Summary
Background
Observational studies have investigated the effect of serum lipids on kidney function, but these findings are limited by confounding, reverse causation and have reported conflicting results. Mendelian randomization (MR) studies address this confounding problem. However, they have been conducted mostly in European ancestry individuals. We, therefore, set out to investigate the effect of lipid traits on the estimated glomerular filtration rate (eGFR) based on serum creatinine in individuals of African ancestry.
Methods
We used the two-sample and multivariable Mendelian randomization (MVMR) approaches; in which instrument variables (IV's) for the predictor (lipid traits) were derived from summary-level data of a meta-analyzed African lipid GWAS (MALG, n = 24,215) from the African Partnership for Chronic Disease Research (APCDR) (n = 13,612) & the Africa Wits-IN-DEPTH partnership for Genomics studies (AWI-Gen) dataset (n = 10,603). The outcome IV's were computed from the eGFR summary-level data of African-ancestry individuals within the Million Veteran Program (n = 57,336). A random-effects inverse variance method was used in our primary analysis, and pleiotropy was adjusted for using robust and penalized sensitivity testing. The lipid predictors for the MVMR were high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, and triglycerides (TG).
Findings
We found a significant causal association between genetically predicted low-density lipoprotein (LDL) cholesterol and eGFR in African ancestry individuals β = 1.1 (95% CI [0.411–1.788]; p = 0.002). Similarly, total cholesterol (TC) showed a significant causal effect on eGFR β = 1.619 (95% CI [0.412–2.826]; p = 0.009). However, the IVW estimate showed that genetically predicted HDL-C β = −0.164, (95% CI = [−1.329 to 1.00]; p = 0.782), and TG β = −0.934 (CI = [−2.815 to 0.947]; p = 0.33) were not significantly causally associated with the risk of eGFR. In the multivariable analysis inverse-variance weighted (MVIVW) method, there was evidence for a causal association between LDL and eGFR β = 1.228 (CI = [0.477–1.979]; p = 0.001). A significant causal effect of Triglycerides (TG) on eGFR in the MVIVW analysis β = −1.3 ([−2.533 to −0.067]; p = 0.039) was observed as well. All the causal estimates reported reflect a unit change in the outcome per a 1 SD increase in the exposure. HDL showed no evidence of a significant causal association with eGFR in the MVIVW method (β = −0.117 (95% CI [−1.252 to 0.018]; p = 0.840)). We found no evidence of a reverse causal impact of eGFR on serum lipids. All our sensitivity analyses indicated no strong evidence of pleiotropy or heterogeneity between our instrumental variables for both the forward and reverse MR analysis.
Interpretation
In this African ancestry population, genetically predicted higher LDL-C and TC are causally associated with higher eGFR levels, which may suggest that the relationship between LDL, TC and kidney function may be U-shaped. And as such, lowering LDL_C does not necessarily improve risk of kidney disease. This may also imply the reason why LDL_C is seen to be a poorer predictor of kidney function compared to HDL. In addition, this further supports that more work is warranted to confirm the potential association between lipid traits and risk of kidney disease in individuals of African Ancestry.
Funding
Wellcome (220740/Z/20/Z).
Keywords: Serum lipids, eGFR, Chronic kidney disease, Kidney function, Two-sample Mendelian Randomization
Research in context.
Evidence before this study
A plethora of observational and/or epidemiological studies have reported inconsistent associations between serum lipids and kidney function. Drawing causal inferences from such studies is limited due to confounding, possible reverse causation, and other biases. Mendelian randomization studies which can infer such causality while limiting non-heritable environmental confounders have majorly been conducted in European-ancestry populations. Most of these findings have limited generalizability in African ancestry individuals.
Added value of this study
We employed univariable and multivariable Mendelian randomization methods and revealed evidence of a causal association between serum lipids and estimated glomerular filtration rate based on serum creatinine among individuals from continental Africa. The unexpected positive causal association between bad cholesterol (LDL and TC) suggests a possible U-shaped association between serum lipids and kidney function which should be given further attention by future investigators.
Implications of all the available evidence
Our findings support a causal association between lipid traits and kidney function in continental Africans. However, due to the unexpected causal association observed in this population, our findings warrant future investigations with bigger sample size and individual-level data access to further confirm true causality between serum lipids and kidney function. We also caution generalizing findings from African ancestry individuals in global cohorts to those in continental Africa as there might be notable genetic and environmental differences between these two groups.
Introduction
Chronic kidney disease (CKD) is defined as a reduction in kidney function indicated by estimated glomerular filtration rate (eGFR) <60 ml/min per 1.73 m2 or kidney damage markers or both that persist for at least three months.1 It has a significant impact worldwide, with an estimated prevalence of 10–15% globally as a direct cause of mortality, morbidity, and comorbidity among other complex traits.2 The prevalence of CKD in Africa is equally high with most sub-Saharan African countries showing generally a >10% prevalence. Managing CKD in its advanced stages requires huge amounts of resources, and this is quite cumbersome on most sub-Saharan Africa (SSA) economies.
