Abstract
N6-methyladenosine (m6A) is one of the epigenetic modifications of RNA. The addition of this chemical mark to RNA molecules regulates gene expression by affecting the fate of the RNA molecules. This posttranscriptional RNA modification is reversible and regulated by methyltransferase “writers” and demethylase “erasers”. The fate of m6A-modified RNAs depends on the function of different “readers” that recognize and bind to them. Research on m6A methylation modification has recently increased due to its important role in regulating cancer progression. Noncoding RNAs (ncRNAs) are a class of RNA molecules that are transcribed from the genome but whose roles have been overlooked due to their lack of well-defined potential for translation into proteins or peptides. However, this misconception has now been completely overturned. ncRNAs regulate various diseases, especially tumors, and it has been confirmed that they play either tumor-promoting or tumor-suppressing roles in almost all types of tumors. In this review, we discuss the m6A modification of different types of ncRNA and summarize the mechanisms involved. Finally, we discuss the progress of research on clinical treatment and discuss the important significance of the m6A modification of ncRNAs in the clinical treatment of tumors.
Subject terms: Tumour biomarkers, Targeted therapies
Non-coding RNAs: Modification by adding methyl groups
The reversible addition of a chemical methyl group (CH3) to the base adenine in RNA molecules, creating N6-methyladenosine, regulates the activity of many non-coding RNAs that, unlike messenger RNAs, do not code for the formation of proteins. These RNAs can regulate gene activity. Researchers in China led by Gang Wang at Harbin Medical University, and Qiushi Tang at the Chinese Medical University, Liaoning, review the molecular mechanisms and significance of ‘m6A modification’ of non-coding RNAs. They focus on the progress of research into the role of m6A modification in cancer and its potential relevance for diagnosis and treatment. Approaches to treating cancer by interfering with m6A modification have been explored in mice. Future research will aim to translate the promise of these early animal studies into tests in human cells and clinical trials in patients.
Introduction
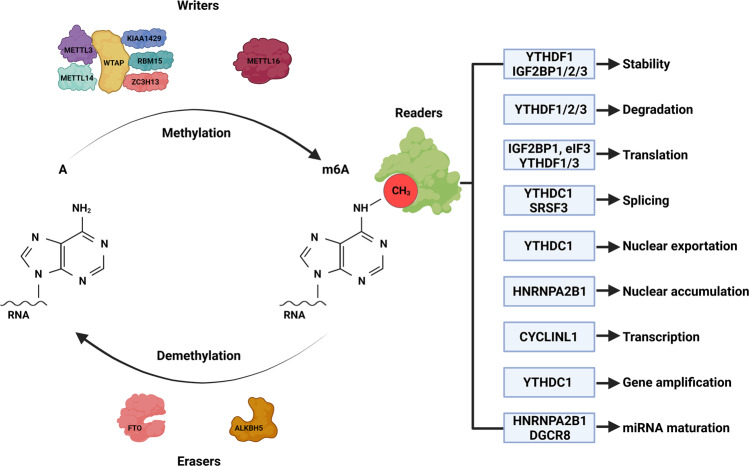
N6-methyladenosine refers to the methylation of the sixth N atom of adenine in RNA molecules (Fig. 1). This modification occurs near stop codons, in 5’- and 3’- untranslated regions, in long internal exons, and in the shared sequence RRACH (R = G/A and H = A/C/U)1. Among the numerous methods of RNA modification discovered to date, m6A methylation is the most common and abundant epigenetic modification of RNA. m6A was first discovered in mRNA in the 1970s2 and has since been found to be widespread in a variety of organisms, such as viruses3, yeast4, and plants5. The m6A modification of RNA is performed by “writers”, including methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), Wilms tumor 1-associated protein (WTAP), KIAA1429, zinc finger CCCH domain-containing protein 13 (ZC3H13), and RNA-binding motif protein 15 (RBM15). METTL3 is the core of the complex and is responsible for catalyzing the methylation of the 6th atoms of adenine (A) in RNA6; METTL14 acts as an RNA binding scaffold to activate and enhance the catalytic activity of METTL37; WTAP has no catalytic activity but promotes m6A methylation by recruiting METTL3 and METTL148; RBM15 binds to METTL3 and WTAP and recruits them to specific RNA sites to promote m6A modification9; ZC3H13 binds to WTAP and allows its retention in nuclear speckles (NSs) to promote m6A modification10; and KIAA1429 guides regioselective methylation by recruiting the m6A methyltransferase complex (MTC) to 3’ UTRs and stop codons11. In addition, methyltransferase-like 15 (METTL15) and methyltransferase-like 16 (METTL16) do not belong to the MTC and independently catalyze m6A methylation. Methyltransferase-like 16 (METTL16), a methylase that was discovered later, promotes m6A modification of U6-snRNA and participates in RNA presplicing12. The erasing of m6A is catalyzed by Human AlkB homolog H5 (ALKBH5) and fat mass and obesity (FTO) of the α-ketoglutarate-dependent dioxygenase family, which are known as “erasers”; thus, this m6A modification is reversible13,14 (Fig. 1). FTO catalyzes demethylation via the oxidation of m6A to N6-hydroxymethyladenosine and N6-formyladenosine, followed by their hydrolysis to adenine; in contrast, ALKBH5 directly removes the m6A modification15. m6A “readers” can be divided into the following categories. Class I readers include those with the YTH domain, including YTHDC1/2 and YTHDF1/2/3. They recognize and bind to m6A-containing transcripts through the YTH domain16. Class II readers are heterogeneous ribonucleic acid proteins (hnRNPs), including hnRNP C and hnRNP A2B1, and they regulate the alternative splicing or processing of transcripts17. The IGF2BP family of proteins, IGF2BP1/2/3, are class III readers, and they share 6 RNA-binding domains, including 2 RNA recognition motifs and 4 KH domains18. Other readers include eukaryotic initiation factor 3 (EIF3)19, which promotes the translation of ncRNAs, and (human antigen R) HuR20, which affects transcript stability (Fig. 1). As mentioned earlier, m6A modification is an important way to regulate gene expression through the modification of multiple steps in mRNA processing. There is evidence that m6A regulates splicing of pre-mRNA21. The mRNA containing m6A was recognized by YTHDC1 and exported to the cytoplasm after splicing22. In the cytoplasm, m6A modification of mRNA affects the stability of mRNA23. In addition, m6A regulates mRNA translation, including translation initiation and translation elongation24,25. Here, we will review the ncRNAs in tumors that are regulated by these three classes of effectors and thus influence tumor progression. With the rapid development of sequencing technology, we found that m6A modification regulates ncRNA fate in many ways. m6A-modified RNAs regulate many pathological and physiological processes by controlling gene expression. Disorders of m6A modification are closely related to the occurrence and development of tumors26. In addition, some other RNA modification methods, such as N1-methyladenosine (m1A), 5-methylcytidine (m5C), N (7)-methylguanosine (m7G), and N4-acetylcytidine (ac4C), will also be briefly described at the end of this review.
Fig. 1. The structure and reversibility of m6A modifications and the function of m6A through recognition by the reader protein.
m6A modification refers to the methylation of the sixth N atom of adenine in RNA molecules, which can be catalyzed by the MTC writing complex consisting of the core components of METTL3-METTL14-WTAP and other regulatory cofactors or by METTL16 alone. The m6A site on RNA can be recognized by the reader protein, causing changes in RNA function, and can also be reversibly removed by the erasure protein FTO or ALKBH5.
ncRNAs are RNAs that are transcribed from the genome and were once thought to be byproducts of meaningless transcription because they are not translated into proteins. However, as research progressed, it was found that these “junk RNAs” are widely involved in important biological functions. ncRNAs comprise many species, many of which have been identified as markers of posttranscriptional regulation. Here, we mainly describe the three types of ncRNA that are most commonly modified by m6A, namely, microRNAs (miRNAs), long noncoding RNAs (LncRNAs) and circular RNAs (circRNAs). miRNAs are a class of ncRNAs that are approximately 20–40 nucleotides in length. The primary transcription product of a miRNA gene, namely, pri-miRNA, is cleaved into precursor miRNA by RNase III Drosha in the nucleus and then transported to the cytoplasm, where it is further cleaved and matured by RNase III Dicer. Mature miRNAs target the 3’ untranslated region of mRNAs to regulate gene expression27. LncRNAs are ncRNAs that are at least 200 nucleotides in length. There are many lncRNAs, and they not only play auxiliary roles as intermediate carriers of genetic information but also perform various regulatory functions, such as genomic imprinting28, chromatin modification29, transcriptional activation30, and transcriptional interference31. Although a large number of studies on lncRNAs have been conducted in recent years, our understanding of them is limited, and the functions of most lncRNAs are still unknown; thus, we are interested in continuing to study these molecules32. CircRNAs are widely expressed in human cells, and pre-mRNAs form this unique ncRNA through reverse splicing. They do not have 5’ end caps and 3’ end poly(A) tails and exist as circular covalently closed structures. CircRNAs perform important regulatory functions, such as acting as competitive endogenous RNAs (ceRNAs)33.
The m6A methylation modification of ncRNAs with regulatory functions is an important part of epigenetic regulation. m6A modification affects various aspects of pathophysiological processes by changing the structure, biogenesis and function of ncRNAs, and it is crucial to the occurrence and progression of many diseases, especially cancer. This research hotspot has been reviewed in previous articles34–38. In recent years, many related studies have elucidated new mechanisms and provided new ideas. This review summarizes the mechanisms involved and discusses the importance of targeting m6A-modified ncRNAs in the treatment of cancer.
Dysregulation of m6A writers in cancer
As mentioned above, the MTC, which primarily mediates m6A modification, consists of several proteins. In addition, some proteins, such as METTL16, function independently of the complex. A reader then recognizes and binds to the ncRNA at the m6A site. Figure 2a shows that ncRNAs are modified by dysregulated writers in tumors and that readers bind to these ncRNAs.
Fig. 2. ncRNAs regulated by writers, erasers, and readers in different cancers.
ncRNAs are modified by writer proteins and recognized by reader proteins (a). ncRNAs targeted by eraser proteins and recognized by reader proteins (b).
METTL3
METTL3 is an S-adenosylmethionine (SAM)-binding protein. Its N-terminal extension includes a leading helix and a nuclear localization signal region, followed by a zinc finger domain (ZFD), a partly ordered linker and a C-terminal MTase domain (MTD). Among these domains, the MTD forms the smallest and most stable complex with the MTD and N-terminal extension of METTL14, which can bind to the cofactor substrate SAM or produce SAH39.
