Abstract
Coronavirus infection induces a variety of cellular antiviral responses either dependent on or independent of type I interferons (IFNs). Our previous studies using Affymetrix microarray and transcriptomic analysis revealed the differential induction of three IFN-stimulated genes (ISGs), IRF1, ISG15 and ISG20, by gammacoronavirus infectious bronchitis virus (IBV) infection of IFN-deficient Vero cells and IFN-competent, p53-defcient H1299 cells, respectively. In this report, the induction kinetics and anti-IBV functions of these ISGs as well as mechanisms underlying their differential induction are characterized. The results confirmed that these three ISGs were indeed differentially induced in H1299 and Vero cells infected with IBV, significantly more upregulation of IRF1, ISG15 and ISG20 was elicited in IBV-infected Vero cells than that in H1299 cells. Induction of these ISGs was also detected in cells infected with human coronavirus-OC43 (HCoV-OC43) and porcine epidemic diarrhea virus (PEDV), respectively. Manipulation of their expression by overexpression, knockdown and/or knockout demonstrated that IRF1 played an active role in suppressing IBV replication, mainly through the activation of the IFN pathway. However, a minor, if any, role in inhibiting IBV replication was played by ISG15 and ISG20. Furthermore, p53, but not IRF1, was implicated in regulating the IBV infection-induced upregulation of ISG15 and ISG20. This study provides new information on the mechanisms underlying the induction of these ISGs and their contributions to the host cell antiviral response during IBV infection.
Keywords: Coronavirus, Infectious bronchitis virus (IBV), IRF1, ISG15, ISG20
1. Introduction
Coronavirus is an important pathogen of human and other animal species. It was the causative agent of severe acute respiratory syndrome (SARS) in 2002–2003, Middle East Respiratory syndrome (MERS) in 2012 and the current COVID-19 pandemic, causing unprecedented damages to the economic and social development (Fung and Liu, 2019, 2021). Belonging to the coronavirus family and the coronavirus subfamily, coronaviruses can be divided into alpha-, beta-, gamma- and deltacoronavirus (Woo et al., 2012). The viral genome encodes 4 structural proteins, namely spike (S), membrane (M), small envelope (E) and nucleocapsid (N) proteins, as well as 15–16 nonstructural proteins (nsp) and some accessory proteins (Liao et al., 2013). Avian infectious bronchitis virus (IBV) is a gammacoronavirus and causes acute respiratory diseases in chickens, with symptoms including dyspnea, cough and runny nose (Liu et al., 2019). IBV infects chickens of all ages, especially young chicks, and poses a constant threat to the poultry industry (Liu et al., 2019). Betacoronavirus human coronavirus OC43 (HCoV-OC43) causes common colds and often occurs in children (Liu et al., 2021), and alphacoronavirus porcine epidemic diarrhea virus (PEDV) causes acute diarrhea, vomiting, dehydration and high mortality in newborn piglets (Jung et al., 2020; Pensaert and de Bouck, 1978).
Virus infection induces the expression of type I interferons (IFN). Together with other proinflammatory cytokines, chemokines and antiviral factors, they form the host innate antiviral response, the first line of host defense against virus infections (Yu et al., 2021). Type I IFNs are induced after recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) in host cells. They then bind to the corresponding cell surface receptor (IFN-α/β receptor 1 and 2, IFNAR1 and IFNAR2), and activate downstream signaling proteins, leading to the activation of JAK/STAT, MAPK, and PI3K signaling pathways, and ultimately inducing the expression of nearly 1000 IFN-stimulated genes (ISGs) (Gürtler and Bowie, 2013). A family of transcription factors, IFN regulatory factor (IRF) family, play a pivotal role in activating and regulating IFN signaling. So far, 10 IRF proteins have been identified, including IRF1, IRF2, IRF3, IRF4/PIP/LSIRF/ICSAT, IRF5, IRF6, IRF7, IRF8/ICSBP, IRF9/ISGF3γ and IRF10 (IRF10 is absent in human and mouse) (Negishi et al., 2018). All IRF proteins have a conserved amino-terminal DNA binding domain (DBD) that recognizes the DNA sequence element, IFN stimulation response element (ISRE), present in the promoters of type I/III IFN and ISG genes (Tanaka et al., 1993). The eponymous member of the IRF family, IRF1, was the first member of the family to be identified (Fujita et al., 1988), and is basically expressed in most human and animal cells. The expression level of IRF1 rapidly increases when stimulated by IFNs and proinflammatory nuclear factor NF-κB as well as virus infection (Carlin et al., 2017).
ISG15, one of the earliest ISGs induced by IFN, comprises two ubiquitin-like domains (UBL1 and UBL2) similar to ubiquitin and also undergoes ISGylation, an enzymatic cascade similar to ubiquitylation (Mirzalieva et al., 2022). The ISRE element in the ISG15 promoter rapidly responds to the virus infection or type I IFN treatment (Der et al., 1998; Meraro et al., 2002; Reich et al., 1987). Other stimuli, such as lipopolysaccharide, exogenous DNA or RNA, retinoic acid and DNA damaging agents can also induce ISG15 expression through various signaling pathways (Perng and Lenschow, 2018). ISG15 plays different roles, both as an effector and a signaling molecule, during various stages of innate immune responses (Freitas et al., 2020, p. 15). For example, ISG15 was shown to prevent the assembly and release of HIV-1 virions in infected cells in an IFN-mediated pathway (Okumura et al., 2006); its binding to p53 in the presence of DNA damage significantly enhanced the binding of p53 to its own gene promoter by promoting phosphorylation/acetylation and target genes, leading to the inhibition of cell growth and tumorigenesis (Park et al., 2016).
ISG20, first discovered in Daudi cells, is an exonuclease capable of cleaving single-stranded RNA and DNA (Gongora et al., 1997; Moser et al., 1997; Nguyen et al., 2001). ISG20 expression is induced by IFN or double-stranded RNA under the control of IRF1 (Espert et al., 2004; Gongora et al., 2000). In the antiviral process, ISG20 can directly degrade viral RNA or inhibit the translation of viral proteins (Deymier et al., 2022; Espert et al., 2003, 2005; Liu et al., 2017). Its C-terminal exonuclease III (ExoIII) domain was found to directly bind to the hepatitis B virus RNA ε stem-loop structure and degrade the intracellular HBV RNA (Liu et al., 2017). In alphavirus infection, IFN-induced ISG20 exerts antiviral effects by upregulating type I IFN-responsive proteins and indirectly regulating RNA virus replication in the cytoplasm (Weiss et al., 2018).
In this report, the induction kinetics of three ISGs, IRF1, ISG15 and ISG20, in cells and their antiviral functions were studied based on previous Affymetrix microarray and transcriptomic analyses, confirming that these three ISGs were indeed differentially induced in H1299 and Vero cells infected with IBV, as well as in cells infected with HCoV-OC43 and PEDV, respectively. IRF1 was shown to play an active anti-IBV role via activation of the type I IFN pathway, however, a minor, if any, antiviral role may be played by ISG15 and ISG20 during IBV infection of culture cells. Furthermore, the induction of ISG15 and ISG20 was demonstrated to be regulated by p53, but not by IRF1.
2. Materials and methods
2.1. Antibodies, chemicals and reagents
Antibodies against β-actin (#HC201-02) and Flag-tag (#HT201-01) were purchased from TransGen Biotech. Antibodies against ISG15 (#P.15981-1-AP), ISG20 (#P.22097-1-AP), IRF1 (#11335-1-AP) were purchased from Proteintech, IRDye® 800CW Goat anti-Rabbit IgG (H + L) (#926–68070), IRDye® 680RD Goat anti-Mouse IgG (H + L) (#926–32211) were purchased from Licor. TansDetect® Cell Counting Kit (CCK) was purchased from TransGen Biotech. Antisera against IBV N protein was prepared in rabbits immunized with bacterially expressed fusion proteins as previously described (Li et al., 2005; Liu and Inglis, 1991). The JAK1/2 inhibitor Ruxolitinib (#S1378) was purchased from Selleckchem, dissolved in DMSO for a 50 mM stock solution, aliquoted and stored at −80 °C.
2.2. Virus, cells, infection and titration
The egg-adapted IBV Beaudette strain (ATCC VR-22) was obtained from the American Type Culture Collection (ATCC) and adapted to Vero cells as previously described (Fang et al., 2005; Lim and Liu, 1998; Shen et al., 2004). HCoV-OC43 (accession No.KU131570.1) was obtained from ATCC(Morfopoulou et al., 2016). PEDV virulent strain DR13 (accession No.JQ023162) was isolated from a suckling pig in 1999, and adapted to growth in Vero cells (Park et al., 2012; Song et al., 2003). The cell lines are preserved by our laboratory. Primary CEF cells are self-produced. Vero cells (#CCL-81), H1299 cells (#CRL-5803) and HEK293T cells (#CRL-3216) were purchased from American Type Culture Collection (ATCC). And DF1 cells (#1101BIR-PUMC000417) were purchased from National Infrastructure of Cell-Line Resource (NICR) from China. Vero, H1299, DF-1, 293T and CEF cells were cultured at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Life Technologies, Carlsbad, CA, USA) supplemented with 6% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin.
To prepare the virus stock, monolayers of Vero cells were infected with IBV at a multiplicity of infection (MOI) of approximately 0.1 and cultured for 24 h. After three freeze/thaw cycles, total cell lysates were clarified, the supernatant was aliquoted and stored at −80 °C as virus stock. UV-inactivation of IBV was performed by exposing the virus stock to 120,000 mJ/cm2 of 254-nm shortwave UV radiation for 30 min with a CL-1000 cross-linker (UVP).
Cells were washed twice with serum-free medium before infected with IBV at an MOI∼2 or incubated with an equal volume of UV-inactivated IBV in serum-free medium. When an inhibitor was used, the inhibitor was diluted to final concentrations using serum-free medium and added. Cells were incubated at 37 °C before harvested. Supernatants were harvested from IBV-infected cells and clarified by centrifugation, and virus titer was determined by the tissue culture infective dose 50 (TCID50) assay using the Reed and Muench method (Yamada and Liu, 2009).
