Abstract
Background:
Diabetic wound healing remains a major challenge due to the impaired functionality of angiogenesis by persistent hyperglycemia. Mesenchymal stem cell exosomes are appropriate candidates for regulating the formation of angiogenesis in tissue repair and regeneration. Here, we explored the effects of exosomes derived from human amniotic mesenchymal stem cell (hAMSC-Exos) on the biological activities of human umbilical vein endothelial cells (HUVECs) treated with high glucose and on diabetic wound healing and investigate lncRNAs related to angiogenesis in hAMSC-Exos.
Methods:
hAMSCs and hAMSC-Exos were isolated and identified by flow cytometry or western blot. A series of functional assays such as cell counting kit-8, scratching, transwell and tube formation assays were performed to evaluate the potential effect of hAMSC-Exos on high glucose-treated HUVECs. The effect of hAMSC-Exos on diabetic wound healing were tested by measuring wound closure rates and immunohistochemical staining of CD31. Subsequently, the lncRNAs profiles in hAMSC-Exos and hAMSCs were examined to screen the lncRNAs related to angiogenesis.
Results:
The isolated hAMSC-Exos had a size range of 30–150 nm and were positive for CD9, CD63 and CD81. The hAMSC-Exos facilitate the functional properties of high glucose-treated HUVECs including the proliferation, migration and the angiogenic activities as well as wound closure and angiogenesis in diabetic wound. hAMSC-Exos were enriched lncRNAs that related to angiogenesis, including PANTR1, H19, OIP5-AS1 and NR2F1-AS1.
Conclusion:
Our findings demonstrated hAMSC-Exos facilitate diabetic wound healing by angiogenesis and contain several exosomal lncRNAs related to angiogenesis, which may represent a promising strategy for diabetic wound healing.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13770-022-00513-w.
Keywords: Mesenchymal stem cells, Amniotic membrane, Exosomes, LncRNAs, Angiogenesis
Introduction
Diabetes mellitus is a multifaceted metabolic disease and has become a rapidly increasing health problem in the worldwide [1]. Approximately 20% of diabetic patients worldwide have developed diabetic wounds. The angiogenesis of diabetic wounds is impaired by persistent hyperglycemia, thus delaying the healing process [2]. Stimulating angiogenesis is an urgent problem for treating diabetic wounds in clinical practice.
Numerous studies have indicated that human amniotic mesenchymal stem cells (hAMSCs) can be isolated from human amniotic tissue of the placenta. They are not only free from ethical or legal concerns and are easily obtained from discarded placentas [3] but also have the merits of self-renewal potential and low immunogenicity and could differentiate into multiple cell types [4, 5], these features make hAMSCs an attractive cell source for wound repair and regeneration. Recently, many studies have demonstrated that the roles of MSC-Exos in promoting wound repair and regeneration mainly lie on paracrine mechanisms, in which exosomes play a major role [6, 7]. It has been demonstrated that exosomes can accelerate the healing of diabetic wounds by angiogenesis [8–10].
Exosomes, as nanoparticles, are secreted by different cell types and have a diameter range of 30–150 nm. They can positively express the protein markers, CD9, CD63 and CD81, as determined by flow cytometry or western blotting [11]. Exosomes are methods of paracrine communication that exist in extracellular environments, such as the conditioned media of cells and various body fluids, from which they were successfully isolated. Further analysis of exosomes revealed that they can carry nucleic acids (e.g., mRNA, miRNA, and lncRNA) and proteins, which can be transferred to target cells to regulate biological processes, which is called intercellular communication [12].
Current research indicates that long noncoding RNAs (lncRNAs, with lengths of more than 200 nucleotides) may play regulatory roles in the healing process of diabetic wounds. For instance, exosomal KLF3-AS1 and HOTAIR derived from MSCs can enhance angiogenesis to promote the healing process of diabetic wounds [13, 14]. To date, there have been no reports regarding the function of hAMSC-Exos in the healing of diabetic wounds and enriched lncRNAs in hAMSC-Exos.
In this study, we sought to investigate whether hAMSC-Exos could facilitate the proliferation, migration and the angiogenic activities of HUVECs and facilitate the diabetic wound healing by angiogenesis. Subsequently, lncRNAs enriched in hAMSC-Exos related to angiogenesis were screened to analyze the underlying mechanisms of angiogenesis facilitated by hAMSC-exos in the healing of diabetic wounds, which may provide novel ideas for further therapy.
Materials and methods
Tissues and ethical approval
Discarded human amniotic tissues were obtained from several puerperae from April 2021 to December 2021 in the Department of Obstetrics and Gynecology, the First Affiliated Hospital of Nanchang University (Jiangxi, China). All patients with hepatitis B, hepatitis C, syphilis and acquired immune deficiency syndrome (AIDS) tested negative from the puerpera, who also had no other infectious diseases. All experiments described in this study were conducted under protocols that were reviewed and approved by the Medical Research Ethics Committees of the First Affiliated Hospital of Nanchang University (no. 20218–008). Written informed consent was signed by the pregnant women or their legal guardians.
Isolation of hAMSCs and hAMSC-Exos
hAMSCs were isolated and cultured as previously described [15]. Human amniotic tissues were separated from maternal placentas under aseptic conditions and rinsed with sterilized PBS (Solarbio, Beijing, China). Then, the amniotic tissues were transferred to 2-mL cryogenic vials and cut into small fragments with ophthalmic scissors. Then digested twice with 0.05% trypsin–EDTA (Gibco, Ottawa, Canada). Subsequently, the cells were digested with 0.75 mg/mL collagenase II (Worthington, OH, USA) containing DNase I (Solarbio) at 37 °C for approximately 1 h. The cells were then digested with Dulbecco's Modified Eagle’s medium (DMEM) (Solarbio) supplemented with 10% fetal bovine serum (FBS) (Solarbio Biotechnology, Berlin, Germany). The filtrate containing cells was collected by a 300-mesh filter, centrifuged and resuspended with DMEM supplemented with 10% FBS and 10 ng/mL basic fibroblast growth factor (bFGF) (Peprotech, Cranbury, NJ, USA) (complete medium) and incubated in an incubator. The culture medium was first replaced after 12 h. Third-generation hAMSCs were collected for further use.
Isolation of hAMSC-Exos was carried out as previously described [16]. The hAMSCs were seeded into 100-mm culture dishes (Corning Inc., Corning, NY, USA) with complete medium for 72 h. Then, washed three times with PBS and incubated with DMEM free of FBS for 48 h. After that, the culture medium was collected, and the hAMSC-Exos were collected by ultracentrifugation. Briefly, the culture medium of hAMSCs was collected and centrifuged at 500 × g for 10 min at 4 °C, 3000 × g for 20 min at 4 °C, and 10,000 × g for 30 min at 4 °C to eliminate cell debris; a 0.22-μm filter (Merck Millipore, Burlington, MA, USA) was used to filter the supernatant. Subsequently, the supernatant was ultracentrifuged at 100,000 × g with a 70Ti rotor (Beckman Coulter, Brea, CA, USA) for 70 min at 4 °C, and the pellet obtained was washed with PBS and ultracentrifuged again. The pellet in which the hAMSC-Exos were enriched was obtained and resuspended in 100μL of PBS, and a BCA protein assay kit (Solarbio) was used to measure the concentrations of hAMSC-Exos proteins. Samples were stored at −80 °C prior to use.
Differentiation potential of hAMSCs
To demonstrate that hAMSCs could differentiate into osteoblasts and adipocytes, hAMSCs at passage 4 were seeded in a 6-well plate according to the manufacturer’s instruction. After the hAMSCs reached 70% confluence, OriCell human MSC osteogenic differentiation medium (Cyagen Biosciences, Jiangsu, China) was added to the 6-well plate and replaced every 4 days. The differentiation potential of osteogenic formation of calcium nodules was assessed by Alizarin Red S (pH 4.2) staining after 26 days of induction. The hAMSCs of passage 4 were seeded in a 6-well plate as before. When the hAMSCs reached 100% confluence, OriCell human MSC adipogenic differentiation medium A was added to the 6-well plate for 3 days, and medium B was then changed to culture for 1 day. The differentiation potential of adipogenesis formation of lipid droplets was assessed the by Oil red O staining after 21 days of induction (All obtained from Cyagen Biosciences).