Long noncoding RNA
METTL3 catalyzes the methylation of lncRNAs and increases their expression level by reducing RNA degradation and stabilizing RNA transcripts40–55. The increased stability of LncRMRP due to m6A modification activates the SMAD2/SMAD3 pathway by enhancing TGFBR1 transcription, ultimately accelerating non-small cell lung cancer (NSCLC) progression42. Conversely, after m6A methylation modification, some lncRNAs are downregulated due to reduced stability. Due to METTL3-mediated m6A modification, LncMEG3 expression is downregulated, its inhibitory effect on miR-544b is weakened, and the decreased expression of BTG2 promotes the progression of hepatocellular carcinoma (HCC)56.
m6A-modified lncRNAs sequester miRNAs, causing endogenous competition. Via METTL3 catalysis, the LncZFAS1 expression level remained unchanged, while the m6A level increased. LncZFAS1 sequesters miR-647 in an m6A-dependent manner to exert an overall cancer-promoting effect57.
Another important function of m6A-modified lncRNAs is binding to other molecules as scaffolds. By binding to CYCLINL1 at the m6A site, NEAT1-1 becomes a bridge that connects CYCLINL1 and CDK19 and then phosphorylates pol II Ser2 in the RUNX2 promoter, ultimately promoting the bone metastasis of prostate cancer (PC)58.
In addition, m6A modification increases the nuclear accumulation of lncRNAs. METTL3-mediated m6A modification leads to increased nuclear accumulation of LncRP11, and then, LncRP11 reduces the Siah1 and Fbxo45 levels and increases Zeb1 stability by forming the RP11-hnRNP A2B1-mRNA complex, ultimately playing an oncogenic role in colorectal cancer (CRC)43.
A recent study also found that METTL3 catalyzes m6A modification to promote lncRNA splicing. In pancreatic cancer (PAC), m6A-modified LncANRIL undergoes splicing with the participation of SRSF3. The spliceosome ANRIL-208 is increased and complexed with RING1b and EZH2 to mediate gemcitabine resistance by enhancing DNA HR repair59.
Circular RNA
circRNAs, a type of ncRNA, usually lack the potential to be translated into proteins or peptides. However, after modification with m6A, they are able to be translated. m6A-methylated circMAP3K4, catalyzed by METTL3, can be translated into circMAP3K4-455aa with the help of IGF2BP1. This protein interacts with AIF to protect it from cleavage and reduce its nuclear distribution, ultimately leading to cisplatin resistance in HCC cells60.
METTL3 catalyzes circRNA methylation and increases its expression, mainly by reducing RNA degradation and stabilizing RNA transcripts61–64. METTL3-mediated m6A modification leads to the upregulation of circSORE, which sequesters miR-103a-2-5p and miR-660-3p. The Wnt/β-catenin pathway is activated, and HCC develops resistance to sorafenib61. Conversely, some m6A-modified circRNAs undergo endoribonucleolytic cleavage that is mediated by the reader, resulting in reduced stability. The protein ASK1-272a.a encoded by circASK1 competitively binds to Akt1, and the ASK1/JNK/p38 signaling pathway is activated to induce apoptosis and chemosensitivity in lung adenocarcinoma (LUAD)65.
m6A modification promotes circRNA back splicing and biosynthesis66–68. METTL3 overexpression mediates the m6A modification of circARL3 and promotes its biosynthesis. circARL3 acts as a ceRNA to absorb miR-1305, a molecule that inhibits target oncogenes, thereby promoting the growth of HBV + liver cancer cells66.
METTL3-mediated m6A modification promoted circRNA nuclear output. METTL3-mediated m6A modification leads to enhanced expression and cytoplasmic transport of circHPS5 and its increased ability to absorb miR-370 and upregulates HMGA2, ultimately accelerating HCC cell tumorigenesis69.
pre-microRNA
The biological significance of m6A modification in pre-microRNAs is mainly the promotion of their biological formation70–72. The current study mainly describes the following mechanisms: METTL3 promotes the splicing and maturation of miRNA precursors. METTL3-catalyzed m6A modification triggers the biogenesis of miR143-3p by promoting the splicing of pre-microRNA, leading to brain metastases in lung cancer71.
METTL14
The catalytic domain of METTL14 does not contain SAM and exists in a closed conformation. METTL14 and METTL3 are connected by a loop near the active site of METTL3 to form a stable 1:1 heterodimer, which performs a more stable and powerful m6A catalytic function. The N-terminus of METTL14 contains a long helix that is approximately 500 amino acids long and interacts with METTL13, and an extension parallel to the N-terminal helix at the C-terminus achieves balance and functional coordination73. METTL14 increases the level of m6A methylation modification in lncRNAs and changes their stability74–80. METTL14 increases the levels of m6A-modified FAM225A and enhances its stability. FAM225A acts as a ceRNA to absorb miR-590-3p and miR-1275, eventually upregulating the target gene ITGB3 to promote nasopharyngeal carcinogenesis (NPC) cell tumorigenesis and metastasis74. When methylated by METTL14, lncRNAs act as molecular scaffolds and bind to m6A readers81,82. METTL14 catalyzes the m6A modification of LncPACERR. LncPACERR acts as a molecular scaffold to bind to IGF2BP2 and synergistically enhance the stability of KLF12 and C-MYC in TAMs82. In addition to IGF2BP2, m6A modifications catalyzed by METTL14 can also be recognized by other reader proteins, such as HuR, YTHDF1 and YTHDF278,79,81.
METTL14 silencing leads to increased ncRNA expression in some tumors. Downregulation of METTL14 increases circORC5 expression in gastric cancer (GC). CircORC5 acts as a miR-30C-2-3p sponge and reverses the METTL14-induced upregulation of miR-30C-2-3p and downregulation of AKT1S1 and EIF4B, promoting GC cell growth and invasion83. The significance of METTL14-mediated m6A modification of pri-microRNAs is the increased maturation of its precursor84,85. When modified by METTL14, pri-microRNAs recruit DGCR8 and promote its maturation. METTL14 mediates the m6A modification of pri-miR-211, which in turn accelerates miR-211 biosynthesis through DGCR8 silencing. Increased miR-211 expression reduces cell proliferation in T-cell lymphoblastic lymphoma (T-LBL) by targeting the miR-211/TCF12 axis85.
METTL3/14
METTL3/14 catalyzes the m6A modification of ncRNAs and stabilizes their transcripts. After modification, LncAROD exhibits enhanced stability and directly binds to SRSF3 to induce the switch from PKM to PKM2. As a ceRNA that inhibits miR-145-5p, LncAROD enhances glycolysis, cell proliferation, migration, invasion and chemical resistance of HCC cells76. When METTL3/14 cooperate to catalyze m6A methylation modification, ncRNAs act as molecular scaffolds to promote the formation of various complexes. For instance, circNDUFB2 forms a ternary complex with TRIM25 and IGF2BPs to promote the ubiquitination and degradation of IGF2BPs, which are tumor-promoting factors86.
WTAP
WTAP is a splicing regulator that contains an extended N-terminal helical coil region and an unstructured C-terminal portion. The N-terminal portion of WTAP interacts with the N-terminal leader helix (LH) of METTL3, allowing METTL3/14 to localize to the nuclear speckle and play an important role in RNA m6A methylation87. WTAP promotes the methylation modification of ncRNAs and favors their stability88–90. WTAP enhances the stability of transcripts by promoting the m6A modification of LncFOXD2-AS1, and FOXD2-AS1 binds to its target FOXM1 mRNA to form a complex that promotes the stability of the latter and ultimately promotes the occurrence of osteosarcoma (OS)89. WTAP binds to the m6A site of LncDLGAP1-AS1 to improve its stability, and DLGAP1-AS1 sponges miR-299-3p to reverse the inhibition of WTAP mRNA, forming a regulatory loop and promoting adriamycin (ADR) resistance in breast cancer (BRC)90.
KIAA1429
KIAA1429, the largest factor related to m6A, is localized to NSs and has been shown to recruit METTL3, METTL14 and WTAP to guide regionally selective methylation. KIAA1429-catalyzed m6A modification typically occurs near 3’-UTRs and stop codons. Similar to METTL3 and METTL14, KIAA1429-mediated m6A methylation may cause changes in the expression of some ncRNAs. In HCC, KIAA1429 catalyzes the m6A methylation modification of circDLC1, reduces its expression level, and promotes tumor progression by altering the circDLC1-HuR-MMP1 axis91.
RBM15
RBM15 recruits MTC to its target transcript by directly binding to U-rich sequences in RNA92. RBM15 specifically targets ncRNA secondary structures, such as the A-repeat region of LncXIST. There is a conserved AUCG tetraloop hairpin in the A repeat region of LncXIST. RBM15 recruits the write complex to XIST for m6A modification by binding to the A-repeat, and the YTH domain recognizes the m6A site in the A-repeat (m6A) UCG tetraloop. The (m6A) UCG tetraloop of the XIST A-repeat hairpin RNA is bound by an arc-like surface of the YTH domain93. In esophageal squamous cell carcinoma (ESCC), RBM15 interacts with METTL3 in a WTAP-dependent manner to deposit m6A on LncMALAT1. LncMALAT1 binds to YTHDC1 as a molecular scaffold to maintain its nuclear localization and related oncogene expression, ultimately promoting metastasis of cancer cells94.
Abnormal m6A modification of ncRNAs that is regulated by writer proteins affects tumor development. We summarize the role of methyltransferase and its target ncRNAs in various cancers (Table 1).
Table 1.
