Abstract
Background
Little is known about the clinical characteristics of talaromycosis with hyper–immunoglobulin E syndrome (HIES).
Methods
We conducted a multicenter retrospective study, which included 7 hospitals from 2016 to 2022. Five consecutive cases of human immunodeficiency virus (HIV)–negative patients with systemic Talaromyces marneffei infections due to STAT3-HIES were identified. A systematic literature review of original articles published in English identified an additional 7 cases. Clinical characteristics and laboratory parameters were collected.
Results
Forty-two percent (5/12) of patients were young adults. The main symptoms of 10 patients were similar: fever (75%), cough (75%) and dyspnea (33%), but two patients mainly had gastrointestinal symptoms. Most patients had a history of infections since infancy. T marneffei was cultured from the bronchoalveolar lavage fluid (50%) and 25% of patients were next-generation sequencing positive. Eight patients had significantly elevated serum immunoglobulin E, increased B cells and decreased natural killer cells. There were ten different STAT3 mutations, three of which were reported for the first time in this study. Chest computed tomography examinations showed multiple exudations with cavities in the lungs. Voriconazole combined with thymosin was effective. Despite given antifungal agents, most had poor outcomes and the case fatality rate was as high as 25%.
Conclusions
STAT3-HIES is most likely a susceptibility factor for T marneffei infections among HIV-negative patients, which has a high case fatality rate. Increased awareness among clinicians is necessary to help in early diagnosis.
Keywords: hyper-IgE syndrome, immunodeficiency, STAT3 mutation, Talaromyces marneffei
Talaromyces marneffei is a dimorphic endemic fungus causing life-threatening mycoses in Southeast Asia and southern China [1]. Talaromycosis was once considered to affect only patients with human immunodeficiency virus (HIV); however, the number of HIV-negative individuals with talaromycosis is increasing [1] and their mortality is higher than that in HIV-positive patients, partially due to the time-consuming pathogen diagnoses and the fact that clinicians are inexperienced in diagnosing and treating the disease [2]. Except for secondary immunodeficiency, patients with primary immunodeficiency due to anti-interferon-gamma (IFN-γ) autoantibodies and genetic variants that lead to susceptibility to T marneffei have been sought and recognized, such as CARD9, STAT1, and STAT3 mutations [3–5].
Heterozygous mutations in STAT3 are the main cause of autosomal dominant hyper–immunoglobulin E syndrome (HIES) [6], whereas biallelic mutations in the gene encoding cell division 8 (DOCK8) have been identified as the most common cause of autosomal recessive HIES [7]; different treatments are required between these subtypes [8, 9]. HIES are a heterogeneous group of inborn errors of immunity sharing manifestations characterized by a triad of eczema, recurrent skin and lung infections, and elevated immunoglobulin E (IgE) levels [10]. Bacterial infections, with Staphylococcus aureus being predominant, and fungal infections, such as Candida, are common in HIES because these infections are promoted by the dysfunction of T-helper 17 cells (Th17) [11, 12], whereas T marneffei was rare in patients with HIES according to previous reports. Little is known about the clinical characteristics of talaromycosis with HIES, so we performed a multicenter retrospective study and systematic literature review to describe the clinical characteristics of 12 HIV-negative patients with T marneffei infections due to STAT3-HIES. This study aimed to provide clinical experience for the early diagnosis of this condition.
METHODS
Multicenter Retrospective Study
A retrospective review was performed of HIV-negative patients diagnosed with talaromycosis from December 2015 through December 2021 in 7 hospitals, and the patients were mainly from the First Affiliated Hospital of Guangzhou Medical University and the First Affiliated Hospital of Guangxi Medical University. Five patients who were enrolled in this study presented with culture, biopsy, and/or next-generation sequencing (NGS). Genetic sequencing of STAT3 exon sequencing was performed. Informed consent was obtained from the patients with the support of the First Affiliated Hospital of Guangzhou Medical University and the First Affiliated Hospital of Guangxi Medical University.
Systematic Review
Search Strategy
We searched PubMed, Web of Science, Embase, and BIOSIS Library for original case reports and cohort studies on T marneffei published in English between 1 January 1985 and 31 December 2021. We used the keywords “penicilliposis,” “Penicillium marneffei,” “talaromycosis,” “Talaromyces marneffei,” and “T marneffei.” The references of the retrieved articles were reviewed for additional relevant citations. The abstracts of all identified articles were viewed, and the full-text versions of relevant articles were retrieved for data extraction and analysis.
Case Definition
Disseminated Talaromycosis
Talaromyces marneffei infection was defined according to the 2020 Revised Definitions of Invasive Fungal Disease published by the European Organization for Research and Treatment of Cancer/Mycoses Study Group [13, 14]. Diagnosis was confirmed by positive culture of T marneffei from the blood or other affected sites or via direct microscopy or histopathology in an individual with an illness consistent with endemic mycoses. Positive T marneffei cultures were characterized by characteristic thermal dimorphism and microscopic morphology and were also diagnosed by serology, molecular techniques, and nucleic acid detection technology [15]. Disseminated infection was defined as the involvement of 2 or more organs.
STAT3-Mutant HIES
STAT3-mutant HIES was defined according to the 2010 guidelines for STAT3-mutant HIES by Woellner et al [10]. “Possible” was defined as IgE 1000 IU/mL plus a weighted score of clinical features >30 based on recurrent pneumonia, newborn rash, pathologic bone fractures, characteristic face, and high palate. “Probable” was defined as these characteristics plus a lack of Th17 cells or a family history for definitive HIES. “Definitive” was defined as these characteristics plus a dominant-negative heterozygous mutation in STAT3 [10].