Serum lipids: high-density lipoprotein (HDL) and low-density lipoprotein (LDL) cholesterol, rank among the highest commonly measured biomarkers in clinical medicine.3 Most epidemiological studies have reported an association between these lipids and kidney function, indicating that low HDL cholesterol is associated with poor kidney function and CKD progression.4, 5, 6 In a well-powered study of 2 million United States veterans who were followed up for a median of 9 years, Bowe et al.,7 reported on the association between HDL cholesterol concentrations and various CKD end points. The authors reported individuals with low HDL cholesterol concentrations (<30 mg/dL) have the highest risk for CKD or CKD progression.5 Other studies have found that higher levels of blood total cholesterol (TC), LDL, TC: HDL ratio, TG: HDL ratio, and lower levels of blood HDL cholesterol, are associated with a higher risk of incident CKD.8 However, evidence from these epidemiological and observational studies is limited by its inability to demonstrate a causal relationship and inconsistencies between several studies.9, 10, 11, 12 Further still, most of such high-powered studies have not only been limited by sample selection bias towards majorly European ancestries, but also confounding from environmental factors.
By fulfilling key assumptions, Mendelian randomization studies can enable causal inference while limiting environmental confounding and reverse causation.3,13, 14, 15 However, similar to observational studies, the association of serum lipids and eGFR has been conflicted, even in the MR studies. Studies like that by Coassin et al., indicated that HDL cholesterol does not influence eGFR, and they further proposed pleiotropic effects on eGFR for some of the associated SNPs.8 Other findings elsewhere conflicted these findings and reported a genetically higher HDL concentration being associated with higher eGFR.16,17 Such studies, however, have been subject to sample selection bias due to the lack of ethnic diversity in the Genome-Wide Association Studies (GWASs) used which are primarily based on European ancestry individuals.18,19 A two-sample Mendelian randomization analysis of data from the most extensive lipid and CKD cohorts supported genetically higher HDL cholesterol concentration as causally associated with better kidney function.20 Further still, a recent study by Humaira et al., found no significant association between serum lipids and kidney function, and reported a weak effect of increased LDL-c levels and higher eGFRcrea.21 This analysis and several others were performed on European ancestry individuals, and the results cannot be confidently generalized to non-European ancestry individuals.19,22, 23, 24, 25
In this study, therefore, we set out to use bi-directional and multivariable MR methods to investigate the causal relationship between serum lipid profiles and kidney function using estimated glomerular filtration rate based on serum creatinine (eGFRcrea) as a marker. We leveraged datasets from individuals of African-ancestry selected from the Million veteran program (MVP) and Meta-analysed continental African Lipid GWASs (APCDR and AWI-Gen), which we called MALG (n = 24,215).
Methods
GWAS data sources
We selected eGFR instruments from GWAS summary statistics of all individuals of African ancestry within the U.S. Veteran's Administration million veteran program, MVP (N = 57,336).26 Genetic instruments for lipid traits were obtained from summary statistics of MALG (n = 24,215)—13,612 African-ancestry participants from the African Partnership for Chronic Disease Research (APCDR) & the Africa Wits-IN-DEPTH partnership for Genomics studies (AWI-Gen).27 More information about the African cohorts (AWI-Gen + APCDR) from which the lipids instrumental variables were obtained are detailed elsewhere.26, 27, 28, 29, 30
Ethics statement
Participant consent and ethical approval were obtained in the original studies.
Univariable Mendelian Randomization
After instrument harmonization and selection, the inverse-weighted variance (IVW) method was used to perform the bi-directional MR analysis. In the absence of directional pleiotropy and heterogeneity between exposure and outcome, the estimates from this method have been reported to be reasonably accurate.28 We checked for the possible presence of horizontal pleiotropy between instrumental variables by including the MR-Egger regression method and MR-PRESSO. Evidence of horizontal pleiotropy was based on the MR-Egger intercept value deviating significantly from zero with a p-value ≤ 0.05.28,31 The weighted median method was used as the method of choice in case of observed pleiotropy.32
Multivariable Mendelian Randomization
The Multivariable Mendelian Randomization method can be applied for multiple genetic instruments regardless of their association with the exposure.33 In this MVMR method, the instrumental variables may be associated with more than one risk factor but they must fulfill the equivalent instrumental-variable assumptions.34 Thus, we applied this method by considering all the instrumental variables for HDL, LDL, and TG to determine their independent effects on eGFR.
