Abstract
Transcription factor NRF2 (NF-E2-related factor 2) is a master regulator of cellular responses against environmental stresses. NRF2 induces expression of detoxification and antioxidant enzymes and suppresses inductions of pro-inflammatory cytokine genes. KEAP1 (Kelch-like ECH-associated protein 1) is an adaptor subunit of CULLIN 3 (CUL3)-based E3 ubiquitin ligase. KEAP1 regulates the activity of NRF2 and acts as a sensor for oxidative and electrophilic stresses. NRF2 has been found to be activated in many types of cancers with poor prognosis. Therapeutic strategies to control NRF2-overeactivated cancers have been considered not only by targeting cancer cells with NRF2 inhibitors or NRF2 synthetic lethal chemicals, but also by targeting host defense with NRF2 inducers. Understanding precise molecular mechanisms how the KEAP1-NRF2 system senses and regulates the cellular response is critical to overcome intractable NRF2-activated cancers.
Keywords: cancer therapy, defense system, KEAP1, NRF2, stress sensing
INTRODUCTION
Our body is equipped with defense systems that upregulate expression of cytoprotective enzymes and NRF2 (NF-E2-related factor 2) is the central player in the inducible expression of cellular defense enzymes (Ishii et al., 2000; Itoh et al., 1997). NRF2 belongs to the CNC (cap-n-collar) subfamily of basic region-leucine zipper (bZIP)-type transcription factors (Itoh et al., 1995). NRF2 dimerizes with one of the small MAF (musculoaponeurotic fibrosarcoma) proteins (sMAF). The NRF2-sMAF heterodimer binds to antioxidant or electrophile response element (ARE/EpRE), which is now collectively referred to as the CNC-sMaf binding element (CsMBE), located in the regulatory regions of the cytoprotective enzyme genes (Friling et al., 1990; Otsuki et al., 2016; Rushmore et al., 1991).
Kelch-like ECH-associated protein 1 (KEAP1) was identified as a negative regulator of NRF2 (Itoh et al., 1999). KEAP1 acts as a substrate-recognition subunit of the CULLIN 3 (CUL3)-based E3 ubiquitin ligase complex that specifically targets NRF2 (Itoh et al., 2003; Kobayashi et al., 2004). In the absence of stress stimuli, NRF2 is efficiently ubiquitinated by the KEAP1-CUL3 E3 ligase and degraded rapidly through the proteasome pathway, such that cellular NRF2 activity is constitutively suppressed in unstressed conditions. Upon exposure to oxidative or electrophilic stresses, KEAP1 loses the ability to ubiquitinate NRF2, allowing NRF2 to accumulate in the nucleus and activate transcription of its target genes. It should be noted that recent studies expand extraordinary our knowledge about the KEAP1-NRF2 system and its relation to intractable cancers with massive NRF2 activation.
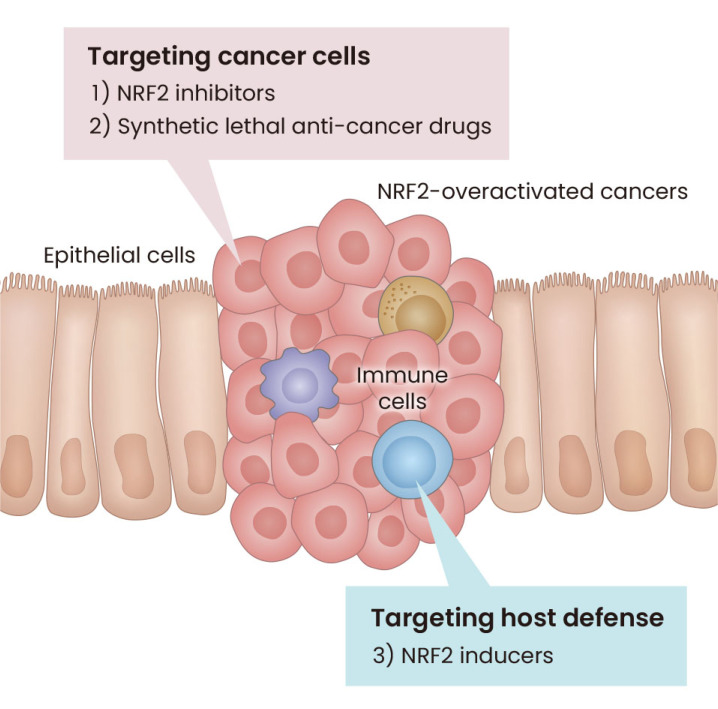
It has been shown that in many types of cancers NRF2 activity is overactivated with high frequency (Padmanabhan et al., 2006; Romero et al., 2017; Shibata et al., 2008). These NRF2 overactivations are brought by somatic mutations, epigenomic errors, exon skipping, etc., which destroy protein-protein interactions (PPI) of NRF2 and KEAP1 and make NRF2 stable (Adam et al., 2011; DeNicola et al., 2011; Goldstein et al., 2016; Komatsu et al., 2010; Ooi et al., 2011; 2013; Padmanabhan et al., 2006; Shibata et al., 2010; Wang et al., 2008). The NRF2-overactivated cancers manifest strong resistance to currently standard therapies of cancers and brings about poor prognosis. Therefore, as summarized in Fig. 1, therapeutic strategies to control malignant NRF2-overeactivated cancers have been designed and developed. These approaches include not only targeting cancer cells with NRF2 inhibitors or NRF2 synthetic lethal chemicals, but also by targeting host defense with NRF2 inducers (Hayashi et al., 2020; Hiramoto et al., 2014; Satoh et al., 2010). Of note, these strategies are supported by in-depth understanding the mechanistic basis of the KEAP1-NRF2 system function as well as understanding the system’s contribution to the fundamental cellular defense machinery. In this review, we especially introduce molecular mechanisms underpinning how this system senses various environmental stresses and potentiates our defense systems.
Fig. 1. Therapeutic strategies to control NRF2-overeactivated cancers.