Identification of biomarkers of hAMSCs by flow cytometry
The standard flow cytometry method was used to analyze the hAMSC phenotypic. The primary hAMSCs were cultured until passage 5 and when the hAMSCs reached 80% confluence, they were resuspended with PBS at a concentration of 1 × 106 cells/mL and stained with fluorescein isothiocyanate (FITC)-conjugated antibodies mouse anti-human CD44, CD45, CD90, CD105, phycoerythrin (PE)-conjugated antibodies mouse anti-human CD34, CD73, HLA-DR (all obtained from BD Biosciences, San Jose, CA, USA) and their isotype controls, normal mouse IgG1-FITC, IgG2b-FITC, IgG1-PE, IgG2a-PE (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4 °C for 30 min. Then, the cells were washed twice with 2 mL of PBS. The cells were resuspended in 300 μL of PBS and analyzed by a FACSCanto II (BD Biosciences). FLOWJO V10 software was used to analyze the data (TreeStar Inc., Ashland, OR, USA).
Morphological and size analyses of hAMSC-Exos
For transmission electron microscope (TEM) detections, 10 μL of hAMSC-Exos were fixed using glutaraldehyde and dropped onto the copper mesh of an electron microscope (Hitachi, HT-7700, Tokyo, Japan). The cells were then subjected to negative staining with 10 μL of uranyl acetate two times. An HT-7700 electron microscope was used to analyze the hAMSC-Exos at 100 kV.
For the size distribution analysis of the hAMSC-Exos, 10 μL of hAMSC-Exos was diluted to 30 μL. The standard product was used to test the high sensitivity flow cytometry for nanoparticle tracking analysis (NTA) (NanoFCM Co. Ltd., Xiamen, China). After
passing that, hAMSC-Exos were loaded, and the particle sizes and concentrations of hAMSC-Exos were then detected by high-sensitivity flow cytometry for NTA.
Western blotting analysis of hAMSC-Exos marker proteins
hAMSC-Exos were lysed with protein extraction reagent (Applygen Technologies Inc., Beijing, China) containing a complete protease inhibitor tablet, according to the manufacturer’s instructions. The protein lysates were subjected to SDS-PAGE and separated by electrophoresis. Then, they were transferred to a polyvinylidene difluoride membrane (PVDF, Merck Millipore). The membranes were blocked with 5% nonfat milk and then incubated with rabbit monoclonal anti-human CD9, CD63, and CD81 antibodies (1:1000 dilution; Abcam, Cambridge, MA, USA) at 4 °C overnight. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h. Finally, the proteins were visualized by an enhanced chemiluminescence system (Clinx Science Instruments, Shanghai, China).
Labeling and uptake of hAMSC-Exos
The hAMSC-Exos were labeled with PKH26 (Sigma-Aldrich, St. Louis, MO, USA) fluorescent solution at room temperature for 20 min. Then, they were centrifuged at 100, 000 × g for 70 min to remove the supererogatory fluorescent solution at 4 °C. HUVECs were incubated with PKH26-labeled hAMSC-Exos for 8 h in the dark at 37 °C. Subsequently, the cells were fixed with 4% paraformaldehyde, and the cell nuclei were stained with 0.5 μg/mL DAPI (Invitrogen, Carlsbad, CA, USA). Finally, the cells were observed with an inverted fluorescence microscope (Leica, Wetzlar, Germany).
Cell proliferation assay
HUVECs were purchased from the American Type Culture Collection and cultured in endothelial cell medium (ECM) with 33 mmol/L glucose to mimic a hyperglycemic environment, which was supplemented with 5% FBS and 1% penicillin/streptomycin. Mannitol (5.5 mM glucose + 27.5 mM D-mannitol) was used as an osmotic control. HUVECs were cultured in a hyperglycemic environment for 7 days, and the media were replaced every 48 h. A total of 1 × 104 cells were seeded into 96-well plates, and cultured in exosome-free low-serum (2% FBS, VivaCell Biotechnology, Denzlingen, Germany) medium containing PBS or 100 μg/mL hAMSC-Exos for 0, 12, 24, 36, 48 and 60 h. Cell counting kit-8 reagent (CCK-8; 10 μL per well; Dojindo, Kumamoto, Japan) was used to assay the proliferation. After incubation at 37 °C for 2 h, the absorbances were evaluated at 450 nm using a Varioskan Flash (Thermo Fisher Scientific, Waltham, MA, USA).
Scratch assay
After culturing with 33 mmol/L glucose for 7 days, the HUVECs were seeded into 6-well plates. The cells were scratched with a pipette tip (200 μL) and washed twice with PBS when the cells reached 100% confluence. Then, the cells were cultured in exosome-free low-serum (2% FBS) ECM containing PBS or 100 μg/mL hAMSC-Exos. Images were obtained at 0 h, 12 h, and 24 h. ImageJ (National Institutes of Health, Bethesda, MD, USA) software was used to measure the wound areas. The following formula: migration area (%) = (A0 − At)/A0 × 100% was used determine the migration rates, where A0 represents the area of the initial wound scratch and At represents the area of the remaining wound at the measurement point.
Transwell assay
HUVECs (4 × 104 cells) were resuspended in serum-free ECM and seeded into the upper chamber with 8-μm pore-size polycarbonate membranes of a 24-well transwell chamber (Corning). The lower chamber was filled with exosome-free low-serum (2% FBS) ECM containing PBS or 100 μg/mL hAMSC-Exos. After incubation at 37 °C with 5% CO2 for 24 h, the cells migrated to the lower surface of the membrane. The migrated cells on the lower surface were fixed with 4% formaldehyde for 15 min. Then the cells were stained with 0.1% crystal violet (Solarbio) for 30 min. Images of the migrated cells were captured with a microscope, and the numbers of migrated cells were counted using ImageJ software. The numbers of cells that had migrated through the membranes were counted in at least three randomly selected microscopic fields.
Tube-formation assay
A 96-well plate was covered with 100 μL of Matrigel (Corning) and incubated for 30 min. The HUVECs (2 × 104 cells/well) were cultured in exosome-free low-serum (2% FBS) ECM containing PBS or 100 μg/mL hAMSC-Exos. After incubation at 37 °C for 8 h, the capillary-like structures were observed with a microscope, and the total lengths of the network structures were measured using ImageJ software.
The effects of hAMSC-Exos on diabetic wounds
Twenty male Sprague Dawley (SD) rats (7–8 weeks) were purchased from SJA Laboratory Animal Co, Ltd. (Hunan, China). All procedures were approved by the Experimental Animal Welfare Ethics Committee of the First Affiliated Hospital of Nanchang University (Jiangxi, China) (CDYFY-IACUC-202208QR040). Streptozocin (50 mg/kg) was intraperitoneally injected after 16 SD rats were fed a high-fat diet for one month. The criterion for inclusion was exceeding 16.7 mmol/L glucose. Then, the rats were anesthetized by intraperitoneal injections of 1% 35 mg/kg pentobarbital sodium (Sigma–Aldrich), and a full-thickness cutaneous wounds (2.5 cm in diameter) were created on the dorsum of each rat. All the rats were divided into three groups and subcutaneously injected with hAMSC-Exos or PBS: (1) PBS + NC group (normal rats treated with 100 μL PBS), (2) PBS + DM group (diabetic rats treated with 100 μL of PBS), and (3) hAMSC-Exos + DM group (diabetic rats treated with 100 μL of 100 μg/mL hAMSC-Exos). The wounds were photographed at Days 0, 7, 10 and 14 postoperation. The wound closure rates were calculated as scratch assay. The wound areas were evaluated with ImageJ software.
