Abstract
Background and objectives
Obesity is a global health problem with few pharmacologic options. Semaglutide is a glucagon-like peptide-1 (GLP-1) analogue that induces weight loss. Yet, the role of semaglutide in adipose tissue has not yet been examined. The following study investigated the mechanism of semaglutide on lipid metabolism by analyzing proteomics of epididymal white adipose tissue (eWAT) in obese mice.
Methods
A total of 36 C57BL/6JC mice were randomly divided into a normal-chow diet group (NCD, n = 12), high-fat diet (HFD, n = 12), and HFD+semaglutide group (Sema, n = 12). Mice in the Sema group were intraperitoneally administered semaglutide, and the HFD group and the NCD group were intraperitoneally administered an equal volume of normal saline. Serum samples were collected to detect fasting blood glucose and blood lipids. The Intraperitoneal glucose tolerance test (IPGTT) was used to measure the blood glucose value at each time point and calculate the area under the glucose curve. Tandem Mass Tag (TMT) combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) were used to study the expression of eWAT, while cellular processes, biological processes, corresponding molecular functions, and related network molecular mechanisms were analyzed by bioinformatics.
Results
Compared with the model group, the semaglutide-treated mice presented 640 differentially expressed proteins (DEPs), including 292 up-regulated and 348 down-regulated proteins. Bioinformatics analysis showed a reduction of CD36, FABP5, ACSL, ACOX3, PLIN2, ANGPTL4, LPL, MGLL, AQP7, and PDK4 involved in the lipid metabolism in the Sema group accompanied by a decrease in visceral fat accumulation, blood lipids, and improvement in glucose intolerance.
Conclusion
Semaglutide can effectively reduce visceral fat and blood lipids and improve glucose metabolism in obese mice. Semaglutide treatment might have beneficial effects on adipose tissues through the regulation of lipid uptake, lipid storage, and lipolysis in white adipose tissue.
Keywords: semaglutide, white adipose tissue, obesity, proteomics, lipid metabolism
1. Introduction
Obesity is a multifactorial chronic disease characterized by excessive fat accumulation in adipose tissue, which can lead to insulin resistance, hypertension, and dyslipidemia (1). Dyslipidemia is an important link between obesity and the development of type 2 diabetes mellitus(T2DM), cardiovascular disease (CVD), and certain types of cancer, such as breast cancer and colon adenomas (2). Semaglutide, a glucagon-like peptide-1 (GLP-1) analogue, has been reported to induce weight loss among overweight or obese adults, as well as to have a beneficial effect on cardiometabolic health in these populations (3). Gabery et al. reported that semaglutide lowers rodent body weight via distributed neural pathways (4). Moreover, Pontes-da-Silva et al. discovered that semaglutide reduces insulin resistance, liver inflammation, and endoplasmic reticulum stress in obese mice (5). Another study reported that semaglutide has beneficial effects on a pro-inflammatory pathway, PDX1, and PPAR-alpha and gamma, by reducing the lesion on the islet. However, the impact of semaglutide on weight loss was found to have little relevance in the pancreatic islet caused by insulin resistance (6).
There are evidences shows GLP-1 analogues directly signal to adipose tissue. Previous study provided evidence for the presence of GLP-1 receptor in adipose tissue (7). The beneficial effects of GLP-1 have been found to be associated with changes in the adipogenesis, lipolysis, thermogenesis and anti-inflammation process. A recent study discovered that GLP-1 down-regulated the expression of adipogenic/lipogenic genes on adipose tissue in vivo and in vitro, while increasing that of lipolytic markers and adiponectin (8). Zhang proved that GLP-1 analogue liraglutide decreased adipocyte size, increased secretion of FGF21, and promoted phosphorylation of LKB1, AMPK and Acetyl coenzyme A carboxylase 1 (ACC1) in white adipose tissue of DM mice (9). Similarly, Shao also found that with liraglutide treatment, visceral adipose tissue of mice was reduced with AMPK activation and Akt suppression, which was associated with reduction of lipogenetic process (10). GLP-1 analogues may also target epicardial adipose tissue GLP-1R and therefore reduce local adipogenesis, improve fat utilization and induce brown fat differentiation (11). Moreover, Wan showed that GLP-1 analogue supaglutide reduces HFD-induced obesity, which is associated with increased Ucp-1 in white adipose tissue of mice (12). Absalon discovered the anti-diabetic effects of liraglutide was mediated by transient upregulation of IL-6, which activates canonical IL-6R signaling resulted in adipose tissue browning and thermogenesis linked with STAT3 activation (13). GLP-1 also has anti-inflammatory effects on adipose tissue, it reduces macrophage infiltration and directly inhibits inflammatory pathways in adipocytes and adipose tissue macrophages, possibly contributing to the improvement of insulin sensitivity (14). Administration of GLP-1 analogues exenatide or liraglutide reduced inflammatory marker mRNA in adipose tissue of T2DM obese subjects (8). However, no study has examined the role of semaglutide on adipose tissue so far.
In mammals, the white adipose tissue (WAT) is the major organ that stores extra energy from diets in the form of triglycerides (TG) or fat, which can be mobilized to meet energy demands (15). Therefore, dysfunction in white adipose tissue metabolism is a cardinal event in the development of insulin resistance and metabolic disorders (16). In this study, we investigated changes in lipid metabolism proteomes in epididymal white adipose tissue (eWAT) of diet-induced obese (DIO) mice in response to semaglutide intervention by TMT combined with LC-MS/MS to provide greater insight into the mechanism of lipid metabolism by semaglutide.
2. Methods
2.1. Mice
A total of 36 male C57BL/6JC mice (7-week-old, 16−20 g) purchased from Hebei INVIVO Laboratory Animal Technology Co., Ltd. (Hebei, China) were housed (3–5 mice per cage) in a pathogen-free facility in a temperature-controlled room (22°C) with a 12-h light/dark cycle, and were given free access to food and water. All animal studies (including the mice euthanasia procedure) were done in compliance with the regulations and guidelines of Hebei General Hospital institutional animal care and conducted according to the AAALAC and the IACUC guidelines.
The current study only investigated male mice since they are more susceptible to diet-induced obesity and diet-induced insulin resistance than female mice (17). After one week of acclimatization, mice were randomly distributed into two groups and fed with either a normal-chow diet (NCD, n = 12) or a high-fat diet (60% fat, 20% carbohydrate, 20% protein, total calories 524kcal/100 g) (n = 24). After 12 weeks of feeding, the high-fat diet group was further divided into the HFD+saline group (HFD, n = 12) and HFD+semaglutide group (Sema, n = 12). Mice in the Sema group were intraperitoneally administered semaglutide (30nmol/kg/d, Novo Nordisk, Bagsværd, Denmark), whereas the NCD and HFD groups were treated with saline. Body weight were measured once a week.
After 12 weeks of treatment, glucose tolerance tests and metabolic measurements were carried out. Mice were fasted for 12 h prior to sacrifice. At the end of the experiment, mice were anesthetized with 1% sodium pentobarbital (60 mg/kg) intraperitoneal injection. Blood was collected from the retro-orbital sinus and placed into sterile tubes containing 1 mm EDTA, after which the mice were euthanized. Interscapular BAT (iBAT) and epididymal WAT (eWAT) were collected, weighted, and subjected to hematoxylin and eosin (H&E) staining or snap-frozen in liquid nitrogen and stored at 80°C until analysis.
2.2. Glucose tolerance tests and AUC measurements
Blood glucose was monitored by examining tail vein blood using the Roche blood glucose monitoring system. The intraperitoneal glucose tolerance test (i.p. GTT) was carried out after 12 weeks of sumaglutide treatment. For i.p. GTT, mice were fasted overnight and were given 2 g of 50% glucose/kg body weight via intraperitoneal injection. Tail vein blood glucose levels were measured at 0, 15, 30, 60, 90, and 120 min, and the area under the curve (AUC) was obtained.
2.3. Serum analysis
Serum samples were separated by centrifugation at 4°C and stored at -80°C until used for measurements. Insulin levels were detected by enzyme-linked immunosorbent assay (ELISA) using the Mouse INS (Insulin) ELISA Kit (Elabscience Biotechnology Co., Ltd). Triglyceride (TG), total cholesterol (TC), LDL-C, and HDL-C assay kits were purchased from Jiancheng Biology Institution PeproTech (Nanjing, China). The above assays were conducted according to the manufacturers’ instructions.
2.4. Histopathological analysis
Both sides of epididymal white adipose tissues and interscapular brown adipose tissues were removed, weighed, fixed in 4% paraformaldehyde for 48 h, and immersed in the dehydration box for dehydration and wax leaching. The wax-soaked tissues were embedded. The paraffin blocks were cut into 4 µm. After de-paraffinization, they were stained with hematoxylin and eosin (H&E). The target area of the tissue was selected for 200x imaging using an Eclipse Ci-L photomicroscope (Nikon Eclipse E100), and the tissue was imaged to fill the entire field of view as much as possible to ensure consistent background light in each photograph. After imaging was completed, Image-Pro Plus 6.0 (Nikon DS-U3) analysis software was used to uniformly measure 5 muscle fiber diameters in each section with mm as the standard unit; the number of adipocytes was counted in 3 fields of view in each section and the total area of adipocytes was measured in the field of view as well as the area of the field of view; the average area of adipocytes was calculated as = total area of adipocytes/number of adipocytes, and the adipose cell density was calculated as = number of adipocytes/area of field of view.
2.5. Protein digestion and peptide labelling
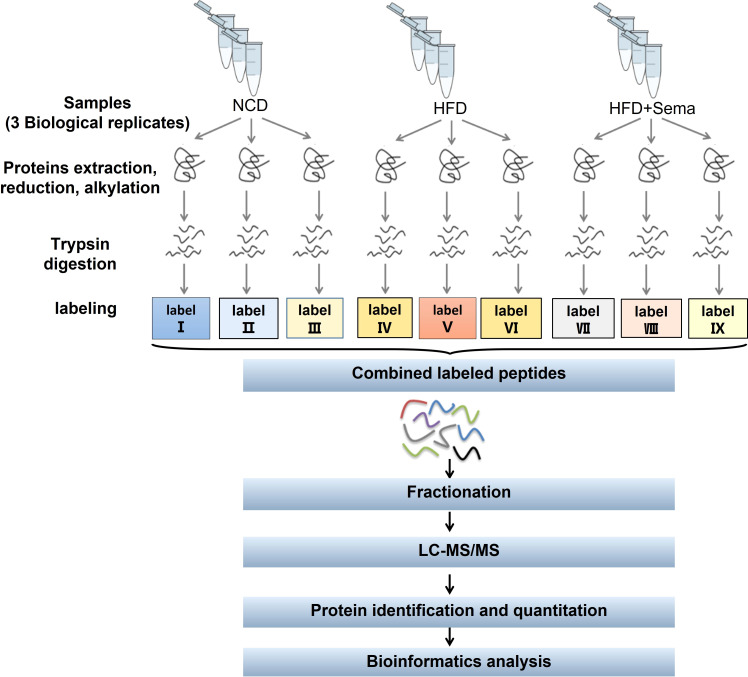
The flowchart of proteomics and bioinformatics analysis is shown in Figure 1 . Nine epididymal white adipose tissues (3 tissues/group) were ground by liquid nitrogen into cell powder, lysed, and extracted in SDT (4%SDS, 100mM Tris-HCl, 1mM DTT, pH7.6) buffer. The amount of protein was quantified with the BCA Protein Assay Kit (Bio-Rad, USA). Protein digestion by trypsin was performed according to the filter-aided sample preparation (FASP) procedure described by Matthias Mann (18). The digest peptides of each sample were desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), concentrated by vacuum centrifugation, and reconstituted in 40 µl of 0.1% (v/v) formic acid. The purity of proteins was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system, after which 100 μg peptide mixture of each sample was labeled using iTRAQ reagent (Applied Biosystems)/TMT reagent (Thermo Scientific) according to the manufacturer’s instructions.
