Abstract
The reversible acetylation of histone lysine residues is controlled by the action of acetyltransferases and deacetylases (HDACs) which regulate chromatin structure and gene expression. The sirtuins are a family of NAD-dependent HDAC enzymes and one member, sirtuin 6, Sirt6, influences DNA repair, transcription, and aging. Here we demonstrate that Sirt6 is efficient at deacetylating several histone H3 acetylation sites, including its canonical site Lys9, in the context of nucleosomes but not free acetylated histone H3 protein substrates. By installing a chemical warhead at the Lys9 position of histone H3, we trap a catalytically poised Sirt6 in complex with a nucleosome and employ this in cryo-EM structural analysis. The structure of Sirt6 bound to a nucleosome reveals extensive interactions between distinct segments of Sirt6 and the H2A/H2B acidic patch and nucleosomal DNA, which accounts for the rapid deacetylation of nucleosomal H3 sites and the disfavoring of histone H2B acetylation sites. These findings provide a new framework for understanding how HDACs target and regulate chromatin.
Keywords: Sirtuin 6, Histone, Nucleosome, Cryo-EM, NAD
Graphical Abstract
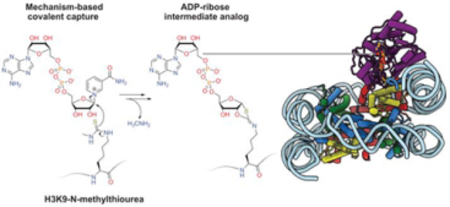
INTRODUCTION
Reversible lysine acetylation modifications on histones and other proteins are a major and critical class of post-translational modifications that regulate chromatin structure, gene expression, and numerous biological processes (Figure 1A).1 In humans, there are 18 established histone deacetylases (HDACs, aka KDACs), which include 11 metallohydrolase HDACs2 and 7 NAD-dependent HDACs known as sirtuins.3 The dysregulation of HDACs in cancer and other diseases has led to drug discovery efforts to identify HDAC modulators and several metallohydrolase HDAC inhibitors are approved for the treatment of cutaneous T cell lymphoma and other malignancies.4 Sirtuin inhibitors5 and activators6 have also been intensively pursued for various therapeutic applications.7
Figure 1.
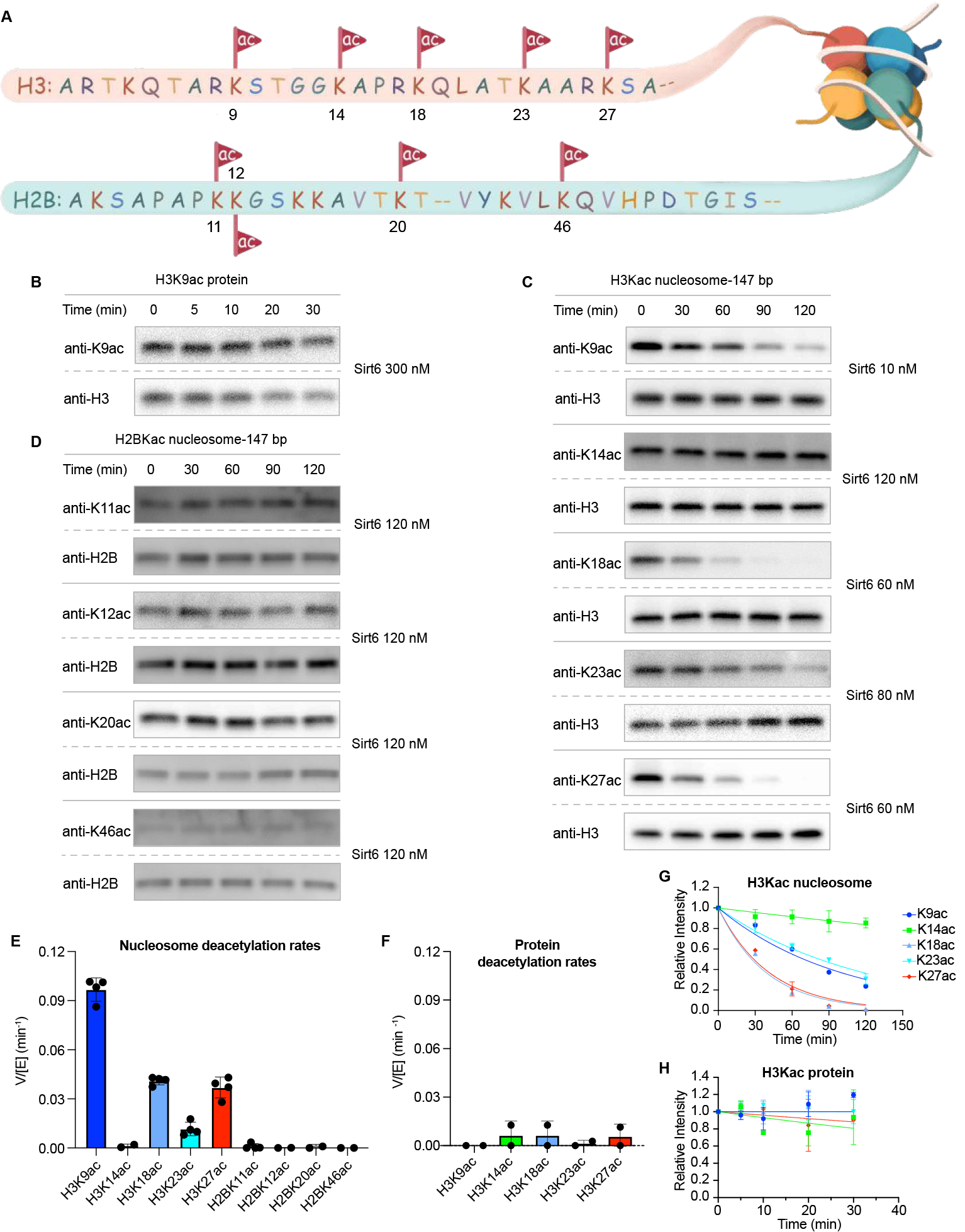
Sirt6 nucleosome deacetylation kinetics. (A) Illustration of histone H3 and H2B N-terminal acetylation sites and surrounding sequence. (B) Western blot analysis of Sirt6 deacetylation of free H3K9ac protein. Enzyme concentrations (right) were selected to maintain diminishing H3K9ac intensity in the antibody linear range. (C) Western blot analysis of Sirt6 deacetylation of multiple nucleosome H3Kac sites. Enzyme concentrations (right) were selected to maintain diminishing H3K9ac intensity in the antibody linear range. (D) Western blot analysis of Sirt6 deacetylation of multiple nucleosome H2BKac sites. Enzyme concentrations (right) were selected to maintain diminishing H2BK9ac intensity in the antibody linear range. (E) Comparison of Sirt6 V/[E] for different nucleosome H3Kac and H2BKac sites. (F) Comparison of Sirt6 V/[E] for different free protein H3Kac sites (G) Deacetylation rate curve fitting for different nucleosome H3Kac sites. (H) Deacetylation rate curve fitting for different free protein H3Kac sites (n=2–4).
Although the site-specificities of particular HDACs for various histone acetylation sites has been identified, the mechanistic details of how deacetylation occurs in the context of chromatin are generally lacking.8 In this study, we focus on sirtuin 6, Sirt6. Sirt6 is a 355 amino acid-long enzyme that has been shown to play a critical role in DNA damage response, modulating transcriptional pause release, and has been found to show altered activity in various diseases.9 Increased Sirt6 activity has been shown to enhance lifespan in Drosophila10 and mice,11 and DNA sequence polymorphisms in humans have been correlated with altered longevity.12 As a result, there has been an effort to develop Sirt6 allosteric activator small molecules such as MDL-80013 that are hypothesized to impede biological aging.
Sirt6 can catalyze at least three distinct reaction types: Lys deacetylation, long chain Lys deacylation14, and ADP-ribosylation.15 However Sirt6’s specific activity and selectivity for different histone lysine sites,16 how these activities are regulated,17 and the connections between enzymatic activities and biological function18 are unclear.19 Sirt6 has been reported to be poor at deacetylation of free histone tail peptides14, 20 but in cellular studies, Sirt6 has a documented preference for cleavage of Lys9 acetylation of histone H3 (H3K9ac),21 leading to the hypothesis that Sirt6 can deacetylate histones more efficiently in chromatin substrates.22 Several X-ray structures of histone peptides in complex with Sirt6 have been reported,14, 23 but the molecular recognition of nucleosome substrates by Sirt6 has been poorly understood.24 Sirt6 appears to have relatively high affinity for nucleosomes (nanomolar KD values) which is partially mediated by the H2A/H2B “acidic patch” as well as nucleosomal DNA, though the structural details of these interactions are lacking.24 Here we have analyzed site-specifically acetylated nucleosomes as substrates to assess the rate of deacetylation of a range of Lys sites in histone H3 and histone H2B. Moreover, we have used protein semisynthesis to prepare a Lys9-thiocarbonyl derivatized nucleosome to trap a Sirt6-nucleosome complex. This approach facilitated the structural determination of Sirt6 bound to nucleosome in a catalytically poised conformation.