Immunohistochemical staining
Skin specimens around wounds were harvested after the rats were sacrificed on postoperative Day 14. Then the sections were cut into 5-mm paraffin sections after fixation with 4% paraformaldehyde. Three sections from each group were subjected to immunohistochemistry after the slides were prepared. Primary antibodies against CD31 (Abcam) were used for the immunohistochemistry determinations.The cells were incubated with the primary antibody against CD31 overnight at 4 °C. Images were captured by microscopy (Leica) and the capillary densities were evaluated using ImageJ software.
LncRNA/mRNA microarray analysis of hAMSC-Exos and hAMSCs
Total RNA samples from hAMSC-Exos and hAMSCs were prepared using TRIzol. An Arraystar Human LncRNA Microarray V5.0 was used to detect the profiles of exosomal lncRNAs in these samples. In this study, we selected the differentially expressed lncRNAs and mRNAs between hAMSCs and hAMSC-Exos with fold changes ≥ 5. Then, GO and KEGG pathway function analyses of the differentially expressed mRNAs were performed using in-house scripts written by Kang Cheng Biology (Shanghai, China).
Gene ontology (GO) and KEGG pathway analyses
GO covers three domains, namely biological processes, molecular functions and cellular component, and their analysis was used to analyze the main functions of the differentially expressed genes and gene products (http://www.geneontology.org). Fisher’s exact tests in the Bioconductor topGO package were used to classify the GO categories. The significance of the functions was measured by using the GO term enrichments of the differentially expressed genes. p < 0.05 is recommended as the threshold of significance. Pathway analysis is a functional analysis that maps genes to KEGG pathways (http://www.genome.jp/kegg). Fisher’s exact tests were used to identify the significantly enriched pathways. p < 0.05 was considered statistically significant.
The reliability of the lncRNA microarray results was verified by real-time quantitative polymerase chain reaction (RT–qPCR)
RT–qPCR was used to verify the reliability of the microarray results. Four differentially expressed lncRNAs and four mRNAs were randomly selected for analysis. The primers for the lncRNAs and mRNAs are shown in Table 1. Gradient dilution DNA templates were used to draw standard curves. The PCR conditions were as follows: 95 °C, 10 min; 40 PCR cycles at 95 °C, 10 s; and 60 °C, 60 s. After that, the fluorescence signals were collected. U6 was used as an internal reference. The 2−∆∆Ct method was used to calculate the expression levels of lncRNAs and mRNAs 17.
Table 1.
Sequences of the primers used in RT-qPCR
| LncRNA and mRNA | Primer sequence (5′–3′) | Size(bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| HOTAIR | CTCCGTGGGGTCCTGCT | AGGCTTCTAAATCCGTTCCAT | 53 |
| H19 | GGTCTGTTTCTTTACTTCCTCCA | CATATTCATTTCCAAGCTTTCAT | 94 |
| LINC01857 | AACCTGCTGGTGCCTTCATCT | AGAGTGCGGGCTCCATAGTTC | 660 |
| MALAT1 | GCTCGCCTGAAGGTGGTAA | CTTCCGCCGCCTCTTAAA | 75 |
| STAT4 | CTTTGGATTGATGGGTATGTC | CTTTGTAGTCTCGCAGGATGT | 238 |
| MYC | CCTACCCTCTCAACGACAGC | CTCTGACCTTTTGCCAGGAG | 248 |
| VEGFD | GCTGCCTGATGTCAACTGCT | GTGGACTGAGATGATCGCTTC | 157 |
| MAPK6 | CATCTTTGCTGAAATGCTGACT | AAACTGGAATTACGCTGAGAAG | 137 |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT | 89 |
Construction of the ceRNA network of differentially expressed lncRNAs
To understand the underlying functions of the differentially expressed lncRNAs, a ceRNA network was constructed to identify the potential miRNA response elements (MREs). In the ceRNA hypothesis, lncRNAs acting as ceRNAs can modulate miRNA target gene expressions by harboring miRNAs. This means that both lncRNAs and mRNAs have the same miRNA-binding sites that are the foundations of those interactions. The lncRNA–miRNA–mRNA interactions were predicted using miRDB (http://www.mirdb.org/) and TargetScan (www.targetscan.org/).
Statistical analysis
All the data are presented as the mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism 8.0 software. T-tests were used to evaluate the significant differences between two groups. One-way analysis of variance (ANOVA) and two-way ANOVA were used to analyze the differences among three groups and differences between two or three groups at different time points. p < 0.05 was considered statistically significant.
Results
Isolation and identification of hAMSCs and hAMSC-Exos
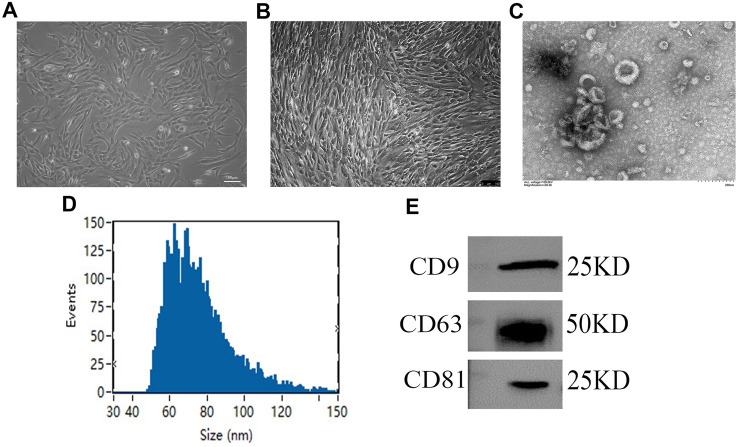
The hAMSCs showed spindle-shaped and fibroblast-like morphologies (Fig. 1A). When the hAMSCs were passaged until passage 3, we observed all of the hAMSCs (Fig. 1B). Flow cytometry showed that the hAMSCs strongly expressed the MSC marker proteins, CD44, CD73, CD90, and CD105, but did not express the hematopoietic stem cell marker proteins, CD34, and CD45, or the histocompatibility protein, HLA-DR, (Supplemental Fig. 1A). After culturing with osteogenic and adipogenic differentiation media, hAMSCs could be induced to differentiate into osteoblasts and adipocytes, respectively, in vitro (Supplemental Fig. 1B and C). The above results demonstrate that hAMSCs express specific marker proteins of MSCs and possess low immunogenicity as well as multidifferentiation potential, which may mean that hAMSCs have potential as a therapy for use in diabetic wounds.
Fig. 1.
Characterization of hAMSC-Exos. A Primary hAMSCs with spindle shapes on culture dishes. B The hAMSCs at passage 3. C The morphology of hAMSC-Exos was captured by TEM. D The diameters and particle concentrations of hAMSC-Exos were detected via NTA. E The specific surface marker proteins (e.g., CD9, CD63, CD81) of hAMSC-Exos were assessed by western blotting
To evaluate the characteristics of nanoparticles derived from the supernatant of the hAMSC culture medium, transmission electron microscopy (TEM) and high sensitivity flow cytometry for nanoparticle analysis were employed for these determinations. TEM showed that hAMSC-Exos exhibited circular, bilayer membrane structures with different sizes (Fig. 1C). The Flow NanoAnalyzer revealed that the hAMSC-Exos had a mean diameter of 75.72 nm, and their diameters were within the range of 30–150 nm (Fig. 1D). Western blot analysis demonstrated that the hAMSC-Exos expressed the exosomal surface markers, CD9, CD63 and CD81 (Fig. 1E). All data indicated that the hAMSC-Exos were successfully harvested from the supernatant of hAMSC culture medium.
The hAMSC-Exos facilitated the proliferation of HUVECs
As shown in Fig. 2A, PKH26-labeled hAMSC-Exos exhibiting red fluorescence were transferred to the cytoplasm of HUVECs after co-incubation for 12 h. The CCK-8 assay demonstrated that the hAMSC-Exos could accelerate the proliferation of HUVECs from 12 h (Fig. 2B). These data revealed that hAMSC-Exos were successfully absorbed by HUVECs and could accelerate their proliferation.
Fig. 2.