Sensitivity analyses
We performed analyses using only IVs that met the three MR assumptions: The relevance assumption, the independence assumption, and the exclusion restriction assumption. All the exposure instrumental variables used in subsequent sensitivity analyses were significantly associated with the exposure for both the UVMR and MVMR at p < 5E-8. This ensured validity of the first MR assumption. Furthermore, we ensured validity of the second MR assumption by selecting only IVs with L.D < 0.01 after clumping at a 500 kb window. The downstream sensitivity analyses were then performed on these instruments that fulfilled the first 2 MR assumptions. We performed a sensitivity analysis using the penalization method in which the contribution of some of the instrumental variables (e.g., heterogeneous or outlying IVs) to the analysis is down-weighted (or penalized).32 We also performed the systematic leave-one-out approach to determine potential pleiotropy per SNP so as to test adherence to the exclusion restriction assumption. The resultant effect was assessed using the robust penalized IVW estimate. The change in results before and after SNP removal was then assessed. We then checked for heterogeneity between instrumental variables determined by Q statistics at p-value ≤ 0.05. We assessed for our instrument-strength by calculating the F-statistic as detailed elsewhere.35 We further tested for adherence to the third assumption of MR; the exclusion restriction assumption, by using various methods like the PhenoScanner to eliminate any instruments that showed horizontal pleiotropy.36 We also assessed for horizontal pleiotropy using the MR-Egger regression based on its intercept terms and the Mendelian randomization pleiotropy RESidual sum and outlier (MR-PRESSO).37 We further conducted a power analysis to estimate the smallest effect we could assess per exposure.38
Statistical analysis
We performed the MR analyses using the two-sample random-effects inverse-variance weighted (IVW) method implemented in the Mendelian Randomization R package.39 This method determines the causal estimates for instruments that meet the instrumental variable assumptions reported elsewhere.14 To account for the documented horizontal pleiotropy between lipids, we conducted a multivariable MR (MVMR) including instrumental variables from HDL, LDL, and TG at p-value < 5 × 10−8. We further checked for reverse causality by conducting an MR analysis considering eGFRcrea from MVP as exposure and lipid traits from MALG as outcome. The genetic instruments included in this study for all analyses were selected as those significantly associated with the risk of lipid traits at p-value < 5 × 10−8 in the MALG dataset with clumping at 500 kb. Statistical significance for causal associations was considered at p-value < 0.05. In multiple testing, an adjusted p-value after Bonferroni correction, p-value < 0.05/3 = 0.016 was considered statistically significant. All analyses were performed using Mendelian Randomization packages in R.
Role of funding source
Funding sources had no role in the conduct or reporting of the research.
Results
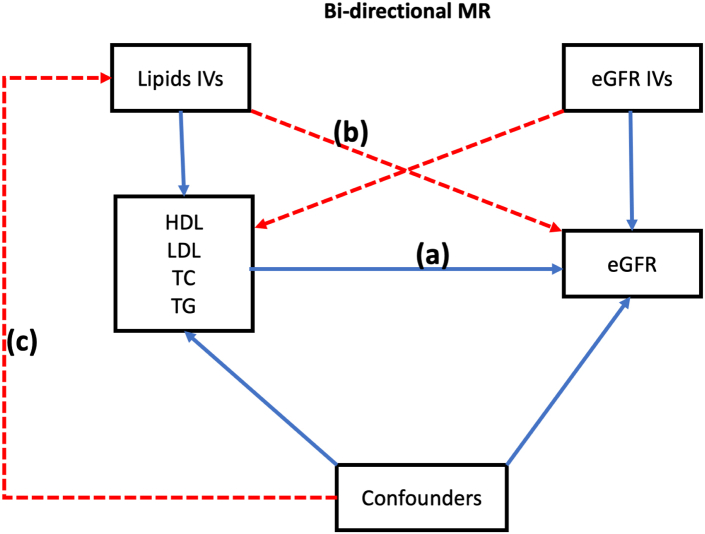
The bi-directional MR analysis was performed as shown in Fig. 1. Further details on the instrumental variables chosen can be found in Supplementary Data.
Fig. 1.
A schematic representation of bi-directional MR analyses: (a) Forward univariable MR; (b) IVs for lipid traits should not have an association with eGFR; (c) IVs for lipid traits are not related to measured or unmeasured confounding. HDL, high-density lipoprotein; LDL, low-density lipoprotein; TG, triglycerides; eGFR, estimated glomerular filtration rate; SNP, single-nucleotide polymorphism; MR, Mendelian Randomization; F/R, Forward/Reverse; IVs, Instrumental variables.