RESULTS
Sirt6 deacetylation of acetylated nucleosome substrates.
To understand the site- and substrate-selectivity of Sirt6 required access to chemically defined protein substrates for enzymology. Scarless semisynthetic histone H3 and H2B containing known acetylations at specific sites (Figure 1A) were prepared using engineered sortase enzymes and synthetic tail peptides as previously described (Figure S1).25 For histone H3, we produced K9ac, K14ac, K18ac, K23ac, and K27ac forms and for H2B, we generated K11ac, K12ac, K20ac, and K46ac forms (Figure S1). The modified histone H3s were assembled into octamers with unmodified recombinant histones H2A, H2B, and H4, and modified H2Bs incorporated into octamers with unmodified recombinant H2A, H3, and H4. Octamers were assembled into nucleosomes with either 147 bp or 185 bp DNA (nucleosome-147 and nucleosome-185),26 and purified by anion exchange chromatography.27 Acetylated histone protein and nucleosomes were analyzed as substrates of purified Sirt6 (Figure S2) using quantitative Western blot time course assays with the corresponding site-specific anti-acetyl-Lys antibodies (Figure 1B,C,D, Figure S3–S8).25 The Sirt6 deacetylation rates, calculated as velocity/enzyme concentration (V/[E]), are shown in Figure 1E and 1F. Three of the H3 acetylated nucleosome forms revealed robust deacetylation including H3K9ac, H3K18ac, and H3K27ac with V/[E] of 0.1, 0.04, and 0.04 min−1, respectively. The rate of nucleosome H3K9ac deacetylation is comparable to the reported rate of one of the faster class I HDAC1 complexes, CoREST, and faster than recently reported for Sirt1 (V/[E]<0.001 min−1), Sirt2 (0.03 min−1), Sirt3 (0.006 min−1), and Sirt5 (<0.001 min−1).25 In contrast to H3K9ac, H3K18ac, and H3K27ac substrates, Sirt6-mediated deacetylation of H3K23ac (V/[E] 0.011 min−1) and H3K14ac (0.0012 min−1) nucleosome substrates, while detectable, was considerably slower (Figure 1E,G, Table S1). Surprisingly, acetylated H2B nucleosomes were not observed to undergo Sirt6-mediated deacetylation at any of the four sites investigated (V/[E] <0.001 min−1) (Figure 1D,E). Moreover, neither histone H3 nor H2B free protein were detectably deacetylated by Sirt6 (V/[E] <0.01 min−1) (Figure 1F,H, Figure S7,S8, Table S1). The marked preference of Sirt6 for H3 acetylated nucleosomes over either H2B nucleosomes or free histone proteins starkly contrasts with other sirtuins (Sirt1, Sirt2, Sirt3, and Sirt5)25 and HDAC complexes (CoREST, MIDAC, Sin3, NuRD, MIER, RERE, and SMRT)28, which generally deacetylate free histone protein faster than nucleosomes, and show little preference between nucleosomes with acetylated H3 or H2B.
To further characterize H3K9ac nucleosome deacetylation by Sirt6, we examined the substrate concentration dependence and derived an apparent Km in the range of 17–19 nM, independent of DNA length (Figure 2A,B, Figure S9,10). This apparent Km is in the same range as the previously reported Kd value of nucleosome binding to Sirt6.24 It has been reported that sirtuin family members29 exhibit different NAD binding modes30 with NAD affinities depending on the specific acyl-Lys substrate31. We determined the NAD concentration dependence here with nucleosomal substrate, which showed an apparent Km of ~150 μM (Figure 2C, Figure S11), significantly larger than the Km of NAD obtained with a peptide substrate (~15 μM).32 We also explored the influence of ionic strength by varying the reaction NaCl concentration. We observed a sharp decline in Sirt6-catalyzed deacetylation rate with increasing NaCl, indicating the importance of ionic interactions in the binding and/or deacetylation process (Figure S12). Prior reports that DNA stimulates sirtuin activity prompted our evaluation of linker DNA outside of the core 147 bp Widom 601 sequence. We assessed the deacetylation of H3K9ac nucleosomes containing 185 bp DNA and found that Sirt6 deacetylatese 147 bp and 185 bp nucleosomes at similar rates (Figure S10).As mentioned, MDL-800 is reported to be an allosteric activator of Sirt6 and as such we evaluated that here. Using H3K9ac 147 bp nucleosomes at 20 nM (close to the apparent Km), we observed an MDL-800 concentration dependent stimulation of Sirt6 deacetylation activity with a ~4-fold maximal effect and an EC50 of ~3.5 μM relative to DMSO vehicle control (Figure 2D–G, Figure S13). These results indicate that MDL-800 is an allosteric activator for both peptide substrates and physiologically relevant acetylated nucleosome substrates (Figure 2, Table S1).
Figure 2.
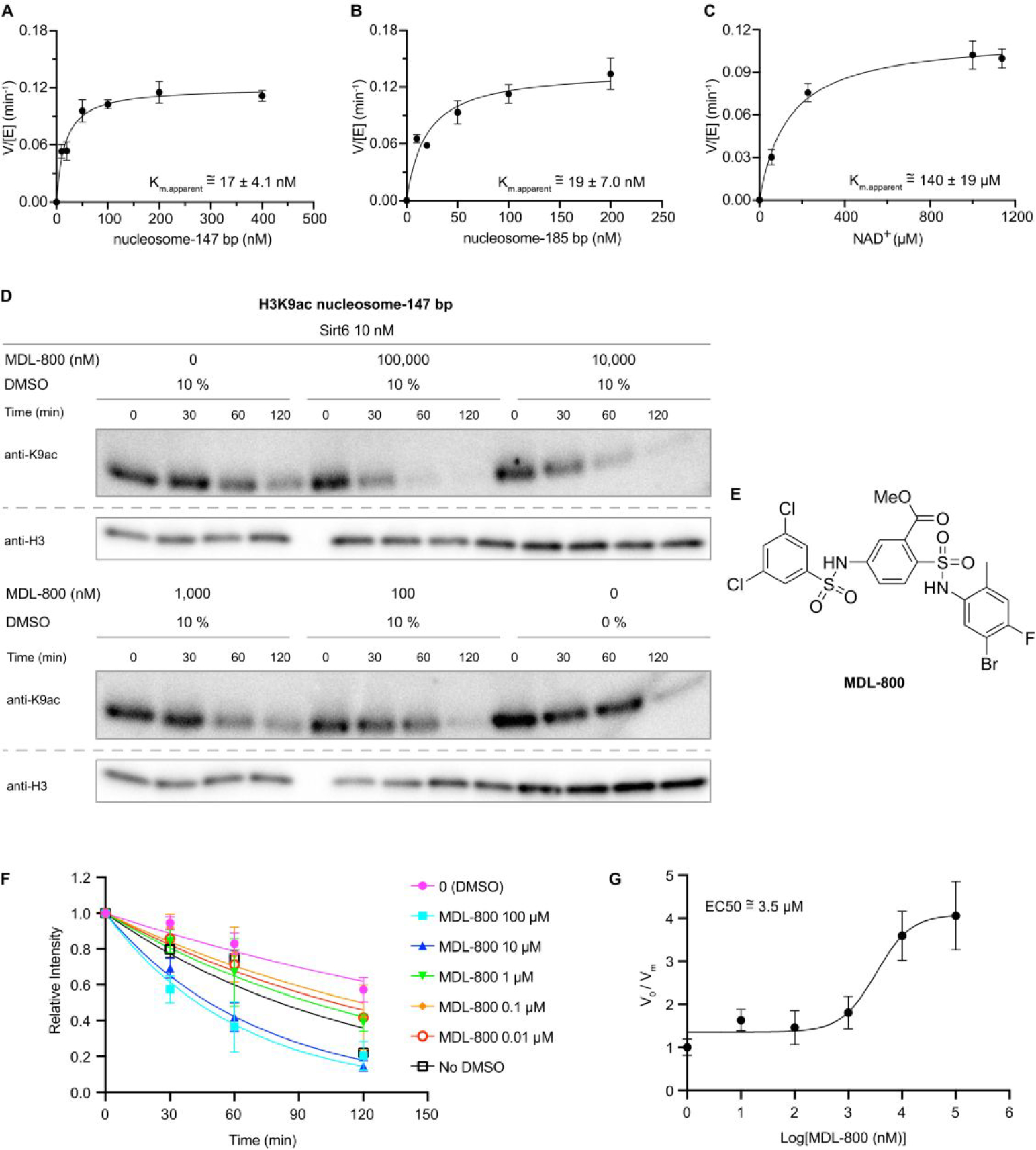
Determination of Sirt6 apparent substrate Km values and allosteric activation by MDL-800. (A) Sirt6 V/[E] vs. 147 bp H3K9ac nucleosome substrate. (B) Sirt6 V/[E] vs. 185 bp H3K9ac nucleosome substrate. (C) Sirt6 V/[E] vs. NAD with 147 bp H3K9ac nucleosome substrate. (D) Western blot analysis of the effect of MDL-800 on Sirt6 deacetylation of H3K9ac 147 bp nucleosome substrates with DMSO as the vehicle control, as DMSO caused a slight rate reduction. (E) MDL-800 structure. (F) Fitting Sirt6 deacetylation rates at different concentrations of MDL-800. (G) MDL-800 activation curve (n=2–3).