2.3. Microarray hybridization, image analysis and transcriptomic analysis
Microarray hybridization and image analysis were carried out as previously described (Nasirudeen and Liu, 2009; Zhong et al., 2012). Briefly, RNA was independently prepared from IBV-infected Vero cells harvested at 24 h post-infection and hybridized to GeneChip® Human Genome U133A Array (Affymetrix, USA), according to the manufacturer's instructions (Affymetrix). GeneChip arrays were scanned on an Affymetrix probe array scanner, and data were analyzed using the statistics software Microarray Suite version 5.0 (MAS5.0) from Affymetrix.
Transcriptomic analysis was carried out by the Biomarker Technologies Co, LTD, Beijing, China, as previously described (Yuan et al., 2022). Briefly, RNA was independently prepared from IBV-infected H1299 cells harvested at 20 h post-infection and sequenced using Illumina HiSeq sequencing technology platform to construct transcriptome libraries and obtain sequencing data.
2.4. Cell proliferation assay after treatment with the JAK1/2 inhibitor Ruxolitinib
Vero and H1299 cells were seeded on 96-well plates, incubated at 37 °C for 16 h and treated with different concentrations of Ruxolitinib. At indicated times post-treatment, 10 μl of CCK solution was added to the plates and incubated at 37 °C for 2 h. The absorbance at 450 nm was detected by an automatic microplate reader.
2.5. RNA extraction and RT-qPCR analysis
Total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Briefly, cells were lysed with 1 ml TRIzol per 10 cm2 effective growth area, and lysates were vigorously mixed with one-fifth volume of chloroform. The mixture was then centrifuged at 12,000 g at 4 °C for 15 min, and the aqueous phase was mixed with an equal volume of isopropanol. The RNA was precipitated by centrifugation at 12,000 g at 4 °C for 15 min, washed twice with 75% ethanol, and dissolved in 30–50 μl RNase-free water.
Total RNA was reverse-transcribed using the FastKing gDNA Dispelling RT SuperMix kit (Tiangen) according to the manufacturer's instructions. The cDNA was then diluted 20-fold with RNase-free water for quantitative PCR (qPCR) analysis using the Talent qPCR PreMix SYBR Green kit (Tiangen), generating the cycle threshold (CT) values. Using the ΔΔCT method, the relative abundance of a transcript was calculated using GAPDH as an internal control and normalized to the respective reference sample in each experiment.
The qPCR primers for IBV: gRNA, 5′-GTTCTCGCATAAGGTCGGCTA-3′ and 5′-GCTCACTAAACACCACCAGAAC-3’; sgRNA2, 5′-GCCTTGCGCTAGATTTTTAACTG-3′ and 5′-AGTGCACACAAAAGAGTCACTA-3’. And the qPCR primers for HCoV-OC43/PEDV gRNA: OC43 gRNA, 5′-CTATCTGGGAACAGGACCGC-3′ and 5′-TTGGGTCCCGATCGACAATG-3’; PEDV gRNA, 5′-AGTAGCCATCGCAAGTGCTG-3′ and 5′-AACCGGAGGAAGGCTGTTTG-3’. Primer pairs for human cells and Vero cells: GAPDH, 5′-CTGGGCTACACTGAGCACC-3′ and 5′-AAGTGGTCGTTGAGGGCAATG-3’; IRF1, 5′- ATGCCCATCACTCGGATGC -3′ and 5′-CCCTGCTTTGTATCGGCCTG-3’; ISG15(H), 5′- TGGACAAATGCGACGAACCTC-3′ and 5′-TCAGCCGTACCTCGTAGGTG-3’; ISG15(V), 5′- TGGACAGATGCGATGAACCTC-3′ and 5′-GTCAGCTGTACCTCGTAGGTG-3’; ISG20, 5′- CTCGTTGCAGCCTCGTGAA-3′ and H-5′-CGGGTTCTGTAATCGGTGATCTC-3′, V-5′- CCGTGTTCTGTAATCGGTGATCTC-3’; JAK2, 5′-ATCCACCCAACCATGTCTTCC-3′ and 5′- ATTCCATGCCGATAGGCTCTG-3’; IFN-β, 5′-TCTCCTGTTGTGCTTCTCCAC-3′ and 5′- GCCTCCCATTCAATTGCCAC-3’. Primers for DF1 and CEF cells: GAPDH, 5′-GACCACTGTCCATGCCATCA-3′ and 5′-TTTCCCCACAGCCTTAGCAG-3’; IRF1, 5′-AACCAGCGTTGAGGGGAAAG -3′ and 5′-GTCCATCATGCGGAACTCCA-3’; ISG20, 5′-AGCATTGTGGGCTATGAGGG-3′ and 5′-AATGTCCGCTTTTGCCAACC-3’.
2.6. SDS-PAGE and Western blot analysis
Whole cell lysates were prepared from cells harvested at indicated time points and lysed in RIPA buffer. After clarified by centrifugation and determined the protein concentration, the cell lysate was then mixed with 5x Laemmli sample buffer. To analyze proteins in the culture supernatant, the supernatant was collected, clarified by brief centrifugation and mixed with 5x Laemmli sample buffer. Equal amounts of protein samples were loaded to each well and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The resolved proteins were then transferred to a nitrocellulose membrane, incubated with 5% skim milk in TBST buffer at room temperature for 1 h. The membrane was then incubated with 1 μg/ml specific primary antibody dissolved in TBST with 3% BSA (w/v) at 4 °C overnight, washed three times with TBST, and incubated with 1:10000 diluted goat anti-rabbit or goat anti-mouse IgG secondary antibodies (Licor) at room temperature for 2 h. Fluorescence images were obtained using the Azure c600 Imager, and densitometric measurement was performed using the AzureSpot software. All experiments were repeated at least three times with similar results, and one of the representative results was shown.
2.7. RNA interference
The siRNA duplexes for IRF1 were purchased from Sangon Biotech (Shanghai, China). The sequence of the siRNA sense strand: GGAAAUUACCUGAGGACAUdTdT; negative control was provided by Sangon Biotech. Transfection of siRNA was carried out using transIntroTM EL Transfection Reagent (Transgen biotech) as follows. H1299 or Vero cells were plated to a 12-well plate the day before transfection, 5 μl of 20 μM siRNA duplex and 2.5 μl TransIntro EL were diluted with 100 μl Opti-MEM (Gibco) per well and incubated for 20 min. Cells were replenished with 400 μl Opti-MEM containing 5% FBS, and the transfection mixture was added to each well dropwise. Virus infection was performed at 24 h post-transfection.
2.8. Plasmid constructions
Expression plasmids XJ40-Flag-P53, XJ40-Flag-P53-R175H, XJ40-Flag-P53-R273H, XJ40-Flag-ISG15, XJ40-Flag-ISG20 and XJ40-Flag-IRF1 were constructed by inserting corresponding PCR products into a pXJ40-based plasmid. The nucleotide sequences of primers:P53/R175H/R273H, 5′- ATTAGGATCCATGGAGGAGCCGCAGTCA-3′ and 5′- ATTACTCGAGCCGTCTGAGTCAGG CCCTTCTGT-3’; ISG15, 5′- CGCGGATCCATGGGCTGGGACCTGAC-3′ and 5′- CCGCTCGA GTTAGCTCCGCCCGCCAG-3’; ISG20, 5′- CGCGGATCCATGGCTGGG AGCCGTGA-3′ and 5′- CCGCTCGAGTCAGTCTGACACAGCCAGGC-3′, IRF1(Vero), 5′- CGCGGATCCATGCCCAT CACTCGGATGCG-3′ and 5′- CCCAAGCTTCTACGGTGCACAGGG AATGGC-3’; IRF1(H1299), 5′- CGC GGATCCATGCCCATCACTCGGATG-3′ and 5′- CCCAAGCTT CTACGGTGCACAGG GAATG-3’.
Plasmid X459-IRF1 used to knockout IRF1 in Vero cell was constructed by inserting the two complementary oligonucleotides (5′-CACCGTCATGCGCATCCGAGTGAT-3′ and 5′-AAACATCACTCGGATGCGCATGAC-3′), coding for the small guide RNA with Bbs1 ends, into pX459. The small guide RNA was designed using the online program CRISPOR.
2.9. Northern blot analysis
Vero cells were infected with IBV at an MOI∼1 and total RNA was extracted from the infected cells. Total RNA (10 μg) was added to a mixture of 1 × MOPS, 37% formaldehyde and incubated at 65 °C for 20 min before subjected to gel electrophoresis. The separated RNA bands were transferred onto a Hybond N+ membrane (Amersham Biosciences) via capillary action overnight and fixed by UV crosslinking (Stratalinker). Hybridization of Dig-labeled DNA probes was carried out at 50 °C in hybridization oven overnight. Membranes were washed 3 times for 15 min each with the probe buffer, before proceeding to detection with CDP-Star (Roche) according to the manufacturer's instructions.
2.10. Statistical analysis
The one-way ANOVA method was used to analyze the significant difference between the indicated sample and the respective control sample. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
3. Result
3.1. IBV infection of IFN-β-deficient Vero and IFN-β -competent H1299 cells differentially regulates the expression of IRF1, ISG15 and ISG20
In our previous studies, the general expression profiles of differentially regulated host antiviral genes in IBV infection of IFN-β-deficient Vero and IFN-β-competent H1299 cells were probed by Affymetrix array and transcriptomic analyses (Liao et al., 2011; Yuan et al., 2021, 2022). Among hundreds of IFN-stimulated genes, mRNA levels of IRF1, ISG15 and ISG20 were 4-, 6.5- and 64-fold upregulated, respectively, in IBV-infected Vero cells at 24 h post-infection (hpi)(Liao et al., 2011). However, a different induction pattern of these three genes in IBV-infected H1299 cells was revealed by transcriptomic analysis: the mRNA level of IRF1 was 2.9-fold upregulated, but ISG15 and ISG20 were 0.794- and 1.098-fold regulated (Yuan et al., 2021, 2022).
