Abstract
Pallas's cat, or the manul cat (Otocolobus manul), is a small felid native to the grasslands and steppes of central Asia. Population strongholds in Mongolia and China face growing challenges from climate change, habitat fragmentation, poaching, and other sources. These threats, combined with O. manul’s zoo collection popularity and value in evolutionary biology, necessitate improvement of species genomic resources. We used standalone nanopore sequencing to assemble a 2.5 Gb, 61-contig nuclear assembly and 17097 bp mitogenome for O. manul. The primary nuclear assembly had 56× sequencing coverage, a contig N50 of 118 Mb, and a 94.7% BUSCO completeness score for Carnivora-specific genes. High genome collinearity within Felidae permitted alignment-based scaffolding onto the fishing cat (Prionailurus viverrinus) reference genome. Manul contigs spanned all 19 felid chromosomes with an inferred total gap length of less than 400 kilobases. Modified basecalling and variant phasing produced an alternate pseudohaplotype assembly and allele-specific DNA methylation calls; 61 differentially methylated regions were identified between haplotypes. Nearest features included classical imprinted genes, non-coding RNAs, and putative novel imprinted loci. The assembled mitogenome successfully resolved existing discordance between Felinae nuclear and mtDNA phylogenies. All assembly drafts were generated from 158 Gb of sequence using seven minION flow cells.
INTRODUCTION
Pallas's cat (Otocolobus manul), or the manul cat, is a small-bodied carnivore native to the montane grassland and shrubland steppe habitats of Central Asia (1). O. manul's wide geographic range spans varied climatic regions including the Caucuses Mountains, Mongolia and the Tibetan Plateau, but populations are concentrated at higher altitudes (2,3). Deemed the ‘the grumpiest cat in the world’ by the BBC documentary series Frozen Planet II (4), manuls are solitary hunters and subsist on a diet of small mammals (5). Prussian zoologist Peter Simon Pallas assigned the manul cat's taxonomic name as Felis manul in 1776 (6); the genus name Otocolobus was proposed in 1842 by Johann Fiedrich von Brandt (7) and formalized in 1907 based on skull morphology divergence from Felis (8).
Because of its wide home range and low population density, the conservation status of O. manul varies geographically. The International Union for Conservation of Nature (IUCN) Red List classifies the species as ‘Least Concern’ as of 2020 after being ‘Near Threatened’ in assessments from 2002 to 2016 (1, 9). However, O. manul is locally classified as endangered in several countries and either likely or confirmed extinct in some parts of its former range. Populations are also severely geographically fragmented, raising concerns for reduced effective population size and genome heterozygosity (10,11). The main threats to manul cats are anthropogenic, including poaching, habitat destruction, and the widespread use of rodenticides, which reduce prey populations and result in secondary poisoning of predators (12). A successful captive breeding program has led to O. manul being widespread in zoo collections around the world (13).
The manul cat is the sole extant species in the monotypic genus Otocolobus; phylogenetic analyses surrounding the relationship of this genus to other small-bodied cats are ongoing. A phylogenetic study using select nuclear genes across the felid radiation placed Otocolobus as a sister lineage to the genus Prionailurus, a taxonomic group that includes the fishing cat (P. viverrinus) and leopard cat (P. bengalensis) among others (14). In contrast, a whole-mitogenome phylogeny placed Otocolobus closer to Felis, which includes the domestic house cat (F. catus) (15). Both studies are concordant with skull morphology data suggesting a Miocene divergence between Pantheridae and other cat lineages (16). Improved genomic resources for the manul cat will increase resolution for phylogenetic analyses within Felidae.
High-quality genome assemblies and comparative genomics experiments increasingly include data on the epigenome and repetitive DNA content (17–19). DNA methylation, the most well-studied epigenetic mark, is critical in embryonic development, genomic imprinting, and X-inactivation with additional influence on gene expression and transposable element (TE) suppression (20–25). Methyl-cytosines also exhibit a 2–3-fold increase in mutation rate relative to unmethylated cytosines via accelerated deamination (26), providing a molecular substrate for phenotypic change across evolutionary time. Long-read sequencing permits haplotype phasing based on DNA sequence variants, meaning that DNA methylation can also be segregated by parent of origin (27). This development is of paramount importance for genome-wide measurement of allele-specific DNA methylation, a signature of imprinted genes. Despite its biological significance, imprinting is chronically understudied due to technical limitations such as bisulfite fragmentation, multi-mapping, and reduced information complexity (28,29).
Widespread improvements in genome assembly quality reflect reduced sequencing costs, greater local computing power, and improved contiguity via long-read sequencing (30). Now within reach even for low-resource laboratories, these powerful tools can support conservation efforts for diverse animal species. Here, we provide a high-quality diploid nuclear genome assembly, updated mitogenome, and allele-specific methylation analysis for O. manul, all generated exclusively from Oxford Nanopore sequencing reads.
MATERIALS AND METHODS
DNA sample
A 5-year-old male manul cat residing at the Utica Zoo (Utica, NY, USA), Tater, was chosen for genome sequencing (Figure 1). Whole blood was collected by jugular venipuncture and shipped frozen, then thawed and combined with 2 volumes of DNA Shield (Zymo Research, Irvine, CA, USA). Genomic DNA was extracted using a Quick DNA Miniprep Plus kit (Zymo Research) yielding 25 μg of DNA from 200 ml of blood. DNA quality was checked using a nano-spectrophotometer (Implen N60, Munich, Germany) and run out on a 1% gel to visualize fragmentation and RNA contamination.
Figure 1.

Otocolobus manul holotype photo depicting Tater, the 5-year-old male manul cat sampled for genome assembly.
Library preparation
Seven gDNA library preparations were performed using the SQK-LSK110 kit (Oxford Nanopore Technologies, OX4 4DQ, UK) following a modified version of the manufacturer's instructions (Nanopore Protocol version GDE_9108_v110_revJ_10Nov2020). In the authors’ modified version, each library prep used 3–5 μg of input DNA and was eluted with 45 μl of elution buffer (EB). Each elution was then split into three 15 μl aliquots to permit one initial flow cell loading step plus two reloads per library prep.
Sequencing
Sequencing was performed on seven R9.4.1 minION flow cells using Oxford Nanopore's minKNOW software (v22.05.5) and Guppy basecalling (v6.2.1). A total of three 15 μl DNA aliquots were loaded onto each flow cell with nuclease flushes between reloads per the SQK-LSK110 kit manual's ‘Priming and loading’ section. Each DNA aliquot was sequenced for 24 h for a total of 72 h of sequencing time per flow cell (21 total sequencing days). Guppy was set to fast basecalling during sequencing; post-hoc basecalling was performed using the ‘super accuracy’ model (r941_min_sup_g507). All assemblies were built using basecalls from the r941_min_sup_g507 model. Sequencing summary statistics (Table 1) and a histogram of read length data (Supplemental File 1) are provided.
Table 1.
O. manul nanopore sequencing read summary. Reads were generated from 25 ug of DNA on seven minION flow cells over 21 total sequencing days
| Number of reads | 15349224 |
|---|---|
| Number of bases | 158 248 024 728 |
| N50 read length (bp) | 16 601 |
| Longest read (bp) | 379 006 |
| Shortest read (bp) | 38 |
| Mean read length (bp) | 10 309 |
| Median read length (bp) | 7099 |
| Mean read quality (QV) | 16.26 |
| Median read quality (QV) | 14.51 |
Computational methods
Detailed pipeline information including software versions is provided in Supplemental File 2. The final assembly required the use of cluster computing resources with 2TB of allocated random access memory (RAM); downstream analyses were performed on two local computers: one running Linux Ubuntu 20.04.1 with 128GB RAM, an NVIDIA 3090Ti graphics processing unit (GPU), and a 16-core, 32-thread AMD Ryzen 9 5950x central processing unit (CPU), and another running Linux Ubuntu (20.04.1) with 64GB RAM, two NVIDIA graphics cards (3080Ti and 2080Ti), and a 12-core, 24-thread Ryzen 9 3900x CPU. Abbreviated computational methods are included below.
Genome assembly and polishing
Because there is no single best assembly pipeline for mammalian genomes, we generated draft sequences using a variety of assemblers and polishers. Draft assemblies were generated locally or via institutional cluster computing using Flye (31), NextDenovo (github.com/Nextomics/NextDenovo), Shasta (32), and Raven (33). Polishing was performed with Medaka (github.com/nanoporetech/medaka) for consensus assemblies and Racon (34) or NextPolish (35) for non-consensus assemblies. Assembly quality was assessed by searching for Carnivora-specific genes with benchmarking universal single-copy orthologs (BUSCO) version 5.3.2 (36,37). The two highest-scoring assemblies from Flye and NextDenovo, respectively, were combined using the Quickmerge metassembler (38) to increase contiguity while preserving quality. The Purge Haplotigs pipeline was applied for reassignment and removal of allelic contigs (39,40). Software versions and calls to the assemblers are provided in Supplementary File 2.
Contamination detection
Unmapped raw reads were scanned for microbial DNA with Kraken2 (41) and Pavian (42). We manually inspected the final assembly for spurious contaminant contigs using GC content, unusually high (>1000×) or low (<1×) coverage depth, lack of BUSCO genes, and sequence similarity as metrics. Sequence similarity contamination screening was performed using Megablast versus NCBI’s nucleotide (nt) database (43). Blobtools2 (44) was used to visualize GC content and read coverage (Supplemental Figure 1). An apparently chimeric 420 kb contig with >1000× coverage and no Carnivora-specific BUSCO genes matching Felidae by identity, ctg001250, was removed to generate the final primary assembly. Megablast hits to ctg001250 are provided in Supplemental File 3.
Coverage and quality statistics
Assembly coverage was assessed using Mosdepth version 0.3.345. K-mer quality value estimates were generated with Inspector, a tool built for long read data (45). We also applied Merqury, which was built for high accuracy short reads, to demonstrate its behavior on less accurate data (46).
Repeat identification
Repetitive DNA masking and classification was performed with RepeatMasker version 4.1.0 (47). Annotation using existing repeat classes, families and subfamilies was elected based on good representation of Carnivora in the Dfam version 3.5 open source repeat library.
