Abstract
Cancer stem cells (CSCs) are a small population of tumor cells characterized by self-renewal and differentiation capacity. CSCs are currently postulated as the driving force that induces intra-tumor heterogeneity leading to tumor initiation, metastasis, and eventually tumor relapse. Notably, CSCs are inherently resistant to environmental stress, chemotherapy, and radiotherapy due to high levels of antioxidant systems and drug efflux transporters. In this context, a therapeutic strategy targeting the CSC-specific pathway holds a promising cure for cancer. NRF2 (nuclear factor erythroid 2-like 2; NFE2L2) is a master transcription factor that regulates an array of genes involved in the detoxification of reactive oxygen species/electrophiles. Accumulating evidence suggests that persistent NRF2 activation, observed in multiple types of cancer, supports tumor growth, aggressive malignancy, and therapy resistance. Herein, we describe the core properties of CSCs, focusing on treatment resistance, and review the evidence that demonstrates the roles of NRF2 signaling in conferring unique properties of CSCs and the associated signaling pathways.
Keywords: antioxidant system, cancer plasticity, cancer stem cell, NRF2/NFE2L2, therapy resistance
INTRODUCTION
Despite the increase in the success rate of treatment, cancer remains a disease with high recurrence and mortality rates. The main cause of failure of cancer treatment is attributed to the heterogeneous cell population within the tumor mass that induces differential sensitivity of individual cancer cells to chemotherapy (Prasetyanti and Medema, 2017; Yung et al., 1982). Along with genetic mutations and epigenetic alterations, intratumoral heterogeneity is ascribed to cancer stem cells (CSCs), a small population of quiescent self-renewing cells within tumors (Lapidot et al., 1994; Prasetyanti and Medema, 2017). CSCs drive metastasis, chemotherapy resistance, and radiation resistance in cancers, which eventually leads to cancer relapse after successful initial treatment (Batlle and Clevers, 2017; Lytle et al., 2018). Currently, extensive efforts are being made to develop novel treatment strategies to target CSCs by identifying key factors and signaling pathways involved in the survival and maintenance of CSCs.
NRF2 (nuclear factor erythroid 2-like 2; NFE2L2) is a master regulator of the expression of genes involved in reactive oxygen species (ROS)/electrophile detoxification, glutathione (GSH) production/regeneration, heme/iron metabolism, NAD(P)H generation, cell metabolism, and drug efflux (Itoh et al., 1997; Tebay et al., 2015; Tonelli et al., 2018). NRF2 activity is primarily regulated by Kelch-like ECH-associated protein 1 (KEAP1). Under normal conditions, two molecules of the KEAP1 protein interact with an NRF2 protein, which leads to continuous degradation of NRF2 through the Cullin 3-dependent ubiquitin ligase and proteasome system (Baird and Yamamoto, 2020; Itoh et al., 1999). Additionally, NRF2 levels are regulated not only by KEAP, but also by KEAP-independent mechanisms such as, such as the negative regulation by β transducin repeats-containing protein (β-TrCP)/glycogen synthase kinase-3 (GSK3) and positive regulation by competing protein p62 (Tebay et al., 2015). Although NRF2 plays a critical role in cytoprotection against carcinogenesis and chemicals/oxidant-induced tissue injuries, a deleterious role of NRF2 in cancer growth and progression has been observed in numerous types of cancer (Choi et al., 2021; Kitamura and Motohashi, 2018; Lee et al., 2022; Rojo de la Vega et al., 2018). Furthermore, since the expression of antioxidant genes and drug efflux transporters, which are under the control of NRF2, is frequently increased in CSCs, the potential implication of NRF2 signaling in CSCs physiology has been raised (Kahroba et al., 2019; Rojo de la Vega et al., 2018; Ryoo et al., 2016b). Herein, we review current evidence of the role of NRF2 in CSC maintenance, survival, and therapy resistance.
CSC CONCEPT
CSCs were initially described as a cancer cell subpopulation that can increase the tumor CSC pool and differentiate into progenitor cancer cells (Bjerkvig et al., 2005). When asymmetric and symmetric division of CSCs is balanced, tumors are composed of CSCs and most cancer cells; however, when there is a shift to symmetric division, the proportion of CSCs increases and cancer transforms to a more aggressive and undifferentiated high-grade state (Batlle and Clevers, 2017; Lytle et al., 2018). Other terms for CSCs include tumor-initiating cells, tumor-progenitor cells, and cancer stem-like cells (Nguyen et al., 2012). CSCs are currently postulated to be key drivers in initiating tumorigenesis and causing cancer metastasis and recurrence (Lytle et al., 2018). The CSC concept was first tested in acute myeloid leukemia. To identify leukemic-initiating cells, leukemic cells were transplanted into SCID (severe combined immune-deficient) mice, and CD34+CD38- fraction of cells was found to be tumor-initiating cells (Lapidot et al., 1994). Subsequently, it was shown that CD34+CD38- leukemic cells have the capacity for differentiation, proliferation, and self-renewal (Bonnet and Dick, 1997). Since these studies, surface marker-based cell purification and subsequent transplantation in immunodeficient mice have confirmed functional CSCs in various solid tumors, such as glioma, breast, and colon cancers (Medema, 2013).
The surface markers for CSCs isolated from solid tumors include CD29, CD24, CD44, CD133, leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5), and epithelial cell adhesion molecule (EpCAM) (Yang et al., 2020). CSC marker expression differs for each type of cancer and varies between patient tumors. CD133 was identified as a marker for glioblastoma CSCs (Singh et al., 2004), whereas CD44+CD24- cell populations were identified as breast CSCs (Ponti et al., 2005). CD44+CD49f+CD133+ cells showed higher tumorigenic capacity than CD44+CD24- cells in estrogen receptor-negative breast tumors (Meyer et al., 2010). In addition, the side population (SP) cancer cells, which do not accumulate Hoechst 33343 dye due to high expression of ATP-binding cassette (ABC) transporter ABCG2, possess self-renewal and differentiation capacities (Zhou et al., 2001). Cancer cells with high expression of aldehyde dehydrogenase 1 (ALDH1), a cytosolic enzyme oxidizing aldehyde, correlates with high tumorigenic and metastatic characteristics (Ricardo et al., 2011). The sphere-forming assay has also been used to enrich CSCs; tumor sphere-forming ependymoma cells are multipotent and initiate tumors in mice (Taylor et al., 2005).
Signaling pathways contributing to the survival, maintenance, self-renewal, and differentiation of CSCs are complex; however, some transcription factors and signaling pathways are shared between normal stem cells and CSCs (Table 1). These signaling pathways include the Wnt/β-catenin, neurogenic locus notch homolog protein (NOTCH), and Sonic Hedgehog pathways, which are involved in the self-renewal of CSCs (Yang et al., 2020). Wnt signaling is aberrantly activated in many cancers, such as colorectal cancer and invasive ductal breast cancer, and its activation induces the conversion of dormant CSCs to active CSCs through β-catenin-mediated cell cycle progression and MYC elevation (Giancotti, 2013). Overactivation of NOTCH4 signaling was found in breast CSCs, and NOTCH4 inhibition completely blocked the tumor-initiating ability of CSCs (Harrison et al., 2010). Major transcription factors for pluripotent stem cell conversion, including octamer-binding transcription factor 4 (OCT4), homeobox protein NANOG (NANOG), Krüppel-like factor 4 (KLF4), MYC, and transcription factor SOX-2 (SOX2), are also utilized in CSCs (Yang et al., 2020). The expression of OCT4, a master regulator of cell pluripotency, is high in hepatocellular carcinoma (HCC) CSCs and breast CSCs, and high OCT4 levels are associated with self-renewal, tumorigenicity, and chemoresistance of these CSCs (Murakami et al., 2015; Ponti et al., 2005).
Table 1.
