Abstract
The rapid advance in deep sequencing technologies has identified numerous long non-coding RNAs (lncRNA) and their biological functions are increasingly being recognized as important regulators of gene expression and cell signaling pathways. H19 is the first lncRNA identified and characterized as the first imprinted gene in the pre-genomic era. During the last three decades, H19 has been extensively investigated as a multitasking lncRNA. H19 plays a crucial role in regulating many biological functions and is intimately involved in the pathogenesis of various human diseases. Here, we highlight the recent findings related to H19 in liver diseases. The unique features of H19 biogenesis and regulation make it an attractive diagnostic and prognostic biomarker and potential therapeutic target for certain liver diseases.
Conclusions:
The roles of H19 in liver disease remains obscure. The rapid advance in new technologies offers promise for the understanding of the mechanisms of lncRNA H19 in physiological and pathological processes of liver diseases and for the development novel therapeutics.
Keywords: non-coding RNA, cholestasis, nonalcoholic fatty liver disease, hepatocellular carcinoma
Introduction
The completion of human genome sequencing in the early 2000s identified that most of the human genome (~93%) is transcribed, but less than 2% of transcripts encode proteins. Most of the transcripts represent non-coding RNAs (ncRNAs). Long non-coding RNAs (lncRNAs) are ncRNAs with more than 200 nucleotides. LncRNAs are further classified into sense, antisense, bidirectional, intronic, and intergenic lncRNA based on their topographic relation to the nearest protein-coding gene (1). Recent advance in next-generation sequencing has identified thousands of lncRNA loci and the number of lncRNAs which are linked to human diseases, including liver disease, is rapidly growing (2). However, compared to protein-coding genes and microRNAs (small ncRNAs with 20-24 nucleotides), lncRNAs are poorly characterized, and their biological functions remain largely unknown.
LncRNA H19 was the first lncRNA and first imprinted gene identified in eukaryotes as a hepatic fetal-specific non-translatable mRNA in the late 1980s (3). Its role in embryogenesis has been well characterized. The biological function of H19 as an RNA molecule remained a mystery until the identification of another lncRNA X-inactive-specific transcript in the early 1990s (4). During the last three decades, H19 has been well characterized as a multitasking lncRNA. The aberrant expression of H19 has been linked to various human cancers, including gastric, liver, and pancreatic cancers (5, 6). Recent studies also reported that H19 is involved in chronic liver diseases such as nonalcoholic fatty liver disease (NAFLD) and cholestatic liver disease, which are major global health issues. The high mortality and morbidity associated with hepatocellular carcinoma (HCC) and cholangiocarcinoma, the end stages of most chronic liver diseases, have imposed huge financial burdens on individuals and the health care systems (7). Therefore, there is an unmet need to identify novel diagnostic biomarkers and therapeutic targets for chronic liver diseases. In this concise review, we discussed the current understanding of H19 in the pathogenesis of chronic liver diseases.
Expression and regulation of H19 and functional mechanisms
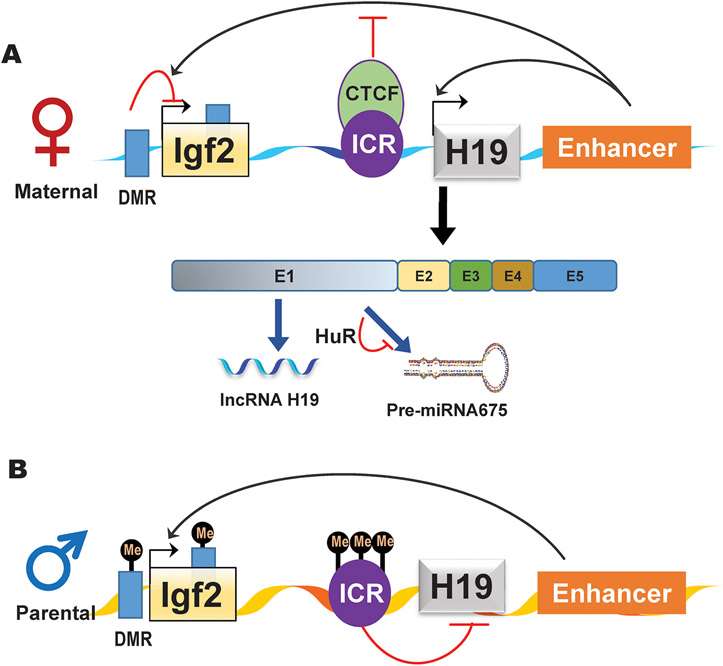