Gene annotation
GeMoMa v1.8 and 1.9 (48,49) were applied to the primary assembly for homology-based protein prediction. First, version 1.8 was run with the F. catus gene annotation (GCF_018350175.1) used as the single reference; with the release of version 1.9, ‘GeMoMaPipeline’ was re-run with four felid gene annotations (F. catus, GCF_018350175.1; P. viverrinus, GCF_022837055.1; P. uncia GCF_023721935.1; O. geoffroyi, GCF_018350155.1) as a combined reference. Specific parameters are available in Supplemental File 2. ‘GeMoMaPipeline’ was also run on the P. viverrinus reference nucleotide assembly to assess the comparability of our O. manul output to NCBI’s reference annotations. All predicted proteins were scored using BUSCO’s protein mode and the carnivora_odb10 lineage dataset. Liftoff (50) was used with the F. catus (GCF_018350175.1) reference annotation to display gene features on differential DNA methylation plots for the O. manul assembly; DMR annotations were sanity checked by overlapping the GeMoMa and Liftoff annotation intervals. Proteins were not predicted for the Liftoff output.
Variant calling
Variant calling and phasing was performed using the PEPPER-Margin-DeepVariant pipeline (27). Variant statistics were generated using WhatsHap version 1.4 (51) and VCFtools version 0.1.17 (52). A consensus FASTA for the secondary haplotype was generated by applying all biallelic variants to the final assembly (considered the primary haplotype) with the consensus module of BCFtools version 1.15.1 (53). Runs of homozygosity and genomic demographics were assessed with SMC++ (54).
DNA methylation
DNA methylation (5 mC) at cytosine guanine dinucleotides (CpGs) was determined by re-basecalling QC-passed FAST5 files with a modified base configuration of GPU-mode Guppy (dna_r9.4.1_450bps_modbases_5mc_cg_sup). The final primary assembly was used as a reference. The resulting modified BAMs (modBAMs) were concatenated together into a single file, then sorted and indexed with Samtools (53). PEPPER-Margin-DeepVariant was then re-run to generate a haplotagged modBAM; this file and the primary assembly were used as input for ModBAM2BED version 0.6.2 (github.com/epi2me-labs/modbam2bed), which aggregates modified base counts to generate bedMethyl files. These files were then used as input for global and allele-specific methylation analysis.
Allele-specific DNA methylation was analyzed using the R package DSS developer version 2.43.2 (55–58). The package's two-group statistical comparison module ‘DMLtest’ was used to identify differentially methylated loci between the two haplotypes. The ‘callDMR’ module was then used with strict parameters (Pthreshold = 0.001, delta = 0.5, minlen = 100, minCG = 15, dis.merge = 1500) to identify multi-CpG differentially methylated regions (DMRs).
DMR sequences were annotated via lifting of F. catus reference (GCF_018350175.1) gene features onto the O. manul assembly using Liftoff (50). The nearest feature to each DMR was then identified using AGAT (https://github.com/NBISweden/AGAT) and the closest module of BEDTools version 2.30.0 (59,60). DMRs annotated near genes with a ‘LOC’ symbol were additionally annotated with an alias or gene description, when available, via manual look up in NCBI’s Gene database (https://www.ncbi.nlm.nih.gov/gene). Visualizations were generated using MethylArtist version 1.2.3 (61), which required haplotagging the PEPPER output with Longphase (62), and JBrowse 2 (63) to validate each DMR.
Mitochondrial genome
Nanopore reads were aligned to the F. catus mitogenome using Minimap2 (v2.22-r1101) (64,65). Aligned reads were then downsampled to 10 000 reads using Seqtk version 1.3-r106 (https://github.com/lh3/seqtk) and Flye (v2.9-b1768) in metagenome mode was used for assembly. Mitogenome annotation and manual rearrangement to start the circular mitogenome at COX1 was done previously (66). A mitogenome phylogeny was built using MUSCLE version 3.8 (67) for alignment and IQ-TREE version 1.6.12 (68) for bootstrapped maximum-likelihood tree inference.
Scaffolding
The contig-level assembly was scaffolded onto the fishing cat (P. viverrinus) reference genome with the scaffold module of RagTag (69). Alignment of the two genomes was assessed with Dot (github.com/marianattestad/dot) visualizations of Nucmer alignments (70). An ideogram of contig positions in the scaffolds was generated with the R pakcage chromoMap (CRAN.R-project.org/package = chromoMap).
RESULTS
Sequencing
We isolated DNA for the assembly from the whole blood of Tater, a captive-bred 5-year-old male manul cat residing at the Utica Zoo (Utica, NY, USA). A holotype photo is provided (Figure 1). DNA extraction yielded approximately 25 μg of total DNA. DNA was sequenced over 21 days using seven minION flow cells and base calling was performed using the GPU version of Guppy (v6.2.1). After default quality filtering, sequencing runs generated a total of 15.3 million reads and 158 Gb of sequence with a read N50 of 16.6 kb (Table 1). Of these reads, 66% had a quality score greater than Q20 (i.e. error rate of 1 in 100 base calls) and 20% were greater than Q30 (one error in 1000 base calls).
Initial assembly
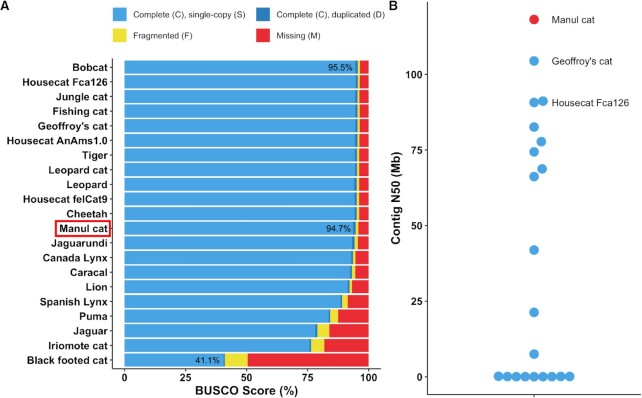
We generated multiple de novo assemblies and chose the highest-quality result for further analysis based on BUSCO score (37) (Table 2). Twenty other Felidae reference genomes, including three domestic house cat (F. catus) assemblies, provided benchmarking (64,71–81). Other felid genomes ranged in sequencing coverage from 17× to 159× with an average BUSCO score of 89.56% and a median score of 95.1% (Figure 2); full felid genome assembly statistics used in this comparison are available in Supplemental File 3.
Table 2.
Assembly statistics for selected O. manul drafts and reference genomes for fishing cat (P. viverrinus) and domestic house cat (F. catus). House cat is the felid species with the most high-quality genome assemblies; fishing cat is more closely related to O. manul by nuclear DNA phylogeny (14,15)
| Assembly | Size (Gb) | Contigs/scaffolds/ chromosomes | Contig N50 (Mb) | BUSCO (% complete) | Depth | Technique |
|---|---|---|---|---|---|---|
| F. catus Fca126_mat1.0 (GCF_018350175.1) | 2.42 | 110/71/19 | 90.7 | 95.5 | 76× | PacBio Sequel 2 |
| P. viverrinus UM_Priviv_1.0 (GCF_022837055.1) | 2.46 | 255/192/19 | 68.8 | 95.4 | 30× | PacBio Sequel |
| O. manul NextDenovo + Medaka | 2.49 | 175/–/– | 36.2 | 94.6 | 56× | Nanopore |
| O. manul Shasta + Medaka | 2.47 | 714/–/– | 26.3 | 94.1 | 56× | Nanopore |
| O. manul Flye + Medaka | 2.49 | 1046/–/– | 28.6 | 94.8 | 56× | Nanopore |
| O. manul Raven + Racon x2 + Medaka | 2.50 | 568/–/– | 9.2 | 94.7 | 56× | Nanopore |
| O. manul Quickmerge (Flye + NextDenovo) + NextDenovo | 2.49 | 99/–/– | 118.2 | 94.7 | 56× | Nanopore |
| O. manul Quickmerge + Purge Haplotigs | 2.49 | 62/–/– | 118.2 | 94.7 | 56× | Nanopore |
| OtoMan1.0 | 2.49 | 61/23/19 | 118.2 | 94.7 | 56× | Nanopore |
Figure 2.
OtoMan1.0 is a near-complete and highly contiguous genome assembly for Pallas's cat (Otocolobus manul). (A) Felid reference assembly completeness was compared to our O. manul assembly by locally searching Carnivora-specific genes (carnivora_odb10) with BUSCO v5.3.2. OtoMan1.0, a nanopore-only assembly, ranked within 1% of the highest felid assembly BUSCO scores. (B) Contig N50 distribution for O. manul and felid reference assemblies. The O. manul assembly exhibits high contiguity (N50 = 118.2 Mb) when compared to other Felidae genomes, which differ minimally (∼3 Mb) in size.
We ran multiple de novo assemblers with relatively low computational memory requirements (i.e. runnable with 64–128GB local RAM) including Flye, NextDenovo, Shasta, and Raven. Running Flye with the full sequencing read set on a mammal-sized genome required remote high-powered computing resources (≥400GB RAM), so Flye was also run locally with filtered read sets (Supplemental File 3). Filtering reads by length or quality reduced computational resources enough to run assembly software locally but resulted in slightly lower BUSCO scores, even after polishing with the full read set. Therefore, the full QC-passed read set was applied for both drafts used in the final assembly.
The two highest quality draft assemblies were generated by Flye and NextDenovo, both polished once with Medaka (https://github.com/nanoporetech/medaka). Of the two, the Flye sequence achieved the highest BUSCO completeness score for Carnivora-specific genes at 94.8% across 1000 contigs (Table 2). The NextDenovo assembly had fewer contigs (175) but a lower BUSCO score of 94.6%. Both sequences were appropriate in size for a felid genome at 2.49 Gb. A merged assembly was generated with Quickmerge to combine the best traits of both drafts (i.e. highest BUSCO score and largest contigs, respectively) (38). Merging parameters were set to reduce the likelihood of perpetuating errors or misjoining contigs, and the quality of each draft was assessed with Inspector (Supplemental File 2) (45). The Flye-Medaka draft's QV was 31.2 and the NextDenovo-Medaka draft's QV was 31.4. Visualizing alignments of the draft assemblies to each other and to the merge result additionally indicated that (i) the drafts were similar to one another pre-merge and (ii) the merge favored the Flye draft over the NextDenovo draft (Supplemental Figure 3). After merging, reassignment of allelic contigs was performed on the merge result with the Purge Haplotigs pipeline (39,40), bringing the number of contigs in the assembly down to 62.
