Abstract
The molecular mechanism of the pathological impact of COVID-19 in lung cancer patients remains poorly understood to date. In this study, we used differential gene expression pattern analysis to try to figure out the possible disease mechanism of COVID-19 and its associated risk factors in patients with the two most common types of non-small-cell lung cancer, namely, lung adenocarcinoma and lung squamous cell carcinoma. We also used network-based approaches to identify potential diagnostic and molecular targets for COVID-19-infected lung cancer patients. Our study showed that lung cancer and COVID-19 patients share 36 genes that are expressed differently and in common. Most of these genes are expressed in lung tissues and are mostly involved in the pathogenesis of different respiratory tract diseases. Additionally, we also found that COVID-19 may affect the expression of several cancer-associated genes in lung cancer patients, such as the oncogenes JUN, TNC, and POU2AF1. Moreover, our findings suggest that COVID-19 may predispose lung cancer patients to other diseases like acute liver failure and respiratory distress syndrome. Additionally, our findings, in concert with published literature, suggest that molecular signatures, such as hsa-mir-93-5p, CCNB2, IRF1, CD163, and different immune cell-based approaches could help both diagnose and treat this group of patients. Altogether, the scientific findings of this study will help formulate appropriate management measures and guide the development of diagnostic and therapeutic measures for COVID-19-infected lung cancer patients.
Keywords: COVID-19, Lung cancer, Risk factors, Diagnosis, Treatment
1. Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease-2019 (COVID-19), has infected more than 590 million people and resulted in the death of 6.4 million infected individuals worldwide since its emergence (Source: Johns Hopkins University Coronavirus Resource Centre). Although SARS-CoV-2 can affect almost every part of the body subsequent to infection, lung damage remains the leading cause of COVID-19-associated mortality [1]. Furthermore, the presence of any co-existing disease may increase the susceptibility to infection and the severity of illness in COVID-19 patients [2]. For example, patients with hypertension, hepatitis, diabetes, cardiovascular disease, chronic renal disease, lung cancer (LC), blood cancer, and infectious diseases remain the most vulnerable groups to COVID-19 [3]. Among them, LC is one of the most commonly diagnosed and leading causes of cancer-related mortality and is accountable for approximately 1.8 million deaths worldwide [4,5]. Studies have found that LC patients are more likely to develop COVID-19 (frequency: 25%) than other types of cancer [6]. LC can be divided into two vital groups known as small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). Majority of the LC falls into the NSCLC group which involves the two most frequently diagnosed types of LC i.e., lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) [7]. For example, more than 500,000 annual deaths are recorded from lung adenocarcinoma making it the most frequent type of LC worldwide [8]. On the contrary, the number of mortalities by LUSC is over 400,00 annually throughout the world which makes it the second most widespread type of LC [9].
Previous studies suggest that intensive care unit (ICU) and mechanical ventilation supports are highly required for such patients with pre-existing lung injuries from cancer when compared to COVID-19 patients without comorbidities [10,11]. It is hypothesized that LC patients are at increased risk for COVID-19-related medical complications due to the abnormality in the pulmonary and alveolar structure that is generated by therapeutic procedures and LC itself. These modifications in the alveolar and pulmonary epithelium create a new milieu that promotes the production of proinflammatory cytokines including interleukin-1 (IL-1), IL-6, and tumor necrosis factor-, which in turn help the invading virus develop pneumonitis [12]. Moreover, the immunocompromised conditions in LC patients with immune cells fighting against cancer cells also leave them vulnerable to SARS-CoV-2-related medical manifestations [13]. It is also hypothesized that the virus manifests its pathogenicity with a higher level of macrophage activation, interferons, T cell lymphocytes, and interleukins which make LC patients vulnerable to medical consequences like developing acute respiratory distress syndrome (ARDS). However, the specific pathophysiology of SARS-CoV-2 in LC patients remains poorly understood to date [14].
Bioinformatics-based meta-analysis could assist in the understanding of the pathology of different complex diseases including cancer with multiple comorbidities [15,16]. However, previous studies aimed to analyze the pathological impact of COVID-19 in NSCLC patients are limited by looking at one type of NSCLC histology (LUAD more specifically), analyzing an inadequate transcriptome (i.e., artificially induced infection in LC cells grown in culture medium) and ignoring the risk of other comorbidities [[17], [18], [19]].
In this study, we explored the pathological mechanism of COVID-19 in two most common forms of LC i.e., LUAD and LUSC by investigating transcriptome expression patterns in the lung tissues collected from LC and COVID-19 patients. We also assessed the risk of comorbidities in LC patients infected with SARS-CoV-2 and identified the molecular signatures that could help proper screening and treatment of this group of patients posing a higher risk of COVID-19. The scientific findings of this study will allow a better understanding of the pathophysiological mechanism to help make diagnostic and therapeutic decisions in SARS-CoV-2-infected LC patients.
2. Materials and methods
2.1. Dataset selection and retrieval
In this step, we explored the National Centre for Biotechnology Information Gene Expression Omnibus (NCBI-GEO) database to select the datasets for this study. For the COVID-19 samples, we retrieved the GSE151764 dataset which contains the high-throughput sequencing data of the whole transcriptome (RNA-seq) collected from 34 to 16 postmortem specimens of LUNG tissues from the patients who died of COVID-19 and non-infectious diseases, respectively, with an average sample collection time of 20 h preceding the decease [20]. We retrieved the GSE116959 (LUAD: 57, Normal: 11) and GSE115002 (LUAD: 52, Normal: 52) microarray datasets that contain the whole transcriptome expression data from the lung and adjacent normal tissue from LUAD patients [21,22]. For the LUSC samples, we collected the GSE30219 (LUSC: 60, Normal: 14) and GSE2088 (LUSC: 48, Normal: 30) microarray datasets incorporating the total mRNA expression profile from LUSC and adjacent normal lung tissue samples [23,24]. We omitted the data from samples cultured in medium and received any treatment.
2.2. Differential gene expression analysis and identification of shared DEGs between COVID-19 and LC patients
The raw read count matrix of the RNA-seq data was preprocessed and normalized in R studio with DESeq2 package [25,26]. Initially, we removed the items containing low total read counts (<50) in the whole sample. Since the dataset contains multiple samples from the same patient, we collapsed the technical replicates and summed the gene expression count values. Thereafter, the count matrix was normalized and the differentially expressed genes were identified by performing the automated Wald t-test with the help of DESeq2 package.
For the microarray datasets, we employed the LIMMA (Linear Models for Microarray Data) package to identify the DEGs [27]. Primarily, the expression matrix was checked and log2 transformation was applied to the datasets where applicable. The datasets were then normalized using the Quantile normalization method with Bioconductor package [28]. Finally, the DEGs were identified by performing moderated t-test with the help of LIMMA package. Most significant DEGs were then selected based on the Benjamini–Hochberg applied multitest adjusted p-value<0.01 and |log2FC|>1.0 cutoffs for all the datasets. Finally, we utilized the InteractiVenn online tool (http://www.interactivenn.net/) to identify the shared DEGs between COVID-19 and LUAD (COVID-19vsLUAD) and LUSC (COVID-19vsLUSC) patients [29].
2.3. Examining the molecular characteristics of the shared DEGs and their translational products
In this step, we queried the total shared DEGs in ShinyGO server (http://bioinformatics.sdstate.edu/go/) to examine their chromosomal location [30]. Thereafter, we explored the Annotation feature of the Metascape server (https://metascape.org/gp/index.html#/main/step1) to optimize the tissue-specific distribution and the classification of the translated proteins of the DEGs [31]. Thereafter, we visualized the relevant items using the ggplot2 package in R studio and CIRCOS online tool (http://circos.ca/) [32,33]. Lastly, we utilized the NetworkAnalyst web-based server (https://www.networkanalyst.ca/) to understand the lung tissue-specific protein-protein interaction (PPI) network of the shared DEGs [34] where we searched our genes of interest against the integrated STRING database with high confidence cutoff threshold of >0.9 to construct the PPI [35]. After that, we applied a degree cutoff of 3.0 to identify the most connected nodes. We then downloaded the network and customized it in Cytoscape (v 3.7.2) app [36].
2.4. Analyzing the correlation between the expression of DEGs and different diseases and comorbidities
Initially, we utilized the DisGeNET plugin of the Cytoscape tool to examine the association between the DEGs and different respiratory tract diseases with default parameters [37]. After that, we explored the DisGeNET online server with help of the Metascape web-utility tool to assess the association of the DEGs with the top 20 diseases. We used a p-value cutoff of 0.01 and a minimum enrichment score cutoff of 1.5 for the associations to be established. Later, the Expression Atlas server (https://www.ebi.ac.uk/gxa/home) was used to evaluate the expression pattern of DEGs in different human disease studies including sample data without treatment and/or stimulus [38]. Finally, we investigated The Network of Cancer Genome database (http://ncg.kcl.ac.uk/) to identify the known cancer-related genes from our gene pool (DEGs) keeping the parameter values at default [39]. The data generated from the aforementioned analyses were then visualized utilizing the ggplot2 package in R studio.
2.5. Analysis of the COVID-19-related ontology terms and functional relevance of the DEGs
In this step, we first searched our genes of interest against the Coronascape, a curated database that enlists the top 300 deregulated genes from different COVID-19-related omics studies, through the Metascape server to observe the expression pattern of the DEGs in different COVID-19 studies [31]. Later, the KEGG pathway analysis on the identified DEGs was performed using the Clusterprofiler package in R studio [40]. We also analyzed the top gene ontology terms i.e., biological processes (BP), molecular function (MF), and cellular component (CC) of the DEGs with the help of the Clusterprofiler package. The results were then analyzed based on multi-test corrected p-value and the significant top 15 gene ontology terms were then visualized using enrichplot and ggplot2 package [40].
2.6. Construction of the protein-protein interaction network of DEGs and identification of hub proteins
We constructed the generic PPI of the proteins expressed by the DEGs using the NetworkAnalyst server and selecting the STRING database with a high overall confidence cutoff (0.900) [41]. Thereafter, we employed the most accurate local (i.e., MCC) and global-based (i.e., EPC, Closeness) methods for the extraction of the top 10 most connected nodes (hub proteins) from the generic PPI using the cytoHubba plugin in Cytoscape tool [42]. Finally, we considered the overlapping hub proteins from the 3 hub networks as the most significant hubs. Later, we analyzed the top biological processes terms on the overlapping hubs using the Cluterprofiler package in R studio.
2.7. Identification of transcriptional and post-transcriptional regulatory signatures and drug targets of the DEGs
In this step, we initially searched the DEGs against the miRTARbase database which is a publicly available repository of the experimentally validated miRNA-gene interaction database using the NetworkAnalyst web-based server [43]. After that, we explored the ENCODE, an experimentally validated and curated database, to identify the gene-transcription factor (TF) interaction network via the NetworkAnalyst online tool [44]. In both cases, a degree cutoff of <3.0 was applied to generate the mostly connected gene-miRNA and gene-TFs targets of the DEGs. Later, the networks were downloaded and customized using the Cytoscape server. Finally, the DEGs were quired against the DrugBank database to identify the protein targets and their respective drug candidates [45].
