Abstract
This work aimed to develop a modified stack-of-stars golden-angle radial sampling scheme with variable-density acceleration along the slice (kz) dimension (referred to as VD-stack-of-stars) and to test this new sampling trajectory with multi-coil compressed sensing reconstruction for rapid motion-robust 3D liver MRI. VD-stack-of-stars sampling implements additional variable-density undersampling along the kz dimension, so that slice resolution (or volumetric coverage) can be increased without prolonging scan time. The new sampling trajectory (with increased slice resolution) was compared with standard stack-of-stars sampling with fully sampled kz (with standard slice resolution) in both non-contrast-enhanced free-breathing liver MRI and dynamic contrast-enhanced MRI (DCE-MRI) of the liver in volunteers. For both sampling trajectories, respiratory motion was extracted from the acquired radial data, and images were reconstructed using motion-compensated (respiratory-resolved or respiratory-weighted) dynamic radial compressed sensing reconstruction techniques. Qualitative image quality assessment (visual assessment by experienced radiologists) and quantitative analysis (as a metric of image sharpness) were performed to compare images acquired using the new and standard stack-of-stars sampling trajectories. Compared to standard stack-of-stars sampling, both non-contrast-enhanced and DCE liver MR images acquired with VD-stack-of-stars sampling presented improved overall image quality, sharper liver edges and increased hepatic vessel clarity in all image planes. The results have suggested that the proposed VD-stack-of-stars sampling scheme can achieve improved performance (increased slice resolution or volumetric coverage with better image quality) over standard stack-of-stars sampling in free-breathing DCE-MRI without increasing scan time. The reformatted coronal and sagittal images with better slice resolution may provide added clinical value.
Keywords: variable-density stack-of-stars, free-breathing, DCE-MRI, golden-angle radial, GRASP
1. Introduction
Stack-of-stars sampling has become an attractive hybrid 3D MRI acquisition scheme combining advantages of radial and Cartesian trajectories [1–3]. For stack-of-stars acquisition, radial sampling is employed in the kx-ky plane, which enables reduced sensitivity to motion [4–9] and allows for acceleration of k-space acquisitions along both the kx and ky dimensions with incoherence [9–11]. The Cartesian encoding is employed along the slice dimension (kz), which allows for flexible selection of volumetric coverage/slice resolution and also ensures effective fat suppression [1,2]. As a result, this acquisition scheme is well-suited for rapid motion-robust MRI in combination with sparsity-based image reconstruction methods, and it has been demonstrated and evaluated in different clinical applications [3,5,9,12–20].
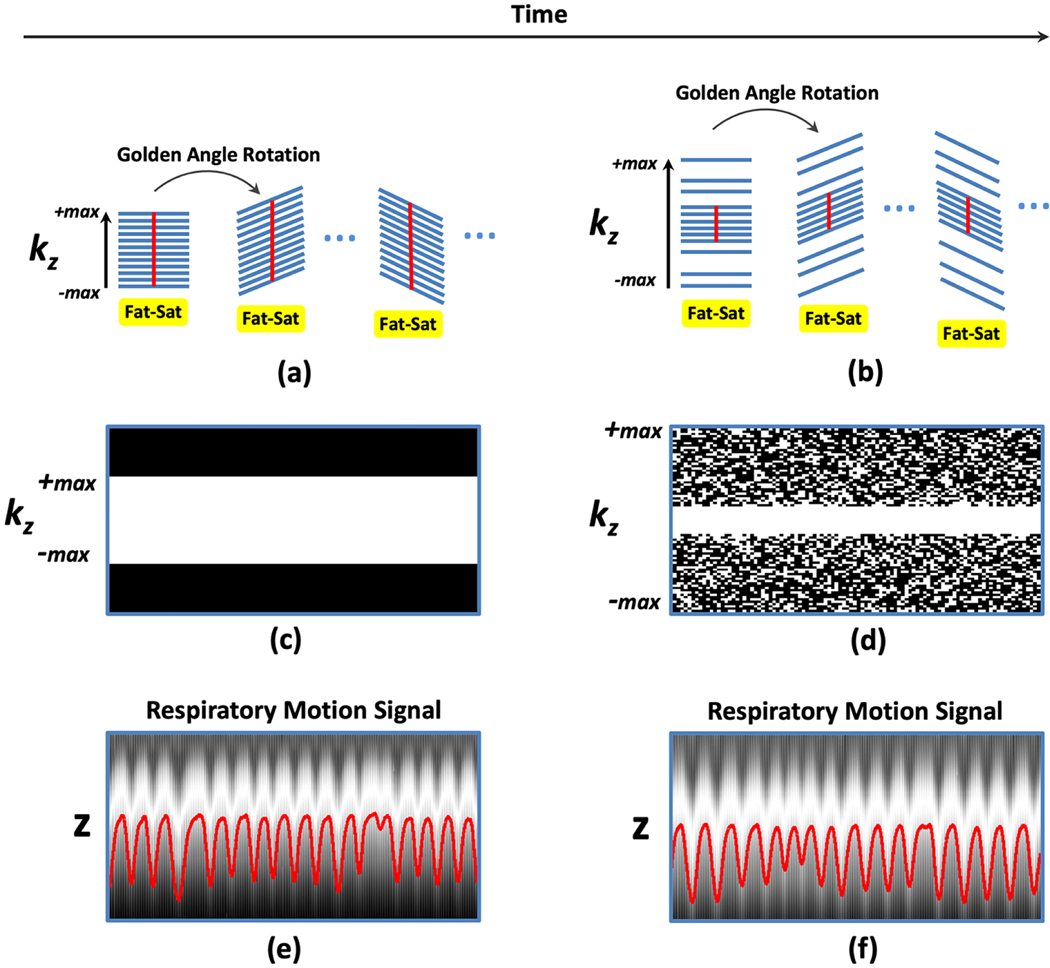
A typical sampling scheme illustrating how conventional stack-of-stars trajectory is implemented is shown in Figure 1a. In this example, the rotation angle of the trajectory follows a golden angle defined as 180°/1.618=111.25° [21]. Specifically, for each rotation angle, imaging normally starts with a fat saturation module (not always required, but essential for avoiding fat-water blurring in radial sampling due to off-resonance) and is followed by acquisition of spokes at different kz positions (referred to as a stack hereafter) without changing the rotation angle. Depending on the fat saturation scheme and other associated parameters in the protocol, the acquisition of each stack can be performed with a linear ordering scheme (sampling from −kzmax to +kzmax as the case in Figure 1a) or with a center-out ordering scheme (sampling from kz=0 to ±kzmax, not shown in the figure). Meanwhile, the kz dimension is typically fully sampled to meet the Nyquist requirement. Such a simple sampling strategy gives a few advantages, including i) parallel reconstruction of different slices after disentangling the Fourier-encoding along kz with a fast Fourier transform (FFT) and ii) smooth change of gradient from partition to partition within each stack to avoid artifacts (e.g., eddy current effects) caused by large gradient jumps. It should be noted that when a golden-angle rotation is employed for stack-of-stars sampling, gradient still changes dramatically when moving from the last spoke of one stack to the first spoke of the next stack. However, this is generally not a problem for two reasons. First, the acquisition of each stack is typically preceded by a fat-saturation module, which causes a delay/gap between two stacks and thus greatly minimize the effects of gradient jumps. Second, in case of linear kz sampling ordering, the gradient jump occurs in the outer k-space region, which has reduced influence to the reconstructed image. When center-out kz sampling order is used, additional dummy spokes can be inserted before the start of the first acquisition of each radial stack.
Figure 1.
(a-b) Comparison of conventional stack-of-stars sampling with fully sampled kz (a) with VD-stack-of-stars sampling with varying kz-t random undersampling (b) for the same number of k-space measurements. (c-d) Corresponding kz-t sampling masks. A VD-stack-of-stars pattern enables larger kz coverage without increasing scan time, which results in increased slice resolution. For stack-of-stars, respiratory motion can be extracted from the fully sampled k-space centers (red straight lines in a) along kz (e). For VD-stack-of-stars sampling, a central k-space region is fully sampled, and the fully sampled central-k-space points (red straight lines in b) can be used for motion detection from corresponding z-direction projection profiles, as shown in (f).
Although fully-sampling the slice dimension facilitates both acquisition and reconstruction in conventional stack-of-stars sampling, it suffers from a few limitations. First, slice resolution is often sacrificed in order to maintain sufficient volumetric coverage and in-plane resolution in dynamic MRI applications. As a consequence, the imaging performance of stack-of-stars sampling is reduced for clinical applications that require high slice resolution (for delineation of small lesions) or large volumetric coverage. Second, sparsity and coil sensitivity encoding along the slice dimension are not exploited, limiting achievable acceleration rates. A few variants of the stack-of-stars trajectory have been previously proposed in several prior works. For example, Peters et al described a radial sampling pattern in which the density of distributing radial lines along the kz dimension is varied (i.e., the sampling density is higher in the center than the periphery). This leads to a reduced level of streaking artifacts without affecting scan time compared to conventional stack-of-stars sampling with uniform kz sampling [22]. Berman and Li et al later demonstrated the performance of combining a similar sampling pattern with compressed sensing reconstruction for improved imaging performance [23,24]. Later, Zhou et al also proposed to use a stack-of-stars trajectory with varying rotation angle along the kz dimension for brain imaging [25]. Despite potential increase of incoherence by reordering all the measurements and jittering all the acquired spokes within a radial stack, these proposed stack-of-stars sampling schemes could be more sensitive to eddy current effects. Meanwhile, implementation of fat saturation pulses in these trajectories was not discussed but is important for radial imaging. Thus, the overall imaging performance of their trajectories, particularly in free-breathing abdominal imaging with self-motion navigation where the stack-of-stars sampling is used a lot, remains to be further explored. Furthermore, these variants all maintain the same spatial resolution and volumetric coverage compared to the standard stack-of-stars trajectory.
