Ductal-derived pancreatic tumors have elevated epithelial and CD8+GZM+ T-cell CD73 expression that confers sensitivity to small-molecule inhibition of CD73 or Adora2b to promote CD8+ T-cell–mediated tumor regression.
Abstract
The microenvironment that surrounds pancreatic ductal adenocarcinoma (PDAC) is profoundly desmoplastic and immunosuppressive. Understanding triggers of immunosuppression during the process of pancreatic tumorigenesis would aid in establishing targets for effective prevention and therapy. Here, we interrogated differential molecular mechanisms dependent on cell of origin and subtype that promote immunosuppression during PDAC initiation and in established tumors. Transcriptomic analysis of cell-of-origin–dependent epithelial gene signatures revealed that Nt5e/CD73, a cell-surface enzyme required for extracellular adenosine generation, is one of the top 10% of genes overexpressed in murine tumors arising from the ductal pancreatic epithelium as opposed to those rising from acinar cells. These findings were confirmed by IHC and high-performance liquid chromatography. Analysis in human PDAC subtypes indicated that high Nt5e in murine ductal PDAC models overlaps with high NT5E in human PDAC squamous and basal subtypes, considered to have the highest immunosuppression and worst prognosis. Multiplex immunofluorescent analysis showed that activated CD8+ T cells in the PDAC tumor microenvironment express high levels of CD73, indicating an opportunity for immunotherapeutic targeting. Delivery of CD73 small-molecule inhibitors through various delivery routes reduced tumor development and growth in genetically engineered and syngeneic mouse models. In addition, the adenosine receptor Adora2b was a determinant of adenosine-mediated immunosuppression in PDAC. These findings highlight a molecular trigger of the immunosuppressive PDAC microenvironment elevated in the ductal cell of origin, linking biology with subtype classification, critical components for PDAC immunoprevention and personalized approaches for immunotherapeutic intervention.
Significance:
Ductal-derived pancreatic tumors have elevated epithelial and CD8+GZM+ T-cell CD73 expression that confers sensitivity to small-molecule inhibition of CD73 or Adora2b to promote CD8+ T-cell–mediated tumor regression.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is predicted to become the second leading cause of cancer-related deaths by the year 2030 in the United States and emerging clinical trials aim to target various components of the immunosuppressive tumor microenvironment (1–3). CD73 is a key ectoenzyme involved in dampening inflammatory responses in inflamed and hypoxic microenvironments that can be soluble but is predominantly localized on the cell surface and generates extracellular adenosine (4, 5). Extracellular adenosine is involved in the resolution stage of inflammation and pancreas tissue repair by signaling through four adenosine receptors (6–8). In the absence of CD73, increased extracellular adenosine triphosphate (ATP) mediates inflammation by binding to purinergic receptors (9–11). In the presence of ectonucleotidase triphosphate diphosphohydrolase-1 (CD39), a cell-surface enzyme with catalytic activity, ATP is rapidly converted to AMP, which is the substrate for catalytic conversion to adenosine by CD73. Preclinical models have shown targeting adenosine signaling has potent antitumor effects (12–19).
Subtypes of human PDAC have been defined using comprehensive whole transcriptomic profiling of large cohorts of patients (20–22). In this article, we found that murine ductal-derived PDAC had elevated CD73 implicating a strong adenosine signaling-dependent immunosuppressive microenvironment. To expand upon these findings, we evaluated which adenosine receptors were important in PDAC. Using The Cancer Genome Atlas (TCGA) analysis and cBioPortal comparisons, we determined high ADORA2B is associated with reduced survival in human PDAC. To test the hypothesis that paracrine signaling through Adora2b is important for PDAC, we implanted KPC cells into wild-type (WT) and Adora2b−/− mice, which resulted in a significant reduction in tumor growth rates in Adora2b−/− mice and a significant increase in CD8+ cells. We then evaluated the requirement for Adora2b-mediated antitumor immunity by performing a study in CD8KO mice. Treatment with an Adora2b small-molecule inhibitor alone significantly reduced the growth of KPC tumors in WT mice; however, the effect was significantly reduced in the CD8KO mice, indicating paracrine adenosine signaling through the Adora2b receptor on CD8+ T cells is a critical mediator of immunosuppression in PDAC.
Materials and Methods
RNA preparation and sequencing
Samples were rinsed in PBS and immediately frozen in liquid nitrogen. Total RNA was extracted using a miRNeasy Mini Kit (74104, Qiagen) and submitted to the Cancer Genomics Center at the University of Texas Health Science Center. Total RNA quality was measured using Agilent RNA 6000 Pico kit (#5067-1513) by Agilent Bioanalyzer 2100 (Agilent Technologies). The samples with an RNA integrity number greater than 7 were used for library preparation. Libraries were prepared following the manufacturer's instructions for the Roche KAPA mRNA HyperPrep Kit (KK8581) and the KAPA Unique Dual-indexed Adapter Kit (KK8727). The quality of the final libraries was examined using the Agilent High Sensitive DNA Kit (#5067-4626) by Agilent Bioanalyzer 2100 (Agilent Technologies), and the library concentrations were determined by qPCR using Collibri Library Quantification kit (#A38524500, Thermo Fisher Scientific). The pooled libraries were sequenced on the Illumina NextSeq 550 platform using the pair-ended 75 bp by a 150-cycle High Output v2.5 Kit (#20024907, Illumina, Inc.). We used ultrafast universal RNA-seq aligner STAR (v2.5.3a) to map the RNA-seq reads to mouse reference genome GRCm38 (23). To obtain the uniquely mapped reads per gene from the GencodeM15 (GRCm38) reference, we set the argument –quantMode to “GeneCounts.” We filtered out those genes with < 5 reads in all samples and conducted the differential expression analysis for the remaining genes by DESeq2 software (24). The P values of genes were adjusted using the Benjamini and Hochberg's procedure to control the false discovery rate (FDR). And the differentially expressed genes were defined as the genes with absolute log2 (fold change) > 0.58 and FDR < 0.05. Nonredundant Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis were performed using WebGestalt (v0.4.3) software (25). All raw data and processed read counts have been submitted to the Gene Expression Omnibus (GSE189130).
Basal and acinar signature genes derived from mouse in human homologs
We adapted the differential expressed genes analysis from our recent cell-of-origin genetically engineered mouse (GEM) models between ductal tumor and acinar tumor in mice (26). We defined those genes (304) with log2 (fold change) > 2 and FDR < 0.01 as the signature of ductal signature genes (917). We further defined acinar signature genes as those genes with log2 (fold change) < −2 and FDR < 0.01. We then downloaded the latest Mouse Human homolog gene symbol (v7.4) file from https://data.broadinstitute.org/gsea-msigdb/msigdb/annotations_versioned/ (accessed on April 20, 2021). We obtained the mouse-derived human homologs signatures for ductal (271) and acinar tumor genes (877), respectively.
Gene set variation analysis
We obtained RNA-seq data sets and differentially expressed gene signatures from two independent PDAC patient cohorts, primary PDACs of high cellularity from the Australian International Cancer Genome Initiative (ICGC; accessed on April 21, 2021) and primary PDACs from TCGA Research Network (accessed through TCGA data portal on April 22, 2021; refs. 21, 22). Specifically, immunogenic, squamous, classic, and basal subtypes classification were considered as previously described by TCGA (Supplementary Table S1 in ref. 22; GUID: C0853F1D-79F0-4125-BA86-8545A54FA572). We constructed the signature genes using the aforementioned human homolog Ductal and Acinar tumor gene sets. Then, we conducted the gene set variation analysis (GSVA) with default setting for immunogenic and squamous samples and basal and classic samples, respectively (27). The GSVA score indicates the relative variation of signature gene activity over the AU and TCGA samples.
Venn diagram analysis
Differentially expressed genes between immunogenic versus squamous and classic versus basal subtypes were obtained from (21) and TCGA (Supplementary Table S1; GUID: 3DD81EAF-3FD4-48CE-A9DA-2454820DAB10; ref. 22). Upregulated genes in human squamous and basal subtypes were compared with upregulated genes in the KPCDuct mouse model and shown as a Venn diagram analysis.
Mass cytometry
Tumor tissues were harvested and digested with 1 mg/mL collagenase P (MilliporeSigma) and 0.5 mg/mL DNase I (MilliporeSigma). Single-cell suspensions were stained with 5 μmol/L Cell-ID Cisplatin (Fluidigm Corp.) and incubated with Fc block (BD Biosciences), followed by a surface antibody cocktail. Antibody details including final concentrations can be found in Supplementary Table S1. Next, cells were washed and fixed in Maxpar Fix I buffer (Fluidigm Corp.) and barcoded using the Cell-ID 20-Plex Pd Barcoding Kit (Fluidigm Corp.). Next, the cells were stained with 1.25 μmol/L Cell-ID Intercalator-Ir (Fluidigm Corp.) overnight. Sample acquisition was performed on a Helios mass cytometer (Fluidigm Corp.). The analysis was performed using R package CyTOF Workflow (28) and FlowJo version 10 software (FlowJo LLC).
Cell lines
HPNE, Capan-2, and ASPC1 cells were purchased from ATCC and were maintained following the vendor's instructions. Murine pancreatic adenocarcinoma cells derived from KrasG12D/+; Trp53LsL-R172H/+; Pdx1-Cre mice (KPC) were a generous gift from Dave Tuveson (Cold Spring Harbor Laboratory) and were maintained in DMEM (Thermo Fisher Scientific-10567014) with 10% fetal bovine serum. All cell lines were maintained at low passage numbers and were cultured at 37°C and 5% CO2 in a humified incubator. Mycoplasma test was performed on HPNE, Capan-2, and ASPC1 cells, immediately before the experiment and no cell line manipulations were performed for this study.
