Abstract
Cremanthodium Benth. is an endemic genus in the Himalayas and adjacent areas. Some plants of the genus are traditional medicinal plants in Tibetan medicine. In this study, the chloroplast genomes of five species (Cremanthodium arnicoides (DC. ex Royle) Good, Cremanthodium brunneopilosum S. W. Liu, Cremanthodium ellisii (Hook. f.) Kitam., Cremanthodium nervosum S. W. Liu, and Cremanthodium rhodocephalum Diels) were collected for sequencing. The sequencing results showed that the size of the chloroplast genome ranged from 150,985 to 151,284 bp and possessed a typical quadripartite structure containing one large single copy (LSC) region (83,326–83,369 bp), one small single copy (SSC) region (17,956–18,201 bp), and a pair of inverted repeats (IR) regions (24,830–24,855 bp) in C. arnicoides, C. brunneopilosum, C. ellisii, C. nervosum, and C. rhodocephalum. The chloroplast genomes encoded an equal number of genes, of which 88 were protein-coding genes, 37 were transfer ribonucleic acid genes, and eight were ribosomal ribonucleic acid genes, and were highly similar in overall size, genome structure, gene content, and order. In comparison with other species in the Asteraceae family, their chloroplast genomes share similarities but show some structural variations. There was no obvious expansion or contraction in the LSC, SSC or IR regions among the five species, indicating that the chloroplast gene structure of the genus was highly conserved. Collinearity analysis showed that there was no gene rearrangement. The results of the phylogenetic tree showed that the whole chloroplast genomes of the five species were closely related, and the plants of this genus were grouped into one large cluster with Ligularia Cass. and Farfugium Lindl.
Keywords: Cremanthodium Benth., Chloroplast genomes, Endemic genera, Sequencing
Introduction
The genus of Cremanthodium Benth. belongs to perennial herbaceous plants of Asteraceae, is distributed primarily in the Tibetan Plateau and southwest mountain area, and contains approximately 64 species in China. The Cremanthodium genus is an endemic genus of the Himalayas and adjacent areas, growing in alpine bushwood, grassy marshland, and screes (Flora Reipublicae Popularis Sinicae 1989). Some plants of this genus are traditional medicinal plants in Tibetan medicine.
At present, studies on this genus have mainly focused on chemicals and pharmacology. Chemical constituents of C. ellisii (Hook. f.) Kitam., C. potaninii C. Winkl., C. discoideum Maxim., C. rhodocephalum Diels, C. lineare Maxim., C. helianthus (Franch.) W. W. Smith, C. stenactinium Diels ex Limpr., and C. brunneopilosum S. W. Liu were studied. Sesquiterpenoids (Chen et al. 1996; Saito et al. 2012; Tori et al. 2012), phenylpropanoids (Zhu et al. 2001), lignin (Yang et al. 1995; Su et al. 1999, 2020; Wang et al. 2004) and steroids (Zhu et al. 2000) are the main constituents of the Cremanthodium genus, as well as fatty acids (Tu et al. 2006) and volatile oils. Recent studies have tapped into the potential of the Cremanthodium genus as antibacterial and antitumour plants (Li et al. 2007). Specifically, the volatile oils of C. discoideum have been well known for their ability to relieve coughing and repell mosquitoes (Wu et al. 2003), while the ether extract of C. humile exhibits significant activities for inducing HeLa cell apoptosis (Li et al. 2007).
Although the Cremanthodium genus has many species, their chloroplast genome has not been used for species identification. This study focused on characterizing the chloroplast genome sequences of C. arnicoides, C. brunneopilosum, C. ellisii, C. nervosum, and C. rhodocephalum (A-B-E-N-R) by the Illumina sequencing platform and discussing the structures and features of the five newly sequenced chloroplast genomes to provide evidence for species identification of the Cremanthodium genus.
Materials and methods
Plant material
Fresh leaves from five species of Cremanthodium were collected for DNA extraction (Fig. 1). The leaf material of C. nervosum (Voucher No. JXZY187) was collected in Yadong County, Tibet, China (27° 36′ 28.7″ N, 89° 02′ 28.65″ E, 3520 m). The leaf material of C. rhodocephalum (Voucher No. JXZY113) was collected from Jiangzi County, Tibet, China (28° 57′ 29.83″ N, 89° 30′ 4.15″ E, 4630 m), and the leaf material of C. ellisii (Voucher No. JXZY496) was collected from Nyalam, Tibet, China (28° 19′ 06.2″ N, 86° 02′ 19.5″ E, 4172 m). The leaf material of C. arnicoides (Voucher No. JXZY286) and C. brunneopilosum (Voucher No. JXZY491) were collected from Chefei Township, Bailang County, Tibet, China (29° 20′ 50.0″ N, 89° 38′ 30.3″ E, 3776 m).
Fig. 1.
Five species of Cremanthodium genus. a C. arnicoides, b C. brunneopilosum, c C. ellisii, d C. nervosum, e C. rhodocephalum
DNA extraction and sequencing
The quality of isolated genomic DNA was verified using two methods: (1) DNA degradation and contamination were monitored on 1% agarose gels; and (2) DNA concentration was measured in a Qubit® 3.0 Fluorometer with a Qubit® DNA Assay Kit (Invitrogen, USA).
A total amount of 0.2 μg DNA per sample was used as input material for the DNA library preparations. The sequencing library was generated using the NEB Next® Ultra™ DNA Library Prep Kit for Illumina sequencing (NEB, USA) following the manufacturer’s recommendations, and index codes were added to each sample. Briefly, genomic DNA samples were fragmented by sonication to a size of 350 bp. Then, DNA fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing, followed by further PCR amplification. After the PCR products were purified by an AMPure XP system (Beckman Coulter, Beverly, USA), the DNA concentration was measured by a Qubit®3.0 Fluorometer (Invitrogen, USA) and the libraries were analysed for size distribution by NGS3K/Calliper and quantified by real-time PCR (3 nM).
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using an Illumina PE Cluster Kit (Illumina, USA) according to the manufacturer’s instructions. After cluster generation, the DNA libraries were sequenced on an Illumina platform, and 150 bp paired-end reads were generated.