Figure 1.
Study workflow. eWAT samples in each group (≥ 600 mg per group) were subjected to three biological replicates (each replicate was a mixture of eWAT from 5–7 mice). LC-MS/MS, liquid chromatography-tandem mass spectrometry; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
2.6. LC-MS/MS analysis
Labeled peptides were fractionated by High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific). The collected fractions were desalted on C18 Cartridges and concentrated by vacuum centrifugation. LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Thermo Fisher Scientific) for 60/90 min. The peptides were loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm*2 cm, nanoViper C18) connected to the C18-reversed-phase analytical column (Thermo Scientific Easy Column) in buffer A (0.1% Formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% Formic acid) at a flow rate of 300 nl/min controlled by IntelliFlow technology. The MS raw data for each sample were searched using the MASCOT engine (Matrix Science, London, UK; version 2.2) embedded into Proteome Discoverer 1.4 software for identification and quantitation analysis.
2.7. Bioinformatics analysis
Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and Java Treeview software (http://jtreeview.sourceforge.net) were used to perform hierarchical clustering analysis. CELLO (http://cello.life.nctu.edu.tw/), a multi-class SVM classification system, was used to predict protein subcellular localization. Protein sequences were searched using the InterProScan software to identify protein domain signatures from the InterPro member database Pfam. The protein sequences of the selected differentially expressed proteins were locally searched using the NCBI BLAST+ client software (ncbi-blast-2.2.28+-win32.exe) and InterProScan to find homologue sequences, after which gene ontology (GO) terms were mapped, and sequences were annotated using the software program Blast2GO. Following annotation steps, the studied proteins were blasted against the online Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://geneontology.org/) to retrieve their KEGG orthology identifications, after which they were mapped to pathways in KEGG. Enrichment analysis was applied based on Fisher’ exact test, considering the whole quantified proteins as the background dataset. Benjamini- Hochberg correction for multiple testing was further applied to adjust derived p-values. Functional categories and pathways with p-values < 0.05 were considered statistically significant.
2.8. Statistical analysis
All mice experiments were analyzed using Prism 8 GraphPad Software (San Diego, California). All data were analyzed using ordinary or repeated-measures one-way or two-way ANOVA; when indicated, Tukey’s or Dunnett’s were used for multiple comparison tests. All data are expressed as mean ± SEM (Standard error of mean). Differences with a p-value of < 0.05 were considered statistically significant.
3. Results
3.1. Semaglutide reduces body weight and improves metabolic profiles
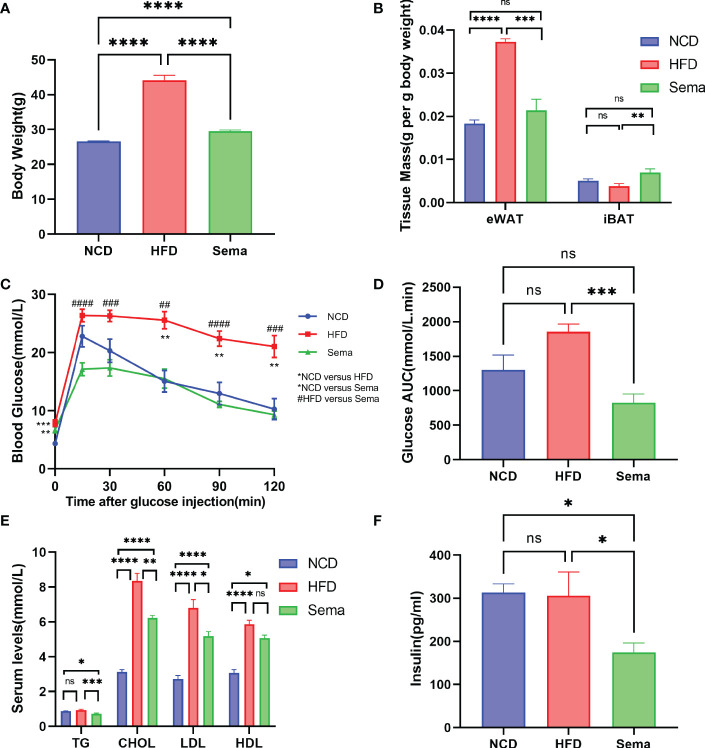
At the end of the treatment, the body weight of mice in the Sema group was significantly lower than that of the HFD group mice ( Figure 2A ), and the eWAT weight/body weight ratio showed differences consistent with body weight, while the iBAT weight/body weight ratio was significantly higher (p<0.05) in Sema group ( Figure 2B ), suggesting that semaglutide led to a weight-loss effect by lowering the proportion of visceral fat mass and increasing the brown fat mass relative to total body mass.
Figure 2.
The effects of semaglutide on reducing body weight and visceral obesity and improving metabolic profiles. (A) Average body weight of each group before being sacrificed (n = 8). (B) Fat mass/body weight ratio (n = 8). (C, D) i.p. GTT after semaglutide treatment and area of the curve (AOC) generated by the area under the curve (AUC), with subtraction of the basal glucose (n = 8). (E, F) Determination of serum TC, TG, LDL-C, HDL-C, and insulin concentration (n =8); Data were presented as mean ± SEM. “ns” p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001. NCD, normal control; HFD, high-fat diet-induced mice; Sema, HFD diet treated by semaglutide.
As shown in Figures 2C, D , there were no significant differences in fasting blood glucose levels between the Sema and HFD groups (P > 0.05). However, compared with the HFD group, semaglutide significantly reduced its blood glucose concentration at 15 min, 30 min, 60 min, 90 min, and 120 min, whilst the area under the blood glucose curve significantly decreased (P < 0.001). The fluctuation of blood glucose levels in HFD-fed mice was alleviated by semaglutide treatment, suggesting that semaglutide could improve the rate of glucose clearance and insulin sensitivity.
Next, we measured the content of TC, TG, LDL-C, HDL-C, and insulin ( Figures 2E, F ) in the serum obtained on the day the mice were sacrificed. TC, LDL-C, and HDL-C levels were significantly elevated in HFD-fed mice compared with NCD-fed mice, while the elevation of TC and LDL was significantly suppressed (p<0.05) after semaglutide treatment. In addition, the insulin and TG levels were comparable between HFD group mice and the control group but were significantly decreased with semaglutide treatment. However, no significant difference in HDL-C level was observed between HFD and Sema groups.
3.2. Semaglutide decreases the size of adipocytes in eWAT and iBAT
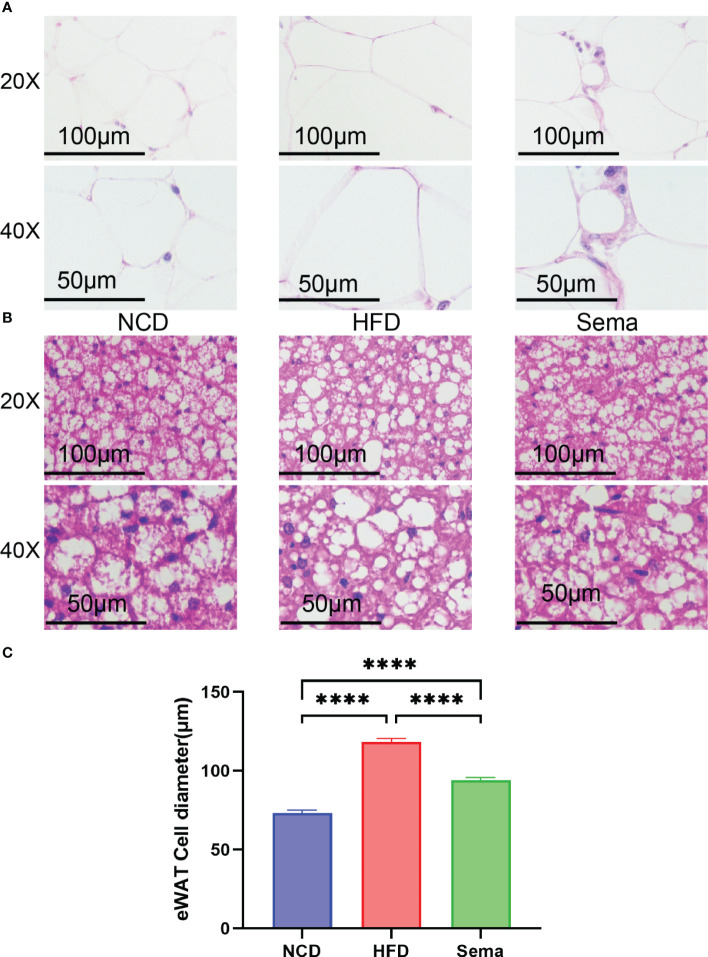
H&E staining showed that the adipocytes in eWAT of NCD group were similar in size and regular in shape, same patterns were observed in iBAT. Yet, compared with NCD group, the adipocytes in eWAT and iBAT of HFD group mice were significantly larger, with distinct morphologies and more observable in lipid droplets, which could be alleviated by the treatment of semaglutide ( Figures 3A, B ). Also, the diameter of adipocytes in eWAT was markedly decreased in Sema group, compared with HFD group ( Figure 3C ).
Figure 3.
Representative image of histological changes of eWAT and iBAT. The size of adipocyte evaluated by staining with H&E. (A) Representative H&E staining from eWAT of mice. Scale bar, 100 µm, and 50 µm. (n = 3). (B) Representative H&E staining from iBAT of mice. Scale bar, 100 µm, and 50 µm. (n = 3). (C) The fat diameter of the adipocyte was quantified by Image J (n = 5). Statistical results are presented as the mean ± SEM. **** p < 0.0001 is an indicative significance compared with vehicle-treated right-side fat as self-control.
