Abstract
Cuproptosis is a novel type of copper-induced cell death that primarily occurs in cells that utilize oxidative phosphorylation as the main metabolic pathway to produce energy. Copper directly associates with the lipoylated proteins of the tricarboxylic acid cycle, leading to the disulfide-bond-dependent aggregation of these lipoylated proteins, destabilization of the iron-sulfur cluster proteins, and consequent proteotoxic stress. Cancer cells prefer glycolysis (Warburg effect) to oxidative phosphorylation for producing intermediate metabolites and energy, thereby achieving resistance to cuproptosis. Interestingly, the tumor suppressor p53 is a crucial metabolic regulator that inhibits glycolysis and drives a metabolic switch towards oxidative phosphorylation in cancer cells. Additionally, p53 regulates the biogenesis of iron-sulfur clusters and the copper chelator glutathione, which are two critical components of the cuproptotic pathway, suggesting that this tumor suppressor might play a role in cuproptosis. Furthermore, the possible roles of mutant p53 in regulating cuproptosis are discussed. In this essay, we review the recent progress in the understanding of the mechanism underlying cuproptosis, revisit the roles of p53 in metabolic regulation and iron-sulfur cluster and glutathione biosynthesis, and propose several potential mechanisms for wild-type and mutant p53-mediated cuproptosis regulation.
Subject terms: Tumour-suppressor proteins, Cancer metabolism
Copper homeostasis and cuproptosis
Copper is an essential cofactor for enzymes and it is required for biological processes—including oxidative phosphorylation, reactive oxygen species (ROS) detoxification, iron homeostasis, connective tissue cross-linking, and signal transmission—in almost all organisms [1, 2]. Copper deficiency is associated with a wide range of genetic, neurological, cardiovascular, and metabolic diseases [3, 4]. However, beyond the threshold maintained by an evolutionarily conserved homeostatic mechanism, copper ions can become toxic [5]. Several copper ionophores, such as elesclomol and disulfiram, were found to promote copper-dependent regulated cell death (RCD), namely cuproptosis, in cells with high rates of oxidative phosphorylation [6]. However, it remained unclear how excessive copper induces cell death until Tsvetkov et al. revealed that copper directly targets lipoylated proteins of the tricarboxylic acid (TCA) cycle to induce cuproptosis [7].
Tsvetkov et al. [7] investigated whether copper is required for elesclomol-induced cell death. As serum is the main source of copper for cultured cells, withdrawal of serum from the culture medium increased the resistance of cells to elesclomol. In contrast, supplementation with copper ions in the culture medium dramatically sensitized cells to elesclomol. In addition to elesclomol, a panel of copper ionophores, such as disulfiram and NSC319726, also induced cell death in the presence of copper, whereas the addition of other metals, including iron, cobalt, zinc, and nickel, did not affect this type of cell death. Copper chelators were also included in this study to verify the importance of copper in this process. Indeed, depletion of the endogenous copper chelator glutathione (GSH) promoted, while the addition of the exogenous copper chelator tetrathiomolybdate repressed, elesclomol-induced cell death. Further studies have shown that copper ionophore-induced cell death cannot be remediated by inhibitors of apoptosis, ferroptosis, necroptosis, or oxidative stress. Together, these results suggest that copper-dependent cell death, termed cuproptosis [6], differs from other known types of cell death.
In addition, it was found that cells addicted to mitochondrial metabolism are more sensitive to copper ionophores than those using glycolysis as the main source of energy [7]. Consistently, treatment of cells with inhibitors of complexes I and II of the electron transport chain (ETC) or mitochondrial pyruvate uptake alleviated cuproptosis induced by elesclomol. The mitochondrial uncoupling agent, FCCP, which reduces mitochondrial membrane potential and ATP biogenesis, did not affect cuproptosis, suggesting that ATP production is not required for this process. Cells grown under hypoxic conditions were resistant to cuproptosis, but forced activation of the HIF pathway alone did not affect cuproptosis of cells under normoxic conditions, again underscoring the important role of mitochondrial respiration in mediating copper-induced cell death. Interestingly, elesclomol dramatically reduced the spare capacity of respiration but not basal or ATP-linked respiration. These results indicate that elesclomol-copper may directly target the components of the TCA cycle without affecting ETC activity and ATP synthesis.
To elucidate the mechanistic basis underlying cuproptosis, genome-wide CRISPR-Cas9 screening was performed [7]. Seven genes have been identified as regulators of cuproptosis. Among them, FDX1 encodes a reductase that converts Cu2+ to Cu+ which is a more cytotoxic form of copper; LIPT1, LIAS, and DLD encode the components of the lipoic acid pathway; and DLAT, PDHA1, and PDHB encode the lipoylated components of the pyruvate dehydrogenase (PDH) complex, suggesting that the lipoic acid pathway and protein lipoylation may be important for cuproptosis. Interestingly, copper was found to directly bind to lipoylated TCA cycle proteins, leading to disulfide-bond-dependent aggregation of these proteins and degradation of iron-sulfur (Fe-S) cluster proteins. More importantly, FDX1 was considered a central regulator of cuproptosis, because FDX1 depletion led to complete loss of protein lipoylation, marked decrease in cellular respiration, accumulation of pyruvate and α-ketoglutarate, reduction of succinate, and stabilization of Fe-S cluster proteins. Altogether, excessive copper promotes aggregation of lipoylated TCA cycle proteins and destabilization of Fe-S cluster proteins, both of which are mediated by FDX1, consequently resulting in cuproptosis.
Finally, the authors investigated the effects of intracellular copper fluctuation on cuproptosis by experimentally manipulating the genes encoding the copper importer SLC31A1 and exporter ATP7B in vitro and in vivo [7]. Overexpression of SLC31A1 in cells dramatically increased their sensitivity to CuCl2-induced aggregation of lipoylated proteins and degradation of Fe-S cluster proteins. Consistently, SLC31A1-sensitized, copper-induced cell death was partially rescued by copper chelators and depletion of FDX1 or LIAS. Remarkably, these in vitro findings were further validated in a Wilson’s disease mouse model with Atp7b depletion (Atp7b−/−). Compared with the livers of Atp7b+/− and wild-type mice, those of Atp7b−/− mice showed significant loss of lipoylated and Fe-S cluster proteins, demonstrating that accumulation of intracellular copper leads to cuproptosis in vivo.