Thiocarbonyl nucleosomes and Sirt6 interactions.
To investigate the structural basis for the selective Sirt6 deacetylation of H3Kac nucleosome substrates, we prepared nucleosomes modified with thiocarbonyl groups at the H3K9 position.36 Thioacetyl (Sac) and methyl-thiourea (MTU) functionalities were installed using sortase-mediated histone H3 semisynthesis.37 Both Sac38 and MTU39 groups have previously been installed in synthetic peptides, leading to potent sirtuins inhibitors40. Thiocarbonyls can trap enzyme-substrate complexes due to slow turnover of ADPthiogylcoside intermediates (Figure 3A,B, Figure S14)41. Such chemical traps have aided in the structural determination of sirtuins bound to the peptide substrate analogs. Unexpectedly, the H3K9-Sac nucleosomes were efficiently cleaved by Sirt6, rendering them ineffective for covalent capture (Figure 3C,D, Figure S15). In contrast, H3K9-MTU appeared more resistant to removal by Sirt6, and nucleosome electrophoretic mobility shift assays (EMSAs) revealed the formation of two sharp, higher molecular weight bands that were presumed to have one or two Sirt6 polypeptides tightly bound to the nucleosome (Figure 3E). This EMSA behavior was dependent on the presence of NAD, consistent with the predicted formation of a covalent adduct between the MTU group and ADP-ribose. Characterization of the four histones by intact mass spectrometry revealed that the H3 alone displayed a 511 Da increase compared to the H3K9-MTU starting material (Figure 3F, Figure S16). Based on the increased molecular weight of the modified H3 and the generally accepted sirtuin catalytic mechanism, the deconvoluted product mass of 15,822 Da corresponds to the chemical structure of a Lys Nε−1,3-oxathiolan-2-yliden amine ADP-ribose adduct (ADP-ribose intermediate analog) formed after elimination of methylamine (Figure 3G). This adduct appears to be an analog of a cyclic intermediate proposed for the catalytic steps of the deacetylation process (Figure 3A,B) and the capacity to eliminate methylamine allows for the stable modified H3 to be isolable. It is noteworthy that there was little unreacted H3K9-MTU protein visible suggesting a robust conversion to the ADP-ribose intermediate analog.
Figure 3.
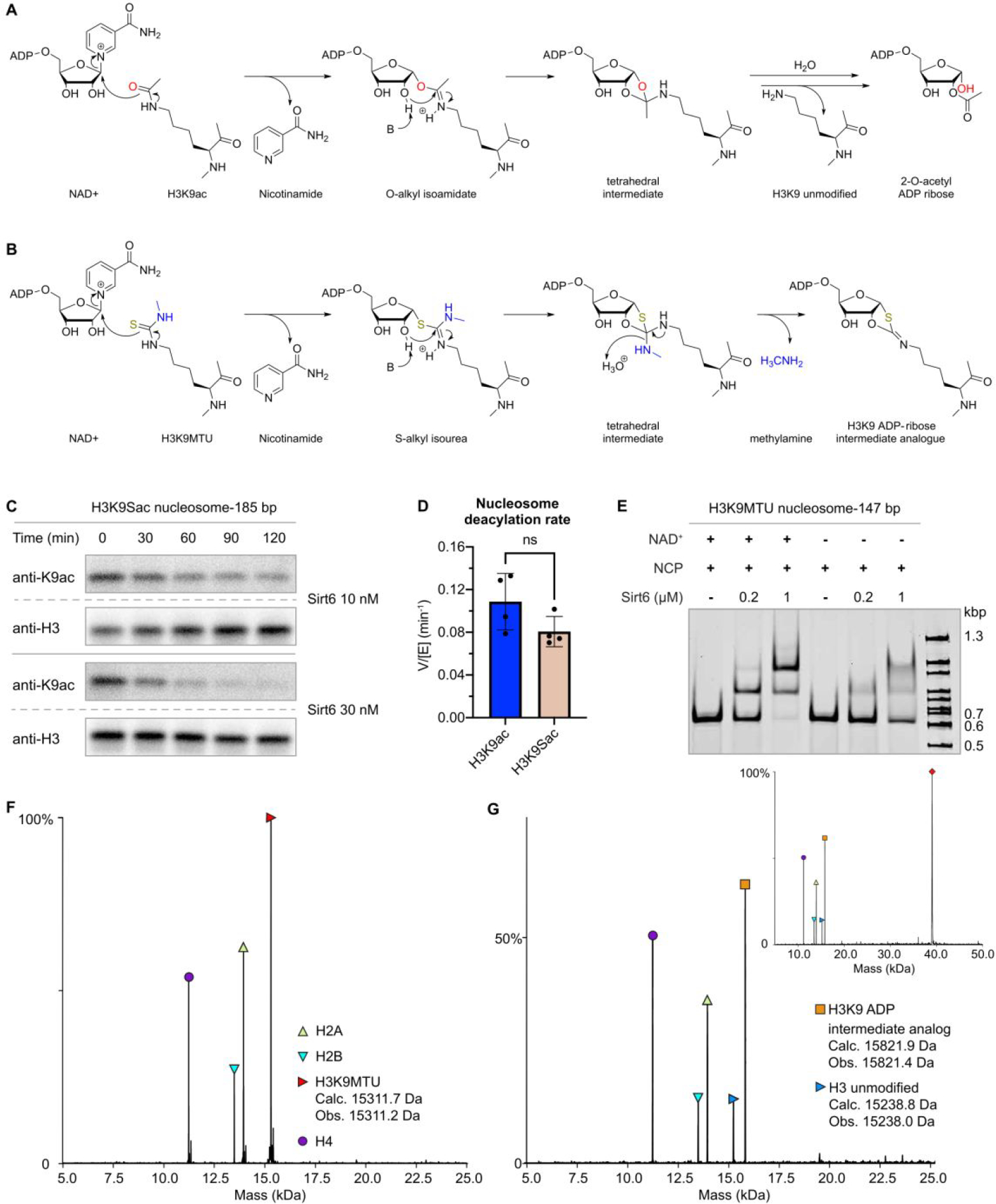
Covalent capture of Sirt6 by cofactor crosslinking with ADP-ribose intermediate analogues. (A) Sirtuin deacylation mechanism illustrating proposed O-alkyl isoamidate (PDBID: 4BVE)33 and tetrahedral intermediates (PDBID: 4F56),34 and observed products (PDBID: 2H59).35 (B) Proposed mechanism of N-methyl thiourea reaction and ADP-ribose intermediate analog. (C) Dethioacetylation of 185 bp H3K9 thioacetyl nucleosomes. (D) Comparison of H3K9 deacetylation and dethioacetylation rates employed an unpaired t-test with a two-tailed P value (ns p>0.1) (n=4). (E) Electrophoretic mobility shift assay of 185 bp H3K9 N-methyl thiourea nucleosome substrates with increasing concentrations of Sirt6, and in the presence or absence of NAD+(n=2). Shifted bands likely correspond to either one (middle) or two (upper) molecules of sirtuins binding to one nucleosome. (F) Deconvoluted intact ESI-MS of H3K9MTU (red triangle; Calculated: 15311.72 Da; Observed: 15311.2 Da) nucleosomes including H4 (purple circle; Calculated: 11236.15 Da; Observed: 11235.5 Da), H2A (upward-pointed yellow green triangle; Calculated 13950.20; Observed: 13949.6 Da) and H2B (downward-pointed cyan triangle; Calculated: 13493.68 Da; Observed: 13493.1 Da) prior to Sirt6 treatment. (G) Deconvoluted intact ESI-MS of H3K9MTU nucleosomes including H4, H2A and H2B following treatment with Sirt6 in the presence of NAD. Masses corresponding to deacylation product (rightward-pointed green triangle) and Nε−1,3-oxathiolan-2-ylidene amine product (orange square) are illustrated, along with the Sirt6 mass (red diamond, inset; Calculated: 39090.82 Da; Observed: 39087.1 Da).
Cryo-EM structural analysis of the Sirt6 nucleosome complex.
