Summary
Diagnostic tests usually need to operate at a high sensitivity or specificity level in practice. Accordingly, specificity at the controlled sensitivity, or vice versa, is a clinically sensible performance metric for evaluating continuous biomarkers. Meanwhile, the performance of a biomarker may vary across sub-populations as defined by covariates, and covariate-specific evaluation can be informative. In this article, we develop a novel modeling and estimation method for covariate-specific specificity at a controlled sensitivity level. Unlike existing methods which typically adopt elaborate models of covariate effects over the entire biomarker distribution, our approach models covariate effects locally at a specific sensitivity level of interest. We also extend our proposed model to handle the whole continuum of sensitivities via dynamic regression and derive covariate-specific ROC curves. We provide the variance estimation through bootstrapping. The asymptotic properties are established. We conduct extensive simulation studies to evaluate the performance of our proposed methods in comparison with existing methods, and further illustrate the applications in two clinical studies for aggressive prostate cancer.
Keywords: Continuous biomarker, Dynamic regression, Logistic regression, Quantile regression, Receiver operating characteristic curve, Sensitivity, Specificity, Specificity at controlled sensitivity
1 ∣. INTRODUCTION
Evaluation of biomarkers for their diagnostic ability is a common task in biomedical research. It is relatively straightforward to evaluate binary biomarkers using metrics such as sensitivity and specificity. The evaluation of continuous biomarkers, however, is more complicated as a threshold is needed to define the normal and abnormal ranges of the measurement for disease diagnosis. The threshold for a diagnostic test usually needs to attain a high sensitivity or specificity level to keep false negatives or positives, respectively, to minimal e.g., Sanda (2017).1 As a result, specificity at a controlled sensitivity (or vice versa) has been used as a clinically sensible metric.2,3
Meanwhile, covariates, such as age, race, and sample collection conditions, may influence biomarkers. Given a desired sensitivity or specificity level, the diagnostic threshold of a biomarker may change in sub-populations as defined by these covariates. Moreover, the diagnostic ability of a biomarker at a fixed threshold may also associate with or be influenced by covariates. Consequently, covariates can confound the assessment of continuous biomarkers, biasing the results if ignored. For example, if the covariates affect the distribution of the biomarker but not the covariate-specific ROC curves, Pepe (2003)4 showed that ignoring covariate effects may lead to underestimated diagnostic ability of a biomarker comparing to its actual performance. Therefore, as already recognized and discussed by many existing studies5,6,7,4, it is important to adjust for covariate effects in the evaluation of continuous biomarkers.
Many existing methods imposed models on both the case and the control biomarker distributions to subsequently induce the covariate effects on the ROC curves.8,9,10,11,12 For example, Faraggi (2003) adopted the normal linear regression models for both the case and control biomarker distributions.11 The approach of Pepe (1998) was more general by adopting semiparametric models.9 Additionally, Inácio de Carvalho et al. (2013)12 and Inácio and Rodriguez-Álvarez (2021)13 developed Bayesian methods based on dependent dirichlet process mixtures to target the whole conditional distribution. Nevertheless, all these methods modeled the covariate effects on the ROC curves in an indirect fashion. Thus their coefficients cannot be directly interpreted with respect to the ROC curve. To address that, several parametric distribution-free (PDF) methods that directly model the ROC curve have been proposed.5,14,15,16 These PDF methods can accommodate multiple test types and continuous covariates, and they may also target restricted portions of the ROC curve that are of interest. In particular, Alonzo and Pepe (2002) and Cai and Pepe (2002) developed generalized linear models for covariate effects on the ROC curve.15,16 Even with these PDF methods, the models are still restrictive because they presume covariate effects, as measured in regression coefficients, to be constant over the ROC curve of interest.
As a related problem, covariate adjustment has been developed for test thresholds so as to keep a controlled sensitivity or specificity level uniform across patient sub-populations. Janes and Pepe (2009) developed a non-parametric estimator in the circumstance of discrete covariates.6 Our previous work generalized the method by imposing a parsimonious quantile regression model for the thresholds.17 These methods may provide a biomarker evaluation at covariate-adjusted thresholds for the overall population, but do not permit subpopulation-specific evaluation, as focused on in this article.
In this work, we develop a novel modeling and estimation method for covariate-specific specificity at a controlled sensitivity level. It generalizes the PDF methods by targeting the particular controlled sensitivity level of interest only or accommodating potential varying covariate effects at different sensitivity levels. At the same time, The proposed approach extends our previous work17 to provide covariate-specific biomarker assessment. We first model the covariate effects among the diseased population by quantile regression, locally at a sensitivity of interest. Subsequently, the covariate-specific specificity is modeled among the non-diseased population by logistic regression. This formulation uses covariate-adjusted thresholds to equally control the sensitivity among sub-populations, meanwhile providing flexibility to estimate specificity for given covariate values. The proposed method starts with a local model for specificity at a controlled sensitivity level, and it extends naturally to covariate-specific ROC curves by addressing the continuous spectrum of sensitivity levels. It is worthwhile to point out that the same method directly applies to covariate-specific sensitivity at controlled specificity by switching the roles of cases and controls.
The subsequent sections are organized as follows. Section 2 considers the covariate-specific specificity locally at a controlled sensitivity level. Inference and asymptotic properties are established. Section 3 extends the proposal to covariate-specific ROC curve with related inference and asymptotic properties. We evaluate the performance of our proposed estimator and inference in the simulation studies presented in Section 4. Section 5 illustrates our proposals with applications to aggressive prostate cancer. Discussions and remarks are presented in Section 6. Technical proofs are relegated to the Appendix. The software of our proposed methods is available through R/CRAN package caROC.
2 ∣. COVARIATE-ADJUSTED SPECIFICITY AT A CONTROLLED SENSITIVITY LEVEL
Denote the continuous biomarker of interest by and for cases and controls, respectively. Let their associated covariates be and , respectively. The covariates could be discrete or continuous. Write the conditional biomarker distribution for cases as and for controls as . The corresponding conditional quantile function for the cases is . To control the sensitivity level at , we adopt a quantile regression model on the cases as follows:
| (1) |
where is the regression coefficient. One is added to the covariate vector to incorporate an intercept. Denote the true value of by .
Since the covariate-specific performance of the biomarker is of interest, we further model specificity over covariates in the control population. A logistic regression model is adopted, with the threshold imposed on the biomarker to control sensitivity at uniformly among the subpopulations:
| (2) |
where is the regression coefficient of interest. A logit link function is used here but it can be replaced by other link functions, e.g., probit link. Write the true value of as . The measure gauges the covariate-adjusted specificity at the controlled sensitivity level for the subpopulation with covariate value .