In this work, a golden-angle stack-of-stars sampling scheme with variable-density (VD) random undersampling of the kz dimension was designed. This new trajectory does not perform spoke jittering within a stack, maintains the same scheme for fat saturation as employed in conventional stack-of-stars trajectory, enables respiratory motion detection for self-navigation, and is suitable for multicoil sparse reconstruction that exploits additional sparsity and coil sensitivity encoding along the slice dimension. More importantly, this new trajectory enables additional acceleration along kz that can be leveraged to increase slice resolution or volumetric coverage without affecting scan time. The new sampling scheme was first tested for motion-resolved free-breathing non-contrast-enhanced liver MRI using XD-GRASP (Golden-angle RAdial Sparse Parallel MRI with eXtra Dimensions) reconstruction [26]. It was then further evaluated in free-breathing dynamic contrast-enhanced MRI (DCE-MRI) of the liver in combination with respiratory-weighted GRASP reconstruction [27]. With these experiments, we aimed to show that this new sampling scheme enables increased slice resolution and better overall image quality compared to standard stack-of-stars sampling without increasing scan time in freebreathing liver MRI, an application that can benefit from the use of this sampling trajectory. For simplicity, the new sampling scheme is referred to as VD-stack-of-stars and conventional stack-of-stars is referred to as stack-of-stars in the following sections.
2. Methods
2.1. Extension of Stack-of-Stars Sampling to VD-Stack-of-Stars Sampling
Figure 1 shows the extension of stack-of-stars trajectory with fully sampled kz (a) to the VD-stack-of-stars trajectory (b) with the same number of radial spokes and thus the same scan time. The kz-t random undersampling in the VD-stack-of-stars trajectory enables i) larger kz coverage (thus higher slice resolution) without increasing scan time, and ii) incoherent undersampling along both kz and time dimension, which is suitable for compressed sensing reconstruction. Corresponding kz-t sampling masks are shown in Figure 1c-d. For VD-stack-of-stars sampling, a central k-space region is fully sampled in each stack in order to extract respiratory motion from the corresponding low-resolution z-direction projection profiles. As in the case of stack-of-stars trajectory, respiratory motion signal can be obtained by performing a FFT along the fully sampled kz points (red straight lines in Figure 1a-b) followed by the procedures described previously using principal component analysis (PCA) [26,27]. Figure 1e-f shows the extracted respiration motion signals superimposed in underlying z projection profiles from stack-of-stars sampling (e) and VD-stack-of-stars sampling (f) in the same subject. As an important feature of this new trajectory, the number of spokes acquired for each radial stack in Figure 1b is the same as in Figure 1a, and the acquisition is performed without jittering the spokes. This maintains the performance of fat saturation as in conventional stack-of-stars sampling, avoids the risk of introducing artifacts due to large gradient jumps, while the acquisition can reach a higher slice resolution.
2.2. Imaging Experiments
This Health Insurance Portability and Accountability Act (HIPAA) compliant study was approved by Institutional Review Board (IRB) and written informed consent was obtained from all subjects before MR scans. All the imaging studies were performed on a 3.0T MR scanner (MAGNETOM Prisma, Siemens Healthineers, Erlangen, Germany). Two types of MRI experiments were performed, including non-contrast-enhanced free-breathing 3D liver MRI and free-breathing DCE-MRI of the liver.
2.2.1. Non-Contrast-Enhanced Free-Breathing Liver MRI
Four healthy volunteers (females, age=32.8±13.6 years) were first recruited for free-breathing 3D liver MRI without contrast injection to demonstrate the performance of VD-stack-of-stars sampling compared to stack-of-stars sampling. Imaging was performed in transverse orientation using a prototype golden-angle radial gradient-echo sequence. In the first three volunteers, non-contrast-enhanced free-breathing 3D liver MRI using both stack-of-stars and VD-stack-of-stars sampling was compared to demonstrate the increase of slice resolution with the same scan time. Relevant imaging parameters for stack-of-stars sampling included: FOV = 350×350×240 mm3, matrix size = 256×256×48, voxel size = 1.37×1.37×5 mm3, TR/TE=3.6/1.6 ms, flip angle = 12°. Data were continuously acquired for 102 seconds with a total of 650 golden-angle rotations. With 80% partial Fourier sampling applied along the slice dimension, 38 fully-sampled partition lines (out of the total 48 slice locations) were acquired at each rotation angle and were preceded by spectral fat saturation. Imaging parameters for VD-stack-of-stars sampling were the same except for a larger matrix size (256×256×80) resulting in higher slice resolution (1.37×1.37×3 mm3). With 80% partial Fourier sampling and additional incoherent undersampling applied along the slice dimension, 38 undersampled partition lines were acquired at each rotation angle preceded by spectral fat saturation, leading to the same scan time as the stack-of-stars sampling (the same number of spokes for each rotation angle but with larger kzmax as shown in Figure 1b). For VD-stack-of-stars imaging, the central 12 kz partitions (out of the total 38 acquired partitions, see the implementation details below) were fully sampled for respiratory motion signal extraction. In the third volunteer, an additional MRI scan was performed using VD-stack-of-stars sampling with further push of slice resolution. In this scan, the matrix size was increased to 256×256×120 to reach a voxel size of 1.37×1.37×2 mm3 with the same scan time by acquiring 38 undersampled partition lines at each rotation angle.
In the fourth volunteer, non-contrast-enhanced free-breathing 3D abdominal MRI using stack-of-stars and VD-stack-of-stars sampling was compared to demonstrate the increase of volumetric coverage with the same scan time. The imaging protocol for stack-of-stars was the same as above. Relevant imaging parameters for VD-stack-of-stars sampling were the same as the stack-of-stars acquisition except for a larger volumetric coverage (FOV = 350×350×400 mm3) with a matrix size of 256×256×80. For this comparison, both acquisitions had the same voxel size of 1.37×1.37×5 mm3. With 80% partial Fourier sampling and additional incoherent undersampling applied along the slice dimension, 38 undersampled partition lines were acquired at each rotation angle preceded by spectral fat saturation, with 12 of them fully sampled in the center for respiratory motion signal extraction.
2.2.2. Free-Breathing DCE-MRI of the Liver
Five healthy volunteers (age=34.8±12.9 years, 4 females, 1 male) were recruited for free-breathing DCE-MRI of the liver. Due to the need to perform two contrast injections in the same subject, two separate DCE-MR scans were performed in each subject in two separate visits with an average of 15.8±5.0 days between the visits. One visit was for DCE-MR scan with stack-of-stars sampling and the other visit was for DCE-MR scan with VD-stack-of-stars sampling. Free-breathing imaging was performed in transverse orientation using the same imaging sequence as in the non-contrast-enhanced imaging. Relevant imaging parameters for stack-of-stars sampling included: FOV = 350×350×240 mm3, matrix size = 256×256×48, voxel size = 1.37×1.37×5 mm3, TR/TE=3.6/1.6 ms, flip angle = 12°. Data were continuously acquired for 190 seconds with a total of 1222 spokes acquired in each partition. With 80% partial Fourier applied along the slice dimension, 38 fully-sampled partition lines were acquired at each rotation angle and were preceded by spectral fat saturation. Imaging parameters for VD-stack-of-stars sampling was the same except for a larger matrix size of 256×256×80 and a smaller voxel size of 1.37×1.37×3 mm3. With 80% partial Fourier and additional incoherent undersampling applied along the slice dimension, 38 undersampled partition lines were acquired at each rotation angle preceded by spectral fat saturation. The central 12 spokes (out of the total 38 acquired partitions) were fully sampled for respiration motion signal extraction. For each MR scan, half dose injection of weight-based (0.1 mmol per kg of body weight) gadopentetate dimeglumine (Magnevist; Bayer Healthcare) was performed 20 seconds after the start of data acquisition, at a rate of 2 mL/sec.
2.2.3. Implementation of Sampling Patterns
This section provides more details about our implementation of both stack-of-stars and VD-stack-of-stars sampling. For stack-of-stars sampling, 80% slice partial Fourier was implemented in our sequence, which is commonly used in body MRI acquisitions. Specifically, the reconstructed images for stack-of-stars sampling have a total of 48 slices and a slice FOV of 240 mm (slice thickness = 5 mm). With 80% slice partial Fourier, the first 10 slices were not acquired and only slice 11 to slice 48 were acquired without undersampling.