In the tumor, NRF2 inhibitors help suppress tumor growth and metastasis by decreasing NRF2 protein levels. In addition, synthetic lethality anti-cancer drugs are transformed into strongly active drugs by NRF2-targeted drug-metabolizing enzymes. In contrast, activation of NRF2 in the microenvironment inhibits the progression of NRF2-overactivated malignant tumors. NRF2, NF-E2-related factor 2.
EXPANSION OF NRF2 TARGET GENES
Various NRF2 target genes have been identified through gene expression profiling analysis and chromatin immunoprecipitation (ChIP) analysis (Chorley et al., 2012; Hirotsu et al., 2012). Of the target genes, major group is the cytoprotective genes encoding anti-oxidative stress enzymes, detoxifying enzymes and enzymes related to metabolic reprogramming in cancer. For instance, heme oxygenase 1 (HO-1) gene is one of the representative NRF2 target genes, which acts for anti-oxidative stress response. Recent analyses of peritoneal macrophages from HO-1-DsRed knock-in mice lacking NRF2 demonstrate that electrophilic NRF2 inducers, such as diethylmaleate (DEM) and 1-[2-cyano-3,12,28-triox-ooleana-1,9(11)-dien-28-yl]-1H-imidazole (CDDO-Im), activate expression of HO-1 gene in an NRF2-dependent manner, while HO-1 substrate hemin induces expression of HO-1 gene in an NRF2-independent manner (Zhang et al., 2021). NRF2 also regulates glucose metabolism genes, which becomes important in the metabolic reprogramming in cancer (Mitsuishi et al., 2012). In addition, NRF2 has also been shown to attenuate inflammation by suppressing the induction of pro-inflammatory cytokine gene transcription (Kobayashi et al., 2016; Suzuki et al., 2020a).
These NRF2 target genes are regulated by the heterodimer composed of NRF2 and one of the sMAF proteins, e.g., MAFF, MAFG, or MAFK, through CsMBE. Recently, Katsuoka et al. (2019) provided direct and compelling evidence for the NRF2-MAFG heterodimer-mediated transcription of NRF2-target genes. By introducing a tethered NRF2-MAFG heterodimer using a flexible linker peptide into the cells lacking all three sMaf proteins, which ensured no interference from other endogenous CNC-sMAF heterodimers or sMAF homodimers, they showed that the NRF2-MAFG heterodimer acts as a dominant transcriptional activator of NRF2-dependent gene transcription through CsMBE (Katsuoka et al., 2019). Structural analysis conducted by Sengoku et al. (2022), strongly supports this notion. Their crystal structure analysis revealed that the NRF2-MAFG heterodimer has approximately 200-fold stronger affinity for CsMBE than do the canonical bZIP proteins, such as activator protein 1 (AP-1), and this is realized by utilization of the CNC motif in NRF2 protein that forms extensive contact with the DNA backbone phosphates (Sengoku et al., 2022).
FLOODGATE MODEL FOR THE KEAP1-MEDIATED ACTIVATION OF NRF2
KEAP1 interacts with the Neh2 (NRF2-ECH homology domain 2) degron domain of NRF2 (Itoh et al., 1999). The stoichiometry of KEAP1 and NRF2 within the KEAP1-NRF2 complex is 2:1, which is shown by means of isothermal calorimetry analysis (Tong et al., 2006a), nuclear magnetic resonance (NMR) titration analyses (Horie et al., 2021; Tong et al., 2006a), and an analytical centrifugation analysis (Iso et al., 2016). A single NRF2 protein binds to a KEAP1 homodimer using a high-affinity ETGE motif and a low-affinity DLGex motif. This two-site recognition/binding mechanism between the NRF2 Neh2 domain and KEAP1 homodimer is critical for the efficient ubiquitination of NRF2 (Tong et al., 2006a; McMahon et al., 2010; Suzuki et al., 2015).
KEAP1 mainly localize in the cytoplasm loosely attaching perinuclear cytoskeleton network (Watai et al., 2007). Oxidative or electrophilic stresses provoke nuclear accumulation of NRF2 without altering the cytoplasmic localization of KEAP1 (Watai et al., 2007). It should be noted that these stresses appear not to trigger dissociation of the KEAP1-NRF2 complex (Kobayashi et al., 2006), but rather repress strongly the KEAP1-based E3 ubiquitin ligase activity. Thus, reduction of the NRF2 ubiquitination leads to the stabilization and nuclear accumulation of de novo synthesized NRF2 (Kobayashi et al., 2006).
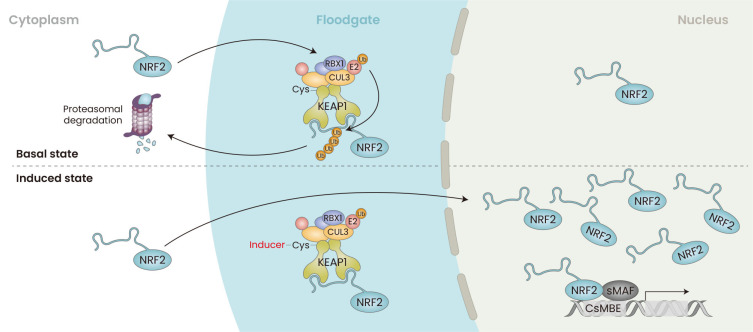
Quantitative biochemical studies revealed that in the basal state, NRF2 protein is maintained at a level significantly lower than the levels of KEAP1. When challenged with the electrophilic NRF2 inducer DEM, abundance and localization of KEAP1 protein does not change, whereas nuclear NRF2 protein rises to the level considerably higher than that of KEAP1 (Iso et al., 2016). Thus, in the basal uninduced conditions the KEAP1-based E3 ubiquitin ligase acts as a “floodgate” that degrades NRF2 efficiently in collaboration with the proteasome system (Fig. 2) (Iso et al., 2016). On the contrary, in response to oxidative or electrophilic stimuli, i.e., in response to inducers, the KEAP1-CUL3 complex loses the floodgate function, i.e., the ubiquitin ligase activity, and NRF2 accumulates in the nucleus and activates the target gene expression (Iso et al., 2016; Suzuki et al., 2017; Yamamoto et al., 2018).
Fig. 2. Floodgate Model for the KEAP1-mediated Activation of NRF2.
