Mass photometry is a technique that is less than five years old, yet it has already experienced widespread laboratory adoption, is sold commercially, and has been involved in hundreds of publications. This nondestructive and label-free technique has unprecedented sensitivity for probing the mass of molecules, which is valuable for studying nucleic acids, single membrane proteins, or protein– protein interactions. Despite its youth, mass photometry relies on the well-known and extensively applied concept of light scattering, which dates to the 17th century. Mass photometry makes use of clever ways to circumvent the ubiquitous problem of single molecules producing barely detectable scattering signals, and manages to accurately probe the mass of molecules that can hide in plain sight.
SINGLE MOLECULE AND UNIVERSAL ANALYSIS: THEBEST OF BOTH WORLDS
On one hand, single molecules have been detectable since the late 1980s using light microscopes and fluorescent imaging. The advantage of this technique is its specificity, which permits detection of only the molecules of interest via refined fluorescent dyes and optimized optronics. However, a main drawback of this approach is its nonuniversality. It cannot be used to visualize the majority of objects, either because they are not themselves fluorescent or because they cannot be marked by fluorescent probes.
On the other hand, other techniques can be used for universal molecular characterization. Mass spectroscopy is both universal and specific, and scattering-based techniques, including X-ray crystallography and electron microscopy, are suitable for structural analysis of very tiny objects. Light scattering has proved its usefulness and sensitivity for the study of biological objects, but it requires either powerful amplification (Graciani et al., 2022) or scattering markers (Choi et al., 2019) to function at all.
While light scattering is in theory applicable to all biological structures, its ultra-low intensity has remained a practical obstacle for experimentalists. For instance, while gold is an excellent scattering material and has been used in the design of scattering-based microscopes (Lindfors et al., 2004), proteins typically produce a signal that is weaker by a factor of 10−6. It was only in 2009 that Philipp Kukura developed a technique for using scattering microscopy to examine biologically relevant objects (Kukura et al., 2009). Later, this technique would be refined into interferometric scattering microscopy or iSCAT (Ortega-Arroyo and Kukura, 2012).
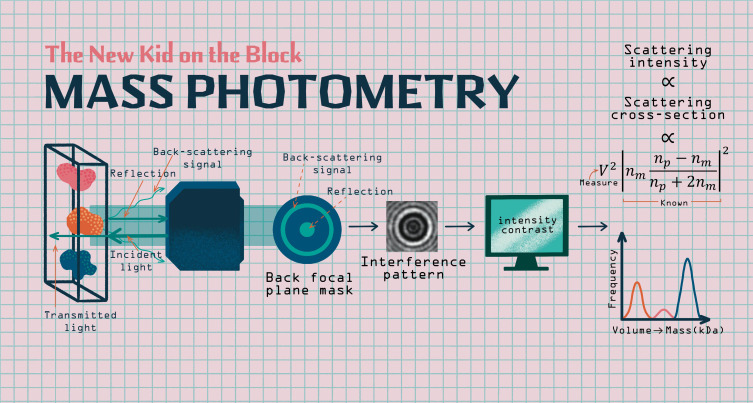
The concept is straightforward: coherent light illuminates a sample through a microscope objective. While the light transmitted through the sample is lost, a small fraction is reflected back from the sample coverslip. An even smaller fraction is collected as backscattering from the molecules sitting close to the lower interface of the coverslip. These two radiations interfere with one another, yielding a dim interference pattern that can be visualized on camera. Given the low intensity of the signal, this technique requires systematic noise reduction corrections.
A NEEDLE IN A HAYSTACK
How can this noise barrier be overcome? The reflections from the coverslip interface constitute a background illumination, which is no different in wavelength than the backscattering signal from the molecules under study. Consequently, color filters—such as those used in fluorescent microscopy—are not useful. Fortunately, the two radiation sources differ in their angular emission profiles and can therefore be spatially filtered. When placed at the back focal plane of the objective, a partially mirrored mask attenuates the background—which propagates collinearly with the illumination—and transmits the backscattering, which is emitted at a higher angle. Unlike a dark-field microscope, both radiations must remain phase-locked and not be eliminated entirely, since they must still be able to interfere with each other (Liebel et al., 2017).