Signaling pathways implicated in CSC property
| Signaling pathways | Property | References |
|---|---|---|
| NOTCH | Self-renewal, stemness | (Takebe et al., 2015) |
| Wnt/β-catenin | Self-renewal, stemness | (Vermeulen et al., 2010) |
| Hedgehog | Self-renewal, stemness | (Justilien and Fields, 2015) |
| BMI-1 | Self-renewal, stemness | (Kreso et al., 2014) |
| TWIST | EMT, stemness | (Beck et al., 2015) |
| ZEB1 | EMT | (Caramel et al., 2013) |
| TGF-β | EMT, stemness | (Mani et al., 2008) |
| TAZ | Stemness, EMT | (Cordenonsi et al., 2011) |
| Nestin | Self-renewal | (Neradil and Veselska, 2015) |
| p62 | Self-renewal | (Li et al., 2021; Umemura et al., 2016) |
CSC, cancer stem cell; NOTCH, neurogenic locus notch homolog protein; TGF-β, transforming growth factor β; EMT, epithelial-to-mesenchymal transition.
CSC AND CANCER PLASTICITY
Earlier studies assumed that the intra-tumor hierarchy was rigid and unidirectional, with CSCs being viewed as the exclusive source of self-renewal and pluripotency (Chaffer et al., 2011). However, recent studies have suggested that both CSCs and non-CSCs possess plasticity. Isolated stem cell-, basal-, and luminal-like cells from breast cancer cell lines generate two other types of cells and can recompose the original phenotypic equilibrium through stochastic transition (Gupta et al., 2011). Among the three phenotypic cell types, only cells with a stem-like phenotype acquired tumorigenic capacity, which enabled to predict de novo CSC generation from non-CSCs. Selective deletion of LGR5+ CSCs in colorectal organoids could initially restrict tumor growth; however, the organoids regained tumorigenicity due to the re-emergence of LGR5+ CSCs (Shimokawa et al., 2017). LGR5- cancer cells have been shown to continuously replenish the LGR5+ CSCs pool, which is critical for liver metastasis from colorectal cancers (de Sousa e Melo et al., 2017).
Epithelial-to-mesenchymal transition (EMT), a phenotypic conversion of epithelial cancer cells to gain mesenchymal properties, such as migration and invasion (Batlle et al., 2000), has garnered substantial attention due to its association with CSCs. The forced EMT phenotype can exacerbate CSC traits, such as tumor-initiating capacity, and CSC-like populations show an increased EMT phenotype in many types of cancer (Batlle and Clevers, 2017). In breast cancer, an increase in the EMT-related transcription factors SNAIL and TWIST not only accelerates the EMT, but also increases the CD44+/CD24- population and mammosphere forming capacity (Mani et al., 2008). In particular, the observations of reversible transition between epithelial and mesenchymal cells and the hybrid EMT phenotype led to the idea that CSC plasticity is attributed to EMT (Gupta et al., 2019; Nieto et al., 2016). The EMT activator zinc finger E-box-binding homeobox 1 (ZEB1) drives the conversion of non-CSCs of basal breast cancers to CSCs, and this phenotypic plasticity is mediated by the change in chromatin configuration of the ZEB1 promoter (Chaffer et al., 2013). In addition to experimental observations, a recent simulation study with mechanism-based mathematical modeling showed that the hybrid epithelial/mesenchymal (hybrid E/M) phenotype appears to have more stemness traits than pure mesenchymal cells (Pasani et al., 2020).
CSC AND THERAPY RESISTANCE
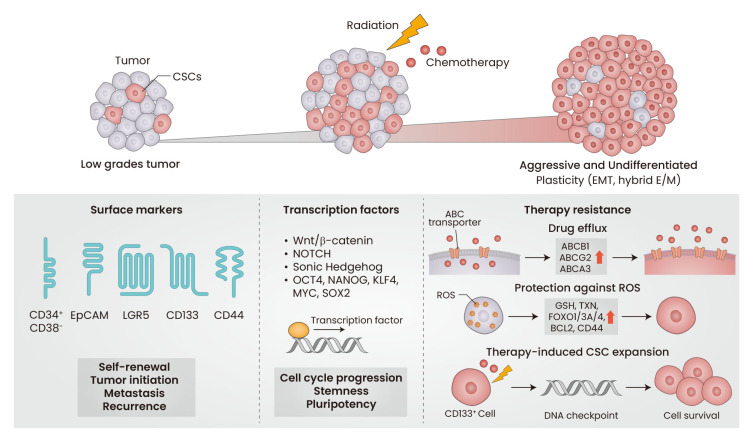
The biggest challenge in cancer treatment is differential sensitivity of intra-tumor cells to anticancer drugs, and the chemotherapy-resistant cells that drive tumor recurrence after the initial treatment are considered CSCs (Fig. 1). This notion is supported by several characteristics of CSCs, including dormancy, upregulation of drug efflux transporters, and enhanced capacity for ROS protection (Nassar and Blanpain, 2016).
Fig. 1. Core properties of cancer stem cells (CSCs).
The increase of CSCs in tumor mass changes the low grades tumors into aggressive and undifferentiated tumors. Several surface markers, including CD34+/CD38- in leukemic cancer, CD44+/CD24- in breast cancer, CD133+ in glioblastoma, LGR5+, and EpCAM+ have been used to isolate CSC-enriched population from tumors. Signaling pathways involved in Wnt/β-catenin, NOTCH, and Sonic Hedgehog play roles in cell cycle progression, stemness, and pluripotency of CSCs. CSCs-associated transcription factors include OCT4, NANOG, KLF4, MYC, and SOX2. Therapy resistance is one of core properties of CSCs. The increase of drug efflux transporters, protection against ROS by antioxidant genes upregulation, and DNA checkpoint elevation lead to survival and maintenance of CSCs. EMT, epithelial-to-mesenchymal transition; E/M, epithelial/mesenchymal; EpCAM, epithelial cell adhesion molecule; LGR5, leucine-rich repeat-containing G-protein coupled receptor 5; NOTCH, neurogenic locus notch homolog protein; OCT4, octamer-binding transcription factor 4; NANOG, homeobox protein NANOG; KLF4, Krüppel-like factor 4; ABC, ATP-binding cassette; ROS, reactive oxygen species; GSH, glutathione; TXN, thioredoxin.
Upregulation of ABC transporters
Drug efflux transporters, including ABCB1 (P-glycoprotein; P-gp) and ABCG2 (breast cancer resistance protein; BCRP), are highly expressed in CSCs to maintain a high efflux capacity for anticancer drugs (Dean et al., 2005). Tumor cells with high ABCG2 and ABCA3 levels, isolated from neuroblastoma patients, showed sustained expansion ex vivo and higher survival after cytotoxic drug treatment (Hirschmann-Jax et al., 2004). The lung cancer SP population, which expresses high levels of ABCG2, is highly tumorigenic and resistant to multiple cancer drugs (Ho et al., 2007). Based on these results, ABCB1 and ABCG2 are often used to isolate CSC-enriched cell populations from tumor tissues (Hadnagy et al., 2006; Ho et al., 2007).
Enhanced protection against ROS
Many conventional chemotherapeutic drugs have been reported to increase ROS and electrophile levels lethally, and CSCs cope with these treatments by upregulating the antioxidant system (Nassar and Blanpain, 2016). The relationship between ROS and stem cell quiescence has been demonstrated in normal stem cells. ROS levels were lower in murine embryonic stem cells (ESC) when compared with those in differentiated cells due to high expression of GSH biosynthesis enzymes and thioredoxin (Saretzki et al., 2004). Knockout of Foxo1/Foxo3a/Foxo4 in mice resulted in the depletion of hematopoietic stem cells (HSC) due to ROS elevation (Tothova et al., 2007).
Considerable evidence has shown that ROS is also involved in CSCs physiology. Breast CSCs retain lower levels of ROS than non-tumorigenic cells due to high levels of GSH synthesis enzymes, and the radiation resistance of CSCs was attributed to low ROS levels (Diehn et al., 2009). Treatment with a BCL-2 inhibitor ablated leukemic stem cells by increasing ROS levels through mitochondrial dysfunction (Lagadinou et al., 2013). FOXO3 activation contributes to low ROS levels in leukemic stem cells; therefore, deletion of FOXO3 blocked the leukemia-initiating capacity of leukemic stem cells (Naka et al., 2010). The mammosphere-derived CD44+/CD24- subpopulation maintained lower levels of ROS following radiation, and thus showed higher viability (Phillips et al., 2006). These results indicate that an enhanced ROS coping system could be a critical determinant of CSCs survival upon radiation therapy and chemotherapy.