Collectively, the study reported by Tsvetkov et al. [7] provides mechanistic insight into cuproptosis and suggests an important role for copper homeostasis in human diseases, including cancer. However, numerous questions remain regarding this understudied field. Is there a threshold for the copper concentration in a cell to induce cuproptosis? Does cuproptosis respond to or is it regulated by cellular stress signals? How do energy metabolic pathways, such as glycolysis, oxidative phosphorylation, and Fe-S cluster metabolism, coordinate with cuproptosis in cancer? Could modulating the copper-induced cell death be exploited for the development of new anti-cancer therapies? These questions warrant further investigation.
Perspective on mechanisms underlying p53 regulation of cuproptosis
The tumor suppressor p53 inhibits cancer progression mainly by promoting different types of RCD, such as apoptosis, ferroptosis, parthanatos, programmed necrosis, and autophagic cell death, in response to various cellular stresses [8–10]. The representative mechanisms underlying p53-mediated RCD are outlined in Fig. 1. Copper dyshomeostasis was previously found to be correlated with p53-regulated apoptosis [11]. Copper at physiological concentrations can directly interact with p53 and inhibit its DNA-binding capacity [12], whereas high levels of copper induce p53 activation and consequent apoptosis in cancer cells [13]. Copper accumulation may result in non-cancer diseases, such as Wilson disease, neurodegenerative diseases, and cardiovascular disorders [14, 15]. It is likely that p53 activation may be partially associated with these diseases by enhancing apoptosis of hepatocytes, neuronal cells, or cardiomyocytes [16–18]. However, it remains elusive whether p53 is involved in cuproptosis. Recently, growing evidence has shown that p53 plays a crucial role in reshaping cancer energy metabolism, including glycolysis and oxidative phosphorylation [19, 20]. As these two tightly coupled metabolic processes are closely associated with cell sensitivity to cuproptosis [7], it is reasonable to speculate that p53 may play a role in the regulation of cuproptosis. To support this perspective, multiple lines of evidence have been discussed (Fig. 2).
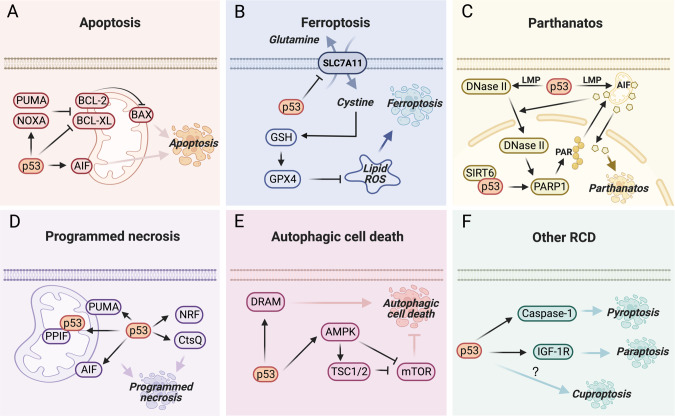
Fig. 1. The representative mechanisms for p53-mediated regulated cell death.
A p53 plays a key role in mitochondrial apoptosis. p53 transactivates the expression of the pro-apoptotic BCL-2 members, such as PUMA and NOXA, to inactivate anti-apoptotic BCL-2 and BCL-XL, resulting in the activation of the pore-forming protein, BAX, and the consequent release of numerous apoptogenic factors [86], such as AIF that is also encoded by a p53-target gene [52]. B p53 can either promote or inhibit ferroptosis in the context of different cancers [87]. One of the well-characterized mechanisms is that p53 represses the expression of SLC7A11, reducing GSH biogenesis, increasing lipid ROS, and thus promoting ferroptosis [9]. C Parthanatos is dependent on PARP1 activation, which leads to generation of PAR, nuclear translocation of AIF, and chromatinolysis [88]. p53 promotes LMP to facilitate nuclear translocation of DNase II with the help of AIF. DNase II then cleaves the DNA to activate the PARP1-AIF axis [54]. Also, p53, together with SIRT6, can directly activate PARP1 to induce parthanatos [89]. D Programmed necrosis is executed via both RIPK-dependent and independent pathways [90]. p53 promotes programmed necrosis by inducing the expression of AIF [91, 92], PUMA [93], CtsQ [94], and the long noncoding RNA NRF [95], or physically interacting with PPIF [96]. E Persistent activation of autophagy results in autophagic cell death. p53 activates AMPK signaling [36, 37] or DRAM expression [97, 98] to enhance this type of RCD. F p53 may also promote pyroptosis and paraptosis by regulating Caspase-1 [99] and IGF-1R [100], respectively.
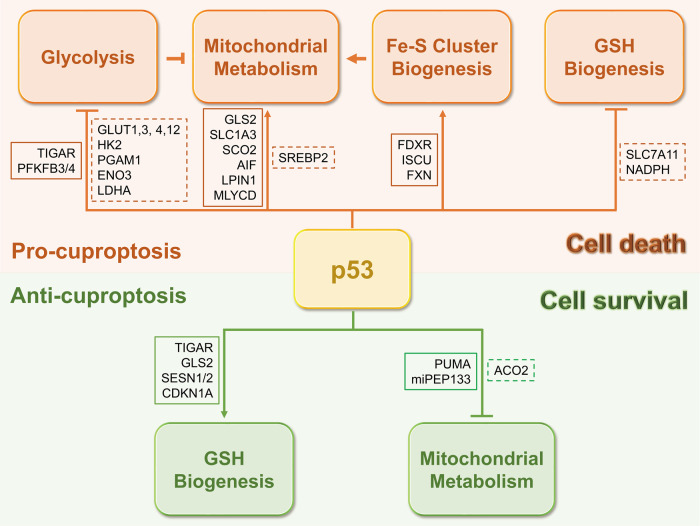
Fig. 2. p53 may regulate cuproptosis via different signaling pathways.