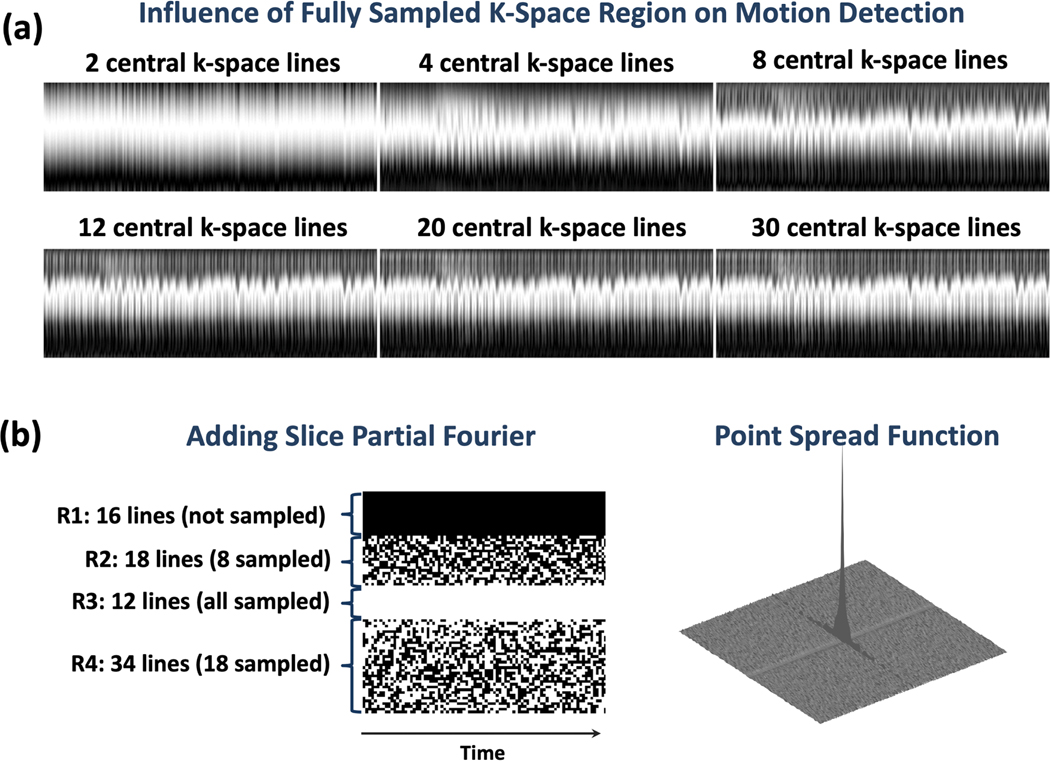
For VD-stack-of-stars sampling, the total number of image slices was increased to 80 with a slice thickness of 3 mm (same FOV = 240 mm). To maintain the same scan time as in stack-of-stars sampling, 38 slices were acquired from all the 80 slice locations. To ensure reliable respiratory motion detection, a small region of central k-space lines along kz were fully sampled. This number of fully sampled central k-space lines was empirically selected by comparing different values as shown in Figure 2a. It can be seen that 12 central k-space lines are sufficient to delineate respiratory motion pattern in the resulting z projection profiles with no visible difference compared to 20 lines or 30 lines. On the other side, while less k-space lines provide less information as indicated by the z projection profiles. As such, 12 central k-space lines were fully sampled in our experiments. Figure 2b shows how the variable-density kz-t undersampling mask was generated for VD-stack-of-stars sampling. The first 16 lines (denoted as region 1 or R1) were not sampled with 80% slice partial Fourier; the 8 k-space lines were randomly sampled above the central region (denoted as region 2 or R2); and 18 k-space lines were randomly sampled below the central region (denoted as region 4 or R4). The fully sampled central k-space region is then denoted as region 3 or R3. This leads to a total of 38 sampled k-space lines from all the 80 slice locations. The sampling pattern was varied along the time dimension, so that compressed sensing can be applied to exploit the temporal correlations. Figure 2b also shows the corresponding point spread function (PSF) for the kz-t variable-density undersampling pattern.
Figure 2.
(a) Influence of fully sampled region with different number of k-space lines on motion detection. 12 central k-space lines are sufficient to delineate respiratory motion pattern in the resulting z projection profiles with no visible difference compared to 20 lines or 30 lines, while less k-space lines provide less information as indicated by the z projection profiles. (b) Design of variable-density kz-t undersampling mask for VD-stack-of-stars sampling in our current implementation. Data acquisition has a total of 80 slices. The first 16 lines (denoted as region 1 or R1) were not sampled with 80% slice partial Fourier; the 8 k-space lines were randomly sampled above the central region (denoted as region 2 or R2); and 18 k-space lines were randomly sampled below the central region (denoted as region 4 or R4). The fully sampled central k-space region is then denoted as region 3 or R3. This leads to a total of 38 sampled k-space lines from all the 80 slice locations. The sampling pattern was varied along the time dimension, so that compressed sensing can be applied to exploit the temporal correlations. Corresponding point spread function (PSF) for the kz-t variable-density undersampling pattern is also shown.
2.3. Image Reconstruction
2.3.1. Reconstruction of Free-Breathing Non-Contras-Enhanced MRI Data
All the non-contrast-enhanced MRI datasets, including both stack-of-stars and VD-stack-of-stars acquisitions, were reconstructed using XD-GRASP [26]. Specifically, given the respiratory motion signal obtained from the z projection profiles as shown in Figure 1 and Figure 2, XD-GRASP reconstruction was performed after sorting the acquired data into four respiratory phases spanning from end-expiration to end-inspiration, resulting in a four-dimensional image-set (x-y-z-respiration). In addition, the coil-unstreaking technique, as described in [28], was incorporated to suppress residual streaking artifacts to improve image quality. XD-GRASP reconstruction was performed by solving the following cost function:
| [1] |
Here, y is sorted dynamic radial k-space data, D is a density compensation function for radial sampling, F is the non-uniform FFT (NUFFT) operator mapping between radial k-space domain and image domain, C represents coil sensitivities, and x is the corresponding motion-resolved dynamic image-series to be reconstructed. S is a temporal finite differences operator applied along the new respiratory dimension to minimize temporal total variation and λ is corresponding regularization parameter. The density compensation was separated into two square root parts as shown in the cost function [29]. R is a coil weighting matrix controlling the contribution of each coil element, in a way that coil elements containing a high level of streaking artifacts (thus with a high artifact weight) contribute less to the final reconstruction, as previously described in [28].
2.3.2. Reconstruction of Free-Breathing DCE-MRI Data
All the DCE-MRI datasets, including both stack-of-stars and VD-stack-of-stars acquisitions, were reconstructed using the RACER-GRASP approach [27]. Specifically, k-space data are automatically sorted at temporal positions of [−30, −15, 0, +5, +10, +15, +22, +30, +45, +60, +90, +120, +150] seconds with a temporal footprint of ~15 seconds, and the data sorting was guided by a contrast enhancement curve extracted from the acquired data. The temporal resolution of 15 seconds was selected for multiphasic assessment reflecting our clinical practice. A total of 13 contrast phases were reconstructed for each dataset. Here, 0s indicates the peak aortic enhancement position and −/+ indicates whether the current phase is before or after the peak aortic enhancement. The phases at −30s and −15s are both pre-contrast phases, the phases at +5s, +10s and +15s are early to late arterial phases, the phase at +60s is the portal venous phase, and the phase at +150s is the delayed phase.
After k-space sorting and motion detection, respiratory-weighted GRASP reconstruction was performed by solving the following cost function:
| [2] |
The variables in Equation 2 are similar as that in Equation 1 above. In addition, W is a motion weighting matrix controlling the contribution of each motion state to the final reconstructed images (i.e., expiratory phases have higher weights and inspiratory phases have lower weights) [27]. In our current implementation, k-space data from each contrast phase were separated into four motion phases based on the detected respiratory motion signa, representing end-expiration to the end-inspiration. The motion weight for the four motion phases was assigned as [1, 0.25, 0.252, 0.253], respectively. This effectively led to less contribution from the inspiratory phases to the final reconstructed images. As in the XD-GRASP reconstruction in Equation 1, R is also a coil weighting matrix controlling the contribution of each coil element based on the associated streaking artifact.
All the non-contrast-enhanced MRI and DCE-MRI reconstruction tasks were performed using a non-linear conjugate gradient algorithm. Coil sensitivity maps were estimated from a static 3D image volume reconstructed using all the acquired radial spokes using the adaptive combination approach [30]. Regularization parameters were empirically selected separately for stack-of-stars and VD-stack-of-stars acquisitions. All the reconstructions were performed using a server equipped with two 16-core CPUs, 256 GB RAM, and two 6-Gb NVIDIA GPU cards.
2.4. Image Quality Assessment and Statistical Analysis
Visual image quality assessment was performed to compare the reconstructed liver MR images acquired using stack-of-stars and VD-stack-of-stars sampling. Before the visual assessment, stack-of-stars images are interpolated to have the same number of slices with the VD-stack-of-stars. The assessment was performed by two abdominal radiologists with 3-year and 1-year of post-fellowship experience who were blinded to the acquisition schemes. For DCE-MRI, scoring was performed for a total of 6 contrast phases from each dataset, including the phases at +5 and +15 seconds (arterial phases), the phase at +45 seconds, the phase at +60 seconds (venous phase), and the phases at +120 and +150 seconds (delayed phases). Image quality assessment was also performed in coronal and sagittal planes, which were reformatted from the axial images from the same dataset. The two radiologists independently scored the overall image quality (including consideration of the residual artifact/noise level), liver edge sharpness and hepatic vessel conspicuity of each image volume based on a 5-point Likert-type scale (5: excellent; 4: good; 3: adequate; 2: borderline; 1: non-diagnostic).
For each assessment category (e.g., overall image quality, liver edge sharpness and hepatic vessel conspicuity), the reported scores were averaged over the two readers and all the evaluated contrast phases to represent mean ± standard deviation for axial, coronal and sagittal image planes, separately. A nonparametric paired two-tailed Wilcoxon signed-rank test was used to compare the image quality scores, and a P value less than 0.05 indicates statistical significance. In addition, weighted kappa coefficients were calculated to assess the inter-observer variability in image quality assessment, where a higher coefficient indicates better inter-observer agreement.
