Abstract
Apoptosis is a process of programmed cell death in which a cell commits suicide while maintaining the integrity and architecture of the tissue as a whole. Apoptosis involves activation of one of two major pathways: the extrinsic pathway, where extracellular pro-apoptotic signals, transduced through plasma membrane death receptors, activate a caspase cascade leading to apoptosis. The second, the intrinsic apoptotic pathway, where damaged DNA, oxidative stress, or chemicals, induce the release of pro-apoptotic proteins from the mitochondria, leading to the activation of caspase-dependent and independent apoptosis. However, it has recently become apparent that proteins involved in apoptosis also exhibit non-cell death-related physiological functions that are related to the cell cycle, differentiation, metabolism, inflammation or immunity. Such non-conventional activities were predominantly reported in non-cancer cells although, recently, such a dual function for pro-apoptotic proteins has also been reported in cancers where they are overexpressed. Interestingly, some apoptotic proteins translocate to the nucleus in order to perform a non-apoptotic function. In this review, we summarize the unconventional roles of the apoptotic proteins from a functional perspective, while focusing on two mitochondrial proteins: VDAC1 and SMAC/Diablo. Despite having pro-apoptotic functions, these proteins are overexpressed in cancers and this apparent paradox and the associated pathophysiological implications will be discussed. We will also present possible mechanisms underlying the switch from apoptotic to non-apoptotic activities although a deeper investigation into the process awaits further study.
Keywords: Apoptosis, Cancer, Metabolism, Mitochondria, SMAC/Diablo, VDAC1
Introduction
In recent years, it has become clear that many proteins have more than their main well-characterized functions, possessing additional functions. Among these are pro-apoptotic proteins which can also perform non-apoptotic functions Apoptosis is a tightly regulated process of programmed cell death that plays an important role in tissue regeneration, in the development of normal organs in the embryonic stage, and in many other cellular processes. A number of pro-apoptotic proteins reside in the mitochondrial intermembrane space (IMS) and are released into the cytosol in response to an apoptotic signal, leading to caspase activation, where caspase activity is considered a hallmark of apoptosis [1].
In recent years, it has become clear that in addition to their well-characterized activities, many pro-apoptotic proteins also possess additional non-apoptotic functions [2–4] and, apparently paradoxically, many are overexpressed in a variety of cancers.
Here, we describe about 15 apoptotic proteins that also exhibit non-apoptotic activity, while focusing on 2 mitochondrial proteins that are overexpressed in cancer. The first is VDAC1, which is a mitochondrial gatekeeper and multifunctional protein involved in energy production and metabolism, Ca2+ homeostasis, and intracellular communication between the endoplasmic reticulum (ER) and mitochondria, as well as mitochondrial fission, autophagy, inflammation, and apoptosis [5–10]. Depletion of VDAC1was found to re-program cell energy and alter the expression and functions of several pro-apoptotic proteins with no activation of apoptosis, suggesting a cell energy level-controlled shift from a pro-apoptotic to a non-apoptotic state [11].
The second protein is the second mitochondria-derived activator of caspase/inhibitor of apoptosis-binding protein with low pI (SMAC/Diablo), which is released from the mitochondria to the cytosol in response to diverse apoptotic stimuli and activates apoptosis. However, SMAC is also overexpressed in cancers where it participates in non-apoptotic processes including cell proliferation in the tumor microenvironment, inflammation, the immune system, and lipid/phospholipid synthesis [12–14].
The duality of pro-apoptotic proteins also being involved in multiple processes essential for tumor growth and progression, suggests a potential as druggable treatment targets.
Apoptosis pathways
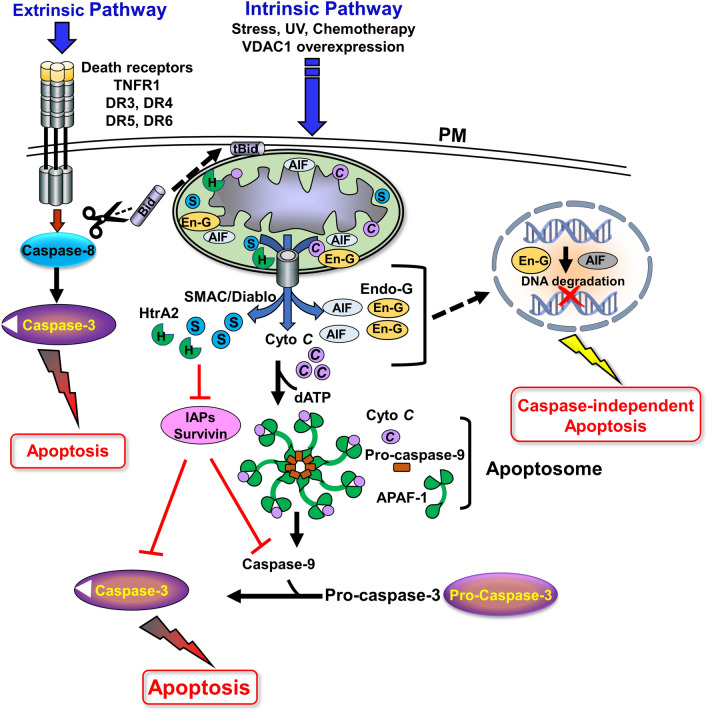
There are two major apoptotic signaling pathways [1]. The extrinsic pathway is induced by external ligands that bind to cell surface death receptors, such as Fas/CD95, tumor necrosis factor (TNFα), tumor necrosis factor receptor 1 and 2 (TNFR1, TNFR2), the TNF-related apoptosis-inducing ligand (TRAIL) receptors, and death receptors 4 and 5 (DR4 and DR5). Activation of these receptors promotes the formation of a death signaling complex (DISC), leading to activation of pro-caspase-8, which activates caspases-3 and -7 for the final execution of cell death [15, 16]. Caspase-8 can also activate mitochondria-mediated apoptosis by cleaving Bid and the cleaved Bid (tBid) then activates Bax and/or Bak, to mediate the release of pro-apoptotic proteins from the mitochondria, thereby, activating apoptosis.
The second pathway, the intrinsic mitochondrial-dependent apoptotic pathway, can be activated by intracellular signals such as DNA damage, Ca2+ overload, increased reactive oxygen species (ROS) levels, chemotherapy drugs [1], and VDAC1 overexpression [17]. The apoptotic signals change the permeability of the outer mitochondrial membrane (OMM), leading to the release of the apoptotic factors residing in the IMS. These released apoptotic proteins include cytochrome c (Cyto c), SMAC/Diablo, high-temperature requirement protein A2 (HtrA2/Omi), endonuclease G (Endo-G), and apoptosis-inducing factor (AIF) [18]. The released factors trigger a downstream cascade of cell death protease (caspase) activity. The released Cyto c binds to the apoptotic protease activating factor 1 (Apaf-1), which in the presence dATP forms an oligomeric structure that recruits cytoplasmic inactive pro-caspase-9 to form the apoptosome, where pro-caspase-9 undergoes activation and then activates caspase-3, and -7, leading to apoptosis [19] (Fig. 1).
Fig. 1.
Apoptosis pathways; Activation and regulation
A schematic presentation of the two apoptosis signaling pathways: (a) extrinsic apoptosis—binding of death ligands to their receptors at the plasma membrane (PM) induce the assembly of DISC, which consists of an adaptor protein and the relevant ligand, and recruits and activates pro-caspase-8. Active caspase-8 activates the executioner caspase-3/7, which in turn, cleaves cellular substrates and brings about the formation of apoptotic bodies. (b) Intrinsic apoptosis—mitochondria-mediated pathway in which multiple stimuli such as UV, DNA damage, oxidants, chemotherapies, ROS, and Ca2+ overload, trigger the formation of the protein-conducting channel at the OMM, allowing the release of apoptogenic factors (Cyto c, AIF, SMAC/Diablo, HtrA2/Omi, Endo-G) from the IMS into the cytosol. The released Cyto c, together with the cytosolic Apaf-1 and procaspase-9 form the apoptosome in the presence of dATP, leading to caspase-9 activation, which cleaves effector caspases, such as caspase-3/7. The activated effector caspases cleave cellular substrates and are responsible for destroying the cell from within, while apoptotic bodies are formed. AIF is cleaved by calpains and/or cathepsins and translocated to the nucleus, where it activates chromatin degradation and condensation. Endo-G also translocated to the nucleus where it cleaves chromatin DNA into nucleosomal fragments. The released SMAC in the cytosol interacts with the inhibitors of caspase IAPs (including survivin), thereby antagonizing caspase inhibition. The crosstalk of extrinsic apoptosis and mitochondrial intrinsic apoptosis is mediated by caspase-8, which cleaves Bid to form the truncated protein (tBid). This protein is then translocated to the mitochondria and activates the intrinsic apoptotic pathway.
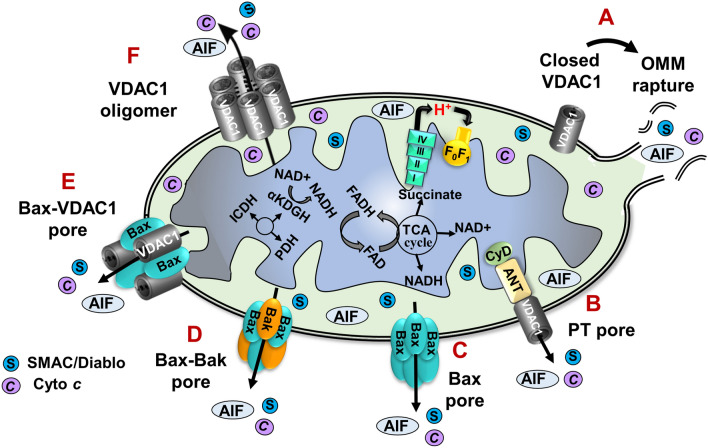
There are several possible mechanisms that could explain how the apoptotic initiators cross the OMM. These include non-specific release of IMS proteins due to OMM rupture [20], opening of the permeability transition pore (PTP) in response to overproduction of ROS or Ca2+ overload [21], formation of a large channel composed of Bax and/or Bak oligomers [22, 23], and the possible formation of hetero-oligomers of VDAC1 and Bax or VDAC1 oligomers [7, 24–34] (Fig. 2). Notably, multiple pathways of Cyto c release may co-exist in the cell [17].
Fig. 2.
Proposed mechanisms for apoptotic initiators crossing the OMM. A schematic representation of different models for the release of apoptogenic proteins. Apoptotic stimuli, stress conditions, or Ca2+ overload induce the assembly of a protein-conducting channel in the OMM, thereby allowing the release of Cyto c, AIF, HtrA2, Endo-G, and SMAC from the IMS to the cytosol where they trigger apoptotic cell death: (A) Prolonged VDAC1 closure leads to mitochondrial matrix swelling and OMM rupture, thereby providing a non-specific method of release of apoptogenic proteins; (B) Mitochondrial swelling leading to the assembly of a permeability transition pore (PTP); (C) Activation of the pro-apoptotic protein Bax leads to its association with the OMM and oligomerization to provide a protein-conducting channel; (D) Bax/Bak oligomerization and activation, provides a route for the release of pro-apoptotic proteins; (E) Bax and VDAC1 form hetero-oligomers that mediate the release; (F) VDAC1 oligomers serve as a protein-conducting channel that can mediate the release of apoptogenic proteins to the cytosol
Mitochondria-mediated apoptosis and VDAC1
Mitochondria are a common site for multiple metabolic, biosynthetic, and bioenergetic reactions as well as apoptosis, mitophagy, NETosis, pyroptosis, necroptosis, parthanatos, ferroptosis, alkaliptosis, and oxeiptosis, and they also regulate innate immunity, and epigenetics, among others [35]. Moreover, mitochondrial dysfunction is associated with many diseases [36].
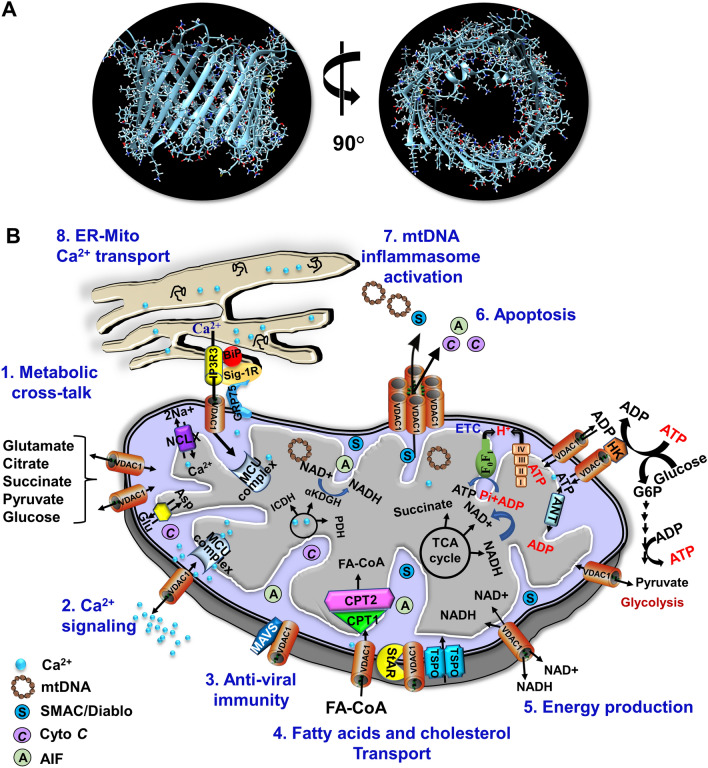
VDAC1 is one of the key controllers of mitochondrial function and is located in the OMM. VDAC1 is composed of 19 transmembrane β-strands that are connected through flexible loops with strands β1 and β19 in parallel conformation. This forms a β-barrel [6] with a 25-residue-long N-terminal region that lies inside the pore and can translocate from the internal pore to the channel surface [37–40] (Fig. 3A). The mobility of the N-terminal region is important for channel gating, interaction with anti-apoptotic proteins, and for VDAC1 dimer formation [37]. In addition, the region serves as the site of interaction for apoptosis-regulating members of the Bcl-2 family (i.e., Bax, Bcl-2, and Bcl-xL) [26, 37, 41–43] and hexokinase (HK) [26, 44].
Fig. 3.