Observe that our model is more general than existing methods in many aspects. Pepe (1998) estimated the biomarker distribution and using semiparametric location-scale regression models9, whereas Faraggi (2003) adopted normal linear regression models for both distributions.11 It is easy to see that both models on are more restrictive than our quantile regression model (1). The normal linear regression model on of Faraggi (2003) implies the probit counterpart of our model (2).11 Thus, our model (2) is also more general than the model of Faraggi (2003) on .11 In comparison with the PDF methods of Alonzo et al. (2002)15 and Cai and Pepe (2002)16, our model (2) is much less restrictive in that the covariate effects are modeled for the controlled sensitivity level only, rather than assumed the same across various sensitivity levels.
2.1 ∣. Estimation
Consider a case cohort study. Suppose the data contain i.i.d. case samples, , and i.i.d. control samples, , . The point estimator for could be obtained using the standard quantile regression method by Koenker and Bassett (1978).18 After is estimated by , a binary diagnostic result based on the estimated threshold is computed for every control sample, , . The logistic regression is then performed with the binary result over the covariates in the control sample to obtain the point estimation for . The estimator is the solution to the following set of estimating equations:
where and . To estimate the variance for the proposed estimators, the standard non-parametric bootstrap can be applied to cases and controls separately. That is, within cases or controls, the pairs of biomarker and covariates are resampled.
2.2 ∣. Asymptotic study
We study the asymptotic properties of the estimators and . The regularity conditions are given as follows:
Condition 1. The control and case size ratio approaches a constant as .
Condition 2. Covariates and are bounded.
Condition 3. Both and are nonsingular, where for vector .
Condition 4a. Both and are differentiable at the threshold with derivative bounded away from 0 and ∞ uniformly in over the supports of and , respectively.
All these conditions are standard and mild. Previous works using quantile regression have adopted similar assumptions.19,17
Theorem 1. Suppose that the quantile regression model for the cases given in (1) and the logistic regression model for the controls given in (2) hold locally at the controlled sensitivity level , along with Conditions 1, 2, 3, and 4a. Then, is consistent almost surely for . In addition, , converges to a bivariate normal distribution with mean 0 and variance
where , ,
and , , , and .
Note that has two components. The second component, , is the additional variabilities in , due to the estimation of . For given covariate , since is a continuous function of , the asymptotic properties for can be established by applying the continuous mapping theorem and delta method.
3 ∣. COVARIATE-SPECIFIC ROC CURVE
The local model in (1) and (2) pertains to a given sensitivity level . This can be naturally extended to the whole spectrum of sensitivity values to obtain a global model. For cases, the quantile regression model for any sensitivity becomes
| (3) |
and for controls, the coefficients of logistic regression also vary with
| (4) |
Again, the logit link here can be replaced by other link functions. Since and are distribution functions for all , there are natural constraints on the coefficient processes, and in the preceding models. Obviously, needs to be non-increasing in for all in Equation (3). With being differentiable, that is equivalent to for all . For the controls with any ,
Plugging the right-hand side of (4), we have , . Given (3) holds, the constraint simplifies to for all .
Of course, the above general model is more restrictive than the earlier local model. Nevertheless, the covariate effects are allowed to vary over various sensitivity levels. Thus, it remains to be more general than the existing methods9,11,15,16, just like the local model as discussed before.
We could apply the estimation procedure developed for local model to estimate the parameters of (3) and (4) in a pointwise way based on the estimating equations:
The computational burden may seem heavy as the solutions may be needed for each and every . However, the estimator is actually a step function and can be efficiently solved by the parametric programming algorithm described in Koenker (2005).19 Portnoy (1991)20 showed that the number of breakpoints is , where is the number of covariates and is the sample size. For logistic regression, one only needs to solve the estimator when changes, which is a subset of the breakpoints in quantile regression. Our R/CRAN package, caROC, provides efficient implementations for both local and global models.
3.1 ∣. An asymptotic analysis
To derive the asymptotic properties of the global model, we strengthen Condition 4a.
Condition 4b. Both and have density functions and , respectively, which are continuous in for given and bounded uniformly in and over the supports of and , respectively. Meanwhile, is continuously differentiable on for any and such that .
This condition is also standard and has been used before. For example, Janes and Pepe (2009) used similar conditions for the existence of density function when the ROC curve was of interest.6 Similarly, the differentiability of the quantile regression estimand has been adopted in Koenker (2005).19
Theorem 2. Suppose that the quantile regression model for the cases given in (3) and the logistic regression model for controls given in (4) holds globally over sensitivity levels through with , along with Conditions 1, 2, 3, and 4b. Then, converges almost surely to uniformly over . Furthermore, , converges weakly to a Gaussian process over .
3.2 ∣. Monotonization and inference
There is inherent monotonicity in covariate-specific ROC curves for all , and accordingly and are necessarily monotonicity-respecting. However, as both quantile regression and logistic regression are solved in a point-wise fashion, lack of respect for such monotonicity may arise in , , and subsequently the estimated covariate-specific ROC curves and so do illogical results. The monotonicity-respecting restoration method of Huang (2017)21 may be used, targeting either and or the estimated covariate-specific ROC curves. In our related work17, the regression-based and the ROC-based monotonization method demonstrate comparable accuracy in the estimations, but ROC-based method has better computational performance. In this work, we shall adopt ROC-based monotonization. Consider an estimated covariate-specific ROC curve , which is a step function; note that we view an ROC curve as 1-specificity versus sensitivity in this article. Denote the set of break points along with boundary points, i.e., 0 and 1, by . From a starting point , we find the left nearest monotonicity-respecting neighbor in as . Each identified point then has its own left nearest monotonicity-respecting neighbor, and we repeat this procedure until no such neighbor exists. In the opposite direction, we can similarly identify the right nearest monotonicity-respecting neighbor of , , and recursively identify all the right monotonicity-respecting points. We denote the set containing all these points including the starting one by . A monotonized covariate-specific ROC curve is obtained by linear interpolating over the points in . As discussed in Huang (2017)21, the monotonicity-restored estimator is robust to the potential tail instability of the original estimators as long as is selected away from the tails. Additionally, Huang (2017)21 established the asymptotic equivalence between the monotonized and original esitmators. Therefore, our asymptotic theory applies for estimators with monotonicity restoration as well.
For inference, the procedures described for the local model could be adopted if a point on the ROC curve is of interest. When inference of the whole ROC curve is needed, one may construct a confidence band using a non-parametric bootstrap. Conditional on the data, the distribution of is asymptotically the same as . Thus, given a set of interested covariates , the -level equal-precision confidence band of can be constructed by
where with and is the standard error of . is the estimated -percentile of . is the standard error obtained from bootstrap resamples. For a monotonized ROC curve, the confidence band can be similarly obtained by replacing the and with their monotonized versions.
4 ∣. SIMULATIONS
We evaluate the finite sample properties of the proposed method through two simulation studies. In each study, we compare the proposed method with three existing covariate-specific ROC estimation methods: Pepe (1998)9, Faraggi (2003)11, and Inácio de Carvalho et al. (2013)12, which have been implemented in R/CRAN package ROCnReg.22 These existing methods are adapted for covariate-specific specificity at controlled sensitivity levels by switching the roles of cases and controls. Unfortunately, many other methods do not have their software readily available and thus are not included for comparison.