3.3. TMT-based quantitative proteomics analysis of eWAT
Quantitative proteomic analysis, based on TMT labeling, was conducted on the eWAT of NCD, HFD, and Sema groups (n = 3 mice per each group). A total of 53,911 peptide fragments were used, of which 48,914 were unique peptides corresponding to a total of 7590 proteins ( Figure S1 , Table S1 ). A satisfactory quality deviation was obtained during the data acquisition process using a high-quality Q Exactive mass spectrometer. The mass deviations of all the identified peptides were primarily distributed within 10 ppm, indicating that the identification results were accurate and reliable ( Figure S2A ). A great score with a median of 29.19 was attained, and more than 68.52% of peptides scored higher than 20 when evaluating each MS2 spectrogram ( Figure S2B ). The protein ratio distribution in the Sema/HFD groups is shown in Figure S2C . A 1.2-fold change cut-off, with P < 0.05, was used to indicate significant changes in the abundance of the DEPs in the Sema/HFD groups ( Table S2 ).
3.4. The identification of differentially expressed proteins
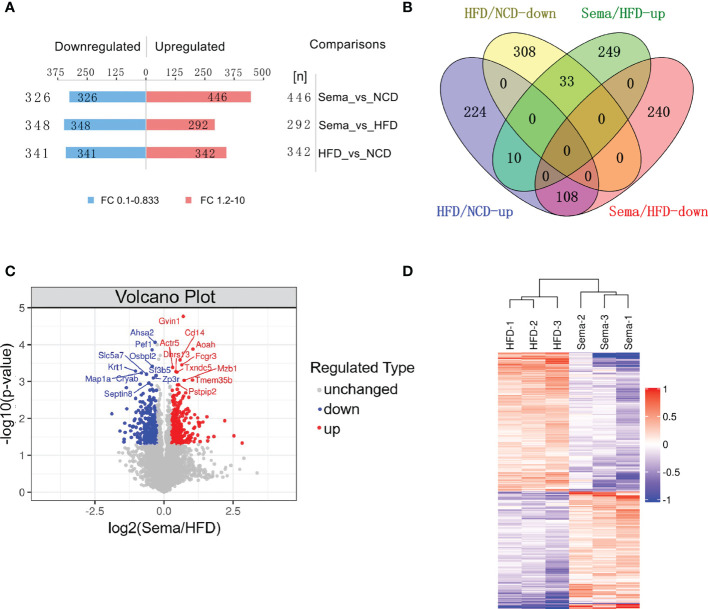
A total of 683 DEPs, 342 up-regulated and 341 down-regulated, were identified in the HFD group versus the NCD group. Semaglutide treatment resulted in 640 DEPs, 292 up-regulated and 348 down-regulated, compared with the HFD group, and semaglutide induced 772 DEPs, 446 up-regulated and 326 down-regulated, compared with the NCD group (n = 3 per each group) ( Figure 4A ). Fold Changes ratios > 1.2 or < 0.83 and P-values (T-test) < 0.05 were considered to be DEPs. The list of the up- and down-regulated proteins between the Sema and HFD groups is shown in Tables S3 and S4 . In addition, a volcano plot and K-means clustering heatmaps were used to show the distribution of significance and fold change of the DEPs between the Sema and HFD groups ( Figures 4C, D ). As shown in Table 1 , several proteins among the top 10 up-regulated DEPs were involved in the lipopolysaccharide catabolic process, cellular response to diacyl bacterial lipopeptide, antibody-dependent cellular cytotoxicity, apoptotic cell clearance, positive regulation of cell proliferation, cell migration including AOAH, CD14, FCGR3, TXND5, MZB1, and PPIP2. The top 10 down-regulated proteins participated in the ER to Golgi vesicle-mediated transport and cholesterol transport, and were the integral component of the membrane. There were 141 overlapping DEPs between Sema/HFD and HFD/NCD in Venn diagram, including 33 down-regulated and 108 up-regulated proteins with HFD reversed by Sema. ( Figure 4B ). The list of the DEPs reversed by semaglutide between HFD/NCD and Sema/HFD ( Table 2 ).
Figure 4.
Identification and quantitative analysis of proteins. (A) The number of DEPs among each group. (B) The number of up-regulated and down-regulated proteins was compared between the two groups in Venn diagram. (C) Volcano plots show the distribution of significance and fold change of identified proteins between the Sema and HFD groups. The blue spots and red spots indicate significantly down-regulated and up-regulated proteins. Red indicates upregulation and blue represents downregulation. (D) Hierarchical clustering of DEPs from the Sema group compared with the HFD group. Red indicates upregulation, and blue represents downregulation.
Table 1.
List of the top 10 DEPs significantly up/down-regulated between the Sema and HFD groups.
| Accession | Protein Name | Gene Name | Sema / HFD |
|---|---|---|---|
| Q80SU7 | Interferon-induced very large GTPase 1 | Gvin1 | 1.626029763 |
| O35298 | Acyloxyacyl hydrolase | Aoah | 2.069990226 |
| P10810 | Monocyte differentiation antigen CD14 | Cd14 | 1.496559752 |
| P08508 | Low affinity immunoglobulin gamma Fc region receptor III | Fcgr3 | 1.566858006 |
| Q80US4 | Actin-related protein 5 | Actr5 | 1.242135008 |
| Q5SS80 | Dehydrogenase/reductase SDR family member 13 | Dhrs13 | 1.349912505 |
| Q91W90 | Thioredoxin domain-containing protein 5 | Txndc5 | 1.381413158 |
| Q3U0Y2 | Transmembrane protein 35B | Tmem35b | 2.04449729 |
| Q9D8I1 | Marginal zone B- and B1-cell-specific protein | Mzb1 | 1.658031446 |
| Q99M15 | Proline-serine-threonine phosphatase-interacting protein 2 | Pstpip2 | 1.443331029 |
| Q8N9S3 | Activator of 90 kDa heat shock protein ATPase homolog 2 | Ahsa2 | 0.801091655 |
| P41242 | Megakaryocyte-associated tyrosine-protein kinase | Matk | 0.568717607 |
| Q9DC63 | F-box only protein 3 | Fbxo3 | 0.722183075 |
| P56375 | Acylphosphatase-2 | Acyp2 | 0.589639337 |
| Q6TCG2 | Membrane progesterone receptor epsilon | Paqr9 | 0.620337924 |
| Q6IME9 | Keratin, type II cytoskeletal 72 | Krt72 | 0.562148059 |
| B1AZA5 | Transmembrane protein 245 | Tmem245 | 0.633014045 |
| Q80WR1 | Tetraspanin-18 | Tspan18 | 0.483043027 |
| Q8BFY6 | Peflin | Pef1 | 0.743245711 |
| Q8BX94 | Oxysterol-binding protein-related protein 2 | Osbpl2 | 0.74768025 |
Table 2.
List of the DEPs reversed by semaglutide between HFD/NCD and Sema/HFD.
| Accession | Protein Name | Gene Name | Sema/HFD | HFD/NCD |
|---|---|---|---|---|
| O35855 | Branched-chain-amino-acid aminotransferase, mitochondrial | Bcat2 | 1.281506424 | 0.687858846 |
| O35930 | Platelet glycoprotein Ib alpha chain | Gp1ba | 1.558792701 | 0.76277469 |
| O88986 | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial | Gcat | 1.282493527 | 0.675599541 |
| P01631 | Ig kappa chain V-II region 26-10 | 2.183937191 | 0.786278394 | |
| P04370 | Myelin basic protein | Mbp | 1.337736286 | 0.66124981 |
| P07934 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | Phkg1 | 1.810779455 | 0.635150402 |
| P08074 | Carbonyl reductase [NADPH] 2 | Cbr2 | 1.422088101 | 0.795488833 |
| P08553 | Neurofilament medium polypeptide | Nefm | 1.538117971 | 0.487675052 |
| P24472 | Glutathione S-transferase A4 | Gsta4 | 1.554097442 | 0.50216426 |
| P27573 | Myelin protein P0 | Mpz | 1.73576827 | 0.507765755 |
| P27931 | Interleukin-1 receptor type 2 | Il1r2 | 1.539524739 | 0.804402067 |
| P51949 | CDK-activating kinase assembly factor MAT1 | Mnat1 | 1.231841048 | 0.717964191 |
| P56565 | Protein S100-A1 | S100a1 | 1.556928633 | 0.560643756 |
| Q05421 | Cytochrome P450 2E1 | Cyp2e1 | 1.481945121 | 0.400138198 |
| Q3TC72 | Fumarylacetoacetate hydrolase domain-containing protein 2A | Fahd2 | 1.346361794 | 0.653543367 |
| Q3U0Y2 | Transmembrane protein 35B | Tmem35b | 2.04449729 | 0.708955297 |
| Q5XJY4 | Presenilins-associated rhomboid-like protein, mitochondrial | Parl | 1.334386893 | 0.758964767 |
| Q61024 | Asparagine synthetase [glutamine-hydrolyzing] | Asns | 1.318686424 | 0.710323152 |
| Q61878 | Bone marrow proteoglycan | Prg2 | 2.936173825 | 0.823299954 |
| Q8BVZ5 | Interleukin-33 | Il33 | 1.346081593 | 0.737888785 |
| Q8C0Q2 | Zinc fingers and homeoboxes protein 3 | Zhx3 | 1.223233196 | 0.698919446 |
| Q8C7H1 | Methylmalonic aciduria type A homolog, mitochondrial | Mmaa | 1.266632979 | 0.738615031 |
| Q8K0T0 | Reticulon-1 | Rtn1 | 1.439219824 | 0.768530397 |
| Q8K3V7 | Major intrinsically disordered Notch2-binding receptor 1 | Minar1 | 1.485521325 | 0.432834834 |
| Q8K4F5 | Protein ABHD11 | Abhd11 | 1.323545844 | 0.778982921 |
| Q91WM2 | Haloacid dehalogenase-like hydrolase domain-containing 5 | Hdhd5 | 1.378265367 | 0.754543827 |
| Q9CR59 | Growth arrest and DNA damage-inducible proteins-interacting protein 1 | Gadd45gip1 | 1.512518484 | 0.783963033 |
| Q9CZ57 | 5-methylcytosine rRNA methyltransferase NSUN4 | Nsun4 | 1.248652722 | 0.829931513 |
| Q9D2R0 | Acetoacetyl-CoA synthetase | Aacs | 1.433225626 | 0.511798207 |
| Q9D2V5 | Protein AAR2 homolog | Aar2 | 1.466674803 | 0.71822847 |
| Q9DCC4 | Pyrroline-5-carboxylate reductase 3 | Pycr3 | 1.245197813 | 0.792313915 |
| Q9ERD7 | Tubulin beta-3 chain | Tubb3 | 1.218067096 | 0.801430453 |
| Q9Z211 | Peroxisomal membrane protein 11A | Pex11a | 1.401647745 | 0.555367229 |
| A2AJ76 | Hemicentin-2 | Hmcn2 | 0.802742454 | 1.288415013 |
| B1AZA5 | Transmembrane protein 245 | Tmem245 | 0.633014045 | 1.349864576 |
| E9Q634 | Unconventional myosin-Ie | Myo1e | 0.62298073 | 1.804548698 |
| E9Q6P5 | Tetratricopeptide repeat protein 7B | Ttc7b | 0.717785347 | 1.220191175 |
| F7BWT7 | Tetraspanin-15 | Tspan15 | 0.647667159 | 1.36638246 |
| O09131 | Glutathione S-transferase omega-1 | Gsto1 | 0.72555588 | 1.231584915 |
| O09164 | Extracellular superoxide dismutase [Cu-Zn] | Sod3 | 0.516482216 | 1.574537595 |
| O35075 | Vacuolar protein sorting-associated protein 26C | Vps26c | 0.608732943 | 1.406154887 |
