruments (Figure S2), which was consistent with previous studies.39,40 LPL is the only drug target associated with a lower T2D risk (OR = 0.64 [95% CI, 0.57–0.71]; p = 3.86 × 10−16) (Figure S2). The F statistics for the respective genetic instruments ranged from 68.0 to 254.3, suggesting that instrument bias was unlikely to affect the analyses. The strength of the genetic instruments for each drug target and the statistical power of the MR analyses are presented in Table S8.
The associations of genetic proxies for the effects of nine lipid-lowering drug classes on NAFLD from two independent datasets were shown in Fig. 2. Genetic mimicry of LPL enhancement equivalent to a 1-mmol/L (88.9 mg/dL) reduction in TG was significantly associated with lower NAFLD risk in the discovery dataset (OR = 0.60 [95% CI 0.50–0.72], p = 2.07 × 10−8) and replication dataset (OR = 0.57 [95% CI 0.39–0.82], p = 3.00 × 10−3). A similar finding was noted for the genetic mimicry of APOC3 inhibition on the protective effect of NAFLD risk in the discovery cohort (OR = 0.85 [95% CI 0.77–0.94], p = 1.00 × 10−3), but this observation was not validated in the replication cohort (OR = 1.23 [95% CI 0.96–1.57], p = 0.11). Other genetic mimicries of drug targets (HMGCR, NPC1L1, PCSK9, APOB, LDLR, ABCG5/ABCG8 and ANGPTL3) were shown to have neutral effects on NAFLD outcomes.
Fig. 2.
Association of genetically proxied drug targets with risk of non-alcoholic fatty liver disease. Forest plot of the association between a 1-mmol/L (LDL-C, 38.7 mg/dL; TG, 88.9 mg/dL) change in the lipid levels of nine lipid-lowering drug targets with NAFLD risk in the discovery dataset (A), and NAFLD risk in the replication dataset (B). Data are represented as odds ratios (ORs) with 95% confidence intervals (error bars). An OR of <1.00 suggests a decreased risk of disease associated with lipid-lowering drug treatment. Associations are significant after correcting for multiple testing (9 genes, p < 0.05/9). Abbreviations: LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; SNP, single-nucleotide polymorphisms; HMGCR, HMG-CoA reductase; NPC1L1, Niemann-Pick C1-like protein 1; PCSK9, proprotein convertase subtilisin/kexin type 9; APOB, Apolipoprotein B-100; ABCG5, ATP Binding Cassette Subfamily G Member 5; ABCG8, ATP Binding Cassette Subfamily G Member 8; LDLR, LDL Receptor; ANGPTL3, angiopoietin-like 3; APOC3, Apolipoprotein C-III; LPL, lipoprotein lipase.
The results of the alternative MR methods were generally consistent (Table S9 and S10). MR-Egger intercept did not find evidence of pleiotropy, which improves causal inferences (Table S11). Besides, these findings were robust in the leave-one-out sensitivity analysis (Table S12). After adjustment of LD correlation between genetic variants (Table S13), the results were consistent with the main findings (Table S14). Additional analysis for the LD threshold with more stringent thresholds (r2 < 0.1, r2 < 0.01 and r2 < 0.001) did not appreciably alter confidence interval widths (Table S15–S18).
