Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2) mediates the cellular antioxidant response, allowing adaptation and survival under conditions of oxidative, electrophilic and inflammatory stress, and has a role in metabolism, inflammation and immunity. Activation of Nrf2 provides broad and long-lasting cytoprotection, and is often hijacked by cancer cells, allowing their survival under unfavorable conditions. Moreover, Nrf2 activation in established human tumors is associated with resistance to chemo-, radio-, and immunotherapies. In addition to cancer cells, Nrf2 activation can also occur in tumor-associated macrophages (TAMs) and facilitate an anti-inflammatory, immunosuppressive tumor immune microenvironment (TIME). Several cancer cell-derived metabolites, such as itaconate, L-kynurenine, lactic acid and hyaluronic acid, play an important role in modulating the TIME and tumor-TAMs crosstalk, and have been shown to activate Nrf2. The effects of Nrf2 in TIME are context-depended, and involve multiple mechanisms, including suppression of pro-inflammatory cytokines, increased expression of programmed cell death ligand 1 (PD-L1), macrophage colony-stimulating factor (M-CSF) and kynureninase, accelerated catabolism of cytotoxic labile heme, and facilitating the metabolic adaptation of TAMs. This understanding presents both challenges and opportunities for strategic targeting of Nrf2 in cancer.
Keywords: anti-tumor immunity, immunosuppression, Keap1, Nrf2, tumor microenvironment
Nrf2 AND ITS REGULATION
Identified in 1994 (Moi et al., 1994) and shown to mediate the transcriptional upregulation of phase 2 drug detoxification enzymes (Itoh et al., 1997), nuclear factor erythroid 2-related factor 2 (Nrf2) is the principal regulator of the cellular antioxidant response, providing adaptation to disturbances in cellular redox homeostasis and protection against oxidative, electrophilic and inflammatory stress. As redox signaling is essential for most physiological processes (Sies and Jones, 2020; Sies et al., 2022), it is not surprising that, in addition to drug metabolism, Nrf2 is involved in regulating numerous cellular processes, including intermediary metabolism, mitochondrial bioenergetics, proliferation and autophagy (Cuadrado et al., 2019; Hayes and Dinkova-Kostova, 2014; Yamamoto et al., 2018).
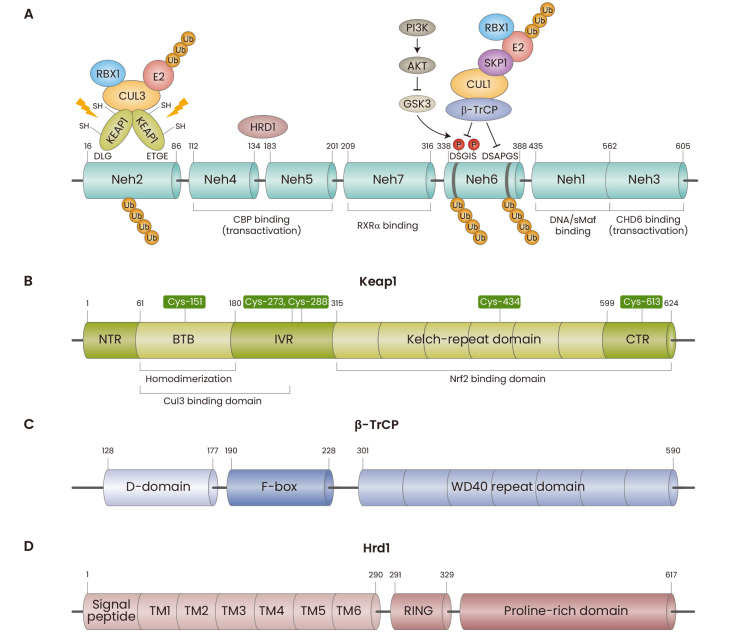
Nrf2 is a modular protein composed of seven Nrf2-ECH homology domains (Neh1-7) (Fig. 1A), which is regulated by a complex cellular machinery. For post-translational regulation, Nrf2 binds to Keap1 (Kelch-like ECH-associated protein 1), a substrate adaptor of the Keap1-Cul3-Rbx1 protein complex (Fig. 1B), β-transducin repeats-containing protein (β-TrCP), a substrate adaptor of the β-TrCP-Skp1-Cul1-Rbx1 complex (Fig. 1C), and/or hydroxymethyl glutaryl-coenzyme A reductase degradation protein 1 (Hrd1), an ER (endoplasmic reticulum)-associated E3 ubiquitin ligase (Fig. 1D), all of which target Nrf2 for ubiquitylation and proteasomal degradation. Inhibition of these ubiquitin ligase complexes by oxidants, electrophiles or protein-protein interaction (PPI) inhibitors (Cuadrado et al., 2019), or the processes involved in ubiquitination, such as neddylation (Soucy et al., 2009), leads to Nrf2 accumulation, its nuclear translocation and transcriptional upregulation of Nrf2-target genes, which encode a network of cytoprotective proteins. Indeed, pharmacological Nrf2 activation has protective effects in numerous animal models of human disease (Liby and Sporn, 2012), and Nrf2 activators are currently in various stages of drug development (Cuadrado et al., 2019).
Fig. 1. Domain structure of Nrf2 and its main negative regulators.
(A) Nrf2 consists of seven Nrf2-ECH homology (Neh) domains. Neh1 is a CNC-bZIP domain that interacts with sMafs proteins and binds DNA antioxidant/electrophile response element (ARE) sequences in DNA. The amino acid motifs DLG and ETGE in Neh2 are responsible for the negative regulation of Nrf2 by Keap1. The amino acid motifs DSGIS and DSAPGS in Neh6 are responsible for the negative regulation of Nrf2 by β-TrCP; the DSGIS motif requires prior phosphorylation by GSK3. Hrd1 interacts with both Neh4 and Neh5 for Nrf2 ubiquitination. Neh3 is a transactivation domain that contains a conserved VFLVPK motif, which is necessary for binding to the chromodomain helicase DNA-binding protein 6 (CHD6). Neh4 and 5 are transactivation domains that interact with the histone-modifying enzyme CBP/p300. The retinoid X receptor α (RXRα) binds Neh7, repressing both basal and inducible expression of Nrf2-target genes. (B) Keap1 contains four domains: Broad complex, Tramtrack and Bric-à-Brac (BTB), intervening region (IVR), double glycine repeat (DGR)/Kelch, and C-terminal region (CTR). The BTB domain mediates homodimer formation and recruitment of Cullin-3. The Kelch-repeat domain and the C-terminal domain (CTR) together form a six-bladed β-propeller structure and bind to the ETGE and DLG motifs in the Neh2 domain of Nrf2, exposing lysine residues in between the motifs for ubiquitination (McMahon et al., 2006; Tong et al., 2007). The cysteine residues (Cys-) indicated in the figure function as sensors for electrophiles and/or oxidants. Keap1 has several critical cysteine residues, including Cys-151 in the BTB domain and Cys-273 and Cys-288, within the IVR domain involved in electrophilic stress sensing, essential in regulating Nrf2 stability and activity (Dayalan Naidu and Dinkova-Kostova, 2020; Dinkova-Kostova et al., 2002; Levonen et al., 2004; McMahon et al., 2010; Zhang and Hannink, 2003). (C) In β-TrCP, the D-domain is responsible for dimerization, forming homo- and heterodimers between β-TrCP1 and β-TrCP2. The F-box region is involved in the recruitment of S-Phase Kinase-Associated Protein 1 (Skp1). The WD40 repeat domain binds to DSGIS and DSAPGS motifs in Neh6 of Nrf2 (Chowdhry et al., 2013; Rada et al., 2011). Notably, the β-TrCP-mediated degradation of Nrf2 requires phosphorylation of the DSGIS motif by glycogen synthase kinase-3 (GSK3) (Chowdhry et al., 2013). GSK3 is a kinase that is constitutively active, but requires prior phosphorylation of its substrates (Robertson et al., 2018), but the identity of this priming kinase(s) that phosphorylates Nrf2 allowing subsequent recognition and phosphorylation by GSK3 is currently unknown. (D) Hrd1 is comprised of a signal peptide, six consecutive transmembrane segments (TMs), a RING-finger region, and a proline-rich cluster (Karamali et al., 2022). Hrd1 interact with the Neh4-5 domains of Nrf2 with its C-terminal domain (Wu et al., 2014).
