Abstract
SARS-CoV-2 can cause acute respiratory distress and death in some patients1. Although severe COVID-19 is linked to substantial inflammation, how SARS-CoV-2 triggers inflammation is not clear2. Monocytes and macrophages are sentinel cells that sense invasive infection to form inflammasomes that activate caspase-1 and gasdermin D, leading to inflammatory death (pyroptosis) and the release of potent inflammatory mediators3. Here we show that about 6% of blood monocytes of patients with COVID-19 are infected with SARS-CoV-2. Monocyte infection depends on the uptake of antibody-opsonized virus by Fcγ receptors. The plasma of vaccine recipients does not promote antibody-dependent monocyte infection. SARS-CoV-2 begins to replicate in monocytes, but infection is aborted, and infectious virus is not detected in the supernatants of cultures of infected monocytes. Instead, infected cells undergo pyroptosis mediated by activation of NLRP3 and AIM2 inflammasomes, caspase-1 and gasdermin D. Moreover, tissue-resident macrophages, but not infected epithelial and endothelial cells, from lung autopsies from patients with COVID-19 have activated inflammasomes. Taken together, these findings suggest that antibody-mediated SARS-CoV-2 uptake by monocytes and macrophages triggers inflammatory cell death that aborts the production of infectious virus but causes systemic inflammation that contributes to COVID-19 pathogenesis.
SARS-CoV-2 causes severe COVID-19 marked by acute respiratory distress that can progress to multiorgan failure and death in older individuals and patients with comorbidities1. Increased chronic inflammation is associated with ageing (inflammaging) and the comorbidities linked to severe disease4, and severe disease is linked to signs of inflammation2. When myeloid cells sense invasive infection, they activate inflammasomes to sound an innate immune alarm3. Inflammasome activation is required to process and release interleukin-1 (IL-1)-family cytokines, arguably the most potent inflammatory mediators5. However, activation of NF-κB, the TNF receptor superfamily and T helper 17 (TH17) cell cytokines can also cause severe inflammation. When inflammasomes sense infection, they recruit the ASC adaptor and assemble into large complexes that recruit and activate caspase-1, which in turn processes IL-1 pro-cytokines and the pore-forming gasdermin D (GSDMD) to disrupt the cell membrane, leading to cell death and cytokine release3. Pyroptotic cell membrane rupture releases cytokines, chemokines and other alarmins that recruit immune cells to infection sites. LDH release is pathognomonic for pyroptosis and other forms of necrotic cell death3 and elevated LDH is one of the best correlates of severe COVID-196.
COVID-19 blood shows signs of pyroptosis
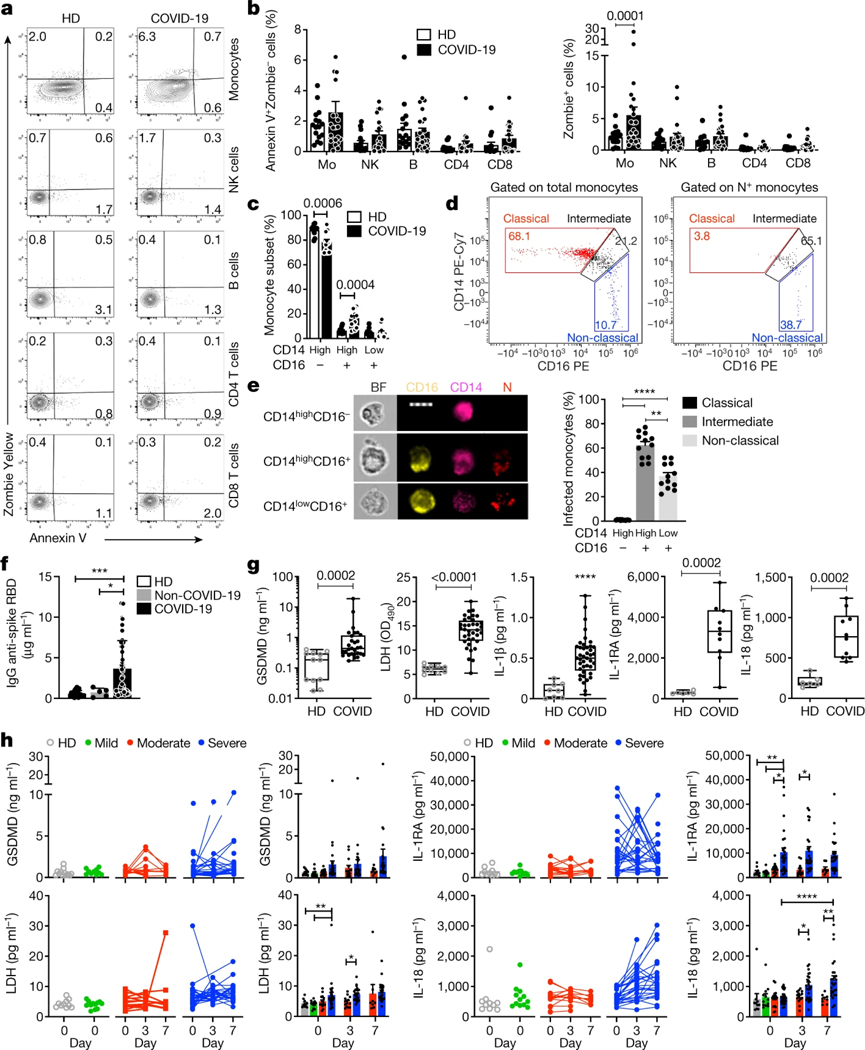
As inflammasome activation is a major mediator of inflammation7, we examined the blood of patients infected with SARS-CoV-2 for inflammasome activation and pyroptosis. Freshly isolated mononuclear cells from 19 healthy donor individuals (HDs) and 22 patients with COVID-19 in the emergency department were stained for haematopoietic cell markers; with a small fixable dye (Zombie Yellow) that enters cells with damaged plasma membranes; and for annexin V, an indicator of programmed cell death (Fig. 1a, b, Extended Data Fig. 1a and Supplementary Table 1). Annexin V+Zombie− apoptotic cells did not increase in any subpopulation in samples from patients with COVID-19. However, around 6% of monocytes of patients with COVID-19 on average took up Zombie dye, a sign of membrane damage consistent with pyroptosis. None of the lymphocyte subsets in samples from patients with COVID-19 showed increased pyroptosis. Monocyte flow cytometry analysis indicated that there was a reduced frequency of classical monocytes (CD14highCD16−) in 15 patients with COVID-19 compared with 13 HDs, whereas intermediate monocytes (CD14highCD16+) were significantly increased, but there was no change in the non-classical subset (CD14lowCD16+) (Fig. 1c and Extended Data Fig. 1b). Many intermediate (about 60%) and non-classical (about 40%), but none of the more abundant classical, monocytes had taken up SARS-CoV-2 virus as they stained for nucleocapsid (N) (Fig. 1d, e). As only monocytes that expressed CD16–an important mediator of antibody-dependent phagocytosis–took up virus, anti-spike RBD IgG plasma titres were measured in plasma samples of 64 patients with COVID-19 that were obtained at presentation at the emergency department, 20 HDs and 5 patients who presented with COVID-19-like symptoms but were SARS-CoV-2 PCR negative (hereafter, non-COVID-19 patients) (Fig. 1f). Most patients with COVID-19, but not HDs or non-COVID-19 controls, had elevated anti-spike RBD IgG, suggesting that they had been infected for approximately a week8. Plasma samples from patients with COVID-19 with diverse disease outcomes and HDs were compared for pyroptosis-specific markers (GSDMD, IL-1β, IL-1RA, IL-18 and LDH activity) (Fig. 1g), inflammatory markers not specific for pyroptosis (inflammatory cytokines IL-6, TNF and IL-17/17A; growth factors IL-7 and G-CSF; and chemokines CCL7, CXCL9 and CXCL10) and interferons (IFNβ and IFNγ). Consistent with published data9,10, all inflammation markers that are not specific for pyroptosis were significantly elevated in the plasma of patients with COVID-19 (except for IL-17/17A) and IFNs were not detected above the baseline (data not shown). All pyroptosis markers were significantly elevated in the plasma of patients with COVID-19 compared with HDs. Although significantly higher in samples from patients with COVID-19, plasma IL-1β was low, which was not surprising as it is rapidly cleared and is usually not detected even in patients with pyroptosis-mediated diseases. However, its antagonist IL-1RA, used as a surrogate5, was greatly increased in samples from patients with COVID-19. Note that IL-1 cytokines and pyroptosis potently activate the other elevated inflammation markers11.
Fig. 1 |. Monocytes of patients with COVID-19 undergo pyroptosis.
a, b, Representative flow cytometry plots (a) and the percentage of lymphocyte subset and monocyte (Mo) staining for annexin V only or Zombie dye (b) in fresh blood from HDs (n = 16) and patients with COVID-19 (n = 22). NK, natural killer cells. c, The frequency of monocyte subsets (classical, CD14highCD16−; intermediate, CD14highCD16+; and non-classical, CD14lowCD16+) in freshly isolated blood from HDs (n = 11) and patients with COVID-19 (n = 12). d, e, Imaging flow cytometry analysis of SARS-CoV-2 infection in monocyte subsets of patients with COVID-19 (n = 12). Monocytes from patients with COVID-19 were enriched by negative selection and stained for CD14, CD16 and SARS-CoV-2 N. d, Representative dot plots of monocyte subsets gated on all monocytes (left) or N+ monocytes. e, Representative images of imaging flow cytometry (left) and quantification of infection (N+) in the monocyte subsets (right). BF, bright field. Scale bar, 7 μm. f, The concentration of anti-spike RBD IgG in the plasma of HDs (n = 20), non-COVID-19 patients (with COVID-19-like symptoms but PCR negative for SARS-CoV-2; n = 5) and patients with COVID-19 (n = 68) at presentation. g, The concentration of pyroptosis biomarkers and cytokines in HD and COVID-19 plasma. GSDMD (n = 12 (HD), n = 29 (COVID-19)); LDH activity (n = 10 (HD), n = 36 (COVID-19)); IL-1β (n = 8 (HD), n = 41 (COVID-19)); IL-1RA and IL-18 (n = 6 (HD), n = 10 (COVID-19)). A description of the samples is provided in Supplementary Table 1. OD490, optical density at 490 nm. h, Plasma pyroptosis biomarkers at presentation (day 0) and during hospitalization (day 3 and 7) in patients with COVID-19 with mild (n = 12), moderate (n = 16) and severe (n = 32) COVID-19 Acuity scores (the samples are described in Supplementary Table 2). Left, individual patient data. Right, grouped data. For b, c, e, f, h, data are mean ± s.e.m. The plots in g show the median (centre line), the interquartile range between the 25th and 75th percentiles (box), and the 25th percentile value − 1.5× the interquartile range (lower whisker) and the 75th percentile value + 1.5× the interquartile range (upper whisker) . Statistical analysis was performed using two-tailed nonparametric unpaired t-tests (b, c), one-way analysis of variance (ANOVA) with Tukey multiple-comparisons test (e, f), two-tailed nonparametric unpaired t-tests (g) and two-way ANOVA with Tukey multiple-comparisons test (h); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine whether pyroptosis biomarkers correlate with COVID-19 disease severity, plasma from 10 HDs and 60 patients with COVID-19 was analysed for GSDMD, LDH, IL-1RA and IL-18 at presentation and on days 3 and 7 for hospitalized patients (Fig. 1h and Supplementary Table 2). The patients were grouped into mild, moderate or severe disease using the MGH COVID Acuity scale12. Plasma GSDMD, LDH, IL-1RA and IL-18 were all elevated in the samples from patients with severe disease compared with those with mild or moderate disease, but the increase in GSDMD was not significant. Taken together, these results suggest ongoing pyroptosis in COVID-19 blood that was more prominent in severe disease.