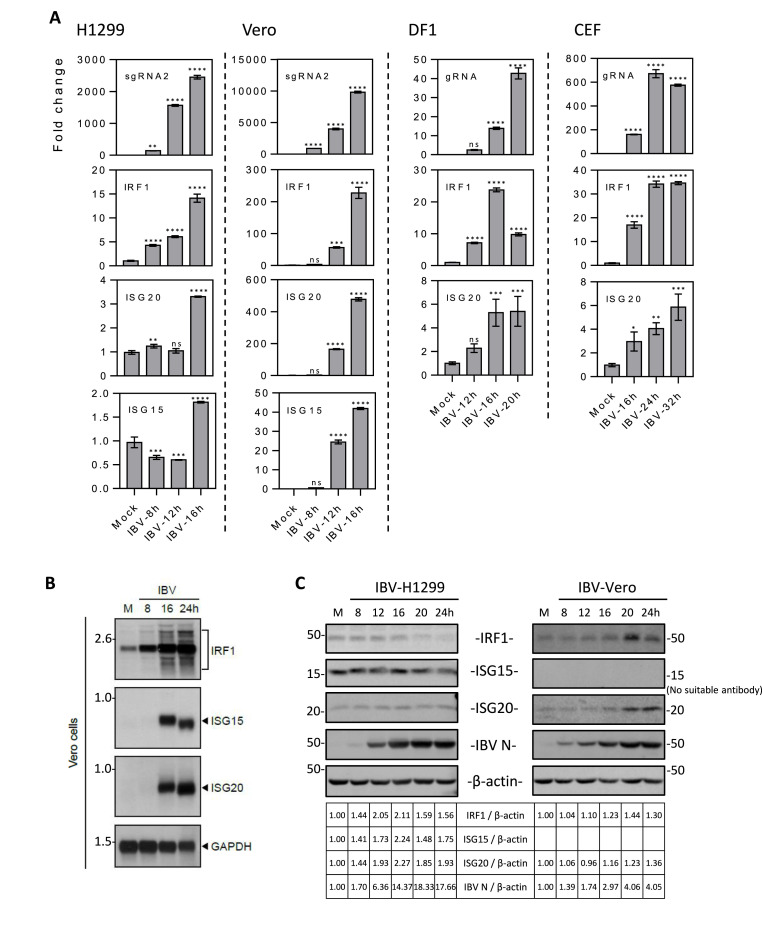
To verify these results, IBV-infected H1299 and Vero cells harvested at indicated times post-infection were subjected to RT-qPCR analysis, revealing variable transcription levels of the three genes. Up to 255-fold induction of IRF1, 43-fold induction of ISG15 and 489-fold induction of ISG20 were detected in IBV-infected Vero cells (Fig. 1 A). However, a mere 4.28- to 14.13-fold induction of IRF1, 0.65- to 1.7-fold induction of ISG15, and 1.24- to 3.3-fold induction of ISG20 were detected in IBV-infected H1299 cells (Fig. 1A). To confirm if the induction of these genes also occurred in chicken cells, their mRNA levels in IBV-infected DF-1 and CEF cells, respectively, were determined, showing a 25- to 35-fold induction of IRF1 and a 5- to 6.5-fold induction of ISG20 (Fig. 1A).
Fig. 1.
Induction of IRF1, ISG20 and ISG15 at RNA and protein levels in IBV-infected cells.
A. RT-qPCR analysis of the induction of IRF1, ISG20 and ISG15 in H1299, Vero, DF1 and CEF cells infected with IBV. Cells were infected with IBV at an MOI∼2 or mock-treated with UV-inactivated IBV, and harvested at indicated time points for RNA extraction. Equal amounts of total RNA were reverse-transcribed. The levels of IBV genomic RNA (IBV gRNA) and IBV subgenomic RNA2 (IBV sgRNA2), and mRNA levels of IRF1, ISG20 and ISG15 were determined by qPCR. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001)
B. Northern blot analysis of the induction of IRF1, ISG20 and ISG15 in Vero cells infected with IBV. Total RNA extracted from Vero cells infected with IBV and mock-treated cells at indicated times were separated on 1% denaturing agarose gel and transferred to a Hybond N+ membrane. IRF1, ISG20 and ISG15 were probed with a Dig-labeled DNA probe corresponding to a region in IRF1, ISG20 and ISG15. Numbers on the left indicate nucleotides in kilobase, and the IRF1, ISG20 and ISG15 RNAs were indicated on the right
C. Western blot analysis of the upregulation of IRF1, ISG20 and ISG15 in IBV-infected H1299 and Vero cells. H1299 and Vero cells were infected with IBV at an MOI∼2 or mock-treated with UV-inactivated IBV, harvested at the indicated time points and subjected to Western blot analysis using antibodies against IRF1, ISG15, ISG20 and IBV N proteins. Beta-actin was included as the loading control. Sizes of protein ladders in kDa were indicated on the left.
The induction of IRF1, ISG15 and ISG20 at the mRNA and protein levels in IBV-infected Vero cells was further studied by Northern and Western blotting. Northern blot analysis of the total RNAs extracted from IBV-infected Vero cells demonstrated that the IRF1 mRNA was at a low level in the mock-treated cells, but markedly increased at 8hpi and accumulated to very high levels at 16 and 24hpi (Fig. 1B). ISG15 and ISG20 mRNAs were not detectable in the mock-treated or 8hpi samples, but were induced to high levels at 16 and 24hpi (Fig. 1B). Western blot analysis of IBV-infected Vero cells showed a low basal level of IRF1 protein in mock-treated cells and in infected cells from 8 to 16 hpi, with a sudden increase to high level at 20hpi and slight reduction at 24hpi (Fig. 1C). The expression of ISG20 protein was similar to that of IRF1 protein, being increased rapidly at 20hpi, and reached the highest level at 24hpi (Fig. 1C). Attempts made to determine the expression levels of ISG15 protein in IBV-infected Vero cells were not successful due to the lack of reactivity of the anti-ISG15 antibodies to monkey proteins. Meanwhile, the expression of these three genes in IBV-infected H1299 cells was also detected, showing minimal to moderate induction at 16hpi and declining afterwards (Fig. 1C). These results reveal a similar induction pattern of these genes at the protein level as their induction kinetics at the mRNA level in the two cell lines. Taken together, these results verified the Affymetrix array and transcriptomic data that the expression of these anti-viral genes was differentially regulated in IBV-infected Vero and H1299 cells, and further demonstrated that upregulation of these gene expression is a common phenomenon during IBV infection of different cell types.
3.2. HCoV-OC43 and PEDV infection of H1299 and/or Vero cells regulates the expression of IRF1, ISG15 and ISG20
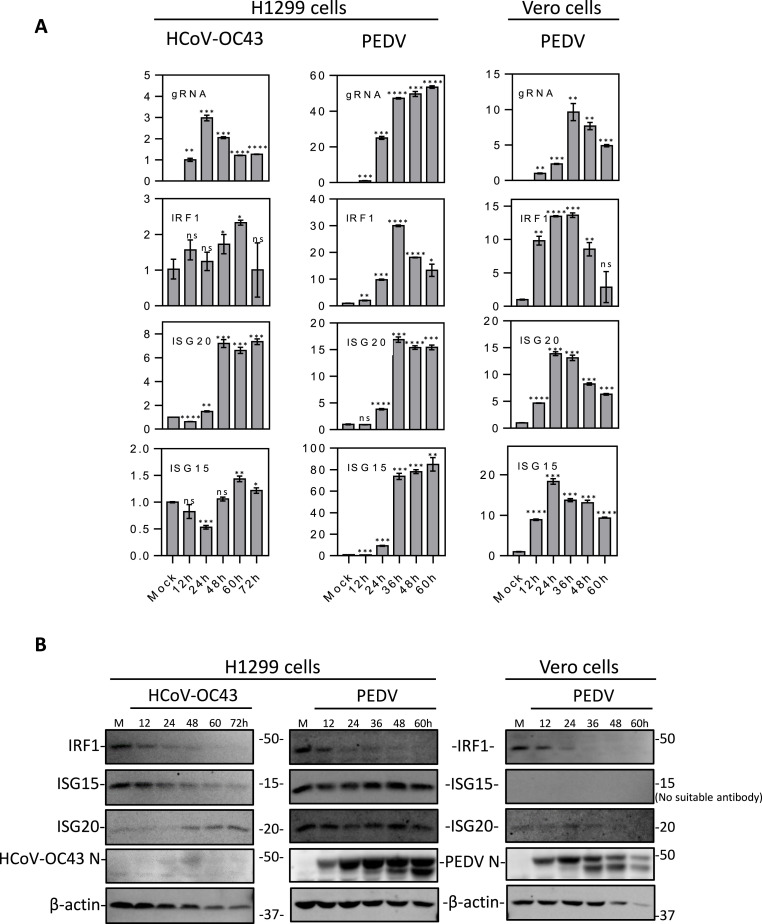
To verify whether the upregulation of IRF1, ISG15 and ISG20 is a common mechanism shared by other coronaviruses, similar time course experiments were conducted in H1299 and/or Vero cells infected with HCoV-OC43and PEDV, respectively. At the mRNA level, IRF1, ISG15 and ISG20 were 2.4-, 1.4- and 7.3-fold, respectively, upregulated in HCoV-OC43-infected H1299 cells (Fig. 2 A). When H1299 cells were infected with PEDV, the induction of IRF1, ISG20 and ISG15 reached 30-, 16.8- and 84.9-fold, respectively (Fig. 2A). In Vero cells infected with PEDV, IRF1, ISG20 and ISG15 were all up-regulated 13.6-,13.8- and 18.3-fold, respectively (Fig. 2A).
Fig. 2.
Induction of IRF1, ISG20 and ISG15 at mRNA and protein levels in HCoV-OC43- and PEDV-infected cells.
A. RT-qPCR analysis of the induction of IRF1, ISG20 and ISG15 in H1299 and/or Vero cells infected with HCoV-OC43 and PEDV, respectively. Cells were infected with HCoV-OC43/PEDV at an MOI∼2 or mock-treated with UV-inactivated HCoV-OC43/PEDV, and harvested at indicated time points for RNA extraction. Equal amounts of total RNA were reverse-transcribed. The levels of HCoV-OC43 genomic RNA (HCoV-OC43 gRNA) and PEDV genomic RNA (PEDV gRNA), and mRNA levels of IRF1, ISG20 and ISG15 were determined by qPCR. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
B. Western blot analysis of the upregulation of IRF1, ISG20 and ISG15 in HCoV-OC43- and PEDV-infected H1299 and/or Vero cells. H1299 and Vero cells were infected and harvested as described in A and subjected to Western blot analysis using antibodies against IRF1, ISG15, ISG20 and HCoV-OC43/PEDV N proteins. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the right.