To validate the improved image sharpness from the higher slice resolution, a previously described image blurring metric [31] was used to quantitatively assess the image sharpness of the reconstructed DCE-MRI images. This metric has a range of 0 to 1. The higher the score, the more blurring an image has, and the lower the score, the higher sharpness an image has. In the analysis, a score reflecting the sharpness is assigned for each of the reconstructed slices. Scores were also quantified for the reformatted coronal and sagittal images. The same analysis was repeated for all the reconstructed phases, and they were averaged to represent mean ± standard deviation.
3. Results
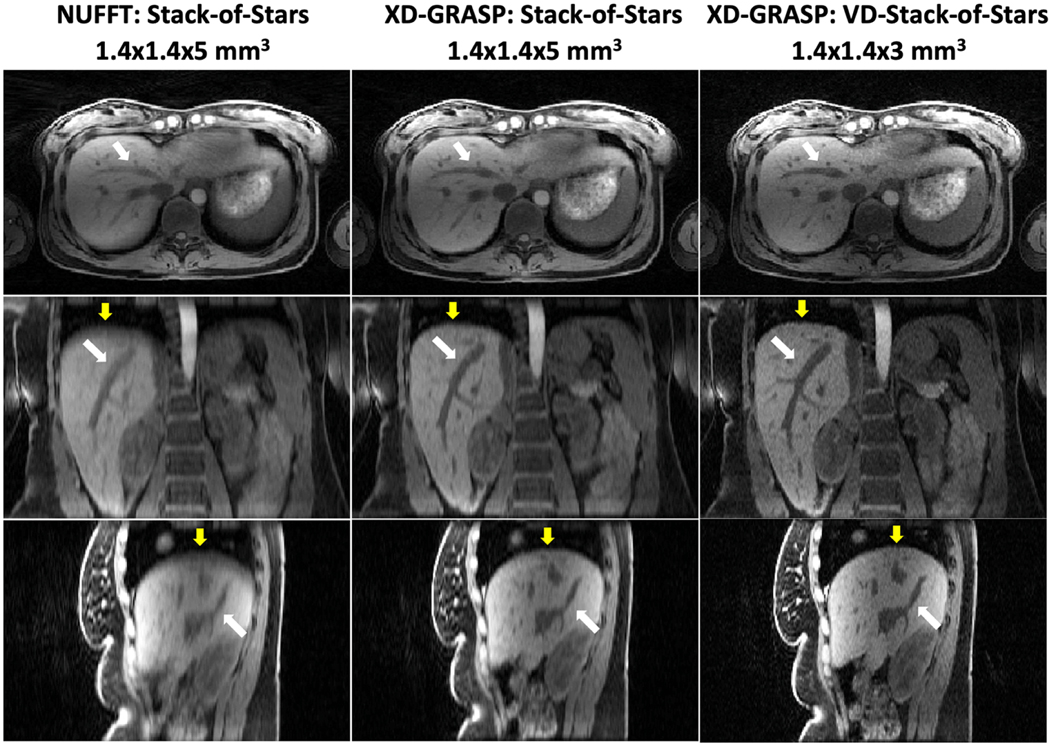
All the reconstruction tasks were performed successfully. Images were reconstructed in axial plane and were reformatted to generate coronal and sagittal images. The left and middle columns of Figure 3 first present non-contrast-enhanced liver images from stack-of-stars sampling in one volunteer reconstructed with standard non-iterative NUFFT reconstruction using all the acquired spokes without motion compensation and with XD-GRASP reconstruction (end-expiratory phase displayed). The right column shows matching liver images from VD-stack-of-stars sampling reconstructed with XD-GRASP (end-expiratory phase). Overall, XD-GRASP reconstruction was able resolve respiratory motion and achieve higher sharpness in liver edge and hepatic vessel (yellow and white arrows), particularly in coronal and sagittal image planes. Further improved sharpness and clarity can be appreciated in XD-GRASP results with VD-stack-of-stars sampling, which present better delineation of both liver edge and hepatic vessels (yellow and white arrows) for the same scan time.
Figure 3.
Left: Non-contrast 3D liver images acquired with stack-of-stars sampling and reconstructed with NUFFT using all spokes without motion sorting. Middle: Corresponding XD-GRASP images resolved respiratory motion and achieved notable increase in image sharpness and vessel clarity (yellow and white arrows). (c) Non-contrast 3D liver images acquired with VD-stack-of-stars sampling and reconstructed with XD-GRASP present further improved delineation of liver edge and hepatic vessel (yellow and white arrows) with the same scan time.
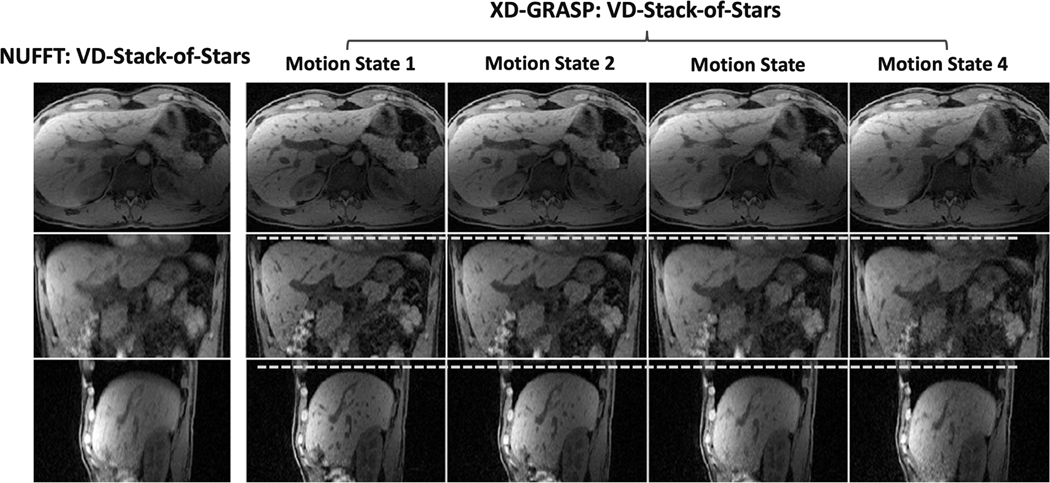
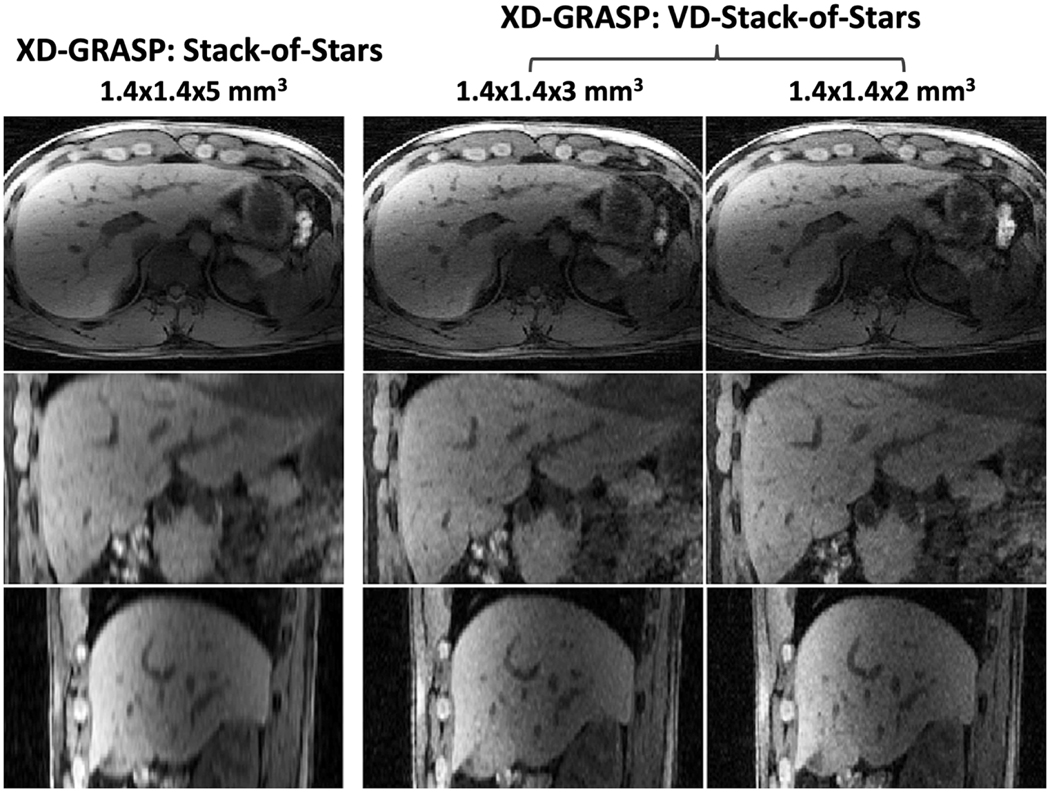
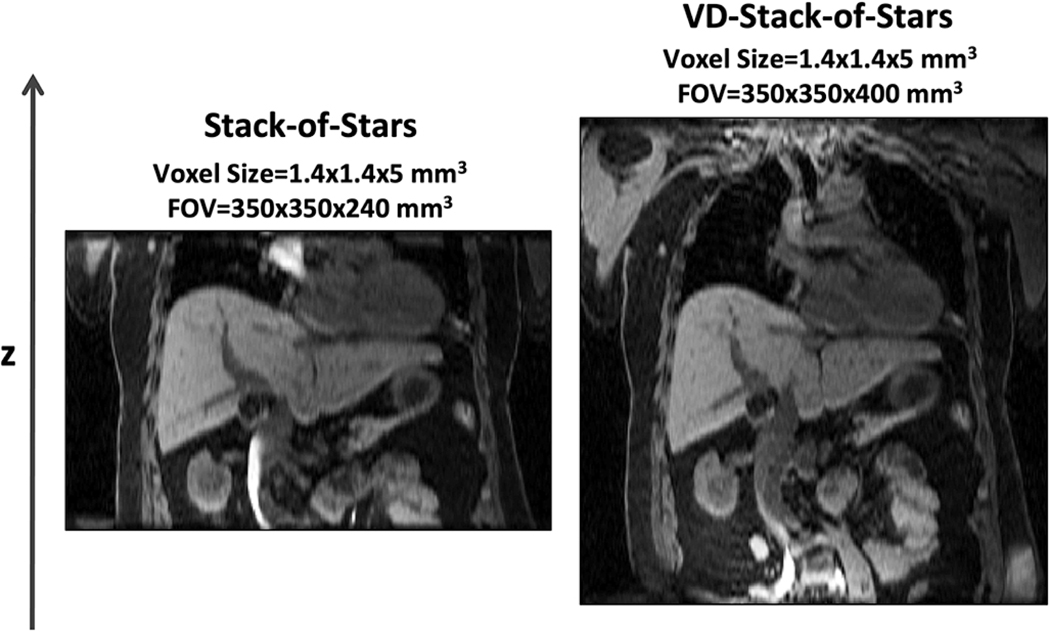
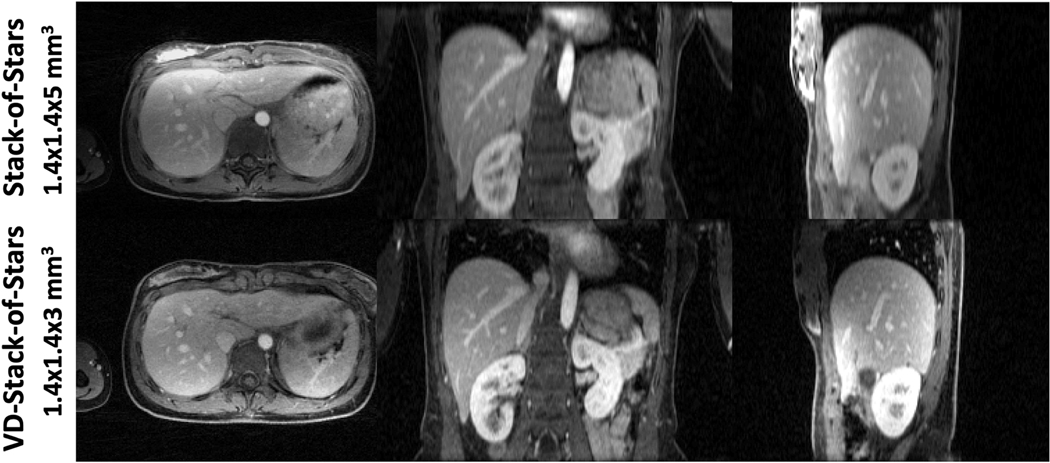
Figure 4 shows non-contrast-enhanced XD-GRASP results with VD-stack-of-stars sampling at different respiratory states (note the changing positions of the diaphragm with respect to the white dashed line) in the second volunteer. When compared to omtion-averaged NUFFT reconstruction, XD-GRASP achieved clearly improved image quality, image sharpness and vessel clarity in all image planes. Figure 5 compares non-contrast-enhanced XD-GRASP results from stack-of-stars sampling with non-contrast-enhanced XD-GRASP results from VD-stack-of-stars sampling with different slice resolutions in the third volunteer. Better