Association of estimated glomerular filtration rate with lipid levels
Univariable MR
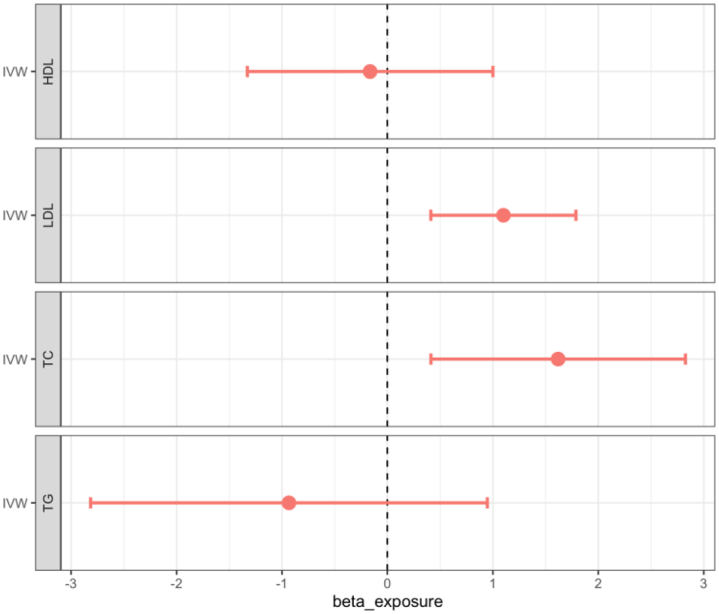
The associations between genetically predicted lipid traits and eGFR are shown in Table 1, Fig. 2, Supplementary Figure S2. We found no evidence of a statistically significant causal association between genetically predicted HDL-C and eGFR (β = −0.164, 95% CI = −1.329 to 1.00; p = 0.782). The effect estimates (β) [95% confidence intervals (CIs)] for the other lipid traits on eGFR were 1.1 ([0.411–1.788]; 0.002), 1.619 ([0.412–2.826]; 0.009) and −0.934 ([−2.815 to 0.947]; 0.33) for LDL, TC, and TG respectively. There was evidence of a significant causal association between genetically predicted LDL cholesterol and eGFR. Similarly, TC showed a significant causal effect on eGFR (Fig. 2). Genetically predicted Triglycerides (TG) were not significantly associated with eGFR as well. Details of the exposure instrumental variables are shown in Supplementary Table S1.
Table 1.
Univariable IVW Mendelian Randomization results.
| Exposure | Outcome | BETA | SE | 95% CI | p-value |
|---|---|---|---|---|---|
| HDL | eGFR | −0.164 | 0.594 | −1.329 to 1 | 0.782 |
| LDL | eGFR | 1.1 | 0.351 | 0.411 to 1.788 | 0.002a |
| TC | eGFR | 1.619 | 0.616 | 0.412 to 2.826 | 0.009a |
| TG | eGFR | −0.934 | 0.96 | −2.815 to 0.947 | 0.33 |
LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TC, Total Cholesterol; TG, Triglycerides; IVW, Inverse Variance Weighted; SE, standard error.
Statistically significant (p < 0.05).
Fig. 2.
Forest plot of the beta estimates and their 95% confidence intervals between genetically predicted lipid traits and eGFR using the IVW univaribale MR method. IVW, inverse-variance weighted; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; TC, total cholesterol.
The reverse MR analysis showed no significant causal association between eGFR and all four lipid traits, as shown in the Supplementary Figure S1 and Supplementary Table S1. For the reverse MR, the effect estimate ([95% CI]) for HDL, LDL, TC, and TG was 0.01 ([−0.011 to 0.012]; p = 0.873), 0.007 ([−0.005 to 0.018]; p = 0.265), 0.008 ([−0.005 to 0.021]; p = 0.225) and 0.00 ([−0.011 to 0.011]; p = 0.984) respectively. eGFR showed no evidence of a reverse causal effect on this population's genetically predicted lipid traits.
Multivariable MR
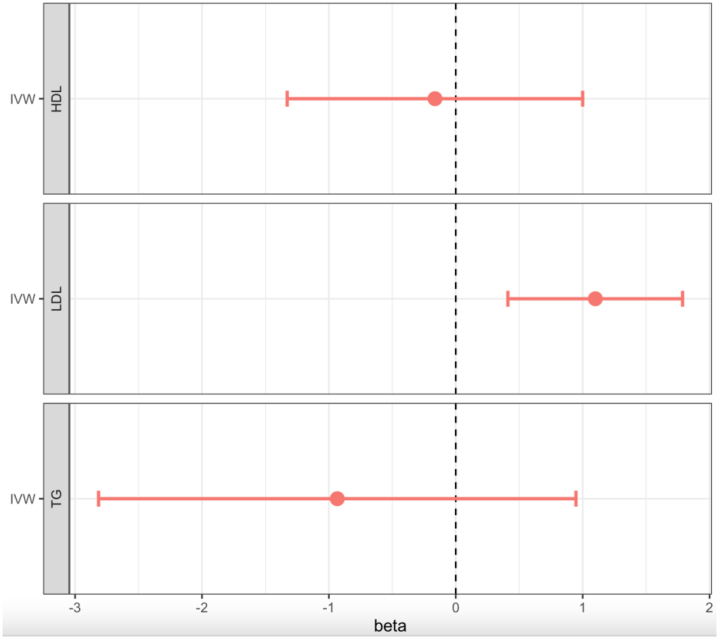
The MVMR analysis showed statistically significant causal associations for genetically predicted lipid traits; LDL and TG on eGFR (Fig. 3; Supplementary Table S2). LDL cholesterol had a significant positive causal effect on eGFR, consistent with that observed in the forward univariable analysis (β = 1.228 ([0.477–1.979]; p = 0.001)). There was evidence of a significant causal effect of genetically predicted TG on eGFR (β = −1.3 ([−2.533 to 0.067]; p = 0.039)). HDL was not significantly associated with eGFR, just like in prior analyses.