The role of lncRNA H19 in the regulation of liver development has been well-documented (8). Genetic and molecular studies have shown that H19 is a parentally imprinted and maternally expressed gene, which is localized to chromosome 7 in mice and chromosome 11p15.5 in humans, respectively; downstream of another maternally imprinted and paternally expressed protein-coding gene, called insulin-like growth factor 2 (Igf2) (9). The H19 gene contains five exons and four small introns and encodes a ~2.3 kb fully capped, spliced, and polyadenylated transcript (8). The expression of H19 is controlled by a promoter and an imprinting control region (ICR), also called a differentially methylated domain (DMD) or a differentially methylated region (DMR). H19 and IGF2 are expressed in the same tissues, and their reciprocal expression is controlled by the zinc-finger protein, CCCTC binding factor (CTCF), which binds to unmethylated maternal ICR and prevents the activation of Igf2 by downstream enhancers. It also has been reported that H19 is the precursor of miRNA675, which is embedded in exon 1 of H19. The excision process of miRNA675 from H19 is regulated by RNA-binding protein (RBP) human antigen R (HuR) (Fig.1) (10). H19 is highly expressed during fetal development and down-regulated after birth, except in skeletal muscle. Aberrant expression of H19 has been linked to various human diseases, especially tumorigenesis (11). It also has been reported that H19 expression is upregulated by estrogen, c-Myc, hypoxia, and oxidative stress (12-15). The hepatic H19 expression level is very low under normal physiological conditions, but it can be upregulated under pathological conditions (16). The relative expression levels of H19 in different types of hepatic cells depend on the pathophysiological conditions. Despite the discrepancy found in the previous studies, there is consensus that H19 impacts various hepatic cells as a multitask regulator of gene expression. The mechanisms by which H19 regulates cellular functions include epigenetic regulation, sponge of miRs, production of miR-675, and regulation of target gene expression via binding to RBPs (17).
Fig.1. Regulation of H19 and Igf2 expression.
A) H19 is expressed from the maternal allele. The binding of CTCF to the unmethylated imprinting control region (ICR) prevents the down-stream enhancer from interacting with the promoter region of Igf2 but allows the enhancer to interact with the H19 promoter. The H19 transcript contains five exons. In exon 1, there is the coding region for miR675. The production of pre-miR675 is inhibited by RBP HuR. B) In the parental allele, the ICR is methylated, which prevents the binding of CTCF and allows the enhancer to interact with the promoter region of Igf2. DMR; DNA methylation region; ICR: imprinting control region; CTCF: CCCTC binding factor.
LncRNA H19 in the nonalcoholic fatty liver disease (NAFLD)
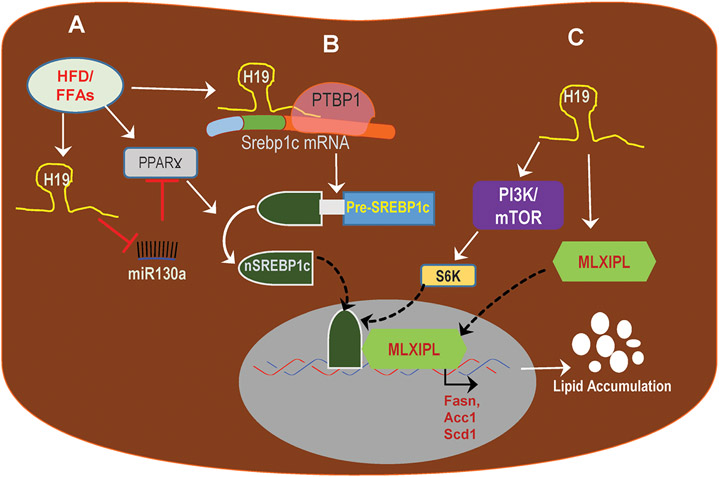
The liver is the most important metabolic organ and plays many vital life functions. The incidence of NAFLD has rapidly increased during the last two decades due to the global pandemic of obesity. NAFLD can progress to steatohepatitis (NASH) and is the second most common indication for liver transplant and a major cause of HCC. Due to the complexity of disease pathology, no reliable diagnostic biomarkers and regulatory-approved drugs are available (18). There is compelling evidence supporting the “Multi-hit” over the “Two-hit” hypothesis of NAFLD pathogenesis (19). NAFLD is not a single organ disease. The progression from simple steatosis to NASH is correlated with systemic and adipose tissue inflammation, dysbiosis, and disruption of gut barrier function. It has been reported that overexpression of H19 disrupts the intestinal barrier function via miR675 (20). A recent study further showed that H19 inhibited the function of Paneth and goblet cells via suppressing autophagy (21). The role of H19 in NAFLD remains unexplored until the identification of its aberrant expression in the livers of NASH patients (22). By using H19−/− and AAV8-mediated overexpression of H19 mouse models, Liu, C et al reported that H19 