VDAC1 as a multi-functional channel mediates the transport of metabolites, ions, lipids, and Ca2+ signaling, and regulates energy production, ER-mitochondria crosstalk, and apoptosis. A Side- and top views of the crystal structure of VDAC1 (PDB code: 3EMN). The β-barrel is formed by 19 β-strands, and the N-terminal domain is folded into the pore interior. B. (2) VDAC1 in the OMM controls transport of various metabolites and ions in and out of the IMS; (2) Transport of Ca2+ to the IMS, where it is transported to the matrix Ca2+ uniporter (MCU) complex; (3) MAVS (mitochondria anti-viral signaling), associated with VDAC1, enables anti-viral signaling Anti-viral activity via interaction with MAVS; (4) VDAC1 mediates the transfer of acyl-CoAs across the OMM to the IMS, where they are converted into acylcarnitine by CPT1a for further processing by β-oxidation. VDAC1 is involved in cholesterol transport as a constituent of the multi-protein complex termed the transduceosome, which contains Star/TSPO/VDAC1. (5) Mediation of cellular energy production by transportation of ATP/ADP and NAD+/NADH and regulation of glycolysis via association with HK; The role of Ca2+ in regulating energy production is mediated via activation of the TCA cycle enzymes pyruvate dehydrogenase (PDH), isocitrate dehydrogenase (ICDH), and α-ketoglutarate dehydrogenase (α-KGDH) leading to enhanced activity of the TCA cycle. The electron transport chain (ETC) and the ATP synthase (FoF1) are also presented. (6) apoptotic signaling induced VDAC1 oligomerization leads to the formation of a large channel conducting apoptogenic proteins (e.g., Cyto c and AIF) transport from the mitochondrial IMS to the cytosol, leading to apoptosis; (7) VDAC1 oligomers from a large channel mediate the release of mtDNA, which is associated with inflammation; (8) VDAC1 structurally and functionally contributes to the mitochondria-associated membranes, where the inositol 3 phosphate receptor type 3 (IP3R3), the sigma 1 receptor (Sig1R), binding immunoglobulin protein (BiP), the glucose-regulated protein 75 (GRP75/HSP70), and VDAC1 mediate Ca2+ transport from the ER to mitochondria
VDAC1 controls the metabolic and energetic crosstalk between the mitochondria and the rest of the cell by transporting metabolites such as pyruvate, malate, and succinate, nucleotides such as ATP/ADP and NADH/NAD+, and lipids, and ions including Ca2+ [5, 6, 17] (Fig. 3B). The protein serves as a hub that interacts with over 100 proteins involved in key cellular functions including metabolism, signal transduction, anti-oxidation, regulation of DNA/RNA structure, and apoptosis [5, 45].
One important function of VDAC1 involves the release of mitochondrial pro-apoptotic proteins from the IMS to the cytosol where they interact with apoptosis regulatory proteins such as IAPs [5, 6, 17] (Fig. 2). It is now accepted that apoptosis induction leads to VDAC1 oligomerization, and results in the formation of a large channel within a VDAC1 homo-oligomer that serves as a release route for Cyto c and other pro-apoptotic proteins, leading to apoptosis activation [7, 24–34, 37, 46] (Fig. 2). Oligomeric VDAC1 also mediates mitochondrial DNA (mtDNA) release into the cytosol, thereby triggering the type-Ι interferon responses that cause systemic lupus erythematosus [24, 47].
Although VDAC1 overexpression is associated with apoptosis [17], it is somewhat unexpectedly also overexpressed in cancers [17, 36, 45, 48–50], as well as in other pathologies [36] such as Alzheimer’s disease [51–54], in β-cells in type 2 diabetes [55], and in autoimmune diseases such as lupus [24], inflammatory bowel diseases [56], non-alcoholic steatohepatitis [57], acute liver injury [58], rheumatoid arthritis [59] cardiovascular diseases [60], and in the T cells of COVID-19 patients [61].
This apparent paradox may be explained by the finding that, while inhibition of apoptosis may lead to cancer and other severe disorders [62], activation of apoptosis may cause neurodegenerative and [54] autoimmune diseases [24, 56]. The latter can be targeted by inhibiting VDAC1 oligomerization, and thereby apoptosis [31, 52].
The involvement of VDAC1 in the control of pro- and non-apoptotic proteins is described below.
Apoptotic proteins with non-apoptotic function
The pro-apoptotic proteins caspases-3, -7, -8, and -9, AIF, APAF, survivin, SMAC/Diablo, Endo-G, HtrA2/Omi, TSPO, VDAC1, HK1, the Bcl2 family, and Cyto c [2, 3, 63] (Table 1) have all been shown to possess additional non-apoptosis related functions.
Table 1.
Pro-apoptotic proteins with non-apoptotic functions
| No | Protein | Apoptosis related function | Non-apoptosis function | References |
|---|---|---|---|---|
| 1 |
Cytochrome c (Cyto c) |
Upon release from the mitochondria, forms part of the apoptosome leading to caspase-9 activation and apoptosis induction | An electron transfer protein, function in the respiration chain. Influences cell differentiation and localizes to the nucleus under non-apoptotic conditions | [2, 11] |
| 2 | Caspase-3 | Caspase-3 is the most important executioner caspase and is activated by both the intrinsic and extrinsic pathways. When activated, it cleaves multiple structural and regulatory proteins that are critical for cell survival and maintenance | Plays a role in cell proliferation and migration during neuronal differentiation, keratinocyte differentiation, in mammalian red blood cell nuclear extrusion, brain development, liver regeneration, and skin wound healing | [64, 74] |
| 3 | Caspase-7 | Activated by initiator caspases and acts as an executioner caspase to cleave multiple proteins | Plays a pivotal role in bone formation and exhibits different functions depending on the developmental history of the bone | [82] |
| 4 | Caspase-8 | Activated by the extrinsic apoptosis pathway, which itself is triggered by death receptors. When activated, it activates caspase-3 and Bid | Performs multiple non-apoptotic roles in cells, such as activation of NF-κB signaling, alteration of endosomal trafficking, and enhancement of cellular adhesion and migration | [93, 100] |
| 5 | Caspase-9 | Activated by the mitochondrial apoptotic pathway in the apoptosome and activates other caspases such as -3 and -7 | Induces differentiation of leukemic cancer cells, granulocytic cells, myoblasts, neuronal cells, and mouse embryonic stem cells | [103–106] |
| 6 | AIF | Is released from the IMS in response to apoptotic signals, then translocates to the nucleus, and activates DNA fragmentation and chromatin condensation to initiate a caspase-independent apoptosis | Transduces DNA-damage-induced signals, controlling both adipocyte differentiation and mitochondrial metabolism | [2] |
| 7 | APAF | A key factor in the apoptosome, that acts at the DNA damage checkpoint and is required for chromosomal stability | Protein functions in the DNA damage-induced cell cycle | [107–110] |
| 8 | Survivin | Inhibits caspase activation, thereby leading to negative regulation of apoptosis | Survivin in the cytosol and nucleus is associated with chromosome alignment and segregation during mitosis and cytokinesis. The protein is involved in the regulation of cell division and DNA-double-strand repair | [111–114] |
| Direct and indirect binding of Survivin to initiator or effector caspases contributes to its inhibition of apoptosis [50]. Survivin binds to SMAC/DIABLO and prevents caspase activation [52] | ||||
| 9 | SMAC | Is released from the mitochondria in response to apoptotic signals and interacts with IAPs in the cytosol to prevent their inhibition of caspases, thereby, initiating apoptosis | Regulates phospholipid synthesis by controlling PSD activity. It also modulates inflammation and immunity | [12, 14] |
| 10 | HtrA2/Omi | High-temperature requirement protein A2. This serine protease is localized to mitochondria and is released to the cytosol following apoptotic insult, where it interacts with XIAP, thereby allowing caspase activation | Regulates the cell cycle, inactivates premature aging in mammals, and promotes hepatic fibrogenesis. It is responsible for quality control in mitochondrial homeostasis | [115] |
| 11 | Endo-G | Endo-G is a mitochondrial protein, a DNA/RNA non-specific nuclease. Once released from mitochondria, it functions in the caspase-independent apoptotic pathway to induce nuclear DNA fragmentation | Regulates mitochondrial bioenergetics by cleaving DNA–RNA hybrids to yield the RNA primers necessary for mtDNA replication. It may play an important role in differentiated cardiomyocytes | [116, 117] |
| 12 | VDAC1 | Participates in the release of mitochondrial pro-apoptotic proteins from the IMS to the cytosol and interacts with apoptosis regulatory proteins such as Bcl-2 | Multifunctional protein involved in energy production and metabolism, Ca2+ homeostasis, intracellular communication, autophagy, and inflammation | [5–9, 17] |
| 13 | TSPO | Proposed to mediate ROS production and formation of PTP to mediate Cyto c release, thus, inducing apoptosis | Involved in cholesterol import, porphyrin transport, heme synthesis, regulation of cell proliferation, inflammation, quality control, and mitochondrial metabolism | [11, 118] |
| 14 | Bcl2 family | Pro- and anti-apoptotic proteins | BH3-only proteins inhibit autophagy, others induce autophagy, or modulate Ca2+ signaling through the ER | [119–129] |
| 15 | Hexokinase | Function as anti-apoptotic proteins when bound to VDAC1 | Hexokinase catalyzes the phosphorylation of glucose | [44, 46, 130–137] |
Caspases: Non-apoptotic functions of caspases
As already described, caspase activity is the hallmark of apoptosis. Caspases are a family of cysteinyl aspartate-specific proteases that usually exist in the cell as inactive proenzymes. However, in response to apoptosis initiation, the initiator caspases are activated and activate the executioner caspases to cleave selected target proteins and execute cell death [64].
Recent studies have now reported caspase participation in multiple non-apoptotic cellular processes, including removal of protein aggregates, cell differentiation or de-differentiation, and rebuilding tissue [65–67]. Caspases are required in the early stages of differentiation [67] and for the terminal differentiation of cell types such as haematopoietic, epithelial, sperm, muscle, and trophoblast cells [2], as well as in the differentiation from human monocytes to macrophages [68]. Caspase activation is also required for terminal erythroid differentiation [69, 70]. The non-apoptotic functions of the pro-apoptotic proteins are summarized below and in Table 1.
Caspase-3 pro-apoptotic and non-apoptotic activities
Caspase-3 is an executioner caspase and a key mediator of both intrinsic and extrinsic pathway activated apoptosis (Fig. 1). Following activation by initiator caspases, casepase-3 cleaves proteins, leading to apoptotic cell death.
However, accumulating evidence now indicates that caspase-3 also possesses non-apoptotic functions, and is involved in regulating the growth and homeostasis of both normal and malignant cells and tissues in multicellular organisms [2, 3]. Caspase-3 activation has also been implicated in the differentiation of multiple cell types, including osteoblastic differentiation, and caspase-3-deficient mice accordingly exhibit delayed ossification and decreased bone mineral density [71–73]. The protein also affects the differentiation and maturation of human and mouse cell types [65], and is involved in cell proliferation and migration during neuronal, keratinocyte, and neuroendocrine cell differentiation, and the movement of neurons and neural and glial progenitor cells [2, 74]. Inhibition of caspase-3 prevents the differentiation of erythroid precursors [70] and affects brain development, liver regeneration, and skin wound healing [75]. The effects are seen in patient-derived glioma cells, where caspase-3 knockdown reduced the ability to form tumor spheres in culture [76].
Similarly, caspase-3-deficient mice are characterized by perinatal lethality with surviving newborns exhibiting various neural phenotypes [77], including specific aspects of cognition and behavior [78]. In contrast, caspase-3 overexpression has been reported in diverse types of malignant tumors [79] and high levels of caspase-3 mRNA expression have been found in neuroblastomas, Hodgkin’s disease, and in hepatoma cell lines [80]. These findings support the theory that the non-apoptotic functions of caspase-3 may be beneficial for cancer.
Caspase-7 pro-apoptotic and non-apoptotic activities
Pro-caspase-7 is activated by caspase-9 and functions in apoptosis execution (Fig. 1). Non-apoptotic activities of caspase-7 have been described in hard tissue formation and in differentiating cells during odontogenesis [81], and the protein also plays a role during osteogenesis [82] and spermatogenesis [83]. Caspase-7 has been reported to regulate the cell cycle and differentiation of certain cell types, and it is essential for proliferation and cell growth [84], where caspase-7 deficiency has an adverse effect on cell cycle progression resulting in the reduction of cell proliferation [85].
Caspase-3/-7 double-deficient mice exhibit embryonic lethality, whereas mice singly deficient in either caspase are born at normal Mendelian ratios and display no gross abnormalities when maintained on a C57BL/6 genetic background [86].
Caspase-7 is downregulated in some cancers [87]. In breast cancer carcinoma, the protein is abnormally expressed and contributes to cell growth and proliferation, with the active forms localized in the nucleus apart from the cytosol [88]. Elevated levels of caspase-7 are associated with well-differentiated tumors in oral squamous cell carcinomas [89].
Caspase-8 pro-apoptotic and non-apoptotic activities
Caspase-8 is an initiator caspase in the extrinsic apoptosis pathway that activates caspases-3 and -7 for the final execution of cell death [90] (Fig. 1).
The protein was recently suggested to serve as molecular switch for apoptosis, necroptosis, and pyroptosis, and prevents tissue damage during embryonic development and adulthood [91], while caspase-8 auto-cleavage plays a role in regulating necroptosis and maintaining lymphocytes homeostasis [92].
The non-apoptotic functions of caspase-8 involve nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-mediated pro-survival pathways, T- and B-cell proliferation, macrophage differentiation [68, 93, 94], and heart muscle differentiation [64]. Accordingly, loss-of-function mutations in the human caspase-8 gene cause defects in the activation of T, B and NK cells, culminating in immunodeficiency [95, 96].
In-vitro studies have indicated that caspase-8 induces the production of cytokines by acting as a scaffolding protein [97, 98]. In addition, caspase-8 enhances cleavage of focal adhesion substrates and cell migration, and its knockdown disrupts metastasis in neuroblastoma in vivo [99].
Caspase-8 deficiency has been reported in several cancer types, where it may contribute to tumorigenic transformation by interfering with cell differentiation [100]. Malignant neuroendocrine tumors, such as in neuroblastoma, primitive neuro-ectoderm tumors, medulloblastoma, glioblastoma, and small cell lung carcinoma frequently show a loss of caspase-8 [100].
Caspase 9 pro-apoptotic and non-apoptotic activities
Caspase-9 is an initiator of intrinsic apoptosis and when activated in the apoptosome, the protein stimulates downstream executioner caspases such as caspase-3 and caspase-7 (Fig. 1).
The non-apoptotic functions of caspase-9 include regulation of cellular differentiation/maturation, innate immunity, mitochondrial homeostasis, and autophagy, and activation of the NF-kB pro-survival pathway by interacting with cellular inhibitor of apoptosis protein-1 (cIAP1/BIRC2) [101]. Caspase-9 also promotes myocyte differentiation and proliferation, and its suppression may impede muscle [102] and neuronal cell differentiation [103].
The expression of caspase-9b is upregulated in several cancers, including non-small-cell lung cancer, thereby providing potential as a target for tumor treatment [101]. Caspase-9 is also involved in differentiating granulocytic and leukemic cancer cells [104].
Overall, the non-apoptotic functions of caspases are diverse, ranging from involvement in immune responses, cell proliferation, cellular remodeling, and cell fate determination, to cytoskeletal reorganization, with implications for cancer, neurodegeneration, wound repair, and regeneration.
Cytochrome c apoptotic and non-apoptotic functions
Cyto c is a key player in respiration, where it shuttles electrons between complex III and complex IV of the respiratory chain [138], but it also performs multiple other functions associated with cell life and death [139]. Cyto c release is a hallmark of the intrinsic apoptotic pathway in mitochondria-mediated apoptosis, and it forms the apoptosome in the presence of dATP and Apaf-1, thereby acting as an allosteric activator of caspase-9 [140, 141] (Fig. 1). The activity depends on the subcellular compartment in which the Cyto c resides, with electron transport activity associated with the IMS, and the pro-apoptotic functions seen when the protein is released into the cytosol.
Interestingly, in response to stress, such as DNA damage, Cyto c migrates to the nucleus before the cytoplasmic caspase are activated [142, 143].
Non-apoptotic functions of Cyto c include accumulation in the nucleus and induction of chromatin remodeling [144]. This localization of Cyto c to the cytoplasm or nucleus is associated with nitration, which thereby controls the cellular functions [145].