Monocytes have activated inflammasomes
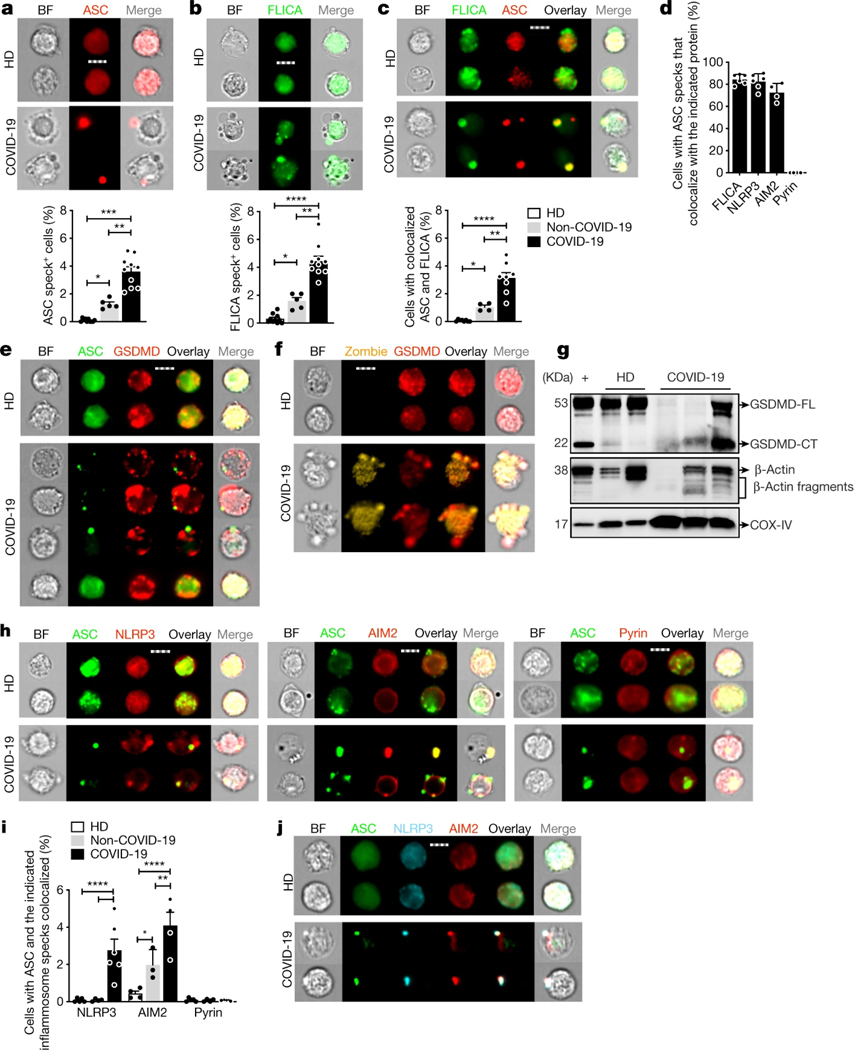
These data suggested that monocytes in patients with COVID-19 might die of pyroptosis and release inflammatory cytokines to contribute to poor outcome. Not much is known about how viruses interact with the 27 potential human canonical inflammasome sensors3. The NLRP3 inflammasome, which detects K+ efflux generated by a variety of stimuli, could be activated by specific viral proteins13,14. Three SARS-CoV-2 proteins–Orf3a, Orf8 and envelope (E)–are thought to be ‘viroporins’ (ion channels) that potentially activate K+ efflux, as previously described for SARS-CoV15. Orf3 and Orf8 are encoded only by pathogenic human coronaviruses. Interestingly, bats, which are the natural hosts of SARS-CoV and SARS-CoV-2, have a dampened NLRP3 response to multiple viruses, including MERS-CoV, which might explain their toleration of these infections despite high viral loads16. To examine whether monocytes of patients with COVID-19 undergo pyroptosis, freshly isolated, enriched monocytes from HDs, patients with COVID-19 of mixed disease severity (Supplementary Table 1) and non-COVID-19 patients were analysed using imaging flow cytometry for the expression and intracellular distribution of the common inflammasome adaptor ASC, activated caspase-1 (using the fluorochrome-labelled inhibitor of caspases assay (FLICA)) and GSDMD. Activated canonical inflammasomes form large micrometre-sized inflammasome–ASC–caspase-1 specks3. About 4% of monocytes from patients with COVID-19, 1% of monocytes from non-COVID-19 patients, but no monocytes from HDs, had caspase-1 and ASC specks (Fig. 2a–c and Extended Data Fig. 2a, b). These results suggest that other causes of respiratory distress activate monocyte inflammasomes, but activation is more extensive in SARS-CoV-2 infection. Most cells with ASC specks (about 80%) from patients with COVID-19 also had colocalized caspase-1 specks (Fig. 2d).
Fig. 2 |. Monocytes of patients with COVID-19 have activated inflammasomes, caspase-1 and GSDMD.
Monocytes from HDs, non-COVID-19 patients or patients with COVID-19 at the time of presentation were analysed by imaging flow cytometry for ASC, GSDMD, caspase-1 activation (FLICA) and/or Zombie dye uptake. a–c, The percentage of monocytes with activated ASC (a) or caspase-1 (b) (n = 8 (HD), n = 5 (non-COVID-19), n = 10 (COVID-19)) or colocalized ASC/caspase-1 specks (c) (n = 8 (HD), n = 4 (non-COVID-19), n = 8 (COVID-19)) (c). Representative images (top) and quantification of all samples (bottom) are shown. d, The percentage of ASC-speck-containing monocytes with colocalized activated caspase-1, NLRP3, AIM2 or pyrin specks. n = 6. e, f, Representative images of ASC (e) or Zombie dye (f) and GSDMD co-stained monocytes. n = 4 independent experiments. g, Lysates of purified monocytes of HDs and patients with COVID-19, and of LPS- and nigericin-treated monocytes of HDs (+) probed with a monoclonal antibody that recognizes full length GSDMD (GSDMD-FL) and the C-terminal of GSDMD (GSDMD-CT) (top), β-actin (middle) and COX-IV (bottom). Representative of n = 4 independent experiments. h, i, Representative images of ASC co-staining with NLRP3 (left; n = 5 (HD), n = 4 (non-COVID-19), n = 6 (COVID-19)), AIM2 (middle; n = 4 (HD), n = 3 (non-COVID-19), n = 4 (COVID-19)) and pyrin (right; n = 4 (HD), n = 4 (non-COVID-19), n = 5 (COVID-19)) (h), and quantification of monocytes showing ASC specks colocalized with the indicated inflammasomes (i). j, Representative images of co-staining of ASC, NLRP3 and AIM2. n = 3 independent experiments. For a–c, e, f, h, j, scale bars, 7 μm. For a–d, i, data are mean ± s.e.m. Statistical analysis was performed using one-way ANOVA with Tukey multiple-comparisons test (a–d) and two-way ANOVA with Tukey multiple-comparisons test (i); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
COVID-19 monocytes with ASC specks showed ballooning plasma membranes, GSDMD redistribution from the cytoplasm to cell membrane puncta and Zombie dye uptake, consistent with GSDMD pore formation and pyroptosis, but cells without ASC specks did not (Fig. 2e, f and Extended Data Fig. 2b, e). Most Zombie+ cells had ASC specks (62 ± 9%), suggesting that most COVID-19 monocyte death is due to inflammasome activation. However, only 28 ± 5% of cells with ASC specks had taken up Zombie dye. This difference could be because cell membrane permeabilization is delayed after ASC activation and dying cells with damaged membranes are rapidly removed from the blood. Immunoblots of monocyte lysates of HDs and patients with COVID-19 were probed for full-length GSDMD (GSDMD-FL) and its C-terminal fragment (GSDMD-CT) and housekeeping proteins, β-actin and COX-IV (Fig. 2g and Extended Data Fig. 2g). During pyroptosis, cleaved GSDMD and actin are released and the actin cytoskeleton disintegrates, whereas membrane-bound proteins, such as COX-IV, are mostly retained3,17. GSDMD-FL was detected in all of the HD samples, but in only 1 out of 3 samples from patients with COVID-19. GSDMD-CT was detected in monocytes of patients with COVID-19 and the positive control (LPS + nigericin-treated HD monocytes). Although COX-IV was detected in all of the samples, full-length β-actin was not detected in one COVID-19 sample, but β-actin fragments were detected in all of the samples from patients with COVID-19 and in nigericin-activated HD monocytes. Thus, monocytes of patients with COVID-19 are undergoing pyroptosis.
To identify the activated inflammasome, monocytes of HDs and patients with COVID-19 were co-stained for ASC and three canonical inflammasomes (NLRP3, AIM2 (activated by cytoplasmic DNA) and pyrin (activated by bacterial toxins))14 (Fig. 2d, h–j and Extended Data Fig. 2c–f). In monocytes of patients with COVID-19, ASC specks colocalized with NLRP3 and AIM2, but there were no pyrin specks. AIM2 activation was unexpected, although AIM2 is activated by RNA viruses in rare cases by an unclear mechanism18. AIM2 might sense host mitochondrial DNA as mitochondrial membranes are damaged during pyroptosis19. Almost all ASC-speck-positive monocytes had colocalized NLRP3 and AIM2 specks (Fig. 2d), and ASC, NLRP3 and AIM2 colocalized (Fig. 2j). We did not expect to find more than one inflammasome stimulated in the same cell, although colocalization of two distinct inflammasomes has been reported20. Confocal microscopy confirmed ASC, caspase-1, NLRP3 and AIM2 colocalization in inflammasomes selectively in COVID-19 monocytes (Extended Data Fig. 2f). These data showing inflammasome specks and GSDMD membrane localization and cleavage, together with the detection of dying annexin V−Zombie+ monocytes and plasma GSDMD and IL-1 cytokines (Fig. 1), indicate that COVID-19 monocytes die of pyroptosis.
