Abstract
Immune checkpoint inhibitors (ICIs) have transformed the treatment of melanoma. However, the majority of patients have primary or acquired resistance to ICIs, limiting durable responses and patient survival. Interferon-gamma (IFNγ) signaling and the expression of IFNγ-stimulated genes correlate with either response or resistance to ICIs, in a context-dependent manner. While IFNγ-inducible immunostimulatory genes are required for response to ICIs, chronic IFNγ signaling induces the expression of immunosuppressive genes, promoting resistance to these therapies. Here, we show that high levels of ULK1 correlate with poor survival in melanoma patients and overexpression of ULK1 in melanoma cells enhances IFNγ-induced expression of immunosuppressive genes, with minimal effects on the expression of immunostimulatory genes. In contrast, genetic or pharmacological inhibition of ULK1 reduces expression of IFNγ-induced immunosuppressive genes. ULK1 binds IRF1 in the nuclear compartment of melanoma cells, controlling its binding to the PD-L1 promoter region. Additionally, pharmacological inhibition of ULK1 in combination with anti-PD-1 therapy further reduces melanoma tumor growth in vivo. Our data suggest that targeting ULK1 represses IFNγ-dependent immunosuppression. These findings support the combination of ULK1 drug-targeted inhibition with ICIs for the treatment of melanoma patients to improve response rates and patient outcomes.
Implications:
This study identifies ULK1, activated downstream of IFNγ signaling, as a druggable target to overcome resistance mechanisms to ICI therapy in metastatic melanoma.
Introduction
Immunotherapies designed to overcome tumor immune escape have been emerging as a promising strategy to treat melanoma, with the immune checkpoint inhibitors (ICIs) anti-CTLA-4 monoclonal antibody (mAb) ipilimumab and the anti-PD-1 mAbs pembrolizumab and nivolumab approved by the U.S. Food and Drug Administration in 2011 and 2014, respectively (1). However, primary and acquired resistance to ICIs occurs in the majority of patients, with only ~25% of the patients achieving a durable response (2, 3).
Surprisingly, interferon-gamma (IFNγ) signaling pathways and expression of IFN-stimulated genes (ISGs) in tumor cells correlate with either response or resistance to ICIs in melanoma in a context-dependent manner (4). Using a genome-scale CRISPR/Cas9 library, Patel et al. demonstrated that expression of genes involved in IFNγ signaling in melanoma cells is essential for the effector function of tumor-reactive CD8+ T cells and response to ICIs (5). Moreover, loss-of-function mutations in the IFNγ-receptor-associated Janus kinase 1 (JAK1) and JAK2 genes are associated with primary and acquired resistance to ICI therapy (6, 7). However, ICI-induced sustained IFNγ signaling in melanoma cells favors expression of immunosuppressive genes, such as IDO1, PD-L1 and PD-L2, driving an immunosuppressive tumor microenvironment and ICI resistance (8-10). These findings suggest that targeting IFNγ signaling could reverse resistance to ICIs. However, it could also negatively affect the immunostimulatory effects of IFNγ required for response to ICI therapies (5, 11). This dual and opposing role of IFNγ signaling in melanoma supports the need to identify means to selectively modulate its cellular effects to enhance anti-tumor immune cell activity without promoting immunosuppression or abrogating an immune response. I.e., identify means to reduce expression of IFNγ-induced immunosuppressive genes that inhibit an immune response/promote resistance to immunotherapies, without affecting the expression of IFNγ-dependent immunostimulatory genes that have been shown to be required for response to immunotherapies.
In earlier studies, we identified Unc-51 like kinase 1 (ULK1) as a key regulator of specific IFN-induced signaling pathways, controlling expression of a subset of ISGs (12, 13). Previously known to play important roles in autophagy and axon growth, ULK1 has more recently been found to play a central role in controlling IFN-regulated immune responses independently of its autophagy-related functions (14, 15). Here, we report that increased expression levels of ULK1 correlate with poor overall survival in melanoma patients. Our results show that ULK1 activity primarily controls the expression of IFNγ-induced immunosuppressive genes in melanoma cells and pharmacological inhibition of ULK1 reduces expression of these genes induced by anti-PD-1 therapy in a melanoma in vivo model, improving anti-tumor immune responses.
Materials and Methods
Correlation between ULK1 and ULK2 expression and survival in melanoma patients
Gene expression of ULK1 and ULK2 measured by RNA Sequencing and melanoma patient survival data were extracted from the GDC TCGA Melanoma (SKCM) dataset using the University of California Santa Cruz (UCSC) Xena (RRID:SCR_018938) browser (https://xena.ucsc.edu/) (16). The method of Kaplan–Meier was used to estimate overall survival, and groups were compared using the logrank test. Plots and statistical analysis were generated using GraphPad Prism 8 software (RRID:SCR_002798).
Comparison of ULK1 and ULK2 gene expression in melanoma cell lines
Gene expression data for ULK1 and ULK2 (Expression 22Q1 Public) were downloaded from the public Cancer Dependency Map Portal (RRID:SCR_017655) (https://depmap.org/portal/) for a total of 77 melanoma cell lines. Data were graphed and statistical analysis was performed using GraphPad Prism 8 software (RRID:SCR_002798).
Assessment of ULK1 gene expression before and after anti-PD-1 therapy in melanoma patients
Data from RNA-sequencing analysis of gene expressions on biopsies from melanoma patients treated with anti-PD-1 therapy were downloaded from dbGaP (https://www.ncbi.nlm.nih.gov/gap/) (study accession: phs001919.v1.p1) (17). Differential expression analysis was performed to evaluate the expression levels of ULK1 prior and post anti-PD-1 therapy in melanoma patients. We used edgeR's trimmed mean of M values (TMM) to normalize data (18). Next, we implemented the limma/voom pipeline (19-21) with duplicateCorrelation (22) to account for correlated samples between baseline and after-treatments. We applied voom and duplicateCorrelation two times to improve the estimations of precision weights and correlations. Then we fitted a linear model to ULK1 and conducted the empirical Bayes moderated t-test.
Animals
C57BL/6J mice were obtained from The Jackson Laboratory (Strain #:000664; RRID:IMSR_JAX:000664) and were housed at Northwestern University in a pathogen-free barrier facility. All animal studies were performed within the parameters of the Guidelines for the Care and Use of Laboratory Animals and were approved by the Northwestern University Institutional Animal Care and Use Committee (ID: IS00014582). Upon arrival, male (6-7-week-old) C57BL/6J mice were allowed to acclimate for one week and then were used to assess the effects of ULK inhibition and/or anti-PD-1 therapy on mouse melanoma tumor growth. Group sample sizes of 5 and 5 achieve 80% power to reject the null hypothesis of equal gene expression means between two groups when the population mean difference is 60.7 with a standard deviation for both groups of 30.0 and with a significance level (alpha) of 0.05 using a two-sided two-sample equal-variance t test. Thus, for the in vivo studies we used at least 5 animals per treatment group.
Cell lines
A375 (CRL-1619, ATCC; RRID:CVCL_0132) cells were grown in Dulbecco's Modified Eagle's Medium (DMEM), SK-MEL-2 (HTB-68, ATCC; RRID:CVCL_0069) cells were grown in Eagle's Minimum Essential Medium (EMEM) and YUMMER1.7 cells (23) (RRID:CVCL_A2AX) were grown in DMEM/F12 (1:1) with GlutaMAX supplement medium. All media was supplemented with 10% fetal bovine serum (FBS) and antibiotics. All three cell lines were maintained in culture at low passage numbers (≤12 passages) at 37°C and 5% CO2. Low passage stocks were maintained in liquid nitrogen. Testing for mycoplasma contamination was performed using a MycoAlert PLUS mycoplasma detection kit, following the manufacturer’s instructions (Lonza) every 3-6 months. Cell line authentication was performed annually using Short Tandem Repeat (STR) analyses.