Inclusion and Exclusion Criteria
Original studies reporting patients with talaromycosis and STAT3-HIES as defined above were included for analysis. The following exclusion criteria were applied: (1) articles on mycology, diagnostics, antifungal susceptibility, and experimental animal studies, (2) general review articles on penicilliosis, and (3) case reports or series involving HIV-positive individuals. For duplicate publications, the most recent article was used for data extraction.
RESULTS
Demographic Data and Clinical Characteristics of T marneffei With STAT3-HIES
In this multicenter retrospective study, 12 patients with STAT3-HIES and infection with T marneffei were evaluated in this study, with 5 from the multicenter retrospective study and 7 patients from 6 original articles that were retrieved and filtered from the review of the literature [16–21] after elimination of duplicate reports. All of the patients were Chinese: 2 patients resided in Hong Kong, whereas 10 patients were from mainland China and lived in Guangdong (6/12 [50%]), Guangxi (2/12 [17%]), Hunan (1 [8%]), or Hangzhou (1 [8%]). Fifty-eight percent (7/12) of the patients were male, and the median age of the 12 patients was 14 years (1–34 years). Forty-two percent (5/12) were young adults (age range, 18–34 years); 2 of them were teenagers (Table 1).
Table 1.
National Institutes of Health Score of the Patients
| Clinical Features | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 |
|---|---|---|---|---|---|
| IgE score (max kU/L) | 8 (1159) | 10 (>5000) | 8 (1063.7) | 4 (814) | … |
| Skin abscess (times) | 4 (3–4) | 4 (3–4) | 0 | 0 | 0 |
| Pneumonia (times) | 8 (>3) | 6 (3) | 6 (3) | 2 | 0 |
| Lung anomalies | 6a | 6a | 0 | 0 | 0 |
| Retained primary teeth | 0 | 0 | 0 | 0 | 0 |
| Scoliosis | 0 | 0 | 0 | 0 | 0 |
| Fractures | 0 | 0 | 0 | 0 | 0 |
| EOS (max 109/L) | 6 (3.69) | 0 (0.14) | 6 (4.96) | 0 (0.10) | 0 |
| Characteristic asymmetric face | 0 | 5 | 0 | 0 | 0 |
| Newborn rash | 0 | 0 | 0 | 0 | 0 |
| Eczema | 4 | 2 | 0 | 2 | 0 |
| URIs per year | 2 | 1 | 1 | 1 | 0 |
| Candidiasis | 4 | 4 | 0 | 0 | 0 |
| Serious infections | 4 | 4 | 4 | 4 | 4 |
| Hyperextensibility of joints | 0 | 0 | 0 | 0 | 0 |
| Lymphoma | 0 | 0 | 0 | 0 | 0 |
| High palate | 0 | 2 | 0 | 0 | 0 |
| Age-adjusted | 0 | 0 | 0 | 0 | 5 |
| NIH score | 46 | 44 | 25 | 13 | 9 |
Abbreviations: EOS, eosinophils; IgE, immunoglobulin E; NIH, National Institutes of Health; URI, upper respiratory infection.
Fever (10/12 [83%]), cough (9/12 [75%]), dyspnea (4/12 [33%]), and lymphadenopathy (4/12 [33%]) were the most common clinical presentations, followed by skin lesions (3/12 [25%]) and hepatosplenomegaly (3/12 [25%]). In particular, 2 patients (2/12 [17%]) presented with gastrointestinal symptoms, such as abdominal distension. Five of them were in poor living conditions due to recurrent infections. Four patients had no medical history, while 2 of them were <4 years old. Seven patients had previous infections or coinfections, including S aureus (3/12 [25%]), Aspergillus (3/12 [25%]), and other pathogens such as Stenotrophomonas maltophilia, Streptococcus pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Talaromyces marneffei was cultured from bronchoalveolar lavage fluid (BALF) (6/12 [50%]), sputum (4/12 [33%]), blood (3/12 [25%]), bone marrow (3/12 [25%]), abscess fluid, and lung tissue. Twenty-five percent (3/12) of patients were NGS-positive and 17% (2/12) of the patients were biopsy-positive (Tables 1 and 2; Figure 1A).
Table 2.
Clinical Features
| Case | Area of Residence | Age | Sex | Presentation | Previous OIs or Coinfections | Medical History | Antifungal Therapy | Outcome |
|---|---|---|---|---|---|---|---|---|
| P1 | Guangdong | 21 y | Male | Fever, cough, skin lesion (pustule), cervical lymphadenopathy | Staphylococcus aureus, Aspergillus, and tuberculosis | Tuberculosis at the age of 5 and 18 y | Amphotericin B; 6 mo | Relapsed; 5 y |
| P2 | Guangdong | 18 y | Female | Fever, cough, sputum | S aureus, Pseudomonas aeruginosa | 1–2 lung infections per year | Amphotericin B and voriconazole; 6 mo | Relapsed; 6 mo |
| P3 | Guangxi | 24 y | Male | Fever, cough, sore throat | Tuberculosis, Aspergillus | None | Voriconazole; 12 mo | Cured; 12 mo |
| P4 | Guangdong | 15 y | Male | Fever, cough, eczema | No | None | Voriconazole; 9 mo | Improved; 9 mo |
| P5 | Guangxi | 37 mo | Male | Fever, abdominal pain, lymphadenopathy, hepatomegaly, anemia | No | None | Voriconazole; 7 mo | Cured; 18 mo |
| P6 | Hongkong | 30 y | Male | Hemoptysis, dyspnea | Stenotrophomonas maltophilia | Cutaneous infections and recurrent pneumonia since childhood | Amphotericin B; NA | Dead |
| P7 | Hongkong | 12 mo | Female | Fever, cough | Aspergillosis, coagulase negative, S aureus bacteremia | None | Itraconazole; NA | Improved; 17 mo |
| P8 | Hangzhou | 34 y | Female | Fever, cough, dyspnea, anemia | No | Immunocompromised (detail was not clear) since infancy | Itraconazole; 3 mo | Improved; NA |
| P9 | Guangdong | 26 mo | Female | Fever, cough, skin lesion, lymphadenopathy, hepatomegaly | NA | NA | NA | Dead |
| P10 | Guangdong | 19 mo | Male | Cough, trachyphonia, dyspnea | NA | NA | Voriconazole and itraconazole; NA | Cured; NA |
| P11 | Guangdong | 156 mo | Male | Fever, trachyphonia, dyspnea, skin lesion, lymphadenopathy | Acinetobacter baumannii | NA | Amphotericin B, voriconazole and itraconazole; 2 mo | Improved; NA |
| P12 | Hunan | 2 y | Female | Abdominal distension, fever, jaundice, cough, hepatosplenomegaly | Streptococcus pneumoniae | Poor living conditions | Amphotericin B, and voriconazole | Dead |
Abbreviations: NA, not available; OI, opportunistic infection.