‘THE DARK SIDE’ OF Nrf2
Although overwhelming evidence clearly shows that Nrf2 is cytoprotective and in the early stages of carcinogenesis suppresses disease progression, the pro-tumorigenic effects of its constitutive activation in cancer are now well recognized (Rojo de la Vega et al., 2018). This dichotomy is not surprising considering that Nrf2 is the master regulator of the cellular redox homeostasis and that, depending on concentration, reactive oxygen species (ROS) affect cancer evolution in multiple ways, including initiating and facilitating the carcinogenesis process, supporting cancer cell proliferation, or causing senescence and/or cancer cell death (Hayes et al., 2020). However, Nrf2 activation in cancer also creates metabolic and redox vulnerabilities and enhances the bioactivation of certain prodrugs (Baird et al., 2022; Sayin et al., 2017; Torrente and DeNicola, 2022). This sets forth both challenges and opportunities for targeting Nrf2.
THE HETEROGENEOUS TUMOR MICROENVIRONMENT (TME)
Tumors exhibit complex characteristics in multiple dimensions which results in inter- and intra-tumoral heterogeneity. Factors such as the extracellular matrix, cytokines, chemokines, neurotrophic factors, morphogenic factors, specific nutrient and metabolic conditions, and expression of non-coding RNAs, add to the complexities of the TME. The immune landscape within tumors is diverse and complex (Fig. 2). The various cell types within the tumor immune microenvironment (TIME) range from cells derived in the periphery to specialised organ-resident cells, each contributing to cancer formation, progression and treatment responses (Quail and Joyce, 2017). Cancer cells recruit immune cells, such as resident and infiltrating monocyte-derived macrophages (MDMs), dendritic cells, myeloid-derived suppressor cells (MDSCs), CD4+ T cells, and regulatory T cells (Treg) into the tumor by secreting chemokines, cytokines and growth factors.
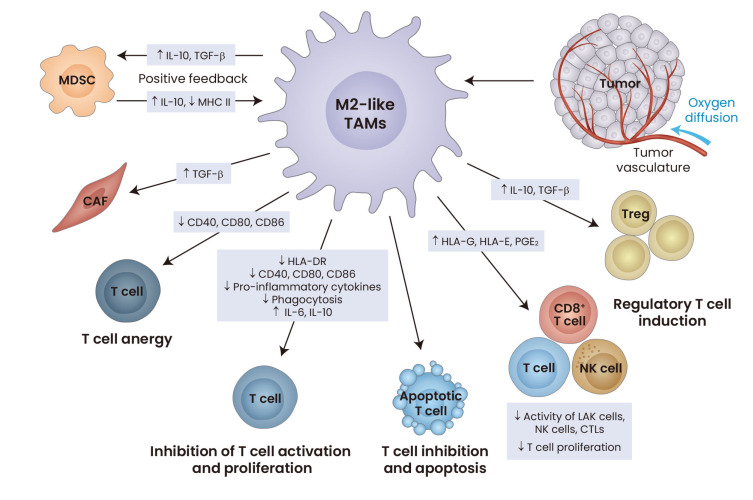
Fig. 2. The immunosuppressive properties of tumor-associated macrophages (TAMs) in the tumor immune microenvironment (TIME).
Numerous tumor-derived factors, some of which have been shown to activate Nrf2, induce the development of the TAMs into ‘alternatively activated M2’ phenotype, exhibiting immunosuppressive and pro-invasive characteristics. These M2-like TAMs promote immune suppression in various ways, including induction of regulatory T (Treg) cells (through secretion of IL-10 and TGF-β), inhibition of T-cell activation and proliferation, reduction of the activity of natural killer (NK) cells and CD8+ T cells, and promoting T-cell apoptosis. Increased levels of the MHC class I antigens HLA-G, HLA-E and prostaglandin E2 (PGE2) in TAMs may affect the activity of lymphokine-activated killer (LAK) cells, NK cells and cytotoxic T cells (CTLs), and inhibit the proliferation of T cells further. Increased levels of IL-6 and IL-10 and decreased levels of the MHC class II cell surface receptor HLA-DR, co-stimulatory factors, such as CD40, CD80, CD86, and pro-inflammatory cytokines, as well as diminished phagocytic activity of TAMs also contribute to T-cell inhibition. Reduced expression of CD40, CD80 and CD86 promotes T cell anergy and apoptosis. Additionally, TAMs induce differentiation of MDSCs into M2-like macrophages and increase recruitment of cancer-associated fibroblasts (CAFs). MDSC, myeloid-derived suppressor cell; IL-10, interleukin 10; TGF-β, transforming growth factor β; MHCII, MHC class II. Adapted from Wurdinger et al. (2014).
A large portion (up to 30%-50%) of the tumor mass can be comprised of tumor-associated macrophages (TAMs), which include resident and infiltrating MDMs. Macrophages are multifunctional cells that exhibit considerable plasticity in response to extracellular cues and are influenced by both innate and adaptive immune signals (Mosser and Edwards, 2008; Okabe and Medzhitov, 2016). Based on in vitro polarization studies, macrophages are broadly classified into two types: the ‘classically activated M1’ ‘Fight’ type that preferentially produces nitric oxide and initiates a Th1 immune response; the ‘alternatively activated M2’ ‘Fix’ type, which produces ornithine, promoting proliferation and repair through polyamines and collagen (Mills, 2012). During inflammation, macrophages adopt M1-like phenotype, exhibiting increased capacity to migrate, phagocytose, secrete cytotoxic factors and express MHC class II and co-stimulatory molecules for T cell activation. By contrast, TAMs polarize towards an M2-like phenotype, exhibiting immunosuppressive and tumor-promoting characteristics (Wurdinger et al., 2014), and accumulate with higher tumor grade (Komohara et al., 2008). However, in vivo this M1/M2 classification is not absolute as many TAMs express both M1 and M2 phenotypic markers, suggesting that the phenotype of macrophages is context-specific (Ginhoux et al., 2016; Muller et al., 2017).
THE ANTI-INFLAMMATORY ROLE OF Nrf2
Shortly after its recognition as the main transcription factor mediating the cellular antioxidant response, it was noted that Nrf2 plays a role in adaptation to oxidative stress in macrophages. Experiments using thioglycolate-elicited peritoneal macrophages isolated from Nrf2-knockout mice revealed that their response to electrophilic and ROS-producing agents was impaired (Ishii et al., 2000). It was subsequently shown that the electrophilic 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), which accumulates in macrophages from pleural lavage, activates Nrf2 by forming adducts with Keap1 (Itoh et al., 2004; Kim et al., 2017), whereas the non-electrophilic analog 9,10-dihydro-15d-PGJ2 had no effect (Kim et al., 2017). Furthermore, the upregulation of the Nrf2 transcriptional targets cluster of differentiation 36 (CD36) (Maruyama et al., 2008) and heme oxygenase 1 (HO-1) (Prestera et al., 1995) was essential for the anti-inflammatory activity of 15d-PGJ2 (Kim et al., 2017). In addition to CD36, a scavenger receptor for oxidized low-density lipoproteins, Nrf2 activates the transcription of other genes important for macrophage function, such as macrophage receptor with collagenous structure (MARCO), a receptor required for bacterial phagocytosis (Harvey et al., 2011) and interleukin-17D (IL-17D), a virus- and tumor cell-surveillance mediator (Saddawi-Konefka et al., 2016). Conversely, Nrf2 is often dysregulated in inflammation, including in lungs from patients with COVID-19 (Olagnier et al., 2020).