Animal models
All animal procedures were performed in compliance with UTHealth's and The University of Texas MD Anderson Cancer Center's Center for Laboratory Animal Medicine and Care and are approved on Dr. Bailey's and Dr. McAllister's IACUC and Animal Welfare Committee protocols. C57BL/6 (000664) mice were purchased from Jackson Laboratories. Hnf1b:CreERTM and Ptf1a:CreERTM mice were purchased from Jackson Laboratories. KPCDuct and KPCAcinar mice were generated by crossing to LSL-KrasG12D and LSL-TP53R172H as described previously (26). For obtaining Kras expression alone, transgenic mice with CAG-lox-GFP-stop-lox-KrasG12V (29) were received from Craig Logsdon, MD Anderson Cancer Center, Houston, TX. Strains of Hnf1b:CreERTM mice were crossed with cLGL-KRASG12V to generate KCDuct mice to obtain mutant Kras expression in adult pancreatic ductal cells. Similarly, Ptf1a:CreERTM were crossed with cLGL-KRASG12V mice to generate CKAcinar mice to obtain Kras expression in mature acinar cells. These mice express GFP in the whole body and lose GFP after Cre-mediated recombination. Mice were genotyped by PCR or Transnetyx. To achieve mutant Kras expression in mature ductal or acinar cells, mice at an age of 6 to 8 weeks were injected subcutaneously with 5 mg tamoxifen (Sigma, T5648) for 2 consecutive days (10 mg dose). For subcutaneous xenograft models, 2×105 KPC cells in PBS: Matrigel mix (1:1) were injected in the left flank of C57BL/6 or Adora2b−/− mice. Tumor size was calculated twice a week with a vernier caliper. Tumor volume was calculated as length × width × width/2 in cubic millimeters. Tumor doubling time was calculated using the method described previously (30).
Histopathology
Formalin-fixed and sectioned pancreas tissue was deparaffinized with histoclear followed by hydration with ethanol and water and staining with hematoxylin. Sections were next counterstained with eosin and dehydrated stepwise with ethanol and histoclear. Slides were mounted with a coverslip using a mounting medium. All pancreatic pathologies in the genetically engineered models were classified by pathologists at the University of Texas Health Science Center.
CD73 inhibitor administration
APCP (αβ-methylene ADP, Sigma-Aldrich; catalog no. M3763) was purchased from Sigma. AB680 was purchased from MedChemExpress (cat. no. HY-125286). Mice bearing subcutaneous KPC tumors or mice with spontaneous PDAC tumors were treated with CD73 inhibitors. 20 mg/kg APCP in PBS or vehicle control (PBS) was administered IP for the spontaneous model and peritumor in the subcutaneous model. Mice were given APCP on alternate days until the end of the experiment. For AB680 treatment, the stocks were prepared in 100% DMSO. For oral gavage, AB680 was diluted in 10% DMSO + 90% SBE beta cyclodextrin (SBE-b-CD) in 0.9% saline. 10 mg/kg AB680 or vehicle control (10% DMSO + 90% SBE-b-CD in 0.9% saline) were given by oral gavage on alternate days until the end of the experiment.
Adora2b inhibitor administration
PSB1115 was purchased from Tocris. C57BL/6 or CD8KO mice (Jax) were implanted with 100K KPC cells subcutaneously and treated with PSB1115 at a concentration of 1 mg/kg for 5 consecutive days per week throughout the duration of the experiment. Vehicle was PBS.
Western blot analysis
Cell and tissue extracts were prepared using cell lysis buffer (Cell Signaling Technology; #9803S) with protease inhibitor cocktail tablets (Cell Signaling Technology; #5871). The BCA method (Thermo Fisher Scientific) was used for protein quantification. Protein (20 μg) was separated using SDS-PAGE (Bio-Rad). Trans-blot Turbo Transfer kit (Bio-Rad) was used for semi-dry transfer. After transfer, membranes were blocked using 5% skimmed milk (Bio-Rad) in TBST (TBS buffer containing 0.5% Tween-20) for 1 hour, followed by overnight incubation with primary antibodies (in 5% milk and dilution as per the manufacturer's instructions) at 4°C. On day −2, the membrane was washed 4 times with TBST buffer and incubated with the respective HRP-conjugated secondary antibody (1:5,000) for 1 hour. Further, membranes were washed four times with TBST buffer and developed using ClarityTM Western ECL Substrate (Bio-Rad; #1705061). Primary antibodies used in this study are described in Supplementary Table S2.
Primary pancreatic duct culture
Pancreatic ducts were cultured as defined previously (31) from Tam0 (WT) and Tam10 mice 4 days after tamoxifen administration. Briefly, pancreata were collected, minced to 1-mm pieces, and digested for 30 minutes at 37°C in a digestive solution (0.1% soybean trypsin inhibitor and 0.1% collagenase). Cells were filtered through a 40-μm filter, washed an additional two times with culture medium and plated on collagen-coated plates in complete medium (DMEM/F12; Life Technologies 11330-032) 500 mL, penicillin–streptomycin (100×; Life Technologies 15140-122) 5 mL, 1×Nu-serum IV (BD Biosciences 355104) 25 mL, 5% bovine pituitary extract (3 mg/mL; BD Biosciences 354123) 4.2 mL, 25 μg/mL ITS+ Premix (BD Biosciences 354352) 2.5 mL, epidermal growth factor (100 μg/mL; BD Biosciences 354001) 100 μL, 20 ng/mL cholera toxin (1 mg/mL; Sigma-Aldrich C8052) 50 μL, 100 ng/mL3,3′,5-Triiodo-L-thyronine (50 μmol/L; Sigma-Aldrich T2877) 50 μL, 5 nmol/L dexamethasone (100 mmol/L; Sigma-AldrichD1756) 5 μL, 1 μmol/L D-glucose (Sigma-Aldrich G5400) 2.5 g 4.7 mg/mL, nicotinamide (Sigma-Aldrich N3376) 0.66 g 1.22 mg/mL and Soybean trypsin inhibitor (type I; Sigma-Aldrich T6522) 50 mg 0.1 mg/mL. The cultures grew to confluency in 1 week and fibroblast contamination was reduced using the differential trypsinization method. Cell lysates were collected, and equal amounts of protein were subjected to Western analysis.
IHC
Paraformaldehyde fixed and sectioned tissue were baked at 60°C for 30 minutes. Deparaffinization and rehydration were performed using histoclear followed by subsequent incubation with 100%, 70%, 30% ethanol, and deionized water. Sections were permeabilized using PBST (PBS containing 0.25% Tween-20) for 10 minutes and endogenous peroxidases were blocked with 0.3% H2O2 in PBS for 15 minutes. Antigen retrieval was performed by antigen unmasking solution (Vector Laboratories, H-3300) using heat-mediated microwave method. Sections were blocked with 10% FBS in PBST followed by incubation with primary antibodies overnight at 4°C (primary antibodies used in this study are described in Supplementary Table S2). The next day, sections were washed three times with PBST and then incubated with secondary antibodies (1:500) at room temperature for 2 hours. The sections were washed three times with PBST and detection was performed using Vectastain Elite ABC kit (Vector Laboratories, PK-6100) and DAB Peroxidase (HRP) Substrate kit (Vector Laboratories, SK- 4100). Sections were counterstained with hematoxylin, mounted with a coverslip using mounting media, and visualized under light microscopy.
ImageJ quantification of IHC
IHC quantification was performed using ImageJ (http://imagej.nih.gov/ij/) software. Color intensity threshold was used to identify positive staining and normalize all tissues. Five to six representative fields per tissue were selected for quantification. For the CD73 epithelial versus stroma comparison, a combination of color intensity threshold and freehold selection tool was used to isolate positive staining by tissue subtype.
Nucleoside/nucleotide extraction and quantification
Cell culture supernatants, mouse serum, tumor tissues, or ex vivo cultured tissue from GEM models were collected at the end of the study. As previously described (15), serum used for adenosine analysis was frozen with an adenosine inhibitor cocktail containing 10 μmol/L APCP (CD73 inhibitor; αβ-methylene ADP, Sigma-Aldrich; cat. no. M3763), 10 μmol/L dipyridamole (equilibrative nucleoside transporter inhibitor; Sigma-Aldrich; cat. no. D9766), and 10 μmol/L deoxycoformycin (adenosine deaminase inhibitor; R&D Systems; cat. no. 2033) to preserve nucleosides. Tumors were flash frozen with liquid nitrogen and stored at −80°C. Cell supernatants, serum, or tumor protein lysate was extracted with perchloric acid and neutralized with KHCO3/KOH. Samples were acidified with ammonium dihydrogen phosphate and phosphoric acid. Reaction supernatant was collected by centrifugation. Extracted samples were analyzed by reversed-phase high-performance liquid chromatography (RP-HPLC; ref. 32). Representative AMP and adenosine peaks were identified and measured using the respective standard HPLC curve. For tumors, adenosine levels were normalized to the protein levels in the tumor lysate. To determine if PDAC cell lines had increased CD73 activity, 200 μL of cell line supernatant was analyzed by HPLC or supernatant was replaced with HBSS and 250 ng AMP was added. To evaluate CD73 activity, we extracted 100 μL of HBSS supernatant at times 0,10, 30, 60, and 120 minutes.
RNA isolation and quantitative RT-PCR
As previously described (15), total RNA was extracted with the RNeasy RNA isolation kit (Qiagen) and reverse transcribed with a cDNA Reverse Transcription Kit (Bio-Rad). Quantitative RT-PCR was performed with SYBR Green Master Mix (Bio-Rad) on a Bio-Rad real-time PCR system. PCR primer sequences used in the study were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) and were synthesized at Integrated DNA Technologies. GAPDH was used as a housekeeping gene, and the expression levels of mRNA of interest were normalized to GAPDH.
TCGA analysis
The Kaplan–Meier curves were generated using TCGA RNA-seq data (FPKM_UQ) for PDAC samples, after excluding PNETs and non-PDAC samples. Higher and lower expression levels were stratified on the basis of average expression. Statistical analysis was performed using log-rank tests, and HRs were calculated using Prism software (GraphPad Software, Inc.).
Immune cell distribution by Quantiseq algorithm
The distribution of immune cells in TCGA data sets was extracted from publicly available The Cancer Immunome Database https://tcia.at/home. Data show immune cell abundance as analyzed by the Quantiseq algorithm.
Multiplex immunofluorescence
Assessment of the murine tumor microenvironment by multiplex immunofluorescence was performed and analyzed using Opal and Vectra as previously published (33). Antibodies include Anti-Ms CK19 (TROMA-III), Anti-Ms Granzyme B (E5V2L), Anti-Ms CD73 (D7F9A), Anti-Ms CD8 (D4W2Z), and Anti-Ms F4/80 (D2S9R). Staining and quantification of multiplex images were calculated as previously published (34).