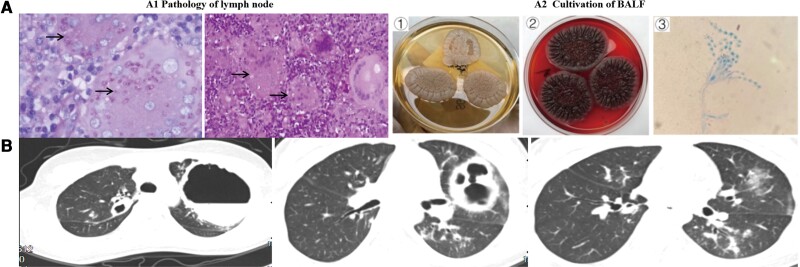
Figure 1.
Chest computed tomography (CT) and etiological diagnosis of Talaromyces marneffei. A, Pathology of the lymph node (A1). A large number of epithelioid granulomas can be seen in the lymph nodes, including many multicellular giant cells and some caseous necrosis (hematoxylin and eosin staining, ×200 magnification). Periodic acid-Schiff staining (×600 magnification) revealed multiple fungal yeast cells in the cytoplasm of multicellular giant cells, as indicated by the arrows. A2 describes the isolation of T marneffei from bronchoalveolar lavage fluid (BALF) at 35°C (①), 25°C (②). The specific morphological features of T marneffei were detected by an oil immersion lens (③). A typical symmetrical bipolar broom-like branch can be seen under high magnification. B, CT scan showing multiple cystic thick-walled cavities in both lungs. The fluid plane of the cavity is visible, and the lungs had scattered patches of tree bud–like blurred shadows (case 2).
Laboratory Examination and Image Features of T marneffei With STAT3-HIES
Some laboratory test data were not available. We tested 5 patients for HIV antibodies and IFN-γ autoantibodies, and the results were negative. Routine blood tests showed that 43% (3/7) of the patients had normal leukocyte counts, 57% (4/7) had elevated leukocyte counts, 67% (4/6) had normal neutrophil counts, 50% (3/6) had normal eosinophil counts, and 50% (3/6) had elevated eosinophil counts. Fifty-seven percent (4/7) of the patients had normal CD4+ T-cell levels, and 43% (3/7) had decreased levels, 2 of whom had only a mild decrease in their CD4+ T cells. Eighty-six percent (6/7) of the patients had normal CD8+ T-cell levels, and 14% (1/7) had decreased levels. B cells were elevated in 63% (5/8) of the patients and natural killer (NK) cells were decreased in 83% (5/6). IgE was significantly elevated in 91% (10/11) of the patients and were normal in 9% (1/11). Immunoglobulin G (IgG) was normal in 50% (3/6) of the patients and elevated in 50% (3/6), but 2 patients received intravenous γ-globulin prior to testing. Immunoglobulin A (IgA) was normal in 82% (9/11) and elevated in 18% (2/11) of patients. Immunoglobulin M (IgM) was normal in 45% (5/11), elevated in 45% (5/11), and decreased in 10% (1/11) of patients (Table 3). By comparison, we found that the percentages of B cells and IgE in HIES patients were higher than those in non-HIES patients, whereas IgA was lower than that in non-HIES patients, and the difference was statistically significant (Table 4).
Table 3.
