Abstract
Viruses are obligate intracellular parasites that depend on host cellular machinery for performing even basic biological functions. One of the many ways they achieve this is through molecular mimicry, wherein the virus mimics a host sequence or structure, thereby being able to hijack the host's physiological interactions for its pathogenesis. Such adaptations are specific recognitions that often confer tissue and species-specific tropisms to the virus, and enable the virus to utilise previously existing host signalling networks, which ultimately aid in further steps of viral infection, such as entry, immune evasion and spread. A common form of sequence mimicry utilises short linear motifs (SLiMs). SLiMs are short-peptide sequences that mediate transient interactions and are major elements in host protein interaction networks. This work is aimed at providing a comprehensive review of current literature of some well-characterised SLiMs that play a role in the attachment and entry of viruses into host cells, which mimic physiological receptor-ligand interactions already present in the host. Considering recent trends in emerging diseases, further research on such motifs involved in viral entry can help in the discovery of previously unknown cellular receptors utilised by viruses, as well as help in the designing of targeted therapeutics such as vaccines or inhibitors directed towards these interactions.
Keywords: Host–pathogen interactions, Short linear motifs (SLiMs), Viral entry, Molecular mimicry, Cell surface biomolecules, Viral entry receptors
Introduction
Viruses are obligate intracellular parasites that require host cell machinery for performing even the most basic biological functions, such as nucleic acid replication and protein synthesis [1]. The first major step in a viral infection cycle is the attachment and entry of the virus into a suitable host cell, after which further pathogenesis occurs [2, 3]. Viruses interact with host cell surface biomolecules such as proteins or carbohydrates, to attach to, and enter host cells. These interactions are generally specific recognitions that often confer tissue and species-specific tropisms to these viruses [2, 4, 5].
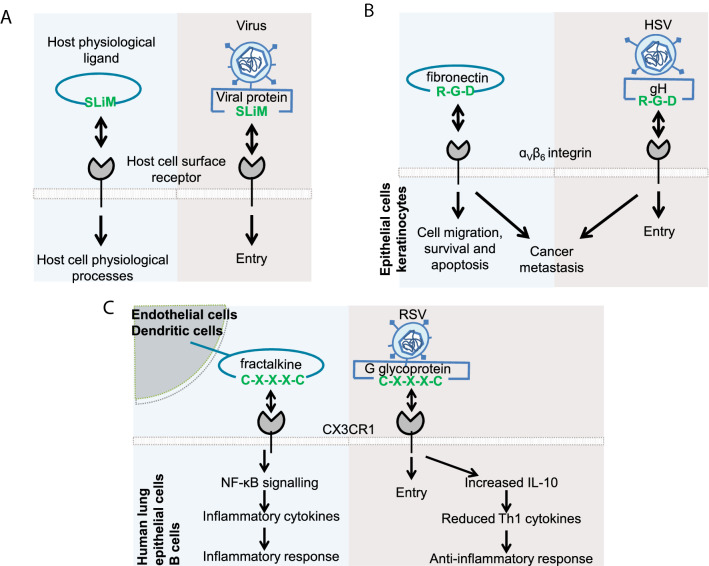
As viruses are dependent on host cellular machinery for their basic biological processes, many viruses have evolved to mimic host interactions for these processes [6]. These adaptations, referred to as molecular mimicry, enable the virus to mimic host interactions to hijack previously existing host signalling networks (Fig. 1A) [6]. Studies have shown that such mimicry can confer various advantages to the virus, which ultimately aids in further viral infection and spread: entry, immune modulation and evasion, replication, etc. [7]. At the molecular level, the mimicry can be either structural or sequence mimicry [6]. In this article, we review some well-characterised sequence motifs involved in host–virus interactions that play a role in the attachment and entry of the virus into the host cells, that mimic some physiological receptor–ligand interactions present in host cells.
Fig. 1.
Viral mimicry of host SLiMs for entry into host cells. A Viruses often utilise existing host cellular network for carrying out their own cellular functions. One such mechanism is the mimicry of the host cell’s short linear motif-mediated physiological receptor-ligand interactions for attachment and entry of the virus into the host cell. B and C The subsequent downstream signalling triggered by such mimicry can follow the exact same pathway as the physiological interaction (B), or may follow a different pathway (C)
Viral mimicry of host short linear motif-mediated interactions
Short linear motifs (SLiMs) or mini motifs are short ~ 3–10 amino acid sequences that mediate specific molecular functions [6, 8]. They are generally present in relatively unstructured or disordered regions of proteins, and can easily appear or disappear during evolution because of point mutations. They also typically mediate transient interactions with low affinities, thus, contributing greatly to protein interaction networks [6, 8, 9].
Due to the properties mentioned above, mimicking SLiMs is a widespread molecular mimicry mechanism utilised by many pathogens, including viruses [6, 7]. Moreover, viral mimicry directly disrupts the host signalling mechanisms by competitively binding to host surface receptors and preventing physiological interactions [7]. Furthermore, they may trigger further downstream responses that are similar to the physiological ones (Fig. 1B), or lead to an entirely different response (Fig. 1C).
In this article, we review some well-characterised SLiMs involved in host–virus interactions that play a role in the attachment and entry of the virus into the host cells, which mimic some physiological receptor–ligand interactions present in host cells (Table 1).
Table 1.