The role of methyltransferases and their regulated ncRNAs in various cancers.
| Writer | Role in cancer | Type | Target ncRNAs | Mechanism |
|---|---|---|---|---|
| METTL3 | Oncogene | CC | LncFOXD2-AS150 | Stability |
| circ000006963 | Stability | |||
| LncZFAS157 | Adsorb microRNAs | |||
| HCC | LncAROD49 | Stability | ||
| LNC127355 | Stability | |||
| circMAP3K460 | Translation | |||
| circSORE61 | Stability | |||
| circARL366 | Circularization | |||
| circHPS569 | Cytoplasmic output | |||
| CRC | LncPTTG3P41 | Stability | ||
| LncRP1143 | Nuclear accumulation | |||
| circALG164 | Stability | |||
| circ166267 | N/A | |||
| LUAD | LncLCAT347 | Stability | ||
| pri- miR-143-3p71 | Maturation | |||
| pri-miR-106b133 | Maturation | |||
| NSCLC | LncRMRP41 | Stability | ||
| circIGF2BP367 | Circularization | |||
| PAC | LncLIFR-AS151 | Stability | ||
| LncANRIL58 | Splicing | |||
| pri-miR-2570 | Maturation | |||
| BRC | LNC0095846 | Stability | ||
| LncMALAT1122 | Stability | |||
| PC | LncNEAT1-158 | Scaffold | ||
| LncSNHG754 | Stability | |||
| GC | LncTHAP7-AS148 | Stability | ||
| EOC | LncRHPN1-AS144 | Stability | ||
| TET | LncMALAT151 | Stability | ||
| RCC | LncDUXAP945 | Stability | ||
| HNSCC | LncAROD76 | Stability | ||
| HPSCC | circCUX162 | Stability | ||
| ESCC | pri-miR-320b72 | Maturation | ||
| Tumor suppressor | LUAD | circASK165 | Stability | |
| GBM | LncMALAT153 | Stability | ||
| HCC | LncMEG356 | N/A | ||
| METTL14 | Oncogene | HNSCC | LncAROD76 | Stability |
| NPC | LncFAM225A74 | Stability | ||
| HCC | LncMIR155HG79 | Stability | ||
| GC | LNC0132075 | Stability | ||
| circORC583 | N/A | |||
| TLBL | pri-miR-21185 | Maturation | ||
| BRC | miR-146a-5p84 | Reshape the miRNA profile | ||
| CRC | LncXIST78 | Stability | ||
| PAC | LncPACERR82 | Scaffold | ||
| RCC | LncLSG181 | Scaffold | ||
| Tumor suppressor | RCC | LncNEAT180 | Stability | |
| Tumor suppressor | LUAD | LncHCG1177 | Stability | |
| METTL16 | Tumor suppressor | HCC | LncRAB11B-AS197 | Stability |
| METTL3/14 | Oncogene | HCC | LncAROD49 | Stability |
| Oncogene | NSCLC | circNDUFB286 | Scaffold | |
| WTAP | Oncogene | NPC | LncDIAPH1-AS188 | Stability |
| Oncogene | OS | LncFOXD2-AS150 | Stability | |
| Oncogene | BRC | LncDLGAP1-AS190 | Stability | |
| KIAA1429 | Oncogene | GC | LNC00958149 | Stability |
| Tumor suppressor | HCC | circDLC191 | N/A | |
| RBM15 | Oncogene | ESCC | LncMALAT194 | Scaffold |
CC cervical cancer, HCC hepatocellular carcinoma, CRC colorectal cancer, LUAD lung adenocarcinoma, NSCLC non-small cell lung cancer, PAC pancreatic cancer, BRC breast cancer, PC prostate cancer, GC gastric cancer, EOC epithelial ovarian cancer, TET thymic epithelial tumor, RCC renal cell carcinoma, HNSCC head and neck squamous cell carcinoma, HPSCC hypopharyngeal squamous cell carcinoma, ESCC esophageal squamous cell carcinoma, GBM glioblastoma multiforme, NPC nasopharyngeal carcinogenesis, TLBL T-cell lymphoblastic lymphoma.
METTL16
The conserved core of the SAM methyltransferase METTL16 consists of β-sheets, α-helices and 310(η) helices. The order of the 7-strand β-sheets is 3214576, located between the α and 310 (η) helical clusters. SAM and RNA binding sites are created within the N-terminal and C-terminal segments of the β-sheet, respectively95. In some ncRNAs, such as MALAT1, the U-rich internal loop at the 3’ end of MALAT1 associates with a downstream genomically encoded A-rich stretch to form a bipartite triple helix composed of canonical triples: nine U•A-U, one C•G-C, and a C-G doublet. The triple helix structure stabilizes MALAT1 and allows it to accumulate in cells96. METTL16 recognizes the RNA triple helix at the 3’ end of MALAT195. Similar to METTL3 and METTL14, METTL16 also affects the stability of ncRNA by inducing the catalytic modification of ncRNAs with m6A. METTL16 catalyzes the m6A modification of LncRAB11B-AS1 to reduce its stability. When LncRAB11B-AS1 expression is downregulated, the malignant phenotype of HCC cells is inhibited97.
Dysregulation of m6A erasers in cancer
To date, two types of erasers have been identified, namely, FTO and ALKBH5. Although there are few types of eraser proteins, their dysfunction has been observed in a variety of tumors. In light of their important role in cancer, we list demethylated ncRNAs (Fig. 2b) and explain their mechanisms of action.
FTO
FTO was the first RNA demethylase to be discovered, and its dysregulation plays tumor-promoting or tumor-suppressing roles13. There are two pairs of positively charged residues that bind to oligonucleotide-like pincers in two loops near the oligonucleotide-binding region of FTO. A hydrophobic pocket is formed in the inner chain of the catalytic pocket of FTO, and the N6 methyl group is stably bound in this pocket for oxidation to Fe(II) and α-KG98. FTO erases the m6A methylation modification of ncRNAs, ultimately inhibiting the degradation of ncRNAs through m6A readers. In ESCC, FTO removes the m6A modification of LNC00022, leading to the inhibition of LNC00022 degradation via the m6A reader YTHDF2 and promoting tumor proliferation by accelerating P21 ubiquitination and degradation99. FTO increases the stability of ncRNAs by reducing methylation modification. When targeted by FTO, LncHOXC13-AS is upregulated and interacts with CBP to induce FZD6 upregulation, promoting Wnt/β-catenin activation and ultimately leading to increased cervical cancer (CC) invasiveness100.
ALKBH5
Similar to all eukaryotic ALKBH family members, the basic scaffold of ALKBH5 consists of eight antiparallel β-sheets, and the secondary structure motifs are essential for the catalytic function of ALKBH5. For example, motif 1 is associated with the activity of the ALKBH5 enzyme, and its substrate specificity is conferred by several motifs, including important active site coordinating residues HxD. H motif and motif 2, which specifically bind to the m6A single-chain substrate body101. ALKBH5 increases the stability of ncRNAs102–104. Under hypoxic conditions, the stability of LncNEAT1 is increased because its m6A modification is eliminated by ALKBH5, and the posttranscriptional repressor SFPQ is relocated from its position in the CXCL8 promoter to paraspeckles, ultimately upregulating CXCL8/IL8 and promoting glioblastoma multiforme (GBM) progression103. Conversely, ALKBH5 also reduces the stability of ncRNAs105,106. In PAC, downregulation of ALKBH5 allows demethylation and decreased expression of LncKCNK15-AS1, which inhibits epithelial–mesenchymal transition, resulting in tumor invasion and metastasis105. ALKBH5 upregulates the level of catalyzed ncRNAs107–110. ALKBH5 upregulates circCPSF6 through demethylation, and circCPSF6 competitively interacts with PCBP2 to stabilize YAP1 mRNA and promote the malignant phenotype of HCC110. ALKBH5 upregulates LncNEAT1 expression through demethylation, leading to the overexpression of a subunit of the polycomb repressive complex EZH2 and ultimately promoting the malignant phenotype of GC107. ncRNAs modified by ALKBH5 act as molecular scaffolds. ALKBH5 catalyzes the demethylation of LncTRERNA1 and upregulates its expression. LncTRERNA1 acts as a molecular scaffold to recruit EZH2 and silence its expression through H3K27me3 modification of the promoter region of P21, which is a cyclin-dependent kinase inhibitor, thus promoting cell proliferation and cell cycle progression in diffuse large B-cell lymphoma (DLBCL)111. ncRNA molecules that are targeted by ALKBH5 have silencing potential. ALKBH5 deficiency increases the level of m6A modification of miR-21-5p and increases the expression level of miR-21-5p target genes, such as MAPK10, PTEN, SMAD7, PDCD4 and SOX7, in NSCLC112.
The dysregulation of ncRNA m6A modification catalyzed by eraser proteins plays an important role in tumorigenesis and progression. We summarize the role of demethylases and their target ncRNAs in various cancers (Table 2).
Table 2.
The role of demethylases and their regulated ncRNAs in various cancers.
| Eraser | Role in cancer | Type | Target ncRNAs | Mechanism |
|---|---|---|---|---|
| FTO | Oncogene | ESCC | LNC0002299 | Stability |
| CC | LncHOXC13-AS100 | Stability | ||
| ALKBH5 | Oncogene | OS | LncPVT1104 | Stability |
| GBM | LncNEAT1103 | Stability | ||
| LSCC | LncKCNQ1OT1117 | Stability | ||
| DLBCL | LncTRERNA1111 | Scaffold | ||
| CRC | circ3823102 | Stability | ||
| HCC | circCPSF6110 | Stability | ||
| NSCLC | miR-21–5p112 | Silencing potency | ||
| GC | LncNEAT1107 | N/A | ||
| CRC | LncNEAT1109 | N/A | ||
| LUAD | LncRMRP108 | N/A | ||
| Tumor suppressor | ESCC | LNC00278106 | Translation | |
| Tumor suppressor | PAC | LncKCNK15-AS1105 | N/A |
ESCC esophageal squamous cell carcinoma, CC cervical cancer, OS osteosarcoma, GBM glioblastoma multiforme, LSCC laryngeal squamous cell carcinoma, DLBCL diffuse large B-cell lymphoma, CRC colorectal cancer, HCC hepatocellular carcinoma, NSCLC non-small cell lung cancer, GC gastric cancer, LUAD lung adenocarcinoma, PAC pancreatic cancer.