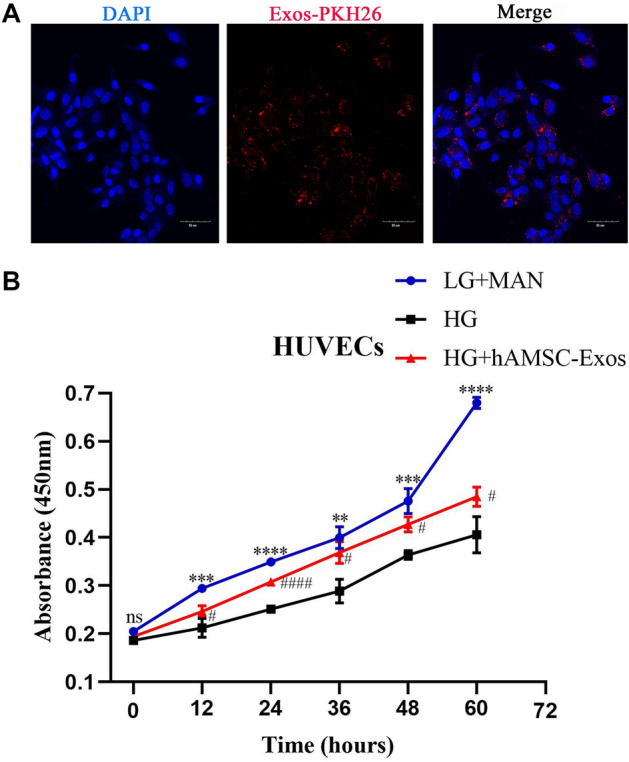
hAMSC-Exos facilitated the proliferation of HUVECs. A PKH26-labeled hAMSC-Exos with red fluorescence were transferred to the cytoplasm of HUVECs. B Cell counting kit-8 assays were used to evaluate the proliferation of HUVECs treated with hAMSC-Exos. *p < 0.05, low glucose with mannitol compared with high glucose group; #p < 0.05, hAMSC-Exos compared with high glucose group, n.s. no significance
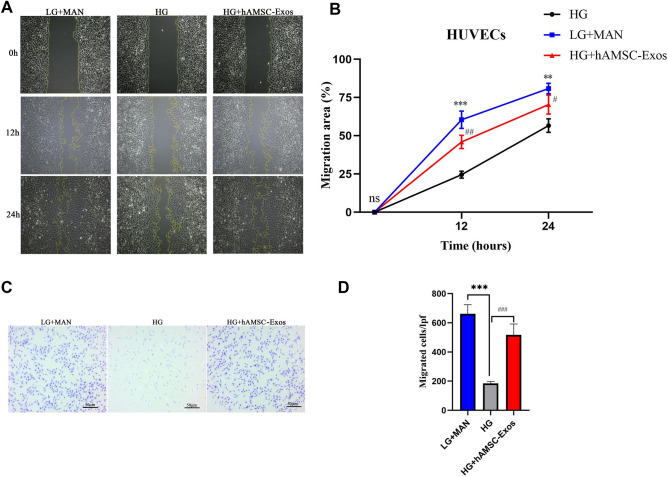
The hAMSC-Exos facilitated the migration of HUVECs
Migration of HUVECs is important for angiogenesis to accelerate the healing of diabetic wounds. Here, scratch and transwell assays were conducted to evaluate the migration of HUVECs by hAMSC-Exos (Fig. 3A–D). Both transwell and scratch assays demonstrated that hAMSC-Exos significantly increased migration of HUVECs (p < 0.05).
Fig. 3.
hAMSC-Exos facilitated migration of HUVECs. A Representative images of scratch assays in HUVECs treated with hAMSC-Exos or PBS. B Quantitative analysis of the migration rates in (A). C Representative images of the transwell assay in HUVECs treated with hAMSC-Exos or PBS. D Quantitative analysis of the migrated cells in (C). *p < 0.05, low glucose with mannitol compared with high glucose group; #p < 0.05, hAMSC-Exos compared with high glucose group, n.s. no significance
hAMSC-Exos enhanced the angiogenic activities of HUVECs
Angiogenesis can provide nutritional support to wound sites, thereby promoting the healing of diabetic wounds. Tube-formation assays on Matrigel are recognized as a model of angiogenesis in vitro. After treatment with hAMSC-Exos, the HUVECs showed a greater number of capillary-like structures compared with the control group (Fig. 4A and B) (p < 0.05).
Fig. 4.
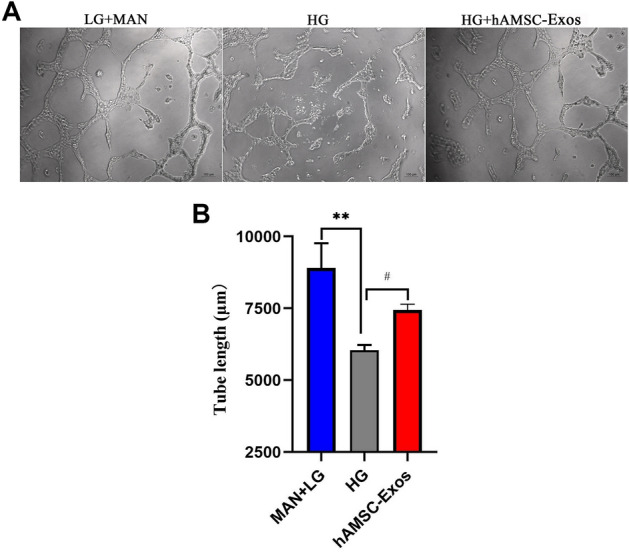
A Representative images of tube formation assays in HUVECs treated with hAMSC-Exos or PBS. B Quantitative analysis of the tube lengths of in (A). *p < 0.05, low glucose with mannitol compared with high glucose group; #p < 0.05, hAMSC-Exos compared with high glucose group, n.s. no significance
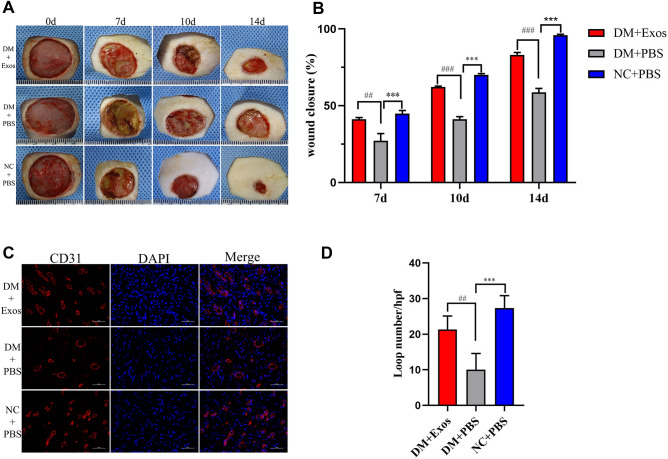
hAMSC-Exos accelerated the healing of diabetic wounds
To evaluate the effects of hAMSC-Exos on the cutaneous healing process of diabetic wounds, PBS or hAMSC-Exos were injected subcutaneously around the wound. The wounds images at Days 0, 7, 10 and 14 postoperation showed that the wounds of normal rats were healing faster than the wounds of diabetic rats. However, diabetic rats injected with hAMSC-Exos showed much faster wound closure rates than those injected with PBS (Fig. 5A and B) (p < 0.05).