Assembly curation and scaffolding
Recent nanopore-only genome assemblies have used anomalous coverage depth and expected GC content as a means of filtering alien contigs (82). Here, these parameters were assessed using Blobtools2 (44). There was one 420 kb contig with 1178× average coverage depth in the initial O. manul assembly, ctg001250. NCBI megablast matched the sequence to Felidae, making a contaminant or endoparasite source unlikely (Supplemental File 3). Examining sequencing read alignment to ctg001250 in JBrowse 2(63) revealed regions of typical (∼60×) coverage interrupted by stretches covered by up to 4000 reads (Supplemental Figure 2). We concluded that ctg001250 was falsely contiguous based on a low number of reads spanning the high and low coverage areas, perhaps due to misplacement of interspersed repeats. Fifty percent of bases on ctg001250 were masked, and 40% were classified as LINE1 elements by RepeatMasker (Supplemental File 3). BUSCO score was intact following removal of ctg001250 (Table 2).
The final contig-level assembly, OtoMan1.0, was 2487293883 bp in length with 61 contigs, an N50 of 118.2 Mb, and a 94.7% BUSCO completeness score for Carnivora genes (Figure 2). Total genomic GC content was 41.87%. For comparison, 20 other Felidae reference genomes were locally reanalyzed with BUSCO v5.3.2 (carnivora_odb10). Our score of 94.7% places this nanopore-only assembly within 1% of the highest quality reference assemblies in Felidae. Additionally, the assembly's contig N50 was the highest among all assessed felid genomes. Half of OtoMan1.0 was covered by 8 contigs (L50) and the largest contig was 218.2 Mb (Supplemental Figure 4; Supplemental File 3). For reference, chromosome A1 in the current F. catus assembly is 239.4 Mb (RefSeq NC_058368.1). The shortest O. manul contig was 133 kb.
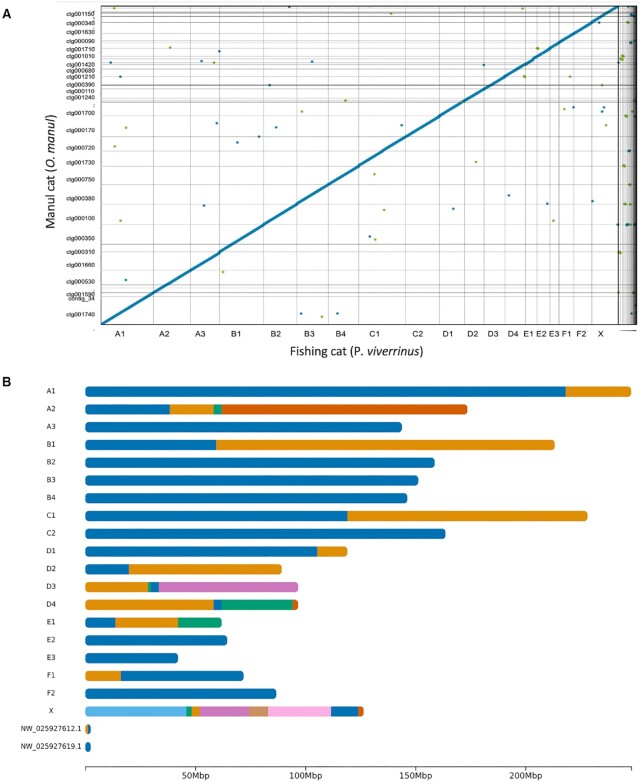
OtoMan1.0 was scaffolded onto the high-BUSCO fishing cat (P. viverrinus) reference genome (GCF_022837055.1) with the scaffold module of RagTag (69) (Figure 3). Fishing cat was chosen for alignment-based scaffolding due to (i) a lack of congenial species in Otocolobus, (ii) the genus sharing its most recent common ancestor with Prionailurus rather than Felis based on nuclear DNA (14,15) and (iii) the high BUSCO score of the P. viverrinus assembly. RagTag joined the 61 O. manul contigs into 23 scaffolds with 38 gaps, a total inferred gap length of 371685 bp, and a chromosome-level N50 of 151.9 Mb (Supplemental File 3). The scaffolds covered all 18 fishing cat autosomes, the X chromosome, and two unplaced BUSCO-containing fishing cat scaffolds (NW_025927612.1 and NW_025927619.1) (Supplemental Figure 3). Eight of the 18 cat autosomes (44%) were captured by single contigs in our assembly (Figure 3B), but two O. manul contigs were left unplaced. One of the two unplaced contigs was 593 kb in length and had 98.5% identity to a fragment of the 1.9 Mb F. catus Y chromosome (51% query coverage) when assessed with NCBI Megablast (43); the animal sampled to build UM_Priviv_1.0 was female. The second unplaced O. manul contig was 187 kb in length and exhibited high but discontiguous BLAST identity to F. catus, P. bengalensis and other felid sequences.
Figure 3.
OtoMan1.0 cross-genus alignment and scaffolding. (A) Genome sequence alignment of the fishing cat (P. viverrinus) reference genome (GCF_022837055.1) and the contig-level nanopore assembly of O. manul, OtoMan1.0, was generated using Nucmer (70) and Dot (https://github.com/marianattestad/dot). The alignment quality reflects gross genome collinearity present across Felidae (80,83). (B) Ideogram of OtoMan1.0 scaffolds generated via alignment to the fishing cat (P. viverrinus) reference assembly (GCF_022837055.1). Color transitions represent breakpoints between contigs on the scaffold. All 19 felid chromosomes were covered by the OtoMan1.0 assembly, and eight autosomes (A3, B2, B3, B4, C2, E2, E3 and F2) were captured by single contigs. Manul contigs were also aligned to two BUSCO-containing fishing cat unplaced scaffolds while two of the 61 O. manul contigs were unplaced. Correlations between the fishing cat chromosome lengths and O. manul scaffold lengths are provided in Supplemental Figure 5.
Repetitive DNA
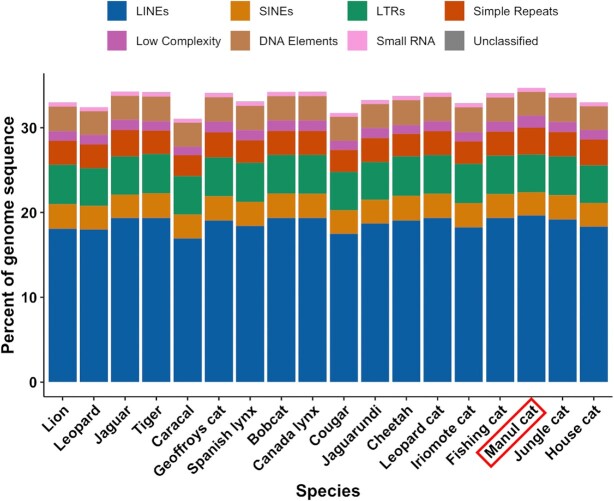
Repetitive DNA, particularly interspersed repeats, are valuable drivers of genetic diversity in vertebrates (84,85). Since carnivores are well-represented in transposable element (TE) databases, we classified and annotated O. manul repetitive DNA with RepeatMasker (Table 3) (47). Overall, 34.72% of the assembly was classified as repetitive DNA, with a majority of retroelements (26.83%) and fewer DNA elements (2.8%), simple repeats (3.2%), and low-complexity regions (1.4%). These results are consistent with the dominance of retroelements in mammalian genomes (86,87). Local analysis of other cat assemblies with RepeatMasker indicated that OtoMan1.0’s repeat complement was consistent with other members of Felidae (Figure 4). Conservation of global repeat content in this clade was high: the 18 analyzed species varied by 3.67% for total bases masked, 2.72% for LINE content, 0.16% for SINE content, and 0.15% for DNA element content (Supplemental File 3).
Table 3.
Repetitive DNA classification results for the manul cat (Otocolobus manul) genome assembly, OtoMan1.0, generated with RepeatMasker
| Element count | Total length (bp) | % of Genome covered | |
|---|---|---|---|
| Total bases masked | 863500886 | 34.72 | |
| Total interspersed elements | 737829702 | 29.66 | |
| Retroelements | 1596531 | 667340949 | 26.83 |
| SINE: | 470064 | 69243802 | 2.78 |
| MIR | 462472 | 68339639 | 2.75 |
| LINE: | 836683 | 488479415 | 19.64 |
| LINE1 | 468806 | 391204655 | 15.73 |
| LINE2 | 312177 | 84936852 | 3.41 |
| L3/CR1 | 41423 | 8915823 | 0.36 |
| RTE | 13010 | 3218808 | 0.13 |
| LTR: | 289784 | 109787884 | 4.41 |
| ERVL | 88371 | 39831330 | 1.60 |
| ERVL-MaLRs | 148224 | 51463432 | 2.07 |
| ERV_classI | 28861 | 12430326 | 0.50 |
| DNA elements: | 356331 | 69720741 | 2.80 |
| hAT-Charlie | 198573 | 36753878 | 1.48 |
| TcMar-Tigger | 55938 | 14630791 | 0.59 |
| Unclassified | 3794 | 597860 | 0.02 |
| Small RNA | 146099 | 11047204 | 0.44 |
| Simple repeats | 1677548 | 79649218 | 3.20 |
| Low complexity | 647428 | 34786006 | 1.40 |
Figure 4.
Repetitive element content of the assembled O. manul genome and 17 other felid species assessed with RepeatMasker (47). Manul cat repeat content was highly similar to other members of Felidae and dominated by retroelements.
Diploid genome
After read phasing OtoMan1.0 with the PEPPER-Margin-DeepVariant pipeline (27), approximately 83% of the assembly was covered by 6613 phase blocks with a block NG50 of 0.53 Mb (Supplemental File 3). As expected, most variants were single nucleotide variants (SNVs) and small insertions and deletions, with fewer large indels. Of 2045611 heterozygous variants detected, 1 561 420 (76.3%) were phased and 1 087 364 (53.2%) were phased, biallelic SNVs. The transition-transversion ratio (Ts/Tv) for biallelic SNVs was 2.12, in line with highly methylated mammalian genomes (Supplemental File 4). The software employed for variant calling does not detect inversions. The scaffolded primary assembly was deposited as BioProject PRJNA885133 and the contig-level alternate haplotype assembly as BioProject PRJNA889808.
An alternate pseudohaplotype assembly was generated by switching biallelic variants from the PEPPER variant call file into the primary, contig level assembly using the ‘consensus’ module of BCFtools (53). The resulting sequence was 2 484 282 002 bp in length with a contig N50 of 118.0 Mb and 94.8% BUSCO score.
We estimated diploid assembly quality using Inspector, a reference-free, k-mer-based tool built for long reads (45). Inspector's estimated QV for the primary contig-level assembly was 31.3 (99.93% accuracy) with a k-mer completeness score of 97.9% (Supplemental File 3). Inspector additionally scored the pre-merge assembly drafts from Flye and NextDenovo at Q31.2 and Q31.4, respectively. Inspector also includes a correction module for structural and base-level errors, but running it reduced OtoMan1.0’s BUSCO from 94.7% to 93.6% and inflated the reported number of structural errors.