delineation of hepatic vessels can be achieved with higher slice resolution, but this also results in increased residual noise in the images with 2-mm slice resolution. Figure 6 shows non-contrast-enhanced XD-GRASP results from stack-of-stars and VD-stack-of-stars sampling with the same voxel size but with different slice coverage in the fourth volunteer, in which VD-stack-of-stars sampling enabled larger coverage along the slice dimension with the same scan time.
Figure 4.
Comparison of NUFFT images acquired with VD-stack-of-stars sampling and reconstructed without motion sorting using all spokes with images reconstructed using XD-GRASP reconstruction at different respiratory motion states. White dashed lines indicate resolved respiratory motion phases in XD-GRASP.
Figure 5.
Comparison of XD-GRASP images with stack-of-stars sampling with XD-GRASP images with VD-Stack-of-Stars sampling at different slice resolutions. Better delineation of hepatic vessels can be achieved with higher slice resolution, but this also results in increased residual noise level particularly in the images with 2mm slice resolution.
Figure 6.
XD-GRASP results (end-expiratory phase) using stack-of-stars sampling (left) and VD-stack-of-stars sampling (right) with the same scan time and same voxel size, but with different slice coverage. VD-stack-of-stars sampling enabled increased volumetric coverage without increasing scan time.
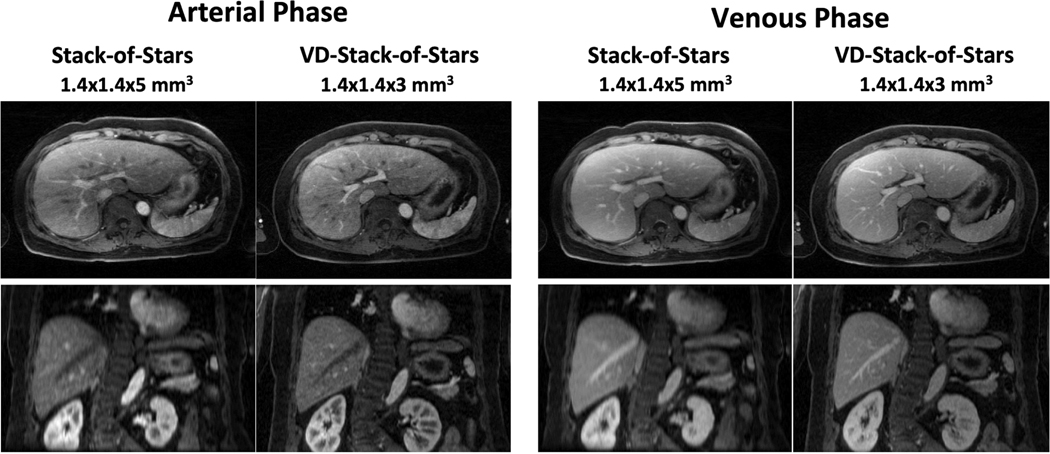
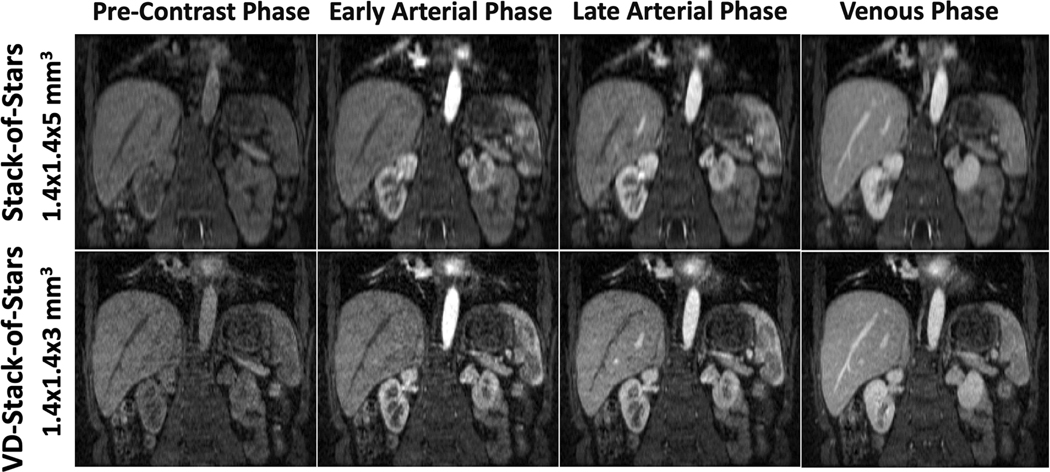
Figure 7 compares DCE-MR images acquired using both stack-of-stars and VD-stack-of-stars sampling in one volunteer. VD-stack-of-stars sampling enabled improved delineation of hepatic vessels and sharper liver edges in both arterial and venous phases. The improvement of slice resolution using VD-stack-of-stars can be better appreciated in the coronal images, in which a smaller voxel size allows for better delineation of small vessels and more detailed structure about the liver.
Figure 7.
Comparison of liver DCE-MR images acquired with both stack-of-stars and VD-stack-of-stars sampling and reconstructed with respiratory-weighted GRASP in one subject. VD-stack-of-stars enabled improved delineation of hepatic vessels and sharper liver edges in both arterial and venous phases. The improvement of slice resolution using VD-stack-of-stars sampling can be better appreciated in the coronal images, in which a smaller voxel size allows for better delineation of small vessels and more detailed structures about the liver.
Figure 8 shows the same comparison in the second DCE-MRI volunteer in axial and sagittal planes. Similar to that in Figure 7, the results with VD-stack-of-stars sampling demonstrated increased image sharpness and vessel clarity in both arterial phase and venous phase, particularly in the reformatted sagittal plane. A similar comparison in the third DCE-MRI volunteer is shown in Figure 9 for different image planes in the venous phase, where the same improvement of image quality, sharpness and conspicuity can be observed in the VD-stack-of-stars results.
Figure 8.