Transfection of melanoma cell lines and drug treatments
The ULK1 (pRK5/myc-hULK1) plasmid was purchased from Addgene (a gift from Do-Hyung Kim; Addgene plasmid #31961; http://n2t.net/addgene:31961; RRID:Addgene_31961) (24). The pRK5 control empty vector (EV) plasmid was generated as previously reported by Saleiro et al. (25). A375 and SK-MEL-2 cells were transfected with ULK1 or EV plasmids using Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were either left untreated or were treated with human IFNγ (Gibco, # PHC4033) for 6 hours at 2,500 IU/mL. Cell pellets were collected and processed for RNA isolation.
Control and ULK1 targeting siRNAs were obtained from Santa Cruz Biotechnology (control siRNA-B #sc-44230 and ULK1 siRNA (h) #sc-44182). A375 and SK-MEL-2 cells were transfected with siRNAs using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were either left untreated or were treated with human IFNγ for 6 hours at 2,500 IU/mL. Cell pellets were collected and processed for RNA isolation.
To assess the effects of ULK1 drug-targeted inhibition on IFNγ-induced gene expression in melanoma cells, human A375 and SK-MEL-2 and mouse YUMMER1.7 melanoma cells were pre-treated with either vehicle-control (DMSO) or 10μM SBI-0206965 (SBI) (Cayman Chemical, # 18477) for 1 hour, followed by 6 hours treatment with either vehicle-control (DMSO), 10μM SBI and/or 2,500 IU/mL IFNγ (Gibco, human IFNγ # PHC4033 and mouse IFNγ # PMC4033). Cell pellets were collected and processed for RNA isolation. To assess the effects of ULK1 drug-targeted inhibition on IFNγ-induced protein expression in melanoma cells, human A375 and SK-MEL-2 and mouse YUMMER1.7 melanoma cells were treated for twenty-four hours with either vehicle-control (DMSO), SBI (10μM for A375, 5μM for SK-MEL-2, 1μM for YUMMER1.7 cells) and/or 1,000 IU/mL IFNγ. Cell pellets were collected and processed for immunoblotting analyses.
To identify novel ULK1-protein complexes in melanoma cells, A375 cells were either left untreated or were treated with IFNγ for 10 minutes (5000 IU/mL) or 4 hours (2500 IU/mL), followed by cytoplasmic and nuclear cell fractionation, co-immunoprecipitation of endogenous ULK1-protein complexes and mass spectrometry analysis or immunoblotting analysis. To validate the binding between ULK1 and IRF1 in the nuclear compartment of melanoma cells in the presence of SBI and/or IFNγ treatment, A375 cells were pre-treated with either vehicle-control (DMSO) or 10μM SBI for 1 hour, followed by 6 hours treatment with either vehicle-control (DMSO), 10μM SBI and/or 2,500 IU/mL IFNγ, followed by cytoplasmic and nuclear cell fractionation, co-immunoprecipitation of endogenous ULK1-protein complexes and immunoblotting analysis.
YUMMER1.7 mouse melanoma in vivo model
YUMMER1.7 cells were injected subcutaneously in the right flank of C57BL/6J male mice (600,000 cells in 100 μl of Hank’s Balanced Salt Solution/mouse). Tumors were allowed to grow for seven days until becoming palpable and then mice were randomized by tumor volume into four treatment groups: 1- vehicle-isotype control, 2- ULK inhibitor, 3- anti-PD-1 and 4- ULK inhibitor plus anti-PD-1. It was established a priori that mice that did not present/develop palpable tumors initially would be excluded from the experiment/analysis. InVivoPlus anti-mouse PD-1 (CD279) antibody (clone RMP1-14, BioXCell #BP0146, RRID:AB_10949053) or InVivoPlus rat IgG2a isotype control (clone 2A3, BioXCell #BP0089, RRID:AB_1107769) were injected intraperitoneally at 10mg/kg body weight (BW) twice per week for four weeks. The ULK inhibitor SBP-7455 (TargetMol, #T8850) or its vehicle (5% DMSO, 10% Tween, 85% water) were administered by oral gavage at 10mg/kg BW five times per week for four weeks. Mice BW and tumor size were recorded three times per week throughout the study. Tumor length (L) and width (W) were measured using calipers, and tumor volume was calculated using the formula: (L x W2)/2. To assess the effects of the drug treatments on gene expression and immune cell infiltration of YUMMER1.7 tumors grown in vivo, mice were treated as described above and on day 35, mice were treated once by oral gavage with either SBP-7455 (10 mg/kg BW) or its vehicle and tumors were collected twenty-four hours later and processed for qRT-PCR, flow cytometry and immunofluorescence analyses.
Statistical Analysis
GraphPad Prism 8 (RRID:SCR_002798) was used to perform all statistical analyses, except for the studies assessing ULK1 expression before and after anti-PD-1 therapy in melanoma patients (detailed above) and the analysis of tumor growth, which was done using Statistical Analysis System (SAS) software (RRID:SCR_008567), Version 9.4 (26) as described in the figure legend. Plots shown represent at least three independent biological replicates or data from tissue isolated from at least five different mice. Sample sizes are reported in each respective figure legend. For the mouse experiments each data point represents one mouse. Two-sample two-tailed t-tests were used when comparing normally distributed outcomes between two groups. One-way ANOVA models were used when comparing more than two groups, and pairwise group comparisons were adjusted using Tukey’s post-hoc test and multiple comparisons correction. Two-way ANOVA was used to compare groups in experiments with two independent variables (factors) with Tukey’s post-hoc test for multiple comparisons correction. The figure legends contain the statistical analysis used for each experiment. p-values were considered statistically significant when less than 0.05.
Data availability
The data generated in this study are available upon request from the corresponding author. Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the project accession #: PXD035347.
Supplementary materials and methods:
The following are included in the supplementary materials and methods due to space constraints: quantitative RT-PCR analysis, immunoblotting analysis, cytoplasmic and nuclear cell fractionation, co-immunoprecipitation of ULK1-protein complexes, mass spectrometry analysis, chromatin-immunoprecipitation assay, flow cytometric immunophenotyping of mouse melanoma tumors, and immunofluorescence staining of mouse melanoma tumors.