Genome sequencing, assembly, and annotation
Pair-end Illumina raw reads were cleaned, adaptors and barcodes were removed, and then quality filtering was performed using Trimoraic. Individual bases with Phred quality score < 20 were removed from both ends of reads, as well as more than three consecutive uncalled bases. Entire reads with a median quality score lower than 21 or less than 40 bp in length after trimming were discarded. After quality filtering, reads were mapped to the chloroplast genome of the closest species with a chloroplast genome available (NCBI download) using Bowtie2 v.2.2.6 (https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.2.6/) to exclude reads of nuclear and mitochondrial origins. All putative chloroplast reads mapped to the reference sequence above were then used for de novo assembly to reconstruct the chloroplast genomes using Get Organelle (Jin et al. 2020). Automatic annotation of the chloroplast genomes was generated by CpGAVAS2 (Shi et al. 2019), and a circular representation of both sequences was drawn using the online tool OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw. html). The draft annotations given by CpGAVAS2 were then manually corrected using Artemis software and other plastid genomes for comparison.
Analysis of chloroplast genomic characteristics
Relative synonymous codon usage (RSCU) analysis was used to express the ratio of the actual codon usage value to the theoretical codon usage value of five species. The characteristics of scattered repeat sequences were analysed by Reputer software (Kurtz et al. 2001). Simple repeat sequences were identified by the MISA tool (http://pgrc.ipk-gatersleben.de/misa/) (parameters: 1-10 2-53-4 4-3-3 6-3).
Comparative genomic analysis
Based on the genome map, the known genes and genome structures were compared to reveal gene function, expression regulation mechanisms, species evolution and other aspects. IR expansion and contraction were analysed by online IR scope (http://irscrope.shinyapps.io/irapp/). The Mauve tool (http://darlinglab.org/mauve/mauve.html) was used to analyse multiple sequences to determine the local collinearity between genomes. The chloroplast genomes of 25 species of Asteraceae were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/), and the chloroplast genomes were constructed by RAxML software (Stamatakis 2014).
Results
Genomic characteristics of chloroplasts
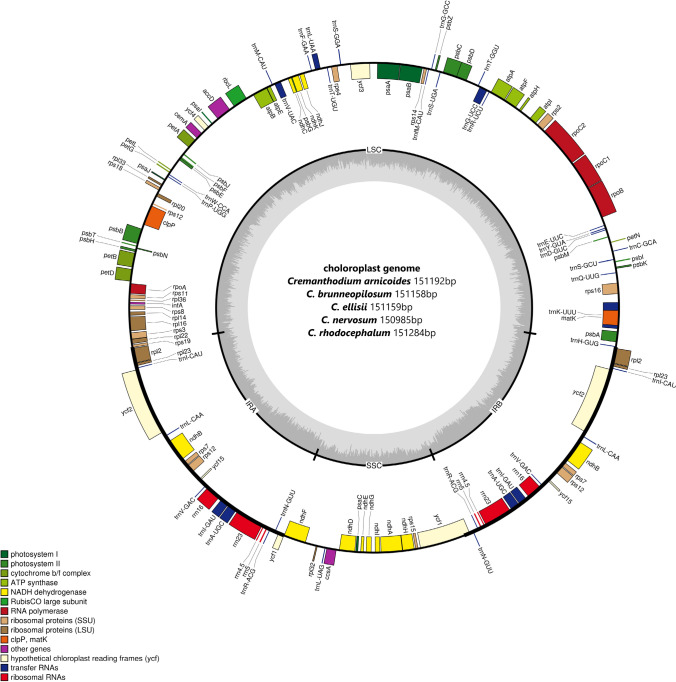
After assembly, the lengths of the A-B-E-N-R chloroplast genomes were 151,192 bp, 151,158 bp, 151,159 bp, 150,985 bp, and 15,1284 bp, respectively. The chloroplast genomes of five Cremanthodium species (A-B-E-N-R) all had a typical quadripartite structure: a large single copy region (LSC), a small single copy region (SSC), and two inverted repeat regions (IRs) (Fig. 2). In the A-B-E-N-R chloroplast genomes, the lengths of the LSC region were 83,357 bp, 83,351 bp, 83,326 bp, 83,369 bp, and 83,423 bp; the lengths of the SSC regions were 18,125 bp, 18,107 bp, 18,173 bp, 17,956 bp, and 18,201 bp; and the lengths of the pair of inverted repeats (IRs) were 24,855 bp, 24,850 bp, 24,830 bp, 24,830 bp, and 24,830 bp, respectively (Fig. 2). The GC content in the A-B-E chloroplast genome was 37.45%, that in the N chloroplast genome was 37.48%, and that in the R chloroplast genome was 37.46%. The chloroplast genomes were highly conserved in structure, which was basically the same among species. All the chloroplast genome sequences have been uploaded to NCBI (GenBank: OM386855, OM386856, OM386857, OM386858, OM386859).
Fig. 2.
The assembly, size, and features of A-B-E-N-R chloroplast genomes (Cremanthodium). The genes outside the circle are transcribed in the counter clockwise direction, and the genes inside the circle are transcribed in the clockwise direction. Different colors in genes represent different functions. The dark gray area and light gray area of the inner circle represent the GC content and AT content of the genome, respectively
Through gene annotation, we found that the chloroplast genomes of five Cremanthodium species showed similar genome structures, containing 133 unique genes (88 protein coding genes, 37 tRNA genes, and 8 rRNA genes) (Table 1). There was no significant difference in gene sequence, gene type or quantity among the five chloroplast genomes. All five chloroplast genomes had 19 genes containing two copies distributed in the IR region, including eight protein editing genes (ndhB, rpl2, rpl23, rps12, rps7, ycf1, ycf15, and ycf2), seven tRNA genes (trnA-UGC, trnG-UCC, trnI-CAU, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5). A total of 18 genes had introns, of which nine protein-coding genes (ndhA, ndhB, petB, petD, atpF, rpl16, rpl2, rps16, and rpoC1) and six tRNA genes contained one intron (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC), and three protein-coding genes (rps12, clpP, and ycf3) contained two introns (Table 2). In all five chloroplast genomes, the ycf1 gene spanned the SSC and IRb junction.