Data availability statement
All raw RNA-seq data and processed read counts have been submitted to Gene Expression Omnibus (GSE189130). We obtained RNA-seq data sets and differentially expressed genes signatures from two independent PDAC patient cohorts, primary PDACs of high cellularity from the Australian International Cancer Genome Initiative (ICGC; accessed on April 21, 2021) and primary PDACs from TCGA Research Network (accessed through TCGA data portal on April 22, 2021; refs. 21, 22). The Cancer Immunome Database was used to identify immune cell distributions. The data generated in this study are available within the article and its Supplementary Data files or from the corresponding author upon reasonable request.
Results
Ductal-derived PDAC tumors have a distinct immunosuppressive gene signature inclusive of high expression of NT5E/CD73
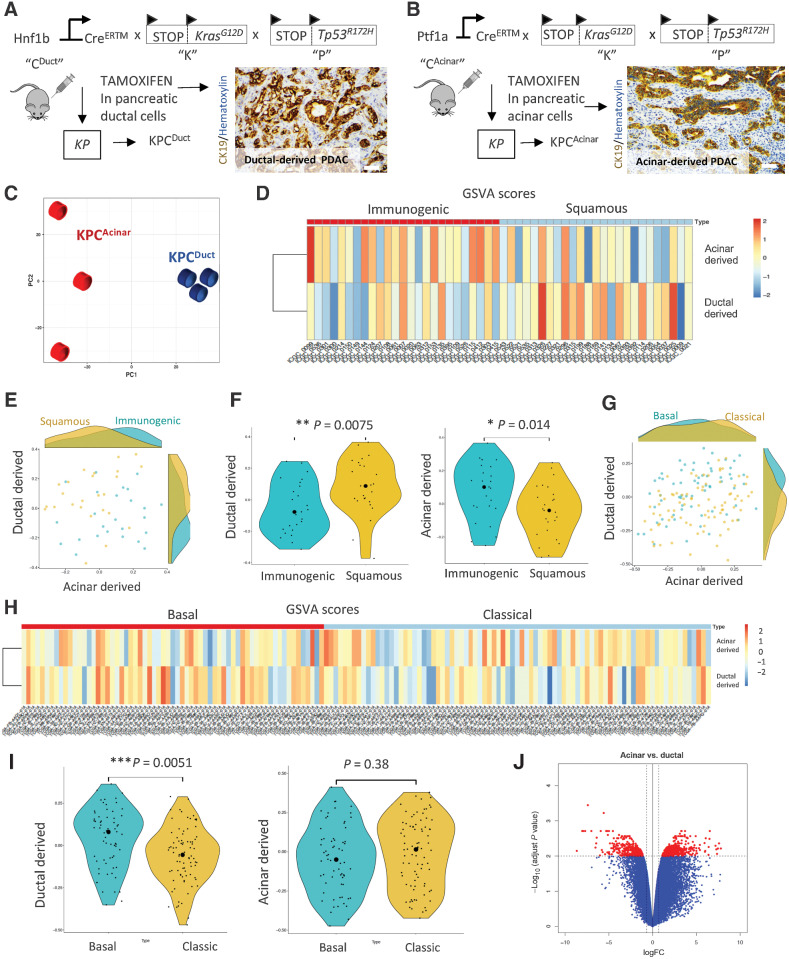
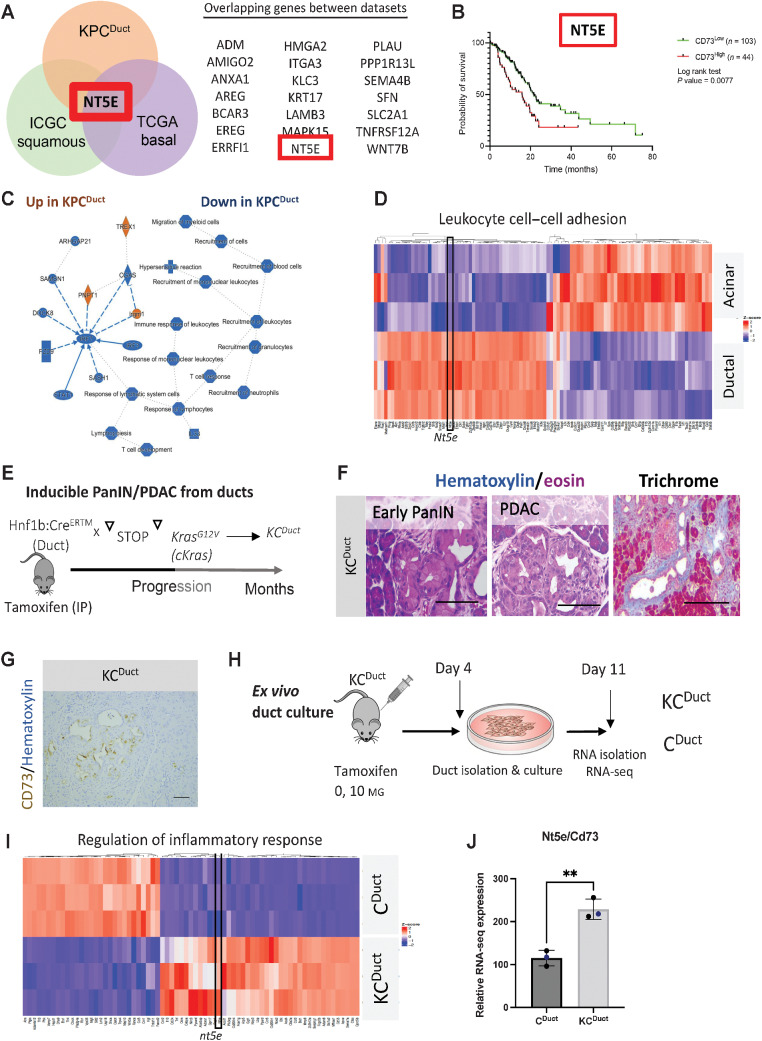
Cell-of-origin studies in inducible murine PDAC models have shown that tumors arising in pancreatic ducts have overlapping transcriptomic signatures with high concordance to basal and squamous human PDAC subtypes and tumors arising in acinar cells have transcriptomic signatures that overlap with high concordance to classic and immunogenic human PDAC subtypes (35). Analyses of bulk tumors generated from acinar (LsL-KrasG12D; LsL-Trp53R127H; Ptf1a:CreERTM (KPCAcinar)) or ductal (LsL-KrasG12D; LsL-Trp53R127H; Hnf1b:CreERTM (KPCDuct)) epithelium previously published by our lab (Fig. 1A and B; ref. 26) resulted in similar findings with tumors arising in acinar cells having RNA-seq signatures that align with immunogenic and classic subtypes and tumors arising in ductal cells having RNA-seq signatures that align with squamous and basal human PDAC subtypes (Fig. 1C–J). By analyzing genes with elevated expression in Squamous and Basal when compared with ductal-derived murine tumors, we identified 21 overlapping genes (Fig. 2A). Notably, NT5E/CD73 was one of the top overlapping genes, as shown by Venn diagram analysis (Fig. 2A; Supplementary Table S3). Using the TCGA data set, we determined high NT5E is associated with a significant reduction in the probability of survival in human PDAC patients (Fig. 2B). Ingenuity Pathway Analysis of top altered pathways increased (orange) or decreased (blue) in murine cell-of-origin tumors revealed an immunosuppressive signature in ductal-derived tumors with immune response to leukocytes, T-cell response and T-cell development all significantly downregulated in ductal-derived murine PDAC compared with acinar-derived PDAC (Fig. 2C; Supplementary Fig. S1A–S1C). We also evaluated differentially expressed genes using GO. Leukocyte cell–cell adhesion was the highest GO category identified (Benjamini–Hochberg procedure corrected PBH = 0.0004; Fig. 2D) and Nt5e was significantly elevated in this category (Fig. 2D; Supplementary Table S4). We recently published another cell-of-origin model system using an LsL-KrasG12V allele, which generates histologically relevant early and advanced PanIN and PDAC (Fig. 2E and F; ref. 36). Using IHC to label for CD73 in paraffin-embedded tissue sections from this model, we observed expression of CD73 in ductal-derived PanIN and PDAC (Fig. 2G). We performed RNA-seq on ex vivo–cultured ducts from the KCDuct model, which confirmed a significant increase in Nt5e expression in Kras-mutant ducts compared with WT ducts (Fig. 2H–J). To determine the cellularity of CD73 in human PDAC, we used IHC to stain for CD73 in human PanIN and intraductal pancreatic mucinous neoplasia (IPMN), another ductal precursor lesion. We observed epithelial CD73 expression in the neoplastic epithelium in 42% of PanIN (n = 12) and 75% of malignant IPMN (n = 4) and 54% of PDAC we analyzed (n = 44; Supplementary Fig. S2A–S2B).
Figure 1.
Comparison of ductal and acinar cell–derived tumor signatures with human molecular subtypes of PDAC. A and B, Schematic of the transgenic mouse breeding scheme to generate mutant Kras and Tp53 tumors from acinar and ductal cells and IHC analysis of CK19 to show ductal adenocarcinoma arising in ductal and acinar mouse models of PDAC. C, Principal component analysis of KPCDuct and KPCAcinar RNA-seq samples. Tumors arising in ductal and acinar cells have distinct profiles. D, Heat map shows the GSVA scores for human homolog genes derived from mouse ductal and acinar signatures in different samples from ICGC immunogenic and squamous subtypes. The color represents the relative GSVA score. E, Scatter plot of the GSVA scores for human homolog genes derived from mouse ductal and acinar signature genes in ICGC immunogenic and squamous subtypes. F, Violin plot of the GSVA scores for human homolog genes derived from mouse ductal and acinar signature genes in ICGC immunogenic and squamous subtypes. G, Scatter plot of the GSVA scores for human homolog genes derived from mouse ductal and acinar signature genes in TCGA basal and classic subtypes. The big round dots represent the medium of the GSVA scores, while the small round dots represent the score for each sample. We used a nonparametric Wilcoxon rank sum test for both groups. *, P < 0.05; **, P < 0.01. H, Heat map shows the GSVA scores for human homolog genes derived from mouse ductal and acinar signatures in different samples from TCGA basal and classic subtypes. I, Violin plot of the GSVA scores for human homolog genes derived from mouse ductal and acinar signature genes in TCGA basal and classic subtypes. J, Volcano plot showing distinct transcriptomic signatures in the duct and acinar tumors.
Figure 2.