| O35382 | Exocyst complex component 4 | Exoc4 | 0.824478971 | 1.220650116 |
| O54998 | Peptidyl-prolyl cis-trans isomerase FKBP7 | Fkbp7 | 0.79323375 | 1.245320989 |
| O55186 | CD59A glycoprotein | Cd59a | 0.39357848 | 1.585667381 |
| O70318 | Band 4.1-like protein 2 | Epb41l2 | 0.689916081 | 1.248056386 |
| O70325 | Phospholipid hydroperoxide glutathione peroxidase | Gpx4 | 0.731072157 | 1.281299472 |
| O70571 | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial | Pdk4 | 0.5890158 | 1.589726035 |
| O88495 | Melatonin-related receptor | Gpr50 | 0.592898971 | 1.576243268 |
| O88822 | Lathosterol oxidase | Sc5d | 0.73134654 | 1.251946776 |
| P00493 | Hypoxanthine-guanine phosphoribosyltransferase | Hprt1 | 0.637009885 | 1.455198993 |
| P02468 | Laminin subunit gamma-1 | Lamc1 | 0.626512243 | 1.213661846 |
| P04925 | Major prion protein | Prnp | 0.708954669 | 1.245567387 |
| P09541 | Myosin light chain 4 | Myl4 | 0.67612683 | 1.210510072 |
| P10107 | Annexin A1 | Anxa1 | 0.455736333 | 2.177574731 |
| P13020 | Gelsolin | Gsn | 0.768408546 | 1.318531459 |
| P14069 | Protein S100-A6 | S100a6 | 0.724108013 | 1.470089004 |
| P14824 | Annexin A6 | Anxa6 | 0.7645763 | 1.323836591 |
| P16546 | Spectrin alpha chain, non-erythrocytic 1 | Sptan1 | 0.735131675 | 1.220197343 |
| P20152 | Vimentin | Vim | 0.662676811 | 1.382356005 |
| P23298 | Protein kinase C eta type | Prkch | 0.657262069 | 1.56266634 |
| P23927 | Alpha-crystallin B chain | Cryab | 0.549626596 | 1.555245656 |
| P24288 | Branched-chain-amino-acid aminotransferase, cytosolic | Bcat1 | 0.651778664 | 1.697720252 |
| P25911 | Tyrosine-protein kinase Lyn | Lyn | 0.79279044 | 1.442995613 |
| P28798 | Progranulin | Grn | 0.645125472 | 1.339155655 |
| P41242 | Megakaryocyte-associated tyrosine-protein kinase | Matk | 0.568717607 | 1.70906332 |
| P43883 | Perilipin-2 | Plin2 | 0.610554981 | 1.72825748 |
| P48036 | Annexin A5 | Anxa5 | 0.687029392 | 1.435505226 |
| P49446 | Receptor-type tyrosine-protein phosphatase epsilon | Ptpre | 0.79828216 | 1.636672446 |
| P50427 | Steryl-sulfatase | Sts | 0.695983487 | 1.526770237 |
| P50543 | Protein S100-A11 | S100a11 | 0.682927439 | 1.511632053 |
| P54763 | Ephrin type-B receptor 2 | Ephb2 | 0.665503893 | 1.284773575 |
| P56375 | Acylphosphatase-2 | Acyp2 | 0.589639337 | 1.344562254 |
| P62983 | Ubiquitin-40S ribosomal protein S27a | Rps27a | 0.773990343 | 1.21193522 |
| P68254 | 14-3-3 protein theta | Ywhaq | 0.785198761 | 1.210454097 |
| P70180 | Atrial natriuretic peptide receptor 3 | Npr3 | 0.593193139 | 1.487836231 |
| P70387 | Hereditary hemochromatosis protein homolog | Hfe | 0.768285731 | 1.349846034 |
| P97298 | Pigment epithelium-derived factor | Serpinf1 | 0.747353637 | 1.305666984 |
| P97467 | Peptidyl-glycine alpha-amidating monooxygenase | Pam | 0.579600387 | 1.552475558 |
| Q00612 | Glucose-6-phosphate 1-dehydrogenase X | G6pdx | 0.653932733 | 1.300257577 |
| Q00724 | Retinol-binding protein 4 | Rbp4 | 0.567152695 | 1.364799047 |
| Q00941 | Granulocyte-macrophage colony-stimulating factor receptor subunit alpha | Csf2ra | 0.647337505 | 1.368877519 |
| Q03350 | Thrombospondin-2 | Thbs2 | 0.733103206 | 1.350827671 |
| Q05793 | Basement membrane-specific heparan sulfate proteoglycan core protein | Hspg2 | 0.666620077 | 1.26311188 |
| Q3TTY5 | Keratin, type II cytoskeletal 2 epidermal | Krt2 | 0.551401154 | 1.33451487 |
| Q3UQ28 | Peroxidasin homolog | Pxdn | 0.560483414 | 2.667881936 |
| Q3UV17 | Keratin, type II cytoskeletal 2 oral | Krt76 | 0.526516721 | 1.362560015 |
| Q4ZJN1 | Complement C1q and tumor necrosis factor-related protein 9 | C1qtnf9 | 0.703123667 | 1.397849658 |
| Q562D6 | tRNA (adenine(37)-N6)-methyltransferase | Trmo | 0.269978943 | 2.993587642 |
| Q5SXY1 | Cytospin-B | Specc1 | 0.628059005 | 1.868882268 |
| Q5U4D9 | THO complex subunit 6 homolog | Thoc6 | 0.770677586 | 1.207621793 |
| Q60675 | Laminin subunit alpha-2 | Lama2 | 0.61438 | 1.520674813 |
| Q60870 | Receptor expression-enhancing protein 5 | Reep5 | 0.608420449 | 1.317769204 |
| Q61699 | Heat shock protein 105 kDa | Hsph1 | 0.798913398 | 1.462266446 |
| Q62048 | Astrocytic phosphoprotein PEA-15 | Pea15 | 0.682596546 | 1.34870236 |
| Q62261 | Spectrin beta chain, non-erythrocytic 1 | Sptbn1 | 0.740102084 | 1.214602281 |
| Q64449 | C-type mannose receptor 2 | Mrc2 | 0.63460548 | 1.459026095 |
| Q6IRU5 | Clathrin light chain B | Cltb | 0.762439764 | 1.217178585 |
| Q6TCG2 | Membrane progesterone receptor epsilon | Paqr9 | 0.620337924 | 1.985498298 |
| Q71LX4 | Talin-2 | Tln2 | 0.721065361 | 1.205132269 |
| Q80V53 | Carbohydrate sulfotransferase 14 | Chst14 | 0.666580674 | 1.367212788 |
| Q8BG73 | SH3 domain-binding glutamic acid-rich-like protein 2 | Sh3bgrl2 | 0.755024856 | 1.533962261 |
| Q8BGA2 | LHFPL tetraspan subfamily member 2 protein |
Lhfpl2 | 0.495844074 | 1.439941209 |
| Q8BGY9 | High affinity choline transporter 1 | Slc5a7 | 0.644645782 | 1.980018733 |
| Q8BHL5 | Engulfment and cell motility protein 2 | Elmo2 | 0.797536093 | 1.259214907 |
| Q8BHZ0 | CYFIP-related Rac1 interactor A | Cyria | 0.790382516 | 1.222435043 |
| Q8BI08 | Protein MAL2 | Mal2 | 0.786294133 | 1.441881996 |
| Q8BPS4 | Integral membrane protein GPR180 | Gpr180 | 0.331242807 | 1.361768073 |
| Q8BZ03 | Serine/threonine-protein kinase D2 | Prkd2 | 0.652307623 | 1.231827765 |
| Q8C142 | Low density lipoprotein receptor adapter protein 1 | Ldlrap1 | 0.703602688 | 1.392901081 |
| Q8C147 | Dedicator of cytokinesis protein 8 | Dock8 | 0.789642671 | 1.517773691 |
| Q8CGA4 | Maturin | Mturn | 0.491145 | 1.578948145 |
| Q8CHH9 | Septin-8 | Septin8 | 0.686151817 | 1.372095604 |
| Q8JZK9 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | Hmgcs1 | 0.641253478 | 1.406928093 |
| Q8K3K8 | Optineurin | Optn | 0.680041902 | 1.365399359 |
| Q8R238 | Serine dehydratase-like | Sdsl | 0.70692237 | 1.290868637 |
| Q8VD33 | Small glutamine-rich tetratricopeptide repeat-containing protein beta | Sgtb | 0.472949058 | 1.371492625 |
| Q91XD7 | Protein disulfide isomerase Creld1 | Creld1 | 0.6104103 | 1.326438835 |
| Q91YX5 | Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 | Lpgat1 | 0.701753748 | 1.380005096 |
| Q91ZF0 | DnaJ homolog subfamily C member 24 | Dnajc24 | 0.731687837 | 1.305430535 |
| Q921Q3 | Chitobiosyldiphosphodolichol beta-mannosyltransferase | Alg1 | 0.60958506 | 1.247437542 |
| Q923D4 | Splicing factor 3B subunit 5 | Sf3b5 | 0.821544189 | 1.219278931 |
| Q99J47 | Dehydrogenase/reductase SDR family member 7B | Dhrs7b | 0.71252128 | 1.269246863 |
| Q99PJ0 | Neurotrimin | Ntm | 0.659723271 | 1.360605816 |
| Q9CW42 | Mitochondrial amidoxime-reducing component 1 | Mtarc1 | 0.547778041 | 1.312522546 |
| Q9CWS0 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | Ddah1 | 0.38989088 | 2.640649238 |
| Q9CXT7 | Transmembrane protein 192 | Tmem192 | 0.811599619 | 1.447067984 |
| Q9CY64 | Biliverdin reductase A | Blvra | 0.760695509 | 1.40052152 |
| Q9D379 | Epoxide hydrolase 1 | Ephx1 | 0.700642139 | 1.425222882 |
| Q9D3P8 | Plasminogen receptor (KT) | Plgrkt | 0.660636171 | 1.344838373 |
| Q9D7G0 | Ribose-phosphate pyrophosphokinase 1 | Prps1 | 0.708687594 | 1.652020885 |
| Q9EPL9 | Peroxisomal acyl-coenzyme A oxidase 3 | Acox3 | 0.774813018 | 1.314661776 |
| Q9ES57 | Cell surface glycoprotein CD200 receptor 1 | Cd200r1 | 0.477497904 | 1.726018437 |
| Q9JKX3 | Transferrin receptor protein 2 | Tfr2 | 0.441377047 | 2.209652728 |
| Q9JL62 | Glycolipid transfer protein | Gltp | 0.732512513 | 1.432249415 |
| Q9QYR6 | Microtubule-associated protein 1A | Map1a | 0.567535959 | 1.750284473 |
| Q9R001 | A disintegrin and metalloproteinase with thrombospondin motifs 5 | Adamts5 | 0.635456846 | 1.324770169 |
| Q9R013 | Cathepsin F | Ctsf | 0.613397598 | 1.407441745 |
| Q9Z0F7 | Gamma-synuclein | Sncg | 0.654712098 | 1.337344502 |
| Q9Z0J0 | NPC intracellular cholesterol transporter 2 | Npc2 | 0.549065572 | 1.397981367 |
| Q9Z0Z4 | Hephaestin | Heph | 0.72666994 | 1.281808528 |
| Q9Z138 | Tyrosine-protein kinase transmembrane receptor ROR2 | Ror2 | 0.628599362 | 1.370936865 |
3.5. Functional classification of DEPs
We used the subcellular structure prediction software CELLO to analyze the subcellular location of all the DEPs. DEPs were mainly located in nuclear (34.72% in Sema/HFD, 38.19% in total). The remaining DEPs were mainly located in cytoplasmic (22.73% in Sema/HFD, 25.96% in total), extracellular (16.29% in Sema/HFD, 11.47% in total), plasma membrane (15.15% in Sema/HFD, 11.47% in total) and mitochondrial (9.22% in Sema/HFD, 10.5% in total; Figures 5A, B ).