Fig. 5.
hAMSC-Exos facilitated cutaneous wound healing and increased the capillary density in rats. A Gross views of wounds treated with NC + PBS, DM + PBS and DM + hAMSC-Exos at Days 0, 7, 10 and 14 postoperation. B Quantitative analysis of the wound closure rates for each group at the indicated times. C Representative images of immunohistochemical staining for CD31 in wound sections treated with NC + PBS, DM + PBS and DM + hAMSC-Exos. D Quantitative analysis of the loop numbers for each field in every group. NC normal control, DM diabetes mellitus. *p < 0.05, NC + PBS compared with DM + PBS; #p < 0.05, DM + hAMSC-Exos compared with DM + PBS, n.s. no significance
hAMSC-Exos promoted the healing process of diabetic wounds by improving angiogenesis
CD31 expressions are widely used to evaluate the extent of angiogenesis in wound sites. To explore the impact of hAMSC-Exos, immunohistochemical staining for CD31 was performed. The results showed that greater numbers of CD31-stained cells appeared in the wounds of normal rats than in diabetic rats, which demonstrated that the wound healing process could be delayed by hyperglycemia. However, among the diabetic rats, more CD31-stained positive cells appeared around wounds treated with hAMSC-Exos in contrast to those treated with PBS (Fig. 5C and D). The above results suggested that hAMSC-Exos were able to facilitate the angiogenesis of diabetic wounds, which demonstrated the potential of hAMSC-Exos in promoting angiogenesis.
Differential expression of lncRNAs and mRNAs
MSC-Exos can carry lncRNAs [12], which can be transferred to target cells to regulate the biological processes. To explore the potential contents of hAMSC-Exos, we conducted a comprehensive analysis of the lncRNA expressions between hAMSCs and hAMSC-Exos by using a microarray. We screened the differential expressions of lncRNAs and mRNAs (fold changes ≥ 5) in hAMSC-Exos. The results showed that 2686 lncRNAs were significantly upregulated and 2388 lncRNAs were significantly downregulated. Concurrently, 857 mRNAs were significantly upregulated and 3979 mRNAs were significantly downregulated. These results suggest that hAMSCs could release exosomes in a specifific manner, and that lncRNAs and mRNAs may be selectively packaged into hAMSC-Exos. We selected the partially differentially expressed exosomal lncRNAs (Table 2) and mRNAs (Table 3).
Table 2.
Partially differentially expressed exosomal lncRNAs ( hAMSCs-Exos vs hAMSCs)
| Transcript_ID | Fold change | Regulation | Gene symbol |
|---|---|---|---|
| ENST00000429730 | 90.9142162 | Up | LINC01857 |
| ENST00000443988 | 40.5410851 | Up | PANTR1 |
| ENST00000634576 | 38.2345874 | Up | LINC00466 |
| ENST00000559368 | 20.0761399 | Up | OIP5-AS1 |
| ENST00000439545 | 8.489689 | Up | HOTAIR |
| ENST00000412712 | 8.4132814 | Up | ID2-AS1 |
| ENST00000442037 | 8.1966398 | Up | H19 |
| ENST00000606233 | 7.0520906 | Up | NR2F1-AS1 |
| ENST00000473846 | 6.9437933 | Up | GLIS3 |
| ENST00000433869 | 5.4139862 | Up | ERLNC1 |
| ENST00000619449 | 219.8320917 | Down | MALAT1 |
| ENST00000498411 | 41.5729725 | Down | FLNA |
| ENST00000572915 | 30.4030594 | Down | PAFAH1B1 |
| ENST00000487310 | 21.1927544 | Down | JAK2 |
| ENST00000462077 | 17.9850517 | Down | FLRT3 |
| ENST00000585639 | 16.7170572 | Down | LINC00907 |
| ENST00000645313 | 16.6357361 | Down | CDKN2B-AS1 |
| ENST00000431243 | 12.0066769 | Down | MTHFR |
| ENST00000512932 | 10.036883 | Down | THAP9-AS1 |
| ENST00000422420 | 6.0660912 | Down | CDKN2B-AS1 |
Table 3.
Partially differentially expressed exosomal mRNAs ( hAMSCs-Exos vs hAMSCs)
| Transcript_ID | Fold CHANGE | Regulation | Gene symbol |
|---|---|---|---|
| ENST00000382333 | 61.8289843 | Up | FGFBP1 |
| ENST00000392320 | 16.5734116 | Up | STAT4 |
| ENST00000392870 | 14.9090414 | Up | GRK5 |
| ENST00000380874 | 8.5155086 | Up | FOXC1 |
| ENST00000225275 | 7.1874011 | Up | MPO |
| ENST00000222390 | 6.9418515 | Up | HGF |
| ENST00000345617 | 6.705519 | Up | HDAC4 |
| ENST00000222572 | 6.4455032 | Up | PON2 |
| ENST00000297904 | 5.5418924 | Up | VEGFD |
| ENST00000360648 | 5.3705867 | Up | PAK3 |
| ENST00000621592 | 102.3147973 | Down | MYC |
| ENST00000361099 | 66.3645547 | Down | STAT1 |
| ENST00000613296 | 51.7064087 | Down | ALMS1 |
| ENST00000551116 | 42.4786968 | Down | DDIT3 |
| ENST00000261845 | 33.5184144 | Down | MAPK6 |
| ENST00000545279 | 24.077475 | Down | SMAD5 |
| ENST00000218388 | 22.5360599 | Down | TIMP1 |
| ENST00000298139 | 21.2347851 | Down | WRN |
| ENST00000221930 | 18.3312543 | Down | TGFB1 |
| ENST00000412585 | 11.8659524 | Down | HLA-B |
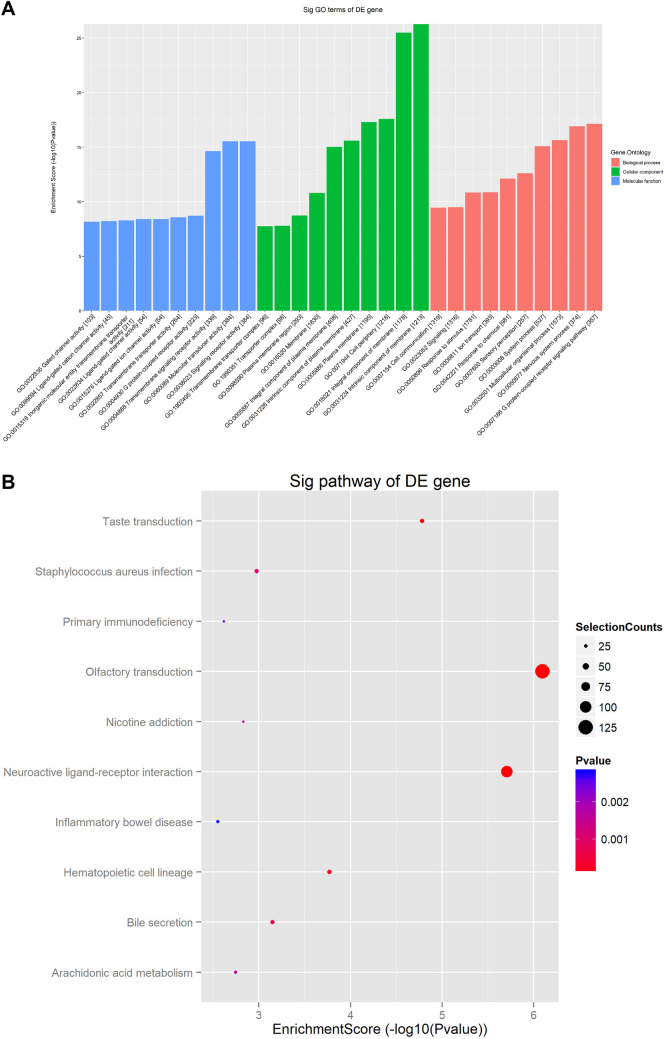
GO and KEGG pathway analyses of the differential expression of genes from the hAMSC-Exos
TopGO was used to perform a GO analysis of the upregulated differential expressions of genes to infer the cellular components, molecular functions and biological processes in which they were involved. Enrichment of the top 10 items is shown in Fig. 6A. The biological processes included the positive regulation of transcription from multicellular organismal process and response to stimulus and cell communication. Molecular functions were involved in signaling receptor activity, molecular transducer activity and transmembrane signaling receptor activity. Cellular components were involved in intrinsic component of membrane, transporter complex, and transmembrane transporter complex. These results suggest that hAMSC-Exos may regulate the construction of cell membrane, recognition of cell molecules and receptors and transfer of information between cells through the bioactive molecules they contain.
Fig. 6.