Lipid-lowering drug targets and risk of liver function traits
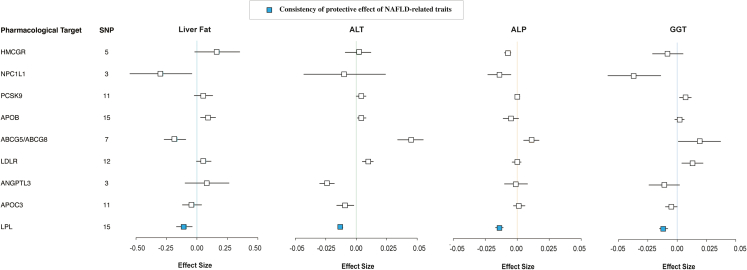
Fig. 3 illustrates the association of genetic proxies for the effects of nine lipid-lowering drug targets on liver function traits. The genetic mimicry of LPL enhancement presented protective effects on liver fat (β = −0.104 [95% CI −0.168, −0.039], p = 1.71 × 10−3), ALT (β = −0.013 [95% CI −0.015, −0.011], p = 8.46 × 10−35), ALP (β = −0.014 [95% CI −0.017, −0.011], p = 1.09 × 10−19), GGT (β = −0.012 [95% CI −0.015, −0.008], p = 1.39 × 10−10) (Table S19). Similar findings were observed in the genetic mimicry of NPC1L1 inhibition. By contrast, APOB, LDLR, and PCSK9 showed adverse effects on liver enzymes and/or liver fat. Genetically proxied inhibition of HMGCR, APOC3, and ANGPTL3 were identified to have neutral effects on liver fat and protective effects on liver enzymes. ABCG5/ABCG8 showed a protective effect on liver fat, but a harmful effect on liver enzymes.
Fig. 3.
Association of genetically proxied drug targets with risk of liver function traits. Forest plot of the association between a 1-mmol/L (LDL-C, 38.7 mg/dL; TG, 88.9 mg/dL) change in the lipid levels of nine lipid-lowering drug targets on the effect of liver fat, ALT, ALP and GGT. Data are represented as effect sizes (β) with 95% confidence intervals (error bars). A β of <0.00 suggests a decreased effect of trait associated with lipid-lowering drug treatment. Blue boxes represent the consistency of the protective effect of the drug target on liver fat and three liver enzymes. Abbreviations: NAFLD, non-alcoholic fatty liver disease; SNP, single-nucleotide polymorphisms; ALT, alanine aminotransferase; ALP, alkaline phosphatase; GGT, γ-glutamyl transferase; TG, triglycerides; LDL-C, low-density lipoprotein cholesterol; HMGCR, HMG-CoA reductase; NPC1L1, Niemann-Pick C1-like protein 1; PCSK9, proprotein convertase subtilisin/kexin type 9; APOB, Apolipoprotein B-100; ABCG5, ATP Binding Cassette Subfamily G Member 5; ABCG8, ATP Binding Cassette Subfamily G Member 8; LDLR, LDL Receptor; ANGPTL3, angiopoietin-like 3; APOC3, Apolipoprotein C-III; LPL, lipoprotein lipase; OR, odds ratio.
Gene expression and NAFLD risk
Because the TG-lowering genetic variants in the LPL gene showed a unique association with a lower NAFLD risk, genetic variants related to LPL expression in whole blood and subcutaneous adipose tissues where the gene was highly expressed were used as instrumental variables for further validation. Findings from a 1-SD increase in blood tissue LPL expression and adipose tissue LPL expression were associated with a lower risk of NAFLD (blood tissue: OR = 0.95 [95% CI, 0.91–0.99]; p = 0.01; adipose tissue: OR = 0.71 [95% CI, 0.58–0.87]; p = 1.20 × 10−3) (Table 3). When more stringent LD thresholds (r2 < 0.1, r2 < 0.01 and r2 < 0.001) were used, the results of LPL expression in adipose tissue remained stable (Table S20 and S21). Moreover, given the inconsistent findings of the association between TG-lowering genetic variants in the APOC3 gene and NAFLD risk, the relationship was further explored using APOC3 expression in liver tissue and blood tissue. This finding was close to null in secondary analyses (Table S22).
Table 3.
