Abstract
Cancer progresses by evading the immune system. Elucidating diverse immune evasion strategies is a critical step in the search for next-generation immunotherapies for cancer. Here we report that cancer cells can hijack the mitochondria from immune cells via physical nanotubes. Mitochondria are essential for metabolism and activation of immune cells. By using field-emission scanning electron microscopy, fluorophore-tagged mitochondrial transfer tracing and metabolic quantification, we demonstrate that the nanotube-mediated transfer of mitochondria from immune cells to cancer cells metabolically empowers the cancer cells and depletes the immune cells. Inhibiting the nanotube assembly machinery significantly reduced mitochondrial transfer and prevented the depletion of immune cells. Combining a farnesyltransferase and geranylgeranyltransferase 1 inhibitor, namely, L-778123, which partially inhibited nanotube formation and mitochondrial transfer, with a programmed cell death protein 1 immune checkpoint inhibitor improved the antitumour outcomes in an aggressive immunocompetent breast cancer model. Nanotube-mediated mitochondrial hijacking can emerge as a novel target for developing next-generation immunotherapy agents for cancer.
The current understanding of tumour immune evasion primarily relies on the capacity of cancer cells to express ligands that engage immune checkpoints1-3. For example, cancer cells express programmed cell death ligand 1 and ‘eat-me-not’ CD47, which bind to immune checkpoints, such as programmed cell death protein 1 (PD1) on T cells and SIRPα on macrophages, respectively, leading to immune suppression4-6. Blocking these checkpoints can exert durable clinical benefits, but only a subset of cancer patients respond to such immunotherapies5. One possible explanation for the restricted effectiveness of existing immune checkpoint inhibitors (ICIs) is that the cancer cells deploy several immune evasion strategies. Elucidating these alternative immune evasion strategies, as well as inhibiting them, is critical towards improving antitumour outcomes7.
Nanotube-mediated mitochondrial trafficking
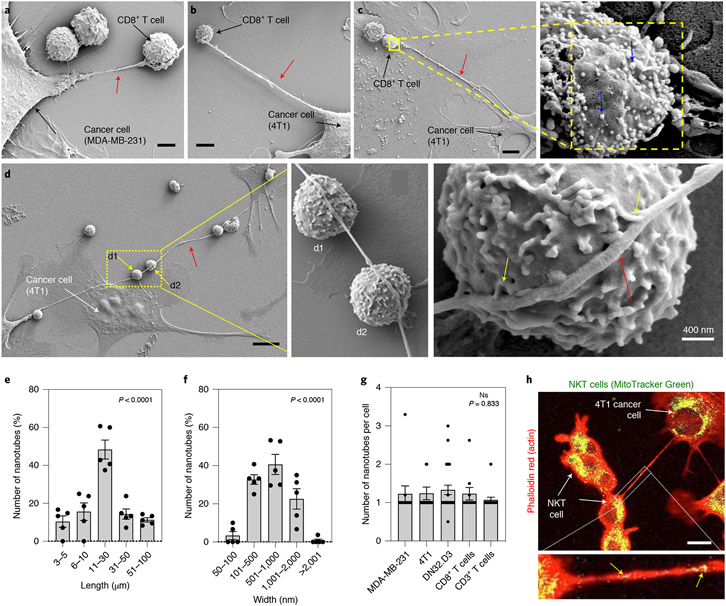
We rationalized that investigating the communication between cancer cells and immune cells at the nanoscale could shed newer insights into mechanisms underlying immune evasion. We set up a simple experiment coculturing different murine and human breast cancer cells with effector immune cells, such as natural killer T (NKT) or CD3+/CD8+ T cells, to create both syngeneic (self) and xenogeneic (non-self) conditions, and examined the intercellular interactions in the coculture using field-emission scanning electron microscopy (FESEM). Interestingly, FESEM image analysis revealed that cancer cells and immune cells physically connect via nanoscale tube-like structures (Fig. 1a-d and Supplementary Figs. 1-2). In certain cases, a single nanotube arising from a cancer cell was found to connect with several immune cells in series and forming multiple contacts with the immune cell membrane (Fig. 1d). The nanotubes have lengths in a broad range of ~3–100 μm (mostly within 10–30 μm) (Fig. 1e and Supplementary Figs. 1 and 2) and widths ranging from 50 nm to 2 μm (mostly in the 100–1,000 nm range) (Fig. 1f and Supplementary Figs. 1 and 2). In some cases, nanotubes aggregated to form a thicker nanotube (Supplementary Fig. 1d). Although the average number of heterotypic nanotubes was approximately one per cell (Fig. 1g), this observation is likely an underestimation of the actual numbers as many of the fragile nanotubes are lost during the sample preparation stage of the experiment (Supplementary Fig. 2m).
Fig. 1 ∣. Cancer cells and effector immune cells connect via physical nanotubes.
a,b, FESEM images showing nanotubes (red arrow) between breast cancer cells and immune cells. CD8+/CD3+ T cells were added to either MDA-MB-231 (a) or 4T1 cells (b). In a, the nanotube appears to branch around the T cell. Scale bars, 2 μm (a) and 4 μm (b). c, Left: FESEM image showing a nanotube (red arrow) between immunogold-labelled CD8+ T cells and 4T1 cancer cells. Scale bar, 4 μm. Right: gold nanoparticles (diameter, 10 nm) are visible (blue arrows) on the surface of the T cells at higher magnification. d, Left: FESEM image showing that a single nanotube can connect a cancer cell (4T1) with multiple T cells (yellow arrows). Scale bar, 10 μm. Middle: magnified view showing the interaction between the nanotube and immune cells. Right: yellow arrows show the buds from the nanotube fusing with the immune cells. e,f, Graphs showing the distribution of lengths and widths of nanotubes connecting the cancer cells and immune cells, as calculated from the FESEM images. Data shown are mean ± s.e.m. (ANOVA followed by Tukey’s multiple comparisons test). Each data point represents a cell pair, that is, CD3+ T/4T1 (n = 7), CD8+ T/MDA-MB-231 (n = 18), TALL/MDA-MB-231 (n = 11), NKT/MDA-MB-231 (n = 28) and NKT/4T1 (n = 11). g, Graph showing the number of nanotubes between cancer cells and immune cells formed per cell (calculated using the FESEM images). Data are represented as mean ± s.e.m. (ANOVA followed by Tukey’s multiple comparisons test; NS, not significant). h, Top: representative confocal image shows a nanotube connecting the NKT and 4T1 cells. Scale bar, 10 μm. The mitochondria in NKT cells (DN32.D3) were labelled with MitoTracker Green dye before coculture with cancer cells (4T1). Rhodamine phalloidin (red) was used to label the actin filaments in all cells. The presence of the green signal (appears yellow in the merged image) in the cancer cell represents the transfer of MitoTracker-Green-tagged mitochondria from the immune cell to the cancer cell. Bottom: MitoTracker localization (yellow arrows) with actin in the nanotube.