In non-cancerous systems, Cyto c appears to modulate differentiation across a wide spectrum of cell types [2], including differentiation associated with caspase-3 and -9 activation [4]. In cancers, elevated levels of Cyto c have been reported in patients with hepatocellular cancer [146], with downregulation in renal cell carcinoma tissues [147]. Cyto c has also been found in secretory granules in the pancreas and anterior pituitary [148].
Cyto c activities are tightly regulated by posttranslational modifications [149]. Specifically, phosphorylation can serve as the switch between the multiple activities of the protein [150].
AIF non-apoptotic functions
AIF is a pro-apoptotic protein that is released from the mitochondria upon apoptosis induction following its cleaved by calpains and/or cathepsins to yield the pro-apoptotic truncated AIF form, that translocates to the nucleus, where it participates in chromatin condensation and chromatinolysis [151–154].
Non-apoptotic pro-survival activities of AIF are related to oxidative phosphorylation. Murine aif knockout causes a defect in oxidative phosphorylation [155, 156].
AIF has been proposed to have an antioxidant function, which is important for optimal oxidative phosphorylation, as well as for the maintenance of mitochondrial morphology and cell-cycle regulation [154, 156]. An absence of AIF in murine cardiac tissue leads to the development of dilated cardiomyopathy and heart failure [157]. These mice also exhibit increased glucose tolerance, higher insulin sensitivity, decreased fat mass, and resistance to high-calorie-diet-induced obesity [158]. In this context AIF has been shown to control adipocyte differentiation [159].
AIF also possesses a pro-survival function as a component of the coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHD4)-dependent import pathway by which a set of nuclear-encoded cysteine-motif-carrying protein substrates are translocated to the IMS [160].
AIF levels are elevated in chronic lymphocytic leukemia (CLL) [161] and in prostate cancer, where the protein supports the growth and survival of aggressive prostate cancer cells [162]. The levels are also increased in colorectal cancer where AIF protect the cells from stress-induced apoptosis [163]. Thus, elevated AIF protein levels benefit tumorigenesis.
SMAC as a non-apoptotic protein
SMAC is a pro-apoptotic protein that is released from the IMS upon apoptosis induction and interacts and antagonizes inhibitors of apoptosis proteins (IAPs) in the cytosol, thereby promoting the activation of caspases and apoptosis [164].
SMAC was found to be upregulated in a variety of tumors [12, 14, 161]. The non-apoptotic functions of overexpressed SMAC in cancer were recently identified [12, 14], and are presented below.
HtrA2/Omi as a non-apoptotic protein
The high temperature requirement protein A 2 (HtrA2) also known as stress-activated/regulated endoprotease (Omi), belongs to a family of serine proteases [165, 166]. HtrA2/Omi is released from the IMS to the cytosol in response to apoptosis signals, and once there, proteolytically removes IAPs and promotes both caspase-dependent and independent apoptosis [167, 168]. However, HtrA2/Omi has non-apoptotic activities include regulating the cell cycle by cleaving the large tumor suppressor kinase 1 (LATS1) protein and generating LATS1 fragments that inhibit the G1–S transition [169]. HtrA2/Omi inactivation has been reported to increase the accumulation of mtDNA deletions and deficiency of the protein induces mtDNA damage, DNA mutation, accumulation of unfolded proteins in the mitochondria, oxidative stress, and defective mitochondrial respiration, suggesting an important role in mitochondrial homeostasis [170]. An inactivating point mutation leads to premature aging in mammals [171].
HtrA2/Omi expression has been found to promote hepatic fibrogenesis and is suggested to be responsible for quality control in mitochondrial homeostasis [166].
Reduced Omi/HtrA2 activity has been connected to neurodegeneration. The protease activity of Omi/HtrA2 was shown to be required for the physiological processing of amyloid-β precursor protein and it is therefore suggested as a possible therapeutic target for Alzheimer’s disease [172–174]. Omi/HtrA2 has also been linked to Parkinson’s disease [175, 176].
Omi/HtrA2 has also been suggested to act as a negative regulator of cell cycle progression through the proteolytic processing of the mitotic kinase WTS/large tumor suppressor 1 (WARTS) [169].
Endo-G as an apoptotic and non-apoptotic protein
Endo-G is a mitochondrial endonuclease that is situated in the IMS and plays a role in mitochondrial DNA replication [177]. However, during apoptosis Endo-G is released from mitochondria and translocates to the nucleus, and induces nuclear DNA fragmentation cleaving chromatin DNA into nucleosomal fragments and induce cell death in caspase-independent pathways [116]. It was also proposed that Endo-G activates DNA damage response and triggers autophagy through its endonuclease activity [178].
Endo-G knockout mice are viable and grow to maturity with no evident defects, and fibroblasts derived from Endo-G-deficient animals showed no difference in sensitivity to a variety of intrinsic and extrinsic apoptotic stimuli, suggesting that Endo-G is not essential for early development or apoptosis [179].
The non-apoptotic functions of Endo-G include a role in recombination and repair during mitochondrial DNA replication [179]. The protein appears to be required for proliferation, as Endo-G-depleted cells accumulate at the G2–M transition [180]. In addition, the protein plays an important role in differentiated cardiomyocytes [116]. Endo-G is a novel determinant of cardiac hypertrophy and mitochondrial function and is involved in regulating the estrogen-related receptor alpha (ERRα) and the peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α) [181]. Endo-G knockdown causes the death of polyploid tumor cells [182].
Endo-G was detected in tumor than non-tumor parts of liver tissue of the patient. of the patient [183].
Survivin, BIRC5 as a non-apoptotic protein
Survivin, first identified in 1997, is a member of the IAP family of proteins with only one baculoviral IAP repeat (BIR) domain [111]. It has a dual function in preventing cell death by inhibiting apoptosis while playing an essential role in mitosis [111, 112]. Survivin, like other IAP family members, is thought to be a negative regulator of apoptosis [111, 112] and has been shown to interact with both caspase-3 and caspase-7 and to inhibit their activity [113]. In addition, the survivin suppression frequently reported in cancer cells is associated with caspase-3 activation and apoptosis enhancement [184–188]. Moreover, survivin has been shown to control the nuclear translocation of AIF, and hence negatively modulates the activation of caspase-independent apoptosis [20, 189–191].
Unlike the other IAP family members, survivin is not only involved in apoptosis regulation but also in promoting mitosis by encouraging the formation of the chromosomal passenger complex [189, 192, 193]. Survivin in the nucleus is associated with chromosome alignment and segregation during mitosis and also controls cytokinesis, thereby regulating cell division, and the protein is also involved in DNA-double-strand repair [114, 194].
Survivin is highly expressed in most cancers and high levels are associated with chemotherapy resistance and increased tumor recurrence [190].
TSPO functions in apoptotic and non-apoptotic processes
TSPO, previously known as the peripheral-type benzodiazepine receptor (PBR), is located in the OMM [195]. The receptor is involved in the regulation of cellular proliferation, apoptosis, and mitochondrial functions [196].
Several studies suggest that TSPO functions in mitochondrial apoptosis [11, 118] by interacting with mitochondrial PTP, and with several other apoptosis-related proteins [197].
However, TSPO’s main activities are unrelated to apoptosis and include cholesterol import, regulation of mitochondrial metabolism, cell proliferation, inflammation, porphyrin transport, heme synthesis [198], and mitochondrial quality control [118]. TSPO deregulates mitochondrial Ca2+ signaling, leading to activation of Ca2+-dependent NADPH oxidase, and thereby, increasing ROS levels [199]. TSPO also regulates mitochondrial energy homeostasis through the modulation of fatty acid oxidation in steroidogenic cells [200].
TSPO is overexpressed in a wide variety of malignant human cells and in colon, brain, prostate, breast, ovarian, esophageal, gliomas, endometrial, and hepatic carcinomas [196], 201. Moreover, the TSPO expression level correlated positively with disease progression of some cancers suggesting that the multiple functions are advantageous to cancer growth and development [196].
Certain TSPO functions are thought to involve interaction with VDAC1 [202–204].
As presented below, the expression of most of these proteins is altered upon VDAC1 or SMAC depletion, suggesting that their expression is linked to cell metabolism.
Hexokinase (HK) dual functions in metabolism and apoptosis
Hexokinase catalyzes the phosphorylation of glucose to glucose-6-phosphate, in an ATP dependent reaction that serves as the entry point for glucose to glycolysis. The four isozymes of HK present in mammalian tissues, termed HK-I to HK-IV, differ in their subcellular localization and metabolic function [205]. HK-I and HK-II are primarily mitochondrial and predominantly dock on to the cytosolic surface of the OMM by binding to VDAC [206]. Binding of HK to VDAC1 has been proposed to allow direct access to newly produced ATP in the mitochondria [207], thereby facilitating the maintenance of a high glycolytic flux rate in tumors with the accompanying increase in energy and metabolite production [130].
Interestingly, the results of both in vitro and in vivo studies suggest that HK-I and HK-II can function as anti-apoptotic proteins when bound to VDAC1, with their detachment enabling activation of apoptosis [44, 46, 130–137]. HK prevents Cyto c release and subsequent apoptosis [44, 46, 130–137] and also protects against Bax- or Bak-mediated apoptosis [131, 132, 208]. Thus, HK not only plays an important metabolic role, but also acts to inhibit apoptosis.
HK-I and HK-II are over expressed in many types of cancer, including colon, prostate, lymphoma, glioma, gastric adenomas, carcinomas, and breast cancers [209–213], as well as in hepatomas [214] and in aggressive tumors such as gliomas and colorectal cancer [215]. The observation of these elevated levels of HK-I and HK-II in cancer cells provide evidence that a high rate of glycolysis allows tumor cells to evade apoptosis, thereby allowing proliferation to continue [216].
In addition to promoting energy production and acting as an anti-apoptotic protein, HK binding to VDAC1 also regulates ROS production/efflux from the mitochondria [217] and increases the synthesis and uptake of cholesterol [131]. Thus, the multifunctional HK is beneficial for cancer cells and make HK dissociation from the mitochondria an attractive target for anti-cancer therapy [218, 219].
Non-apoptotic functions of other cell death regulatory proteins
Other proteins associated with apoptosis such as APAF1, Bax, XIAP, and the Bcl-2 family have also been shown to have non-apoptotic functions [220].
Bcl-2 family proteins: Bcl-2 family anti-apoptotic proteins such as Bcl-2, Bcl-XL, Bim, Bcl-w, and Mcl-1 [121, 126, 127], were found to inhibit autophagy via interaction with Beclin 1 [128, 129]. Conversely, pro-apoptotic BH3-only members of the Bcl-2 family such as BNIP3, Bad, Noxa, Puma, and others have been shown to induce autophagy by competing with Bcl-2 and Bcl-XL, and by interaction with Beclin 1 [121, 122]. Interestingly, both anti-apoptotic and pro-apoptotic proteins from the Bcl-2 family were shown to modulate Ca2+ signaling in the ER [123–125].
Bcl-2 and Bcl-XL were found to lower luminal Ca2+ levels by enhancing Ca2+ leakage from the ER, through modulation of the gating of the inositol-1,4,5-trisphosphate receptor (IP3R) by its ligand [221, 222]. In contrast, Bcl-2, Bcl-xL, and Mcl-1 modulate Ca2+ homeostasis via interaction with IP3Rs at the mitochondria-associated membranes (MAM) [223]. It has been suggested that Bcl-xL interacts with IP3Rs to promote Ca2+ release, thereby regulating mitochondrial metabolism and neuronal function and development [224].
Conversely, Bax and Bak enhance the release of Ca2+ from ER stores, by involving the SERCA [119, 120].
Interestingly, Bcl-xL, but not Bcl2 can strongly suppress the mitophagy receptor FUN14 domain-containing 1 (FUNDC1)-mediated mitophagy by inhibiting the mitochondrial Ser/Thr phosphatase PGAM5, thereby preventing the dephosphorylation of FUNDC1 and blocking hypoxia-induced mitophagy [225]. When FUNDC1-mediated mitophagy is blocked by the microtubule inhibitor vinblastine, PGAM5 dephosphorylates FUNDC1 and mediates mitochondrial fission that aggravates vinblastine-induced cell death [226].
Bcl2 family members can affect the morphology of mitochondria. The activation of pro-apoptotic Bax and Bak promotes fragmentation of the mitochondrial network during apoptosis. This is not inhibited by the expression of Bcl-xL, MCL1, or other members of the Bcl2 subfamily [226].
Over expression of anti-apoptotic Bcl-2 members (e.g., Bcl-2, Bcl-XL, Mcl-1) is quite frequent in newly diagnosed cancer as well as after developing resistance to therapy [227]. Bcl2 and Bcl-xL are expressed at high levels in many types of cancer, such as colon, prostate, lymphoma, glioma, gastric adenomas, carcinomas, and breast cancer [155, 156]. When Bcl-2 is overexpressed in cancer cells, it may inhibit the pro-apoptotic signals, allowing the cancer cell to survive under stress [228].
Decreased Bak expression associated with increased expression of anti-apoptotic components has been shown in tumors of colon cancer patients [132]. It is now accepted that the ratio of expression of anti- and pro-apoptotic proteins in the cell determines the resistance to apoptosis induction.
APAF-1: Apaf-1, which is the main component of the apoptosome, a platform required for caspase 9 activation (Fig. 1), has been reported to function in the DNA damage checkpoint and is required for chromosomal stability [107–110]. The results of Apaf-1 knockdown in human cells and knockout in mice suggest that the protein functions in DNA damage-induced cell cycle arrest [107, 108, 110]. Apaf-1 mutants lacking the N-terminal CARD domain can replace endogenous Apaf-1 in the control of the DNA damage-induced cell-cycle blockade, suggesting that the cycle-arresting function of Apaf-1 is independent of caspase activation [107, 108, 110]. Apaf-1 depletion was also found to decrease the DNA damage induced, activating phosphorylation of the checkpoint kinase Chk1 [107, 108, 110], and knockdown of Chk1 abrogates Apaf-1-mediated cell-cycle arrest. A study of biopsies from non-small cell lung cancer patients revealed a correlation between the nuclear translocation of Apaf-1 induced by DNA damaging agents, and the endogenous activation of Chk1.
In summary, the findings presented above suggest that proteins categorized as apoptotic proteins may also possess non-apoptotic functions ranging from regulating cell proliferation to affecting differentiation and modulating the immune response. Many of these apoptotic proteins are overexpressed in cancer, suggesting their importance for cancer development and survival.
The paradox of VDAC1 overexpression of in cancer
Exogenous overexpression of VDAC1 was found to induce apoptosis in the absence of any apoptosis stimuli and regardless of cell type [30, 46, 229]. Overexpressed VDAC1 undergoes oligomerization and forms a large channel that allows passage of IMS-located apoptotic proteins such as Cyto c, SMAC, and AIF, to cross the OMM and be released to the cytosol, where they induce apoptosis [7, 25–30, 230] (Fig. 3).
Moreover, VDAC1 expression could be increased by stimuli such as chemotherapy drugs, ROS, Ca2+, and stress conditions, which activate mitochondria-mediated apoptosis via different mechanisms, but all lead to VDAC1 oligomerization [7, 29, 36].