Association of LPL expression in the subcutaneous adipose and whole blood tissue with risks of NAFLD and liver function traits.
| Drug target | Trait | OR/effect size (95% CI) | P value |
|---|---|---|---|
| LPL (subcutaneous adipose tissue) | NAFLD | 0.711 (0.579, 0.874) | 1.20 × 10−3 |
| Liver fat | −0.117 (−0.213, −0.021) | 0.017 | |
| ALT | −0.004 (−0.009, 0.001) | 0.090 | |
| ALP | −0.009 (−0.015, −0.003) | 5.00 × 10−3 | |
| GGT | −0.004 (−0.009, 0.000) | 0.049 | |
| LPL (whole blood tissue) | NAFLD | 0.946 (0.906, 0.989) | 0.014 |
| Liver fat | −0.117 (−0.213, −0.021) | 1.17 × 10−4 | |
| ALT | −0.002 (−0.003, −0.001) | 4.01 × 10−4 | |
| ALP | −0.004 (−0.004, −0.003) | 2.17 × 10−11 | |
| GGT | −0.002 (−0.004, −0.001) | 3.73 × 10−3 |
Abbreviations: OR, odds ratio; NAFLD, nonalcoholic fatty liver disease; ALT, alanine aminotransferase; ALP, alkaline phosphatase; GGT, γ-glutamyl transferase.
We further performed colocalisation analyses to identify the probability that genetic variants associated with LPL or APOC3 expression in relevant tissues and NAFLD shared causal SNPs. LPL expression in subcutaneous adipose tissue and NAFLD shared a causal variant (PP.H4 = 0.85) (Table S23), whereas the colocalisation finding of APOC3 expression was poorly identified (liver tissue: PP.H4 = 0.01; blood tissue: PP.H4 = 0.01) (Table S24). A causal variant (rs326) was associated with both LPL expression in adipose tissue and NAFLD within the LPL locus (Figure S3), providing evidence against the association driven by distinct SNPs that are in LD.
Mediation analysis
Given that BMI, waist circumference, T2D and systolic blood pressure are well-established risk factors for NAFLD, they could be mediators underlying the effect of LPL on the NAFLD risk. A two-step MR analysis was performed to investigate the mediating pathway from LPL to NAFLD. Among the four potential mediators, we only identified a causal relationship between LPL and T2D risk (Table S25). The indirect effect of LPL on NAFLD via T2D was 0.95 (95% CI, 0.92–0.99; p = 0.03) (Fig. 4A). After adjusting for T2D, the direct effect (β) of LPL on NAFLD decreased from 0.52 (95% CI, 0.31–0.74) to 0.47 (95% CI, 0.26–0.69), suggesting that lowering the risk of NAFLD derived from LPL enhancement was partially mediated by decreasing the risk of T2D (Table S26). No evidence of heterogeneity was found in the two-step cis-MR analysis (p = 0.31).
Fig. 4.
Mediation analysis of the effect of lipoprotein lipase on non-alcoholic fatty liver disease via potential mediators under a two-step Mendelian randomisation analysis framework. ‘Direct effect’ indicates the effect of LPL on NAFLD risk after adjusting for the mediator (type 2 diabetes [A] or fasting insulin [B]). ‘Indirect effect’ indicates the effect of LPL on NAFLD risk through the mediator (type 2 diabetes [A] or fasting insulin [B]).
Additional analyses were carried out to investigate the potential role of glycaemic traits (i.e. fasting insulin, fasting glucose, and homeostasis model assessment of β-cell function) as mediators in the associations between LPL activation and NAFLD risk. We discovered that lowering fasting insulin levels moderately mediated the association (mediation proportion: 7.40% [95% CI, 1.70%–14.63%], p = 0.03) (Fig. 4B and Table S27 and S28).
Discussion
In this MR analysis of 9,917 NAFLD cases and 787,961 controls, LPL was the only drug target that significantly lowered the risk of NAFLD. This finding was validated by different approaches of constructing genetic instruments (TG-lowering genetic variants in the LPL gene or genetic variants related to LPL gene expression) and two independent NAFLD datasets. Our study provided strong evidence that LPL is a promising drug target for NAFLD. Evidence regarding the effect of lipid traits and eight lipid-lowering drug targets on lowering the NAFLD risk was not found, suggesting that the mechanism of action of LPL in NAFLD risk is independent of its lipid-lowering effects. Mediation analysis showed that decreasing the risk of NAFLD derived from LPL was partially mediated by lowering the risk of T2D, and further analyses of glycaemic traits revealed that the mechanism of action of LPL on NAFLD may be partially through the regulation of insulin levels.