Dependence on Nrf2 and the Keap1 sensor cysteine Cys-151 for the ability to suppress pro-inflammatory responses in peritoneal murine macrophages has been shown for a range of cyclic cyanoenones, the most potent Nrf2 activators known to date (Dayalan Naidu et al., 2018). Pre-treatment with CDDO-Im, an imidazole derivative of 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid, attenuated lipopolysaccharide (LPS)-induced cytokine expression in human peripheral blood mononuclear cells (PBMCs) (Thimmulappa et al., 2007). Mechanistically, Nrf2 inhibits LPS-induced expression of some proinflammatory cytokines (e.g., IL-6 and IL-1b) in macrophages by disrupting recruitment of RNA polymerase II, in a ROS- and antioxidant/electrophile response element (ARE)-independent manner (Kobayashi et al., 2016), as well as by ARE-dependent induction of activating transcription factor 3 (ATF3), a negative regulator of IL-6 transcription (Hoetzenecker et al., 2012).
Two structure-activity studies in murine cells identified suppression of inflammation as a consistent property of Nrf2 activators, suggesting it as a central aspect of the cytoprotective actions of these compounds (Dinkova-Kostova et al., 2005; Liu et al., 2008). Correlation between the ability to activate Nrf2-dependent transcription and inhibit markers of inflammation was also observed in PBMCs isolated from blood of human subjects following oral administration of preparations delivering the classical Nrf2 activator sulforaphane (Liu et al., 2020).
Compared to wild-type mice, Nrf2-knockout mice are more sensitive (Thimmulappa et al., 2006), whereas mice with Keap1 deletion in myeloid leukocyte cells are more resistant to endotoxin- and cecal ligation and puncture-induced septic shock (Kong et al., 2011). Under conditions of Nrf2 deficiency, NADPH oxidase-dependent ROS amplify Toll-like receptor 4 (TLR4) signaling and sepsis-induced mortality (Kong et al., 2010). Additionally, Nrf2 negatively regulates the type I interferon response (Gunderstofte et al., 2019; Olagnier et al., 2017; 2018). A recent high-resolution proteomics study confirmed that Nrf2 suppresses the type I interferon response and reprograms macrophage intermediary metabolism, and further revealed that, upon LPS stimulation, Nrf2 activation enhances the metabolic switch from oxidative phosphorylation to glycolysis whilst promoting mitochondrial fusion (Baker et al., 2022; Ryan et al., 2022). Overall, it is now recognized that Nrf2 is not only a regulator of the cellular redox homeostasis, but also has a role in regulating inflammation and immunity (van der Horst et al., 2022).
Nrf2 IN THE TUMOR IMMUNE MICROENVIRONMENT (TIME)
Human KEAP1-mutant tumors have high levels of Nrf2, low infiltration of T cells, and unfavorable immunotherapy responses (Baird et al., 2022). Single-cell RNA sequencing (scRNA-seq) analysis of ten human lung adenocarcinomas and ten normal control tissues has confirmed that KEAP1-mutant tumors display high levels of TAMs and exhausted CD8+ T cells and associate with poor patient prognosis (Bischoff et al., 2021). An immunosuppressive microenvironment was documented in a lung cancer model in mice with lung-specific reduced expression of Keap1 and phosphatase and tensin homolog deleted on chromosome 10 (Pten) and partly attributed to Nrf2 activation (Best et al., 2018), suggesting that Nrf2 has a role in TIME immunosuppression.
Lung tumors with high Nrf2 levels display high PD-L1 positivity (Best et al., 2018; Fahrmann et al., 2022) and limited responses to anti-PD-L1 treatment (Singh et al., 2021), whereas immune checkpoint inhibition leads to a decrease in lung tumor burden and an increase in infiltrating lymphoid cells (Best et al., 2018). Additionally, Nrf2 has been implicated in the LKB1/AMPK-mediated increase of PD-L1 expression in lung cancer cells (Shen et al., 2020) and in the UVB radiation-induced PD-L1 expression in primary human keratinocytes and melanocytes (Zhu et al., 2018). Conversely, PD-L1 expression is lower in distal colon tissues of Nrf2-knockout in comparison with wild-type female mice following azoxymethane/dextran sodium sulfate treatment (Kang et al., 2021). In models of melanoma, Nrf2 depletion suppresses PD-L1 expression, increases tumor-infiltrating T cell activation and inhibits tumor growth (Zhu et al., 2018). Together, these studies implicate induction of PD-L1 as one mechanism by which Nrf2 contributes to immunosuppression in cancer.
Another way by which Nrf2 activation enhances immunosuppression in TIME could be through increased HO-1 expression (and consequently, accelerated catabolism of cytotoxic labile heme) in subsets of TAMs recruited to the invasive tumor margin. Indeed, interfering with recruitment of such TAMs, or pharmacologic inhibition or myeloid-specific deletion of HO-1 increased the antitumor immunity and the efficacy of anti–PD-L1 immunotherapy (Consonni et al., 2021). In this context, it is noteworthy that macrophage colony-stimulating factor (M-CSF), which plays a role in the expansion of TAMs, is a transcriptional target of Nrf2 (Liu et al., 2022). Interestingly, a role for Nrf2 in the reduced responses to anti-cytotoxic T-lymphocytes-associated protein 4 therapy was also recently described (Ahmed et al., 2022).
However, not all experimental models have led to identical conclusions. In contrast with the immunosuppressive effects of the lung-specific simultaneous knockdown of Keap1 and Pten described above, Nrf2 activation in global Keap1-knockdown mice attenuated the activity of MDSCs and decreased lung cancer metastasis; this correlated with lower ROS levels in Keap1-knockdown MDSCs of tumor-bearing mice (Satoh et al., 2010), although by lowering ROS, Nrf2 sustains MDSCs survival (Ostrand-Rosenberg et al., 2020). In a model of urethane-induced lung cancer, the adenomas that formed in Keap1-knockdown mice were smaller compared to those in wild-type animals; however, they grew faster when transplanted into immunodeficient mice (Satoh et al., 2016). The use of the Kras-induced lung adenocarcinoma mouse model confirmed that Keap1 knockdown in cells of the hematopoietic lineage suppressed development of Keap1-mutant tumors (Hayashi et al., 2020). Although the effect of Nrf2 activation on natural killer (NK) cells was not examined in this study, one way by which TAMs may contribute to the immunosuppressive TIME is through inhibition of the activation of NK cells and lowering their cytotoxicity against tumor cells (Krneta et al., 2017). In this context, it has been shown that pharmacologic Nrf2 activation (with RTA-408) in NK cells within TIME restores anti-tumor activity (Poznanski et al., 2021).
Lung carcinogenesis is exacerbated in Nrf2-deficient mice treated with vinyl carbamate, which induces invasive adenocarcinomas, and tumor multiplicity and size were increased in comparison with wild-type mice (Zhang et al., 2018). Consistent with higher tumor burden, there were more TAMs in lungs of Nrf2-deficient mice, and the MDSCs were also elevated in the spleens of these animals, whereas the CD8+ cytotoxic T cells and CD4+ helper T cells in the lung, were reduced. RNA sequencing revealed that numerous cytokines and chemokines, such as Cxcl1, Csf1, Ccl9, Cxcl12, as well as major histocompatibility complex antigens that promote tumor growth, were upregulated in tumors from Nrf2-knockout mice. Notably, the expression of CXCL10 increases upon knockdown of Nrf2 in human macrophages (THP1) and decreases following pharmacologic Nrf2 activation (Ryan et al., 2022).