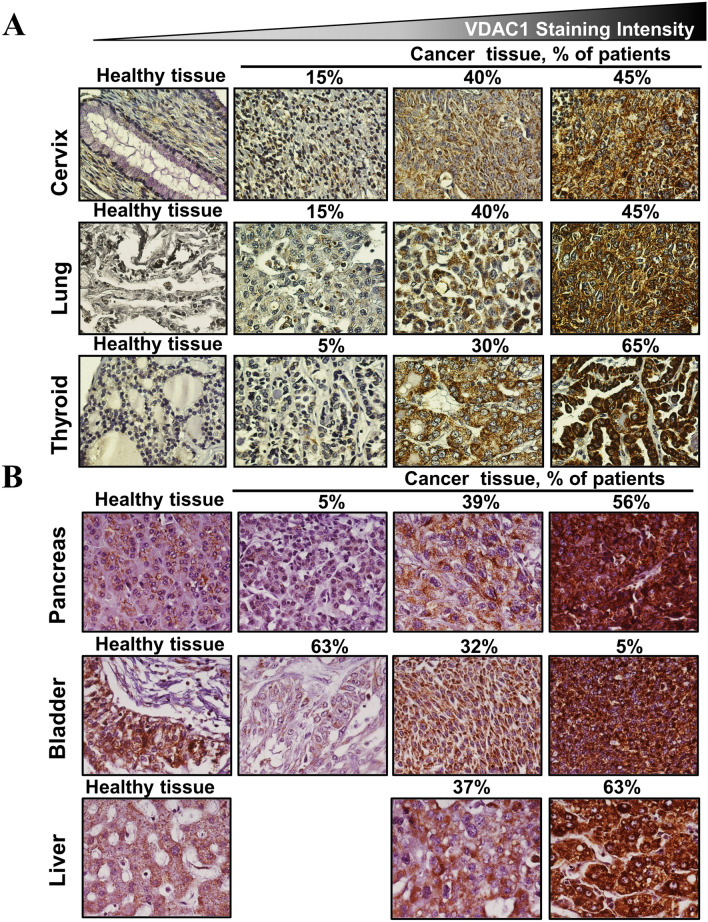
In cancers, although VDAC1 is overexpressed [17, 36, 45, 48–50] (Fig. 4), there is no induction of apoptosis. This is due to the prevention of apoptosis by the concomitant over expression of anti-apoptotic proteins such as hexokinase and Bcl2 [17, 36, 45, 49].
Fig. 4.
VDAC1 overexpression in cancers. A, B Immunohistochemical staining of VDAC1 on tissue microarray slides obtained from Biomax: US slide (MC5003) comprising cancer tissues (n = 18–20 patients for each cancer type) and normal tissue (n = 5). Representative sections of cervix, lung, thyroid, pancreas, bladder, and liver are shown. The slides were incubated overnight at 4 °C with anti-VDAC1 antibodies (1:1000) in PBS containing 1% BSA and then with the appropriate secondary antibody (1:5000) diluted in PBS containing 1% BSA. The slides were subsequently treated with 3′3-diaminobenzidine tetra-hydrochloride (DAB) and counter-stained with hematoxylin. Negative controls lacked the incubation with the primary antibody. Sections of tissue were observed under an Olympus microscope and images were recorded at 200× magnification with the same light intensity and exposure time. The percentage of patients with positive staining at the intensity indicated by the scale at the top is shown
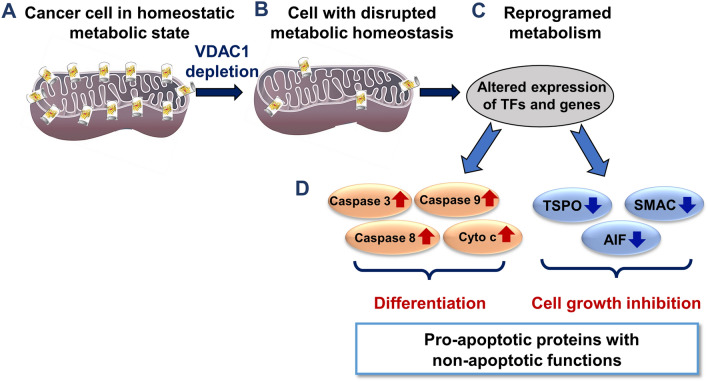
Why do cancer cells overexpress VDAC1 despite its pro-apoptotic activity? Cancer cells reprogram their metabolism to support the increased biosynthetic and energetic demands required for their growth and motility. In these cells, VDAC1 is overexpressed to support cell energy and metabolism homeostasis [5, 6, 17]. Accordingly, downregulation of VDAC1 expression decreases metabolite exchange between the mitochondria and the cytosol and inhibits cell growth [49, 231]. Moreover, we demonstrated that depleting VDAC1 in tumor cells leads to metabolic reprogramming, with a resultant inhibition of cell proliferation and tumor growth, elimination of cancer stem cells, alteration of the inflammation in the tumor microenvironment, and induction of differentiation of the malignant cells to become normal-like cells [11, 49, 232–236].
Thus, VDAC1 can be considered as a pro-survival protein with pro-apoptotic activity that is controlled by the expression level and the interacting proteins.
Reducing VDAC1 expression in cancer alters the expression of pro-apoptotic proteins performing a non-apoptotic role
Silencing VDAC1 expression in tumors and cells in culture resulted in metabolic reprogramming [49, 231, 233] and alteration in the levels of expression of pro-apoptotic proteins [11]. Our results indicate that VDAC1 deletion leads to an upregulation of the pro-apoptotic proteins, caspases-3, -8, and -9, p53, and Cyto c, albeit with no induction of apoptosis, but rather, an association with cell differentiation [11, 232–236]. The increase in levels of Cyto c and the protein translocation to the nucleus in VDAC1-depleted tumors with no apoptotic cell death could be related to its role in cell differentiation [2]. The increase in caspase-3, -8 and -9 expression levels with no apoptosis seen when VDAC1 is depleted in a tumor [11, 235, 236] may be related to the proposed non-apoptotic functions of the caspases, for example in differentiation [2, 64].
Notably, the expression of other apoptosis-regulatory proteins such as SMAC, AIF, and TSPO are also decreased in VDAC1-silenced cells [11].
Thus, we can conclude that reprogramming cancer cell energy and metabolism shifts protein functions from pro-apoptotic to non-apoptotic, with effects on processes ranging from bioenergetics and metabolism to inflammation and differentiation.
Indeed, metabolic reprogramming via silencing VDAC1 expression encourages differentiation of cancer cells, both in culture and in mouse models [233–235, 237]. In glioblastoma (GBM), and in lung and breast cancers, cell metabolism rewired by VDAC1 depletion results in cell growth arrest and differentiation [49, 231–234, 238]. Similarly, in GBM tumors and in cells in culture, silencing VDAC1 expression results in differentiation of astroglia U-87MG cell into astrocyte- and neuron-like cells [233, 234]. In triple negative breast cancer, MDA-MB-231 cells, which correspond to a poorly differentiated tumor that does not express the estrogen, progesterone, or ERBB2/Her2 receptors [239], silencing VDAC1 expression in tumors and cells in culture increased the expression levels of all these receptors as well as that of prolactin [234, 237].
VDAC1 silencing also downregulated other pro-apoptotic proteins, including SMAC, AIF, and TSPO, which are associated with cell growth support [11].
AIF may offer advantages to cancer cells through non-apoptotic functions associated with pro-survival, oxidative phosphorylation, ROS detoxification, cell cycle regulation, and others [154]. Since VDAC1 depletion leads to a dramatic decrease in the levels of AIF [11], this should also reduce the AIF functions.
Similarly, a marked decrease in TSPO levels was observed in VDAC1-depleted tumors [11]. This decrease will affect the multiple functions of TSPO in cholesterol import, mitochondrial metabolism, cell proliferation, Ca2+ signaling, and oxidative stress regulation [240]. As TSPO binds to and acts via VDAC1 [202–204], the decrease in TSPO levels may result from VDAC1 depletion.
These findings reveal that metabolic reprogramming of tumors following silencing of VDAC1 expression both in cultured cells and in mouse models, results in changes in the levels of pro-apoptotic proteins, which then fulfil non-apoptotic functions, such as acting as regulators of cell growth and differentiation [233–235, 237] (Fig. 5).
Fig. 5.
Reprograming tumor metabolism via VDAC1 depletion alters the expression of pro-apoptotic proteins and shifts their activities towards arrested cell growth and induction of cell differentiation. A schematic presentation of cancer cell VDAC1-depletion leading to altered levels of apoptotic proteins and expression of their non-apoptotic functions associated with arrested cell growth and cell differentiation induction. Cells expressing VDAC1 maintain a homeostatic metabolic state supporting cell growth and survival (A). Following VDAC1 depletion, cells have impaired energy and metabolite generation (B). This results in altered expression of thousands of genes and transcription factors (TFs) such as metabolism regulators, increased p53 levels supporting cell differentiation, and decreased HIF1-α, c-Myc, and NF-kB/RelA levels, resulting in cell growth inhibition [11] (C). Metabolism reprogrammed tumors overexpress pro-apoptotic proteins that possess cell differentiation promotion functions, while pro-apoptotic proteins whose non-apoptotic function associated with cell growth promotion are downregulated (D)
To conclude, the results presented here suggest that players in apoptosis pathways may have a dual function in cancer and be both apoptotic and non-apoptotic, with the switch between these activities being controlled by the cell energy level on the metabolism-epigenetics axis. Accordingly, depletion of the mitochondria gatekeeper VDAC1 leads to a re-programming of energy and shifts the activity of these proteins from a pro-apoptotic to a non-apoptotic mode. This is accompanied by translocation of some of these proteins from the mitochondria/cytosol to the nucleus. Possible mechanisms that regulate the pro-apoptotic or non-apoptotic functions are discussed below.
The paradox of overexpression of the pro-apoptotic protein SMAC in cancer
SMAC is a 25-kDa protein requiring post-translational modification for maturation, activation, and translocation to IMS. Smac/DIABLO at 2.2 A resolution reveals that it homodimerizes through an extensive hydrophobic interface [241] (Fig. 7A). SMAC is a pro-apoptotic IMS-located protein, that, upon apoptosis induction, is released into the cytosol where it interacts with members of the IAP family (cIAP1, cIAP2, and XIAP) to neutralize their inhibition of caspases, thereby initiating apoptosis [164, 242, 243] (Fig. 1).
Fig. 7.
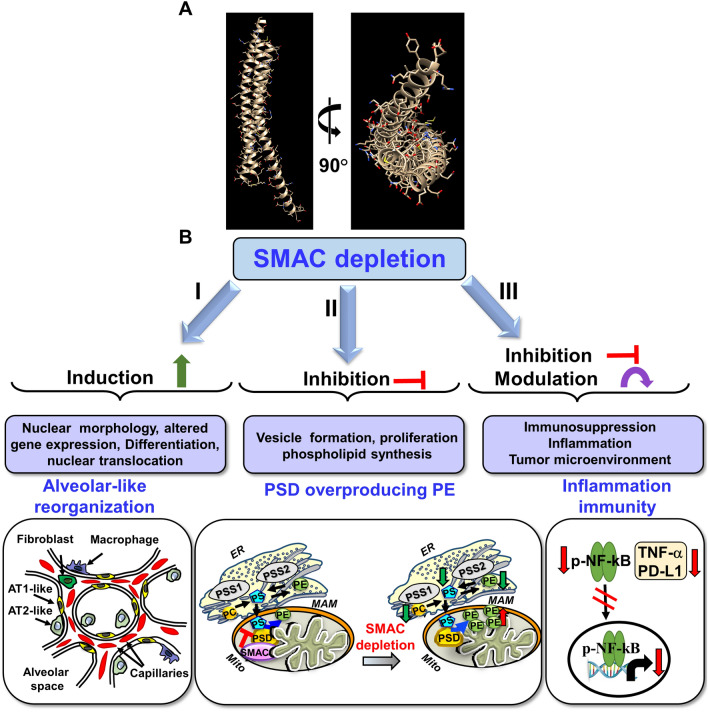
SMAC depletion effects on tumor morphology and properties. A Crystal structure of SMAC, PDB https://doi.org/10.2210/pdb1FEW/pdb. B SMAC depletion leads to: (I) Induction of cell differentiation and reorganization into glandular/alveoli-like structures. Pulmonary alveoli cross-section showing the major cell types with the tumor implemented AT2-like cells were differentiation into AT1-like cells, and changes in blood capillary organization; (II) Inhibition of vesicle formation, cell proliferation in the tumor microenvironment, and phospholipid synthesis. (b) Induction of altered expression of genes associated with the cell membrane, exosomes, and ER- and Golgi-related proteins, nuclear translocation, and cell and tumor morphological changes. This is due to SMAC functions in the regulation and maintenance of phospholipid synthesis [12]. In mammalian cells, distinct pools of PE are synthesized in either mitochondria or ER membranes [252]. The major site of phospholipid synthesis is in the ER [261], where PS is transferred from the ER to mitochondria [262]. In the ER, PS is produced from PC and PE by PSS1 and PSS2, respectively, and then transferred to the mitochondria, where it is converted into PE by PSD [250]. (III) Inhibition or modulation of inflammation and immunity in the small tumors developed from CRISPER/Cas9 SMAC/Diablo-depleted A549 lung cancer cells showed reduced expression of inflammation-related proteins such as NF-kB and TNF-α and of the programmed death-ligand 1 PD-L1 associated with additive immune system suppression [13]
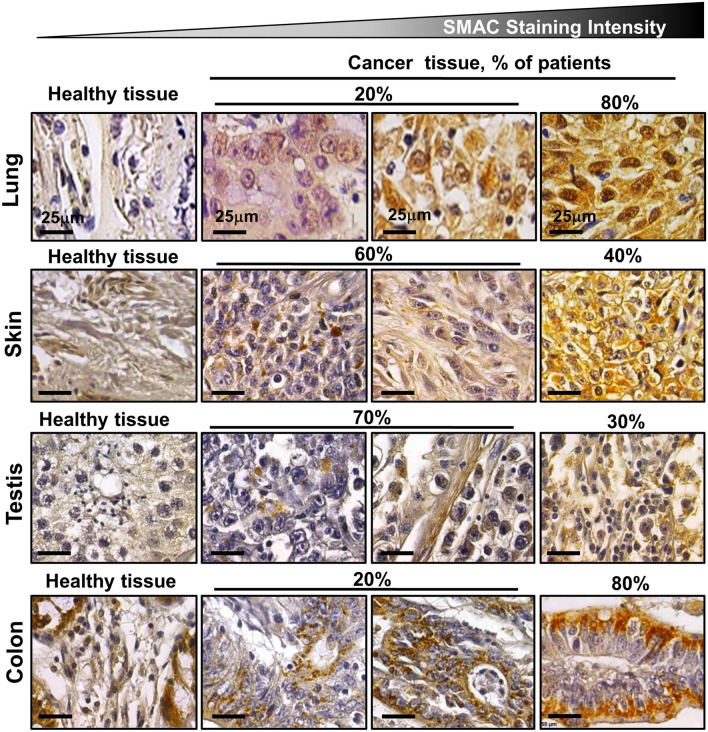
Unexpectedly, we and others have demonstrated overexpression of SMAC in many cancer types and cell lines, including cervical cancer, lung, ovarian, prostate, gastric and renal cell carcinomas, and different types of sarcomas (Fig. 6) [12, 14]. This may suggest that SMAC overexpression is beneficial to cancer cells, through the manifestation of non-apoptotic functions. Indeed, our results indicate that SMAC is required for the growth of cancerous, but not of non-cancerous cells in culture, and that silencing SMAC expression inhibits lung cancer-derived tumor growth, restores altered lipid metabolism, and modulates a number of oncogenic properties [12, 14] (Fig. 7).