In the basal state, the ubiquitin ligase KEAP1-CUL3 complex acts as a floodgate and ubiquitinates NRF2. Ubiquitinated NRF2 degrades through the proteasome system. In response to oxidative and electrophilic stimuli, NRF2 accumulates significantly in the nucleus. These stimuli do not affect the abundance and subcellular localization of KEAP1 and CUL3. NRF2 dimerizes with one of the small MAF proteins (sMAF) to form an NRF2-sMAF heterodimer that recognizes the CNC-sMAF binding element (CsMBE) and activates target gene expression. KEAP1, Kelch-like ECH-associated protein 1; NRF2, NF-E2-related factor 2; CUL3, CULLIN 3; CNC, cap-n-collar.
CYSTEINE CODE FOR THE KEAP1 SENSING OF ELECTROPHILIC NRF2 INDUCERS
A variety of chemical NRF2 inducers are reported, and the majority are electrophiles that react with nucleophilic thiols, including cysteine sulfhydryl groups (Dinkova-Kostova and Talaly, 2008). Specific patterns of KEAP1 cysteine modifications by individual chemical NRF2 inducers have been identified by mass spectrometry analyses (Eggler et al., 2005; Kobayashi et al., 2009) and these modifications affect the KEAP1-based E3 ubiquitin ligase activity.
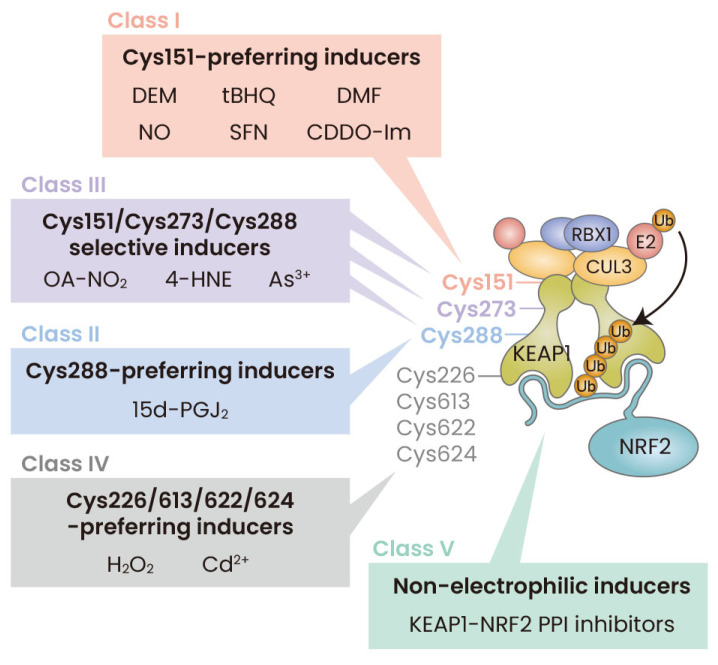
Functional significance of the cysteine residues in KEAP1 has been examined by means of site-directed mutagenesis (Saito et al., 2016; Suzuki et al., 2019; Takaya et al., 2012; Yamamoto et al., 2008; Zhang and Hannink, 2003). The chemicals triggering NRF2 activation appear to modify KEAP1 cysteine residues in a multiple pattern. This unique use of cysteine residues as sensors has led to the “cysteine code” concept (Suzuki et al., 2013). The concept explains the unique feature of the KEAP1-NRF2 system, which responds to a diverse array of chemical and oxidative insults (Suzuki et al., 2013). Our recent results revealed molecular basis as to how Keap1 employs multiple cysteine residues as sensors enabling activations of various NRF2-mediated cytoprotective responses (Suzuki et al., 2019). Based on the analyses, we classified the Nrf2 inducers into five classes (Fig. 3), which are going to be introduced below.
Fig. 3. Model for the multiple stress-sensing mechanisms acting through KEAP1.
There are five classes of NRF2 inducers: Class I, Cys151-preferring; Class II, Cys288-preferring; Class III, Cys151/Cys273/Cys288-selective; Class IV, Cys226/613/622/624-preferring; and Class V, Non-electrophilic KEAP1-NRF2 PPI inhibitors. Chemicals representing each class are shown. KEAP1, Kelch-like ECH-associated protein 1; NRF2, NF-E2-related factor 2; DEM, diethylmaleate; tBHQ, tert-butyl hydroquinone; DMF, dimethylfumarate; NO, nitric oxide; SFN, sulforaphane; CDDO-Im, 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]-imidazol; OA-NO2, nitro-octadec-9-enoic acid; 4-HNE, 4-hydroxy-nonenal; 15d-PGJ2, 15-deoxy-∆12,14-prostaglandin J2; H2O2, hydrogen peroxide; CUL3, CULLIN 3; PPI, protein-protein interactions.
Class I; KEAP1-Cys151-preferring inducers
The importance of KEAP1-Cys151 as a sensor has been verified by means of knock-in or transgenic mice expressing a KEAP1 mutant in which Cys151 of KEAP1 is substituted with serine, which are referred to as KEAP1C151S (Eggler et al., 2009; Kobayashi et al., 2009; McMahon et al., 2010; Saito et al., 2016; Takaya et al., 2012; Yamamoto et al., 2008; Zhang and Hannink, 2003). In KEAP1C151S mouse embryonic fibroblasts (MEFs) and peritoneal macrophages, the Cys151 residue was found to be indispensable for the accumulation of NRF2 in response to an array of electrophilic chemicals. These NRF2 inducers include DEM, tBHQ (tert-butyl hydroquinone), DMF (dimethylfumarate), nitric oxide (NO), sulforaphane (SFN), and CDDO-Im (Saito et al., 2016; Takaya et al., 2012). These are categorized into Class I or Cys151-preferring inducers (Fig. 3).