Suppose that the biomarker in cases and controls rely on two continuous covariates and , both of which follow uniform distribution in region [0, 1]. In the first simulation setting, the biomarker in cases is associated with the two covariates under the quantile regression model (1) with coefficients , , , . The biomarker in controls is associated with the two covariates under the logistic regression model (2) with coefficients , , , , where and is the cumulative density function for standard normal distribution. The true specificities at controlled sensitivity levels for given observation could be obtained from .
The first simulation setting uses the modeling assumptions that our proposed model holds but not the three existing ones. To provide a fair comparison with the three existing methods, we design the second simulation setting that all three models hold. In the second setting, the case biomarkers is associated with the two covariates through normal distribution and control biomarkers . Denote the distribution functions for and by and . For a given sensitivity and covariates , the covariate-specific specificity is .
The estimation accuracy of the proposed method and the performance of the bootstrap inference are evaluated at three different covariate values and four sensitivity levels. Table 1 reports the performance using the proposed method with bootstrap-based inference as well as using the semiparametric method proposed by Pepe (1998)9 and Faraggi (2003)11, respectively, under covariate value . The four blocks of rows correspond to the results under different specified sensitivity levels. All presented results are summarized over 5000 Monte Carlo datasets. First, our proposed method overall achieves good accuracy for all covariate values. The estimation accuracy is higher when the controlled sensitivity is away from the 1 () compared to near the border (). Our confidence intervals have good coverage rates in most sample sizes and covariate settings. A logit-transformation based 95% confidence interval is adopted here, as the logit-transformation based confidence interval is more stable than the regular confidence interval when sensitivity is near 0 or 1. Second, the bootstrap inference has a stable and good variance estimation as well as coverage rate even when sample size is relatively small. The standard errors are very close to the empirical standard deviations. Lastly, compared with the proposed method, Pepe (1998)9 and Faraggi (2003)11 overall have larger bias, worse standard error estimations and lower coverage probabilities. The semiparametric method by Pepe (1998)9 has much better performance than the results by Faraggi (2003)11.
TABLE 1.
Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at covariate value .
| n1 = n0 | Proposed method | Pepe (1998) | Faraggi (2003) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bias | SD | SE | LCov | Bias | SD | SE | LCov | Bias | SD | SE | LCov | |
| 100 | 174 | 1061 | 1126 | 96.0 | 348 | 992 | 936 | 91.4 | 1250 | 866 | 782 | 59.0 |
| 200 | 78 | 710 | 779 | 96.4 | 181 | 687 | 660 | 92.7 | 1250 | 622 | 571 | 41.3 |
| 500 | 22 | 435 | 469 | 96.1 | 47 | 429 | 415 | 94.1 | 1230 | 391 | 371 | 12.2 |
| 1000 | 21 | 306 | 323 | 95.2 | 6 | 298 | 292 | 94.0 | 1230 | 275 | 264 | 1.22 |
| 5000 | 3 | 136 | 140 | 95.1 | −22 | 135 | 130 | 93.6 | 1230 | 124 | 119 | 0 |
| 100 | 134 | 1086 | 1184 | 97.2 | 256 | 1030 | 963 | 92.1 | 876 | 769 | 704 | 68.1 |
| 200 | 71 | 752 | 807 | 96.0 | 123 | 728 | 693 | 93.1 | 886 | 543 | 506 | 55.4 |
| 500 | 20 | 457 | 492 | 96.0 | 19 | 459 | 443 | 94.3 | 872 | 342 | 326 | 27.9 |
| 1000 | 19 | 329 | 343 | 95.2 | −11 | 326 | 313 | 93.8 | 871 | 240 | 231 | 6.32 |
| 5000 | 2 | 147 | 150 | 95.3 | −34 | 144 | 140 | 93.8 | 874 | 108 | 104 | 0 |
| 100 | 93 | 1035 | 1113 | 97.2 | 173 | 985 | 901 | 92.9 | 406 | 665 | 614 | 81.6 |
| 200 | 53 | 711.4 | 766 | 96.6 | 85 | 692 | 659 | 93.8 | 415 | 464 | 437 | 76.9 |
| 500 | 17 | 441 | 473 | 96.0 | 2 | 440 | 424 | 93.9 | 406 | 292 | 280 | 64.9 |
| 1000 | 15 | 320 | 329 | 94.8 | −23 | 314 | 303 | 94 | 405 | 206 | 198 | 45.8 |
| 5000 | −1 | 142 | 145 | 95.1 | −38 | 141 | 136 | 92.8 | 408 | 92.4 | 88.7 | 0.8 |
| 100 | 63 | 928 | 998 | 97.4 | 106 | 869 | 799 | 93.3 | −19 | 573 | 532 | 90.9 |
| 200 | 44 | 648 | 685 | 96.5 | 41 | 622 | 587 | 93.9 | −11 | 397 | 377 | 92.2 |
| 500 | 10 | 399 | 424 | 96.0 | −12 | 391 | 381 | 94.6 | −18 | 250 | 240 | 93.0 |
| 1000 | 12 | 286 | 297 | 95.2 | −31 | 282 | 272 | 93.7 | −19 | 177 | 170 | 93.2 |
| 5000 | −2 | 128 | 131 | 95.3 | −39 | 126 | 122 | 92.5 | −16 | 79.1 | 75.9 | 93.3 |
Bias, ; SD, standard deviation ×104; SE, standard error estimated using bootstrap ×104; LCov (%), coverage rates of logit transformation-based 95% confidence interval.
Table S1-S3 presents the performance of all three methods with three different covariate selections. The results are also summarized over the same 5000 simulation datasets as in Table 1. As discussed in the introduction, Pepe (1998)9 and Faraggi (2003)11 adjusted covariate effects using a general model over the entire ROC curve, which may not be able to handle the changing covariate effects well. We find the proposed method has good accuracy and superior coverage probabilities compared to the two existing methods in all covariate selections. This comparison may not be completely fair since the data is generated based on our model. However, it shows that the methods by Pepe (1998)9 and Faraggi (2003)11 may not provide accurate ROC estimation and inference when the covariate effect changes with specificity levels, as is the situation in our simulation setting.
Table 2 presents the performance of the three methods at covariate value in the second simulation setting. Table S4-S6 present the simulation results using our proposed method and the two existing methods in this setting at the other two covariate values. The model assumption holds for all methods in this setting. All the results are summarized over 5000 Monte Carlo datasets. As expected, we observe that the two existing methods show improved biases results compared to the ones from the previous setting. Meanwhile, our proposed method demonstrates comparable and sometimes even better results in comparison to the two existing methods, suggesting the favorable performance and robustness of the proposed method. The existing methods of Pepe (1998)9 and Faraggi (2003)11 have slightly higher efficiency compared to the proposed method. This is not surprising as our proposal uses a non-parametric approach to model the covariate effect on the case population.