Fig. 6.
SMAC overexpression in cancer. Immunohistochemical staining for SMAC was performed on tissue microarray slides obtained from Biomax US: MC5003 comprising cancer tissues (n = 10–20 patients for each cancer type) and normal tissues (n = 5). Representative lung, skin, testes, and colon are shown. The slides were incubated overnight at 4 °C with anti-SMAC antibodies (1:1000) in PBS containing 1% BSA and then with the appropriate secondary antibody (1:5000) diluted in PBS containing 1% BSA. The slides were subsequently treated with 3′3-diaminobenzidine tetra-hydrochloride (DAB) and counter-stained with hematoxylin. Negative controls lacked the incubation with the primary antibody. Sections of tissue were visualized under an Olympus microscope and images were recorded at 200× magnification with the same light intensity and exposure time. The percentage of patients with positive staining at the intensity indicated by the scale at the top is shown
This is particularly interesting, since mice lacking SMAC are viable, and grow, and mature normally, without any histological abnormalities, and exhibit wild-type responses to all types of apoptotic stimuli [244]. This suggests that cancer cells require novel non-apoptosis-related functions.
SMAC possesses non-apoptotic functions required for cancer survival and development
The non-apoptotic functions of SMAC in cancer range from regulating cell proliferation to differentiation [12, 14].
The non-apoptotic function of SMAC was demonstrated by silencing its expression using specific siRNA in cells in culture or in sub-cutaneous lung cancer xenografts in mice [14, 245]. The non-apoptotic functions of SMAC in cancer range from inhibiting cell proliferation and tumor growth to differentiation [12, 14]. The si-hSMAC-treated residual tumor demonstrated morphological changes, including cell differentiation and reorganization into glandular/alveoli-like structures and elimination of surfactant-producing organs, the lamellar bodies [12, 14]. Similarly, CRISPER/Cas9 SMAC/Diablo-depleted A549 lung cancer cells displayed inhibited cell proliferation and cell migration [13]. Using the proteomic approach of LC–MS/MS analysis, these SMAC/Diablo depleted cells, showed altered expression of proteins associated with lipid- and lipid signaling, vesicular transport and trafficking, metabolism, epigenetic, extracellular matrix, cell signaling and neutrophil-mediated immunity [13].
Moreover, tumors established from SMAC-KO A549 cells showed, altered expression of several components of the tumor microenvironment, reduced expression of inflammation-related proteins such as NF-kB and TNF-α and of the programmed death-ligand 1 PD-L1 associated with additive immune system suppression [13].
These results suggest that SMAC is involved in multiple processes that are essential for tumor growth and progression.
Next generation sequencing (NGS) analysis of lung cancer-derived tumors that were silenced for SMAC expression revealed altered expression of genes associated with transporters of metabolites, lipids, and ions, and of enzymes involved in the metabolism of cholesterol, lipids, and nucleotides [12, 14].
SMAC depletion altered expression of genes associated with cholesterol, lipid transport and synthesis, and lipid degradation and regulation [12, 14]. In addition, the expression of many genes associated with the formation of vesicles that mediate intra- and extra-cellular transport and that are related to organelles, such as the ER, Golgi and exosomes, was modified [14]. In addition morphologically, the tumors exhibit differentiation and reorganization into glandular/alveoli-like structures. (Fig. 7B).
These non-apoptotic functions may explain the SMAC overexpression observed in multiple cancers.
SMAC regulates lipid metabolism in cancer via modulation of mitochondrial PSD activity
Metabolism reprogramming is a hallmark of cancer, with cancer cells demonstrating a high dependence on synthesis of the lipids [246], and phospholipids (PLs) that are essential for cancer growth, development, migration, invasion, and immune escape [247, 248]. PLs are essential building-block components of cellular membranes, but also function as regulators of various cellular functions such as cell adhesion and migration, signal transduction, vesicular trafficking, apoptosis, metabolism, and post-translational modifications [249]. As presented above, lipid and phospholipid synthesis pathways are highly affected by depletion of SMAC in a tumor [12, 14].
We identified SMAC interaction with and modulation of the mitochondrial enzyme phosphatidylserine decarboxylase (PSD), which is also present in the IMS, as SMAC, and converts phosphatidylserine (PS) to phosphatidyl-ethanol amine (PE) [12, 250]. Accordingly, SMAC deletion, resulted in about a 2-fold decrease in total PLs and phosphatidyl choline (PC) levels both in tumors and in CRISPR/Cas9 SMAC-KO cancer cells, while PE levels were increased 2-fold in the mitochondria [12]. Purified SMAC directly interacts with purified PSD and inhibited its activity, and SMAC downregulation resulted in enhanced PSD activity, thereby, overproducing mitochondrial PE, leading to cell depletion of PC and PS and inhibition of cancer cell proliferation [12] (Fig. 7D).
Such changes reflect significant differences in cellular physiology since PSD and PE are essential components. PSD-KO is embryonic lethal [251], and loss of PSD causes defects in mitochondrial morphology and function and disrupts electron transport chain complex formation [252]. PE is not only a membrane component, but also functions in the nucleus as a lipid chaperone signaling molecule where it modulates the structural organization of chromatin, nucleic acid synthesis, and DNA replication [253], and has a role in autophagy [254]. PE levels are elevated in several cancers, and it is a target of potent anticancer natural products [255].
SMAC-interacting network
SMAC has multiple effects on cancers which may be related to the interaction of SMAC with a variety of non-apoptotic proteins located in the IMS, cytosol, and nucleus [12, 14]. These include Bcl-2, mitogen-activated protein kinase [256], c-Jun N-terminal kinase [257], and protein kinase C (PKCδ) [258]. SMAC also interacts with NADE (p75NTR-associated cell death), thereby promoting TRAIL-induced apoptosis [259] SMAC interacts with two nuclear proteins, the aryl hydrocarbon receptor nuclear translocator, ARNT/HIF-1β, and the transcriptional co-activator the mastermind like transcriptional coactivator 2 truncated poly Q MAML2, [260].
We recently identified the binding sites of SMAC with ten SMAC-interacting proteins: baculoviral IAP repeat containing two (BIRC2), TNF receptor associated factor 2 (TRAF2), MAML2(ARENT/HIF-1β), the ubiquitin conjugating enzyme E2K (UBE2K), baculoviral IAP2 repeat containing five (BIRC5, survivin), HtrA2, and PSD [12, 14].
The diverse proteins that interact with SMAC, and their functions and subcellular localizations clearly explain the multiple non-apoptotic functions of SMAC that are essential for cancer cells.
In summary, the pro-apoptotic SMAC is overexpressed in many cancers, probably because of additional non-apoptotic functions that contribute to cancer cell survival. Reducing SMAC expression inhibits cancer cell proliferation, both in culture and in tumors. The hundreds of genes shown to be differentially expressed in SMAC-depleted tumors are associated with a wide variety of cellular and tissue morphological traits and functions, which probably reflects the non-apoptotic functions of SMAC that are essential for tumorigenesis. These results highlight the significance of SMAC for tumor growth and progression, and as a potential target for the development of new therapeutic approaches for cancer treatment.
What controls whether apoptotic proteins promote pro- or non-apoptotic functions?
A major question is how, when, and where is the transformation from a pro-apoptotic to a non-apoptotic protein initiated and regulated?
The signal or condition(s) that determine the pro- or non-apoptotic outcomes are unclear, although several mechanisms have been postulated [263]. One mechanism proposes that whether caspases fulfil apoptotic or non-apoptotic functions depends on the activation of substrates or cofactors that are unique to each event [264], or that are influenced by the timing and intensity of the signaling pathway [264]. Another proposed mechanism suggests that the caspases induce the expression of specific genes [265, 266]. Indeed, microinjection of active caspase-3 into cells was found to induce the expression of muscle-specific genes [266]. In caspase-8-deficient neuroblastoma cells, death effector domain expression attenuated tumor growth by a mechanism involving upregulation of p53 and differentiation markers [265].
Our results indicate that silencing VDAC1 expression, reduces cellular ATP levels and cell proliferation, induces metabolic reprogramming, inhibits tumor development and angiogenesis, and alters the tumor microenvironment and the expression of over 4000 genes, including epigenetic-related enzymes and transcription factors [49, 231–234, 238, 267]. These findings suggest that the rewiring of cancer cell metabolism induced by VDAC1 depletion, alters the expression of caspases, Cyto c, AIF, SMAC, and TSPO [11], most probably via an interplay between metabolism and epigenetics.
The result of pro-apoptotic versus non-apoptotic activity may also depend on the sub-cellular localization of the protein. Indeed, caspases, AIF, and Cyto c can be found either in the nucleus or the cytosol [144, 268], but in VDAC1-depleted tumors, Cyto c, AIF, caspase-8 and -9, and SMAC are present in the nucleus [11, 12, 14]. However, the molecular basis of protein sub-cellular localization remains unknown.
A further option is that other apoptotic regulatory proteins modulate the activity of the pro-apoptotic proteins. For example, ubiquitination-mediated XIAP degradation may be responsible for some caspase-3 non-apoptotic functions [269].
Finally, other suggested mechanisms for controlling protein pro- or non-apoptotic function include spatial restriction, as shown for caspase-9 activation during thrombopoiesis only in perinuclear granular structures, or temporal restriction as seen for caspase-3 activation only during early erythropoiesis, or by substrate specificity, as demonstrated in several models of cellular differentiation [2].
Perspectives
Numerous proteins have additional functions, over and their main and well-characterized activities.
Many pro-apoptotic proteins fulfil non-apoptotic, non-lethal physiological functions in the cell cycle, differentiation, metabolism, and inflammation.
Several of these apoptotic proteins are overexpressed in cancers where they perform non-apoptotic functions and thus, are potential targets for treating cancer.
Some of these pro-apoptotic proteins translocate to the nucleus to perform non-apoptotic functions that include regulating gene expression and modulating DNA and RNA structures.
The mechanisms orchestrating the transformation from a pro-apoptotic to a non-apoptotic protein are not clear. They may involve the timing and intensity of the signaling pathway, substrate or cofactor specificity, metabolism-epigenetics cross-talk, and sub-cellular localization, although the exact mechanisms await further study.
Author contributions
TA: Writing the draft of some sections. AS-K: Prepared the figures. VSB: Writing, reviewing and editing, VSB: Supervision. All authors read and approved the submitted version.
Funding
This research was funded by The Israel Science Foundation, grant No 974/19.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Data availability
All data generated or analyzed during this study are included in this published articles and their supplementary information files.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17(7):395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L, et al. Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep. 2012;13(4):322–330. doi: 10.1038/embor.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskandari E, Eaves CJ (2022) Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J Cell Biol 221(6):e202201159 [DOI] [PMC free article] [PubMed]
- 4.Galluzzi L, et al. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 2008;15(7):1113–1123. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoshan-Barmatz V, Maldonado EN, Krelin Y. VDAC1 at the crossroads of cell metabolism, apoptosis and cell stress. Cell Stress. 2017;1(1):11–36. doi: 10.15698/cst2017.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoshan-Barmatz V, et al. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med. 2010;31(3):227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Keinan N, Tyomkin D, Shoshan-Barmatz V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol Cell Biol. 2010;30(24):5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchi S, Patergnani S, Pinton P. The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim Biophys Acta. 2014;1837(4):461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Zhou R, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 10.Hu H, et al. Mitochondrial VDAC1: a potential therapeutic target of inflammation-related diseases and clinical opportunities. Cells. 2022;11(9):3174. doi: 10.3390/cells11193174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arif T, Krelin Y, Shoshan-Barmatz V. Reducing VDAC1 expression induces a non-apoptotic role for pro-apoptotic proteins in cancer cell differentiation. Biochim Biophys Acta. 2016;1857(8):1228–1242. doi: 10.1016/j.bbabio.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Pandey SK, et al. SMAC/Diablo controls proliferation of cancer cells by regulating phosphatidylethanolamine synthesis. Mol Oncol. 2021;15(11):3037–3061. doi: 10.1002/1878-0261.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey SK, et al. Non-apoptotic activity of the mitochondrial protein SMAC/Diablo in lung cancer: novel target to disrupt survival, inflammation, and immunosuppression. Front Oncol. 2022;12:992260. doi: 10.3389/fonc.2022.992260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul A, et al. A new role for the mitochondrial pro-apoptotic protein SMAC/diablo in phospholipid synthesis associated with tumorigenesis. Mol Ther. 2018;26(3):680–694. doi: 10.1016/j.ymthe.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashkenazi A. Targeting the extrinsic apoptotic pathway in cancer: lessons learned and future directions. J Clin Invest. 2015;125(2):487–489. doi: 10.1172/JCI80420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider P, Tschopp J. Apoptosis induced by death receptors. Pharm Acta Helv. 2000;74(2–3):281–286. doi: 10.1016/S0031-6865(99)00038-2. [DOI] [PubMed] [Google Scholar]
- 17.Shoshan-Barmatz V, et al. (2015) The mitochondrial voltage-dependent anion channel 1 in tumor cells. Biochim Biophys Acta. 1848;1848(10 Pt B):2547–2575. doi: 10.1016/j.bbamem.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 18.van Loo G, et al. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 2002;9(1):20–26. doi: 10.1038/sj.cdd.4400970. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Lai Y, Hua ZC (2019) Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep 39(1). 10.1042/BSR20180992 [DOI] [PMC free article] [PubMed]
- 20.Vander Heiden MG, et al. Outer mitochondrial membrane permeability can regulate coupled respiration and cell survival. Proc Natl Acad Sci USA. 2000;97(9):4666–4671. doi: 10.1073/pnas.090082297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biasutto L, et al. (2016) The mitochondrial permeability transition pore in AD 2016: an update. Biochim Biophys Acta Mol Cell Res. 1863;10:2515–2530. doi: 10.1016/j.bbamcr.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol. 2006;18(6):685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135(6):1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019;366(6472):1531–1536. doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalk R, et al. Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem J. 2005;386(Pt 1):73–83. doi: 10.1042/BJ20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abu-Hamad S, et al. The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J Cell Sci. 2009;122(Pt 11):1906–1916. doi: 10.1242/jcs.040188. [DOI] [PubMed] [Google Scholar]