Monocyte infection triggers pyroptosis
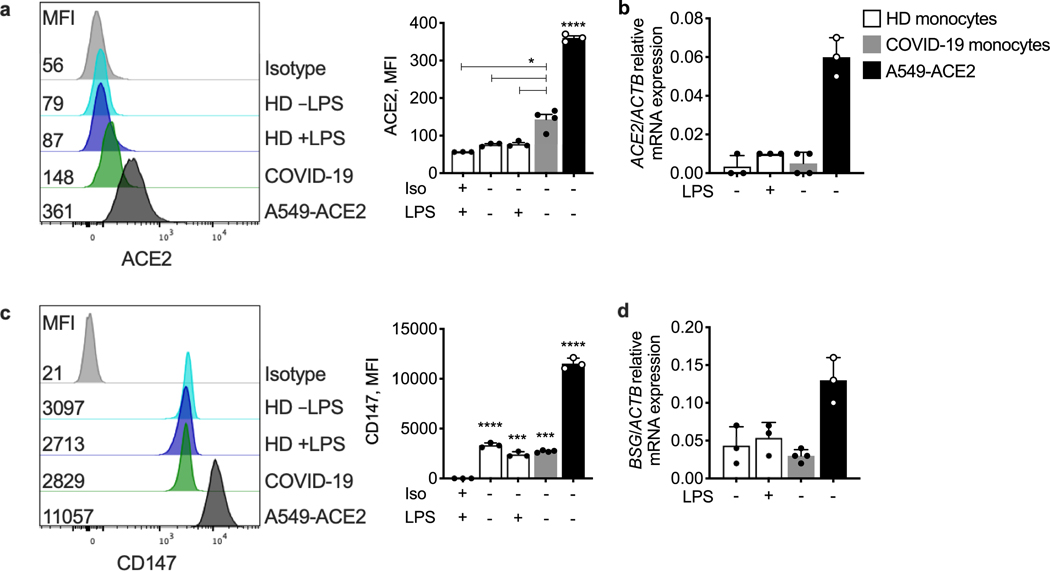
We next examined what activates inflammasomes in COVID-19 monocytes. As inflammasomes sense invasive infection, monocyte infection might be the trigger. A few reports suggest that monocytes10,21 and macrophages can be infected by SARS-CoV-2, and we detected nucleocapsid in patient monocytes (Fig. 1d, e). However, monocytes do not express ACE2, the viral entry receptor22. Indeed, ACE2 was undetected or barely detected by flow cytometry and quantitative PCR with reverse transcription (RT–qPCR) analysis of monocytes of patients with COVID-19 and HDs (Extended Data Fig. 3a, b). Monocytes of HDs and patients with COVID-19 expressed similar levels of CD147 (also known as basigin and EMMPRIN), which is reported to bind to the SARS-CoV-2 spike protein and facilitate viral uptake, although this finding is controversial23–25 (Extended Data Fig. 3c, d). Monocytes express three Fcγ receptors—CD64 (FcγRI) and CD32 (FcγRII), which is expressed on most blood monocytes, and CD16 (FcγRIIIa), which is expressed on a small minority of blood monocytes (around 10% in HDs)26,27—that are increased in COVID-199. These receptors could recognize antibody-opsonized virions and mediate uptake through antibody-dependent phagocytosis28. Anti-SARS-CoV-2 spike antibodies are detected early in SARS-CoV-2 infection, about when patients develop inflammatory symptoms8,29, as in our cohort (Fig. 1f). To examine whether monocytes of patients with COVID-19 are infected, we co-stained monocytes of HDs and patients with COVID-19 for nucleocapsid (N) (Fig. 3a–d) or double-stranded RNA (dsRNA) (anti-J2 antibodies) (Fig. 3e–h) and ASC. N staining indicates virus internalization, but J2 staining indicates active infection30. Monocytes of HDs did not stain for N, dsRNA or ASC. About 10% of monocytes from patients with COVID-19 stained for N or dsRNA (Fig. 3b, f) and around 95% of N+ monocytes were also J2 positive, indicating viral replication. Almost all infected cells showed ASC specks (Fig. 3c, g) and all ASC-speck-positive cells were infected (Fig. 3d, h). Thus, SARS-CoV-2 monocyte infection activates inflammasomes and pyroptosis.
Fig. 3 |. SARS-CoV-2-infected monocytes and lung macrophages have activated inflammasomes.
a–h, Monocytes of HDs and patients with COVID-19 were stained for SARS-CoV-2 N (n = 5) (a–d) or dsRNA (anti-J2 antibodies) (n = 4) (e–h) and ASC. a, e, Representative imaging flow cytometry images. b, f, Quantification of infected cells on the basis of N (b) or J2 (f) staining. c, g, Uninfected or infected cells that showed ASC specks. d, h, The percentage of cells with or without ASC specks that were infected. For a, e, scale bars, 7 μm. i–k, Lung autopsies from five patients with COVID-19 (the samples are described in Supplementary Table 3) and three control individuals who have experienced trauma were stained for N (green), ASC (red) and CD14 (magenta), and with DAPI (blue). i, Digital scanner images of a representative patient who experienced trauma (left) and a patient with COVID-19 (middle), showing a magnified image of representative infected CD14+ (top) and CD14− (bottom) cells from the lungs of the patient with COVID-19 (right). Scale bars, 50 μm (left), 100 μm (middle). j, k, Representative confocal microscopy COVID-19 lung images of infected CD14+ (j) and CD14− (k) cells (left). Right, quantification of CD14+ (j) and CD14− (k) cells that are N positive and/or have ASC specks in the lungs of patients with COVID-19 (n = 5) and control individuals (n = 3). In k, representative images of CD14−N+ cells (left) were co-stained for ASC and E-cadherin, an epithelial marker (top), or CD31, an endothelial marker (bottom). For j, k, scale bars, 7 μm. For b–d, f–h, j, k, data are mean ± s.e.m. Statistical analysis was performed using two-tailed nonparametric unpaired t-tests (Mann–Whitney U-tests) (b–d, f–h) and two-way ANOVA with Tukey multiple-comparisons test (j, k); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Lung macrophages have inflammasome specks
As the respiratory tract is the main infection site, we next assessed whether macrophages in lung autopsies were infected with SARS-CoV-2 and had active inflammasomes. Fixed lung slides from five individuals with SARS-CoV-2 infection (Supplementary Table 3) and three uninfected individuals who have experienced trauma were co-stained for CD14, ASC, N and DAPI (Fig 3i–k). In the lungs of patients with COVID-19, 15.1 ± 2.9% of CD14− cells and 8.3 ± 4.2% of CD14+ cells stained for N, but N was not detected in the unaffected individuals who have experienced trauma (Fig 3i–k). As expected, both E-cadherin+ epithelial and CD31+ endothelial CD14− cells stained for N (Fig 3k). However, ASC specks were detected only in CD14+, but not in CD14−, COVID-19 lung cells, indicating that tissue-resident macrophages have activated ASC-containing inflammasomes, but infected lung epithelial and endothelial cells do not. Most CD14+N+ cells had ASC specks (Fig. 3j). ASC specks were not seen in control autopsies. About a quarter of CD14+ lung cells had ASC specks, although only approximately 8% were N positive, suggesting that danger-associated molecular patterns, released from infected or otherwise damaged lung cells, may have activated inflammasomes in uninfected macrophages.
CD16 mediates infection of opsonized virus
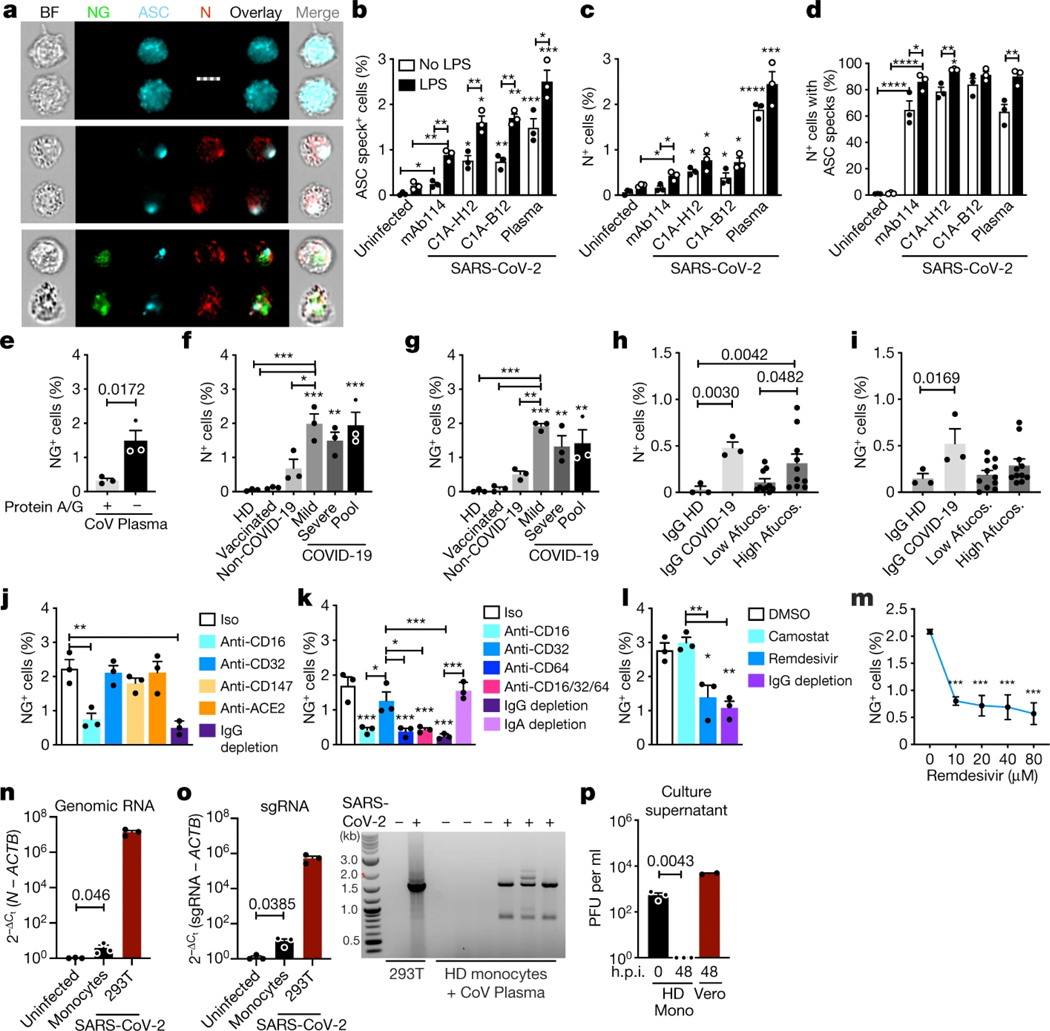
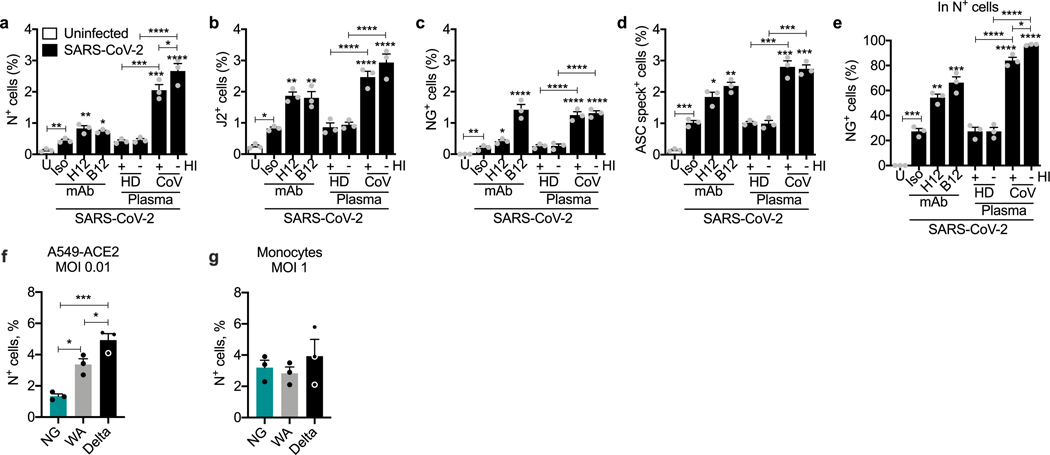
To confirm that monocytes can be infected, monocytes of HDs were infected with an engineered infectious clone (icSARS-CoV-2-mNG) encoding a Neon Green (NG) fluorescent reporter of viral replication31. Monocytes, primed or not with LPS, were infected (multiplicity of infection of 1) with reporter virus preincubated with IgG1 isotype control antibodies (mAb114), anti-spike monoclonal antibodies (non-neutralizing (C1A-H12) or neutralizing (C1A-B12))32 or pooled plasma (heat-inactivated or not) from HDs or patients with COVID-19. Antibodies and plasma were also present during culture. After 48 h, monocytes were analysed for N, dsRNA and ASC by imaging flow cytometry (Fig. 4a–g and Extended Data Fig. 4). Without LPS, anti-spike antibodies or COVID-19 pooled plasma, few monocytes of HDs took up or replicated the virus, but infection increased significantly in the presence of anti-spike monoclonal antibodies or plasma from patients with COVID-19. Antibody-neutralizing activity and plasma heat-inactivation did not affect infection (Extended Data Fig. 4a–e), suggesting that complement was not involved. IgG-depletion of plasma from patients with COVID-19 nearly abrogated viral infection, assessed by NG fluorescence, but IgA depletion had no effect on infection (Fig. 4e, j, k). These results suggest that infection is mediated by virus opsonized by anti-spike antibodies. Nonetheless, N-, J2- and NG-positive monocytes were detected at low levels after infection of HD monocytes with virus preincubated with isotype control monoclonal antibodies or with HD plasma, suggesting possible inefficient anti-SARS-Cov-2-antibody-independent monocyte infection. The highest in vitro infection rate was around 3% in HD monocytes that were pretreated with LPS and incubated with patient plasma. N and J2 staining were comparable, with a low background of around 0.1% in uninfected samples; fewer cells were NG fluorescent (about half as many) and there was no background NG fluorescence. More J2+ or N+ cells in samples with the highest infection rates (treated with LPS and patient plasma or anti-spike antibodies) were also NG fluorescent, indicating viral replication (Extended Data Fig. 4e). NG may be detected less often than N or dsRNA because it is expressed late in the viral lifecycle and/or is more difficult to detect. ASC specks were barely detected in uninfected HD monocytes but increased with SARS-CoV-2 infection (Fig. 4c and Extended Data Fig. 4d). ASC-speck-positive cells increased when SARS-CoV-2 was preincubated with anti-spike antibodies and still more when preincubated with patient plasma. HD monocyte infection with the fluorescent molecular clone was similar to infection with the parental Washington (WA) strain or a Delta variant clinical isolate but, as expected, the molecular clone less efficiently infected A549-ACE2 cells compared with the WA strain or the more infectious Delta variant (Extended Data Fig. 4f, g). The similarity of HD monocyte infection for all three viruses suggested that monocyte viral entry might be ACE2-independent.