TABLE 2.
Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods9,11 are estimated and presented.
| Proposed method | Pepe (1998) | Faraggi (2003) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bias | SD | SE | LCov | Bias | SD | SE | LCov | Bias | SD | SE | LCov | |
| 100 | 136 | 1025 | 1073 | 93.5 | 468 | 1000 | 932 | 91.1 | 130 | 695 | 654 | 91.1 |
| 200 | 63 | 658 | 734 | 95.7 | 242 | 672 | 644 | 93.4 | 68 | 475 | 456 | 92.7 |
| 500 | −16 | 401 | 432 | 96.7 | 88 | 406 | 389 | 94.1 | 21 | 294 | 282 | 93.3 |
| 1000 | −40 | 281 | 297 | 95.9 | 49 | 280 | 275 | 94.2 | 13 | 206 | 200 | 93.5 |
| 5000 | −50 | 125 | 128 | 94.0 | 8 | 126 | 122 | 93.4 | 2 | 91.7 | 89 | 93.3 |
| 100 | 87 | 1377 | 1495 | 97.4 | 389 | 1300 | 1180 | 92.6 | 76 | 1030 | 944 | 91.3 |
| 200 | 37 | 957 | 1039 | 97.1 | 210 | 915 | 861 | 93.5 | 46 | 727 | 686 | 92.8 |
| 500 | −42 | 622 | 642 | 95.5 | 68 | 578 | 557 | 94.2 | 8 | 457 | 439 | 93.1 |
| 1000 | −55 | 432 | 449 | 95.3 | 42 | 409 | 394 | 94.2 | 9 | 322 | 313 | 93.5 |
| 5000 | −60 | 193 | 196 | 94.0 | 9 | 184 | 178 | 93.3 | 1 | 145 | 141 | 93.0 |
| 100 | 90 | 1395 | 1514 | 97.8 | 244 | 1270 | 1160 | 92.8 | 9 | 1060 | 959 | 91.5 |
| 200 | 54 | 976 | 1045 | 96.7 | 135 | 911 | 857 | 93.5 | 15 | 746 | 701 | 92.9 |
| 500 | 11 | 625 | 652 | 95.5 | 48 | 577 | 558 | 94.1 | −6 | 468 | 451 | 93.1 |
| 1000 | 11 | 440 | 454 | 95.5 | 27 | 407 | 398 | 94.4 | 3 | 330 | 321 | 93.1 |
| 5000 | 8 | 195 | 200 | 95.0 | 3 | 187 | 179 | 93.6 | 0 | 149 | 144 | 93.3 |
| 100 | 78 | 1206 | 1328 | 98.2 | 139 | 1100 | 1010 | 93.4 | −33 | 931 | 844 | 91.9 |
| 200 | 82 | 845 | 909 | 96.6 | 87 | 801 | 750 | 93.4 | −7 | 656 | 614 | 93.0 |
| 500 | 58 | 540 | 562 | 95.4 | 22 | 504 | 483 | 93.8 | −13 | 410 | 395 | 93.1 |
| 1000 | 63 | 381 | 393 | 94.9 | 16 | 353 | 346 | 94.1 | −1 | 289 | 281 | 93.2 |
| 5000 | 63 | 170 | 171 | 93.3 | 3 | 160 | 156 | 93.8 | −1 | 131 | 126 | 93.1 |
Bias, ; SD, standard deviation ×104; SE, standard error estimated using bootstrap ×104; LCov (%), coverage rates of logit transformation-based 95% confidence interval.
The comparison between our proposed method and Inácio de Carvalho et al. (2013)12 is presented in Table S5-10 and Figure S1-8. The implementation of Inácio de Carvalho et al. (2013) is slower than our proposed and two other existing methods for the construction of confidence interval. Thus we only perform 100 Monte Carlo iterations and summarize the results in supplementary materials, not 5000 Monte Carlo iterations as shown in Table 1 and 2. In the first simulation setting (Table S5-7), we observe that the proposed method generally has smaller bias and better coverage probability for the setting (0.5,0.5)T, (0.25,0.75)T, and (0.75,0.75)T. In settings of (0.25,0.25)T and (0.75,0.25)T, Inácio de Carvalho et al. (2013) are better in some sample sizes. In the second simulation setting with Gaussian data (Table S8-10), the two methods are mostly comparable in bias and covarage probability. However, Inácio de Carvalho et al. (2013) has a smaller estimation variance compared to our proposed method, as our method has less assumptions. The advantage of the proposed method in computational performance is substantial. For the sample size of 5000, the Inácio de Carvalho et al. (2013) method takes about 3.8 minutes to construct confidence interval at a given sensitivity level (TimeCI) while the proposed method with bootstrap confidence interval construction only takes about 3 seconds. However, the construction of confidence band using bootstrap by our method (TimeCB) is slower than Inácio de Carvalho et al. (2013) when the sample size is very large (e.g., 1000 or 5000).
We also evaluate the performance our method with monotonicity restoration under the two simulations. Table S11 shows the bias, standard deviation, standard error, and the coverage probability. As shown in Huang (2017)21, the monotonized and the original estimators are asymptotically equivalent. Comparing Table S11 versus Table 1 and 2 in the main manuscript, we find that the results using the proposed method with monotonicity restoration have similar performance as the method without monotonicity restoration, which is consistent with the previous findings21. Overall, the results using our method with monotonicity restoration show good variance estimation and coverage probability.
5 ∣. ILLUSTRATION WITH TWO CLINICAL DATASETS
Many previous studies have reported improved outcomes from treating aggressive prostate cancer patients at an early stage.23,24 However, such survival benefits can be undermined by harms from treating over-diagnosed indolent prostate cancer patients. To improve the diagnostic accuracy, biomarkers for aggressive prostate cancer usually need to achieve high specificity while maintaining sensitivity at a desirable level to provide clinical utility. Below, we illustrate the usage of the proposed method through a multi-center clinical study for aggressive prostate cancer.
The whole NCI-EDRN dataset was collected by researchers from Harvard University, Cornell University and Michigan University over the past two decades.1,25 It enrolled a total of 2261 men and collected their pre-diagnosis biomarkers, characteristics and the biopsy-confirmed diagnosis. Among them, 615 were aggressive prostate cancer patients with Gleason scores ≥ 7 and the rest had indolent prostate cancer or were normal controls. We provide evaluations of two biomarkers, prostate-specific antigen (PSA) and prostate health index (phi). In the first part of the analysis, we will use all patients and apply the proposed method to evaluate PSA. Since phi is a much newer biomarker than the PSA, only a subset of 502 men in the data has phi measurement and will be included in the second part of the analysis.