It is not possible to directly compare the various models summarized here due to differences in genetic background of the experimental animals, types of carcinogens, treatment protocols and timepoints during tumor development. Nonetheless, the results show that the role of Nrf2 in TIME is complex and context-dependent, and requires further investigation.
ENDOGENOUS Nrf2 ACTIVATORS IN THE TUMOR IMMUNE MICROENVIRONMENT
Recent experimental evidence suggests that several metabolites can activate Nrf2 in the TIME. These include the TCA cycle-derived immunometabolite itaconate, the tryptophan metabolite L-kynurenine (and its downstream electrophilic metabolites), lactate and hyaluronic acid (Table 1).
Table 1.
Endogenous Nrf2 activators in the tumor immune microenvironment
| Metabolite | Chemical structure | Nrf2 activation in: | Reference |
|---|---|---|---|
| Itaconate |
|
human keratinocytes (HaCaT) | (Chen et al., 2022) |
| L-Kynurenine |
|
human cervical cancer cells (HeLa) human macrophage cells (U937) |
(Fiore et al., 2022) (Li et al., 2022) |
| Lactic acid |
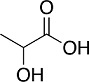
|
human macrophage cells (THP1 and peripheral blood-derived) human neuroblastoma cells (SH-SY5Y) Caenorhabditis elegans |
(Feng et al., 2018) (Tauffenberger et al., 2019) |
| Hyaluronic acid |
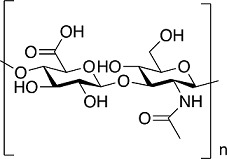
|
bovine chondrocytes frostbite-wounded skin tissues of Wistar rats doxorubicin-resistant human breast cancer cells (MCF7) |
(Onodera et al., 2015) (Joshi et al., 2021) (Choi et al., 2022) |
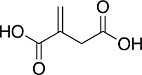
Itaconate
Itaconate is a Krebs cycle-derived anti-inflammatory immunometabolite with diverse roles in inflammation and immunity, including inhibition of succinate dehydrogenase, glycolysis, the NLRP3 inflammasome, and activation of Nrf2 and ATF3 (Bambouskova et al., 2018; Mills et al., 2018; Peace and O'Neill, 2022). Itaconate accumulates during the metabolic reprogramming in activated macrophages due to upregulation of immune-responsive gene 1 (IRG1). Thus, gene expression profiling of inflammatory murine macrophages has shown that Irg1 is one of the most upregulated genes (Basler et al., 2006; Thomas et al., 2006). IRG1 catalyzes the production of itaconate from cis-aconitate, connecting cellular metabolism to immunity (Michelucci et al., 2013). Moreover, itaconate is important in macrophage-mediated immune responses (Strelko et al., 2011). Consistent with IRG1 upregulation, markedly increased levels of itaconate (reaching a concentration of 5 mM) were detected in human and murine macrophages activated by LPS (Mills et al., 2018). A zebrafish (Danio rerio) line expressing an Irg1-EGFP fusion protein has been developed as a reporter of macrophage activation (Sanderson et al., 2015).
Itaconate contains an electrophilic α,β-unsaturated carboxylic acid, and can alkylate protein cysteines by Michael addition. As a cysteine-based sensor for electrophiles, Keap1 is one such candidate (Dayalan Naidu and Dinkova-Kostova, 2020), and alkylation of its cysteine sensors leads to Nrf2 accumulation, its nuclear translocation and activation of antioxidant and anti-inflammatory responses. Indeed, 4-octyl itaconate (4-OI), a cell-permeable analog of itaconate, covalently binds to cysteines of Keap1 and robustly activates Nrf2 (Mills et al., 2018). In turn, Nrf2 mediates some, although not all, of the anti-inflammatory activities of 4-OI (Liao et al., 2019; Mills et al., 2018; Ryan et al., 2022; Sun et al., 2020). Nrf2 activation by itaconate was recently observed in human keratinocytes (HaCaT) (Chen et al., 2022). In a model of hepatic fibrosis, the levels of Nrf2 and its downstream targets are lower in CCl4-treated Irg1-deficient mice in comparison with their wild-type counterparts and restored by 4-OI (Fan et al., 2022).
In addition to its function in inflammation and immunity, a role for IRG1 in cancer and anti-tumor immunity has also been described. Thus, exposure of RAW264.7 cells to tumor-conditioned medium increases the levels of IRG1 and itaconate secretion (Zhao et al., 2022), and xenografted human cancer cells activate the Irg1-EGFP reporter in zebrafish (Sanderson et al., 2015). Furthermore, itaconate is secreted from tumor-associated MDSCs and suppresses the proliferation, cytotoxic activity and immune effector functions of CD8+ T-cells, whereas loss of IRG1 (and the corresponding decrease in itaconate levels) enhances their anti-tumor activity and increases the sensitivity to anti-PD-1 immune checkpoint blockade in mice (Zhao et al., 2022). The levels of IRG1 are increased in monocytes isolated from ascites fluid of ovarian carcinoma patients (Weiss et al., 2018) and in glioma tumors, where this increase is associated with disease progression and poor prognosis (Pan et al., 2014). Knockdown of IRG1, on the other hand, leads to a decrease in migration and invasion (Pan et al., 2014) and reduced tumor growth (Weiss et al., 2018). In agreement, the survival of metastatic melanoma patients treated with anti-PD-1 immune checkpoint blockade is longer if they express low levels of IRG1 compared to those where IRG1 expression is high (Zhao et al., 2022). Additionally, the levels of microRNA-378, which targets IRG1, are decreased in glioma (Shi et al., 2018). This suggests IRG1 plays a role in the progression of at least some tumors. Considering that, as discussed above, itaconate, activates Nrf2, it is possible that at least some of the tumor-promoting effects of IRG1 are mediated by Nrf2. Such possibility is likely as compared to wild-type, the levels of IRG1 are higher in bone-marrow derived macrophages from Keap1-knockdown mice and lower in Nrf2-deficient cell (Ryan et al., 2022), suggesting that the expression of IRG1 could be in part regulated by Nrf2 and/or its transcriptional target(s). In this context, it has been shown that HO-1 induction upregulates IRG1 expression and inhibits inflammation (Jamal Uddin et al., 2016).
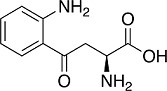
Kynurenine
Another endogenous metabolite associated with tumor immunosuppression is the tryptophan metabolite L-kynurenine. The metabolic reprogramming in cancer cells leads to an increase in L-kynurenine biosynthesis and secretion due to increased oxidation of tryptophan to N-formyl L-kynurenine, the rate limiting step in the kynurenine pathway. The immunosuppressive effects of L-kynurenine in the TME, many of which have been attributed to activation of the aryl hydrocarbon receptor (AhR), include suppression of T cells and NK cells, as well as activation of MDSCs and Treg cells (Prendergast et al., 2014). Although the precise mechanism(s) has not been established, L-kynurenine and/or its metabolites upregulate Nrf2 in several cell types (Carreno et al., 2022; Fiore et al., 2022; Pae et al., 2006), including macrophages (Li et al., 2022 and our unpublished observations), and in mice (Carreno et al., 2022). A recent study suggests that one potential mechanism for Nrf2 activation by L-kynurenine is through activation of the AhR, which in turn binds to the promoter of NFE2L2 and transcriptionally upregulates Nrf2 (Li et al., 2022). Proteomic analysis of 47 lung adenocarcinoma cell lines (11 with and 36 without mutations in KEAP1) has revealed kynureninase, the enzyme that catalyzes the hydrolysis of L-kynurenine, as the most highly overexpressed protein associated with Nrf2 activation (Fahrmann et al., 2022). Nrf2 activation by genetic and pharmacologic means confirmed the dependence of kynureninase expression on Nrf2. High levels of kynureninase are associated with suppressed antitumor immunity, including increases in PD-L1 positivity and Treg cells. Notably, Nrf2 and its transcriptional target, the cystine/glutamate antiporter solute carrier SLC7A11 are important determinants of Treg cell proliferation in response to pseudo-starvation (Procaccini et al., 2021). Moreover, it has been shown that Treg cells co-cultured with ovarian cancer ascites undergo apoptosis due to low levels of Nrf2 and antioxidant defences, whereas the pharmacologic Nrf2 activator sulforaphane reduced tumor Treg cells apoptosis, increased effector T cell cytokine expression and inhibited tumor growth (Maj et al., 2017).