Results
Overexpression of ULK1 correlates with poor overall survival in melanoma patients and enhances expression of IFNγ-induced immunosuppressive genes
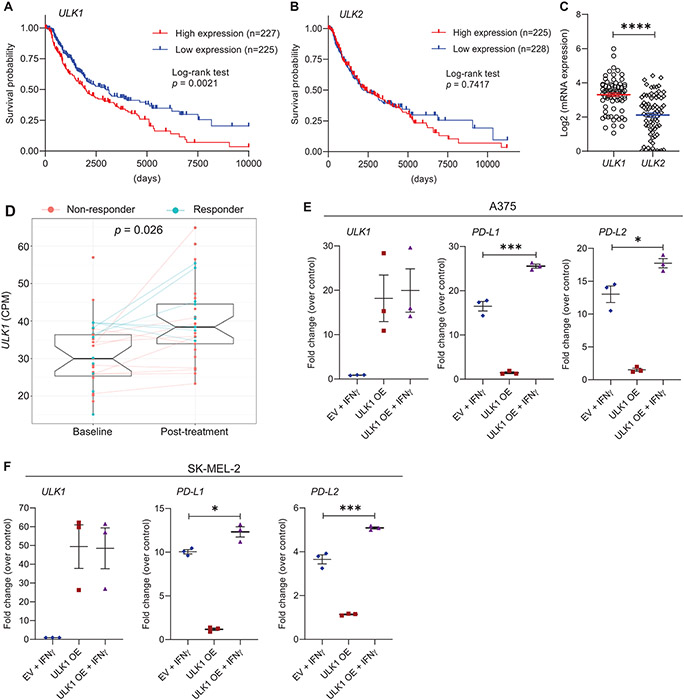
In initial studies, we assessed how ULK1 expression correlates with survival in melanoma patients using the GDC TCGA Melanoma (SKCM) dataset via the UCSC Xena Data Portal (16). We found that patients expressing high levels of ULK1 had shorter overall survival compared with patients expressing low levels of ULK1 (Fig. 1A). In contrast, expression of ULK2, the closest ULK1 homologous kinase, showed no clear correlation with survival in melanoma patients (Fig. 1B) and ULK1 was found to be expressed at higher levels in melanoma cells compared to ULK2 (Fig. 1C). Additionally, analysis of a previously published dataset (17) revealed that ULK1 expression is increased in melanoma patients post anti-PD-1 therapy (Fig. 1D). Thus, we next sought to evaluate the effects of ULK1 overexpression in regulation of IFNγ-induced gene expression in melanoma cells. Overexpression of ULK1 in the A375 and SK-MEL-2 melanoma cell lines resulted in a significant increase in IFNγ-mediated expression of the immunosuppressive genes PD-L1 and PD-L2 (Fig. 1E-F), with minimal effects on the expression of the IFNγ-induced immunostimulatory genes HLA-B, IFI30 and TAP1 (Supplementary Fig. S1).
Figure 1. Overexpression of ULK1 correlates with poor survival in melanoma patients and increased expression of IFNγ-induced immunosuppressive genes in melanoma cells.
(A-B) Kaplan–Meier survival plots show survival probability for melanoma patients with (A) high (n = 227) versus low (n = 225) expression levels of ULK1 (cut-point 17.17) and (B) high (n = 225) versus low (n = 228) expression levels of ULK2 (cut-point 15.95). Data were extracted from the GDC TCGA Melanoma (SKCM) dataset using the UCSC Xena browser (https://xena.ucsc.edu/). Statistical analyses were performed using Log-rank test and p values are shown. (C) Scatter dot plot with means ± SEM shows Log2 of mRNA expression of ULK1 and ULK2 in melanoma cell lines measured by RNA-sequencing (n = 77, Expression 22Q1 Public). Data were extracted from DepMap Portal (https://depmap.org/portal/). Statistical analysis was performed using two-sample two-tailed t test. ****, p < 0.0001. (D) Differential expression analysis of ULK1 in melanoma patient biopsies prior and post anti-PD-1 therapy (dbGaP study accession: phs001919.v1.p1). Box-plots show counts per million (CPM) of ULK1 expression, on average 30.7 (± 9.17 SD) for 27 patients prior to treatment (baseline) and 39.6 (± 10.18 SD) for 33 patients post-treatment. Patients who did not respond to therapy are colored in red (non-responder) and patients who responded are colored in blue (responder). Lines connect data available for paired samples. Empirical Bayes moderated t-test was used to calculate statistically significant differences between baseline and post-treatment groups as described in the Methods section (adjusted p-value = 0.026). (E-F) (E) A375 and (F) SK-MEL-2 melanoma cells were transfected with empty vector (EV) or ULK1 expression plasmid (ULK1 OE). The next day, transfected cells were either left untreated or were treated with IFNγ (2500 IU/mL) for 6 hours and then processed for qRT-PCR analysis. Scatter dot plots show fold change of mRNA expression for the indicated genes over untreated EV-transfected cells (control). Means ± SEM from three independent experiments are shown. Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *, p < 0.05, ***, p < 0.001.
Targeted inhibition of ULK1 suppresses expression of IFNγ-induced immunosuppressive genes
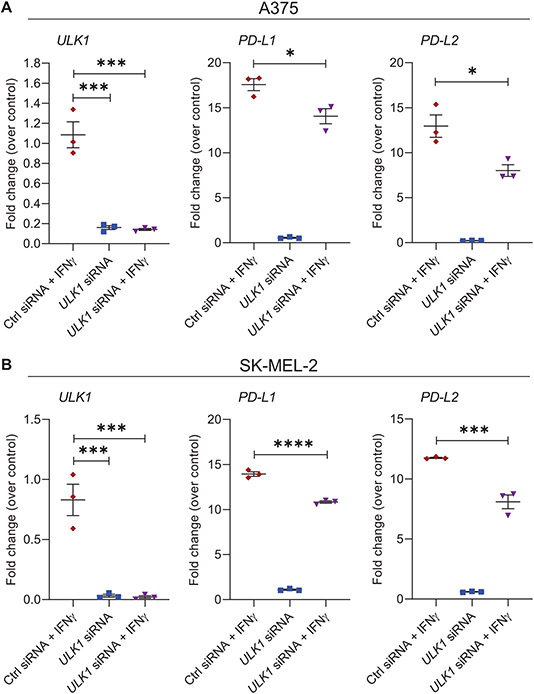
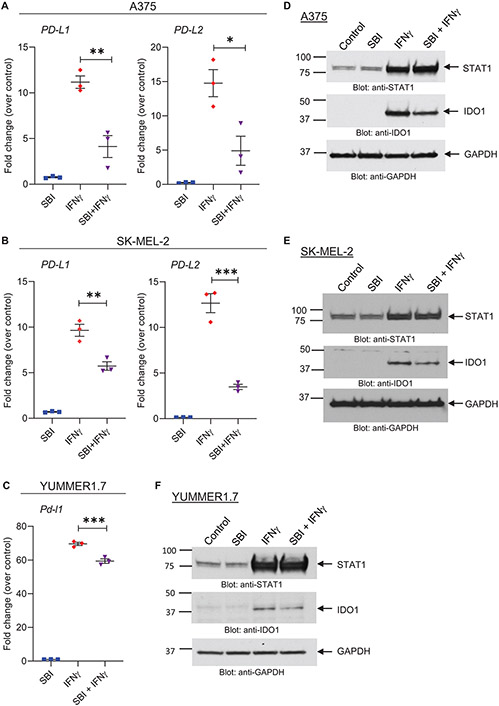
Given that overexpression of ULK1 promotes IFNγ-mediated expression of immunosuppressive genes, we sought to determine the effects of targeted inhibition of ULK1 on the expression of IFNγ-induced genes in melanoma cells. We observed that siRNA-mediated knockdown of ULK1 in A375 and SK-MEL-2 melanoma cells reduced induction of PD-L1 and PD-L2 expression following IFNγ treatment compared to IFNγ-treated control siRNA-transfected cells (Fig. 2). Minimal or negligible differences were observed in the expression of the IFNγ-induced immunostimulatory genes HLA-B, IFI30 and TAP1 (Supplementary Fig. S2). Additionally, small molecule drug targeted inhibition of ULK1 kinase activity using SBI-0206965 (27) was found to decrease IFNγ-mediated expression of the immunosuppressive molecules PD-L1, PD-L2 and IDO1 in A375 and SK-MEL-2 human melanoma cells and in YUMMER1.7 mouse melanoma cells (Fig. 3A-F). In contrast, co-treatment of SBI-0206965 with IFNγ showed minimal or no effects on IFNγ-induced expression of the immunostimulatory genes HLA-B, IFI30, TAP1 and STAT1, as compared to IFNγ treatment alone in all three cell lines (Fig. 3D-F and Supplementary Fig. S3). Together these findings support a selective role for ULK1 in the expression of IFNγ-dependent genes in melanoma cells.