p53 inhibits glucose uptake and glycolysis, and promotes a metabolic switch towards the TCA cycle and oxidative phosphorylation, resulting in cuproptosis sensitization. In addition, p53 regulates the biogenesis of iron-sulfur clusters and glutathione, which are two critical components of the cuproptosis pathway, thereby promoting or preventing cuproptosis.
p53 might increase sensitization to cuproptosis by inhibiting glycolysis
Cancer cells tend to utilize glycolysis, but not the TCA cycle, to produce intermediate metabolites and energy, even under oxygen-rich conditions, which is known as aerobic glycolysis or the Warburg effect. Inhibition of glucose uptake or glycolysis may induce a metabolic shift from glycolytic metabolism to mitochondrial metabolism by using alternate energy sources, such as galactose or glutamine, which sensitizes cells to cuproptosis [7, 21]. p53 inhibits glucose metabolism through various mechanisms. First, p53 reduces glucose uptake by repressing the expression or activity of the glucose transporters GLUT1, GLUT3, GLUT4, and GLUT12 [22–25]. In addition, p53 perturbs several critical catalytic reactions in glycolysis. For example, p53 suppresses the conversion of glucose to glucose-6-phosphate by promoting the decay of HK2 mRNA [26], fructose-6-phosphate to fructose-1,6-bisphosphate by inhibiting PFK1 activity via its target genes, PFKFB3/4 and TIGAR [27–29], 3-phosphoglycerate to 2-phosphoglycerate by reducing the protein level of PGAM1 [30], and 2-phosphoglycerate to phosphoenolpyruvate by repressing the expression of ENO3 [31]. In the final step of glycolysis, phosphoenolpyruvate is converted to pyruvate, which either produces lactate under the action of LDHA/B or fuels the TCA cycle. p53 also inhibits the expression of LDHA by directly binding to its promoter [32] or promoting the degradation of HIF-1α, a transcription factor of LDHA [33]. This may result in the accumulation of intracellular pyruvate, which is available for the TCA cycle and oxidative phosphorylation. p53 can boost mitochondrial metabolism in many ways, which will be described in the next section. Glucose is the main source of ATP for mammalian cells. Upon glucose restriction, the intracellular ATP level declines, resulting in the increased ratio of AMP to ATP and consequent activation of the AMPK signaling [34]. AMPK activation induces p53 phosphorylation and activation [35], thus forming a feedback circuit between p53 and glucose metabolism, which further prompts a switch from glycolysis to oxidative phosphorylation. It was also noted that persistent activation of AMPK triggers autophagic cell death through, for example, inhibition of mTOR [36, 37]. Thus, cuproptosis might coincide with autophagic cell death in response to limitation of glucose supply. Collectively, p53 may sensitize cells to cuproptosis by inhibiting glycolysis and driving a shift towards mitochondrial metabolism, as discussed below.
p53 might promote cuproptosis by enhancing mitochondrial metabolism
p53 activity has been found to maintain mitochondrial integrity and performance, as mutation or deficiency of p53 leads to a significant reduction in mitochondrial content, cytochrome c oxidase (COX) activity, and respiratory metabolism [38–40], which also suggests a possible role for p53 in cuproptosis. p53 promotes the TCA cycle and oxidative phosphorylation via numerous mechanisms. In addition to maintaining a high level of pyruvate by inhibiting LDHA as mentioned above, p53 facilitates the conversion of pyruvate to acetyl-CoA by promoting dephosphorylation and thus activation of the PDH complex [41, 42], a critical component of the TCA cycle required for cuproptosis [7]. It is unclear whether p53 regulates the biosynthesis of lipoic acid or mediates lipoylation of the PDH complex, including DLAT, PDHA1, and PDHB, though lipoic acid can modulate p53 protein stability [43]. Also, p53 increases acetyl-CoA production by enhancing fatty acid oxidation [44, 45] and suppressing lipid synthesis [46], thereby boosting the initiation of the TCA cycle. Glutamine, an important source of nutrients in the TCA cycle, is converted to glutamate and subsequently to α-ketoglutarate. p53 promotes the conversion of glutamine into glutamate by transcriptionally activating GLS2 [47, 48]. Interestingly, upon glutamine starvation, p53 activates the expression of SLC1A3, which encodes an aspartate/glutamate transporter, to sustain aspartate metabolism, consequently fueling the TCA cycle [49]. Thus, these findings suggest that p53 may induce cuproptosis sensitization by promoting the TCA cycle. However, p53 suppresses mitochondrial pyruvate uptake through the activation of PUMA in hepatocarcinoma [50] and the catalytic activity of ACO2, an Fe-S cluster protein responsible for the conversion of citrate into isocitrate, in prostate cancer [51], suggesting that p53 may prevent cuproptosis in the context of different cancers (Fig. 3). Additionally, p53 might increase cuproptotic cell death by augmenting oxidative phosphorylation. For example, p53 was found to promote mitochondrial respiration by inducing the expression of SCO2, which is essential for the assembly of the COX complex, whereas depletion of SCO2 induces a metabolic switch towards glycolysis, which is manifested in p53-deficient cells [39]. Another p53-target gene, AIF, which encodes a mitochondrial membrane protein, is required for efficient oxidative phosphorylation as it is involved in the proper assembly of the mitochondrial respiratory complex I [52]. Interestingly, AIF seems a key node in the p53-mediated RCD network, as apoptosis, parthanatos, and programmed necrosis could be connected with one another through this mitochondrial protein [53, 54] (Fig. 1). However, this does not necessarily mean that cuproptosis can be accompanied by apoptosis, parthanatos, or programmed necrosis. For instance, the anti-apoptotic BCL-2 members are often overexpressed in cancers, leading to the impaired apoptotic pathway and chemoresistance [55]. Under this scenario, other types of RCD, such as cuproptosis, might be alternately activated in response to cancer therapies. Taken together, these findings strongly suggest that p53 may support cuproptosis by enhancing mitochondrial metabolism via different mechanisms.