Lactate
Cancer cells produce high levels of lactate, which is secreted into the extracellular space and contributes to the immunosuppressive TME and cancer progression (Certo et al., 2021). Exposure to lactate increased the intracellular ROS and activated Nrf2 in neuroblastoma cells (SH-SY5Y) and Caenorhabditis elegans (Tauffenberger et al., 2019). Interestingly, the same study showed that lactate also caused activation of PI3K/AKT, but the possible involvement of AKT in the mechanism of Nrf2 activation by lactate has not been investigated. In co-culture experiments, cancer cell-derived lactate activates Nrf2 in macrophages, and polarizes them towards anti-inflammatory phenotype (Feng et al., 2018).
Hyaluronic acid
Hyaluronic acid (HA) is a negatively charged polysaccharide, which is present inside the cell, at the cell surface, and in the extracellular matrix (ECM), where it functions as a microenvironmental cue in the co-regulation of cell behaviour during embryogenesis, inflammation, healing and tumor growth and development (Toole, 2004). It is generally accepted that high molecular-weight HA in the ECM has anti-inflammatory and wound-healing roles, whereas low-molecular-weight HA promotes tumor growth and metastasis, and supports the development of immunosuppressive PD-L1+ macrophages in the TIME (Donelan et al., 2022). However, it was recently shown that ovarian cancer cell-derived high molecular-weight HA drives cholesterol efflux and depletion of lipid rafts from macrophages, which in turn drives IL-4 signaling and inhibition of IFNγ-induced gene expression (Goossens et al., 2019). Treatment with HA activates Nrf2 and induces Nrf2-target genes in bovine chondrocytes and human breast cancer cells, with increased phosphorylation of AKT or higher levels of p62, which competes with Nrf2 for binding to Keap1 (Komatsu et al., 2010), correlating with Nrf2 activation (Choi et al., 2022; Onodera et al., 2015; Ryoo et al., 2018). In addition, a hydrogel formulation of HA increased the levels of Nrf2 in frostbite-wounded skin tissues of Wistar rats (Joshi et al., 2021). To our knowledge, the possibility that Nrf2 activation may also occur in macrophages exposed to HA has not been explored.
CONCLUDING REMARKS
With the high rate of immune cell infiltration into the tumor, understanding of the TIME could shed light on new therapeutic targets and strategies. There are extensive interactions between TAMs and tumor cells within the TIME, culminating in an immunosuppressive environment that promotes tumor growth and invasion. Emerging evidence suggests that Nrf2 activation triggered by cancer cell-derived metabolites, such as itaconate, L-kynurenine, lactic acid and hyaluronic acid, and/or other secreted factors, plays an important role in modulating the TIME and tumor-TAMs crosstalk. The results from the models summarized here show that Nrf2 has a complex role in the TIME, which is not fully understood and merits further investigation. This is particularly important considering that several Nrf2 activators are currently in clinical trials for a number of disease indications.
ACKNOWLEDGMENTS
We thank the Medical Research Council (MR/W023806/1) and the Ninewells Cancer Campaign for funding our research.
Footnotes
AUTHOR CONTRIBUTIONS
J.F., O.J.R., and A.T.D.-K. wrote the manuscript. A.T.D.-K. secured funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahmed K.M., Veeramachaneni R., Deng D., Putluri N., Putluri V., Cardenas M.F., Wheeler D.A., Decker W.K., Frederick A.I., Kazi S., et al. Glutathione peroxidase 2 is a metabolic driver of the tumor immune microenvironment and immune checkpoint inhibitor response. J. Immunother. Cancer. 2022;10:e004752. doi: 10.1136/jitc-2022-004752.3658e8d79c574173b33bffd98561e7d7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird L., Kensler T.W., Yamamoto M. Novel NRF2-activated cancer treatments utilizing synthetic lethality. IUBMB Life. 2022;74:1209–1231. doi: 10.1002/iub.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.P., Phair I.R., Brenes A.J., Atrih A., Ryan D.G., Bruderer R., Dinkova-Kostova A.T., Lamont D.J., Arthur J.S.C., Howden A.J.M. DIA label-free proteomic analysis of murine bone-marrow-derived macrophages. STAR Protoc. 2022;3:101725. doi: 10.1016/j.xpro.2022.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambouskova M., Gorvel L., Lampropoulou V., Sergushichev A., Loginicheva E., Johnson K., Korenfeld D., Mathyer M.E., Kim H., Huang L.H., et al. Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature. 2018;556:501–504. doi: 10.1038/s41586-018-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler T., Jeckstadt S., Valentin-Weigand P., Goethe R. Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopolysaccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J. Leukoc. Biol. 2006;79:628–638. doi: 10.1189/jlb.0905520. [DOI] [PubMed] [Google Scholar]
- Best S.A., De Souza D.P., Kersbergen A., Policheni A.N., Dayalan S., Tull D., Rathi V., Gray D.H., Ritchie M.E., McConville M.J., et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metab. 2018;27:935–943.e4. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Bischoff P., Trinks A., Obermayer B., Pett J.P., Wiederspahn J., Uhlitz F., Liang X., Lehmann A., Jurmeister P., Elsner A., et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene. 2021;40:6748–6758. doi: 10.1038/s41388-021-02054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno M., Pires M.F., Woodcock S.R., Brzoska T., Ghosh S., Salvatore S.R., Chang F., Khoo N.K.H., Dunn M., Connors N., et al. Immunomodulatory actions of a kynurenine-derived endogenous electrophile. Sci. Adv. 2022;8:eabm9138. doi: 10.1126/sciadv.abm9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo M., Tsai C.H., Pucino V., Ho P.C., Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2021;21:151–161. doi: 10.1038/s41577-020-0406-2. [DOI] [PubMed] [Google Scholar]
- Chen F., Elgaher W.A.M., Winterhoff M., Bussow K., Waqas F.H., Graner E., Pires-Afonso Y., Casares Perez L., de la Vega L., Sahini N., et al. Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism. Nat. Metab. 2022;4:534–546. doi: 10.1038/s42255-022-00577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.H., Ryoo I., Sim K.H., Ahn H.J., Lee Y.J., Kwak M.K. High levels of hyaluronic acid synthase-2 mediate NRF2-driven chemoresistance in breast cancer cells. Biomol. Ther. (Seoul) 2022;30:368–379. doi: 10.4062/biomolther.2022.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni F.M., Bleve A., Totaro M.G., Storto M., Kunderfranco P., Termanini A., Pasqualini F., Ali C., Pandolfo C., Sgambelluri F., et al. Heme catabolism by tumor-associated macrophages controls metastasis formation. Nat. Immunol. 