Table 1.
Chloroplast genome characteristics of five species in Cremanthodium genus
| Species | Size (bp) | No. of PCGs | No. of tRNAs | No. of rRNAs | No. of genes | GC content (%) | LSC size (bp) | SSC size (bp) | IR size (bp) |
|---|---|---|---|---|---|---|---|---|---|
| C.arnicoides | 151,192 | 88 | 37 | 8 | 133 | 37.45 | 83,357 | 18,125 | 24,855 |
| C. brunneopilosum | 151,158 | 88 | 37 | 8 | 133 | 37.45 | 83,351 | 18,107 | 24,850 |
| C. ellisii | 151,159 | 88 | 37 | 8 | 133 | 37.45 | 83,326 | 18,173 | 24,830 |
| C. nervosum | 150,985 | 88 | 37 | 8 | 133 | 37.48 | 83,369 | 17,956 | 24,830 |
| C. rhodocephalum | 151,284 | 88 | 37 | 8 | 133 | 37.46 | 83,423 | 18,201 | 24,830 |
Table 2.
Encoded genes in chloroplast genome of five Cremanthodium species
| Category | Group | Genes |
|---|---|---|
| Photosynthetic | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbG, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA†, ndhB (× 2)†, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB†, petD†, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF†, atpH, atpI | |
| Large subunit of RubisCO | rbcL | |
| Self-replication | Large subunit of ribosomal | rpl14, rpl16†, rpl2 (× 2)†, rpl20, rpl22, rpl23 (× 2), rpl32, rpl33, rpl36 |
| Samll subunit of ribosomal | rps11, rps12 (× 2)†, rps14, rps15, rps16†, rps18, rps19, rps2, rps3, rps4, rps7 (× 2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1†, rpoC2 | |
| Ribosomal RNAs | rrn16 (× 2), rrn23 (× 2), rrn4.5 (× 2), rrn5 (× 2) | |
| Transfer RNAs | trnA-UGC (× 2)†, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC†, trnH-GUG, trnI-CAU (× 2), trnI-GAU (× 2)†, trnK-UUU†, trnL-CAA (× 2), trnL-UAA†, trnL-UAG, trnM-CAU, trnN-GUU (× 2), trnP-UGG, trnQ-UUG, trnR-ACG (× 2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (× 2), trnV-UAC†, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Tanslational initiation factor | infA | |
| Other | Protease | clpP† |
| Maturase | matK | |
| Envelope membrane protein | cemA | |
| c-type cytochrome synthesis gene | ccsA | |
| Subunit of Acetyl-CoA- carboxylase | accD | |
| Hypothetical chloroplast reading frames | ycf1 (× 2), ycf15 (× 2), ycf2 (× 2), ycf3†, ycf4 |
(× 2) indicates that the gene has two copies. †Indicate genes containing introns
The codon-anticodon recognition pattern and codon usage
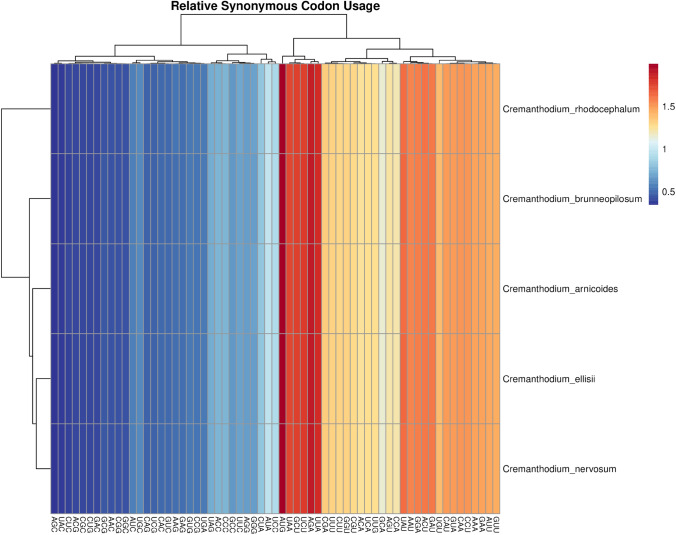
The codon preference analysis of the five chloroplast genomes showed that there was no significant difference in the overall size, base composition, or AT/GC content of the protein coding region. There were few differences in codon usage preferences among the chloroplast genomes of the five species (Table 3, Fig. 3).
Table 3.