TG may not be recognised as a causal risk factor for NAFLD with the available evidence. Two MR studies investigated the overall effect of lipid traits on NAFLD. Yuan et al.41 reported strong association between genetically indexed TG and increased NAFLD risk (OR = 1.23 [95% CI, 1.15–1.33]; p = 3.08 × 10−8) using the TG GWAS dataset from UKB. However, their finding may lead to potential bias due to overlapping information between two sample datasets. Only the finnGen NAFLD dataset (894 cases and 217,898 non-cases) revealed a positive association between TG and NAFLD risk (OR = 1.22, p = 0.022), according to Xie et al.,42 whereas the three combined datasets revealed no association at all. Consistent with previous findings, we did not find strong piece of evidence to support a positive association in the largest NAFLD pooled dataset at present.
It has been proposed that lipid-lowering drugs may protect against NAFLD by lowering TG levels.43,44 However, only TG-lowering genetic variants in LPL were associated with a lower NAFLD risk in this study; the association between TG-lowering genetic variants in APOC3 and ANGPTL3 with a lower NAFLD risk was not validated. The lack of a causal association between TG, APOC3 and ANGPTL3 with NAFLD suggests that modulating LPL could have physiological effects other than TG metabolism. In recent studies, LPL genetic variants showed metabolic effects different from those of ANGPTL3 genetic variants.40,45 They were found to improve insulin resistance and increase insulin sensitivity. Insulin resistance is a critical mechanism that leads to hepatic steatosis and steatohepatitis, and there is a stepwise increased resistance to insulin-mediated suppression of peripheral lipolysis and hepatic glucose output from steatosis to steatohepatitis.46 Our mediation analysis revealed that LPL activation's protective effect on lowering NAFLD risk was partially mediated by lower fasting insulin levels, implying that improved insulin resistance and lower insulin levels may act as a mediating mechanism in lowering NAFLD risk.
The clinical relevance of LPL and NAFLD has been confirmed by several epidemiological, laboratory and genetic studies. Lower LPL activity is closely linked with the development of NAFLD. An observational study conducted by Maltais et al. showed that 42% of patients with familial chylomicronemia syndrome (inherited LPL deficiency) and 74% of multifactorial chylomicronemia syndrome (functional LPL deficiency) met the criteria for NAFLD.47 Concordantly, Shirakawa et al. found patients with NAFLD but no other metabolic disorders were associated with a significant reduction in LPL mass.48 Enhancing LPL enzymatic activity reduces liver lipogenesis and the risk of liver inflammation. Direct LPL activators, NO-188649 and C10d,50 were found to improve the high-fat diet-induced NAFLD in animal models. LPL was recently identified as a susceptibility locus for NAFLD in a GWAS meta-analysis by Ghodsian et al.,28 and a negative association between LPL expression and NAFLD risk was reported. Our study supported this causal association using drug target MR analysis and further elucidated the potential mediators.
The pharmacological properties of LPL modulation can be observed in various medications, such as fibrates, omega-3 fatty acid, thiazolidinediones and metformin; however, it is not the primary mechanism of action of these drugs.51 Recently, there is growing interest in developing new drugs targeting LPL activation as their central mechanistic effect. Several new approaches, such as C10d and 50F10 by directly activating LPL, are currently being investigated.51 The American Association for the Study of Liver Diseases recommends that any candidate drug for NAFLD should be neutral from a cardiovascular risk perspective or ideally reduce cardiovascular risks.52 In addition to NAFLD risk reduction, our study linked LPL activation to lower risks of common NAFLD comorbidities (T2D, CAD, and dyslipidemia). These findings highlight a significant pleiotropic benefit of LPL activation. Given LPL's superior contribution to metabolic improvement, our findings, in conjunction with previous pharmacological and genetic studies,40,45,51,53 indicate that there is a high clinical interest in the development of LPL-enhancing drugs.