Fig. 4 |. HD monocytes take up antibody-opsonized SARS-CoV-2 through an FcγR but viral replication is aborted.
a–d, HD monocytes (n = 3) were primed (black bars) or not (white bars) with LPS, infected with icSARS-CoV-2-mNG and stained 48 h later for N and ASC. Virus was preincubated with IgG1 control mAb114, non-neutralizing anti-spike (C1A-H12) or neutralizing anti-RBD (C1A-B12), or with pooled plasma from patients with COVID-19, and these were retained throughout culture. a, Representative imaging flow cytometry images of uninfected (top), N+NG− (middle) or N+NG+ (bottom) monocytes. Scale bar, 7 μm. b–d, Quantification of the percentage of ASC speck+ (b) or N+ (c) monocytes, and of N+ monocytes with ASC specks (d). n = 3. e–i, LPS-activated HD monocytes were infected with icSARS-CoV-2-mNG preincubated with pooled COVID-19 plasma, depleted or not depleted of immunoglobulins using protein A/G beads (n = 3; e), or preincubated with pooled plasma from HDs, recipients of a COVID-19 mRNA vaccine, non-COVID-19 patients or patients with COVID-19 with mild and/or severe disease (n = 3; f, g); or with purified IgG from HDs (n = 3), pooled from patients with COVID-19 of mixed severity (n = 3) or patients with COVID-19 with low (about 8%) or high (about 30%) afucosylated (Afucos.) anti-spike IgG (n = 11) (h, i). Infection was quantified by N staining (f, h) or NG fluorescence (e, g, i). j–m, LPS-treated HD monocytes were infected with icSARS-CoV-2-mNG, preincubated with pooled plasma from patients with COVID-19, depleted or not depleted of IgG or IgA as indicated, in the presence of the indicated blocking or isotype control (Iso) antibodies (n = 3; j, k) or antiviral drugs (l (10 μM remdesivir), m), and infection was assessed 48 h later by NG fluorescence. The statistical analysis in m compared drug with no drug. n, o, RT–qPCR analysis of genomic SARS-CoV-2 N RNA (n) and sgRNA (o, left) in uninfected or infected HD monocytes (n = 3), normalized to ACTB mRNA. Infected HEK293T cells were used as a positive control (n = 3). Agarose gel electrophoresis of ethidium-bromide-stained RT–qPCR-amplified sgRNA is shown (o, right). The approximately 1,600-bp band in the samples from patients with COVID-19 was sequenced and confirmed to be N sgRNA. p, SARS-CoV-2 plaque-forming units (PFU) in culture supernatants of infected monocytes (Mono) or Vero E6 cells collected at the indicated hours post-infection (h.p.i.). For b–p, data are mean ± s.e.m. Statistical analysis was performed using two-way ANOVA with Sidak multiple-comparisons test (b–d), two-tailed nonparametric unpaired t-tests (e) and one-way ANOVA with Tukey multiple-comparisons test (f–p); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Data are representative of n = 3 replicate experiments.
To assess whether disease severity or antibodies raised by vaccination increased monocyte virus uptake, LPS-activated monocytes were infected in the presence of pooled plasma from uninfected donors, mRNA vaccine recipients or patients with COVID-19 with mild or severe disease. Importantly, uninfected HD and post-vaccination plasma did not facilitate virus uptake or replication, even though plasma anti-RBD IgG was around twofold higher in HD vaccine recipients (6.5 ± 1.1 μg ml−1) than in patients with COVID-19 (3.6 ± 0.5 μg ml−1) (Fig. 4f, g). However, pooled plasma from non-COVID-19 patients slightly increased infection, but the increase was not significant, suggesting possible inefficient viral uptake by some non-COVID plasma component. Disease severity did not affect infection by the plasma of patients with COVID-19 as pooled mild and/or severe plasma similarly facilitated infection.
Patients with severe acute COVID-19 have increased antiviral IgGs that are afucosylated in their Fc region and bind better to CD1633–35. To test whether afucosylation affects HD monocyte infection, HD monocyte infection by virus preincubated with purified IgG from pooled plasma from HDs or patients with COVID-19, or from patients with COVID-19 with relatively low (about 8%) or high (about 30%) afucosylation (2 patients of each) was compared (Fig. 4h, i). As expected, purified HD plasma IgG did not lead to N staining or NG fluorescence, whereas IgG from pooled plasma from patients with COVID-19 did. Low afucosylated IgG did not significantly increase infection compared to HD IgG, but more highly afucosylated COVID-19 IgGs modestly, but significantly, increased N+ cells. However, NG fluorescence did not increase significantly after adding either low- or high-afucosylated IgG from patients with COVID-19 compared to HD IgG, perhaps because this assay is less sensitive than N staining. Purified IgG enhanced HD monocyte infection less than patient plasma (compare Fig. 4l, m with Fig. 4f, g), suggesting that an Ig-independent plasma component might facilitate infection.
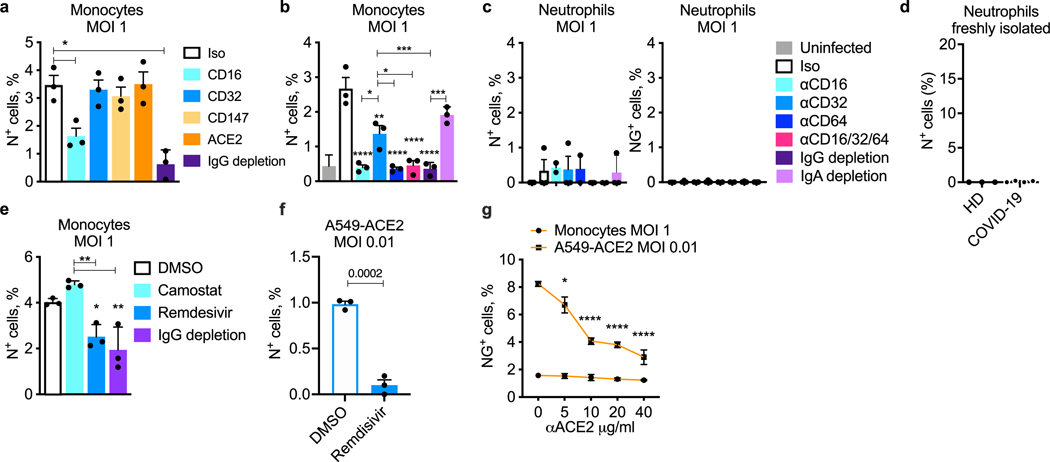
To identify the viral receptor on monocytes, purified HD monocytes were infected with the reporter virus in the presence of plasma of patients with COVID-19 that was or was not depleted of IgG or in the presence of blocking antibodies to potential monocyte receptors—ACE2, CD147 and the three monocyte FcγRs, CD16, CD32 and CD64 (Fig. 4j, k and Extended Data Fig. 5a, b). Blocking CD16 or CD64 or IgG depletion strongly inhibited infection, whereas blocking the other receptors had no significant effect. The combination of anti-CD16 and anti-CD64 blocking antibodies did not inhibit virus uptake more than either blocking antibody on its own. Thus, SARS-CoV-2 infection of monocytes is mostly mediated by CD16 and/or CD64 uptake of opsonized virus.
CD16 is also expressed on neutrophils and cytotoxic T and natural killer cells, which could be infected by a similar antibody-dependent mechanism. We did not observe increased cell death in patient lymphocytes (Fig. 1a) and therefore did not study them further. However, neutrophils contribute to SARS-CoV-2 immunopathology and inflammation36. To determine whether neutrophils are infected, HD neutrophils and monocytes were infected side by side in the presence of COVID-19 plasma (Extended Data Fig. 5b, c). Infection of HD neutrophils was low compared with monocyte infection (around 0.2% versus almost 3% in monocytes) and not significantly increased above background. To assess whether neutrophils are infected in vivo, the frequency of in vivo neutrophil infection in samples from COVID-19 patients of mixed disease severity and HDs was assessed by N staining negatively selected, fresh blood neutrophils (Extended Data Fig. 5d). Infection was not detected in neutrophils of patients with COVID-19.