Some well-characterised sequence motifs involved in viral molecular mimicry for attachment and entry into host cells
| Motif | Appearance of sequence motifs | Interacting host cell surface receptor | References | |
|---|---|---|---|---|
| Viral glycoprotein | Host physiological ligand | |||
| CX3C | Glycoprotein G in respiratory syncytial virus | Fractalkine | CX3CR1 | [10, 11] |
| GPR | VP7 in human rotaviruses | Fibrinogen | ɑxꞵ2 integrins | [12–14] |
| RGD | gB glycoprotein in Kaposi’s sarcoma-associated herpesvirus | Vitronectin and Fibronectin | ɑVꞵ3, ɑVꞵ1, and ɑVꞵ5 integrins | [15, 16] |
| BMRF-2 in Epstein-Barr virus | Vitronectin and Fibronectin | β1, α3, α5, and αv integrins | [15, 16] | |
| Penton base in type C adenoviruses(Ad5 and Ad2) | Vitronectin and Fibronectin | αvβ3 and αvβ5 | [15, 17] | |
| gH in herpes simplex virus | Fibronectin | αvβ6- and αvβ8 | [15, 18] | |
| VP1 in foot-and-mouth disease virus (A12) | Vitronectin and Fibronectin | αvβ3 | [15, 19] | |
| VP1 in coxsackievirus A9 | Fibronectin | αvβ6 | [15, 20] | |
| KGD | gH/gL in Epstein-Barr virus | Collagen XVII | αVβ5, αVβ6, and αVβ8 | [21–25] |
| DGE | VP5 in rotaviruses | Type I collagen | α2β1 | [14, 26, 27] |
| R/K-X-X-R/K | S1 subunit in SARS-CoV-2 | VEGF165 | Neuropilin-1 | [28–31] |
| gB in Epstein-Barr virus | VEGF165 | Neuropilin-1 | [28, 29, 32] | |
| Surface (SU) subunit human T-cell lymphotropic viruses | VEGF165 | Neuropilin-1 | [28, 29] | |
| LDV and LDI | VP7 in rotavirus group A | Fibronectin (CS-1), VCAM-1, MAdCAM-1 | α4β1 and α4β7 | [13, 33–35] |
| IDA | VP4 in rotavirus group A | VCAM-1 | α4β1 and α4β7 | [13, 33–35] |
| YGL | VP4 in rotavirus group A | Osteopontin | α4β1 | [13, 36] |
| VP37 in white spot syndrome virus | Osteopontin | β integrins | [23, 37] | |
CX3C motif
The CX3C (Cys-Xxx-Xxx-Xxx-Cys) motif present in the chemokine fractalkine (CX3C ligand 1—CX3CL1) mediates its interaction with CX3C receptor 1 (CX3CR1) [10]. Physiologically, interaction of fractalkine with the CX3CR1 is important for leukocyte migration across endothelial barriers and chemotaxis. It triggers further downstream signalling pathways that induce NF-kB activation and expression of CX3CR1 as well as inflammatory cytokines such as IL-8 and MIG (monokine induced by interferon gamma) [11, 38, 39].
One mode of entry of respiratory syncytial virus (RSV) into the host regulatory B-cells, and airway and lung epithelial cells is through the interaction of RSV glycoprotein G with CX3C receptor 1 (CX3CR1) present on the host cells, through the CX3C motif of glycoprotein G [11, 40]. This interaction mimics the physiological interaction between CX3CR1 and the CX3C motif present in the chemokine fractalkine (the physiological ligand for CX3CR1) [10]. Thus, RSV glycoprotein G competes with fractalkine to interact with CX3CR1. Meanwhile, RSV glycoprotein G interaction with CX3CR1 also triggers various cellular responses that are distinct from the physiological fractalkine–CX3CR1 interaction (Fig. 1C) [10, 38].
GPR motif
The GPR (Gly-Pro-Arg) motif present in the N-terminal domain of ɑ chain of fibrinogen is crucial for its interaction with ɑxꞵ2integrin. This fibrinogen—ɑxꞵ2 integrin (CD11c/CD18) interaction plays an important role in the adhesion of tumour necrosis factor-stimulated polymorphonuclear leukocytes (PMNs) to fibrinogen/fibrin thrombi, especially during clot formation [12].
After human rotaviruses attach to major entry receptor ɑ2ꞵ1 integrin through viral spike protein VP4, the interaction of viral outer capsid protein VP7 with ɑxꞵ2 integrin in the early endosomes mediate the entry of the virus into the host human kidney or intestinal epithelial cells, resulting in further infection [13]. Interestingly, mucosal PMNs and inflammation increase during rotavirus infection, suggesting a possible interaction between the virus and the PMNs [41]. Subsequent studies have shown that VP7 also contains a conserved GPR motif that is involved in its recognition by integrin [14, 33, 41].
RGD motif
The RGD (Arg-Gly-Asp) motifs present in many extracellular interacting partners of integrins (such as vitronectin, fibronectin, von Willebrand factor, fibrinogen, osteopontin, and thrombospondin) mediate their interactions with integrins, thus regulating vital cellular processes such as cell migration, proliferation, survival, and apoptosis [42–45]. For example, vitronectin interacts with ɑVꞵ1, ɑVꞵ3, ɑVꞵ5, and ɑIIꞵ1 integrins, while fibronectin interacts with ɑ3ꞵ1, ɑVꞵ1, ɑ8ꞵ1, ɑVꞵ1, ɑVꞵ3, ɑVꞵ5, ɑVꞵ8, ɑVꞵ6 and ɑIIꞵ3 integrins, and cadherin-17 and VE-cadherin can recognise α2β1 integrin in an RGD-dependent manner [15, 46]. Studies also show that many of the RGD-binding integrins are important in mediating cancer metastasis [46].