Cys151 is located in the BTB domain of KEAP1, which is responsible for the interaction of KEAP1 with CUL3. Consistent with these observations, several reports have shown that modification of Cys151 inhibits the KEAP1-CUL3 interaction and prevents ubiquitination of NRF2 (Cleasby et al., 2014; Eggler et al., 2009; Zhang et al., 2004). It should be noted that there are a couple of reports that disagree with this model (Baird et al., 2013; Li et al., 2012). In this regard, pull-down and analytical centrifugation analyses show that many of the Cys151-targeting electrophilic inducers do not provoke dissociation of the KEAP1-CUL3 complex, except in the case of CDDO-Im (Iso et al., 2016). Chemical inducers produce conformational changes in KEAP1, as shown by a hydrophobicity probe (Dinkova-Kostova et al., 2005). We surmise that cysteine modification elicits structural alterations in KEAP1 by affecting the complex status of KEAP1 and CUL3 without causing their dissociation. This structural alteration in KEAP1 prevents NRF2 ubiquitination. It is possible that Cys151 modification affects the angle of orientation of the KEAP1-CUL3 association, thereby significantly altering the distance between ubiquitin and the target lysine residues in the Neh2 domain.
Class II & III; NRF2 inducers consisting of Cys273/288 on KEAP1-IVR domain
It has been assumed that other cysteine residues, especially Cys273 and Cys288 in the intervening region (IVR), may also contribute to the stress-sensor function of KEAP1. The Cys273 and Cys288 residues have been suggested to react with 15-deoxy-∆12,14-prostaglandin J2 (15d-PGJ2) (Kobayashi et al., 2009). However, substitution of Cys273 and Cys288 with serine or alanine failed to repress NRF2 activity in both a reporter co-transfection transactivation assay (Wakabayashi et al., 2004; Zhang and Hannink, 2003) and in transgenic complementation rescue mouse experiments (Yamamoto et al., 2008), precluding a simple validation of this notion.
To overcome this problem, we systematically introduced amino acid substitutions into Cys273 and Cys288 and identified amino acids that do not affect the ability of KEAP1 to repress NRF2 accumulation (Saito et al., 2016). Finally, we found that KEAP1 mutants in which Cys273 was replaced with tryptophan and Cys288 was replaced with glutamate (Saito et al., 2016) retained the ability to repress NRF2 accumulation. In unstressed conditions, it seems that Cys273 and Cys288 are kept in hydrophobic and hydrophilic conditions, respectively. These characteristics of Cys273 and Cys288 might be critical for the structural integrity of KEAP1 for maintaining ubiquitin ligase activity. However, conclusive insights into these structural requirements await further elucidation of the structure surrounding the IVR that includes Cys273 and Cys288. It was shown in MEFs from KEAP1-Cys288Glu (Keap1C288E) knock-in mice that 15d-PGJ2 is recognized by KEAP1 Cys288. Therefore, 15d-PGJ2 belongs to Class II chemicals known as Cys288-preferring inducers (Fig. 3).
On the other hand, all three cysteine residues (Cys151, Cys273, and Cys288) are indispensable for the activity of KEAP1 that senses 9-nitro-octadec-9-enoic acid (9-OA-NO2), 4-hydroxy-2-nonenal (4-HNE) and sodium arsenite (NaAsO2). These chemicals therefore belong to Class III and are referred to as Cys151/Cys273/Cys288-selective inducers (Fig. 3).
Class IV; KEAP1-Cys226/613/622/624-selective NRF2 inducers
Oxidative stress is involved in the development and progression of many diseases, including Alzheimer’s disease, atherosclerosis, and cancer (Sies et al., 2022). Hydrogen peroxide (H2O2) is classified into the Cys151/Cys273/Cys288-independent inducer (Saito et al., 2016). NRF2 accumulation by H2O2 was not affected in KEAP1C151S&C273W&C288E-expressing MEFs, indicating that the three cysteine residues are dispensable for NRF2 activation in response to H2O2. This suggests that alternative cysteine residues are critical for stress recognition by KEAP1.
To address this hypothesis, we generated a KEAP1 mutant protein with 11 cysteine residue substitution along with various individual and combination mutations of these cysteine residues. Functional analyses of an additional range of KEAP1 mutants showed that the H2O2 sensor actually resides in the 11 cysteine residues, and finally revealed that Cys226, Cys613, Cys622, and Cys624 are involved in the sensing of H2O2 by KEAP1 (Suzuki et al., 2019). Of note, despite the fact that our results delineate that the four cysteine residues are involved in the H2O2 sensor activity, no individual cysteine residue is critical for the sensor function, as the single mutation of any of the four cysteine residues does not inactivate the oxidative stress sensor. These observations support our contention that disulfide bond between Cys226, Cys613, and Cys622/624 is formed in response to oxidative stress, and the sensor for the H2O2 contains a robust compensatory mechanism. Thus, these findings indicate that an elaborate fail-safe mechanism consisting of Cys226, Cys613, and Cys622/Cys624 ensures the KEAP1’s response to H2O2 across a range of conditions.
These mechanistic studies revealed that various electrophiles are sensed by unique multiple cysteine residues, including Cys151, Cys273, and Cys288. Electrophiles make thiol-alkylation covalent bondage with these cystines. On the other hand, H2O2 forms a disulfide bond between Cys226, Cys613, Cys622, and Cys624. A pair between these residues is formed between two of these cysteine residues. These results thus indicate that electrophiles and ROS employs distinct cysteine residues of KEAP1. However, precise mechanisms as to how these cysteine modifications alter ubiquitin ligase activity of KEAP1 remain to be elucidated.
Class V; non-electrophilic KEAP1-NRF2 PPI inhibitor or NRF2 inducers
Classic inducers of NRF2 interact with the cysteine residues of KEAP1 by virtue of their electrophilic nature and can therefore inherently react with glutathione or thiol in proteins. Since an over-dose of such thiol-reactive chemicals could cause electrophilic damage in cells, chemicals that directly inhibit the PPI of KEAP1 and NRF2 are emerging as candidates of attractive novel NRF2 inducers. We refer these small chemical inducers to as Class V (Fig. 3). For instance, the isoquinoline PRL-295 (Lazzara et al., 2020) was developed with the safety and physicochemical properties of the earlier naphthalene- and isoquinoline-based KEAP1 inhibitors (Richardson et al., 2018). The potency of PRL-295 in cell-based assays is similar to that of sulforaphane (Dayalan et al., 2022). We have utilized PRL-295 recently to test the “Hinge-Latch” model of NRF2 activation. The results revealed that the PPI inhibitors disrupt the DLGex-KEAP1 interaction preferentially over the ETGE-KEAP1 interaction (Horie et al., 2021). The “Hinge-Latch” model will be discussed more in detail in the next section. Another example is that Scohia Pharma, Inc. claimed a patent of WO2020116660 that described macrocyclic KEAP1-NRF2 inhibitors harboring the tetrahydroisoquinoline scaffold.