SARS-CoV-2 monocyte infection is aborted
dsRNA and NG detection strongly suggested that monocytes replicate SARS-CoV-2. To confirm viral replication and further assess whether uptake is ACE2 mediated, HD monocytes were infected in the presence of plasma from patients with COVID-19 and the antiviral drugs remdesivir, an inhibitor of the viral RNA-dependent RNA polymerase, and camostat mesylate, an inhibitor of TMPRSS2, which primes the spike protein for ACE2-mediated entry37 (Fig. 4l, m and Extended Data Fig. 5e–g). Monocyte infection, assessed by N or NG positivity, was unaffected by camostat, but significantly and comparably inhibited by Ig depletion or remdesivir, confirming antibody-dependent entry and viral replication. A lack of inhibition by camostat and anti-ACE2 antibodies suggests that ACE2 is unlikely to be a dominant receptor for viral entry into monocytes but does not rule out a small role in monocyte infection or a more prominent role in the infection of ACE2+ macrophages. Early in viral replication, a series of positive-strand subgenomic RNAs (sgRNAs) is transcribed with a common leader sequence that specifically indicates viral replication16. RT–qPCR was used to detect SARS-CoV-2 genomic RNAs (gRNAs) and sgRNAs using primers targeting the N1 region of the N gene and the shared leader sequence and 3′ UTR sequences of the sgRNAs, respectively. gRNA and sgRNA were detected only in SARS-CoV-2-infected HD monocytes (Fig. 4n, o). The most abundant amplified sgRNA fragment migrated on agarose gels at the size of the N sgRNA (1,560 nucleotides), and its identity was confirmed by sequencing.
Although multiple assays indicated monocytes begin viral replication, we next assessed whether infected monocytes produce infectious virus. Infectious SARS-CoV-2 is detected in plasma of patients with COVID-19 only with especially sensitive assays, and we did not detect infectious virus by plaque assay in plasma samples from nine patients with COVID-19. Although infected HD monocyte culture supernatants formed plaques in Vero cells when culture supernatants were collected immediately after infection (probably detecting input virus), no infectious virus was detected when culture supernatants were collected 48 h after infection (Fig. 4p). By contrast, plaques were easily detected in culture supernatants from infected Vero cells collected at 48 h after infection. Thus, monocyte infection did not produce infectious virus.
Discussion
Here we show antibody-opsonized SARS-CoV-2 infects and replicates in blood monocytes and lung macrophages. About 10% of monocytes and 8% of lung macrophages in patients with COVID-19 were SARS-CoV-2-infected. We found a one-to-one correspondence between monocyte infection and inflammasome caspase-1 activation and pyroptosis. Most dying monocytes in the blood of patients with COVID-19 had activated inflammasomes, suggesting that monocytes are dying of pyroptosis. This is a large number, considering that dying cells are rapidly eliminated in vivo. It may be surprising that monocyte infection and cell death has not been widely recognized. However, this may be because (1) many COVID-19 studies use thawed, frozen cells, and dying cells do not survive freeze–thawing; (2) investigation of whether circulating mononuclear cells are dying is lacking in published studies; and (3) few researchers have looked for monocyte infection because monocytes do not express ACE2. A few previous studies have shown increased IL-1 cytokines in the plasma of patients with COVID-19, in vitro SARS-CoV-2 entry in myeloid cells or NLRP3 inflammasome caspase-1 activation in blood cells of patients with COVID-199,10,21,38. However, no previous study showed that SARS-CoV-2 infection of monocytes is antibody mediated, identified the monocyte receptor, showed that viral replication does not produce infectious virions, identified monocyte infection as the cause of inflammasome activation or showed evidence of pyroptosis. However, two previous studies suggested that monocyte-derived macrophages can be abortively infected38. In contrast to our findings, monocyte-derived macrophages weakly express ACE2 and their infection may be partly mediated by ACE2, as in vitro infection in the absence of anti-spike is blocked by anti-ACE238.
FcγR-mediated uptake of antibody-coated virus into monocytes is a double-edged sword. Pyroptosis, which occurs rapidly, probably aborts viral infection before infectious virions are fully assembled. Monocyte/macrophage infection is a dead end for the virus—it removes virions from the extracellular milieu, blocks them from producing infectious progeny and prevents them from disseminating. Pyroptosis in infected monocytes/macrophages also sounds a potent immune alarm to recruit and activate innate and adaptive immune cells to infection sites to mobilize immune defence. By contrast, the inflammatory mediators released from pyroptotic monocytes and macrophages can cause a cytokine storm. It may not be a coincidence that clinical deterioration coincides temporally with the detection of SARS-CoV-2 antibody responses8,29,39. In fact, some recent studies suggest that higher antibody titres correlate with disease severity29,39.
Pyroptotic myeloid cells are probably a major cause of the serious inflammatory sequelae that lead to acute lung injury, multiorgan damage, vascular leak and respiratory distress in patients with severe disease. In particular, patients with severe COVID-19 had increased plasma biomarkers of pyroptosis compared with patients with mild or moderate COVID-19. However, neither antibody titres nor the proportion of infected ASC-speck-positive monocytes at presentation correlated with severe disease, perhaps because of the small number of samples. Larger cohorts are needed to better assess the relative importance of monocyte/macrophage pyroptosis in severe COVID-19 pathogenesis. The large numbers of infected monocytes and macrophages, the fact that a quarter of lung macrophages have activated inflammasomes, and that myeloid cells are the major source of IL-1 and other inflammatory cytokines make it probable that monocyte/macrophage infection and inflammasome activation are important in severe COVID-19 pathogenesis. Although neutrophils could potentially be infected, infection of freshly isolated COVID-19 neutrophils or in vitro-infected HD neutrophils was not detected. Thus, neutrophil infection is probably not a major contributor to pathogenesis, although neutrophil activation of GSDMD-dependent NETosis (a cell death process involving neutrophil extracellular traps (NETs)) or other features of neutrophil activation may well be important drivers. It will be worthwhile to study other infected cells as potential sources of inflammation, and to understand what aspects of monocyte/macrophage activation enhance infection.
Four times as many lung-resident macrophages had activated inflammasomes as were infected. Further studies are needed to identify what stimulates inflammation in uninfected macrophages, but alarmins released by lung tissue damage are probably culprits. Although inflammasome activation was detected in almost every infected monocyte and macrophage, it was not detected in lung epithelial cells. Why lung epithelial cells resist inflammasome activation will require further study. It is worth examining whether infection might activate inflammasome-independent pyroptosis by other gasdermins in non-myeloid cells in the lungs. NLRP3 and AIM2 inflammasomes that recognize cell membrane damage and cytosolic DNA, respectively, formed in SARS-CoV-2-infected monocytes. Further work is needed to understand how SARS-CoV-2 activates these inflammasomes, whether activation is restricted to virulent coronaviruses, and whether other inflammasomes are activated, such as NLRP1 and NLRP6, which sense dsRNA40,41.
In this study, blocking antibodies against two FcγRs, CD16 and CD64, inhibited monocyte infection. CD64 is expressed on all monocytes, including the dominant classical subtype that is not infected, whereas CD16 is more selectively expressed, and all the infected patient monocytes are CD16 positive. This means that CD16 is probably the major Fc receptor that mediates viral entry into monocytes. Blocking infection by anti-CD64 antibodies may be indirect, as CD64 and CD16 use the same signalling adaptors and associate on the cell surface.
At diagnosis, plasma biomarkers of pyroptosis, including IL-1RA, IL-18, LDH and GSDMD, were increased in patients who developed severe disease—suggesting that they might help to predict prognosis—and who would benefit from immune-modulating therapy. Repurposing FDA-approved drugs that inhibit inflammatory cytokines or GSDMD is worth assessing but, so far, controlled clinical trials evaluating inhibiting inflammatory cytokines (anti-IL-1β (canakinumab), anti-IL-1RA (anakinra), anti-IL-6 and anti-IL-6R) have shown at best weak protection, which may be due to suboptimal timing or because any cytokine is only one of many inflammatory mediators. Two FDA-approved inhibitors of GSDMD, disulfiram (antabuse)42 and dimethyl fumarate (tecfidera)43 are currently being evaluated in clinical studies (NCT04485130, NCT04594343 and NCT04381936). In mouse models of sepsis, which has overlapping features with severe COVID-19 disease, these drugs strongly improved survival and reduced plasma IL-6 and TNF.
Our findings, which implicate opsonizing antibodies in monocyte infection and inflammasome activation, suggest that antibodies may contribute to deleterious immune reactions associated with severe disease44. FcγR-mediated monocyte infection is an example of antibody-mediated enhancement of infection. Nonetheless, overwhelming evidence shows that vaccine-generated neutralizing antibodies prevent infection and improve the clinical outcome of breakthrough infections, suggesting that anti-spike antibodies are highly beneficial. Plasma from vaccinated individuals did not promote monocyte infection, indicating that antibody-mediated enhancement is not a concern with respect to vaccination. However, therapeutically administered anti-spike neutralizing monoclonal antibodies only improve the clinical outcome if given early, before hospitalization45,46, and antibody-containing convalescent sera have not shown clinical benefit47. Thus, it is worth considering whether some antibodies might have both protective and deleterious effects48. Antibodies are clearly beneficial for blocking infection of ACE2-expressing lung and airway epithelia, in which the virus completes replication to produce infectious progeny. However, antibody properties that affect Fc-receptor-mediated cellular uptake, phagocytosis, cytotoxicity and complement activation can affect disease pathogenesis28.
Early development of afucosylated anti-spike antibodies promotes alveolar macrophage inflammation and is associated with COVID-19 severity33–35. Afucosylated antibodies are increased during acute infection with enveloped viruses like SARS-CoV-2 but are not abundant after COVID-19 vaccination49 or other types of antigen exposure34. IgG isolated from patients with COVID-19 with a higher proportion of afucosylated antibodies significantly, but weakly, increased in vitro monocyte infection but IgG from patients with fewer afucosylated antibodies did not. The increased pathogenicity of afucosylated antibodies could be secondary to antibody-mediated infection and downstream inflammasome activation in monocytes and macrophages. However, our findings about afucosylation are preliminary and more work is needed to make this association. Characterizing how antibody features, such as afucosylation, sialylation and choice of constant region, alter protective versus deleterious functions of anti-spike antibodies will be important not only for understanding SARS-CoV-2 pathogenesis, but also for choosing the best preparations of convalescent patient plasma and monoclonal antibodies for therapy and/or prevention of severe disease.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-04702-4.
Methods
Human participants
Fresh PBMCs and plasma cohort.