2021;22:595–606. doi: 10.1038/s41590-021-00921-5. [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- Dayalan Naidu S., Dinkova-Kostova A.T. KEAP1, a cysteine-based sensor and a drug target for the prevention and treatment of chronic disease. Open Biol. 2020;10:200105. doi: 10.1098/rsob.200105.c5acd0ef8f23493ba09ecad96169f537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayalan Naidu S., Muramatsu A., Saito R., Asami S., Honda T., Hosoya T., Itoh K., Yamamoto M., Suzuki T., Dinkova-Kostova A.T. C151 in KEAP1 is the main cysteine sensor for the cyanoenone class of NRF2 activators, irrespective of molecular size or shape. Sci. Rep. 2018;8:8037. doi: 10.1038/s41598-018-26269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova A.T., Liby K.T., Stephenson K.K., Holtzclaw W.D., Gao X., Suh N., Williams C., Risingsong R., Honda T., Gribble G.W., et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelan W., Dominguez-Gutierrez P.R., Kusmartsev S. Deregulated hyaluronan metabolism in the tumor microenvironment drives cancer inflammation and tumor-associated immune suppression. Front. Immunol. 2022;13:971278. doi: 10.3389/fimmu.2022.971278.ddb7b3f9d565427fb3f0665aae8e6ae2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrmann J.F., Tanaka I., Irajizad E., Mao X., Dennison J.B., Murage E., Casabar J., Mayo J., Peng Q., Celiktas M., et al. Mutational activation of the NRF2 pathway upregulates kynureninase resulting in tumor immunosuppression and poor outcome in lung adenocarcinoma. Cancers (Basel) 2022;14:2543. doi: 10.3390/cancers14102543.7da46032bbd349bea7f5437e7dee2a8e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K., Zan X., Zhi Y., Yang Y., Hu K., Zhang X., Zhang X., Zhao S., Chen K., Gong X., et al. Immune response gene 1 deficiency impairs Nrf2 activation and aggravates liver fibrosis in mice. Biochem. Biophys. Res. Commun. 2022;607:103–109. doi: 10.1016/j.bbrc.2022.03.110. [DOI] [PubMed] [Google Scholar]
- Feng R., Morine Y., Ikemoto T., Imura S., Iwahashi S., Saito Y., Shimada M. Nrf2 activation drive macrophages polarization and cancer cell epithelial-mesenchymal transition during interaction. Cell Commun. Signal. 2018;16:54. doi: 10.1186/s12964-018-0262-x.a1a4347d39394e1685299d302a0cfe1a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A., Zeitler L., Russier M., Gross A., Hiller M.K., Parker J.L., Stier L., Kocher T., Newstead S., Murray P.J. Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling. Mol. Cell. 2022;82:920–932.e7. doi: 10.1016/j.molcel.2022.02.007. [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Schultze J.L., Murray P.J., Ochando J., Biswas S.K. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2016;17:34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- Goossens P., Rodriguez-Vita J., Etzerodt A., Masse M., Rastoin O., Gouirand V., Ulas T., Papantonopoulou O., Van Eck M., Auphan-Anezin N., et al. Membrane cholesterol efflux drives tumor-associated macrophage reprogramming and tumor progression. Cell Metab. 2019;29:1376–1389.e4. doi: 10.1016/j.cmet.2019.02.016. [DOI] [PubMed] [Google Scholar]
- Gunderstofte C., Iversen M.B., Peri S., Thielke A., Balachandran S., Holm C.K., Olagnier D. Nrf2 negatively regulates type I interferon responses and increases susceptibility to herpes genital infection in mice. Front. Immunol. 2019;10:2101. doi: 10.3389/fimmu.2019.02101.2c4c7ade5fba4afea79db4b7aa15877d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey C.J., Thimmulappa R.K., Sethi S., Kong X., Yarmus L., Brown R.H., Feller-Kopman D., Wise R., Biswal S. Targeting Nrf2 signaling improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model. Sci. Transl. Med. 2011;3:78ra32. doi: 10.1126/scitranslmed.3002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Kuga A., Suzuki M., Panda H., Kitamura H., Motohashi H., Yamamoto M. Microenvironmental activation of Nrf2 restricts the progression of Nrf2-activated malignant tumors. Cancer Res. 2020;80:3331–3344. doi: 10.1158/0008-5472.CAN-19-2888. [DOI] [PubMed] [Google Scholar]
- Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Hayes J.D., Dinkova-Kostova A.T., Tew K.D. Oxidative stress in cancer. Cancer Cell. 2020;38:167–197. doi: 10.1016/j.ccell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoetzenecker W., Echtenacher B., Guenova E., Hoetzenecker K., Woelbing F., Bruck J., Teske A., Valtcheva N., Fuchs K., Kneilling M., et al. ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-associated immunosuppression. Nat. Med. 2012;18:128–134. doi: 10.1038/nm.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K., Mochizuki M., Ishii Y., Ishii T., Shibata T., Kawamoto Y., Kelly V., Sekizawa K., Uchida K., Yamamoto M. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Delta(12,14)-prostaglandin j(2) Mol. Cell. Biol. 2004;24:36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal Uddin M., Joe Y., Kim S.K., Oh Jeong S., Ryter S.W., Pae H.O., Chung H.T. IRG1 induced by heme oxygenase-1/carbon monoxide inhibits LPS-mediated sepsis and pro-inflammatory cytokine production. Cell. Mol. Immunol. 2016;13:170–179. doi: 10.1038/cmi.2015.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K., Mazumder B., Chattopadhyay P., Goyary D., Das M., Dwivedi S.K. Exploring the frostbite healing potential of hyaluronic acid based hydrogel of Manuka honey through in-silico antithrombotic and anti-platelet studies of major phytoconstituents and in-vivo evaluation in Wistar rat model. Drug Dev. Ind. Pharm. 2021;47:1326–1334. doi: 10.1080/03639045.2021.1989459. [DOI] [PubMed] [Google Scholar]
- Kang C., Song C.H., Kim N., Nam R.H., Choi S.I., Yu J.E., Nho H., Choi J.A., Kim J.W., Na H.Y., et al. The enhanced inhibitory effect of estrogen on PD-L1 expression following Nrf2 deficiency in the AOM/DSS model of colitis-associated cancer. Front. Oncol. 2021;11:679324. doi: 10.3389/fonc.2021.679324.8e9848fc1ad44b7da4ed93e25b105aaa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamali N., Ebrahimnezhad S., Khaleghi Moghadam R., Daneshfar N., Rezaiemanesh A. HRD1 in human malignant neoplasms: Molecular mechanisms and novel therapeutic strategy for cancer. Life Sci. 2022;301:120620. doi: 10.1016/j.lfs.2022.120620. [DOI] [PubMed] [Google Scholar]
- Kim W., Lee H.N., Jang J.H., Kim S.H., Lee Y.H., Hahn Y.I., Ngo H.K., Choi Y., Joe Y., Chung H.T., et al. 15-Deoxy-Delta(12,14)-prostaglandin J2 exerts proresolving effects through nuclear factor E2-related factor 2-induced expression of CD36 and heme oxygenase-1. Antioxid. Redox Signal. 2017;27:1412–1431. doi: 10.1089/ars.2016.6754. [DOI] [PubMed] [Google Scholar]
- Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624.64f68c1a2df449c4a3c17e7079f6efb6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Komohara Y., Ohnishi K., Kuratsu J., Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008;216:15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- Kong X., Thimmulappa R., Craciun F., Harvey C., Singh A., Kombairaju P., Reddy S.P., Remick D., Biswal S. Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am. J. Respir. Crit. Care Med. 2011;184:928–938. doi: 10.1164/rccm.201102-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Thimmulappa R., Kombairaju P., Biswal S. NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J. Immunol. 2010;185:569–577. doi: 10.4049/jimmunol.0902315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krneta T., Gillgrass A., Poznanski S., Chew M., Lee A.J., Kolb M., Ashkar A.A. M2-polarized and tumor-associated macrophages alter NK cell phenotype and function in a contact-dependent manner. J. Leukoc. Biol. 2017;101:285–295. doi: 10.1189/jlb.3A1215-552R. [DOI] [PubMed] [Google Scholar]