Western blot analysis showed that in H1299 and/or Vero cells infected with HCoV-OC43 and PEDV, the protein expression of ISG15 and ISG20 was similar to their mRNA expression levels, reaching the highest level in the middle to later stage of infection (Fig. 2B). However, the expression level of IRF1 protein appeared to be gradually reduced, probably due to the prolonged duration of the time course experiments for these two viruses (Fig. 2B). In general, these results demonstrate that induction of IRF1, ISG15 and ISG20 is a common mechanism shared by different coronaviruses, suggesting that they play important regulatory roles in coronavirus replication and virus-host interactions.
3.3. Inhibition of type I/III IFN pathways does not significantly affect the upregulation of IRF1, ISG15 and ISG20 by IBV-infected Vero cells
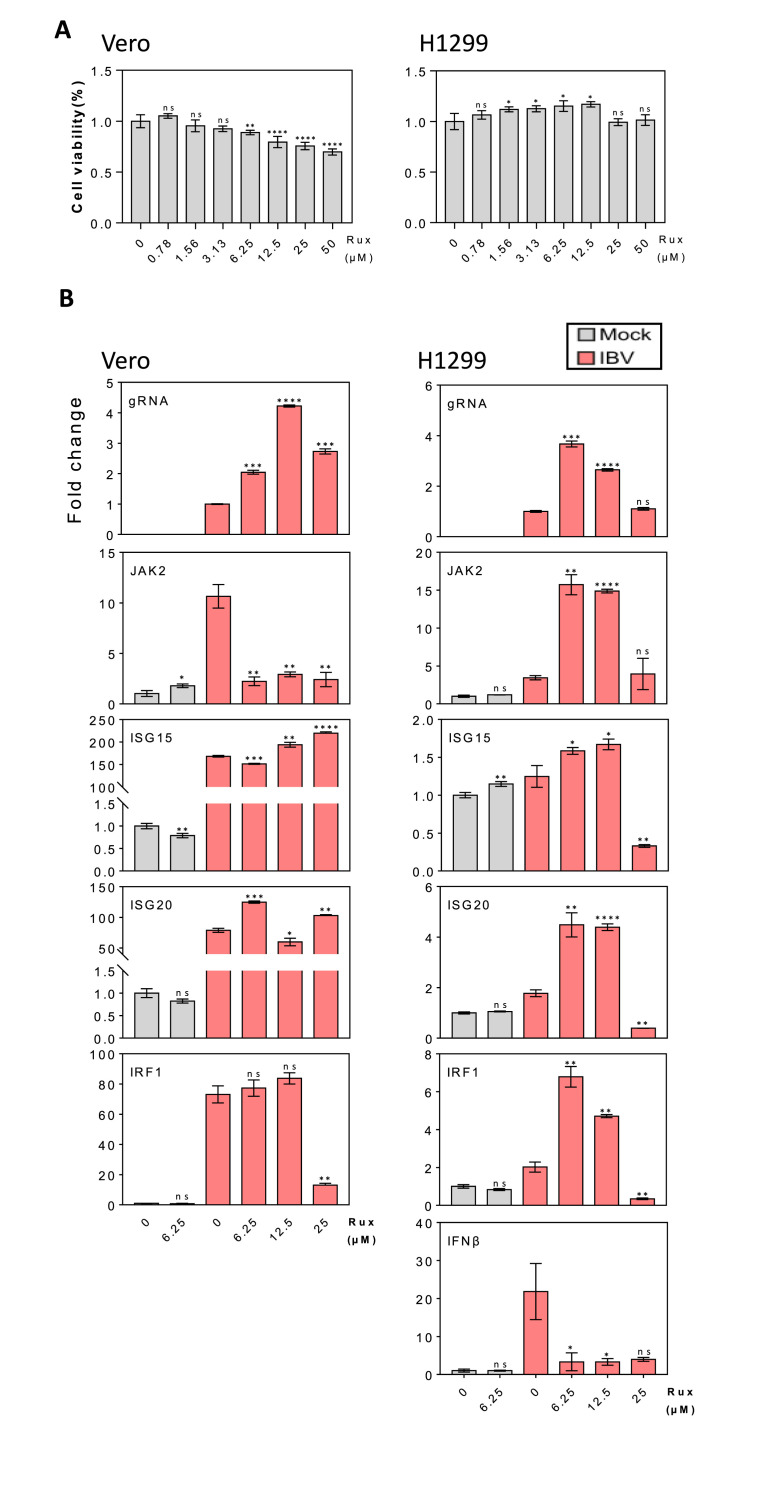
As Vero cells are known to be IFN-β-deficient, the highly efficient induction of IRF1, ISG15 and ISG20 by IBV infection of this cell line would be IFN-β-independent. To further confirm this and to investigate if other type I/III IFNs might be involved in the activation of these three genes, Ruxolitinib, an effective JAK2 inhibitor, was used to explore the effect of IFN inhibition on IBV-induced upregulation of IRF1, ISG15 and ISG20 in Vero and H1299 cells. The effects of different concentrations of Ruxolitinib on cell proliferation were first tested, revealing no significant difference in the proliferation rate of Vero cells in the presence of 0.78, 1.56 and 3.13 μM of Ruxolitinib, as compared with the DMSO control (Fig. 3 A). Cell proliferation rates were significantly reduced when the concentrations of Ruxolitinib were raised to 12.5–50 μM (Fig. 3A). The growth and proliferation rates of H1299 cells were not much affected when 0.78–50 μM of Ruxolitinib were used (Fig. 3A).
Fig. 3.
Effect of JAK1/2 inhibitor Ruxolitinib on the induction of IRF1, ISG15 and ISG20 in IBV-infected Vero and H1299 cells.
A. Effect of different concentrations of Ruxolitinib on the viability of Vero and H1299 cells. Vero or H1299 cells were plated to a 96-well plate and cultured for 16–20 h, treated with Ruxolitinib at indicated concentrations or the same volume of DMSO for 24 h. Cell viability was determined by measuring the absorbance value at 450 nm after adding the CCK solution for 1–4 h. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
B. RT-qPCR analysis of levels of IBV gRNA, JAK2, IRF1, ISG15, ISG20 and IFNβ in IBV-infected Vero (IFNβ-deficient) and H1299 cells in the presence of different concentrations of Ruxolitinib. At 1 hpi, cells infected with MOI∼2 of IBV were treated with Ruxolitinib at the indicated concentrations or same volume of DMSO, and harvested at 20hpi for RNA extraction. Equal amounts of total RNA were reverse-transcribed, and the level of IBV gRNA and mRNA levels of JAK2, ISG15, ISG20, IRF1 and IFNβ were determined by qPCR. Significance levels were presented by the p-value as in A.
The effects of Ruxolitinib on IBV-induced upregulation of JAK2, IRF1, ISG15 and ISG20 were then analyzed in IBV-infected Vero cells in the presence of different concentrations of the inhibitor. Treatment of the infected Vero cells with 6.25, 12.5 and 25 μM of Ruxolitinib, respectively, showed 2-, 4- and 3-fold enhancement of IBV replication (Fig. 3B). Interestingly, the induction of JAK2 was totally inhibited by the addition of Ruxolitinib, but further upregulation of IRF1, ISG15 and ISG20 was observed in most cases in the presence of these concentrations of the inhibitor (Fig. 3B). The effects of Ruxolitinib on IBV-induced upregulation of JAK2, IRF1, ISG15 and ISG20 were then analyzed in IBV-infected H1299 cells, an IFN-β-competent cell line. In the presence of all concentrations of the inhibitor used, the induction of IFN-β by IBV infection was significantly inhibited (Fig. 3B), confirming that treatment with this inhibitor suppressed the induction of IFN-β. Similar to the results observed in Vero cells, treatment of H1299 cells with 6.25 and 12.5 μM of Ruxolitinib, respectively, significantly increased IBV replication, and meanwhile, significant upregulation of JAK2, IRF1, ISG15 and ISG20 was also detected in these cells (Fig. 3B). This enhancement effect on IBV replication and induction of the three ISGs was ablated when the concentration of the inhibitor was increased to 25 μM (Fig. 3B). These results support that induction of IRF1, ISG15 and ISG20 in IBV-infected cells may be controlled by an IFN-β-independent mechanism.
3.4. Overexpression of IRF1 suppresses IBV replication mainly through the induction of IFN pathways
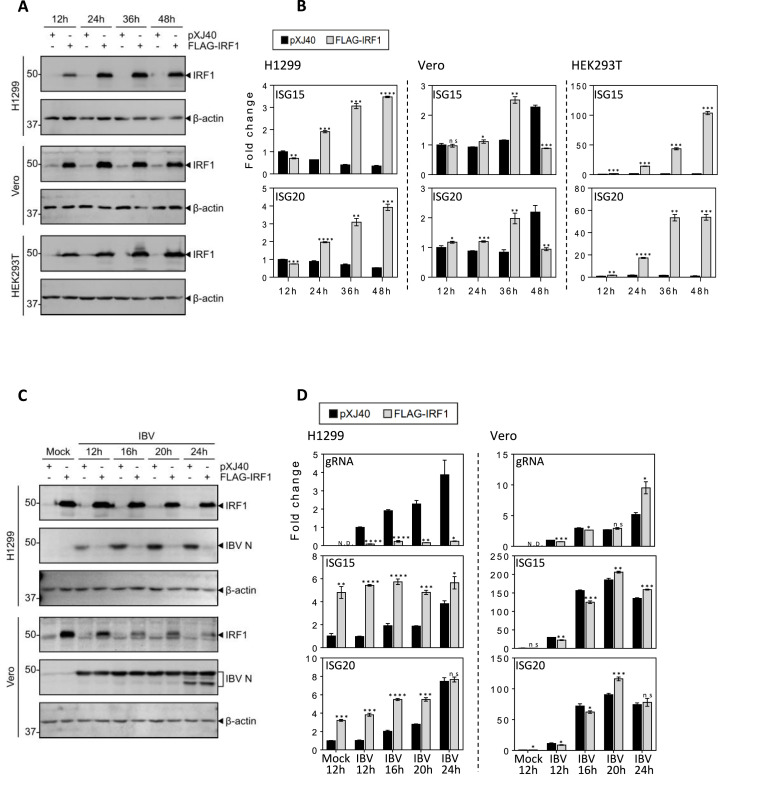
The functional significance of IRF1 upregulation in the replication of IBV was then studied by overexpression of the protein in H1299, Vero and HEK293T cells. Cells were transfected with either an empty plasmid or a Flag-tagged IRF1, and harvested at 12, 24, 36 and 48hpi, respectively, for analysis by Western blotting and the transcription levels of ISG15 and ISG20 by RT-qPCR. Efficient expression of the Flag-tagged IRF1 at the protein level was observed in all the three transfected cell lines (Fig. 4 A). Its overexpression in Vero and H1299 cells resulted in a moderate 2- to 4-fold induction of ISG15 and ISG20, but a 10- to 100-fold induction of the two genes was observed in HEK293T cells, reflecting a generally much higher transfection efficiency of this cell line (Fig. 4A and B).