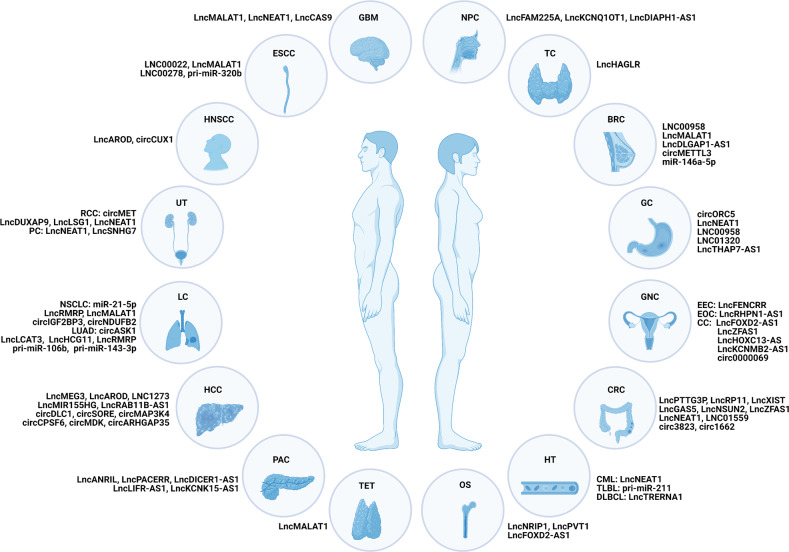
Thus far, it is not difficult to find that dysregulated m6A modification of ncRNAs, including increased m6A modification caused by methyltransferases and decreased m6A modification by demethylases, affects the development of various cancers. These changes have been observed in more than 20 tumors (Fig. 3).
Fig. 3. Dysregulation of m6A-modified ncRNAs in human cancer.
GBM glioblastoma multiforme, ESCC esophageal squamous cell carcinoma, LSCC laryngeal squamous cell carcinoma, HNSCC head and neck squamous cell carcinoma, UT urinary tract, RCC renal cell carcinoma, BC bladder cancer, PC prostate cancer, LC lung cancer, NSCLC non-small cell lung cancer, LUAD lung adenocarcinoma, HCC hepatocellular carcinoma, PAC pancreatic cancer, TET thymic epithelial tumor, OS osteosarcoma, HT hematological system, CML chronic myeloid leukemia, TLBL T-cell lymphoblastic lymphoma, DLBCL diffuse large B-cell lymphoma, CRC colorectal cancer, GNC genital system, EEC endometrioid endometrial carcinoma, EOC epithelial ovarian cancer, CC cervical cancer, GC gastric cancer, BRC breast cancer, TC thyroid cancer, NPC nasopharyngeal carcinogenesis.
Dysregulation of m6A readers in cancer
YTHDF1
The YTH domain of YTHDF1 consists of 5 α helices (α0-α4), 6 parallel β chains (β1-β6), and a 310 helix following the β5 chain. In this YTH domain, the rings between β4β5 and β1α1β2 at the C-terminus and α2 at the N-terminus form the m6A-binding pocket. The aromatic cage composed of Trp411, Trp465 and Trp470 specifically recognizes m6A-modified sites113. YTHDF1 is the most versatile and robust of all reader proteins; it recognizes G (m6A) C and A (m6A) C RNA sequences without sequence selectivity114. YTHDF1 binds to m6A sites on ncRNAs and recruits translation machinery to increase translation efficiency. Yin Yang 1 (YY1)-binding micropeptide is translated by m6A-modified LNC00278 with the help of YTHDF1, which blocks the interaction between YY1 and the androgen receptor AR, reduces the expression of eEF2K through the AR signaling pathway, increases the level of caspase-3, and causes ESCC cells to undergo apoptosis. ALKBH5-mediated demethylation in patients who smoke inhibits this process and ultimately mediates the malignant phenotype of ESCC106.
YTHDF2
The YTH region of YTHDF2 has a spherical shape overall, with 4 β-strands (β1-β4) in the center, 4 α-helices (α1-α4) around it, and flanking regions on each side. The YTH surface has a structure that is rich in basic residues, in which the K416 residue on the β1 strand and the R527 residue on the linker helix α3α4 are essential for binding m6A methyl groups. In the nearby hydrophobic pocket, residues W432 and W486 are involved in the specific recognition of m6A modifications115. According to high-throughput sequencing, YTHDF1 and YTHDF2 have no sequence preference and recognize the m6A site equally116. YTHDF2 binds to m6A-modified sites in lncRNAs to promote their transcriptional stability. In laryngeal squamous cell carcinoma (LSCC), decreased ALKBH5 levels lead to increases in m6A modification of LncKCNQ1OT1 and enhanced LncKCNQ1OT1 stability mediated by YTHDF2. LncKCNQ1OT1 directly binds to HOXA9 to further regulate the proliferation, invasion and metastasis of LSCC cells117. When m6A is recognized by YTHDF2, ncRNAs are degraded65,78,99,118. METTL14 induces m6A methylation modification of LncXIST and reduces XIST expression through YTHDF2-dependent RNA degradation, which plays an important role in CRC progression78. Similarly, FTO decreases the m6A modification of LNC00022, and the weakened binding of YTHDF2 accelerates the ubiquitination and degradation of the P21 protein by inhibiting the degradation of LNC00022, thus promoting the proliferation of ESCC99.
YTHDF3
YTHDF3, the third member of the YTH family, functions as a partner of YTHDF1 and YTHDF2 to promote translation by recognizing m6A sites and affecting the cytoplasmic metabolism of RNAs119,120. By recognizing m6A sites, YTHDF3 changes the stability of ncRNAs118,121,122. LncDICER1-AS1 promotes the transcription of its sense gene DICER1 by recruiting the YY1 transcription factor to the DICER1 promoter. DICER1 promotes the maturation of miR-5586-5p, thereby inhibiting the expression of the glycolytic genes LDHA, HK2, PGK1 and SLC2A1. YTHDF3 binds to the m6A site of LncDICER1-AS1 to promote its degradation, which ultimately enhances the glycolysis of PAC cells118. YTHDF3 recognizes the m6A site of LncGAS5 and promotes its ubiquitin-mediated degradation. After the content of LncGAS5 decreases, its ability to promote the degradation of YAP decreases, and the level of YAP increases. YTHDF3 is the target of YAP, and its level increases with increasing YAP levels. The three form a regulatory loop that promotes the progression of CRC105.
There are strong debates regarding whether DF1/DF2 have distinct roles or redundant roles. Previously, different YTHDF paralogs were thought to be bound to different m6A sites to perform different functions24,119,123,124. However, with advances in research, new studies have proposed that the YTH domain and all m6A sites are equally bound and that the three DF paralogs act redundantly to mediate mRNA degradation and cellular differentiation125–127. These studies reveal a unified model of m6A function in which all m6A-modified mRNAs are subjected to the combined action of YTHDF proteins in proportion to the number of m6A sites126.
YTHDC1
The YTH domain consists of 5 α-helices (α0-α4), 6 β-chains (β1-β6) and 310 helices. The m6A binding pocket consists of a β1 residue, a ring between α1 and β1, α1, β1 and a partial ring connecting the β4 and β5 chains, while the aromatic pocket that recognizes m6A modification sites consists of W377, W428 and L439128. YTHDC1 regulates RNA metabolic processes in a m6A-dependent manner16,129. YTHDC1 locates oncogenes in nuclear spots by recognizing the m6A site in ncRNAs, amplifying the expression of oncogenes. The m6A site in MALAT1 acts as a scaffold for recruiting YTHDC1 to NSs and promotes the upregulation of key oncogenes in ESCC94. YTHDC1 binds to the m6A site of ncRNAs to promote their reverse splicing and biosynthesis66,68. METTL3 overexpression mediates the m6A modification of circARL3, and YTHDC1 binds to the m6A site and promotes its biosynthesis. circARL3 acts as a ceRNA to absorb miR-1305 and causes downregulation of WNT2, UBE2T, MDM2, TGF-β2 and POLR3G, thereby promoting the growth of HBV + HCC cells66. Another example is YTHDC1, which recognizes the site of METTL3-mediated m6A modification in circIGF2BP3 and promotes circIGF2BP3 cyclization. circIGF2BP3 acts as a ceRNA to absorb miR-328-3p and miR-3173-5p and upregulates the expression of PKP3, thereby destroying tumor immunity in NSCLC68. YTHDC1 promotes the nuclear output of ncRNA69,130. YTHDC1 promotes the nuclear output of ncRNAs. With the help of YTHDC1, m6A-modified circMET is transported to the cytoplasm, where it promotes the growth of NONO-TFE3 tRCC by enhancing CDKN2A mRNA attenuation, competitively absorbing miR1197 and blocking SMAD3 mRNA expression130. METTL3-mediated m6A modification increases the expression of circHPS5, while YTHDC1 promotes the transport of circHPS5 to the cytoplasm. These two act together to increase the ability of circHPS5 to function as a ceRNA to absorb miR-370, upregulate HMGA2, and promote the progression of HCC69.
YTHDC2
YTHDC2 affects tumor progression by regulating RNA metabolism. YTHDC2 affects tumor progression by regulating RNA metabolism. YTHDC2 binds to m6A-modified mRNA and promotes its degradation. m6A-modified SLC7A11 mRNA binds to YTHDC2, which enhances the degradation of YTHDC2 and reduces cystine uptake, thereby blocking the downstream antioxidant program and ultimately suppressing the progression of LUAD131. In addition, YTHDC2 binds to mRNA to promote its translation initiation. YTHDC2 binds to IGF1R mRNA and promotes its translation initiation to activate the IGF1R-AKT/S6 axis, which ultimately leads to NPC radioresistance132. However, no study has shown the role of YTHDC2 in ncRNA m6A modification.
IGF2BP1
IGF2BP1, the first IGF2BP family member to be described, consists of six typical binding domains, including four C-terminal K homology (KH) domains that promote RNA binding and two N-terminal RNA recognition domains (RRMs) that promote the stability of IGF2BP-RNA complexes133. IGF2BP1 binds to m6A sites in ncRNAs, increasing their stability. IGF2BP1 recognizes the m6A modification site in circMDK to increase the stability of its transcripts. circMDK sponges miR-346, and miR-874-3p upregulates autophagy-related 16-like 1 (ATG16L1), thereby activating the PI3K/AKT/mTOR signaling pathway and promoting HCC cell proliferation, migration and invasion134. METTL3 and IGF2BP1 act together to increase the stability of LncAROD through m6A modification. Elevated LncAROD increases the levels of pyruvate kinase isoform M2 (PKM2) in hypoxic environments or HIF1α and promotes HCC progression49.