- Levonen A.L., Landar A., Ramachandran A., Ceaser E.K., Dickinson D.A., Zanoni G., Morrow J.D., Darley-Usmar V.M. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 2004;378:373–382. doi: 10.1042/bj20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yuan Y., Chen H., Dai H., Li J. Indoleamine 2,3-dioxygenase mediates the therapeutic effects of adipose-derived stromal/stem cells in experimental periodontitis by modulating macrophages through the kynurenine-AhR-NRF2 pathway. Mol. Metab. 2022;66:101617. doi: 10.1016/j.molmet.2022.101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S.T., Han C., Xu D.Q., Fu X.W., Wang J.S., Kong L.Y. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 2019;10:5091. doi: 10.1038/s41467-019-13078-5.2b66b3e7c6704dc1a4818c1e7d684e87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liby K.T., Sporn M.B. Synthetic oleanane triterpenoids: multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol. Rev. 2012;64:972–1003. doi: 10.1124/pr.111.004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Dinkova-Kostova A.T., Talalay P. Coordinate regulation of enzyme markers for inflammation and for protection against oxidants and electrophiles. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15926–15931. doi: 10.1073/pnas.0808346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhao D., Li H., Zhang W., Lin Q., Wang X., Zheng S., Zhang L., Li L., Hu S., et al. Blocking iASPP/Nrf2/M-CSF axis improves anti-cancer effect of chemotherapy-induced senescence by attenuating M2 polarization. Cell Death Dis. 2022;13:166. doi: 10.1038/s41419-022-04611-4.e43648bab7cf4183b91c36e80139afdb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zimmerman A.W., Singh K., Connors S.L., Diggins E., Stephenson K.K., Dinkova-Kostova A.T., Fahey J.W. Biomarker exploration in human peripheral blood mononuclear cells for monitoring sulforaphane treatment responses in autism spectrum disorder. Sci. Rep. 2020;10:5822. doi: 10.1038/s41598-020-62714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maj T., Wang W., Crespo J., Zhang H., Wang W., Wei S., Zhao L., Vatan L., Shao I., Szeliga W., et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017;18:1332–1341. doi: 10.1038/ni.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama A., Tsukamoto S., Nishikawa K., Yoshida A., Harada N., Motojima K., Ishii T., Nakane A., Yamamoto M., Itoh K. Nrf2 regulates the alternative first exons of CD36 in macrophages through specific antioxidant response elements. Arch. Biochem. Biophys. 2008;477:139–145. doi: 10.1016/j.abb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- McMahon M., Lamont D.J., Beattie K.A., Hayes J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a "tethering" mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A., et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C.D. M1 and M2 macrophages: oracles of health and disease. Crit. Rev. Immunol. 2012;32:463–488. doi: 10.1615/CritRevImmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M.P., Costa A.S.H., Higgins M., Hams E., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Kohanbash G., Liu S.J., Alvarado B., Carrera D., Bhaduri A., Watchmaker P.B., Yagnik G., Di Lullo E., Malatesta M., et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18:234. doi: 10.1186/s13059-017-1362-4.b3ec0e0d5c1c43f3ad2d248d295ddd67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y., Medzhitov R. Tissue biology perspective on macrophages. Nat. Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- Olagnier D., Brandtoft A.M., Gunderstofte C., Villadsen N.L., Krapp C., Thielke A.L., Laustsen A., Peri S., Hansen A.L., Bonefeld L., et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 2018;9:3506. doi: 10.1038/s41467-018-05861-7.5fc54bf6fbd8427c94401b47231854b6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagnier D., Farahani E., Thyrsted J., Blay-Cadanet J., Herengt A., Idorn M., Hait A., Hernaez B., Knudsen A., Iversen M.B., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3.7eb1ed3741cc4fbfa366937b0037cc35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagnier D., Lababidi R.R., Hadj S.B., Sze A., Liu Y., Naidu S.D., Ferrari M., Jiang Y., Chiang C., Beljanski V., et al. Activation of Nrf2 signaling augments vesicular stomatitis virus oncolysis via autophagy-driven suppression of antiviral immunity. Mol. Ther. 2017;25:1900–1916. doi: 10.1016/j.ymthe.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Teramura T., Takehara T., Fukuda K. Hyaluronic acid regulates a key redox control factor Nrf2 via phosphorylation of Akt in bovine articular chondrocytes. FEBS Open Bio. 2015;5:476–484. doi: 10.1016/j.fob.2015.05.007.6481c50207494948b790650f2534062e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S., Beury D.W., Parker K.H., Horn L.A. Survival of the fittest: how myeloid-derived suppressor cells survive in the inhospitable tumor microenvironment. Cancer Immunol. Immunother. 2020;69:215–221. doi: 10.1007/s00262-019-02388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pae H.O., Oh G.S., Lee B.S., Rim J.S., Kim Y.M., Chung H.T. 3-Hydroxyanthranilic acid, one of L-tryptophan metabolites, inhibits monocyte chemoattractant protein-1 secretion and vascular cell adhesion molecule-1 expression via heme oxygenase-1 induction in human umbilical vein endothelial cells. Atherosclerosis. 2006;187:274–284. doi: 10.1016/j.atherosclerosis.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Pan J., Zhao X., Lin C., Xu H., Yin Z., Liu T., Zhang S. Immune responsive gene 1, a novel oncogene, increases the growth and tumorigenicity of glioma. Oncol. Rep. 2014;32:1957–1966. doi: 10.3892/or.2014.3474. [DOI] [PubMed] [Google Scholar]
- Peace C.G., O'Neill L.A. The role of itaconate in host defense and inflammation. J. Clin. Invest. 2022;132:e148548. doi: 10.1172/JCI148548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznanski S.M., Singh K., Ritchie T.M., Aguiar J.A., Fan I.Y., Portillo A.L., Rojas E.A., Vahedi F., El-Sayes A., Xing S., et al. Metabolic flexibility determines human NK cell functional fate in the tumor microenvironment. Cell Metab. 2021;33:1205–1220.e5. doi: 10.1016/j.cmet.2021.03.023. [DOI] [PubMed] [Google Scholar]
- Prendergast G.C., Smith C., Thomas S., Mandik-Nayak L., Laury-Kleintop L., Metz R., Muller A.J. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol. Immunother. 2014;63:721–735. doi: 10.1007/s00262-014-1549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestera T., Talalay P., Alam J., Ahn Y.I., Lee P.J., Choi A.M. Parallel induction of heme oxygenase-1 and chemoprotective phase 2 enzymes by electrophiles and antioxidants: regulation by upstream antioxidant-responsive elements (ARE) Mol. Med. 1995;1:827–837. doi: 10.1007/BF03401897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccini C., Garavelli S., Carbone F., Di Silvestre D., La Rocca C., Greco D., Colamatteo A., Lepore M.T., Russo C., De Rosa G., et al. Signals of pseudo-starvation unveil the amino acid transporter SLC7A11 as key determinant in the control of Treg cell proliferative potential. Immunity. 2021;54:1543–1560.e6. doi: 10.1016/j.immuni.2021.04.014. [DOI] [PubMed] [Google Scholar]
- Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H., Hayes J.D., Sutherland C. A partnership with the proteasome; the destructive nature of GSK3. Biochem. Pharmacol. 2018;147:77–92. doi: 10.1016/j.bcp.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D.G., Knatko E.V., Casey A.M., Hukelmann J.L., Dayalan Naidu S., Brenes A.J., Ekkunagul T., Baker C., Higgins M., Tronci L., et al. Nrf2 activation reprograms macrophage intermediary metabolism and suppresses the type I interferon response. iScience. 2022;25:103827. doi: 10.1016/j.isci.2022.103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo I.G., Choi B.H., Ku S.K., Kwak M.K. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: implications for cancer stem cell resistance. Redox Biol. 2018;17:246–258. doi: 10.1016/j.redox.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddawi-Konefka R., Seelige R., Gross E.T., Levy E., Searles S.C., Washington A., Jr., Santosa E.K., Jr., Liu B., Jr., O'Sullivan T.E., Jr., Harismendy O., Jr., et al. Nrf2 induces IL-17D to mediate tumor and virus surveillance. Cell Rep. 2016;16:2348–2358. doi: 10.1016/j.celrep.2016.07.075.ac468b8017164efb8c90cff21f7339e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson L.E., Chien A.T., Astin J.W., Crosier K.E., Crosier P.S., Hall C.J. An inducible transgene reports activation of macrophages in live zebrafish larvae. Dev. Comp. Immunol. 2015;53:63–69. doi: 10.1016/j.dci.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Satoh H., Moriguchi T., Saigusa D., Baird L., Yu L., Rokutan H., Igarashi K., Ebina M., Shibata T., Yamamoto M. NRF2 intensifies host defense systems to prevent lung carcinogenesis, but after tumor initiation accelerates malignant cell growth. Cancer Res. 2016;76:3088–3096. doi: 10.1158/0008-5472.CAN-15-1584. [DOI] [PubMed] [Google Scholar]
- Satoh H., Moriguchi T., Taguchi K., Takai J., Maher J.M., Suzuki T., Winnard P.T., Jr., Raman V., Jr., Ebina M., Jr., Nukiwa T., Jr., et al. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis. 2010;31:1833–1843. doi: 10.1093/carcin/bgq105. [DOI] [PubMed] [Google Scholar]
- Sayin V.I., LeBoeuf S.E., Singh S.X., Davidson S.M., Biancur D., Guzelhan B.S., Alvarez S.W., Wu W.L., Karakousi T.R., Zavitsanou A.M., et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife. 2017;6:e28083. doi: 10.7554/eLife.28083.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Zhao Y., Liu G., Zhou H.L., Fan J., Zhang L., Li Y.L., Wang Y., Liang J., Xu Z.X. Upregulation of programmed death ligand 1 by liver kinase B1 and its implication in programmed death 1 blockade therapy in non-small cell lung cancer. Life Sci. 2020;256:117923. doi: 10.1016/j.lfs.2020.117923. [DOI] [PubMed] [Google Scholar]
- Shi H.Z., Wang D., Sun X.N., Sheng L. MicroRNA-378 acts as a prognosis marker and inhibits cell migration, invasion and epithelial-mesenchymal transition in human glioma by targeting IRG1. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3837–3846. doi: 10.26355/eurrev_201806_15268. [DOI] [PubMed] [Google Scholar]
- Sies H., Belousov V.V., Chandel N.S., Davies M.J., Jones D.P., Mann G.E., Murphy M.P., Yamamoto M., Winterbourn C. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 2022;23:499–515. doi: 10.1038/s41580-022-00456-z. [DOI] [PubMed] [Google Scholar]
- Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- Singh A., Daemen A., Nickles D., Jeon S.M., Foreman O., Sudini K., Gnad F., Lajoie S., Gour N., Mitzner W., et al. NRF2 activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin. Cancer Res. 2021;27:877–888. doi: 10.1158/1078-0432.CCR-20-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S., et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Strelko C.L., Lu W., Dufort F.J., Seyfried T.N., Chiles T.C., Rabinowitz J.D., Roberts M.F. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 2011;133:16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K.A., Li Y., Meliton A.Y., Woods P.S., Kimmig L.M., Cetin-Atalay R., Hamanaka R.B., Mutlu G.M. Endogenous itaconate is not required for particulate matter-induced NRF2 expression or inflammatory response. Elife. 2020;9:e54877. doi: 10.7554/eLife.54877.sa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauffenberger A., Fiumelli H., Almustafa S., Magistretti P.J. Lactate and pyruvate promote oxidative stress resistance through hormetic ROS signaling. Cell Death Dis. 2019;10:653. doi: 10.1038/s41419-019-1877-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa R.K., Fuchs R.J., Malhotra D., Scollick C., Traore K., Bream J.H., Trush M.A., Liby K.T., Sporn M.B., Kensler T.W., et al. Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid. Redox Signal. 2007;9:1963–1970. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa R.K., Lee H., Rangasamy T., Reddy S.P., Yamamoto M., Kensler T.W., Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.M., Francescutti-Verbeem D.M., Kuhn D.M. Gene expression profile of activated microglia under conditions associated with dopamine neuronal damage. FASEB J. 2006;20:515–517. doi: 10.1096/fj.05-4873fje. [DOI] [PubMed] [Google Scholar]
- Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole B.P. Hyaluronan: from extracellular glue to pericellular cue. Nat. Rev. Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- Torrente L., DeNicola G.M. Targeting NRF2 and its downstream processes: opportunities and challenges. Annu. Rev. Pharmacol. Toxicol. 2022;62:279–300. doi: 10.1146/annurev-pharmtox-052220-104025. [DOI] [PubMed] [Google Scholar]
- van der Horst D., Carter-Timofte M.E., van Grevenynghe J., Laguette N., Dinkova-Kostova A.T., Olagnier D. Regulation of innate immunity by Nrf2. Curr. Opin. Immunol. 2022;78:102247. doi: 10.1016/j.coi.2022.102247. [DOI] [PubMed] [Google Scholar]
- Weiss J.M., Davies L.C., Karwan M., Ileva L., Ozaki M.K., Cheng R.Y., Ridnour L.A., Annunziata C.M., Wink D.A., McVicar D.W. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 2018;128:3794–3805. doi: 10.1172/JCI99169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Zhao F., Gao B., Tan C., Yagishita N., Nakajima T., Wong P.K., Chapman E., Fang D., Zhang D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28:708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T., Deumelandt K., van der Vliet H.J., Wesseling P., de Gruijl T.D. Mechanisms of intimate and long-distance cross-talk between glioma and myeloid cells: how to break a vicious cycle. Biochim. Biophys. Acta. 2014;1846:560–575. doi: 10.1016/j.bbcan.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Rennhack J., Andrechek E.R., Rockwell C.E., Liby K.T. Identification of an unfavorable immune signature in advanced lung tumors from Nrf2-deficient mice. Antioxid. Redox Signal. 2018;29:1535–1552. doi: 10.1089/ars.2017.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.D., Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Teng D., Yang L., Xu X., Chen J., Jiang T., Feng A.Y., Zhang Y., Frederick D.T., Gu L., et al. Myeloid-derived itaconate suppresses cytotoxic CD8(+) T cells and promotes tumour growth. Nat. Metab. 2022 Nov 1; doi: 10.1038/s42255-022-00676-9. [Epub]. https://doi.org/10.1038/s42255-022-00676-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Tang L., Chen S., Yin C., Peng S., Li X., Liu T., Liu W., Han C., Stawski L., et al. Targeting the upstream transcriptional activator of PD-L1 as an alternative strategy in melanoma therapy. Oncogene. 2018;37:4941–4954. doi: 10.1038/s41388-018-0314-0. [DOI] [PubMed] [Google Scholar]