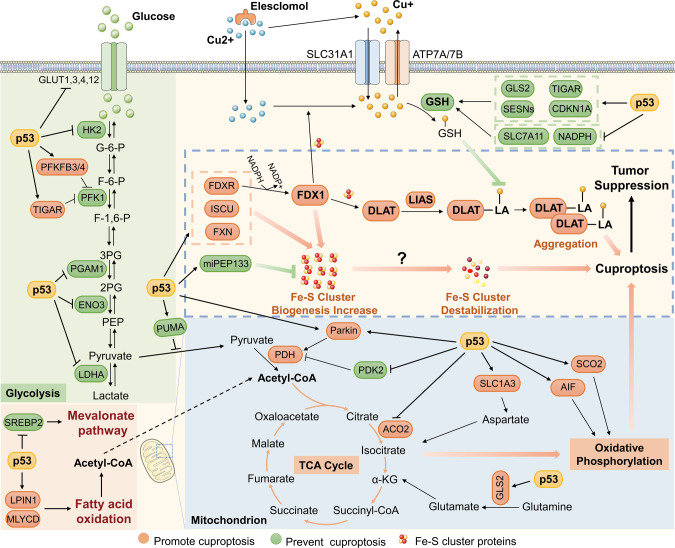
Fig. 3. p53 may have divergent roles in cuproptosis.
p53 may maintain a proper physiological copper level to prevent copper-induced cytotoxicity (lower panel), while also promote cuproptosis when the copper level is pathologically or experimentally upregulated (upper panel). Solid boxes indicate genes that are positively regulated by p53, and dashed boxes indicate genes that are negatively regulated by p53.
p53 regulates genes involved in the biogenesis of iron-sulfur clusters
Fe-S clusters are ubiquitous proteins that act as cofactors involved in a wide range of biological processes, including enzymatic catalysis, electron transport, and metabolic stress sensing [56]. Tsvetkov et al. found that copper-induced cell death is accompanied by the destabilization of Fe-S cluster proteins [7], although it remains unclear whether Fe-S cluster degradation is a causative factor or a consequence of cuproptosis. Considering that p53 regulates the expression of multiple genes involved in Fe-S cluster biogenesis, we speculate that this may be an additional mechanism underlying p53-regulated cuproptosis that would be worth testing in the future. One of these genes is FDXR, which encodes a ferredoxin reductase responsible for electron transport from NADPH to FDX1/2 and then to cytochrome P450 for Fe-S cluster biogenesis [57]. FDXR was originally identified as a p53-target gene required for p53-induced oxidative stress and apoptosis by two independent groups [58, 59]. It was later shown that FDXR is involved in p53-mediated iron metabolism and possibly Fe-S cluster homeostasis [60]. In addition, p53 induces the expression of ISCU [61] and FXN [62, 63], both of which encode scaffold components critical for Fe-S cluster assembly. Recently, the tumor suppressor microprotein miPEP133, which is encoded by the primary transcript of miR-34a activated by p53, was found to interact with HSPA9 to impair its function in the mitochondria [64]. Inhibition of HSPA9 results in a decrease in mitochondrial membrane potential and mitochondrial content and, probably, the perturbation of Fe-S cluster biogenesis, as HSPA9 serves as a chaperone protein for Fe-S cluster assembly [65]. These findings suggest that p53 may regulate cuproptosis by coordinating biogenesis and homeostasis of Fe-S clusters, although whether p53 plays a role in the destabilization of Fe-S clusters remains to be determined.
p53 controls the level of the endogenous copper chelator glutathione
GSH not only acts as a central antioxidant mediator but also as a copper chelator, as GSH deficiency results in increased labile copper levels [66]. Consistently, Tsvetkov et al. showed that depletion of GSH by buthionine sulfoximine promotes copper-induced cell death [7]. Interestingly, several studies have shown that p53 plays a role in GSH biogenesis. p53 was found to transcriptionally repress the expression of SLC7A11, which is critical for the import of the GSH precursor cystine, resulting in reduced levels of GSH, increased lipid ROS, and ferroptosis [9]. In addition, p53 suppresses the production of NADPH, a powerful reducing agent that provides electrons to regenerate GSH, by inhibiting malic enzymes [67] or G6PD and thus the pentose phosphate pathway [68]. These findings, together with those of Tsvetkov et al. [7], imply that iron- and copper-induced cell death may share a common surveillance mechanism involving GSH, which can be counteracted by p53. Interestingly, p53 also enhances GSH biosynthesis to protect cells from oxidative toxicity through the activation of several metabolic genes, including TIGAR [27], GLS2 [47, 48], and SESN1/2 [69]. Additionally, the p53-target gene CDKN1A, also known as p21, triggers the NRF2 antioxidant pathway, resulting in increased GSH and NADPH synthesis [70]. These studies suggest that p53 may play both positive and negative roles in cuproptosis by differentially regulating GSH biogenesis (Fig. 3). One possible interpretation is that p53 may act as a monitor of the intracellular copper concentration, on the one hand, to maintain a proper physiological copper level by preventing accumulation of excessive labile copper, and on the other hand, to eliminate copper-damaged cells when the uptake or level of copper is pathologically or experimentally upregulated. This statement is partially supported by previous studies documenting that p53 has either pro-survival or pro-death activity in response to varying degrees of stress signals [20, 21]. For instance, p53 facilitates DNA damage repair and ROS clearance to help cells survive mild genotoxic and oxidative stresses. In contrast, p53 can also promote apoptosis and ferroptosis to eliminate cells with irreparable damage caused by drastic stress. Altogether, p53 may prevent or promote cuproptosis through divergent regulation of the biogenesis of the copper chelator GSH under the physiological or copper-excessive condition.