5.1 ∣. Covariate-specific evaluation of PSA
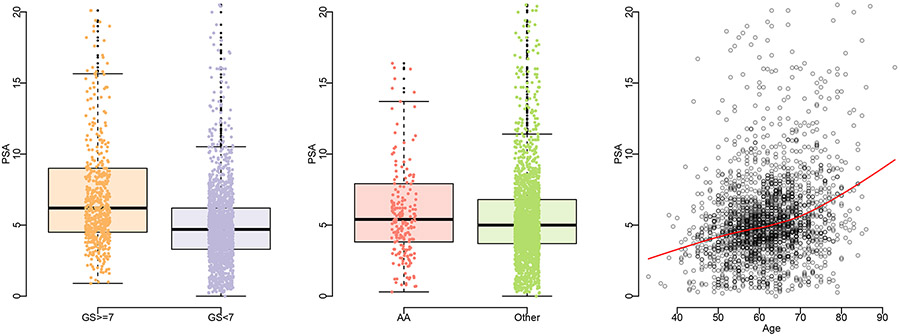
The data from the total 2261 men are used for the evaluation of PSA. The left panel of Figure 1 shows that the subjects with Gleason score equal to or greater than 7 tend to have higher PSA values. The impacts of patient characteristics on PSA have been reported before in many publications. For example, it is known that elder men and African American men tend to have elevated PSA values.26,27 The right panel of Figure 1 demonstrates a clear trend of higher PSA for elder men. Although we have a very unbalanced distribution of being African American (AA) - only 219 out of 2261 men are African American men, we still observe consistent covariate effects of AA on PSA as shown in the middle panel of Figure 1. As a nonlinear trend of age on PSA can be observed in Figure 1, we also consider including squared age into the modeling. However, the term is not significant in both the quantile regression and logistic regression, and thus we exclude the squared term from the final analysis. Motivated by all the observations, we include age and being African American (AA) as covariates in the following evaluation of PSA.
FIGURE 1.
Exploratory plots of PSA versus patient characteristics in the NCI-EDRN cohort.
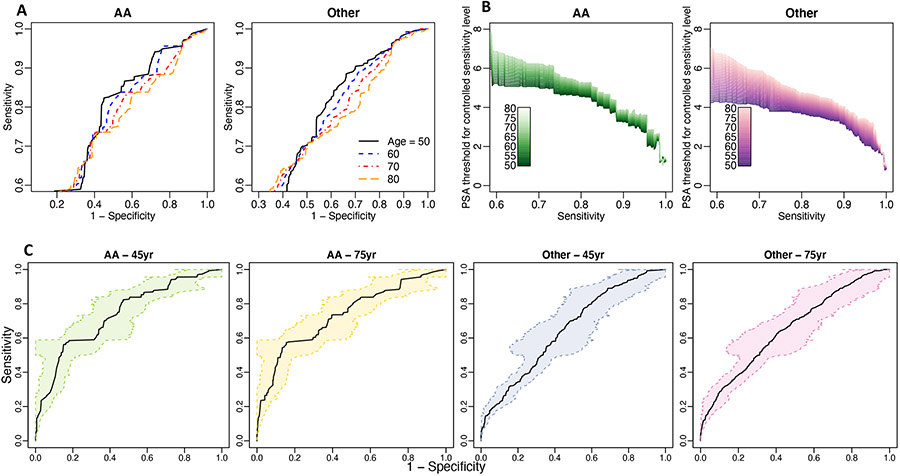
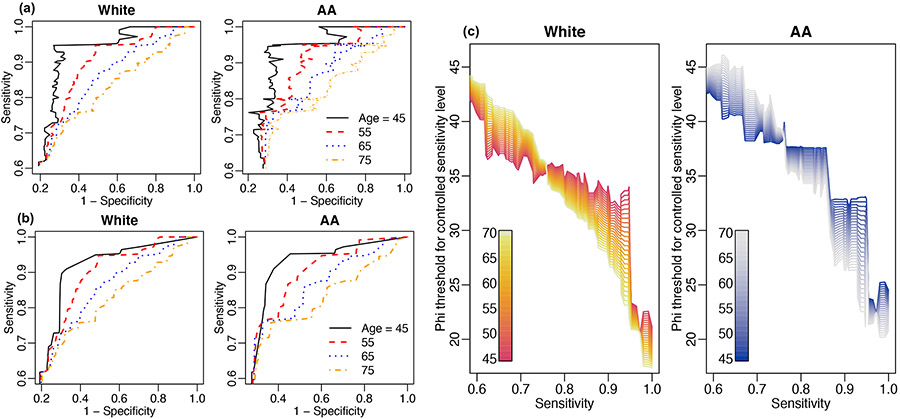
Figure 2 presents the results obtained from applying the proposed methods. Panel A demonstrates that PSA has better specificity at high controlled sensitivity levels for younger patients (age = 50) than older patients (age = 80) in both African American and other races. Our results indicate that older and African American men need higher PSA thresholds to achieve the same controlled sensitivity level (Figure 2 Panel B). This figure could help clinicians to make diagnostic decisions for patients in different age and race groups controlling sensitivity at the same high level. We then obtain the 95% bootstrap-based confidence bands of the monotonized ROC curves for age 45 and 75 years old in both African American and other subpopulations (Figure 2 Panel C).
FIGURE 2.
Results from applying the proposed method to PSA of the NCI-EDRN cohort. Panel A is the monotonized ROC curve. Panel B is the PSA threshold for controlled sensitivity level for different race and age groups. Panel C is the 95% bootstrap-based confidence band for the monotonized ROC curve in different subpopulations.
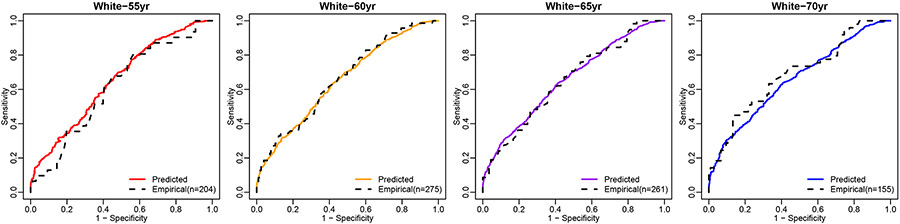
We also perform a model check with this relatively large dataset. Without model specification, calculating covariate-specific ROC curve requires sub-setting the data at each of the covariate values. When datasets have limited sample sizes, sub-setting data to each age level generally results in too few data points to construct an ROC curve. The large sample size of this NCI-EDRN dataset provides an opportunity for us to scrutinize our model fitting by comparing the model-based ROC curves with empirical ROC curves. Figure 3 shows that the predicted ROC curves using our proposed method are very close to the empirical ROC curves, especially when a good number of data points for the specific covariate are available. For example, there are a total of 275 patients being both White and around 60 years old (59 ≤ age ≤ 61). The constructed empirical ROC curve aligns well with our predicted ROC curve for this subpopulation (yellow curve). These results confirm that the proposed method provides a good fit for the data.