Figure 5.
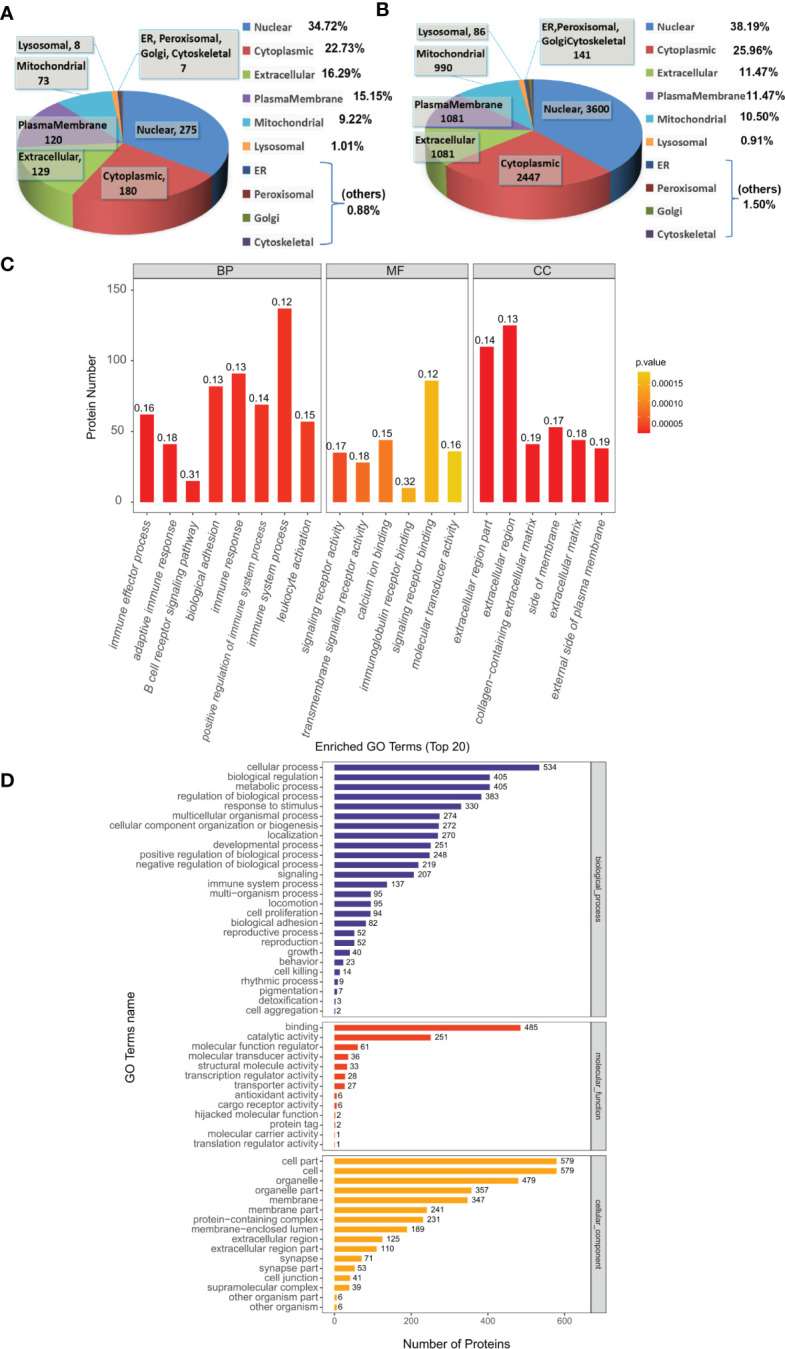
Functional analysis of DEPs. (A, B) Subcellular localization analysis of DEPs in Sema/HFD group and total proteome. The numbers show the proteins located in different subcellular components. (C) Overall enriched GO terms of DEPs between the Sema and HFD groups. BP: biological processes; MF: molecular functions; CC: cellular components. (D) Top 20 enriched GO analyses of DEPs. The color gradient from orange to red represents the p-value. The numbers above the bar charts represent the rich factor (rich factor ≤ 1).
The DEPs were then analyzed against the GO database using three sets of ontologies: biological process (BP), molecular function (MF), and cellular component (CC). In the Sema/HFD group, the most enriched GO terms of BP, MF, and CC were annotated as immune effector process, signaling receptor activity, and extracellular region part, respectively ( Figures 5C, D , Table 3 ). Other important BPs were included in adaptive immune response, B cell receptor signaling pathway, biological adhesion, immune response, positive regulation of immune system process and immune system process, and leukocyte activation ( Figure 5C ). Other important MFs included transmembrane signaling receptor activity, calcium ion binding, immunoglobulin receptor binding, and signaling receptor binding molecular transducer activity ( Figure 5C ). Other important CCs included extracellular region, collagen-containing extracellular matrix, side of membrane, extracellular matrix, and external side of the plasma membrane ( Figure 5C ).
Table 3.
Distribution of proteins and signaling pathways between Sema and HFD groups, based on GO and KEGG analysis.
3.6. KEGG pathways
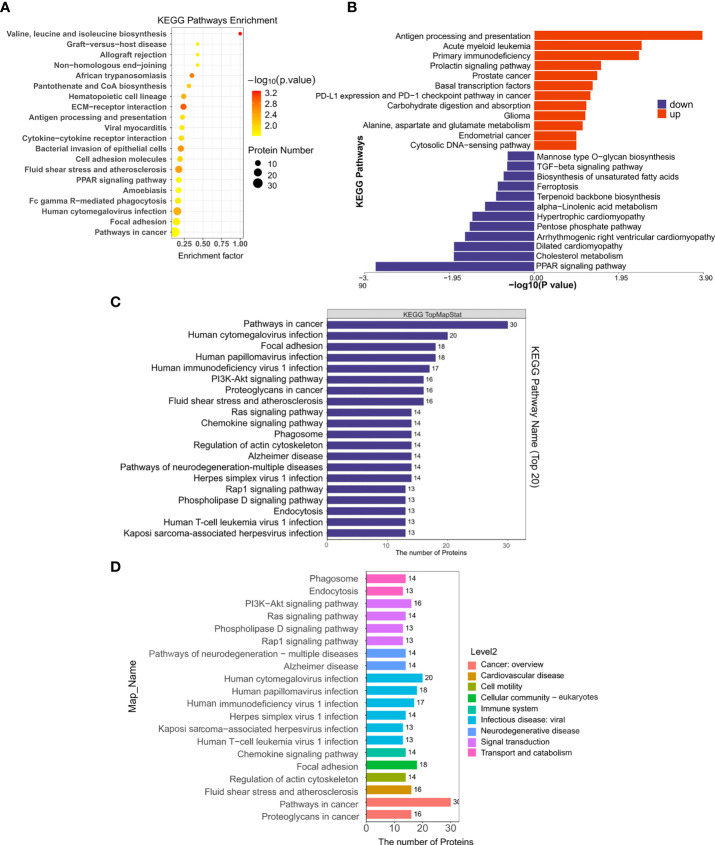
By searching the major biological pathways and relevant regulatory processes involved in the KEGG, we analyzed all of the DEPs in the Sema and HFD groups. Valine, leucine, and isoleucine biosynthesis, ECM-receptor interaction, African trypanosomiasis, fluid shear stress and atherosclerosis, bacterial invasion of epithelial cells, human cytomegalovirus infection, hematopoietic cell lineage, focal adhesion, Fc gamma R-mediated phagocytosis, and cell adhesion molecules, resulted as significant enrichment pathways ( Figure 6A , Table 3 ). Most of the DEPs were enriched in pathways related to cancer and human cytomegalovirus infection ( Figures 6C, D ).
Figure 6.
The KEGG pathway enrichment of DEPs. (A) KEGG pathway enrichment bubble chart. The horizontal axes represent the enrichment factor (enrichment factor ≤ 1). The vertical axis represents the statistical results of DEPs under the Top 20 KEGG pathways. The color gradient from green to red represents the p-value; the closer to the red color, the lower the p-value and the higher the significance level corresponding to the enrichment. (B) Significantly enriched pathways in up/down-regulated DEPs in the Sema/HFD group. (C) Numbers of DEPs under Top 20 KEGG pathways. (D) Numbers of DEPs under level 2 KEGG pathways.