UPDATED “HINGE-LATCH” MODEL OF THE KEAP1-NRF2 SYSTEM
Various structure-function analyses of the KEAP1-NRF2 interaction revealed the importance of the two-site binding model between the KEAP1 homodimer (i.e., two DC domains) and the Neh2 domain of NRF2 (via DLGex and ETGE motifs) (Fukutomi et al., 2014; Tong et al., 2006a). The Hinge-Latch model was proposed as a plausible mechanism for the KEAP1-mediated NRF2 activation (Tong et al., 2006b). This model is based on the fact that the DLGex and ETGE motifs show approximately two-orders of magnitude difference in the binding affinity to the KEAP1-DC domain (Tong et al., 2007). Various lines of evidence including somatic mutation analyses in clinical cancer studies support the presence of this model (Shibata et al., 2008). However, the model was not validated to date due to technical difficulties in examining the DLGex-KEAP1 binding and ETGE-KEAP1 binding simultaneously.
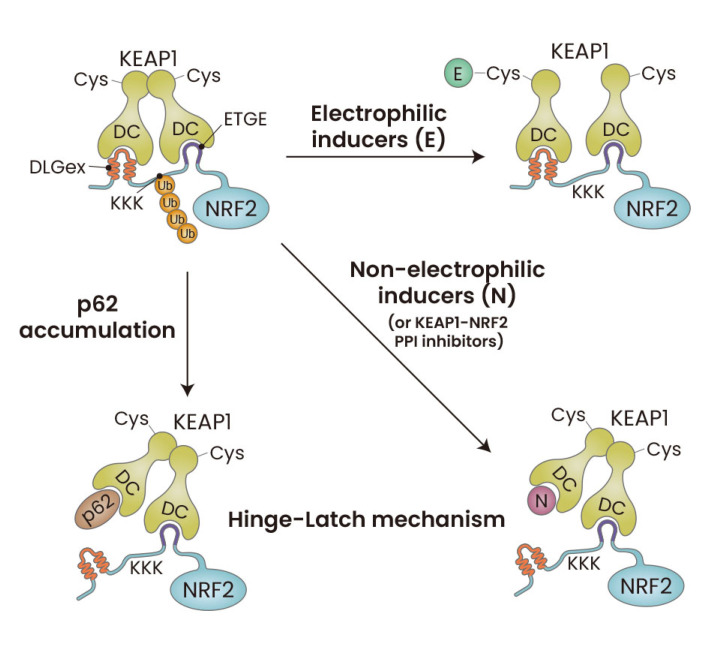
To overcome this problem, Horie et al. (2021), utilized NMR titration, which is capable of simultaneous and sequences-specific assignment of the DLGex and ETGE motifs. By utilizing this method, Horie et al. (2021), found that the Hinge-Latch mechanism indeed operates during the activation of NRF2 by p62 accumulation, which can be induced by autophagy deficiency, as well as pharmacological KEAP1-NRF2 PPI inhibitors such as PRL-295, but not by electrophilic NRF2 inducers such as CDDO-Im, SFN or 15d-PGJ2 (Fig. 4).
Fig. 4. Updated Hinge-Latch model.
Autophagy chaperone p62 accumulation disrupts the DLGex-Keap1 interaction and activates NRF2, and this mechanism conforms to the concept of the Hinge-Latch model. Similarly, pharmacological KEAP1-NRF2 PPI inhibitors disrupt the DLGex-KEAP1 interaction preferentially to the ETGE-KEAP1 interaction, and molecular basis for the efficacy of these chemicals is the utilization of the Hinge-Latch mechanism. In contrast, electrophilic NRF2-activating compounds activate NRF2 without disrupting the Neh2-KEAP1 interaction, implying that these KEAP1 thiol-modifying chemicals utilize mechanisms distinct from the Hinge-Latch mechanism. KEAP1, Kelch-like ECH-associated protein 1; NRF2, NF-E2-related factor 2; PPI, protein-protein interactions.
THERAPEUTIC STRATEGIES TO CONTROL NRF2-OVERACTIVATED CANCERS
NRF2 plays diverse role in the context of cancer. As the KEAP1-NRF2 pathway is an essential cell defense system against environmental stresses, NRF2 provides important anti-cancer ability (Aoki et al., 2001; Ohkoshi et al., 2013; Ramos-Gomez et al., 2003). On the other hand, aberrant NRF2 overactivation has been found in several types of cancer cells, and such NRF2 overactivation in cancer cells correlates strongly with poor clinical outcomes (Padmanabhan et al., 2006; Romero et al., 2017; Shibata et al., 2008). Aberrant NRF2 activation in cancer cells induces expression of cellular defense enzymes and metabolic reprogramming conferring therapeutic resistance and aggressive proliferation properties (Homma et al., 2009; Mitsuishi et al. 2012). Previous studies have shown multiple mechanisms of NRF2 activation in cancer cells, such as somatic mutations (Ooi et al., 2013; Padmanabhan et al., 2006; Shibata et al., 2010), loss of NRF2 gene exon 2 (Goldstein et al., 2016), enhanced transcription of NRF2 (DeNicola et al., 2011), p62 accumulation (Komatsu et al., 2010), epigenetic modification of KEAP1 gene (Wang et al., 2008) and production of oncometabolites (Adam et al., 2011; Ooi et al., 2011).