Figure 2. Gene-targeted inhibition of ULK1 reduces transcription of IFNγ-induced immunosuppressive genes in melanoma cells.
(A-B) qRT-PCR analysis of ULK1, PD-L1 and PD-L2 mRNA expression in control (Ctrl) siRNA or ULK1 siRNA-transfected (A) A375 and (B) SK-MEL-2 melanoma cells, either left untreated or treated with IFNγ (2500 IU/mL) for 6 hours. Scatter dot plots, with means ± SEM from three independent experiments, show fold change of mRNA expression for each gene compared to Ctrl siRNA-transfected untreated cells (control) for each independent experiment. Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *, p < 0.05, ***, p < 0.001, ****, p < 0.0001.
Figure 3. Pharmacological inhibition of ULK1 activity reduces transcription of IFNγ-induced immunosuppressive genes in melanoma cells.
(A-C) qRT-PCR analysis of PD-L1 and PD-L2 mRNA expression in (A) A375 and (B) SK-MEL-2 and of Pd-l1 in (C) YUMMER1.7 cells treated with either vehicle (DMSO, control), the ULK kinase inhibitor SBI-0206965 (SBI) (10 μM), IFNγ (2500 IU/mL) or combination therapy with SBI-0206965 and IFNγ (SBI + IFNγ) for 6 hours. Scatter dot plots, with means ± SEM from three independent experiments, show fold change of mRNA expression for each gene compared to vehicle-treated cells (control) for each independent experiment. Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *, p < 0.05, **, p < 0.01, ***, p < 0.001. (D-F) Immunoblotting analysis of STAT1 and IDO1 in (D) A375, (E) SK-MEL-2 and (F) YUMMER1.7 cells treated with either vehicle (DMSO, control), the ULK kinase inhibitor SBI-0206965 (SBI) (A375: 10μM, SK-MEL-2: 5μM, YUMMER1.7: 1μM), IFNγ (1000 IU/mL) or combination therapy with SBI-0206965 and IFNγ (SBI + IFNγ) for 24 hours. GAPDH expression was used as loading control. Immunoblots shown are representative of three independent experiments.
ULK1 interacts with IRF1 in the nucleus controlling its binding to the PD-L1 promoter region
Next, to examine whether inhibition of ULK1 expression does not affect IFNγ-induced phosphorylation of STAT1, we performed siRNA-mediated knockdown of ULK1 in A375 melanoma cells and treated control and ULK1 siRNA-transfected cells with IFNγ for 10 minutes. Our results show that IFNγ treatment induces STAT1 phosphorylation on serine 727 and on tyrosine 701 on both control and ULK1 siRNA-transfected A375 cells (Supplementary Fig. S4). These results are consistent with our previous published studies using Ulk1/2 double knockout mouse embryonic fibroblasts (13).
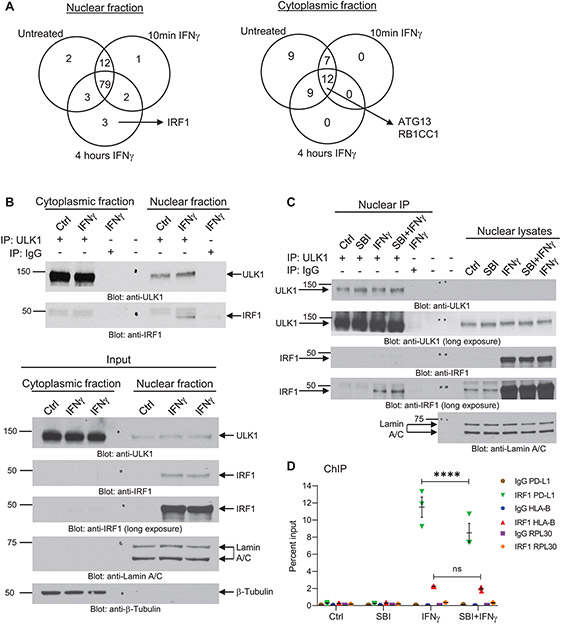
Given that ULK1 is expressed in both nuclear and cytoplasmic compartments (15, 28), we next performed nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) analysis to identify novel interactor proteins of ULK1 in each compartment in the absence and/or presence of IFNγ treatment that could be part of a complex regulating IFNγ-induced transcription of ISGs. For this, we performed nuclear and cytoplasmic fractionation of A375 cells that were either left untreated or treated with IFNγ for 10 minutes or 4 hours (Supplementary Fig. S5A), followed by co-immunoprecipitation of ULK1-protein complexes (Supplementary Fig. S5B). These complexes were resolved by SDS-PAGE and then submitted for nLC-MS/MS analysis. Proteins identified in the rabbit IgG negative control group were removed from the analysis. Consistent with previous reports (24), our results showed that ULK1 binds the autophagy-related proteins ATG13 and RB1CC1 (also known as FIP200) in the cytoplasmic compartment independently of IFNγ treatment (Fig. 4A, cytoplasmic fraction and Supplementary Table S1). Notably, ULK1 was found to interact with IRF1 in the nucleus after 4 hours of IFNγ treatment (Fig. 4A, nuclear fraction and Supplementary Table S1), suggesting a role for ULK1 in mediating IFNγ signaling independent of its autophagy-related function. As IRF1 has been previously shown to act as a transcription factor for ISGs (29) and to bind the PD-L1 promoter region (30), we next sought to validate the binding between ULK1 and IRF1 in A375 cells treated with IFNγ and/or the ULK1 inhibitor SBI-0206965. Initially, we observed that IFNγ treatment induces expression of IRF1 in the nuclear compartment only (Fig. 4B, input lower panel) and validated that ULK1 binds IRF1 in this compartment in A375 melanoma cells treated with IFNγ (Fig. 4B, upper panel). Additionally, we show that IFNγ-induced expression of IRF1 in the nucleus is not affected by co-treatment with SBI-0206965 (Fig. 4C, nuclear lysates) and that ULK1 interacts with IRF1 under both IFNγ alone and IFNγ plus SBI-0206965 treatment conditions (Fig. 4C, nuclear IP). Thus, we next performed ChIP assays to determine whether drug-targeted inhibition of ULK1 kinase activity affects the binding of IRF1 to the PD-L1 promoter and/or HLA-B promoter regions in A375 cells. Co-treatment with IFNγ and SBI-0206965 significantly decreased the binding of IRF1 to the PD-L1 promoter region as compared to IFNγ treatment alone (Fig. 4D). In contrast, no significant differences were observed between IFNγ and IFNγ plus SBI-0206965 treatment conditions for the binding of IRF1 to the HLA-B promoter region (Fig. 4D). These results suggest that ULK1 activity may selectively regulate the affinity of IRF1 to bind specific DNA promoter regions.
Figure 4. ULK1 binds IRF1 in the nucleus and controls IFNγ-induced IRF1 binding to the promoter region of PD-L1.