promoted lipogenesis by facilitating polypyrimidine tract-binding protein 1 (PTBP1), a RBP, to stabilize sterol regulatory element-binding protein 1c mRNA and increase protein cleavage and nuclear translocation (23). Recently, two groups reported that H19 expression is induced by free fatty acids in hepatocytes and high-fat diet feeding in vivo (24, 25). Mechanistically, H19 promotes hepatic lipogenesis by downregulating miR130a, an inhibitor of peroxisome proliferator-activator receptor γ; or upregulating transcription factors, MLX-interacting protein-like (MLXIPL, also called carbohydrate-responsive element-binding protein), and PI3K/mTOR pathways (24, 25). These studies identified H19 as an essential player in diet-induced hepatic steatosis (Fig.2). However, it remains unclear whether and how H19 is involved in NAFLD/NASH disease progression.
Fig.2. Potential mechanisms of H19-induced hepatic lipid accumulation.
HFD/FFAs induce upregulation of H19 in hepatocytes. A) H19 inhibits miR130a expression, an inhibitor of PPARƔ, and results in activation of PPARƔ and hepatic lipogenesis. B) H19 facilitates RBP, PTBP1, to stabilize the Srebp1c mRNA and promotes SREBP1c protein cleavage and nuclear translocation of the activated nuclear form, nSREBP1c and results in the increase of transcription of lipogenic genes. C) H19 induces activation of PI3K/mTOR pathway and upregulates lipogenic transcription factor, MLXIPL, resulting in increased lipid accumulation. HFD: High-fat diet; FFAs: free fatty acids; PPARƔ: peroxisome proliferator-activated receptor Ɣ; RBP: RNA binding protein; PTBP1: Polypyrimidine Tract Binding Protein 1; Srebp1c: Sterol regulatory element-binding protein 1c; Mlxipl: MLX interacting protein-like; nSREBP1, the nuclear form of SREBP1c; PI3K/mTOR: phosphoinositide 3-kinase/mammalian target of rapamycin.
LncRNA H19 in cholestatic liver disease
Cholestatic liver diseases, such as primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC) in adults and biliary atresia (BA) and Alagille syndrome in children, are a significant cause of morbidity and mortality and liver transplant. Cholestasis is defined as the impairment of bile flow due to disruption of bile acid formation or excretion or obstruction of bile ducts (26). Bile acids are exclusively formed in hepatocytes and play critical roles in nutrient absorption via intrahepatic circulation. More importantly, bile acids function as signaling molecules in regulating lipid and glucose metabolism (27). Disruption of intrahepatic bile acid circulation or accumulation of bile acids in the liver can cause hepatocyte injury, cholangiocyte proliferation, ductal reaction, activation of hepatic stellate cells, and inflammation. Although cholestatic liver diseases are relatively rare compared to NAFLD, the incidence and prevalence are increasing. The available therapeutic agents are limited to Ursodeoxycholic acid and obeticholic acid for PBC, which are largely nonspecific and often ineffective. There is an urgent need to identify diagnostic biomarkers and new therapeutic targets for cholestatic liver diseases. LncRNAs are increasingly recognized as promising potential therapeutic targets for cholestatic liver diseases (28). Zhang Y, et al. first identified H19 as a key player in BDL-induced cholestatic liver injury (22). Several studies have reported upregulation of H19 in different hepatic cells, cholestatic mouse models, and human PSC, PBC and BA patients (29-39). H19 not only functions as a sponge of miRNAs but also activates different signaling pathways involved in the activation of macrophages, cholangiocytes, and hepatic stellate cells (HSCs). Table 1 summarizes the key findings of the most recent studies, which assessed the roles of H19 in different hepatic cells and cholestatic animal models.
Table 1.
