Abstract
Introduction
The overall prevalence of Klebsiella spp., a group of important zoonotic pathogens, in the global dairy herds and the risk of cross-species transmission between humans and dairy cows remain to be clarified. This systematic review aimed to determine the prevalence of Klebsiella spp. in milk samples from dairy cows with mastitis worldwide and to assess the factors influencing the prevalence of these strains.
Methods
Qualified studies published from 2007 to 2021 were retrieved from ScienceDirect, Web of Science, PubMed, WanFang Database, China National Knowledge Infrastructure (CNKI), and VIP Chinese Journal Database. Calculations of prevalence and their 95% confidence intervals (CIs) were performed for all the studies using the Freeman-Tukey double arcsine transformation (PFT).
Results
A total of 79,852 milk samples from 55 manuscripts were examined in this meta-analysis, and 2,478 samples were found to be positive for Klebsiella spp. The pooled prevalence estimates worldwide were 7.95% (95% CI: 6.07%–10.06%), with significant heterogeneity (I2 = 98.8%, p = 0). The sampling period of 2013–2020 had a higher (p < 0.05) Klebsiella-positive proportion of milk samples (12.16%, 95% CI: 8.08%–16.90%) than that of 2007–2012 (3.85%, 95% CI: 2.67%–5.21%), indicating that bovine mastitis caused by Klebsiella may become increasingly prevalent. The risk factors for the high prevalence of Klebsiella in milk samples mainly included: economic development level (developing countries; 11.76%, 95% CI: 8.25%–15.77%), mastitis type (CM; 11.99%, 95% CI: 8.62%–15.79%), and population density (>500 per sq km; 10.28%, 95% CI: 2.73%–21.58%). Additionally, a bivariate meta-regression analysis revealed that the multidrug-resistance (MDR) rate of the epidemic strains was also closely related to economic development level (R2 = 78.87%) and population density (R2 = 87.51%).
Discussion
Due to the potential risk of cross-species transmission between humans and cows, the prevalence of mastitis milk-derived Klebsiella and its high MDR rate need to be monitored, especially in developing countries with high population densities.
Keywords: meta-analysis, bovine mastitis, Klebsiella, multidrug-resistance (MDR), prevalence
Introduction
As one of the common diseases in dairy farming, bovine mastitis can lead to decreased milk production, poor milk quality, reproductive barriers and even culling of dairy cows, causing great economic losses to the global dairy industry (1). Bovine mastitis is classified as clinical mastitis (CM) and subclinical mastitis (SCM) according to symptoms and milk characteristics. In general, CM is defined as a condition showing swelling of the infected quarter and in some cases systemic signs such as fever and anorexia. However, it is difficult to detect SCM visually unless through diagnostic techniques such as the California Mastitis Test (CMT), Wisconsin Mastitis Test (WMT) or Somatic Cell Count (SCC) (2). Due to the complexity of the bacteria that cause intramammary infection (IMI) in dairy cows, the control of bacterial infection is mainly performed with antibiotic management and assisted by the treatment of some bacteriophages and natural compounds at present. Moreover, some vaccines have been developed to prevent bovine mastitis (1).
As a common zoonotic pathogen that is widely prevalent in hospitals and communities, Klebsiella pneumoniae mainly causes pneumonia, liver abscess, urinary diseases, toxemia, septicaemia and other infection symptoms (3). Due to the long-term irrational use of antibiotics in the medical field, a wide variety of carbapenemase [such as K. pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM) and oxacillinase (OXA)]-producing and extended spectrum β-lactamase (ESBL, such as CTX-M, SHV and TEM)-producing multidrug-resistant (MDR) Klebsiella have emerged in an endless number of cases (4). Klebsiella spp., as vehicles of multiple drug-resistance genes, are monitored by the World Health Organization (WHO) (5). Under the pressure of antibiotic selection, drug-resistance genes are transferred to other strains through mobile genetic elements (MGEs) such as plasmids, transposons, and insertion elements, thus increasing the challenge of mitigating bacterial infections (6). Klebsiella spp. is also an important cause of bovine mastitis, with K. pneumoniae and K. oxytoca as the most prevalent species (7, 8). In dairy cattle, Klebsiella spp. are transmitted by contact with teats, mainly through manure, bedding and other farm equipment, and invade mammary epithelial cells and persist for a long time (9). Moreover, these strains not only seriously affect the milk quality and performance of adult cows, but also pose a fatal threat to the survival of newborn calves (10). To the best of our knowledge, bovine mastitis caused by K. pneumoniae was first publicly reported in 1954 (11), but in the following decades, the prevalence of Klebsiella in milk samples appeared to be nonsignificant compared to that of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli. However, over the past decade or so, the number of cases of Klebsiella spp. detected in milk samples has risen dramatically worldwide. In some countries, these bacteria are second only to E. coli in the incidence of gram-negative bacteria in dairy cow udders (8).
The global spread and distribution of carbapenemase-producing and hypervirulent Klebsiella spp. from human sources in the past decade have been revealed (12, 13). During the same period, reports of Klebsiella spp. being detected in raw milk samples from dairy farming regions (especially Holstein-Friesian dairy farms) around the world have increased. Although K. pneumoniae isolated from companion animals shows zoonotic potential (14, 15), there is currently no assessment of risk factors for cross-species transmission of these strains in humans and dairy herds due to the lack of systematic analysis of the global distribution of milk-derived Klebsiella spp. To fully evaluate the global prevalence of Klebsiella spp. in milk samples from dairy cows with mastitis and to assess risk factors for influencing the prevalence of these strains, this systematic review and meta-analysis was conducted on the prevalence of Klebsiella spp. in milk samples from major Holstein dairy farming regions using articles published from 2007 to 2021 based on subgroups (sampling years, detection methods, geographic information, level of economic development, mastitis type, population density, Klebsiella species, and MDR rate of Klebsiella isolates).
Materials and methods
Study design and search strategies
We conducted a systematic literature search for studies that examined the prevalence of Klebsiella spp. in milk samples from dairy cows worldwide following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (16). On January 12, 2022, six databases were searched: ScienceDirect, Web of Science (all databases), PubMed, WanFang Database, China National Knowledge Infrastructure (CNKI), and VIP Chinese Journal Database. We found that the search items “Klebsiella” and “Mastitis” produced the most qualified studies by presearch.
The keywords “Klebsiella” and “Mastitis” were used in the process of screening ScienceDirect, and “Research articles” was chosen as the article type. Web of Science was searched by combining two queries #1 “TS = (Mastitis)” and #2 “TS = (Klebsiella)” with “AND”. In PubMed, we used the MeSH terms “Klebsiella” and “Mastitis” in an advanced search to generate the search formula (Klebsiella) AND (Mastitis). In the WanFang database, CNKI, and the VIP Chinese Journal database, the topics were defined as “Mastitis” AND “Klebsiella” in Chinese in the advanced search. Considering that some qualified studies may have not been included in the electronic database we built, all the references cited in relevant studies were carefully checked. Subsequently, the supplementary search was conducted using Google Scholar, which allowed those missing articles with available data to be included in this meta-analysis. In addition to English and Chinese studies, a Spanish study and a Korean study (both with English abstracts) were included through a supplementary search. Records identified in the search process were uploaded into EndNote (version X 9.3.3).
Selection criteria
We preliminarily screened articles based on duplication, and those with duplicate titles and abstracts were excluded with the help of Endnote. After title/abstract screening, full texts were screened afterwards. Qualified studies were selected according to the following criterion: articles which were published between 2007 and 2021 that examined the prevalence of Klebsiella spp. in milk samples. Studies were excluded if they met the following criteria: (1) the full text was unavailable, (2) sample totals or prevalence were not provided, (3) the sampling time was unclear, and (4) the samples collected were not milk.