(A) A375 cells were either left untreated or were treated with IFNγ for 10 min (5000 IU/mL) or 4 hours (2500 IU/mL), followed by cytoplasmic and nuclear cell fractionation (Supplementary Fig. S5A). ULK1-protein complexes were co-immunoprecipitated using an anti-ULK1 monoclonal specific antibody conjugated to sepharose beads for each compartment/treatment condition. As negative control, the same procedure was followed, but using a rabbit monoclonal antibody IgG isotype control conjugated to sepharose beads instead of the anti-ULK1 antibody for the 4 hours (240 min) IFNγ treatment condition (see also Supplementary Fig. S5B). Protein complexes were eluted from the beads and submitted for nano-liquid chromatography-tandem mass spectrometry analysis. Venn diagrams show the number of putative ULK1 interacting proteins in the nuclear and cytoplasmic fractions under the three different treatment conditions. Proteins found on the negative control group were removed from the analysis. (B) A375 cells were either left untreated (Ctrl) or were treated with IFNγ for 4 hours (2500 IU/mL), followed by cytoplasmic and nuclear cell fractionation, as indicated. (Top panel) ULK1-protein complexes were co-immunoprecipitated using an anti-ULK1 monoclonal specific antibody and protein G sepharose beads for each compartment/treatment condition (IP: ULK1). As negative control, the same procedure was followed, but using a rabbit monoclonal antibody IgG isotype control instead of the anti-ULK1 antibody for the 4 hours IFNγ treatment condition (IP: IgG). Protein complexes were eluted from the beads and resolved by SDS-PAGE and immunoblotting analysis was performed for ULK1 and IRF1. (Bottom panel) Equal amounts of nuclear and cytoplasmic protein lysates for each treatment condition (Input) were resolved by SDS-PAGE and immunoblotting analysis was performed for ULK1 and IRF1. Lamin A/C and β-tubulin were used as loading control for the nuclear and cytoplasmic compartments, respectively. (C) A375 cells were pre-treated for 1 hour with either DMSO (Ctrl and IFNγ groups) or SBI-0206965 (SBI) (10 μM) followed by 6 hours of treatment with either DMSO (Ctrl), SBI (10 μM), IFNγ (2500 IU/mL) or SBI + IFNγ, as indicated. After treatment, cells were separated into nuclear and cytoplasmic fractions and ULK1-protein complexes were co-immunoprecipitated using an anti-ULK1 monoclonal specific antibody and protein G sepharose beads for each nuclear fraction treatment condition (IP: ULK1). As negative control, the same procedure was followed, but using a rabbit monoclonal antibody IgG isotype control instead of the anti-ULK1 antibody for the IFNγ treatment condition (IP: IgG). Protein complexes were eluted from the beads and resolved by SDS-PAGE and immunoblotting analysis was performed for ULK1 and IRF1. Equal amounts of nuclear protein lysates for each treatment condition were resolved on the same gel and immunoblotted for ULK1, IRF1 and Lamin A/C (loading control). (D) A375 cells were pre-treated for 1 hour with either DMSO (Ctrl and IFNγ groups) or SBI-0206965 (SBI) (10 μM) followed by 6 hours of treatment with either DMSO (Ctrl), SBI (10 μM), IFNγ (2500 IU/mL) or SBI + IFNγ, as indicated. ChIP assay was performed in A375 cells at the PD-L1 promoter, the HLA-B promoter, and the RPL30 promoter (negative control) for IRF1 binding. IgG antibody was used for each promoter region as negative control. Scatter dot plot shows data as percent enrichment relative to input ± SEM for three independent experiments. Statistical analyses were performed using two-way ANOVA followed by Tukey’s multiple comparisons test between treatment conditions for each antibody binding to each promoter region (IgG to PD-L1, HLA-B or RPL30 and IRF1 to PD-L1, HLA-B or RPL30). ****, p < 0.0001; ns, p > 0.05 between IFNγ and SBI + IFNγ treatment conditions.
Drug targeted inhibition of ULK1 reduces anti-PD-1 therapy-induced Pd-l1 and Pd-l2 expression in a melanoma in vivo model
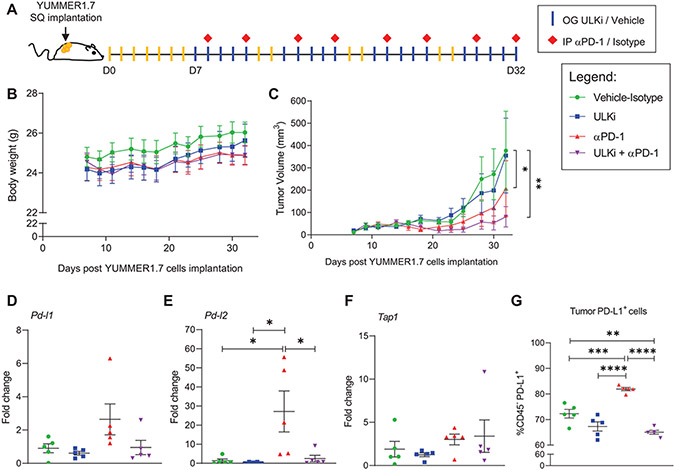
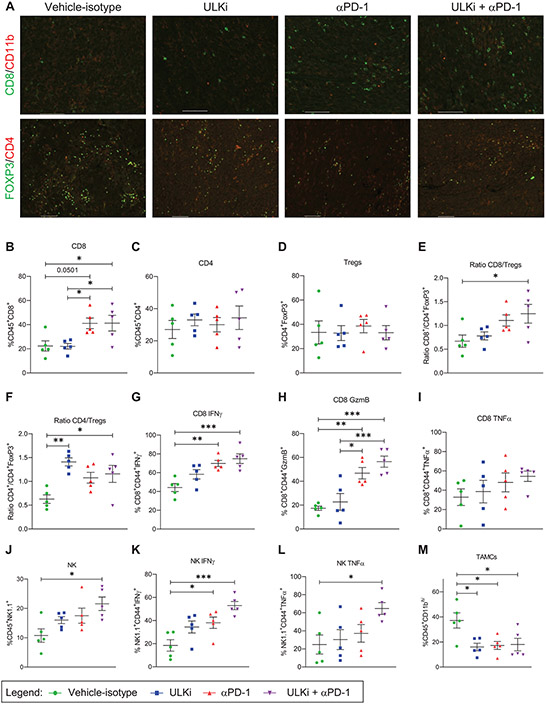
As our results show that ULK1 controls expression of specific IFNγ responsive genes in melanoma cells in vitro, we next sought to evaluate the effects of drug-targeted inhibition of ULK1 activity alone and in combination with anti-PD-1 therapy using the YUMMER1.7 immunogenic mouse melanoma in vivo model (23). YUMMER1.7 mouse melanoma cells carry the three driver mutations: BrafV600E, Pten−/−, and Cdkn2a−/− and present a high somatic mutation burden induced by UV-radiation characteristic of human melanoma (23). These cells were implanted subcutaneously into syngeneic C57BL/6J mice and one week later mice were randomized by tumor size into four treatment groups: 1- Vehicle-Isotype, 2- ULK inhibitor (ULKi), 3- anti-PD-1 and 4- ULKi plus anti-PD-1 (Fig. 5A). For these studies, we used SBP-7455 drug, which was recently reported as a potent orally bioavailable specific ULK1/2 inhibitor in mice (31). Mice were treated by oral gavage with either SBP-7455 (ULKi) or its vehicle five times per week and by intraperitoneal injection with either anti-PD-1 antibody or isotype control antibody twice per week for four weeks (Fig. 5A). Mice were found to tolerate the different drug treatments well, as monitored by body weight (Fig. 5B). Tumor volume was monitored three times per week (Fig. 5C and Supplementary Fig. S6) and at the end of the study (day 32), tumor volume was significantly reduced in both the anti-PD-1 and combination therapy groups (Fig. 5C). Importantly, the degree of growth suppression was most significant in the mice treated with combination therapy compared with the vehicle-isotype treatment group (Fig. 5C). In further studies, we sought to determine the in vivo effects of each treatment condition on expression of IFNγ-induced genes and on tumor-infiltrating immune cells. YUMMER1.7-tumor bearing mice were treated as described above (Fig. 5A) and, on day 35, mice were treated once more by oral gavage with