Moreover, up-regulated DEPs were found to be enriched in antigen processing and presentation, human immunodeficiency virus 1 infection, herpes simplex virus 1 infection, acute myeloid leukemia, etc. ( Figure 6B , Table 4 ). Down-regulated DEPs were found to be enriched in the PPAR signaling pathway and cholesterol metabolism ( Figure 6B , Table 4 ). Interestingly, we found 10 proteins involved in the fatty acid uptake, lipid storage, unsaturated fatty acid synthesis, lipid peroxidation, and glycerol efflux down-regulated in Sema/HFD group, including CD36 FABP5, ACSL, ACOX3, PLIN2, ANGPTL4, LPL, MGLL, AQP7, and PDK4 ( Figure 7 , Table 5 ). In addition, the expression of PDK4, ACOX3, PLIN2 were up-regulated with HFD and significantly reversed by Sema. Taken together, these findings suggested that semaglutide treatment might have beneficial effects on adipose tissues through the regulation of lipid uptake, lipid storage, and lipolysis in white adipose tissue ( Figures S3–6 ).
Table 4.
Significantly enriched pathways in up/down regulated DEPs between Sema and HFD groups identified through KEGG pathway enrichment analysis.
| Terms | Count | p Value | FDR | richFactor | Accession | Sema/HFD DEPs Regulated Type |
|---|---|---|---|---|---|---|
| Antigen processing and presentation | 7 | 0.00013769 | 0.033596295 | 0.22580645 | P01899 P97372 P36371 P21958 P06339 Q9R233 P06332 | up |
| Human immunodeficiency virus 1 infection | 14 | 0.000609053 | 0.054097938 | 0.10447761 | P01899 Q60710 P36371 P21958 Q8CIH5 Q05144 P06339 Q9R233 P31938 P31750 Q3TBT3 Q9DB50 P06332 O35904 | up |
| Herpes simplex virus 1 infection | 12 | 0.000665139 | 0.054097938 | 0.11428571 | P01899 P36371 P21958 P06339 Q9R233 Q8R5F7 P31750 Q3TBT3 Q9EQU3 Q8VI93 O35904 Q9EP53 | up |
| Human cytomegalovirus infection | 13 | 0.001234466 | 0.075302447 | 0.1015625 | P01899 P36371 P21958 Q05144 P06339 Q9R233 P31938 P31750 P51675 Q3TBT3 O35904 A3KGF7 Q9EP53 | up |
| Acute myeloid leukemia | 6 | 0.003441545 | 0.13284056 | 0.15384615 | P42232 P31938 P10810 P31750 P17433 O35904 | up |
| Cell adhesion molecules | 7 | 0.003512205 | 0.13284056 | 0.13461539 | P01899 P06339 P13597 P15702 P06332 Q9Z261 P27573 | up |
| Primary immunodeficiency | 3 | 0.003983193 | 0.13284056 | 0.33333333 | P36371 P21958 P06332 | up |
| Natural killer cell mediated cytotoxicity | 7 | 0.004355428 | 0.13284056 | 0.12962963 | P01899 Q8CIH5 Q05144 P06339 P31938 P13597 O35904 | up |
| Human T-cell leukemia virus 1 infection | 10 | 0.005680549 | 0.150034667 | 0.09803922 | P01899 P42232 P06339 P31938 P13597 P31750 P27931 P17433 P06332 O35904 | up |
| Fc gamma R-mediated phagocytosis | 8 | 0.006148962 | 0.150034667 | 0.11111111 | Q8CIH5 Q05144 P08101 P31938 P28667 P31750 Q7TQF7 O35904 | up |
| ECM-receptor interaction | 10 | 3.35E-05 | 0.008284126 | 0.2173913 | Q05793 Q61292 P97927 P02469 Q60675 P09055 Q08857 P43406 O70309 Q03350 | down |
| PPAR signaling pathway | 10 | 0.000226586 | 0.025015808 | 0.1754386 | P41216 Q62417 Q05816 Q08857 P43883 Q9EPL9 Q8JZK9 P11152 Q9Z1P8 O54794 | down |
| Bacterial invasion of epithelial cells | 10 | 0.000303836 | 0.025015808 | 0.16949153 | P09055 P49817 Q9WVC3 Q8CHH9 P42208 Q8BZ98 Q8BHL5 Q6IRU5 Q61140 Q9QYY0 | down |
| Focal adhesion | 14 | 0.001675795 | 0.103480359 | 0.112 | Q71LX4 Q61292 P97927 P02469 P57780 Q60675 P09055 P49817 Q9WVC3 P43406 O70309 Q61140 Q03350 P70182 | down |
| Valine, leucine and isoleucine biosynthesis | 2 | 0.006098131 | 0.301247675 | 0.66666667 | Q8R238 P24288 | down |
| Small cell lung cancer | 7 | 0.011087266 | 0.44441595 | 0.12962963 | Q61292 P97927 P02469 Q60675 P09055 P43406 O89106 | down |
| Cholesterol metabolism | 5 | 0.014282383 | 0.44441595 | 0.15625 | Q08857 Q9Z0J0 P11152 Q9Z1P8 Q8C142 | down |
| Dilated cardiomyopathy | 6 | 0.014394039 | 0.44441595 | 0.13636364 | P48678 Q60675 P09055 P43406 O70309 P97490 | down |
| Arrhythmogenic right ventricular cardiomyopathy | 5 | 0.025679591 | 0.704762112 | 0.13513514 | P48678 Q60675 P09055 P43406 O70309 | down |
| Pentose phosphate pathway | 4 | 0.033096704 | 0.8174886 | 0.14814815 | P05064 Q00612 Q9D7G0 Q8R1Q9 | down |
Figure 7.
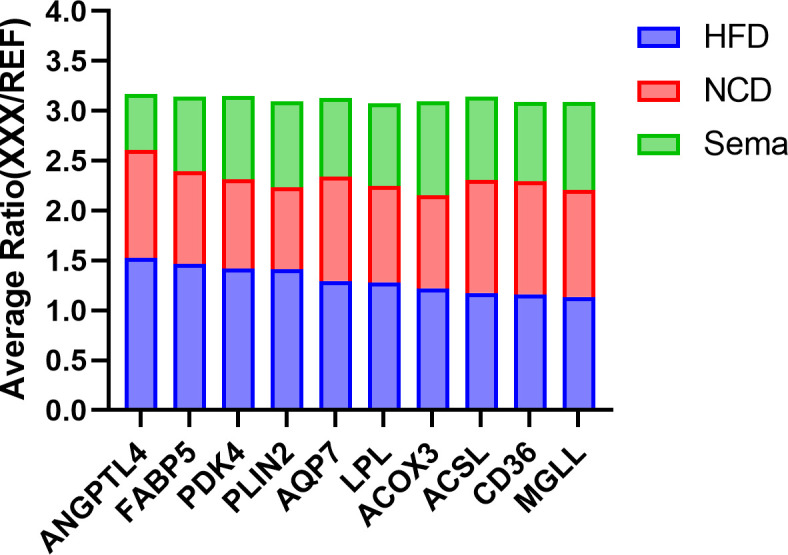
The Average Ratio (XXX/REF) of lipid metabolism proteins in HFD, NCD, and Sema groups. Mean relative protein expression of the sample group relative to the internal reference (REF).
Table 5.
DEPs down -regulated in the Sema/HFD group modulating the lipid metabolism.
| Accession | Protein Name | Gene Name | Sema / HFD |
|---|---|---|---|
| P41216 | Long-chain-fatty-acid--CoA ligase 1 | Acsl1 | 0.714539589 |
| Q05816 | Fatty acid-binding protein 5 | Fabp5 | 0.513802071 |
| O54794 | Aquaporin-7 | Aqp7 | 0.6124858 |
| P43883 | Perilipin-2 | Plin2 | 0.610554981 |
| O35678 | Monoglyceride lipase | Mgll | 0.775676329 |
| Q9EPL9 | Peroxisomal acyl-coenzyme A oxidase 3 | Acox3 | 0.774813018 |
| Q9Z1P8 | Angiopoietin-related protein 4 | Angptl4 | 0.366305511 |
| P11152 | Lipoprotein lipase | Lpl | 0.649312671 |
| Q08857 | Platelet glycoprotein 4 | Cd36 | 0.691987249 |
| O70571 | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial | Pdk4 | 0.5890158 |
4. Discussion
Obesity is a highly prevalent, chronic, relapsing disease requiring long-term management (19–21). Semaglutide is a potent long-acting glucagon-like peptide-1 (GLP-1) analogue that requires once-weekly administration (22). It regulates blood glucose via the incretin pathway, stimulating insulin and inhibiting glucagon secretion in a glucose-dependent manner, leading to lower blood glucose levels with low risk for hypoglycaemia (23). Semaglutide provides weight-loss effects, and greater reduction in fat mass than that reported with liraglutide (3). The Semaglutide Treatment Effect in People with obesity (STEP) clinical trial programme provides evidences that improvements in cardiometabolic risk factors, including high blood pressure, atherogenic lipids and benefits on physical function and quality of life were seen with semaglutide 2.4 mg (24).
To determine the role of semaglutide in obese mice, we performed intraperitoneally injections of semaglutide to HFD-induced obese mice, whereas the NCD and HFD groups were treated with saline for 12 weeks. As expected, we observed that semaglutide exerted a weight-loss effect by lowering the proportion of visceral fat mass and increasing the brown fat mass relative to total body mass. Both plasma lipids (cholesterol and triglycerides) and plasma lipoprotein (LDL) levels were significantly decreased after semaglutide treatment. Semaglutide significantly reduced blood glucose concentrations in glucose tolerance tests. However, plasma levels of fasting insulin decreased significantly in the semaglutide treated mice, when it is known that semaglutide does the opposite in humans (25). A potential factor explaining this observation may be the loss of body weight during the course of the experiment, possibly impacting insulin resistances. A large board of studies and clinical correlations has suggested that hyperinsulinemia is associated with obesity, and there is a close relationship between hyperinsulinemia and dyslipidemia (26, 27). Recent in vivo evidence has also shown that reducing circulating insulin levels may protect and reverse adiposity, insulin resistance, and hyperglycemia that is associated with obesity (28). A previous study discovered that plasma fasting insulin and leptin levels decreased in the GLP-1 (rhGLP-1) Beinaglutide (BN) treated mice, suggesting that BN could reduce hyperinsulinemia and hyperleptinemia associated with obesity (29). Based on the above findings, in this study we indicate that semaglutide could reduce hyperinsulinemia and promote the insulin sensitivity in obese mice.
Adipose tissues are important regulators of whole-body energy homeostasis. In this study, we focused on the role of semaglutide on adipose metabolism and found that it has several beneficial effects on the adipose tissues of obese mice.