The study was approved by the Investigation Review Boards of Boston Children’s Hospital and Massachusetts General Hospital (MGH), and all of the enrolled patients signed an informed consent. A total of 73 patients aged 18 years or older with clinical symptoms suggestive of COVID-19 infection were enrolled at the time of presentation to the MGH emergency department (ED) from 9 July 2020 to 15 October 2021. A 10-ml EDTA blood sample was transported to Boston Children’s Hospital and processed within 2 h of collection. Samples from patients with COVID-19 were all RT–qPCR verified for SARS-CoV-2 infection on the day on which blood was drawn. Patients who presented to the ED with COVID-19-like symptoms, but were PCR negative, were used as non-COVID-19 samples. Patients who had received SARS-CoV-2 vaccination before presentation were excluded from the study. A summary of demographic and clinical data is provided in Supplementary Table 1. HD samples were processed and analysed in parallel with the patient samples. The participants were enrolled from 9 July 2020 to 10 January 2021 at Boston Children’s Hospital (BCH) with IRB-approved waiver of informed consent. Vaccinated HDs (n = 6), who received two doses of the Pfizer-BioNtech mRNA vaccine, were enrolled 3 weeks after the second dose and their plasma was pooled to evaluate whether it promoted monocyte infection.
Frozen plasma cohort.
A total of 60 patients aged 18 years or older with clinical symptoms suggestive of COVID-19 infection were enrolled in the MGH ED from 15 March 2020 to 15 April 2020 with an IRB-approved waiver of informed consent. The enrolled patients had at least one of the following: (1) tachypnea, ≥22 breaths per minute; (2) oxygen saturation, ≤92% on room air; (3) requirement for supplemental oxygen; and (4) positive-pressure ventilation. A 10-ml EDTA tube was obtained with the initial clinical blood draw in the ED (n = 60). Blood was also obtained on days 3 (n = 42) and 7 (n = 35) if the patient was hospitalized on those dates. Clinical course was followed for 28 days after enrolment or until hospital discharge if after 28 days. SARS-CoV-2-confirmed patients (by RT–qPCR) were assigned a maximum acuity score (A1–A5) (A1, died; A2, required mechanical ventilation; A3, hospitalized requiring supplemental oxygen; A4, hospitalized but not requiring supplemental oxygen; and A5, discharged and not requiring hospitalization)12. Patients were grouped on the basis of their worst acuity score over 28 days and divided into three groups for comparison (A1 and A2, severe disease; A3, moderate disease; and A4 and A5, mild disease). Only 1 patient was in A4; most of the mild patients therefore represent those who were discharged immediately from the ED and therefore have only a day-0 sample. A summary of the demographic and clinical data for each outcome group is provided in Supplementary Table 2.
Lung tissue samples.
Lung samples from five individuals who died from COVID-19 (Supplementary Table 3) and three individuals who died from trauma and without lung disease were obtained from MGH. The study was approved by the institutional review board of MGH IRB 2020P001147. Informed consent was obtained from the relatives of study participants. Lung tissue specimens were obtained within 24 h of autopsy and immediately formalin-fixed and embedded in paraffin.
Reagents and antibodies
A list of reagents and antibodies and their sources is provided in Supplementary Table 4.
Plasma, PBMC, neutrophil and monocyte isolation
Samples were processed using the recommended safety precautions in a BSL-2+ facility. Blood tubes were centrifuged at 2,000 rpm for 10 min to separate the plasma from blood cells. The plasma was collected in a new tube and incubated or not with 1% Triton X-100 for 1 h on ice before aliquoting and freezing at −80 °C. Blood cells were resuspended in PBS and layered over Ficoll for density centrifugation. PBMCs were collected from the interface and subjected to red blood cell lysis (if necessary) with Red Blood Cell Lysing Buffer Hybri-Max for 5 min on ice, followed by quenching with RPMI medium supplemented with 10% FBS and 1% penicillin–streptomycin. PBMCs were washed once more with RPMI and one fraction was stained for flow cytometry, while the remaining cells were used for monocyte purification by negative selection using the RosetteSep Human Monocyte Enrichment Cocktail. Neutrophils of patients with COVID-19 were isolated from the whole blood by immunomagnetic negative selection using the EasySep Direct Human Neutrophil Isolation Cocktail, according to the manufacturer’s instructions. HD monocytes for in vitro infection were purified from PBMCs by positive selection with CD14+ magnetic beads. The red blood cell pellet from the Ficoll density centrifugation was used to isolate neutrophils from the same HD samples. Neutrophils were separated from the RBC pellet by hypotonic lysis.
Cell lines
The THP-1 monocytic cell line and Vero E6 cells were obtained from ATCC. A549 cells and HEK293T cells overexpressing ACE2 were obtained from the MassCPR variants repository at Ragon Institute. ACE2 expression was validated by RT–qPCR and anti-ACE2 flow cytometry. All cells were tested for mycoplasma contamination.
Multiplex luminex, immunoassay and LDH activity assay
IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12, IL-13, IL-17, IL-18, IL-21, IL-23, CCL3, CCL7, CCL9, CXCL10, G-CSF, TNF, IFNβ and IFNγ were measured in plasma samples using a custom Luminex assay (R&D Systems) according to the manufacturer’s instructions. Sample data were acquired using the Luminex xPONENT 4.2 for MAGPIX Analyzer at the Analytical Instrumentation Core Lab of Boston University and analysed with Milliplex Analyst v5. The plasma levels of IL-1β were measured using the Simple Plex cartridge Ella (ProteinSimple) according to the manufacturer’s instructions at the BCH. All of the samples were diluted 1:3 with the dilution buffer and the analytical performance was conducted on the ProteinSimple Ella automated immunoassay platform (Bio-Techne). The samples were acquired using the Simple Plex Runner v.3.7.2.0 software and analysed using Simple Plex Explorer 3.7.2.0. GSDMD was measured in the same samples using the Human GSDMD ELISA kit (MyBiosource) according to the manufacturer’s instructions and LDH activity was measured using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega). Results from the latter assays were analysed using the Biotek Synergy 2 analyzer; GSDMD absorbance was measured at 450 nm and LDH absorbance was measured at 490 nm. Absorbance levels were quantified by linear regression based on the standard curve.
Anti-spike RBD ELISA
The enzyme-linked immunosorbent assay (ELISA) anti-spike RBD kit (BioLegend) was used to quantify antigen-specific IgG in the plasma from HDs, non-COVID-19 patients and patients with COVID-19. ELISA was performed according to the manufacturer’s instructions. Anti-spike RBD absorbance was measured at 450 nm and 570 nm and quantified by linear regression based on the standard curve.
Intracellular staining for imaging flow cytometry and confocal microscopy
Fixed monocytes were permeabilized with 0.1% Triton X-100 for 10 min and washed twice with PBS + 3% FBS. Monocytes were then blocked for 30 min with PBS + 5% FBS, washed twice and then stained with unconjugated primary antibodies against ASC (1:200, mouse or rabbit), NLRP3 (1:200, goat), AIM2 (1:200, mouse), GSDMD (1:200, mouse), pyrin (1:200, rabbit), dsRNA (J2, mouse) (1:500) or SARS-CoV-2 nucleocapsid protein (1:500, rabbit) for 2 h, followed by three washes with PBS + 3% FBS. The cells were then stained with secondary antibodies (donkey anti-mouse, rabbit or goat conjugated with Alexa Fluor 488, 546 or 647, at 1:1,000) for 1 h in PBS + 3% FBS, followed by three washes. Untreated THP-1 cells, THP-1 cells treated with LPS + nigericin or transfected with Poly(dA:dT) using Lipofectamine 2000, and HEK293T cells (negative control) were stained with anti-NLRP3 and anti-AIM2 antibodies for antibody validation.
For microscopy analysis, cells were fixed and then stained with DAPI (1:1,000) for 10 min, washed three times and cytospun onto glass slides (VWR), and sealed using polyvinyl alcohol and 1.5 mm coverslips (VWR). Confocal images were acquired using the Zeiss LSM 800 system with 405-nm, 488-nm, 561-nm and 633-nm lasers (emission filters, 465 nm, 509 nm, 561 nm and 668 nm, respectively) and a ×40 or ×63 1.4 NA oil-immersion objective. Images were acquired using Zen Black 2.0 and processed using Zen Blue 3.2.
For imaging flow cytometry, cells were resuspended in PBS + 3% FBS for analysis. Data were acquired using the ImageStream X MKII system with ×60 magnification (Amnis), the INSPIRE v.2 acquisition software and were analysed using IDEAS v.6.2 (Amnis). Monocytes were gated based on area/aspect ratio. ASC, NLRP3, AIM2 and pyrin specks were gated and quantified on the basis of fluorophore intensity/maximum pixels.
Flow cytometry
PBMCs were washed and stained for viability with Zombie Yellow in PBS (1:200) for 15 min on ice. Cells were washed with PBS, centrifuged and then stained with anti-annexin V PE (1:200) antibodies in 1× annexin buffer for 15 min on ice. After washing with 1× annexin V buffer, cells were blocked for 10 min with anti-CD32 (1:100) in PBS + 3% FBS, and then stained for 15 min on ice with a cocktail of antibodies to identify lymphocyte and myeloid cell subsets (all 1:200 except CD19 BV650, CD123 PerCP-Cy5.5 and CD56 APC-Cy7, 1:100). Purified monocytes and an A549 cell line overexpressing ACE2 were blocked with anti-CD32, then stained with primary antibodies for ACE2 (1:100) for 15 min on ice. The secondary anti-goat AF488 antibody was co-incubated with anti-CD14 PE-Cy7 (1:200) and anti-CD147 APC (1:100) antibodies. After the last wash, cells were resuspended in 2% PFA and kept at 4 °C until flow cytometry analysis. In vitro-infected monocytes were fixed and permeabilized with 0.1% Triton X-100, then blocked with PBS + 5% FBS. Cells were stained with primary antibodies for dsRNA (J2, mouse) (1:500), then stained with secondary antibodies (donkey anti-mouse conjugated with Alexa Fluor 647, at 1:500) and anti-CD14 PE-Cy7 antibodies. Cells were acquired using the FACS Canto II or LSR II using the FACSDiva v7 acquisition software, and data were analysed using FlowJo v.10.7.1.
FLICA assay
Freshly isolated monocytes were washed and resuspended in RPMI 10% FBS with FLICA substrate (BioRad FAM-FLICA Caspase-1 kit) and cultured for 1 h at 37 °C. Cells were then washed twice with 1× apoptosis buffer (from the kit) and fixed with 1× fixative (from the kit). Cells were kept at 4 °C until further staining and analysis.
Immunoblot analysis
Lysates of enriched monocytes from HDs and patients with COVID-19, the former treated or not for 16 h at 37 °C with 100 ng ml−1 LPS and 20 μM nigericin, were resolved on 12% SDS–PAGE gels, transferred to nitrocellulose membranes and blotted to detect GSDMD using (Abcam ab210070) primary rabbit monoclonal antibodies and secondary anti-rabbit IgG. The membranes were also blotted for β-actin and COX-IV.
Immunofluorescence analysis of lung samples
Formalin-fixed and paraffin-embedded lung parenchymal samples were stained for SARS-CoV-2 N, ASC and CD14, and immunofluorescence was analysed on the Leica Bond RX automated staining platform using the Leica Biosystems Refine Detection Kit (Leica). The antibody for SARS nucleocapsid (Novus) was run with citrate antigen retrieval and tagged with Alexa Fluor 488 Tyramide (Life). After citrate stripping, the antibody for CD14 (Cell Signaling) was incubated and tagged with Alexa Fluor 594 Tyramide (Life). After EDTA stripping, staining for ASC (Santa Cruz) was analysed using antibodies tagged with Alexa Fluor 647 Tyramide (Life). EDTA stripping was performed before anti-CD31 or anti-E-cadherin staining tagged to Alexa Fluor 555 Tyramide (Life). The samples were counterstained with DAPI. The slides were scanned using the Aperio Versa Digital Pathology Scanner (Leica) and analysed using Aperio ImageScope v.12.4.3 (Leica). The slides were also analysed by confocal microscopy as described above.