Comparison of liver DCE-MR images acquired using both stack-of-stars and VD-stack-of-stars sampling and reconstructed with respiratory-weighted GRASP in another subject for both axial and sagittal planes. Same as in Figure 6, VD-stack-of-stars sampling achieved increased image sharpness and vessel clarity in both arterial and venous phases, particularly in the reformatted sagittal plane.
Figure 9.
Comparison of liver DCE-MR images with stack-of-stars sampling and VD-stack-of-stars sampling in a venous phase in the third subject. Similar improvement of image quality, sharpness and conspicuity can be observed in the VD-stack-of-stars results.
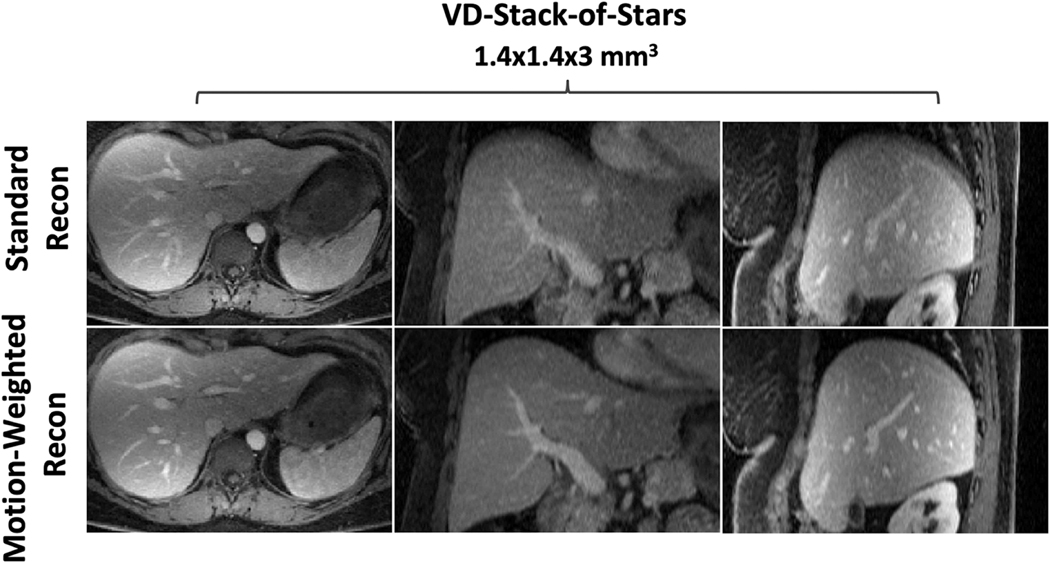
Figure 10 shows results from the fourth DCE-MRI volunteer comparing contrast-enhanced liver images with both sampling trajectories at different contrast phases in coronal image plane. VD-stack-of-stars sampling enabled better delineation of contrast enhancement, increased image sharpness and vessel conspicuity in both the liver and the kidney. Lastly, Figure 11 compares the same DCE-MR data acquired with VD-stack-of-stars sampling but different reconstruction schemes. Specifically, images in the top row of Figure 11 were reconstructed using standard GRASP reconstruction without respiratory motion weighting, while images in the bottom row was reconstructed with respiratory-weighted GRASP reconstruction. With respiratory-weighted sparse reconstruction, improved overall image quality, increased liver edge sharpness and hepatic vessel conspicuity can be achieved.
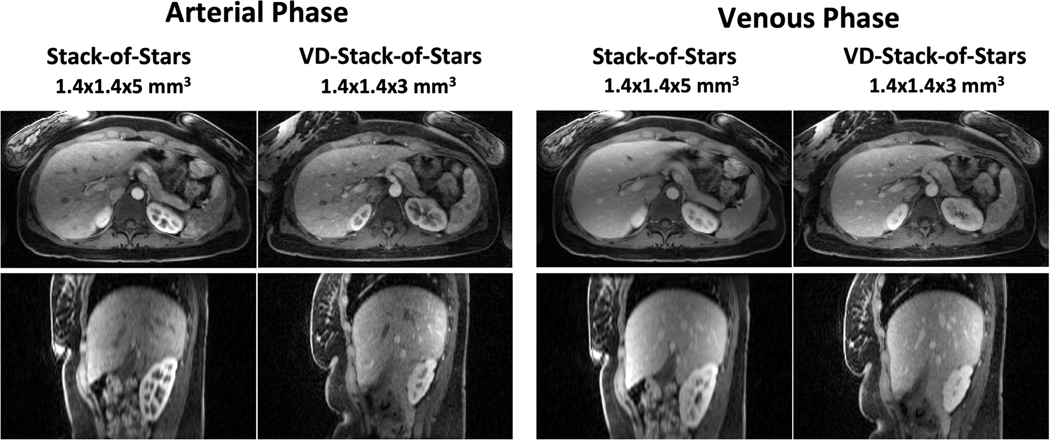
Figure 10.
Comparison of liver DCE-MR images with stack-of-stars sampling and VD-stack-of-stars sampling at different contrast phases. VD-stack-of-stars enabled better delineation of contrast enhancement, increased image sharpness and vessel conspicuity in both the liver and kidney.
Figure 11.
Comparison of the same DCE-MR data acquired with VD-stack-of-stars sampling but reconstructed with and without respiratory motion compensation/weighting. Without respiratory motion compensation (top row), images suffer from residual blurring, particularly in the reformatted coronal and sagittal image planes. With respiratory-weighted sparse reconstruction (bottom row), improved overall image quality, increased liver edge sharpness and hepatic vessel conspicuity can be achieved.
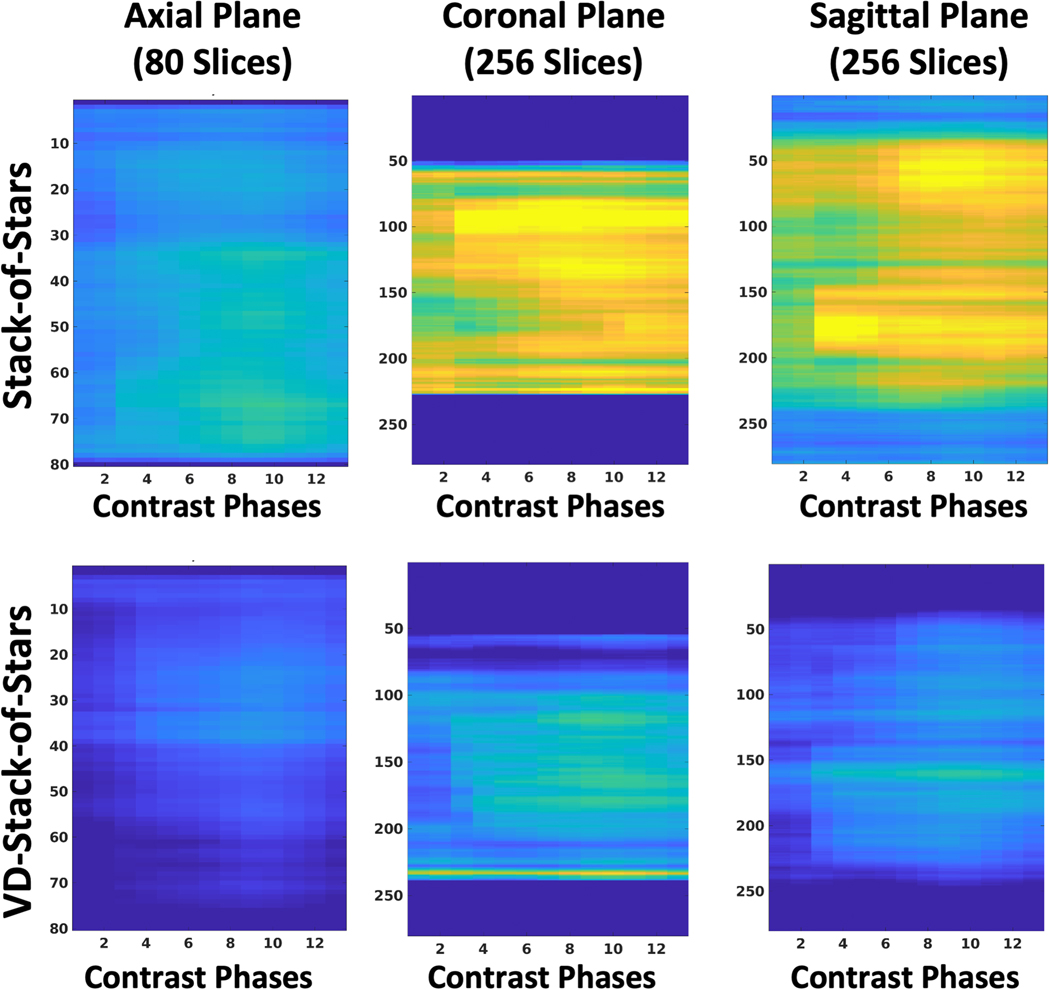
Figure 12 shows the results of the quantitative sharpness analysis from one subject for all the reconstructed DCE-MRI contrast phases. The proposed VD-stack-of-stars sampling enabled higher image sharpness (low quantitative scores) than standard stack-of-stars sampling. The differences are more obvious in the reformatted coronal and sagittal comparison, as the increased resolution is along the slice (kz) dimension. The averaged blurring metrics from all the subjects are summarized in Table 1.
Figure 12.
Scores of the quantitative sharpness analysis in a representative DCE-MR dataset. The sharper the image, the lower the resulting score. Results from the proposed VD-stack-of-stars are consistently sharper than the standard stack-of-stars for all three image planes. The stack-of-stars results were interpolated to have the same number of slices as the VD-stack-of-stars results.