The codon-anticodon recognition pattern and codon usage for five Cremanthodium species
| Amino acid | Codon | C. arnicoides | C. brunneopilosum | C. ellisii | C. nervosum | C. rhodocephalum | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No | RSCU | No | RSCU | No | RSCU | No | RSCU | No | RSCU | ||
| Ter | UAA | 51 | 1.7385 | 51 | 1.7385 | 51 | 1.7385 | 51 | 1.7385 | 51 | 1.7385 |
| UAG | 21 | 0.7158 | 21 | 0.7158 | 21 | 0.7158 | 21 | 0.7158 | 21 | 0.7158 | |
| UGA | 16 | 0.5454 | 16 | 0.5454 | 16 | 0.5454 | 16 | 0.5454 | 16 | 0.5454 | |
| Ala | GCA | 409 | 1.144 | 409 | 1.144 | 409 | 1.1448 | 409 | 1.144 | 412 | 1.1524 |
| GCC | 231 | 0.646 | 232 | 0.6488 | 232 | 0.6496 | 232 | 0.6488 | 231 | 0.646 | |
| GCG | 160 | 0.4476 | 160 | 0.4476 | 160 | 0.448 | 160 | 0.4476 | 159 | 0.4448 | |
| GCU | 630 | 1.7624 | 629 | 1.7596 | 628 | 1.758 | 629 | 1.7596 | 628 | 1.7568 | |
| Cys | UGC | 91 | 0.6066 | 90 | 0.598 | 91 | 0.6066 | 91 | 0.6046 | 91 | 0.6108 |
| UGU | 209 | 1.3934 | 211 | 1.402 | 209 | 1.3934 | 210 | 1.3954 | 207 | 1.3892 | |
| Asp | GAC | 213 | 0.4068 | 213 | 0.4064 | 213 | 0.4064 | 212 | 0.4046 | 213 | 0.405 |
| GAU | 834 | 1.5932 | 835 | 1.5936 | 835 | 1.5936 | 836 | 1.5954 | 839 | 1.595 | |
| Glu | GAA | 986 | 1.473 | 986 | 1.4738 | 985 | 1.4724 | 986 | 1.475 | 988 | 1.4768 |
| GAG | 352 | 0.262 | 352 | 0.5262 | 353 | 0.5276 | 351 | 0.525 | 350 | 0.5232 | |
| Phe | UUC | 517 | 0.6888 | 515 | 0.688 | 517 | 0.688 | 517 | 0.6898 | 518 | 0.6944 |
| UUU | 984 | 1.3112 | 982 | 1.312 | 986 | 1.312 | 982 | 1.3102 | 974 | 1.3056 | |
| Gly | GGA | 704 | 1.582 | 704 | 1.582 | 704 | 1.582 | 704 | 1.582 | 702 | 1.58 |
| GGC | 193 | 0.4336 | 194 | 0.436 | 194 | 0.436 | 194 | 0.436 | 196 | 0.4412 | |
| GGG | 302 | 0.6788 | 301 | 0.6764 | 301 | 0.6764 | 301 | 0.6764 | 301 | 0.6776 | |
| GGU | 581 | 1.3056 | 581 | 1.3056 | 581 | 1.3056 | 581 | 1.3056 | 578 | 1.3012 | |
| His | CAC | 155 | 0.5008 | 155 | 0.5024 | 155 | 0.5024 | 155 | 0.5024 | 154 | 0.5 |
| CAU | 464 | 1.4992 | 462 | 1.4976 | 462 | 1.4976 | 462 | 1.4976 | 462 | 1.5 | |
| Ile | AUA | 720 | 0.9711 | 723 | 0.9753 | 721 | 0.9726 | 722 | 0.9738 | 720 | 0.9717 |
| AUC | 428 | 0.5772 | 425 | 0.5733 | 425 | 0.5733 | 424 | 0.5718 | 425 | 0.5736 | |
| AUU | 1076 | 1.4514 | 1076 | 1.4514 | 1078 | 1.4541 | 1078 | 1.4541 | 1078 | 1.4547 | |
| Lys | AAA | 1042 | 1.4738 | 1042 | 1.4738 | 1043 | 1.4742 | 1042 | 1.4728 | 1031 | 1.4708 |
| AAG | 372 | 0.5262 | 372 | 0.5262 | 372 | 0.5258 | 373 | 0.5272 | 371 | 0.5292 | |
| Leu | CUA | 387 | 0.8184 | 388 | 0.8196 | 387 | 0.819 | 388 | 0.8202 | 383 | 0.813 |
| CUC | 188 | 0.3978 | 187 | 0.3948 | 187 | 0.396 | 187 | 0.3954 | 187 | 0.3972 | |
| CUG | 192 | 0.4062 | 192 | 0.4056 | 192 | 0.4062 | 192 | 0.4056 | 191 | 0.4056 | |
| CUU | 616 | 1.3026 | 617 | 1.3038 | 617 | 1.3056 | 617 | 1.3038 | 612 | 1.2996 | |
| UUA | 859 | 1.8168 | 861 | 1.8192 | 859 | 1.818 | 861 | 1.8198 | 856 | 1.8174 | |
| UUG | 595 | 1.2582 | 595 | 1.257 | 593 | 1.2552 | 594 | 1.2552 | 597 | 1.2678 | |
| Met | AUG | 639 | 1.9938 | 639 | 1.9938 | 641 | 1.9938 | 639 | 1.9938 | 638 | 1.9968 |
| GUG | 2 | 0.0062 | 2 | 0.0062 | 2 | 0.0062 | 2 | 0.0062 | 1 | 0.0032 | |
| Asn | AAC | 285 | 0.4388 | 287 | 0.4416 | 287 | 0.4412 | 286 | 0.44 | 285 | 0.4378 |
| AAU | 1014 | 1.5612 | 1013 | 1.5584 | 1014 | 1.5588 | 1014 | 1.56 | 1017 | 1.5622 | |
| Pro | CCA | 328 | 1.206 | 329 | 1.2096 | 328 | 1.206 | 328 | 1.206 | 325 | 1.198 |
| CCC | 203 | 0.7464 | 203 | 0.7464 | 203 | 0.7464 | 203 | 0.7464 | 202 | 0.7448 | |
| CCG | 146 | 0.5368 | 145 | 0.5332 | 146 | 0.5368 | 146 | 0.5368 | 149 | 0.5492 | |
| CCU | 411 | 1.5112 | 411 | 1.5112 | 411 | 1.5112 | 411 | 1.5112 | 409 | 1.508 | |
| Gln | CAA | 712 | 1.5116 | 714 | 1.5144 | 713 | 1.5122 | 713 | 1.5122 | 714 | 1.5144 |
| CAG | 230 | 0.4884 | 229 | 0.4856 | 230 | 0.4878 | 230 | 0.4878 | 229 | 0.4856 | |
| Arg | AGA | 504 | 1.89 | 503 | 1.8888 | 504 | 1.8912 | 503 | 1.8888 | 501 | 1.881 |
| AGG | 177 | 0.6636 | 177 | 0.6648 | 177 | 0.6642 | 177 | 0.6648 | 177 | 0.6648 | |
| CGA | 354 | 1.3278 | 353 | 1.3254 | 353 | 1.3248 | 352 | 1.3218 | 353 | 1.3254 | |
| CGC | 108 | 0.405 | 108 | 0.4056 | 108 | 0.405 | 109 | 0.4092 | 108 | 0.4056 | |
| CGG | 116 | 0.435 | 116 | 0.4356 | 116 | 0.435 | 116 | 0.4356 | 116 | 0.4356 | |
| CGU | 341 | 1.2786 | 341 | 1.2804 | 341 | 1.2798 | 341 | 1.2804 | 343 | 1.2876 | |