First, cell size and turnover of adipocytes are major determinants of fat tissue metabolism and mass, the alterations of which are associated with pathological conditions (16). Semaglutide-treated mice not only show dramatically reduced adipose tissue weight, H&E staining also showed that the large lipid droplets with distinct morphologies in the adipocytes of obese mice were alleviated by semaglutide. Hypertrophy, an increase in cell size of adipocyte is closely associated with dyslipidemia and insulin resistance in humans (30, 31). A reduction in the diameter of adipocytes induced by semaglutide may indicate improvement of insulin sensitivity in the fat tissues of obese mice.
Second, although the changes of adipose tissue respond to GLP-1 analogues treatment has been studied from the gene expression, lipidomic perspectives (29, 32),comprehensive protein level profiling is rare. Gene expression and protein expression diverge (33). Profiling the adipose tissue proteomic signatures identified pathways hat were not available through gene expression studies (34).
To provide greater insight into the role of semaglutide in obese white adipose tissue, we applied TMT proteomic approach to analyze a total of 7553 proteins quantifiable in the three experimental groups. There were 683 DEPs in the HFD/NCD group and 640 DEPs in the HFD/Sema group, with a total of 141 significant overlapping DEPs. Previous study provided evidence for the presence of GLP-1 receptor in adipose tissue and show that its mRNA and protein expressions increased in visceral adipose depots from morbidly obese patients with a high degree of Insulin resistances (IR) (7). Interestingly, protein O35659 was not identified by TMT LC-MS in our study, which was consistent with another study of h rhGLP-1 Beinaglutide (BN) on adipose tissue (29). Potential explanations of this observation may be that the fat loss effect by semaglutide during the course of the experiment, might lower the degree of IR, leading to a lower expression of Glpr in mice, or the beneficial effects of GLP-1 are associated with the activation of not only the canonical GLP-1 receptor but also an additional, as yet unknown, receptor (8), the effects of semaglutide on adipose tissues might not be through its regulation on expression of GLP-1R. This phenomenon may also be due to technical limitations that the protein extraction does not efficiently extract membrane proteins, the hydrophobic properties of these proteins make full structural and functional characterization challenging because of the need to use detergents or other solubilizing agents when extracting them from their native lipid membranes (35).
It is well established that GLP-1/GLP-1R functions through activation of down-stream PKA-, MAPK-, or AMPK-related signaling pathways in various cells. Xu et al. investigated the contribution of brown remodelling of WAT to the weight-lowering-effect induced by the GLP-1R agonist exenatide, and researched the role of SIRT1 in this process (32). Their findings indicated that exenatide induces the phosphorylation of AMPK which, in turn, activates SIRT1 by regulating NAD+ concentration, triggering a lipolytic cycle. Our data have also shown that semaglutide stimulates the expression of Q8C078 (Camkk2) up-regulated in the obese adipose tissue ( Table S3 ). AMPKs can be activated by a Ca2+-dependent pathway mediated by CaMKKb (36, 37). It was indicated that the actions of GLP-1 on VAT should be modulated by sympathetic tone to activate AMPK, leading to decreased lipogenesis and reduced triglyceride content (38). We indicate that semaglutide can activate cAMP-associated signaling. The KEGG analyses of the proteomic data have also shown that PI3K-AKT and PPAR signaling pathways and several lipid-metabolism-related pathways are among the top enriched pathways in the obese adipose tissue with semaglutide treatment.
Notably, the KEGG analyses have shown that down-regulated DEPs in Sema/HFD group were found to be enriched in the cholesterol metabolism and PPAR signaling pathways. These results suggest that semaglutide down-regulates several lipid-metabolism-related proteins in adipose tissues, it might be reducing PPAR pathway activity when PPAR gamma (39) is good for metabolic health. A potential explanation of this observation may be that semaglutide could also be acting on other signals resulted in reducing PPAR activity in the white adipose tissue, but is associated with reducing visceral fat accumulation and plasma lipid level and increasing insulin sensitivity. Shao et al. found that with liraglutide treatment, the lipogenetic transcription factors PPARγ and C/EBPα expressions were both reduced with AMPK activation and Akt suppression in visceral adipose tissue (VAT), which was associated with reduction of lipogenetic process in VAT (10). A recent study also found that GLP‐1 significantly decreased the expression levels of two key markers of adipocyte differentiation, PPARγ and FABP4, as well as increased that of adiponectin in VAT explants from morbidly obese patients, compared with untreated controls (8). This can be interpreted as the reduction of VAT after weight loss with semaglutide treatment might be related to the adipogenesis decreases and lipolysis increases to avoid more fat accumulation and improve metabolic health, which deserves further investigation.
Based on the results of the study, we found 10 proteins significantly concentrated in lipid metabolism down-regulated in Sema/HFD group, including fatty acid transport and oxidation (CD36 FABP5, ACSL, ACOX3). CD36 is a multifunctional glycoprotein that has a critical role in LCFA uptake and transport in adipocytes (40–42). A previous study indicated that during fat depot expansion, CD36 deficiency negatively affects preadipocyte recruitment in mature adipocytes (43). Fatty acid-binding protein 5 (FABP5) delivers specific fatty acids from the cytosol to the nucleus, wherein they activate nuclear receptors (44) and modulate inflammation by regulating PTGES induction via NF-kappa-B activation. Long-chain-fatty-acid–CoA ligase 1 (ACSL) plays important roles in lipid metabolism for accelerating fatty acid biosynthesis by both synthesis of cellular lipids, and degradation via beta-oxidation (45). Peroxisomal acyl-coenzyme A oxidase 3 (ACOX3) positively regulates fatty acid oxidation (46).
Besides, LPL, PLIN2, ANGPTL4 which positively regulate the process of preadipocyte differentiation (46–48), involved in the adipocyte hypertrophy. Hypertrophic adipocytes are associated with increased insulin resistance (49–51). And MGLL, AQP7, PDK4 all positively regulate energy use in maintaining metabolic balance between carbohydrates and lipids (52–54). All of these 10 proteins had significantly higher expression in the HFD group than in the Sema group and NCD group. In addition, the expression of several proteins was significantly lower in Sema group than that in NCD group ( Figure 7 ). It indicates that sema might alleviate the HFD-induced adipose tissue expansion by down-regulated proteins related to the lipid metabolism, both biosynthesis and degradation.
Conclusion: we indicate that semaglutide treatment might have beneficial effects on adipose tissues through the regulation of lipid uptake, lipid storage, and lipolysis in white adipose tissue.
5. Conclusion
In the present study, we used a quantitative proteomics approach to investigate alterations of eWAT protein expression in obese mice and examined the therapeutic effect of semaglutide on peripheral metabolic health. Our results suggested that semaglutide could remarkably improve glucose and lipid metabolism, and we indicate that semaglutide treatment might have beneficial effects on adipose tissues through the regulation of lipid uptake, lipid storage, and lipolysis in white adipose tissue.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Hebei General Hospital institutional animal care and conducted according to the AAALAC and the IACUC guidelines.
Author contributions
RZ and SC carried out the studies, participated in collecting data, and drafted the manuscript. RZ and SC performed the statistical analysis and participated in its design. RZ and SC participated in the acquisition, analysis, or interpretation of data and drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the Hebei Provincial Central-leading Local Science and Technology Development funds Project (206Z7702G).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1095432/full#supplementary-material
References
- 1. Lustig RH, Collier D, Kassotis C, Roepke TA, Kim MJ, Blanc E, et al. Obesity I: Overview and molecular and biochemical mechanisms. Biochem Pharmacol (2022) 199:115012. doi: 10.1016/j.bcp.2022.115012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koene RJ, Prizment AE, Blaes A, Konety SH. Shared risk factors in cardiovascular disease and cancer. Circulation (2016) 133(11):1104–14. doi: 10.1161/circulationaha.115.020406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wilding JPH, Batterham RL, Calanna S, Davies M, Van Gaal LF, Lingvay I, et al. Once-weekly semaglutide in adults with overweight or obesity. New Engl J Med (2021) 384(11):989–1002. doi: 10.1056/NEJMoa2032183 [DOI] [PubMed] [Google Scholar]
- 4. Gabery S, Salinas CG, Paulsen SJ, Ahnfelt-Rønne J, Alanentalo T, Baquero AF, et al. Semaglutide lowers body weight in rodents via distributed neural pathways. JCI Insight (2020) 5(6). doi: 10.1172/jci.insight.133429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pontes-da-Silva RM, de Souza Marinho T, de Macedo Cardoso LE, Mandarim-de-Lacerda CA, Aguila MB. Obese mice weight loss role on nonalcoholic fatty liver disease and endoplasmic reticulum stress treated by a glp-1 receptor agonist. Int J Obes (2005) (2022) 46(1):21–9. doi: 10.1038/s41366-021-00955-7 [DOI] [PubMed] [Google Scholar]
- 6. Marinho TS, Martins FF, Cardoso LEM, Aguila MB, Mandarim-de-Lacerda CA. Pancreatic islet cells disarray, apoptosis, and proliferation in obese mice. Role Semaglutide Treat Biochimie (2022) 193:126–36. doi: 10.1016/j.biochi.2021.10.017 [DOI] [PubMed] [Google Scholar]
- 7. Vendrell J, El Bekay R, Peral B, García-Fuentes E, Megia A, Macias-Gonzalez M, et al. Study of the potential association of adipose tissue glp-1 receptor with obesity and insulin resistance. Endocrinology (2011) 152(11):4072–9. doi: 10.1210/en.2011-1070 [DOI] [PubMed] [Google Scholar]
- 8. El Bekay R, Coín-Aragüez L, Fernández-García D, Oliva-Olivera W, Bernal-López R, Clemente-Postigo M, et al. Effects of glucagon-like peptide-1 on the differentiation and metabolism of human adipocytes. Br J Pharmacol (2016) 173(11):1820–34. doi: 10.1111/bph.13481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang N, Liu C, Zhang Y, Xu D, Gui L, Lu Y, et al. Liraglutide regulates lipid metabolism via FGF21- LKB1- AMPK- ACC1 pathway in white adipose tissues and macrophage of type 2 diabetic mice. Biochem Biophys Res Commun (2021) 548:120–6. doi: 10.1016/j.bbrc.2021.02.065 [DOI] [PubMed] [Google Scholar]