Laboratory Tests of the Patients
| Case | IgG, g/La | IgA, g/La | IgM, g/La | IgE, kU/La | Leukocytes, 109/La | Neutrophils, 109/La/% | Eosinophils, 109/La | CD4%, Cells/µL | CD8%, Cells/µL | CD19%, Cells/µL | NK%, Cells/µL | Radiography | Sites of Positive Culture, Histology, or NGS | Genetic Test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 43.8 (6.0–16.0) | 0.84 (0.7–5.0) | 0.84 (0.6–2.0) | 1159 (<100) | 7.96 (4.00–10.00) | 2.5 (1.8–8.0)/31.6% (40%–70%) | 3.69 (0.05–0.30) | 33.20% (30%–60%) | 30.4% (13%–41%) | 13% (5%–18%) | 3.8% (7%–40%) | Multiple infiltrates, bronchiectasis, cavity | Blood cultures, lymph node biopsy, CSF NGS | STAT3 mutation (c.1784G > A p. Arg595Gln) heterozygous mutations |
| P2 | 21.8 (6.0–16.0) | 2.83 (0.7–5.0) | 2.84 (0.6–2.0) | 5000 (<100) | 13.23 (4.00–10.00) | 11 (1.8–8.0)/82.8% (40%–70%) | 0.14 (0.05–0.30) | 44.7% (30%–60%) | 23.3% (13%–41%) | 24.8% (5%–18%) | 3% (7%–40%) | Multiple cystic thick-walled cavities | Sputum and BALF cultures | STAT3 mutation (c.1909G > A p. Val637Met) heterozygous mutations |
| P3 | … | 2.74 (2.01–2.69) | 2.29 (0.84–1.32) | 1406.5 (<100) | 18.11 (4.00–10.00) | … | 4.96 (0.02–0.52) | … | … | … | … | … | Sputum and BALF cultures | STAT3 mutation (c.1138_1139 + 1del p.?) heterozygous mutations |
| P4 | 12.0 (6.0–16.0) | 1.67 (0.7–5.0) | 0.21 (0.6–2.0) | 814 (<100) | 5.60 (4.00–10.00) | 2.4 (1.8–8.0)/42.3% (40%–70%) | 0.10 (0.05–0.30) | 27.5% (29%–61%) | 21.3% (15%–44%) | 486 (90–560) | 491 (150–110)/17.9% (7%–40%) | Multiple patchy and nodular hyperdense lesions, small cavities | BALF NGS | STAT3 mutation (c.1422G > A (p.Trp474*) heterozygous mutations |
| P5 | Normal | Normal | Slightly elevated | … | 16.9 (4.00–10.00) | 4.4 (…) | 0.22 (0.02–0.52) | … | … | … | … | Multiple infiltrates | Bone marrow cultures, intestinal biopsy | STAT3 mutation (c.1673G > A,p.G558D) heterozygous mutations |
| P6 | Normal | Normal | Normal | 2510 (<100) | 16.4 (4–11) | 85.1% (…) | 9.5% (…) | … | … | … | … | Pneumatoceles, cavity | Sputum, abscess fluid cultures | Not done |
| P7 | 14.6 (3.46–9.32) | 1.52 (0.13–0.86) | 0.94 (0.38–1.82) | 748 (<100) | … | … | … | 17% (30%–60%) | 9% (13%–41%) | 70% (5%–18%) | … | Bilateral consolidative and cystic changes | Blood, bone marrow cultures | STAT3 mutation (c.1121A > G,p.D374G) heterozygous mutation |
| P8 | Normal | Normal | Normal | Normal | … | … | … | … | … | … | … | Multiple ground glass opacities with multiple bullae | BALF NGS, BALF, and lung tissue cultures | STAT3 mutation (c.92G > A p.R31Q) heterozygous mutation |
| P9 | 23.9b (3.82–10.58) | 0.57 (0.14–1.14) | 1.99 (0.4–1.28) | 6440 (<60) | … | … | … | 869.13 (410–1590) | 403.87 (190–1140) | 1049.56 (90–660) | 33.8 (90–590) | … | Blood cultures | Not done |
| P10 | 8.49 (3.82–10.58) | 0.16 (0.14–1.14) | 1.36 (0.4–1.28) | 422 (<60) | … | … | … | 664.87 (410–1590) | 418.42 (190–1140) | 789.31 (90–660) | 10.91 (90–590) | … | BALF cultures | STAT3 mutation (c.1679_1681del) heterozygous mutations |
| P11 | 17.1 (6.36–13.24) | 1.07 (0.49–2.29) | 0.61 (0.42–1.46) | 7660 (<200) | … | … | … | 233.91 (240–1317) | 0.56 (0.47–2.05) | 75.91 (210–1514) | 75.91 (210–1514) | Thin-walled cystic lesion | Airway mucosal biopsy, BALF cultures | STAT3 mutation (c.A1593T) heterozygous mutations |
| P12 | … | … | … | 1620 (…) | 7.34 ( …) | 0.807 (…) | … | … | … | 50.21% (NA) | … | Multiple patchy high-density shadows in both lungs | Bone marrow, blood, BALF, and sputum cultures | STAT3 mutation heterozygous mutations |
Abbreviations: BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; IgA, immunoglobulin A; IgE, immunoglobulin E; IgG, immunoglobulin G; IgM, immunoglobulin M; NA, not available; NGS, next-generation sequencing; NK, natural killer.
Normal range is presented in parentheses.
They were treated with intravenous immunoglobulin (IVIg) administration before the examination.
Table 4.
Comparison in Laboratory Tests Between Patients With Versus Without Hyper–Immunoglobulin E Syndrome
| Laboratory Measure | HIES | Non-HIES | P Value |
|---|---|---|---|
| IgG, g/L | 17.10 (12.00–23.90) | 15.10 (11.00–21.10) | .5506 |
| IgA, g/L | 1.30 (0.63–2.47) | 2.69 (1.55–3.59) | .0198a |
| IgM, g/L | 1.15 (0.67–2.22) | 1.08 (0.90–1.50) | >.9999 |
| IgE, kU/L | 1513.25 (797.50–5360) | 67.20 (13.80–210.0) | .0001a |
| Leukocytes, 109/L | 13.23 (7.34–16.9) | 8.60 (5.92–11.08) | .2344 |
| Neutrophils, 109/L | 2.50 (1.60–7.70) | 5.90 (3.88–8.40) | .0866 |
| Neutrophil percentage | 0.63 (0.34–0.85) | 0.71 (0.64–0.78) | .6253 |
| Eosinophils, 109/L | 0.22 (0.12–4.33) | 0.18 (0.04–0.22) | .3856 |
| CD4, cells/µL | 767 (341.65–938.78) | 565.00 (162.25–849.75) | .4275 |
| CD4 percentage | 0.30 (0.20–0.42) | 0.29 (0.20–0.44) | .8996 |
| CD8, cells/µL | 595.21 (407.51–820.75) | 340.50 (164.50–448.00) | .0773 |
| CD8 percentage | 0.22 (0.12–0.29) | 0.25 (0.18–0.32) | .3023 |
| CD19, cells/µL | 637.66 (178.43–637.66) | 157.00 (81.00–309.50) | .074 |
| CD19 percentage | 0.6 | 0.15 (0.04–0.19) | .0205a |
| NK, cells/µL | 54.86 (16.63–387.23) | 241.00 (47.00–366.75) | .6571 |
| NK percentage | 0.04 | 0.18 (0.06–0.25) | .2744 |
Data are presented as median range unless otherwise indicated.