Nanotube-like structures have been implicated in mediating intercellular communication between immune cells8, transfer of HIV virus between T cells9, transfer of prions between neurons10, conferring survival advantage to cancer cells11 and metastasis12. Nanotubes also enable intercellular organelle trafficking13-15, including the transfer of mitochondria between epithelial cells15,16. Mitochondrial function is essential for the expansion of antigen-specific CD8+ T cells, cytokine production and effective memory response17. NKT cells are exquisitely dependent on oxidative phosphorylation in the mitochondria18. Furthermore, mitochondrial gain and increased oxidative phosphorylation has been implicated in cancer progression and resistance development towards chemotherapy19,20. We, therefore, tested for nanotube-mediated transfer of mitochondria between cancer cells and immune cells. We labelled the mitochondria in the NKT cells with MitoTracker Green, which covalently binds to the free thiol groups of cysteine residues on mitochondrial proteins21. The immune cells were washed to remove any unbound dye and then added to a culture of metastatic breast cancer cells, which did not have any labelled mitochondria in the beginning. At 16 h post-coculture, we observed substantial levels of punctate green fluorescence in the cancer cells (Fig. 1h), consistent with the transfer of mitochondria from the immune cell to cancer cell. Using phalloidin red to stain F-actin in the nanotube8,12, we observed the colocalization of MitoTracker-labelled mitochondria within the nanotubes (Fig. 1h). To control the possibility of non-specific leakage of the dye into the media from immune cells resulting in labelling of the mitochondria in the cancer cells, in a separate experiment, we collected the conditioned media from a 16 h monoculture of MitoTracker-Green-labelled immune cells, and added it to the cancer cell culture. Simultaneously, the labelled immune cells were collected, washed and added to a parallel culture of cancer cells. We quantified the fraction of MitoTracker-Green-positive cancer cells after 16 h of incubation under both conditions using fluorescence-activated cell sorting (FACS; Supplementary Fig. 3). A non-specific leakage of MitoTracker from the immune cells and not the media would have meant that cancer cells in both culture conditions should exhibit notable levels of mitochondrial labelling. However, as shown in Supplementary Fig. 3, the cancer cells did not show any notable MitoTracker Green labelling when cultured with the conditioned media. In contrast, a substantial fraction of the cancer cells exhibited MitoTracker Green labelling when cocultured with the immune cells, validating the fact that labelled mitochondria signal in the cancer cell was not due to dye leakage from the immune cells. Nanotube-mediated mitochondrial transfer was also visualized when CD8+ T or NKT cells were cocultured with MDA-MB-231 breast cancer cells (Fig. 2a and Supplementary Fig. 4). When the coculture was treated with Hoechst 33342 dye to label DNA (both nuclear and mitochondrial), we observed the colocalization of MitoTracker Green and Hoechst 33342 within the nanotubes (Fig. 2a), supporting mitochondrial trafficking via the nanotube.
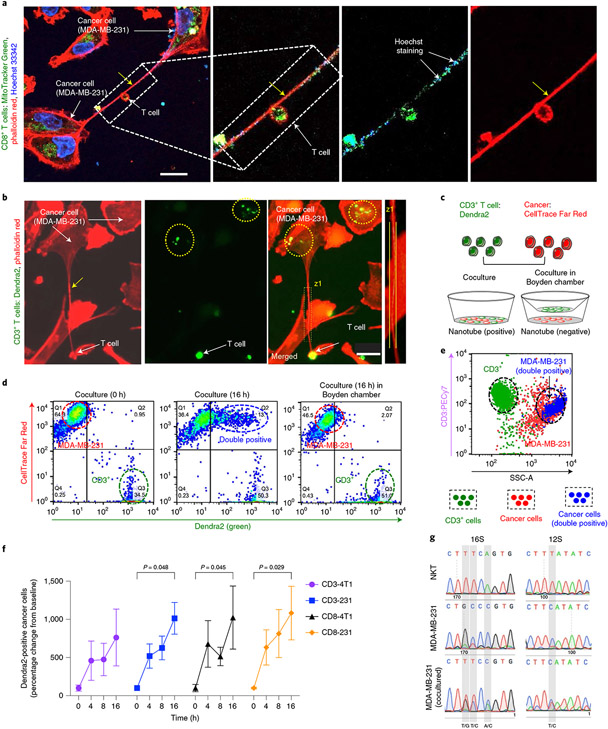
Fig. 2 ∣. Nanotubes mediate organelle transfer between immune cells and cancer cells.
a, Confocal image showing nanotube-mediated transfer of MitoTracker-Green-tagged mitochondria from CD8+ T cell to MDA-MB-231 cells. Scale bar, 20 μm. The colocalization of Hoechst 33342 (which stains mitochondrial DNA) and MitoTracker in the merged image supports nanotube-mediated mitochondrial trafficking. b, Fluorescence image showing the nanotube and transfer of Dendra2-positive mitochondria in the coculture of CD3+ T cell (from PhAMexcised mice expressing a mitochondria-specific version of Dendra2) and MDA-MB-231 cells. Scale bar, 20 μm. Actin was stained with rhodamine phalloidin. c, Experimental design to quantify mitochondrial hijacking by cancer cells from immune cells. CTFR-labelled MDA-MB-231 cells were cultured with Dendra2-positive CD3+ T cell in direct contact or separated in a Boyden chamber by a porous membrane. d, Dot plots showing the transfer of Dendra2-positive mitochondria from CD3+ T cell to MDA-MB-231 cells. At time 0 h (T0h), the MDA-MB-231 cells and CD3+ T cells are displayed as two distinct populations corresponding to their fluorescent labelling. After 16 h, a new dual red and green population (top right) appears, consistent with the transfer of mitochondria from the immune cells to cancer cells. Minimal transfer of mitochondria was observed in the coculture in the Boyden chamber. e, Representative plot of the above coculture at 16 h after staining the T cells with anti-CD3 (PE-Cy7) antibody and sorting the cells based on side scattering (SSC). The green population represents Dendra2-positive CD3+ T cells and the red population represents CTFR-stained cancer cells. The blue population indicates cells having both red and green fluorescence. The blue population superimposed with the population of the cancer cells as seen in the SSC signify the transport of mitochondria from the immune cell to the cancer cell. f, Graph showing the time-dependent transfer of mitochondria from Dendra2-positive CD3+/CD8+ T cells to cancer cells. The data are normalized to the T0h values, and presented as mean ± s.e.m. (n = 3; P = 0.048, 0.045, 0.029; two-way ANOVA with Tukey’s multiple comparisons test). g, Mitochondrial genotyping using species-specific SNPs show mitochondrial transfer from mouse DN32.D3 cells into human MDA-MB-231 cancer cells.
We further validated our observations using T cells isolated from PhAMexcised (photo-activatable mitochondria) mice, which ubiquitously express a mitochondria-localized version of the fluorescent protein Dendra2 (ref. 22). We observed the nanotube-mediated transfer of Dendra2-labelled mitochondria from these T cells to cancer cells (Fig. 2b and Supplementary Fig. 5). To quantify the transfer of mitochondria between immune cells and cancer cells, we labelled the cancer cells with CellTrace Far Red (CTFR) before establishing a coculture with Dendra2-positive (green) mitochondria-containing T cells, and then quantified the transfer of tracers between the cells using flow cytometry. As a control, we set up a coculture in a Boyden chamber, where the cancer cells and immune cells were separated by a membrane with 400 nm pores that allowed the secreted signalling molecules and exosomes to pass through but prevented direct cellular communication via physical nanotubes (Fig. 2c). As shown in Fig. 2d and Supplementary Figs. 6-8, although the cancer cells and immune cells appear as separate red and green populations at the start, a new dual red- and green-positive population of cells appears by 16 h, consistent with the transfer of Dendra2-positive mitochondria from the immune cells to CTFR-labelled cancer cells. We did not observe any notable transfer in the Boyden chamber (Fig. 2d), indicating that the transfer seen in the coculture assay was due to direct physical communication. To confirm that the new dual-positive population is indeed cancer cells, we labelled the Dendra2-positive immune cells with an anti-CD3 antibody and the cancer cells with CTFR, and then sorted the cocultured cells at different time points. Cancer cells and T cells appear as distinct pools in the side-scatter (SSC) analysis. As shown in Fig. 2e (and Supplementary Fig. 6d), the cells initially sorted into two main populations: (1) immune cells that were CD3 (high), CTFR (low), Dendra2 (high) and SSC (low); (2) cancer cells that were CTFR (high), CD3 (low) and SSC (high). A third population emerged in a temporal manner, which was Dendra2 (high) and CTFR (high), but CD3 (low) and SSC (high), consistent with cancer cells that have received Dendra2-labelled mitochondria from the immune cells (Fig. 2e,f and Supplementary Figs. 6-8). Similar observations were made using MitoTracker-Green-labelled CD3+/CD8+ T or NKT cells cocultured with 4T1 or MDA-MB-231, where SSC analysis revealed the new dual-positive population overlaid with the cancer cells (Supplementary Figs. 9 and 10). To test for any non-specific transfer of Dendra2-labelled mitochondria, we cultured the cancer cells in the conditioned media of Dendra2-labelled CD3+ T cells. Both flow cytometry and microscopy revealed negligible population of cancer cells with green fluorescence, signifying minimal non-specific transfer of mitochondria from CD3+ T cells to cancer cells (Supplementary Fig. 11). We further validated the mitochondrial transfer from murine immune cells (DN32.D3) to human cancer cells (MDA-MB-231) by mitochondrial genotyping using species-specific single-nucleotide polymorphisms (SNPs) (Fig. 2g, Supplementary Fig. 12 and Supplementary Table 1).