NT5E/CD73 is highly expressed in murine ductal-derived PDAC. A, Venn diagram showing the number of top overlapping genes in murine KPCDuct and human squamous and basal subtypes. NT5E/CD73 is one of the top overlapping genes expressed between KPCDuct, squamous, and basal subtypes. B, TCGA analysis revealed high expression of NT5E/CD73 in human PDAC significantly correlates with worse prognosis. C, Ingenuity Pathway Analysis of top altered pathways increased (orange) or decreased (blue) in KPC murine cell-of-origin tumors. D, Leukocyte cell–cell adhesion was one of the top differentially expressed GO categories and nt5e was a significantly elevated gene in ductal-derived PDAC in this category. E, Schematic of a mouse model to generate PanIN and PDAC from ductal cells using an inducible KrasG12V allele. F, Representative hematoxylin and eosin and trichrome staining from KCDuct mice. G, IHC analysis of CD73 expression in KCDuct mice. H, Schematic of the experimental setup to generate whole transcriptomic profiles of Kras-mutant pancreatic ducts. I, GO heat map of highly enriched pathways in Kras-mutant ducts. Regulation of inflammatory response is one of the top GO pathways. Nt5e is elevated in GO: Regulation of inflammatory response. J, Relative RNA-seq signature of Nt5e/CD73 in ex vivo–cultured pancreatic ducts. CD73 is significantly increased in Kras-mutant pancreatic ducts. **, P < 0.01. Student t test. n = 3 per group analyzed by RNA-seq.
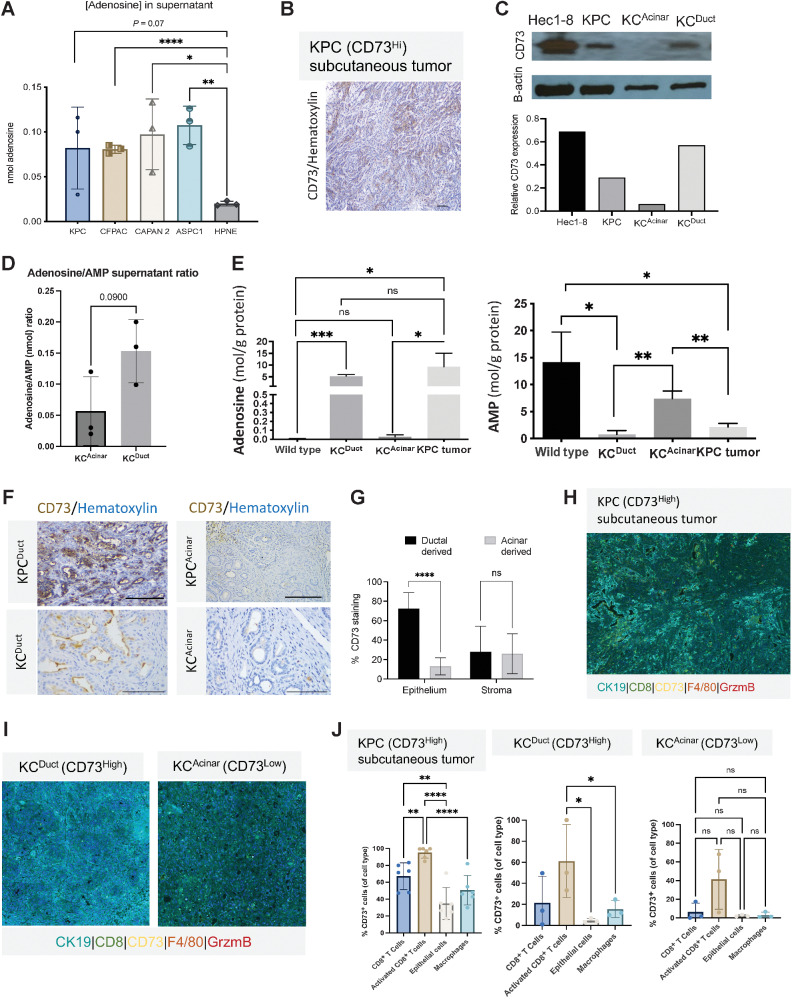
To assess CD73 activity, we analyzed adenosine levels by HPLC in the supernatant of human and murine PDAC cell lines compared with normal human pancreatic ductal HPNE cells (37). Adenosine concentrations were significantly elevated in CFPAC, CAPAN-2, and ASPC1 supernatant compared with HPNE cells (Fig. 3A; Supplementary Fig. S2C).
Figure 3.
CD73 expression levels on epithelial cells and immune cells correlate with adenosine concentrations in supernatant and tissue. KCDuct and KPC subcutaneous tumors present the highest intratumoral levels of adenosine. A, Supernatant adenosine levels are significantly increased in PDAC cell lines relative to control pancreatic cells (HPNE). B, IHC images of CD73+ KPC tumor. C, Western blot image and quantification of CD73 levels in Hec1-8 cell lines and lysates from KPC subcutaneous tumors, KCAcinar and KCDuct pancreata. D, HPLC analysis of the adenosine:AMP ratio from ex vivo–cultured KCDuct and KCAcinar pancreata at the time of euthanasia (n = 3 mice per group). Adenosine:AMP ratios are elevated in cultured KCDuct pancreata compared with KCAcinar pancreata (P = 0.09). E, HPLC analysis of adenosine and AMP levels in WT pancreas, KCDuct, or KCAcinar. N = 3 samples per group. Intrapancreatic adenosine levels are significantly elevated in KCDuct pancreata, consistent with high CD73 expression in KCDuct neoplasia and intrapancreatic AMP levels are highest in WT and KCAcinar tissue, indicating lack of CD73 activity in these tissues. F, Representative CD73 IHC showing high percentage expression of CD73 in ductal-derived PDAC from both KPCDuct and KCDuct when compared with acinar-derived PDAC from KPCAcinar and KCAcinar. G, ImageJ quantification of CD73 expression in the epithelium compared with stroma in ductal-derived compared with acinar-derived PDAC (n = 5). H and I, Representative composite multiplex immunofluorescent image from a KPC subQ tumor (H) and KCDuct and KCAcinar pancreata (I). J, Quantitative assessment of KPC subQ tumor, KCDuct and KCAcinar pancreata using multiplex immunofluorescence. In all groups, activated CD8+ T cells have the highest fluorescence intensity compared with CD8+ T cells, epithelial cells, and macrophages. The overall percentage of CD73 cells was the highest in KPC and KCDuct samples compared. Scale bars, 50 μm. A Student t test was used for statistical comparison. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
CD73 overexpression results in increased production of adenosine
To directly compare stromal and epithelial mechanisms of ductal versus acinar-derived Kras-dependent development of PanIN, we crossed the CAcinar allele to the KrasG12V allele to generate KCAcinar mice (Supplementary Fig. S3A). Similar to what we recently observed and published in pancreatic ducts expressing KrasG12V (KCDuct), KCAcinar display PanIN, fibrosis, and PDAC (Supplementary Fig. S3B). Immunolabeling for CD73 on sections from these mice revealed epithelial expression of CD73 in an average of 5% of acinar-derived PanIN and PDAC analyzed by ImageJ compared with 62% of ductal-derived PanIN and PDAC (Supplementary Fig. S3C and S3D).
IHC staining was used to show KPC subcutaneous tumors expressed CD73 (Fig. 3B). Western blots of Hec1-8 cells and lysates from KPC subcutaneous tumors or pancreata from KCAcinar and KCDuct GEM models revealed CD73 was expressed most abundantly in KPC tumors and KCDuct pancreata (Fig. 3C). We analyzed the adenosine:AMP ratios in supernatant from ex vivo–cultured KCAcinar and KCDuct tissue and quantified elevated adenosine:AMP ratio in KCDuct compared with KCAcinar pancreata (P = 0.09; Fig. 3D). To determine if elevated CD73 resulted in increased parenchymal adenosine, intrapancreatic adenosine levels from KCDuct and KCAcinar pancreata were analyzed by HPLC. Adenosine levels in KCDuct pancreata were significantly higher than WT or KCAcinar pancreata (Fig. 3D) and levels of adenosine monophosphate (AMP), the substrate for CD73, were significantly increased in WT and KCAcinar pancreata compared with KCDuct (Fig. 3E). Using ImageJ convolution software, we quantified a significant difference in epithelial, but not stromal expression of CD73 in the GEM models. Tumors arising in ductal cells had significantly higher expression of tumor-specific staining for CD73 than tumors arising in acinar cells (Fig. 3F and G). Recent studies have shown CD39 and CD73 cooperate to promote PDAC, so we evaluated the cellularity of CD39 in our cell-of-origin GEM models (38). In contrast to what we observed with CD73, for both models, we observed predominantly stromal expression of CD39 (Supplementary Fig. S4).
Activated CD8+ T cells have the highest expression of CD73 by multiplex immunofluorescent analysis
To further characterize the cellularity of CD73 expression in PDAC, we used a second method and analyzed CD73 expression on various cellular compartments using Multiplex Immunofluorescence of KPC subQ tumors and pancreata from KCDuct and KCAcinar GEM models (33). We used a stringent threshold Immunofluorescent setting to determine which cell types expressed the highest levels of CD73. These data revealed in all three GEM models that granzyme B (GZM) + CD8+ (activated) T cells presented the highest expression of CD73 (Fig. 3H–J). When comparing the overall %CD73+ cells of each cell type, we observed CD8+ T cells (nonproducers of GZM), epithelial cells and macrophages also express CD73, though at lower levels. Congruent with our adenosine HPLC measurements, KPC subQ tumors and KCDuct GEM overall had the highest %CD73+ cells of each cell type analyzed (Fig. 3J) and highlight a significant role for CD8+ T cells in the generation of adenosine within the tumor microenvironment.