Abbreviations: HIES, hyper–immunoglobulin E syndrome; IgA, immunoglobulin A; IgE, immunoglobulin E; IgG, immunoglobulin G; IgM, immunoglobulin M; NK, natural killer.
Significant difference from non-HIES patients (P < .05).
Chest computed tomography (CT) showed that 80% (8/10) of the patients suffered from lesions in both lungs, and 20% (2/10) suffered from lesions in only the right lobes. Eighty percent (8/10) of the patients showed cavities, including 3 thick cavities and 5 cystic changes, 2 of which showed air and liquid levels. Forty percent (4/10) showed multiple patchy shadows (Table 2; Figure 1B).
Genetic Test of T marneffei With STAT3-HIES
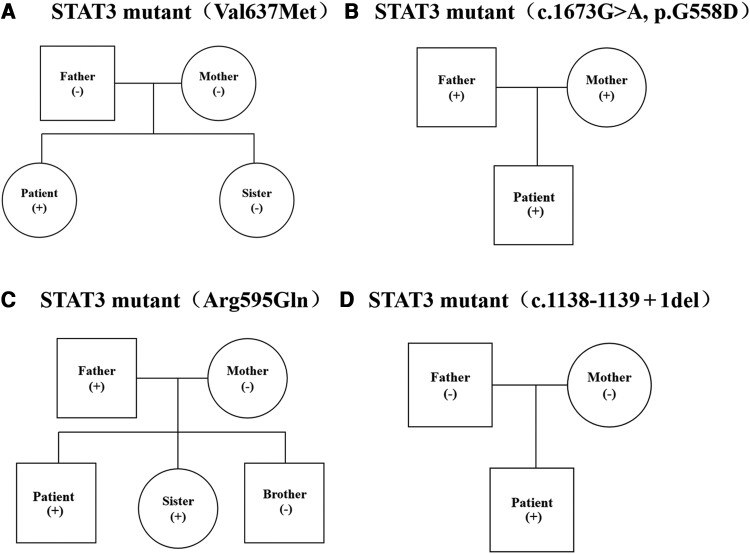
STAT3 gene detection was performed in our 5 patients and 6 of the patients that were included through a literature review, and STAT3 gene mutations were detected in all of them. Among the STAT3 mutations in our 5 cases, 1 STAT3 mutation (c.1909G > A p. Val637Met) (Figure 2A), a sporadic mutation, has been shown to inhibit STAT3 phosphorylation in previous studies. STAT3 mutation (c.1673G > A, p. G558D) (Figure 2B) was reported previously, while the other mutations have not been reported before. STAT3 mutation (c.1784G > A p. Arg595Gln) (Figure 2C), a missense mutation, was found in 1 patient by direct sequencing of the exon regions of 170 hereditary immunodeficiency-related genes and was compared with the reference sequences. Bioinformatics software predicts the disease-causing potential of mutations. This mutation was transmitted from his father and also appeared in his sister who also suffered from talaromycosis as well. Direct sequencing of the exon regions of 171 hereditary immunodeficiency-related genes was compared with the reference sequences. The sequences were compared, and a STAT3 mutation (c.1422G > A p. Trp474*) was found in the patient. This mutation is a nonsense mutation. It is expected that the 474th amino acid of the encoded protein will have Trp changed to a stop codon, which is expected to truncate the expressed protein, and the bioinformatics software predicted that this mutation may be pathogenic. One STAT3 mutation (c.1138_1139 + 1del p.? ) (Figure 2D) was a sporadic mutation detected by whole-exome sequencing by high-throughput sequencing technology (NGS), and the mutation was intron and exon deletion mutations at the junction, which are expected to cause splice site changes and disrupt the encoded protein to lose its normal function. Four different STAT3 mutations were detected in the 7 patients from the literature review, except 1 whose site of mutation was not available and 2 who did not undergo STAT3 detection.
Figure 2.
Pathogenetic diagnosis of hyper–immunoglobulin E syndrome. A, STAT3 gene mutation (V637 M) in case 2 and the family members. B, STAT3 gene mutation (c.1673G > A, p. G558D) in case 5 and the family members. C, STAT3 gene mutation (A595G) in case 1 and the family members. D, STAT3 gene mutation (c.1138_1139 + 1del p.?) in case 3 and the family members.
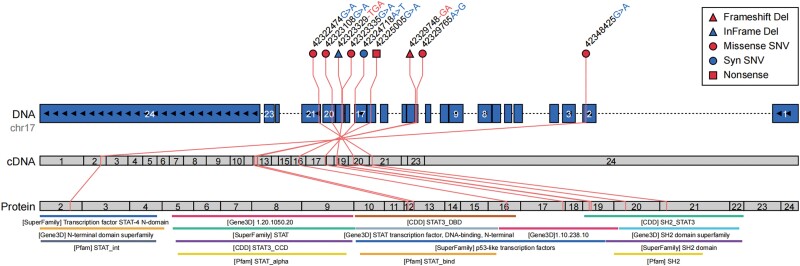
One hundred percent (9/9) of the STAT3 mutations occurred in exons. Forty-four percent (4/9) of the patients carried STAT3 mutations affecting the SH2 domain, 33% (3/9) affected the DNA-binding domain, 11% (1/9) affected the N-terminal domain, and 11% (1/9) affected the linker domain. Fifty-six percent (5/9) of the patients carried a missense mutation, and other mutations included frameshift deletion, in-frame deletion, synonymous mutation, and nonsense mutation (11% [1/9]) (Figure 3; Table 2).