IGF2BP2
IGF2BP2 is widely expressed in a variety of tissues and is an independent prognostic factor for various cancer types; it functions by recognizing the m6A sites of different types of RNAs, such as miRNAs, mRNAs and LncRNAs, that are involved in the occurrence and growth of cancer135. IGF2BP2 increases the stability of ncRNAs41,45,77,136–140. For example, IGF2BP2 recognizes and stabilizes LncDANCR harboring m6A methylation, and then, these molecules work together to promote the progression of PC137. IGF2BP2 also contributes to the formation of multiplex complexes. The m6A methylation-modified circNDUFB2 functions as a scaffold that forms a ternary complex with TRIM25 and IGF2BPs to promote the ubiquitination and degradation of the NSCLC-promoting factor IGF2BPs86. When circNSUN2 undergoes m6A modification by METTL3, its transport to the cytoplasm increases with the help of YTHDF2. CircNSUN2 enters the cytoplasm to form a circNSUN2/IGF2BP2/HMGA2 RNA protein ternary complex, increasing the stability of HMG mRNA and enhancing CRC malignancy141.
IGF2BP3
Similar to IGF2BP1, IGF2BP3 is expressed in only a few tissues and cells of healthy adults but is resynthesized or substantially upregulated in various tumors. Therefore, to some extent, they can be characterized as oncofetal genes142. IGF2BP3 recognizes and binds to the m6A site of ncRNAs to improve their stability. IGF2BP3 increases the expression of LncKCNMB2-AS1 by binding to the m6A methylation modification site. Upregulated LncKCNMB2-AS1 acts as a competing endogenous RNA of miR-130b-5p and miR-4294 and ultimately facilitates CC progression143.
hnRNPs
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a class of RNA-binding proteins (RBPs). There are four unique RNA-binding domains (RBDs) in hnRNP proteins: RNA recognition mods (RRM), quasi-RRM, glycine rich domains that constitute RGG boxes, and KH domains. RRM is the most common RBD, and it includes two conserved RNP1 octamer and RNP2 hexacomer sequences and four β-sheets and two α-helices (βαβαβ) that are critical for RNA binding specificity. hnRNPs play an important role in many aspects of nucleic acid metabolism144. hnRNP A2B1 promotes the maturation of miR-106b-5p by reading the m6A site in pri-miR-106b, thereby reducing the level of secreted frizzled-related protein 2 (SFRP2) to activate the Wnt/β-catenin signaling pathway and promote LUAD tumor occurrence145. hnRNP A2B1 binds to the m6A site in LncRP11 to increase its nuclear accumulation and recruit Siah1 and Fbxo45 mRNA. Enhanced mRNA degradation of two E3 ligases, Siah1 and Fbxo45, prevents Zeb1 proteasome degradation and ultimately promotes the CRC malignant phenotype43.
HuR
HuR is an RNA-binding protein that recognizes U/AU-rich elements in different RNAs through two RNA recognition motifs, namely, RRM1 and RRM2. The RRM domain adopts a β1-α1-β2-β3-α2-β4 topology with two α-helices stacked in an antiparallel four-stranded β-sheet. Residues located at conserved positions on β strands 1 and 3 are critical for RN binding. HuR tightly regulates target RNA fate at the posttranscriptional level146. HuR binds to the m6A sites of ncRNAs to stabilize their transcripts53,79. Lipopolysaccharide (LPS) of intestinal bacteria upregulates METTL14, promotes LncMIR155HG methylation and stabilizes it in a HuR-dependent manner. LncMIR155HG acts as a ceRNA to induce PD-L1 expression through the miR-223/STAT1 axis and plays an important role in immune escape in HCC79.
EIF3
Translation initiation in eukaryotes relies on a number of eukaryotic initiation factors (eIFs) that stimulate the recruitment of initiating tRNA, Met-TRNAiMet, and mRNA to the 40S ribosomal subunit and subsequently scan the mRNA for the AUG initiation codon. EIF3, one of the first initiating factors to be discovered in the 1970s, organizes a network of interactions among several eIFs. Studies have found that circRNAs, as ncRNAs, do not have the ability to be translated into proteins or peptides, but when they undergo m6A modification and EIF3 binds to the m6A site, some ncRNAs can be translated. When EIF3 binds to the m6A modification site in the initiation codon of circARHGAP35, a truncated protein that contains four FF domains and lacks the Rho GAP domain is produced, and this protein promotes tumor progression by interacting with the TFII-I protein in the nucleus19.
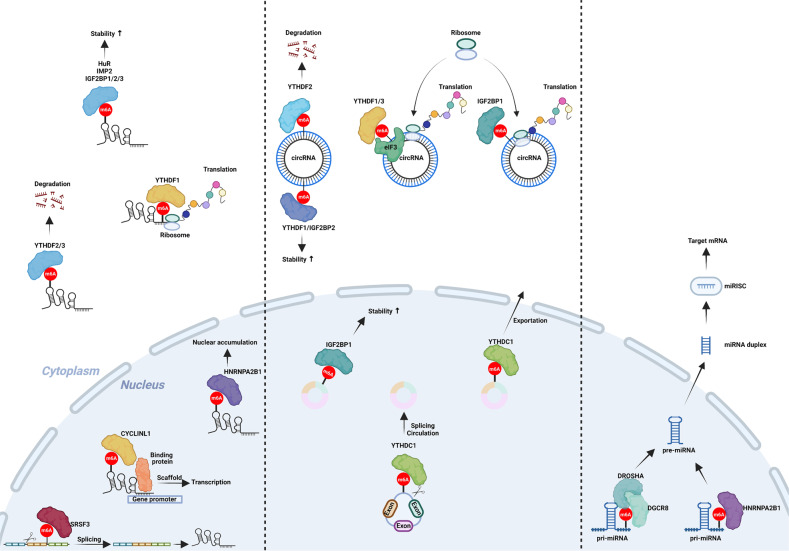
After the m6A modifications of ncRNAs are installed by writers or removed by erasers, the ability of reader proteins to bind to these molecules changes, and the metabolic fate of these ncRNAs is determined by different mechanisms (Fig. 4). We summarize the roles of reader proteins and their target ncRNAs in a variety of tumors (Table 3).
Fig. 4. Functions of m6A modifications in noncoding RNAs.
The functions of m6A modifications in circRNAs (left) include reverse shearing, changes in stability, and enhanced cytoplasmic transport; these circRNAs are also able to be translated. The functions of m6A modification in lncRNAs (middle) include splicing, acting as molecular scaffolds, nuclear accumulation, and stability changes, and these lncRNAs can also be translated. The functions of m6A modifications (right) are mainly to promote their maturation.
Table 3.
Dysregulation of m6A-binding proteins in cancer.
| Reader | Role in cancer | Type | Target ncRNAs | Mechanism |
|---|---|---|---|---|
| YTHDF1 | Oncogene | CRC | circALG164 | Stability |
| Tumor suppressor | ESCC | LNC00278106 | Translation | |
| YTHDF2 | Oncogene | CRC | lncXIST78 | Stability |
| ESCC | LNC0002299 | Stability | ||
| LSCC | LncKCNQ1OT1117 | Stability | ||
| HCC | LNC127355 | Stability | ||
| circCPSF6110 | Stability | |||
| Tumor suppressor | EEC | LncFENDRR155 | Stability | |
| LUAD | circASK165 | Stability | ||
| YTHDF3 | Oncogene | BRC | LncMALAT1122 | Stability |
| Tumor suppressor | CRC | LncGAS5121 | Stability | |
| PAC | LncDICER1-AS1118 | Stability | ||
| YTHDC1 | Oncogene | ESCC | LncMALAT194 | Scaffold |
| HCC | circARL366 | Circularization | ||
| circHPS569 | Cytoplasmic output | |||
| RCC | LncLSG181 | Scaffold | ||
| circMET130 | Cytoplasmic output | |||
| NSCLC | circIGF2BP368 | Circularization | ||
| IGF2BP1 | Oncogene | HCC | LncAROD49 | Stability |
| circMDK134 | Stability | |||
| circMAP3K460 | Translation | |||
| IGF2BP2 | Oncogene | CRC | LncPTTG3P41 | Stability |
| circNSUN2141 | Cytoplasmic export | |||
| LncZFAS1139 | Stability | |||
| GBM | lncCASC9136 | Stability | ||
| NPC | LncDIAPH1-AS188 | Stability | ||
| RCC | LncDUXAP945 | Stability | ||
| PAC | LncDANCR137 | Stability | ||
| LncPACERR82 | Scaffold | |||
| TC | LncHAGLR138 | Stability | ||
| NSCLC | LncMALAT1140 | Stability | ||
| Tumor suppressor | NSCLC | circNDUFB286 | Scaffold | |
| LUAD | lncHCG1177 | Stability | ||
| IGF2BP3 | Oncogene | CC | lncKCNMB2-AS1143 | Stability |
| HNRNPA2B1 | Oncogene | LUAD | pri-miR-106b145 | Maturation |
| CRC | LncRP1143 | Nuclear accumulation | ||
| EIF3 | Oncogene | HCC | circARHGAP3519 | Translation |
| HuR | Oncogene | HCC | LncMIR155HG79 | Stability |
| GBM | LncMALAT153 | Stability | ||
| Tumor suppressor | HCC | circDLC191 | N/A |
CRC colorectal cancer, ESCC esophageal squamous cell carcinoma, LSCC laryngeal squamous cell carcinoma, HCC hepatocellular carcinoma, EEC endometrioid endometrial carcinoma, LUAD lung adenocarcinoma, BRC breast cancer, PAC pancreatic cancer, RCC renal cell carcinoma, NSCLC head and neck squamous cell carcinoma, GBM glioblastoma multiforme, NPC nasopharyngeal carcinogenesis, TC thyroid cancer, CC cervical cancer.