We next tested the directionality of mitochondrial transfer. We labelled the mitochondria of immune cells with MitoTracker Green and the cancer cells with CTFR, and established that the two cell types are indeed two distinct pools when sorted based on forward scatter (FSC) and SSC using flow cytometry (Supplementary Fig. 13a,b). When we cocultured these cells for 16 h and then sorted the cells, consistent with previous observations, a dual-positive, that is, MitoTracker Green (high) and CTFR (high), population appeared, comprising ~45% of the total population of the cells. Analysis of this dual-positive (high) subset using FSC and SSC revealed that ~90% of dual-positive cells fell in the FSC and SSC gates for cancer cell population, and ~5% fell within the gates for NKT cells. Next, we reversed the experimental condition, that is, the mitochondria in cancer cells were labelled with MitoTracker Green and the immune cells were labelled with CTFR before establishing the cocultures. At 16 h post-coculture, we observed three pools of cells in this case. The newly emerged population was dual positive, that is, MitoTracker Green (high) and CTFR (high), and it comprised only ~19% of the total population of the cells; again, ~92% of the these cells were within the FSC and SSC gates set for cancer cells, whereas ~5% were in the gates set for NKT cells. This translates to cancer cells that received labelled mitochondria from immune cells comprising ~41.00% of the total cells in the coculture, whereas NKT cells that received the labelled mitochondria from cancer cells in the reverse culture formed only 0.99% of the total cells in the coculture (Supplementary Fig. 13c). This study highlights three conclusions: (1) the trafficking of mitochondria clearly is predominantly unidirectional—from immune cells to cancer cells; (2) leakage or random diffusion of the MitoTracker dye is unlikely to substantially contribute to dual-positive labelling; if that was the case, we would see similar fractions of dual-positive cells in both culture conditions; and (3) the fact that the dual-positive cells in the reverse culture are predominantly cancer cells means that there is a transfer of CFTR-labelled cytoplasmic content from immune cells to cancer cells; however, this was less than the transfer of mitochondria, which could be potentially due to the well-known active transport of mitochondria via nanotubes14.
Mitochondrial trafficking alters the metabolic state
We next studied the impact of mitochondrial transfer on cancer cells and immune cells by metabolic profiling using the Seahorse platform (Fig. 3a)23,24. Cancer cells isolated from the coculture assay exhibited higher basal respiration and spare respiratory capacity than control cancer cells that were either monocultured or cocultured with immune cells in a Boyden chamber (Fig. 3b-d and Supplementary Fig. 14). In contrast, a significant reduction in basal respiration and spare respiratory capacity was observed in the case of cocultured immune cells (Fig. 3e-g). To test that the metabolic augmentation seen in cancer cells is indeed due to the gain of mitochondria from immune cells, we partially damaged the mitochondria of immune cells with antimycin A and rotenone (10.0 μM each) or ethidium bromide (0.5 μM) before coculture with cancer cells (Supplementary Fig. 15a). As shown in Supplementary Fig. 15b, these treatments blocked the gain of metabolic functions, in terms of basal respiration and spare respiratory capacity, in the cancer cells (Supplementary Fig. 15b-d), validating the fact that metabolic augmentation in cancer cells is due to mitochondrial hijacking from immune cells. Consistent with the rationale that the loss of mitochondria is catastrophic for immune cells and hence detectable early, we observed a significant reduction in immune cell population in the coculture assay by 16 h (Fig. 3h and Supplementary Fig. 16). We did not observe any notable change in the population of cancer cells at this early time point. Interestingly, monitoring cell proliferation over several days revealed that cancer cells isolated from the cocultures exhibited significantly higher growth compared with control cancer cells that were separately either monocultured or cocultured in a Boyden chamber (Fig. 3i and Supplementary Fig. 16).
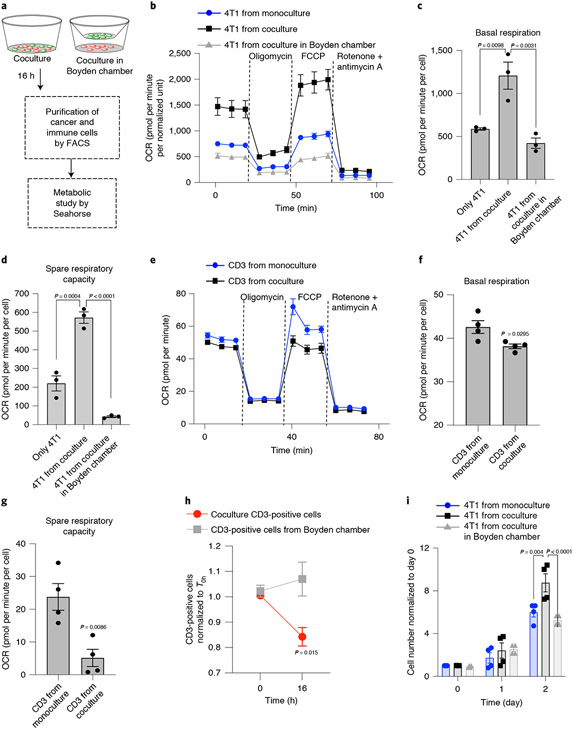
Fig. 3 ∣. Metabolic effect of mitochondrial hijacking.
a, Schematic showing the experimental design. After 16 h of coculture, the cancer cells and immune cells were separated using FACS, and the metabolic state of each cell type was measured using the Seahorse XFe24 platform. b, Oxygen consumption rate (OCR) from Seahorse XF Mito stress test of cancer cells isolated from a coculture in which they form direct contact with immune cells shows increased mitochondrial respiration. Data are normalized according to the total protein concentration in each well after the assay and represented as mean ± s.e.m. (n = 3). c,d, Graphs showing basal respiration (c) and spare respiratory capacity (d) in 4T1 cells. Data are represented as mean ± s.e.m. (n = 3; P = 0.0098 and P = 0.0031 for basal respiration and P = 0.0004 and P < 0.0001 for spare respiratory capacity; one-way ANOVA with Tukey’s multiple comparisons test). e, Seahorse XF Mito stress test profile shows decreased mitochondrial respiration in CD3+ T cells when they are cultured in the presence of cancer cells. The cells were separated by FACS after 10 h and used for the metabolism assay using Seahorse XFe96. Data are represented as mean ± s.e.m. (n = 3). f,g, Graphs showing basal respiration (f) and spare respiratory capacity (g) of cocultured and monocultured CD3+ T cells. Data are shown as mean ± s.e.m. (n = 4; P = 0.0295 for basal respiration and P = 0.0086 for spare respiratory capacity; unpaired two-tailed t-test). h, Graph showing the reduction in T cell population when cocultured with 4T1 cancer cells versus when they are cocultured in a Boyden assay. Data are normalized to the T0h population (n = 3; P = 0.004 and P < 0.0001 at 48 h; two-way ANOVA with Bonferroni’s multiple comparisons test). i, Graph showing the proliferation of cancer cells post-coculture with immune cells. The 4T1 cells were isolated using FACS after 16 h of coculture and further maintained as monocultures for an additional 48 h. Data are normalized according to the population at 0 h for each condition. Data are shown as mean ± s.e.m. (n = 4; P = 0.0032 and 0.0021 at 48 h; two-way ANOVA with Bonferroni’s multiple comparisons test).