Fig. 3.
Forest plot showing the beta estimates and 95% confidence intervals of Multivariate MR of lipids vs eGFR traits. HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, Triglycerides.
Sensitivity analyses
We accounted for the pleiotropic effects between instrumental variables using MR-Egger, penalized robust MR-Egger, leave-one-out analysis, simple median, and weighted median analyses. We found no evidence of horizontal pleiotropy between IVs using the MR-Egger regression intercept analysis. All associations had p-values > 0.005 for the MR-Egger intercept, as shown in Fig. 2 and Supplementary Table S4. This was the same case for the reverse MR analysis (Supplementary Table S1). We further estimated any horizontal pleiotropy using the leave-one-out approach and found no evidence of any confounding due to pleiotropy between SNPs with all p-value > 0.05 (Table 2). The Phenoscanner analysis indicated no association between the SNPs and any other traits that could confound the exposure-outcome relationship, as the instrumental variables were only associated with lipid traits. The Phenoscanner analysis indicated no association between the SNPs and any other traits that could confound the exposure-outcome relationship. A Steiger filtering analysis to test for directionality confirmed that the lipid traits have a causal effect on eGFR. We tested for weak instruments by calculating the F-statistics. We found no difference in associations between lipid traits and eGFRcrea after dropping weak instruments as shown in Supplementary Table S2. MR-Egger analysis and weighted median analysis showed no evidence of horizontal pleiotropy for both the forward and reverse analyses. The power calculation for the forward MR analysis ranged from 55 to 100% while that of the reverse was 100% (Supplementary Table S5).
Table 2.
Leave-one-out sensitivity analyses for all SNPs in the multivariable MR.
| SNP | MR-Egger intercept | SE | 95% CI | p-value |
|---|---|---|---|---|
| rs1800588 | −0.058 | 0.084 | −0.222 to 0.107 | 0.493 |
| rs17111732 | −0.038 | 0.098 | −0.229 to 0.153 | 0.698 |
| rs116513376 | −0.061 | 0.085 | −0.227 to 0.106 | 0.476 |
| rs59523416 | −0.056 | 0.084 | −0.220 to 0.108 | 0.503 |
| rs12740374 | −0.028 | 0.073 | −0.171 to 0.115 | 0.703 |
| rs143375141 | −0.070 | 0.085 | −0.236 to 0.097 | 0.413 |
| rs35804417 | −0.057 | 0.084 | −0.221 to 0.107 | 0.497 |
| rs75143493 | −0.073 | 0.075 | −0.220 to 0.075 | 0.334 |
| rs73015020 | −0.095 | 0.079 | −0.250 to 0.060 | 0.229 |
| rs10416720 | −0.076 | 0.089 | −0.251 to 0.099 | 0.393 |
| rs7412 | −0.084 | 0.095 | −0.271 to 0.102 | 0.375 |
| rs3810308 | −0.107 | 0.083 | −0.270 to 0.056 | 0.199 |
| rs326 | −0.073 | 0.088 | −0.246 to 0.100 | 0.406 |
| rs2070895 | −0.054 | 0.088 | −0.227 to 0.119 | 0.538 |
| rs12721054 | −0.035 | 0.069 | −0.170 to 0.101 | 0.613 |
| rs114139997 | −0.054 | 0.077 | −0.204 to 0.096 | 0.477 |
Discussion
In this African-ancestry MR study, we investigated the causal effect of genetically predicted lipid traits on eGFRcrea using a two-sample and multivariable MR approach. In the primary MR-IVW forward analysis, LDL-C and TC showed evidence of a significant causal association with eGFR. Therefore, we report significant evidence that genetically predicted lipids; LDL and TC are causally associated with eGFRcrea in this African population. However, the reverse MR-IVW analysis indicated a non-significant causal association between eGFRcrea and either of the genetically predicted lipids.