Encouraged by these findings with the H3K9-MTU nucleosome/Sirt6 reaction, we used single-particle cryo-EM to analyze the structure of the macromolecular complex (Figure 4). We reconstituted a H3K9-MTU nucleosome with 20 base pairs of extranucleosomal DNA on both sides for a total construct length of 185 base pairs for the nucleosome and prepared the H3K9-MTU nucleosome-Sirt6 complex in the presence of NAD using size exclusion chromatography (Figure S17). Peak fractions containing the H3K9-MTU nucleosome-Sirt6 complex were mildly cross-linked with glutaraldehyde and then prepared for single-particle cryo-EM. Single-particle cryo-EM data were collected on a Titan Krios microscope with a K3 direct electron detector and energy filter (Methods). The cryo-EM data was subsequently processed and analyzed (Figure S18,S19). The structure of Sirt6 bound to the H3K9-MTU nucleosome was determined from 95,205 particles with an overall resolution of 3.1 Å (Figure 4B, Table S2). The nucleosome was resolved to a resolution of 3–5 Å and Sirt6 to resolutions of 3–6 Å (Figure S19). A known crystal structure of the nucleosome42 and a previously reported crystal structure of Sirt613 were docked into the density and locally adjusted. Additional extranucleosomal DNA was only visible on the Sirt6 bound site and the DNA was extended to accommodate an additional three base pairs. We docked the N-terminus of Sirt6 from an AlphaFold2 model43, which sits on top of the Sirt6 hydrophobic pocket that is formed by the Sirt6 Rossman fold and the zinc-binding domain (Figure 4C). Only a small part of the Sirt6 C-terminus could not be resolved (Figure 4). In our structure, Sirt6 sits on top of the nucleosomal disc above the H2A C-terminus docking domain and the H3 ɑN helix next to the nucleosomal DNA at SHL 6 to 7. Sirt6 is relatively unaltered compared to published crystal structures with a root mean square deviation of 1.723 Å. After completing the structural model of the nucleosome and Sirt6, we observed additional density that allowed us to build the ADP-ribose intermediate analog and H3 tail residues 3–13 into our density (Figure 5A). The ADP-ribose intermediate analog moiety is bound in the Sirt6 Rossman fold and the histone tail is positioned between nucleosomal DNA at superhelical location 6.5 and Sirt6, positioning the substrate H3K9 lysine for insertion into the Sirt6 active site proximal to Sirt6 residue His133. The nucleosome-Sirt6 structure was refined and showed good stereochemistry (Table S2,S3).
Figure 4.
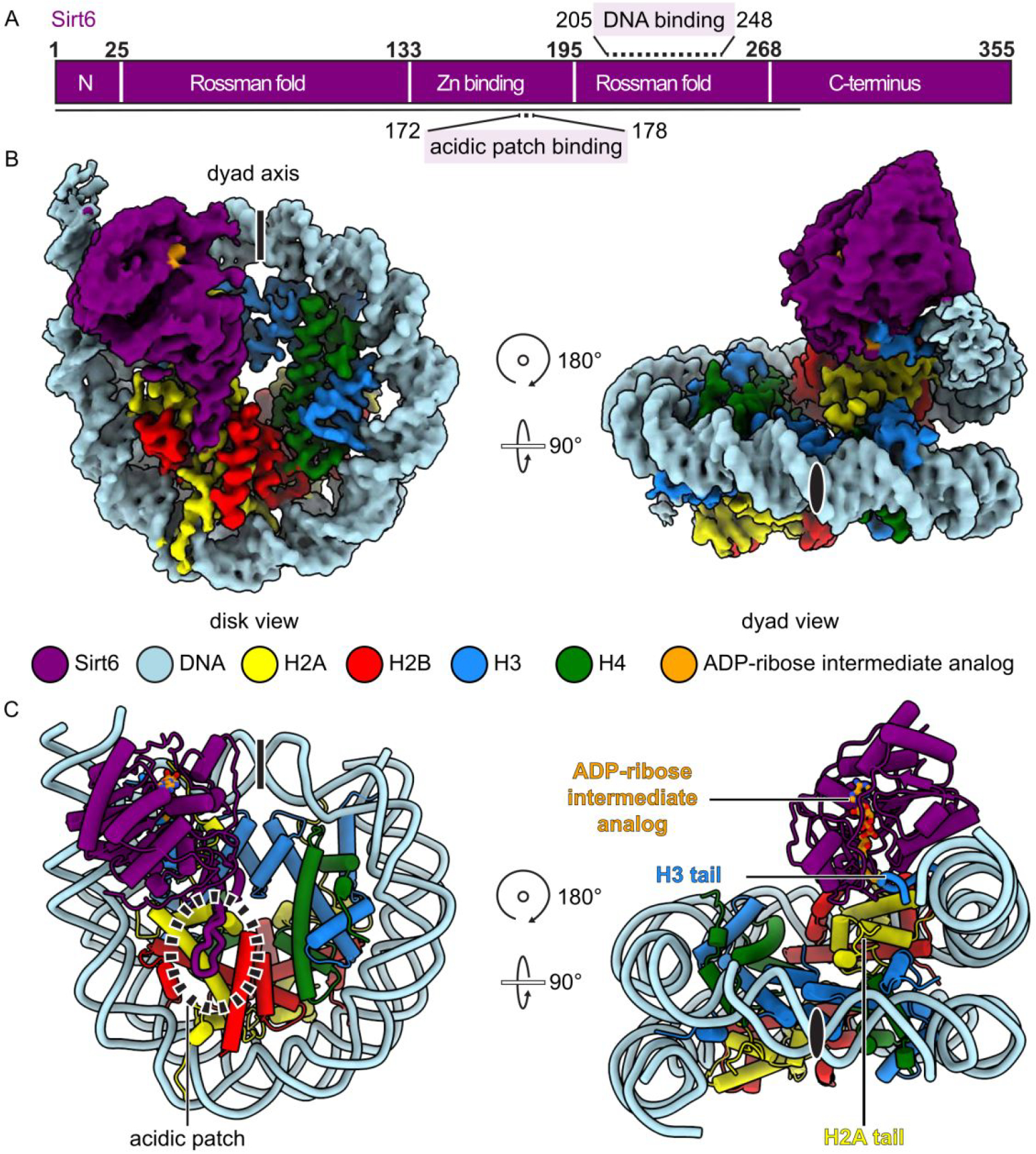
Cryo-EM structure of Sirt6-Nucleosome complex. (A) Domain map of Sirt6 with acidic patch-binding loop (170–178) and DNA-binding (205–248) regions highlighted. Modeled region is indicated as black line. (B) Complex density map. Dyad axis indicated as line or black oval. (C) Structure of Sirt6-nucleosome complex, showing the H2A C-terminal tail.
Figure 5.