Table 1.
| Axial Plane | Sagittal Plane | Coronal Plane | ||
|---|---|---|---|---|
| Dataset 1 | Stack-of-Stars | 0.2631±0.0089 | 0.3122±0.0096 | 0.3260±0.0110 |
| VD-Stack-of-Stars | 0.2296±0.0093 | 0.2420±0.0089 | 0.2637±0.0130 | |
| Dataset 2 | Stack-of-Stars | 0.2574±0.0099 | 0.3260±0.0122 | 0.3262±0.0145 |
| VD-Stack-of-Stars | 0.2310±0.0115 | 0.2425±0.0129 | 0.2576±0.0154 | |
| Dataset 3 | Stack-of-Stars | 0.2654±0.0092 | 0.3293±0.0123 | 0.3213±0.0088 |
| VD-Stack-of-Stars | 0.2318±0.0086 | 0.2563±0.0137 | 0.2779±0.0130 | |
| Dataset 4 | Stack-of-Stars | 0.2330±0.0101 | 0.3287±0.0130 | 0.3481±0.0123 |
| VD-Stack-of-Stars | 0.2161±0.0110 | 0.2418±0.0097 | 0.2779±0.0154 | |
| Dataset 5 | Stack-of-Stars | 0.2641±0.0106 | 0.3157±0.0114 | 0.3532±0.0154 |
| VD-Stack-of-Stars | 0.2304±0.0104 | 0.2397±0.0098 | 0.2888±0.0154 |
Quantitative scores for assessing image sharpness and blurring averaged over different contrast phases. The score has a range between 0–1. The sharper the image, the lower the resulting score (e.g., 1 indicates the most image blurring and 0 indicates the highest image sharpness). Results from VD-stack-of-stars sampling are consistently sharper than the standard stack-of-stars sampling for all the image planes.
Table 2 summarizes the reader’s scores for different assessment categories comparing liver MR images acquired with stack-of-stars and VD-stack-of-stars sampling. Although the scores of stack-of-stars were around or above 3 (adequate) in axial orientation, its scores were only around 2.5 (between borderline and adequate) in both coronal and sagittal orientations mostly due to poor slice resolution. On the other hand, the scores of VD-stack-of-stars were above 4 (good) in axial orientation and were around 3.5 (between adequate and good) in both coronal and sagittal orientations. For all the imaging planes, VD-stack-of-stars sampling achieved superior image quality and statistical significance was reached in all assessment categories (P<0.05). The kappa coefficients were 0.93, 0.91 and 0.88 for the scores from the two readers to assess overall image quality, liver edge sharpness and hepatic vessel conspicuity, respectively, indicating excellent inter-observer agreement.
Table 2.
| Overall Image Quality | Liver Edge Sharpness | Hepatic Vessel Conspicuity | ||
|---|---|---|---|---|
| Axial Plane | Stack-of-Stars | 2.95±0.20 | 3.02±0.28 | 3.10±0.33 |
| VD-Stack-of-Stars | 4.03±0.18 a | 4.03±0.18 a | 4.05±0.15 a | |
| Coronal Plane | Stack-of-Stars | 2.00±0.13 | 2.00±0.13 | 2.02±0.09 |
| VD-Stack-of-Stars | 2.98±0.09 a | 3.00±0.01 a | 3.02±0.09 a | |
| Sagittal Plane | Stack-of-Stars | 2.00±0.01 | 2.00±0.01 | 2.00±0.01 |
| VD-Stack-of-Stars | 2.98±0.09 a | 2.98±0.09 a | 3.00±0.01 a |
Visual image quality comparison between free-breathing liver DCE-MRI with stack-of-stars sampling and VD-stack-of-stars sampling in different image planes. VD-stack-of-stars sampling achieved improvement, and statistical significance was reached in all the assessment categories.
The scoring criteria for all the assessment categories were as follows: 5=excellent; 4=good; 3=adequate; 2=borderline; 1=non-diagnostic.
The difference between stack-of-stars and VD-stack-of-stars was statistically significant, P<0.001.
4. Discussion
This study proposes a VD-stack-of-stars sampling scheme that incorporates additional variable-density undersampling along slice dimension for hybrid radial-Cartesian acquisition, so that increased slice resolution (or volumetric coverage) can be achieved without prolonging scan time and additional sparsity and coil sensitivity encoding can be exploited along the slice dimension in multicoil compressed sensing reconstruction. This new imaging trajectory maintains the advantages of standard stack-of-stars sampling, including the performance of fat saturation, low sensitivity to effects associated with large gradient jumps, flexible selection of slice resolution, and self-navigation. The VD-stack-of-stars sampling trajectory was combined with the XD-GRASP and RACER-GRASP reconstruction methods for free-breathing liver MRI to demonstrate better image quality, increased liver edge sharpness and higher hepatic vessel clarity over standard stack-of-stars sampling, particularly in the reformatted coronal and sagittal image planes. This can provide additional information for improved clinical use in liver MRI where detection and characterization of small lesions are important.
In the VD-stack-of-stars trajectory, 12 central k-space partitions are fully sampled in our current implementation, so that a respiratory motion signal can be extracted from the corresponding projections for motion-weighted or motion-resolved reconstruction. The acquisition of 12 fully sampled center spokes was empirically determined to achieve a good balance between the performance of motion detection and incoherence along kz as shown in Figure 2. Despite reduced resolution in the self-navigation projections compared to stack-of-stars sampling, as can be seen in Figure 1a-d, these projections are only used for extracting motion signals, and the performance was found to be comparable with standard stack-of-stars as shown in Figure 1e-f. Meanwhile, the fully sampled central k-space region naturally results in a variable density sampling pattern, which is favorable in compressed sensing reconstruction.
In routine clinical practice, slice partial Fourier is commonly employed for body MRI. This is known as VIBE imaging (Volumetric Interpolated Breath-hold Examination) on the Siemens platform. As such, 80% slice partial Fourier was also implemented in all the experiments in our study. The effect of the slice partial Fourier is expected to be similar for both standard stack-of-stars sampling and VD-stack-of-stars sampling, which which could result in certain blurring along the slice dimension.
In addition to increasing slice resolution, the VD-stack-of-stars sampling can also be used to increase volumetric coverage for the same scan times. This can be achieved by sampling more densely (in an undersampling scheme) along the slice dimension while keeping the same kzmax as in standard stack-of-stars sampling, which may be particularly useful for clinical applications requiring a large volumetric coverage, such as MR Enterography [32], which typically covers both the upper and lower abdomen.
The VD-stack-of-stars sampling shares similarity with kooshball-type 3D radial sampling, which is often applied for cardiovascular MR applications [33–36]. They both allow for acceleration along all spatial dimensions (kx-ky-kz), thus enabling exploitation of sparsity and incoherence along all dimensions. However, challenges associated with kooshball-like 3D radial sampling include reduced performance in fat saturation and increased sensitivity to eddy currents due to the fact that each spoke passes through the center of k-space and carries equal amount of information. Besides, 3D radial sampling can result in a lower signal to noise ratio (SNR) because of the intrinsically acquired isotropic spatial resolution, which is not required in many applications. In contrast, the proposed VD-stack-of-stars sampling allows for selection of flexible spatial resolution along kz. For example, the spatial resolution of VD-stack-of-stars sampling was empirically set as 1.4×1.4×3mm3 in this study by an experienced radiologist to maintain a good balance between image sharpness and SNR in DCE-MR scans.
A major limitation of this study is the small number of subject (n=5) in our experiments. This was because of the need to perform two separate contrast-enhanced DCE-MR exams for each subject, which substantially increase the difficulty in recruiting subjects. Meanwhile, only half dose of contrast agent was given in each MR scan, which limited achievable performance and SNR. Further studies in a larger number of subjects with clinical standard contrast dose will be necessary to fully assess the clinical utility of the proposed VD-stack-of-stars trajectory in different applications.
5. Conclusion
This study has demonstrated the initial performance of VD-stack-of-stars sampling that can be used to improve slice resolution and image quality in rapid and motion-robust non-contrast-enhanced liver MRI and DCE-MRI MRI without increasing scan times. The combination of VD-stack-of-stars with the GRASP MRI (and its variants) is a promising method for free-breathing body MRI.
Acknowledgement
This work was supported in part by the NIH (Grant Support: R01 EB030549, R21EB032917 and P41 EB017183). The authors thank Dr. Tiejun Zhao for helping implement the VD-Stack-of-Stars MRI sequence, thank Dr. Lihua Chen for help with statistical analyses, and thank Drs. Daniel Sodickson and Ricardo Otazo for helping edit the early version of this paper.