Evidence regarding the beneficial effects of other lipid-lowering drug targets on NAFLD was not found in the present study. In contrast, we discovered that genetic mimicry of APOB inhibition increases the risk of liver fat accumulation and elevated hepatic enzyme levels. Mipomersen is an antisense oligonucleotide drug used to treat homozygous familial hypercholesterolemia by inhibiting apolipoprotein B synthesis. A meta-analysis of 13 RCTs found that mipomersen was associated with increased risks of hepatic steatosis and an increase in liver enzymes, which was consistent with our findings.54 Therefore, pharmacovigilance for adverse effects on liver function among mipomersen users is required.
Some limitations should be considered when interpreting the results of this study. First, genetic variants reflect the effect of lifelong changes in lipid levels on NAFLD risk, and the magnitude of the effect may not be comparable with the short-term effects of lipid-lowering drugs.55 MR analysis is more helpful in determining the direction of associations rather than quantifying the magnitude of the association. Second, original GWAS data are not stratified by certain subtypes (NAFL or NASH). Therefore, stratified analysis was unable to be performed in the present study. It remains a research subject and should be considered when specific datasets become available in the future. Third, environmental responses to genetic risk of metabolic diseases may bias effect estimates.55 Fourth, GWAS data for liver fat and liver enzymes from UKB may lead to potentially “healthy volunteer” bias and may restrict the generalizability of our results. Fifth, our study only predicts the on-target effects of specific drug targets, and these models do not estimate potential off-target effects. Sixth, horizontal pleiotropy cannot be completely excluded, although various sensitivity analyses were performed to test the assumptions of MR analyses. Last, since our findings were limited to individuals of European ancestry, these findings are not necessarily valid for other ethnic groups.
In summary, this study does not support lipid traits (i.e. TG, LDL-C and TC) as causal risk factors for NAFLD, and the beneficial effect of the seven-class lipid-lowering drugs (i.e. statins, ezetimibe, proprotein convertase subtilisin/kexin type 9 inhibitors, bile acid sequestrants, angiopoietin-like 3 inhibitors and antisense oligonucleotide targeting apolipoprotein C-III mRNA) on NAFLD was not found. Larger GWAS datasets and additional potentially associated genetic variants are required to validate this finding. Moreover, this study demonstrates that LPL is a promising candidate drug target for NAFLD. Part of the mechanism may be through the regulation of insulin-glucose metabolism other than lipid metabolism. The underlying mechanisms should be elucidated in further research, and the role of LPL activators in NAFLD in basic or even clinical trials might be worth assessing.
Contributors
All authors read and approved the final version of the manuscript. Z.L. and B.Z. proposed the idea, performed the MR analyses and drafted the manuscript. Q.L., E.Z., Z.M., H.Z., S.G., Y.C., Z.T., C.W., J.P., L.D., B.G., J.L., and Y.H. checked the integrity and plausibility of data analysis. H.X. and Y.W. revised the manuscript and was responsible for the integrity of data acquisition and statistical analyses. Z.L., B.Z., and Y.W. verified the underlying data.
Data sharing statement
All data are publicly available. Detailed information for these datasets is summarised in Table S2.
Declaration of interests
Nothing to disclosure.
Acknowledgements
This work was supported by the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS, grant number: 2021-I2M-C&T-A-010) and Capital’s Funds for Health Improvement and Research (CFH 2022-4-4037). Dr. Zhang holds a State Scholarship Fund from China Scholarship Council (No. 201806210439) and a fund from Capital’s Funds for Health Improvement and Research (CFH 2022-4-4037). Dr. Wu holds a fund from Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS, grant number: 2021-I2M-C&T-A-010). The authors thank the participants and investigators for providing publicly available summary statistics. The images used in the figure were created by Figdraw. The authors would like to thank Enago for the language polishing.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104543.
Contributor Information
Haiyan Xu, Email: xuhaiyan@fuwaihospital.org.