Viruses such as Kaposi’s sarcoma-associated herpesvirus (KSHV), Epstein-Barr virus (EBV), adenoviruses, herpes simplex virus (HSV), foot-and-mouth disease virus (FMDV) and coxsackievirus A9 (CAV9) exploit the RGD motif for interacting with RGD-dependent integrins, mediating their entry into host cells [16–20, 47]. KSHV forms complex with integrins (ɑVꞵ3, ɑVꞵ1 and ɑVꞵ5) and CD98/xCT through the glycoprotein B containing an RGD motif to enter into human dermal microvascular endothelial cells [16]. Antibodies against RGD motif, fibronectin or ɑV integrins reduce KSHV infection by ~ 50 per cent, highlighting the requirement of RGD motif for its entry. EBV glycoprotein BMRF-2 contains an RGD motif in its extracellular domain essential for its entry into oral epithelial cells by interacting with ꞵ1 and ɑV family of integrins [47].Similarly, after attaching to its primary entry receptor CAR in human melanoma cell lines, adenovirus type 5 (Ad5) binds to ɑVꞵ3 and ɑVꞵ5 integrins through the RGD motif of the penton base protein [17]. The glycoprotein H of HSV interacts specifically with ɑVꞵ6 and ɑVꞵ8 integrins through an RGD motif which helps them to get endocytosed in epithelial cells and keratinocytes, mediating its entry (Fig. 1B) [18]. Mutational studies also highlighted that FMDV (A12) containing an RGD motif in G-H loop of VP1 and CAV9 containing an RGD motif near the C terminus of VP1, bind to αvꞵ3 and αvꞵ6, respectively, for their entry [19, 20]. It is interesting to note that several of these viruses, such as KSHV and EBV are also generally involved in viral-induced tumours [48–50].
KGD motif
KGD motif is closely related to the RGD motif [21]. Physiologically, multiple KGD (Lys-Gly-Asp) motifs present in the COL15 domain of collagen XVII are important for its interaction with α5β1 and αVβ1integrins. These interactions mediate keratinocyte spreading and migration [21]. Similarly, the KGD motif of avian tenascin-W (present in the fibronectin type III domain) is important for its interaction with integrins, which regulates developmental patterning [51]. KGD motif binds more specifically to the αIIbβ1 integrin [52].
KGD motifs present on the surface-exposed loop of the glycoproteins H and L of EBV are important for the membrane fusion of EBV to epithelial as well as B-cells, the primary infection sites of EBV [22, 23]. Mutation of the KGD motif or down-regulation of αV integrins (αVβ6 and αVβ8) leads to reduced EBV infection in epithelial cells. These data together suggest that EBV interaction with these integrins through the KGD motif is important for its entry [23–25]. Furthermore, EBV infections also cause severe thrombocytopenia (decreased platelet counts), in a manner similar to the mode of action of several viper venoms which contain disintegrins (small proteins found in viper venom with RGD or KGD motifs that disrupt platelet aggregation and integrin-dependent cell adhesion through competitive binding) [52–54].
DGE motif
DGE (Asp-Gly-Glu) motifs present in type I collagen are important for its interaction with α2β1 integrin present on platelets and fibroblasts [26]. This interaction is important for platelet activation and thrombosis [55].
Rotavirus spike protein VP4 is proteolytically cleaved into VP5 and VP8 [27]. The VP5 is responsible for its attachment to α2β1 integrins which is essential for viral entry into intestinal cells [14, 27]. This interaction is mediated by a DGE present on the VP5 of rotaviruses [14]. Monoclonal antibodies against α2 integrins inhibit the VP5-α2β1 interaction, similar to the physiological collagen-α2β1 interaction, thus, reducing rotavirus infection [33]. On the other hand, clinical studies suggest that acute human rotavirus infection decreases the mean platelet volume, especially in children [56], suggesting a possible interaction between the platelets and human rotaviruses.
(R/K)XX(R/K)
The (R/K)XX(R/K) (Arg/Lys-Xxx-Xxx-Arg/Lys) motif present in vascular endothelial growth factor (VEGF) mediates its interaction with neuropilin-1 [28]. The C-terminal region of VEGF-A containing the polybasic motif R/K-X-X-R/K must be proteolytically cleaved to become active, thus, named as C-end rule (CendR) motif [57]. Neuropilin-1, a physiological co-receptor for VEGF-A, is a transmembrane protein expressed on various immune cells, and is involved in regulating VEGF-A dependent biological processes such as axon guidance, angiogenesis, and endothelial and vascular permeability, development and leakage [29, 58]. Studies have shown that certain peptides and nanoparticles containing a C-terminal arginine (or rarely lysine) in the CendR motif can interact with neuropilin-1, and internalise into the cells [57]. Due to this property, many viruses exploit the CendR motif: neuropilin-1 interaction for their entry into host cells [28, 30–32, 59–61].
After the CendR motif of EBV glycoprotein B (RRRR) becomes exposed by furin protease cleavage, it interacts with neuropilin-1, which is essential in mediating EBV entry into human nasopharyngeal epithelial cells [32]. Furthermore, this interaction activates the downstream signalling pathways (such as EBV-activated EGFR/RAS/ERK and neuropilin-1-dependent receptor tyrosine kinase pathways) that further facilitates EBV infection.
Human T-cell lymphotropic virus type 1 (HTLV-1) usesneuropilin-1 as a receptor for entering CD4+ T cells and dendritic cells [28]. The HTLV-1 surface (SU) subunit of Env protein consists of a KPXR motif that is involved in its interaction with neuropilin-1 present on these host cells. This region is also highly conserved in various HTLVs (1, 2 and 3), as well as in related simian viruses.
Similarly, furin protease cleavage of CendR site present in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein S results in the formation of RRAR in the S1 subunit, important for enhanced fusion capacity and increased infectivity [30, 31]. This interaction with neuropilin-1, when co-expressed with ACE-2 and TMPRSS2, increases infectivity of the virus. Furthermore, as VEGF-A164-neuropilin-1 interaction also regulates nociception, viral alteration of this physiological interaction has been hypothesised as a reason for VEGF-A-driven pain in SARS-CoV-2 patients [59, 62].