| Ser | AGC | 117 | 0.3468 | 118 | 0.3504 | 118 | 0.3498 | 118 | 0.3498 | 120 | 0.3576 |
| AGU | 413 | 1.2252 | 412 | 1.2228 | 412 | 1.221 | 412 | 1.2222 | 410 | 1.2222 | |
| UCA | 424 | 1.2576 | 424 | 1.2582 | 425 | 1.2594 | 425 | 1.2606 | 425 | 1.2666 | |
| UCC | 308 | 0.9132 | 307 | 0.9108 | 308 | 0.9126 | 307 | 0.9108 | 303 | 0.903 | |
| UCG | 164 | 0.4866 | 165 | 0.4896 | 165 | 0.489 | 165 | 0.4896 | 164 | 0.489 | |
| UCU | 597 | 1.7706 | 596 | 1.7688 | 597 | 1.7688 | 596 | 1.7676 | 591 | 1.7616 | |
| Thr | ACA | 413 | 1.2524 | 413 | 1.2524 | 413 | 1.2524 | 413 | 1.2524 | 412 | 1.254 |
| ACC | 247 | 0.7492 | 247 | 0.7492 | 246 | 0.746 | 247 | 0.7492 | 243 | 0.7396 | |
| ACG | 133 | 0.4032 | 133 | 0.4032 | 133 | 0.4032 | 133 | 0.4032 | 132 | 0.402 | |
| ACU | 526 | 1.5952 | 526 | 1.5952 | 527 | 1.598 | 526 | 1.5952 | 527 | 1.6044 | |
| Val | GUA | 525 | 1.4968 | 525 | 1.4968 | 525 | 1.4988 | 525 | 1.4968 | 523 | 1.4976 |
| GUC | 175 | 0.4988 | 175 | 0.4988 | 175 | 0.4996 | 175 | 0.4988 | 175 | 0.5012 | |
| GUG | 195 | 0.556 | 195 | 0.556 | 193 | 0.5512 | 195 | 0.556 | 194 | 0.5556 | |
| GUU | 508 | 1.4484 | 508 | 1.4484 | 508 | 1.4504 | 508 | 1.4484 | 505 | 1.446 | |
| Trp | UGG | 455 | 1 | 455 | 1 | 454 | 1 | 454 | 1 | 451 | 1 |
| Tyr | UAC | 181 | 0.3668 | 181 | 0.366 | 181 | 0.366 | 182 | 0.368 | 181 | 0.3676 |
| UAU | 806 | 1.6332 | 808 | 1.634 | 808 | 1.634 | 807 | 1.632 | 804 | 1.6324 | |
Fig. 3.
Codon RSCU clustering in chloroplast genome of five Cremanthodium species
The protein coding sequences of C. arnicoides and C. brunneopilosum were 79,005 bp, and 88 protein coding genes encoded 26,335 codons. The protein coding sequence of the C. ellisii was 79,017 bp, and 88 protein coding genes encoded 26,339 codons. The protein coding sequence of the C. nervosum was 79,008 bp, and 88 protein coding genes encoded 26,336 codons. The protein coding sequence of the C. rhodocephalum was 78,807 bp, and 88 protein coding genes encoded 26,269 codons. There were three stop codons (UAA, UAG and UGA) in the protein coding sequences of five chloroplast genomes (Table 3). UAA appeared 51 times, with more than 50% frequency; UAG appeared 21 times, and UGA appeared 16 times.
In the coding protein sequences of the A-B-E-N-R chloroplast genomes, the most frequent amino acid encoded by codons was leucine (Leu), which appeared 2837, 2840, 2835, 2839 and 2826 times, respectively, and the most frequents codon was AUU of isoleucine (Ile), which appeared 1076 times, 1076 times, 1078 times, 1078 times and 1078 times, respectively. Only tryptophan (Trp) has one codon, and other amino acids have 2–6 synonymous codons. RSCU > 1 indicates codon preference, RSCU < 1 indicates low usage rate, and RSCU = 1 indicates no codon preference.
Detection of chloroplast repeat sequences and SSRs
The scattered repeat sequences were analysed by Repeater software. In this study, we identified 38, 39, 36, 38, and 39 interspersed repeat sequences (LTRs) and 62, 63, 62, 60 and 61 simple repeated sequences (SSRs) in the A-B-E-N-R chloroplast genomes (Table 4), respectively. There were two main types of LTRs: forwards LTRs, accounting for 43.6–50.0% of all repeats, and palindromic LTRs, accounting for 50.0–56.4% of all repeats.
Table 4.
Distribution of chloroplast genomes LTR in chloroplast genome of five Cremanthodium species
| Type | C. arnicoides | C. brunneopilosum | C. ellisii | C. nervosum | C. rhodocephalum |
|---|---|---|---|---|---|
| Forward | 19 | 18 | 17 | 18 | 17 |
| Palindromic | 19 | 21 | 19 | 20 | 22 |
| Reverse | 0 | 0 | 0 | 0 | 0 |
| Complement | 0 | 0 | 0 | 0 | 0 |
All single nucleotide repeats were A/T homopolymers. Single nucleotide repeats accounted for 62.3–65.0% of the SSR, and 10–14 bp repeats accounted for 80.0–89.5% of the single nucleotide repeats. There were 6–7 dinucleotide repeats, accounting for 9.5–11.3% of the SSRs (Table 5). All dinucleotide repeats were AT/TA. The number of trinucleotides in all repeats was 5. The types of trinucleotides were ATG, ATT, TTA, and TTC. The number of tetranucleotides in all repeats was 11. The types of trinucleotides were ATAA, AAAT, ACTA, TATT, TTTC, AATT, ATAG, AATA, AAAT, and AATC. The only hexanucleotide was ACTCCT, and it was detected in the chloroplast genome of C. rhodocephalum.
Table 5.