Therapy-induced CSC expansion
Cancer cells that survive chemotherapy and radiation therapy are the primary cause of tumor relapse, and CSCs are found to be enriched in residual tumors after treatment. The levels of CD133+ glioma CSCs are highly increased following radiation in vitro and in human glioma xenografts, and activation of the radiation-induced DNA damage checkpoint pathway was found to be a mechanism of CD133+ CSCs enrichment (Bao et al., 2006). Carboplatin treatment induced CSC-like properties in normal HCC, and silencing OCT3/4 and SOX2 blocked this change (Hu et al., 2012). Cancer plasticity and conversion to CSCs following radiation and chemotherapy have been demonstrated. Patient-derived non-breast CSCs can be converted to CSCs following radiation exposure, with concomitant increases in OCT4 and NANOG expression (Lagadec et al., 2012). During EMT in squamous cell carcinoma CSCs, chemoresistance is acquired through conversion to a slow proliferative state (Oshimori et al., 2015). Breast CSCs became more abundant in residual tumor tissue after chemotherapy or endocrine therapy, and an increase in mesenchymal gene expression was accompanied (Creighton et al., 2009).
NRF2 SIGNALING AND CANCER
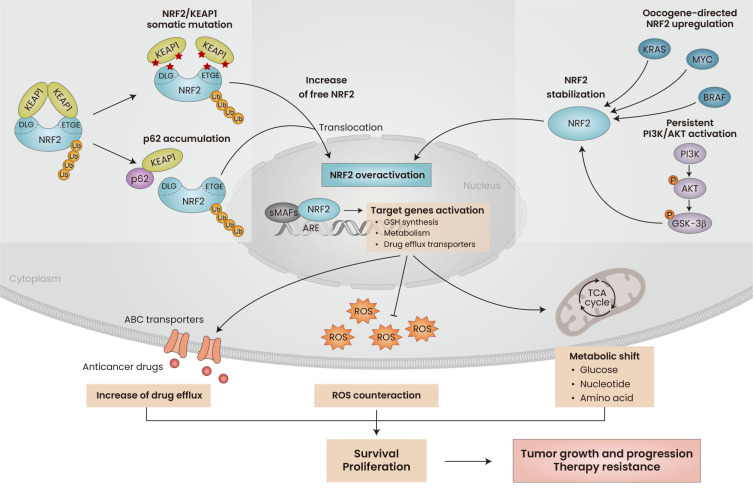
In normal and healthy cells, NRF2 is accepted as a central cytoprotective factor that counteracts ROS/electrophile stress by promoting the removal of deleterious cytotoxic challenges (Baird and Yamamoto, 2020; Cho and Kleeberger, 2020; Taguchi and Kensler, 2020; Tsushima et al., 2020). Extensive efforts have been made to develop therapeutic interventions that increase NRF2 activity to prevent or treat multiple chronic diseases, such as respiratory, cardiovascular, and neurodegenerative diseases, as well as cancer (Cuadrado et al., 2019; Taguchi and Yamamoto, 2020). On the contrary, NRF2 is often overactivated in cancer cells and associated with tumor growth, cancer progression, development of therapy resistance, and poor patient prognosis (Murakami and Motohashi, 2015; Rojo de la Vega et al., 2018). Aberrant NRF2 activation in cancers results from multiple molecular events: (i) somatic mutations and genomic alterations in NRF2 or KEAP1 (Goldstein et al., 2016; Kitamura and Motohashi, 2018; Wang et al., 2008), (ii) oncogene (K-RAS, B-RAS, MYC)-directed upregulation of NRF2 (DeNicola et al., 2011), (iii) phosphoinositide 3-kinases (PI3K)/AKT activation-induced NRF2 stabilization (Best et al., 2018), and (iv) elevation of p62/SQSTM1 and competition with NRF2 for KEAP1 binding (Ichimura et al., 2013).
The notion that cancer cells hijack NRF2 signaling to enhance their survival and growth under a stress-rich tumor microenvironment has been consistently supported by numerous reports that show NRF2 inhibition via pharmacological or genetic methods could suppress tumor growth and progression and improve therapy resistance (Zhu et al., 2016). The beneficial effects of persistent activation of NRF2 on cancer cells can be explained in several ways. NRF2-driven cancer survival is attributed to the increased ROS counteracting system (Singh et al., 2010), increased tumor growth as a result of a metabolic shift to facilitate cell proliferation (Mitsuishi et al., 2012; Romero et al., 2017), and inhibition of mRNA translational regulating factors (Chio et al., 2016). In addition, resistance in NRF2-activated cancers is mediated by the elevated levels of multiple detoxifying enzymes and ABC transporters (Maher et al., 2007) (Fig. 2).
Fig. 2. Role of NRF2 in cancer.
Aberrant NRF2 activation in cancer is often resulted from NRF2/KEAP1 somatic mutation and binding competition by p62 that results in liberation of NRF2 from KEAP1-mediated degradation system. As KEAP1-independent regulation, NRF2 stabilization is achieved by oncogenes-directed upregulation and persistent activation of PI3K/AKT signaling pathway. NRF2 overactivation increases its target genes expression to counteract ROS imbalance, enhance anticancer drug efflux, and re-direct metabolism to increase survival and proliferation, thus supports tumor growth, progression, and therapy resistance. ROS, reactive oxygen species; ABC, ATP-binding cassette; KEAP1, Kelch-like ECH-associated protein 1; DLG, DLG motif; ETGE, ETGE motif; sMAFs, small MAF proteins; ARE, antioxidant response element; GSH, glutathione; TCA, tricarboxylic acid; PI3K, phosphoinositide 3-kinases; GSK-3, glycogen synthase kinase-3.
NRF2 SIGNALING IN CSC
NRF2 signaling in normal stem cells
Evidence indicates an association between NRF2 and stem-like traits in normal stem cells. NRF2 was shown to maintain the balance of quiescence and self-renewal of HSCs, and Nrf2 knockout showed an expanded HSCs pool and progenitor cells in mouse bone marrow (Tsai et al., 2013). NRF2 expression was higher in human ESCs than in non-stem cells, and NRF2 levels decreased in the differentiated state (Jang et al., 2014). Knockdown of NRF2 in hESCs has shown that constitutive NRF2 activity is necessary for the self-renewal and pluripotency of ESCs.
In line with these findings, a link between NRF2 and stem cell-related NOTCH signaling has been demonstrated. NOTCH1 expression was reduced in Nrf2-null cells, and the functional antioxidant response element, an enhancer recognized by the NRF2/sMAF complex, was identified in the murine Notch1 gene. This correlation was further strengthened by an animal study showing delayed liver regeneration in partially hepatectomized Nrf2-knockout mice and the rescue phenotype by NOTCH expression (Wakabayashi et al., 2010). Administration of the NRF2 activator led to NOTCH signaling activation and HPSC reconstitution in irradiated NOTCH reporter mice (Kim et al., 2014). Squamous epithelial cells from the tongue tissues of Keap1-knockout mice showed increased NOTCH and hyperproliferative signaling, confirming a positive correlation between NRF2 and NOTCH (Fan et al., 2017). Interestingly, reciprocal relationship between NRF2 and NOTCH was also observed. Overexpression of the NOTCH intracellular domain (NICD) in mice increases NRF2 expression in enlarged livers, which can be reversed by Nrf2 disruption. NOTCH-mediated NRF2 elevation was found to be a direct effect of NICD on the Nrf2 promoter (Wakabayashi et al., 2014). ROS flux promotes NRF2-mediated self-renewal and proliferation of airway basal stem cells (ABSCs), and NOTCH signaling was a downstream mediator of NRF2 in ABSCs self-renewal regulation (Paul et al., 2014).
NRF2 activation in CSC
Recent accumulating evidence has shown that NRF2 is activated in CSCs, and this increase is crucial for maintaining CSC phenotype. In glioma CSCs, NRF2 expression was negatively correlated with the differentiation state, and the knockdown of NRF2 suppressed tumor growth by differentiating glioma CSCs (Zhu et al., 2014). NRF2 was upregulated in de-differentiated breast epithelial cells, which were established by TWIST overexpression, and the protein kinase R-like endoplasmic reticulum kinase (PERK)-mediated NRF2 activation was responsible for chemotherapy resistance of these cells (Del Vecchio et al., 2014). NRF2 levels were highly upregulated in breast cancer spheres, and treatment with brusatol, a chemical inhibitor of NRF2, sensitized breast cancer spheres to taxol treatment (Wu et al., 2015). Similarly, NRF2/ABCG2 expression was increased in mammospheres and colonospheres, and silencing NRF2 gene suppressed sphere growth and enhanced doxorubicin cytotoxicity (Ryoo et al., 2015; 2016a). Activated NRF2 in ROSlow head and neck CSCs promotes a metabolic shift to glycolysis and maintains CSC stemness by maintaining low ROS levels (Chang et al., 2018).