LDV and LDI motifs
Alternatively spliced domains of fibronectin contains an LDV (Leu-Asp-Val) and LDI (Leu-Asp-Ile) motifs, and VCAM-1 consists of an LDV motif, respectively, which is essential for their interaction with α4β1, mediating leukocyte adhesion and plays important role in modulating inflammatory responses [34, 35, 63].
Rotavirus group A expresses α4 integrin-binding motifs LDV and LDI in its outer capsid protein VP7 [13]. Cell-based studies showed that some of the rotaviruses use α4 integrins (α4β1 and α4β7) as their entry receptor or co-receptor. White spot syndrome virus (WSSV) possesses an LDV motif in its outer surface glycoproteins VP26 and VP31 [64]. Also, small peptides or antibodies of the LDV sequence reduces α4 integrin interactions with rotaviruses and decreases their infectivity [13, 33]. Peptides containing LDV sequences partially inhibit their infection by reducing their interaction with β integrins [33, 37, 64]. Disrupting these physiological interactions may help rotaviruses to re-infect the host cells. Also, many α4 integrins are present in immune cells like T cells and B-cells, suggesting a possible explanation for immune modulation exhibited by rotaviruses [65]. Furthermore, the interactions of LDV motifs present in fibronectin connecting segment-1 region with α4β1 also leads to oral cancer cell adhesion, migration and invasion [66]. Rotavirus infection shows a similar type of B-cells accumulation in mesenteric lymph nodes leading to lymph node hypertrophy and increased cytokine levels in oral mucosa [67].
IDA motif
Rotavirus group A expresses α4 integrin-binding motif IDA (Ile-Asp-Ala) in their spike protein VP4 [13]. Cell-based studies showed that some of the rotaviruses use α4 integrins (α4β1 and α4β7) as their entry receptor or co-receptor. Physiologically, alternatively spliced domains of fibronectin contain an IDA motif which is essential for its interaction with α4β1, thus, mediating leukocyte adhesion and modulating inflammatory responses [34, 35, 63].
YGL motif
YGL (Tyr-Gly-Leu) motif present in osteopontin is important for its interaction with α4β1 and α9β1, and signals Th-1 cytokine-mediated immune responses sometimes leading to chronic inflammation.
Rotaviruses also contain an YGL motif in their VP4 [13]. In the rotavirus infection assay, peptides containing YGL motifs tend to inhibit the binding of α4β1 to MAdCAM-1 and fibronectin, and partially inhibit the interaction with VCAM-1. WSSV possesses a YGL motif in outer surface glycoprotein VP37 [36, 37, 64].
Structural perspectives
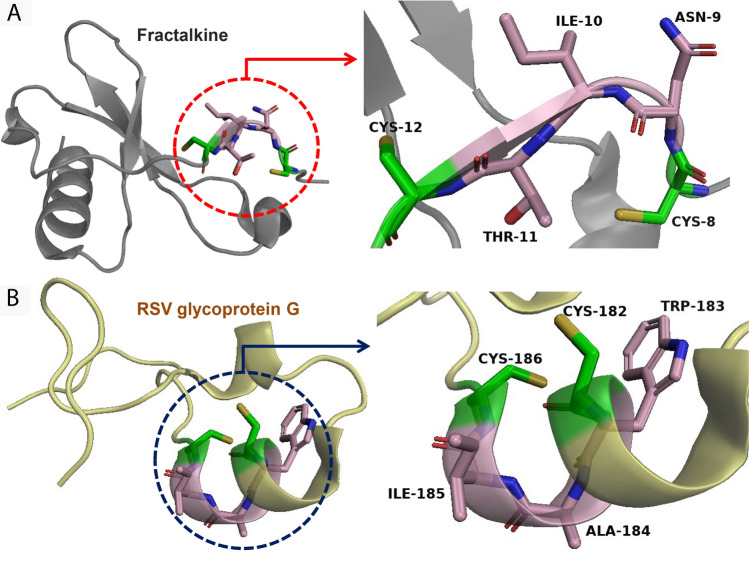
As SLiMs are short-peptide sequences that are generally found in relatively unstructured regions of proteins, their three-dimensional conformation may vary drastically between the host physiological ligand and the viral ligand [6]. For instance, the CX3C motif present in RSV glycoprotein G does not take the same structural conformation as the CX3C motif of the physiological ligand fractalkine, although they both recognise the same cellular receptor CX3CR1 (Fig. 2). Furthermore, due to their short nature, SLiMs have been found to appear, mutate, and disappear relatively easily during the course of normal evolution [1, 68–70]. Hence, these properties are hypothesised to confer some flexibility to these peptides to adapt, mutate and evolve as necessary, thus, making them indispensable during the evolution of protein interaction networks within the host, as well as in host–pathogen interactions.
Fig. 2.
The structural conformation of SLiMs may differ between the host physiological ligand and the viral ligand, as SLiMs are short-peptide sequences that are generally found in relatively unstructured regions of proteins. For example, the CX3C motifs (shown in red) of RSV glycoprotein G (PDB id: 1F2L) (A) and the physiological ligand fractalkine (PDB id: 6BLH) (B) do not exhibit the same structural conformation, although they both recognise the same cellular receptor CX3CR1
Conclusion and future directions
Viruses are a unique category of pathogens that are dependent completely on their hosts for performing basic cellular processes, and have, thus, evolved to utilise existing host interaction and signalling networks for various steps during their pathogenesis [1–3]. One of the many ways they achieve this is by molecular mimicry, wherein the virus mimics a host sequence or structure, thereby being able to hijack the host physiological interactions for their own pathogenesis. Common sequence mimicry utilises SLiMs, short-peptide sequences that are major elements in host protein interaction networks [6]. In this article, we have reviewed some well-characterised sequence mimicry by viruses that are vital for the entry of the virus into host cells.