Our findings in the main analysis on HDL and TC differ from those reported on MR analyses in European ancestry cohorts by Lanktree et al. and other groups.16,17,40 They reported a significant association between higher HDL levels with higher eGFR. Here, we report no evidence of association between genetically-proxied HDL cholesterol and better kidney function in this African cohort. However, our findings tally with those from other studies conducted on European ancestry individuals which reported a non-significant effect of HDL on eGFR levels, and hence kidney function.8,21 Notably, elevated HDL has been shown to lower the mortality rate of CKD within observed ranges.41
Our causal association between LDL and eGFR differs with findings from elsewhere.4,10 The Chronic Renal Insufficiency Cohort Study reported no association between LDL-C levels and the change rate of eGFR in low proteinuria individuals at baseline.42 However, similar to our findings, a study conducted in European ancestry individuals reported a weak effect of LDL-c and eGFR based on serum creatinine, and similarly observed no association between other serum lipids and kidney function except apolipoprotein B.21 We, therefore, suggest better powered future studies within the same African ancestry to clarify the true association between serum lipids and kidney function as measured by eGFR in this ancestry. Importantly, high LDL levels have been linked to poor kidney function by observational and Mendelian randomization studies, we therefore further suggest future studies to explore a U-shaped association in a stratified MR study to better resolve this association.
The reverse univariable analysis showed no evidence of a significant causal association between eGFR and lipid traits. Our findings from the reverse association between eGFR and serum lipids are consistent with findings elsewhere.21
In the main univariable analysis, we report that high LDL and TC levels had a strong significant causal effect on eGFR levels. In the multivariable MR analysis, low TG levels had a protective effect on eGFR. Unlike TC, genetically predicted low TG levels showed a consistent causal effect on eGFR between the MVMR and the main forward univariable analysis, showing significance in the latter. Findings from other studies have reported a conflicting association between TG and eGFR, but these have been based on European ancestry populations.16,21,43,44 Evidence from observational studies supports a greater triglyceride to HDL cholesterol ratio as associated with a decline in eGFR.20,45 These observational studies are, however, limited by confounding and inability to determine direction of effect.
The respective directions of effect from the MVMR analysis were quite similar to those observed in the forward univariabe MR analysis. In this MVMR analysis, both LDL and TG had protective causal effects on eGFR. The un-expected direction of effect of genetically predicted LDL and TG on eGFR reported in this study might be due to the low statistical power in this study. Noteworthy, a recent study reported an inconsistent evidence between higher atherogenic lipids including LDL-C, TG, and Apo B and weak increase in eGFR.21 A higher eGFR association with higher LDL-C and TG has been previously associated with glomerular hyperfiltration rates that occur in individuals with cardiometabolic conditions.46 We couldn't verify the role of underlying cardiometabolic conditions towards the observations in this study. We recommend a more powered study on African-ancestry individuals, accounting for such clinical parameters to further clarify our findings.
Our study strengths were in the use of continental African-derived GWAS summary statistics (MALG) and assessing a possibility for a reverse causation between eGFR and serum lipids. We also performed sensitivity analyses including multi-variable MR-Egger to determine reliability of our instrumental variables as detailed under the methods section.
Study limitations
The study was limited by a lack of access to individual-level data as we only had access to GWAS summary statistics. This meant that we couldn't verify a possible U-shaped relationship between serum lipid traits and eGFRcrea. We were also not able to correct for sample overlap. We also did not assess for ancestral differences in the instrumental variables with other ancestries, as suggested by Graham et al.47 Our study cohorts couldn't be expanded in this study to achieve a larger sample size to correct for some of the instruments that exhibited low power. Furthermore, this made it difficult to explore potential non-linearity of some of the associations.
Conclusions
This Mendelian Randomization study suggests a causal association between LDL cholesterol and higher eGFR, but not HDL cholesterol. We report that genetically elevated LDL cholesterol levels are associated with developing higher eGFR. Our findings suggest that the relationship between non-HDL cholesterol and kidney function may be U-shaped. This may be a reason why LDL is seen to be a poor predictor of renal function compared to HDL, and as such lowering LDL does not necessarily improve risk of kidney disease. Therefore, our findings highlight the need for bigger MR studies focused on African ancestry individuals to accurately determine the association between serum lipid traits and kidney function measured by eGFRcrea in continental Africans.
Contributors
SF conceptualised the study. CK performed the analyses. OS and SF verified the underlying data. CK, OS, TC and SF wrote the first draft of the manuscript.
SF, TC, DG, JF, DN and MN interpreted results. ABK, AK, RM, RK, BBS and DJ interpreted the data. All authors provided critical revisions and edits to the manuscript. All authors read and approved of the final manuscript. SF is the guarantor of this work.
All authors read and approved the final version of the manuscript.
Data sharing statement
All scripts for the analysis are available from the authors upon request.
Declaration of interests
DG is employed part-time by Novo Nordisk and has received consultancy fees from Policy Wisdom. No potential conflicts of interest relevant to this article were reported by all other authors.