Over the course of analyses for molecular features of the KEAP1-NRF2 signaling pathway, we have shown some promising compounds to treat NRF2-activated cancers (Fig. 1). Initially, halofuginone was identified as an NRF2 inhibitor using a high-throughput screening (Tsuchida et al., 2017). Halofuginone suppresses NRF2 protein accumulation by inhibition of prolyl-tRNA synthetase. Recently, we developed novel compound “halofuginone-micelle” which attacks NRF2-activated cancers with less adverse effect (Panda et al., 2022). Another promising approach is to isolate synthetic lethal drugs including mitomycin C and the geldanamycin-derived HSP90 inhibitor (Baird and Yamamoto, 2021; Baird et al., 2020) (Fig. 1). These compounds are anticancer prodrugs and bioactivated by NRF2-dependent drug metabolizing enzymes. Targeting the NRF2 transcriptome enables to reduce toxicity in normal cells and provide specificity to NRF2 activated cancers.
In addition, NRF2 activation in cancer microenvironment can also be an effective approach against NRF2-activated cancers (Hayashi et al., 2020) (Fig. 1). In a mouse model experiment with systemic and myeloid cell specific NRF2 knocked out as well as transplanted 3 Lewis lung carcinoma (3LL) cells, higher ROS accumulation in the myeloid-derived suppressor cells (MDSCs) was observed (Hiramoto et al., 2014; Satoh et al., 2010). These mice also displayed a higher rate of cancer incidence and spontaneous lung metastasis, demonstrating that high NRF2 levels in the tumor microenvironment suppress tumor cell development by reducing the ROS levels in MDSCs. These observations indicate that NRF2 activation in host myeloid cells can be a therapeutic target in clinical settings. Several NRF2 inducers have been provided and some of them have been already used in clinical setting, so further studies of NRF2 inducer will lead to novel anticancer drug development (Gold et al., 2012; Szczesny-Malysiak et al., 2020; de Zeeuw et al., 2013).
FUTURE PERSPECTIVES
Accumulating lines of evidence have proven that KEAP1 senses a wide range of NRF2-inducing chemicals (Saito et al., 2016; Suzuki et al., 2019). Nonetheless, how KEAP1 senses a variety of chemicals utilizing multiple and distinct sets of cysteine residues remains to be clarified. Additional studies on the sensing mechanisms will advance our understanding of the environmental stress response.
NRF2 activators targeting the cysteine thiols of KEAP1 have been extensively developed as discussed above and NRF2 activators targeting the KEAP1-NRF2 PPI are also entering the stage of extensive development. It is interesting to note that Cuadrado (2015) has been developing β-transduction repeat-containing protein (β-TRCP)-NRF2 PPI inhibitor in patent (WO2022152800) as [β-TRCP-S-phase kinase-associated protein 1-CULLIN1 (β-TRCP-SKP1-CUL1) E3 ligase complex is also responsible for ubiquitination of NRF2 (Cuadrado, 2015; Kuga et al., 2022). Novel NRF2 activators targeting other sites will also be developed.
Of the many interesting recent topics in the NRF2 field, one highly interesting topic is the Nrf2-knockout mouse travel of international space station. The Nrf2 knockout mice stayed there for 31 days and all the mice returned safely to the ground (Suzuki et al., 2020b). Analyses of the mice revealed that NRF2 acts to preserve homeostasis of the skeletal muscle, kidney, and epididymal white adipose tissue (Hayashi et al., 2021; Suzuki et al., 2022; Uruno et al., 2021). Next challenge of the space mouse study is to verify that the NRF2 activation indeed acts to maintain homeostasis of whole body against the space stress and the study will continue.
ACKNOWLEDGMENTS
This work was supported in part by MEXT/JSPS KAKENHI (19H05649 to M.Y. and 22K06876 to T.S.), the Takeda Science Foundation (to T.S.), the Suzuken Memorial Foundation (to T.S.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to T.S.), the Foundation for Promotion of Cancer Research (to T.S.), the Gonryo Medical Foundation (to T.S.), the Life Science Foundation of Japan (to T.S.), and Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED (JP22ama121038 to M.Y.).
Footnotes
AUTHOR CONTRIBUTIONS
T.S., J.T., and M.Y. wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Adam J., Hatipoglu E., O'Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K., et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Sato H., Nishimura N., Takahashi S., Itoh K., Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol. Appl. Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- Baird L., Dinkova-Kostova A.T. Diffusion dynamics of the Keap1-Cullin3 interaction in single live cells. Biochem. Biophys. Res. Commun. 2013;433:58–65. doi: 10.1016/j.bbrc.2013.02.065. [DOI] [PubMed] [Google Scholar]
- Baird L., Suzuki T., Takahashi Y., Hishinuma E., Saigusa D., Yamamoto M. Geldanamycin-derived HSP90 inhibitors are synthetic lethal with NRF2. Mol. Cell. Biol. 2020;40:e00377–20. doi: 10.1128/MCB.00377-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird L., Yamamoto M. NRF2-dependent bioactivation of mitomycin C as a novel strategy to target KEAP1-NRF2 pathway activation in human cancer. Mol. Cell. Biol. 2021;41:e00473–20. doi: 10.1128/MCB.00473-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleasby A., Yon J., Day P.J., Richardson C., Tickle I.J., Williams P.A., Callahan J.F., Carr R., Concha N., Kerns J.K., et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9:e98896. doi: 10.1371/journal.pone.0098896.e0c4767540824921bd96b680725f39ab [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/β-TrCP. Free Radic. Biol. Med. 2015;88(Pt B):106–109. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Dayalan Naidu S., Suzuki T., Dikovskaya D., Knatko E.V., Higgins M., Sato M., Novak M., Villegas J.A., Moore T.W., Yamamoto M., et al. The isoquinoline PRL-295 increases the thermostability of Keap1 and disrupts its interaction with Nrf2. iScience. 2022;25:103703. doi: 10.1016/j.isci.2021.103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova A.T., Holtzclaw W.D., Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova A.T., Talalay P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol. Nutr. Food Res. 2008;52(Suppl 1):S128–S138. doi: 10.1002/mnfr.200700195. [DOI] [PubMed] [Google Scholar]