Furthermore, mimicking host motifs can also confer other advantages to the virus: SLiMs can evolve and mutate easily, and can be easily integrated into existing protein interaction networks. This can also lead to immune evasion, where the host immune system recognises these imposters as self, and fails to act [1, 68–70]. Hence, various pathogens (including viruses, as well as prokaryotic and eukaryotic) have evolved to utilise SLiMs for their pathogenesis through molecular mimicry [6].
Although similar sequence motifs present on both the viruses and the physiological ligands mediate these interactions, some of these interactions between viral glycoproteins and their cognate receptors on the host cells mimic physiological downstream processes triggered by the physiological ligand, while others induce signalling cascades that are different [7, 10, 48]. Moreover, the three-dimensional conformation of these motifs may also differ in the viral and physiological ligand, which can be explained by the properties of SLiMs such as their short lengths and occurrence in disordered regions of proteins [6]. The exact molecular and structural determinants involved in many of these interactions remain under explored.
SLiMs are also known for their integral role in a wide range of physiological processes, including cell signalling. Thus, finding the whole repertoire of SLiMs and understanding the mechanisms involved in regulating these diverse processes are of at most importance. However, their low complexity, small size and periodic mutations impart a challenge in identifying new SLiMs from the complex human proteome. In recent years, viral mimicry has been suggested as one of the integral part of a viral infection, and researchers have been able to identify a number of novel viral SLiMs essential for their entry into hosts. Convergent evolution of the human SLiMs in viruses as well as the existence of simpler proteome compared to the humans can help us to identify novel SLiMs in humans [71, 72].
Cell surface biomolecules are exploited by viruses for attachment and entry, thus, dictating host specificity and tissue tropism. Research on such motifs can predict the cellular receptors of various viruses, as well as help in the designing of targeted therapeutics such as vaccines or inhibitors designed specifically for modifying these interactions [73]. Recent studies have shown that SLiM-based peptide inhibitors and drugs such as nutlin, venetoclax and cilengitide can reduce the infectivity of many viruses. However, the broad spectrum antiviral capability of these therapeutic strategies remain to be explored [74, 75]. Thus, elucidating the structural and molecular basis of the interactions of viral SLiMs with host cell surface receptors can help us to design newer intervention techniques against viral infections in future.
Perspectives
While the research on emerging diseases has been gaining focus in the past two decades, there is renewed interest in this field due to the ongoing pandemic.
As entry receptors present on the host cell surface play a vital role in the viral pathogenesis process, targeting these interactions has been a standard strategy for preventing further spread of the virus.
Considering recent trends in emerging diseases, further research on such motifs involved in viral entry can help in the discovery of previously unknown cellular receptors utilised by viruses, as well as help in the designing of targeted therapeutics such as vaccines or inhibitors directed towards these interactions.
Abbreviations
- Ad5
Adenovirus type 5
- CAV9
Coxsackievirus A9
- CD
Cluster of differentiation
- CendR
C-end rule
- CX3CR1
CX3C receptor 1
- CX3CL1
CX3C ligand 1
- EBV
Epstein-Barr virus
- ECM
Extracellular matrix
- FMDV
Foot-and-mouth disease virus
- HSV
Herpes simplex virus
- HTLV
Human T-cell lymphotropic virus type 1
- IL
Interleukin
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- MIG
Monokine induced by interferon gamma
- NF-kB
Nuclear factor kappa B
- PMN
Polymorphonuclear leukocytes
- RSV
Respiratory syncytial virus
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SLiMs
Short linear motifs
- VEGF
Vascular endothelial growth factor
- WSSV
White spot syndrome virus
Author contributions
Conceptualization, K.D. and D.S.; literature review, S.D. and K.D.; writing—original draft, review and editing, S.D., K.D. and D.S.; supervision, D.S. All authors have read and agreed to the submission of the article.
Funding
Not applicable.
Declarations
Conflict of interest
Saumyadeep Goswami declares that he has no conflict of interest. Kheerthana Duraivelan declares that she has no conflict of interest. Dibyendu Samanta declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dibyendu Samanta, Email: dibyendu.samanta@iitkgp.ac.in.
Kheerthana Duraivelan, Email: kheerthana.duraivelan@einsteinmed.edu.