GO and KEGG pathway analyses of differentially expressed mRNAs. A The top 10 enriched GO terms involved in biological processes, cellular components and molecular functions. B The top 10 most enriched pathways are presented
In terms their functions, the differentially expressed mRNAs were used for pathway analysis to infer the pathways in which they were involved. The annotations of the microarray genes for pathways were downloaded from KEGG. The differentially expressed genes that were upregulated in hAMSC-Exos play roles in several different pathways, and the most enriched pathways were olfactory transduction, neuroactive ligand-receptor interaction, and hematopoietic cell lineage (Fig. 6B), which may play dominant roles in wound repair and tissue regeneration.
RT–qPCR verification
The reliability of the microarray results was verified by RT–qPCR. LINC01857, H19, HOTAIR, MALAT1, VEGFD, STAT4, MAPK6, and MYC were selected for RT–qPCR verification. LINC01857, H19, HOTAIR, VEGFD and STAT4 were upregulated and MALAT1, MAPK6 and MYC were downregulated in hAMSC-Exos, which is consistent with the microarray data. This means that the data acquired by the microarray were reliable. The RT–qPCR results are shown in Fig. 7 (p < 0.05).
Fig. 7.
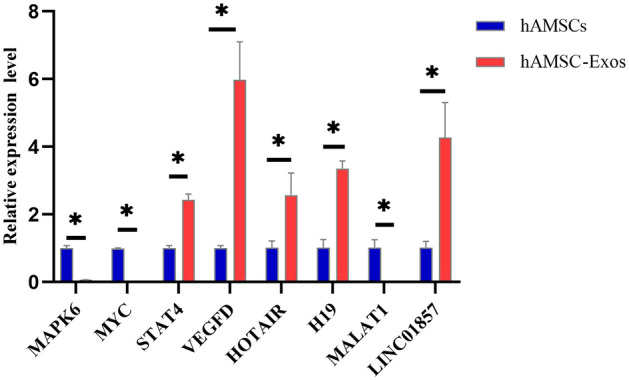
Relative expression levels of LINC01857, H19, HOTAIR, MALAT1, VEGFD, STAT4, MAPK6, and MYC in hAMSC-Exos. They were verified by RT–qPCR, *p < 0.05, hAMSC-Exos compared with hAMSCs
Construction of ceRNA networks
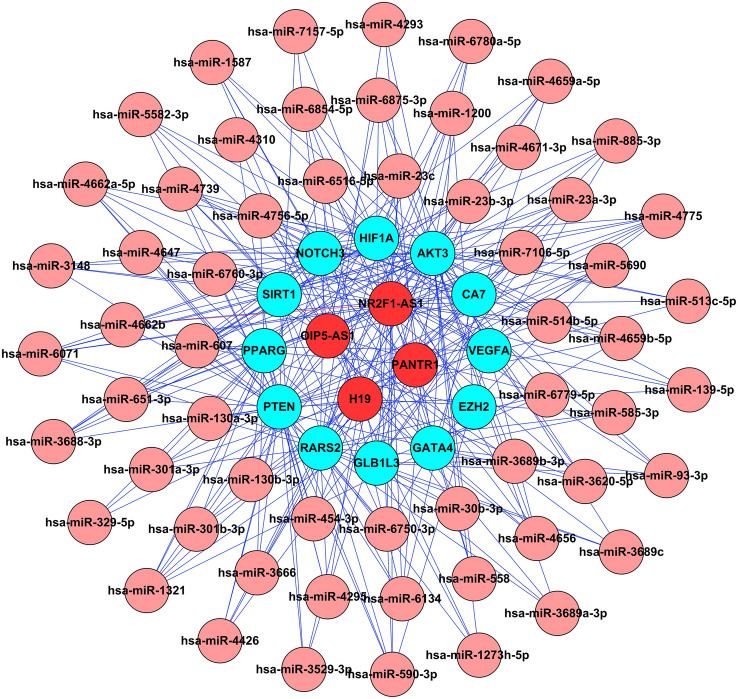
In terms of the ceRNA hypothesis, miRNAs can silence mRNAs expressions by binding to mRNA, and lncRNAs can competitively bind to miRNAs to regulate the expressions of mRNAs. The lncRNAs and mRNAs exhibited coexpression patterns in the networks. Therefore, lncRNAs can be considered and may function as ceRNAs. Here, ceRNA networks were constructed, that could directly exhibit lncRNAs–miRNA–mRNA interactions. The angiogenesis-related lncRNAs, PANTR1, H19, OIP5-AS1 and NR2F1-AS1, were selected to construct the ceRNA networks (Fig. 8). They can act as ceRNAs and bind to miRNAs to regulate the expressions of the miRNA targets, VEGFA, PTEN, HIF1A, and SIRT1, which may participate in regulating the healing process of diabetic wounds.
Fig. 8.
The ceRNA interaction network of lncRNA–miRNA–mRNA. The lncRNAs were linked to mRNAs via miRNAs in this network. Red nodes represent lncRNAs, pink nodes represent miRNAs, and cyan nodes represent mRNAs
Discussion
In this study, we demonstrated that exosomes secreted by hAMSCs could facilitate the proliferation, migration, and tube formation of HUVECs in vitro as well as facilitate diabetic wound healing by angiogenesis in vivo. Previous studies have indicated that the MSCs could affect the healing process of diabetic wounds by transplantation [18, 19]. For example, intracutaneous injection of hAMSCs around the wound can promote diabetic wound healing by angiogenesis [20].The effects by which hAMSCs promote angiogenesis lie on paracrine mechanism in which soluble growth factors and exosomes play a major role. However, there are many unpredictable risk factors in the direct transplantation of the MSCs, such as potential tumorigenicity, undesired immune responses and storage limitations [21]. Exosomes secreted by MSCs through paracrine mechanisms not only have the same effects as MSCs but also exhibit great potential for angiogenesis formation during wound healing due to their high stability, low immunogenicity, nontumorigenicity and ease to storeage [22–24]. MSC-Exos have been shown to have positive treatment effects for cutaneous wounds, and provide more efficient and safe cell-free therapy [25].
Many studies have indicated that exosomes originating from adipose-derived mesenchymal stem cells (ADMSCs), bone marrow mesenchymal stem cells (BMSCs) and umbilical cord mesenchymal stem cells (UCMSCs) can accelerate cutaneous wound healing. For example, ADMSC-Exos could promote vascularization to accelerate the healing of diabetic cutaneous wounds [26]. Exosomes derived from atorvastatin-pretreated BMSCs can accelerate the healing of diabetic skin defects by promoting the formation of blood vessels in diabetic rats [8]. UCMSC-Exos facilitate diabetic wound repair by regulating oxidative stress, and promoting angiogenesis [9, 27]. However, there have been no reports regarding the application of hAMSC-Exos in diabetic wound healing thus far. The present study reported only that the application of conditioned medium from hAMSCs could promote wound healing in diabetic mice [28]. To explore the role of hAMSC-Exos in diabetic wound healing, we successfully isolated and identified hAMSCs and hAMSC-Exos and observed the effects of hAMSC-exos on the biological activities of HUVECs treated with high glucose levels and diabetic wound healing. Our results demonstrated that hAMSC-Exos could facilitate the proliferation, migration and angiogenic activities of HUVECs in vitro and facilitate diabetic wound healing by angiogenesis in vivo.
Although the hAMSC-Exos have already exhibited therapeutic value in diabetic wounds through angiogenesis, the underlying mechanisms of hAMSC-Exos in diabetic wound healing remain poorly understood. Exosomes contain a variety of bioactive molecules, and lncRNA is one of the important molecules secreted by MSC-Exos [29, 30]. It has been reported that exosomal lncRNAs can promote angiogenesis via ceRNAs [31, 32]. To explore whether the hAMSC-Exos contain lncRNAs associated with angiogenesis, the differential expressions of lncRNAs in hAMSCs and hAMSC-Exos were compared by using a microarray; subsequently, lncRNAs related to angiogenesis were screened. The results showed that PANTR1 (also known as linc-POU3F3), H19, OIP5-AS1 and NR2F1-AS1 were enriched in hAMSC-Exos, which could regulate angiogenesis [33–36].