Exocyst complex involved in mitochondrial transfer
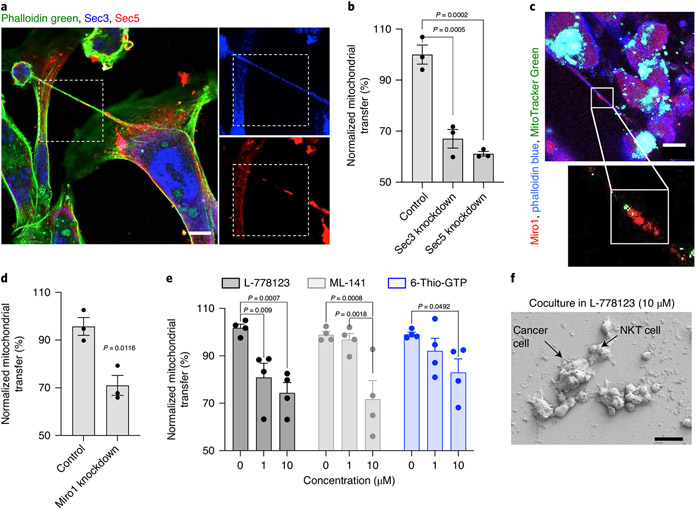
Previous studies have implicated the exocyst complex in nanotube formation25,26. The exocyst complex is an octameric protein complex made up of Sec proteins25 and interacts with the Rho and Ral GTPase family for actin remodelling during nanotube formation27,28. Indeed, immunolabelling the coculture of breast cancer cells and immune cells revealed the colocalization of Sec3 and Sec5 at the site of actin cytoskeletal recruitment in the nanotube (Fig. 4a and Supplementary Fig. 17a)29. Small interfering RNA (siRNA)-mediated knockdown of either Sec3 or Sec5 reduced mitochondria trafficking between immune cells and cancer cells (Fig. 4b and Supplementary Fig. 17b,c). Furthermore, immunolabelling revealed the colocalization of MitoTracker-Green-labelled mitochondria and mitochondrial Rho GTPase (Miro1) within the nanotubes (Fig. 4c). A significant reduction in mitochondrial transfer was observed with an siRNA-based partial knockdown of Miro1 (Fig. 4d and Supplementary Fig. 17d,e), supporting a Miro1-mediated active mitochondrial transport from immune cells to cancer cells30,31. The Ras/Rho GTPase signalling has been implicated in nanotube-mediated intercellular communication32. These GTPases are prenylated by geranylgeranyltransferase for membrane association and biological activity33. Indeed, pharmacological inhibitors, namely, ML-141 (inhibits Cdc42), 6-Thio-GTP (inhibits Rac1 GTPase) and L-778123 (inhibits farnesyltransferase and geranylgeranyltransferase type 1), significantly reduced nanotube formation and mitochondrial hijacking at non-cytotoxic concentrations (Fig. 4e,f and Supplementary Fig. 18). Taken together, these observations are consistent with previous reports that nanotubes are formed via Ras-GTPase-exocyst-enabled cytoskeletal rearrangements, and that Miro1 is implicated in the active trafficking of mitochondria via the nanotube.
Fig. 4 ∣. Mechanism underlying nanotube formation and mitochondrial transfer.
a, Representative confocal image showing the localization of exocyst proteins, namely, Sec3 (blue) and Sec5 (red), at the base and within a nanotube between MDA-MB-231 cells and NKT cells. Actin was stained with phalloidin green. Scale bar, 10 μm. b, Graph showing the induced transfer of mitochondria after siRNA-mediated partial knockdown of Sec3 and Sec5 in MDA-MB-231 cells. Data are normalized to mitochondrial transfer in controls and represented as mean ± s.e.m. (n = 3; P = 0.0005 and 0.0002 for Sec3 and Sec5, respectively; one-way ANOVA with Dunnett’s multiple comparisons test). c, Immunofluorescence image showing the colocalization of mitochondrial Rho GTPase-1 (Miro1) with MitoTracker-labelled mitochondria. NKT cells loaded with MitoTracker Green were cocultured with MDA-MB-231 cells. The coculture was fixed at 16 h, and immunolabelled with anti-Miro1 antibody (red). Phalloidin blue was used to stain actin. The image shows the colocalization of mitochondria (green) and Miro1 (red) in the nanotube. Scale bar, 10 μm. d, Graph showing the decrease in transfer of mitochondria after siRNA-targeting partial knockdown of Miro1 in MDA-MB-231 cells. The transfected cancer cells were labelled with CTFR and cocultured with MitoTracker-Green-loaded NKT cells. Data are normalized to the control and represented as mean ± s.e.m. (n = 3; P = 0.0116; unpaired two-tailed t-test). e, Graph showing the drug-concentration-dependent reduction in mitochondrial transfer from immune cells to cancer cells. The transfer of mitochondria was monitored by flow cytometry and the data are normalized according to mitochondrial transfer in the control condition. Data are shown as mean ± s.e.m. (n = 4; P = 0.009; P = 0.0007 for L-778123; P = 0.0008; P = 0.0018 for ML-141; P = 0.0492 for 6-Thio-GTP; two-way ANOVA with Tukey’s multiple comparisons test). f, Scanning electron microscopy images of cocultures of NKT and MDA-MB-231 cell culture in the presence of L-778123 (10 μM) for 16 h showing the distinct change in interactions between the cells and the absence of nanotubes. Scale bar, 20 μm.
Inhibition of mitochondrial transfer and antitumour outcome
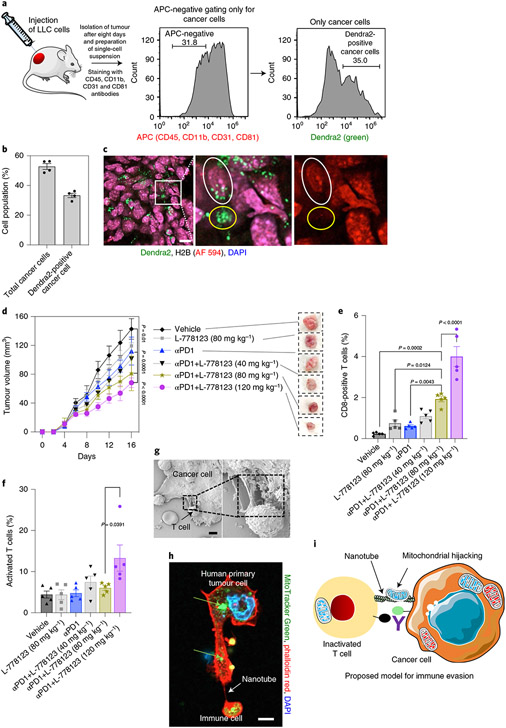
To test for mitochondrial hijacking by cancer cells in an in vivo setting, we subcutaneously injected Lewis lung carcinoma (LLC) cells into syngeneic C57BL/6 PhAMexcised mice. After eight days, we isolated the tumours from the animals, and tested for Dendra2-positive mitochondria in the cancer cells. As shown in Fig. 5a-c, both flow cytometry and confocal imaging revealed Dendra2-labelled mitochondria in the cancer cells, consistent with mitochondrial hijacking. To further validate the mitochondrial transfer from T cells to cancer cells, we mixed CTFR-labelled cancer cells (LLC or B16F10 melanoma) and Dendra2-labelled CD3+ T cells in Matrigel, and subcutaneously injected them into PhAMexcised mice. At 60 h postimplantation, 17.7% and 11.6% of the CTFR (high) LLC or B16/F10 cells, respectively, were found to be Dendra2-positive (Supplementary Fig. 19), validating the mitochondrial trafficking from T cells to cancer cells in vivo. We next tested if the efficacy of pharmacological inhibitors could be replicated in vivo. We observed lethality, in vivo, with ML-141. Therefore, we used L-778123 in further studies. Confocal imaging and flow cytometry analysis of a single-cell suspension revealed the presence of dual-positive cells when CTFR-labelled 4T1 and MitoTracker-Green-labelled CD3+ T cells were mixed in a Matrigel plug and subcutaneously injected (Supplementary Fig. 20) into BALB/c mice, consistent with mitochondrial transfer. This dual-positive cell population was reduced after treatment with L-778123 (10 μM) in vivo. In a separate experiment, we implanted 4T1 breast cancer cells into the flank of immunocompetent BALB/c mice, and treated the animals with either vehicle (control) or L-778123 (120 mg kg−1), administered by intraperitoneal injection every alternate day. A significant reduction in tumour growth was observed in the L-778123-treated group (Supplementary Fig. 21). The animals were killed after six cycles of treatment, and a single-cell suspension was prepared from the excised tumour and the mitochondria were labelled with MitoTracker Green. Gating for cancer cells using FSC and SSC via FACS and then quantifying the level of MitoTracker-labelled mitochondria in the cancer cells revealed a reduced fraction of cancer cells with high mitochondria signature in the L-778123-treated group compared with the vehicle controls (Supplementary Fig. 22). Treating 4T1 cells with L-778123 did not reveal any direct drug effect on mitochondrial mass (Supplementary Fig. 23). Taken together, these results suggest that L-778123 inhibits mitochondrial hijacking from immune cells to cancer cells.