Flow cytometry-based CSC isolation also suggests a correlation between CSC and NRF2 activation. Proteome analysis showed that HCC CSCs with CD44 variant 9 (CD44v9) express high levels of NRF2 compared to CD44v9- HCC (Kakehashi et al., 2016). The levels of NRF2 and its target genes are high in the CD44+/CD24- breast CSCs population, and NRF2 activation is necessary for CSCs survival and therapy resistance (Ryoo et al., 2018). Colorectal CSCs with CD133+ showed high levels of NRF2-driven ABCB1 and ABCG2 expression resulting in drug resistance (Goto et al., 2020; Park et al., 2022). Ovarian CSCs populations with high ALDH expression showed NRF2 signaling activation, and NRF2 silencing blocked CSC traits, such as anchorage-independent growth, migration, sphere formation, therapy resistance, and tumor growth (Kim et al., 2018a). The ALDH+ breast CSCs are resistance to radiotherapy and NRF2-mediated ALDH elevation has been reported to contribute to this resistance (Kamble et al., 2021). The EpCAMhigh cell population is enriched in cisplatin-resistant head and neck squamous cell carcinoma, and high levels of NRF2, mediated by IL-6 elevation and p62 accumulation, are responsible for treatment resistance (Noman et al., 2020). These results indicate that the NRF2 pathway is upregulated in multiple CSC models and plays a crucial role in survival, maintenance, and therapy-resistant CSCs.
Association of NRF2 with CSC signaling
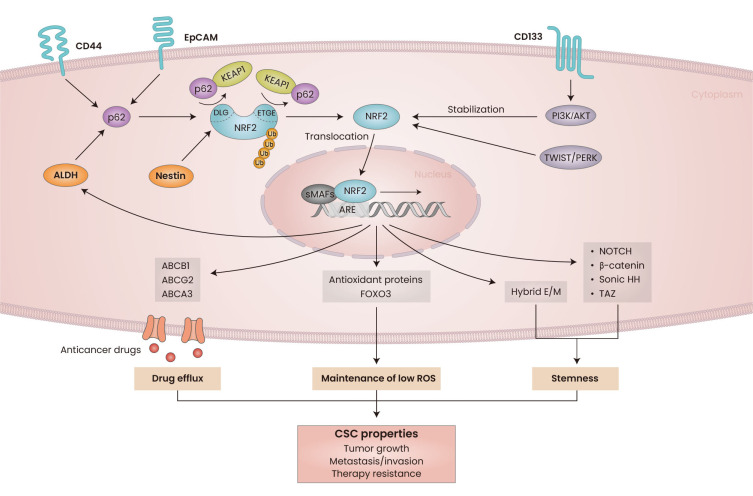
The molecular events that define the role of NRF2 in CSCs signaling have been identified (Fig. 3). In particular, the expression levels of key stemness molecule NOTCH have been linked to NRF2 in multiple types of cancer. Loss of KEAP1, which results in persistent NRF2 activation, increases the self-renewal capacity of head and neck CSCs through NRF2 elevation and subsequently NOTCH signaling activation (Islam et al., 2022). Radiation-induced lung cancer migration depends on NOTCH1 signaling, and NRF2 inhibition suppresses metastasis with concomitant NOTCH1 level reduction (Zhao et al., 2017). In breast cancer cells, carbon monoxide (CO), derived from NRF2/heme oxygenase-1 elevation, stimulates mammosphere formation through upregulation of NOTCH1 expression (Kim et al., 2018b). Constitutive NRF2 activation in KEAP1-mutated lung cancer cells induced unique remodeling of the enhancer of NOTCH3 gene, and the cooperation between CCAAT/enhancer-binding protein β (CEBPB) and NRF2 promoted NOTCH3 elevation, which resulted in enhanced tumor-initiating capacity (Okazaki et al., 2020a). In a subsequent study, NRF2 was found to increase CEBPB expression directly, and NRF2-CEBPB cooperation regulated additional gene sets involved in chemoresistance in NRF2-active lung cancers (Okazaki et al., 2022).
Fig. 3. Association of NRF2 with cancer stem cells (CSCs) properties.
NRF2 signaling is activated in CSCs and contributes to CSC properties, such as tumor initiation, metastatic malignancy, and therapy resistance. NRF2 is activated by CSC markers such as CD44, EpCAM, and ALDH, and p62 accumulation is associated with NRF2 activation. CD133 leads to NRF2 stabilization through PI3K/AKT pathway activation. TWIST-mediated PERK activation directly induce NRF2 accumulation. Competitive binding of Nestin with KEAP1 induces NRF2 liberation and translocation into the nucleus. NRF2 upregulates multiple antioxidant defense genes and FOXO3 to maintain low ROS levels. ABC transporters, including ABCB1, ABCG2, and ABCA3, are upregulated by NRF2 and contribute to chemotherapy resistance. High level of NRF2 is also associated with the upregulation of transcription factors, including NOTCH1/3, Sonic Hedgehog, β-catenin, and TAZ to maintain stemness of cancer cells. NRF2 activation stabilizes cells in hybrid epithelial/mesenchymal (hybrid E/M) state to support phenotypic conversion to CSCs. ROS, reactive oxygen species; EpCAM, epithelial cell adhesion molecule; PI3K, phosphoinositide 3-kinases; sMAFs, samll MAF proteins; ARE, antioxidant response element; KEAP1, Kelch-like ECH-associated protein 1; DLG, DLG motif; ETGE, ETGE motif; ALDH, aldehyde dehydrogenase; NOTCH, neurogenic locus notch homolog protein; HH, hedgehog homolog.
In addition to NOTCH, NRF2 has been reported to contribute to the regulation of other stemness-related molecules in cancers. Activated NRF2 signaling in breast CSCs upregulates forkhead box protein O3 (FOXO3) and downstream BMI-1 expression via reductive stress, resulting in an enhanced self-renewal capacity (Kim et al., 2020). NRF2/FOXM1-mediated upregulation of sulfiredoxin-peroxiredoxin contributes to the stemness and survival of colon CSCs (Escoll et al., 2020; Song et al., 2021). β-Catenin expression was directly enhanced by NRF2; therefore, persistent activation of NRF2/β-catenin promotes hepatic stem cell proliferation and subsequently initiates tumorigenesis (Fragoulis et al., 2022). NRF2, which is activated in liver tumor-initiating cells, can directly upregulate sonic hedgehog homolog to activate the sonic hedgehog pathway for tumorigenesis (Leung et al., 2020). Nestin, a type IV intermediate filament protein highly expressed in stem cells and cancer cells, competitively interacts with the Kelch domain of the KEAP1 protein, thus, stabilizing the NRF2 protein and leading to oxidative stress resistance and malignancy initiation in non-small cell lung cancer (Wang et al., 2019). High NRF2 levels in glioblastoma CSCs induce tumorigenesis by directly elevating the expression of TAZ, a Hippo pathway effector participating in cancer migration, invasion, and stemness.
p62 (encoded by SQSTM1 gene), an autophagy adaptor protein involved in selective autophagy, has been suggested to act as an oncoprotein (Ichimura et al., 2013; Li et al., 2013). Recent evidence indicates the role of p62 in CSCs: selective p62 inhibition attenuated the cancer-initiating capacity of acute myeloid leukemia cells (Li et al., 2021). Furthermore, there are considerable reports indicating that the increase of NRF2 in CSCs is associated with p62. Accumulation of p62 was found to be necessary for HCC-initiated cell survival and expansion by increasing NRF2 activity (Umemura et al., 2016). High levels of p62 are directly associated with NRF2 signaling activation in multiple CSCs models, including the CD44+/CD24- breast CSCs, ALDH+ ovarian CSCs, and EpCAM+ HNSC CSCs (Kim et al., 2018a; Noman et al., 2020; Ryoo et al., 2018). The interplay between CSCs and the niche microenvironment is important for tumor progression and therapy resistance. Tumor-initiating cells from NRF2-high squamous cell carcinoma stimulated the release of IL-33, which promotes the differentiation of macrophages to secrete transforming growth factor β (TGF-β). CSCs-mediated paracrine effect on TGF-β signaling consequently induces cancer invasion and resistance (Taniguchi et al., 2020).