Fig. 4.
Overexpression of IRF1 increased the induction of ISG15 and ISG20 in H1299, Vero and HEK293T cells and suppressed IBV replication.
A. Western blot analysis of the expression efficiency of IRF1 in H1299, Vero and HEK293T cells transfected with IRF1. Cells were transfected with pXJ40, or pXJ40-FLAG-IRF1, harvested at the indicated time points. Cells were subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left.
B. RT-qPCR analysis of the induction of ISG15 and ISG20 in H1299, Vero and HEK293T cells transfected with IRF1. Cells were transfected and harvested as in (A), total RNA were extracted and equal amounts of total RNA were reverse-transcribed. The mRNA expression levels of ISG15 and ISG20 were determined by qPCR. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
C. Western blot analysis of IRF1 and IBV N protein in IRF1-overexpressed H1299 and Vero cells infected with IBV. Cells were transfected with pXJ40, or pXJ40-FLAG-IRF1, before being infected with IBV at MOI∼2 or mock-treated with UV-inactivated IBV, harvested at the indicated time points. Cells were subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left
D. RT-qPCR analysis of the induction of ISG15 and ISG20 in IRF1-overexpressed H1299 and Vero cells infected with IBV. Cells were transfected, infected and harvested as in (C), and total RNA extracted. Equal amounts of total RNA were reverse-transcribed, and the levels of IBV genomic RNA (IBVgRNA), and the mRNA expression levels of ISG15 and ISG20 were determined by qPCR. Significance levels were presented by the p-value as in B.
IBV infection of IRF1-transfected H1299 cells showed almost completely inhibition of viral replication at both mRNA and protein levels, and did not induce a higher ISG15 induction (Fig. 4C and D). A moderate induction of ISG20 was observed in the same infected cells (Fig. 4D), reflecting a different induction mechanism of the two ISGs. Infection of IRF1-transfected Vero cells with IBV did not show any inhibitory effect on viral replication (Fig. 4D). In fact, higher viral replication was detected in the IRF1-transfected cells than did in cells transfected with the empty vector alone (Fig. 4D). Furthermore, very similar induction kinetics of ISG15 and ISG20 were observed in the infected cells transfected with or without IRF1 (Fig. 4D). As Vero cells are IFN-β-deficient, these results demonstrate that the antiviral effects of IRF1 on the replication of IBV are mainly mediated through activation of the type I IFN pathways in the transfected cells. In addition, ISG15 was not significantly induced in p53-deficient H1299 cells transfected with IRF1, pointing to the involvement of p53 in the induction of this ISG during IBV infection.
3.5. The endogenous IRF1 plays a minor role in the inhibition of IBV replication and in the induction of ISG15 and ISG20
To assess the contribution of the endogenous IRF1 to the host antiviral response in IBV-infected cells, transient knockdown of IRF1 with siRNA and establishment of IRF1-knockout stable cells with CRISPR-cas9 were carried out. Knockdown of IRF1 with siRNA was first performed in Vero cells, followed by infection with IBV, showing efficient knockdown of the endogenous IRF1 at both protein and mRNA levels (Fig. 5 A and B). IBV infection of the knockdown cells showed only slightly reduced ISG20 induction and even higher induction of ISG15 at the mRNA level, compared with the siNC control cells (Fig. 5B). Knockdown of IRF1 in Vero cells also rendered a very minor, if any, effect on IBV replication, as revealed by Western blot analysis of IBV N protein and RT-qPCR analysis of viral genomic RNA (Fig. 5B). In contrast, IBV infection of IRF1-knockdown H1299 cells showed significantly higher levels of viral genomic RNA replication and slightly higher synthesis of IBV N protein (Fig. 5A and B). Correspondingly, the expression of ISG15 was also upregulated in the IRF1-knockdown H1299 cells, probably reflecting the higher levels of viral replication (Fig. 5A and B).
Fig. 5.
Effect of knockdown/knockout of IRF1 on the replication of IBV and the induction of ISG15 and ISG20 in H1299 and Vero cells.
A. Western blot analysis of IBV N and IRF1 in IRF1-knockdown Vero and H1299 cells infected with IBV. Cells were transfected with siNC or siIRF1 before being infected with IBV at MOI∼2 or mock-treated with UV-inactivated IBV, harvested at indicated time points. Cells were subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left.
B. RT-qPCR analysis of IBV gRNA, IRF1, ISG15 and ISG20 in IRF1-knockdown cells infected with IBV. Cells were infected and harvested as in (A), and total RNA extracted. Equal amounts of total RNA were reverse-transcribed, levels of IBV gRNA and mRNA levels of IRF1, ISG15 and ISG20 were determined by RT-qPCR. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001)
C. Construction of an IRF1-knockout clone based on Vero cells using CRISPR-cas9 technique. Sequence analysis of a selected IRF1-knockout clone, showing the insertion of an “A” at the 5′-terminal region of the IRF1 gene.
D. Western blot analysis of IBV N and IRF1 in IRF1-knockout Vero cells infected with IBV. Wild type and IRF1-knockout Vero cells were infected with IBV at an MOI∼2 or mock-treated with UV-inactivated IBV, harvested at the indicated time points. Cells were subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left.
E. RT-qPCR analysis of IBV gRNA, IRF1, ISG15 and ISG20 in IRF1-knockout cells infected with IBV. Cells were infected and harvested as in (D), and total RNA extracted. Equal amounts of total RNA were reverse-transcribed, levels of IBV gRNA and mRNA levels of IRF1, ISG15 and ISG20 were determined by qPCR. Significance levels were presented by the p-value as in B.
Knockout of IRF1 in Vero cells by CRISPR-cas9 was also attempted, isolating a knockout cell clone with an “A” insertion between nucleotide position A10 and C11 in the IRF1-coding region (Fig. 5C). This “A” insertion would result in the translation of a truncated IRF1 protein terminated at 17th amino acid. Western blot analysis of the isolated clone showed much reduced detection of the IRF1 protein (Fig. 5D). However, it appears that low levels of IRF1 expression were still detectable, suggesting that the ‘A’ insertion may occur in one chromosome only. Infection of wild type and knockout cells with IBV showed minimal effects on the expression of IBV N protein (Fig. 5E). Unexpectedly, the viral gRNA transcription was slightly reduced in the knockout cells (Fig. 5E), suggesting that the partial knockout of IRF1 may render a certain effect on the growth and proliferation of the cells, which, in turn, inhibits the replication of IBV. The induction of ISG20 was also suppressed in a similar pattern as the viral gRNA, however, significantly more induction of ISG15 was observed in the partial knockout cells at 20 and 24hpi (Fig. 5E). Taken together, these results are consistent with data generated from the overexpression of IRF1, reinforcing the conclusions that the IRF1-mediated anti-IBV response is mainly type I IFN-dependent and that the induction of ISG15 and IRG20 in IBV-infected Vero cells is not dependent on the induction of IRF1.