Possible roles of mutant p53 in the regulation of cuproptosis
TP53 is the most frequently mutated gene in human cancers. The majority of cancer-associated p53 mutations are missense mutations that often occur in the DNA-binding domain. Many of these mutant p53 (mtp53) proteins acquire “gain-of-function” (GOF) activities to promote cancer development [71, 72] independently of the function of wild-type p53 as discussed above. Mtp53 was found to enhance glycolysis and repress mitochondrial metabolism through different mechanisms [19, 20]. For instance, mtp53 increases glucose uptake via the RhoA-ROCK-GLUT1 cascade [73] and promotes glycolysis by transcriptionally inducing the expression of HK2 [74] and PLA2G16 [75]. Mtp53 also facilitates mTOR signaling to mediate phosphorylation of PKM2, leading to enhanced glycolysis [76, 77]. In addition, p53 mutants attenuate oxidative phosphorylation, as they can interact with and suppress the function of PGC-1α [78], or downregulate PCK2 via the miR-200c-ZEB1/BMI1 axis [79]. It is also worth noting that distinct mtp53 proteins may have different roles in regulating glycolysis and mitochondrial metabolism [80]. Moreover, mtp53 augments lipid synthesis and inhibits fatty acid oxidation, thus reducing the availability of acetyl-CoA to the TCA cycle. Mechanistically, mtp53 cooperates with SREBPs to activate the mevalonate pathway [81, 82], transactivates PLA2G16 expression to increase phospholipid synthesis [83], and suppresses AMPK signaling to support anabolic metabolism [76, 84, 85]. Together, these studies suggest that mtp53 may protect cancer cells from cuproptosis (Fig. 4).
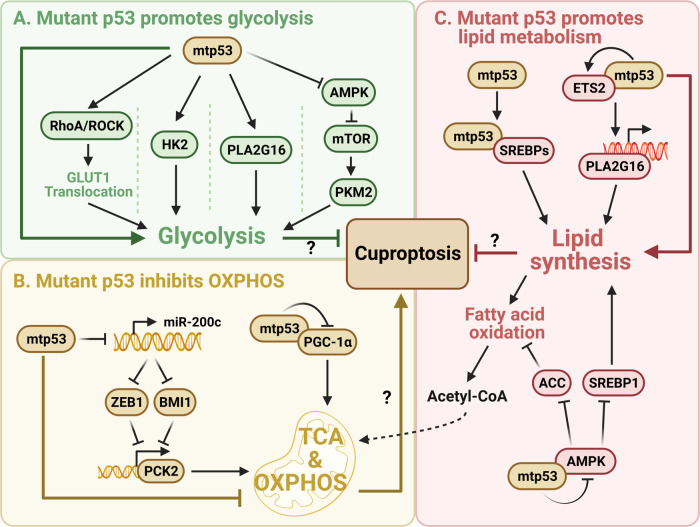
Fig. 4. Possible roles of mutant p53 in regulating cuproptosis.
Mutant p53 may prevent cuproptosis by enhancing glycolysis (A), inhibiting mitochondrial metabolism (B), and reducing the availability of acetyl-CoA to the TCA cycle through the regulation of lipid metabolism (C).
Concluding remarks
Cuproptosis is a novel form of copper-induced cell death. Cells that utilize glycolysis as the main metabolic pathway to produce energy are resistant to cuproptosis, whereas those that are dependent on the TCA cycle and oxidative phosphorylation are sensitive to cuproptosis. Excessive labile copper can directly associate with lipoylated TCA cycle proteins, leading to aggregation of these lipoylated proteins, destabilization of Fe-S cluster proteins, and consequent cell death. p53 acts as an important metabolic regulator that mediates the transition from glycolysis to oxidative phosphorylation, enhances Fe-S cluster biogenesis and orchestrates the level of the copper chelator GSH. The p53- or mtp53-regulated genes, which may be associated with cuproptosis as discussed above, are illustrated in Tables 1 and 2. In light of these findings, it is rational to propose that p53 may participate in cuproptosis in a context-dependent manner, and activation of p53-mediated cuproptosis could be an attractive strategy to eradicate cancer cells.
Table 1.
p53-regulated genes possibly associated with cuproptosis.
| Functional category | p53-regulated gene | Function of gene | p53 may promote (+) or prevent (−) cuproptosis by regulating the gene | Reference |
|---|---|---|---|---|
| Glycolysis | GLUT1/3/4/12 | Transport glucose into cells | + | [22–25] |
| HK2 | Convert glucose to glucose-6-phosphate | + | [26] | |
| PFKFB3/4 | Inhibit PFK1 activity to prevent the conversion of fructose-6-phosphate to fructose-1,6-bisphosphate | + | [28, 29] | |
| TIGAR | Inhibit PFK1 activity to prevent the conversion of fructose-6-phosphate to fructose-1,6-bisphosphate | + | [27] | |
| PGAM1 | Convert 3-phosphoglycerate to 2-phosphoglycerate | + | [30] | |
| ENO3 | Convert 2-phosphoglycerate to phosphoenolpyruvate | + | [31] | |
| LDHA | Convert pyruvate to lactate, thus diminishing pyruvate entry into the mitochondria | + | [32, 33] | |
| Mitochondrial metabolism | GLS2 | Promote the conversion of glutamine to glutamate and subsequently to α-KG | + | [47, 48] |
| SLC1A3 | Promote aspartate uptake or aspartate-glutamate exchange at the mitochondria | + | [49] | |
| SCO2 | Sustain the assembly of the COX complex | + | [39] | |
| AIF | Sustain the assembly of the Complex I | + | [52] | |
| ACO2 | Promote the conversion of citrate to isocitrate | − | [51] | |
| PUMA | Disrupt the function of mitochondrial pyruvate carrier (MPC) to inhibit pyruvate uptake | − | [50] | |
| Lipid metabolism | LPIN1 | Promote fatty acid oxidation | + | [44] |
| MLYCD | Convert malonyl-CoA to acetyl-CoA | + | [45] | |
| SREBP2 | Promote the mevalonate pathway | + | [46] | |
| Fe-S cluster biogenesis | FDXR | Promote electron transport from NADPH to FDX1/2 and then to cytochrome P450 for Fe-S cluster biogenesis | + | [58–60] |
| ISCU | Encode scaffold components critical for Fe-S cluster assembly | + | [61] | |
| FXN | Encode scaffold components critical for Fe-S cluster assembly | + | [62, 63] | |
| Pri-miR-34a | Encode miPEP133 to interact with HSPA9 and impair Fe-S cluster assembly | − | [64] | |
| GSH biogenesis | SLC7A11 | Encode a component of the cystine/glutamate antiporter to import cystine and promote GSH biosynthesis | + | [9] |
| ME1/2 | Produce NADPH to facilitate GSH regeneration | + | [67] | |
| G6PD | Produce NADPH to facilitate GSH regeneration | + | [68] | |
| TIGAR | Promote PPP to produce NADPH and then GSH | − | [27] | |
| GLS2 | Convert glutamine to glutamate for GSH biosynthesis | − | [47, 48] | |
| SESN1/2 | Promote GSH biosynthesis | − | [69] | |
| CDKN1A | Trigger the NRF2 antioxidant pathway to promote GSH biosynthesis | − | [70] |
Table 2.