References
- 1.Hraber P, O’Maille PE, Silberfarb A, Davis-Anderson K, Generous N, McMahon BH, Fair JM. Resources to discover and use short linear motifs in viral proteins. Trends Biotechnol. 2020;38(1):113–127. doi: 10.1016/j.tibtech.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimitrov DS. Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2(2):109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrell CJ, Howard CR, Murphy FA. Pathogenesis of virus Infections. In: Burrell CJ, Howard CR, Murphy FA, editors. Fenner and white’s medical virology. 5. London: Academic Press; 2017. pp. 77–104. [Google Scholar]
- 4.Brito AF, Pinney JW. Protein-protein interactions in virus-host systems. Front Microbiol. 2017;8:1557. doi: 10.3389/fmicb.2017.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grove J, Marsh M. The cell biology of receptor-mediated virus entry. J Cell Biol. 2011;195(7):1071–1082. doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Via A, Uyar B, Brun C, Zanzoni A. How pathogens use linear motifs to perturb host cell networks. Trends Biochem Sci. 2015;40(1):36–48. doi: 10.1016/j.tibs.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Sobhy H. A review of functional motifs utilized by viruses. Proteomes. 2016;4(1):3. doi: 10.3390/proteomes4010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey NE, van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella H, Gibson TJ. Attributes of short linear motifs. Mol BioSyst. 2012;8(1):268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 9.Elkhaligy H, Balbin CA, Gonzalez JL, Liberatore T, Siltberg-Liberles J. Dynamic, but not necessarily disordered, human-virus interactions mediated through slims in viral proteins. Viruses. 2021;13(12):2369. doi: 10.3390/v13122369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson CS, Chu CY, Wang Q, Mereness JA, Ren Y, Donlon K, Bhattacharya S, Misra RS, Walsh EE, Pryhuber GS, Mariani TJ. CX3CR1 as a respiratory syncytial virus receptor in pediatric human lung. Pediatric Res. 2020;87(5):862–867. doi: 10.1101/19002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizoue LS, Bazan JF, Johnson EC, Handel TM. Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry. 1999;38(5):1402–1414. doi: 10.1021/bi9820614. [DOI] [PubMed] [Google Scholar]
- 12.Loike JD, Sodeik B, Cao L, Leucona S, Weitzt JI, Detmers PA, Wright SD, Silverstein SC. CD11c/CD18 on neutrophils recognizes a domain at the N terminus of the Aa chain of fibrinogen. Proc Natl Acad Sci U S A. 1991;88:1044–1048. doi: 10.1073/pnas.88.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham KL, Fleming FE, Halasz P, Hewish MJ, Nagesha HS, Holmes IH, et al. Rotaviruses interact with α4β7 and α4β1 integrins by binding the same integrin domains as natural ligands. J Gen Virol. 2005;86(12):3397–3408. doi: 10.1099/vir.0.81102-0. [DOI] [PubMed] [Google Scholar]
- 14.Graham KL, Halasz P, Tan Y, Hewish MJ, Takada Y, Mackow ER, et al. Integrin-using rotaviruses bind α2β1 integrin α2 I domain via VP4 DGE sequence and RECOGNIZE αXβ2 and αVβ3 by using VP7 during cell entry. J Virol. 2003;77(18):9969–9978. doi: 10.1128/JVI.77.18.9969-9978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275(29):21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 16.Veettil MV, Sadagopan S, Sharma-Walia N, Wang FZ, Raghu H, Varga L, et al. Kaposi’s sarcoma-associated herpesvirus forms a multimolecular complex of integrins (αVβ5, αVβ3, and α3β1) and CD98-xCT during infection of human dermal microvascular endothelial cells, and CD98-xCT is essential for the postentry stage of infection. J Virol. 2008;82(24):12126–12144. doi: 10.1128/JVI.02052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyle C, Mccormick F. Integrin αvβ5 is a primary receptor for adenovirus in CAR-negative cells. Virol J. 2010;7(1):1–13. doi: 10.1186/1743-422X-7-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gianni T, Salvioli S, Chesnokova LS, Hutt-Fletcher LM, Campadelli-Fiume G. αvβ6- and αvβ8-integrins serve as interchangeable receptors for HSV gH/gL to promote endocytosis and activation of membrane fusion. PLoS Pathog. 2013;9(12):1–14. doi: 10.1371/journal.ppat.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson T, Sharma A, Ghazaleh RA, Blakemore WE, Ellard FM, Simmons DL, et al. Arginine-glycine-Aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin avb3 in vitro. J Virol. 1997;71(11):8357–8361. doi: 10.1128/jvi.71.11.8357-8361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams ÇH, Kajander T, Hyypiä T, Jackson T, Sheppard D, Stanway G. Integrin αvβ6 is an RGD-dependent receptor for coxsackievirus A9. J Virol. 2004;78(13):6967–6973. doi: 10.1128/JVI.78.13.6967-6973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nykvist P, Tasanen K, Viitasalo T, Käpylä J, Jokinen J, Bruckner-Tuderman L, et al. The cell adhesion domain of type XVII collagen promotes integrin-mediated cell spreading by a novel mechanism. J Biol Chem. 2001;276(42):38673–38679. doi: 10.1074/jbc.M102589200. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Rowe CL, Jardetzky TS, Longnecker R. The KGD motif of Epstein-Barr virus gH/gL is bifunctional, orchestrating infection of B cells and epithelial cells. MBio. 2012;3(1):e00290–e00311. doi: 10.1128/mBio.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Zhang X, Jardetzky TS, Longnecker R. The Epstein-Barr virus (EBV) glycoprotein B cytoplasmic C-terminal tail domain regulates the energy requirement for EBV-induced membrane fusion. J Virol. 2014;88(20):11686–11695. doi: 10.1128/JVI.01349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, et al. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol. 2018;3(2):172–180. doi: 10.1128/mBio.02892-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Longnecker R. Epithelial cell infection by Epstein-Barr virus. FEMS Microbiol Rev. 2019;43(6):674–683. doi: 10.1093/femsre/fuz023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staatzs WD, Fokg KF, Zutters MM, Adamss SP, Rodriguez BA, Santoros SA. Identification of a tetrapeptide recognition sequence for the alpha 2 beta 1 integrin in collagen. J Biol Chem. 1991;266(12):7363–7367. doi: 10.1016/S0021-9258(20)89455-1. [DOI] [PubMed] [Google Scholar]