Emerging evidence has shown that PANTR1, H19, OIP5-AS1 and NR2F1-AS1 can regulate angiogenesis by promoting the migration, proliferation, and tube formation of endothelial cells [36–39]. One of the mechanisms underlying angiogenesis facilitated by lncRNAs is that they act as ceRNAs to regulate mRNA expressions by sponging miRNAs. For example, H19 can increase VEGFA expressions by sponging miR-29c to regulate the progression of corneal neovascularization [40]. OIP5-AS1 affects hepatocellular carcinoma angiogenesis by regulating VEGFA expressions through sponging miR-3163 [38]. To analyze the mechanisms of exosomal lncRNAs that are involved in promoting angiogenesis, ceRNA networks were constructed. The networks showed that VEGFA, PTEN, HIF1A, and SIRT1 were the target genes of exosomal lncRNAs, the lower expression of them are associated with delaying the healing process of diabetic wounds [41–43]. Our next study explored the effect of lncRNAs on diabetic wound healing through the ceRNA mechanism.
In summary, our study demonstrated that hAMSC-Exos could facilitate the proliferation, migration and angiogenic activities of HUVECs treated with high glucose and facilitate diabetic wound healing by angiogenesis in rats and contained several exosomal lncRNAs related to angiogenesis such as PANTR1, H19, OIP5-AS1 and NR2F1-AS1. These findings suggested that hAMSC-Exos may represent a promising strategy for diabetic wound healing.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplemental Fig. 1. A Flow cytometry analysis of the CD44, CD73, CD90, CD105, CD34, CD45 and HLA-DR expressions in hAMSCs. Blue lines represent the isotype control, and red lines represent the level of surface markers. B and C The differentiation potential of hAMSCs. The hAMSCs could be induced to differentiate into osteocytes and adipocytes with osteogenic and adipogenic differentiation medium, respectively, and the hAMSCs were stained with Alizarin Red S and Oil Red O after osteocytes and adipocytes differentiated (DOCX 734 kb)
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant No. 81460293).
Declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Ethical statement
This study approved by the Medical Research Ethics Committees of the First Affiliated Hospital of Nanchang University (no. 20218-008) and the written informed consents were signed by the pregnant women or their legal guardians.
The animal experiment were approved by the Experimental Animal Welfare Ethics Committee of the First Affiliated Hospital of Nanchang University (Jiangxi, China) (CDYFY-IACUC-202208QR040).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shangfeng Fu, Hongyan Zhang have contributed equally to this work.
References
- 1.An T, Chen Y, Tu Y, Lin P. Mesenchymal stromal cell-derived extracellular vesicles in the treatment of diabetic foot ulcers: application and challenges. Stem Cell Rev Rep. 2021;17:369–378. doi: 10.1007/s12015-020-10014-9. [DOI] [PubMed] [Google Scholar]
- 2.Burgess JL, Wyant WA, Abdo AB, Kirsner RS, Jozic I. Diabetic wound-healing science. Medicina (Kaunas) 2021;57:1072. doi: 10.3390/medicina57101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniadou E, David AL. Placental stem cells. Best Pract Res Clin Obstet Gynaecol. 2016;31:13–29. doi: 10.1016/j.bpobgyn.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Abbasi-Kangevari M, Ghamari SH, Safaeinejad F, Bahrami S, Niknejad H. Potential therapeutic features of human amniotic mesenchymal stem cells in multiple sclerosis: immunomodulation, inflammation suppression, angiogenesis promotion, oxidative stress inhibition, neurogenesis induction, mmps regulation, and remyelination stimulation. Front Immunol. 2019;10:238. doi: 10.3389/fimmu.2019.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parolini O, Alviano F, Bagnara GP, Bilic G, Buhring HJ, Evangelista M, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first international workshop on placenta derived stem cells. Stem Cells. 2008;26:300–311. doi: 10.1634/stemcells.2007-0594. [DOI] [PubMed] [Google Scholar]
- 6.Azari Z, Nazarnezhad S, Webster TJ, Hoseini SJ, Brouk MP, Baino F, et al. Stem cell-mediated angiogenesis in skin tissue engineering and wound healing. Wound Repair Regen. 2022;30:421–435. doi: 10.1111/wrr.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mankuzhy PD, Ramesh ST, Thirupathi Y, Mohandas PS, Chandra V, Sharma TG. The preclinical and clinical implications of fetal adnexa derived mesenchymal stromal cells in wound healing therapy. Wound Repair Regen. 2021;29:347–369. doi: 10.1111/wrr.12911. [DOI] [PubMed] [Google Scholar]
- 8.Yu M, Liu W, Li J, Lu J, Lu H, Jia W, et al. Exosomes derived from atorvastatin-pretreated MSC accelerate diabetic wound repair by enhancing angiogenesis via AKT/eNOS pathway. Stem Cell Res Ther. 2020;11:350. doi: 10.1186/s13287-020-01824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan C, Xv Y, Lin Z, Endo Y, Xue H, Hu Y, et al. Human umbilical cord mesenchymal stem cell-derived exosomes accelerate diabetic wound healing via ameliorating oxidative stress and promoting angiogenesis. Front Bioeng Biotechnol. 2022;10:829868. doi: 10.3389/fbioe.2022.829868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Cai Y, Zhang Q, Zhang Y. Pharmaceutical Activation of Nrf2 Accelerates Diabetic Wound Healing by Exosomes from Bone Marrow Mesenchymal Stem Cells. Int J Stem Cells. 2022;15:164–172. doi: 10.15283/ijsc21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha DH, Kim HK, Lee J, Kwon HH, Park GH, Yang SH, et al. Mesenchymal stem/stromal cell-derived exosomes for immunomodulatory therapeutics and skin regeneration. Cells. 2020;9:1157. doi: 10.3390/cells9051157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casado-Diaz A, Quesada-Gomez JM, Dorado G. Extracellular vesicles derived from mesenchymal stem cells (MSC) in regenerative medicine: applications in skin wound healing. Front Bioeng Biotechnol. 2020;8:146. doi: 10.3389/fbioe.2020.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han ZF, Cao JH, Liu ZY, Yang Z, Qi RX, Xu HL. Exosomal lncRNA KLF3-AS1 derived from bone marrow mesenchymal stem cells stimulates angiogenesis to promote diabetic cutaneous wound healing. Diabetes Res Clin Pract. 2022;183:109126. doi: 10.1016/j.diabres.2021.109126. [DOI] [PubMed] [Google Scholar]
- 14.Born LJ, Chang KH, Shoureshi P, Lay F, Bengali S, Hsu A, et al. HOTAIR-loaded mesenchymal stem/stromal cell extracellular vesicles enhance angiogenesis and wound healing. Adv Healthc Mater. 2022;11:e2002070. doi: 10.1002/adhm.202002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu QW, Liu QY, Li JY, Wei L, Ren KK, Zhang XC, et al. Therapeutic efficiency of human amniotic epithelial stem cell-derived functional hepatocyte-like cells in mice with acute hepatic failure. Stem Cell Res Ther. 2018;9:321. doi: 10.1186/s13287-018-1063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purushothaman A. Exosomes from cell culture-conditioned medium: isolation by ultracentrifugation and characterization. Methods Mol Biol. 2019;1952:233–244. doi: 10.1007/978-1-4939-9133-4_19. [DOI] [PubMed] [Google Scholar]
- 17.Adnan M, Morton G, Hadi S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2(-DeltaDeltaCT) method. Mol Cell Biochem. 2011;357:275–282. doi: 10.1007/s11010-011-0898-y. [DOI] [PubMed] [Google Scholar]