Potential targets of lncRNA H19 in cholestatic liver fibrosis
| Animal models or Human Samples |
In vitro models | Targets | Effects | Ref # (Year) |
|---|---|---|---|---|
| C57BL/6 & H19−/− mice. 2-week BDL | HepG2, Huh7, Hep3B, H69, Mz-Cha-1, CCLP-1, HuCCT1, SG231. Mouse Hepa1, MLC, and MSC. | ZEB1 EpCAM SOX9 | BDL-induced H19 suppressed ZEB1 expression, which resulted in the de-repression of EpCAM by ZEB1 and cholestatic liver fibrosis. | (32)(2017) |
| C57 male mice 3 and 4-week CCl4 | Primary mouse hepatocytes, AML12 cell line | Sox9 | Sox9-mediated upregulation of H19 is responsible for CCl4-induced liver fibrosis. | (5)(2017) |
| Sprague-Dawley male rats, 12-week CCl4 model | HSC-T6 cell line | DNMT1 ERK | DNMT1-mediated epigenetic regulation of H19 and H19-mediated activation. ERK1/2 promoted HSC activation and liver fibrosis | (38)(2018) |
| Mdr2−/−, H19−/− (Δ Exon1/+) PSC liver samples (n=16) 8-week CCL4 mouse model | Primary mouse hepatocytes, cholangiocytes, and Kupffer cells; MLC cell line | FXR/SHP S1PR2/ERK1/2 | Bile acid/estrogen-induced H19 expression in cholangiocytes is responsible for gender disparity of cholestatic liver injury in Mdr2−/− mice by downregulation of SHP and activation of S1PR2 /ERK1/2 signaling pathways. | (16, 29)(2017, 2018) |
| C57/BL6 male mice 8-week CCl4 mouse model | Primary mouse HSCs, human LX2 and L02 cell lines | miR148a, USP4 TGF-β/SMAD | H19 promoted hepatic fibrosis by activating HSCs via sponging miR148a and upregulating USP4, which enhanced the TGF-β-mediated activation of SMAD in HSCs. | (37)(2018) |
| C57/BL6 Mdr2−/−, H19−/− (Δ Exon1/+) and DKO mice; 2-week BDL model | Primary mouse hepatocytes, HSCs, cholangiocytes, and Kupffer cells; MLC, H69 and LX2 cell lines | CyclinD1/p21 CCL2/CCR2 | Cholangiocyte-derived exosomal H19 is preferentially taken up by HSCs and Kupffer cells significantly promoted the activation of HSCs and macrophages in liver fibrotic progression. | (30,31)(2019, 2020) |
| C57/BL6 male mice BDL for 2, 4, and 6 weeks. | JS-1 murine HSC cell line | PI3K/AKT/mTOR | H19 promoted autophagy by interacting with the PI3K/AKT/mTOR pathway, which was responsible for IGFBPrP1-induced activation of HSC and liver fibrosis. | (39)(2019) |
| Human BA patient liver samples (n=57) Mdr2−/−, H19−/− and DKO 2-week BDL model | MLC cell line HUCCT1 | S1PR2 Let7/HMGA2 | H19 expression level is correlated to disease severity in BA patients. H19 promotes cholangiocyte proliferation and fibrotic liver injury by regulating S1PR2 and let-7/ HMGA2–mediated pathways. | (35)(2019) |
| ICR male mice 8-week CCl4 mouse model | Primary mouse HSCs and human LX2 cell lines | ADH3/ALDH1 RARα/RXRβ AMPKα/LKB1 | H19 induced HSC activation by upregulation of ADH3/ALDH1 and retinoic acid signaling pathways and by activation of AMPKα via facilitating the formation of AMPKα/LKB1 complex. | (33,34)(2020) |
| C57/BL6j H19−/−male mice 1-week BDL model | Human Huh7, Mouse Hepa1, MSC, and MLC cell lines | PTBP1 and Let7 | H19 suppressed the expression of PTBP1, which inhibited the biogenesis of Let7, but enhanced the bioavailability to their targets. | (36)(2020) |
LncRNA H19 in HCC
HCC is the most common primary liver cancer and is often diagnosed at late stages due to the lack of diagnostic and prognostic biomarkers. Liver transplant remains the only therapeutic option. Aberrant up-regulation of H19 in tumorigenesis has been well established in different types of human cancers (11). However, the role of H19 in HCC is more complicated and remains controversial. Most studies with human HCC samples were limited by small sample size and variations in patient populations and tissue sampling. The major findings in the past three decades related to H19 in HCC and the potential reasons for the conflicting results are summarized and discussed in an excellent recent review (40). Studies with cultured HCC cell lines, in vivo animal models and human HCC patient samples indicate that H19 can be an oncogene or tumor suppressor by regulating different miRNAs, RBPs, and diverse signaling pathways (41-46). The most recent studies related to H19 in HCC are summarized in Table 2. The functions of lncRNAs are linked to their intracellular localization. More mechanistic and comprehensive studies are needed to define the role of H19 in the progression of HCC.