Two authors (JS and WX) performed the selection process according to the eligibility criteria.
Data extraction
Standardized forms generated by Microsoft Excel 2019 (version 2203) were used to extract the following information from the eligible studies: first author, publication year, sampling years, sample size, number of Klebsiella-positive milk samples, detection methods, geographic information, mastitis type, economic development level and population density of the country where the study was conducted, and the MDR rate of Klebsiella spp. isolated from milk samples. The milk samples included were all individual quarter samples.
In 2013, MDR K. pneumoniae was listed by the Centers for Disease Control and Prevention (CDC) as an urgent threat to public health, along with other carbapenem-resistant Enterobacteriaceae (CRE) (17). Therefore, we chose 2013 as a time point to divide the timeline into two parts to conduct a systematic review and meta-analysis.
The economic development levels of the countries were established according to World Economic Situation and Prospects 2022—UN (18). A list of countries by population density was derived from World Population Prospects 2022—UN (19).
Quality assessment
The quality of each eligible article was estimated by scoring (20, 21). In brief, the following items were given 1 point when they were present in a study: random sampling; explicit detection method; detailed sampling procedures; sampling year; and risk factors ≥4. The articles were scored with a range of 0 to 5, and they were divided into three intervals: 0–1 points, 2–3 points, and 4–5 points. The scoring criteria were only applicable to this meta-analysis and did not represent the research level of the included studies.
Statistical analysis
All the data were analyzed by the package “meta” (version 4.0.0) in R software version 4.0.0 (R Foundation for Statistical Computing, Vienna, Austria). The prevalence of Klebsiella-positive milk samples was estimated as the number of milk samples in which Klebsiella spp. was detected divided by the number of total tested milk samples. Calculations of prevalence and their 95% confidence intervals (CIs) were performed for all the studies. The Freeman-Tukey double arcsine transformation (PFT) showed better variance stabilization performance by normalizing the data with different meta packages previously. Therefore, to make the distribution in accordance with Gaussian distribution, we performed rate conversion with PFT. The formulates for PFT were as follows (22):
Note: t = transformed prevalence; n = sample size; r = positive number; se = standard error.
The transformed summary proportion and its confidence interval were reconverted for better readability. The I2, Cochran's Q, and χ2 tests were used to quantify the variation (23, 24). Because of the high degree of heterogeneity, we conducted the meta-analysis using a random effects model. Publication bias (indicated by symmetry of the funnel plot) was evaluated by performing a funnel plot, trim and fill method, and Egger's test. A p < 0.05 was considered to be the statistical significance threshold (25, 26). Funnel plots were generated to further assess every subgroup. To determine the stability of the study, we also conducted sensitivity analysis, which evaluated the effect of each study on the overall results by sequentially excluding single studies. The R code for this meta-analysis is shown in Supplementary Table 1.
Subgroup analysis and univariate meta-regression analysis were used to reveal factors that may contribute to heterogeneity among studies. The independent factors that we selected were sampling years (comparison of 2013 or later with before 2013), detection methods (comparison of automatic analysis systems with other methods), continents (comparison of Oceania with other continents), longitude (comparison of 0–20°E with other intervals), latitude (comparison of 20–30°N with other intervals), economic development level (comparison of developing countries with developed countries), mastitis type (comparison of CM with SCM), population density (comparison of “ < 50 per sq km” with other groups), and Klebsiella species (comparison of K. pneumoniae with K. oxytoca). In addition, we performed a bivariate meta-regression analysis to analyze the correlation of the MDR rate of Klebsiella spp. isolated from milk samples with population density and economic development level, and R2 was used to explain the heterogeneity of each term by indicating the proportion (27).
Results
Search results and eligible studies
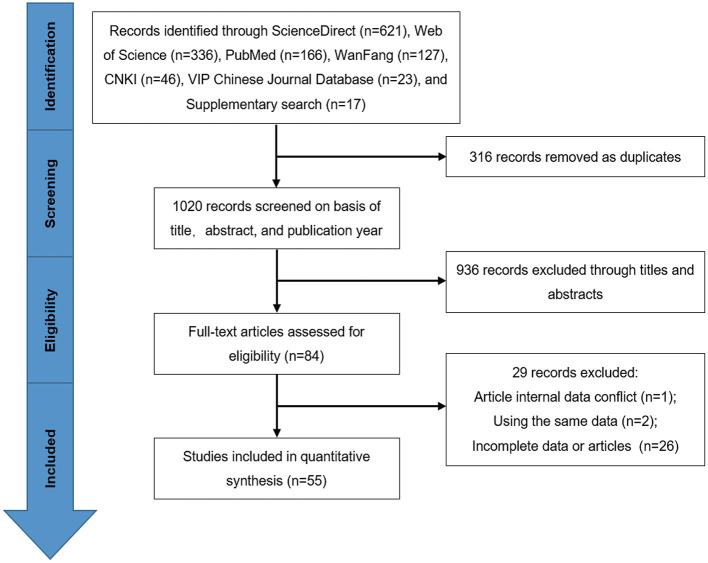
A total of 1,319 records were identified by searching the databases. First, 316 duplicate studies were removed. According to the previously established inclusion criterion, 936 studies were excluded because failed to meet publication years (2007–2021) or targeted objectives (Klebsiella spp. in milk samples from dairy cows with mastitis) through screening of the titles and abstracts. After that, the full texts of the remaining 84 studies were derived for further assessment based on the inclusion criterion. Finally, we included 55 studies in this meta-analysis (Figures 1, 2). In terms of quality, 14 studies scored 2 or 3 points, and 41 studies scored 4 or 5 points (Supplementary Table 2).
Figure 1.
Flow diagram of search and selection process of eligible studies.
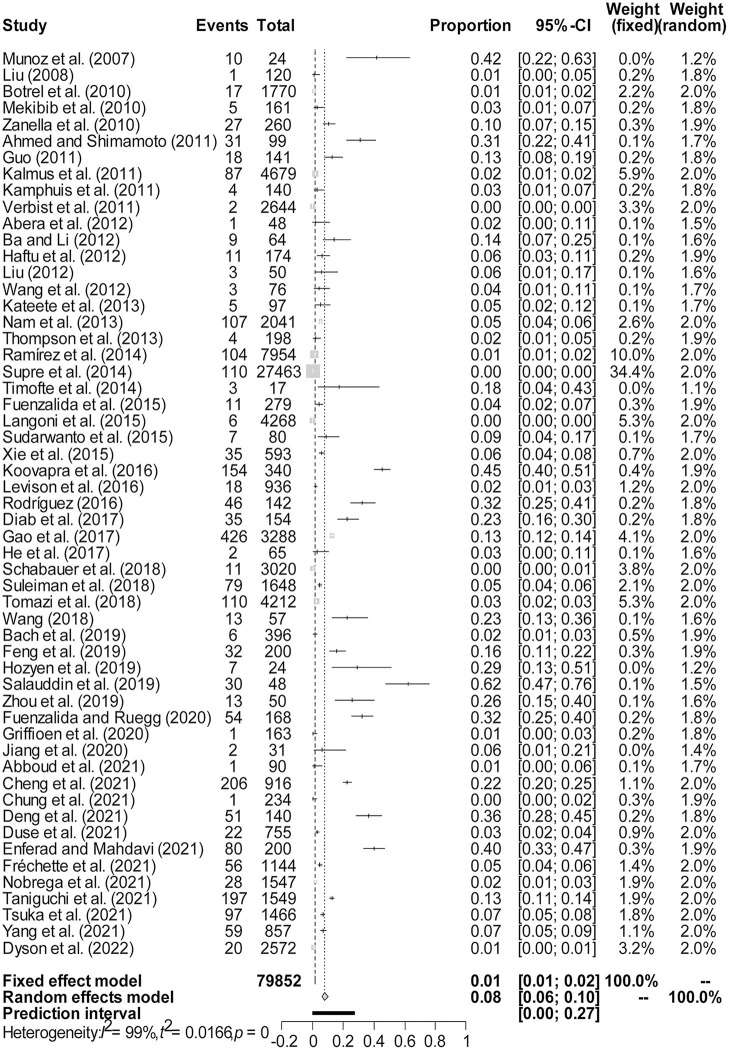
Figure 2.