Acknowledgements
This work was supported by the Wellcome Trust [grant number: 220740/Z/20/Z] awarded to Segun Fatumo. TC is an international training fellow supported by the Wellcome Trust grant (214205/Z/18/Z). DG was supported by the British Heart Foundation Centre of Research Excellence (RE/18/4/34215) at Imperial College, and a National Institute for Health Research Clinical Lectureship (CL-2020-16-001) at St. George's, University of London. This work was supported by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement, through core funding to the MRC/UVRI and LSHTM Uganda Research Unit. Funding sources had no role in the conduct or reporting of the research.
The authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/for more details). The citation for MVP is Gaziano, J.M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Veterans Administration (VA) Cooperative Studies Program (CSP) award #G002.
Dr. Segun Fatumo is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104537.
Appendix A. Supplementary data
References
- 1.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. 2013;3:1–150. doi: 10.1038/ki.2013.243. [DOI] [PubMed] [Google Scholar]
- 2.Webster A.C., Nagler E.V., Morton R.L., Masson P. Chronic kidney disease. Lancet. 2017;389(10075):1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 3.Voight B.F., Peloso G.M., Orho-Melander M., et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaeffner E.S., Kurth T., Curhan G.C., et al. Cholesterol and the risk of renal dysfunction in apparently healthy men. J Am Soc Nephrol. 2003;14:2084–2091. doi: 10.1681/ASN.V1482084. [DOI] [PubMed] [Google Scholar]
- 5.Cases A., Coll E. Dyslipidemia and the progression of renal disease in chronic renal failure patients. Kidney Int Suppl. 2005;99:S87–S93. doi: 10.1111/j.1523-1755.2005.09916.x. [DOI] [PubMed] [Google Scholar]
- 6.Morton J., Zoungas S., Li Q., et al. Low HDL cholesterol and the risk of diabetic nephropathy and retinopathy: results of the ADVANCE study. Diabetes Care. 2012;35(11):2201–2206. doi: 10.2337/dc12-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowe B., Xie Y., Xian H., Balasubramanian S., Al-Aly Z. Low levels of high-density lipoprotein cholesterol increase the risk of incident kidney disease and its progression. Kidney Int. 2016;89(4):886–896. doi: 10.1016/j.kint.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Coassin S., Friedel S., Kottgen A., Lamina C., Kronenberg F. Is high-density lipoprotein cholesterol causally related to kidney function? Evidence from genetic epidemiological studies. Arterioscler Thromb Vasc Biol. 2016;36(11):2252–2258. doi: 10.1161/ATVBAHA.116.308393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae J.C., Han J.M., Kwon S., et al. LDL-C/apoB and HDL-C/apoA-1 ratios predict incident chronic kidney disease in a large apparently healthy cohort. Atherosclerosis. 2016;251:170–176. doi: 10.1016/j.atherosclerosis.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 10.Kuma A., Uchino B., Ochiai Y., et al. Impact of low-density lipoprotein cholesterol on decline in estimated glomerular filtration rate in apparently healthy young to middle-aged working men. Clin Exp Nephrol. 2018;22(1):15–27. doi: 10.1007/s10157-017-1407-8. [DOI] [PubMed] [Google Scholar]
- 11.Obermayr R.P., Temml C., Knechtelsdorfer M., et al. Predictors of new-onset decline in kidney function in a general middle-European population. Nephrol Dial Transplant. 2008;23(4):1265–1273. doi: 10.1093/ndt/gfm790. [DOI] [PubMed] [Google Scholar]
- 12.Zuo P.Y., Chen X.L., Liu Y.W., Zhang R., He X.X., Liu C.Y. Non-HDL-cholesterol to HDL-cholesterol ratio as an independent risk factor for the development of chronic kidney disease. Nutr Metab Cardiovasc Dis. 2015;25(6):582–587. doi: 10.1016/j.numecd.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Burgess S., Scott R.A., Timpson N.J., Davey Smith G., Thompson S.G., Consortium E.-I. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30(7):543–552. doi: 10.1007/s10654-015-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawlor D.A., Harbord R.M., Sterne J.A., Timpson N., Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 16.Lanktree M.B., Theriault S., Walsh M., Pare G. HDL cholesterol, LDL cholesterol, and triglycerides as risk factors for CKD: a mendelian randomization study. Am J Kidney Dis. 2018;71(2):166–172. doi: 10.1053/j.ajkd.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Miao L., Min Y., Qi B., et al. Causal effect between total cholesterol and HDL cholesterol as risk factors for chronic kidney disease: a mendelian randomization study. BMC Nephrol. 2021;22(1):35. doi: 10.1186/s12882-020-02228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J., Zhang Y., Rasheed H., et al. Trans-ethnic Mendelian-randomization study reveals causal relationships between cardiometabolic factors and chronic kidney disease. Int J Epidemiol. 2021;50(6):1995–2010. doi: 10.1093/ije/dyab203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soremekun O., Karhunen V., He Y., et al. Lipid traits and type 2 diabetes risk in African ancestry individuals: a Mendelian Randomization study. eBioMedicine. 