NRF2 AND CANCER PLASTICITY
Given that EMT contributes to the plasticity of CSC and non-CSCs, the potential relationship between NRF2 and EMT is an intriguing question. Several lines of evidence indicate the involvement of NRF2 in EMT; however, this correlation does not seem to be consistent. E-cadherin, a marker of epithelial cells, was shown to inhibit NRF2 accumulation through direct binding and KEAP1-dependent proteasomal degradation; thus, loss of E-cadherin in EMT could increase NRF2 levels, which results in cancer resistance to chemotherapy (Kim et al., 2012). In another study, NRF2 activation suppressed E-cadherin expression through an unidentified mechanism and increased the invasion of pancreatic ductal carcinoma cells (Arfmann-Knübel et al., 2015). In lung cancer cells, TGF-β, a potent inducer of EMT, increases ROS and activates NRF2 signaling. This event is necessary for NOTCH4 induction and TGF-β-induced EMT (Yazaki et al., 2021). However, loss of NRF2 can enhance HCC motility with a concomitant decrease in E-cadherin and an increase in the EMT transcription factor zinc finger protein SNAI2 (SLUG) (Rachakonda et al., 2010). Nrf2-knockout mice develop a higher degree of lung metastasis following inoculation with mouse lung carcinoma cells, and Kepa1-knockdown mice are resistant to cancer cell migration to the lungs (Satoh et al., 2010). TGF-β-induced cancer migration and associated signaling activation were higher in NRF2-silenced lung cancer cells (Ryu et al., 2020).
Conflicting observations suggest that the association between NRF2 and cancer plasticity might be a phase-specific phenomenon. Several recent studies have emphasized the role of hybrid E/M cells, in cancer stemness, which have both epithelial and mesenchymal features. The computational-experimental approach revealed that NRF2 could prevent complete EMT, while stabilizing a hybrid E/M phenotype by upregulating E-cadherin and ZEB-1 in lung cancer and bladder cancer cells (Bocci et al., 2019). In particular, this study showed that the NRF2 levels are maximally increased in the hybrid E/M state, and NRF2 knockdown destabilized the hybrid E/M phenotype. Consistently, a simulation study of network dynamics suggested that NRF2 is a stabilizing factor for the hybrid E/M phenotype (Pasani et al., 2020). In silico-in vitro analysis, NRF2 activation enhances the hybrid E/M phenotype at the migrating edge in a wound healing assay, and the involvement of NOTCH signaling was also confirmed in experimental settings (Vilchez Mercedes et al., 2022). Although sufficient evidence is lacking, these reports suggest the possibility that NRF2 can change and function during the plastic phase of cancer. Therefore, the functional identification of NRF2 in cancer plasticity is expected to provide clues to control the emergence of CSC properties.
CONCLUSION AND PERSPECTIVES
In this review, we examine recent evidence that demonstrates NRF2 signaling is activated in CSCs and contributes to CSC properties, such as tumor initiation, metastatic malignancy, and therapy resistance. To date, multiple CSC markers and transcription factors have been reported to be associated with the upregulation of NRF2 expression in CSCs. In particular, the reciprocal regulation of NRF2 and NOTCH signaling could explain the changes and functions of NRF2 in CSCs. Activated NRF2 signaling in CSCs has been primarily attributed to maintaining intracellular ROS levels low, which leads to enhanced CSC survival after ROS-generating therapy. NRF2 activation also induces resistance to chemotherapy by increasing ABC transporter expression.
Considering the multifaceted role of NRF2 in cancer cells, the function of NRF2 in CSCs is expected to expand. For instance, CSCs are believed to favor glycolysis and pentose phosphate pathway (PPP) metabolism, which aids in maintaining low ROS levels by inhibiting mitochondrial oxidative phosphorylation and NADPH generation (Tuy et al., 2021). From this perspective, activation of the PPP pathway or changes in amino acid metabolic pathways, which are observed in NRF2-activated cancers, may contribute to metabolic changes in CSCs (Hayes and Dinkova-Kostova, 2014; Okazaki et al., 2020b). The immune evasion property is another hallmark of CSCs. The immune response of CSCs is reprogrammed to promote tumor immune escape, resulting in immunotherapy failure (Bayik and Lathia, 2021). As NRF2 was found to upregulate programmed cell death ligand 1 (PD-L1), an immune checkpoint protein for the inhibition of adaptive immune response, high levels of NRF2 in CSCs may contribute to the immune evasion traits of CSCs (Shen et al., 2020; Zhu et al., 2018). The CSC microenvironment is an important contributor to the regulation of CSC fate. In particular, secretory factors derived from the tumor niche have been shown to activate self-renewal and differentiation of CSCs (Batlle and Clevers, 2017). Proteome analysis revealed that the NRF2 antioxidant system is enriched in conditioned media from colorectal CSCs (Emmink et al., 2013). NRF2 is also involved in the secretion of cytokines from non-cancerous and cancerous cells (Kitamura et al., 2017; Taniguchi et al., 2020). Considering these reports, it is probable that NRF2 signaling contributes to the regulation of the crosstalk between CSCs and their microenvironment.
Based on the context discussed above, a novel strategy for targeting NRF2 and CSCs can be developed. EpCam antibodies, such as adecatumumab and γ-secretase inhibitors targeting NOTCH signaling, have been developed as CSC targeting therapies (Yang et al., 2020). Considering the increase in NRF2 in EpCAM+ CSCs and NOTCH-activated cells (Noman et al., 2020; Wakabayashi et al., 2014), it would be interesting to determine whether these drugs improve CSC resistance by suppressing NRF2 signaling. NRF2 inhibitors are being developed to control tumorigenesis and malignant progression. Although the issue of selectivity between normal cells and cancer cells remains, several synthetic and natural compounds, including brusatol, chrysin, and trigonelline, have been investigated to demonstrate their inhibitory effects on tumor growth, malignant progression, and therapy resistance (Cuadrado et al., 2019; Panieri et al., 2020; Taguchi and Yamamoto, 2020). The development of NRF2 inhibitors that selectively act on CSCs is expected to be a promising strategy for suppressing CSC survival and malignant properties.
ACKNOWLEDGMENTS
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2022R1A2C2011866, 2018R1A6A1A03025108). This study was also supported by The Catholic University of Korea, Research Fund 2021.