Classification of cp SSR in five Cremanthodium species
| type | C. arnicoides | C. brunneopilosum | C. ellisii | C. nervosum | C. rhodocephalum |
|---|---|---|---|---|---|
| Mono- | 40 | 41 | 39 | 38 | 38 |
| Di- | 6 | 6 | 7 | 6 | 6 |
| Tri- | 5 | 5 | 5 | 5 | 7 |
| Tetra- | 11 | 11 | 11 | 11 | 11 |
| Penta- | 0 | 0 | 0 | 0 | 0 |
| Hexa- | 0 | 0 | 0 | 0 | 1 |
IR expansion and contraction
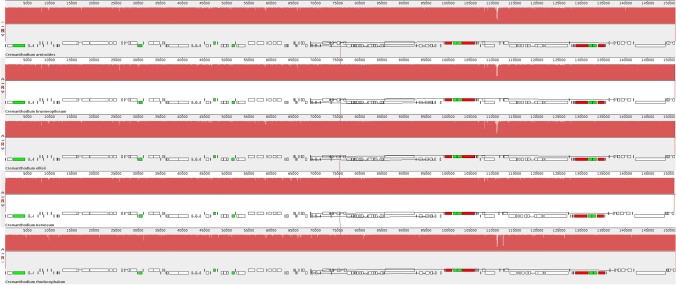
There was no obvious expansion or contraction in the LSC, SSC or IR regions among the five species, indicating that the chloroplast gene structure of the genus was highly conserved (Fig. 4).
Fig. 4.
Chloroplast boundary characteristics of five Cremanthodium species
Collinearity analysis
The chloroplast genomes of five species of the Cremanthodium genus were compared. The results showed that the chloroplast genomes of the five species were collinear, and there was no gene rearrangement (Fig. 5). There still are some differences in their chloroplast genomes which the code gene near the site of 110,000. It can be used as a mutation hotspot, and this area was in the middle between ndhF and rpl32.
Fig. 5.
Chloroplast boundary characteristics of five Cremanthodium species. Colinear analysis of chloroplast genomes of five Cremanthodium species. Color bands represent genes, and different colors represent different blocks. Blocks with the same color between different genes represent homologous regions. Two rows of small blocks below the color band represent genes. The top one is on the positive chain, and the below one is on the complementary chain. Of which, the white block represents the coding genes, a thin line inside the white blocks denotes intron. Green and red blocks represent tRNA and rRNA, respectively
Phylogenetic tree
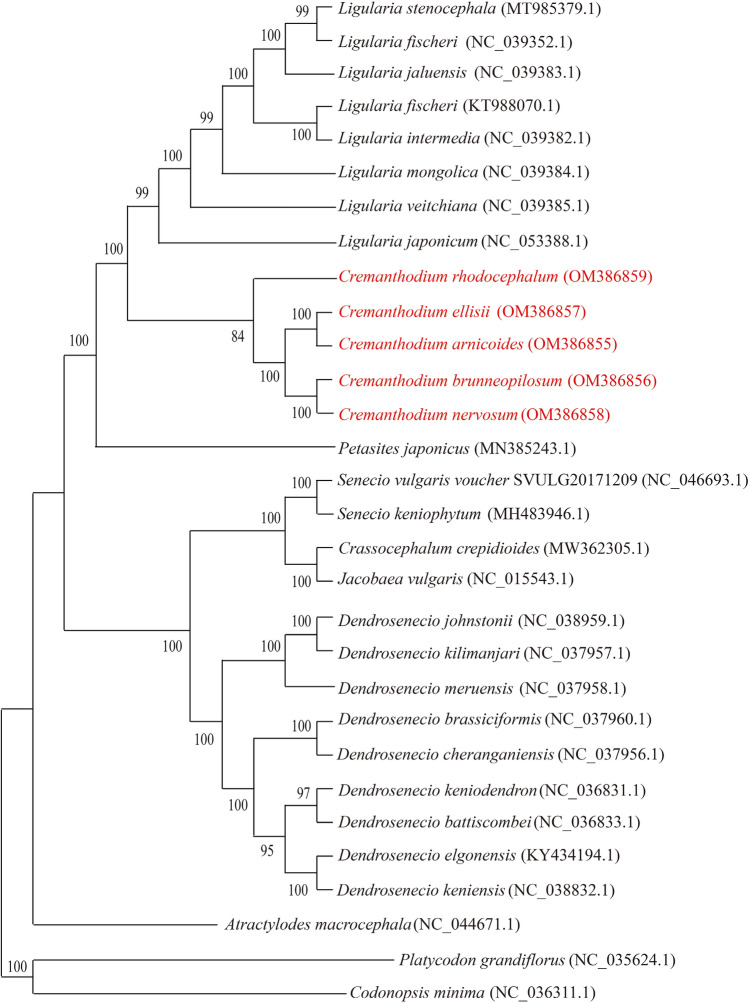
In addition to the five species of Cremanthodium (A-B-E-N-R) in this study, 25 published chloroplast genomes of the Compositae family were selected to construct phylogenetic trees using the maximum likelihood (ML) method to explore phylogenetic relationships. From the ML tree, C. arnicoides and C. ellisii formed a sister group, and C. brunneopilosum and C. nervosum formed a sister group. Five species of Cremanthodium (A-B-E-N-R) formed a monophyletic group (Fig. 6). C. rhodocephalum was the first differentiated species among the five species of Cremanthodium (A-B-E-N-R). Based on the ML tree, there was a closer genetic relationship between the five species of Cremanthodium (A-B-E-N-R), and their next closest genetic relationships were with Ligularia stenocephala, L. fischeri, L. jaluensis, L. intermedia, L. mongolica, L. veitchiana, Farfugium japonicum, and Petasites japonicus.
Fig. 6.