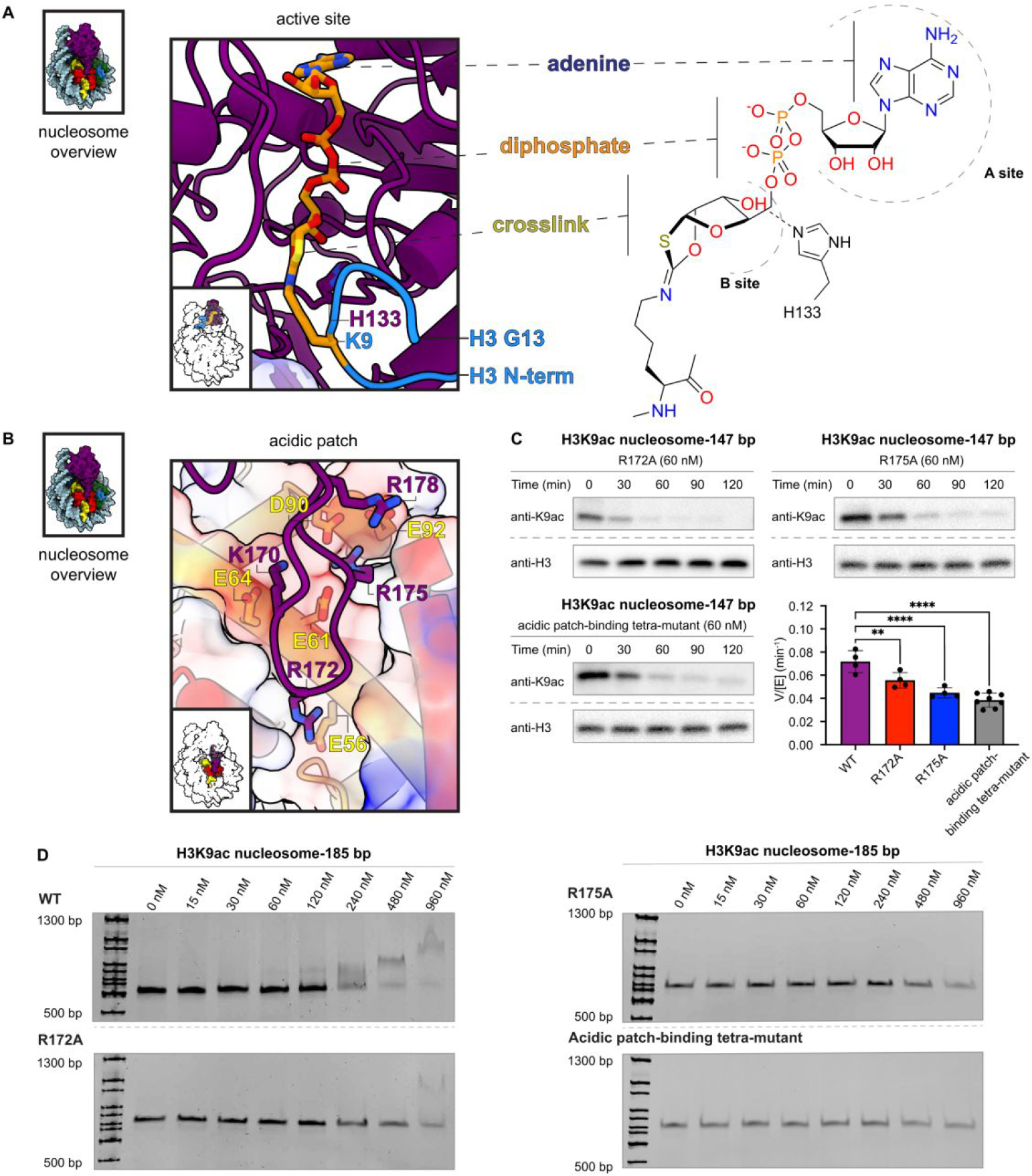
Cofactor crosslink and interactions between Sirt6 and the H2A/H2B acidic patch. (A) Sirt6 active site with modelled K9 Nε−1,3-oxathiolan-2-ylidene amine linkage to ADP-ribose. Histone H3 T3 through G13 are modelled (blue) based on an existing crystal structure (PDBID: 5Y2F).13 (B) Residues involved in interaction between Sirt6 (K170, R172, R175, R178) and H2A (E56, E61, E64, D90, E92). (C) Effect of point mutations to the acidic patch-binding loop on Sirt6 deacetylation of H3K9ac nucleosomes. R172A (top left, n=4), R175A (top right, n=4), and K170A/R172A/R175A/R178A (bottom left, n=8) each significantly decreased (** p=0.0089; **** p<0.0001) using an ordinary one-way ANOVA with multiple comparisons with the WT Sirt6 deacetylation rate (bottom right, n=4). (D) Electrophoretic mobility shift assay of acidic patch-binding point mutants. WT Sirt6 (top left) induces a change in nucleosome migration at the final two concentrations (480 nM and 960 nM). R172A (lower left), R175A (upper right) and K170A/R172A/R175A/R178A (bottom right) show only a slight change in nucleosome intensity at the final concentration (960 nM).
Multivalent interactions of Sirt6 with the nucleosome.
In our structure, a single copy of Sirt6 sits on top of the nucleosomal disk and interacts with histones and nucleosomal DNA to form multiple contacts with the nucleosome (Figure 5,6). First, Sirt6 interacts with the acidic patch of the nucleosome (Figure 5B). Specifically, Sirt6 has a zinc-binding domain with a unique insertion between the third and fourth cysteines of the zinc cluster.44 This insertion is unique to Sirt6 among all sirtuins (Figure S20).44 The ten amino acid long extension forms an approximately 15 Å long loop and is enriched in basic amino acids (Sirt6 residues K170, R172, R175, R178). In our structure, the loop binds the acidic patch of the nucleosome, confirming biochemical observations.24 The interactions are mediated via Sirt6 residues K170, R172, R175. K170 contacts H2A residue E64, Sirt6 R172 inserts into the acidic patch and contacts H2A E56, and Sirt6 R175 binds H2A residues E61, D90, and E92 (Figure 5B). This configuration is similar to other arginine anchors that bind the acidic patch.45 To test the importance of this interaction, we generated multiple Sirt6 mutants that disrupt the binding to the acidic patch. These mutations include a R172A mutant, a R175A mutant, and a mutant with K170A, R172A, R175A, and R178A. All three mutants showed a decreased deacetylase activity and reduced ability to bind the nucleosome (Figure 5C,D, Figure S21,S23).45
Figure 6.
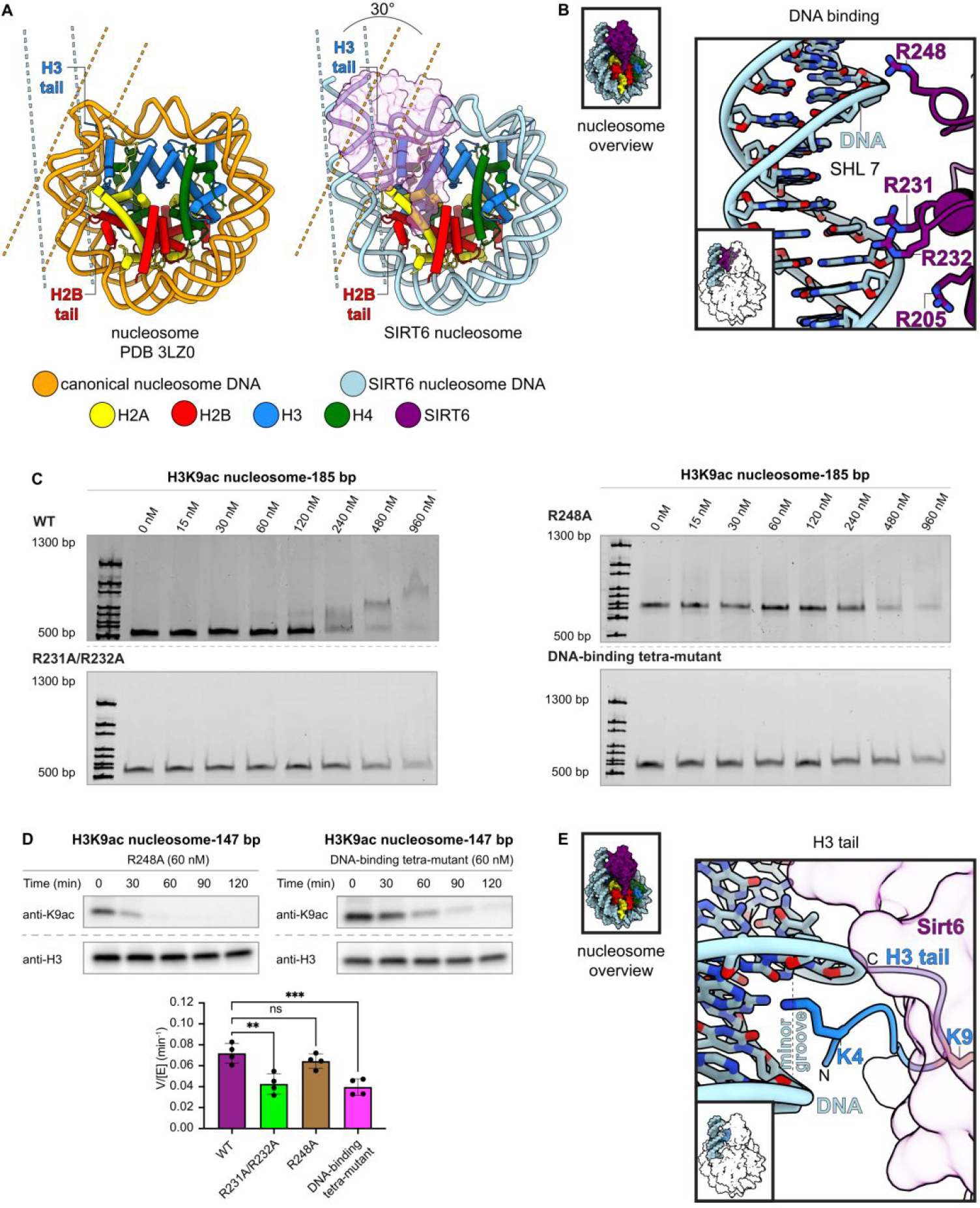
Interactions between Sirt6 and nucleosome entry site DNA. (A) Comparison of canonical nucleosome structure (left) and Sirt6-bound nucleosome (right) illustrating the unwinding of entry site DNA that accompanies Sirt6 binding. The different positions of H3 and H2B tails are illustrated. (B) Residues involved in interaction between Sirt6 (R205, R231, R232, R248) and DNA. (C) Electrophoretic mobility shift assay of DNA-binding point mutants. WT Sirt6 (top left) induces a change in nucleosome migration at the highest two concentrations (480 nM and 960 nM). R231A/R232A (lower left), R248A (upper right) and R205A/R231A/R232A/R248A (bottom right) show little change in migration up to 960 nM. (D) Effect of point mutations to the DNA-binding residues on Sirt6 deacetylation of H3K9ac nucleosomes. R231A/R232A (supplement, n=4) and R205A/R231A/R232A/R248A (top right, n=4), each significantly decreased (** p=0.0011; *** p<0.0005), while R248A (top left, n=4) does not significantly decrease the rate (ns p>0.1) using an ordinary one-way ANOVA with multiple comparisons with the WT Sirt6 deacetylation rate (bottom, n=4). (E) Interaction between H3K4 and the minor groove of DNA at SHL 6.5.