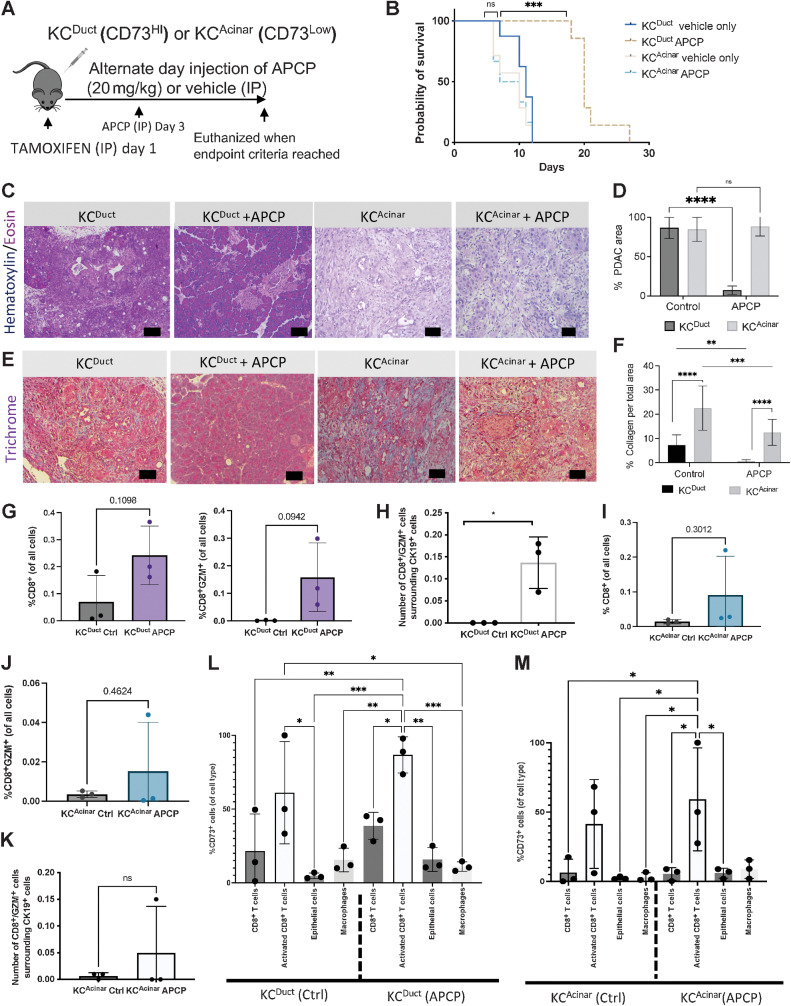
CD73 small-molecule inhibition significantly reduced spontaneous aggressive ductal-derived PDAC
To determine if overall CD73 activity was important for spontaneous ductal-derived PDAC initiation, we treated KCDuct (CD73High) and KCAcinar (CD73Low) GEM mice with adenosine 5′-(α,β-methylene) diphosphate (APCP), a small-molecule inhibitor of CD73, which began 3 days after tamoxifen injections (Fig. 4A) and continued administration every other day for the duration of the experiments. We observed a significant increase in survival in APCP-treated KCDuct, but not KCAcinar GEM mice, indicating tumors with higher levels of CD73-generated adenosine may be more sensitive to inhibition of CD73 (Fig. 4B). Standard hematoxylin and eosin evaluations were used to quantify the percentage of pancreata occupied by PDAC in vehicle-treated compared with APCP-treated mice. Inhibition of CD73 resulted in a significant reduction in surface occupied by PDAC in KCDuct pancreata, but not KCAcinar pancreata, confirming the functional relevance of CD73 in ductal cell transformation (Fig. 4C and D). In addition to hematoxylin and eosin staining, we evaluated collagen-rich stroma using trichrome staining. We quantified a significantly more percentage of collagen staining per field in KCAcinar compared with KCDuct pancreata, and we observed APCP treatment significantly reduced collagen abundance in both KCDuct and KCAcinar pancreata (Fig. 4E and F) implicating adenosine signaling regulates collagen deposition, fibrosis, and stromal remodeling in PDAC. We further characterized neoplastic epithelium by staining for CK19, pAktT308, and pErkT202/TH204. Quantification of this histology determined APCP treatment significantly reduced the percentage of the CK19+ area in KCDuct but not in KCAcinar pancreata as well as Kras downstream pathways, including PI3K and MAPK signaling (Supplementary Fig. S5A–S5D), consistent with recent in vitro studies showing CD73 activity promotes PI3K and MAPK signaling in PDAC cell lines (39). Using IHC, we also observed a significant increase in GZM+ or CD8α+ cells in APCP-treated KCDuct mice and, while we observed an increase in GZM+ cells after APCP treatment, the increase did not reach significance in KCAcinar mice (Supplementary Fig. S5E and S5F).
Figure 4.
Inhibition of CD73 using intraperitoneal delivery of APCP significantly reduces spontaneous aggressive ductal-derived PDAC. A, Schematic of the preclinical model to evaluate the requirement for CD73 in a spontaneous GEM model of ductal-derived PDAC. B, APCP significantly improved survival only in KCDuct GEM mice (n = 5 mice per group). C, Representative hematoxylin and eosin staining of vehicle-treated compared with APCP-treated KCDuct and KCAcinar pancreata. D, Inhibition of CD73 significantly reduced PDAC area in KCDuct (n = 2) compared with control (n = 3) but not KCAcinar GEM mice (n = 2) compared with control (n = 2). E, Trichrome images showing PDAC areas in KCDuct and KCAcinar GEM mice in both vehicle and APCP-treated groups. F, APCP treatment significantly reduced the percentage of collagen in both KCDuct and KCAcinar pancreata. A two-way ANOVA was used to compare groups. G, Quantification of multiplex immunofluorescent analysis of %CD8+ cells or %CD8+GZM+ cells per whole tissue on the section from KCDuct Ctrl- and APCP-treated mice (n = 3). H, Spatial quantification showing a significant increase in CD8+GZM+ cells within 80 μm radius of CD19+ cells in APCP-treated KCDuct pancreata compared with Ctrl KCDuct (n = 3). I and J, Quantification of multiplex immunofluorescent analysis of %CD8+ cells (I) or %CD8+GZM+ cells (J) per field in pancreata from KCAcinar Ctrl- and APCP-treated mice (n = 3). K, Spatial quantification showing an increase in CD8+GZM+ cells within 80 μm radius of CD19+ cells in APCP-treated KCAcinar pancreata compared with Ctrl KCAcinar (n = 3). L and M, Quantification of %CD73+ cells per cell type in Ctrl versus APCP-treated samples. In KCDuct, but not KCAcinar, we quantified a 2-fold increase in CD73+CD8+ T cells, 1.5-fold increase in CD73+CD8+GZM+ T cells, and a 4-fold increase in CD73+ epithelium in pancreata from APCP-treated mice (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Ductal and acinar–derived tumors have divergent changes in CD73+ immune and epithelial cell expression in response to APCP treatment
Multiplex immunofluorescent data were again generated to immune profile the GEM models either with vehicle (Ctrl) treatment or APCP treatment. APCP treatment increased the %CD8a+ and %CD8a+GZM+ T cells in APCP-treated KCDuct and KCAcinar tumors (Fig. 4G–J), increased the %F4/80+ cells and decreased the %CK19+ cells (Supplementary Fig. S6). Using spatial immunoprofiling on the pancreatic sections, we quantified a significant increase in the number of CD8+GZM+ T cells surrounding CK19+ cells, within 80-μm radius, in APCP-treated KCDuct (Fig. 4H), but not APCP-treated KCAcinar pancreata compared with pancreata from vehicle-treated mice (Fig. 4K). We also evaluated differences in %CD73+ cells after APCP treatment in these mice. In the APCP-treated KCDuct pancreata compared with Ctrl-treated KCDuct pancreata, we quantified a 1.7-fold increase in CD73+CD8+ cells, a 1.4-fold increase in CD73+CD8+GZM+ cells and a 4-fold increase in CD73+ epithelial cells and no change in CD73+F4/80+ macrophages (Fig. 4L). We quantified a similar 1.4-fold increase in CD73+CD8+GZM+ cells in APCP-treated KCAcinar compared with Ctrl-treated pancreata, a 1.8-fold increase in CD73+ epithelium and a 9-fold increase in CD73+F4/80+ macrophages (Fig. 4M). These data implicate a mechanism of therapeutic resistance to CD73 inhibitors as well as changes in the tumor microenvironment that may be dependent on the PDAC subtype.
In KPC subQ tumors, inhibition of CD73 significantly elevates intratumoral-activated CD4+ and CD8+ T cells
We tested two CD73 small-molecule inhibitors using syngeneic models with murine PDAC cells generated from Pdx:Cre; LsL-KrasG12D; LsL-Trp53R173H (KPC) mice. Our data revealed that peritumor APCP treatment significantly decreases the final tumor volume (Supplementary Fig. S7A–S7D), intratumoral adenosine levels in vivo (Supplementary Fig. S7E–S7G). Using flow-cytometry analysis, we observed significantly increased CD8α+TCR αβ+ cells in the spleens of APCP-treated mice (Supplementary Fig. S7H). Quantitative analysis of IHC to label CD8α+ T cells in control- compared with APCP-treated tumors showed a significant increase in CD8α+ T cells in APCP-treated compared with control-treated tumors (Supplementary Fig. S7I–7S7J) and flow-cytometry analysis revealed a significant increase in infiltration of CD45+ and T cells in tumors upon APCP treatment versus control (Supplementary Fig. S7K and S7L). Alanine transaminase (AST) and aspartate aminotransferase (AST) analysis showed no evidence of liver toxicity in AB680-treated mice (Supplementary Fig. S7M).