In vitro SARS-CoV-2 infection
icSARS-CoV-2-mNG (a molecular clone of SARS-CoV-2 expressing Neon Green (NG) fluorescent protein) was a gift to A.E.G. from S. P. Yong and the World Reference Center for Emerging Viruses and Arboviruses)31. The NG fusion protein is expressed only during viral replication. The SARS CoV-2 US-WA1/2020 ancestral (WA) variant was obtained from BEI Resources. The B.1.617.1/Delta variant isolate was obtained from the MassCPR variant repository. In brief, the variant was isolated at the Ragon BSL3 by rescue on Vero-E6 cells from primary clinical specimens. The whole genome of subsequent viral stocks was sequenced to confirm that no additional mutation arose during virus expansion. HD monocytes/neutrophils were purified from apheresis leukoreduction collars collected at Brigham and Women’s Hospital. Monocytes were incubated overnight with medium or 100 ng ml−1 LPS, and then infected with icSARS-CoV-2-mNG, SARS-CoV-2 (WA) and SARS CoV-2 B.1.617.1/Delta (multiplicity of infection (MOI) = 1) in a BSL-3 facility. Infection of A549-ACE2 cells at an MOI of 0.01 was used as a control. The viral inoculum was treated with 10 μg ml−1 of antibody (isotype control mAb114, anti-spike C1A-H12, or anti-spike C1A-B12), or 5% pooled plasma (heat-inactivated or not; Ig-depleted or not, as indicated) from HDs (n = 3), patients with COVID-19 of mixed disease severity (n = 12 (total), n = 4 (mild), n = 4 (moderate), n = 4 (severe)) or vaccinated HDs (n = 6) before infection with SARS-CoV-2 for 30 min at room temperature. Treated virus (100 μl) was added to monocytes (2 × 106 cells per well) in 48-well plates. Infected cells were incubated at 37 °C under 5% CO2 with gentle shaking every 10 min for 1 h, after which the culture volume was increased to 500 μl with RPMI supplemented with 5% heat-inactivated normal AB human serum and 10 μg ml−1 of the aforementioned antibodies, or 5% pooled plasma from HDs or patients with COVID-19. Cultures were then incubated at 37 °C under 5% CO2 for 48 h, at which time the cells were collected and fixed for 20 min with 4% PFA and then stained.
IgG from the pooled plasma of patients with COVID-19 was depleted by protein A/G agarose resin and IgA depleted by peptide M agarose. Control samples were incubated with agarose resin without coupled protein. C1A-B12 and C1A-H12, two SARS-CoV-2 spike-targeting human monoclonal antibodies, were produced as previously described32. For blocking experiments, cells were incubated with 10 μg ml−1 monoclonal antibodies, anti-CD16, anti-CD32 (clone IV.3 (Fig. 4j and Extended Data Fig. 5a), clone 6C4 (Fig. 4k and Extended Data Fig. 5b, c)), anti-CD64, anti-ACE2 and anti-CD147 for 30 min, before virus infection. For antiviral drug treatment, monocytes were incubated at 37 °C under 5% CO2 for 1 h with 10 μM remdesivir (GS-5734) or camostat mesylate before infection. To find an appropriate remdesivir concentration, serial dilutions between 10 and 80 μM were analysed. To compare plasma obtained from patients with different disease severity, plasma was pooled on the basis of the MGH acuity score (A1–A5), as described above.
To test the role of IgG afucosylation, IgG purified from serum samples of patients with COVID-19 was analysed by mass spectrometry to define the percentage of afucosylation as described previously33. Low afucosylated samples, provided by T. Wang, contained 8.4 ± 0.7% afucosylated IgG and high afucosylated samples, 30.1 ± 1.5% afucosylated IgG. IgG was also purified from pooled plasma from HDs and patients with COVID-19 using the Melon gel IgG Spin Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Virus was preincubated with 10 μg ml−1 of purified IgG and the infection was performed as described above.
RT–qPCR
RNA was extracted using Trizol reagent (Invitrogen) from monocytes of patients with COVID-19 or from uninfected or infected HD monocytes (stimulated or not with LPS (100 ng ml−1 for 16 h)), then reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Random primers were used to generate cDNA for detection of cellular RNAs (ACE2, BSG and ACTB) and SARS-CoV-2-specific primers were used to generate cDNA to detect viral genomic RNAs (N1 region of N gene)50. cDNA was analysed by RT– qPCR using the SsoFast EvaGreen Supermix (BioRad) (30 s at 95 °C; then 40 cycles of 3 s at 95 °C and 3 s at 54 °C) in the CFX96 Touch Real-Time PCR Detection System (BioRad) using the CFX Manager v.1.6 acquisition/analysis software. To detect SARS-CoV-2 sgRNA, RT–qPCR was performed using a primer pair with the forward primer annealing to the 5′ leader region of the viral genome and the reverse primer annealing to the 3′ UTR. With the cycling conditions used (30 s at 95 °C; then 40 cycles of 30 s at 95 °C, 30 s at 60 °C and 90 s at 72 °C), full-length gRNA was not amplified, but small sgRNA segments (<3 kb) could be amplified16,51,52. For each sample, Ct values were normalized to the ACTB Ct value. Primer sequences are provided in Supplementary Table 4. sgRNA qPCR products were also analysed by electrophoresis on 1% agarose gels stained with ethidium bromide and visualized on the Chemidoc imager (BioRad). The approximately 1,600 nucleotide band was excised and sequenced to confirm its origin as the SARS-CoV-2 sgRNA encoding N.
Plaque assays
Vero E6 cells were seeded as monolayers in 24-well plates 1 day before infection. Virus-infected sample culture supernatants were serially diluted in DMEM. The plates were washed once with DPBS and then infected with 100 μl of diluted sample and incubated at 37 °C under 5% CO2 for 1 h with rocking every 15 min. After 1 h, the inoculum was removed and an overlay of 1% methylcellulose (Sigma-Aldrich) in complete MEM (Gibco) was applied to each well. The plates were incubated at 37 °C until plaques were observable in positive control wells. To visualize plaques, the overlay was removed, and the cell monolayer was fixed with 4% PFA and stained with crystal violet. Plaques were then counted to quantify the virus titre in PFU per ml.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v.9.0. Normal distribution of the data was evaluated using the D’Agostino and Pearson normality test before applying statistical methods. Distributions were considered to be normal if P ≤ 0.05. Parametric or nonparametric (Mann–Whitney U-test) two-tailed unpaired t-tests were used to compare two unpaired groups. Multiple-group comparisons were analysed using one-way ANOVA with Sidak or Tukey multiple-comparisons tests, or nonparametric Kruskal–Wallis with Dunn post-test. Multiple groups were compared using two-way ANOVA with additional Sidak or Tukey multiple-comparisons test. Mean plasma values from hospitalized patients with COVID-19 on each day were compared between severity groups by multiple unpaired t-tests. Correlations of plasma levels were determined by simple linear regression and Pearson correlation coefficient.
Extended Data
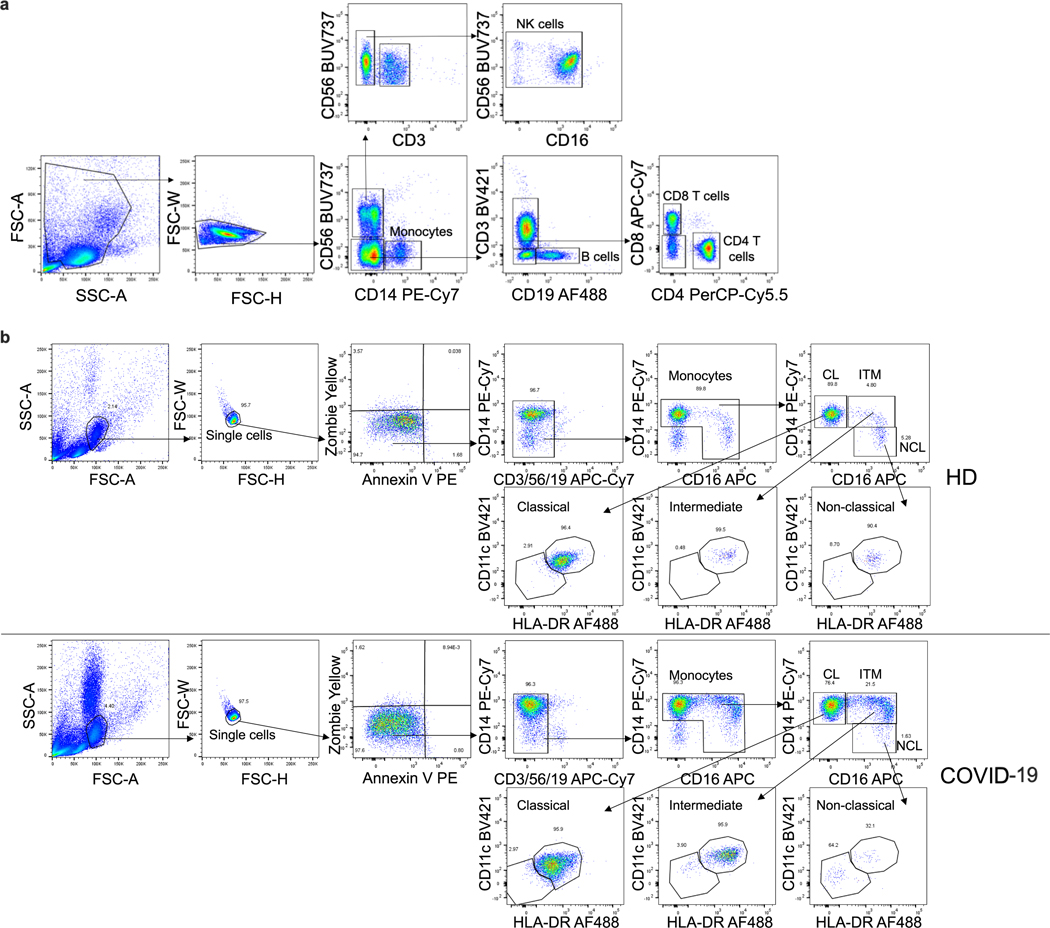
Extended Data Fig. 1 |. Identification of lymphocyte and monocyte subsets in healthy donors and COVID-19 patients.
Flow cytometry gating strategy for identifying lymphocytes and monocytes in Fig. 1a, b (a) and for identifying monocyte subpopulations in Fig. 1c (b). Monocyte subpopulations: CL - classical CD14hiCD16−; ITM - intermediate CD14hiCD16+; NCL - non-classical CD14loCD16+.