either SBP-7455 or its vehicle and tumors were collected 24 hours later. Tumors from each treatment group were harvested and processed separately for qRT-PCR, immunofluorescence and flow cytometry analyses. Tumor Pd-l1, Pd-l2 and Tap1 mRNA expression was analyzed by qRT-PCR and Pd-l2 expression was found to be significantly higher in the anti-PD-1 treatment group compared to all the other three treatment groups (Fig. 5D-F). Additionally, using flow cytometry analysis we observed increased PD-L1 protein expression on the cell surface of tumors from mice treated with anti-PD-1 alone compared to tumors from mice treated with vehicle-isotype, ULKi and ULKi plus anti-PD-1 therapies (Fig. 5G). Notably, PD-L1 expression was also significantly reduced in the tumors from mice treated with combination therapy compared to tumors from mice in the control group (Fig. 5G). Next, we assessed the presence of tumor-infiltrating immune cells for each treatment condition by immunofluorescence (Fig. 6A and Supplementary Fig. S7-S16) and flow cytometry analysis (Fig. 6B-M and Supplementary Fig. S17). The percentage of tumor-infiltrating CD8+ T cells was substantially increased in both anti-PD-1 and ULKi plus anti-PD-1 treatment groups compared to the control and ULKi treatment groups (Fig. 6B), whereas no major differences were observed for the percentage of tumor-infiltrating CD4+ T cells and T regulatory cells (Tregs) between the four treatment groups (Fig. 6C-D). However, the ratio of CD8+ T cells over Tregs and the ratio of CD4+ T cells over Tregs were significantly increased between the combinatorial therapy and the control treatment group (Fig. 6E-F). Interestingly, the ratio of CD4+ T cells over Tregs was also higher in the tumors from ULKi-treated mice compared to the tumors from vehicle-isotype-treated mice (Fig. 6F). Compared to the control-treated mice, tumors from mice treated with ULKi plus anti-PD-1 showed higher percentages of CD8+ effector T cells that produce IFNγ, granzyme B and TNFα (Fig. 6G-I). Moreover, the percentages of tumor-infiltrating natural killer (NK) cells, as well as active NK cells producing IFNγ and TNFα, were significantly increased in the combination treatment group compared to the control treatment group (Fig. 6J-L). Interestingly, the percentage of tumor-associated myeloid cells (TAMCs) was lower in both single dose and combination treatment groups compared to the vehicle-isotype treatment group (Fig. 6M). In summary, these data suggest that pharmacological inhibition of ULK1 in combination with anti-PD-1 treatment could further enhance anti-tumor responses in vivo by, at least in part, decreasing the expression of IFNγ-induced immunosuppressive genes in melanoma cells mediated by treatment with anti-PD-1 antibody.
Figure 5. Drug-targeted inhibition of ULK1 decreases expression of anti-PD-1 therapy-driven IFNγ-induced immunosuppressive genes in an in vivo mouse melanoma model.
(A) Schematic illustration of YUMMER1.7 mouse melanoma in vivo model and therapeutic regimen. Seven days (D7) post subcutaneous (SQ) implantation of YUMMER1.7 mouse melanoma cells into C57BL/6J mice, mice were randomized by tumor size into four treatment groups: 1- Vehicle-Isotype, 2- ULK inhibitor (ULKi), 3- anti-PD-1 (αPD-1) and 4- ULKi + αPD-1. Mice were treated by oral gavage (OG) with either SBP-7455 (ULKi) (10 mg/kg) or its vehicle five times per week and by intraperitoneal injection (IP) with either anti-PD-1 antibody or isotype control antibody (10 mg/kg) twice per week for four weeks, as illustrated. (B) Mice body weight was monitored three times per week throughout the study. Data shown are means ± SEM (n = 6 for vehicle-isotype and ULKi, n = 7 for αPD-1 and ULKi + αPD-1 treatment groups). (C) Tumor volume was measured three times per week throughout the study. Data shown are means ± SEM (n = 6 for vehicle-isotype and ULKi, n = 7 for αPD-1 and ULKi + αPD-1 treatment groups). A linear mixed effects model was fitted with ln(volume) as the outcome and day, treatment group and their interaction as fixed effects. For 0 volume measurements, ln(1) was used. Within-mouse correlation between repeated measurements was modeled using autoregressive order 1 (AR(1)) covariance structure. Pairwise differences between groups at Day 32 were assessed based on the model, and p-values were adjusted for multiple comparisons using the method of Tukey-Kramer. Analyses were done using PROC MIXED in SAS v 9.4 software (26). *, p < 0.05 between vehicle-control and αPD-1 and **, p < 0.01 between vehicle-control and ULKi + αPD-1 treatment groups. p < 0.05 was observed between ULKi and αPD-1 and ULKi + αPD-1 treatment groups. See also Supplementary Fig. S6. (B-C) Results are representative of two independent in vivo studies. (D-G) YUMMER1.7 tumor-bearing mice were treated as illustrated in A, and, on day 35, mice were treated once by oral gavage with either ULKi (10 mg/kg) or its vehicle and tumors were collected 24 hours later. (D-F) qRT-PCR analysis of Pd-l1, Pd-l2 and Tap1 mRNA expression in YUMMER1.7 tumors isolated from mice treated with either vehicle-isotype, ULKi, αPD-1 or ULKi + αPD-1. Scatter dot plots, with means ± SEM (n = 5 for each treatment group), show fold change of mRNA expression for each gene compared to a randomly selected vehicle-isotype-treated mouse tumor. Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *, p < 0.05. (G) Scatter dot plot shows the percentage of tumor cells expressing PD-L1 (CD45− PD-L1+ cells) in tumors isolated from vehicle-isotype, ULKi, αPD-1 and ULKi plus αPD-1-treated mice with means ± SEM (n = 5 for each treatment group). Data were assessed by flow cytometry and the gating strategy used is shown in Supplementary Fig. S17. Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. **, p < 0.01; ***, p < 0.001 and ****, p < 0.0001.
Figure 6. Drug-targeted inhibition of ULK1 increases the frequency of infiltrating anti-tumor immune cells in response to anti-PD-1 therapy in vivo.