We additionally ran Merqury on the diploid assembly (46). Merqury was built for high accuracy short reads, and lower accuracy read sets are more likely to produce QV overestimates. Still, this use case of a popular quality estimation tool is unreported in the literature. On default settings, Merqury gave OtoMan1.0 a k-mer completeness score of 97.5% and quality score of Q45.3 (Supplemental File 2). We also ran Merqury's ‘best_k’ script, which is intended to provide an ideal k-mer length for analysis based on genome size and read error rate. Given our median read error rate of 0.035, ‘best_k’ suggested k = 19; this value further inflated the QV to 53.2 from the original Q45.3 using the default k = 21. Despite these potential QV overestimates, Merqury's histogram output indicated that most small k-mers were excluded from the final assembly, appearing only in the read set (Supplemental Figure 6).
Lastly, genome heterozygosity and effective population size for O. manul were assessed using variant calls. Heterozygosity was 0.048% based on the number of called heterozygous SNVs (1 184 174) divided by the total callable bases in the contig-level primary assembly and 0.082% using all 2045611 heterozygous variants combined (SNVs and indels). Population history inferences for O. manul based on genomic runs of homozygosity were conducted with SMC++ (54). The results suggested an effective population size (Ne) near 10 000 with recovery from a bottleneck approximately 3000 generations ago (Supplemental Figure 7).
Annotation
Homology-based gene annotation was performed on OtoMan1.0 with Gene Model Mapper (GeMoMa) versions 1.8 and 1.9. The F. catus RefSeq annotation (GCF_018350175.1) served as the reference for our initial v1.8 run, generating 21909 amino acid predictions with an 86.2% protein BUSCO score (Supplemental File 2). A second prediction was generated using the P. viverrinus reference annotation as reference (GCF_022837055.1) instead, but BUSCO completeness was lower at 85.3%.
With the release of GeMoMa version 1.9, we re-ran GeMoMaPipeline on OtoMan1.0 using four felid reference annotations (F. catus, GCF_018350175.1; P. viverrinus, GCF_022837055.1; P. uncia GCF_023721935.1; O. geoffroyi, GCF_018350155.1) for a single prediction. The resulting protein BUSCO score was only slightly higher at 88.2% (Supplemental File 2).
To benchmark the GeMoMa software, we also predicted proteins for the P. viverrinus reference nucleotide assembly using F. catus as the annotation reference in GeMoMa v1.8. The resulting protein BUSCO score was 86.8%, within 1% of OtoMan1.0’s original protein score (Supplemental File 2). Both scores fall below our locally calculated BUSCO values for the F. catus (99.6%) and P. viverrinus (99.2%) protein annotations, which were generated with NCBI’s Eukaryotic Gene Annotation Pipeline.
DNA methylation
Nanopore instruments can detect native DNA modifications via modified pore signals compared to the unmodified base. Here, 5-methylcytosine (5mCG) base modifications were called using Guppy with the contig-level assembly as an alignment reference, generating methylation data for 33085506 cytosine-guanine dinucleotides. Similar to other mammals (88), global DNA methylation at CpG dinucleotides was high in O. manul at 78.2%.
Allele-specific methylation, a mechanism that facilitates monoallelic gene expression in genomic imprinting, was assessed by calling differentially methylated regions (DMRs) between O. manul pseudohaplotypes with DSS (55). Strict calling parameters (i.e. methylation delta at least 50%, 100 bp length, at least 15 CpGs) yielded 91 unique loci (Table 4). Visualization of haplotagged read alignments at each DMR facilitated the discovery of 30 false positive loci, where apparent read misalignment for one of the two haplotypes led to a called DMR via a lack of CpGs on one allele. Of the original 91 loci, 61 (67%) were deemed true DMRs following manual inspection (Supplemental File 3). The validated DMRs had a mean length of 969 bp and a mean of 118 CpGs.
Table 4.
Summary statistics and nearest gene feature data for differentially methylated regions (DMRs) called between O. manul pseudohaplotyes. Gene annotation data were lifted onto the OtoMan1.0 assembly from F. catus (F.catus_Fca126_mat1.0) and the nearest feature to each DMR was found with BEDTools (60). DMRs were validated visually using read pileup data. Statistics are shown for validated DMRs. Gene aliases are included in parenthesis
| Initial DMRs | 91 |
| Validated DMRs | 61 |
| Mean length (bp) | 969 |
| Median length (bp) | 705 |
| Mean number of CpGs | 118 |
| Median number of CpGs | 75 |
| Mean absolute difference in methylation (%) | 71.15 |
| Median absolute difference in methylation (%) | 69.67 |
| Mean distance to gene feature (bp) | 3463 |
| Median distance to gene feature (bp) | 0 |
| Nearest genes to DMRs (symbol) | MXRA7, KCNQ1, IGF2, LOC109492247, CTSD, CAMK1G, IGF2R, LOC123383330, LOC109499294, LOC111560210, HERC3, NAP1L5, LOC109500454, BNC1, IGF1R, LOC109500537, UBE2Q2, LOC102900156, KLC1, SOCS6, LOC123378902, MEST, LOC109497613, VWDE, HSPG2, ST8SIA6, LOC111561284, SNU13, ATP4A, LOC123382347, INPP5F, LOC101093666, LOC102899880, LOC101083149, ATP6V1H, FAM110B, LOC109495939, BLCAP, NNAT, LOC102900772 (NESP55), LOC101098453 (GNAS), LOC109497888, CALCB, LOC123380523, GNA12, LOC105261300, TEKT5, ZDBF2, CMKLR2 (GPR1) |
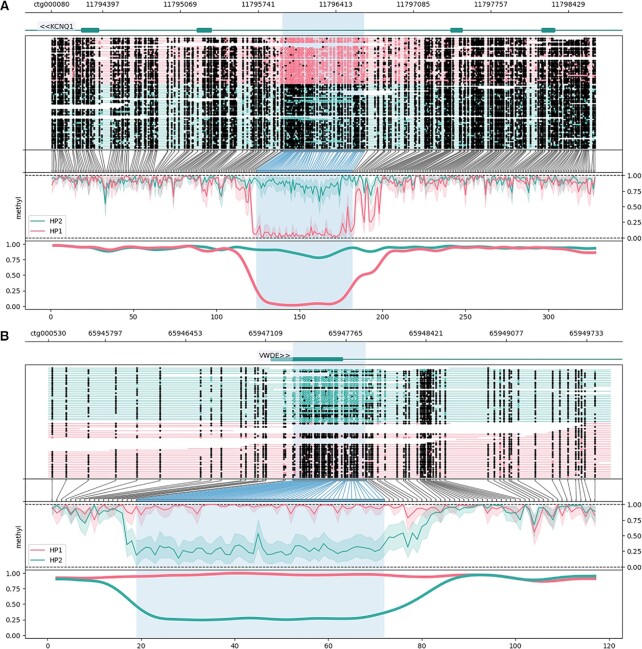
Validated DMRs were annotated by lifting F. catus reference gene features onto the contig-level O. manul assembly and querying the nearest gene with BEDTools (60). This method captured a number of classical imprinted loci described in humans and other animals, including GNAS, NNAT, IGF2, IGF2R, KCNQ1, MEST and ZDBF2 (Table 4) (89–93). In total, 41 validated DMRs directly overlapped 49 genes while non-overlapping DMRs fell between 165 bp and 56.7 kb away from the nearest feature. Putative novel imprinted loci, or DMRs near a feature not previously described as imprinted, including a 595 bp, 106-CpG DMR in the first exon of Von Willebrand factor D and EGF domain-containing protein (VWDE). Representative DMRs are shown in Figure 5. The mean absolute difference in methylation between the two pseudohaplotypes at DMRs was 71.15%. Most validated DMRs (44 of 61, 72.1%) were hypomethylated on the alternate pseudohaplotype, while 17 were hypermethylated.
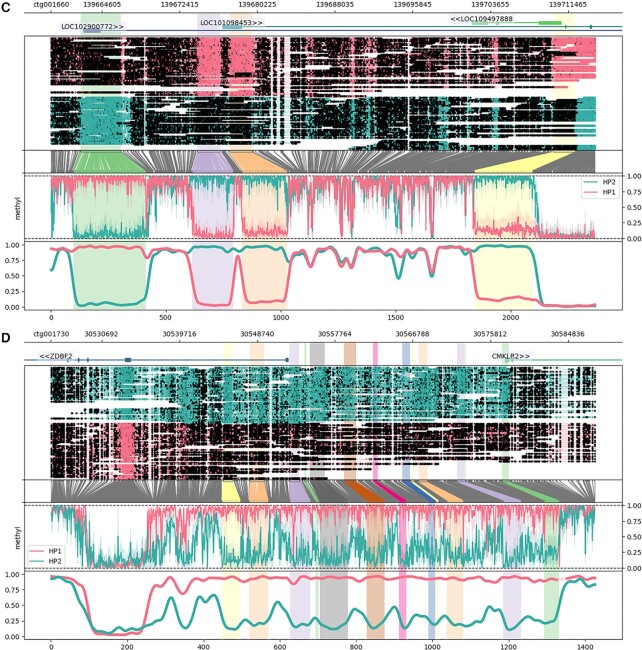
Figure 5.
Allele-specific DNA methylation at differentially located loci (DMRs) called between O. manul assembly pseudohaplotypes assembled from a whole blood sample. DMRs were visualized with the package Methylartist (61). Annotation of the 61 manually validated DMRs was accomplished by lifting gene coordinates from the Felis catus reference assembly (F.catus_Fca126_mat1.0) onto OtoMan1.0 and finding the nearest feature. The results revealed (A, B) singleton DMRs and (C, D) multi-DMR clusters within and near genes. Genes overlapping DMRs included previously described imprinted loci such as (A) potassium voltage-gated channel subfamily Q member 1 (KCNQ1) and (B) the guanine nucleotide-binding protein G(S) subunit alpha (GNAS) complex locus (aliases LOC101098453, LOC102900772). DMRs also overlapped genes not previously reported as imprinted, including (C) von Willebrand factor D and EGF domains (VWDE), a protein with predicted involvement in anatomical development (96). (D) The most notable locus was a 33.2 kb region containing 12 DMRs all hypomethylated on pseudohaplotype 2. The DMRS overlapped the 5’ end of a classical imprinted gene, zinc finger DBF-type containing 2 (ZDBF2), on the negative strand and the 5’ end of chemerin chemokine-like receptor 2 (CMKLR2) on the positive strand. The CMKLR2 antisense RNA (CMKLR2AS; alias GPR1AS), which is not annotated in F. catus, exhibits imprinted expression in the human placenta (97).