- 27.Shoshan-Barmatz V, Mizrachi D. VDAC1: from structure to cancer therapy. Front Oncol. 2012;2:164. doi: 10.3389/fonc.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoshan-Barmatz V, Mizrachi D, Keinan N. Oligomerization of the mitochondrial protein VDAC1: from structure to function and cancer therapy. Prog Mol Biol Transl Sci. 2013;117:303–334. doi: 10.1016/B978-0-12-386931-9.00011-8. [DOI] [PubMed] [Google Scholar]
- 29.Keinan N, et al. The role of calcium in VDAC1 oligomerization and mitochondria-mediated apoptosis. Biochim Biophys Acta. 2013;1833(7):1745–1754. doi: 10.1016/j.bbamcr.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Weisthal S, et al. Ca(2+)-mediated regulation of VDAC1 expression levels is associated with cell death induction. Biochim Biophys Acta. 2014;1843(10):2270–2281. doi: 10.1016/j.bbamcr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Hail D, et al. Begas-Shvartz R, Shalev M, Gruzman A, Shoshan-Barmatz V (2016) The mitochondrial protein VDAC1 as a target for novel anti-apoptotic compounds. Cell Death Dis. 2016;291(48):24986–25003. doi: 10.1074/jbc.M116.744284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geula S, et al. Structure-based analysis of VDAC1 protein: defining oligomer contact sites. J Biol Chem. 2012;287(3):2179–2190. doi: 10.1074/jbc.M111.268920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smilansky A, et al. The voltage-dependent anion channel 1 mediates amyloid beta toxicity and represents a potential target for Alzheimer disease therapy. J Biol Chem. 2015;290(52):30670–30683. doi: 10.1074/jbc.M115.691493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang L, et al. A new fungal diterpene induces VDAC1-dependent apoptosis in Bax/Bak-deficient cells. J Biol Chem. 2015;290(39):23563–23578. doi: 10.1074/jbc.M115.648774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 36.Shoshan-Barmatz V, Shteinfer-Kuzmine A, Verma A (2020) VDAC1 at the intersection of cell metabolism, apoptosis, and diseases. Biomolecules 10(11) [DOI] [PMC free article] [PubMed]
- 37.Geula S, Ben-Hail D, Shoshan-Barmatz V. Structure-based analysis of VDAC1: N-terminus location, translocation, channel gating and association with anti-apoptotic proteins. Biochem J. 2012;444(3):475–485. doi: 10.1042/BJ20112079. [DOI] [PubMed] [Google Scholar]
- 38.Bayrhuber M, et al. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105(40):15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiller S, et al. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ujwal R, et al. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105(46):17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, et al. Identification of the protein-protein contact site and interaction mode of human VDAC1 with Bcl-2 family proteins. Biochem Biophys Res Commun. 2003;305(4):989–996. doi: 10.1016/S0006-291X(03)00871-4. [DOI] [PubMed] [Google Scholar]
- 42.Arbel N, Ben-Hail D, Shoshan-Barmatz V. Mediation of the antiapoptotic activity of Bcl-xL protein upon interaction with VDAC1 protein. J Biol Chem. 2012;287(27):23152–23161. doi: 10.1074/jbc.M112.345918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arbel N, Shoshan-Barmatz V. Voltage-dependent anion channel 1-based peptides interact with Bcl-2 to prevent antiapoptotic activity. J Biol Chem. 2010;285(9):6053–6062. doi: 10.1074/jbc.M109.082990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arzoine L, et al. Voltage-dependent anion channel 1-based peptides interact with hexokinase to prevent its anti-apoptotic activity. J Biol Chem. 2009;284(6):3946–3955. doi: 10.1074/jbc.M803614200. [DOI] [PubMed] [Google Scholar]
- 45.Shoshan-Barmatz V, et al. Voltage-dependent anion channel 1 as an emerging drug target for novel anti-cancer therapeutics. Front Oncol. 2017;7:154. doi: 10.3389/fonc.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abu-Hamad S, et al. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: mapping the site of binding. J Biol Chem. 2008;283(19):13482–13490. doi: 10.1074/jbc.M708216200. [DOI] [PubMed] [Google Scholar]
- 47.Yan J, et al. VDAC oligomer pores: A mechanism in disease triggered by mtDNA release. Cell Biol Int. 2020;44(11):2178–2181. doi: 10.1002/cbin.11427. [DOI] [PubMed] [Google Scholar]
- 48.Shoshan-Barmatz V, Golan M. Mitochondrial VDAC1: function in cell life and death and a target for cancer therapy. Curr Med Chem. 2012;19(5):714–735. doi: 10.2174/092986712798992110. [DOI] [PubMed] [Google Scholar]
- 49.Arif T, et al. Silencing VDAC1 expression by siRNA inhibits cancer cell proliferation and tumor growth in vivo. Mol Ther Nucleic Acids. 2014;3:e159. doi: 10.1038/mtna.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shteinfer-Kuzmine A, et al. Mitochondria and nucleus cross-talk: signaling in metabolism, apoptosis, and differentiation, and function in cancer. IUBMB Life. 2021;73(3):492–510. doi: 10.1002/iub.2407. [DOI] [PubMed] [Google Scholar]
- 51.Cuadrado-Tejedor M, et al. Enhanced expression of the voltage-dependent anion channel 1 (VDAC1) in Alzheimer's disease transgenic mice: an insight into the pathogenic effects of amyloid-beta. J Alzheimers Dis. 2011;23(2):195–206. doi: 10.3233/JAD-2010-100966. [DOI] [PubMed] [Google Scholar]
- 52.Shoshan-Barmatz V, et al. VDAC1, mitochondrial dysfunction, and Alzheimer’s disease. Pharmacol Res. 2018;131:87–101. doi: 10.1016/j.phrs.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Shteinfer-Kuzmine A, et al. A VDAC1-derived N-terminal peptide inhibits mutant SOD1-VDAC1 interactions and toxicity in the SOD1 model of ALS. Front Cell Neurosci. 2019;13:346. doi: 10.3389/fncel.2019.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verma A, et al. Targeting the overexpressed mitochondrial protein VDAC1 in a mouse model of Alzheimer's disease protects against mitochondrial dysfunction and mitigates brain pathology. Transl Neurodegener. 2022;11(1):58. doi: 10.1186/s40035-022-00329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang E, et al. Preserving insulin secretion in diabetes by inhibiting VDAC1 overexpression and surface translocation in β Cells. Cell Metab. 2019;29(1):64–77. doi: 10.1016/j.cmet.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verma A, et al. The role of the mitochondrial protein VDAC1 in inflammatory bowel disease: a potential therapeutic target. Mol Therapy. 2021;30(2):726–744. doi: 10.1016/j.ymthe.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pittala S, et al. A mitochondrial VDAC1-based peptide greatly suppresses steatosis and NASH-associated pathologies in a mouse model. Mol Ther. 2019;27(10):1848–1862. doi: 10.1016/j.ymthe.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Niu B, et al. Protecting mitochondria via inhibiting VDAC1 oligomerization alleviates ferroptosis in acetaminophen-induced acute liver injury. Cell Biol Toxicol. 2021;38(3):505–530. doi: 10.1007/s10565-021-09624-x. [DOI] [PubMed] [Google Scholar]
- 59.Zeng F, et al. Central role of RIPK1-VDAC1 pathway on cardiac impairment in a non-human primate model of rheumatoid arthritis. J Mol Cell Cardiol. 2018;125:50–60. doi: 10.1016/j.yjmcc.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 60.Klapper-Goldstein H, et al. VDAC1 in the diseased myocardium and the effect of VDAC1-interacting compound on atrial fibrosis induced by hyperaldosteronism. Sci Rep. 2020;10(1):22101. doi: 10.1038/s41598-020-79056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson EA, et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34(11):108863. doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian C, et al. Mitochondria related cell death modalities and disease. Front Cell Dev Biol. 2022;10:832356. doi: 10.3389/fcell.2022.832356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olsson M, Zhivotovsky B. Caspases and cancer. Cell Death Differ. 2011;18(9):1441–1449. doi: 10.1038/cdd.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shalini S, et al. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22(4):526–539. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baena-Lopez LA. All about the caspase-dependent functions without cell death. Semin Cell Dev Biol. 2018;82:77–78. doi: 10.1016/j.semcdb.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 66.Su TT (2020) Non-apoptotic roles of apoptotic proteases: new tricks for an old dog. Open Biol 10(8). 10.1098/rsob.200130 [DOI] [PMC free article] [PubMed]
- 67.Schwerk C, Schulze-Osthoff K. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem Pharmacol. 2003;66(8):1453–1458. doi: 10.1016/S0006-2952(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 68.Sordet O, et al. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002;100(13):4446–4453. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- 69.Lamarque M, et al. Role of caspase-10-P13tBID axis in erythropoiesis regulation. Cell Death Differ. 2022;30:208–220. doi: 10.1038/s41418-022-01066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zermati Y, et al. Caspase activation is required for terminal erythroid differentiation. J Exp Med. 2001;193(2):247–254. doi: 10.1084/jem.193.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dehkordi MH, Munn RGK, Fearnhead HO. Non-canonical roles of apoptotic caspases in the nervous system. Front Cell Dev Biol. 2022;10:840023. doi: 10.3389/fcell.2022.840023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miura M, et al. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J Clin Invest. 2004;114(12):1704–1713. doi: 10.1172/JCI20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Svandova E, et al. Activation of pro-apoptotic caspases in non-apoptotic cells during odontogenesis and related osteogenesis. Front Physiol. 2018;9:174. doi: 10.3389/fphys.2018.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Philchenkov A, et al. Caspases and cancer: mechanisms of inactivation and new treatment modalities. Exp Oncol. 2004;26(2):82–97. [PubMed] [Google Scholar]
- 75.Ojha S, Tapadia MG. Nonapoptotic role of caspase-3 in regulating Rho1GTPase-mediated morphogenesis of epithelial tubes of Drosophila renal system. Dev Dyn. 2022;251(5):777–794. doi: 10.1002/dvdy.437. [DOI] [PubMed] [Google Scholar]
- 76.Su TT. Non-apoptotic roles of apoptotic proteases: new tricks for an old dog. Open Biol. 2020;10(8):200130. doi: 10.1098/rsob.200130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le DA, et al. Caspase activation and neuroprotection in caspase-3- deficient mice after in vivo cerebral ischemia and in vitro oxygen glucose deprivation. Proc Natl Acad Sci USA. 2002;99(23):15188–15193. doi: 10.1073/pnas.232473399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lo SC, Scearce-Levie K, Sheng M. Characterization of social behaviors in caspase-3 deficient mice. Sci Rep. 2016;6:18335. doi: 10.1038/srep18335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boudreau MW, Peh J, Hergenrother PJ. Procaspase-3 overexpression in cancer: a paradoxical observation with therapeutic potential. ACS Chem Biol. 2019;14(11):2335–2348. doi: 10.1021/acschembio.9b00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Persad R, et al. Overexpression of caspase-3 in hepatocellular carcinomas. Mod Pathol. 2004;17(7):861–867. doi: 10.1038/modpathol.3800146. [DOI] [PubMed] [Google Scholar]
- 81.Matalova E, et al. Caspase-7 participates in differentiation of cells forming dental hard tissues. Dev Growth Differ. 2013;55(5):615–621. doi: 10.1111/dgd.12066. [DOI] [PubMed] [Google Scholar]
- 82.Svandova E, et al. Non-apoptotic functions of caspase-7 during osteogenesis. Cell Death Dis. 2014;5:e1366. doi: 10.1038/cddis.2014.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lei B, et al. Apoptotic and nonapoptotic function of caspase 7 in spermatogenesis. Asian J Androl. 2017;19(1):47–51. doi: 10.4103/1008-682X.169563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hashimoto T, Kikkawa U, Kamada S. Contribution of caspase(s) to the cell cycle regulation at mitotic phase. PLoS ONE. 2011;6(3):e18449. doi: 10.1371/journal.pone.0018449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Safari F, Akbari B. Knockout of caspase-7 gene improves the expression of recombinant protein in CHO cell line through the cell cycle arrest in G2/M phase. Biol Res. 2022;55(1):2. doi: 10.1186/s40659-021-00369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lakhani SA, et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311(5762):847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palmerini F, et al. Caspase 7 downregulation as an immunohistochemical marker of colonic carcinoma. Hum Pathol. 2001;32(5):461–467. doi: 10.1053/hupa.2001.24328. [DOI] [PubMed] [Google Scholar]
- 88.Chaudhary S, et al. Overexpression of caspase 7 is ERalpha dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip) Oncogenesis. 2016;5:e219. doi: 10.1038/oncsis.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coutinho-Camillo CM, et al. Caspase expression in oral squamous cell carcinoma. Head Neck. 2011;33(8):1191–1198. doi: 10.1002/hed.21602. [DOI] [PubMed] [Google Scholar]
- 90.Denicourt C, Dowdy SF. Targeting apoptotic pathways in cancer cells. Science. 2004;305(5689):1411–1413. doi: 10.1126/science.1102974. [DOI] [PubMed] [Google Scholar]
- 91.Fritsch M et al (2019) Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 575(7784): 683 [DOI] [PubMed]
- 92.Li X, et al. Caspase-8 auto-cleavage regulates programmed cell death and collaborates with RIPK3/MLKL to prevent lymphopenia. Cell Death Differ. 2022;29(8):1500–1512. doi: 10.1038/s41418-022-00938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maelfait J, Beyaert R. Non-apoptotic functions of caspase-8. Biochem Pharmacol. 2008;76(11):1365–1373. doi: 10.1016/j.bcp.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 94.Xia J, et al. Non-apoptotic function of caspase-8 confers prostate cancer enzalutamide resistance via NF-kappaB activation. Cell Death Dis. 2021;12(9):833. doi: 10.1038/s41419-021-04126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chun HJ, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419(6905):395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 96.Barnabei L, et al. NF-kappaB: at the borders of autoimmunity and inflammation. Front Immunol. 2021;12:716469. doi: 10.3389/fimmu.2021.716469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Henry CM, Martin SJ (2017) Caspase-8 acts in a non-enzymatic role as a Scaffold for assembly of a pro-inflammatory “FADDosome” complex upon TRAIL stimulation. Mol Cell 65(4): 715–729 e5 [DOI] [PubMed]
- 98.Philip NH, et al. Activity of uncleaved Caspase-8 controls anti-bacterial immune defense and TLR-induced cytokine production independent of cell death. PLoS Pathog. 2016;12(10):e1005910. doi: 10.1371/journal.ppat.1005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao R, et al. Novel roles of apoptotic caspases in tumor repopulation, epigenetic reprogramming, carcinogenesis, and beyond. Cancer Metastasis Rev. 2018;37(2–3):227–236. doi: 10.1007/s10555-018-9736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stupack DG. Caspase-8 as a therapeutic target in cancer. Cancer Lett. 2013;332(2):133–140. doi: 10.1016/j.canlet.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Avrutsky MI, Troy CM. Caspase-9: a multimodal therapeutic target with diverse cellular expression in human disease. Front Pharmacol. 2021;12:701301. doi: 10.3389/fphar.2021.701301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murray TV, et al. A non-apoptotic role for caspase-9 in muscle differentiation. J Cell Sci. 2008;121(Pt 22):3786–3793. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- 103.Pistritto G, et al. Divergent modulation of neuronal differentiation by caspase-2 and -9. PLoS ONE. 2012;7(5):e36002. doi: 10.1371/journal.pone.0036002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Madadi Z, et al. The non-apoptotic role of caspase-9 promotes differentiation in leukemic cells. Biochim Biophys Acta Mol Cell Res. 2019;1866(12):118524. doi: 10.1016/j.bbamcr.2019.118524. [DOI] [PubMed] [Google Scholar]
- 105.Fernando P, Brunette S, Megeney LA. Neural stem cell differentiation is dependent upon endogenous caspase 3 activity. FASEB J. 2005;19(12):1671–1673. doi: 10.1096/fj.04-2981fje. [DOI] [PubMed] [Google Scholar]