Fig. 5 ∣. Targeting nanotube-mediated mitochondrial hijacking augments antitumour immune response in vivo.
a, Left: LLC cells were subcutaneously injected into syngeneic C57BL/6 J PhAMexcised mice. The infiltrating cells were stained with CD45, CD11b, CD31 and CD81 antibodies (all are APC labelled). The cancer cells were selected by gating the APC-negative cell population. Middle: histogram showing the selection of cancer cells that are not labelled with CD45, CD11b, CD31 and CD81 antibodies. Right: histogram showing the fraction of cancer cells containing Dendra2-positive mitochondria. b, Bar graph showing the total cancer cells, and the fraction of Dendra2-positive cancer cells. Data are represented as mean ± s.e.m. (n = 4). c, Confocal image showing Dendra2-positive mitochondria in cancer cells. Scale bar, 10 μm. Tumour sections were labelled with cancer-cell-specific H2B antibody and DAPI. The green signal in H2B-antibody-labelled cells signifies the presence of trafficked Dendra2-positive mitochondria in the cancer cell. Yellow circle shows Dendra2-positive mitochondria in infiltrated stromal cells, whereas the white outline highlights an H2B cell that has acquired Dendra2-positive mitochondria. d, Tumour growth curves and representative images of tumours showing the effect of eight cycles of L-778123, PD1 inhibitor, or a combination of these two on 4T1 tumours in immunocompetent BALB/c female mice. Data represent mean tumor volume ± s.e.m. (n = 5). e, Graph showing the immunomodulatory effect of L-778123 and PD1 inhibitors on the population of CD8+ T cells in the tumour, as measured by flow cytometry. Data are shown as mean ± s.e.m. (two-way ANOVA followed by Tukey’s multiple comparisons test). f, Graph showing the effect of treatments on the population of activated CD69+ T cells. Data are shown as mean ± s.e.m. (two-way ANOVA followed by Tukey’s multiple comparisons test). g, FESEM image showing the nanotube between a primary human tumour explant and an autologous immune cell. Scale bar, 2 μm. h, Representative confocal image showing nanotube formation and MitoTracker-Green-labelled mitochondrial transfer (green arrows) from PBMCs occurs to the tumour explant. Scale bar, 5 μm. i, In our proposed model, the cancer cell can communicate with the immune cell via physical nanotubes, and metabolically impair the immune cell by hijacking the mitochondria. Next-generation immunotherapies will need to target immune checkpoints as well as mitochondrial hijacking. Panel i adapted from Servier Medical Art under a Creative Commons licence (https://creativecommons.org/licenses/by/3.0/).
Currently, a key goal in cancer immunotherapy is to identify combinations that can improve treatment outcomes. Based on our in vivo observations, we rationalized that L-778123 could improve the outcome with ICIs. We tested this in the syngeneic aggressive 4T1 breast tumour model, which responds poorly to monotherapy with ICIs and thus fits the profile of a tumour where a combination therapy is needed. The animals were randomized into six treatment groups: (1) vehicle; (2) PD1 inhibitor (10 mg kg−1) alone; (3) L-778123 (40 mg kg−1) + PD1 inhibitor (10 mg kg−1); (4) L-778123 (80 mg kg−1) + PD1 inhibitor (10 mg kg−1); (5) L-778123 (120 mg kg−1) + PD1 inhibitor (10 mg kg−1); and (6) L-778123 (80 mg kg−1). The animals were given eight cycles of treatments, with L-778123 and anti-PD1 administered on alternate days. As shown in Fig. 5d, PD1 inhibition resulted in a statistically significant reduction in the tumour volume compared with vehicle controls (P = 0.01). Interestingly, we observed a dose-dependent increase in antitumour efficacy with L-778123 in combination with PD1 inhibitor. Although monotherapy with L-778123 (80 mg kg−1) did not exert statistically significant tumour inhibition compared with the vehicle, the combination of L-778123 (80 mg kg−1) with PD1 inhibitor (10 mg kg−1) resulted in a statistically significant improvement over L-778123 (80 mg kg−1) monotherapy (P = 0.004). Similarly, the combination of L-778123 (120 mg kg−1) with PD1 inhibitor (10 mg kg−1) was found to exert statistically significant tumour growth inhibition over PD1 inhibitor monotherapy (P = 0.0003). Furthermore, both L-778123 (120 mg kg−1) or a combination of L-778123 (120 mg kg−1) and PD1 inhibitor (10 mg kg−1) resulted in substantial tumour inhibition compared with vehicle-treated controls (Supplementary Fig. 21). The combination resulted in a statistically significant improvement in tumour inhibition compared with the GTPase inhibitor alone (P < 0.05). The combination of L-778123 (120 mg kg−1) and PD1 inhibitor (10 mg kg−1) exhibited a significant increase in survival versus PD1 inhibitor monotherapy (logrank P = 0.018; HR (logrank) = 0.29) (Supplementary Fig. 24a). The treatments were well tolerated, and we did not observe any body weight change in individual mice with the treatments (Supplementary Figs. 21b and 24b). The combination of L-778123 (80 mg kg−1) and PD1 inhibitor (10 mg kg−1) exhibited a significant increase in survival versus L-778123 (80 mg kg−1) monotherapy (logrank P = 0.024; HR (logrank) = 0.23). Isolating the tumour after eight cycles of treatments and analysing the single-cell suspension using flow cytometry revealed that the combination of L-778123 (80 mg kg−1) and PD1 inhibitor increased the total CD8+ T cells in the tumour versus PD1 alone (10 mg kg−1) (n = 5; P = 0.0043), L-778123 alone (80 mg kg−1) (n = 5; P = 0.0124) or vehicle controls (n = 5; P = 0.0002). The combination of L-778123 (120 mg kg−1) and PD1 inhibitor significantly increased the total CD8+ T cells in the tumour versus the combination of L-778123 (80 mg kg−1) and PD1 inhibitor (n = 5; P < 0.0001) (Fig. 5e,f and Supplementary Figs. 25-27). Taken together, our in vivo studies indicate that the combination of an ICI with a pharmacological agent perturbing nanotube-mediated mitochondrial hijacking can improve antitumour efficacy.
Finally, in a preliminary analysis, we generated primary tumour explants from three different human tumour biopsies, which were cocultured with autologous immune cells from the same patient34. FESEM imaging of the coculture of the patient’s immune cells with the tumour explant confirmed the presence of heterotypic nanotubes (Fig. 5g and Supplementary Fig. 28). Furthermore, labelling the mitochondria of the immune cells with MitoTracker Green before addition to the cancer explant culture revealed the transfer of MitoTracker to the cancer cells (Fig. 5h), indicating that nanotube-mediated mitochondrial hijacking is conserved in humans.