3.6. Overexpression of ISG15 and ISG20 does not significantly suppress IBV replication
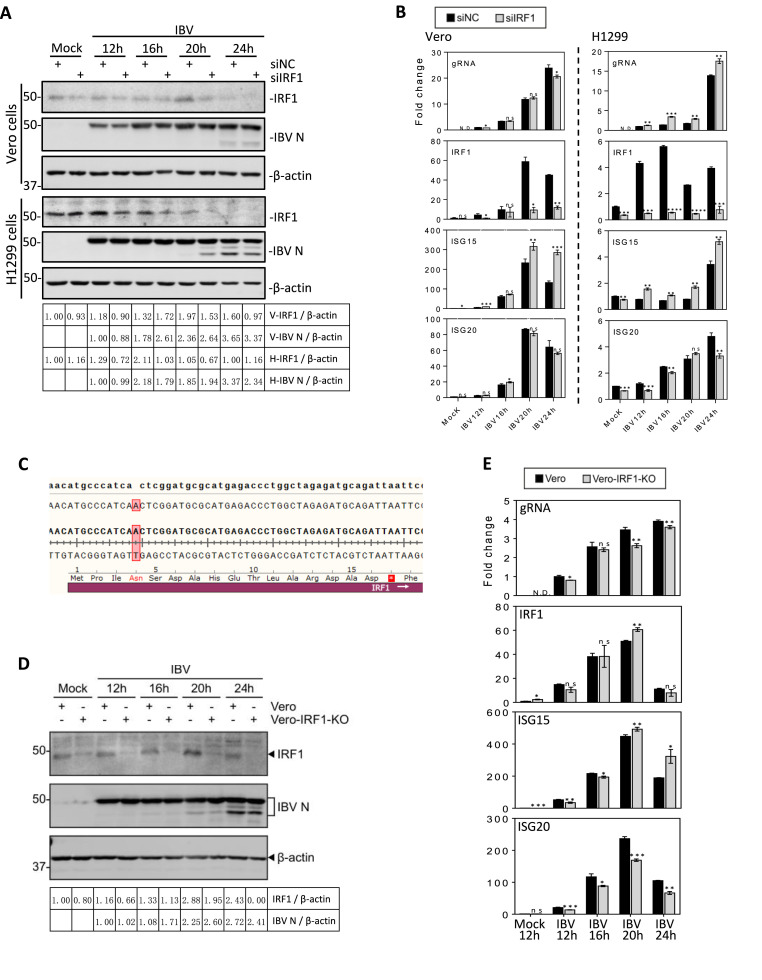
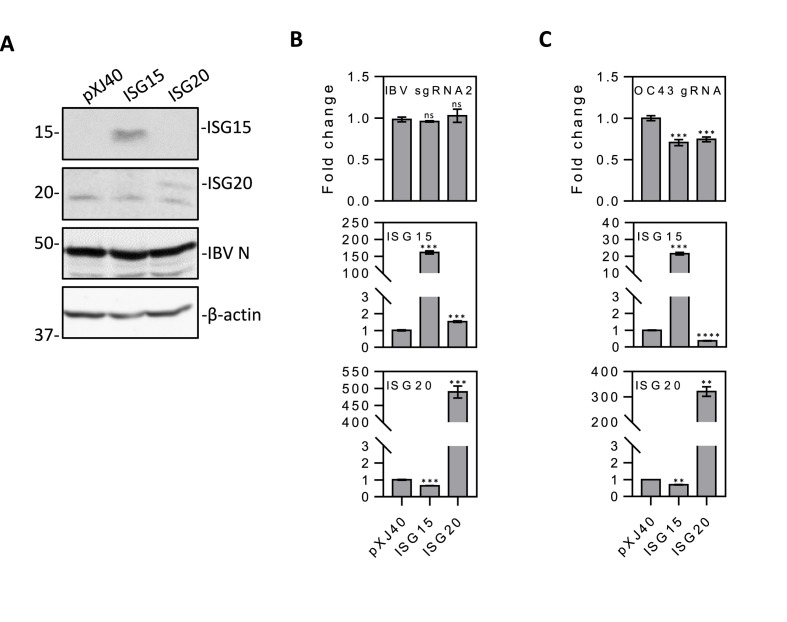
The functional significance of ISG15 and ISG20 induction in IBV replication was then studied by overexpression of the two genes, respectively, in H1299 cells. Analysis of cells transfected with either an empty plasmid, Flag-tagged ISG15 or Flag-tagged ISG20 by Western blot and RT-qPCR demonstrated efficient expression of the two ISGs at protein and mRNA levels (Fig. 6 A and B). Infection of the transfected cells showed that overexpression of either ISG15 or ISG20 did not impose a significantly suppressive effect on IBV replication (Fig. 6B). However, overexpression of ISG15 or ISG20 in H1299 cells infected with HCoV-OC43 significantly inhibited its replication (Fig. 6C), demonstrating that these two ISGs may play a relatively minor role against IBV replication in culture cells.
Fig. 6.
The effect of overexpression of ISG15 and ISG20 on the replication of IBV and HCoV-OC43.
A. Western blot analysis of IBV N, ISG15 and ISG20 in H1299 cells transfected with either ISG15 or ISG20 and infected with IBV. H1299 cells were transfected with pXJ40, pXJ40-FLAG-ISG15, or pXJ40-FLAG-ISG20, before being infected with IBV at MOI∼2. Cell lysates were harvested at 24hpi, and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left
B. RT-qPCR analysis of IBV sgRNA2, ISG15 and ISG20 in H1299 cells transfected with either ISG15 or ISG20 and infected with IBV. Cells were infected and harvested as in (A), and total RNA extracted. Equal amounts of total RNA were reverse-transcribed, and the level of IBV sgRNA2 and mRNA levels of ISG15 and ISG20 were determined by qPCR. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).C. RT-qPCR analysis of HCoV-OC43 gRNA, ISG15 and ISG20 in H1299 cells transfected with either ISG15 or ISG20 and infected with HCoV-OC43. H1299 cells were transfected with pXJ40, pXJ40-FLAG-ISG15 or pXJ40-FLAG-ISG20, before being infected with HCoV-OC43 at MOI∼2. Cell lysates were harvested at 48hpi for RNA extraction. Equal amounts of total RNA were reverse-transcribed, and the level of OC43 gRNA and mRNA levels of ISG15 and ISG20 were determined by qPCR. Significance levels were presented by the p-value as in B.
3.7. IBV-induced ISG15 and ISG20 expression is partially p53-dependent
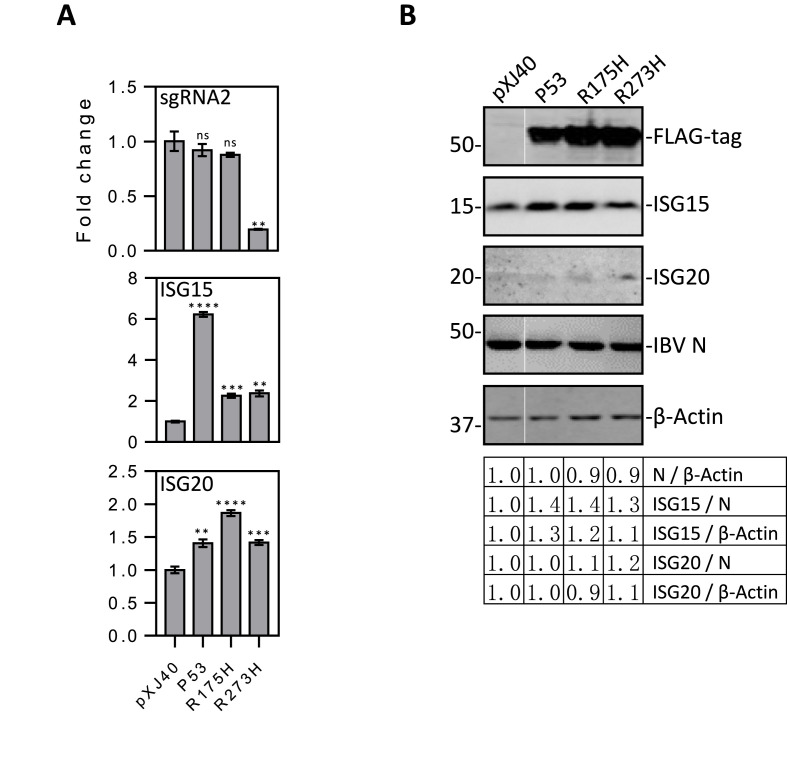
As H1299 cells are p53-deficient, the weak induction of ISG15 and ISG20 by IBV infection of this cell line would point to the potential involvement of p53 in regulating these ISGs during IBV infection. This possibility was studied by transfection of H1299 cells with wild type and two gain-of-function mutant p53, R175H and R273H, prior to infection with IBV. Compared with the vector control, overexpression of R273H suppressed IBV replication as revealed by the much lower level of IBV sgRNA2, but IBV RNA synthesis was not significantly affected in cells transfected with p53 and R175H (Fig. 7 A). Overexpression of p53 and the two mutants moderately enhanced (1.5- to 2-fold) ISG20 induction at the mRNA level (Fig. 7A). A more drastic induction (6-fold) of ISG15 mRNA expression was detected in cells overexpressing wild type p53 (Fig. 7A). However, in cells overexpressing the two mutants, only a moderate induction of ISG15 (about 2-fold) was observed (Fig. 7A). In addition to the much lower IBV replication in R273H-transfected cells, additional functions gained by the two mutants may modulate apoptosis and proliferation of the transfected H1299 cells, limiting the induction of this ISG.
Fig. 7.
The effect of p53 overexpression on the induction of ISG15 and ISG20 in p53-deficient H1299 cells infected with IBV.
A. RT-qPCR analysis of IBV sgRNA2, ISG15 and ISG20 in p53-overexpressed H1299 cells infected with IBV. Cells were transfected with pXJ40, pXJ40-FLAG-P53, pXJ40-FLAG-P53-R175H or pXJ40-FLAG-P53-R273H, before being infected with IBV at MOI∼2. Cells were harvested at 24hpi, and total RNA were extracted. Equal amounts of total RNA were reverse-transcribed, and the levels of IBV sgRNA2 and mRNA levels of ISG15 and ISG20 were determined by qPCR. Significance levels were presented by the p-value (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
B. Western blot analysis of IBV N, ISG15, ISG20 and the expression efficiency of wild type and mutant p53 in H1299 cells infected with IBV. Cells were transfected and infected as in (A). Cells were harvested at 24hpi and subjected to Western blot analysis using the indicated antibodies. Beta-actin was included as the loading control. Sizes of protein ladders in kDa are indicated on the left.
Western blot analysis confirmed the efficient expression of the ectopic wild type and mutant p53 proteins at comparable levels (Fig. 7B). Compared with the vector control, IBV-induced ISG15 protein expression was elevated in cells overexpressing p53 and R175H (Fig. 7B). Whereas no ISG20 protein was detected in the vector control, low levels of IBV-induced ISG20 protein expression could be detected in the transfected cells (Fig. 7B). Taken together, these data suggest that the induction of ISG15 and ISG20 during IBV infection was partially dependent on p53.
4. Discussion
IFN is involved in a variety of immune mechanisms during viral infection, playing a key role in inducing antiviral response and inhibiting the replication of many DNA and RNA viruses (Malmgaard, 2004; Samuel, 2001). In this study, three ISGs, IRF1, ISG15 and ISG20, were shown to be differentially expressed in IBV-infected Vero and H1299 cells, confirming the initial findings by Affymetrix microarray and transcriptomic analyses. Characterization of their functional roles during IBV infection demonstrated that IRF1 played an active role in suppressing IBV replication, mainly through the activation of the IFN pathway. However, a minor, if any, anti-IBV function was played by ISG15 and ISG20 in culture cells. Furthermore, p53, but not IRF1, was shown to be involved in the induction of ISG15 and ISG20 in IBV-infected cells.
The robust induction of IRF1, ISG15 and ISG20 in IFN-β-deficient Vero cells infected with IBV suggests an IFN-independent mechanism controlling the induction of these genes. This was reinforced by the use of JAK1/2 inhibitor Ruxolitinib, which did not significantly inhibit the upregulation of ISG15 and ISG20 in IBV-infected Vero cells, ruling out the potential involvement of other type I/III IFNs in regulating the expression of these ISGs. Our observations demonstrated that high concentration of Ruxolitinib may inhibit IBV replication and, consequently, the induction of these ISGs. This is consistent with a previous report that JAK inhibitors (Baricitinib) may target host factors required for SARS-COV-2 entry into cells, leading to the suppression of type I IFN-driven ACE2 upregulation and viral infection (Jorgensen et al., 2020). Furthermore, the observation that addition of 6.25 and 12.5 μM of Ruxolitinib rendered a differential effect on the induction of JAK2, IRF1, ISG15 and ISG20, lending further support to the conclusion that the induction of IRF1, ISG15 and ISG20 in IBV-infected cells is controlled by a type I IFN-independent mechanism.