Yongjian Wu, Email: wuyongjian@fuwaihospital.org.
Appendix A. Supplementary data
References
- 1.Estes C., Razavi H., Loomba R., Younossi Z., Sanyal A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67(1):123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell E.E., Wong V.W., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397(10290):2212–2224. doi: 10.1016/S0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 3.Jarvis H., Craig D., Barker R., et al. Metabolic risk factors and incident advanced liver disease in non-alcoholic fatty liver disease (NAFLD): a systematic review and meta-analysis of population-based observational studies. PLoS Med. 2020;17(4) doi: 10.1371/journal.pmed.1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.I., Lee H.W., Lee K.S., Lee H.S., Park J.Y. Effects of statin use on the development and progression of nonalcoholic fatty liver disease: a nationwide nested case-control study. Am J Gastroenterol. 2021;116(1):116–124. doi: 10.14309/ajg.0000000000000845. [DOI] [PubMed] [Google Scholar]
- 6.Brunt E.M., Kleiner D.E., Carpenter D.H., et al. NAFLD: reporting histologic findings in clinical practice. Hepatology. 2021;73(5):2028–2038. doi: 10.1002/hep.31599. [DOI] [PubMed] [Google Scholar]
- 7.Rinella M.E., Tacke F., Sanyal A.J., Anstee Q.M. Report on the AASLD/EASL joint workshop on clinical trial endpoints in NAFLD. J Hepatol. 2019;71(4):823–833. doi: 10.1016/j.jhep.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 8.Sanyal A.J., Neuschwander-Tetri B.A., Tonascia J. End points must be clinically meaningful for drug development in nonalcoholic fatty liver disease. Gastroenterology. 2016;150(1):11–13. doi: 10.1053/j.gastro.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Walker V.M., Davey Smith G., Davies N.M., Martin R.M. Mendelian randomization: a novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol. 2017;46(6):2078–2089. doi: 10.1093/ije/dyx207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arsenault B.J. From the garden to the clinic: how Mendelian randomization is shaping up atherosclerotic cardiovascular disease prevention strategies. Eur Heart J. 2022;43(42):4447–4449. doi: 10.1093/eurheartj/ehac394. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt A.F., Finan C., Gordillo-Marañón M., et al. Genetic drug target validation using Mendelian randomisation. Nat Commun. 2020;11(1):3255. doi: 10.1038/s41467-020-16969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill D., Georgakis M.K., Koskeridis F., et al. Use of genetic variants related to antihypertensive drugs to inform on efficacy and side effects. Circulation. 2019;140(4):270–279. doi: 10.1161/CIRCULATIONAHA.118.038814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reay W.R., Cairns M.J. Advancing the use of genome-wide association studies for drug repurposing. Nat Rev Genet. 2021;22(10):658–671. doi: 10.1038/s41576-021-00387-z. [DOI] [PubMed] [Google Scholar]
- 14.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 15.Sabatine M.S., Giugliano R.P., Keech A.C., et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376(18):1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 16.Williams D.M., Finan C., Schmidt A.F., Burgess S., Hingorani A.D. Lipid lowering and Alzheimer disease risk: a mendelian randomization study. Ann Neurol. 2020;87(1):30–39. doi: 10.1002/ana.25642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarmolinsky J., Bull C.J., Vincent E.E., et al. Association between genetically proxied inhibition of HMG-CoA reductase and epithelial ovarian cancer. JAMA. 2020;323(7):646–655. doi: 10.1001/jama.2020.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison S.C., Holmes M.V., Burgess S., et al. Genetic association of lipids and lipid drug targets with abdominal aortic aneurysm: a meta-analysis. JAMA Cardiol. 2018;3(1):26–33. doi: 10.1001/jamacardio.2017.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrivankova V.W., Richmond R.C., Woolf B.A.R., et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA. 2021;326(16):1614–1621. doi: 10.1001/jama.2021.18236. [DOI] [PubMed] [Google Scholar]