2.8. Determination of the immunophenotypes of the DEGs in LUAD and LUSC patients
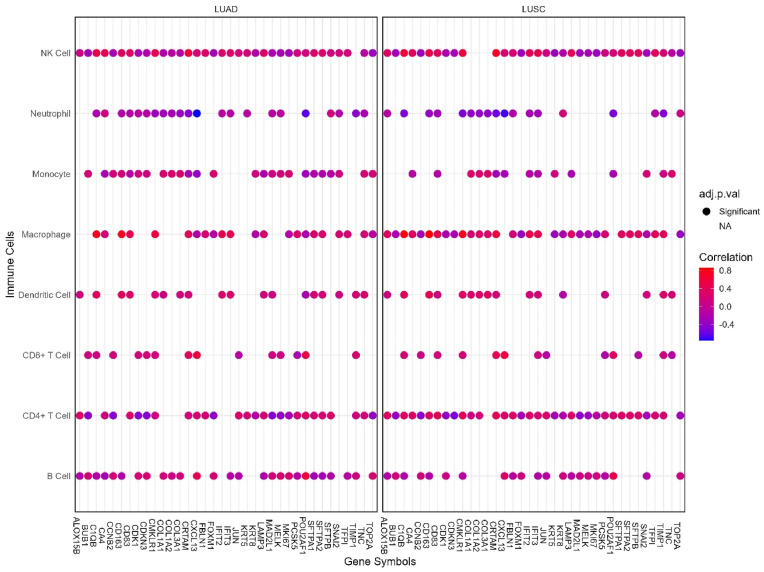
We examined the association between the mRNA level expression of the DEGs and the infiltration levels of different immune cells i.e., B cell, T cell, Macrophage, Neutrophil, Monocyte, Natural Killer (NK) cells in LUAD and LUSC microenvironment by investigating the immune module of the Gene Set Cancer Analysis (GSCA) server (http://bioinfo.life.hust.edu.cn/GSCA/#/) [46]. The association was then analyzed based on a significant adjusted p-value cutoff (<0.05) and spearman correlation coefficient.
2.9. Validation of the DEG expression on TCGA and independent cohorts and their association analysis with LC patients’ survival
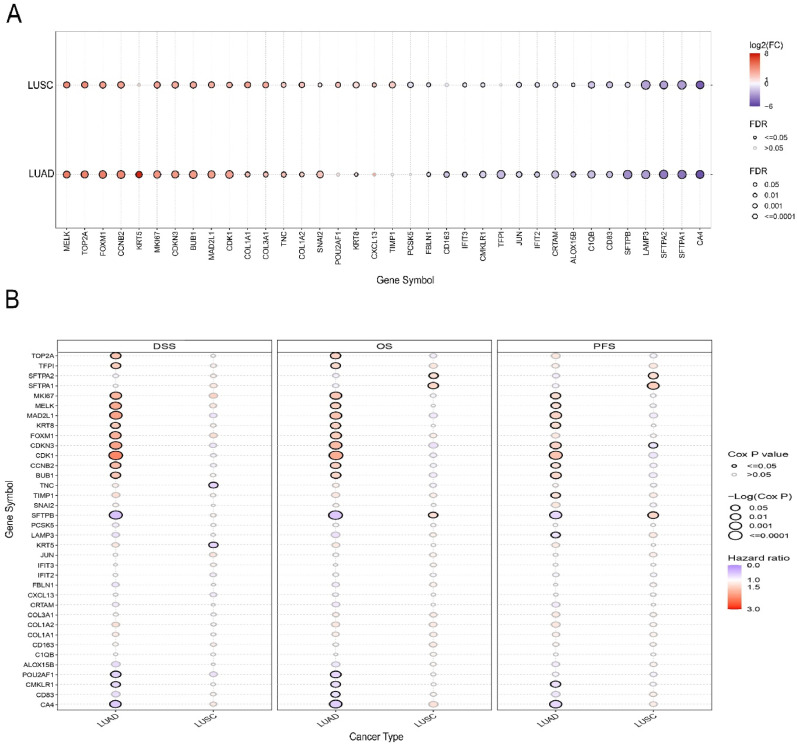
Herein, we again utilized the GSCA server to evaluate the differential gene expression pattern of the DEGs in TCGA LUAD and LUSC cohorts. The GSCA server collects and preprocesses the raw cancer tissue and adjacent normal tissue RNA-seq data the TCGA cohorts and allows DEG analysis with statistical methods [46]. Additionally, it also collects clinical data and assesses the association between gene expression and cancer patients’ survival rates by performing a log-rank t-test and measuring the cox-proportional hazard ratio. We searched our differentially expressed gene pools against the LUAD and LUSC cohorts and observed the pattern of DEG expression and their impact on disease-specific survival (DSS), overall survival (OS) and progression-free survival (PFS) of LUD and LUSC patients using the expression module of GSCA server. Alongside this, we also investigated the expression pattern of the shared DEGs in one COVID-19 (GSE171110) and one lung cancer (GSE74706) dataset. The GSE171110 dataset contains the whole blood transcriptome (RNA-seq) data of 44 COVID-19 patients and 10 age and sex-matched healthy individuals [47]. On the other hand, GSE74706 is a GeneChip dataset that contains the transcriptome profile of cancer tissues (LUSC: 8; LUAD: 10) and adjacent normal tissues (n = 18) of NSCLC patients [48]. The DEG analysis was performed utilizing the same protocol of the mainstream analysis as in step 2.2 and significant DEGs were identified using FDR-adjusted p-value cutoff of 0.05 and absolute log2FC cutoff of 1. The metagenomic pipeline utilized in this paper has been summarized in Fig. 1 .
Fig. 1.
Strategic flowchart of the experimental procedures utilized in the study.
3. Results
3.1. The molecular characteristics of the common DEGs between COVID-19 and LC patients
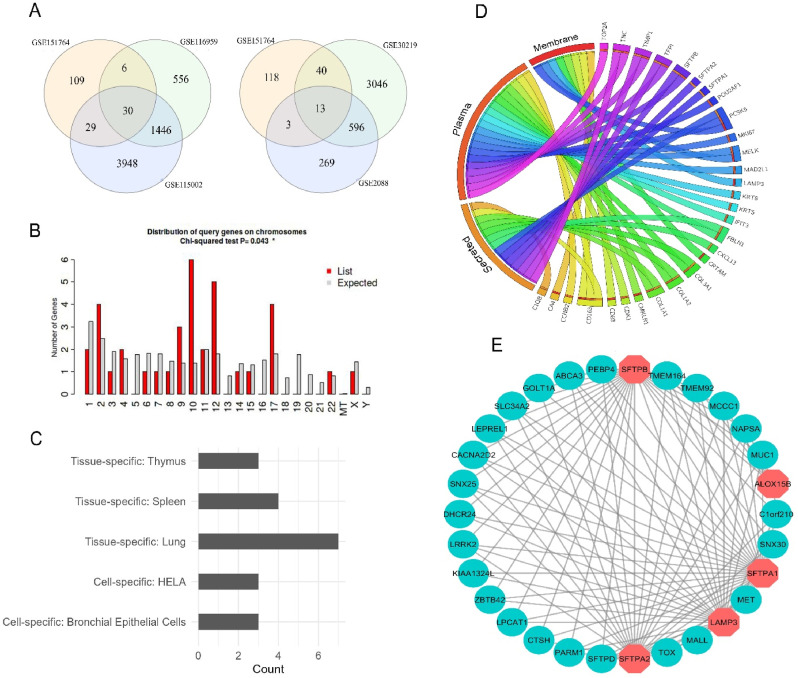
DEG analysis revealed that COVID-19 samples shared a total of 30 and 13 DEGs with LUAD and LUSC datasets, respectively (Fig. 2 A). Further inspection revealed that COVID vs LUAD and COVID vs LUSC datasets again shared 7 genes (COL1A1, MAD2L1, TOP2A, COL3A1, CDKN3, IFIT2 and CXCL13) in common. This resulted in a total of 36 final DEGs that were utilized in the next phases of the experiment. Chromosomal distribution analysis on the common DEGs revealed that most of the genes are located in chromosomes 2, 10, 12, and 17 (Fig. 2B). Furthermore, DEGs were not found on chromosomes 5, 13, 16, 18–21, or the mitochondrial (MT) or Y chromosomes. More information, such as gene description and specific chromosomal location of the DEGs, is available in Supplementary Table S1. In addition, tissue-specific expression analysis revealed that the majority of DEGs were mostly expressed in lung tissues. In particular, several genes were discovered to exhibit thymus tissue and bronchial epithelial cell-specific expression, while the remaining genes were revealed to be spleen tissue and HELA cell-specific. (Fig. 2C). The classification analysis of the translational products of shared DEGs based on their functional position inside the human body revealed that most of the proteins were plasma proteins (20) followed by secreted (14) and membrane proteins (8) (Fig. 2D). The lung-specific PPI network analysis of the common DEGs revealed that five of the deregulated genes (SFTPB, ALOX15B, SFTPA1, LAMP3 and SFTPA2) interact with a total of 25 different proteins that are specifically expressed in lung tissues (Fig. 2E).
Fig. 2.
(A) The venn diagram representation of the shared DEGs between COVID-19 and LUAD patients (left panel) and COVID-19 and LUSC patients (right panel). (B) The histogram representation of the chromosomal distribution of the shared DEGs. (C) Bar diagram reflecting the number of DEGs specifically expressed in different tissues. (D) Chord diagram delineating the functional distribution of the DEGs in different locations inside the human body. (E) The PPI network of the lung tissue-specific interactions of the translated proteins of DEGs with different proteins. The octagonal nodes represent the proteins expressed by the DEGs and the circular nodes represent interacting proteins that are exclusively expressed in lung tissues.