Footnotes
AUTHOR CONTRIBUTIONS
M.K.K. supervised the overall process and wrote the manuscript. J.M.K. and S.P.H. contributed manuscript writing and figure preparation.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Arfmann-Knübel S., Struck B., Genrich G., Helm O., Sipos B., Sebens S., Schäfer H. The crosstalk between Nrf2 and TGF-β1 in the epithelial-mesenchymal transition of pancreatic duct epithelial cells. PLoS One. 2015;10:e0132978. doi: 10.1371/journal.pone.0132978.31ef7273707649fab7aa4e81fd788d71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell. Biol. 2020;40:e00099–20. doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Batlle E., Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Bayik D., Lathia J.D. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer. 2021;21:526–536. doi: 10.1038/s41568-021-00366-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B., Lapouge G., Rorive S., Drogat B., Desaedelaere K., Delafaille S., Dubois C., Salmon I., Willekens K., Marine J.C., et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Best S.A., De Souza D.P., Kersbergen A., Policheni A.N., Dayalan S., Tull D., Rathi V., Gray D.H., Ritchie M.E., McConville M.J., et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metab. 2018;27:935–943.e4. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Bjerkvig R., Tysnes B.B., Aboody K.S., Najbauer J., Terzis A.J. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat. Rev. Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- Bocci F., Tripathi S.C., Vilchez Mercedes S.A., George J.T., Casabar J.P., Wong P.K., Hanash S.M., Levine H., Onuchic J.N., Jolly M.K. NRF2 activates a partial epithelial-mesenchymal transition and is maximally present in a hybrid epithelial/mesenchymal phenotype. Integr. Biol. (Camb.) 2019;11:251–263. doi: 10.1093/intbio/zyz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Caramel J., Papadogeorgakis E., Hill L., Browne G.J., Richard G., Wierinckx A., Saldanha G., Osborne J., Hutchinson P., Tse G., et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Chaffer C.L., Brueckmann I., Scheel C., Kaestli A.J., Wiggins P.A., Rodrigues L.O., Brooks M., Reinhardt F., Su Y., Polyak K., et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer C.L., Marjanovic N.D., Lee T., Bell G., Kleer C.G., Reinhardt F., D'Alessio A.C., Young R.A., Weinberg R.A. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.W., Chen Y.S., Tsay Y.G., Han C.L., Chen Y.J., Yang C.C., Hung K.F., Lin C.H., Huang T.Y., Kao S.Y., et al. ROS-independent ER stress-mediated NRF2 activation promotes warburg effect to maintain stemness-associated properties of cancer-initiating cells. Cell Death Dis. 2018;9:194. doi: 10.1038/s41419-017-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio I.I.C., Jafarnejad S.M., Ponz-Sarvise M., Park Y., Rivera K., Palm W., Wilson J., Sangar V., Hao Y., Öhlund D., et al. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell. 2016;166:963–976. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.Y., Kleeberger S.R. Mitochondrial biology in airway pathogenesis and the role of NRF2. Arch. Pharm. Res. 2020;43:297–320. doi: 10.1007/s12272-019-01182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.H., Kim J.M., Kwak M.K. The multifaceted role of NRF2 in cancer progression and cancer stem cells maintenance. Arch. Pharm. Res. 2021;44:263–280. doi: 10.1007/s12272-021-01316-8. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A.R., Poletti A., et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Creighton C.J., Li X., Landis M., Dixon J.M., Neumeister V.M., Sjolund A., Rimm D.L., Wong H., Rodriguez A., Herschkowitz J.I., et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- Dean M., Fojo T., Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Del Vecchio C.A., Feng Y., Sokol E.S., Tillman E.J., Sanduja S., Reinhardt F., Gupta P.B. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol. 2014;12:e1001945. doi: 10.1371/journal.pbio.1001945.fea5fd64e6e84ad395b3ca4c79149cf3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmink B.L., Verheem A., Van Houdt W.J., Steller E.J., Govaert K.M., Pham T.V., Piersma S.R., Borel Rinkes I.H., Jimenez C.R., Kranenburg O. The secretome of colon cancer stem cells contains drug-metabolizing enzymes. J. Proteomics. 2013;91:84–96. doi: 10.1016/j.jprot.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Escoll M., Lastra D., Pajares M., Robledinos-Antón N., Rojo A.I., Fernández-Ginés R., Mendiola M., Martínez-Marín V., Esteban I., López-Larrubia P., et al. Transcription factor NRF2 uses the Hippo pathway effector TAZ to induce tumorigenesis in glioblastomas. Redox Biol. 2020;30:101425. doi: 10.1016/j.redox.2019.101425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Paiboonrungruan C., Zhang X., Prigge J.R., Schmidt E.E., Sun Z., Chen X. Nrf2 regulates cellular behaviors and Notch signaling in oral squamous cell carcinoma cells. Biochem. Biophys. Res. Commun. 2017;493:833–839. doi: 10.1016/j.bbrc.2017.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoulis A., Schenkel J., Schröder N., Brandt E.F., Weiand M., Neu T., Ramadori P., Caspers T., Kant S., Pufe T., et al. Nrf2 induces malignant transformation of hepatic progenitor cells by inducing β-catenin expression. Redox Biol. 2022;57:102453. doi: 10.1016/j.redox.2022.102453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti F.G. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L.D., Lee J., Gnad F., Klijn C., Schaub A., Reeder J., Daemen A., Bakalarski C.E., Holcomb T., Shames D.S., et al. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 2016;16:2605–2617. doi: 10.1016/j.celrep.2016.08.010.e594afbe1e7541f5bfa7c4dffdee5106 [DOI] [PubMed] [Google Scholar]
- Goto S., Kawabata T., Li T.S. Enhanced expression of ABCB1 and Nrf2 in CD133-positive cancer stem cells associates with doxorubicin resistance. Stem Cells Int. 2020;2020:8868849. doi: 10.1155/2020/8868849.4ad26161cdc3410a8c5897f26daa22f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C., Lander E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Gupta P.B., Pastushenko I., Skibinski A., Blanpain C., Kuperwasser C. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell. 2019;24:65–78. doi: 10.1016/j.stem.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadnagy A., Gaboury L., Beaulieu R., Balicki D. SP analysis may be used to identify cancer stem cell populations. Exp. Cell Res. 2006;312:3701–3710. doi: 10.1016/j.yexcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Harrison H., Farnie G., Howell S.J., Rock R.E., Stylianou S., Brennan K.R., Bundred N.J., Clarke R.B. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Hirschmann-Jax C., Foster A.E., Wulf G.G., Nuchtern J.G., Jax T.W., Gobel U., Goodell M.A., Brenner M.K. A distinct "side population" of cells with high drug efflux capacity in human tumor cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M.M., Ng A.V., Lam S., Hung J.Y. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- Hu X., Ghisolfi L., Keates A.C., Zhang J., Xiang S., Lee D.K., Li C.J. Induction of cancer cell stemness by chemotherapy. Cell Cycle. 2012;11:2691–2698. doi: 10.4161/cc.21021. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Islam S.S., Qassem K., Islam S., Parag R.R., Rahman M.Z., Farhat W.A., Yeger H., Aboussekhra A., Karakas B., Noman A.S.M. Genetic alterations of Keap1 confers chemotherapeutic resistance through functional activation of Nrf2 and Notch pathway in head and neck squamous cell carcinoma. Cell Death Dis. 2022;13:696. doi: 10.1038/s41419-022-05126-8.63807b26d5284eb1ba2fb9405a2b8a18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J., Wang Y., Kim H.S., Lalli M.A., Kosik K.S. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2014;32:2616–2625. doi: 10.1002/stem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V., Fields A.P. Molecular pathways: novel approaches for improved therapeutic targeting of Hedgehog signaling in cancer stem cells. Clin. Cancer Res. 2015;21:505–513. doi: 10.1158/1078-0432.CCR-14-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahroba H., Shirmohamadi M., Hejazi M.S., Samadi N. The role of Nrf2 signaling in cancer stem cells: from stemness and self-renewal to tumorigenesis and chemoresistance. Life Sci. 2019;239:116986. doi: 10.1016/j.lfs.2019.116986. [DOI] [PubMed] [Google Scholar]