- 20.Willer C.J., Schmidt E.M., Sengupta S., et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mach F., Baigent C., Catapano A.L., et al. 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. 2020;41(1):111–188. doi: 10.1093/eurheartj/ehz455. [DOI] [PubMed] [Google Scholar]
- 22.Grundy S.M., Stone N.J., Bailey A.L., et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139(25):e1082–e1143. doi: 10.1161/CIR.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borén J., Taskinen M.R., Björnson E., Packard C.J. Metabolism of triglyceride-rich lipoproteins in health and dyslipidaemia. Nat Rev Cardiol. 2022;19(9):577–592. doi: 10.1038/s41569-022-00676-y. [DOI] [PubMed] [Google Scholar]
- 24.Ridker P.M. LDL cholesterol: controversies and future therapeutic directions. Lancet. 2014;384(9943):607–617. doi: 10.1016/S0140-6736(14)61009-6. [DOI] [PubMed] [Google Scholar]
- 25.Ross S., D'Mello M., Anand S.S., et al. Effect of bile acid sequestrants on the risk of cardiovascular events: a mendelian randomization analysis. Circ Cardiovasc Genet. 2015;8(4):618–627. doi: 10.1161/CIRCGENETICS.114.000952. [DOI] [PubMed] [Google Scholar]
- 26.Rosoff D.B., Bell A.S., Jung J., Wagner J., Mavromatis L.A., Lohoff F.W. Mendelian randomization study of PCSK9 and HMG-CoA reductase inhibition and cognitive function. J Am Coll Cardiol. 2022;80(7):653–662. doi: 10.1016/j.jacc.2022.05.041. [DOI] [PubMed] [Google Scholar]
- 27.The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghodsian N., Abner E., Emdin C.A., et al. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep Med. 2021;2(11) doi: 10.1016/j.xcrm.2021.100437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anstee Q.M., Darlay R., Cockell S., et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol. 2020;73(3):505–515. doi: 10.1016/j.jhep.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Haas M.E., Pirruccello J.P., Friedman S.N., et al. Machine learning enables new insights into genetic contributions to liver fat accumulation. Cell Genom. 2021;1(3) doi: 10.1016/j.xgen.2021.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pazoki R., Vujkovic M., Elliott J., et al. Genetic analysis in European ancestry individuals identifies 517 loci associated with liver enzymes. Nat Commun. 2021;12(1):2579. doi: 10.1038/s41467-021-22338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emdin C.A., Khera A.V., Kathiresan S. Mendelian randomization. JAMA. 2017;318(19):1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 33.Xue A., Wu Y., Zhu Z., et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9(1):2941. doi: 10.1038/s41467-018-04951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikpay M., Goel A., Won H.H., et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giambartolomei C., Vukcevic D., Schadt E.E., et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5) doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z., Zhang Y., Graham S., et al. Causal relationships between NAFLD, T2D and obesity have implications for disease subphenotyping. J Hepatol. 2020;73(2):263–276. doi: 10.1016/j.jhep.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolf B., Zagkos L., Gill D. TwoStepCisMR: a novel method and R package for attenuating bias in cis-mendelian randomization analyses. Genes. 2022;13(9) doi: 10.3390/genes13091541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson E. Multivariable mendelian randomization and mediation. Cold Spring Harb Perspect Med. 2021;11(2) doi: 10.1101/cshperspect.a038984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson T.G., Leyden G.M., Wang Q., et al. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol. 2022;20(2) doi: 10.1371/journal.pbio.3001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q., Oliver-Williams C., Raitakari O.T., et al. Metabolic profiling of angiopoietin-like protein 3 and 4 inhibition: a drug-target Mendelian randomization analysis. Eur Heart J. 2021;42(12):1160–1169. doi: 10.1093/eurheartj/ehaa972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan S., Chen J., Li X., et al. Lifestyle and metabolic factors for nonalcoholic fatty liver disease: mendelian randomization study. Eur J Epidemiol. 2022;37(7):723–733. doi: 10.1007/s10654-022-00868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie J., Huang H., Liu Z., et al. The associations between modifiable risk factors and nonalcoholic fatty liver disease: a comprehensive Mendelian randomization study. Hepatology. 2022;77(3):949–964. doi: 10.1002/hep.32728. [DOI] [PubMed] [Google Scholar]