3.2. Association and expression pattern of the DEGs in different diseases and comorbidities
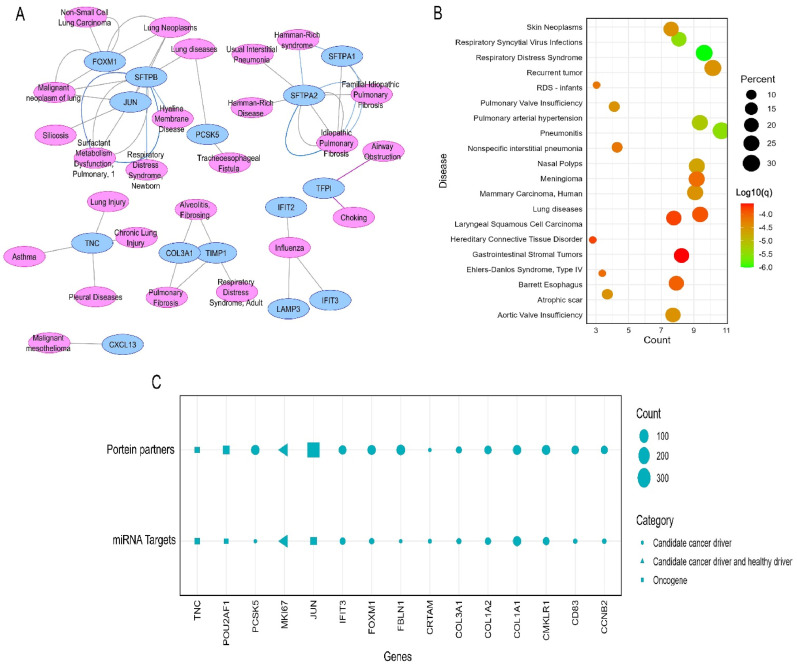
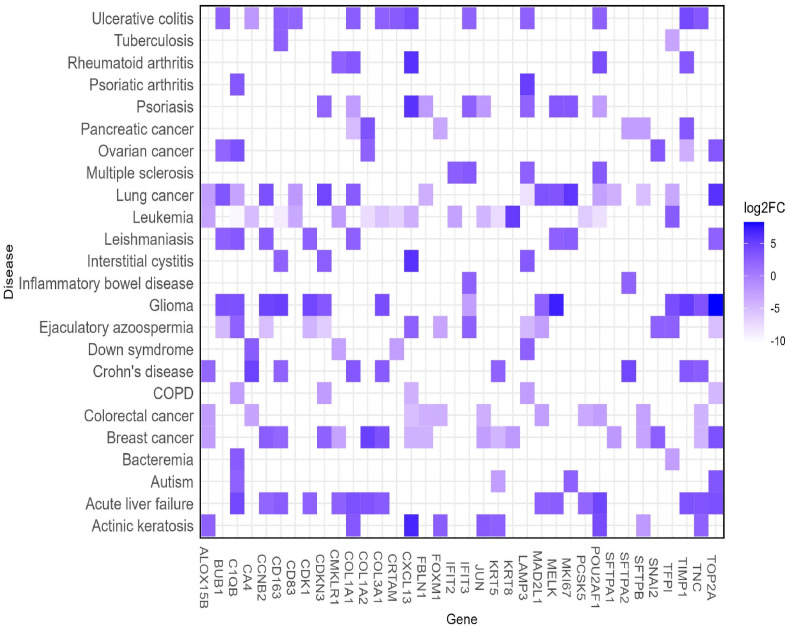
The association analysis between the shared DEGs and other respiratory tract diseases reported that many different genes i.e., FOXM1, SFTPB, JUN, PCSK5, SFTPA2 were involved in numerous respiratory tract diseases such as lung neoplasm, asthma, respiratory distress syndrome (RDS), IPF and chronic lung injury (Fig. 3 A). The inspection of the DEGs in DisGenet irrespective of specific disease groups unsurprisingly revealed that the highest proportion of the genes is involved in critical lung diseases like RDS, respiratory syncytial virus infection, and pulmonary valve insufficiency (Supplementary Table S2) (Fig. 3B). According to the results of our query in the NCG dataset, 15 of the 36 DEGs were cancer-associated genes, including a total of three oncogenes i.e., JUN, POU2AF1, and TNC. (Fig. 3C). The analysis of the shared DEGs in the Expression Atlas dataset reported that the majority of the genes are also differentially expressed in different diseases including carcinoma as the predominant one (Fig. 4 ). More specifically, the DEGs were found to be mostly deregulated in LC (n = 17) followed by leukemia (n = 16), breast cancer (n = 16), glioma (n = 13), and other cancer types. A noticeable number of the DEGs (n = 15) were found to be upregulated in acute kidney failure. Chronic obstructive pulmonary disease (COPD), tuberculosis, bacteremia, and leishmaniasis were mentionable among respiratory diseases other than carcinoma and other infectious disease types. The DEGs were also discovered to be differentially expressed in a few autoimmune disorders, such as multiple sclerosis and rheumatoid arthritis (Fig. 4).
Fig. 3.
(A) The interaction network delineating the association between the proteins expressed by the DEGs and different respiratory tract diseases. (B) The dot-plot representation of the enrichment of the DEGs among different diseases was obtained from the DisGenet database. (C) The dot-plot representation of enrichment of the 15 cancer-related genes identified among the total 36 differentially expressed genes in this study.
Fig. 4.
Heatmap reflecting differential expression pattern of the 36 DEGs in different diseases and comorbidities. The color scale represents the log2FC of the DEGs in those respective disease types. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. The ontology terms and functional relevance of the DEGs in relation to COVID-19
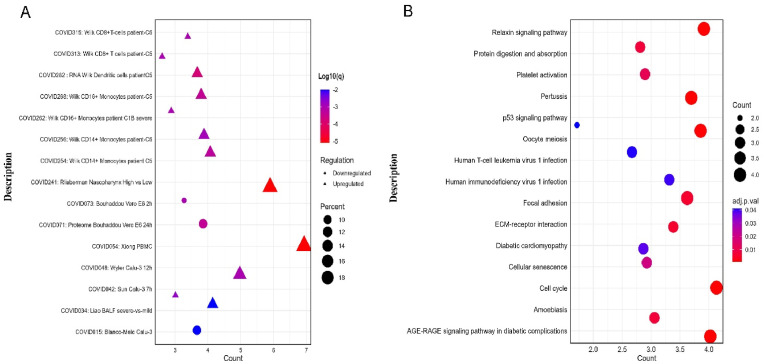
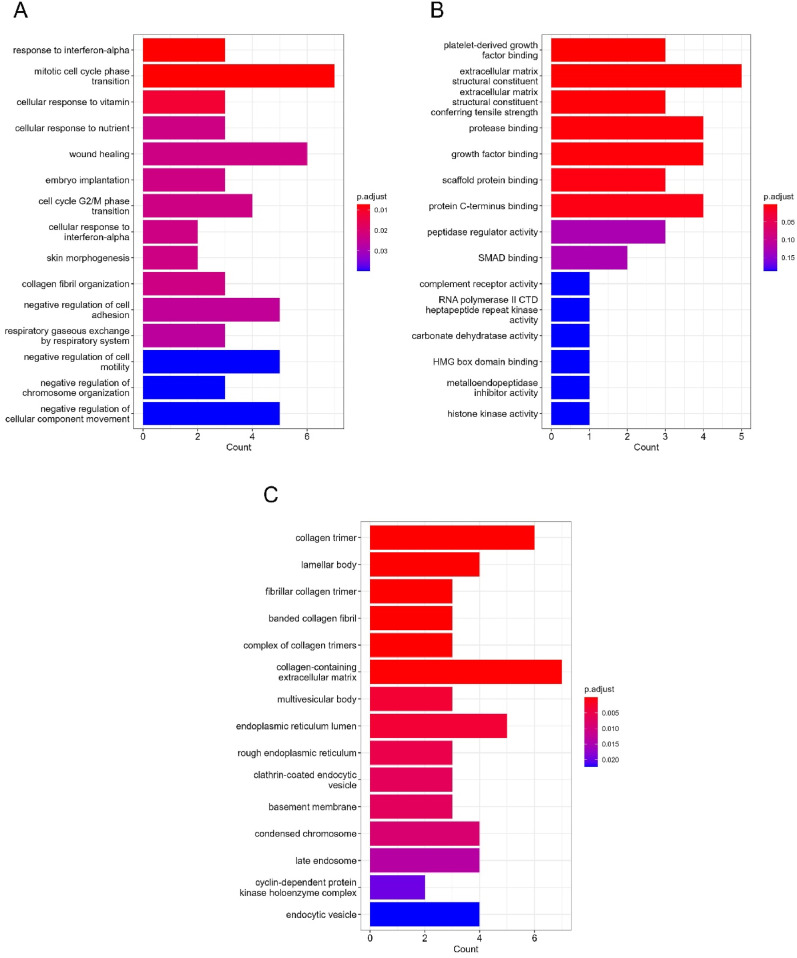
COVID-19 ontology term analysis revealed that the DEGs are disproportionately upregulated and/or downregulated in all the selected COVID-19 transcriptomic and proteomic studies (Supplementary Table S3) (Fig. 5 A). The KEGG pathway analysis on the common DEGs revealed that the majority of the genes were involved in crucial biological pathways like the relaxin signaling pathway, protein digestion and absorption, cell cycle, platelet activation, and cellular senescence (Fig. 5B). Several genes were also found to be associated with several other disease processes, including diabetic cardiomyopathy and pertussis. We evaluated the enrichment of the common DEGs in different biological function categories by examining their gene ontology terms based on BP, MF, and CC. Response to interferon-alpha (IFN-), mitotic cell cycle phase transition, control of chromosomal structure, negative regulation of cell adhesion, cellular response to vitamins and minerals, and so on were identified as the main biological processes shared by the DEGs (Supplementary Table S4) (Fig. 6 A). Platelet-derived growth factor binding, protease binding, peptidase regulator activity, complement receptor activity, and histone kinase activity were the most important molecular functions of the shared DEGs, followed by extracellular matrix organization, growth factor binding, and scaffold protein binding. (Supplementary Table S5) (Fig. 6B). The cellular component ontology term analysis on the DEGs revealed that the majority of the DEGs were functional in biological compartments like collagen trimers, multivesicular body, endoplasmic reticulum lumen, fibrillar collagen trimmer, banded collagen fibril, lamellar body, and collagen-containing extracellular matrix (Supplementary Table S6) (Fig. 6C).
Fig. 5.
(A) Dot-plot representing the regulation pattern of the common DEGs in different COVID-19-related studies from the Metascape server. The y-axis represents the COVID-19-related studies with authors' names. (B) The dot-plot representation of the signaling pathway involved with the shared DEGs. The x-axis represents the overlapping gene counts with the enrichment terms. The color gradient indicates the adjusted p-value distribution of the DEGs within those ontology terms (low: red, high: blue).
Fig. 6.
The bar plot representation of the top 15 gene ontology terms of the shared DEGs: (A) biological processes, (B) molecular function and (c) cellular component. The y-axis represents the ontology terms and the x-axis represents the counts shared with the DEGs. The color gradient indicates the adjusted p-value distribution of the DEGs within those ontology terms (low: red, high: blue).
3.4. The PPI network of the proteins expressed by shared DEGs and its relative hub proteins
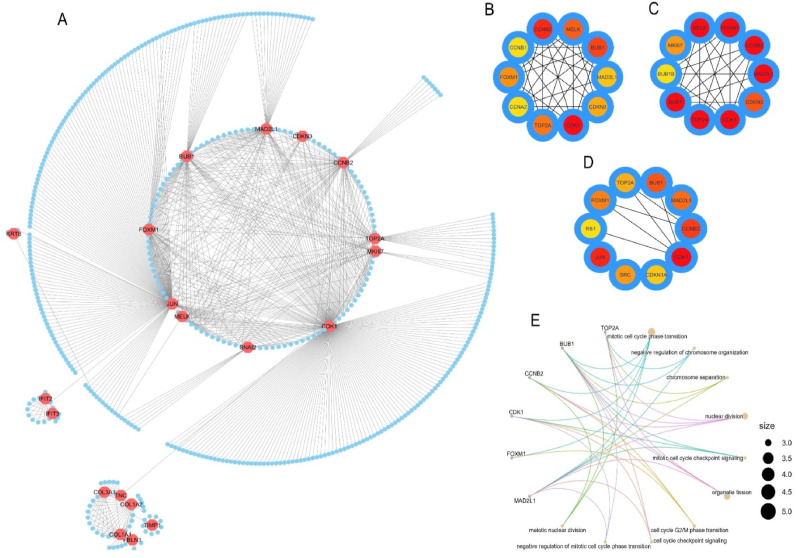
The shared DEGs were utilized to produce a generic PPI network which resulted in a network containing 469 nodes and 771 edges (Fig. 7 A). We then applied different global and local-based algorithms i.e., MCC (Fig. 7B), EPC (Fig. 7C) and Closeness (Fig. 7D) to identify the top 10 most connected nodes in the generic PPI. Each of the hub protein identification methods constructed a network containing the top 10 most connected nodes from the initial PPI network. Later we discovered that all 3 generated hub networks shared 6 proteins in common (data not shown here). Therefore, we considered the 6 overlapping proteins among the 3 hub networks i.e., CDK1, BUB1, TOP2A, CCNB2, FOXM1, and MAD2L1 as the most significant hub proteins. The major three biological functions of the most significant hub proteins were mitotic cell cycle transition, nuclear division, and organelle fission as observed from subsequent BP analysis (Fig. 7E). Additionally, the hub proteins were also involved in other biological functions like negative regulation of chromosome organization, chromosome separation, mitotic cell cycle checkpoint signaling, cell cycle G2/M phase transition.