FIGURE 3.
The covariate-specifc ROC curve estimated from the proposed method and the empirical ROC curve based on NCI-EDRN data for the biomarker PSA.
5.2 ∣. Covariate specific evaluation of phi
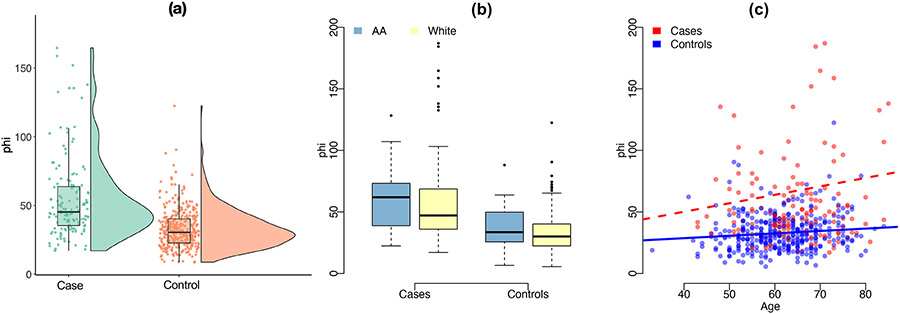
The Beckman Coulter® Prostate Health Index or phi is an FDA-approved multi-analyte blood test for more accurate prostate cancer detection. Proposed in 2010, phi combines three measurements, total prostate-specific antigen (PSA), free PSA and p2PSA, into a mathematical formula .28 It has been reported that the men with a higher total PSA and p2PSA as well as a lower free PSA are more likely to have clinically significant prostate cancer.29,30,31 As a result, a larger phi value indicates more risk for aggressive prostate cancer. The prostate health index may be more accurate in detecting prostate cancer than PSA.32,33,34 Among the 502 patients, a total of 352 patients are biopsy-confirmed aggressive prostate cancer patients. Figure 4(a) shows the distributions of phi in cases and controls, respectively. It can clearly be seen that aggressive prostate cancer patients tend to have higher phi values than the controls.
FIGURE 4.
Exploratory plots for the aggressive prostate cancer clinical study with the biomarker phi.
The covariates under consideration here are again age and being AA. The subjects analyzed in this part also have very unbalanced distributions in both covaraites. The majority of the patients are between age 50 and 70. Only 49 of the subjects are African Americans. Nonetheless, the observations in Figure 4(b) and (c) confirm the covariate effect of age and AA on phi. We observe that African-American men have higher phi values than White men in both cases and controls. In addition, elder men are more likely to have higher phi, especially in the case subjects (p=0.0498 for the interaction of age and disease status in linear regression model).
Figure 5 presents the results of applying the proposed method to this clinical data. Controlling equal sensitivity levels among different covariate groups, we evaluate the diagnostic accuracy of phi for specific sub-populations. Figure 5(a) shows the covariate-specific ROC curves and (b) is the smoothed covariate-specific ROC curves after applying ROC-based monotonization. The presented ROC curves are truncated to sensitivity levels greater than 0.6, as high sensitivity levels are usually desired for clinical utility. We observe phi has better diagnostic performance in younger patients, for example, around age 45 years old, than older patients around age 75. The raw ROC curves of AA men are bumpier than the curves in White men, because this study has fewer AA men than White men, as discussed above. The trend is similar for both raw and monotonized ROC curves. Figure 5(c) is the estimated phi threshold at controlled sensitivity levels for different age groups in White and AA men, respectively.
FIGURE 5.
Covariate-adjusted ROC curve for specific sub-populations from the aggressive prostate cancer clinical trial data by controlling sensitivity levels for the biomarker phi.
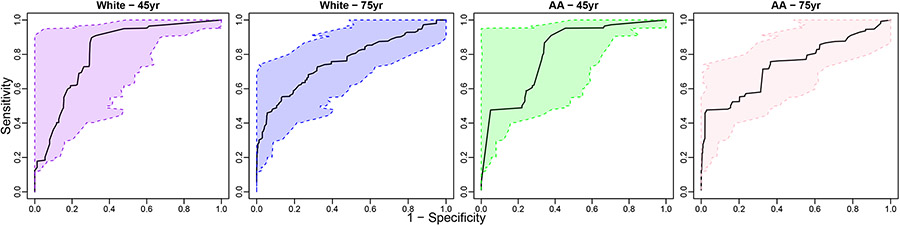
We also obtain the 95% bootstrap-based confidence band for the covariate-specific ROC curves (Figure 6). The presented ROC curves and the related confidence intervals have been monotonized by ROC-based monotonization methods. Compared to the first part, the confidence bands are wider in the current application due to the limited sample size.
FIGURE 6.
The estimated covariate-adjusted ROC curve (black solid curves) with ROC-based monotonization and bootstrap-based confidence bands (colored areas between the dashed lines) for the biomarker phi.
6 ∣. DISCUSSION
In this work, we develop an approach to evaluate the performance of continuous biomarkers at specific covariate levels. It extends our previous work on pooled evaluation with covariate-adjusted threshold17. Although the modeling for the diseased population under quantile regression framework is similar to Li et al. 202117, the covariate-specific evaluation requires further modeling on the controls, which substantially increases the model complexity.
Compared with existing methods, our contribution is twofold. First, by adopting a combined framework of quantile regression and logistic regression, our method allows flexible local covariate adjustment and covariate-specific evaluation for continuous biomarkers. The proposed method is more general than previous methods in many aspects and demonstrates favorable performance. Second, the establishment of asymptotic properties and inference procedures lays a solid foundation to the applications of the proposed method. Our R implementations, wrapped in the R/CRAN package caROC, contain efficient estimation procedures and graphical functions. These allow researchers to easily apply the proposed method for clinical biomarker evaluation. The package provides options for users to control sensitivity or specificity, as well as to specify covariate values of interest. In this era of precision medicine, our method offers a useful tool to improve subpopulation-specific diagnosis.
Supplementary Material
Table S1. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S2. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S3. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S4. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S5. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 500 Monte Carlo iterations.
Table S6. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S7. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S8. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S9. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S10. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S11. Evaluation of the proposed method with monotonicity restoration under the first and second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method with monotonization are estimated and presented. Results were summarized over 5000 Monte Carlo iterations.
Figure S1. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.8. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S2. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.85. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S3. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.90. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S4. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.95. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S5. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.80. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S6. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.85. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S7. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.90. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S8. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.95. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
ACKNOWLEDGMENTS
This project was partly supported by the National Institutes of Health grants R01CA230268, CA113913, and R03CA270725. The authors also thank Dr. David Howard from Emory University and Dr. Yu Liu from MD Anderson Cancer Center for their helpful discussions during the real data analysis.