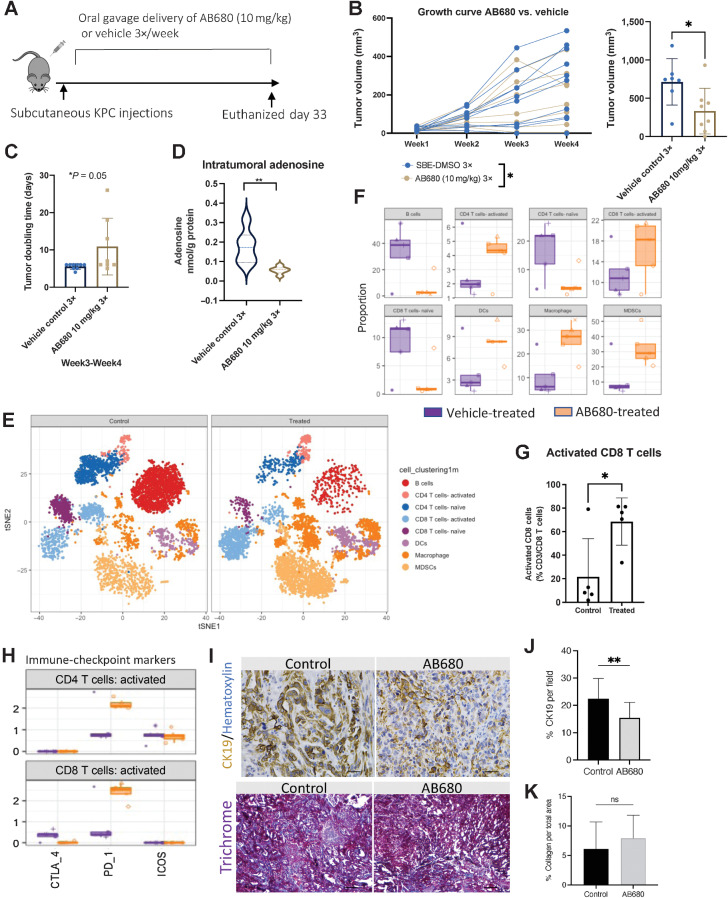
Clinical trials using AB680 from Arcus Bioscience have recently shown AB680 in combination with NP/GEM plus zimberelimab had an overall response rate of 41% (40). Using the KPC subcutaneous model, we initiated oral gavage delivery of AB680 3 days per week until mice needed to be euthanized (Fig. 5A). AB680 treatment significantly reduced tumor growth rates and tumor volume compared with vehicle-treated alone (Fig. 5B) and significantly increased tumor doubling time (Fig. 5C). HPLC analysis confirmed a significant decrease in intratumoral adenosine levels in AB680-treated mice (Fig. 5D). Cytometry time-of-flight (CyTOF) analysis revealed a significant increase in activated CD8+ T cells, activated CD4+ T cells, dendritic cells, macrophages, and myeloid-derived suppressor cells (MDSC; Fig. 5E–G; Supplementary Fig. S8A and S8B). Analysis of immune-checkpoint markers indicated that activated CD4+ and CD8+ T cells increased expression of PD-1 in AB680-treated mice (Fig. 5H). We used IHC to stain for CK19 and quantified a significant reduction in CK19+ staining per field in AB680-treated compared with vehicle-treated tumors (Fig. 5I and J). Trichrome staining did not show a difference in collagen levels in AB680-treated compared with vehicle-treated tumors (Fig. 5I–K).
Figure 5.
AB680 oral gavage treatment reduces tumor KPC tumor growth rate and elevates intratumoral activated CD8+ T cells. A, KPC subcutaneous tumors were analyzed weekly (n = 10 per group). B, AB680 treatment significantly decreased KPC subcutaneous growth rates and at the conclusion of this experiment, AB680-treated mice had significantly smaller tumor volume than vehicle control-treated mice. *, P < 0.05. Statistical analysis was performed using a Student t test in Prism GraphPad software. C, Graphical representation of subcutaneous tumor growth rates for analysis of individual tumor doubling time. n = 6. *, P < 0.05. D, HPLC analysis shows a significant decrease in adenosine levels in tumors from AB680-treated mice versus vehicle-treated mice. **, P < 0.01 compared with vehicle controls (n = 8 per group). E, CyTOF vSNE plots by group show elevated clusters of activated CD8 T cells, CD4 T cells, MDSCs, and macrophages. F, Quantitative global population analysis reveals increased activated CD4 T cells, CD8 T cells, macrophages, and MDSCs. G, Quantitative analysis of significantly increased activated CD8 T cells. *, P < 0.05. A Student test using Prism GraphPad software was used to calculate statistics. H, Activated CD4 and CD8 T cells increased expression of PD-1. I, Representative CK19 and trichrome staining of tumors from control- (vehicle) and AB680-treated mice. J and K, Tumors from AB680-treated mice had significantly reduced expression of CK19 (**, P < 0.01; J) and no significant difference (ns) in % collagen per total area (K). Scale bars, 50 μm.
Adenosine signaling through Adora2b promotes PDAC progression
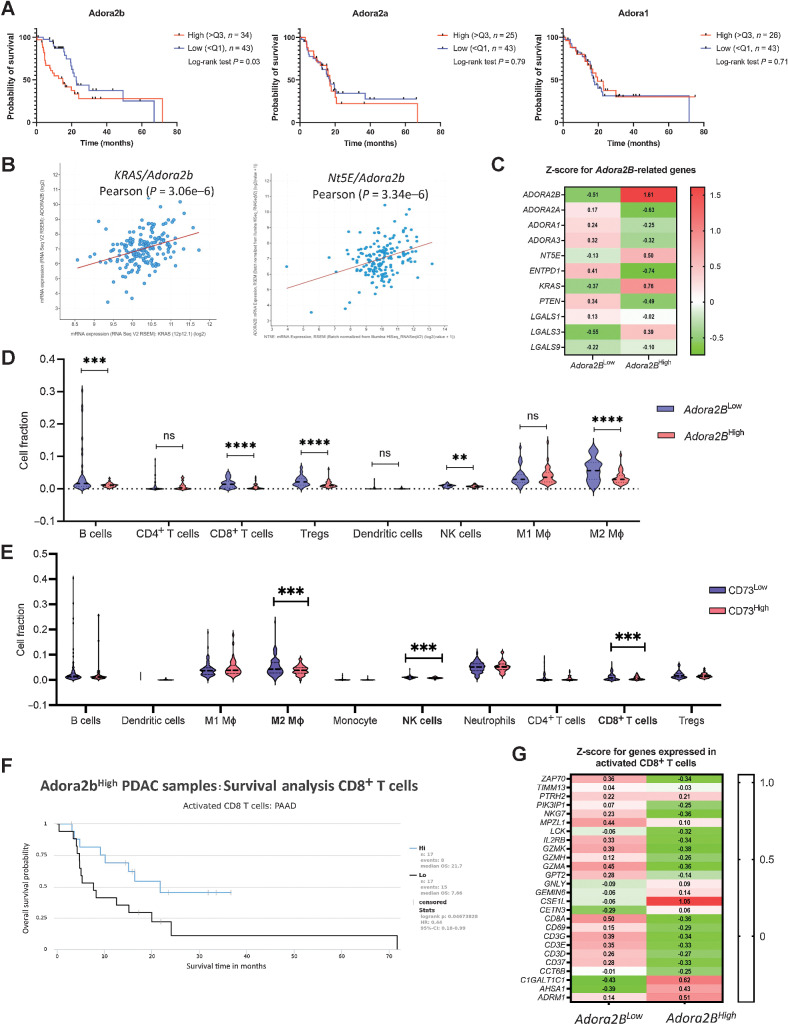
Adenosine signaling through Adora receptors on immune cells has been shown to regulate their function (41). Expression of Adora2b on myeloid cells and antigen-presenting cells indirectly suppresses CD8+ T-cell responses and promotes metastasis in syngeneic ectopic solid tumors (42). We analyzed TCGA data for ADORA1, ADORA2a, and ADORA2b. High expression of ADORA2b was correlated with a significant reduction in overall survival in human PDAC (Fig. 6A). We used cBioPortal to define the correlation of KRAS/Adora2b and NT5E/Adora2b, and we observed both KRAS and NT5E expression levels are correlated with high expression of Adora2b (Fig. 6B). These data were confirmed by heat map analysis of Adora2b-related genes in patients with Adora2bHigh and Adora2bLow expression levels (Fig. 6C). Protein expression as analyzed by IHC and in a tissue microarray with samples from patients with high and low Adora2b expression, and we evaluated correlative levels of CD73 and Adora2b in human PDAC cell lines by Western blot (Supplementary Fig. S9A–S9C). To analyze the distribution of immune cells in these two groups, the Quantiseq algorithm was utilized, and data were extracted from The Cancer Immunome Database. Patients with high Adora2b expression had a significantly lower abundance of B cells, CD8+ T cells, T regulatory cells, NK cells, and M2 macrophages, consistent with a potent immunosuppressive TME (Fig. 6D). We observed similar data for patients with high NT5E expression (Fig. 6E). In addition, patients with High Adora2b expression who have a lower abundance of activated CD8+ T cells also have a worse prognosis, indicating the Adora2b receptor regulates CD8+ T-cell activation (Fig. 6F). Analysis of genes expressed in activated CD8+ T cells confirmed lower overall expression of activated CD8+ T-cell gene expression in patients with high Adora2b expression (Fig. 6G).
Figure 6.