Conclusion
Our results suggest that nanotube-mediated mitochondrial hijacking from immune cells could emerge as a novel mechanism of immune evasion by cancer cells. Although currently approved immunotherapy agents can activate the immune cells by blocking the immune checkpoints, maximal efficacy can be limited if the immune cells are metabolically hindered. Our results indicate that pharmacological inhibitors of nanotube-mediated mitochondrial hijacking could lead to increased anticancer efficacy and emerge as next-generation immunotherapy (Fig. 5i). We recognize that the pharmacological inhibitors used in this proof-of-concept study are not specific inhibitors of nanotube formation and that their anticancer effects could be due to other pathways being perturbed besides the inhibition of nanotube-mediated mitochondrial transfer. We also note that the inhibitors were not able to fully block mitochondrial hijacking, which indicates that there are additional mechanisms driving the process. Clinical translation will require novel targeted therapeutics that specifically inhibit the machinery of nanotube formation. Indeed, our study highlights the exocyst complex and Miro GTPase as attractive therapeutic targets for cancer immunotherapy. Many open questions remain, for example, what drives the formation of these nanotubes towards specific cells or what causes the mitochondria to preferentially move from immune cells to cancer cells. Although these questions are beyond the scope of the present study, answering them can lead to a new generation of immunotherapies.
Methods
Cell culture.
MDA-MB-231 (ATCC; HTB-26), 4T1 (ATCC, CRL-2539), LLC (ATCC; CRL-1642) and B16F10 (ATCC; CRL-6475) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific), supplemented with 10% v/v heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific) and 1% v/v penicillin–streptomycin–glutamine (PSG) (Thermo Fisher Scientific). The DN32.D3 cells (gifted by A. Kulkarni, UMass Amherst) were maintained in Iscove’s modified Dulbecco’s medium (IMDM) (Thermo Fisher Scientific) with 10% FBS and 1% PSG. Primary T cells were cultured either in IMDM or RPMI-1640 (Thermo Fisher Scientific) supplemented with 10% FBS, 1% HEPES, 1% PSG and 50 nM BME. All the cells were free of mycoplasma. Cells were stained with 0.5–1.0 μM CellTrace Red (or CTFR) cell proliferation dye (Invitrogen), 0.5–1.0 μM CellTrace Green CFSE cell proliferation dye (Invitrogen) or 0.5–1.0 μM of MitoTracker Green FM (Invitrogen) at 37 °C in a 5% CO2 incubator as per the manufacturer’s protocols. The MitoTracker-stained cells were washed twice followed by incubation in complete media for 1 h to ensure the removal of excess unbound MitoTracker dyes. Cocultures were established by incubating cancer cells and immune cells in a 1:1 ratio of their respective media for defined time periods according to the experiments. Primary CD3+ T or CD8+ T cells were isolated from mice spleen by a negative selection method using MojoSort Mouse CD3+ (or CD8+) T cell isolation kit (BioLegend), following the supplier’s protocol. After isolation, the purity of CD3+ or CD8+ T cells was validated by co-staining with anti-mouse CD8 and CD3 antibodies followed by flow cytometry analysis. The spleens were obtained either from 6–8-week-old BALB/c mice (Charles River Laboratories) or from PhAMexcised mice (B6;129S-Gt(ROSA)26Sortm1.1(CAG-COX8A/Dendra2)Dcc) (gifted by M. Haigis, Harvard Medical School, and D. Chan, Caltech).
Isolation of primary T cells.
Primary T cells were isolated from 8–10-week-old female BALB/c mice. Female BALB/c mice (6–8 weeks old) were purchased from Charles River Laboratories and housed in specific pathogen-free conditions at the Center of Comparative Medicine in Brigham and Women’s Hospital (BWH) by following the guidelines of BWH’s Institutional Animal Care and Use Committee. The animals were purchased and housed in the facility for at least a week before starting any experiment. Splenocyte was prepared by harvesting the spleen from healthy 8–10-week-old mice. CD3+ and CD8+ T cells were isolated by immunomagnetic negative selection following the manufacturer’s protocol. The purity and viability of the isolated cells were tested before proceeding with further experiments. Splenocyte from PhAMexcised mice (B6;129S-Gt(ROSA)26Sortm1.1(CAG-COX8A/Dendra2)Dcc) was supplied by M. Haigis’ group.
Drug treatments.
For pharmacological inhibition, both cancer cells and immune cells were treated with varying concentrations of farnesyltransferase/geranylgeranyltransferase 1 inhibitor (L-778123) (BioVision), Cdc42/Rac1 GTPase inhibitor (ML-141) (Sigma) or Rac1 GTPase inhibitor (6-Thio-GTP) (Abcam), and separately incubated in basal media for 7 h before coculture in complete media. To test the effect of different treatments on cell viability, MDA-MB-231 and 4T1 cells (breast cancer cell lines) were seeded in a 96-well flat-bottom tissue-culture-treated plates (Corning) at a density of 104 cells per well (per 100 μl) and treated with increasing concentrations of ML-141, 6-Thio-GTP and L-778123 for 7 h in basal media followed by complete media for 24 h at 37 °C in a 5% CO2 incubator. The maximum concentration of 0.2% DMSO was used as the vehicle control. Cell viability was quantified using the MTT (thiazolyl blue tetrazolium bromide) assay. Absorbance was recorded in a BioTek Epoch microplate spectrophotometer (using Gen5 software (v. 1.11)) at a wavelength of 562 nm. All the experiments were performed at least thrice, and the relative cell viability (%) was expressed as a percentage mean ± standard error of the mean (s.e.m.) of untreated cells. Three replicates were performed per condition. Calculations were tabulated using Microsoft Excel 2011 (v. 14.7.3).
Fluorescence microscopy.
Cells grown on coverslips (VWR; diameter, 12 mm) were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature for 2 h. The fixed cells were washed three times with 1× PBS for 20 min. The cells were further permeabilized by incubation with 0.5% Triton X-100 at 4 °C for 10 min. Then, the cells were washed three times with 1× PBS-T (1× PBS + 0.05% Tween 20), blocked with 10% bovine serum albumin (BSA) solution at room temperature for 1 h, and stained with the desired primary antibody for 12–16 h at 4 °C followed by secondary antibody staining (1:300 dilution) for 1 h at room temperature. For actin staining, the cells were incubated with 50 μg ml−1 of rhodamine phalloidin/phalloidin AF 488/phalloidin AF 405 (Thermo Fisher Scientific) and incubated at room temperature for 1 h. For nuclear staining, the cells were incubated with DAPI/Hoechst 33342 (Thermo Fisher Scientific) for 15–20 min. The cells were washed with 1× PBS-T three more times. The coverslips were mounted on glass slides and imaged using a Nikon Eclipse Ti camera (Nikon Instruments) with NIS-Elements imaging software (v. 3.10). Confocal imaging was done on Zeiss LSM 800 and Airyscan confocal laser scanning microscope with ZEN 2.3 software. Post-processing of the images was done using either ImageJ or ZEN lite software. The following antibodies were used for immunostaining: Sec3 (Novus Biological or Proteintech; 1:200), Sec5 (Santa Cruz Biotechnology or Proteintech; 1:200) and Miro1 (Santa Cruz Biotechnology or Sigma; 1:200).
Scanning electron microscopy.
Cells were seeded on 12-mm-diameter glass coverslips (VWR) and incubated for the desired time (16 h in general) according to the experiment. The sample was fixed by 2.5% glutaraldehyde (Sigma) in 0.1 M sodium cacodylate buffer (Electron Microscopy Sciences). Fixed cells were washed 3 × 15 min with 0.1 M sodium cacodylate buffer, post-fixed in 0.1% OsO4 (Sigma) in water for 1 h at room temperature and washed 2 × 10 min with water before dehydration. The dehydration step was performed as follows: 35% ethanol for 5 min, 50% ethanol for 5 min, 70% ethanol for 10 min, 90% ethanol for 10 min and 2 × 100% ethanol for 10 min. After fixation and dehydration, the coverslips were dried and placed on FESEM stubs for sputter coating by EMS 300T D dual-head sputter coater with Au or Pt/Pd (5 nm). Imaging was done on a Zeiss FESEM Supra 55-VP microscope. Images were processed using ImageJ software. In the case of immunogold labelling, the sample was fixed by 1.5% glutaraldehyde (Sigma) and 2% paraformaldehyde in PBS. Then, the cells were washed three times using PBS, blocked with 10% BSA solution at room temperature for 1 h, and probed with the primary antibody for 12–16 h at 4 °C followed by gold nanoparticle (10 nm diameter)-conjugated secondary antibody staining (1:100 dilution) for 1 h at room temperature. The cells were washed with PBS followed by 3 × 15 min with 0.1 M sodium cacodylate buffer. The sample was treated with 0.1% OsO4 in water for 1 h at room temperature and washed for 2 × 10 min. A silver enhancement step was followed to increase the size of the gold nanoparticle for easy visualization.