Extended Data Fig. 2 |. Inflammasome imaging and GSDMD cleavage analysis.
a, Gating strategy for imaging flow cytometry analysis of isolated monocytes. b, Representative imaging flow cytometry images of GSDMD and ASC staining in COVID-19 patient monocytes that lacked ASC specks. c, d, Representative imaging flow cytometry images of HEK293T cells (negative control) and THP-1 cells untreated or treated with LPS+nigericin or transfected with poly(dA:dT), then stained with anti-NLRP3 (c) and anti-AIM2 (d). e, Single staining controls for antibody staining. Representative images of monocytes from COVID-19 patients shown were stained with 1° ASC - 2° AF488; 1° NLRP3 – 2°AF568; 1° GSDMD, Pyrin, AIM2, J2, N - 2° AF647; or FAM FLICA Caspase-1 fluorescence, and Zombie Yellow dye. FLICA+ and Zombie+ cells in cells undergoing pyroptosis; GSDMD, Pyrin, AIM2 and NLRP3 in non-pyroptotic cells (diffuse staining); J2+ and N+ in infected monocytes. Scale bar, 7 μm (b–e). f, Representative confocal image z-stacks and plane projections of monocytes of HD and COVID-19 patients, stained for the same markers as in Figure 2. Scale bars, 5 μm. g, Full scan images for blots shown in Fig. 2g.
Extended Data Fig. 3 |. ACE2 and CD147 expression on circulating monocytes.
Purified blood monocytes from HD (n = 3), COVID-19 patients (n = 4) and A549-ACE2 (n = 3) were analysed by flow cytometry (a, c) and RT–qPCR (b, d) for expression of ACE2 (a, b) or CD147 (BSG) (c, d). HD monocytes were treated or not with LPS before analysis. A549-ACE2 cells were used as positive control. Mean ± S.E.M. is shown. *p<0.05, ***p<0.001, ****p<0.0001 relative to isotype (Iso) control antibody-stained, LPS-activated HD monocytes (a,c) by one-way ANOVA with Tukey’s multiple comparisons test. Data are representative of 2 independent experiments.
Extended Data Fig. 4 |. Effect of anti-spike monoclonal antibodies or pooled COVID-19 plasma on in vitro infection of healthy donor purified monocytes with icSARS-CoV-2-mNG.
a–e, HD monocytes (n = 3) were primed with LPS, infected with icSARS-CoV-2-mNG (MOI, 1), then stained 48 h later for nucleocapsid (N) or dsRNA (J2) and ASC and analysed by imaging flow cytometry. Before infection, virus was preincubated with indicated monoclonal antibodies (IgG1 isotype control mAb114 (Iso)), non-neutralizing anti-spike (C1A-H12 (H12)) or neutralizing anti-RBD (C1A-B12 (B12)) or with pooled HD or COVID-19 patient plasma that had been heat-inactivated (HI) or not. U, uninfected. Quantification of HD monocyte staining for N (a), J2 (b), NG (c, e) or ASC specks (d). (e) Shows the percentage of N+ cells that were also NG fluorescent. f, g, A5490-ACE2 (n = 3) (f) or LPS-primed HD monocytes (n = 3) (g) were infected at the indicated MOI with icSARS-CoV-2-mNG (NG), a molecular clone of the Washington (WA) strain, or with clinical WA and Delta strains. Infection was measured by N staining and flow cytometry. Mean ± S.E.M. is shown. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by one-way ANOVA with Tukey’s multiple comparisons test, relative to Iso or as indicated (a–g). Data are representative of 2 independent experiments.
Extended Data Fig. 5 |. In vitro infection of healthy donor monocytes and neutrophils.
a–c, LPS-primed HD monocytes (n = 3) (a, b) or purified HD neutrophils (n = 3) (c) were infected with icSARS-CoV-2-mNG (MOI, 1), then stained 48 h later for nucleocapsid (N) or analysed for NG fluorescence (c, right). Before infection, virus was preincubated with COVID-19 plasma, depleted or not of IgG as indicated, and infection was carried out in the presence of indicated blocking or isotype (Iso) control antibodies (a–c). The monocyte and neutrophil infections in (b) and (c) were performed with cells isolated from the same HDs. d, Freshly isolated neutrophils, enriched by negative selection, from HD (n = 3) and COVID-19 patients of mixed disease severity (n = 4) were stained for N and analysed by flow cytometry to assess in vivo infection. e, Infection of LPS-primed HD monocytes (n = 3) with icSARS-CoV-2-mNG in the presence of pooled COVID-19 patient plasma, depleted or not of IgG as indicated, and antiviral drugs, Camostat and Remdesivir. f, Infection of A549-ACE2 (n = 3) with icSARS-CoV-2-mNG to verify the inhibitory activity of 10 μM Remdesivir. Infection was measured by N staining and flow cytometry. g, Infection of A549-ACE2 (n = 3) and HD monocytes (n = 3) with icSARS-CoV-2-mNG in the presence of anti-ACE2 blocking antibody at different concentrations. Infection was measured by NG fluorescence. Mean ± S.E.M. is shown. *p<0.05, **p<0.01, ***p<0.001, by one-way ANOVA with Tukey’s multiple comparisons test (a–c, g), nonparametric unpaired t-test (d, f) and two-tailed nonparametric unpaired multiple t-test (e). Data are representative of 2 replicate experiments.
Supplementary Material
Acknowledgements
We thank members of the MGH COVID-19 collection and the processing team (K. Lavin-Parsons, B. Lilley, C. Lodenstein, B. McKaig, N. Charland, H. Khanna, A. Gonye, I. Gushterova, T. Lasalle, N. Sharma, B. C. Russo, M. Rojas-Lopez, M. Sade-Feldman, K. Manakongtreecheep, J. Tantivit and M. Fisher Thomas) for plasma samples; T. Wang for providing the high and low afucosylated IgGs from donors with COVID-19; the members of the Analytical Instrumentation Core Lab of Boston University for running and analysing the Luminex Multiplex assay; D. Briscoe and L. Sheward for analysing IL-1β; the staff at the Specialized Histopathology Core of the Dana-Farber/Harvard Cancer Center (supported in part by an NIH P30CA06516) for histology and immunohistochemistry; the staff at the MassCPR variants repository (funded by the Massachusetts Consortium on Pathogen Readiness) for providing viral strains and ACE2-overexpressing cell lines; and the staff at the Ragon Insitute BSL3 laboratory (funded by the Harvard Center for AIDS Research (NIH P30AI060354)) and the Massachusetts Consortium on Pathogen Readiness. This research was supported by the Lemann Brazil Research Fund (to J.L. and C.J.); the National Institutes of Health grant R01AI124491 (to H.W.); the National Institutes of Health grant U19AI131135 (to L.G.); the Annenberg Foundation and FAST grants and a gift from J. Sullivan (to A.E.G.); the American Lung Association (to M.B.G. and M.R.F.); the British Heart Foundation Programme grant RG/16/4/32218 (to S.B.); a Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) fellowship (to C.J.); a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) fellowship (to L.B.d.L.); and the National Institutes of Health training grant T32AI007245-31A1 (to M.L.).
Footnotes
Competing interests The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-04702-4.
Peer review information Nature thanks Stanley Perlman, Gestur Vidarsson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The data and materials supporting the findings of this study are available from the corresponding authors on request.
References
- 1.Hu B, Guo H, Zhou P. & Shi ZL Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Valle DM et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Xia S, Zhang Z, Wu H. & Lieberman J. Channelling inflammation: gasdermins in physiology and disease. Nat. Rev. Drug Discov. 20, 384–405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbar AN & Gilroy DW Aging immunity may exacerbate COVID-19. Science 369, 256–257 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Dinarello CA Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117, 3720–3732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C. et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 180, 934–943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vora SM, Lieberman J. & Wu H. Inflammasome activation at the crux of severe COVID-19. Nat. Rev. Immunol. 21, 694–703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long QX et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Hadjadj J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369, 718–724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues TS et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 218, e20201707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan AH & Schroder K. Inflammasome signaling and regulation of interleukin-1 family cytokines. J. Exp. Med. 217, e20190314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filbin MR et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep. Med. 2, 100287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan P. et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 12, 4664 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma D. & Kanneganti TD The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J. Cell Biol. 213, 617–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung S-Y, Yuen K-S, Ye Z-W, Chan C-P & Jin D-Y A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg. Microbes Infect. 9, 558–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irving AT, Ahn M, Goh G, Anderson DE & Wang LF Lessons from the host defences of bats, a unique viral reservoir. Nature 589, 363–370 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Davis MA et al. Calpain drives pyroptotic vimentin cleavage, intermediate filament loss, and cell rupture that mediates immunostimulation. Proc. Natl Acad. Sci. USA 116, 5061–5070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spel L. & Martinon F. Detection of viruses by inflammasomes. Curr. Opinion Virol. 46, 59–64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers C. et al. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat. Commun. 10, 1689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson KV et al. A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med. 214, 3611–3626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J. et al. Severe acute respiratory syndrome coronavirus 2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis. 223, 785–795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song X. et al. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry A, 10.1002/cyto.a.24285 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ragotte ,RJ et al. l. Human basigin (CD147) does not directly interact with SARS-CoV-2 spike glycoprotein. mSphere 6, e0064721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shilts J, Crozier TWM, Greenwood EJD, Lehner PJ & Wright GJ No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 11, 413 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K. et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 5, 283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruhns P. & Jonsson F. Mouse and human FcR effector functions. Immunol. Rev. 268, 25–51 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Ong S-M et al. A novel, five-marker alternative to CD16–CD14 gating to identify the three human monocyte subsets. Front. Immunol. 10, 1761 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bournazos S, Gupta A. & Ravetch JV The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 20, 633–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li K. et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat. Commun. 11, 6044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber F, Wagner V, Rasmussen SB, Hartmann R. & Paludan SR Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80, 5059–5064 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X. et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark SA et al. Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2. Preprint at bioRxiv 10.1101/2020.11.13.381533 (2020). [DOI] [Google Scholar]
- 33.Chakraborty S. et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 22, 67–73 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen MD et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science 371, eabc8378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoepel W. et al. High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci. Transl. Med. 13, eabf8654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ackermann M. et al. Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ. 28, 3125–3139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hui KPY et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 8, 687–695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Beltran WF et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell 184, 476–488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen C. et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell 184, 5759–5774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauernfried S, Scherr MJ, Pichlmair A, Duderstadt KE & Hornung V. Human NLRP1 is a sensor for double-stranded RNA. Science 371, eabd0811 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Hu JJ et al. FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol. 21, 736–745 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphries F. et al. Succination inactivates gasdermin D and blocks pyroptosis. Science 369, 1633–1637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwasaki A. & Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 20, 339–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinreich DM et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N. Engl. J. Med. 384, 238–251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta A. et al. Early treatment for COVID-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 385, 1941–1950 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Korley FK et al. Early convalescent plasma for high-risk outpatients with COVID-19. N. Engl. J. Med. 385, 1951–1960 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y. et al. Enhancement versus neutralization by SARS-CoV-2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 34, 108699 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty S. et al. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci. Transl. Med. 14, eabm7853 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Division of Viral Diseases, NCIRD. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primer and Probes, https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (CDC, 2020). [Google Scholar]
- 51.Kim D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perera R. et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg. Infect. Dis. 26, 2701–2704 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials supporting the findings of this study are available from the corresponding authors on request.