IRF1 has long been known to mediate the host defense system, participating in a broad antiviral response by coordinating multiple cellular signaling cascades. In a variety of cell types, overexpression of IRF1 induced the expression of type I IFN and other ISGs, and its deletion led to the limitation of these ISG inductions (Song et al., 2021). IRF1 can directly bind to the promoter of VIPERIN and induce its expression in the absence of IFN signaling (Stirnweiss et al., 2010). Infection of cells with many viruses has been shown to upregulate several ISGs without producing type I IFNs and initiating the JAK-STAT signal pathway. These include the transcriptional upregulation of ISG15 and some other ISGs mediated by IRF3 independent of type I IFNs during human cytomegalovirus (HCMV) infection (Ashley et al., 2019), and the upregulation of ISG56 in JAK1-defecient p2.1 cells infected with vesicular stomatitis virus, encephalomyocarditis virus or Sendai virus (Guo et al., 2000). IRF1 was also shown to activate STAT1 transcription and stimulate ISG expression in hepatitis E virus (HEV) infection without triggering the IFN production (Xu et al., 2016). On the other hand, IRF1 was actively participated in host defense processes in an IFN-β-dependent manner against respiratory pathogen human metapneumovirus (HMPV) infection (Loevenich et al., 2021). The ISRE presented in the ISG20 promoter was reported to mediate the induction of IRF1-dependent ISG20 expression in the absence of functional γ-activating sequence (Gongora et al., 2000). However, as observed in this study, knockdown/out of IRF1 in Vero cells did not reduce the upregulation of ISG15 and ISG20, ruling out the possible involvement of IRF1 in the induction of the two ISGs. Furthermore, overexpression or knockdown/out of IRF1 in Vero cells rendered a minor, if any, effect on IBV replication, indicating that the major anti-IBV activity of IRF1 is IFN-dependent.
The antiviral function of ISG15 is host species-specific. ISG15-deficient human patients did not show an increased susceptibility to virus infection, which is markedly different from mice lacking ISG15 (Hermann and Bogunovic, 2017). Lung cells from ISG15-deficient mice were found to be more suitable for the replication of influenza B virus and a higher viral titer was produced (Lai et al., 2009). In addition to participating in the antiviral response by directly inhibiting virus replication, ISG15 may also directly or indirectly regulate cytokine responses and play a role in cell autophagy and metabolism. In human cells, ISG15 upregulated by IFN promotes the movement of ATG mediators to parasitic vacuoles and regulates IFN-γ-dependent autophagy in Toxoplasma gondii-infected cells (Bhushan et al., 2020). ISG20 is significantly up-regulated by IFNs, but its basal levels are varying in different cell types even in the absence of persistent IFN response. The varying basal levels of ISG20 in different cell types may reflect the fact that its expression is regulated by different transcription factors in different cells, such as specific protein 1 (SP-1) or upstream stimulator 1 (USF-1) (Gongora et al., 2000). As an antiviral protein, ISG20 was considered to mediate the antiviral action through direct degradation of viral RNA owned to its RNase activity. ISG20 was also able to directly degrade deaminated viral DNA and inhibit the translation of viral RNA and non-self RNA (Deymier et al., 2022). In this study, we showed that these two ISGs may play a minimal inhibitory role in IBV replication, suggesting that IBV may have evolved mechanisms to evade their anti-viral actions. As ISG15 is naturally absent in avian species, the functional irrelevance of this ISG to IBV infection of chickens appears to be appreciable. However, further studies would be required to illuminate the strategies exploited by IBV to evade the antiviral functions of ISG20.
Overexpression of p53 and two gain-of-function mutants in p53-deficient H1299 cells enhanced the induction of ISG15 and ISG20 following IBV infection, suggesting the involvement of p53 in the upregulation of these two ISGs. As a well-studied tumor suppressor and the most common mutated gene in human cancers (Oren, 2003), activation of p53 promotes cell cycle arrest, apoptosis and a number of other cellular activities (Ding et al., 2014; Hao et al., 2019). p53 is also known as a central regulator of the innate immune response (Hao et al., 2019; Rivas et al., 2010). In response to viral infection, p53 may promote trans-activation of ISGs as well as IFN production in cells (Muñoz-Fontela et al., 2011). Previous studies reported the presence of a p53 response element in the ISG15 promoter and a coupling relationship between the two genes. In response to DNA damage, ISG15 may positively regulate the tumor suppressor function of p53 and the expression of ISG15 depends on p53 (Park et al., 2016). In HIV-1 infection, ISG15 can modify p53 and the absence of ISG15 leads to the non-degradation of misfolded p53 accumulated by USP18 and enhanced virus replication (Osei Kuffour et al., 2019).
Throughout the study, it was also noted that the protein levels of these ISGs in cells overexpressing the corresponding construct without IBV infection was much higher than those in the same transfected cells infected with IBV. Similarly, in IBV-infected cells, a high-level induction of their expression at the mRNA level was detected, but only minor upregulation at the protein level was usually detected. Our previous studies have demonstrated that the interaction between IBV S protein and eIF3f inhibits host cell protein translation, resulting in strong expression of the same genes at the transcriptional level, but only minimal to moderate induction at the translational level (Xiao et al., 2008). Other studies have also shown that coronavirus infection may inhibit host translation and de nova protein synthesis to efficiently translate viral mRNAs (Waisner et al., 2023). For example, SARS-CoV and SARS-CoV-2 nsp 1 protein can suppress host gene expression by promoting host mRNA degradation, alter the cellular transcriptome and bias the host protein synthesis mechanism towards viral RNA translation (Narayanan et al., 2008; Yuan et al., 2020). Furthermore, induction of ER stress response, especially the activation of the PKR/PERK-eIF2a pathway, may also render an inhibitory effect on host gene expression at the protein level (Fung et al., 2016; Wang et al., 2009). Suppression of these ISG expression at the protein level would be an effective strategy evolved by IBV to evade host antiviral response. Our Northern blot analysis also showed that the migration of ISG15 and ISG20 mRNAs collected at 24 hpi was slightly faster compared to the samples collected at 16 hpi. In addition to the gel artifact, it is possible that an alternative polyadenylation site might be preferentially selected at late stage of IBV infection. This possibility will be investigated in future studies.
5. Conclusion
In conclusion, this study characterizes the relative contribution of IRF1, ISG15 and ISG20, the three prominently upregulated ISGs in IFN-β-deficient Vero cells, to host anti-IBV response in the absence of type I IFN activation. IRF1 plays an active anti-IBV function in an IFN-dependent manner, but very minor, if any, anti-IBV function is played by ISG15 and ISG20 in the presence and absence of IFN signaling. The induction of ISG15 and ISG20 by IBV infection is partially dependent on p53, but not on IRF1. This study provides new information on the mechanisms underlying the induction of ISG15, ISG20 and IRF1, and their relative contribution to host anti-IBV response.
CRediT authorship contribution statement
Si Ying Liu: Data were analyzed by, wrote the manuscript draft, the experiments were performed by. Mei Huang: Data were analyzed by. To Sing Fung: Data were analyzed by. Rui Ai Chen: Data were analyzed by, this study was conceived by. Ding Xiang Liu: This study was conceived by, data were analyzed by, wrote the manuscript draft, did critical revision.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
This work was partially supported by National Natural Science Foundation of China grants (31972660 and 32170152) and Zhaoqing Xijiang Innovative Team Foundation of China (grant number P20211154-0202).