Abbreviations:
- ROC
Receiver Operating Characteristic
parametric distribution-free
- PSA
prostate-specific antigen
- phi
prostate health index
Biographies

Ziyi Li is an Assistant Professor in the Department of Biostatistics at The University of Texas MD Anderson Cancer Center, Houston, TX. She is a a statistician and data scientist who develops statistical and machine learning methods and applies them to different high-dimensional biomedical data. She is also interested in collaboration projects with Biologist and Physicians. Her previous collaboration project involves the study of cancer, Alzheimer’s disease, autism, obesity and cardiovascular diseases.

Yijian (Eugene) Huang is Professor in the Department of Biostatistics and Bioinformatics at Rollins School of Public Health of Emory University. His methodological research interests include survival analysis, measurement errors in covariates, and disease diagnosis using biomarkers. He has collaborated in clinical research of HIV/AIDS, cancer, renal disease, and cardiovascular diseases.

Dattatraya Patil is a senior Biostatistician in the Department of Urology at Emory University School of Medicine. He is the co-author of more than 200 manuscripts for Urological cancers, biomarker detections, and health services research.

Mark Rubin is Professor and Director, Department for BioMedical Research, University of Bern, Switzerland; Project Leader for Precision Medicine, University Hospital of Bern, Switzerland; Professor, Pathology and Laboratory Medicine, Weill Cornell Medical College, New York, NY. His laboratory focuses on understanding prostate cancer disease progression, taking a functional genomics approach. Most recently, he has been trying to understand therapy resistance in the context of lineage plasticity which implicates epigenetic as well as genomic alterations.

Martin G Sanda is an internationally recognized prostate cancer surgeon and scientist. He is the appointed Chair of the Department of Urology at Emory University School of Medicine and service chief for Emory Healthcare. Dr. Sanda’s clinical practice, which includes robotic prostatectomy and robotic cystectomy, is focused on developing new surgical and non-surgical approaches to cancer care and to improving the quality of life among cancer survivors.
APPENDIX
A PROOF FOR THEOREM 1
The asymptotic properties of the quantile regression estimator have been established before, e.g. Koenker (2005, Section 4.1.1 and Theorem 4.1),
| (A1) |
where and are defined in Theorem 1.
Write
Note that is the solution to
By the Glivenko-Cantelli theorem, converges to almost surely and uniformly in . Then it follows that converges to almost surely under Condition 4a.
On the other hand, converges to almost surely by Strong Law of Large Numbers for a fixed . This convergence holds uniformly in because of the monotonicity of the function.
Combining the two results, we have shown that converges to almost surely and uniformly in . Since has a unique solution at , converges to almost surely.
Meanwhile, define
Since is Donsker, converges weakly to a Gaussian process. Under Conditions 2 and 4a, is asymptotically uniformly equicontinuous in probability using an argument similar to Huang (2017, appendix). Together with the consistency result of , it follows that
| (A2) |
By component-wise Taylor Expansion, one then obtains
Thus
Note that the left hand side is equal to 0. We apply the component-wise Taylor expansion on the part involving and obtain
By the Central Limit Theorem,
The asymptotic normality of has been established in (A1). Meanwhile, is independent of . Therefore,
B PROOF FOR THEOREM 2
Write
With the cases, the consistency of has been shown previously, e.g., Li et al. (2021+). Turning to the controls, note that is Donsker. By the Glivenko-Cantelli theorem, almost surely,
| (B3) |
With the consistency of and the continuity of , we have
almost surely. As a result, almost surely,
| (B4) |
The uniform convergence of to holds following the same arguments in the proof of Theorem 1. Thus, almost surely,
| (B5) |
By definition, for any ,
Results (B4) and (B5) then lead to, almost surely,
By component-wise Taylor Expansion, almost surely,
Since the minimum eigen value of is bounded away from 0 and is also bounded by Condition 4b,
almost surely.
Now we prove the weak convergence of the proposed estimators. The weak convergence of has been obtained in Li et al. (2021+):
| (B6) |
uniformly in .
With the afore given Donsker result, converges weakly to a Gaussian process. Under Conditions 2 and 4b, is asymptotically uniformly equicontinuous in probability using arguments similar to those given by Huang (2017, appendix). Therefore,
| (B7) |
Since and ,
| (B8) |
Using the similar uniform equicontinuous argument for , we have
| (B9) |
Results (B8) and (B9) togerther lead to
| (B10) |
To build the connection between and , and , respectively, we apply the component-wise Taylor expansion. Almost surely,
| (B11) |
and
| (B12) |
Combining results (B10), (B11) and (B12) leads to
Together with (A1), we have
uniformly in . Then over converges weakly to a Gaussian process.
Footnotes
Conflict of interest
The authors declare no potential conflict of interests.
Financial disclosure
None reported.
SUPPORTING INFORMATION
The following supporting information is available as part of the online article:
Data availability statement
The proposed methods together with sample simulation data have been wrapped in R/CRAN package caROC. This software is freely available from the CRAN website https://cran.r-project.org/web/packages/caROC/index.html. The prostate data that were analyzed in this study are not publicly available due to privacy or ethical restrictions. The data are available upon reasonable request from Dr. Martin Sanda at the Department of Urology of Emory University.