Fig. 7.
(A) The PPI network of the proteins expressed by the shared DEGs between COVID-19 and lung cancer patients. The nodes represent the proteins and the edges represent the interactions. Octagonal nodes represent the proteins expressed by the DEGs and circular nodes represent the interacting partners. The network of top 10 connected interacting proteins (hub proteins) was identified by (B) MCC, (C) EPC, and (D) Closeness methods. (E) The cnet-plot representation of the top 10 ontology terms (biological processes) of the overlapping hub proteins. The color gradient scale indicates the strength of interactions with partner proteins (i.e., higher intensity of color refers to a greater number of interactions and vice versa).
3.5. The transcriptional and post-transcriptional regulatory signatures and interacting drug molecules of the DEGs
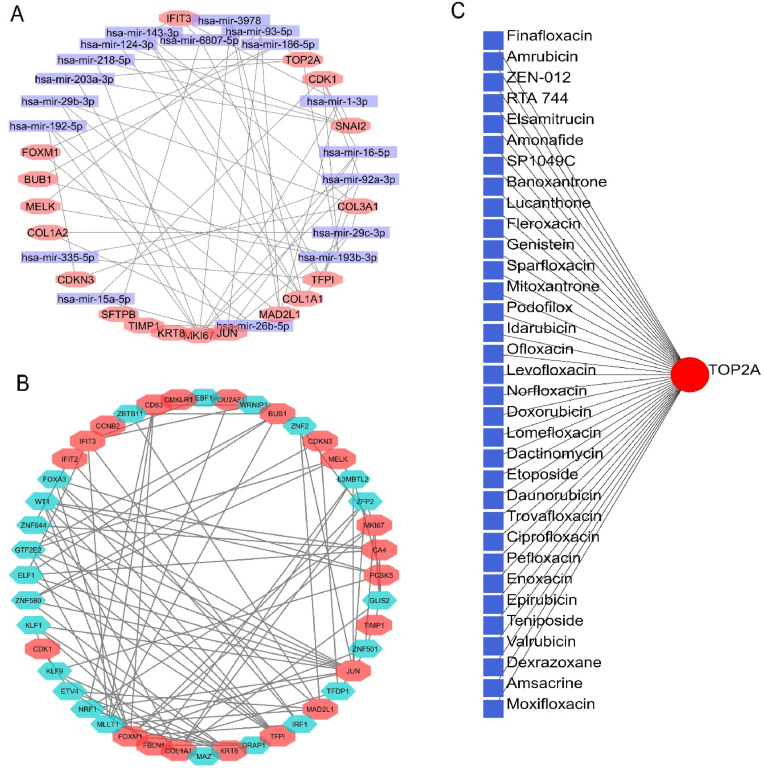
The gene-miRNA interaction analysis of the shared DEGs stated that a total of 18 different DEGs shared interactions with 19 different miRNAs (hsa-mir-192-5p, hsa-mir-29b-5p, hsa-mir-203a-3p, hsa-mir-218-5p, hsa-mir-6807-5p, hsa-mir-192-5p, hsa-mir-3978, hsa-mir-93-5p, hsa-mir-186-5p, hsa-mir-1-3p, hsa-mir-16-5p, hsa-mir-92a-3p, hsa-mir-29c-3p, hsa-mir-193b-3p, hsa-mir-26b-5p, hsa-mir-15a-5p, hsa-mir-335-5p) (Fig. 8 A). Additionally, we recorded several different transcription factor targets for multiple DEGs by gene-TF interaction analysis. More specifically, out of the 36 identified DEGs, we reported a total of 23 DEGs that are targeted by 21 different TFs (ZBTB11, EBF1, WRNIP1, ZNF2, L3MBTL2, ZFP2, GLIS2, ZNF501, TFDP1, IRF1, DRAP1, MAZ, MLLT1, NRF1, ETV4, KLF9, KLF1, ZNF580, ELF1, GTF2E2, ZNF644, WT1, FOXA3) (Fig. 8B). Finally, the protein-drug interaction analysis reported TOP2A as a potential protein target whose activity could be regulated by different currently available drugs in the markets. The TOP2A protein interacted with 33 different drug molecules including many antineoplastic agents i.e., Amrubicin, Amonafide, Banoxantrone, Doxorubicin, chemotherapeutic agents i.e., Elsamitrucin, Etoposide and anti-infectious drugs i.e., Finafloxacin, Lucanthone, Moxifloxacin, and few others (Fig. 8C).
Fig. 8.
(A) The gene-miRNA network of the shared DEGs. The rectangular nodes represent individual miRNA and octagonal nodes represent DEGs. (B) The gene-TF interaction network of the DEGs. The hexagonal nodes correspond to interacting proteins whereas the octagonal nodes indicate shared DEGs. (C) The protein-drug interaction network of the proteins encoded by shared DEGs.
3.6. The association between the DEGs expression and the level of immune abundance in LUAD and LUSC patients
According to our findings, most of the DEGs are linked to the infiltration levels of the chosen immune cells (B cells, T cells, NK cell, Neutrophil, Monocytes, and Macrophages) in LUAD and LUSC patients. (Fig. 9 ). Overall, the infiltration level of NK cells showed a significant correlation with most of the DEGs followed by CD4+ T cell, B cell and macrophage infiltration levels in both LUAD and LUSC patients. However, a greater number of DEGs were reported to be associated with the abundance level of NK cells in LUAD microenvironment than in LUSC patients. CD4+ T cells and macrophages, on the contrary, were observed to be associated with the mRNA level expression of a higher number of DEGs in LUSC patients compared to the LUAD patients (Fig. 9). Other immune cells showed a significant correlation with a similarly small number of DEGs both in LUAD and LUSC microenvironments. Importantly, the DEGs maintained a similar projection of association (i.e., either positively or negatively correlated) with the infiltration levels of different immune cells in both cancer types.
Fig. 9.
The association between the mRNA level expression of DEGs and different immune cell infiltration levels in LUAD and LUSC tissues. The color gradient indicates (low: blue, high: red) the spearman correlation coefficient. The circle represents a significant correlation (adjusted p-value<0.05) and the blank space indicates an insignificant correlation.
3.7. The expression pattern of the DEGs in TCGA database and their impact on LUAD and LUSC Patient's survival
Most of the genes were found to be significantly upregulated and/or downregulated in LUAD and LUSC samples than in adjacent normal lung tissues. The shared DEGs were observed to exhibit a similar pattern of deregulation (i.e., either upregulated or downregulated) in both cancer types. The log2FC value of the deregulated genes varied between −6 and 8 as evidenced by the experiment carried out on the TCGA cohorts (Fig. 10 A). The association analysis between the expression of the DEGs and different survival rates revealed that the mRNA level expression of most of the genes was associated with both favorable and unfavorable survival of LUAD and LUSC patients in a different manner (Fig. 10B). In summary, MKI67, MELK, MAD2L1, KRT8 (except for PFS), FOXM1, CDKN3, CDK1, CCNB2, and BUB1 expression was discovered to be associated with the unfavorable DSS, OS and PFS of LUAD patients as suggested by higher HR values. Additionally, SFTPA2 and SFTPA1 expression was found to be able to predict poor prognosis in LUSC patients. In contrast, POU2AF1, CMLRK1, CD83 (associated with DSS only) and CA4 expression showed significant association with the better DSS, OS and PFS of LUAD patients (Fig. 10B). The impact of the expression of the DEGs on LUAD and LUSC patients’ survival trend can be further explored on the TCGA dataset and other independent datasets from GEPIA 2.0 (http://gepia2.cancer-pku.cn/#survival) [49] and PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/) databases [50], respectively, in the form of Kaplan-Meier plot. Additionally, we also investigated the expression pattern of the shared DEGs in one COVID-19 (GSE171110) and one lung cancer (GSE74706) datasets. And at least 28 of the 36 DEGs were found to show significant differential expression (|log2FC|>1; p < 0.05) in those datasets as well (Supplementary Fig. S1).
Fig. 10.
(A) The dot-plot representation of the expression pattern of the DEGs in LUAD and LUSC tissue samples in comparison with normal lung tissue samples from TCGA database. The color gradient scale corresponds to the log2FC (low: blue; high: red). (B) The association between the expression level of DEGs and different survival rates of LUAD and LUSC patients from TCGA database. The color gradient scale indicates the distribution of hazard ratio (low: blue; high: red). DSS: disease-specific survival; OS: overall survival; PFS: progression-free survival.
4. Discussion
In this study, we employed a comprehensive bioinformatics meta-analysis pipeline to assess the pathological impact of COVID-19 on two most common forms of NSCLC i.e., LUAD and LUSC. Meta-analysis in genomic study refers to a method that uses a series of statistical analysis modules to combine the results of multiple studies to increase the analysis power and provide a more comprehensive and accurate summary of the evidence [51]. With the advent of advanced sequencing and computational techniques, meta-analysis has become an important research protocol in predicting the mechanism and clinical outcome of different genetic and infectious diseases by combining the findings from multiple studies [52]. In genomic studies, meta-analysis methods have several advantages over other conventional methods including the ability to pool data from different studies to increase sample size and reduce the risk of false negative results, synthesize evidence from multiple sources to overcome the limitations of individual studies, identify patterns and trends in the data that are not apparent from individual studies, and provide more precise estimates of the true effect size by combining the results of multiple studies [[53], [54], [55]]. Thus, meta-analysis methods in combination with other approaches like pathway and network-based strategies play an important role in synthesizing the available evidence and providing a more comprehensive and accurate understanding of the effects of various interventions and identifying potential molecular targets in different diseases like cancer [56].
Differential gene expression pattern analysis can help understand the involvement and possible roles of different genes in various complex diseases like cancer [[57], [58], [59]]. We identified 36 differentially expressed genes that are common in COVID-19, LUAD and LUSC patients suggesting that the SARS-CoV-2 infection may modulate the disease progression of LC patients. We also discovered that most of the deregulated genes are expressed specifically in lung tissues which suggests that the SARS-CoV-2 infection may affect the overall lung homeostasis and hasten the LUAD and LUSC-associated lung injury further. Additionally, a large number of proteins expressed by the DEGs were classified as secreted and plasma proteins which signifies the relevance of those proteins in the non-invasive diagnosis of COVID-19-affected LUAD and LUSC patients.