- Kakehashi A., Ishii N., Sugihara E., Gi M., Saya H., Wanibuchi H. CD44 variant 9 is a potential biomarker of tumor initiating cells predicting survival outcome in hepatitis C virus-positive patients with resected hepatocellular carcinoma. Cancer Sci. 2016;107:609–618. doi: 10.1111/cas.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamble D., Mahajan M., Dhat R., Sitasawad S. Keap1-Nrf2 pathway regulates ALDH and contributes to radioresistance in breast cancer stem cells. Cells. 2021;10:83. doi: 10.3390/cells10010083.00e1a97e69a647c8bc4ca51560353b7d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Choi B.H., Ryoo I.G., Kwak M.K. High NRF2 level mediates cancer stem cell-like properties of aldehyde dehydrogenase (ALDH)-high ovarian cancer cells: inhibitory role of all-trans retinoic acid in ALDH/NRF2 signaling. Cell Death Dis. 2018a;9:896. doi: 10.1038/s41419-018-0903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Jang J.H., Kwon O.S., Cha H.J., Youn H.J., Chun K.S., Surh Y.J. Nuclear factor erythroid-derived 2-like 2-induced reductive stress favors self-renewal of breast cancer stem-like cells via the FoxO3a-Bmi-1 axis. Antioxid. Redox Signal. 2020;32:1313–1329. doi: 10.1089/ars.2019.7730. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Yoon H.J., Cha Y.N., Surh Y.J. Role of heme oxygenase-1 and its reaction product, carbon monoxide, in manifestation of breast cancer stem cell-like properties: Notch-1 as a putative target. Free Radic. Res. 2018b;52:1336–1347. doi: 10.1080/10715762.2018.1473571. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Thimmulappa R.K., Kumar V., Cui W., Kumar S., Kombairaju P., Zhang H., Margolick J., Matsui W., Macvittie T., et al. NRF2-mediated Notch pathway activation enhances hematopoietic reconstitution following myelosuppressive radiation. J. Clin. Invest. 2014;124:730–741. doi: 10.1172/JCI70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.D., Kim Y.W., Cho I.J., Lee C.H., Kim S.G. E-cadherin inhibits nuclear accumulation of Nrf2: implications for chemoresistance of cancer cells. J. Cell Sci. 2012;125:1284–1295. doi: 10.1242/jcs.095422. [DOI] [PubMed] [Google Scholar]
- Kitamura H., Motohashi H. NRF2 addiction in cancer cells. Cancer Sci. 2018;109:900–911. doi: 10.1111/cas.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura H., Onodera Y., Murakami S., Suzuki T., Motohashi H. IL-11 contribution to tumorigenesis in an NRF2 addiction cancer model. Oncogene. 2017;36:6315–6324. doi: 10.1038/onc.2017.236. [DOI] [PubMed] [Google Scholar]
- Kreso A., van Galen P., Pedley N.M., Lima-Fernandes E., Frelin C., Davis T., Cao L., Baiazitov R., Du W., Sydorenko N., et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 2014;20:29–36. doi: 10.1038/nm.3418. [DOI] [PubMed] [Google Scholar]
- Lagadec C., Vlashi E., Della Donna L., Dekmezian C., Pajonk F. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30:833–844. doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou E.D., Sach A., Callahan K., Rossi R.M., Neering S.J., Minhajuddin M., Ashton J.M., Pei S., Grose V., O'Dwyer K.M. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Kwon Y.W., Kim H.R., Shin N., Son H.J., Cheong C.S., Kim D.J., Hwang O. A novel pyrazolo[3,4-d]pyrimidine induces heme oxygenase-1 and exerts anti-inflammatory and neuroprotective effects. Mol. Cells. 2022;45:134–147. doi: 10.14348/molcells.2021.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H.W., Lau E.Y.T., Leung C.O.N., Lei M.M.L., Mok E.H.K., Ma V.W.S., Cho W.C.S., Ng I.O.L., Yun J.P., Cai S.H., et al. NRF2/SHH signaling cascade promotes tumor-initiating cell lineage and drug resistance in hepatocellular carcinoma. Cancer Lett. 2020;476:48–56. doi: 10.1016/j.canlet.2020.02.008. [DOI] [PubMed] [Google Scholar]
- Li L., Shen C., Nakamura E., Ando K., Signoretti S., Beroukhim R., Cowley G.S., Lizotte P., Liberzon E., Bair S., et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell. 2013;24:738–750. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li Y., Yin J., Wang C., Yang M., Gu J., He M., Xu H., Fu W., Zhang W., et al. A mitophagy inhibitor targeting p62 attenuates the leukemia-initiation potential of acute myeloid leukemia cells. Cancer Lett. 2021;510:24–36. doi: 10.1016/j.canlet.2021.04.003. [DOI] [PubMed] [Google Scholar]
- Lytle N.K., Barber A.G., Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer. 2018;18:669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J.M., Dieter M.Z., Aleksunes L.M., Slitt A.L., Guo G., Tanaka Y., Scheffer G.L., Chan J.Y., Manautou J.E., Chen Y., et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema J.P. Cancer stem cells: the challenges ahead. Nat. Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- Meyer M.J., Fleming J.M., Lin A.F., Hussnain S.A., Ginsburg E., Vonderhaar B.K. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70:4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Murakami S., Motohashi H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic. Biol. Med. 2015;88(Pt B):168–178. doi: 10.1016/j.freeradbiomed.2015.06.030. [DOI] [PubMed] [Google Scholar]
- Murakami S., Ninomiya W., Sakamoto E., Shibata T., Akiyama H., Tashiro F. SRY and OCT4 are required for the acquisition of cancer stem cell-like properties and are potential differentiation therapy targets. Stem Cells. 2015;33:2652–2663. doi: 10.1002/stem.2059. [DOI] [PubMed] [Google Scholar]
- Naka K., Hoshii T., Muraguchi T., Tadokoro Y., Ooshio T., Kondo Y., Nakao S., Motoyama N., Hirao A. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- Nassar D., Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- Neradil J., Veselska R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015;106:803–811. doi: 10.1111/cas.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.V., Vanner R., Dirks P., Eaves C.J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Noman A.S.M., Parag R.R., Rashid M.I., Islam S., Rahman M.Z., Chowdhury A.A., Sultana A., Jerin C., Siddiqua A., Rahman L., et al. Chemotherapeutic resistance of head and neck squamous cell carcinoma is mediated by EpCAM induction driven by IL-6/p62 associated Nrf2-antioxidant pathway activation. Cell Death Dis. 2020;11:663. doi: 10.1038/s41419-020-02907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K., Anzawa H., Katsuoka F., Kinoshita K., Sekine H., Motohashi H. CEBPB is required for NRF2-mediated drug resistance in NRF2-activated non-small cell lung cancer cells. J. Biochem. 2022;171:567–578. doi: 10.1093/jb/mvac013. [DOI] [PubMed] [Google Scholar]
- Okazaki K., Anzawa H., Liu Z., Ota N., Kitamura H., Onodera Y., Alam M.M., Matsumaru D., Suzuki T., Katsuoka F., et al. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat. Commun. 2020a;11:5911. doi: 10.1038/s41467-020-19593-0.e2fd0ce97bcb4b3aabb6a909e9e673dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K., Papagiannakopoulos T., Motohashi H. Metabolic features of cancer cells in NRF2 addiction status. Biophys. Rev. 2020b;12:435–441. doi: 10.1007/s12551-020-00659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., Oristian D., Fuchs E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160:963–976. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panieri E., Buha A., Telkoparan-Akillilar P., Cevik D., Kouretas D., Veskoukis A., Skaperda Z., Tsatsakis A., Wallace D., Suzen S., et al. Potential applications of NRF2 modulators in cancer therapy. Antioxidants (Basel) 2020;9:193. doi: 10.3390/antiox9030193.232290a69e0a4704a98074c0afcb06a9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Kim S.K., Hallis S.P., Choi B.H., Kwak M.K. Role of CD133/NRF2 axis in the development of colon cancer stem cell-like properties. Front. Oncol. 2022;11:808300. doi: 10.3389/fonc.2021.808300.1f0b1ac739704af0a6d2816c4bb15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasani S., Sahoo S., Jolly M.K. Hybrid E/M phenotype(s) and stemness: a mechanistic connection embedded in network topology. J. Clin. Med. 2020;10:60. doi: 10.3390/jcm10010060.efe89c58175f4ba8925d39c8f76a119d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.K., Bisht B., Darmawan D.O., Chiou R., Ha V.L., Wallace W.D., Chon A.T., Hegab A.E., Grogan T., Elashoff D.A. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent notch signaling. Cell Stem Cell. 2014;15:199–214. doi: 10.1016/j.stem.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T.M., McBride W.H., Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J. Natl. Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M.A., Daidone M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Prasetyanti P.R., Medema J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer. 2017;16:41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachakonda G., Sekhar K.R., Jowhar D., Samson P.C., Wikswo J.P., Beauchamp R.D., Datta P.K., Freeman M.L. Increased cell migration and plasticity in Nrf2-deficient cancer cell lines. Oncogene. 2010;29:3703–3714. doi: 10.1038/onc.