Figure 3.
Mutations in STAT3. The mutations in STAT3, as visualized by Oviz-Bio (https://academic.oup.com/nar/article/48/W1/W415/5835823) and showing the single-nucleotide variants (SNVs) and InDels with positions and functional annotations on 3 layers: genome (DNA), cDNA, and protein. The top panel specifies the positions and content changes of the variants, with shapes and colors indicating the different types of variants. The bottom panel gives the protein domains annotated by the following databases: SuperFamily (https://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/publications.html), Gene3D (https://pubmed.ncbi.nlm.nih.gov/22139938/), CDD (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6943070/), and Pfam (https://academic.oup.com/nar/article/49/D1/D412/5943818).
Treatment and Outcome of T marneffei With STAT3-HIES
One patient improved with thymosin in combination with antifungal drugs but relapsed and developed an intracranial infection after the discontinuation of thymosin. A total of 25% (3/12) of the patients had voriconazole as an antifungal treatment and were cured or improved. Seventeen percent (2/12) of the patients improved after receiving itraconazole, 8% (1/12) recovered after the itraconazole was combined with voriconazole, 8% (1/12) improved after the itraconazole was combined with amphotericin B, and 8% (1/12) received amphotericin B combined with voriconazole for 2 weeks and itraconazole orally for 2 months as antifungal agents with good clinical and imaging response. Seventeen percent (2/12) of the patients relapsed or died after receiving amphotericin B, and 17% (2/12) of the patients relapsed or died after receiving amphotericin B combined with voriconazole. The case fatality rate from our study was 25% (3/12) (Table 2).
DISCUSSION
STAT3 plays an important role in regulating proliferation, differentiation, migration, cell survival, and apoptosis by mediating responses to multiple cytokines and growth factors [22]. Ligands that are transduced through STAT3 include interleukin (IL) 6, IL-10, IL-11, IL-21, IL-22, and IL-23 [23]. STAT3-HIES patients could not respond well to these cytokines [24] and had difficulties generating Th17 lymphocytes [24], IL-22–producing T cells, and circulating memory B cells [25]. Th17 lymphocytes release antimicrobial peptides and produce the IL-17 cytokine family involved in the response to bacteria, typically S aureus, as well as fungi such as Aspergillus and Candida [26]. Therefore, the tests of Th17 and memory B cells may provide proof of STAT3-HIES. Mutations in STAT3 abrogate the ability of human CD8+ T cells to differentiate into granzyme B–expressing effector cells in response to IL-21 [27]. In this study, we described 12 patients with disseminated talaromycosis who were suffering from STAT3-HIES. Furthermore, due to a lack of insight into talaromycosis in HIV-negative populations and routine screening for gene mutation testing, there may be more cases that have been missed, which indicates that STAT3-HIES is a susceptibility factor for disseminated T marneffei infection in HIV-negative hosts.
In this study, most of these patients were from southern China. They had a median age of 14 years, and nearly half of the patients were young adults, whereas it was previously believed that HIES mostly occurred in infants and children [26]. There were no typical symptoms different from other HIV-negative patients with talaromycosis [28], but the rate of gastrointestinal symptoms was higher than previously reported [29]. Most HIES patients are prone to infections from childhood onward. The most common pathogens of patients were S aureus and Aspergillus, similar to previous reports, while Candida albicans was less common in this study [26]. The most significant abnormality in the laboratory tests was a markedly elevated IgE level, but some patients had normal IgE levels, so this could not be used as a basis for excluding HIES. IgA, IgM, and IgG were normal or mildly abnormal. These patients had elevated or normal white blood cell counts, neutrophils, and eosinophils and normal or mildly abnormal numbers of CD4+ T cells and CD8+ T cells, which is significantly different from HIV-positive patients with talaromycosis. In most patients, their B-cell counts were increased, which may account for the elevated IgE, and NK cells showed a downward trend, which may contribute to dysfunction of Th17 [30]. Multiple cavities and patchy shadows in both lungs were the most common features of the CT findings in this study. Therefore, when a patient has a history of repeated S aureus, C albicans, or Aspergillus infections, significantly increased IgE, increased B cells, and decreased IgA, as well as multiple cavities and patchy shadows in both lungs, it is necessary to consider whether the patient has HIES, and the diagnosis can be confirmed by STAT3 gene detection. In patients with STAT3-HIES, especially those living in talaromycosis-endemic areas, clinicians should consider the possibility of T marneffei infection and vice versa. The positive rates of BALF and sputum cultures were the highest in this study, whereas the positive rates of blood cultures were lower than those reported previously [2]. Therefore, NGS could also be used to assist in the identification of pathogens [31]. Therefore, the detection of T marneffei can be improved by using blood culture combined with other specimens and supplemented by NGS.
The National Institutes of Health (NIH) score plays an important role in diagnosing HIES [32]. However, the NIH scores of 2 patients in this study were both <30, so the diagnosis of HIES cannot be ruled out based on the NIH score. Both patients carried mutations in the DNA-binding domain, similar to those found in previous studies. Also, 1 of the mutations also occurred in exon 16 [33], but the cause was unclear. The patients in this study were mainly young people, not children, which may be determined by the location and type of STAT3 mutation in these patients. Previous studies have shown that the same mutation of STAT3 may also be heterogeneous in different patients, so the reason is still unclear. Three novel STAT3 mutations were identified in this study, but the effect of these mutations on function could not be confirmed. STAT3 mutations can be inherited, so it is important information for the patients’ relatives.