- 10. Shao Y, Yuan G, Zhang J, Guo X. Liraglutide reduces lipogenetic signals in visceral adipose of Db/Db mice with ampk activation and akt suppression. Drug design Dev Ther (2015) 9:1177–84. doi: 10.2147/dddt.S79175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dozio E, Vianello E, Malavazos AE, Tacchini L, Schmitz G, Iacobellis G, et al. Epicardial adipose tissue GLP-1 receptor is associated with genes involved in fatty acid oxidation and white-to-brown fat differentiation: A target to modulate cardiovascular risk? Int J Cardiol (2019) 292:218–24. doi: 10.1016/j.ijcard.2019.04.039 [DOI] [PubMed] [Google Scholar]
- 12. Wan Y, Bao X, Huang J, Zhang X, Liu W, Cui Q, et al. Novel GLP-1 analog supaglutide reduces HFD-induced obesity associated with increased ucp-1 in white adipose tissue in mice. Front Physiol (2017) 8:294. doi: 10.3389/fphys.2017.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gutierrez AD, Gao Z, Hamidi V, Zhu L, Saint Andre KB, Riggs K, et al. Anti-diabetic effects of GLP1 analogs are mediated by thermogenic interleukin-6 signaling in adipocytes. Cell Rep Med (2022) 3(11):100813. doi: 10.1016/j.xcrm.2022.100813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee YS, Park MS, Choung JS, Kim SS, Oh HH, Choi CS, et al. Glucagon-like peptide-1 inhibits adipose tissue macrophage infiltration and inflammation in an obese mouse model of diabetes. Diabetologia (2012) 55(9):2456–68. doi: 10.1007/s00125-012-2592-3 [DOI] [PubMed] [Google Scholar]
- 15. Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature (2006) 444(7121):847–53. doi: 10.1038/nature05483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morigny P, Boucher J, Arner P, Langin D. Lipid and glucose metabolism in white adipocytes: Pathways, dysfunction and therapeutics. Nat Rev Endocrinol (2021) 17(5):276–95. doi: 10.1038/s41574-021-00471-8 [DOI] [PubMed] [Google Scholar]
- 17. Stubbins RE, Najjar K, Holcomb VB, Hong J, Núñez NP. Oestrogen alters adipocyte biology and protects female mice from adipocyte inflammation and insulin resistance. Diabetes Obes Metab (2012) 14(1):58–66. doi: 10.1111/j.1463-1326.2011.01488.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods (2009) 6(5):359–62. doi: 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 19. Frühbeck G, Busetto L, Dicker D, Yumuk V, Goossens GH, Hebebrand J, et al. The abcd of obesity: An easo position statement on a diagnostic term with clinical and scientific implications. Obes facts (2019) 12(2):131–6. doi: 10.1159/000497124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet (London England) (2016) 387(10026):1377–96. doi: 10.1016/s0140-6736(16)30054-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bray GA, Kim KK, Wilding JPH. Obesity: A chronic relapsing progressive disease process. a position statement of the world obesity federation. Obes Rev (2017) 18(7):715–23. doi: 10.1111/obr.12551 [DOI] [PubMed] [Google Scholar]
- 22. Blundell J, Finlayson G, Axelsen M, Flint A, Gibbons C, Kvist T, et al. Effects of once-weekly semaglutide on appetite, energy intake, control of eating, food preference and body weight in subjects with obesity. Diabetes Obes Metab (2017) 19(9):1242–51. doi: 10.1111/dom.12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Donath MY, Burcelin R. Glp-1 effects on islets: Hormonal, neuronal, or paracrine? Diabetes Care (2013) 36 Suppl 2(Suppl 2):S145–8. doi: 10.2337/dcS13-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bergmann NC, Davies MJ, Lingvay I, Knop FK. Semaglutide for the treatment of overweight and obesity: A review. Diabetes Obes Metab (2023) 25(1):18–35. doi: 10.1111/dom.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kapitza C, Dahl K, Jacobsen JB, Axelsen MB, Flint A. Effects of semaglutide on beta cell function and glycaemic control in participants with type 2 diabetes: A randomised, double-blind, placebo-controlled trial. Diabetologia (2017) 60(8):1390–9. doi: 10.1007/s00125-017-4289-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corkey BE. Banting lecture 2011: Hyperinsulinemia: Cause or consequence? Diabetes (2012) 61(1):4–13. doi: 10.2337/db11-1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Erion KA, Corkey BE. Hyperinsulinemia: A cause of obesity? Curr Obes Rep (2017) 6(2):178–86. doi: 10.1007/s13679-017-0261-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page MM, Johnson JD. Mild suppression of hyperinsulinemia to treat obesity and insulin resistance. Trends Endocrinol metabolism: TEM (2018) 29(6):389–99. doi: 10.1016/j.tem.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 29. Zhang F, Chen Z, Wu D, Tian L, Chen Q, Ye Y, et al. Recombinant human glp-1 beinaglutide regulates lipid metabolism of adipose tissues in diet-induced obese mice. iScience (2021) 24(12):103382. doi: 10.1016/j.isci.2021.103382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffstedt J, Arner E, Wahrenberg H, Andersson DP, Qvisth V, Löfgren P, et al. Regional impact of adipose tissue morphology on the metabolic profile in morbid obesity. Diabetologia (2010) 53(12):2496–503. doi: 10.1007/s00125-010-1889-3 [DOI] [PubMed] [Google Scholar]
- 31. Veilleux A, Caron-Jobin M, Noël S, Laberge PY, Tchernof A. Visceral adipocyte hypertrophy is associated with dyslipidemia independent of body composition and fat distribution in women. Diabetes (2011) 60(5):1504–11. doi: 10.2337/db10-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu F, Lin B, Zheng X, Chen Z, Cao H, Xu H, et al. Glp-1 receptor agonist promotes brown remodelling in mouse white adipose tissue through Sirt1. Diabetologia (2016) 59(5):1059–69. doi: 10.1007/s00125-016-3896-5 [DOI] [PubMed] [Google Scholar]
- 33. Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, et al. Comparative analysis of proteome and transcriptome variation in mouse. PloS Genet (2011) 7(6):e1001393. doi: 10.1371/journal.pgen.1001393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plubell DL, Wilmarth PA, Zhao Y, Fenton AM, Minnier J, Reddy AP, et al. Extended multiplexing of tandem mass tags (Tmt) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol Cell Proteomics MCP (2017) 16(5):873–90. doi: 10.1074/mcp.M116.065524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rawlings AE. Membrane proteins: Always an insoluble problem? Biochem Soc Trans (2016) 44(3):790–5. doi: 10.1042/bst20160025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Investigating the mechanism for amp activation of the amp-activated protein kinase cascade. Biochem J (2007) 403(1):139–48. doi: 10.1042/bj20061520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin F, Ribar TJ, Means AR. The Ca2+/Calmodulin-dependent protein kinase kinase, Camkk2, inhibits preadipocyte differentiation. Endocrinology (2011) 152(10):3668–79. doi: 10.1210/en.2011-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nogueiras R, Pérez-Tilve D, Veyrat-Durebex C, Morgan DA, Varela L, Haynes WG, et al. Direct control of peripheral lipid deposition by cns glp-1 receptor signaling is mediated by the sympathetic nervous system and blunted in diet-induced obesity. J Neurosci Off J Soc Neurosci (2009) 29(18):5916–25. doi: 10.1523/jneurosci.5977-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marion-Letellier R, Savoye G, Ghosh S. Fatty acids, eicosanoids and ppar gamma. Eur J Pharmacol (2016) 785:44–9. doi: 10.1016/j.ejphar.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 40. Glatz JF, Luiken JJ. From fat to fat (Cd36/Sr-B2): Understanding the regulation of cellular fatty acid uptake. Biochimie (2017) 136:21–6. doi: 10.1016/j.biochi.2016.12.007 [DOI] [PubMed] [Google Scholar]
- 41. Pohl J, Ring A, Korkmaz U, Ehehalt R, Stremmel W. Fat/Cd36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Mol Biol Cell (2005) 16(1):24–31. doi: 10.1091/mbc.e04-07-0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coburn CT, Knapp FF, Jr., Febbraio M, Beets AL, Silverstein RL, Abumrad NA. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of Cd36 knockout mice. J Biol Chem (2000) 275(42):32523–9. doi: 10.1074/jbc.M003826200 [DOI] [PubMed] [Google Scholar]
- 43. Vroegrijk IO, van Klinken JB, van Diepen JA, van den Berg SA, Febbraio M, Steinbusch LK, et al. Cd36 is important for adipocyte recruitment and affects lipolysis. Obes (Silver Spring Md) (2013) 21(10):2037–45. doi: 10.1002/oby.20354 [DOI] [PubMed] [Google Scholar]
- 44. Kaczocha M, Vivieca S, Sun J, Glaser ST, Deutsch DG. Fatty acid-binding proteins transport n-acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J Biol Chem (2012) 287(5):3415–24. doi: 10.1074/jbc.M111.304907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, et al. Structure and regulation of rat long-chain acyl-coa synthetase. J Biol Chem (1990) 265(15):8681–5. doi: 10.1016/S0021-9258(19)38942-2 [DOI] [PubMed] [Google Scholar]
- 46. Weinstock PH, Bisgaier CL, Aalto-Setälä K, Radner H, Ramakrishnan R, Levak-Frank S, et al. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest (1995) 96(6):2555–68. doi: 10.1172/jci118319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3-12, adipophilin, and Tip47 package lipid in adipocytes. J Biol Chem (2005) 280(19):19146–55. doi: 10.1074/jbc.M500978200 [DOI] [PubMed] [Google Scholar]
- 48. Yin W, Romeo S, Chang S, Grishin NV, Hobbs HH, Cohen JC. Genetic variation in Angptl4 provides insights into protein processing and function. J Biol Chem (2009) 284(19):13213–22. doi: 10.1074/jbc.M900553200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arner E, Westermark PO, Spalding KL, Britton T, Rydén M, Frisén J, et al. Adipocyte turnover: relevance to human adipose tissue morphology. Diabetes (2010) 59(1):105–9. doi: 10.2337/db09-0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type ii diabetes independent of insulin resistance. Diabetologia (2000) 43(12):1498–506. doi: 10.1007/s001250051560 [DOI] [PubMed] [Google Scholar]
- 51. Lundgren M, Svensson M, Lindmark S, Renström F, Ruge T, Eriksson JW. Fat cell enlargement is an independent marker of insulin resistance and 'Hyperleptinaemia'. Diabetologia (2007) 50(3):625–33. doi: 10.1007/s00125-006-0572-1 [DOI] [PubMed] [Google Scholar]
- 52. Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. Cdna cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem (1997) 272(43):27218–23. doi: 10.1074/jbc.272.43.27218 [DOI] [PubMed] [Google Scholar]
- 53. Maeda N, Funahashi T, Hibuse T, Nagasawa A, Kishida K, Kuriyama H, et al. Adaptation to fasting by glycerol transport through aquaporin 7 in adipose tissue. Proc Natl Acad Sci United States America (2004) 101(51):17801–6. doi: 10.1073/pnas.0406230101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeoung NH, Wu P, Joshi MA, Jaskiewicz J, Bock CB, Depaoli-Roach AA, et al. Role of pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) in glucose homoeostasis during starvation. Biochem J (2006) 397(3):417–25. doi: 10.1042/bj20060125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.