Adenosine signaling through Adora2b on CD8+ T cells correlates with reduced survival and PDAC progression. A, TCGA analysis revealed that high expression of Adora2b human PDAC significantly correlates with worse prognosis (left). No differences were found in Adora2a (middle) or Adora1 (right). B,Adora2b expression in human PDAC positively correlates with Kras and Nt5E. C, Z score comparison of Adora2b-related genes shows a positive correlation with Kras and Nt5E, whereas a negative correlation was observed for Adora2a, Adora1, Adora3, Entpd1/CD39, and Pten. Z scores were downloaded from TCGA–PAAD cBioPortal. D, To analyze the distribution of immune cells in patients with high (n = 26) and low (n = 46) Adora2b expression in two groups, data analyzed with the Quantiseq algorithm were downloaded from The Cancer Immunome Database. Patients with high Adora2b expression presented a decreased abundance of B cells, CD8+ T cells, Tregs, NK cells, and M2 macrophages. Student t test was used to analyze significance. E, Similar results were observed when analyzing Immunome atlas data from patients with high (n = 44) and low (n = 103) expression levels of NT5E. Student t test was used to analyze significance. F, Patients with high Adora2b expression who have a lower abundance of activated CD8+ T cells present a worse prognosis. G, Analysis of genes expressed in activated CD8+ T cells confirmed lower overall expression of activated CD8+ T-cell gene expression in patients with high Adora2b expression. Z scores were extracted from The Cancer Immunome Database and TCGA–PAAD cBioPortal. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Antitumoral effect of Adora2b inhibition is CD8+ T-cell dependent
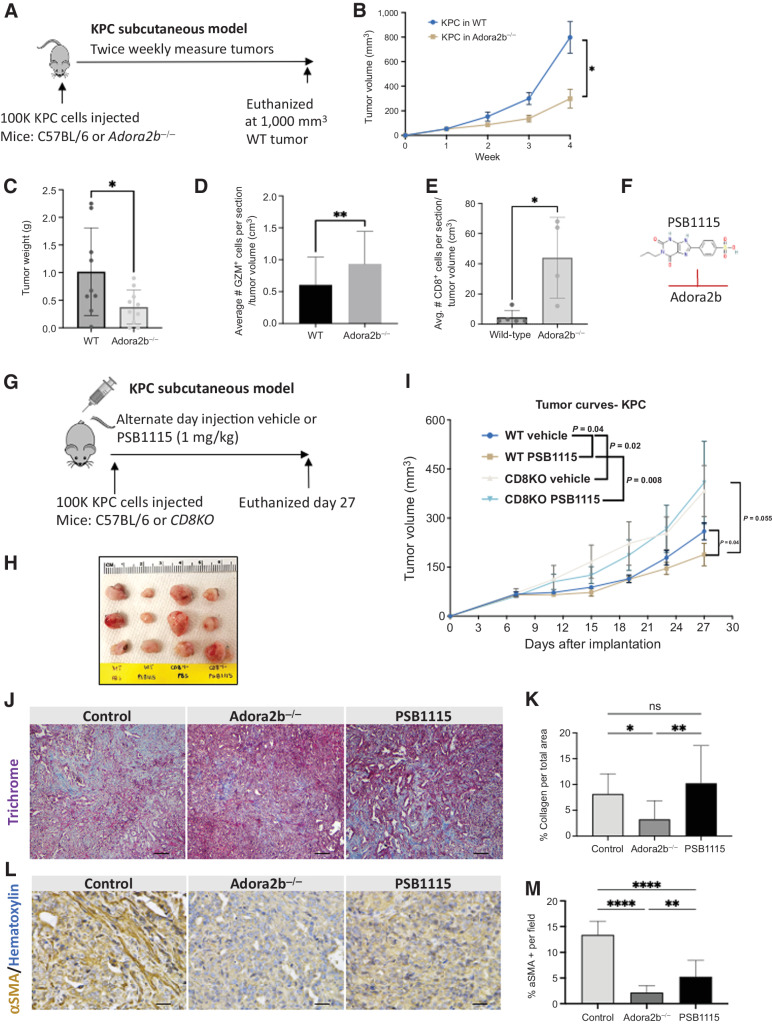
To study the role of paracrine adenosine signaling, we implanted KPC cells into WT and Adora2b−/− mice and monitored tumor growth rates (Fig. 7A). KPC subcutaneous tumors grew significantly slower in Adora2b−/− mice compared with WT mice (Fig. 7B). At euthanasia, analysis of final tumor weight confirmed a significant difference in tumors grown in Adora2b−/− compared with WT mice (Fig. 7C). KPC tumors grown in Adora2b−/− mice had significantly more CD8+ T cells and GZM+ cells than tumors in WT mice (Fig. 7D and E). To determine if activated CD8+ T cells were important for Adora2b-mediated immunosuppression, we implanted KPC cells into WT or CD8KO mice and treated both groups with either a vehicle control or an Adora2b small-molecule inhibitor, PSB1115 (1 mg/kg; Fig. 7F and G). PSB1115 treatment significantly reduced the growth rate of KPC tumors in WT mice; however, the effect of the inhibitor was lost in the CD8KO mice, indicating reduced tumor growth by inhibiting adenosine/Adora2b signaling is dependent on activated CD8+ T cells (Fig. 7H and I).
Figure 7.
Antitumoral effect of Adora2b inhibition is CD8+ T-cell dependent. A, Schematic of KPC subcutaneous preclinical model to evaluate the role of Adora2b in PDAC tumor development. B, Mice lacking Adora2b (Adora2b−/−) presented delayed KPC subQ tumor growth compared with WT mice and (C) decreased tumor weight compared with WT mice. D and E, Tumors that developed in mice lacking Adora2b (Adora2b−/−) presented significantly increased GZM staining per tumor volume compared with WT mice (n = 14 fields/group; D) and presented significantly increased CD8+ T cells (E). F and G, Chemical structure of PSB1115 (F), Adora2b inhibitor, and schematic of KPC subcutaneous preclinical model (G) to evaluate the role of Adora2b in PDAC tumor development. H and I, Inhibition of Adora2b with PSB1115 reduces tumor growth rate only when injected in WT mice. The effect is lost when KPC cells are injected in CD8KO mice (n = 8 mice per group). J, Trichrome staining to evaluate collagen abundance in control, Adora2b−/−, and PSB1115-treated tumors. K, KPC tumors growing in Adora2b−/− had significantly reduced collagen compared with control tumors; however, there was no difference in the percentage of collagen per field in tumors from PSB1115-treated mice (n = 12 fields analyzed per group). L, IHC for αSMA to evaluate stromal changes in KPC tumors from Adora2b−/− and tumors from PSB1115-treated mice. M, The percentage of αSMA+ staining was significantly decreased in KPC tumors grown in Adora2b−/− mice and in tumors from PSB1115-treated mice. n = 15 sections per group. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, nonsignificant.
To determine if adenosine signaling through the Adora2b receptor altered the stromal composition of KPC subQ tumors, we stained for trichrome and α smooth muscle actin (αSMA). Tumors from Adora2b−/− had significantly reduced collagen and αSMA compared with Control (vehicle-treated) tumors (Fig. 7J–M). Tumors from PSB1115-treated mice presented a significant reduction in %αSMA + per field (Fig. 7K). These data indicate that the adenosine/Adora2b axis also promotes fibrosis in pancreatic cancer.
Discussion
In normal pancreas, studies have reported normal pancreas does not express CD73, indicating in physiologic conditions, CD73 levels are low in exocrine pancreatic cells (39, 43, 44). In contrast, in PDAC specimens, CD73 is highly expressed in a subset of patients and correlates with immunosuppression and poor prognosis (43). In our GEM models, we observe high immunolabeling for CD73 in the luminal side of PDAC arising in ductal cells, but less common in metaplastic acinar cells. Using multiplex immunofluorescence on sections from KPC subQ tumors and ductal and acinar-derived PDAC, we observe infiltration of CD73+CD8+, CD73+F4/80+ macrophages, and we quantified CD73+CD8+GZM+ cells have the highest expression of CD73. These data indicate in PDAC that CD8+ T cells also generate adenosine in the tumor microenvironment. CD8+ T cells express adenosine receptors, suggesting a cell-autonomous immunosuppressive mechanism on CD8+ T cells. CD73+CD8+ T cells have previously been characterized as having reduced antitumor capacity and, through adenosine-enriched exosomes, may reduce responses to immunotherapy (45, 46). However, in other contexts, CD73+T cells define a subset of long-lived immune memory T cells that may be critical for the adaptive response and augment immunotherapy in PDAC patients (47). Overall, our preclinical data indicate patients with higher CD73 may respond better clinically to CD73 inhibitors.
These data are timely as the role of extracellular adenosine signaling in pancreatic cancer immunosuppression has only recently been evaluated in orthotopic KPC tumor models (13, 38). In the studies by King and colleagues, Nt5e knockdown KPC tumors had decreased levels of GM-CSF and elevated infiltration of CD4+ and CD8+ T cells expressing IFNy. In these studies, CD4+ T-cell depletion, but not CD8+ T-cell depletion, abrogated the effects of Nt5e knockdown. In contrast, in our GEM and KPC models and experiments using multiplex immunofluorescent and CyTOF, we consistently observed elevated CD8+ T cells, and we show adenosine signaling through Adora2b is a critical determinant of the CD8+ T-cell antitumor response. In addition, we show that Adora2b promotes fibrosis and collagen deposition in the tumor microenvironment.
Activation of immune cells after CD73 inhibition, concomitant with reduced spontaneous PDAC arising in pancreatic ducts, highlights cell-autonomous and non–cell-autonomous mechanisms of extracellular adenosine signaling are critical to the development of PDAC. Our studies show that Adora2b high expression positively correlates with both KRAS and NT5E expression in PDAC patients, and elevated Adora2b expression in tumors is associated with poor prognosis. We observe patients with elevated Adora2b present a lower abundance of B cells, CD8+ T cells, T regulatory cells, NK cells, and M2 macrophages, reinforcing Adora2b plays a key role in mediating PDAC immunosuppression. In addition, by analyzing KPC tumor growth in Adora2b−/− mice and using small-molecule inhibitors in CD8KO mice we find Adora2b mediates antitumor immunity, which is dependent, at least in part, on activated CD8+ T cells. Future experiments will determine the role of CD73 on CD8+GZM+ T cells in response to immunotherapy. These data indicate coinhibition of CD73 and Adora2b may provide a stronger CD8+ T-cell–mediated antitumor response for patients with PDAC.
Supplementary Material
Supplementary Figures S1-9
Table S2
Table S1
Table S4
Table S3
Acknowledgments
F. McAllister received support from the V Foundation (Translational Award), NCI R37 (CA237384), and the Cancer Prevention and Research Institute of Texas (RP200173). This work was partially supported by the Cancer Prevention and Research Institute of Texas (CPRIT RP180734), National Cancer Institute (NCI) Division of Cancer Prevention PREVENT Program Base Contract No. 75N91019D00021 “Preclinical Drug Development Program: Preclinical Efficacy and Intermediate Biomarkers” (J.M. Bailey-Lundberg, F. McAllister, and P.H. Brown), Texas Medical Center Digestive Disease Center Pilot Award 2P30DK056338-16 (J.M. Bailey-Lundberg), and the work was partially supported by a 2011 Pancreatic Cancer Action Network–AACR Pathway to Leadership Grant, grant number 11-70-25-BAIL (J.M. Bailey-Lundberg). This project was also supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine, with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672) and the NIH (CA125123 and R01LM012806). The authors thank Joel M. Sederstrom for assistance. H.K. Eltzschig received support from NIH grants R01HL154720, R01DK122796, and R01HL133900.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
N.C. Thosani reports being a consultant for Boston Scientific Corp., Ambu Inc., and Pentax America, a speaker for AbbVie, and has received royalty from UpToDate. D. Bar-Sagi reports grants from NCI during the conduct of the study; grants from Pancreatic Cancer Action Network, TEZCAT Laboratories, Lustgarten Foundation, personal fees from Starr Cancer Consortium, Wistar Institute, and Samumed LLC outside the submitted work. K.L. Poulsen reports grants from NIH during the conduct of the study. M.I. Savage reports grants from NCI during the conduct of the study. Z. Zhao reports grants from the Cancer Prevention and Research Institute of Texas and NIH during the conduct of the study. P.H. Brown reports support from Genetex outside the submitted work. F. McAllister reports being a paid SAB member at Neologics Bio. No disclosures were reported by the other authors.