Forest plot of the prevalence of Klebsiella spp. in milk samples among studies conducted worldwide. The length of the horizontal line represents the 95% confidence interval, and the diamond represents the summarized effect.
Publication bias and sensitivity analysis
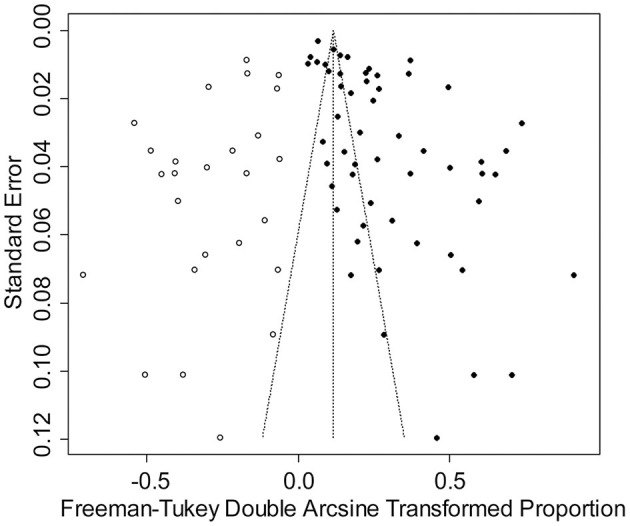
The rate was converted using PFT to ensure that the distribution was more similar to a normal distribution (Table 1). The forest plot revealed a high heterogeneity among the included studies (I2 = 98.8%, p = 0; Figure 2). The funnel plot was not symmetrical, indicating publication bias or small sample size bias in the studies (Figure 3). Egger's test was used to further test the sources of funnel plot asymmetry and provided a p-value of < 0.001 (Figure 4, Supplementary Table 3), indicating that publication bias did exist. The publication bias disappeared after adding 26 studies (the point estimate was 0%) when evaluating publication bias by the trim and fill method (Figure 5). Furthermore, we performed a sensitivity analysis to determine the effect of each study on the pooled prevalence of Klebsiella spp. There was no significant change in the result after excluding individual studies, which indicated the stability of our meta-analysis (Figure 6).
Table 1.
Normal distribution test for the normal rate and the different conversions of the normal rate.
| Conversion form | W | P |
|---|---|---|
| PRAWa | 0.77945 | 1.12e-07 |
| PLNb | 0.96893 | 0.1648 |
| PLOGITc | 0.98594 | 0.7657 |
| PASd | 0.90654 | 0.0004236 |
| PFTe | 0.90207 | 0.0002939 |
aPRAW: original rate.
bPLN: logarithmic conversion.
cPLOGIT: logit transformation.
dPAS: arcsine transformation.
ePFT: double-arcsine transformation.
Figure 3.

Funnel plot with pseudo 95% confidence interval limits for the examination of publication bias.
Figure 4.
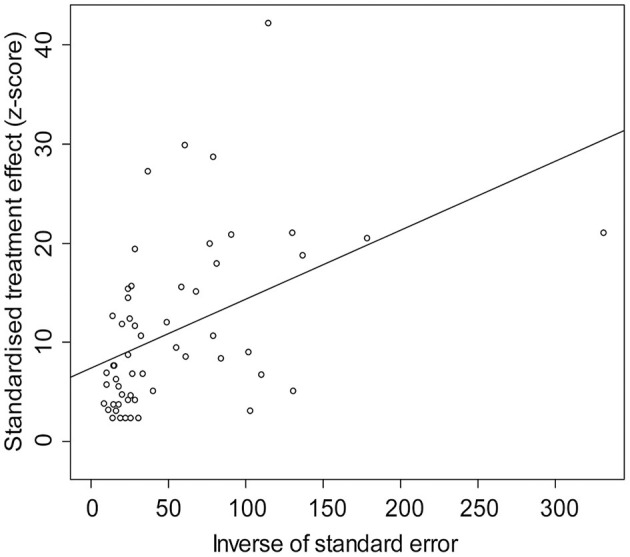
Egger's test for publication bias.
Figure 5.
The trim and fill method for publication bias.
Figure 6.
Sensitivity analysis. After removing one study at a time, the remaining studies were recombined using a random effects model to verify the effect of a single study on the overall results.
The meta-analysis results and publication bias of each subgroup are shown in Supplementary Figures 1–8.
Pooled prevalence of Klebsiella spp. in milk samples
There were 79,852 milk samples tested in our meta-analysis, and 2,478 of them were found to contain Klebsiella spp. The majority of Klebsiella species isolated from the milk of cows with mastitis is K. pneumoniae (25/55), followed by K. oxytoca (6/55), and only one case of K. ozaenae has been reported (Supplementary Table 4). At the global level, the random effect estimated pooled prevalence was 7.95% (95% CI: 6.07%−10.06%), and there was substantial heterogeneity (x2 = 4584.55, I2 = 98.8%, p = 0). The prevalence of Klebsiella-positive milk samples is shown in Figure 2.
The pooled Klebsiella spp. prevalence estimates and the 95% CIs for corresponding subgroups are all reported in Table 2. The pooled prevalence of Klebsiella-positive samples detected in 2013 or later was 12.16% (95% CI: 8.08%−16.90%), which was higher (p < 0.05) than that obtained before 2013 (3.85%, 95% CI: 2.67%−5.21%). By comparing various detection methods, automatic analysis systems (including the API 20E System and VITEK 2 Automated Identification System) showed the highest prevalence (17.14%, 95% CI: 10.33%−25.17%). The pooled prevalence estimates of K. oxytoca (3.22%, 95% CI: 0.73%−7.15%) in milk samples were lower than that of K. pneumoniae (11.51%, 95% CI: 6.72%−17.31%). However, considering p > 0.05 and small number of studies and samples of K. oxytoca, there existed a small sample size bias which indicated the results of this subgroup were not robust.
Table 2.
Pooled prevalence of Klebsiella spp. in milk samples from dairy cows with mastitis worldwide including risk factors and meta-regression.