- Eggler A.L., Liu G., Pezzuto J.M., van Breemen R.B., Mesecar A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler A.L., Small E., Hannink M., Mesecar A.D. Cul3-mediated Nrf2 ubiquitination and antioxidant response element (ARE) activation are dependent on the partial molar volume at position 151 of Keap1. Biochem. J. 2009;422:171–180. doi: 10.1042/BJ20090471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friling R.S., Bensimon A., Tichauer Y., Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutomi T., Takagi K., Mizushima T., Ohuchi N., Yamamoto M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014;34:832–846. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold R., Kappos L., Arnold D.L., Bar-Or A., Giovannoni G., Selmaj K., Tornatore C., Sweetser M.T., Yang M., Sheikh S.I., et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N. Engl. J. Med. 2012;367:1098–1107. doi: 10.1056/NEJMoa1114287. [DOI] [PubMed] [Google Scholar]
- Goldstein L.D., Lee J., Gnad F., Klijn C., Schaub A., Reeder J., Daemen A., Bakalarski C.E., Holcomb T., Shames D.S., et al. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 2016;16:2605–2617. doi: 10.1016/j.celrep.2016.08.010.e594afbe1e7541f5bfa7c4dffdee5106 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Kuga A., Suzuki M., Panda H., Kitamura H., Motohashi H., Yamamoto M. Microenvironmental activation of Nrf2 restricts the progression of Nrf2-activated malignant tumors. Cancer Res. 2020;80:3331–3344. doi: 10.1158/0008-5472.CAN-19-2888. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Kudo T., Fujita R., Fujita S.I., Tsubouchi H., Fuseya S., Suzuki R., Hamada M., Okada R., Muratani M., et al. Nuclear factor E2-related factor 2 (NRF2) deficiency accelerates fast fibre type transition in soleus muscle during space flight. Commun. Biol. 2021;4:787. doi: 10.1038/s42003-021-02334-4.54b77403fca14ea68b052687eebe637e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto K., Satoh H., Suzuki T., Moriguchi T., Pi J., Shimosegawa T., Yamamoto M. Myeloid lineage-specific deletion of antioxidant system enhances tumor metastasis. Cancer Prev. Res. (Phila.) 2014;7:835–844. doi: 10.1158/1940-6207.CAPR-14-0094. [DOI] [PubMed] [Google Scholar]
- Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K., Engel J.D., Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma S., Ishii Y., Morishima Y., Yamadori T., Matsuno Y., Haraguchi N., Kikuchi N., Satoh H., Sakamoto T., Hizawa N., et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin. Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- Horie Y., Suzuki T., Inoue J., Iso T., Wells G., Moore T.W., Mizushima T., Dinkova-Kostova A.T., Kasai T., Kamei T., et al. Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism. Commun. Biol. 2021;4:576. doi: 10.1038/s42003-021-02100-6.b744783a8c5b4f278122faa72d2167f4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- Iso T., Suzuki T., Baird L., Yamamoto M. Absolute amounts and status of the Nrf2-Keap1-Cul3 complex within cells. Mol. Cell. Biol. 2016;36:3100–3112. doi: 10.1128/MCB.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K., Igarashi K., Hayashi N., Nishizawa M., Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995;15:4184–4193. doi: 10.1128/MCB.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., O'Connor T., Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- Katsuoka F., Otsuki A., Takahashi M., Ito S., Yamamoto M. Direct and specific functional evaluation of the Nrf2 and MafG heterodimer by introducing a tethered dimer into small Maf-deficient cells. Mol. Cell. Biol. 2019;39:e00273–19. doi: 10.1128/MCB.00273-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Kang M.I., Watai Y., Tong K.I., Shibata T., Uchida K., Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624.64f68c1a2df449c4a3c17e7079f6efb6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kuga A., Tsuchida K., Panda H., Horiuchi M., Otsuki A., Taguchi K., Katsuoka F., Suzuki M., Yamamoto M. The β-TrCP-mediated pathway cooperates with the Keap1-mediated pathway in Nrf2 degradation in vivo. Mol. Cell. Biol. 2022;42:e0056321. doi: 10.1128/mcb.00563-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzara P.R., David B.P., Ankireddy A., Richardson B.G., Dye K., Ratia K.M., Reddy S.P., Moore T.W. Isoquinoline Kelch-like ECH-associated protein 1-nuclear factor (erythroid-derived 2)-like 2 (KEAP1-NRF2) inhibitors with high metabolic stability. J. Med. Chem. 2020;63:6547–6560. doi: 10.1021/acs.jmedchem.9b01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Paonessa J.D., Zhang Y. Mechanism of chemical activation of Nrf2. PLoS One. 2012;7:e35122. doi: 10.1371/journal.pone.0035122.7686f76406424919a655490b93b7f482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M., Lamont D.J., Beattie K.A., Hayes J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Ohkoshi A., Suzuki T., Ono M., Kobayashi T., Yamamoto M. Roles of Keap1-Nrf2 system in upper aerodigestive tract carcinogenesis. Cancer Prev. Res. (Phila.) 2013;6:149–159. doi: 10.1158/1940-6207.CAPR-12-0401-T. [DOI] [PubMed] [Google Scholar]
- Ooi A., Dykema K., Ansari A., Petillo D., Snider J., Kahnoski R., Anema J., Craig D., Carpten J., Teh B.T., et al. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013;73:2044–2051. doi: 10.1158/0008-5472.CAN-12-3227. [DOI] [PubMed] [Google Scholar]
- Ooi A., Wong J.C., Petillo D., Roossien D., Perrier-Trudova V., Whitten D., Min B.W., Tan M.H., Zhang Z., Yang X.J., et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- Otsuki A., Suzuki M., Katsuoka F., Tsuchida K., Suda H., Morita M., Shimizu R., Yamamoto M. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic. Biol. Med. 2016;91:45–57. doi: 10.1016/j.freeradbiomed.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M.I., Kobayashi A., Yokoyama S., Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Panda H., Suzuki M., Naito M., Saito R., Wen H., Baird L., Uruno A., Miyata K., Yamamoto M. Halofuginone micelle nanoparticles eradicate Nrf2-activated lung adenocarcinoma without systemic toxicity. Free Radic. Biol. Med. 2022;187:92–104. doi: 10.1016/j.freeradbiomed.2022.05.017. [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M., Dolan P.M., Itoh K., Yamamoto M., Kensler T.W. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24:461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- Richardson B.G., Jain A.D., Potteti H.R., Lazzara P.R., David B.P., Tamatam C.R., Choma E., Skowron K., Dye K., Siddiqui Z., et al. Replacement of a naphthalene scaffold in Kelch-like ECH-associated protein 1 (KEAP1)/nuclear factor (erythroid-derived 2)-like 2 (NRF2) inhibitors. J. Med. Chem. 2018;61:8029–8047. doi: 10.1021/acs.jmedchem.8b01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sánchez-Rivera F.J., et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. doi: 10.1016/S0021-9258(18)99004-6. [DOI] [PubMed] [Google Scholar]