Second, we observe that 15 base pairs of nucleosomal DNA from SHL 5.5 to SHL 7 are unwrapped from the histone octamer by approximately 30° compared to a canonical nucleosome (Figure 6A). Sirt6 interacts with the unwrapped DNA via multiple arginine residues. Sirt6 arginine residues 205, 231, and 232 contact the phosphate backbone of the unwrapped DNA at SHL 6.5 and Sirt6 R248 is located in close proximity to the phosphate backbone at SHL 7 (Figure 6B). Confirming our structural observations, mutation of R231 and R232 to alanines or mutation of all identified DNA interacting Sirt6 residues to alanines results in a significant reduction of deacetylase activity and a reduced affinity to the nucleosomal substrate. Notably, disruption of just Sirt6 R248 does not show a significant reduction of H3K9 Sirt6 deacetylase activity but does appear to weaken nucleosome affinity. (Figure 6C and 6D, Figure S22,S23). Because the H3 histone tail is clamped between the two DNA gyres in canonical nucleosome conformations, it is likely that the unwrapping of nucleosomal DNA and the stabilization of this unwrapped conformation by Sirt6 plays an important role in facilitating access of Sirt6 to the acetylated H3 tail. Due to the positioning of Sirt6 on the nucleosomal disk, unwrapping of the DNA is likely required to release the H3 tail from its DNA clamp between the two DNA gyres, which then allows H3K9ac to enter the Sirt6 active site.
Third, Sirt6 is located above the C-terminus of H2A and H2A residues 119–122 reach toward the N-terminal region of Sirt6 (residues 8–11), forming an additional interaction surface. Together, Sirt6 recognizes the nucleosomal substrate through multiple interactions, allowing Sirt6 to be optimally positioned to catalyze the deacylation of acetylated histone H3 lysine residues. Our structural findings do not rule out binding of a second Sirt6 copy on the opposite side of the nucleosome as suggested biochemically.
Interaction of SIRT6 with a catalytic intermediate analog of H3K9ac deacylation
We observe density for the ADP-ribose intermediate analog formed during the deacylation of H3K9 including H3 histone tail residues 3–13. Similar to other sirtuins and as previously observed, the adenosine moiety of the cofactor analog binds in the A site and the reactive ribose moiety binds in the B site46 in close proximity to the important catalytic Sirt6 residue His1336 (Figure 5A). The resolved part of the H3 histone tail extends from the unwrapped DNA towards the active site of Sirt6 to insert H3 residue Lys9 into the Sirt6 active site, facilitating deacylation. Similar to previous crystal structures,14 our structure confirms that Sirt6 can accommodate larger lysine acylations in its hydrophobic pocket and is likely able to complete their deacylation.
We observe the H3 tail residues T3-R8 projecting from the unwrapped DNA to the Sirt6 active site. Notably, H3 residue K4 inserts itself into the minor groove of DNA at SHL 6.5 (Figure 6E). It is likely this interaction with DNA helps stabilize the H3 tail in the proper orientation to ensure placement of H3K9 into the active site of SIRT6.
We do not observe density for H3 tail residues K14 through P38 in our reconstruction, likely due to their inherent flexibility. However, the distance between resolved H3 residues 13 and 39 is only ~20 Å. As such, our model suggests that Sirt6 binding and deacetylation of other residues within the H3 tail such as H3K27 can reach the active site of Sirt6 in the observed conformation, consistent with our biochemical observations (Fig. 1)
DISCUSSION
As critical enzymes that regulate gene regulation and established therapeutic targets, HDACs have been intensively studied since their original molecular identification over two decades ago.47 Nevertheless, how the metallohydrolase HDACs or the NAD-dependent sirtuins target acetyl-Lys sites for removal from chromatin has been unclear. In this study, we employed a series of site-specifically H3- and H2B-acetylated nucleosomes to assess how nucleosome substrates compare to free histone protein as Sirt6 substrates. Two principal features of selectivity were observed for Sirt6 mediated deacetylation. First, H3 tail acetylated nucleosome substrates are far superior to histone N-tail H2B acetylated nucleosome substrates, and second, H3 tail acetylated nucleosome substrates are much more rapidly processed than free histone H3 and H2B protein acetylated substrates. Our cryo-EM analysis of the Sirt6-nucleosome complex readily explains both of these selectivity features. The extensive and intimate interactions between the nucleosome and Sirt6 position the H3 tail in a suitable orientation for active site entry whereas the H2B tail appears to extend away from the Sirt6 catalytic site. This also explains why Sirt6 only exhibits deacylation activity with H2B peptide substrates and not on nucleosomal substrates.14
The cryo-EM structure reconstructed here is effectively modeled with X-ray crystal structures that include both H3K9 modified tail peptide as well as MDL-800 allosteric ligand. This structure can therefore be used to discern the basis for the increased cleavage rates of Sirt6 at H3K9ac, H3K18ac, and H3K27ac relative to H3K23ac and H3K14ac, agreeing with the general selectivity observed in cell experiments as a H3K9ac and H3K18ac deacetylase.48 A common element of H3K9, H3K18, and H3K27 is an Arg residue preceding the targeted Lys. The Sirt6 X-ray structure shows that Arg8 is nicely accommodated by the peptide substrate binding site whereas a Thr preceding Lys23 and especially the small residue Gly preceding Lys14 would appear to be poorer fits. It is interesting that Lys14 acetylation is generally the most difficult to remove among metallohydrolase HDACs and sirtuins and this could account for its high prevalence across chromatin. H3K27ac is an apparently novel Sirt6 deacetylation site that should be considered in future studies.8 Notably, we also observe that H3 residue K4 inserts into the DNA minor groove at SHL 6.5. Because H3K4 can be post-translationally modified (e.g., methylated), future studies should consider how these modifications impact Sirt6 deacylation activity of H3K9.
That MDL-800 can accelerate nucleosome deacetylation in vitro reinforces the idea that it acts to stimulate Sirt6 in vivo. Agents that activate Sirt6 and other sirtuins are proposed to slow down the aging process, and we would suggest that examining their behavior in a purified system with nucleosome substrates as employed here can help select for the most promising agents.
Although to our knowledge this study describes the first high resolution structure of an HDAC in complex with a nucleosome, there are several enzyme-nucleosome complexes previously determined.45, 49–51 A common theme for nucleosome binding proteins is that they involve the H2A/H2B acidic patch by employing one or more basic residues.52, 53 That is certainly the case with Sirt6. Some specialized features regarding DNA binding and H2A C-terminal contact also appear to be facets of Sirt6/nucleosome interaction and allow a targeted recruitment to the nucleosome and positioning of Sirt6 on the substrate to facilitate histone deacylation. Though unexplored in this study, the proximity of the H2A C-tail to the Sirt6 active site suggests a possible H2A deacylase role that fits the function of Sirt6 in safeguarding genome integrity.54
Chemical trapping functionalities like norleucine for histone methyltransferases55, methionine for LSD1 histone demethylase51, and CoA for histone acetyltransferases56 have proven effective for structural analysis of these enzymes. The use of a thiourea as performed in this work rather than a thioacetyl to trap Sirt6-nucleosome interactions may be of general advantage for the sirtuin family in analyzing nucleosome interactions. We also propose that this approach of incorporating warheads like hydroxamic acids8 for metallohydrolase HDACs may facilitate structural determination of these enzymes bound to nucleosomes.
EXPERIMENTAL METHODS
Cloning, expression, and purification of Sirt6.