Authors' Contributions
E.Y. Faraoni: Conceptualization, data curation, formal analysis, writing–review and editing. K. Singh: Conceptualization, data curation, formal analysis, writing–review and editing. V. Chandra: Data curation, formal analysis, writing–review and editing. O. Le Roux: Data curation, formal analysis. Y. Dai: Data curation, formal analysis. I. Sahin: Data curation, formal analysis. B.J. O'Brien: Data curation. L.N. Strickland: Data curation, writing–review and editing. L. Li: Data curation. E. Vucic: Data curation, writing–review and editing. A.N. Warner: Data curation. M. Pruski: Data curation. T. Clark: Data curation. G. Van Buren: Resources. N.C. Thosani: Writing–review and editing. J.S. Bynon: Resources. C.J. Wray: Writing–review and editing. D. Bar-Sagi: Formal analysis. K.L. Poulsen: Writing–review and editing. L.A. Vornik: Project administration. M.I. Savage: Project administration, writing–review and editing. S. Sei: Project administration, writing–review and editing. A. Mohammed: Project administration, writing–review and editing. Z. Zhao: Formal analysis, writing–review and editing. P.H. Brown: Writing–review and editing. T. Mills: Data curation. H.K. Eltzschig: Resources, writing–review and editing. F. McAllister: Conceptualization, resources, data curation, supervision, funding acquisition, writing–review and editing. J.M. Bailey-Lundberg: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, writing original draft.
References
- 1. Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G, et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell 2021;39:1342–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol Hepatol 2020;17:487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katz MHG, Shi Q, Meyers J, Herman JM, Chuong M, Wolpin BM, et al. Efficacy of preoperative mFOLFIRINOX vs mFOLFIRINOX plus hypofractionated radiotherapy for borderline resectable adenocarcinoma of the pancreas: the A021501 phase 2 randomized clinical trial. JAMA Oncol 2022;8:1263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grenz A, Zhang H, Eckle T, Mittelbronn M, Wehrmann M, Köhle C, et al. Protective role of ecto-5'-nucleotidase (CD73) in renal ischemia. J Am Soc Nephrol 2007;18:833–45. [DOI] [PubMed] [Google Scholar]
- 5. Synnestvedt K, Furuta GT, Comerford KM, Louis N, Karhausen J, Eltzschig HK, et al. Ecto-5'-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest 2002;110:993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Brien BJ, Faraoni EY, Strickland LN, Ma Z, Mota V, Mota S, et al. CD73-generated extracellular adenosine promotes resolution of neutrophil-mediated tissue injury and restrains metaplasia in pancreatitis. FASEB J 2023;37:e22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burnstock G, Novak I. Purinergic signalling in the pancreas in health and disease. J Endocrinol 2012;213:123–41. [DOI] [PubMed] [Google Scholar]
- 8. Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature 2014;509:310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MA, Muskens F, et al. Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nat Med 2007;13:913–9. [DOI] [PubMed] [Google Scholar]
- 10. Cowen DS, Lazarus HM, Shurin SB, Stoll SE, Dubyak GR. Extracellular adenosine triphosphate activates calcium mobilization in human phagocytic leukocytes and neutrophil/monocyte progenitor cells. J Clin Invest 1989;83:1651–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Warburton D, Buckley S, Cosico L. P1 and P2 purinergic receptor signal transduction in rat type II pneumocytes. J Appl Physiol (1985) 1989;66:901–5. [DOI] [PubMed] [Google Scholar]
- 12. Hay CM, Sult E, Huang Q, Mulgrew K, Fuhrmann SR, McGlinchey KA, et al. Targeting CD73 in the tumor microenvironment with MEDI9447. Oncoimmunology 2016;5:e1208875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King RJ, Shukla SK, He C, Vernucci E, Thakur R, Attri KS, et al. CD73 induces GM-CSF/MDSC-mediated suppression of T cells to accelerate pancreatic cancer pathogenesis. Oncogene 2022;41:971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faraoni EY, Ju C, Robson SC, Eltzschig HK, Bailey-Lundberg JM. Purinergic and adenosinergic signaling in pancreatobiliary diseases. Front Physiol 2022;13:849258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faraoni EY, Strickland LN, O'Brien BJ, Barraza JF, Thosani NC, Wray CJ, et al. Radiofrequency ablation in combination with CD73 inhibitor AB680 reduces tumor growth and enhances anti-tumor immunity in a syngeneic model of pancreatic ductal adenocarcinoma. Front Oncol 2022;12:995027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen S, Fan J, Zhang M, Qin L, Dominguez D, Long A, et al. CD73 expression on effector T cells sustained by TGF-β facilitates tumor resistance to anti-4–1BB/CD137 therapy. Nat Commun 2019;10:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J, Lupo KB, Chambers AM, Matosevic S. Purinergic targeting enhances immunotherapy of CD73. J Immunother Cancer 2018;6:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sek K, Mølck C, Stewart GD, Kats L, Darcy PK, Beavis PA. Targeting adenosine receptor signaling in cancer immunotherapy. Int J Mol Sci 2018;19:3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma SR, Deng WW, Liu JF, Mao L, Yu GT, Bu LL, et al. Blockade of adenosine A2A receptor enhances CD8. Mol Cancer 2017;16:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research Network. Electronic address aadhe, cancer genome atlas research n. integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017;32:185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao Y, Wang J, Jaehnig EJ, Shi Z, WebGestalt ZB. 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019;47:W199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 2016;35:4282–8. [DOI] [PubMed] [Google Scholar]
- 27. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FJ, Guglietta S, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 2017;6:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ji B, Tsou L, Wang H, Gaiser S, Chang DZ, Daniluk J, et al. Ras activity levels control the development of pancreatic diseases. Gastroenterology 2009;137:1072–82, 82.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mehrara E, Forssell-Aronsson E, Ahlman H, Bernhardt P. Specific growth rate versus doubling time for quantitative characterization of tumor growth rate. Cancer Res 2007;67:3970–5. [DOI] [PubMed] [Google Scholar]
- 31. Reichert M, Rhim AD, Rustgi AK. Culturing primary mouse pancreatic ductal cells. Cold Spring Harb Protoc 2015;2015:558–61. [DOI] [PubMed] [Google Scholar]
- 32. Knudsen TB, Winters RS, Otey SK, Blackburn MR, Airhart MJ, Church JK, et al. Effects of (R)-deoxycoformycin (pentostatin) on intrauterine nucleoside catabolism and embryo viability in the pregnant mouse. Teratology 1992;45:91–103. [DOI] [PubMed] [Google Scholar]
- 33. Quesada PR, Riquelme E, Burks J, Rakoski A, Sahin I, McAllister F. Assessment of the murine tumor microenvironment by multiplex immunofluorescence. Methods Mol Biol 2022;2435:107–27. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y, Chandra V, Riquelme Sanchez E, Dutta P, Quesada PR, Rakoski A, et al. Interleukin-17-induced neutrophil extracellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J Exp Med 2020;217:e20190354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flowers BM, Xu H, Mulligan AS, Hanson KJ, Seoane JA, Vogel H, et al. Cell of origin influences pancreatic cancer subtype. Cancer Discov 2021;11:660–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh K, Pruski M, Bland R, Younes M, Guha S, Thosani N, et al. Kras mutation rate precisely orchestrates ductal derived pancreatic intraepithelial neoplasia and pancreatic cancer. Lab Invest 2021;101:177–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res 2007;67:2098–106. [DOI] [PubMed] [Google Scholar]
- 38. Jacoberger-Foissac C, Cousineau I, Bareche Y, Allard D, Chrobak P, Allard B, et al. CD73 inhibits cGAS-STING and cooperates with CD39 to promote pancreatic cancer. Cancer Immunol Res 2023;11:56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou L, Jia S, Chen Y, Wang W, Wu Z, Yu W, et al. The distinct role of CD73 in the progression of pancreatic cancer. J Mol Med (Berl) 2019;97:803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blocking CD73 can shrink pancreatic tumors. Cancer Discov 2021;11:OF4. [DOI] [PubMed] [Google Scholar]
- 41. Cekic C, Day YJ, Sag D, Linden J. Myeloid expression of adenosine A2A receptor suppresses T and NK cell responses in the solid tumor microenvironment. Cancer Res 2014;74:7250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen S, Akdemir I, Fan J, Linden J, Zhang B, Cekic C. The expression of adenosine A2B receptor on antigen-presenting cells suppresses CD8. Cancer Immunol Res 2020;8:1064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao J, Soto LMS, Wang H, Katz MH, Prakash LR, Kim M, et al. Overexpression of CD73 in pancreatic ductal adenocarcinoma is associated with immunosuppressive tumor microenvironment and poor survival. Pancreatology 2021;21:942–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sciarra A, Monteiro I, Ménétrier-Caux C, Caux C, Gilbert B, Halkic N, et al. CD73 expression in normal and pathological human hepatobiliopancreatic tissues. Cancer Immunol Immunother 2019;68:467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shevchenko I, Mathes A, Groth C, Karakhanova S, Müller V, Utikal J, et al. Enhanced expression of CD39 and CD73 on T cells in the regulation of anti-tumor immune responses. Oncoimmunology 2020;9:1744946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider E, Winzer R, Rissiek A, Ricklefs I, Meyer-Schwesinger C, Ricklefs FL, et al. CD73-mediated adenosine production by CD8 T cell-derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nat Commun 2021;12:5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fang F, Cao W, Zhu W, Lam N, Li L, Gaddam S, et al. The cell-surface 5'-nucleotidase CD73 defines a functional T memory cell subset that declines with age. Cell Rep 2021;37:109981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-9
Table S2
Table S1
Table S4
Table S3
Data Availability Statement
All raw RNA-seq data and processed read counts have been submitted to Gene Expression Omnibus (GSE189130). We obtained RNA-seq data sets and differentially expressed genes signatures from two independent PDAC patient cohorts, primary PDACs of high cellularity from the Australian International Cancer Genome Initiative (ICGC; accessed on April 21, 2021) and primary PDACs from TCGA Research Network (accessed through TCGA data portal on April 22, 2021; refs. 21, 22). The Cancer Immunome Database was used to identify immune cell distributions. The data generated in this study are available within the article and its Supplementary Data files or from the corresponding author upon reasonable request.