Grant Support:
NIH R01 EB030549, R21EB032917, P41 EB017183
Footnotes
Author Statement
Zhitao Li: Methodology, Image Analysis, Manuscript Writing and Editing
Chenchan Huang: Image Analysis
Angela Tong: Image Analysis
Hersh Chandarana: Data Acquisition
Li Feng: Conceptualization, Methodology, Image Reconstruction Design, Data Acquisition, Manuscript Writing and Editing
Disclosure
Li Feng and Hersh Chandarana are co-inventors of a patent (Patent number 9921285) on the GRASP and XD-GRASP MRI technique.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Block KT, Chandarana H, Milla S, Bruno M, Mulholland T, Fatterpekar G, et al. Towards Routine Clinical Use of Radial Stack-of-Stars 3D Gradient-Echo Sequences for Reducing Motion Sensitivity. J Korean Soc Magn Reson Med 2014;18:87. 10.13104/jksmrm.2014.18.2.87. [DOI] [Google Scholar]
- [2].Feng L. Golden-Angle Radial MRI: Basics, Advances, and Applications. J Magn Reson Imaging 2022. 10.1002/JMRI.28187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Feng L, Benkert T, Block KT, Sodickson DK, Otazo R, Chandarana H. Compressed sensing for body MRI. J Magn Reson Imaging 2017;45:966–87. 10.1002/jmri.25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Glover GH, Pauly JM. Projection Reconstruction Techniques for Reduction of Motion Effects in MRI. Magn Reson Med 1992;28:275–89. 10.1002/mrm.1910280209. [DOI] [PubMed] [Google Scholar]
- [5].Chandarana H, Feng L, Block TK, Rosenkrantz AB, Lim RP, Babb JS, et al. Free-breathing contrast-enhanced multiphase MRI of the liver using a combination of compressed sensing, parallel imaging, and golden-angle radial sampling. Invest Radiol 2013;48:10–6. 10.1097/RLI.0b013e318271869c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chandarana H, Feng L, Ream J, Wang A, Babb JS, Block KT, et al. Respiratory motion-resolved compressed sensing reconstruction of free-breathing radial acquisition for dynamic liver magnetic resonance imaging. Invest Radiol 2015;50:749–56. 10.1097/RLI.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chandarana H, Block KT, Winfeld MJ, Lala SV., Mazori D, Giuffrida E, et al. Free-breathing contrast-enhanced T1-weighted gradient-echo imaging with radial k-space sampling for paediatric abdominopelvic MRI. Eur Radiol 2013 242 2013;24:320–6. 10.1007/S00330-013-3026-4. [DOI] [PubMed] [Google Scholar]
- [8].Chandarana H, Block TK, Rosenkrantz AB, Lim RP, Kim D, Mossa DJ, et al. Free-breathing radial 3D fat-suppressed T1-weighted gradient echo sequence: A viable alternative for contrast-enhanced liver imaging in patients unable to suspend respiration. Invest Radiol 2011;46:648–53. 10.1097/RLI.0B013E31821EEA45. [DOI] [PubMed] [Google Scholar]
- [9].Feng L, Grimm R, Block KT obia., Chandarana H, Kim S, Xu J, et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med 2014;72:707–17. 10.1002/mrm.24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magn Reson Med 2007. 10.1002/mrm.21236. [DOI] [PubMed] [Google Scholar]
- [11].Adluru G, McGann C, Speier P, Kholmovski EG, Shaaban A, Dibella EVR. Acquisition and reconstruction of undersampled radial data for myocardial perfusion magnetic resonance imaging. J Magn Reson Imaging 2009;29:466–73. 10.1002/JMRI.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feng L, Wen Q, Huang C, Tong A, Liu F, Chandarana H. GRASP-Pro: imProving GRASP DCE-MRI through self-calibrating subspace-modeling and contrast phase automation. Magn Reson Med 2020;83:94–108. 10.1002/mrm.27903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Feng L. 4D GRASP MRI at Sub-Second Temporal Resolution. Accept NMR Biomed 2022. 10.48550/arxiv.2208.05508. [DOI] [Google Scholar]
- [14].Adluru G, Chen L, Kim SE, Burgon N, Kholmovski EG, Marrouche NF, et al. Three-dimensional late gadolinium enhancement imaging of the left atrium with a hybrid radial acquisition and compressed sensing. J Magn Reson Imaging 2011;34:1465–71. 10.1002/jmri.22808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen L, Liu D, Zhang J, Xie B, Zhou X, Grimm R, et al. Free-breathing dynamic contrast-enhanced MRI for assessment of pulmonary lesions using golden-angle radial sparse parallel imaging. J Magn Reson Imaging 2018;48:459–68. 10.1002/jmri.25977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhou Z, Han F, Yu S, Yu D, Rapacchi S, Song HK, et al. Accelerated noncontrast-enhanced 4-dimensional intracranial MR angiography using golden-angle stack-of-stars trajectory and compressed sensing with magnitude subtraction. Magn Reson Med 2018;79:867–78. 10.1002/mrm.26747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rosenkrantz AB, Geppert C, Grimm R, Block TK, Glielmi C, Feng L, et al. Dynamic contrast-enhanced MRI of the prostate with high spatiotemporal resolution using compressed sensing, parallel imaging, and continuous golden-angle radial sampling: Preliminary experience. J Magn Reson Imaging 2015;41:1365–73. 10.1002/JMRI.24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim SG, Feng L, Grimm R, Freed M, Block KT, Sodickson DK, et al. Influence of temporal regularization and radial undersampling factor on compressed sensing reconstruction in dynamic contrast enhanced MRI of the breast. J Magn Reson Imaging 2016;43:261–9. 10.1002/JMRI.24961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Feng L, Liu F, Soultanidis G, Liu C, Benkert T, Block KT, et al. Magnetization-prepared GRASP MRI for rapid 3D T1 mapping and fat/water-separated T1 mapping. Magn Reson Med 2021;86:97–114. 10.1002/mrm.28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Z, Xu X, Yang Y, Feng L. Repeatability and robustness of MP-GRASP T1 mapping. Magn Reson Med 2022;87:2271–86. 10.1002/MRM.29131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the golden ratio for time-resolved MRI. IEEE Trans Med Imaging 2007;26:68–76. 10.1109/TMI.2006.885337. [DOI] [PubMed] [Google Scholar]
- [22].Peters D, Nezafat R, Manning R. Radial Undersampling that is Variable in kz. In Proceedings of the 15th Annual Meeting of ISMRM, Berlin, Germany, 2007. p. 304., n.d. [Google Scholar]
- [23].Li Z, Berman B, Altbach M, Galons J, Martin D, Dong B et al. Highly Accelerated 3D Dynamic Imaging with Variable Density Golden Angle Stack-Of-Stars Sampling. In Proceedings of the 21th Annual Meeting of ISMRM, Salt Lake City, Utah, USA, 2013. p. 3797., n.d. [Google Scholar]
- [24].Berman B, Li Z, Altbach MI, Galons J, Martin DR, Dong B. How to Stack the Stars: A Variable Center-Dense K-Space Trajectory for 3D MRI. In Proceedings of the 21th Annual Meeting of ISMRM, Salt Lake City, Utah, USA, 2013. p. 3829., n.d. [Google Scholar]
- [25].Zhou Z, Han F, Yan L, Wang DJJ, Hu P. Golden-ratio rotated stack-of-stars acquisition for improved volumetric MRI. Magn Reson Med 2017;78:2290–8. 10.1002/MRM.26625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med 2016;75:775–88. 10.1002/mrm.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Feng L, Huang C, Shanbhogue K, Sodickson DK, Chandarana H, Otazo R. RACER-GRASP: Respiratory-weighted, aortic contrast enhancement-guided and coil-unstreaking golden-angle radial sparse MRI. Magn Reson Med 2018;80:77–89. 10.1002/mrm.27002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Feng L, Chandarana H, Sodickson DK, Otazo R. Unstreaking: Radial MRI with Automatic Streaking Artifact Reduction. In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, HI, USA, 2017. p. 4001., n.d. [Google Scholar]
- [29].Benkert T, Tian Y, Huang C, DiBella EVR, Chandarana H, Feng L. Optimization and validation of accelerated golden-angle radial sparse MRI reconstruction with self-calibrating GRAPPA operator gridding. Magn Reson Med 2018;80:286–93. 10.1002/mrm.27030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Walsh DO, Gmitro AF, Marcellin MW. Adaptive Reconstruction of Phased Array MR Imagery 2000. 10.1002/(SICI)1522-2594(200005)43:5. [DOI] [PubMed] [Google Scholar]
- [31].Crete F, Dolmiere T, Ladret P, Nicolas M. The blur effect: perception and estimation with a new no-reference perceptual blur metric. Https://DoiOrg/101117/12702790 2007;6492:196–206. 10.1117/12.702790. [DOI] [Google Scholar]
- [32].Ream JM, Doshi A, Lala SV., Kim S, Rusinek H, Chandarana H. High Spatiotemporal Resolution Dynamic Contrast-Enhanced MR Enterography in Crohn Disease Terminal Ileitis Using Continuous Golden-Angle Radial Sampling, Compressed Sensing, and Parallel Imaging. Http://DxDoiOrg/102214/AJR1413674 2015;204:W663–9. 10.2214/AJR.14.13674. [DOI] [PubMed] [Google Scholar]
- [33].Stehning C, Börnert P, Nehrke K, Eggers H, Stuber M. Free-breathing whole-heart coronary MRA with 3D radial SSFP and self-navigated image reconstruction. Magn Reson Med 2005;54:476–80. 10.1002/mrm.20557. [DOI] [PubMed] [Google Scholar]
- [34].Piccini D, Monney P, Sierro C, Coppo S, Bonanno G, Van Heeswijk RB, et al. Respiratory self-navigated postcontrast whole-heart coronary MR angiography: Initial experience in patients. Radiology 2014;270:378–86. 10.1148/radiol.13132045. [DOI] [PubMed] [Google Scholar]
- [35].Feng L, Coppo S, Piccini D, Yerly J, Lim RP, Masci PG, et al. 5D whole-heart sparse MRI. Magn Reson Med 2018;79:826–38. 10.1002/mrm.26745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Feng L, Delacoste J, Smith D, Weissbrot J, Flagg E, Moore WH, et al. Simultaneous Evaluation of Lung Anatomy and Ventilation Using 4D Respiratory-Motion-Resolved Ultrashort Echo Time Sparse MRI. J Magn Reson Imaging 2019;49:411–22. 10.1002/jmri.26245. [DOI] [PMC free article] [PubMed] [Google Scholar]