Of the 61 validated DMRs, 12 (19.67%) occurred in one contiguous 33.2 kb region on feline chromosome C1 (Figure 5). All DMRs in the region were hypomethylated on pseudohaplotype 2. The region fell directly upstream of two genes, chemerin chemokine-like receptor 2 (CMKLR2) on the positive strand and zinc finger DBF-type containing 2 (ZDBF2) on the negative strand. ZDBF2 is a canonical imprinted gene expressed paternally in most human and mouse tissues (94); imprinted expression of CMKLR2, a paralog of opioid receptor kappa 1 (OPRK1), has not been reported, but expression of its antisense transcript (GPR1AS) is imprinted in human and mouse placenta (95).
Mitochondrial genome
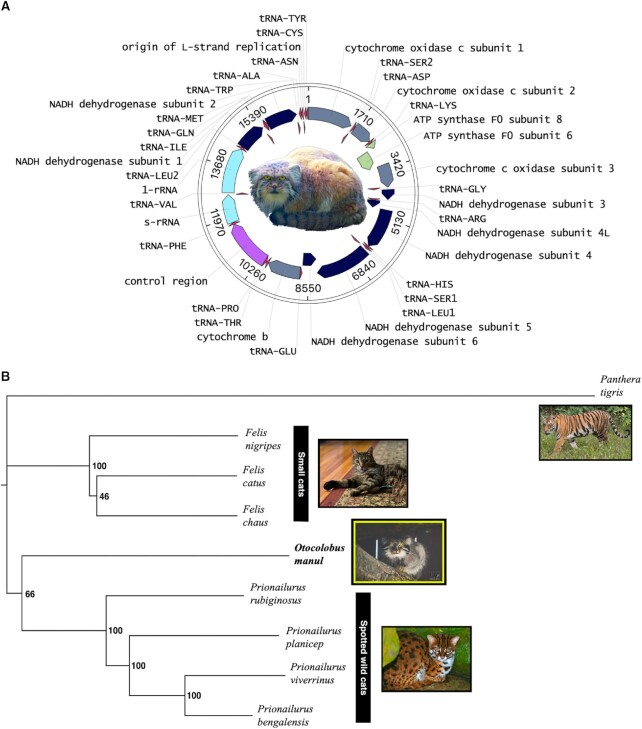
The mitochondrial genome assembly built using Flye in metagenome mode contained one circular contig 17 097 bp in length with 800× read coverage (Figure 6). Megablast revealed a 99.7% nucleotide identity match to a previously assembled O. manul mitogenome (Supplemental File 3) (98). However, our assembly was longer than this previous manul mitogenome assembled from Sanger sequencing reads in 2019, which was 16 672 bp (GenBank MH978908.1). The 400 bp gap identified between our assembly and the 2019 version was located in the non-coding D-loop region of the mitogenome, making misassembly less likely than in the case of a large genic insertion. A 2015 Illumina mitogenome assembly for O. manul was closer to our assembly's length at 17 009 bp (KR132585.1), but identity and query coverage were lower. From our mitogenome, a new phylogeny was built using tiger (Panthera tigris) as the outgroup. The resulting phylogeny matched previous nuclear DNA trees, with Otocolobus sharing its most recent ancestor with Prionailurus (14) in contradiction of a 2016 mitochondrial phylogeny that placed Otocolobus closer to Felis (15). The mitogenome was deposited as part of the scaffolded primary assembly at BioProject PRJNA885133.
Figure 6.
Nanopore mitochondrial genome assembly of O. manul and felid mtDNA phylogeny. (A) The circular length of the built O. manul mitogenome assembly was 17 097 bp; read coverage was 800×. (B) Phylogenetic relationships of O. manul were derived through bootstrapped maximum likelihood comparison of the assembled mitogenome to existing Felidae references. In contrast to previously published mtDNA for O. manul (98), the current tree matches those generated through analysis of nuclear DNA (14).
DISCUSSION
Here, we have produced a highly contiguous diploid nuclear genome assembly, mitogenome, and allele-specific DNA methylation dataset for the manul cat (Otocolobus manul) via standalone nanopore sequencing. With 61 contigs and a contig N50 of 118.2 Mb, the assembly was more contiguous than any other Felidae reference genome to date before scaffolding. The felid clade is particularly well-suited for cross-genus scaffolding due to conserved genome collinearity among its members; our scaffolding strategy may not yield such unambiguous results in other animal families (80,83). Comparing our primary assembly to the original read set with Inspector yielded a k-mer completeness score of 97.9% and QV of 31.3 (99.93% accuracy). Using a single sequencing technology without parental data, this QV score approaches the ‘platinum-quality’ reference genome standard of Q40 (99.99% accuracy) established by the Vertebrate Genomes Project (VGP) (77,99,100). Our assembly exceeds VGPs other benchmarks of contig N50 ≥ 1 Mb and chromosomal scaffold N50 ≥10 Mb by 118-fold and 15-fold, respectively. Merqury yielded a higher QV estimate of 45.3 on default settings, but the score is likely an overestimate due to a relatively high read error rate. Still, Merqury's k-mer spectra histograms indicated that most small k-mers were excluded from the final assembly, suggesting that our coverage depth was able to overcome the read set error rate to produce a high-quality genome.
Our assembly's BUSCO score for Carnivora-specific genes, 94.7%, also fell within 1% of the most complete felid genomes. Importantly, BUSCO values were re-calculated locally using an updated version of the program (v5.3.2), leading to lower scores overall compared to those currently displayed on NCBI’s Genome pages (v4.1.4). The described genome, OtoMan1.0, reflects the continued, rapid improvement of assembly pipelines and the necessity of long-read sequencing for generating contiguous, high-quality reference genomes (101–104). Our results also provide evidence against concerns for low nanopore-only assembly quality due to sequencing error rates.
Repetitive elements have been minimally explored in felids despite their high genome occupancy and role in phenotype evolution (105–108). SINE element insertions are major sources of genomic diversity in canids, with links to multiple phenotypes in domestic dogs (Canis lupus familiaris) including early retinal degeneration, polyneuropathy, myopathy, and merle coat color (109–115). While global repeat content is strongly conserved across Felidae based on our analysis, individual repeat family activity is potentially variable and relevant to intra- and inter-species conservation. In a biomedically relevant example, domestic cats but not large cats possess an infectious endogenous retrovirus (ERV), RD-114, which can contaminate vaccines manufactured with feline cell lines and infect experimentally inoculated dogs (116–118). Long-read sequencing enables more accurate investigation of genomic repeat content; single reads can span full TEs and capture unique flanking host sequences, reducing multi-mapping issues inherent in short read assembly (119–121).
Nanopore sequencing permits native DNA methylation detection alongside DNA base calls, combining genome assembly and epigenome analysis into one step at the bench top. The epigenome is responsive to an animal's environment and shifts in a predictable manner across the life span (122,123). Here, we present a phased methylome and allele-specific DNA methylation analysis for O. manul. Our differentially methylated region (DMR) detection criteria were relatively strict in an effort to reduce false positives and yielded 61 validated loci. We successfully captured a number of classical imprinted genes including NESP55/GNAS, KCNQ1, NNAT, IGF1R and IGF2/H19 (124–128). IGF2R contained a DMR in this study suggestive of imprinted expression; this gene is maternally expressed in dogs (129,130) while monoallelic expression appears to have been lost in the primate lineage (131). Putative novel imprinting loci were also identified in genes such as Von Willebrand factor D and EGF domain-containing protein (VWDE), a paralog of fibulin 2 (FBLN2) required for multi-tissue limb regeneration in the axolotl (Ambystoma mexicanum) (132). A 2012 study found that 40% of human genomes deposited by the 1000 Genomes project (www.1000genomes.org) carried at least one loss-of-function mutation in VWDE (133). This locus and others were evaluated as distinct from 30 false DMRs, where misassembly resulted in a lack of CpGs on one pseudohaplotype. This phenomenon is not detectable to the DMR calling software DSS, necessitating manual curation; humans have over 200 imprinted genes (127,134) meaning our DMR calling parameters likely also introduced false negatives. Although imprinted gene expression cannot be predicted based on methylation data alone, this assembly provides a high-quality alignment reference for future transcriptomic analyses in O. manul. Future comparative investigations in diverse species will enrich our functional understanding of the allele-specific methylation mechanism.
Detection of low genome heterozygosity in this captive-bred O. manul individual is a concerning but valuable insight for both zoo breeding programs and wild population management. At 0.048%, our estimate of heterozygosity using biallelic pseudohaplotype SNVs was similar to the critically endangered Amur tiger (Panthera tigris altaica), a species that experienced a severe population bottleneck in the 1940s with a current estimated population of 400 animals (135–138). In contrast, the manul cat population is estimated at 58 000 individuals (1). Although captive breeding programs are a crucial tool in global conservation, a small pool of founder animals inevitably leads to reduced genetic diversity. Practices like sperm biobanking have the potential to support captive population genetic quality, but such systems are underutilized (139). Fortunately, our estimate of effective population size using runs of homozygosity was relatively large at Ne = 10 000. Results also suggested that manul cats experienced a crash in genetic diversity, but that this event was in the relatively distant past (∼10000 years ago); effective population size appears to have continued recovering despite anthropogenic challenges to the species. Given the large, low-density geographic range and solitary social strategy of O. manul, it is unclear whether these values are typical across the entire population.
While largely conserved among metazoans, mitogenomes also contain structural and nucleotide variation that can serve as phylogenetic markers (140). Their smaller size, relative to nuclear genomes, make them an attractive tool in phylogenetics. However, incongruities between mitogenome and nuclear DNA phylogenies can occur due to uniparental inheritance of organelles (141), horizontal gene transfer (142), or incomplete lineage sorting (143). Previously, an O. manul mitogenome assembly placed Otocolobus as a sister taxa to the Felis clade (15). The mitogenome assembly presented here contains a previously unidentified 400 bp gap in the D-loop control region; a new phylogenetic analysis placed O. manul closest to Prionailurus in agreement with the nuclear genome phylogeny (14). This finding suggests that O. manul nuclear DNA and mtDNA have similar phylogenetic histories. Recently, felid mitogenomes were used to investigate the contribution of a wild cat to domestication of F. catus (144). Using our mitogenome assembly, a similar study could investigate introgression among wild Asian cats (i.e. O. manul and Prionailurus spp.) and between wild and domestic cats in this region. Hybridization among cat species could have important implications for genetic diversity and conservation. Notably, evidence of hybridization among domestic and wild cats is found in South Africa and Europe, and is related to human population density in proximate spaces to wild cat habitat (145,146).