(A-M) YUMMER1.7 tumor-bearing mice were treated as illustrated in Fig. 5A, and, on day 35, mice were treated once by oral gavage with either ULKi (10 mg/kg) or its vehicle and 24 hours later tumors were collected and processed for immunofluorescence and flow cytometry analyses. (A) Immunofluorescence imaging of CD8+ T cells (CD8), tumor-associated myeloid cells (TAMCs) (CD11b), CD4+ T cells (CD4) and T regulatory cells (Tregs) (FOXP3) in the tumors isolated from mice treated with vehicle-isotype, ULKi, αPD-1 and ULKi plus αPD-1. Representative images of merged staining for the indicated antibodies are shown for each treatment group. Pictures were taken with Nikon Ti2 Widefield microscope equipped with Photometrics IRIS15 sCMOS camera with a 10X panfluorNA03 objective and zoomed in using NIS Elements Imaging software as shown in Supplementary Fig. S7-S14. (B-M) Scatter dot plots show the percentage of tumor-infiltrating (B) CD8+ T cells, (C) CD4+ T cells, (D) Tregs, (E) ratio of CD8+ T cells over Tregs, (F) ratio of CD4+ T cells over Tregs, (G) IFNγ-producing CD8+ T cells, (H) granzyme B-producing CD8+ T cells, (I) TNFα-producing CD8+ T cells, (J) natural killer (NK) cells, (K) IFNγ-producing NK cells, (L) TNFα-producing NK cells and (M) TAMCs in tumors isolated from vehicle-isotype, ULKi, αPD-1 and ULKi plus αPD-1-treated mice with means ± SEM (n = 5 for each treatment group). The flow cytometry gating strategy used is shown in Supplementary Fig. S17. Statistical analyses were performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *, p < 0.05; **, p < 0.01 and ***, p < 0.001.
Discussion
Prior to 2011, few treatment options were available for patients with advanced or metastatic melanoma disease, and median overall survival remained limited to 9 months (32). The field of melanoma research was redrawn with the discovery that re-activation of anti-tumor immune responses through inhibition of immune checkpoints, such as PD-1 and CTLA-4, can induce durable responses in patients. Combination therapy with the ICIs nivolumab (anti-PD-1 antibody) and ipilimumab (anti-CTLA-4 antibody) improved median overall survival to 6 years in patients with advanced melanoma (33). However, despite this progress, resistance to ICIs has become a significant clinical barrier (34). Approximately 55% of melanoma patients present with primary resistance to ICIs and, by three years, at least 40% of responding patients develop secondary resistance (2, 35). Thus, improving the response rate to these therapies by targeting resistance mechanisms is of critical importance.
Paradoxically, IFNγ signaling and IFNγ-responsive genes in melanoma cells have been associated with both response and resistance to ICIs (3, 5, 36). Clinical studies have shown that melanoma tumors carrying genomic alterations of IFNγ pathway genes (e.g., IFNGR, JAK1 and JAK2) do not respond to anti-CTLA-4 and exhibit primary and acquired resistance to anti-PD-1 therapy (6, 7, 37). Thus, intact IFNγ signaling is necessary for immune activation and the anti-tumor activity of ICIs. However, chronic IFNγ signaling mediates downregulation of antigen presentation on the tumor cell surface, resulting in loss of immunogenicity and ICI efficacy (38). In another study, loss of the IFNγ receptor was shown to be associated with decreased expression of ICI resistance genes (39). Additionally, increased CD8+ T cell activation and infiltration are required for an appropriate response to ICIs (40-42). Melanoma tumors resistant to ICIs are often found to be in a T cell excluded state or to have characteristics of an “exhausted” immune state consistent with chronic immune activation (3). This exhausted phenotype is partially driven by IFNγ-mediated upregulation of immunosuppressive genes (36). Moreover, in low tumor burden state, IFNγ secretion was shown to stimulate an immune-intrinsic mechanism of resistance to ICIs (10). Similarly, in healthy tissues, IFNγ plays a critical role in immune activation, but also in the feedback mechanisms that downregulate the immune response, preventing autoimmunity and deleterious tissue damage. These innate negative feedback mechanisms also seem to protect tumor cells from a prolonged immune response and immune-mediate cell death, allowing tumor escape (11, 34). Thus, in order to optimize responses to ICIs, timely and selective control of IFNγ signaling is required to inhibit IFNγ-driven immunosuppressive responses without affecting activation of immunostimulatory ones.
IFNγ binds to its receptor composed of two IFNGR1 and two IFNGR2 subunits bound to JAK1 and JAK2 proteins, respectively (43). Engagement of IFNGR/JAKs activates STAT-dependent and independent-signaling pathways that tightly control transcription and translation of ISGs (43). Previously, blocking IFNγ signaling at the receptor level by pharmacologic inhibition of JAK1/2 was shown to reverse resistance to ICIs in vivo (8), but this approach can also negatively impact the initial IFNγ-immunostimulatory effects required for response to ICIs, as previously shown (5-7, 37). Additionally, JAK proteins are required for activation of signaling pathways by other cytokines, such as GM-CSF, and growth factors (44, 45), whose inhibition could lead to off-target effects and negatively impact immune responses (46, 47).
In a previous study, using RNA-sequencing and gene ontology analyses we have shown that targeted deletion of Ulk1/2 genes in mouse embryonic fibroblasts does not affect IFNγ-induced expression of genes involved in activation of innate immune responses and antigen presentation, but increases expression of IFNγ-induced genes associated with immune cell proliferation, cytokine production and T cell activation (13). These results suggest that targeted inhibition of ULK1 may enhance IFNγ-mediated immunostimulatory effects. In the present study, we show that increased levels of ULK1 correlate with poor survival of melanoma patients and observed an increased expression of ULK1 in melanoma patients post anti-PD-1 therapy supporting a potential role for this kinase on anti-PD-1-driven immune responses. We show that ULK1 interacts with IRF1 transcription factor in the nucleus of melanoma cells and inhibition of its kinase activity reduces IRF1 binding to the immunosuppressive gene PD-L1 promoter region, but not to the immunostimulatory gene HLA-B promoter region. Consistently, overexpression of ULK1 in melanoma cells promotes IFNγ-induced expression of immunosuppressive genes, while genetic or pharmacologic inhibition of ULK1 decreases it, with minimal or no effects on the expression of immunostimulatory genes. Together with other studies, our data suggest that JAK/STAT signaling mediates a primary response to IFNγ inducing expression of IFNγ-primary response genes, including expression of IRF1 in the nucleus (11) and nuclear ULK1 then selectively controls the binding of IRF1 to DNA promoter regions of IFNγ secondary response genes. Future studies will explore the precise mechanism by which ULK1 regulates IRF1 binding to DNA promoter regions and other potential mechanisms through which ULK1 may specifically modulate IFNγ-responses both in melanoma and immune cells. Our results suggest that ULK1 can directly phosphorylate IRF1 controlling its binding affinity to the promoter region of specific IFNγ-induced genes, depending on the sequence of the IRF1 binding site on those genes. It is also plausible that the presence of other transcription factor binding sites, such as those of other IRF proteins or STAT proteins near the IRF1 site in specific genes could enhance or downregulate IRF1 binding to their promoters in an IFNγ-induced ULK1-dependent manner. In vivo, combination of drug-targeted inhibition of ULK1 with anti-PD-1 therapy further reduced melanoma tumor growth, decreased Pd-l2 and PD-L1 expression in tumor cells and further enhanced infiltration and activation of anti-tumor immune cells within the tumor microenvironment. Interestingly, in another study (48) murine IRF1-deficient tumors were found to have lost the capacity to increase PD-L1 expression, becoming more susceptible to T-cell cytotoxicity in vivo. These results suggest that IRF1 could promote tumor immune evasion through PD-L1 expression (48).
Inhibition of ULK1 has also been shown to inhibit induction of autophagy in several types of cancer leading to either cancer cell death or survival, depending on the type of cancer (49, 50). In LKB1-mutant non-small lung cancer models, inactivation of ULK1 kinase function restores antigen presentation through increasing immunoproteasome activity and synergizes with anti-PD-1 therapy in vivo by enhancing CD8+ T cell expansion (51). Together these reports suggest that ULK1 plays distinct autophagy-dependent and -independent roles that control tumor growth and anti-tumor immune responses. Our data support specific targeting of ULK1 to suppress IFNγ-dependent immunosuppressive effects, while still promoting IFNγ-mediated immune cell activity against melanoma, to overcome resistance mechanisms to ICIs.