Phylogenetic tree for 30 species in Asteraceae family using maximum likelihood (ML), based on alignments of complete chloroplast genomes
Discussion
Senecioneae Cass. belongs to subfamily Asteroideae (Asteraceae), which contains about 3500 species and 152 genera, and is widely distributed around the world (Nordenstam 2007; Nordenstam et al. 2009). However, due to the possible rapid diversification in the early evolution of Asteraceae, the specific system location of Senecioneae Cass. has not been determined (Kim et al.2005; Panero and Funk 2008). Different genus of Senecioneae Cass. formed a large complex (Ligularia-Cremanthodium-Parasenecio complex; L-C-P complex) except for Tussilago L. and Petasites Mill.. Ligularia Cass., Cremanthodium Benth., Parasenecio W. W. Sm. and J. Small, and Sinosenecio B. Nord. which are defined according to morphological characters instead of monophyletic groups, and the boundaries between genera need to be revised (Liu et al. 2006). The position and genetic relationship of Ligularia Cass., Cremanthodium Benth., and Parasenecio W. W. Sm. in the system are not clear, which needs more experimental verification at the molecular level. In this study, the whole chloroplast genomes of five Cremanthodium species were sequenced and annotated for the first time, which enriched the chloroplast genome data of L-C-P complex and provided a basis for their intergeneric boundaries. According to the expeimental results, the chloroplast genomes length of five Cremanthodium species are similar to that of other species of Asteraceae. Due to the narrow distribution of Cremanthodium plants, the chloroplast genome showed obvious conservation. The assembled chloroplast genome sequences of C. arnicoides, C. brunneopilosum, C. ellisii, C. nervosum, and C. rhodocephalum, with lengths of 150,985–151,284 bp (Fig. 2), were similar to the most sequenced chloroplast genomes: Chrysanthemum L. (151,010–151,098 bp) (Tyagi et al. 2020a, b), Senecio L. (150,000–151,000 bp) (Gichira et al. 2019), Ligularia Cass. (151,118–151,253 bp) (Chen et al. 2018; Lee et al. 2016), Farfugium Lindl. (151,222 bp) (Gu et al. 2016), Taraxacum F. H. Wigg. (151,307 and 151,451 bp) (Kim et al. 2016), Saussurea involucrate (152,490 bp) (Wang et al. 2020), Aster flaccidus (151,329 bp) (Tyagi et al. 2020a, b), and Carpesium abrotanoides L. (151,394 bp) (He et al. 2022). The chloroplast genomes encoded equal number of 133 unique genes, of which 88 were protein-coding genes, 37 were transfer ribonucleic acid genes, and eight were ribosomal ribonucleic acid genes (Table 1), and were highly similar in overall size, genome structure, gene content, and order. The GC content is also similar, indicating that there is little variation among the species in the genus.
Codon usage analysis showed that there were 31 codons with RSCU > 1 in the five species, of which 16 ended in U, 13 in A, and 2 in G, which indicated that more codons ended with U or A (Table 3, Fig. 3). In Cremanthodium plants, the identification of microsatellite loci in the intergenic spacer region and introns can show a potential polymorphism since coding regions were conserved across other genomes. There is a predominance of mononucleotides, followed by tetranucleotides. The number of SSRs identified for these five Cremanthodium chloroplast genomes ranged from 60 to 63.
Comparing the junction of four parts of chloroplast genome of five Cremanthodium species, it was found that the composition and distribution of genes at the boundary were highly similar (Fig. 4). The gene rps19 is located at the junction of the LSC region and the IRb region, and the sequence length distribution in the two regions is stable. The gene length of rps19 entering the IRb region is 60 bp, and the length retained in the LSC region is 219 bp. The gene ycf1 is located at the junction of IRb region and SSC region and between SSC region and IRa region, while ycf1 gene is highly conserved at the boundary.
The collinearity of chloroplast genes (Fig. 5) showed that the sequences of protein coding genes, tRNA and rRNA genes were similar, and the gene structure was conservative. There is an obvious difference in 112 kb between Cremanthodium rhodocephalum and other species, and it is speculated that there is a relatively distant relationship between this and other species, which is also consistent with the relationship on ML tree.
Based on the chloroplast genome sequences of five Cremanthodium species, 23 species of Asteraceae, and 2 species of Campanulaceae were compared and analyzed, and the taxonomic position and evolutionary relationship of the plants sequenced in this study were evaluated. According to ML tree (Fig. 6), as part of the L-C-P complex, five Cremanthodium species are grouped into one group, which C. arnicoides is resolved as sister to C. ellisii, C. brunneopilosum is resolved as sister to C. nervosum, and C. rhodocephalum is resolved as a separate group. This is different from the results of classical morphological classification, and the taxonomic position of C. nervosum is determined by chloroplast genome data. According to the characteristics of leaf veins, C. nervosum, C. arnicoides and C. ellisii were divided into a sister group (pinnate vein group), but the whole chloroplast genome sequence showed that C. nervosum did not belong to this sister group. Five Cremanthodium species and eight Ligularia species constituted a morphologically distinct with high support rate, the existing chloroplast genomes data support the location of genera, and show that the evolutionary relationship between the two genera is relatively close, which may be derived from the same ancestor (Liu et al. 2006).
Conclusion
The complete chloroplast genome sequences of five Cremanthodium Benth. and their phylogenetic relationship were reported to provide evidence for species identification of the Cremanthodium genus. The structure and composition of the chloroplast genomes are highly similar and their overall sequence, gene content and gene order were conserved. Phylogenetic analyses using other Compositae species and other species supported the taxonomic status of the Cremanthodium within the tribe. This study provides invaluable data for species identification, allowing for future studies on phylogenetic evolution, as well as for further biological discoveries.
Acknowledgements
We thank Prof. Xiang Liu and Prof. Huarong Zhou for their assistance during leaves collection.
Author contributions
WZ performed the experiments, data processing and manuscript draft preparation, XD contributed to analyzing the data, LC and ZM performed sample collection, XW and GZ designed the project and approved the final manuscript version.
Funding
This work was supported by National Key Research and Development Program of China (No. 2019YFC1712300) and Jiangxi University of Chinese Medicine Science and Technology Innovation Team Development Program (CXTD22002).
Data Availability
The data that support the findings of this study have been deposited in the NCBI database (GenBank accession: OM386855, OM386856, OM386857, OM386858, OM386859) (http://www.ncbi.nlm.nih.gov/).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaolang Du, Email: 20131059@jxutcm.edu.cn.
Guoyue Zhong, Email: zgy1037@163.com.