| No. studies | No. tested | No. positive | Positive percentage (95% CIa) | Heterogeneity | Univariate meta-regression | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | p -value b | I2 (%) | p -value | Coefficient (95% CI) | ||||||
| Sampling years | 0.0004 | 0.1339 (0.0599–0.2079) | ||||||||
| Before 2013 | 25 | 26,171 | 536 | 3.85% (2.67–5.21%) | 505.76 | < 0.01 | 94.9% | |||
| 2013 or later | 28 | 24,752 | 1,735 | 12.16% (8.08–16.90%) | 2,725.28 | 0 | 99.0% | |||
| Detection methods | 0.0099 | 0.1565 (0.0376–0.2754) | ||||||||
| Biochemical tests | 32 | 64,699 | 1,626 | 6.97% (4.93–9.31%) | 2,705.81 | 0 | 98.9% | |||
| 16SrDNA identification | 9 | 12,770 | 806 | 11.04% (4.01–20.82%) | 1,550.65 | 0 | 99.5% | |||
| MALDI-TOF MS | 8 | 4,154 | 315 | 4.56% (1.51–9.01%) | 188.45 | < 0.01 | 96.3% | |||
| Automatic analysis systems | 6 | 5,090 | 615 | 17.14% (10.33–25.17%) | 126.76 | < 0.01 | 96.1% | |||
| Continents | 0.00240 | −0.2169 (−0.4053 to −0.0285) | ||||||||
| Africa | 7 | 2,251 | 139 | 8.72% (3.75–15.32%) | 67.02 | < 0.01 | 91.0% | |||
| Asia | 24 | 12,616 | 1,581 | 14.26% (10.49–18.48%) | 789.51 | < 0.01 | 97.1% | |||
| Europe | 9 | 40,651 | 257 | 0.73% (0.20–1.49%) | 159.78 | < 0.01 | 95.0% | |||
| North America | 7 | 3,145 | 159 | 7.52% (2.99–13.73%) | 160.46 | < 0.01 | 96.3% | |||
| South America | 6 | 18,383 | 321 | 4.48% (2.14–7.61%) | 325.69 | < 0.01 | 98.5% | |||
| Oceania | 2 | 2,806 | 21 | 0.67% (0.38–1.03%) | 0.10 | 0.76 | 0.0% | |||
| Longitude | < 0.0001 | −0.2229 (−0.3097 to −0.1362) | ||||||||
| 0–20°E | 7 | 35,955 | 167 | 0.70% (0.28–1.26%) | 64.72 | < 0.01 | 90.7% | |||
| 20–40°E | 10 | 7,174 | 262 | 7.72% (4.00–12.42%) | 199.63 | < 0.01 | 95.5% | |||
| 40–60°E | 1 | 200 | 80 | 40.00% (33.30–46.89%) | 0.00 | — | — | |||
| 80–100°E | 1 | 48 | 30 | 62.50% (48.27–75.75%) | 0.00 | — | — | |||
| 100–120°E | 18 | 7,580 | 649 | 10.11% (7.43–13.13%) | 209.11 | < 0.01 | 91.9% | |||
| 140–160°E | 1 | 2,572 | 20 | 0.78% (0.47–1.16%) | 0.00 | — | — | |||
| 0–20°W | 2 | 447 | 65 | 15.23% (0.00–50.98%) | 67.24 | < 0.01 | 98.5% | |||
| 40–60°W | 3 | 6,075 | 61 | 2.60% (0.15–7.67%) | 113.26 | < 0.01 | 98.2% | |||
| 60–80°W | 4 | 8,516 | 166 | 12.59% (2.29–28.79%) | 168.56 | < 0.01 | 98.2% | |||
| 80–100°W | 2 | 953 | 21 | 6.22% (0.00–29.58%) | 6.81 | < 0.01 | 85.3% | |||
| Latitude | 0.0001 | 0.2421 (0.1189–0.3652) | ||||||||
| 0–10°S | 3 | 1,870 | 132 | 13.37% (1.30–34.48%) | 78.94 | < 0.01 | 97.5% | |||
| 10–20°S | 1 | 1,547 | 28 | 1.81% (1.20–2.54%) | 0.00 | — | — | |||
| 20–30°S | 2 | 4,528 | 33 | 3.23% (0.00–20.37%) | 83.49 | < 0.01 | 98.8% | |||
| 30–40°S | 1 | 2,454 | 19 | 0.77% (0.46–1.16%) | 0.00 | — | — | |||
| 40–50°S | 1 | 118 | 1 | 0.85% (0.00–3.60%) | 0.00 | — | — | |||
| 0–10°N | 4 | 8,260 | 115 | 2.33% (0.63–4.86%) | 9.52 | 0.02 | 68.5% | |||
| 10–20°N | 1 | 174 | 11 | 6.32% (3.12–10.48%) | 0.00 | — | — | |||
| 20–30°N | 4 | 415 | 84 | 24.13% (6.02–48.96%) | 73.14 | < 0.01 | 95.9% | |||
| 30–40°N | 16 | 7,375 | 719 | 12.91% (8.93–17.46%) | 349.68 | < 0.01 | 95.7% | |||
| 40–50°N | 10 | 6,918 | 155 | 6.20% (2.81–10.68%) | 299.14 | < 0.01 | 97.0% | |||
| 50–60°N | 6 | 35,106 | 207 | 0.60% (0.01–1.73%) | 122.70 | < 0.01 | 95.9% | |||
| Economic development level | < 0.0001 | 0.1561 (0.0840–0.2282) | ||||||||
| Developed countries | 21 | 51,658 | 838 | 3.31% (1.89–5.08%) | 1,296.66 | < 0.01 | 98.5% | |||
| Developing countries | 34 | 28,194 | 1,640 | 11.76% (8.25–15.77%) | 2,596.12 | 0 | 98.7% | |||
| Mastitis type | 0.0447 | 0.0987 (0.0023–0.1951) | ||||||||
| Clinical | 31 | 16,990 | 1,233 | 11.99% (8.62–15.79%) | 1,222.44 | < 0.01 | 97.5% | |||
| Subclinical | 13 | 14,627 | 420 | 6.42% (2.97–10.98%) | 770.04 | < 0.01 | 98.4% | |||
| Population density | 0.0141 | −0.1021 (−0.1837 to −0.0206) | ||||||||
| < 50 | 17 | 29,768 | 610 | 4.13% (2.70–5.83%) | 595.21 | < 0.01 | 97.3% | |||
| 50–100 | 2 | 1,848 | 159 | 19.00% (0.00–62.04%) | 153.85 | < 0.01 | 99.4% | |||
| 100–250 | 24 | 12,121 | 968 | 9.58% (5.64–14.37%) | 1,222.15 | < 0.01 | 98.1% | |||
| 250–500 | 6 | 33,479 | 563 | 8.88% (2.18–19.07%) | 1,336.36 | < 0.01 | 99.6% | |||
| >500 | 6 | 2,636 | 178 | 10.28% (2.73–21.58%) | 149.23 | < 0.01 | 96.6% | |||
| Scores of study quality | 0.8368 | −0.0087 (−0.0920 to 0.0745) | ||||||||
| 2–3 | 14 | 39,846 | 572 | 8.20% (4.72–12.49%) | 1,222.17 | < 0.01 | 98.9% | |||
| 4–5 | 41 | 40,006 | 1,906 | 7.88% (5.67–10.39%) | 2,643.54 | 0 | 98.5% | |||
| Total | 55 | 79,852 | 2,478 | 7.95% (6.07–10.06%) | 4,584.55 | 0 | 98.8% | |||
aCI, confidence interval.
bp-value: p < 0.05 is statistically significant.
The 55 eligible studies were from 25 countries on six continents (Asia: 24; Europe: 9; Africa: 7; North America: 7; South America: 6; Oceania: 2). The prevalence of Klebsiella spp. in Asia was the highest (14.26%, 95% CI: 10.49%−18.48%), particularly in the longitude interval 40–100°E. The 0–20°E interval, where most European countries included in this meta-analysis were located, had the lowest positive proportion. In regard to latitude, there was a low Klebsiella prevalence at high latitudes. Almost all countries in the European (0.73%, 95% CI: 0.20–1.49%) and Oceanian (0.67%, 95% CI: 0.38–1.03%) regions are developed countries, which indicates that the distribution of the Klebsiella-positive proportion may be related to the level of national economic development.
The pooled prevalence of Klebsiella spp. samples detected in developing countries was 11.76% (95% CI: 8.25–15.77%), which was higher (p < 0.05) than that in developed countries (3.31%, 95% CI: 1.89–5.08%). There were more (p < 0.05) Klebsiella-positive samples found in clinical mastitis samples (11.99%, 95% CI: 8.62–15.79%) than in subclinical mastitis samples (6.42%, 95% CI: 2.97–10.98%).