Handling Editor: Ms. J Jasmine Tomar
References
- Ashley C.L., Abendroth A., McSharry B.P., Slobedman B. Interferon-independent upregulation of interferon-stimulated genes during human cytomegalovirus infection is dependent on IRF3 expression. Viruses. 2019;11:246. doi: 10.3390/v11030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan J., Radke J.B., Perng Y.-C., Mcallaster M., Lenschow D.J., Virgin H.W., Sibley L.D. ISG15 connects autophagy and IFN-γ-Dependent control of Toxoplasma gondii infection in human cells. mBio. 2020;11:e00852. doi: 10.1128/mBio.00852-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A.F., Plummer E.M., Vizcarra E.A., Sheets N., Joo Y., Tang W., Day J., Greenbaum J., Glass C.K., Diamond M.S., Shresta S. An IRF-3-, IRF-5-, and IRF-7-independent pathway of dengue viral resistance utilizes IRF-1 to stimulate type I and II interferon responses. Cell Rep. 2017;21:1600–1612. doi: 10.1016/j.celrep.2017.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der S.D., Zhou A., Williams B.R., Silverman R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deymier S., Louvat C., Fiorini F., Cimarelli A. ISG20: an enigmatic antiviral RNase targeting multiple viruses. FEBS Open Bio. 2022;12:1096–1111. doi: 10.1002/2211-5463.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Huang Y., Du Q., Dong F., Zhao X., Zhang W., Xu X., Tong D. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem. Biophys. Res. Commun. 2014;445:497–503. doi: 10.1016/j.bbrc.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L., Degols G., Gongora C., Blondel D., Williams B.R., Silverman R.H., Mechti N. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 2003;278:16151–16158. doi: 10.1074/jbc.M209628200. [DOI] [PubMed] [Google Scholar]
- Espert L., Degols G., Lin Y.-L., Vincent T., Benkirane M., Mechti N. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 2005;86:2221–2229. doi: 10.1099/vir.0.81074-0. [DOI] [PubMed] [Google Scholar]
- Espert L., Rey C., Gonzalez L., Degols G., Chelbi-Alix M.K., Mechti N., Gongora C. The exonuclease ISG20 is directly induced by synthetic dsRNA via NF-kappaB and IRF1 activation. Oncogene. 2004;23:4636–4640. doi: 10.1038/sj.onc.1207586. [DOI] [PubMed] [Google Scholar]
- Fang S.G., Shen S., Tay F.P.L., Liu D.X. Selection of and recombination between minor variants lead to the adaptation of an avian coronavirus to primate cells. Biochem. Biophys. Res. Commun. 2005;336:417–423. doi: 10.1016/j.bbrc.2005.08.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas B.T., Scholte F.E.M., Bergeron É., Pegan S.D. How ISG15 combats viral infection. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Sakakibara J., Sudo Y., Miyamoto M., Kimura Y., Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-beta gene regulatory elements. EMBO J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liao Y., Liu D.X. Regulation of stress responses and translational control by coronavirus. Viruses. 2016;8:E184. doi: 10.3390/v8070184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Similarities and dissimilarities of COVID-19 and other coronavirus diseases. Annu. Rev. Microbiol. 2021;75:19–47. doi: 10.1146/annurev-micro-110520-023212. [DOI] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gongora C., David G., Pintard L., Tissot C., Hua T.D., Dejean A., Mechti N. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 1997;272:19457–19463. doi: 10.1074/jbc.272.31.19457. [DOI] [PubMed] [Google Scholar]
- Gongora C., Degols G., Espert L., Hua T.D., Mechti N. A unique ISRE, in the TATA-less human Isg20 promoter, confers IRF-1-mediated responsiveness to both interferon type I and type II. Nucleic Acids Res. 2000;28:2333–2341. doi: 10.1093/nar/28.12.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Peters K.L., Sen G.C. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- Gürtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Fu F., Cao L., Guo L., Liu J., Xue M., Feng L. Tumor suppressor p53 inhibits porcine epidemic diarrhea virus infection via interferon-mediated antiviral immunity. Mol. Immunol. 2019;108:68–74. doi: 10.1016/j.molimm.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann M., Bogunovic D. ISG15: in sickness and in health. Trends Immunol. 2017;38:79–93. doi: 10.1016/j.it.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Jorgensen S.C.J., Tse C.L.Y., Burry L., Dresser L.D. Baricitinib: a review of pharmacology, safety, and emerging clinical experience in COVID-19. Pharmacotherapy. 2020;40:843–856. doi: 10.1002/phar.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J., Wang Q. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Struckhoff J.J., Schneider J., Martinez-Sobrido L., Wolff T., García-Sastre A., Zhang D.-E., Lenschow D.J. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 2009;83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Xiao H., Tam J.P., Liu D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005;579:2387–2396. doi: 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Fung T.S., Huang M., Fang S.G., Zhong Y., Liu D.X. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013;87:8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Wang X., Huang M., Tam J.P., Liu D.X. Regulation of the p38 mitogen-activated protein kinase and dual-specificity phosphatase 1 feedback loop modulates the induction of interleukin 6 and 8 in cells infected with coronavirus infectious bronchitis virus. Virology. 2011;420:106–116. doi: 10.1016/j.virol.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Liu D.X. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology. 1998;245:303–312. doi: 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Liang J.Q., Fung T.S. Encyclopedia of Virology. Elsevier; 2021. Human coronavirus-229e, -OC43, -NL63, and -HKU1 (coronaviridae) pp. 428–440. [DOI] [Google Scholar]
- Liu D.X., Ng Y., Fung T.S. Infectious Bronchitis Virus. In: Avian Virology: Current Research and Future Trends. Caister Academic Press; Norfolk, UK: 2019. pp. 133–178. [DOI] [Google Scholar]
- Liu Y., Nie H., Mao R., Mitra B., Cai D., Yan R., Guo J.-T., Block T.M., Mechti N., Guo H. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loevenich S., Spahn A.S., Rian K., Boyartchuk V., Anthonsen M.W. Human metapneumovirus induces IRF1 via TANK-binding kinase 1 and type I IFN. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.563336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmgaard L. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- Meraro D., Gleit-Kielmanowicz M., Hauser H., Levi B.-Z. IFN-stimulated gene 15 is synergistically activated through interactions between the myelocyte/lymphocyte-specific transcription factors, PU.1, IFN regulatory factor-8/IFN consensus sequence binding protein, and IFN regulatory factor-4: characterization of a new subtype of IFN-stimulated response element. J. Immunol. Baltim. Md. 2002;168:6224–6231. doi: 10.4049/jimmunol.168.12.6224. 1950. [DOI] [PubMed] [Google Scholar]
- Mirzalieva O., Juncker M., Schwartzenburg J., Desai S. ISG15 and ISGylation in human diseases. Cells. 2022;11:538. doi: 10.3390/cells11030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfopoulou S., Brown J.R., Davies E.G., Anderson G., Virasami A., Qasim W., Chong W.K., Hubank M., Plagnol V., Desforges M., Jacques T.S., Talbot P.J., Breuer J. Human coronavirus OC43 associated with fatal encephalitis. N. Engl. J. Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- Moser M.J., Holley W.R., Chatterjee A., Mian I.S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C., Pazos M., Delgado I., Murk W., Mungamuri S.K., Lee S.W., García-Sastre A., Moran T.M., Aaronson S.A. p53 serves as a host antiviral factor that enhances innate and adaptive immune responses to influenza A virus. J. Immunol. Baltim. Md. 2011;187:6428–6436. doi: 10.4049/jimmunol.1101459. 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.-T.K., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirudeen A.M.A., Liu D.X. Gene expression profiling by microarray analysis reveals an important role for caspase-1 in dengue virus-induced p53-mediated apoptosis. J. Med. Virol. 2009;81:1069–1081. doi: 10.1002/jmv.21486. [DOI] [PubMed] [Google Scholar]
- Negishi H., Taniguchi T., Yanai H. The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harbor Perspect. Biol. 2018;10:a028423. doi: 10.1101/cshperspect.a028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.H., Espert L., Mechti N., Wilson D.M. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry. 2001;40:7174–7179. doi: 10.1021/bi010141t. [DOI] [PubMed] [Google Scholar]
- Okumura A., Lu G., Pitha-Rowe I., Pitha P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Osei Kuffour E., König R., Häussinger D., Schulz W.A., Münk C. ISG15 deficiency enhances HIV-1 infection by accumulating misfolded p53. mBio. 2019;10:e01342. doi: 10.1128/mBio.01342-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Yang S.W., Park J.M., Ka S.H., Kim J.-H., Kong Y.-Y., Jeon Y.J., Seol J.H., Chung C.H. Positive feedback regulation of p53 transactivity by DNA damage-induced ISG15 modification. Nat. Commun. 2016;7 doi: 10.1038/ncomms12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-J., Kim H.-K., Song D.-S., An D.-J., Park B.-K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012;86:5964. doi: 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng Y.-C., Lenschow D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N., Evans B., Levy D., Fahey D., Knight E., Darnell J.E. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. USA. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C., Aaronson S.A., Munoz-Fontela C. Dual role of p53 in innate antiviral immunity. Viruses. 2010;2:298–313. doi: 10.3390/v2010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Law Y.C., Liu D.X. A single amino acid mutation in the spike protein of coronavirus infectious bronchitis virus hampers its maturation and incorporation into virions at the nonpermissive temperature. Virology. 2004;326:288–298. doi: 10.1016/j.virol.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Yang J.S., Oh J.S., Han J.H., Park B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21:1833–1842. doi: 10.1016/s0264-410x(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Gao Y., Dozmorov I., Malladi V., Saha I., McDaniel M.M., Parameswaran S., Liang C., Arana C., Zhang B., Wakeland B., Zhou J., Weirauch M.T., Kottyan L.C., Wakeland E.K., Pasare C. IRF1 governs the differential interferon-stimulated gene responses in human monocytes and macrophages by regulating chromatin accessibility. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnweiss A., Ksienzyk A., Klages K., Rand U., Grashoff M., Hauser H., Kröger A. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. Baltim. Md 1950. 2010;184:5179–5185. doi: 10.4049/jimmunol.0902264. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Kawakami T., Taniguchi T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531-4538.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waisner H., Grieshaber B., Saud R., Henke W., Stephens E.B., Kalamvoki M. SARS-CoV-2 harnesses host translational shutoff and autophagy to optimize virus yields: the role of the envelope (E) protein. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.03707-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liao Y., Yap P.L., Png K.J., Tam J.P., Liu D.X. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J. Virol. 2009;83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C.M., Trobaugh D.W., Sun C., Lucas T.M., Diamond M.S., Ryman K.D., Klimstra W.B. The interferon-induced exonuclease ISG20 exerts antiviral activity through upregulation of type I interferon response proteins. mSphere. 2018;3:e00209–e00218. doi: 10.1128/mSphere.00209-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B.-J., Chan K.-H., Yuen K.-Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Xu L.H., Yamada Y., Liu D.X. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhou X., Wang W., Wang Y., Yin Y., van der Laan L.J.W., Sprengers D., Metselaar H.J., Peppelenbosch M.P., Pan Q. IFN regulatory factor 1 restricts hepatitis E virus replication by activating STAT1 to induce antiviral IFN-stimulated genes. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016;30:3352–3367. doi: 10.1096/fj.201600356R. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Bruneau R.C., Brennan G., Rothenburg S. Battle royale: innate recognition of poxviruses and viral immune evasion. Biomedicines. 2021;9:765. doi: 10.3390/biomedicines9070765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L.X., Liang J.Q., Zhu Q.C., Dai G., Li S., Fung T.S., Liu D.X. A gammacoronavirus, avian infectious bronchitis virus, and an alphacoronavirus, porcine epidemic diarrhea virus, exploit a cell survival strategy by upregulating cFOS to promote virus replication. J. Virol. 2021;95 doi: 10.1128/JVI.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L.X., Yang B., Fung T.S., Chen R.A., Liu D.X. Transcriptomic analysis reveals crucial regulatory roles of immediate-early response genes and related signaling pathways in coronavirus infectious bronchitis virus infection. Virology. 2022;575:1–9. doi: 10.1016/j.virol.2022.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Shen Q., Wu S., Chen S., Lomakin I.B., Xiong Y. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol. Cell. 2020;80:1055–1066.e6. doi: 10.1016/j.molcel.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Liao Y., Fang S., Tam J.P., Liu D.X. Up-regulation of Mcl-1 and Bak by coronavirus infection of human, avian and animal cells modulates apoptosis and viral replication. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]