Regarding the treatment of patients with T marneffei infection and HIV, the accepted therapeutic regimen was amphotericin B induction therapy and itraconazole maintenance therapy [34]. When the immune status of HIV-positive patients was improved after receiving antiretroviral therapy, antifungal treatments can be effective. However, there are no guidelines for the treatment of T marneffei infection in patients who are HIV negative. We tried to treat the 3 patients using voriconazole, which showed a good therapeutic effect in the initial treatment. However, as to why the infection recurred, we think it is related to the underlying disease, HIES. Current research shows that patient populations may benefit from hematopoietic stem cell transplantation, omalizumab, dupilumab, and adenine base editing; however, there is a lack of evidence-based practice [35–37], so we did not give any specific therapeutic intervention for STAT3-HIES. Thymosin may be effective when combined with antifungal treatment [38].
There are several limitations to this study. First, the number of patients included in this retrospective study was small, and some clinical data were incomplete. Second, this study lacked a non-HIES cohort of patients with talaromycosis as a control group. In addition, the effects of 3 novel STAT3 mutations in this study on function were not verified. However, this study is the first to systematically review the clinical features of T marneffei infection in patients with STAT3-HIES of all ages and provides a basis for the early diagnosis of such patients.
In summary, T marneffei infections with STAT3-HIES have poor prognosis and high mortality, while the knowledge of clinicians on these infections remains insufficient. These patients usually present with obvious elevated serum IgE, increased B cells, decreased IgA cells, and cavity lesions on CT.
Notes
Author contributions. All authors participated in the conception and design of the study. L. Z. T., Y. J. L., Q. Y., F. Yang, T. M. X., and F. Ye analyzed the information and data. L. S. Q., Z. Y. Q., L. Y. M., T. S. F., and J. C. collected and tested the clinical information and specimens. All authors provided suggestions that improved this study. L. Z. T. and F. Ye wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments. The authors thank Drs Su Danhong, Gu Yingying, and Zeng Qingsi from the Department of Microbiology Laboratory, the Pathology Laboratory, the Imaging Service of the First Affiliated Hospital of Guangzhou Medical University, and King Med Center for Clinical Laboratory, respectively. In addition, we appreciate Dr Sook-San Wong and Dr Mark Zanin for their help in editing the manuscript. We also thank Springer Nature (http://authorservices.springernature.com/) for providing English-language editing of the manuscript.
Patient consent. Ethical approval of the study protocol was granted from the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (number 2019–20; Guangzhou, China). Written informed consent was obtained from each participant and their families for publication of this case report and any accompanying images.
Financial support. This work was funded by the independent fund of the National Natural Science Foundation of China (82270007); the National Natural Science Foundation of China (82202544); the State Key Laboratory of Respiratory Diseases (SKLRD-Z-202019); the Guangzhou Institute of Respiratory Health Open Project (2019GIRHZ06); and the Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020003). An open-access licence has been selected.
Contributor Information
Zhengtu Li, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Jinglu Yang, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Ye Qiu, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China; The First Affiliated Hospital of Guangxi Medical University, Nanning, China.
Feng Yang, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Mengxin Tang, The First Affiliated Hospital of Guangxi Medical University, Nanning, China.
Shaoqiang Li, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Yangqing Zhan, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Yongming Li, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Sufang Tang, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Cheng Jing, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Feng Ye, State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
References
- 1. Vanittanakom N, Cooper CJ, Fisher MC, et al. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev 2006; 19:95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kawila R, Chaiwarith R, Supparatpinyo K. Clinical and laboratory characteristics of Penicilliosis marneffei among patients with and without HIV infection in northern Thailand: a retrospective study. BMC Infect Dis 2013; 13:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ba H, Peng H, Cheng L, et al. Case report: Talaromyces marneffei infection in a Chinese child with a complex heterozygous CARD9 mutation. Front Immunol 2021; 12:685546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen K, Tan J, Qian S, et al. Case report: disseminated Talaromyces marneffei infection in a patient with chronic mucocutaneous candidiasis and a novel STAT1 gain-of-function mutation. Front Immunol 2021; 12:682350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pan M, Qiu Y, Zeng W, et al. Disseminated Talaromyces marneffei infection presenting as multiple intestinal perforations and diffuse hepatic granulomatous inflammation in an infant with STAT3 mutation: a case report. BMC Infect Dis 2020; 20:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holland SM, DeLeo FR, Elloumi HZ, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 2007; 357:1608–19. [DOI] [PubMed] [Google Scholar]
- 7. Engelhardt KR, McGhee S, Winkler S, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol 2009; 124:1289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma CA, Stinson JR, Zhang Y, et al. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat Genet 2017; 49:1192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yanagimachi M, Ohya T, Yokosuka T, et al. The potential and limits of hematopoietic stem cell transplantation for the treatment of autosomal dominant hyper-IgE syndrome. J Clin Immunol 2016; 36:511–6. [DOI] [PubMed] [Google Scholar]