In addition, the prevalence of Klebsiella spp. was related to population density. The analysis of population density in different countries showed that the lowest Klebsiella spp. prevalence was in countries where there were < 50 per sq km (4.13%, 95% CI: 2.70–5.83%) and revealed that the higher the population density of the sampling area, the higher the prevalence of Klebsiella spp. in milk samples.
To further explore the MDR rate of Klebsiella spp. isolated from milk samples, an MDR rate correlation analysis was conducted for economic development level and population density. The R2 values were 78.87 and 87.51% (Table 3), respectively, which revealed that the MDR rate was closely related to the level of economic development and population density.
Table 3.
The MDR rate of Klebsiella spp. isolated from milk samples: a bivariate meta-regression analysis.
| No. studies | No. isolatesa | No. MDRb | Positive percentage (95% CI) | Heterogeneity | Univariate meta-regression | Bivariate meta-regression | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | p -value | I 2 | p -value c | Coefficient (95% CI) | R 2 –economic development level | R 2 –population density | |||||
| MDR rate | 0.0214 | −0.5828 (-1.0793 to−0.0863) | 78.87% | 87.51% | |||||||
| 0 | 4 | 33 | 0 | 0.00% (0.00–0.00%) | 0.89 | 0.83 | 0.0% | ||||
| >0, < 0.5 | 8 | 860 | 129 | 15.02% (5.85–26.97%) | 103.38 | < 0.01 | 93.2% | ||||
| > 0.5, < 1 | 2 | 112 | 73 | 63.13% (46.19–78.61%) | 2.76 | 0.10 | 63.8% | ||||
| 1 | 6 | 81 | 81 | 100.0% (100.0–100.0%) | 1.38 | 0.93 | 0.0% | ||||
aNo. isolates: the number of Klebsiella isolates.
bNo. MDR: the number of MDR Klebsiella isolates.
cp-value: p < 0.05 is statistically significant.
Discussion
This study investigated the prevalence of Klebsiella spp. in milk samples from dairy cows with mastitis worldwide by analyzing articles published from 2007 to 2021 to assess risk factors for influencing the prevalence of these strains and to improve solutions for bovine intramammary infections. The studies included in this analysis from 2007 to 2013 mostly came from European and North American developed countries. However, the prevalence of Klebsiella-positive milk samples from the European Union (EU) has decreased dramatically after 2013 (Table 4). For example, the prevalence of Klebsiella in CM milk samples from the Netherlands during 2017–2018 was lower than that in 2006–2009 (28, 29), which might be due to strict health management regulations by the EU member countries (30). Because Klebsiella spp. in bovine mastitis mostly originates from the manure attached to the surrounding bedding and farming equipment, especially the organic bedding materials such as sawdust (31, 32). In developed countries, frequent changes in bedding, along with cow cleanliness and the hygiene of housing facilities greatly reduce the contact transmission of these bacteria (33). Although the Klebsiella-positive rate of CM milk samples from the United States and Japan was comparatively prominent after 2013, cases were mainly reported in developing countries and showed a global distribution, especially in Asia. For instance, the prevalence of K. pneumoniae in mastitis milk samples in West Bengal, Jharkhand and Mizoram in India was 45.29% (95% CI: 40.03–50.61%) (34). Random sampling of free-range dairy herds in northwestern Iran revealed that the K. pneumoniae-positive proportion was 40.00% (95% CI: 33.30–46.89%) (35). The prevalence in CM milk samples in Rangpur Division, Bangladesh was up to 62.50% (95% CI: 48.27–75.75%) (36). One distinguishing feature of these regions is that they have extremely high population densities, and Bangladesh and some regions in India each have a population density of over 1,000 per sq km (19). In addition, in these regions, milk is usually produced in rural settings where the majority of the population resides (37). We also noted a surge in cases from China in the past decade, which may be partly related to the selection of three Chinese databases for this study and the increasing attention to bovine mastitis in this country. As fecal shedding of K. pneumoniae plays a critical role in pathogen dissemination (38), fecal-oral transmission cycles may perpetuate and amplify the presence of such pathogens (32). Human feces are recognized as a reservoir of reverse transmission (39). Although the evidence for its role in the transmission of Klebsiella from humans to dairy herds is unclear, some latrines in close proximity to dairy herds should be considered a risk factor that can promote human-derived Klebsiella transmission, and this risk may be increased by excessive population density.
Table 4.
Characteristics of the included studies.
| References | Sampling time | No. positive/No. testeda | Prevalence | Mastitis type | Country | Population densityb | Detection methodsc | Klebsiella speciesd | Genotyping methods | Gene detectione | MDR ratef |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Munoz et al. (50) | 2006.04 | 10/24 | 41.67% | Clinical | The United States | 36.39 | — | Kpn | RAPD | — | — |
| Liu (51) | 2008 | 1/120 | 0.83% | Clinical | China | 153.8 | Biochem | Unknown Species | — | — | 0/1 |
| Botrel et al. (52) | 2007.01–2008.03 | 17/1,770 | 0.96% | Bothg | France | 119.5 | Biochem | Unknown Species | — | — | — |
| Mekibib et al. (53) | 2008.11–2009.04 | 5/161 | 3.11% | Subclinical | Ethiopia | 117.9 | Biochem | Kpn | — | — | — |
| Zanella et al. (54) | 2006.01–2007.06 | 27/260 | 10.38% | — | Brazil | 25.6 | Biochem | Kpn, Kox, and Koz | — | — | — |
| Ahmed and Shimamoto (48) | 2008 | 31/99 | 31.31% | — | Egypt | 104.7 | Biochem and Auto | Kpn and Kox | — | Drug resistance | 14/31 |
| Guo (55) | 2010 | 18/141 | 12.77% | Both | China | 153.8 | Biochem | Unknown Species | — | — | — |
| Kalmus et al. (56) | 2007–2009 | 87/4,679 | 1.86% | Both | Estonia | 31.26 | Biochem | Unknown Species | — | — | — |
| Kamphuis et al. (28) | 2011 | 4/140 | 2.86% | Clinical | The Netherlands | 509.3 | Biochem | Unknown Species | — | — | — |
| Verbist et al. (31) | 2008.05–2009.05 | 2/2,644 | 0.08% | Clinical | Belgium | 384.2 | Biochem | Unknown Species | — | — | — |
| Abera et al. (57) | 2008.10–2009.05 | 1/48 | 2.08% | — | Ethiopia | 117.9 | — | Unknown Species | — | — | — |
| Ba and Li (58) | 2011.07–2011.08 | 9/64 | 14.06% | — | China | 153.8 | Biochem | Unknown Species | — | — | 0/9 |
| Haftu et al. (59) | 2009.10–2010.05 | 11/174 | 6.32% | Both | Ethiopia | 117.9 | Biochem | Kpn | — | — | — |
| Liu (60) | 2009.01–2009.12 | 3/50 | 6.00% | Subclinical | China | 153.8 | Biochem | Unknown Species | — | — | — |