Mutant p53-regulated genes possibly associated with cuproptosis.
| Functional category | mtp53-regulated gene | Function of gene | mtp53 may promote (+) or prevent (−) cuproptosis by regulating the gene | Reference |
|---|---|---|---|---|
| Glycolysis | RhoA/ROCK | Stimulate GLUT1 translocation | − | [73] |
| HK2 | Promote glycolysis | − | [74] | |
| PLA2G16 | Promote glycolysis | − | [75] | |
| AMPK | Inhibits glycolysis through multiple mechanisms | − | [76] | |
| Mitochondrial metabolism | PGC-1α | Promote oxidative phosphorylation | − | [78] |
| miR-200c | Inhibit ZEB1 and BMI1 to activate PCK2 | − | [79] | |
| Lipid metabolism | SREBPs | Promote the mevalonate pathway | − | [81, 82] |
| PLA2G16 | Promote phospholipid synthesis | − | [83] | |
| AMPK | Inhibit lipid synthesis and promote fatty acid oxidation | − | [76] |
Supplementary information
Acknowledgements
We thank the innovative research team of the high-level local university in Shanghai.
Author contributions
CX, QH, and XZ drafted the manuscript, HL, QH, and XZ revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82273098 and 82072879 to XZ, and 82173022 to QH).
Competing interests
The authors declare no competing interests.
Footnotes
Edited by G. Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qian Hao, Email: haoqian@fudan.edu.cn.
Xiang Zhou, Email: xiangzhou@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01125-0.
References
- 1.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–62. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz LM, Libedinsky A, Elorza AA. Role of copper on mitochondrial function and metabolism. Front Mol Biosci. 2021;8:711227. doi: 10.3389/fmolb.2021.711227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weihl CC, Lopate G. Motor neuron disease associated with copper deficiency. Muscle Nerve. 2006;34:789–93. doi: 10.1002/mus.20631. [DOI] [PubMed] [Google Scholar]
- 4.Su TA, Shihadih DS, Cao W, Detomasi TC, Heffern MC, Jia S, et al. A Modular ionophore platform for liver-directed copper supplementation in cells and animals. J Am Chem Soc. 2018;140:13764–74. doi: 10.1021/jacs.8b08014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge EJ, Bush AI, Casini A, Cobine PA, Cross JR, DeNicola GM, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer. 2022;22:102–13. doi: 10.1038/s41568-021-00417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsvetkov P, Detappe A, Cai K, Keys HR, Brune Z, Ying W, et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat Chem Biol. 2019;15:681–9. doi: 10.1038/s41589-019-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–61. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471–80. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 9.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranjan A, Iwakuma T. Non-canonical cell death induced by p53. Int J Mol Sci. 2016;17:2068. doi: 10.3390/ijms17122068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Formigari A, Gregianin E, Irato P. The effect of zinc and the role of p53 in copper-induced cellular stress responses. J Appl Toxicol. 2013;33:527–36. doi: 10.1002/jat.2854. [DOI] [PubMed] [Google Scholar]
- 12.Hainaut P, Rolley N, Davies M, Milner J. Modulation by copper of p53 conformation and sequence-specific DNA binding: role for Cu(II)/Cu(I) redox mechanism. Oncogene. 1995;10:27–32. [PubMed] [Google Scholar]
- 13.Narayanan VS, Fitch CA, Levenson CW. Tumor suppressor protein p53 mRNA and subcellular localization are altered by changes in cellular copper in human Hep G2 cells. J Nutr. 2001;131:1427–32. doi: 10.1093/jn/131.5.1427. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Jiang Y, Shi H, Peng Y, Fan X, Li C. The molecular mechanisms of copper metabolism and its roles in human diseases. Pflug Arch. 2020;472:1415–29. doi: 10.1007/s00424-020-02412-2. [DOI] [PubMed] [Google Scholar]
- 15.Ahuja A, Dev K, Tanwar RS, Selwal KK, Tyagi PK. Copper mediated neurological disorder: visions into amyotrophic lateral sclerosis, Alzheimer and Menkes disease. J Trace Elem Med Biol. 2015;29:11–23. doi: 10.1016/j.jtemb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Strand S, Hofmann WJ, Grambihler A, Hug H, Volkmann M, Otto G, et al. Hepatic failure and liver cell damage in acute Wilson’s disease involve CD95 (APO-1/Fas) mediated apoptosis. Nat Med. 1998;4:588–93. doi: 10.1038/nm0598-588. [DOI] [PubMed] [Google Scholar]
- 17.Vanlandingham JW, Tassabehji NM, Somers RC, Levenson CW. Expression profiling of p53-target genes in copper-mediated neuronal apoptosis. Neuromolecular Med. 2005;7:311–24. doi: 10.1385/NMM:7:4:311. [DOI] [PubMed] [Google Scholar]
- 18.Men H, Cai H, Cheng Q, Zhou W, Wang X, Huang S, et al. The regulatory roles of p53 in cardiovascular health and disease. Cell Mol Life Sci CMLS. 2021;78:2001–18. doi: 10.1007/s00018-020-03694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and metabolism. J Mol Cell Biol. 2019;11:284–92. doi: 10.1093/jmcb/mjy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Gu W. The complexity of p53-mediated metabolic regulation in tumor suppression. Semin Cancer Biol. 2021;85:4–32. [DOI] [PMC free article] [PubMed]
- 21.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 22.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–8. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 23.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–33. doi: 10.1158/0008-5472.CAN-03-0846. [DOI] [PubMed] [Google Scholar]