2022;78 doi: 10.1016/j.ebiom.2022.103953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuruya K., Yoshida H., Nagata M., et al. Impact of the triglycerides to high-density lipoprotein cholesterol ratio on the incidence and progression of CKD: a longitudinal study in a large Japanese population. Am J Kidney Dis. 2015;66(6):972–983. doi: 10.1053/j.ajkd.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Rasheed H., Zheng J., Rees J., et al. The causal effects of serum lipids and apolipoproteins on kidney function: multivariable and bidirectional Mendelian-randomization analyses. Int J Epidemiol. 2021;50(5):1569–1579. doi: 10.1093/ije/dyab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiza A.B., Toure S.M., Vujkovic M., et al. Transferability of genetic risk scores in African populations. Nat Med. 2022;28(6):1163–1166. doi: 10.1038/s41591-022-01835-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fatumo S., Karhunen V., Chikowore T., et al. Metabolic traits and stroke risk in individuals of African ancestry: mendelian randomization analysis. Stroke. 2021;52(8):2680–2684. doi: 10.1161/STROKEAHA.121.034747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chikowore T., Ekoru K., Vujkovi M., et al. Polygenic prediction of type 2 diabetes in Africa. Diabetes Care. 2022;45(3):717–723. doi: 10.2337/dc21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fatumo S., Chikowore T., Choudhury A., Ayub M., Martin A.R., Kuchenbaecker K. A roadmap to increase diversity in genomic studies. Nat Med. 2022;28(2):243–250. doi: 10.1038/s41591-021-01672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaziano J.M., Concato J., Brophy M., et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Gurdasani D., Carstensen T., Fatumo S., et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179(4):984–1002.e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhury A., Brandenburg J.T., Chikowore T., et al. Meta-analysis of sub-Saharan African studies provides insights into genetic architecture of lipid traits. Nat Commun. 2022;13(1):2578. doi: 10.1038/s41467-022-30098-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fatumo S., Mugisha J., Soremekun O.S., et al. Uganda genome resource: a rich research database for genomic studies of communicable and non-communicable diseases in Africa. Cell Genom. 2022;2(11) doi: 10.1016/j.xgen.2022.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatumo S., Chikowore T., Kalyesubula R., et al. Discovery and fine-mapping of kidney function loci in first genome-wide association study in Africans. Hum Mol Genet. 2021;30(16):1559–1568. doi: 10.1093/hmg/ddab088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden J., Del Greco M.F., Minelli C., Davey Smith G., Sheehan N., Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36(11):1783–1802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess S., Bowden J., Fall T., Ingelsson E., Thompson S.G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28:30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson E., Davey Smith G., Windmeijer F., Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48(3):713–727. doi: 10.1093/ije/dyy262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess S., Davies N.M., Thompson S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40(7):597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce B.L., Ahsan H., Vanderweele T.J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley J.R., Blackshaw J., Kamat M.A., et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brion M.J., Shakhbazov K., Visscher P.M. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalil R.S., Wang J.H., de-Boer I.H., et al. Effect of extended release niacin on cardiovascular events and kidney function in chronic kidney disease: a post-hoc analysis of the AIM-HIGH trial. Kidney Int. 2015;87(6):1250–1257. doi: 10.1038/ki.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaziri N.D. HDL abnormalities in nephrotic syndrome and chronic kidney disease. Nat Rev Nephrol. 2016;12(1):37–47. doi: 10.1038/nrneph.2015.180. [DOI] [PubMed] [Google Scholar]
- 42.Rahman M., Yang W., Akkina S., et al. Relation of serum lipids and lipoproteins with progression of CKD: the CRIC study. Clin J Am Soc Nephrol. 2014;9(7):1190–1198. doi: 10.2215/CJN.09320913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh A.K., Kari J.A. Metabolic syndrome and chronic kidney disease. Curr Opin Nephrol Hypertens. 2013;22(2):198–203. doi: 10.1097/MNH.0b013e32835dda78. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y.B., Sheng L.T., Wei W., et al. Association of blood lipid profile with incident chronic kidney disease: a Mendelian randomization study. Atherosclerosis. 2020;300:19–25. doi: 10.1016/j.atherosclerosis.2020.03.020. [DOI] [PubMed] [Google Scholar]
- 45.Thompson M., Ray U., Yu R., et al. Kidney function as a determinant of HDL and triglyceride concentrations in the Australian population. J Clin Med. 2016;5(3):35. doi: 10.3390/jcm5030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palatini P. Glomerular hyperfiltration: a marker of early renal damage in pre-diabetes and pre-hypertension. Nephrol Dial Transplant. 2012;27(5):1708–1714. doi: 10.1093/ndt/gfs037. [DOI] [PubMed] [Google Scholar]
- 47.Graham S.E., Clarke S.L., Wu K.H., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600(7890):675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.