The comorbidity profiling of the DEGs revealed that multiple genes are associated with different forms of respiratory tract diseases and patients with such comorbidities are the most vulnerable groups to SARS-CoV-2 infection-related severity and mortality [60]. Furthermore, all of the previously discovered lung-specific DEGs were subsequently shown to be implicated in various lung injuries. Later, our examination of the DEGs versus all disease pools indicated correlations with additional complicated respiratory illnesses such as RDS. Studies have shown that adult patients with a history of lung damage have a greater risk of developing RDS after contracting COVID-19, which increases fatality rates by 35% [61]. Therefore, COVID-19 may further increase the risk of developing RDS in LUAD and LUSC patients by deregulating the genes associated with RDS. Afterward, we observed that the DEGs are also deregulated in other types of diseases including carcinoma with LC as the predominant disease. Among the deregulated genes in LC, TOP2A is associated with tumorigenic progression and unfavorable survival in LUAD patients [62]. Previous studies reported that patients with LUAD and LUSC had worse prognoses when they express high levels of TOP2A. Thus, TOP2A is reported as a marker for predicting patient outcomes, such as disease recurrence and overall survival in patients with these two types of LC [63,64]. TOP2A has also been demonstrated to contribute to the pathogenesis of the disease in COVID-19 patients such as severe symptoms and an elevated risk of death have been linked to high expression of TOP2A [65]. As a result, it is possible that TOP2A might also be used as a therapeutic target in COVID-19 patients. Among the other deregulated genes in LC, MELK promotes the mitotic and metastatic potential of LC cells [66], BUB1 is associated with the division of LC cells and poor prognosis of NSCLC patients [67,68]. Moreover, an in vitro study has recently shown that CCNB2 serves as a prognostic indicator and therapeutic target for LUAD patients infected with COVID-19 [69]. Additionally, other deregulated genes in LC i.e., C1QB, have been found to be overexpressed in both NSCLC and IPF patients and reported to act as an unfavorable survival indicator for both diseases [70]. Furthermore, C1QB expression level has recently been proposed to be able to discriminate the macrophage infiltration levels between healthy subjects and COVID-19 patients [71]. Additionally, increased CD163 expression was associated with the damaged lymph nodes obtained from lung autopsy samples of COVID-19-infected patients [72]. Collectively, these findings bolster the hypothesis that COVID-19-induced expressional changes may predispose LUAD and LUSC patients to LC-related complexities. Furthermore, COVID-19 has been shown to induce acute liver injuries in infected patients irrespective of prior history of liver diseases [73]. In our study, we reported that a total of 15 genes of the 36 DEGs are differentially expressed in acute liver failure. Among those deregulated genes in liver failure, CD163 expression is correlated with the development and progression of chronic liver failure [74]. The genomic instability of POU2AF1 is associated with primary biliary cirrhosis of the liver [75]. Moreover, the optimum expression of TOP2A is required to maintain liver homeostasis by regulating c-Myc [76]. These findings suggest that the presence of COVID-19 in LUAD and LUSC patients may further increase the risk of COVID-19-associated liver injury.
We also observed that the DEGs were differentially expressed in different autoimmune diseases. COVID-19 patients with autoimmune disease are assumed to experience more complications due to the similar immune system-mediated pathomechanism of both diseases [77,78]. Remarkably, few studies have recorded the development of autoimmune diseases in individuals who had no previous medical history of the illness following SARS-CoV-2 infection [79]. Moreover, a case report has recently recorded evidence of ulcerative colitis (newly diagnosed) in a patient after a SARS-CoV-2 infection [80]. We also observed that the DEGs were deregulated in a few infectious diseases i.e., tuberculosis. A meta-analysis on COVID-19 patients with tuberculosis (n = 2765) by Gao et al., concluded that the case fatality rate in those patients was around 4.5% [81]. Therefore, the extant literature suggests that LUAD and LUSC patients with such comorbidities need special medical care and the involvement of experts for emergency clinical decision-making (i.e., chemotherapy, immunotherapy). Finally, we identified that out of 36 DEGs 15 genes are cancer-associated genes involving 3 oncogenes i.e., JUN, POU2AF1, and TNC. JUN signaling pathway is associated with LC development [82]. Studies suggest that high JUN expression might be associated with a poorer prognosis in patients with LUAD and LUSC [[83], [84], [85]], which has led researchers to investigate the use of JUN as a diagnostic marker in the early detection of lung cancer. Studies also showed that the products of the TNC gene seem to help LC spread and thus promote cancer invasion and exacerbation [86]. In addition, high TNC expression has been linked to poor prognosis in LUAD and LUSC, indicating its potential use as a prognostic marker. [87,88]. POU2AF1 has been linked to the development of other types of cancer, like myeloma [89], but its connection to LC or COVID-19-infected LUAD and LUSC patients is still not clear.
The KEGG pathway analysis reported that most of the DEGs are involved in controlling the cell cycle, cellular senescence, extracellular matrix organization, cell cycle phase transition, p53 signaling pathway and the deregulation of such activities by abnormal gene expression can lead to cancer development, progression and distant metastasis [90,91]. We also observed that the deregulated DEGs control different signaling pathways involved in diabetic complications. Diabetic patients were among the hardest hit by SARS-CoV-2 infection and this group of patients had a higher risk of developing ARDS and requires an extended period of ICU support [92]. Response to IFN-α and platelet-derived growth factor binding was among the most significant biological processes and molecular functions of the DEGs. IFN-α has been accused to induce the expression of the Angiotensin Converting Enzyme 2 receptor (a molecule that assists SARS-CoV-2 entering inside the cell) in the human upper respiratory tract [93]. Furthermore, increased IFN- production is linked to morbidity because to COVID-19 [94]. Hematologic platelet counts may provide light on the prognosis of patients with COVID-19 and LC [95,96] since platelets are the first line of defense against lung damage. Patients with LC infected with COVID-19 may benefit from using a variety of immune modulators for diagnosis and treatment.
The interconnection pattern of the PPI network signified that the DEGs may affect the functions controlled by many other genes in COVID-19-affected LUAD and LUSC patients. Moreover, we next identified 6 significant hub proteins from the PPI network and targeting those proteins can provide the most efficacy in the therapeutic intervention approach for this group of patients [97]. Among the identified hub proteins, CDK1 and MAD2L1 serve as potential therapeutic targets and unfavorable prognostic indicators in NSCLC patients [[98], [99], [100]]. Likely, FOXM1 overexpression is associated with epithelial to mesenchymal transition and the poor prognosis of NSCLC patients [101,102]. We previously found that the remaining hub proteins i.e., BUB1, TOP2A and CCNB2 are associated with LC exacerbation and other comorbidities. We later discovered that regulating the cell cycle and cell division were the predominant biological processes of the hub proteins justifying their therapeutic potential.
The miRNA and TF interaction analysis suggested that multiple DEGs and their respective mRNA and proteins are targeted by different miRNA and TFs. Of the identified interacting miRNAs, downregulation of the hsa-mir-203a-3p provides a therapeutic advantage in LC patients [103]. As a prognostic marker, hsa-mir-203a-3p has been found to be associated with poor prognosis in lung cancer patients, with low expression levels linked to increased risk of death [104,105]. As a therapeutic target, hsa-mir-203a-3p has been found to regulate key cancer-related genes, including those involved in cell proliferation, invasion and metastasis, making it a promising target for cancer therapy [106]. Additionally, hsa-mir-203a-3p has been shown to be upregulated in COVID-19 patients, and may play a role in the inflammation and oxidative stress response associated with severe COVID-19 outcomes [107]. Targeting hsa-mir-203a-3p could therefore have therapeutic implications for COVID-19 patients. Another identified miRNA i.e., hsa-mir-218-5p is assumed to play a significant role in the pathogenesis of NSCLC by being upregulated in LUAD and LUSC patients, and thus has been shown to be an independent predictor of poor prognosis in these patients [[108], [109], [110]]. Also, in COVID-19 patients, hsa-mir-218-5p has been found to be associated with severe disease and poor outcomes in patients with COVID-19-related pneumonia [111]. Additionally, the expression of hsa-mir-218-5p has been shown to be regulated by the viral infection, suggesting its potential as a therapeutic target in COVID-19 patients [112]. Moreover, hsa-mir-186-5p promotes the LUAD cell growth and invasion through inhibiting the activity of PTEN [113]. In LUAD, the expression of hsa-mir-186-5p was found to be a predictor of poor survival outcomes and increased drug resistance, making it a potential target for personalized therapies [114]. In LUSC, hsa-mir-186-5p was found to be overexpressed and associated with more aggressive tumors, making it a potential prognostic biomarker [115]. Additionally, recent studies have also shown that hsa-mir-186-5p is involved in the development and progression of COVID-19, where it was found to be overexpressed in severe cases and associated with increased cytokine production and inflammation [116]. This makes hsa-mir-186-5p a potential diagnostic and therapeutic target for the stratification and treatment of this group of patients. Additionally, hsa-mir-193b-3p has been found to regulate the expression of various genes involved in tumor progression and angiogenesis in LUSC, making it a potential therapeutic target for LUSC therapy [117,118]. In LUAD, hsa-mir-193b-3p was found to be a powerful prognostic marker, with low levels associated with poor overall survival and high levels linked to better patient outcomes [119].
Among the identified TFs, EBF1 knockdown with shRNA can block the cell cycle at the G1 phase in LUAD cell lines [120]. In LUAD and LUSC, EBF1 has been found to have a strong association with tumor progression and poor prognosis, with higher EBF1 expression levels being associated with reduced survival and increased metastasis which suggests that EBF1 may be a useful target for therapeutic intervention in lung cancer [[121], [122]].TFDP1 hyperactivity promotes the p53 signaling pathway and progresses the NSCLC cells through the cell cycle [123]. Moreover, TFDP1 has been shown to be upregulated in infected individuals and may play a role in the severity of COVID-19. Studies thus also suggest that targeting TFDP1 may have therapeutic potential for treating severe cases of COVID-19 [124]. Huang et al. showed that IRF1 acts as a tumor suppressor in LC cells and provides a therapeutic target for IRF1-based LC treatment strategies [125]. Moreover, IRF1-mediated inhibition of MHC class I and IFN signaling pathway helps SARS-CoV-2 evade the immune system of the human body [126]. Additionally, ETV4 overexpression promotes NSCLC cell proliferation by upregulating genes involved in maintaining focal adhesion i.e., MMP1 and PXN [127]. Furthermore, ETV4 has been identified as a potential target for the treatment of COVID-19, with studies indicating its involvement in the regulation of the immune response and susceptibility to the virus [128]. Finally, we discovered TOP2A as a potential target for the treatment of COVID-19-infected LC patients which interacted with many approved drugs including several antineoplastic agents. Therefore, the miRNA, TF targets, and different drug molecules could be investigated further for the treatment of LUAD and LUSC patients infected with SARS-CoV-2.