Flow cytometry.
Cell suspensions were prepared in PBS with 2% FBS by maintaining a density of 1–5 million cells ml−1. Depending on the experimental design, fixation, permeabilization and antibody staining were performed as stated above. Each of the antibodies were used at 1:50 dilution for flow cytometry analysis. Samples were examined by Accuri C6 flow cytometer or BD LSRFortessa flow cytometer. All the FACS data were processed with Cflow Plus or FlowJo software. For flow sorting, a single-cell suspension was prepared from coculture; the cells were sorted according to their fluorescence signal. The cells were collected in a 5 ml flow tube containing complete media (DMEM/IMDM). The cells were transferred to fresh media after sorting and counting was performed before the next experiment.
siRNA knockdown.
Cancer cells were transfected with EXOC1 and EXOC2 siRNA (MISSION siRNA, Sigma-Aldrich), RhoT1/Miro1 (Santa Cruz Biotechnology) and GAPDH control siRNA (Sigma-Aldrich) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Briefly, cells at 90% confluence were cultured in Opti-MEM I (Invitrogen). An siRNA–lipid complex, comprising 2.5 μl (0.1 pM) and 4 μl (0.2 pM) of EXOC1, EXOC2 or RhoT1 siRNA (60 nM) in 150 μl Opti-MEM I mixed with 9 μl Lipofectamine RNAiMAX in 150 μl Opti-MEM I, was prepared. After incubation for 15 min at room temperature, the cells were incubated for 12 h with the siRNA–lipid complex and then the cells were only left in complete media for the next 36 h. Knockdown was validated by Western blot analysis.
Mitochondrial genotyping.
Human and mouse mitochondrial 16S and 12S sequences were downloaded from NCBI (accession numbers: NC_012920.1:1671-3229 and NC_005089.1:1094-2675 for human and mouse 16S sequences, respectively; NC_012920.1:648-1601 and NC_005089.1:70-1024 for human and mouse 12S sequences, respectively). Forward and reverse primers were designed using Primer3 software to amplify two different non-overlapping regions in each gene. Forward and reverse primer pairs were constructed in conserved regions so that every primer pair could be used for PCR amplification in both species. DNA was extracted from the mouse NKT cell line DN32.D3 and human cancer cell line MDA-MB-231 (each at approximately 3 × 106 cells) using the Qiagen DNeasy blood and tissue kit (catalogue number 69504) and used as a template for the PCR amplification of listed genes using the designed primers. The PCR product was run on 1% agarose gel; gel bands at the expected sizes were eluted using IBI Scientific’s gel/PCR DNA fragment extraction kit (catalogue number IB47030). The purified PCR product was then sequenced using the primer (in bold). Species-specific (and cell-line-specific) SNPs were identified for further analyses by comparing individual chromatograms using SnapGene software. For testing the hypothesis that human cancer cell line MDA-MB-231 showed a specific uptake of mitochondria from mouse NKT cells, both cell lines were cocultured for 16 h and FACS was used to separate the cancer cells. DNA extraction, PCR amplification, gel elution and sequencing were performed on cocultured cancer cells as described above, and all the three cell lines were then used for comparative SNP analyses. A few representative SNP examples for each gene are shown in Supplementary Table 1 as illustrative examples.
Western blotting.
Cells were seeded at a density of 2–3 × 106 cells in T-25 flasks. Cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (supplemented with protease and phosphatase inhibitor) for 30 min on ice with mild vortexing every 10 min followed by centrifugation at 4 °C. The amount of protein was measured by bicinchoninic acid assay (Thermo Fisher Scientific) (according to the supplier’s protocol) and equal amounts of protein lysates were electrophoresed on a 10–15% polyacrylamide gel at 80–140 V for 1 h. The proteins were transferred to nitrocellulose or polyvinylidene difluoride membrane (pore size, 0.2 μm) through wet blotting for 2 h at 350 mA (Bio-Rad Laboratories). Membranes were blocked by 5% BSA in 1× TBS-T (1× TBS and 0.1% Tween 20) for 1 h and incubated with primary antibodies at 4 °C for 12–14 h. After washing thrice with 1X TBS-T, the membrane was incubated with the secondary HRP-goat-anti-mouse/rabbit antibody (1:3000) (Bio-Rad Laboratories) for 1 h at room temperature. The membranes were washed twice with 1X TBS-T and once with TBS before developed using chemiluminescent Femto Substrate (Thermo Scientific) and imaged using G:BOX bio-imaging system (Syngene) using Quantity One software for image analysis. The following antibodies were used for Western blotting: Sec3/ExoC1 (Proteintech; 1:500), Sec5/ExoC2 (Proteintech; 1:500), RhoT1 (Sigma; 1:500), and normalized to GAPDH (SCBT; 1:2000).
Metabolic studies by Seahorse.
Metabolic analysis was carried out using the XFe24 or XFe96 extracellular flux analyser (Seahorse Bioscience). For cancer cells, 0.04 × 106 cells were seeded onto XFe24-well Seahorse microplates in complete media, which allow the adhesion of cells overnight. The media in each well were replaced with the XF base medium (Agilent) supplemented with 10 mM glucose, 1 mM sodium pyruvate and 2 mM l-glutamine, and adjusted to a pH of 7.4. After that, the plate was equilibrated at 37 °C in an incubator without CO2 for 1 h. For immune cells, 0.2 × 106 cells were seeded onto poly-d-lysin-treated XFe96-well Seahorse microplates, allowing the adhesion of cells for 1 h with the supplemented XF base medium mentioned above. After that, the plate was equilibrated at 37 °C in an incubator without CO2 for 1 h. The standard Mito stress test was performed with the stepwise injection of oligomycin (1 μM), carbonyl cyanide-p-triflouromethoxyphenylhydrazone (FCCP) (1.0 μM for cancer cells and 1.5 μM for immune cells), and a mixture of rotenone (0.5 μM) and antimycin A (0.5 μM). At the end of the experiment, the data were normalized according to the total protein concentration in each well. Cell lysates were prepared with the addition of 50 μl radioimmunoprecipitation assay lysis buffer in each well and the total protein concentration was determined by the bicinchoninic acid assay (the earlier section provides further details). All the chemicals used in this experiment were purchased from Sigma, unless stated otherwise. For immune cells, the data were normalized according to the cell number.
In vivo animal study (mitochondrial transfer).
Syngeneic C57BL/6PhAMexcised mice (~20 g; 8–10 weeks) of both sex were subcutaneously injected with LLC lung cancer cells (1 × 106 cells) in the flanks. The tumour was harvested after eight days. The tumour tissues were washed with cold PBS, thoroughly minced in less than 1 mm size and suspended in type IV collagenase solution (1.0–1.5 mg ml−1). The tumour samples with digestion media were incubated for 1 h at 37 °C and 5% CO2 incubator and finally passed through a 70 μm filter to obtain a single-cell suspension. The cell suspension was washed with RPMI-1640 media by centrifuging at 350 r.c.f. (relative centrifugal force) for 5 min (twice). The single-cell tumour suspension was then used for FACS analysis. The cells (106 cells per 100 μl) were labelled for CD45, CD11b, CD31 and CD81 using antibodies (BioLegend) at 1:100 dilution for 1 h on ice. The cells were washed twice with PBS, and analysed using a BD LSR II flow cytometer. The flow cytometry data were analysed using FlowJo software (v. 10.5.3). All the animal procedures were approved by BWH’s Institutional Animal Care and Use Committee (protocol number 2016N000393). The animals were housed in dedicated institutional centralized animal facility where light cycles of 12 h (12 h light and 12 h dark) were maintained. In the rodent-holding rooms, the temperature was set at 71 F and controlled within ±3 F. Humidity was set at 50% and controlled to a range between 30% and 70%.