- 43.Mato J.M., Alonso C., Noureddin M., Lu S.C. Biomarkers and subtypes of deranged lipid metabolism in non-alcoholic fatty liver disease. World J Gastroenterol. 2019;25(24):3009–3020. doi: 10.3748/wjg.v25.i24.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das Pradhan A., Glynn R.J., Fruchart J.C., et al. Triglyceride lowering with pemafibrate to reduce cardiovascular risk. N Engl J Med. 2022;387(21):1923–1934. doi: 10.1056/NEJMoa2210645. [DOI] [PubMed] [Google Scholar]
- 45.Lotta L.A., Stewart I.D., Sharp S.J., et al. Association of genetically enhanced lipoprotein lipase-mediated lipolysis and low-density lipoprotein cholesterol-lowering alleles with risk of coronary disease and type 2 diabetes. JAMA Cardiol. 2018;3(10):957–966. doi: 10.1001/jamacardio.2018.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanyal A.J., Campbell-Sargent C., Mirshahi F., et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120(5):1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 47.Maltais M., Brisson D., Gaudet D. Non-alcoholic fatty liver in patients with chylomicronemia. J Clin Med. 2021;10(4) doi: 10.3390/jcm10040669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirakawa T., Nakajima K., Yatsuzuka S., et al. The role of circulating lipoprotein lipase and adiponectin on the particle size of remnant lipoproteins in patients with diabetes mellitus and metabolic syndrome. Clin Chim Acta. 2015;440:123–132. doi: 10.1016/j.cca.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 49.Kusunoki M., Tsutsumi K., Iwata K., et al. NO-1886 (ibrolipim), a lipoprotein lipase activator, increases the expression of uncoupling protein 3 in skeletal muscle and suppresses fat accumulation in high-fat diet-induced obesity in rats. Metabolism. 2005;54(12):1587–1592. doi: 10.1016/j.metabol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Geldenhuys W.J., Caporoso J., Leeper T.C., et al. Structure-activity and in vivo evaluation of a novel lipoprotein lipase (LPL) activator. Bioorg Med Chem Lett. 2017;27(2):303–308. doi: 10.1016/j.bmcl.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geldenhuys W.J., Lin L., Darvesh A.S., Sadana P. Emerging strategies of targeting lipoprotein lipase for metabolic and cardiovascular diseases. Drug Discov Today. 2017;22(2):352–365. doi: 10.1016/j.drudis.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanyal A.J., Friedman S.L., McCullough A.J., Dimick-Santos L. Challenges and opportunities in drug and biomarker development for nonalcoholic steatohepatitis: findings and recommendations from an American Association for the Study of Liver Diseases-U.S. Food and Drug Administration Joint Workshop. Hepatology. 2015;61(4):1392–1405. doi: 10.1002/hep.27678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ference B.A., Kastelein J.J.P., Ray K.K., et al. Association of triglyceride-lowering LPL variants and LDL-C-lowering LDLR variants with risk of coronary heart disease. JAMA. 2019;321(4):364–373. doi: 10.1001/jama.2018.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fogacci F., Ferri N., Toth P.P., Ruscica M., Corsini A., Cicero A.F.G. Efficacy and safety of mipomersen: a systematic review and meta-analysis of randomized clinical trials. Drugs. 2019;79(7):751–766. doi: 10.1007/s40265-019-01114-z. [DOI] [PubMed] [Google Scholar]
- 55.Holmes M.V., Richardson T.G., Ference B.A., Davies N.M., Davey Smith G. Integrating genomics with biomarkers and therapeutic targets to invigorate cardiovascular drug development. Nat Rev Cardiol. 2021;18(6):435–453. doi: 10.1038/s41569-020-00493-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.