In LUAD and LUSC patients, DEGs are favorably and adversely correlated with immune cells. High B cell abundance in LC cancer microenvironment predicts good survival, and adoptive B cell transfer may regress cancer development and limit metastasis [129]. Higher T cell and NK cell abundance enhances LC patient survival, and both are prospective candidates for immune-based therapeutic intervention [130,131]. COVID-19 patients generally have lymphopenia with fewer B, T, and NK cells [132,133]. Infiltration of immune cells may act as a therapeutic marker and immunotherapy implication in COVID-19-infected LC patients. The expression profile of differentially expressed genes (DEGs) in the TCGA LUAD and LUSC cohorts, which include a significant number of RNA-seq data from cancer and surrounding normal tissues of respective cancer types. We detected differential expression of the identified DEGs in TCGA cohorts. The survival analysis of DEGs demonstrated that the differential expression of a substantial number of DEGs is associated with the survival conditions of LUAD and LUSC patients, indicating their underlying involvement in the development and progression of LC. Finally, we also investigated the expression pattern of the DEGs in two different independent databases including one that involves the blood transcriptome of COVID-19 patients. We found that at least 28 of the 36 DEGs are differentially expressed in the cells present in the blood circulation system of COVID-19 patients which may further assist in formulating non-invasive diagnosis of this group of patients. At the same time, how COVID-19 may promote the LC invasion and influence the prognosis of LC patients by deregulating these set of genes in cells that may travel to distant organs requires further investigation.
5. Conclusion
This study sought to investigate the potential pathophysiology and risk factors of COVID-19 in LUAD and LUSC patients by differential gene expression pattern analysis. The SARS-CoV-2 infection was reported to impact the course of LC by deregulating many oncogenes and cancer-associated genes. Moreover, it was found that the presence of COVID-19 may affect and manifest distinct respiratory illness phenotypes in LUAD and LUSC patients by deregulating lung-specific genes. Later, we used a network-based strategy to discover critical molecular markers with diagnostic and therapeutic potential for this group of patients. While the comorbidity and functional enrichment profiles of the deregulated genes in COVID-19-affected LUAD and LUSC patients may guide the adoption of necessary management measures, the distinct regulatory signatures identified in this study may facilitate the development of diagnostic and therapeutic options for this vulnerable patient group. Therefore, more laboratory research on regulatory signatures for future clinical development is required.
Author’s contribution Statement
MU: Conceptualization and experimental design. MU, SA, AM: Experiment, Formal analysis, interpretation and data Visualization. MU, SA, AM, TA and AS: Writing, Writing – review & editing. AS: Supervision. All authors have approved and agreed to the publication of the final manuscript.
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Availability of data and material
All the data are provided within the manuscript and the supplementary material. The R codes utilized in this study were taken and adjusted from publicly available CRAN projects (https://cran.r-project.org/) and Bioconductor resources (https://www.bioconductor.org/).
Funding
No specific grant was received for this study.
Declaration of competing interest
All the authors declare that they have no conflict of interest regarding the publication of the paper.
Acknowledgment
The authors are thankful to the members of Swift Integrity Computational Lab, Dhaka, Bangladesh, and, Cell Genetics and Plant Biotechnology Laboratory (www.cgpbl.ac.bd), Jahangirnagar University, Dhaka, Bangladesh, for their support during the preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2023.106855.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bösmüller H., Matter M., Fend F., Tzankov A. The pulmonary pathology of COVID-19. Virchows Arch. 2021;478(1):137–150. doi: 10.1007/s00428-021-03053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang B., Li R., Lu Z., Huang Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging. 2020;12(7):6049. doi: 10.18632/aging.103000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh M.K., Mobeen A., Chandra A., Joshi S., Ramachandran S. A meta-analysis of comorbidities in COVID-19: which diseases increase the susceptibility of SARS-CoV-2 infection? Comput. Biol. Med. 2021;130 doi: 10.1016/j.compbiomed.2021.104219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islami F., Torre L.A., Jemal A. Global trends of lung cancer mortality and smoking prevalence. Transl. Lung Cancer Res. 2015;4(4):327. doi: 10.3978/j.issn.2218-6751.2015.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schabath M.B., Cote M.L. Cancer progress and priorities: lung cancer. Cancer Epidemiol. Biomark. Prev. 2019;28(10):1563–1579. doi: 10.1158/1055-9965.EPI-19-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alkhathami M.G., Advani S.M., Abalkhail A.A., Alkhathami F.M., Alshehri M.K., Albeashy E.E., Alsalamah J.A. Prevalence and mortality of lung comorbidities among patients with COVID-19: a systematic review and meta-analysis. Lung India. Official Organ of Indian Chest Society. 2021;38(Suppl 1):S31. doi: 10.4103/lungindia.lungindia_497_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilad S., Lithwick-Yanai G., Barshack I., Benjamin S., Krivitsky I., Edmonston T.B., Bibbo M., Thurm C., Horowitz L., Huang Y., Feinmesser M. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J. Mol. Diagn. 2012;14(5):510–517. doi: 10.1016/j.jmoldx.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E., Cho J., Suh J., Capelletti M., Sivachenko A., Sougnez C. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J., Rizvi H., Preeshagul I.R., Egger J.V., Hoyos D., Bandlamudi C., McCarthy C.G., Falcon C.J., Schoenfeld A.J., Arbour K.C., Chaft J.E. COVID-19 in patients with lung cancer. Ann. Oncol. 2020;31(10):1386–1396. doi: 10.1016/j.annonc.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provencio M., Gallego J.M., Calles A., Antoñanzas M., Pangua C., Rubio X.M., Nadal E., Castro R.L., López-Martín A., Del Barco E., Dómine M. Lung cancer patients with COVID-19 in Spain: GRAVID study. Lung Cancer. 2021;157:109–115. doi: 10.1016/j.lungcan.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passaro A., Bestvina C., Velez M.V., Garassino M.C., Garon E., Peters S. Severity of COVID-19 in patients with lung cancer: evidence and challenges. J. Immunother. Cancer. 2021;9(3) doi: 10.1136/jitc-2020-002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J., Rizvi H., Preeshagul I.R., Egger J.V., Hoyos D., Bandlamudi C., McCarthy C.G., Falcon C.J., Schoenfeld A.J., Arbour K.C., Chaft J.E. COVID-19 in patients with lung cancer. Ann. Oncol. 2020;31(10):1386–1396. doi: 10.1016/j.annonc.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Addeo A., Obeid M., Friedlaender A. COVID-19 and lung cancer: risks, mechanisms and treatment interactions. J. Immunother. Cancer. 2020;8(1) doi: 10.1136/jitc-2020-000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auwul M.R., Rahman M.R., Gov E., Shahjaman M., Moni M.A. Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Briefings Bioinf. 2021;22(5) doi: 10.1093/bib/bbab120. bbab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ullah M.A., Sarkar B., Akter F. Prediction of biomarker signatures and therapeutic agents from blood sample against Pancreatic Ductal Adenocarcinoma (PDAC): a network-based study. Inform. Med. Unlocked. 2020;19 [Google Scholar]
- 17.Wang B., Huang Y. Which type of cancer patients are more susceptible to the SARS-COX-2: evidence from a meta-analysis and bioinformatics analysis. Crit. Rev. Oncol.-Hematol. 2020;153 doi: 10.1016/j.critrevonc.2020.103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang X., Chen Y., Fan Y. Bioinformatics approach to identify common gene signatures of patients with coronavirus 2019 and lung adenocarcinoma. Environ. Sci. Pollut. Control Ser. 2022;29(15):22012–22030. doi: 10.1007/s11356-021-17321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lou H., Li X., Gao S., Zhang Y., Chen H., Zhai X. Identifying potential gene defect patterns related to COVID-19 based on pharmacological and bioinformatics analysis for lung adenocarcinoma. Int. J. Gen. Med. 2022;15:4285. doi: 10.2147/IJGM.S356444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nienhold R., Ciani Y., Koelzer V.H., Tzankov A., Haslbauer J.D., Menter T., Schwab N., Henkel M., Frank A., Zsikla V., Willi N. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 2020;11(1):1–3. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno Leon L., Gautier M., Allan R., Ilié M., Nottet N., Pons N., Paquet A., Lebrigand K., Truchi M., Fassy J., Magnone V. The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA contributes to an aggressive phenotype in lung adenocarcinoma through regulation of oxidative stress. Oncogene. 2019;38(46):7146–7165. doi: 10.1038/s41388-019-0935-y. [DOI] [PubMed] [Google Scholar]
- 22.Cui Y., Fang W., Li C., Tang K., Zhang J., Lei Y., He W., Peng S., Kuang M., Zhang H., Chen L. Development and validation of a novel signature to predict overall survival in “driver gene–negative” lung adenocarcinoma (LUAD): results of a multicenter StudyA prognostic classifier for “driver gene–negative” LUAD. Clin. Cancer Res. 2019;25(5):1546–1556. doi: 10.1158/1078-0432.CCR-18-2545. [DOI] [PubMed] [Google Scholar]
- 23.Rousseaux S., Debernardi A., Jacquiau B., Vitte A.L., Vesin A., Nagy-Mignotte H., Moro-Sibilot D., Brichon P.Y., Lantuejoul S., Hainaut P., Laffaire J. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 2013;5(186) doi: 10.1126/scitranslmed.3005723. 186ra66- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiwara T., Hiramatsu M., Isagawa T., Ninomiya H., Inamura K., Ishikawa S., Ushijima M., Matsuura M., Jones M.H., Shimane M., Nomura H. ASCL1-coexpression profiling but not single gene expression profiling defines lung adenocarcinomas of neuroendocrine nature with poor prognosis. Lung Cancer. 2012;75(1):119–125. doi: 10.1016/j.lungcan.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014 Dec;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allaire J. 2012. RStudio: Integrated Development Environment for R. Boston, MA; pp. 165–171. 770(394) [Google Scholar]
- 27.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):1–6. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth G.K. InBioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York, NY: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 29.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 2015;16(1):1–7. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36(8):2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019 Apr 3;10(1) doi: 10.1038/s41467-019-09234-6. 1-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham H., Chang W., Wickham M.H. Package ‘ggplot2’. Create elegant data visualisations using the grammar of graphics. Version. 2016;2(1):1–89. [Google Scholar]