| Wang et al. (61) | 2011.03–2011.04 | 3/76 | 3.95% | Subclinical | China | 153.8 | Biochem | Unknown Species | — | — | — |
| Kateete et al. (62) | 2010.02–2011.03 | 5/97 | 5.15% | Clinical | Uganda | 235.8 | Biochem | Kox | — | — | 2/5 |
| Nam et al. (63) | 2012.01–2012.11 | 107/2,041 | 5.24% | — | Korea | 527.7 | Biochem | Unknown Species | — | — | — |
| Thompson et al. (64) | 2007.06–2008.08 | 4/198 | 2.02% | Clinical | Canada | 4.168 | — | Unknown Species | — | — | — |
| Ramírez et al. (65) | 2009.01–2010.12 | 104/7,954 | 1.31% | Subclinical | Colombia | 46.21 | Biochem | Unknown Species | — | — | — |
| Supre et al. (66) | 2012.09–2013.09 | 110/27,463 | 0.40% | — | Belgium | 384.2 | Biochem | Unknown Species | — | — | 0/59 |
| Timofte et al. (67) | 2010 | 3/17 | 17.65% | Clinical | The United Kingdom | 281.9 | Auto | Kpn | — | Drug resistance | 3/3 |
| Fuenzalida et al. (68) | 2011.05–2013.11 | 11/279 | 3.94% | Clinical | The United States | 36.39 | Biochem | Unknown Species | — | — | — |
| Langoni et al. (69) | 2013 | 6/4,268 | 0.14% | — | Brazil | 25.6 | 16S rDNA | Kpn | — | — | — |
| Sudarwanto et al. (43) | 2011.11–2011.12 | 7/80 | 8.75% | — | Indonesia | 152.6 | — | Kpn | PFGE | Drug resistance | 8/8 |
| Xie et al. (70) | 2013 | 35/593 | 5.90% | — | China | 153.8 | Biochem | Kox | — | — | — |
| Koovapra et al. (34) | 2016 | 154/340 | 45.29% | Both | India | 468.7 | — | Unknown Species | — | Drug resistance | 23/291 |
| Levison et al. (71) | 2011.04–2012.05 | 18/936 | 1.92% | Clinical | Canada | 4.168 | Biochem | Unknown Species | — | — | — |
| Rodríguez (72) | 2015.09–2015.12 | 46/142 | 32.39% | — | Peru | 26.06 | — | Unknown Species | — | — | — |
| Diab et al. (49) | 2015.09–2015.11 | 35/154 | 22.73% | — | Lebanon | 661.7 | MALDI-TOF | Kpn | PFGE | Drug resistance | 36/36 |
| Gao et al. (73) | 2014.03–2016.09 | 426/3,288 | 12.96% | Clinical | China | 153.8 | Biochem, 16S rDNA, and Auto | Unknown Species | — | — | — |
| He et al. (45) | 2015 | 2/65 | 3.08% | Clinical | China | 153.8 | MALDI-TOF | Kpn | PFGE | Drug resistance | 2/2 |
| Schabauer et al. (74) | 2015.10–2016.09 | 11/3,020 | 0.36% | — | Austria | 109.7 | 16S rDNA | Kpn | — | — | — |
| Suleiman et al. (75) | 2014.01–2014.07 | 79/1,648 | 4.79% | Subclinical | Tanzania | 69.43 | Biochem | Unknown Species | — | — | — |
| Tomazi et al. (76) | 2014.03–2016.01 | 110/4,212 | 2.61% | Clinical | Brazil | 25.6 | Biochem | Unknown Species | — | — | — |
| Wang (77) | 2017.04 | 13/57 | 22.81% | Clinical | China | 153.8 | Biochem | Kpn | — | — | — |
| Bach et al. (78) | 2017.02–2017.05 | 6/396 | 1.52% | — | The United States | 36.39 | Biochem | Unknown Species | — | — | — |
| Feng et al. (79) | 2017 | 32/200 | 16.00% | Clinical | China | 153.8 | 16S rDNA | Unknown Species | — | — | 17/32 |
| Hozyen et al. (80) | 2017.09–2017.12 | 7/24 | 29.17% | Clinical | Egypt | 104.7 | Biochem | Kpn | — | — | — |
| Salauddin et al. (36) | 2019 | 30/48 | 62.50% | Clinical | Bangladesh | 1278 | Biochem | Unknown Species | — | — | 30/30 |
| Zhou et al. (81) | 2018.06 | 13/50 | 26.00% | Subclinical | China | 153.8 | Biochem and 16S rDNA | Kpn | — | — | — |
| Fuenzalida and Ruegg (82) | 2016.06–2016.12 | 54/168 | 32.14% | Clinical | The United States | 36.39 | — | Kpn | PFGE | — | — |
| Griffioen et al. (29) | 2017.05–2018.07 | 1/163 | 0.61% | Clinical | The Netherlands | 509.3 | MALDI-TOF | Kpn | — | — | — |
| Jiang et al. (83) | 2019.06–2019.10 | 2/31 | 6.45% | Clinical | China | 153.8 | 16S rDNA | Kpn | — | — | 2/2 |
| Abboud et al. (84) | 2019.03–2019.04 | 1/90 | 1.11% | — | Lebanon | 661.7 | MALDI-TOF | Kox | — | — | 0/1 |
| Cheng et al. (85) | 2019 | 206/916 | 22.49% | Both | China | 153.8 | Biochem and 16S rDNA | Kpn | — | Virulence | — |
| Chung et al. (86) | 2020.04–2020.09 | 1/234 | 0.43% | Both | Australia | 3.357 | MALDI-TOF | Kpn | — | — | — |
| Deng et al. (87) | 2019.09–2019.12 | 51/140 | 36.43% | Clinical | China | 153.8 | 16S rDNA and Auto | Kpn | — | Virulence | 51/130 |
| Duse et al. (88) | 2013.08–2018.12 | 22/755 | 2.91% | Clinical | Sweden | 24.76 | MALDI-TOF | Unknown Species | — | — | 0/22 |
| Enferad and Mahdavi (35) | 2018.04–2018.10 | 80/200 | 40.00% | — | Iran | 52.21 | Biochem | Kpn | — | Drug resistance | 56/80 |
| Fréchette et al. (89) | 2018–2019 | 56/1,144 | 4.90% | Clinical | Canada | 4.168 | MALDI-TOF | Unknown Species | — | — | — |
| Nobrega et al. (90) | 2009.12–2011.07 | 28/1,547 | 1.81% | — | Brazil | 25.6 | Biochem | Kpn | — | Drug resistance | 13/81 |
| Taniguchi et al. (8) | 2016.08–2017.07 | 197/1,549 | 12.72% | — | Japan | 345.8 | MALDI-TOF | Kpn and Kox | RAPD | Drug resistance | 22/197 |
| Tsuka et al. (91) | 2012.10–2014.12 | 97/1,466 | 6.62% | Clinical | Japan | 345.8 | Auto | Kpn | PFGE | Drug resistance | — |
| Yang et al. (92) | 2017–2018 | 59/857 | 6.88% | Clinical | China | 153.8 | 16S rDNA | Kpn | — | Drug resistance | 4/66 |
| Dyson et al. (93) | 2011.01–2012.03 | 20/2,572 | 0.78% | — | Australia | 3.357 | Biochem | Unknown Species | — | — | — |
aNo. postive/No. tested: the ratio of Klebsiella-positive milk samples to total tested milk samples.
bPopulation density: unit—per sq km.
cDetection methods: Biochem, biochemical tests; 16S rDNA, 16S rDNA identification; MALDI-TOF, MALDI-TOF MS; Auto, automatic analysis systems.
dKlebsiella species: Kpn, Klebsiella pneumoniae; Kox, Klebsiella oxytoca; Koz, Klebsiella ozaenae.
eGene detection: molecular characterization of drug resistance genes or virulence genes.
fMDR rate: the MDR rate of Klebsiella isolates.
gBoth: studies with both clinical mastitis milk samples and subclinical mastitis milk sample.
Although Klebsiella spp. were occasionally detected in milk samples from EU member countries (Table 5), the drug-resistance rate of these bacteria has declined since 2009 (40), and the occurrence of MDR strains is very rare. This may be attributed to the EU's policy of a total ban on the use of antibiotic growth promoters (AGPs) in animal feeding since 2006 (41), which substantially reduced the probability of Klebsiella carrying drug-resistance genes in the dairy farm environment. In this meta-analysis, China had the highest number of reports of MDR Klebsiella in milk samples, which included seven studies from different regions of the country. In July 2019, Announcement No. 194 was issued by the Chinese Ministry of Agriculture and Rural Affairs to prohibit all antimicrobials from being used as AGPs and was implemented in on July 1, 2020 (42). However, whether this will achieve the desired governance results is pending further investigations in the future.