The clinical potential of ncRNAs and m6A modifiers
With ncRNA methylation playing such an important role in tumor progression, m6A-modified ncRNAs and various m6A effectors, including writers, erasers, and readers, may be potential tumor markers. Chen et al. analyzed the circRNA expression profiles of 6 pairs of CRC tissues and adjacent nontumor tissues by high-throughput sequencing and found that circ1662 expression was increased and correlated with poor prognosis and depth of tumor invasion in CRC. Moreover, the level of METTL3 in CRC tissues was higher than the level of METTL3 in adjacent tissues and predicted poor prognosis. Thus, METTL3 and its modified circ1662 have the potential to serve as biomarkers to judge the prognosis of CRC67. Shen et al., showed through high-throughput methylated RNA immunoprecipitation sequencing of RCC cells that LncLSG1 was upregulated after m6A modification, which was closely related to tumor metastasis. Meanwhile, significantly high expression of METTL14 in RCC tissues resulted in tumor metastasis and poor OS and DFS. In conclusion, METTL14 and its modified lncRNA LncLSG1 can be considered potential markers to predict RCC metastasis81. Decreased expression of circDLC1, which is regulated by KIAA1429, is associated with more advanced HCC stages91. In CRC, high expression of m6A-modified LNC00460 is associated with lower 5-year overall survival and disease-free survival rates147. Increased LncTHAP7-AS1 expression after m6A modification is associated with poor prognosis of GC48. The expression of m6A-modified LncRHPN1-AS1 is an important indicator for assessing the prognosis of epithelial EOC44. Notably, the same m6A-modified ncRNA can play a tumor-promoting role in many tumors, suggesting that this molecule may be a potential tumor marker. For example, increased expression of LncMALAT1 affects the malignant phenotype of various tumors. The increased LncMALAT1 expression observed in TC stimulates the proliferation, migration and invasion of TC cells by competitively binding to miR-204, upregulating IGF2BP2 and enhancing MYC expression148. In ESCC, LncMALAT1 acts as a molecular scaffold to bind to YTHDC1, which is crucial for the maintenance of NSs and the expression of related oncogenes, thereby promoting cancer cell metastasis94. In TET, LncMALAT1 induces high expression of c-MYC protein to promote proliferation and metastasis51. Increased stability of LncMALAT1 in GBM activates the NF-κB signaling pathway to promote tumor growth, and this process is positively associated with higher malignant grade and poorer prognosis53. Moreover, activation of the MALAT1/miR-26b/HMGA2 axis in BC leads to epithelial-mesenchymal transition and enhanced tumor cell invasion122. LNC00958 also plays a role in a variety of tumors. In BC, LNC00958 upregulates YY1, which functions as a ceRNA for miR-378a-3p, promoting tumor progression46. In GC, LNC00958 positively regulates aerobic glycolysis by enhancing the transcriptional stability of GLUT1 mRNA, promoting tumor progression and reducing the survival rate of patients149. LNC00958 sponges miR3619-5p in HCC, leading to the upregulation of hepatoma-derived growth factor (HDGF) and thereby promoting HCC adipogenesis and progression40. LncNEAT1 also plays a role in the initiation and progression of various tumors. LncNEAT1 binds to CYCLINL1 at m6A sites and acts as a bridge between CYCLINL1 and CDK19, promoting the bone metastases of PC by phosphorylating Pol II Ser2 in the RUNX2 promoter58. The increase in LncNEAT1 expression relocates the posttranscriptional repressor SFPQ from its position in the CXCL8 promoter to paraspeckles, which ultimately upregulates CXCL8/IL8 and promotes the progression of GBM103. Upregulated LncNEAT1 expression causes the overexpression of a subunit of the polycomb repressive complex EZH2, which ultimately promotes the malignant phenotype of GC107.
RNA-modifying effectors may be targets for therapeutic intervention. m6A has been studied for decades, but earlier studies focused on DNA methylation. Therefore, both azacytidine and decitabine, which have been approved for the clinical treatment of tumors, are DNA methyltransferase inhibitors150,151. Although RNA methylation was more recently discovered, a number of small molecules that inhibit the RNA demethylation enzyme FTO have been identified152–154. For example, the natural compound rhein has been shown to competitively bind to FTO to inhibit its effects on its ssRNA or ssDNA substrates152. An FTO-specific inhibitor, meclofenamic acid (MA), inhibits the demethylation of m6A-modified ssRNA or ssDNA by inhibiting FTO153. The efficacy of anti-PD-1 therapy in METTL3 or METTL14 KO mice as well as ALKBH5 KO mice is increased.
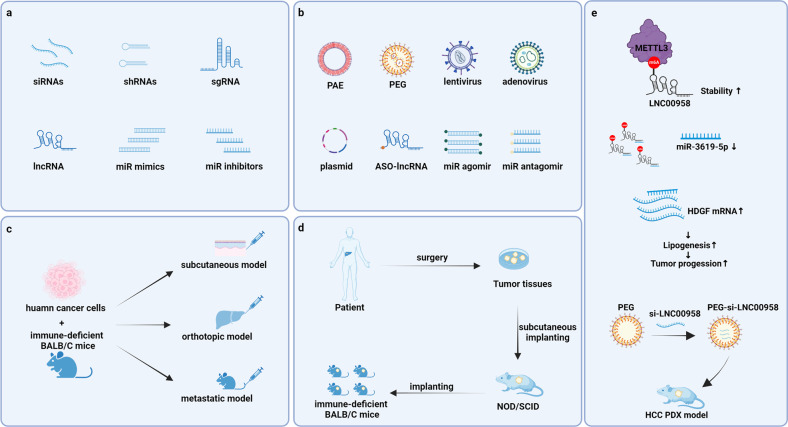
Several ncRNA-based therapies, including siRNAs, shRNAs, sgRNAs, therapeutic lncRNAs, miRNA mimics, and miR inhibitors (Fig. 5a), have been shown to effectively suppress the malignant phenotype of cancer in mice. In recent years, new types of platforms, such as polymeric nanoparticle (NP) platforms and poly (β-amino esters) (PAEs), have been developed as carriers of cancer treatment that effectively deliver siRNA to target cells and tissues (Fig. 5b). The efficacy of targeting m6A effectors or their target ncRNAs for cancer treatment has been demonstrated in a variety of mouse models, including subcutaneous, orthotopic, metastatic and patient-derived tumor xenograft (PDX) models (Fig. 5c, d). For example, Lnc00958 sponges miR3619-5p in HCC, leading to the upregulation of hepatoma-derived growth factor and thereby promoting HCC adipogenesis and progression. In the PDX model, the weight and volume of the xenografts were significantly reduced after tail vein injection of PLGA-PEG (si-LINC00958) NPs (Page 20, Lines 565-569) (Fig. 5e)40. In a cell-derived xenograft (CDX) model, tail vein injection of local short hairpin RNA to stably knock down LCAT3 (shLACT3) was found to inhibit tumor growth in lung cancer47. In vivo delivery of circSORE RNAi via local short hairpin RNA lentiviral injection significantly enhanced the efficacy of sorafenib in animal models61. The growth of endometrioid endometrial carcinoma cells was inhibited in mice transplanted with pcDNA-FENDRR-transfected cells155. In a PDX model, tail vein injection of NEAT1–1 #4 m6A-mut had no positive effect on the distant metastasis of mouse prostate cancer58. Tumor growth was significantly suppressed by in vivo-optimized RNA interference (RNAi) inhibition of LINRIS156. PAEs to assist in the transport of circMDK siRNA (PAE-siRNA) were demonstrated to be effective in inhibiting tumor progression without causing significant adverse effects in four HCC models, including a subcutaneous model, orthotopic model, lung metastasis model and PDX model134. These studies suggest that targeting RNA effector molecules for clinical treatment is a promising research direction. However, in general, our understanding of ncRNAs and RNA modification is not thorough, and more studies are needed to help us explore more accurate and faster detection methods and to develop more effective targeted therapies.
Fig. 5. ncRNA-based therapeutics and delivery systems in a mouse model.
Several ncRNA-based therapeutics exist, including siRNAs, shRNAs, sgRNAs, therapeutic lncRNAs, miRNA mimics and miR inhibitors (a). These therapeutic ncRNAs can be delivered by some vectors, including PAE, PEG, lentivirus, adenovirus and plasmids, and can also be administered in the forms of ASO-lncRNAs, miR agomirs, and miR antagomirs to play a therapeutic role (b). ncRNA-based therapy has been shown to be effective in cell-derived xenograft (CDX) models, including subcutaneous, orthotopic and metastatic models (c). ncRNA-based therapy has been shown to be effective in patient-derived tumor xenograft (PDX) models (d). Lnc00958 sponges miR3619-5p in HCC, leading to the upregulation of hepatoma-derived growth factor and thereby promoting HCC adipogenesis and progression. In the PDX model, the weight and volume of the xenografts were significantly reduced after tail vein injection of PLGA-PEG (si-LINC00958) NPs (e).
Other RNA chemical modifications
In addition to m6A modification, RNA can also undergo more than 170 chemical modifications, including m1A, m5C, m7G, ac4C, etc. m1A refers to the methylation of the first N atom of adenine in RNA molecules. Unlike m6A, m1A mainly dominates tRNA and rRNA, leading to changes in their secondary structure and playing a role in affecting translation157,158. m1A modifications can also occur in mRNAs and affect their stability and translation159,160. m5C is the methylation of the 5th carbon atom of cytosine. m5C is also involved in multiple RNA metabolisms, including mRNA export161, RNA stability162 and translation163. m7G refers to the methylation modification of the 7th N atom of guanine usually located at the 5’ cap of mRNA164, rRNA165, tRNA166 and miRNAs167. At present, a few studies have discussed m7G modification and revealed that m7G and other RNA epigenetic modifications have synergistic effects on different biological processes168. ac4C is the acetylation modification of the 4th N atom of cytosine, which was first discovered on tRNA and helps tRNA read codons correctly169. ac4C modification has also been demonstrated to exist on rRNA, which is related to maintaining the accuracy of translation170. ac4C also widely exists in mRNA and affects its stability171.
Conclusion
In summary, the study of m6A effectors and their target ncRNAs provides new directions for the diagnosis and treatment of cancer. Although the mechanism underlying the effect of m6A modification on ncRNA metabolism has been well studied and progress has been made in research on the contribution of m6A effectors and related ncRNAs to tumor phenotypes, current treatments have only been validated in mouse models, and substantial work remains to render clinical translation feasible. This is the direction of future research on the m6A modification of ncRNAs.
Acknowledgements
This paper was supported by grants from the National Natural Scientific Foundation of China (81770639, 82070657), the Applied Technology Research and Development Project of Heilongjiang Province (GA20C019), Outstanding Youth Funds of the First Affiliated Hospital of Harbin Medical University (HYD2020JQ0006), and Research Projects of the Chinese Research Hospital Association (Y2019FH-DTCC-SB1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ziying Liu, Lei Gao, Long Cheng.
Contributor Information
Gang Wang, Email: wgilu79@163.com.
Qiushi Tang, Email: tangqiushi@vip.163.com.