Table 2.
Potential targets of lncRNA H19 in HCC
| Animal models or Human Samples |
In vitro models | Targets | Potential mechanisms | Ref# (Year) |
|---|---|---|---|---|
| C57/BL6J mice transplanted with TICs from DEN-treated Tgfbr2fl/fl mice by splenic injection followed by ip injection of CCl4 for 3 weeks and tail vein injection of Ad-Cre. | TICs isolated from B6.129S6-Tgfbr2fl/fl mice (male, 14-day-old) injected with DEN (25 mg/kg). | TGFβ/ Tgfbr2-Sox2 | TGFβ regulated H19 expression via suppression of SOX2. Inactivation of Tgfbr2 in TICs simultaneously increased SOX2 and H19 levels, which are responsible for HCC development and progression. | (43)(2019) |
| Human HBV patient liver tissues and matched normal tissue (n=20) | Human L02 cell line | miR675/PPARα Akt/mTOR | HBV x protein-induced H19/miRNA675 is responsible for HBV-associated hepatitis and liver injury by activating PPARα and Akt/mTOR signaling pathways. | (42)(2019) |
| 16-month Female and 17.7-month male C57/BL6 Mdr2−/−and Mdr2−/−/H19−/−DKO mice. Human HCC patient liver tissues (n=242) with matched non-tumor tissues (n=298) | Primary cells from mouse non-tumor liver tissues | H19 is pro-oncogenic in inflammation-mediated HCC. | The single-cell transcriptome analysis of the non-tumor tissue of 18-month old female Mdr2−/− mouse indicated that H19 was mainly expressed in hepatocytes, endothelial cells and macrophages. H19 expression level in HCC tumor inversely correlated to the patient’s survival. | (41)(2020) |
| Human HCC liver tissues | Huh7, Hep3B, SNU-449, and SNU-387 cell lines | miR675 | High H19 expression is negatively related to sorafenib sensitivity by upregulation of miR675 in HCC cells. | (45)(2020) |
| Human HCC liver tissues and matched non-cancerous liver tissues (n=55) | HepG2 cell line | G3BP1 Myc | NSUN2-mediated m5C-modified H19 promotes HCC by recruiting G3BP1 oncoprotein, which leads to MYC accumulation. | (44)(2020) |
| Human HCC liver tissues (n=64) and TCGA cohort (N=393) | Hep-G2, Hep2B2, THP-1, SK-OV-3 and NCI-H520 | miRNA-193b MAPK1 axis | TAM-derived H19 promotes tumor cell migration and invasion and immune cell infiltration by hijacking miR-193b as a sponge and activating MAPK1. | (46)(2020) |
Conclusion and future perspectives
LncRNA H19 has gained increasing attention due to its broad spectrum of physiological and pathological functions. The ease of detection of lncRNAs in serum and other bodily fluids make them attractive biomarkers. Although H19 was discovered more than three decades ago, its potential roles in liver diseases remain largely obscure. Recent studies have solidified the idea that H19 represents a novel diagnostic and prognostic biomarker for various liver diseases. The fundamental role of H19 in promoting hepatic lipogenesis, inflammation, and epithelial-mesenchymal transition makes this lncRNA a promising therapeutic target in liver diseases. Recent advances in technologies for gene profiling and editing, as well as nanotechnology for RNA delivery, have rapidly moved the lncRNA research field forward. Considering the complexity of various liver diseases, it is essential to understand the comprehensive and systemic effects of H19 in different physiological and pathological settings. More mechanistic and translational studies with tissue- and cell type-specific H19−/− animal models are needed in order to validate H19 as a novel therapy target for liver diseases.