- 24.Zawacka-Pankau J, Grinkevich VV, Hunten S, Nikulenkov F, Gluch A, Li H, et al. Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J Biol Chem. 2011;286:41600–15. doi: 10.1074/jbc.M111.240812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama M, Okada S, Nakagomi A, Moriya J, Shimizu I, Nojima A, et al. Inhibition of endothelial p53 improves metabolic abnormalities related to dietary obesity. Cell Rep. 2014;7:1691–703. doi: 10.1016/j.celrep.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Xiong H, Wu F, Zhang Y, Wang J, Zhao L, et al. Hexokinase 2-mediated Warburg effect is required for PTEN- and p53-deficiency-driven prostate cancer growth. Cell Rep. 2014;8:1461–74. doi: 10.1016/j.celrep.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 28.Franklin DA, He Y, Leslie PL, Tikunov AP, Fenger N, Macdonald JM, et al. p53 coordinates DNA repair with nucleotide synthesis by suppressing PFKFB3 expression and promoting the pentose phosphate pathway. Sci Rep. 2016;6:38067. doi: 10.1038/srep38067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ros S, Floter J, Kaymak I, Da Costa C, Houddane A, Dubuis S, et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene. 2017;36:3287–99. doi: 10.1038/onc.2016.477. [DOI] [PubMed] [Google Scholar]
- 30.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–85. doi: 10.1158/0008-5472.177.65.1. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Du J, Lin W, Long Z, Zhang N, Huang X, et al. Regulation of lactate production through p53/beta-enolase axis contributes to statin-associated muscle symptoms. EBioMedicine. 2019;45:251–60. doi: 10.1016/j.ebiom.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Niu W, Luo Y, Li H, Xie Y, Wang H, et al. p53/Lactate dehydrogenase A axis negatively regulates aerobic glycolysis and tumor progression in breast cancer expressing wild-type p53. Cancer Sci. 2019;110:939–49. doi: 10.1111/cas.13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121–35. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–93. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 36.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–52. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- 38.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–92. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 39.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 40.Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci USA. 2011;108:16259–64. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–7. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 43.Farhat D, Lincet H. Lipoic acid a multi-level molecular inhibitor of tumorigenesis. Biochim Biophys Acta Rev Cancer. 2020;1873:188317. doi: 10.1016/j.bbcan.2019.188317. [DOI] [PubMed] [Google Scholar]
- 44.Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, et al. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, He Y, Jin A, Tikunov AP, Zhou L, Tollini LA, et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci USA. 2014;111:E2414–2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon SH, Huang CH, Houlihan SL, Regunath K, Freed-Pastor WA, Morris JPt, et al. p53 represses the mevalonate pathway to mediate tumor suppression. Cell. 2019;176:564–80.. [DOI] [PMC free article] [PubMed]
- 47.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA. 2010;107:7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tajan M, Hock AK, Blagih J, Robertson NA, Labuschagne CF, Kruiswijk F, et al. A role for p53 in the adaptation to glutamine starvation through the expression of SLC1A3. Cell Metab. 2018;28:721–36. doi: 10.1016/j.cmet.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Yu L, Chen W, Xu Y, Wu M, Todorova D, et al. Wild-Type p53 promotes cancer metabolic switch by inducing PUMA-dependent suppression of oxidative phosphorylation. Cancer Cell. 2019;35:191–203. doi: 10.1016/j.ccell.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Xue YN, Liu YN, Su J, Li JL, Wu Y, Guo R, et al. Zinc cooperates with p53 to inhibit the activity of mitochondrial aconitase through reactive oxygen species accumulation. Cancer Med. 2019;8:2462–73. doi: 10.1002/cam4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stambolsky P, Weisz L, Shats I, Klein Y, Goldfinger N, Oren M, et al. Regulation of AIF expression by p53. Cell Death Differ. 2006;13:2140–9. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 53.Delavallee L, Cabon L, Galan-Malo P, Lorenzo HK, Susin SA. AIF-mediated caspase-independent necroptosis: a new chance for targeted therapeutics. IUBMB Life. 2011;63:221–32. doi: 10.1002/iub.432. [DOI] [PubMed] [Google Scholar]
- 54.Tarayrah-Ibraheim L, Maurice EC, Hadary G, Ben-Hur S, Kolpakova A, Braun T, et al. DNase II mediates a parthanatos-like developmental cell death pathway in Drosophila primordial germ cells. Nat Commun. 2021;12:2285. doi: 10.1038/s41467-021-22622-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat Rev Cancer. 2009;9:501–7. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 56.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–8. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 57.Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci USA. 2010;107:11775–80. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, et al. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–7. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu G, Chen X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21:7195–204. doi: 10.1038/sj.onc.1205862. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Qian Y, Zhang J, Yan W, Jung YS, Chen M, et al. Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 2017;31:1243–56. doi: 10.1101/gad.299388.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Funauchi Y, Tanikawa C, Yi Lo PH, Mori J, Daigo Y, Takano A, et al. Regulation of iron homeostasis by the p53-ISCU pathway. Sci Rep. 2015;5:16497. doi: 10.1038/srep16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sawamoto M, Imai T, Umeda M, Fukuda K, Kataoka T, Taketani S. The p53-dependent expression of frataxin controls 5-aminolevulinic acid-induced accumulation of protoporphyrin IX and photo-damage in cancerous cells. Photochem Photobiol. 2013;89:163–72. doi: 10.1111/j.1751-1097.2012.01215.x. [DOI] [PubMed] [Google Scholar]
- 63.Shimizu R, Lan NN, Tai TT, Adachi Y, Kawazoe A, Mu A, et al. p53 directly regulates the transcription of the human frataxin gene and its lack of regulation in tumor cells decreases the utilization of mitochondrial iron. Gene. 2014;551:79–85. doi: 10.1016/j.gene.2014.08.043. [DOI] [PubMed] [Google Scholar]