- 34.Zhou G., Soufan O., Ewald J., Hancock R.E., Basu N., Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piñero J., À Bravo, Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., García-García J., Sanz F., Furlong L.I. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw943. gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papatheodorou I., Fonseca N.A., Keays M., Tang Y.A., Barrera E., Bazant W., Burke M., Füllgrabe A., Fuentes A.M., George N., Huerta L. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46(D1):D246–D251. doi: 10.1093/nar/gkx1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Repana D., Nulsen J., Dressler L., Bortolomeazzi M., Venkata S.K., Tourna A., Yakovleva A., Palmieri T., Ciccarelli F.D. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019;20(1):1–2. doi: 10.1186/s13059-018-1612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(4):1–7. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M., Chien C.H. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res. 2011;39(suppl_1):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ENCODE Project Consortium A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4) doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C.J., Hu F.F., Xia M.X., Han L., Zhang Q., Guo A.Y. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 47.Levy Y., Wiedemann A., Hejblum B.P., Durand M., Lefebvre C., Surenaud M., Lacabaratz C., Perreau M., Foucat E., Dechenaud M., Tisserand P. CD177, a specific marker of neutrophil activation, is associated with coronavirus disease 2019 severity and death. iScience. 2021;24(7) doi: 10.1016/j.isci.2021.102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marwitz S., Depner S., Dvornikov D., Merkle R., Szczygieł M., Müller-Decker K., Lucarelli P., Wäsch M., Mairbäurl H., Rabe K.F., Kugler C. Downregulation of the TGFβ pseudoreceptor BAMBI in non–small cell lung cancer enhances TGFβ signaling and InvasionRole of BAMBI in lung cancer. Cancer Res. 2016;76(13):3785–3801. doi: 10.1158/0008-5472.CAN-15-1326. [DOI] [PubMed] [Google Scholar]
- 49.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizuno H., Kitada K., Nakai K., Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009;2(1) doi: 10.1186/1755-8794-2-18. 1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon C., Daniel R. Metagenomic analyses: past and future trends. Appl. Environ. Microbiol. 2011;77(4):1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borenstein M., Hedges L.V., Higgins J.P., Rothstein H.R. Introduction to Meta-Analysis. John Wiley & Sons; 2009. [Google Scholar]
- 53.Berkey C.S., Hoaglin D.C., Mosteller F., Colditz G.A. A random-effects regression model for meta-analysis. Stat. Med. 1995;14(4):395–411. doi: 10.1002/sim.4780140406. [DOI] [PubMed] [Google Scholar]
- 54.Zeggini E., Ioannidis J.P. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10(2):191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y.H. An overview of meta-analysis for clinicians. Kor. J. Intern. Med. 2018;33(2):277–283. doi: 10.3904/kjim.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Campos M.A., Espinal-Enríquez J., Hernández-Lemus E. Pathway analysis: state of the art. Front. Physiol. 2015;6:383. doi: 10.3389/fphys.2015.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutta S.K., Mitra P.S., Ghosh S., Zang S., Sonneborn D., Hertz-Picciotto I., Trnovec T., Palkovicova L., Sovcikova E., Ghimbovschi S., Hoffman E.P. Differential gene expression and a functional analysis of PCB-exposed children: understanding disease and disorder development. Environ. Int. 2012;40:143–154. doi: 10.1016/j.envint.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang P., Pardee A.B. Analysing differential gene expression in cancer. Nat. Rev. Cancer. 2003;3(11):869–876. doi: 10.1038/nrc1214. [DOI] [PubMed] [Google Scholar]
- 59.Crow M., Lim N., Ballouz S., Pavlidis P., Gillis J. Predictability of human differential gene expression. Proc. Natl. Acad. Sci. USA. 2019;116(13):6491–6500. doi: 10.1073/pnas.1802973116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aveyard P., Gao M., Lindson N., Hartmann-Boyce J., Watkinson P., Young D., Coupland C.A., San Tan P., Clift A.K., Harrison D., Gould D.W. Association between pre-existing respiratory disease and its treatment, and severe COVID-19: a population cohort study. Lancet Respir. Med. 2021;9(8):909–923. doi: 10.1016/S2213-2600(21)00095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibson P.G., Qin L., Puah S.H. COVID‐19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre‐COVID‐19 ARDS. Med. J. Aust. 2020;213(2):54–56. doi: 10.5694/mja2.50674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kou F., Sun H., Wu L., Li B., Zhang B., Wang X., Yang L. TOP2A promotes lung adenocarcinoma cells' malignant progression and predicts poor prognosis in lung adenocarcinoma. J. Cancer. 2020;11(9):2496. doi: 10.7150/jca.41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma W, Wang B, Zhang Y, Wang Z, Niu D, Chen S, Zhang Z, Shen N, Han W, Zhang X, Wei R, Wang C. Prognostic significance of TOP2A in non-small cell lung cancer revealed by bioinformatic analysis. Cancer Cell Int. 2019;19:239. doi: 10.1186/s12935-019-0956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su Q, Zhang S, Ran J. TOP2A Serves as a Prognostic Marker Associated with Immune Infiltration in Hepatocellular Carcinoma. Research Square. 2022 [Google Scholar]
- 65.Tian Y, Carpp LN, Miller HER, Zager M, Newell EW, Gottardo R. Single-cell immunology of SARS-CoV-2 infection. Nat Biotechnol. 2022 Jan;40(1):30–41. doi: 10.1038/s41587-021-01131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Q., Li W., Zheng X., Ren L., Liu J., Li S., Wang J., Du G. MELK is an oncogenic kinase essential for metastasis, mitotic progression, and programmed death in lung carcinoma. Signal Transduct. Targeted Ther. 2020 Dec 2;5(1):1–2. doi: 10.1038/s41392-020-00288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stav D., Bar I., Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. Int. J. Biol. Markers. 2007;22(2):108–113. doi: 10.1177/172460080702200204. [DOI] [PubMed] [Google Scholar]
- 68.Qian X., Song X., He Y., Yang Z., Sun T., Wang J., Zhu G., Xing W., You C. CCNB2 overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed. Pharmacother. 2015;74:222–227. doi: 10.1016/j.biopha.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Yang L., Xiong H., Li X., Li Y., Zhou H., Lin X., Chan T.F., Li R., Lai K.P., Chen X. Network pharmacology and comparative transcriptome reveals biotargets and mechanisms of curcumol treating lung adenocarcinoma patients with COVID-19. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.870370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kou W., Li B., Shi Y., Zhao Y., Yu Q., Zhuang J., Xu Y., Peng W. High complement protein C1q levels in pulmonary fibrosis and non-small cell lung cancer associated with poor prognosis. BMC Cancer. 2022;22(1):1–6. doi: 10.1186/s12885-021-08912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaath H., Vishnubalaji R., Elkord E., Alajez N.M. Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells. 2020;9(11):2374. doi: 10.3390/cells9112374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haslbauer J.D., Matter M.S., Stalder A.K., Tzankov A. Histomorphological patterns of regional lymph nodes in COVID-19 lungs. Pathologe. 2021;42(1):89–97. doi: 10.1007/s00292-021-00945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nardo A.D., Schneeweiss‐Gleixner M., Bakail M., Dixon E.D., Lax S.F., Trauner M. Pathophysiological mechanisms of liver injury in COVID‐19. Liver Int. 2021;41(1):20–32. doi: 10.1111/liv.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nielsen M.C., Hvidbjerg Gantzel R., Clària J., Trebicka J., Møller H.J., Grønbæk H. Macrophage activation markers, CD163 and CD206, in acute-on-chronic liver failure. Cells. 2020;9(5):1175. doi: 10.3390/cells9051175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamura M., Nishida N., Kawashima M., Aiba Y., Tanaka A., Yasunami M., Nakamura H., Komori A., Nakamuta M., Zeniya M., Hashimoto E. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet. 2012;91(4):721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hishida T., Yamamoto M., Hishida-Nozaki Y., Shao C., Huang L., Wang C., Shojima K., Xue Y., Hang Y., Shokhirev M., Memczak S. In vivo partial cellular reprogramming enhances liver plasticity and regeneration. Cell Rep. 2022;39(4) doi: 10.1016/j.celrep.2022.110730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y., Sawalha A.H., Lu Q. COVID-19 and autoimmune diseases. Curr. Opin. Rheumatol. 2021;33(2):155. doi: 10.1097/BOR.0000000000000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodríguez Y., Novelli L., Rojas M., De Santis M., Acosta-Ampudia Y., Monsalve D.M., Ramírez-Santana C., Costanzo A., Ridgway W.M., Ansari A.A., Gershwin M.E. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J. Autoimmun. 2020;114 doi: 10.1016/j.jaut.2020.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saad M.A., Alfishawy M., Nassar M., Mohamed M., Esene I.N., Elbendary A. Covid-19 and autoimmune diseases: a systematic review of reported cases. Curr. Rheumatol. Rev. 2021;17(2):193–204. doi: 10.2174/1573397116666201029155856. [DOI] [PubMed] [Google Scholar]
- 80.Aydın M.F., Taşdemir H. Ulcerative colitis in a COVID-19 patient: a case report. Turk. J. Gastroenterol. 2021;32(6):543–547. doi: 10.5152/tjg.2021.20851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao Y., Liu M., Chen Y., Shi S., Geng J., Tian J. Association between tuberculosis and COVID-19 severity and mortality: A rapid systematic review and meta-analysis. J Med Virol. 2021 Jan;93(1):194–196. doi: 10.1002/jmv.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y., Ikezoe T., Saito T., Kobayashi M., Koeffler H.P., Taguchi H. Proteasome inhibitor PS‐341 induces growth arrest and apoptosis of non‐small cell lung cancer cells via the JNK/c‐Jun/AP‐1 signaling. Cancer Sci. 2004;95(2):176–180. doi: 10.1111/j.1349-7006.2004.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo Z., Han Z., Shou F, Li Y., Chen Y. LINC00958 Accelerates Cell Proliferation and Migration in Non-Small Cell Lung Cancer Through JNK/c-JUN Signaling. Hum Gene Ther Methods. 2019 Dec;30(6):226–234. doi: 10.1089/hgtb.2019.115. [DOI] [PubMed] [Google Scholar]
- 84.Qiao G.B, Wang R.T, Wang S.N, Tao S.L., Tan Q.Y., Jin H. GRP75-mediated upregulation of HMGA1 stimulates stage I lung adenocarcinoma progression by activating JNK/c-JUN signaling. Thorac Cancer. 2021 May;12(10):1558–1569. doi: 10.1111/1759-7714.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao M., Xu P., Liu Z., et al. RETRACTED ARTICLE: Dual roles of miR-374a by modulated c-Jun respectively targets CCND1-inducing PI3K/AKT signal and PTEN-suppressing Wnt/β-catenin signaling in non-small-cell lung cancer. Cell Death Dis. 2018;9:78. doi: 10.1038/s41419-017-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Tang Y.A., Chen C.H., Sun H.S., Cheng C.P., Tseng V.S., Hsu H.S., Su W.C., Lai W.W., Wang Y.C. Global Oct4 target gene analysis reveals novel downstream PTEN and TNC genes required for drug-resistance and metastasis in lung cancer. Nucleic Acids Res. 2015;43(3):1593–1608. doi: 10.1093/nar/gkv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yilmaz A., Loustau T., Salomé N., Poilil Surendran S., Li C., Tucker R.P., Izzi V., Lamba R., Koch M., Orend G.. Advances on the roles of tenascin-C in cancer. J Cell Sci. 2022 Sep 15;135(18):jcs260244. [DOI] [PMC free article] [PubMed]