Long-read sequencing in general has been critical for increased genome assembly quality, but the portability and cost-effectiveness of nanopore sequencing make it particularly valuable for reducing barriers to entry in genomics (82,104). Still, assembly of mammalian-sized genomes (3+ Gb) can exceed computational and financial resources available to small laboratories. Even Flye, an efficient assembler (31), required distributed computing resources when using our full read set (Table 2; Supplemental File 3). Unequal access to such resources can inadvertently perpetuate ‘parachute science’ and limit the participation of researchers from historically colonized countries in endemic species conservation (147–149). However, we were able to build O. manul assemblies locally using a subset of the longest reads with only a small reduction in BUSCO score (Supplemental File 3). On-demand cloud computing resources such as Amazon Web Services (AWS) are also increasingly affordable, eliminating the necessity of access to a university cluster. We hope that the provided data and computational pipeline will be useful resources for O. manul conservation as well as for other small groups assembling mammal genomes.
DATA AVAILABILITY
DNA sequencing reads are available in FASTQ format in the Sequencing Read Archive repository (https://www.ncbi.nlm.nih.gov/sra) under submission SRR22085263. The scaffolded primary assembly (including mitogenome) is available as BioProject PRJNA885133; the contig-level alternate assembly is available as BioProject PRJNA889808. Modified base calls (DNA methylation) are also available under BioProject PRJNA885133. Sample data are available as BioSample SAMN31076064. Gene annotation, variant calling, and DNA methylation data are available by request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Utica Zoo and Tater for providing the whole blood sample used in this study. For Figure 6B, leopard cat photo credit goes to Bernard Dupont and tiger photo credit to Charles J. Sharp, both under Creative Commons licenses.
Contributor Information
Nicole Flack, Department of Veterinary and Biomedical Sciences, University of Minnesota, Saint Paul, MN 55108, USA.
Melissa Drown, Department of Ecology, Evolution, and Behavior, University of Minnesota, Saint Paul, MN 55108, USA.
Carrie Walls, Department of Animal Science, University of Minnesota, Saint Paul, MN 55108, USA.
Jay Pratte, Bloomington Parks and Recreation, Miller Park Zoo, Bloomington, IL 61701, USA.
Adam McLain, Department of Biology and Chemistry, SUNY Polytechnic Institute, Utica, NY 13502, USA.
Christopher Faulk, Department of Animal Science, University of Minnesota, Saint Paul, MN 55108, USA.
Supplementary Data
Supplementary Data are available at NARGAB Online.
FUNDING
United States Department of Agriculture National Institute of Food and Agriculture [HATCH AES MIN-16–12 to C.F.]; Norn Group [Longevity Impetus Grant to C.F.]; National Institutes of Health [R21 AG071908 to C.F., L70 AG079467-01 to N.F., T32 OD010993 to N.F.]; National Science Foundation [IOS 1556396 and IOS 1754437 to M.K.D.]
Conflict of interest statement. None declared.
REFERENCES
- 1. Ross S., Barashkova A., Dhendup T., Munkhtsog B., Smelansky I., Barclay D., moqanaki E. Otocolobus Manul. 2019; 10 February 2023, date last accessedhttp://www.10.2305/iucn.uk.2020-2.rlts.t15640a180145377.en. [Google Scholar]
- 2. Gittleman J.L. Heptner, V.G. and Sludskii, A.A. 1992. Mammals of the soviet union. volume II, part 2. Carnivora (hyaenas and cats). Smithsonian Institution Libraries and National Science Foundation. J. Mammal. 1993; 74:510–511. [Google Scholar]
- 3. Murdoch J., Tserendorj M., Reading R. Pallas’ cat ecology and conservation in the semi-desert steppes of mongolia. CAT News. 2006; 45:18–19. [Google Scholar]
- 4. BBC The grumpiest cat in the world. 2022; 10 February 2023, date last accessedhttps://www.bbc.co.uk/programmes/p0cz5p0y.
- 5. Ross S., Munkhtsog B., Harris S. Dietary composition, plasticity, and prey selection of Pallas's cats. J. Mammal. 2010; 91:811–817. [Google Scholar]
- 6. Pallas P.S. Reise durch verschiedene provinzen des russischen reichs. Vol. Reise aus sibirien zurück an die wolga im 1773ten jahr. 1776; St. Petersburg, Russian Empire: Kayserliche Academie der Wissenschafen. [Google Scholar]
- 7. Brandt J. Observations sur le manoul (felis manul pallas). Bull. Sc. Ac. Imp. Sc. St. Petersb. 1842; 9:37–39. [Google Scholar]
- 8. Thomas O., Wroughton R.C. 3. The rudd exploration of South Africa.—VII. List of Mammals obtained by Mr. Grant at Coguno, Inhambane. Proc. Zoo Soc. Lond. 1907; 77:285–306. [Google Scholar]
- 9. Ross S., Barashkova A., Farhadinia M., Appel A., Riordan P., Sanderson J., munkhtsog B. Otocolobus Manul. 2014; 1 February 2023, date last accessedhttp://www10.2305/iucn.uk.2016-1.rlts.t15640a87840229.en. [Google Scholar]
- 10. Spong G., Johansson M., Björklund M. High genetic variation in leopards indicates large and long-term stable effective population size. Mol. Ecol. 2000; 9:1773–1782. [DOI] [PubMed] [Google Scholar]
- 11. Palstra F.P., Ruzzante D.E. Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence?. Mol. Ecol. 2008; 17:3428–3447. [DOI] [PubMed] [Google Scholar]
- 12. Ross S., Munkhtsog B., Harris S. Dietary composition, plasticity, and prey selection of Pallas's cats. J. Mammal. 2010; 91:811–817. [Google Scholar]
- 13. Barclay D., Smelansky I., Nygren E., Antonevich A. Legal Status, Utilisation, Management and Conservation of Manul. 2019; 37–40. [Google Scholar]
- 14. Johnson W.E., Eizirik E., Pecon-Slattery J., Murphy W.J., Antunes A., Teeling E., O’Brien S.J The late miocene radiation of modern felidae: a genetic assessment. Science. 2006; 311:73–77. [DOI] [PubMed] [Google Scholar]
- 15. Li G., Davis B.W., Eizirik E., Murphy W.J. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 2015; 26:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sakamoto M., Ruta M. Convergence and divergence in the evolution of cat skulls: temporal and spatial patterns of morphological diversity. PLoS One. 2012; 7:e39752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohamed M., Dang N.T.-M., Ogyama Y., Burlet N., Mugat B., Boulesteix M., Mérel V., Salces-Ortiz J., Severac D. et al. A transposon story: from TE content to TE dynamic invasion of drosophila genomes using the single-molecule sequencing technology from Oxford nanopore. Cells. 2020; 9:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moss E.L., Maghini D.G., Bhatt A.S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 2020; 38:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ewing A.D., Smits N., Sanchez-Luque F.J., Faivre J., Brennan P.M., Richardson S.R., Cheetham S.W., Faulkner G.J. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol. Cell. 2020; 80:915–928. [DOI] [PubMed] [Google Scholar]
- 20. Razin A., Cedar H. DNA methylation and gene expression. Microbiol. Rev. 1991; 55:451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993; 3:226–231. [DOI] [PubMed] [Google Scholar]
- 22. Greenberg M.V.C., Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019; 20:590–607. [DOI] [PubMed] [Google Scholar]
- 23. Zemach A., McDaniel I.E., Silva P., Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010; 328:916–919. [DOI] [PubMed] [Google Scholar]
- 24. Doskočil J., Šorm F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim. Biophys. Acta. 1962; 55:953–959. [DOI] [PubMed] [Google Scholar]
- 25. Riggs A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Genome Res. 1975; 14:9–25. [DOI] [PubMed] [Google Scholar]
- 26. Cooper D.N., Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 1989; 83:181–188. [DOI] [PubMed] [Google Scholar]
- 27. Shafin K., Pesout T., Chang P.-C., Nattestad M., Kolesnikov A., Goel S., Baid G., Kolmogorov M., Eizenga J.M., Miga K.H. et al. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat. Methods. 2021; 18:1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Battaglia S., Dong K., Wu J., Chen Z., Najm F.J., Zhang Y., Moore M.M., Hecht V., Shoresh N., Bernstein B.E. Long-range phasing of dynamic, tissue-specific and allele-specific regulatory elements. Nat. Genet. 2022; 54:1504–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akbari V., Garant J.-M., O’Neill K., Pandoh P., Moore R., Marra M.A., Hirst M., Jones S.J Genome-wide detection of imprinted differentially methylated regions using nanopore sequencing. Elife. 2022; 11:e77898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rhie A., McCarthy S.A., Fedrigo O., Damas J., Formenti G. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021; 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019; 37:540–546. [DOI] [PubMed] [Google Scholar]
- 32. Shafin K., Pesout T., Lorig-Roach R., Haukness M., Olsen H.E., Bosworth C., Armstrong J., Tigyi K., Maurer N., Koren S. et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 2020; 38:1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaser R., Šikić M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 2021; 1:332–336. [DOI] [PubMed] [Google Scholar]
- 34. Vaser R., Sović I., Nagarajan N., Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017; 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu J., Fan J., Sun Z., Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2019; 36:2253–2255. [DOI] [PubMed] [Google Scholar]
- 36. Manni M., Berkeley M.R., Seppey M., Zdobnov E.M. BUSCO: assessing genomic data quality and beyond. Curr. Protoc. 2021; 1:e323. [DOI] [PubMed] [Google Scholar]
- 37. Manni M., Berkeley M.R., Seppey M., Simão F.A., Zdobnov E.M. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021; 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Solares E.A., Chakraborty M., Miller D.E., Kalsow S., Hall K., Perera A.G., Emerson J.J., Hawley R.S. Rapid low-cost assembly of the Drosophila melanogaster reference genome using low-coverage, long-read sequencing. G3 (Bethesda). 2018; 8:3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Solares E.A., Chakraborty M., Miller D.E., Kalsow S., Hall K., Perera A.G., Emerson J.J., Hawley R.S. Rapid low-cost assembly of the Drosophila melanogaster reference genome using low-coverage, long-read sequencing. G3 (Bethesda). 2018; 8:3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roach M.J., Schmidt S.A., Borneman A.R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinf. 2018; 19:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019; 20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Breitwieser F.P., Salzberg S.L. Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics. 2019; 36:1303–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000; 7:203–214. [DOI] [PubMed] [Google Scholar]