References
- Chen H, Zhu Y, Shen XM, Jia ZJ. Four new sesquiterpene polyol esters from Cremanthodium ellisii. J Nat Prod. 1996;59:1117–1120. doi: 10.1021/NP9601768. [DOI] [Google Scholar]
- Chen XL, Zhou JG, Cui YX, Wang Y, Duan BZ, Yao H. Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol. 2018;9:1–11. doi: 10.3389/fphar.2018.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora Reipublicae Popularis Sinicae Editorial Board of the Chinese Academy of Sciences (1989) Flora reipublicae popularis sinicae, vol 7. Science Press, Beijing, p 115
- Gichira AW, Avoga S, Li Z, Huang GW, Wang QF, Chen JM. Comparative genomics of 11 complete chloroplast genomes of Senecioneae (Asteraceae) species DNA barcodes and phylogenetics. Bot Stud. 2019;60:1–17. doi: 10.1186/s40529-019-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Ma Q, Lu Y. Characterization of the complete chloroplast genome of Farfugium japonicum (Asteraceae) Mitochondrial DNA B. 2016;6:678–679. doi: 10.1080/23802359.2021.1881928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XY, Dong SJ, Gao CS. The complete chloroplast genome of Carpesium abrotanoides L. (Asteraceae): structural organization, comparative analysis, mutational hotspots, and phylogenetic implications within the tribe Inuleae. Biologia. 2022;77:1861–1876. doi: 10.1007/s11756-022-01038-2. [DOI] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241–272. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Choi KS, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae) Mol Biol Evol. 2005;22:1783–1792. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- Kim JK, Park JY, Lee YS, Woo SM, Park HS, Lee TJ, Sung SH, Yang TJ. The complete chloroplast genomes of two Taraxacum species, T. platycarpum Dahlst. and T. mongolicum Hand. –Mazz. (Asteraceae) Mitochondrial DNA B. 2016;1:412–413. doi: 10.1080/23802359.2016.1176881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee H, Lee SC, Sung SH, Kang JH, Lee TJ, Yang T. The complete chloroplast genome sequence of Ligularia fischeri (Ledeb.) Turcz. (Asteraceae) Mitochondrial DNA B. 2016;1:4–5. doi: 10.1080/23802359.2015.1137793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang LJ, Qiu GF, Yu JQ, Liang SC, Hu XM. Apoptosis of Hela cells induced by extract from Cremanthodium humile. Food Chem Toxicol. 2007;45:2040–2046. doi: 10.1016/j.fct.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Liu JQ, Wang YJ, Wang AL, Hideaki O, Abbott RJ. Radiation and diversification within the Ligularia–Cremanthodium–Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 2006;38:31–49. doi: 10.1016/j.ympev.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Nordenstam B. Tribe senecioneae cass. In: Kadereit JW, Jeffrey C, editors. The families and genera of vascular plants, flowering plants: eudicots, Asterales. Berlin: Springer; 2007. pp. 208–242. [Google Scholar]
- Nordenstam B, Pelser PB, Kadereit JW, et al. Senecioneae. In: Funk VA, Susanna A, Stussey TF, et al., editors. Systematics, Evolution, and biogeography of compositae. Vienna: International Association for Plant Taxonomy; 2009. pp. 503–525. [Google Scholar]
- Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phyl Evol. 2008;47:757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Saito Y, Ichihara M, Okamoto Y, Gong X, Kuroda C, Tori M. Four new eremophilane-type alcohols from Cremanthodium helianthus collected in China. Nat Prod Commun. 2012;7:423–426. doi: 10.1177/1934578x1200700402. [DOI] [PubMed] [Google Scholar]
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;7:65–73. doi: 10.1093/nar/gkz345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su BN, Zhu QX, Jia ZJ. A new nor-neolignan from Cramanthodium ellisii. Chin Chem Lett. 1999;10:129–130. [Google Scholar]
- Su BN, Zhu QX, Jia ZJ. Nor-lignan and sesquiterpenes from Cremanthodium ellisii. Phytochemistry. 2020;53:1103–1108. doi: 10.1016/S0031-9422(99)00584-1. [DOI] [PubMed] [Google Scholar]
- Tori M, Saito Y, Takiguchi K, Gong X, Kuroda C. Three new bisabolanetype sesquiterpenoids from Cremanthodium Rhodocephalum (Asteraceae) Heterocycles. 2012;86:497–503. doi: 10.3987/COM-12-S(N)47. [DOI] [Google Scholar]
- Tu YQ, Yang RP, Shou QY. Study on volatile chemical constituents of Cremanthodium pleurocaulis. China J Chin Mater Med. 2006;6:522–524. [Google Scholar]
- Tyagi S, Jung JA, Kim JS, Won SY. A comparative analysis of the complete chloroplast genomes of three Chrysanthemum boreale strains. PeerJ. 2020;8:e9448. doi: 10.7717/peerj.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Jung JA, Kim JS, Won SY. Comparative analysis of the complete chloroplast genome of mainland Aster spathulifolius and Other aster species. Plants. 2020;9:568. doi: 10.3390/plants9050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AX, Zhong Q, Jia ZJ. Phenylpropanosids, lignans and other constituents from Cremanthodium ellisii. Pharmazie. 2004;59:889–892. [PubMed] [Google Scholar]
- Wang R, Liu JF, Liu SY, Guan SY, Jiao P. Characterization of the complete chloroplast genome of Saussurea involuerata (Compositae), an endangered species endemic to China. Mitochondrial DNA B. 2020;5:511–512. doi: 10.1080/23802359.2019.1705195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QX, Zhu Y, Jia ZJ (2003) Study on volatile chemical constituents of Cremanthodium Discoideum. Lanzhou Daxue Xuebao, Ziran Kexueban 39:107–108. 10.13885/j.issn.0455-2059.2003.01.022
- Yang L, Chen H, Jia ZJ (1995) Lignan and a Coumarin from Cremanthodium ellisii Kitam. Indian J Chem B: Org Chem Incl Med Chem 34:975–977
- Zhu Y, Zhu QX, Jia ZJ. Epoxide sesquiterpenes and steroids from Cremanthodium discoideum. Aust J Chem. 2000;53:831–834. doi: 10.1071/ch00100. [DOI] [Google Scholar]
- Zhu Y, Liang QW, Jia ZJ (2001) Sesquiterpenes and phenolic compounds from Cremanthodium Discoideum. Lanzhou Daxue Xuebao, Ziran Kexueban 37:68–75. 10.13885/j.issn.0455-2059.2001.04.015
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study have been deposited in the NCBI database (GenBank accession: OM386855, OM386856, OM386857, OM386858, OM386859) (http://www.ncbi.nlm.nih.gov/).