In vivo animal study (drug treatment).
4T1 breast cancer cells (1 × 106 cells) were subcutaneously injected in the flanks of syngeneic female BALB/c mice (~20 g; 8–10 weeks old). The drug therapy was started on the day after tumour implantation. Each animal was intraperitoneally injected every alternate day (eight cycles) with the vehicle (for the control group), αPD1 (10 mg kg−1), L-778123 (80 mg kg−1), αPD1 (10 mg kg−1) and L-778123 (40 mg kg−1), αPD1 (10 mg kg−1) and L-778123 (80 mg kg−1), or αPD1 (10 mg kg−1) and L-778123 (120 mg kg−1). The tumours were measured every other day using a Vernier caliper, and the tumour volume (Vt) was calculated as per the following formula: L × B2/2, where L is the longest dimension and B is the shortest dimension. The total body weight was routinely measured as a measure of any gross toxicity. All the tumour tissues were harvested for further studies. All the animal procedures were approved by BWH’s Institutional Animal Care and Use Committee (protocol number 2016N000393). The maximum permitted tumour volume (2 cm3) was not exceeded in any study.
FACS analysis of immune contexture of tumours.
Single-cell suspensions were prepared from each tumour sample following the abovementioned procedure using type IV collagenase. The single-cell tumour suspension was then used for FACS analysis. The cells (106 cells per 100 μl) were labelled for CD3, CD4, CD8, CD45, and CD69 using antibodies (BioLegend) at 1:100 dilution for 1 h on ice. The cells were washed twice with PBS, and analysed using a BD LSR II flow cytometer. The flow cytometry data were analysed using FlowJo software (v. 10.5.3).
Human PBMCs–tumour explant culture.
Cancer cells were isolated from (1) a surgical biopsy of thymoma from an anonymous female patient, (2) a liver biopsy of metastatic breast cancer in an anonymous 42-year-old female having metastatic breast cancer, and (3) peritoneal biopsy from an anonymous 47-year-old female with metastatic breast cancer. The cancer cells were cocultured with autologous peripheral blood mononuclear cells (PBMCs) from the matched patient. At 16 h post-incubation, the explant coculture was fixed with glutaraldehyde and processed for either FESEM or confocal imaging to visualize the nanotube. In parallel, the explant was counter-labelled with phalloidin (for actin) and Hoechst 33342 (for nucleus), and imaged using a confocal microscope to track the transfer of MitoTracker-labelled mitochondria from immune cells to cancer cells. Tissues were collected using an institutional review board–approved sample collection protocol (IRB: 13-416) at the Mass General Hospital Cancer Center, and the subjects were consented for serial blood collections and tissue collections for study. The study was compliant with all the relevant ethical regulations regarding research involving human participants.
Statistical analysis and reproducibility.
All the statistical analyses were performed by Prism 8 software (GraphPad). The experimental data are expressed as mean ± s.e.m. and we used the Student’s t-test, one-way (or two-way) analysis of variance (ANOVA) followed by an appropriate post-test to calculate the statistical significance. Here P < 0.05 was considered significant. Each experiment has been independently repeated at least thrice, unless mentioned otherwise.
Supplementary Material
Acknowledgements
This work is supported by grants from the National Institute of Health (NIH AR073135_ HLJ, CA236702_SS_HLJ, CA214411_SS and CA229772_SS_Co-I), American Lung Association Discovery Grant (LCD-618834_SS) and Department of Defense (DoD PC180355_HLJ and CA201065_HLJ). This work was performed in part at the Center for Nanoscale Systems (CNS), Harvard University, a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), supported by the National Science Foundation. We thank M. Haigis and A. Goldman for help with the PhAM mouse studies. K.K. is a Gilead Sciences Fellow of the Life Sciences Research Foundation.
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
S.S. is a co-founder and owns equity in Vyome Therapeutics, Akamara Therapeutics and Invictus Oncology, and receives fees from Famygen and Advamedica. H.L.J. is a founder and owns equity in Curer. A.B. is involved in the following consulting/advisory boards: Pfizer, Novartis, Genentech, Merck, Radius Health, Immunomedics, Taiho, Sanofi, Daiichi Pharma/AstraZeneca, Puma, Biothernostics, Phillips, Eli Lilly and Foundation Medicine; A.B. is also contracted via research/grant to the following institutions: Genentech, Novartis, Pfizer, Merck, Sanofi, Radius Health, Immunomedics, Daiichi Pharma/AstraZeneca and Natera. T.S., C.D., R.J., S.K., A.K., K.K., J.M. and P.K.M. declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41565-021-01000-4.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41565-021-01000-4.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1.Wei SC et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170, 1120–1133.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P & Allison JP Dissecting the mechanisms of immune checkpoint therapy. Nat. Rev. Immunol 20, 75–76 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Wolchok J Putting the immunologic brakes on cancer. Cell 175, 1452–1454 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Kulkarni A et al. A designer self-assembled supramolecule amplifies macrophage immune responses against aggressive cancer. Nat. Biomed. Eng 2, 589–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei SC, Duffy CR & Allison JP Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Tseng D et al. Anti-CD47 antibody–mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc. Natl Acad. Sci. USA 110, 11103–11108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Önfelt B, Nedvetzki S, Yanagi K & Davis DM Cutting edge: membrane nanotubes connect immune cells. J. Immunol 173, 1511–1513 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Sowinski S et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol 10, 211–219 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Gousset K et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol 11, 328–336 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Osswald M et al. Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Connor Y et al. Physical nanoscale conduit-mediated communication between tumour cells and the endothelium modulates endothelial phenotype. Nat. Commun 6, 8671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rustom A, Saffrich R, Markovic I, Walther P & Gerdes H-H Nanotubular highways for intercellular organelle transport. Science 303, 1007–1010 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Ahmad T et al. Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J. 33, 994–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X & Gerdes HH Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 22, 1181–1191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J et al. Tunneling nanotubes promote intercellular mitochondria transfer followed by increased invasiveness in bladder cancer cells. Oncotarget 8, 15539–15552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sena LA et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A et al. Enhanced oxidative phosphorylation in NKT cells is essential for their survival and function. Proc. Natl Acad. Sci. USA 116, 7439–7448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vyas S, Zaganjor E & Haigis MC Mitochondria and cancer. Cell 166, 555–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman A et al. Targeting tumor phenotypic plasticity and metabolic remodeling in adaptive cross-drug tolerance. Sci. Signal 12, eaas8779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clutton G, Mollan K, Hudgens M & Goonetilleke N A reproducible, objective method using MitoTracker® fluorescent dyes to assess mitochondrial mass in T cells by flow cytometry. Cytometry 95, 450–456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham AH, McCaffery JM & Chan DC Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis 50, 833–843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelletier M, Billingham LK, Ramaswamy M & Siegel RM in Methods Enzymol, Vol. 542 (eds Galluzzi L & Kroemer G) 125–149 (Academic Press, 2014). [DOI] [PubMed] [Google Scholar]
- 24.Kaplon J et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Hase K et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol 11, 1427–1432 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto M et al. Potential role of the formation of tunneling nanotubes in HIV-1 spread in macrophages. J. Immunol 196, 1832–1841 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Moskalenko S et al. The exocyst is a Ral effector complex. Nat. Cell Biol 4, 66–72 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Hanna SJ et al. The role of Rho-GTPases and actin polymerization during macrophage tunneling nanotube biogenesis. Sci. Rep 7, 8547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo W, Tamanoi F & Novick P Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol 3, 353–360 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Fransson Å, Ruusala A & Aspenström P The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun 344, 500–510 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Glater EE, Megeath LJ, Stowers RS & Schwarz TL Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol 173, 545–557 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arkwright PD et al. Fas stimulation of T lymphocytes promotes rapid intercellular exchange of death signals via membrane nanotubes. Cell Res. 20, 72–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bustelo XR, Sauzeau V & Berenjeno IM GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays 29, 356–370 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumder B et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun 6, 6169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.