- 27.Zárate S, Cuadras MA, Espinosa R, Romero P, Juárez KO, Camacho-Nuez M, et al. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J Virol. 2003;77(13):7254–7260. doi: 10.1128/JVI.77.13.7254-7260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert S, Bouttier M, Vassy R, Seigneuret M, Petrow-Sadowski C, Janvier S, et al. HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood. 2009;113(21):5176–5185. doi: 10.1182/blood-2008-04-150342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth L, Prahst C, Ruckdeschel T, Savant S, Weström S, Fantin A, et al. Neuropilin-1 mediates vascular permeability independently of vascular endothelial growth factor receptor-2 activation. Sci Signal. 2016;9(425):ra42. doi: 10.1126/scisignal.aad3812. [DOI] [PubMed] [Google Scholar]
- 30.Daly JL, Simonetti B, Klein K, Chen KE, Kavanagh Williamson M, Antón-Plágaro C, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang HB, Zhang H, Zhang JP, Li Y, Zhao B, Feng GK, et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat Commun. 2015 doi: 10.1038/s41564-017-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coulson BS, Londrigan SL, Lee DJ. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc Natl Acad Sci USA. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis GE, Thomas JS, Madden S. The α4β1 integrin can mediate leukocyte adhesion to casein and denatured protein substrates. J Leukoc Biol. 1997;62(3):318–328. doi: 10.1002/jlb.62.3.318. [DOI] [PubMed] [Google Scholar]
- 35.Mould AP, Humphries MJ. Identification of a novel recognition sequence for the integrin α4β1 in the COOH-terminal heparin-binding domain of fibronectin. EMBO J. 1991;10(13):4089–4095. doi: 10.1002/j.1460-2075.1991.tb04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund SA, Giachelli CM, Scatena M. The role of osteopontin in inflammatory processes. J Cell Commun Signal. 2009;3(3):311–322. doi: 10.1007/s12079-009-0068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng ZS, Xu JY, Liu HP. Cellular entry of white spot syndrome virus and antiviral immunity mediated by cellular receptors in crustaceans. Fish Shellfish Immunol. 2019;93:580–588. doi: 10.1016/j.fsi.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Imai T, Hieshima K, Haskell C, Baba M, Nagira M, Nishimura M, Kakizaki M, Takagi S, Nomiyama H, Schall TH, Yoshie O. Identification and molecular characterization of fractalkine receptor CX 3 CR1, which mediates both leukocyte migration and adhesion. Cell. 1997;91:521–530. doi: 10.1016/S0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhivaki D, Lemoine S, Lim A, Morva A, Vidalain PO, Schandene L, et al. Respiratory syncytial virus infects regulatory B cells in human neonates via chemokine receptor CX3CR1 and promotes lung disease severity. Immunity. 2017;46(2):301–314. doi: 10.1016/j.immuni.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chirkova T, Lin S, Oomens AGP, Gaston KA, Boyoglu-Barnum S, Meng J, Stobart CC, Cotton CU, Hartert TV, Moore ML, Ziady AG, Anderson LJ. CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. J Gen Virol. 2015;96(9):2543–2556. doi: 10.1099/vir.0.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris AP, Estes MK. Microbes and microbial toxins: paradigms for microbial-mucosal interactions VIII. Pathological consequences of rotavirus infection and its enterotoxin. Am J Physiol. 2001;281(1):G303–G3010. doi: 10.1152/ajpgi.2001.281.2.G303. [DOI] [PubMed] [Google Scholar]
- 42.Kapp TG, Rechenmacher F, Neubauer S, Maltsev O, Cavalcanti-Adam EA, Zarka R, et al. A comprehensive evaluation of the activity and selectivity profile of ligands for RGD-binding integrins. Sci Rep. 2017;7(1):1–13. doi: 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludwig BS, Kessler H, Kossatz S, Reuning U. RGD-binding integrins revisited: How recently discovered functions and novel synthetic ligands (re-)shape an ever-evolving field. Cancers (Basel) 2021;13(7):1711. doi: 10.3390/cancers13071711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plow EF, Pierschbachert MD, Ruoslahtit E, Margueriei GA, Ginsberg MH. The effect of Arg-Gly-Asp-containing peptides on fibrinogen and von Willebrand factor binding to platelets. Proc Natl Acad Sci U S A. 1985;82:8057–8061. doi: 10.1073/pnas.82.23.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oldberg A, Franzfn A, Heinegard D. Cloning and sequence analysis of rat bone sialoprotein (osteopontin) cDNA reveals an Arg-Gly-Asp cell-binding sequence. Biochemistry. 1986;83:8819–8823. doi: 10.1073/pnas.83.23.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casal JI, Bartolome RA. RGD cadherins and α2β1 integrin in cancer metastasis: a dangerous liaison. Biochim Biophys Acta Rev Cancer. 2018;2:321–332. doi: 10.1016/j.bbcan.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Xiao J, Palefsky JM, Herrera R, Berline J, Tugizov SM. The Epstein-Barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370(2):430–442. doi: 10.1016/j.virol.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackey JK, Rigden PM, Green M. Do highly oncogenic group A human adenoviruses cause human cancer? Analysis of human tumors for adenovirus 12 transforming DNA sequences. Proc Natl Acad Sci. 1976;73(12):4657–4661. doi: 10.1073/pnas.73.12.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farrell PJ. Epstein-Barr virus and cancer. Annu Rev Pathol. 2019;14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023. [DOI] [PubMed] [Google Scholar]
- 50.Harvala H, Kalimo H, Stanway G, Hyypiä T. Pathogenesis of coxsackievirus A9 in mice: role of the viral arginine-glycine-aspartic acid motif. J Gen Virol. 2003;84(9):2375–2379. doi: 10.1099/vir.0.19246-0. [DOI] [PubMed] [Google Scholar]