- 10. Woellner C, Gertz EM, Schäffer AA, et al. Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J Allergy Clin Immunol 2010; 125:424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorenzini T, Giacomelli M, Scomodon O, et al. Autosomal-dominant hyper-IgE syndrome is associated with appearance of infections early in life and/or neonatal rash: evidence from the Italian cohort of 61 patients with elevated IgE. J Allergy Clin Immunol Pract 2019; 7:2072–5. [DOI] [PubMed] [Google Scholar]
- 12. Conti HR, Baker O, Freeman AF, et al. New mechanism of oral immunity to mucosal candidiasis in hyper-IgE syndrome. Mucosal Immunol 2011; 4:448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Masur H, Brooks JT, Benson CA, et al. Prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: updated guidelines from the Centers for Disease Control and Prevention, National Institutes of Health, and HIV Medicine Association of the Infectious Diseases Society of America. Clin Infect Dis 2014; 58:1308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaplan JE, Benson C, Holmes KK, et al. Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recomm Rep 2009; 58:1–207, E1–4. [PubMed] [Google Scholar]
- 15. Pruksaphon K, Intaramat A, Ratanabanangkoon K, et al. Diagnostic laboratory immunology for talaromycosis (penicilliosis): review from the bench-top techniques to the point-of-care testing. Diagn Microbiol Infect Dis 2020; 96:114959. [DOI] [PubMed] [Google Scholar]
- 16. Lee PP, Chan KW, Lee TL, et al. Penicilliosis in children without HIV infection—are they immunodeficient? Clin Infect Dis 2012; 54:e8–19. [DOI] [PubMed] [Google Scholar]
- 17. Fan H, Huang L, Yang D, et al. Pediatric hyperimmunoglobulin E syndrome: a case series of 4 children in China. Medicine (Baltimore) 2018; 97:e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeng Q, Jin Y, Yin G, et al. Peripheral immune profile of children with Talaromyces marneffei infections: a retrospective analysis of 21 cases. BMC Infect Dis 2021; 21:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang W, Ye J, Qiu C, et al. Rapid and precise diagnosis of T. marneffei pulmonary infection in a HIV-negative patient with autosomal-dominant STAT3 mutation: a case report. Ther Adv Respir Dis 2020; 14:1022303031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma BH, Ng CS, Lam R, et al. Recurrent hemoptysis with Penicillium marneffei and Stenotrophomonas maltophilia in Job’s syndrome. Can Respir J 2009; 16:e50–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan JH, Luo HY, Yang LG, et al. Penicilliosis marneffei in HIV negative children: three case reports. Ann Palliat Med 2021; 10:8437–47. [DOI] [PubMed] [Google Scholar]
- 22. Kane A, Deenick EK, Ma CS, et al. STAT3 is a central regulator of lymphocyte differentiation and function. Curr Opin Immunol 2014; 28:49–57. [DOI] [PubMed] [Google Scholar]
- 23. O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med 2013; 368:161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity 2008; 28:477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Speckmann C, Enders A, Woellner C, et al. Reduced memory B cells in patients with hyper IgE syndrome. Clin Immunol 2008; 129:448–54. [DOI] [PubMed] [Google Scholar]
- 26. Tsilifis C, Freeman AF, Gennery AR. STAT3 hyper-IgE syndrome—an update and unanswered questions. J Clin Immunol 2021; 41:864–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ives ML, Ma CS, Palendira U, et al. Signal transducer and activator of transcription 3 (STAT3) mutations underlying autosomal dominant hyper-IgE syndrome impair human CD8(+) T-cell memory formation and function. J Allergy Clin Immunol 2013; 132:400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chan JF, Lau SK, Yuen KY, et al. Talaromyces (penicillium) marneffei infection in non-HIV-infected patients. Emerg Microbes Infect 2016; 5:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun J, Sun W, Tang Y, et al. Clinical characteristics and risk factors for poor prognosis among HIV patients with Talaromyces marneffei bloodstream infection. BMC Infect Dis 2021; 21:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Babic M, Dimitropoulos C, Hammer Q, et al. NK cell receptor NKG2D enforces proinflammatory features and pathogenicity of Th1 and Th17 cells. J Exp Med 2020; 217:e20190133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Y, Liu Y, Wen Y. Gastrointestinal manifestations of Talaromyces marneffei infection in an HIV-infected patient rapidly verified by metagenomic next-generation sequencing: a case report. BMC Infect Dis 2021; 21:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Al-Shaikhly T, Ochs HD. Hyper IgE syndromes: clinical and molecular characteristics. Immunol Cell Biol 2019; 97:368–79. [DOI] [PubMed] [Google Scholar]
- 33. Wolach O, Kuijpers T, Ben-Ari J, et al. Variable clinical expressivity of STAT3 mutation in hyperimmunoglobulin E syndrome: genetic and clinical studies of six patients. J Clin Immunol 2014; 34:163–70. [DOI] [PubMed] [Google Scholar]
- 34. Le T, Kinh NV, Cuc N, et al. A trial of itraconazole or amphotericin B for HIV-associated talaromycosis. N Engl J Med 2017; 376:2329–40. [DOI] [PubMed] [Google Scholar]
- 35. Alonso-Bello CD, Jiménez-Martínez M, Vargas-Camaño ME, et al. Partial and transient clinical response to omalizumab in IL-21-induced low STAT3-phosphorylation on hyper-IgE syndrome. Case Reports Immunol 2019; 2019:6357256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hashemi H, Mohebbi M, Mehravaran S, et al. Hyperimmunoglobulin E syndrome: genetics, immunopathogenesis, clinical findings, and treatment modalities. J Res Med Sci 2017; 22:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harrison SC, Tsilifis C, Slatter MA, et al. Hematopoietic stem cell transplantation resolves the immune deficit associated with STAT3-dominant-negative hyper-IgE syndrome. J Clin Immunol 2021; 41:934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Serafino A, Pierimarchi P, Pica F, et al. Thymosin α1 as a stimulatory agent of innate cell-mediated immune response. Ann N Y Acad Sci 2012; 1270:13–20. [DOI] [PubMed] [Google Scholar]