- Saito R., Suzuki T., Hiramoto K., Asami S., Naganuma E., Suda H., Iso T., Yamamoto H., Morita M., Baird L., et al. Characterizations of three major cysteine sensors of Keap1 in stress response. Mol. Cell. Biol. 2016;36:271–284. doi: 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H., Moriguchi T., Taguchi K., Takai J., Maher J.M., Suzuki T., Winnard P.T., Raman V., Ebina M., Nukiwa T., et al. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis. 2010;31:1833–1843. doi: 10.1093/carcin/bgq105. [DOI] [PubMed] [Google Scholar]
- Sengoku T., Shiina M., Suzuki K., Hamada K., Sato K., Uchiyama A., Kobayashi S., Oguni A., Itaya H., Kasahara K., et al. Structural basis of transcription regulation by CNC family transcription factor, Nrf2. Nucleic Acids Res. 2022;50:12543–12557. doi: 10.1093/nar/gkac1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Saito S., Kokubu A., Suzuki T., Yamamoto M., Hirohashi S. Global downstream pathway analysis reveals a dependence of oncogenic NF-E2-related factor 2 mutation on the mTOR growth signaling pathway. Cancer Res. 2010;70:9095–9105. doi: 10.1158/0008-5472.CAN-10-0384. [DOI] [PubMed] [Google Scholar]
- Sies H., Belousov V.V., Chandel N.S., Davies M.J., Jones D.P., Mann G.E., Murphy M.P., Yamamoto M., Winterbourn C. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 2022;23:499–515. doi: 10.1038/s41580-022-00456-z. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Iwamura Y., Nakai T., Kato K., Otsuki A., Uruno A., Saigusa D., Taguchi K., Suzuki M., Shimizu R., et al. Gene expression changes related to bone mineralization, blood pressure and lipid metabolism in mouse kidneys after space travel. Kidney Int. 2022;101:92–105. doi: 10.1016/j.kint.2021.09.031. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Hidaka T., Kumagai Y., Yamamoto M. Environmental pollutants and the immune response. Nat. Immunol. 2020a;21:1486–1495. doi: 10.1038/s41590-020-0802-6. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Motohashi H., Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol. Sci. 2013;34:340–346. doi: 10.1016/j.tips.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Muramatsu A., Saito R., Iso T., Shibata T., Kuwata K., Kawaguchi S.I., Iwawaki T., Adachi S., Suda H., et al. Molecular mechanism of cellular oxidative stress sensing by Keap1. Cell Rep. 2019;28:746–758.e4. doi: 10.1016/j.celrep.2019.06.047. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Uruno A., Yumoto A., Taguchi K., Suzuki M., Harada N., Ryoke R., Naganuma E., Osanai N., Goto A., et al. Nrf2 contributes to the weight gain of mice during space travel. Commun. Biol. 2020b;3:496. doi: 10.1038/s42003-020-01227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015;88(Pt B):93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Yamamoto M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J. Biol. Chem. 2017;292:16817–16824. doi: 10.1074/jbc.R117.800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny-Malysiak E., Stojak M., Campagna R., Grosicki M., Jamrozik M., Kaczara P., Chlopicki S. Bardoxolone methyl displays detrimental effects on endothelial bioenergetics, suppresses endothelial ET-1 release, and increases endothelial permeability in human microvascular endothelium. Oxid. Med. Cell. Longev. 2020;2020:4678252. doi: 10.1155/2020/4678252.33dbf1ccd0e046878c69f47532abf8fe [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya K., Suzuki T., Motohashi H., Onodera K., Satomi S., Kensler T.W., Yamamoto M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2012;53:817–827. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong K.I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006a;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong K.I., Kobayashi A., Katsuoka F., Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol. Chem. 2006b;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida K., Tsujita T., Hayashi M., Ojima A., Keleku-Lukwete N., Katsuoka F., Otsuki A., Kikuchi H., Oshima Y., Suzuki M., et al. Halofuginone enhances the chemo-sensitivity of cancer cells by suppressing NRF2 accumulation. Free Radic. Biol. Med. 2017;103:236–247. doi: 10.1016/j.freeradbiomed.2016.12.041. [DOI] [PubMed] [Google Scholar]
- Uruno A., Saigusa D., Suzuki T., Yumoto A., Nakamura T., Matsukawa N., Yamazaki T., Saito R., Taguchi K., Suzuki M., et al. Nrf2 plays a critical role in the metabolic response during and after spaceflight. Commun. Biol. 2021;4:1381. doi: 10.1038/s42003-021-02904-6.4d30a972dc234a618c53b81a1176cb9d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N., Dinkova-Kostova A.T., Holtzclaw W.D., Kang M.I., Kobayashi A., Yamamoto M., Kensler T.W., Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., An J., Ji F., Jiao H., Sun H., Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Watai Y., Kobayashi A., Nagase H., Mizukami M., McEvoy J., Singer J.D., Itoh K., Yamamoto M. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells. 2007;12:1163–1178. doi: 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zeeuw D., Akizawa T., Audhya P., Bakris G.L., Chin M., Christ-Schmidt H., Goldsberry A., Houser M., Krauth M., Lambers Heerspink H.J., et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N. Engl. J. Med. 2013;369:2492–2503. doi: 10.1056/NEJMoa1306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Suzuki T., Adachi S., Naganuma E., Suzuki N., Hosoya T., Itoh K., Sporn M.B., Yamamoto M. Distinct regulations of HO-1 gene expression for stress response and substrate induction. Mol. Cell. Biol. 2021;41:e0023621. doi: 10.1128/MCB.00236-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.D., Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]