- 88.Donovan C., Bai X., Chan Y.L., Feng M., Ho K.-F., Guo H., Chen H., Oliver B.G. Tenascin C in Lung Diseases. Biology. 2023;12(2):199. doi: 10.3390/biology12020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simoes R.D., Shirasaki R., Tang H., Yamano S., Barwick B.G., Gandolfi S., Dhimolea E., Downey-Kopyscinski S.L., Dashevsky O., Glassner B., Sheffer M. POU2AF1 as a master regulator of oncogenic transcription factor networks in myeloma. Blood. 2020;136:18–19. [Google Scholar]
- 90.Oppenheimer S.B. Cellular basis of cancer metastasis: a review of fundamentals and new advances. Acta Histochem. 2006;108(5):327–334. doi: 10.1016/j.acthis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 91.Weinberg R.A. How cancer arises. Sci. Am. 1996;275(3):62–70. doi: 10.1038/scientificamerican0996-62. [DOI] [PubMed] [Google Scholar]
- 92.Singh A.K., Gupta R., Ghosh A., Misra A. Diabetes in COVID-19: prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metabol. Syndr.: Clin. Res. Rev. 2020;14(4):303–310. doi: 10.1016/j.dsx.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ziegler C.G., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369(6504):706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mandaliya H., Jones M., Oldmeadow C., Nordman Prognostic biomarkers in stage IV non-small cell lung cancer (NSCLC): neutrophil to lymphocyte ratio (NLR), lymphocyte to monocyte ratio (LMR), platelet to lymphocyte ratio (PLR) and advanced lung cancer inflammation index (ALI) Transl. Lung Cancer Res. 2019;8(6):886. doi: 10.21037/tlcr.2019.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salamanna F., Maglio M., Landini M.P., Fini M. Platelet functions and activities as potential hematologic parameters related to Coronavirus Disease 2019 (Covid-19) Platelets. 2020;31(5):627–632. doi: 10.1080/09537104.2020.1762852. [DOI] [PubMed] [Google Scholar]
- 97.Shen Y.Z., Ding Y.S., Gu Q., Chou K.C. Identifying the hub proteins from complicated membrane protein network systems. Med. Chem. 2010;6(3):165–173. doi: 10.2174/1573406411006030165. [DOI] [PubMed] [Google Scholar]
- 98.Shi Y.X., Zhu T., Zou T., Zhuo W., Chen Y.X., Huang M.S., Zheng W., Wang C.J., Li X., Mao X.Y., Zhang W. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7(51) doi: 10.18632/oncotarget.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang Z., Shen G., Gao J. CDK1 promotes the stemness of lung cancer cells through interacting with Sox2. Clin. Transl. Oncol. 2021;23(9):1743–1751. doi: 10.1007/s12094-021-02575-z. [DOI] [PubMed] [Google Scholar]
- 100.Zhang C., Elkahloun A.G., Robertson M., Gills J.J., Tsurutani J., Shih J.H., Fukuoka J., Hollander M.C., Harris C.C., Travis W.D., Jen J. Loss of cytoplasmic CDK1 predicts poor survival in human lung cancer and confers chemotherapeutic resistance. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kong F.F., Qu Z.Q., Yuan H.H., Wang J.Y., Zhao M., Guo Y.H., Shi J., Gong X.D., Zhu Y.L., Liu F., Zhang W.Y. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol. Rep. 2014;31(6):2660–2668. doi: 10.3892/or.2014.3129. [DOI] [PubMed] [Google Scholar]
- 102.Xu N., Jia D., Chen W., Wang H., Liu F., Ge H., Zhu X., Song Y., Zhang X., Zhang D., Ge D. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Q.F., Kong J.L., Zou S.C., Gao H., Wang F., Qin S.M., Wang W. LncRNA LINC00342 regulated cell growth and metastasis in non-small cell lung cancer via targeting miR-203a-3p. Eur. Rev. Med. Pharmacol. Sci. 2019;23(17):7408–7418. doi: 10.26355/eurrev_201909_18849. [DOI] [PubMed] [Google Scholar]
- 104.Jiang N., Jiang X., Chen Z., et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2017;36:138. doi: 10.1186/s13046-017-0604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Greither T., Koser F., Holzhausen H.J., Güttler A., Würl P., Kappler M., Wach S., Taubert H.. MiR-155-5p and MiR-203a-3p Are Prognostic Factors in Soft Tissue Sarcoma. Cancers (Basel). 2020 Aug 12;12(8):2254. [DOI] [PMC free article] [PubMed]
- 106.Chen L., Gao , Liang J., Qiao J., Duan J., Shi H., Zhen T., Li H., Zhang F., Zhu Z., Han A. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am. J. Cancer Res. 2018 Dec 1;8(12):2387-2401. [PMC free article] [PubMed]
- 107.Srivastava S., Garg I, Singh Y., Meena R., Ghosh N, Kumari B., Kumar V., Eslavath M.R., Singh S., Dogra V., Bargotya M, Bhattar S., Gupta U., Jain S., Hussain J., Varshney R, Ganju L. Evaluation of altered miRNA expression pattern to predict COVID-19 severity. Heliyon. 2023 Feb;9(2) doi: 10.1016/j.heliyon.2023.e13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jin X., Guan Y., Zhang Z., Wang H. Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer. BMC cancer. 2020 Dec;20(1) doi: 10.1186/s12885-020-06829-x. 1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ye G., Liu Y., Huang L., Zhang C., Sheng Y., Wu B., Wu C., Qi Y. miRNA-218/FANCI is associated with metastasis and poor prognosis in lung adenocarcinoma: a bioinformatics analysis. Ann Transl. Med. 2021 Aug;9(16):1298. doi: 10.21037/atm-21-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gallach S., Jantus-Lewintre E., Calabuig-Fariñas S., Montaner D., Alonso S., Sirera R., Blasco A., Usó M., Guijarro R., Martorell M., Camps C. MicroRNA profiling associated with non-small cell lung cancer: next generation sequencing detection, experimental validation, and prognostic value. Oncotarget. 2017 Jun 22;8(34):56143-56157. [DOI] [PMC free article] [PubMed]
- 111.Karakas Celik S., Cakmak Genc G., Dursun A. A bioinformatic approach to investigating cytokine genes and their receptor variants in relation to COVID-19 progression. Int. J. Immunogenet. 2021 Apr;48(2):211–218. doi: 10.1111/iji.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shan Y., Chen Y., Brkić J., Fournier L., Ma H., Peng C. miR-218-5p Induces Interleukin-1β and Endovascular Trophoblast Differentiation by Targeting the Transforming Growth Factor β-SMAD2 Pathway. Front. Endocrinol. (Lausanne). 2022 Mar 1;13:842587. [DOI] [PMC free article] [PubMed]
- 113.Feng H., Zhang Z., Qing X., French S.W., Liu D. miR-186-5p promotes cell growth, migration and invasion of lung adenocarcinoma by targeting PTEN. Exp. Mol. Pathol. 2019;108:105–113. doi: 10.1016/j.yexmp.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 114.Wang J., Zhang Y., Ge F. MiR-186 Suppressed Growth, Migration, and Invasion of Lung Adenocarcinoma Cells via Targeting Dicer1. J Oncol. 2021 Nov 11;2021:6217469. [DOI] [PMC free article] [PubMed]
- 115.Petkova V., Marinova D., Kyurkchiyan S., Stancheva G., Mekov E., Kachakova-Yordanova D., Slavova Y., Kostadinov D., Mitev V., Kaneva R. MiRNA expression profiling in adenocarcinoma and squamous cell lung carcinoma reveals both common and specific deregulated microRNAs. Medicine (Baltimore). 2022 Aug 19;101(33):e30027. [DOI] [PMC free article] [PubMed]
- 116.Pollet K., Garnier N., Szunerits S., Madder A., Hober D., Engelmann I. Host mirnas as biomarkers of SARS-COV-2 infection: A critical review. Sensors & Diagnostics. 2023;2(1):12–35. [Google Scholar]
- 117.Gastaldi C., Bertero T., Xu N., Bourget-Ponzio I., Lebrigand K., Fourre S., Popa A., Cardot-Leccia N., Meneguzzi G., Sonkoly E., Pivarcsi A. miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis. 2014;35(5):1110–1120. doi: 10.1093/carcin/bgt490. [DOI] [PubMed] [Google Scholar]
- 118.Wang W.L., Yang Z, Zhang Y.J., Lu P., Ni Y.K., Sun C.F., Liu F.Y. Competing endogenous RNA analysis reveals the regulatory potency of circRNA_036186 in HNSCC. Int J Oncol. 2018 Oct;53(4):1529–1543. doi: 10.3892/ijo.2018.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dama E., Melocchi V., Mazzarelli F., Colangelo T., Cuttano R., Di Candia L., Ferretti G.M., Taurchini M., Graziano P., Bianchi F. Non-Coding RNAs as Prognostic Biomarkers: A miRNA Signature Specific for Aggressive Early-Stage Lung Adenocarcinomas. Non-Coding RNA. 2020;6:48. doi: 10.3390/ncrna6040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang L., Feng H, Li D. Effect of shRNA‑mediated knockdown EBF1 gene expression on the proliferation of lung cancer cell line A549 in vitro and in vivo. Oncol. Rep. 2023 May;49(5):90. doi: 10.3892/or.2023.8527. [DOI] [PubMed] [Google Scholar]
- 121.Gao L., Guo Y.N., Zeng J.H., Ma F.C., Luo J., Zhu H.W, Xia S., Wei K.L., Chen G. The expression, significance and function of cancer susceptibility candidate 9 in lung squamous cell carcinoma: A bioinformatics and in vitro investigation. Int J Oncol. 2019;54:1651–1664. doi: 10.3892/ijo.2019.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee B., Lee T., Lee S.H., Choi Y.L., Han J. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget. 2016 Apr 26;7(17):23874-84. [DOI] [PMC free article] [PubMed]
- 123.Zhan W., Wang W., Han T., Xie C., Zhang T., Gan M., Wang J.B. COMMD9 promotes TFDP1/E2F1 transcriptional activity via interaction with TFDP1 in non-small cell lung cancer. Cell. Signal. 2017;30:59–66. doi: 10.1016/j.cellsig.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 124.Xenos A., Malod-Dognin N., Zambrana C., Pržulj N. Integrated Data Analysis Uncovers New COVID-19 Related Genes and Potential Drug Re-Purposing Candidates. Int. J. Mol. Sci. 2023;24:1431. doi: 10.3390/ijms24021431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang J.X., Wu Y.C., Cheng Y.Y., Wang C.L., Yu C.J. IRF1 negatively regulates oncogenic KPNA2 expression under growth stimulation and hypoxia in lung cancer cells. OncoTargets Ther. 2019;12 doi: 10.2147/OTT.S221832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yoo J.S., Sasaki M., Cho S.X., Kasuga Y., Zhu B., Ouda R., Orba Y., de Figueiredo P., Sawa H., Kobayashi K.S. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021;12(1):1–7. doi: 10.1038/s41467-021-26910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y., Ding X., Liu B., Li M., Chang Y., Shen H., Xie S.M., Xing L., Li Y. ETV4 overexpression promotes progression of non–small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol. Carcinog. 2020;59(1):73–86. doi: 10.1002/mc.23130. [DOI] [PubMed] [Google Scholar]
- 128.Zhou Y.W., Xie Y., Tang L.S., et al. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Sig Transduct Target. Ther. 2021;6:317. doi: 10.1038/s41392-021-00733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang S.S., Liu W., Ly D., Xu H., Qu L., Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 2019;16(1):6–18. doi: 10.1038/s41423-018-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pockley A.G., Vaupel P., Multhoff G. NK cell-based therapeutics for lung cancer. Expet Opin. Biol. Ther. 2020;20(1):23–33. doi: 10.1080/14712598.2020.1688298. [DOI] [PubMed] [Google Scholar]
- 131.Wakabayashi O., Yamazaki K., Oizumi S., Hommura F., Kinoshita I., Ogura S., Dosaka‐Akita H., Nishimura M. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non‐small cell lung cancers. Cancer Sci. 2003;94(11):1003–1009. doi: 10.1111/j.1349-7006.2003.tb01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct. Targeted Ther. 2020;5(1):1–8. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are provided within the manuscript and the supplementary material. The R codes utilized in this study were taken and adjusted from publicly available CRAN projects (https://cran.r-project.org/) and Bioconductor resources (https://www.bioconductor.org/).