References
- 1.Zhang H, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48:6251–6264. doi: 10.1093/nar/gkaa347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan B, et al. Viral and cellular N(6)-methyladenosine and N(6),2’-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 2018;3:108–120. doi: 10.1038/s41564-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020;13:117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, et al. Insights into roles of METTL14 in tumors. Cell Prolif. 2021;55:e13168. doi: 10.1111/cpr.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan Q, et al. The critical role of RNA m(6)A methylation in cancer. Cell Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer. 2020;19:88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer. 2020;19:44. doi: 10.1186/s12943-020-01172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, et al. YTH domain proteins: a family of m(6)A readers in cancer progression. Front. Oncol. 2021;11:629560. doi: 10.3389/fonc.2021.629560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Shi Y, Shen H, Xie W. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020;13:35. doi: 10.1186/s13045-020-00872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv. Sci. 2021;8:2001701. doi: 10.1002/advs.202001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin D, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol. Cancer. 2020;19:40. doi: 10.1186/s12943-020-01161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alarcón CR, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roundtree IA, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. ELife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016;23:110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L, et al. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 28.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Böhmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossi E, et al. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 2020;11:936. doi: 10.1038/s41467-020-14623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuman S. Transcriptional interference at tandem lncRNA and protein-coding genes: an emerging theme in regulation of cellular nutrient homeostasis. Nucleic Acids Res. 2020;48:8243–8254. doi: 10.1093/nar/gkaa630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol. Cancer. 2020;19:94. doi: 10.1186/s12943-020-01207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma S, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:310–318. doi: 10.1016/j.bbagrm.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi YC, Chen XY, Zhang J, Zhu JS. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol. Cancer. 2020;19:121. doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49:7239–7255. doi: 10.1093/nar/gkab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo X, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y, et al. N6-methyladenosine modification of PTTG3P contributes to colorectal cancer proliferation via YAP1. Front. Oncol. 2021;11:669731. doi: 10.3389/fonc.2021.669731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin, H. et al. M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway. Cell Death Differ. 10.1038/s41418-021-00888-8 (2021). [DOI] [PMC free article] [PubMed]
- 43.Wu Y, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer. 2019;18:87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging. 2020;12:4558–4572. doi: 10.18632/aging.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan L, et al. N6-methyladenosine modification of LncRNA DUXAP9 promotes renal cancer cells proliferation and motility by activating the PI3K/AKT signaling pathway. Front. Oncol. 2021;11:641833. doi: 10.3389/fonc.2021.641833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rong D, et al. m(6)A-induced LINC00958 promotes breast cancer tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Disco. 2021;7:27. doi: 10.1038/s41420-020-00382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian X, et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J. Hematol. Oncol. 2021;14:112. doi: 10.1186/s13045-021-01123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu HT, et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2021;29:627–641. doi: 10.1038/s41418-021-00879-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia G, et al. LNCAROD enhances hepatocellular carcinoma malignancy by activating glycolysis through induction of pyruvate kinase isoform PKM2. J. Exp. Clin. Cancer Res. 2021;40:299. doi: 10.1186/s13046-021-02090-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Ji F, et al. m(6)A methyltransferase METTL3-mediated lncRNA FOXD2-AS1 promotes the tumorigenesis of cervical cancer. Mol. Ther. Oncolytics. 2021;22:574–581. doi: 10.1016/j.omto.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iaiza A, et al. METTL3-dependent MALAT1 delocalization drives c-Myc induction in thymic epithelial tumors. Clin. Epigenetics. 2021;13:173. doi: 10.1186/s13148-021-01159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen JQ, et al. M(6)A-mediated up-regulation of LncRNA LIFR-AS1 enhances the progression of pancreatic cancer via miRNA-150-5p/ VEGFA/Akt signaling. Cell Cycle. 2021;20:2507–2518. doi: 10.1080/15384101.2021.1991122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang YZ, et al. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021;511:36–46. doi: 10.1016/j.canlet.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Yuan JF, Wang YZ. METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate cancer via SRSF1/c-Myc axis. Exp. Cell Res. 2022;416:113149. doi: 10.1016/j.yexcr.2022.113149. [DOI] [PubMed] [Google Scholar]
- 55.Kong H, Sun J, Zhang W, Zhang H, Li H. Long intergenic non-protein coding RNA 1273 confers sorafenib resistance in hepatocellular carcinoma via regulation of methyltransferase 3. Bioengineered. 2022;13:3108–3121. doi: 10.1080/21655979.2022.2025701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, et al. m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell Through miR-544b/BTG2 Signaling. Onco Targets Ther. 2021;14:3745–3755. doi: 10.2147/OTT.S289198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, et al. ZFAS1 exerts an oncogenic role via suppressing miR-647 in an m(6)A-dependent manner in cervical cancer. Onco Targets Ther. 2020;13:11795–11806. doi: 10.2147/OTT.S274492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen S, et al. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer. 2020;19:171. doi: 10.1186/s12943-020-01293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang ZW, et al. SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep. 2022;39:110813. doi: 10.1016/j.celrep.2022.110813. [DOI] [PubMed] [Google Scholar]
- 60.Duan JL, et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer. 2022;21:93. doi: 10.1186/s12943-022-01537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu J, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol. Cancer. 2020;19:163. doi: 10.1186/s12943-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu P, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12:298. doi: 10.1038/s41419-021-03558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z, et al. circ0000069 promotes cervical cancer cell proliferation and migration by inhibiting miR-4426. Biochem. Biophys. Res. Commun. 2021;551:114–120. doi: 10.1016/j.bbrc.2021.03.020. [DOI] [PubMed] [Google Scholar]
- 64.Lin C, et al. The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer. 2022;21:80. doi: 10.1186/s12943-022-01560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang T, et al. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 2021;520:321–331. doi: 10.1016/j.canlet.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Rao X, et al. N(6) -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life. 2021;73:408–417. doi: 10.1002/iub.2438. [DOI] [PubMed] [Google Scholar]
- 67.Chen C, et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11:4298–4315. doi: 10.7150/thno.51342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z, et al. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol. Cancer. 2021;20:105. doi: 10.1186/s12943-021-01398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rong D, et al. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol. Ther. Nucleic Acids. 2021;26:637–648. doi: 10.1016/j.omtn.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019;10:1858. doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer. 2019;18:181. doi: 10.1186/s12943-019-1108-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Liu T, et al. Exosomal and intracellular miR-320b promotes lymphatic metastasis in esophageal squamous cell carcinoma. Mol. Ther. Oncolytics. 2021;23:163–180. doi: 10.1016/j.omto.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou H, Yin K, Zhang Y, Tian J, Wang S. The RNA m6A writer METTL14 in cancers: roles, structures, and applications. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188609. doi: 10.1016/j.bbcan.2021.188609. [DOI] [PubMed] [Google Scholar]
- 74.Zheng ZQ, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to Sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cell Res. 2019;79:4612–4626. doi: 10.1158/0008-5472.CAN-19-0799. [DOI] [PubMed] [Google Scholar]
- 75.Hu N, Ji H. N6-methyladenosine (m6A)-mediated up-regulation of long noncoding RNA LINC01320 promotes the proliferation, migration, and invasion of gastric cancer via miR495-5p/RAB19 axis. Bioengineered. 2021;12:4081–4091. doi: 10.1080/21655979.2021.1953210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ban Y, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol. Oncol. 2020;14:1282–1296. doi: 10.1002/1878-0261.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao J, Qiu H, Guo L. LncRNA HCG11 mediated by METTL14 inhibits the growth of lung adenocarcinoma via IGF2BP2/LATS1. Biochem. Biophys. Res. Commun. 2021;580:74–80. doi: 10.1016/j.bbrc.2021.09.083. [DOI] [PubMed] [Google Scholar]
- 78.Yang X, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peng L, et al. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol. Toxicol. 2022;38:1159–1173. doi: 10.1007/s10565-022-09718-0. [DOI] [PubMed] [Google Scholar]
- 80.Liu T, et al. Methyltransferase-like 14 suppresses growth and metastasis of renal cell carcinoma by decreasing long noncoding RNA NEAT1. Cancer Sci. 2022;113:446–458. doi: 10.1111/cas.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen D, et al. METTL14-mediated Lnc-LSG1 m6A modification inhibits clear cell renal cell carcinoma metastasis via regulating ESRP2 ubiquitination. Mol. Ther. Nucleic Acids. 2022;27:547–561. doi: 10.1016/j.omtn.2021.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y, et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2022;15:52. doi: 10.1186/s13045-022-01272-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan HN, et al. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer. 2022;21:51. doi: 10.1186/s12943-022-01521-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yi D, et al. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6‑methyladenosine and hsa‑miR‑146a‑5p expression. Oncol. Rep. 2020;43:1375–1386. doi: 10.3892/or.2020.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.An L, Li X, Yang J. MicroRNA-211 attenuates cell proliferation in T-cell lymphoblastic lymphoma through targeting TCF12. Leuk. Res. 2021;110:106653. doi: 10.1016/j.leukres.2021.106653. [DOI] [PubMed] [Google Scholar]
- 86.Li B, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 2021;12:295. doi: 10.1038/s41467-020-20527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schöller E, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li ZX, et al. WTAP-mediated m(6)A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 2022;29:1137–1151. doi: 10.1038/s41418-021-00905-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren Z, et al. N(6)-methyladenosine methyltransferase WTAP-stabilized FOXD2-AS1 promotes the osteosarcoma progression through m(6)A/FOXM1 axis. Bioengineered. 2022;13:7963–7973. doi: 10.1080/21655979.2021.2008218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang T, et al. N(6)-methyladenosine (m(6)A)-mediated lncRNA DLGAP1-AS1enhances breast canceradriamycin resistance through miR-299-3p/WTAP feedback loop. Bioengineered. 2021;12:10935–10944. doi: 10.1080/21655979.2021.2000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. 2021;11:1396–1411. doi: 10.7150/thno.53227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Knuckles P, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones AN, Tikhaia E, Mourão A, Sattler M. Structural effects of m6A modification of the Xist A-repeat AUCG tetraloop and its recognition by YTHDC1. Nucleic Acids Res. 2022;50:2350–2362. doi: 10.1093/nar/gkac080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X, et al. N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell. 2021;56:702–715 e708. doi: 10.1016/j.devcel.2021.01.015. [DOI] [PubMed] [Google Scholar]