References
- 1.Sanda MG, Feng Z, Howard DH, et al. Association between combined TMPRSS2: ERG and PCA3 RNA urinary testing and detection of aggressive prostate cancer. JAMA oncology. 2017;3(8):1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou XH, Qin G. Improved confidence intervals for the sensitivity at a fixed level of specificity of a continuous-scale diagnostic test. Statistics in medicine. 2005;24(3):465–477. [DOI] [PubMed] [Google Scholar]
- 3.Qin G, Davis AE, Jing B. Empirical likelihood-based confidence intervals for the sensitivity of a continuous-scale diagnostic test at a fixed level of specificity. Statistical methods in medical research. 2011;20(3):217–231. [DOI] [PubMed] [Google Scholar]
- 4.Pepe MS. The statistical evaluation of medical tests for classification and prediction. Medicine; 2003. [Google Scholar]
- 5.Pepe MS. A regression modelling framework for receiver operating characteristic curves in medical diagnostic testing. Biometrika. 1997;84(3):595–608. [Google Scholar]
- 6.Janes H, Pepe MS. Adjusting for covariate effects on classification accuracy using the covariate-adjusted receiver operating characteristic curve. Biometrika. 2009;96(2):371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Zhou XH. ROC analysis in biomarker combination with covariate adjustment. Academic radiology. 2013;20(7):874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tosteson ANA, Begg CB. A general regression methodology for ROC curve estimation. Medical Decision Making. 1988;8(3):204–215. [DOI] [PubMed] [Google Scholar]
- 9.Pepe MS. Three approaches to regression analysis of receiver operating characteristic curves for continuous test results. Biometrics. 1998;:124–135. [PubMed] [Google Scholar]
- 10.Dodd LE. Regression methods for areas and partial areas under the receiver-operating characteristic curve.. 2002;. [Google Scholar]
- 11.Faraggi D Adjusting receiver operating characteristic curves and related indices for covariates. Journal of the Royal Statistical Society: Series D (the Statistician). 2003;52(2):179–192. [Google Scholar]
- 12.Inacio-De-Carvalho V, Jara A, Hanson TE, Carvalho M, others. Bayesian nonparametric ROC regression modeling. Bayesian Analysis. 2013;8(3):623–646. [Google Scholar]
- 13.Inacio-De-Carvalho V, Rodríguez-Álvarez MX. The covariate-adjusted ROC curve: the concept and its importance, review of inferential methods, and a new Bayesian estimator. Statistical Science. 2021;in press. [Google Scholar]
- 14.Pepe MS. An interpretation for the ROC curve and inference using GLM procedures. Biometrics. 2000;56(2):352–359. [DOI] [PubMed] [Google Scholar]
- 15.Alonzo TA, Pepe MS. Distribution-free ROC analysis using binary regression techniques. Biostatistics. 2002;3(3):421–432. [DOI] [PubMed] [Google Scholar]
- 16.Cai T, Pepe MS. Semiparametric receiver operating characteristic analysis to evaluate biomarkers for disease. Journal of the American statistical Association. 2002;97(460):1099–1107. [Google Scholar]
- 17.Li Z, Huang Y, Patil D, Sanda MG. Covariate adjustment in continuous biomarker assessment. Biometrics. 2021;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenker R, Bassett G. Regression quantiles. Econometrica: journal of the Econometric Society. 1978;:33–50. [Google Scholar]
- 19.Koenker R Quantile Regression (Econometric Society Monographs). Cambridge university press; 2005. [Google Scholar]
- 20.Portnoy S Asymptotic behavior of the number of regression quantile breakpoints. SIAM journal on scientific and statistical computing. 1991;12(4):867–883. [Google Scholar]
- 21.Huang Y Restoration of monotonicity respecting in dynamic regression. Journal of the American Statistical Association. 2017;112(518):613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodríguez-Álvarez MX, Inacio V. ROCnReg: An R package for receiver operating characteristic curve inference with and without covariate information. arXiv preprint arXiv:2003.13111. 2020;. [Google Scholar]
- 23.Bill-Axelson A, Holmberg L, Garmo H, et al. Radical prostatectomy or watchful waiting in early prostate cancer. New England Journal of Medicine. 2014;370(10):932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Amico AV, Manola J, Loffredo M, Renshaw AA, DellaCroce A, Kantoff PW. 6-month androgen suppression plus radiation therapy vs radiation therapy alone for patients with clinically localized prostate cancer: a randomized controlled trial. Jama. 2004;292(7):821–827. [DOI] [PubMed] [Google Scholar]
- 25.Liss MA, Leach RJ, Sanda MG, Semmes OJ. Prostate Cancer Biomarker Development: National Cancer Institute’s Early Detection Research Network Prostate Cancer Collaborative Group Review. Cancer Epidemiology and Prevention Biomarkers. 2020;29(12):2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson RJ, Eastham JA, Daniel JC, et al. Prostate-specific antigen (PSA) and PSA density: racial differences in men without prostate cancer. Journal of the National Cancer Institute. 1997;89(2):134–138. [DOI] [PubMed] [Google Scholar]
- 27.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nature Reviews Cancer. 2008;8(4):268–278. [DOI] [PubMed] [Google Scholar]
- 28.Jansen FH, Schaik RHN, Kurstjens J, et al. Prostate-specific antigen (PSA) isoform p2PSA in combination with total PSA and free PSA improves diagnostic accuracy in prostate cancer detection. European urology. 2010;57(6):921–927. [DOI] [PubMed] [Google Scholar]
- 29.Djulbegovic M, Beyth RJ, Neuberger MM, et al. Screening for prostate cancer: systematic review and meta-analysis of randomised controlled trials. Bmj. 2010;341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le BV, Griffin CR, Loeb S, et al. [−2] Proenzyme prostate specific antigen is more accurate than total and free prostate specific antigen in differentiating prostate cancer from benign disease in a prospective prostate cancer screening study. The Journal of urology. 2010;183(4):1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guazzoni G, Nava L, Lazzeri M, et al. Prostate-specific antigen (PSA) isoform p2PSA significantly improves the prediction of prostate cancer at initial extended prostate biopsies in patients with total PSA between 2.0 and 10 ng/ml: results of a prospective study in a clinical setting. European urology. 2011;60(2):214–222. [DOI] [PubMed] [Google Scholar]
- 32.Stephan C, Vincendeau S, Houlgatte A, Cammann H, Jung K, Semjonow A. Multicenter evaluation of [−2] proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clinical chemistry. 2013;59(1):306–314. [DOI] [PubMed] [Google Scholar]
- 33.Loeb S, Catalona WJ. The Prostate Health Index: a new test for the detection of prostate cancer. Therapeutic advances in urology. 2014;6(2):74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loeb S, Sanda MG, Broyles DL, et al. The prostate health index selectively identifies clinically significant prostate cancer. The Journal of urology. 2015;193(4):1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S2. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S3. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S4. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and two existing methods (Pepe, 1998; Faraggi, 2003) are estimated and presented.
Table S5. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 500 Monte Carlo iterations.
Table S6. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S7. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S8. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S9. Estimation and inference results under the second simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S10. Estimation and inference results under the first simulation setting. Specificity under controlled sensitivity level at the covariate selection and by our proposed method and an existing Bayesian method (Inácio de Carvalho et al. 2013) are estimated and presented. Results were summarized over 100 Monte Carlo iterations.
Table S11. Evaluation of the proposed method with monotonicity restoration under the first and second simulation setting. Specificity under controlled sensitivity level at the covariate selection by our proposed method with monotonization are estimated and presented. Results were summarized over 5000 Monte Carlo iterations.
Figure S1. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.8. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S2. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.85. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S3. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.90. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S4. Simulation results in the first simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.95. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S5. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.80. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S6. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.85. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S7. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.90. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Figure S8. Simulation results in the second simulation setting of the proposed method (red box) and the Inácio de Carvalho et al. (2013) (blue). From top panel to the bottom panel, the sample sizes are 100, 200, 500, 1000, and 5000. The Sensitivity level is fixed at 0.95. The title of each figure shows the covariate combination for and . All results are summarized over 100 Monte Carlo datasets.
Data Availability Statement
The proposed methods together with sample simulation data have been wrapped in R/CRAN package caROC. This software is freely available from the CRAN website https://cran.r-project.org/web/packages/caROC/index.html. The prostate data that were analyzed in this study are not publicly available due to privacy or ethical restrictions. The data are available upon reasonable request from Dr. Martin Sanda at the Department of Urology of Emory University.