- 18.Chu J, Shi P, Deng X, Jin Y, Liu H, Chen M, et al. Dynamic multiphoton imaging of acellular dermal matrix scaffolds seeded with mesenchymal stem cells in diabetic wound healing. J Biophotonics. 2018;11:e201700336. doi: 10.1002/jbio.201700336. [DOI] [PubMed] [Google Scholar]
- 19.Guo J, Hu H, Gorecka J, Bai H, He H, Assi R, et al. Adipose-derived mesenchymal stem cells accelerate diabetic wound healing in a similar fashion as bone marrow-derived cells. Am J Physiol Cell Physiol. 2018;315:C885–C896. doi: 10.1152/ajpcell.00120.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SW, Zhang HZ, Guo L, Kim JM, Kim MH. Amniotic mesenchymal stem cells enhance wound healing in diabetic NOD/SCID mice through high angiogenic and engraftment capabilities. PLoS ONE. 2012;7:e41105. doi: 10.1371/journal.pone.0041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu WZ, Ma ZJ, Li JR, Kang XW. Mesenchymal stem cell-derived exosomes: therapeutic opportunities and challenges for spinal cord injury. Stem Cell Res Ther. 2021;12:102. doi: 10.1186/s13287-021-02153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shabbir A, Cox A, Rodriguez-Menocal L, Salgado M, Van Badiavas E. Mesenchymal stem cell exosomes induce proliferation and migration of normal and chronic wound fibroblasts, and enhance angiogenesis in vitro. Stem Cells Dev. 2015;24:1635–1647. doi: 10.1089/scd.2014.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y, Zhou Y, Li HJ. Advances in mesenchymal stem cell exosomes: a review. Stem Cell Res Ther. 2021;12:71. doi: 10.1186/s13287-021-02138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong J, Hu H, Guo R, Wang H, Jiang H. Mesenchymal stem cell exosomes as a new strategy for the treatment of diabetes complications. Front Endocrinol (Lausanne) 2021;12:646233. doi: 10.3389/fendo.2021.646233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xunian Z, Kalluri R. Biology and therapeutic potential of mesenchymal stem cell-derived exosomes. Cancer Sci. 2020;111:3100–3110. doi: 10.1111/cas.14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Xie X, Lian W, Shi R, Han S, Zhang H, et al. Exosomes from adipose-derived stem cells overexpressing Nrf2 accelerate cutaneous wound healing by promoting vascularization in a diabetic foot ulcer rat model. Exp Mol Med. 2018;50:1–14. doi: 10.1038/s12276-018-0058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng L, Maqsood M, Zhu M, Zhou Y, Kang M, Zhou J, Chen J. Exosomes derived from human umbilical cord mesenchymal stem cells accelerate diabetic wound healing via promoting M2 macrophage polarization, angiogenesis, and collagen deposition. Int J Mol Sci. 2022;23:10421. [DOI] [PMC free article] [PubMed]
- 28.Takahashi H, Ohnishi S, Yamamoto Y, Hayashi T, Murao N, Osawa M, et al. Topical application of conditioned medium from hypoxically cultured amnion-derived mesenchymal stem cells promotes wound healing in diabetic mice. Plast Reconstr Surg. 2021;147:1342–1352. doi: 10.1097/PRS.0000000000007993. [DOI] [PubMed] [Google Scholar]
- 29.Li KS, Bai Y, Li J, Li SL, Pan J, ChengY Q, et al. LncRNA HCP5 in hBMSC-derived exosomes alleviates myocardial ischemia reperfusion injury by sponging miR-497 to activate IGF1/PI3K/AKT pathway. Int J Cardiol. 2021;342:72–81. doi: 10.1016/j.ijcard.2021.07.042. [DOI] [PubMed] [Google Scholar]
- 30.Su Y, Liu Y, Ma C, Guan C, Ma X, Meng S. Mesenchymal stem cell-originated exosomal lncRNA HAND2-AS1 impairs rheumatoid arthritis fibroblast-like synoviocyte activation through miR-143-3p/TNFAIP3/NF-kappaB pathway. J Orthop Surg Res. 2021;16:116. doi: 10.1186/s13018-021-02248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. MSC-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncRNA-KLF3-AS1/miR-206/GIT1 axis in osteoarthritis. Cell Cycle. 2018;17:2411–2422. doi: 10.1080/15384101.2018.1526603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao Q, Liang XL, Zhang CL, Pang YH, Lu YX. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138-5p/Sirt1 axis. Stem Cell Res Ther. 2019;10:393. doi: 10.1186/s13287-019-1522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seles M, Hutterer GC, Foßelteder J, Svoboda M, Resel M, Barth DA, et al. Long non-coding RNA PANTR1 is associated with poor prognosis and influences angiogenesis and apoptosis in clear-cell renal cell cancer. Cancers (Basel) 2020;12:1200. doi: 10.3390/cancers12051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behera J, Kumar A, Voor MJ, Tyagi N. Exosomal lncRNA-H19 promotes osteogenesis and angiogenesis through mediating Angpt1/Tie2-NO signaling in CBS-heterozygous mice. Theranostics. 2021;11:7715–7734. doi: 10.7150/thno.58410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Lin S, Xie X, Zhu H, Fan T, Wang S. Highly enriched exosomal lncRNA OIP5-AS1 regulates osteosarcoma tumor angiogenesis and autophagy through miR-153 and ATG5. Am J Transl Res. 2021;13:4211–4223. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Li T, Wang Z, Kuang X, Shao N, Lin Y. lncRNA NR2F1-AS1 promotes breast cancer angiogenesis through activating IGF-1/IGF-1R/ERK pathway. J Cell Mol Med. 2020;24:8236–8247. doi: 10.1111/jcmm.15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang HL, Hu GW, Chen Y, Liu Y, Tu W, Lu YM, et al. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci. 2017;21:959–972. [PubMed] [Google Scholar]
- 38.Shi C, Yang Q, Pan S, Lin X, Xu G, Luo Y, et al. LncRNA OIP5-AS1 promotes cell proliferation and migration and induces angiogenesis via regulating miR-3163/VEGFA in hepatocellular carcinoma. Cancer Biol Ther. 2020;21:604–614. doi: 10.1080/15384047.2020.1738908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Z, Bian Y, Ma X, Tang Z, Chen N, Shen M. LncRNA H19 knockdown in human amniotic mesenchymal stem cells suppresses angiogenesis by associating with EZH2 and activating vasohibin-1. Stem Cells Dev. 2019;28:781–790. doi: 10.1089/scd.2019.0014. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z, Ding Y, Jin X, Xu Z, Zhang H. Long non-coding RNA H19 promotes corneal neovascularization by targeting microRNA-29c. Biosci Rep. 2019;39:BSR20182394. doi: 10.1042/BSR20182394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng WX, He PX, Liu LJ, Zhu T, Zhong YQ, Xiang L, et al. LncRNA GAS5 activates the HIF1A/VEGF pathway by binding to TAF15 to promote wound healing in diabetic foot ulcers. Lab Invest. 2021;101:1071–1083. doi: 10.1038/s41374-021-00598-2. [DOI] [PubMed] [Google Scholar]
- 42.Li B, Luan S, Chen J, Zhou Y, Wang T, Li Z, et al. The MSC-derived exosomal lncRNA H19 promotes wound healing in diabetic foot ulcers by upregulating PTEN via MicroRNA-152-3p. Mol Ther Nucleic Acids. 2020;19:814–826. doi: 10.1016/j.omtn.2019.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafez YM, El-Deeb OS, Atef MM. The emerging role of the epigenetic enzyme Sirtuin-1 and high mobility group Box 1 in patients with diabetic foot ulceration. Diabetes Metab Syndr. 2018;12:1065–1070. doi: 10.1016/j.dsx.2018.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. A Flow cytometry analysis of the CD44, CD73, CD90, CD105, CD34, CD45 and HLA-DR expressions in hAMSCs. Blue lines represent the isotype control, and red lines represent the level of surface markers. B and C The differentiation potential of hAMSCs. The hAMSCs could be induced to differentiate into osteocytes and adipocytes with osteogenic and adipogenic differentiation medium, respectively, and the hAMSCs were stained with Alizarin Red S and Oil Red O after osteocytes and adipocytes differentiated (DOCX 734 kb)