- 44. Challis R., Richards E., Rajan J., Cochrane G., Blaxter M. BlobToolKit interactive quality assessment of genome assemblies. G3 (Bethesda). 2020; 10:1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen Y., Zhang Y., Wang A.Y., Gao M., Chong Z. Accurate long-read de novo assembly evaluation with Inspector. Genome Biol. 2021; 22:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rhie A., Walenz B.P., Koren S., Phillippy A.M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020; 21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Storer J., Hubley R., Rosen J., Wheeler T.J., Smit A.F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mobile DNA. 2021; 12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Keilwagen J., Wenk M., Erickson J.L., Schattat M.H., Grau J., Hartung F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016; 44:e89–e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Keilwagen J., Hartung F., Paulini M., Twardziok S.O., Grau J. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinf. 2018; 19:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shumate A., Salzberg S.L. Liftoff: accurate mapping of gene annotations. Bioinformatics. 2021; 37:1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin M., Ebert P., Marschall T.. Peters B.A., Drmanac R. Read-based phasing and analysis of phased variants with WhatsHap. Haplotyping. Methods in Molecular Biology. 2023; 2590:NY: Humana. [DOI] [PubMed] [Google Scholar]
- 52. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V. Twelve years of SAMtools and BCFtools. GigaScience. 2021; 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Terhorst J., Kamm J.A., Song Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2016; 49:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu H., Wang C., Wu Z. A new shrinkage estimator for dispersion improves differential expression detection in RNA-seq data. Biostatistics. 2012; 14:232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Feng H., Conneely K.N., Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014; 42:e69–e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu H., Xu T., Feng H., Chen L., Li B., Yao B., Qin Z., Jin P., Conneely K.N. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 2015; 43:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Park Y., Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 2016; 32:1446–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Quinlan A.R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinform. 2014; 47:11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheetham S.W., Kindlova M., Ewing A.D. Methylartist: tools for visualizing modified bases from nanopore sequence data. Bioinformatics. 2022; 38:3109–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin J.-H., Chen L.-C., Yu S.-C., Huang Y.-T. LongPhase: an ultra-fast chromosome-scale phasing algorithm for small and large variants. Bioinformatics. 2022; 38:1816–1822. [DOI] [PubMed] [Google Scholar]
- 63. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lopez J.V., Cevario S., O’Brien S.J Complete nucleotide sequences of the domestic cat (Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics. 1996; 33:229–246. [DOI] [PubMed] [Google Scholar]
- 65. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wanner N., Larsen P.A., McLain A., Faulk C. The mitochondrial genome and Epigenome of the Golden lion Tamarin from fecal DNA using Nanopore adaptive sequencing. BMC Genom. 2021; 22:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Alonge M., Lebeigle L., Kirsche M., Aganezov S., Wang X., Lippman Z.B., Schatz M.C., Soyk S. Automated assembly scaffolding elevates a new tomato system for high-throughput genome editing. Genome Biol. 2021; 23:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018; 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pontius J.U., Mullikin J.C., Smith D.R., Lindblad-Toh K., Gnerre S., Clamp M., Chang J., Stephens R., Neelam B., Volfovsky N. et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007; 17:1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Buckley R.M., Davis B.W., Brashear W.A., Farias F.H.G., Kuroki K., Graves T., Hillier L.W., Kremitzki M., Li G., Middleton R.P. et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. PLoS Genet. 2020; 16:e1008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Brashear W.A., Bredemeyer K.R., Murphy W.J. Genomic architecture constrained placental mammal X Chromosome evolution. Genome Res. 2021; 31:1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Burger P.A., Steinborn R., Walzer C., Petit T., Mueller M., Schwarzenberger F. Analysis of the mitochondrial genome of cheetahs (Acinonyx jubatus) with neurodegenerative disease. Gene. 2004; 338:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Prost S., Machado A.P., Zumbroich J., Preier L., Mahtani-Williams S., Meissner R., Guschanski K., Brealey J.C., Fernandes C.R., Vercammen P. et al. Genomic analyses show extremely perilous conservation status of African and Asiatic cheetahs (Acinonyx jubatus). Mol. Ecol. 2022; 31:4208–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bredemeyer K.R., Seabury C.M., Stickney M.J., McCarrey J.R., vonHoldt B.M., Murphy W.J. Rapid macrosatellite evolution promotes X-linked hybrid male sterility in a feline interspecies cross. Mol. Biol. Evol. 2021; 38:5588–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rhie A., McCarthy S.A., Fedrigo O., Damas J., Formenti G., Koren S., Uliano-Silva M., Chow W., Fungtammasan A., Kim J. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021; 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Abascal F., Corvelo A., Cruz F., Villanueva-Cañas J.L., Vlasova A. Extreme genomic erosion after recurrent demographic bottlenecks in the highly endangered Iberian lynx. Genome Biol. 2016; 17:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lei W., XiaoBing W., Zhu L., Jiang Z. Mitogenomic analysis of the genus Panthera. Sci. China Life Sci. 2011; 54:917–930. [DOI] [PubMed] [Google Scholar]
- 80. Bredemeyer K.R., Harris A.J., Li G., Zhao L., Foley N.M., Roelke-Parker M., O’Brien S.J., Lyons L.A., Warren W.C., Murphy W.J. Ultracontinuous single haplotype genome assemblies for the domestic cat (Felis catus) and asian leopard cat (Prionailurus bengalensis). J. Hered. 2020; 112:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tamazian G., Dobrynin P., Zhuk A., Zhernakova D.V., Perelman P.L., Serdyukova N.A., Graphodatsky A.S., Komissarov A., Kliver S., Cherkasov N. et al. Draft de novo genome assembly of the elusive jaguarundi, Puma yagouaroundi. J. Hered. 2021; 112:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Faulk C. De novo sequencing, diploid assembly, and annotation of the black carpenter ant, Camponotus pennsylvanicus, and its symbionts by one person for $1000, using nanopore sequencing. Nucleic Acids Res. 2023; 51:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Armstrong E.E., Taylor R.W., Miller D.E., Kaelin C.B., Barsh G.S., Hadly E.A., Petrov D Long live the king: chromosome-level assembly of the lion (Panthera leo) using linked-read, Hi-C, and long-read data. BMC Biol. 2020; 18:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schrader L., Schmitz J. The impact of transposable elements in adaptive evolution. Mol. Ecol. 2018; 28:1537–1549. [DOI] [PubMed] [Google Scholar]
- 85. Böhne A., Brunet F., Galiana-Arnoux D., Schultheis C., Volff J.-N. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008; 16:203–215. [DOI] [PubMed] [Google Scholar]
- 86. Platt R.N., Vandewege M.W., Ray D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018; 26:25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Meredith R.W., Janecka J.E., Gatesy J., Ryder O.A., Fisher C.A., Teeling E.C., Goodbla A., Eizirik E., Simao T.L.L., Stadler T. et al. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011; 334:521–524. [DOI] [PubMed] [Google Scholar]
- 88. Ehrlich M., Gama-Sosa M.A., Huang L.-H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982; 10:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bastepe M., Fröhlich L.F., Linglart A., Abu-Zahra H.S., Tojo K., Ward L.M., Jüppner H. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat. Genet. 2004; 37:25–27. [DOI] [PubMed] [Google Scholar]
- 90. Zaitoun I., Khatib H. Assessment of genomic imprinting of SLC38A4, NNAT, NAP1L5, and H19 in cattle. BMC Genet. 2006; 7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brabazon D.C., Callanan J.J., Nolan C.M. Imprinting of canine IGF2 and H19. Anim. Genet. 2021; 53:108–118. [DOI] [PubMed] [Google Scholar]
- 92. Eßinger C., Karch S., Moog U., Fekete G., Lengyel A., Pinti E., Eggermann T., Begemann M. Frequency of KCNQ1 variants causing loss of methylation of Imprinting Centre 2 in Beckwith-Wiedemann syndrome. Clin Epigenet. 2020; 12:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li X., Song N., Wang D., Han X., Lv Q., Ouyang H., Li Z. Isoform-specific imprinting of the MEST gene in porcine parthenogenetic fetuses. Gene. 2015; 558:287–290. [DOI] [PubMed] [Google Scholar]
- 94. Kobayashi H., Yamada K., Morita S., Hiura H., Fukuda A., Kagami M., Ogata T., Hata K., Sotomaru Y., Kono T. Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics. 2009; 93:461–472. [DOI] [PubMed] [Google Scholar]
- 95. Kobayashi H., Yanagisawa E., Sakashita A., Sugawara N., Kumakura S., Ogawa H., Akutsu H., Hata K., Nakabayashi K., Kono T. Epigenetic and transcriptional features of the novel human imprinted lncRNAGPR1ASsuggest it is a functional ortholog to mouseZdbf2linc. Epigenetics. 2013; 8:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Iwata K., Kawarabayashi K., Yoshizaki K., Tian T., Saito K., Sugimoto A., Kurogoushi R., Yamada A., Yamamoto A., Kudo Y. et al. von Willebrand factor D and EGF domains regulate ameloblast differentiation and enamel formation. J. Cell. Physiol. 2021; 237:1964–1979. [DOI] [PubMed] [Google Scholar]
- 97. Kobayashi H., Yanagisawa E., Sakashita A., Sugawara N., Kumakura S., Ogawa H., Akutsu H., Hata K., Nakabayashi K., Kono T. Epigenetic and transcriptional features of the novel human imprinted lncRNAGPR1ASsuggest it is a functional ortholog to mouseZdbf2linc. Epigenetics. 2013; 8:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu Y., Liu J., Jiang E., Xu Y., Ning F., Du Z., Bai X. The complete mitochondrial genome of Pallas's cat (Otocolobus manul). Mitochondrial DNA B. 2019; 4:658–659. [Google Scholar]
- 99. Paez S., Kraus R.H.S., Shapiro B., Thomas M., Gilbert P., Jarvis E.D.Vertebrate Genomes Project Conservation Group Vertebrate Genomes Project Conservation Group Al-Ajli F O, Ceballos G, Crawford A J, Fedrigo O Reference genomes for conservation. Science. 2022; 377:364–366. [DOI] [PubMed] [Google Scholar]