For small objects, such as single proteins, the interference signal is barely noticeable. Additional software-based background removal methods are necessary, as is a comparison between successive frames designed to identify minute changes in local contrast. Although difficult to detect, predicting the intensity of the scattering signal is simple: it is proportional to the scattering cross-section of the object or, more precisely, the product of its volume squared with a coefficient relative to the contrast in refractive indexes between the object and its environment. Using the methods mentioned above, in 2018, Kukura’s group carefully characterized the distribution of refractive indexes and the densities of thousands of proteins, and were therefore able to infer for the first time the volume—and thus the mass—of single biological macromolecules from their light scattering signal alone (Young et al., 2018).
A WIDE RANGE OF APPLICATIONS
This label-free, in-solution technique has a wide range of potential applications. Protein–protein interactions, for example, have been recently quantified using mass photometry (Soltermann et al., 2020). By quantifying the relative abundance of bound and unbound species, the purity, stoichiometry, and binding affinity were all retrieved in-solution with no labeling required. Kinetic rate constants were also obtained by this technique.
iSCAT applications have been further developed to study membrane organization (Sezgin et al., 2017), and mass photometry of membrane proteins was performed in 2021 (Olerinyova et al., 2021). By showing that distinguishing between different oligomeric and functional states of membrane proteins is possible in various mimetic systems, this work showed that mass photometry shows great promise for the in vitro study of membrane proteins. Moreover, this has many ramifications to other biological systems. Other applications include protein oligomerization, other biomolecular interactions, macromolecular assemblies, as well as assessments of homogeneity, structural integrity, or activity quantification. We envision a bright future for this technique, since it has consistently proven to be a valuable complementary tool for the characterization of single molecules.
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1A3B1071354 to T.-Y.Y.).
Footnotes
AUTHOR CONTRIBUTIONS
G.G. and T-Y.Y. wrote the paper.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Choi H.K., Min D., Kang H., Shon M.J., Rah S.H., Kim H.C., Jeong H., Choi H.J., Bowie J.U., Yoon T.Y. Watching helical membrane proteins fold reveals a common N-to-C-terminal folding pathway. Science. 2019;366:1150–1156. doi: 10.1126/science.aaw8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciani G., King J.T., Amblard F. Cavity-amplified scattering spectroscopy reveals the dynamics of proteins and nanoparticles in quasi-transparent and miniature samples. ACS Nano. 2022;16:16796–16805. doi: 10.1021/acsnano.2c06471. [DOI] [PubMed] [Google Scholar]
- Kukura P., Ewers H., Müller C., Renn A., Helenius A., Sandoghdar V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Methods. 2009;6:923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- Liebel M., Hugall J.T., van Hulst N.F. Ultrasensitive label-free nanosensing and high-speed tracking of single proteins. Nano Lett. 2017;17:1277–1281. doi: 10.1021/acs.nanolett.6b05040. [DOI] [PubMed] [Google Scholar]
- Lindfors K., Kalkbrenner T., Stoller P., Sandoghdar V. Detection and spectroscopy of gold nanoparticles using supercontinuum white light confocal microscopy. Phys. Rev. Lett. 2004;93:037401. doi: 10.1103/PhysRevLett.93.037401. [DOI] [PubMed] [Google Scholar]
- Olerinyova A., Sonn-Segev A., Gault J., Eichmann C., Schimpf J., Kopf A.H., Rudden L.S.P., Ashkinadze D., Bomba R., Frey L., et al. Mass photometry of membrane proteins. Chem. 2021;7:224–236. doi: 10.1016/j.chempr.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Arroyo J., Kukura P. Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Phys. Chem. Chem. Phys. 2012;14:15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- Sezgin E., Levental I., Mayor S., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltermann F., Foley E.D., Pagnoni V., Galpin M., Benesch J.L., Kukura P., Struwe W.B. Quantifying protein-protein interactions by molecular counting with mass photometry. Angew. Chem. Int. Ed. Engl. 2020;59:10774–10779. doi: 10.1002/anie.202001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G., Hundt N., Cole D., Fineberg A., Andrecka J., Tyler A., Olerinyova A., Ansari A., Marklund E.G., Collier M.P., et al. Quantitative mass imaging of single biological macromolecules. Science. 2018;360:423–427. doi: 10.1126/science.aar5839. [DOI] [PMC free article] [PubMed] [Google Scholar]