Supplementary Material
Acknowledgements:
We thank Dr. Antoni Ribas (Jonsson Comprehensive Cancer Center and David Geffen School of Medicine, University of California Los Angeles, Funding Source: R35 CA197633 National Institutes of Health, Bethesda, MD, USA) for his valuable feedback, support on our studies and for depositing the data for study accession: phs001919.v1.p1 on dbGaP. Proteomics services were performed by the Northwestern Proteomics Core Facility of the Robert H. Lurie Comprehensive Cancer Center (supported by P30 CA060553). Flow cytometry work was performed at the Northwestern University Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Facility. All histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory of the Robert H Lurie Comprehensive Cancer Center. Imaging work was performed at the Northwestern University Center for Advanced Microscopy core of the Robert H Lurie Comprehensive Cancer Center. This work was supported by an H Foundation-Robert H. Lurie Comprehensive Cancer Center Basic Insights Award (CMH, DS), grants R50-CA221848 (ETB), P30-CA060553 (MK), R01-CA77816 (LCP) and R21-CA245447 (DS) by NIH-NCI and by a Conquer Cancer Young Investigator Award (SEF), supported by Funder. Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology®, Conquer Cancer®, or the Funder.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Sullivan RJ, Atkins MB, Kirkwood JM, Agarwala SS, Clark JI, Ernstoff MS, et al. An update on the Society for Immunotherapy of Cancer consensus statement on tumor immunotherapy for the treatment of cutaneous melanoma: version 2.0. J Immunother Cancer 2018;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015;373:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minn AJ, Wherry EJ. Combination Cancer Therapies with Immune Checkpoint Blockade: Convergence on Interferon Signaling. Cell 2016;165:272–5. [DOI] [PubMed] [Google Scholar]
- 5.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature 2017;548:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov 2017;7:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016;167:1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 2013;5:200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai CS, Huang JT, Lu X, Simons DM, Park C, Chang A, et al. Clonal Deletion of Tumor-Specific T Cells by Interferon-γ Confers Therapeutic Resistance to Combination Immune Checkpoint Blockade. Immunity 2019;50:477–492.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaidi MR, Merlino G. The two faces of interferon-γ in cancer. Clin Cancer Res 2011;17:6118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saleiro D, Mehrotra S, Kroczynska B, Beauchamp EM, Lisowski P, Majchrzak-Kita B, et al. Central role of ULK1 in type I interferon signaling. Cell Rep 2015;11:605–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleiro D, Blyth GT, Kosciuczuk EM, Ozark PA, Majchrzak-Kita B, Arslan AD, et al. IFN-γ-inducible antiviral responses require ULK1-mediated activation of MLK3 and ERK5. Sci Signal 2018;11:eaap9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013;155:688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleiro D, Kosciuczuk EM, Platanias LC. Beyond autophagy: New roles for ULK1 in immune signaling and interferon responses. Cytokine Growth Factor Rev 2016;29:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abril-Rodriguez G, Torrejon DY, Liu W, Zaretsky JM, Nowicki TS, Tsoi J, et al. PAK4 inhibition improves PD-1 blockade immunotherapy. Nat Cancer 2020;1:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 2015;43:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005;21:2067–75. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Perry CJ, Meeth K, Thakral D, Damsky W, Micevic G, et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res 2017;30:428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009;20:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saleiro D, Wen JQ, Kosciuczuk EM, Eckerdt F, Beauchamp EM, Oku CV, et al. Discovery of a signaling feedback circuit that defines interferon responses in myeloproliferative neoplasms. Nat Commun 2022;13:1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SAS Institute Inc. 2018. SAS/STAT® 15.1 User’s Guide. Cary, NC: SAS Institute Inc. [Google Scholar]
- 27.Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell 2015;59:285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi A, Iyengar R, Joo JH, Li-Harms XJ, Wright C, Marino R, et al. Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death Differ 2016;23:216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forero A, Ozarkar S, Li H, Lee CH, Hemann EA, Nadjsombati MS, et al. Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity 2019;51:451–464.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 2017;19:1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren H, Bakas NA, Vamos M, Chaikuad A, Limpert AS, Wimer CD, et al. Design, Synthesis, and Characterization of an Orally Active Dual-Specific ULK1/2 Autophagy Inhibitor that Synergizes with the PARP Inhibitor Olaparib for the Treatment of Triple-Negative Breast Cancer. J Med Chem 2020;63:14609–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol 2017;14:463–82. [DOI] [PubMed] [Google Scholar]
- 33.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, et al. Long-Term Outcomes With Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients With Advanced Melanoma. J Clin Oncol 2022;40:127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenton SE, Sosman JA, Chandra S. Resistance mechanisms in melanoma to immuneoncologic therapy with checkpoint inhibitors. Cancer Drug Resist 2019;2:744–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, et al. Association of Pembrolizumab With Tumor Response and Survival Among Patients With Advanced Melanoma. JAMA 2016;315:1600–9. [DOI] [PubMed] [Google Scholar]
- 36.Benci JL, Johnson LR, Choa R, Xu Y, Qiu J, Zhou Z, et al. Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell 2019;178:933–948.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016;167:397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda K, Nakayama M, Hayakawa Y, Kojima Y, Ikeda H, Imai N, et al. IFN-γ is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat Commun 2017;8:14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cucolo L, Chen Q, Qiu J, Yu Y, Klapholz M, Budinich KA, et al. The interferon-stimulated gene RIPK1 regulates cancer cell intrinsic and extrinsic resistance to immune checkpoint blockade. Immunity 2022;55:671–685.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleiro D, Platanias LC. Interferon signaling in cancer. Non-canonical pathways and control of intracellular immune checkpoints. Semin Immunol 2019;43:101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 2012;36:542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stark GR, Darnell JE Jr. The JAK-STAT pathway at twenty. Immunity 2012;36:503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwek SS, Kahn J, Greaney SK, Lewis J, Cha E, Zhang L, et al. GM-CSF and ipilimumab therapy in metastatic melanoma: Clinical outcomes and immunologic responses. Oncoimmunology 2015;5:e1101204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaufman HL, Ruby CE, Hughes T, Slingluff CL Jr. Current status of granulocyte-macrophage colony-stimulating factor in the immunotherapy of melanoma. J Immunother Cancer 2014;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao L, Hou W, Scharping NE, Vendetti FP, Srivastava R, Roy C, et al. IRF1 Inhibits Antitumor Immunity through the Upregulation of PD-L1 in the Tumor Cell. Cancer Immunol Res. 2019;7:1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C, Blessing AM, Pulliam TL, Shi Y, Wilkenfeld SR, Han JJ, et al. Inhibition of CAMKK2 impairs autophagy and castration-resistant prostate cancer via suppression of AMPK-ULK1 signaling. Oncogene 2021;40:1690–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu L, Yan L, Liao N, Wu WQ, Shi JL. A Review of ULK1-Mediated Autophagy in Drug Resistance of Cancer. Cancers (Basel) 2020;12:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng J, Thennavan A, Dolgalev I, Chen T, Li J, Marzio A, et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1 mutant lung cancer. Nat Cancer 2021;2:503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author. Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the project accession #: PXD035347.