- 106.Karimzadeh S, et al. Insufficient Apaf-1 expression in early stages of neural differentiation of human embryonic stem cells might protect them from apoptosis. Eur J Cell Biol. 2018;97(2):126–135. doi: 10.1016/j.ejcb.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 107.Shakeri R, Kheirollahi A, Davoodi J. Apaf-1: regulation and function in cell death. Biochimie. 2017;135:111–125. doi: 10.1016/j.biochi.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 108.Shakeri R, Kheirollahi A, Davoodi J. Contribution of Apaf-1 to the pathogenesis of cancer and neurodegenerative diseases. Biochimie. 2021;190:91–110. doi: 10.1016/j.biochi.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 109.Mouhamad S, et al. Apaf-1 deficiency causes chromosomal instability. Cell Cycle. 2007;6(24):3103–3107. doi: 10.4161/cc.6.24.5046. [DOI] [PubMed] [Google Scholar]
- 110.Zermati Y, et al. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol Cell. 2007;28(4):624–637. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 111.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 112.Jaiswal PK, Goel A, Mittal RD. Survivin: A molecular biomarker in cancer. Indian J Med Res. 2015;141(4):389–397. doi: 10.4103/0971-5916.159250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shin S, et al. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40(4):1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 114.Vong QP, et al. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310(5753):1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 115.Liu HR, et al. Role of Omi/HtrA2 in apoptotic cell death after myocardial ischemia and reperfusion. Circulation. 2005;111(1):90–96. doi: 10.1161/01.CIR.0000151613.90994.17. [DOI] [PubMed] [Google Scholar]
- 116.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412(6842):95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 117.Wiehe RS, et al. Endonuclease G promotes mitochondrial genome cleavage and replication. Oncotarget. 2018;9(26):18309–18326. doi: 10.18632/oncotarget.24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Veenman L, Vainshtein A, Gavish M. TSPO as a target for treatments of diseases, including neuropathological disorders. Cell Death Dis. 2015;6:e1911. doi: 10.1038/cddis.2015.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scorrano L, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300(5616):135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 120.Chemaly ER, Troncone L, Lebeche D. SERCA control of cell death and survival. Cell Calcium. 2018;69:46–61. doi: 10.1016/j.ceca.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25(1):65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maiuri MC, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3(4):374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 123.Pihan P, Carreras-Sureda A, Hetz C. BCL-2 family: integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017;24(9):1478–1487. doi: 10.1038/cdd.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pinton P, Rizzuto R. Bcl-2 and Ca2+ homeostasis in the endoplasmic reticulum. Cell Death Differ. 2006;13(8):1409–1418. doi: 10.1038/sj.cdd.4401960. [DOI] [PubMed] [Google Scholar]
- 125.Vervliet T, et al. Modulation of Ca(2+) signaling by anti-apoptotic B-cell lymphoma 2 proteins at the endoplasmic reticulum-mitochondrial interface. Front Oncol. 2017;7:75. doi: 10.3389/fonc.2017.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hardwick JM, Soane L (2013) Multiple functions of BCL-2 family proteins. Cold Spring Harb Perspect Biol 5(2):a008722 [DOI] [PMC free article] [PubMed]
- 127.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Decuypere JP, Parys JB, Bultynck G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells. 2012;1(3):284–312. doi: 10.3390/cells1030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282(17):13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 130.Azoulay-Zohar H, et al. In self-defence: hexokinase promotes voltage-dependent anion channel closure and prevents mitochondria-mediated apoptotic cell death. Biochem J. 2004;377(Pt 2):347–355. doi: 10.1042/bj20031465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pastorino JG, Hoek JB. Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr. 2008;40(3):171–182. doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277(9):7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 133.Shoshan-Barmatz V, et al. Key regions of VDAC1 functioning in apoptosis induction and regulation by hexokinase. Biochim Biophys Acta. 2009;1787(5):421–430. doi: 10.1016/j.bbabio.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 134.Goldin N, et al. Methyl jasmonate binds to and detaches mitochondria-bound hexokinase. Oncogene. 2008;27(34):4636–4643. doi: 10.1038/onc.2008.108. [DOI] [PubMed] [Google Scholar]
- 135.Pastorino JG, Hoek JB, Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer Res. 2005;65(22):10545–10554. doi: 10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- 136.Shoshan-Barmatz V, Keinan N, Zaid H. Uncovering the role of VDAC in the regulation of cell life and death. J Bioenerg Biomembr. 2008;40(3):183–191. doi: 10.1007/s10863-008-9147-9. [DOI] [PubMed] [Google Scholar]
- 137.Shoshan-Barmatz V, et al. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Curr Pharm Des. 2006;12(18):2249–2270. doi: 10.2174/138161206777585111. [DOI] [PubMed] [Google Scholar]
- 138.Perez-Mejias G et al (2022) Novel insights into the mechanism of electron transfer in mitochondrial cytochrome c. Coord Chem Rev 450. 10.1016/j.ccr.2021.214233
- 139.Huttemann M, et al. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: from respiration to apoptosis. Mitochondrion. 2011;11(3):369–381. doi: 10.1016/j.mito.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci. 2010;123(Pt 19):3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li K, van Delft MF, Dewson G. Too much death can kill you: inhibiting intrinsic apoptosis to treat disease. EMBO J. 2021;40(14):e107341. doi: 10.15252/embj.2020107341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gonzalez-Arzola K, et al. Nucleus-translocated mitochondrial cytochrome c liberates nucleophosmin-sequestered ARF tumor suppressor by changing nucleolar liquid-liquid phase separation. Nat Struct Mol Biol. 2022;29(10):1024–1036. doi: 10.1038/s41594-022-00842-3. [DOI] [PubMed] [Google Scholar]
- 143.Gonzalez-Arzola K, et al. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Lett. 2019;593(22):3101–3119. doi: 10.1002/1873-3468.13655. [DOI] [PubMed] [Google Scholar]
- 144.Nur EKA, et al. Nuclear translocation of cytochrome c during apoptosis. J Biol Chem. 2004;279(24):24911–24914. doi: 10.1074/jbc.C400051200. [DOI] [PubMed] [Google Scholar]
- 145.Godoy LC, et al. Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc Natl Acad Sci USA. 2009;106(8):2653–2658. doi: 10.1073/pnas.0809279106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Afify M, et al. The possible role of cytochrome c and programmed cell death protein 4 (PDCD4) on pathogenesis of hepatocellular carcinoma. J Genet Eng Biotechnol. 2015;13(2):157–163. doi: 10.1016/j.jgeb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu Z, et al. Cytochrome C inhibits tumor growth and predicts favorable prognosis in clear cell renal cell carcinoma. Oncol Lett. 2019;18(6):6026–6032. doi: 10.3892/ol.2019.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Soltys BJ, et al. Cytochrome-C localizes in secretory granules in pancreas and anterior pituitary. Cell Biol Int. 2001;25(4):331–338. doi: 10.1006/cbir.2000.0651. [DOI] [PubMed] [Google Scholar]
- 149.Guerra-Castellano A et al (2020) Post-translational modifications of cytochrome c in cell life and disease. Int J Mol Sci 21(22) [DOI] [PMC free article] [PubMed]
- 150.Gomila AMJ, et al. Phosphorylation disrupts long-distance electron transport in cytochrome c. Nat Commun. 2022;13(1):7100. doi: 10.1038/s41467-022-34809-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Artus C, et al. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. EMBO J. 2010;29(9):1585–1599. doi: 10.1038/emboj.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Baritaud M, et al. AIF-mediated caspase-independent necroptosis requires ATM and DNA-PK-induced histone H2AX Ser139 phosphorylation. Cell Death Dis. 2012;3(9):e390. doi: 10.1038/cddis.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Delavallee L, et al. AIF-mediated caspase-independent necroptosis: a new chance for targeted therapeutics. IUBMB Life. 2011;63(4):221–232. doi: 10.1002/iub.432. [DOI] [PubMed] [Google Scholar]
- 154.Sevrioukova IF. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxid Redox Signal. 2011;14(12):2545–2579. doi: 10.1089/ars.2010.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vahsen N, et al. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23(23):4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wischhof L, et al. AIFM1 beyond cell death: An overview of this OXPHOS-inducing factor in mitochondrial diseases. EBioMedicine. 2022;83:104231. doi: 10.1016/j.ebiom.2022.104231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Joza N, et al. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25(23):10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pospisilik JA, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131(3):476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 159.Shen SM, et al. Apoptosis-inducing factor is a target gene of C/EBPalpha and participates in adipocyte differentiation. FEBS Lett. 2011;585(14):2307–2312. doi: 10.1016/j.febslet.2011.05.061. [DOI] [PubMed] [Google Scholar]
- 160.Reinhardt C, et al. AIF meets the CHCHD4/Mia40-dependent mitochondrial import pathway. Biochim Biophys Acta Mol Basis Dis. 2020;1866(6):165746. doi: 10.1016/j.bbadis.2020.165746. [DOI] [PubMed] [Google Scholar]
- 161.Ademoni-Elisha L, Nakdimon I, Shteinfer A, Prezma T, Arif T, Arbel N, Melkov A, Zelichov O, Levi I, Shoshan-Barmatz V, et al. Novel biomarker proteins in chronic lymphocytic leukemia: Impact on diagnosis, prognosis and treatment. PLoS One. 2016;11(4):e0148500. doi: 10.1371/journal.pone.0148500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lewis EM, et al. The enzymatic activity of apoptosis-inducing factor supports energy metabolism benefiting the growth and invasiveness of advanced prostate cancer cells. J Biol Chem. 2012;287(52):43862–43875. doi: 10.1074/jbc.M112.407650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Urbano A, et al. AIF suppresses chemical stress-induced apoptosis and maintains the transformed state of tumor cells. EMBO J. 2005;24(15):2815–2826. doi: 10.1038/sj.emboj.7600746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Du C, et al. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102(1):33–42. doi: 10.1016/S0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 165.Gray CW, et al. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267(18):5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- 166.Hur W et al (2019) Serine protease HtrA2/Omi deficiency impairs mitochondrial homeostasis and promotes hepatic fibrogenesis via activation of hepatic stellate cells. Cells 8(10):1119 [DOI] [PMC free article] [PubMed]
- 167.Vande Walle L, Lamkanfi M, Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15(3):453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- 168.Verhagen AM, et al. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J Biol Chem. 2002;277(1):445–454. doi: 10.1074/jbc.M109891200. [DOI] [PubMed] [Google Scholar]
- 169.Kuninaka S, et al. Serine protease Omi/HtrA2 targets WARTS kinase to control cell proliferation. Oncogene. 2007;26(17):2395–2406. doi: 10.1038/sj.onc.1210042. [DOI] [PubMed] [Google Scholar]
- 170.Goo HG, et al. HtrA2/Omi deficiency causes damage and mutation of mitochondrial DNA. Biochim Biophys Acta. 2013;1833(8):1866–1875. doi: 10.1016/j.bbamcr.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 171.Kang S, Fernandes-Alnemri T, Alnemri ES. A novel role for the mitochondrial HTRA2/OMI protease in aging. Autophagy. 2013;9(3):420–421. doi: 10.4161/auto.22920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vaux DL, Silke J. HtrA2/Omi, a sheep in wolf's clothing. Cell. 2003;115(3):251–253. doi: 10.1016/S0092-8674(03)00851-1. [DOI] [PubMed] [Google Scholar]
- 173.Darreh-Shori T, et al. Increased active OMI/HTRA2 serine protease displays a positive correlation with cholinergic alterations in the Alzheimer's Disease brain. Mol Neurobiol. 2019;56(7):4601–4619. doi: 10.1007/s12035-018-1383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Westerlund M, et al. Altered enzymatic activity and allele frequency of OMI/HTRA2 in Alzheimer's disease. FASEB J. 2011;25(4):1345–1352. doi: 10.1096/fj.10-163402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Plun-Favreau H, et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat Cell Biol. 2007;9(11):1243–U63. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 176.Chung HJ, Jamal M, Hong ST. The function of bacterial HtrA is evolutionally conserved in mammalian HtrA2/Omi. Sci Rep. 2020;10(1):5284. doi: 10.1038/s41598-020-62309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ohsato T, et al. Mammalian mitochondrial endonuclease G. Digestion of R-loops and localization in intermembrane space. Eur J Biochem. 2002;269(23):5765–5770. doi: 10.1046/j.1432-1033.2002.03238.x. [DOI] [PubMed] [Google Scholar]
- 178.Wang W, et al. Endonuclease G promotes autophagy by suppressing mTOR signaling and activating the DNA damage response. Nat Commun. 2021;12(1):476. doi: 10.1038/s41467-020-20780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.David KK, et al. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13(7):1147–55. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- 180.Huang KJ, Ku CC, Lehman IR. Endonuclease G: a role for the enzyme in recombination and cellular proliferation. Proc Natl Acad Sci USA. 2006;103(24):8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McDermott-Roe C, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478(7367):114–8. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Buttner S, et al. Depletion of endonuclease G selectively kills polyploid cells. Cell Cycle. 2007;6(9):1072–1076. doi: 10.4161/cc.6.9.4218. [DOI] [PubMed] [Google Scholar]
- 183.Chao T, et al. Autophagy restricts mitochondrial DNA damage-induced release of ENDOG (endonuclease G) to regulate genome stability. Autophagy. 2021;17(11):3444–3460. doi: 10.1080/15548627.2021.1874209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cheung CH, et al. A cell-permeable dominant-negative survivin protein induces apoptosis and sensitizes prostate cancer cells to TNF-alpha therapy. Cancer Cell Int. 2010;10:36. doi: 10.1186/1475-2867-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lin KY, et al. Delivery of a survivin promoter-driven antisense survivin-expressing plasmid DNA as a cancer therapeutic: a proof-of-concept study. Onco Targets Ther. 2016;9:2601–13. doi: 10.2147/OTT.S101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Montazeri Aliabadi H, et al. Induction of apoptosis by survivin silencing through siRNA delivery in a human breast cancer cell line. Mol Pharm. 2011;8(5):1821–30. doi: 10.1021/mp200176v. [DOI] [PubMed] [Google Scholar]
- 187.Raviv Z, et al. Methyl jasmonate down-regulates survivin expression and sensitizes colon carcinoma cells towards TRAIL-induced cytotoxicity. Br J Pharmacol. 2011;164(5):1433–44. doi: 10.1111/j.1476-5381.2011.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Uchida H, et al. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther. 2004;10(1):162–71. doi: 10.1016/j.ymthe.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 189.Cheung CHA, et al. Anti-apoptotic proteins in the autophagic world: an update on functions of XIAP, Survivin, and BRUCE. J Biomed Sci. 2020;27(1):31. doi: 10.1186/s12929-020-0627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Garg H, et al. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016;16:49. doi: 10.1186/s12935-016-0326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pavlyukov MS, et al. Survivin monomer plays an essential role in apoptosis regulation. J Biol Chem. 2011;286(26):23296–307. doi: 10.1074/jbc.M111.237586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Klein UR, Nigg EA, Gruneberg U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol Biol Cell. 2006;17(6):2547–58. doi: 10.1091/mbc.e05-12-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li D, Hu C, Li H. Survivin as a novel target protein for reducing the proliferation of cancer cells. Biomed Rep. 2018;8(5):399–406. doi: 10.3892/br.2018.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Babkoff A. A direct interaction between survivin and myosin II is required for cytokinesis. J Cell Sci. 2019;132(14):jcs233130. doi: 10.1242/jcs.233130. [DOI] [PubMed] [Google Scholar]