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo S., Vieira A.F., Gerhard R., Leitão D., Pinto R., Cameselle-Teijeiro J.F., Milanezi F., Schmitt F., Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sánchez-Rivera F.J., et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo I.G., Choi B.H., Ku S.K., Kwak M.K. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: implications for cancer stem cell resistance. Redox Biol. 2018;17:246–258. doi: 10.1016/j.redox.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo I.G., Choi B.H., Kwak M.K. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget. 2015;6:8167–8184. doi: 10.18632/oncotarget.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo I.G., Kim G., Choi B.H., Lee S.H., Kwak M.K. Involvement of NRF2 signaling in doxorubicin resistance of cancer stem cell-enriched colonospheres. Biomol. Ther. (Seoul) 2016a;24:482–488. doi: 10.4062/biomolther.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo I.G., Lee S.H., Kwak M.K. Redox modulating NRF2: a potential mediator of cancer stem cell resistance. Oxid. Med. Cell. Longev. 2016b;2016:2428153. doi: 10.1155/2016/2428153.7754a378f0334c8aa6105780a9dfde0e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D., Lee J.H., Kwak M.K. NRF2 level is negatively correlated with TGF-β1-induced lung cancer motility and migration via NOX4-ROS signaling. Arch. Pharm. Res. 2020;43:1297–1310. doi: 10.1007/s12272-020-01298-z. [DOI] [PubMed] [Google Scholar]
- Saretzki G., Armstrong L., Leake A., Lako M., von Zglinicki T. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells. 2004;22:962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- Satoh H., Moriguchi T., Taguchi K., Takai J., Maher J.M., Suzuki T., Winnard P.T., Jr., Raman V., Jr., Ebina M., Jr., Nukiwa T., Jr., et al. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis. 2010;31:1833–1843. doi: 10.1093/carcin/bgq105. [DOI] [PubMed] [Google Scholar]
- Shen X., Zhao Y., Liu G., Zhou H.L., Fan J., Zhang L., Li Y.L., Wang Y., Liang J., Xu Z.X. Upregulation of programmed death ligand 1 by liver kinase B1 and its implication in programmed death 1 blockade therapy in non-small cell lung cancer. Life Sci. 2020;256:117923. doi: 10.1016/j.lfs.2020.117923. [DOI] [PubMed] [Google Scholar]
- Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M., Date S., Sugimoto S., Kanai T., Sato T. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature. 2017;545:187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- Singh A., Bodas M., Wakabayashi N., Bunz F., Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid. Redox Signal. 2010;13:1627–1637. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Song I.S., Jeong Y.J., Jung Y., Park Y.H., Shim S., Kim S.J., Eom D.W., Hong S.M., Lee P.C.W., Kim S.U., et al. The sulfiredoxin-peroxiredoxin redox system regulates the stemness and survival of colon cancer stem cells. Redox Biol. 2021;48:102190. doi: 10.1016/j.redox.2021.102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J., Anderson J.E., Storm E.E., Modrusan Z., Koeppen H., et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- Taguchi K., Kensler T.W. Nrf2 in liver toxicology. Arch. Pharm. Res. 2020;43:337–349. doi: 10.1007/s12272-019-01192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K., Yamamoto M. The KEAP1-NRF2 system as a molecular target of cancer treatment. Cancers (Basel) 2020;13:46. doi: 10.3390/cancers13010046.4d4c99c3899147d589942e1b17428ab5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N., Miele L., Harris P.J., Jeong W., Bando H., Kahn M., Yang S.X., Ivy S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi S., Elhance A., Van Duzer A., Kumar S., Leitenberger J.J., Oshimori N. Tumor-initiating cells establish an IL-33-TGF-β niche signaling loop to promote cancer progression. Science. 2020;369:eaay1813. doi: 10.1126/science.aay1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.D., Poppleton H., Fuller C., Su X., Liu Y., Jensen P., Magdaleno S., Dalton J., Calabrese C., Board J., et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Tebay L.E., Robertson H., Durant S.T., Vitale S.R., Penning T.M., Dinkova-Kostova A.T., Hayes J.D. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015;88(Pt B):108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tsai J.J., Dudakov J.A., Takahashi K., Shieh J.H., Velardi E., Holland A.M., Singer N.V., West M.L., Smith O.M., Young L.F. Nrf2 regulates haematopoietic stem cell function. Nat. Cell Biol. 2013;15:309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima M., Liu J., Hirao W., Yamazaki H., Tomita H., Itoh K. Emerging evidence for crosstalk between Nrf2 and mitochondria in physiological homeostasis and in heart disease. Arch. Pharm. Res. 2020;43:286–296. doi: 10.1007/s12272-019-01188-z. [DOI] [PubMed] [Google Scholar]
- Tuy K., Rickenbacker L., Hjelmeland A.B. Reactive oxygen species produced by altered tumor metabolism impacts cancer stem cell maintenance. Redox Biol. 2021;44:101953. doi: 10.1016/j.redox.2021.101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A., He F., Taniguchi K., Nakagawa H., Yamachika S., Font-Burgada J., Zhong Z., Subramaniam S., Raghunandan S., Duran A., et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E.M.F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- Vilchez Mercedes S.A., Bocci F., Ahmed M., Eder I., Zhu N., Levine H., Onuchic J.N., Jolly M.K., Wong P.K. Nrf2 modulates the hybrid epithelial/mesenchymal phenotype and Notch signaling during collective cancer migration. Front. Mol. Biosci. 2022;9:807324. doi: 10.3389/fmolb.2022.807324.a10c830fc1e94e2190da9ca5b7979327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N., Shin S., Slocum S.L., Agoston E.S., Wakabayashi J., Kwak M.K., Misra V., Biswal S., Yamamoto M., Kensler T.W. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci. Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N., Skoko J.J., Chartoumpekis D.V., Kimura S., Slocum S.L., Noda K., Palliyaguru D.L., Fujimuro M., Boley P.A., Tanaka Y., et al. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol. Cell. Biol. 2014;34:653–663. doi: 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lu Q., Cai J., Wang Y., Lai X., Qiu Y., Huang Y., Ke Q., Zhang Y., Guan Y., et al. Nestin regulates cellular redox homeostasis in lung cancer through the Keap1-Nrf2 feedback loop. Nat. Commun. 2019;10:5043. doi: 10.1038/s41467-019-12925-9.b1b00c42536b4061b50abe0984df2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., An J., Ji F., Jiao H., Sun H., Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wu T., Harder B.G., Wong P.K., Lang J.E., Zhang D.D. Oxidative stress, mammospheres and Nrf2-new implication for breast cancer therapy? Mol. Carcinog. 2015;54:1494–1502. doi: 10.1002/mc.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Shi P., Zhao G., Xu J., Peng W., Zhang J., Zhang G., Wang X., Dong Z., Chen F., et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K., Matsuno Y., Yoshida K., Sherpa M., Nakajima M., Matsuyama M., Kiwamoto T., Morishima Y., Ishii Y., Hizawa N. ROS-Nrf2 pathway mediates the development of TGF-β1-induced epithelial-mesenchymal transition through the activation of Notch signaling. Eur. J. Cell Biol. 2021;100:151181. doi: 10.1016/j.ejcb.2021.151181. [DOI] [PubMed] [Google Scholar]
- Yung W.K., Shapiro J.R., Shapiro W.R. Heterogeneous chemosensitivities of subpopulations of human glioma cells in culture. Cancer Res. 1982;42:992–998. [PubMed] [Google Scholar]
- Zhao Q., Mao A., Guo R., Zhang L., Yan J., Sun C., Tang J., Ye Y., Zhang Y., Zhang H. Suppression of radiation-induced migration of non-small cell lung cancer through inhibition of Nrf2-Notch Axis. Oncotarget. 2017;8:36603–36613. doi: 10.18632/oncotarget.16622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Schuetz J.D., Bunting K.D., Colapietro A.M., Sampath J., Morris J.J., Lagutina I., Grosveld G.C., Osawa M., Nakauchi H., et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- Zhu B., Tang L., Chen S., Yin C., Peng S., Li X., Liu T., Liu W., Han C., Stawski L., et al. Targeting the upstream transcriptional activator of PD-L1 as an alternative strategy in melanoma therapy. Oncogene. 2018;37:4941–4954. doi: 10.1038/s41388-018-0314-0. [DOI] [PubMed] [Google Scholar]
- Zhu J., Wang H., Chen F., Fu J., Xu Y., Hou Y., Kou H.H., Zhai C., Nelson M.B., Zhang Q., et al. An overview of chemical inhibitors of the Nrf2-ARE signaling pathway and their potential applications in cancer therapy. Free Radic. Biol. Med. 2016;99:544–556. doi: 10.1016/j.freeradbiomed.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Zhu J., Wang H., Fan Y., Hu Y., Ji X., Sun Q., Liu H. Knockdown of nuclear factor erythroid 2-related factor 2 by lentivirus induces differentiation of glioma stem-like cells. Oncol. Rep. 2014;32:1170–1178. doi: 10.3892/or.2014.3320. [DOI] [PubMed] [Google Scholar]