- 100. Morin P.A., Archer F.I., Avila C.D., Balacco J.R., Bukhman Y.V., Chow W., Fedrigo O., Formenti G., Fronczek J.A., Fungtammasan A. et al. Reference genome and demographic history of the most endangered marine mammal, the vaquita. Mol. Ecol. Resour. 2020; 21:1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Murigneux V., Rai S.K., Furtado A., Bruxner T.J.C., Tian W., Harliwong I., Wei H., Yang B., Ye Q., Anderson E. et al. Comparison of long-read methods for sequencing and assembly of a plant genome. Gigascience. 2020; 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wick R.R., Holt K.E. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res. 2019; 8:2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen Z., Erickson D.L., Meng J. Benchmarking long-read assemblers for genomic analyses of bacterial pathogens using Oxford Nanopore Sequencing. Int. J. Mol. Sci. 2020; 21:9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hotaling S., Kelley J.L., Frandsen P.B. Toward a genome sequence for every animal: where are we now?. Proc. Nat. Acad. Sci. U.S.A. 2021; 118:e2109019118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bhat A., Ghatage T., Bhan S., Lahane G.P., Dhar A., Kumar R., Pandita R.K., Bhat K.M., Ramos K.S., Pandita T.K. Role of transposable elements in genome stability: implications for health and disease. Int. J. Mol. Sci. 2022; 23:7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., Heidmann T. Paleovirology of ‘ syncytins ’, retroviral env genes exapted for a role in placentation. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013; 368:20120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Stoye J.P. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012; 10:395–406. [DOI] [PubMed] [Google Scholar]
- 108. Chiu E.S., VandeWoude S. Presence of endogenous viral elements negatively correlates with feline leukemia virus susceptibility in Puma and domestic cat cells. J. Virol. 2020; 94:e01274-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Walters-Conte K.B., Johnson D.L.E., Allard M.W., Pecon-Slattery J. Carnivore-specific SINEs (Can-SINEs): distribution, evolution, and genomic impact. J. Hered. 2011; 102:S2–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wiedmer M., Oevermann A., Borer-Germann S.E., Gorgas D., Shelton G.D., Drögemüller M., Jagannathan V., Henke D., Leeb T. A RAB3GAP1 SINE Insertion in Alaskan Huskies with Polyneuropathy, Ocular Abnormalities, and Neuronal Vacuolation (POANV) Resembling Human Warburg Micro Syndrome 1 (WARBM1). G3 (Bethesda). 2016; 6:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Goldstein O., Kukekova A.V., Aguirre G.D., Acland G.M. Exonic SINE insertion in STK38L causes canine early retinal degeneration (erd). Genomics. 2010; 96:362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pelé M., Tiret L., Kessler J.-L., Blot S., Panthier J.-J. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum. Mol. Genet. 2005; 14:1417–1427. [DOI] [PubMed] [Google Scholar]
- 113. Wang W., Kirkness E.F. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005; 15:1798–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Clark L.A., Wahl J.M., Rees C.A., Murphy K.E. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Murphy S.C., Evans J.M., Tsai K.L., Clark L.A. Length variations within the Merle retrotransposon of canine PMEL: correlating genotype with phenotype. Mobile DNA. 2018; 9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Okada M., Yoshikawa R., Shojima T., Baba K., Miyazawa T. Susceptibility and production of a feline endogenous retrovirus (RD-114 virus) in various feline cell lines. Virus Res. 2011; 155:268–273. [DOI] [PubMed] [Google Scholar]
- 117. Okabe H., Gilden R.V., Hatanaka M. RD 114 virus-specific sequences in feline cellular RNA: detection and characterization. J. Virol. 1973; 12:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yoshikawa R., Shimode S., Sakaguchi S., Miyazawa T. Contamination of live attenuated vaccines with an infectious feline endogenous retrovirus (RD-114 virus). Arch. Virol. 2013; 159:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yasir M., Turner A.K., Lott M., Rudder S., Baker D., Bastkowski S., Page A.J., Webber M.A., Charles I.G. Long-read sequencing for identification of insertion sites in large transposon mutant libraries. Sci. Rep. 2022; 12:3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020; 21:597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020; 585:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Raj K., Szladovits B., Haghani A., Zoller J.A., Li C.Z., Black P., Maddox D., Robeck T.R., Horvath S. Epigenetic clock and methylation studies in cats. GeroScience. 2021; 43:2363–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ekström T.J. Parental Imprinting and the IGF2 gene. Horm. Res. 1994; 42:176–181. [DOI] [PubMed] [Google Scholar]
- 125. Tucci V., Isles A.R., Kelsey G., Ferguson-Smith A.C., Tucci V., Bartolomei M.S., Benvenisty N., Bourc’his D., Charalambous M., Dulac C. et al. Genomic imprinting and physiological processes in mammals. Cell. 2019; 176:952–965. [DOI] [PubMed] [Google Scholar]
- 126. Bartolomei M.S., Zemel S., Tilghman S.M. Parental imprinting of the mouse H19 gene. Nature. 1991; 351:153–155. [DOI] [PubMed] [Google Scholar]
- 127. Jima D.D., Skaar D.A., Planchart A., Motsinger-Reif A., Cevik S.E., Park S.S., Cowley M., Wright F., House J., Liu A. et al. Genomic map of candidate human imprint control regions: the imprintome. Epigenetics. 2022; 17:1920–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kanduri C., Fitzpatrick G., Mukhopadhyay R., Kanduri M., Lobanenkov V., Higgins M., Ohlsson R. A differentially methylated imprinting control region within the Kcnq1 locus harbors a methylation-sensitive chromatin insulator. J. Biol. Chem. 2002; 277:18106–18110. [DOI] [PubMed] [Google Scholar]
- 129. O’Sullivan F.M., Murphy S.K., Simel L.R., McCann A., Callanan J.J., Nolan C.M. Imprinted expression of the canine IGF2R, in the absence of an anti-sense transcript or promoter methylation. Evol. Dev. 2007; 9:579–589. [DOI] [PubMed] [Google Scholar]
- 130. Nolan C., O’Sullivan F., Brabazon D., Callanan J. Genomic Imprinting inCanis familiaris. Reprod Domestic Anim. 2009; 44:16–21. [DOI] [PubMed] [Google Scholar]
- 131. Killian J.K. Divergent evolution in M6P/IGF2R imprinting from the jurassic to the quaternary. Hum. Mol. Genet. 2001; 10:1721–1728. [DOI] [PubMed] [Google Scholar]
- 132. Leigh N.D., Sessa S., Dragalzew A.C., Payzin-Dogru D., Sousa J.F., Aggouras A.N., Johnson K., Dunlap G.S., Haas B.J., Levin M. et al. von Willebrand factor D and EGF domains is an evolutionarily conserved and required feature of blastemas capable of multitissue appendage regeneration. Evol. Dev. 2020; 22:297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012; 335:823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tucci V., Isles A.R., Kelsey G., Ferguson-Smith A.C., Tucci V., Bartolomei M.S., Benvenisty N., Bourc’his D., Charalambous M., Dulac C. et al. Genomic imprinting and physiological processes in mammals. Cell. 2019; 176:952–965. [DOI] [PubMed] [Google Scholar]
- 135. Henry P., Miquelle D., Sugimoto T., McCullough D.R., Caccone A., Russello M.A. In situpopulation structure andex siturepresentation of the endangered Amur tiger. Mol. Ecol. 2009; 18:3173–3184. [DOI] [PubMed] [Google Scholar]
- 136. Cho Y., Hu L., Hou H., Lee H., Xu J. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat. Commun. 2013; 4:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Liao W., Reed D.H. Inbreeding-environment interactions increase extinction risk. Animal Conserv. 2009; 12:54–61. [Google Scholar]
- 138. Ning Y., Kostyria A.V., Ma J., Chayka M.I., Guskov V.Y., Qi J., Sheremetyeva I.N., Wang M., Jiang G. Dispersal of Amur tiger from spatial distribution and genetics within the eastern Changbai mountain of China. Ecol. Evol. 2019; 9:2415–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Howell L.G., Frankham R., Rodger J.C., Witt R.R., Clulow S., Upton R.M.O., Clulow J. Integrating biobanking minimises inbreeding and produces significant cost benefits for a threatened frog captive breeding programme. Conserv. Lett. 2021; 14:e12776. [DOI] [PubMed] [Google Scholar]
- 140. Bernt M., Braband A., Schierwater B., Stadler P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 2013; 69:328–338. [DOI] [PubMed] [Google Scholar]
- 141. Birky C.W. Uniparental inheritance of organelle genes. Curr. Biol. 2008; 18:R692–R695. [DOI] [PubMed] [Google Scholar]
- 142. Goremykin V.V., Salamini F., Velasco R., Viola R. Mitochondrial DNA of vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2008; 26:99–110. [DOI] [PubMed] [Google Scholar]
- 143. Folk R.A., Mandel J.R., Freudenstein J.V. Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Syst. Biol. 2016; 66:320–337. [DOI] [PubMed] [Google Scholar]
- 144. Yu H., Xing Y.-T., Meng H., He B., Li W.-J., Qi X.-Z., Zhao J.-Y., Zhuang Y., Xu X., Luo S. Genomic evidence for the Chinese mountain cat as a wildcat conspecific (Felis silvestris bieti) and its introgression to domestic cats. Sci. Adv. 2021; 7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Le Roux J.J., Foxcroft L.C., Herbst M., MacFadyen S. Genetic analysis shows low levels of hybridization between A frican wildcats (Felis silvestris lybica) and domestic cats (F. s. catus) in S outh A frica. Ecol. Evol. 2014; 5:288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Pierpaoli M., Birò Z.S., Herrmann M., Hupe K., Fernandes M., Ragni B., Szemethy L., Randi E. Genetic distinction of wildcat (Felis silvestris) populations in Europe, and hybridization with domestic cats in Hungary. Mol. Ecol. 2003; 12:2585–2598. [DOI] [PubMed] [Google Scholar]
- 147. Asase A., Mzumara-Gawa T.I., Owino J.O., Peterson A.T., Saupe E. Replacing “parachute science” with “global science” in ecology and conservation biology. Conserv Sci Pract. 2022; 4:e517. [Google Scholar]
- 148. Stefanoudis P.V., Licuanan W.Y., Morrison T.H., Talma S., Veitayaki J., Woodall L.C. Turning the tide of parachute science. Curr. Biol. 2021; 31:R184–R185. [DOI] [PubMed] [Google Scholar]
- 149. Li F.-W. Decolonizing botanical genomics. Nat. Plants. 2021; 7:1542–1543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequencing reads are available in FASTQ format in the Sequencing Read Archive repository (https://www.ncbi.nlm.nih.gov/sra) under submission SRR22085263. The scaffolded primary assembly (including mitogenome) is available as BioProject PRJNA885133; the contig-level alternate assembly is available as BioProject PRJNA889808. Modified base calls (DNA methylation) are also available under BioProject PRJNA885133. Sample data are available as BioSample SAMN31076064. Gene annotation, variant calling, and DNA methylation data are available by request.