Table 5.
Estimated pooled prevalence of Klebsiella-positive milk samples by country.
| Country | Klebsiella species*(No. studies) | Continent | No. tested | No. positive | Positive percentage | 95% CI |
|---|---|---|---|---|---|---|
| Australia | Kpn (1); Unknown Species (1) | Oceania | 2,806 | 21 | 0.67% | 0.38–1.03% |
| Austria | Kpn (1) | Europe | 3,020 | 11 | 0.36% | 0.18–0.62% |
| Bangladesh | Unknown Species (1) | Asia | 48 | 30 | 62.50% | 48.27–75.75% |
| Belgium | Unknown Species (2) | Europe | 30,107 | 112 | 0.22% | 0.02–0.64% |
| Brazil | Kpn (3); Kox (1); Koz (1) | South America | 10,287 | 171 | 2.56% | 0.56–5.89% |
| Canada | Unknown Species (3) | North America | 2,278 | 78 | 2.90% | 1.14–5.39% |
| China | Kpn (7); Kox (1); Unknown Species (7) | Asia | 6,648 | 873 | 11.67% | 8.00–15.89% |
| Colombia | Unknown Species (1) | South America | 7,954 | 104 | 1.31% | 1.07–1.57% |
| Egypt | Kpn (2); Kox (1) | Africa | 123 | 38 | 30.72% | 22.70–39.33% |
| Estonia | Unknown Species (1) | Europe | 4,679 | 87 | 1.86% | 1.49–2.27% |
| Ethiopia | Kpn (2); Unknown Species (1) | Africa | 383 | 17 | 4.13% | 2.12–6.68% |
| France | Unknown Species (1) | Europe | 1,770 | 17 | 0.96% | 0.55–1.47% |
| India | Unknown Species (1) | Asia | 340 | 154 | 45.29% | 40.03–50.61% |
| Indonesia | Kpn (1) | Asia | 80 | 7 | 8.75% | 3.39–16.08% |
| Iran | Kpn (1) | Asia | 200 | 80 | 40.00% | 33.30–46.89% |
| Japan | Kpn (2); Kox (1) | Asia | 3,015 | 294 | 9.45% | 4.35–16.24% |
| Korea | Unknown Species (1) | Asia | 2,041 | 107 | 5.24% | 4.32–6.25% |
| Lebanon | Kpn (1); Kox (1) | Asia | 244 | 36 | 9.22% | 0.00–39.36% |
| Peru | Unknown Species (1) | South America | 142 | 46 | 32.39% | 24.92–40.34% |
| Sweden | Unknown Species (1) | Europe | 755 | 22 | 2.91% | 1.82–4.25% |
| Tanzania | Unknown Species (1) | Africa | 1,648 | 79 | 4.79% | 3.81–5.88% |
| The Netherlands | Kpn (1); Unknown Species (1) | Europe | 303 | 5 | 1.52% | 0.04–4.41% |
| The United Kingdom | Kpn (1) | Europe | 17 | 3 | 17.65% | 2.57–39.97% |
| The United States | Kpn (2); Unknown Species (2) | North America | 867 | 81 | 14.61% | 1.94–35.23% |
| Uganda | Kox (1) | Africa | 97 | 5 | 5.15% | 1.48–10.61% |
| Total | — | — | 79,852 | 2,478 | 7.95% | 6.07–10.06% |
*Klebsiella species: Kpn, Klebsiella pneumoniae; Kox, Klebsiella oxytoca; Koz, Klebsiella ozaenae.
We found that reports of MDR Klebsiella spp. in milk samples were also mainly from densely populated developing countries (189/283, 66.78%). Similar to the human carbapenemase-resistant Klebsiella species (12), the studies we included suggest that carbapenemase-resistant Klebsiella spp. in milk samples are also harbor blaKPC, blaNDM or blaOXA. KPC-producing K. pneumoniae has caused serious nosocomial and community infections worldwide, but there are few reports of their detection in milk samples. Among the articles screened in this study, KPC-producing strains were only reported from bulk tank milk in Indonesia (43). Discovered in 2008 (44), the endemic scope of human NDM-producing K. pneumoniae was mainly concentrated in India, Pakistan and Bangladesh. There is also a high prevalence of MDR K. pneumoniae in milk samples from some regions of South Asia, but the detection of blaNDM was ignored by some local reports (36). Accordingly, we recommend that dairy farms in South Asia promote blaNDM detection and strengthen environmental management to take effective measures to constrain the global spread of blaNDM-harboring strains. Although China is not a major epidemic region for NDM-producing K. pneumoniae among humans, the presence of blaNDM − 5-positive K. pneumoniae in both milk and fecal samples was reported in Jiangsu Province. Notably, blaNDM − 5 plasmids carried by these strains are almost identical to the human K. pneumoniae plasmid (pNDM-MGR194) previously reported in India (45), which may be a molecular epidemiological clue of cross-species transmission of blaNDM − 5 plasmids between humans and cows. Therefore, we need to be alert to the risk of cross-species transmission of the blaNDM − 5 gene. OXAs commonly hydrolyze isoxazolylpenicillins (oxacillin, cloxacillin, dicloxacillin, etc.), but the hydrolysis of carbapenems by OXA-48 should not be underestimated (46). OXA-48-producing K. pneumoniae was first identified in Turkey in 2001 (47), and its presence has been reported in Mediterranean countries (12). In this study, we found that milk-derived OXA-48-producing K. pneumoniae also mainly occurred in Mediterranean countries, where their epidemic scope overlaps with that of human strains. According to sampling data from 2008, blaOXA-harboring K. oxytoca strains were detected in milk samples from Egypt (48). Similar reports were subsequently reported in Lebanon, where there was a substantial risk of OXA-48-producing K. pneumoniae being transmitted back to humans due to the local practice of selling raw milk (49). In addition, the endemic scope of these strains has been seen in India and other South Asian countries, and milk-derived variants with both blaOXA − 1 and blaNDM − 5 genes have been identified in China (45). Therefore, to prevent the emergence of more resistant strains due to the combination of drug-resistance genes, the genetic evolution and integration of blaOXA in dairy herds deserves further study.
Among the selected articles, there were only two reports on virulence genes of milk-derived Klebsiella, and both of them were from China (Table 4), a region with a high incidence of hypervirulent K. pneumoniae (hvKP) infection (13). Klebsiella spp. from milk samples also have virulence-related genes associated with siderophore biosynthesis, fimbria proteins, adhesion, and secretion systems, but they are far less abundant than human-derived K. pneumoniae strains in terms of gene variety (94). The ferric uptake operon kfuABC carried by human-derived K. pneumoniae is closely related to colonization and tissue invasion (95). kfuABC of the milk-derived strains also has similar functions, and the gene cluster was more prevalent in CM cases than in SCM cases (94), which explained why the prevalence of Klebsiella in CM milk samples was significantly higher than that in SCM milk samples. Capsular polysaccharides (CPSs) have been identified as the most important virulence factor of K. pneumoniae for assisting bacteria in evading host immune surveillance, and their synthesis is mainly regulated by CPS-regulated genes such as regulator of mucoid phenotype A (rmpA) (96, 97). However, rmpA was detected only in milk-derived K. pneumoniae from China according to this analysis (85). It was found that K. pneumoniae was more likely to generate capsules in milk than in LB medium (98), which may indicate that the nutrient-rich environment reduces the dependence of Klebsiella capsule synthesis on CPS regulatory genes such as rmpA. Despite the extremely low incidence of rmpA in milk-derived Klebsiella strains, this gene can be transferred with pLVPK-like plasmids to MDR K. pneumoniae (especially serotype K47), leading to the emergence of carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP), which has become a more troublesome pathogen of nosocomial infections than MDR KP and hvKP in recent years (99). A study included in our meta-analysis showed that K47 serotype K. pneumoniae had been detected in milk samples from Jiangsu, China (92). Therefore, it is necessary to strengthen the monitoring of the genetic evolution of milk-derived Klebsiella to prevent the emergence and spread of CR-hvKP in dairy herds.