The His-Sirt6 plasmid previously reported57 was employed after a TEV cleavage site was inserted between the His-tag and Sirt6 sequence along with an N-terminal Cys. All subsequent constructs were generated by Q5 site-directed mutagenesis (New England Biolabs). Sirt6 constructs were expressed in LOBSTR E. coli strain derived from Rosetta (DE3) in LB media supplemented with 50 mg/L kanamycin. Cells were grown at 37 °C, 200 rpm to an OD600 of 0.6, and overexpression was induced with the addition of isopropyl β-D-1-thiogalactopyranoside to 0.5 mM. Cells were grown for a further 18 h at 25 °C, 200 rpm. Pelleted cells were resuspended in ice cold lysis buffer (20 mM Tris, 500 mM NaCl, 20 mM imidazole, pH 7.5) and then lysed by passage through a French press. Lysate was clarified by centrifugation at 13,000 × g, 4 °C. Lysate supernatant was applied to pre-equilibrated Ni-NTA resin (2 mL resin bed volume / 1 L culture) and incubated 1 h at 4 °C. The resin was drained and was washed with 10 column volumes of wash buffer (20 mM Tris, 500 mM NaCl, 50 mM imidazole, 0.5 mM TCEP, pH 7.5), followed by 25 column volumes of high-salt wash buffer (20 mM Tris, 2 M NaCl, 20 mM imidazole, pH 7.5), then eluted with 10 column volumes of elution buffer (20 mM Tris, 500 mM NaCl, 200 mM imidazole, 0.5 mM TCEP, pH 7.5). The elution was concentrated by spin concentrator (Amicon, 10 kDa MWCO, 4,000 rpm, 4 °C) and mixed with TEV protease (~1 mg/mL), before overnight dialysis at 4 °C (dialysis buffer: 20 mM Tris, 150 mM NaCl, 20 mM imidazole, 0.5 mM TCEP, pH 7.5. Uncleaved protein was removed by a second passage over Ni-NTA resin (1 h incubation at 4 °C). The flow through was diluted with 10 mL Heparin buffer A (50 mM Tris, 150 mM NaCl, 5 % glycerol, 0.5 mM TCEP, pH 7.5), and concentrated by spin concentrator (10 kDa MWCO, 4,000 rpm, 4 °C). Concentrated sample was applied to a Heparin column (1 mL), and eluted with a linear gradient from 0–50 % Heparin buffer B (50 mM Tris, 2000 mM NaCl, 5 % glycerol, 0.5 mM TCEP, pH 7.5) over 20 column volumes at a flow rate of 0.6 mL/min. Elution fractions were checked by Coomassie-staining following SDS-PAGE (6–20 %), and pure fractions were collected and concentrated. This material was further purified by size exclusion using a Superdex200 increase 10/300 GL column (Cytiva) with Superdex running buffer (50 mM Tris, 150 mM NaCl, 0.5 mM TCEP, pH 7.5). Elution fractions were checked by SDS-PAGE, and the pure fractions were combined and concentrated to ~40 μM. Coomassie-staining after SDS-PAGE was used for densitometry and quality check for WT Sirt6 and all the Sirt6 mutants. WT Sirt6 was characterized by intact ESI-MS (Q Exactive, Thermo Scientific) and deconvoluted using UniDec.58 All Sirt6 constructs were aliquoted and flash-frozen in liquid nitrogen, then stored at −80 °C prior to use.
General deacetylation assay and conditions.
Free histone protein (H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H2BK11ac, H2BK12ac, H2BK20ac, and H2B46ac; final 1.0 μM) or nucleosomes (H3K9ac, H3K9Sac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H2BK11ac, H2BK12ac, H2BK20ac, and H2B46ac; final 100 nM) were diluted in Sirt6 reaction buffer (50 mM HEPES at pH 7.5, 1 mM DTT, 0.2 mg/mL BSA, and 1 mM NAD). This solution was kept on ice until the reaction start. Reactions were commenced by addition of Sirt6, after which samples were incubated at 37 °C. At each indicated time point a small volume was removed from the reaction. For typical nucleosome assays time points were taken at 0, 30, 60, 90, 120 min. Each quenched sample was boiled for 3–5 min at 95 °C and resolved on a 4–20 % gradient SDS-PAGE gel at 180 Volts for ~25 min. Gels were transferred semi dry onto nitrocellulose membrane by iBlot with P3 (20 V) for 5.0 min. Membranes were blocked for 1 h with 5% BSA in TBST, then incubated overnight with primary antibody. Site-specific antibodies for H3, acetylated H3, H2B, acylated H2B are listed in the key materials table. After washing, HRP linked secondary antibody was applied for 1 h at room temperature. Membranes were washed and treated with ECL substrate reagent, then visualized with a G:BOX mini gel imager (Syngene). Band intensity was quantified by ImageJ (imagej.nih.gov/ij/). All intensity values were divided by the intensity value at time 0 to get relative intensity, and then fit to a single-phase exponential decay curve with constraints Y0=1, Plateau=0 (GraphPad Prism 9). For assays with free histone protein substrates the decrease in anti-H3 or anti-H2B signal was accounted for by normalizing the PTM signal relative to the corresponding total histone signal at each time point. Each plotted point represents at least 2 replicates. The kinetic parameter V/[E] was calculated using GraphPad Prism 9. All the Km measurements and activator assays were done with similar procedures.
Cryo-EM structural analysis of the Sirt6 nucleosome complex.
Sirt6-H3K9MTU complexes were mixed with NAD and purified by size exclusion chromatography. Purified complex was mildly cross-linked with glutaraldehyde and dialyzed prior to vitrification onto cryo-EM grids. A 15,192 micrograph dataset and a second dataset with 6,922 micrograph and 30° tilt were both collected on a Titan Krios equipped with a BioQuantum energy filter and Gatan K3 direct electron detector. A total of 2,507,720 particles were picked from the first dataset and 1,556,859 particles were collected from the second dataset. Both datasets were combined and after several rounds of 3D classification in cryoSPARC, 95,205 particles were refined to a final resolution of 3.1 Å (map A). Further classification of the particles resulted in a second map with better resolved Sirt6 and H3 tail features. This second map (map B) was refined to 3.3 Å and aided in model building. The model was built by first rigid-body fitting in the nucleosome (PDB 3LZ0),42 Sirt6 (PDB 5Y2F)13, Sirt6 residues 1–16 (AlphaFold2, AF-Q8N6T7-F1), and the H3 tail (PDB 5Y2F). The ADP-ribose intermediate analog was modeled based on NAD+ (PDB 5Y2F) and placed into the extra density. Local adjustments were made in Coot59 and further refined with Phenix.60
Sirt6 mutants activity and EMSA.
Nucleosome binding of Sirt6 constructs was assessed by electrophoretic mobility shift assay (EMSA). Sirt6 constructs were diluted to a working concentration of 9.6 μM, then serially diluted to 4800, 2400, 1200, 600, 300, 150, 75, and 0 nM. H3K9ac nucleosome-185 bp was diluted in Sirt6 binding buffer, then combined with Sirt6 enzyme to a final nucleosome concentration of 20 nM. NAD was added to a concentration of 150 μM and mixed. Samples were prepared such that the final Sirt6 concentrations were 960, 480, 240, 120, 60, 30, 15, 0 nM. Samples were incubated for 30 min on ice, followed by native electrophoresis on a 4–20 % TBE gel run at 130 Volts for 110 min on ice.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank James Lee from MBCF of DanaFarber Cancer Institute for MALDI, Brad Palanski for technical support, Yi Zheng for illustrations (www.yizhengillustration.com), Marty Taylor and Maggie Chen for advice. The authors acknowledge NIH (GM62437 to P.A.C.), NSF2127882, LLS, American Cancer Society (PF-20–105-01-DMC to S.D.W), American Heart Association (Postdoctoral fellowship award 826614 to K. L) for financial support. L.F. is supported by the Dorsett L. Spurgeon, MD (HMS 1929) Distinguished Research Award at Harvard Medical School, by The Smith Family Awards Program for Excellence in Biomedical Research, and the Damon Runyon-Rachleff Innovation Award. C.S. is supported by the DF/HCC SPARC Program. We thank The Harvard Cryo-EM Center for Structural Biology at Harvard Medical School for support with data collection.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: Additional experimental details, materials, methods, and supporting figures (PDF) Structural coordinates and EM maps are available under accsession codes PDB 8F86 and EMDB-28915.
P.A.C. is a cofounder of Acylin Therapeutics which is involved in developing epigenetic agents and has been a consultant for the pharmaceutical companies Abbvie and Constellation.