- 64.Kang M, Tang B, Li J, Zhou Z, Liu K, Wang R, et al. Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol Cancer. 2020;19:143. doi: 10.1186/s12943-020-01248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schilke B, Williams B, Knieszner H, Pukszta S, D’Silva P, Craig EA, et al. Evolution of mitochondrial chaperones utilized in Fe-S cluster biogenesis. Curr Biol CB. 2006;16:1660–5. doi: 10.1016/j.cub.2006.06.069. [DOI] [PubMed] [Google Scholar]
- 66.Chung CY, Posimo JM, Lee S, Tsang T, Davis JM, Brady DC, et al. Activity-based ratiometric FRET probe reveals oncogene-driven changes in labile copper pools induced by altered glutathione metabolism. Proc Natl Acad Sci USA. 2019;116:18285–94. doi: 10.1073/pnas.1904610116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–93. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–6. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 70.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–73. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang C, Liu J, Xu D, Zhang T, Hu W, Feng Z. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol. 2020;12:674–87. doi: 10.1093/jmcb/mjaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–80. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 75.Xia W, Bai H, Deng Y, Yang Y. PLA2G16 is a mutant p53/KLF5 transcriptional target and promotes glycolysis of pancreatic cancer. J Cell Mol Med. 2020;24:12642–55. doi: 10.1111/jcmm.15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–74. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dando I, Cordani M, Donadelli M. Mutant p53 and mTOR/PKM2 regulation in cancer cells. IUBMB Life. 2016;68:722–6. doi: 10.1002/iub.1534. [DOI] [PubMed] [Google Scholar]
- 78.Basu S, Gnanapradeepan K, Barnoud T, Kung CP, Tavecchio M, Scott J, et al. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1alpha. Genes Dev. 2018;32:230–43. doi: 10.1101/gad.309062.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chao CH, Wang CY, Wang CH, Chen TW, Hsu HY, Huang HW, et al. Mutant p53 attenuates oxidative phosphorylation and facilitates cancer stemness through downregulating miR-200c-PCK2 axis in basal-like breast cancer. Mol Cancer Res MCR. 2021;19:1900–16. doi: 10.1158/1541-7786.MCR-21-0098. [DOI] [PubMed] [Google Scholar]
- 80.Eriksson M, Ambroise G, Ouchida AT, Lima Queiroz A, Smith D, Gimenez-Cassina A, et al. Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism. Mol Cell Biol. 2017;37:e00328–17. doi: 10.1128/MCB.00328-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–66. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 83.Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, et al. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci USA. 2014;111:11145–50. doi: 10.1073/pnas.1404139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hardie DG, Pan DA. Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochem Soc Trans. 2002;30:1064–70. doi: 10.1042/bst0301064. [DOI] [PubMed] [Google Scholar]
- 85.Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–88. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–13. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Gu W. p53 in ferroptosis regulation: the new weapon for the old guardian. Cell Death Differ. 2022;29:895–910. doi: 10.1038/s41418-022-00943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann N. Y Acad Sci. 2008;1147:233–41. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, Zhang C, Li J, Jiang M, Guo S, Yang G, et al. Inhibition of AKT induces p53/SIRT6/PARP1-dependent parthanatos to suppress tumor growth. Cell Commun Signal: CCS. 2022;20:93. doi: 10.1186/s12964-022-00897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Napoletano F, Gibert B, Yacobi-Sharon K, Vincent S, Favrot C, Mehlen P, et al. p53-dependent programmed necrosis controls germ cell homeostasis during spermatogenesis. PLoS Genet. 2017;13:e1007024. doi: 10.1371/journal.pgen.1007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cregan SP, Fortin A, MacLaurin JG, Callaghan SM, Cecconi F, Yu SW, et al. Apoptosis-inducing factor is involved in the regulation of caspase-independent neuronal cell death. J Cell Biol. 2002;158:507–17. doi: 10.1083/jcb.200202130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cabon L, Galan-Malo P, Bouharrour A, Delavallee L, Brunelle-Navas MN, Lorenzo HK, et al. BID regulates AIF-mediated caspase-independent necroptosis by promoting BAX activation. Cell Death Differ. 2012;19:245–56. doi: 10.1038/cdd.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen D, Tong J, Yang L, Wei L, Stolz DB, Yu J, et al. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc Natl Acad Sci USA. 2018;115:3930–5. doi: 10.1073/pnas.1717190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tu HC, Ren D, Wang GX, Chen DY, Westergard TD, Kim H, et al. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci USA. 2009;106:1093–8. doi: 10.1073/pnas.0808173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang K, Liu F, Liu CY, An T, Zhang J, Zhou LY, et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23:1394–405. doi: 10.1038/cdd.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–48. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 98.Hasei J, Sasaki T, Tazawa H, Osaki S, Yamakawa Y, Kunisada T, et al. Dual programmed cell death pathways induced by p53 transactivation overcome resistance to oncolytic adenovirus in human osteosarcoma cells. Mol Cancer Ther. 2013;12:314–25. doi: 10.1158/1535-7163.MCT-12-0869. [DOI] [PubMed] [Google Scholar]
- 99.Gupta S, Radha V, Furukawa Y, Swarup G. Direct transcriptional activation of human caspase-1 by tumor suppressor p53. J Biol Chem. 2001;276:10585–8. doi: 10.1074/jbc.C100025200. [DOI] [PubMed] [Google Scholar]
- 100.Pehar M, O’Riordan KJ, Burns-Cusato M, Andrzejewski ME, del Alcazar CG, Burger C, et al. Altered longevity-assurance activity of p53:p44 in the mouse causes memory loss, neurodegeneration and premature death. Aging Cell. 2010;9:174–90. doi: 10.1111/j.1474-9726.2010.00547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.