Financial Support
This study was supported by VA Merit Award I01BX004033; Research Career Scientist Award (IK6BX004477); VA ShEEP grants (1 IS1 BX004777-01 and 1IS1BX005517-01); National Institutes of Health Grant R01 DK104893, R01DK-057543, and 1R21 AA026629-01.
Abbreviations:
- ncRNAs
non-coding RNAs
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- NAFLD
nonalcoholic fatty liver disease
- HCC
hepatocellular carcinoma
- Igf2
insulin-like growth factor 2
- ICR
imprinting control region
- DMD
differentially methylated domain
- DMR
differentially methylated region
- CTCF
CCCTC binding factor
- RBP
RNA-binding protein
- HuR
human antigen R
- NASH
nonalcoholic steatohepatitis
- PTBP1
polypyrimidine tract-binding protein 1
- PSC
primary sclerosing cholangitis
- PBC
primary biliary cholangitis
- BDL
bile duct ligation
- HSCs
hepatic stellate cells
- EpCAM
epithelial cell adhesion molecule
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nature Reviews Genetics 2018;19:535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Research 2019;47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol 1990;10:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarroux J, Morillon A, Pinskaya M: History, Discovery, and Classification of lncRNAs. In: Springer Singapore, 2017; 1–46. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Deng J, Deng H, Kang Z, Huang Z, Ding Z, Dong L, et al. A Novel Sox9/lncRNA H19 Axis Contributes to Hepatocyte Death and Liver Fibrosis. Toxicological Sciences 2020;177:214–225. [DOI] [PubMed] [Google Scholar]
- 6.Alipoor B, Parvar SN, Sabati Z, Ghaedi H, Ghasemi H. An updated review of the H19 lncRNA in human cancer: molecular mechanism and diagnostic and therapeutic importance. Mol Biol Rep 2020;47:6357–6374. [DOI] [PubMed] [Google Scholar]
- 7.Udompap P, Kim D, Kim WR. Current and Future Burden of Chronic Nonmalignant Liver Disease. Clinical Gastroenterology and Hepatology 2015;13:2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 2010;32:473–480. [DOI] [PubMed] [Google Scholar]
- 9.Rachmilewitz J, Goshen R, Ariel I, Schneider T, De Groot N, Hochberg A. Parental imprinting of the human H19 gene. 1992;309:25–28. [DOI] [PubMed] [Google Scholar]
- 10.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature Cell Biology 2012;14:659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghafouri-Fard S, Esmaeili M, Taheri M. H19 lncRNA: Roles in tumorigenesis. Biomedicine & Pharmacotherapy 2020;123:109774. [DOI] [PubMed] [Google Scholar]
- 12.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 2006;66:5330–5337. [DOI] [PubMed] [Google Scholar]
- 13.Basak P, Chatterjee S, Weger S, Bruce MC, Murphy LC, Raouf A. Estrogen regulates luminal progenitor cell differentiation through H19 gene expression. Endocr Relat Cancer 2015;22:505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, Chen YQ. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol 2016;9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon Y, Kim I, Chang S, Park B, Lee S, Yoo S, Chae S, et al. Hypoxia regulates allele-specific histone modification of the imprinted H19 gene. Biochim Biophys Acta Gene Regul Mech 2020;1863:194643. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, Puri P, et al. The role of long noncoding RNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice. Hepatology 2017;66:869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, Puppo M, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A 2014;111:E5023–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Younossi ZM, Rinella ME, Sanyal A, Harrison SA, Brunt E, Goodman Z, Cohen DE, et al. From NAFLD to MAFLD: Implications of a premature change in terminology. Hepatology 2020. [DOI] [PubMed] [Google Scholar]
- 19.Tilg H, Adolph TE, Moschen AR. Multiple Parallel Hits Hypothesis in NAFLD – Revisited After a Decade. Hepatology 2020;doi: 10.1002/hep.31518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang J-Y, Xu Y, et al. H19Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Molecular and Cellular Biology 2016;36:1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu TX, Chung HK, Xiao L, Piao JJ, Lan S, Jaladanki SK, Turner DJ, et al. Long Noncoding RNA H19 Impairs the Intestinal Barrier by Suppressing Autophagy and Lowering Paneth and Goblet Cell Function. Cell Mol Gastroenterol Hepatol 2020;9:611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, Delker D, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Scientific Reports 2016;6:20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, Tran M, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 2018;67:1768–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Tang T, Wang GD, Liu B. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARgamma axis in non-alcoholic fatty liver disease. Biosci Rep 2019;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Cao Y, Shu L, Zhu Y, Peng Q, Ran L, Wu J, et al. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. Journal of Cellular and Molecular Medicine 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onofrio FQ, Hirschfield GM. The Pathophysiology of Cholestasis and Its Relevance to Clinical Practice. Clinical Liver Disease 2020;15:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong E, Li Y, Hylemon PB, Zhou H. Bile acids and sphingosine-1-phosphate receptor 2 in hepatic lipid metabolism. Acta Pharm Sin B 2015;5:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology 2014;60:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Liu R, Huang Z, Gurley EC, Wang X, Wang J, He H, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 2018;68:599–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Liu R, Wang Y, Zhu W, Zhao D, Wang X, Yang H, et al. Cholangiocyte-Derived Exosomal lncRNA H19 Promotes Macrophage Activation and Hepatic Inflammation under Cholestatic Conditions. Cells 2020;9:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, Gurley EC, et al. Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis. Hepatology 2019;70:1317–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, Pope C, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology 2017;66:1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Yang X, Kai J, Wang F, Wang Z, Shao J, Tan S, et al. HIF-1alpha-upregulated lncRNA-H19 regulates lipid droplet metabolism through the AMPKalpha pathway in hepatic stellate cells. Life Sci 2020;255:117818. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z-M, Xia S-W, Zhang T, Wang Z-Y, Yang X, Kai J, Cheng X-D, et al. LncRNA-H19 induces hepatic stellate cell activation via upregulating alcohol dehydrogenase III-mediated retinoic acid signals. International Immunopharmacology 2020;84:106470. [DOI] [PubMed] [Google Scholar]
- 35.Xiao Y, Liu R, Li X, Gurley EC, Hylemon PB, Lu Y, Zhou H, et al. Long Noncoding RNA H19 Contributes to Cholangiocyte Proliferation and Cholestatic Liver Fibrosis in Biliary Atresia. Hepatology 2019;70:1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Yang Z, Huang W, Wu J. H19 potentiates let-7 family expression through reducing PTBP1 binding to their precursors in cholestasis. Cell Death & Disease 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Luo Z, Pan Y, Zheng W, Li W, Zhang Z, Xiong P, et al. H19/miR-148a/USP4 axis facilitates liver fibrosis by enhancing TGF-beta signaling in both hepatic stellate cells and hepatocytes. J Cell Physiol 2019;234:9698–9710. [DOI] [PubMed] [Google Scholar]
- 38.Yang J-J, She Q, Yang Y, Tao H, Li J. DNMT1 controls LncRNA H19/ERK signal pathway in hepatic stellate cell activation and fibrosis. Toxicology Letters 2018;295:325–334. [DOI] [PubMed] [Google Scholar]
- 39.Huang TJ, Ren JJ, Zhang QQ, Kong YY, Zhang HY, Guo XH, Fan HQ, et al. IGFBPrP1 accelerates autophagy and activation of hepatic stellate cells via mutual regulation between H19 and PI3K/AKT/mTOR pathway. Biomed Pharmacother 2019;116:109034. [DOI] [PubMed] [Google Scholar]
- 40.Tietze L, Kessler SM. The Good, the Bad, the Question–H19 in Hepatocellular Carcinoma. Cancers 2020;12:1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamaev L, Mizrahi L, Friehmann T, Rosenberg N, Pappo O, Olam D, Zeira E, et al. The pro-oncogenic effect of the lncRNA H19 in the development of chronic inflammation-mediated hepatocellular carcinoma. Oncogene 2020. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Xu L, Lu B, Zhao M, Li L, Sun W, Qiu Z, et al. LncRNA H19/microRNA-675/PPARalpha axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mTOR signalling. Mol Immunol 2019;116:18–28. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Han C, Ungerleider N, Chen W, Song K, Wang Y, Kwon H, et al. A Transforming Growth Factor-β and H19 Signaling Axis in Tumor-Initiating Hepatocytes That Regulates Hepatic Carcinogenesis. Hepatology 2019;69:1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, Liu Y, et al. Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 2020;39:6906–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Liu Y, Li Z, Li H, Li X, Yan L, Mao J, et al. Long non-coding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR-675. Oncology Reports 2020;44:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, Yu W, et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett 2020;469:310–322. [DOI] [PubMed] [Google Scholar]