REFERENCES
- (1).Dancy BM; Cole PA Protein lysine acetylation by p300/CBP. Chem Rev. 2015, 115 (6), 2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lee K; Whedon SD; Wang ZA; Cole PA Distinct biochemical properties of the class I histone deacetylase complexes. Curr Opin Chem Biol. 2022, 70, 102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jing H; Lin H Sirtuins in epigenetic regulation. Chem Rev. 2015, 115 (6), 2350–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Shiota H; Alekseyenko AA; Wang ZA; et al. Chemical Screen Identifies Diverse and Novel Histone Deacetylase Inhibitors as Repressors of NUT Function: Implications for NUT Carcinoma Pathogenesis and Treatment. Mol Cancer Res. 2021, 19 (11), 1818–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Jing H; Hu J; He B; et al. A SIRT2-Selective Inhibitor Promotes cMyc Oncoprotein Degradation and Exhibits Broad Anticancer Activity. Cancer Cell. 2016, 29 (3), 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Klein MA; Denu JM Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J Biol Chem. 2020, 295 (32), 11021–11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wang ZA; Cole PA The Chemical Biology of Reversible Lysine Post-translational Modifications. Cell Chem Biol. 2020, 27 (8), 953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wu M; Hayward D; Kalin JH; Song Y; Schwabe JW; Cole PA Lysine-14 acetylation of histone H3 in chromatin confers resistance to the deacetylase and demethylase activities of an epigenetic silencing complex. eLife. 2018, 7, e37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Toiber D; Erdel F; Bouazoune K; et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013, 51 (4), 454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Taylor JR; Wood JG; Mizerak E; et al. Sirt6 regulates lifespan in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2022, 119 (5), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kanfi Y; Naiman S; Amir G; et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012, 483 (7388), 218–221. [DOI] [PubMed] [Google Scholar]
- (12).Hirvonen K; Laivuori H; Lahti J; Strandberg T; Eriksson JG; Hackman P SIRT6 polymorphism rs117385980 is associated with longevity and healthy aging in Finnish men. BMC Med Genet. 2017, 18 (1), 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Huang Z; Zhao J; Deng W; et al. Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol. 2018, 14 (12), 1118–1126. [DOI] [PubMed] [Google Scholar]
- (14).Jiang H; Khan S; Wang Y; et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013, 496 (7443), 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Liszt G; Ford E; Kurtev M; Guarente L Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005, 280 (22), 21313–21320. [DOI] [PubMed] [Google Scholar]
- (16).Tasselli L; Xi Y; Zheng W; et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016, 23 (5), 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rezazadeh S; Yang D; Tombline G; et al. SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res. 2019, 47 (15), 7914–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mao Z; Hine C; Tian X; et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science (80- ). 2011, 332 (6036), 1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Onn L; Portillo M; Ilic S; et al. SIRT6 is a DNA double-strand break sensor. eLife. 2020, 9, e51636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Feldman JL; Baeza J; Denu JM Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by Mammalian Sirtuins. J Biol Chem. 2013, 288 (43), 31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Michishita E; McCord RA; Berber E; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008, 452 (7186), 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang WW; Zeng Y; Wu B; Deiters A; Liu WR A Chemical Biology Approach to Reveal Sirt6-targeted Histone H3 Sites in Nucleosomes. ACS Chem Biol. 2016, 11 (7), 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pan PW; Feldman JL; Devries MK; Dong A; Edwards AM; Denu JM Structure and biochemical functions of SIRT6. J Biol Chem. 2011, 286 (16), 14575–14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Liu WH; Zheng J; Feldman JL; et al. Multivalent interactions drive nucleosome binding and efficient chromatin deacetylation by SIRT6. Nat Commun. 2020, 11 (5244), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wang ZA; Whedon SD; Wu M; et al. Histone H2B Deacylation Selectivity: Exploring Chromatin’s Dark Matter with an Engineered Sortase. J Am Chem Soc. 2022, 144 (8), 3360–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dyer PN; Raji ES; White CL; et al. Reconstitution of Nucleosome Core Particles from Recombinant Histones and DNA. Methods Enzymol. 2004, 375, 23–44. [DOI] [PubMed] [Google Scholar]
- (27).Luger K; Rechsteiner TJ; Richmond TJ Preparation of Nucleosome Core Particle from Recombinant Histones. Methods Enzymol. 1999, 304 (1997), 3–19. [DOI] [PubMed] [Google Scholar]
- (28).Wang ZA; Millard CJ; Lin C-L; et al. Diverse nucleosome siteselectivity among histone deacetylase complexes. eLife. 2020, 9, e57663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Madsen AS; Andersen C; Daoud M; et al. Investigating the Sensitivity of NAD+-dependent Sirtuin Deacylation Activities to NADH. J Biol Chem. 2016, 291 (13), 7128–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Fiorentino F; Mai A; Rotili D Emerging Therapeutic Potential of SIRT6 Modulators. J Med Chem. 2021, 64, 9732–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Feldman JL; Dittenhafer-Reed KE; Kudo N; et al. Kinetic and structural basis for Acyl-group selectivity and NAD+ dependence in sirtuincatalyzed deacylation. Biochemistry. 2015, 54 (19), 3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kugel S; Feldman JL; Klein MA; et al. Identification of and Molecular Basis for SIRT6 Loss-of-Function Point Mutations in Cancer. Cell Rep. 2015, 13 (3), 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gertz M; Fischer F; Nguyen GTT; et al. Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism. Proc Natl Acad Sci U S A. 2013, 110 (30), E2772–E2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhou Y; Zhang H; He B; et al. The bicyclic intermediate structure provides insights into the desuccinylation mechanism of human sirtuin 5 (SIRT5). J Biol Chem. 2012, 287 (34), 28307–28314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hoff KG; Avalos JL; Sens K; Wolberger C Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD+ and Acetylated Peptide. Structure. 2006, 14 (8), 1231–1240. [DOI] [PubMed] [Google Scholar]
- (36).Hirsch BM; Hao Y; Li X; Wesdemiotis C; Wang Z; Zheng W A mechanism-based potent sirtuin inhibitor containing Nε- thiocarbamoyllysine (TuAcK). Bioorganic Med Chem Lett. 2011, 21 (16), 4753–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Piotukh K; Geltinger B; Heinrich N; et al. Directed evolution of sortase A mutants with altered substrate selectivity profiles. J Am Chem Soc. 2011, 133 (44), 17536–17539. [DOI] [PubMed] [Google Scholar]
- (38).Fatkins DG; Monnot AD; Zheng W Nε-Thioacetyl-lysine: A multi-facet functional probe for enzymatic protein lysine Nε-deacetylation. Bioorganic Med Chem Lett. 2006, 16 (14), 3651–3656. [DOI] [PubMed] [Google Scholar]
- (39).Farooqi AS; Hong JY; Cao J; et al. Novel Lysine-Based Thioureas as Mechanism-Based Inhibitors of Sirtuin 2 (SIRT2) with Anticancer Activity in a Colorectal Cancer Murine Model. J Med Chem. 2019, 62 (8), 4131–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cole PA Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008, 4 (10), 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hawse WF; Hoff KG; Fatkins DG; et al. Structural Insights into Intermediate Steps in the Sir2 Deacetylation Reaction. Structure. 2008, 16 (9), 1368–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Vasudevan D; Chua EYD; Davey CA Crystal Structures of Nucleosome Core Particles Containing the “601” Strong Positioning Sequence. J Mol Biol. 2010, 403 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- (43).Jumper J; Evans R; Pritzel A; et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021, 596 (7873), 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sanders BD; Jackson B; Marmorstein R Structural basis for sirtuin function: What we know and what we don’t. Biochim Biophys Acta - Proteins Proteomics. 2010, 1804 (8), 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).McGinty RK; Tan S Principles of nucleosome recognition by chromatin factors and enzymes. Curr Opin Struct Biol. 2021, 71, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).You W; Zheng W; Weiss S; Chua KF; Steegborn C Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci Rep. 2019, 9 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wang M; Lin H Understanding the Function of Mammalian Sirtuins and Protein Lysine Acylation. Annu Rev Biochem. 2021, 90 (1), 245–285. [DOI] [PubMed] [Google Scholar]
- (48).Pastor BM; Mostoslavsky R SIRT6: a new guardian of mitosis. Nat Struct Mol Biol. 2016, 23 (5), 360–362. [DOI] [PubMed] [Google Scholar]
- (49).Song Y; Dagil L; Fairall L; et al. Mechanism of Crosstalk between the LSD1 Demethylase and HDAC1 Deacetylase in the CoREST Complex. Cell Rep. 2020, 30 (8), 2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Worden EJ; Wolberger C Activation and regulation of H2BUbiquitin-dependent histone methyltransferases. Curr Opin Struct Biol. 2019, 59, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kim SA; Zhu J; Yennawar N; Eek P; Tan S Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate. Mol Cell. 2020, 78 (5), 903–914.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Skrajna A; Goldfarb D; Kedziora KM; et al. Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res. 2020, 48 (17), 9415–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).McGinty RK; Henrici RC; Tan S Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014, 514 (7524), 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hao S; Wang Y; Zhao Y; et al. Dynamic switching of crotonylation to ubiquitination of H2A at lysine 119 attenuates transcription-replication conflicts caused by replication stress. Nucleic Acids Res. 2022, 50 (17), 9873–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Worden EJ; Hoffmann NA; Hicks CW; Wolberger C Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell. 2019, 176 (6), 1490–1501.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Qu K; Chen K; Wang H; Li X; Chen Z Structure of the NuA4 acetyltransferase complex bound to the nucleosome. Nature. 2022, 610 (7932), 569–574. [DOI] [PubMed] [Google Scholar]
- (57).Zhang X; Khan S; Jiang H; et al. Identifying the functional contribution of the defatty-Acylase activity of SIRT6. Nat Chem Biol. 2016, 12 (8), 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Marty MT; Baldwin AJ; Marklund EG; Hochberg GKA; Benesch JLP; Robinson CV Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal Chem. 2015, 87 (8), 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Emsley P; Cowtan K Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004, 60 (12 I), 2126–2132. [DOI] [PubMed] [Google Scholar]
- (60).Adams PD; Grosse-Kunstleve RW; Hung LW; et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002, 58 (11), 1948–1954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