- 95.Ruszkowska A. METTL16, methyltransferase-like protein 16: current insights into structure and function. Int. J. Mol. Sci. 2021;22:2176. doi: 10.3390/ijms22042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brown JA, et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dai YZ, et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m(6)A-dependent manner. Cell Mol. Biol. Lett. 2022;27:41. doi: 10.1186/s11658-022-00342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang X, et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl Acad. Sci. USA. 2019;116:2919–2924. doi: 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cui Y, et al. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2021;40:294. doi: 10.1186/s13046-021-02096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang T, et al. FTO-stabilized lncRNA HOXC13-AS epigenetically upregulated FZD6 and activated Wnt/β-catenin signaling to drive cervical cancer proliferation, invasion, and EMT. J. BUON. 2021;26:1279–1291. [PubMed] [Google Scholar]
- 101.Bayoumi M, Munir M. Structural insights into m6A-erasers: a step toward understanding molecule specificity and potential antiviral targeting. Front. Cell Dev. Biol. 2020;8:587108. doi: 10.3389/fcell.2020.587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo Y, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol. Cancer. 2021;20:93. doi: 10.1186/s12943-021-01372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dong F, et al. ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cell Res. 2021;81:5876–5888. doi: 10.1158/0008-5472.CAN-21-1456. [DOI] [PubMed] [Google Scholar]
- 104.Chen S, Zhou L, Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.He Y, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell. Physiol. Biochem. 2018;48:838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 106.Wu S, et al. A novel micropeptide encoded by Y-linked LINC00278 links cigarette smoking and AR signaling in male esophageal squamous cell carcinoma. Cell Res. 2020;80:2790–2803. doi: 10.1158/0008-5472.CAN-19-3440. [DOI] [PubMed] [Google Scholar]
- 107.Zhang J, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiol. Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu H, Zhang Z. ALKBH5-mediated m6A demethylation of lncRNA RMRP plays an oncogenic role in lung adenocarcinoma. Mamm. Genome. 2021;32:195–203. doi: 10.1007/s00335-021-09872-6. [DOI] [PubMed] [Google Scholar]
- 109.Guo T, Liu DF, Peng SH, Xu AM. ALKBH5 promotes colon cancer progression by decreasing methylation of the lncRNA NEAT1. Am. J. Transl. Res. 2020;12:4542–4549. [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Y, et al. Activation of YAP1 by N6-methyladenosine-modified circCPSF6 drives malignancy in hepatocellular carcinoma. Cell Res. 2022;82:599–614. doi: 10.1158/0008-5472.CAN-21-1628. [DOI] [PubMed] [Google Scholar]
- 111.Song W, et al. ALKBH5-mediated N(6)-methyladenosine modification of TRERNA1 promotes DLBCL proliferation via p21 downregulation. Cell Death Disco. 2022;8:25. doi: 10.1038/s41420-022-00819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang H, Song X, Song C, Wang X, Cao H. m(6)A-seq analysis of microRNAs reveals that the N6-methyladenosine modification of miR-21-5p affects its target expression. Arch. Biochem. Biophys. 2021;711:109023. doi: 10.1016/j.abb.2021.109023. [DOI] [PubMed] [Google Scholar]
- 113.Xu C, et al. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Z, Zhong X, Xia M, Zhong J. The roles and mechanisms of the m6A reader protein YTHDF1 in tumor biology and human diseases. Mol. Ther. Nucleic Acids. 2021;26:1270–1279. doi: 10.1016/j.omtn.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu T, et al. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–1496. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arguello AE, Leach RW, Kleiner RE. In vitro selection with a site-specifically modified RNA library reveals the binding preferences of N(6)-methyladenosine reader proteins. Biochemistry. 2019;58:3386–3395. doi: 10.1021/acs.biochem.9b00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Y, et al. ALKBH5-mediated m6A modification of lncRNA KCNQ1OT1 triggers the development of LSCC via upregulation of HOXA9. J. Cell. Mol. Med. 2022;26:385–398. doi: 10.1111/jcmm.17091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu Y, et al. A reciprocal feedback between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1 promotes glycolysis of pancreatic cancer through inhibiting maturation of miR-5586-5p. J. Exp. Clin. Cancer Res. 2022;41:69. doi: 10.1186/s13046-022-02285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shi H, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chang G, et al. YTHDF3 induces the translation of m(6)A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell. 2020;38:857–871.e857. doi: 10.1016/j.ccell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ni W, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao C, Ling X, Xia Y, Yan B, Guan Q. The m6A methyltransferase METTL3 controls epithelial-mesenchymal transition, migration and invasion of breast cancer through the MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int. 2021;21:441. doi: 10.1186/s12935-021-02113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li A, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lasman L, et al. Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 2020;34:1373–1391. doi: 10.1101/gad.340695.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582–1595.e1518. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y, Bedi RK, Moroz-Omori EV, Caflisch A. Structural and dynamic insights into redundant function of YTHDF proteins. J. Chem. Inf. Model. 2020;60:5932–5935. doi: 10.1021/acs.jcim.0c01029. [DOI] [PubMed] [Google Scholar]
- 128.Xu C, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 129.Widagdo J, Anggono V, Wong JJ. The multifaceted effects of YTHDC1-mediated nuclear m(6)A recognition. Trends Genet. 2022;38:325–332. doi: 10.1016/j.tig.2021.11.005. [DOI] [PubMed] [Google Scholar]
- 130.Yang L, et al. CircMET promotes tumor proliferation by enhancing CDKN2A mRNA decay and upregulating SMAD3. Mol. Cancer. 2022;21:23. doi: 10.1186/s12943-022-01497-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ma L, et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021;38:101801. doi: 10.1016/j.redox.2020.101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He JJ, et al. m(6)A reader YTHDC2 promotes radiotherapy resistance of nasopharyngeal carcinoma via activating IGF1R/AKT/S6 signaling axis. Front. Oncol. 2020;10:1166. doi: 10.3389/fonc.2020.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang X, et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J. Hematol. Oncol. 2018;11:88. doi: 10.1186/s13045-018-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Du A, et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol. Cancer. 2022;21:109. doi: 10.1186/s12943-022-01575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J, Chen L, Qiang P. The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int. 2021;21:99. doi: 10.1186/s12935-021-01799-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu H, et al. m(6)A reader IGF2BP2-stabilized CASC9 accelerates glioblastoma aerobic glycolysis by enhancing HK2 mRNA stability. Cell Death Disco. 2021;7:292. doi: 10.1038/s41420-021-00674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hu X, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dong L, et al. IGF2BP2 knockdown suppresses thyroid cancer progression by reducing the expression of long non-coding RNA HAGLR. Pathol. Res. Pract. 2021;225:153550. doi: 10.1016/j.prp.2021.153550. [DOI] [PubMed] [Google Scholar]
- 139.Lu S, et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J. Hematol. Oncol. 2021;14:188. doi: 10.1186/s13045-021-01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Han L, et al. IGF2BP2 regulates MALAT1 by serving as an N6-methyladenosine reader to promote NSCLC proliferation. Front. Mol. Biosci. 2021;8:780089. doi: 10.3389/fmolb.2021.780089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He RZ, Jiang J, Luo DX. M6A modification of circNSUN2 promotes colorectal liver metastasis. Genes Dis. 2021;8:6–7. doi: 10.1016/j.gendis.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bell JL, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Y, Wang D, Wu D, Zhang D, Sun M. Long noncoding RNA KCNMB2-AS1 stabilized by N(6)-methyladenosine modification promotes cervical cancer growth through acting as a competing endogenous RNA. Cell Transpl. 2020;29:963689720964382. doi: 10.1177/0963689720964382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rong L, Xu Y, Zhang K, Jin L, Liu X. HNRNPA2B1 inhibited SFRP2 and activated Wnt-β/catenin via m6A-mediated miR-106b-5p processing to aggravate stemness in lung adenocarcinoma. Pathol. Res. Pract. 2022;233:153794. doi: 10.1016/j.prp.2022.153794. [DOI] [PubMed] [Google Scholar]
- 146.Lal P, et al. Regulation of HuR structure and function by dihydrotanshinone-I. Nucleic Acids Res. 2017;45:9514–9527. doi: 10.1093/nar/gkx623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hou P, et al. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J. Exp. Clin. Cancer Res. 2021;40:52. doi: 10.1186/s13046-021-01857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic role of long noncoding RNAMALAT1 in thyroid cancer progression through regulation of the miR-204/IGF2BP2/m6A-MYC signaling. Mol. Ther. Nucleic Acids. 2021;23:1–12. doi: 10.1016/j.omtn.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang D, et al. m(6) A transferase KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic glycolysis through targeting GLUT1. IUBMB Life. 2021;73:1325–1333. doi: 10.1002/iub.2545. [DOI] [PubMed] [Google Scholar]
- 150.Christman JK. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 151.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 152.Chen B, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 2012;134:17963–17971. doi: 10.1021/ja3064149. [DOI] [PubMed] [Google Scholar]
- 153.Huang Y, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Su R, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90–105.e123. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shen J, et al. N-methyladenosine reader YTHDF2-mediated long noncoding RNA FENDRR degradation promotes cell proliferation in endometrioid endometrial carcinoma. Lab. Invest. 2021;101:775–784. doi: 10.1038/s41374-021-00543-3. [DOI] [PubMed] [Google Scholar]
- 156.Wang Y, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer. 2019;18:174. doi: 10.1186/s12943-019-1105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 158.Sharma S, Watzinger P, Kötter P, Entian KD. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;41:5428–5443. doi: 10.1093/nar/gkt195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dominissini D, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li X, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell. 2017;68:993–1005.e1009. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang X, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen X, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019;21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 163.Janin M, et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053–1074. doi: 10.1007/s00401-019-02062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sloan KE, et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14:1138–1152. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tomikawa C. 7-methylguanosine modifications in transfer RNA (tRNA) Int. J. Mol. Sci. 2018;19:4080. doi: 10.3390/ijms19124080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pandolfini L, et al. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol. Cell. 2019;74:1278–1290.e1279. doi: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen H, et al. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020;30:544–547. doi: 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stern L, Schulman LH. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J. Biol. Chem. 1978;253:6132–6139. doi: 10.1016/S0021-9258(17)34590-8. [DOI] [PubMed] [Google Scholar]
- 170.Sharma S, et al. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dominissini D, Rechavi G. N(4)-acetylation of cytidine in mRNA by NAT10 regulates stability and translation. Cell. 2018;175:1725–1727. doi: 10.1016/j.cell.2018.11.037. [DOI] [PubMed] [Google Scholar]