- 51.Meloty-Kapella CV, Degen M, Chiquet-Ehrismann R, Tucker RP. Avian tenascin-W: Expression in smooth muscle and bone, and effects on calvarial cell spreading and adhesion in vitro. Develop Dynamics. 2006;235(6):1532–1542. doi: 10.1002/dvdy.20731. [DOI] [PubMed] [Google Scholar]
- 52.Yang LJ, Niu B, Zhang D, Yang T. Substitution of the echistatin amino acid motif RGDD with KGDW enhances inhibition of platelet aggregation and thrombogenesis. Int J Pept Res Ther. 2015;21(4):451–458. doi: 10.1007/s10989-015-9475-7. [DOI] [Google Scholar]
- 53.Likic R, Kuzmanic D. Severe thrombocytopenia as a complication of acute Epstein-Barr virus infection. Wien Klin Wochenschr. 2004;116(1):47–50. doi: 10.1007/BF03040424. [DOI] [PubMed] [Google Scholar]
- 54.Lu X, Davies J, Lu D, Xia M, Wattam B, Shang D, et al. The effect of the single substitution of arginine within the RGD tripeptide motif of a modified neurotoxin dendroaspin on its activity of platelet aggregation and cell adhesion. Cell Commun Adhes. 2006;13(3):171–183. doi: 10.1080/15419060600726183. [DOI] [PubMed] [Google Scholar]
- 55.Farndale RW. Collagen-induced platelet activation. Blood Cells Mol Dis. 2006;36(2):162–165. doi: 10.1016/j.bcmd.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 56.Mete E, Akelma AZ, Cizmeci MN, Bozkaya D, Kanburoglu MK. Decreased mean platelet volume in children with acute rotavirus gastroenteritis. Platelets. 2014;25(1):51–54. doi: 10.3109/09537104.2013.764493. [DOI] [PubMed] [Google Scholar]
- 57.Teesalu T, Sugahara KN, Ramana Kotamraju V, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci. 2009;106(38):16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becker PM, Waltenberger J, Yachechko R, Mirzapoiazova T, Sham JSK, Lee CG, et al. Neuropilin-1 regulates vascular endothelial growth factor-mediated endothelial permeability. Circ Res. 2005;96(12):1257–1265. doi: 10.1161/01.RES.0000171756.13554.49. [DOI] [PubMed] [Google Scholar]
- 59.Jobe A, Vijayan R. Neuropilins: C-end rule peptides and their association with nociception and COVID-19. Comput Struct Biotechnol J. 2021;19:1889–1895. doi: 10.1016/j.csbj.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balistreri G, Yamauchi Y, Teesalu T. A widespread viral entry mechanism: the C-end Rule motif-neuropilin receptor interaction. Proc Natl Acad Sci. 2021;118(49):e2112457118. doi: 10.1073/pnas.2112457118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moutal A, Martin LF, Boinon L, Gomez K, Ran D, Zhou Y, et al. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain. 2021;162(1):243–252. doi: 10.1097/j.pain.0000000000002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dominguez-Jimenez C, Sanchez-Aparicio P, Albar JP, Garcia-Pardo A. The α4β1 fibronectin ligands CS-19 HEP II, and RGD induce different intracellular events in B lymphoid cells. Comparison with the effects of the endothelial ligand VCAM-1. Cell Adhesion Commun. 1996;4(4–5):251–267. doi: 10.3109/15419069609010770. [DOI] [PubMed] [Google Scholar]
- 64.Zhang JY, Liu QH, Huang J. Multiple proteins of White spot syndrome virus involved in recognition of β-integrin. J Biosci. 2014;39(3):381–388. doi: 10.1007/s12038-014-9418-z. [DOI] [PubMed] [Google Scholar]
- 65.Youngman KR, Franco MA, Kuklin NA, Rott LS, Butcher EC, Greenberg HB. Correlation of tissue distribution, developmental phenotype, and intestinal homing receptor expression of antigen-specific B cells during the murine anti-rotavirus immune response. J Immunol. 2002;168(5):2173–2181. doi: 10.4049/jimmunol.168.5.2173. [DOI] [PubMed] [Google Scholar]
- 66.Kamarajan P, Garcia-Pardo A, D’silva NJ, Kapila YL. The CS1 segment of fibronectin is involved in human OSCC pathogenesis by mediating OSCC cell spreading, migration, and invasion. BMC Cancer. 2010;10(1):1–8. doi: 10.1186/1471-2407-10-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakawesi J, Konjit GM, DasoveanuDC J-L, Lahl K. Rotavirus infection causes mesenteric lymph node hypertrophy independently of type I interferon or TNF-α in mice. Eur J Immunol. 2021;51(5):1143–1152. doi: 10.1002/eji.202048990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rothenburg S, Brennan G. Species-specific host-virus interactions: implications for viral host range and virulence. Trends Microbiol. 2020;28(1):46–56. doi: 10.1016/j.tim.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hagai T, Azia A, Babu MM, Andino R. Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 2014;7(5):1729–1739. doi: 10.1016/j.celrep.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Acharya D, Dutta TK. Elucidating the network features and evolutionary attributes of intra- and interspecific protein–protein interactions between human and pathogenic bacteria. Sci Rep. 2021 doi: 10.1038/s41598-020-80549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wadie B, Kleshchevnikov V, Sandaltzopoulou E, Benz C, Petsalaki E. Use of viral motif mimicry improves the proteome-wide discovery of human linear motifs. Cell Rep. 2022;39:110764. doi: 10.1016/j.celrep.2022.110764. [DOI] [PubMed] [Google Scholar]
- 72.Shuler G, Hagai T. Rapidly evolving viral motifs mostly target biophysically constrained binding pockets of host proteins. Cell Rep. 2022;40:111212. doi: 10.1016/j.celrep.2022.111212. [DOI] [PubMed] [Google Scholar]
- 73.Corbi-Verge C, Kim PM. Motif mediated protein-protein interactions as drug targets. Cell Commun Signal. 2016;14(1):1–12. doi: 10.1186/s12964-016-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simonetti L, Nilsson J, McInerney G, Ivarsson Y, Davey NE. SLiM-binding pockets: an attractive target for broad-spectrum antivirals. Trends Biochem Sci. 2022 doi: 10.1016/j.tibs.2022.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Gressett TE, Nader D, Robles JP, Buranda T, Kerrigan S, Bix G. Integrins as therapeutic targets for SARS-CoV-2. Front Cell Infect Microbiol. 2022;12:892323. doi: 10.3389/fcimb.2022.892323. [DOI] [PMC free article] [PubMed] [Google Scholar]