- 195.Veenman L, Papadopoulos V, Gavish M. Channel-like functions of the 18-kDa translocator protein (TSPO): regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr Pharm Des. 2007;13(23):2385–405. doi: 10.2174/138161207781368710. [DOI] [PubMed] [Google Scholar]
- 196.Bhoola NH et al (2018) Translocator protein (TSPO) as a potential biomarker in human cancers. Int J Mol Sci 19(8) [DOI] [PMC free article] [PubMed]
- 197.Veenman L, Gavish M. The role of 18 kDa mitochondrial translocator protein (TSPO) in programmed cell death, and effects of steroids on TSPO expression. Curr Mol Med. 2012;12(4):398–412. doi: 10.2174/1566524011207040398. [DOI] [PubMed] [Google Scholar]
- 198.Beinlich A, et al. Relation of cell proliferation to expression of peripheral benzodiazepine receptors in human breast cancer cell lines. Biochem Pharmacol. 2000;60(3):397–402. doi: 10.1016/S0006-2952(00)00325-7. [DOI] [PubMed] [Google Scholar]
- 199.Gatliff J, et al. A role for TSPO in mitochondrial Ca(2+) homeostasis and redox stress signaling. Cell Death Dis. 2017;8(6):e2896–e2896. doi: 10.1038/cddis.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tu LN, et al. Translocator protein (TSPO) affects mitochondrial fatty acid oxidation in steroidogenic cells. Endocrinology. 2016;157(3):1110–21. doi: 10.1210/en.2015-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Batarseh A, Papadopoulos V. Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol Cell Endocrinol. 2010;327(1–2):1–12. doi: 10.1016/j.mce.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shoshan-Barmatz V, Pittala S, Mizrachi D (2019) VDAC1 and the TSPO: expression, interactions, and associated functions in health and disease states. Int J Mol Sci 20(13):3348 [DOI] [PMC free article] [PubMed]
- 203.Joo HK, et al. Peripheral benzodiazepine receptor regulates vascular endothelial activations via suppression of the voltage-dependent anion channel-1. FEBS Lett. 2012;586(9):1349–55. doi: 10.1016/j.febslet.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 204.Veenman L, Shandalov Y, Gavish M. VDAC activation by the 18 kDa translocator protein (TSPO), implications for apoptosis. J Bioenerg Biomembr. 2008;40(3):199–205. doi: 10.1007/s10863-008-9142-1. [DOI] [PubMed] [Google Scholar]
- 205.Wilson JE. Homologous and heterologous interactions between hexokinase and mitochondrial porin: evolutionary implications. J Bioenerg Biomembr. 1997;29(1):97–102. doi: 10.1023/A:1022472124746. [DOI] [PubMed] [Google Scholar]
- 206.Nakashima RA, et al. Hexokinase receptor complex in hepatoma mitochondria: evidence from N, N′-dicyclohexylcarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry. 1986;25(5):1015–21. doi: 10.1021/bi00353a010. [DOI] [PubMed] [Google Scholar]
- 207.Pedersen PL. Voltage dependent anion channels (VDACs): a brief introduction with a focus on the outer mitochondrial compartment's roles together with hexokinase-2 in the “Warburg effect” in cancer. J Bioenerg Biomembr. 2008;40(3):123–6. doi: 10.1007/s10863-008-9165-7. [DOI] [PubMed] [Google Scholar]
- 208.Majewski N, et al. Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol Cell Biol. 2004;24(2):730–40. doi: 10.1128/MCB.24.2.730-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pedersen PL, et al. Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim Biophys Acta. 2002;1555(1–3):14–20. doi: 10.1016/S0005-2728(02)00248-7. [DOI] [PubMed] [Google Scholar]
- 210.Bryson JM, et al. Increased hexokinase activity, of either ectopic or endogenous origin, protects renal epithelial cells against acute oxidant-induced cell death. J Biol Chem. 2002;277(13):11392–400. doi: 10.1074/jbc.M110927200. [DOI] [PubMed] [Google Scholar]
- 211.Brown RS, et al. Expression of hexokinase II and Glut-1 in untreated human breast cancer. Nucl Med Biol. 2002;29(4):443–53. doi: 10.1016/S0969-8051(02)00288-3. [DOI] [PubMed] [Google Scholar]
- 212.Katabi MM, et al. Hexokinase type II: a novel tumor-specific promoter for gene-targeted therapy differentially expressed and regulated in human cancer cells. Hum Gene Ther. 1999;10(2):155–64. doi: 10.1089/10430349950018959. [DOI] [PubMed] [Google Scholar]
- 213.Mathupala SP, Ko YH, Pedersen PL. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta. 2010;1797(6–7):1225–30. doi: 10.1016/j.bbabio.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gelb BD, et al. Targeting of hexokinase 1 to liver and hepatoma mitochondria. Proc Natl Acad Sci USA. 1992;89(1):202–6. doi: 10.1073/pnas.89.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.He XS, et al. Overexpression of Hexokinase 1 as a poor prognosticator in human colorectal cancer. Tumor Biol. 2016;37(3):3887–3895. doi: 10.1007/s13277-015-4255-8. [DOI] [PubMed] [Google Scholar]
- 216.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25(34):4777–86. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.da Silva WS, et al. Mitochondrial bound hexokinase activity as a preventive antioxidant defense: steady-state ADP formation as a regulatory mechanism of membrane potential and reactive oxygen species generation in mitochondria. J Biol Chem. 2004;279(38):39846–39855. doi: 10.1074/jbc.M403835200. [DOI] [PubMed] [Google Scholar]
- 218.Krasnov GS, et al. Targeting VDAC-bound hexokinase II: a promising approach for concomitant anti-cancer therapy. Expert Opin Ther Targets. 2013;17(10):1221–33. doi: 10.1517/14728222.2013.833607. [DOI] [PubMed] [Google Scholar]
- 219.Ganapathy-Kanniappan S (2015) Targeting glycolytic adaptations of cancer cells: from molecular mechanisms to therapeutic opportunities. stress response pathways in cancer. In: Wondrak GT (ed) Chapter 15. Springer, Dordrecht, Heidelberg, New York, London
- 220.Bahatyrevich-Kharitonik B, et al. Cell death related proteins beyond apoptosis in the CNS. Front Cell Dev Biol. 2021;9:825747. doi: 10.3389/fcell.2021.825747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ivanova H, et al. Bcl-2-protein family as modulators of IP(3) receptors and other organellar Ca(2+) channels. Cold Spring Harb Perspect Biol. 2020;12(4):a035089. doi: 10.1101/cshperspect.a035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.White C, et al. The endoplasmic reticulum gateway to apoptosis by Bcl-X(L) modulation of the InsP3R. Nat Cell Biol. 2005;7(10):1021–8. doi: 10.1038/ncb1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nougarede A, et al. Breast cancer targeting through inhibition of the endoplasmic reticulum-based apoptosis regulator Nrh/BCL2L10. Cancer Res. 2018;78(6):1404–1417. doi: 10.1158/0008-5472.CAN-17-0846. [DOI] [PubMed] [Google Scholar]
- 224.Bas J, et al. Involvement of Bcl-xL in neuronal function and development. Int J Mol Sci. 2021;22(6):3202. doi: 10.3390/ijms22063202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ma K, et al. Dynamic PGAM5 multimers dephosphorylate BCL-xL or FUNDC1 to regulate mitochondrial and cellular fate. Cell Death Differ. 2020;27(3):1036–1051. doi: 10.1038/s41418-019-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ma K, et al. Mitophagy, mitochondrial homeostasis, and cell fate. Front Cell Dev Biol. 2020;8:467. doi: 10.3389/fcell.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Um HD. Bcl-2 family proteins as regulators of cancer cell invasion and metastasis: a review focusing on mitochondrial respiration and reactive oxygen species. Oncotarget. 2016;7(5):5193–5203. doi: 10.18632/oncotarget.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Reed JC. Bcl-2 on the brink of breakthroughs in cancer treatment. Cell Death Differ. 2018;25(1):3–6. doi: 10.1038/cdd.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zaid H, et al. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ. 2005;12(7):751–60. doi: 10.1038/sj.cdd.4401599. [DOI] [PubMed] [Google Scholar]
- 230.Shoshan-Barmatz V, Arbel N, Arzoine I. VDAC, the voltage-dependent anion channel: function, regulation & mitochondrial signaling in cell life and death. Cell Science. 2008;4:74–118. [Google Scholar]
- 231.Abu-Hamad S, Sivan S, Shoshan-Barmatz V. The expression level of the voltage-dependent anion channel controls life and death of the cell. Proc Natl Acad Sci USA. 2006;103(15):5787–92. doi: 10.1073/pnas.0600103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Amsalem Z. The mitochondrial protein VDAC1 at the crossroads of cancer cell metabolism: the epigenetic link. Cancers. 2020;12(4):1031. doi: 10.3390/cancers12041031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Arif T, et al. VDAC1 is a molecular target in glioblastoma, with its depletion leading to reprogrammed metabolism and reversed oncogenic properties. Neuro Oncol. 2017;19(7):951–964. doi: 10.1093/neuonc/now297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Arif T et al (2018) Mitochondrial VDAC1 silencing leads to metabolic rewiring and the reprogramming of tumour cells into advanced differentiated states. Cancers (Basel) 10(12) [DOI] [PMC free article] [PubMed]
- 235.Arif T, et al. Rewiring of cancer cell metabolism by mitochondrial VDAC1 depletion results in time-dependent tumor reprogramming: glioblastoma as a proof of concept. Cells. 2019;8(11):1330. doi: 10.3390/cells8111330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zerbib E, et al. VDAC1 Silencing in cancer cells leads to metabolic reprogramming that modulates tumor microenvironment. Cancers. 2021;13(11):2850. doi: 10.3390/cancers13112850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Arif T, Amsalem Z, Shoshan-Barmatz V. metabolic reprograming via silencing of mitochondrial VDAC1 expression encourages differentiation of cancer cells. Mol Ther Nucleic Acids. 2019;17:24–37. doi: 10.1016/j.omtn.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Koren I, Raviv Z, Shoshan-Barmatz V. Downregulation of voltage-dependent anion channel-1 expression by RNA interference prevents cancer cell growth in vivo. Cancer Biol Ther. 2010;9(12):1046–52. doi: 10.4161/cbt.9.12.11879. [DOI] [PubMed] [Google Scholar]
- 239.Rakha EA, et al. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109(1):25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 240.Zeno S, et al. The 18 kDa mitochondrial translocator protein (TSPO) prevents accumulation of protoporphyrin IX. Involvement of reactive oxygen species (ROS) Curr Mol Med. 2012;12(4):494–501. doi: 10.2174/1566524011207040494. [DOI] [PubMed] [Google Scholar]
- 241.Chai J, et al. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406(6798):855–62. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- 242.Shiozaki EN, Shi Y. Caspases, IAPs and Smac/DIABLO: mechanisms from structural biology. Trends Biochem Sci. 2004;29(9):486–94. doi: 10.1016/j.tibs.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 243.Verhagen AM, Vaux DL. Cell death regulation by the mammalian IAP antagonist Diablo/Smac. Apoptosis. 2002;7(2):163–6. doi: 10.1023/A:1014318615955. [DOI] [PubMed] [Google Scholar]
- 244.Okada H, et al. Generation and characterization of Smac/DIABLO-deficient mice. Mol Cell Biol. 2002;22(10):3509–17. doi: 10.1128/MCB.22.10.3509-3517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pandey S et al (2020) SMAC/Diablo controls proliferation of cancer cells by regulating phosphatidylethanolamine synthesis. Mol Oncol 26(3):680–694 [DOI] [PMC free article] [PubMed]
- 246.Baenke F, et al. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6(6):1353–63. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4(7):594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 248.Bandu R, Mok HJ, Kim KP. Phospholipids as cancer biomarkers: mass spectrometry-based analysis. Mass Spectrom Rev. 2018;37(2):107–138. doi: 10.1002/mas.21510. [DOI] [PubMed] [Google Scholar]
- 249.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11(8):593–8. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 250.Di Bartolomeo F, Wagner A, Daum G. Cell biology, physiology and enzymology of phosphatidylserine decarboxylase. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862(1):25–38. doi: 10.1016/j.bbalip.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 251.Steenbergen R, et al. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J Biol Chem. 2005;280(48):40032–40. doi: 10.1074/jbc.M506510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831(3):543–54. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 253.Alessenko AV, Burlakova EB. Functional role of phospholipids in the nuclear events. Bioelectrochemistry. 2002;58(1):13–21. doi: 10.1016/S1567-5394(02)00135-4. [DOI] [PubMed] [Google Scholar]
- 254.Rockenfeller P, et al. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ. 2015;22(3):499–508. doi: 10.1038/cdd.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Patel D, Witt SN. Ethanolamine and phosphatidylethanolamine: partners in health and disease. Oxid Med Cell Longev. 2017;2017:4829180. doi: 10.1155/2017/4829180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zhang XD, et al. Activation of ERK1/2 protects melanoma cells from TRAIL-induced apoptosis by inhibiting Smac/DIABLO release from mitochondria. Oncogene. 2003;22(19):2869–81. doi: 10.1038/sj.onc.1206427. [DOI] [PubMed] [Google Scholar]
- 257.Chauhan D, et al. JNK-dependent release of mitochondrial protein, Smac, during apoptosis in multiple myeloma (MM) cells. J Biol Chem. 2003;278(20):17593–6. doi: 10.1074/jbc.C300076200. [DOI] [PubMed] [Google Scholar]
- 258.Masoumi KC, et al. Identification of a novel protein kinase Cdelta-Smac complex that dissociates during paclitaxel-induced cell death. FEBS Lett. 2012;586(8):1166–72. doi: 10.1016/j.febslet.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 259.Yoon K, Jang HD, Lee SY. Direct interaction of Smac with NADE promotes TRAIL-induced apoptosis. Biochem Biophys Res Commun. 2004;319(2):649–54. doi: 10.1016/j.bbrc.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 260.McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27(38):5138–47. doi: 10.1038/onc.2008.228. [DOI] [PubMed] [Google Scholar]
- 261.Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta. 2014;1841(4):595–609. doi: 10.1016/j.bbalip.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 262.Achleitner G, et al. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264(2):545–53. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 263.Connolly PF, Jager R, Fearnhead HO. New roles for old enzymes: killer caspases as the engine of cell behavior changes. Front Physiol. 2014;5:149. doi: 10.3389/fphys.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fernando P, Megeney LA. Is caspase-dependent apoptosis only cell differentiation taken to the extreme? FASEB J. 2007;21(1):8–17. doi: 10.1096/fj.06-5912hyp. [DOI] [PubMed] [Google Scholar]
- 265.Mielgo A, et al. The death effector domains of caspase-8 induce terminal differentiation. PLoS ONE. 2009;4(11):e7879. doi: 10.1371/journal.pone.0007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Fernando P, et al. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA. 2002;99(17):11025–30. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Maldonado EN, Lemasters JJ. Warburg revisited: regulation of mitochondrial metabolism by voltage-dependent anion channels in cancer cells. J Pharmacol Exp Ther. 2012;342(3):637–41. doi: 10.1124/jpet.112.192153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhao S, Aviles ER, Jr, Fujikawa DG. Nuclear translocation of mitochondrial cytochrome c, lysosomal cathepsins B and D, and three other death-promoting proteins within the first 60 minutes of generalized seizures. J Neurosci Res. 2010;88(8):1727–37. doi: 10.1002/jnr.22338. [DOI] [PubMed] [Google Scholar]
- 269.Khatri N, et al. The autism protein Ube3A/E6AP remodels neuronal dendritic arborization via caspase-dependent microtubule destabilization. J Neurosci. 2018;38(2):363–378. doi: 10.1523/JNEUROSCI.1511-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published articles and their supplementary information files.