The advantages of this meta-analysis are its wide geographical coverage, long time span and clear analysis methods, but we have to acknowledge that this study has some limitations. First, the 55 articles in this study were derived from six large databases (three English databases and three Chinese databases), which may have led to the omission of eligible articles from other databases. Due to the constraints of the databases, the majority of the selected articles were in English or Chinese, with only two in other languages (Korean and Spanish), which may have resulted in studies in other languages being omitted. Possibly due to the language, the studies we have included only covered 25 countries. In addition to one case of K. ozaenae, only six studies reported Klebsiella mastitis caused by K. oxytoca with a relatively small sample which may have led to small sample size bias. About half of the included studies did not identify specific Klebsiella species, so this meta-analysis mainly focused on Klebsiella spp. in dairy milk of cattle with mastitis. Moreover, data was unevenly distributed across countries due to the factors such as research conditions, government attention and trade protection. Unfortunately, we did not find data applicable to this meta-analysis for large milk producing countries such as Russia or New Zealand. However, it is certain that the data cover the characteristics of most dairy farming regions worldwide and can reflect the global distribution and variation trends of Klebsiella in milk samples. Second, bovine mastitis generally occurs in multiparous cows (100), but we were unable to extract available data on the parity of cows to find its correlation with the positive rate of Klebsiella in milk samples. Third, among the studies collected in this meta-analysis, pathogen identification methods for milk samples included biochemical tests, 16S rDNA identification, MALDI-TOF MS, and automatic analysis systems that were widely adopted to identify the pathogen of the milk samples. It is generally accepted that MALDI-TOF MS or automatic analysis systems are considered to have a lower detection error than conventional biochemical tests (101). Reports from developed economies typically involve 2–3 laboratory tests that provide accurate calibration of pathogen incidence in milk samples. In some developing countries, however, only biochemical tests are usually available, which is likely to result in a small number of missed cases. Therefore, we need to prepare for the worsening outbreaks of Klebsiella in dairy herds in some developing countries, and call on local authorities to put more effective measures in place.
Conclusions
This meta-analysis revealed that the prevalence of Klebsiella-positive milk samples and MDR Klebsiella spp. are highly correlated with the economic development level and population density of the country or region. These two factors are often prerequisites for banning AGPs and enforcing environmental management. The milk-derived MDR strains from Asia and the Mediterranean region suggest that these strains may have the potential for interspecies transmission between humans and dairy cows, and that MDR strains in Chinese dairy herds may evolve into CR-hvKP. Our meta-analysis provides a reference for the prevention and treatment of bacterial mastitis in dairy cows and the blocking of the global transmission of zoonotic pathogens.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
RC, RD, and QG conceptualized the study, with funding provided by RC and were responsible for the revision. JS, WX, QW, JY, TT, QY, MZ, GG, JL, ND, FL, and KS performed data extraction. JS and WX established the database and wrote the original draft of the article. QW carried out the data analysis. All authors contributed to the manuscript editing and approved the final manuscript.
Funding Statement
This work was financially supported through grants from the National Natural Science Foundation of China (No. 32102677), the Science and Technology Project of the Jilin Provincial Education Department (No. JJKH20210364KJ), the Natural Science Foundation of Jilin Province (No. YDZJ202201ZYTS449), and the China Postdoctoral Science Foundation (No. 2021M691224).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1143257/full#supplementary-material
References
- 1.Gomes F, Henriques M. Control of Bovine mastitis: old and recent therapeutic approaches. Curr Microbiol. (2016) 72:377–82. 10.1007/s00284-015-0958-8 [DOI] [PubMed] [Google Scholar]
- 2.Ruegg PL, A. 100-year review: mastitis detection, management, and prevention. J Dairy Sci. (2017) 100:10381–97. 10.3168/jds.2017-13023 [DOI] [PubMed] [Google Scholar]
- 3.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. (2017) 41:252–75. 10.1093/femsre/fux013 [DOI] [PubMed] [Google Scholar]
- 5.Organization WH . Antimicrobial Resistance: Global Report on Surveillance. Geneva: WHO. (2014). [Google Scholar]
- 6.Yang X, Dong N, Chan EW, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. (2021) 29:65–83. 10.1016/j.tim.2020.04.012 [DOI] [PubMed] [Google Scholar]
- 7.Massé J, Dufour S, Archambault M. Characterization of Klebsiella isolates obtained from clinical mastitis cases in dairy cattle. J Dairy Sci. (2020) 103:3392–400. 10.3168/jds.2019-17324 [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi T, Latt KM, Tarigan E, Yano F, Sato H, Minamino T, et al. A 1-year investigation of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from bovine mastitis at a large-scale dairy farm in Japan. Microb Drug Resist. (2021) 27:1450–4. 10.1089/mdr.2020.0481 [DOI] [PubMed] [Google Scholar]
- 9.Cheng J, Zhang J, Han B, Barkema HW, Cobo ER, Kastelic JP, et al. Klebsiella pneumoniae isolated from bovine mastitis is cytopathogenic for bovine mammary epithelial cells. J Dairy Sci. (2020) 103:3493–504. 10.3168/jds.2019-17458 [DOI] [PubMed] [Google Scholar]
- 10.Komatsu T, Yoshida E, Shigenaga A, Yasuie N, Uchiyama S, Takamura Y, et al. Fatal suppurative meningoencephalitis caused by Klebsiella pneumoniae in two calves. J Vet Med Sci. (2021) 83:1113–9. 10.1292/jvms.21-0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes LE. Four cases of bovine mastitis caused by Klebsiella pneumoniae. J Am Vet Med Assoc. (1954) 125:50–4. [PubMed] [Google Scholar]
- 12.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. (2016) 7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. (2017) 7:483. 10.3389/fcimb.2017.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marques C, Menezes J, Belas A, Aboim C, Cavaco-Silva P, Trigueiro G, et al. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: Population structure, antimicrobial resistance and virulence genes. J Antimicrob Chemother. (2019) 74:594–602. 10.1093/jac/dky499 [DOI] [PubMed] [Google Scholar]
- 15.Marques C, Belas A, Aboim C, Cavaco-Silva P, Trigueiro G, Gama LT, et al. Evidence of sharing of Klebsiella pneumoniae strains between healthy companion animals and cohabiting humans. J Clin Microbiol. (2019) 57:6. 10.1128/JCM.01537-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prevention CDC. Antibiotic Resistance Threats in the United States. Atlanta: CDC Press. (2013). [Google Scholar]
- 18.United Nations . World Economic Situation Prospects 2022. (2022). Available online at: https://www.un.org/development/desa/dpad/publication/world-economic-situation-and-prospects-2022/?msclkid=5b53c59ec6fd11eca8ee30f679a21fc9 (accessed January 13, 2022).
- 19.United Nations . World Population Prospects 2022. (2022). Available online at: https://population.un.org/wpp/ (accessed July 11, 2022).
- 20.Ding H, Gao YM, Deng Y, Lamberton PH, Lu DB, A. systematic review and meta-analysis of the seroprevalence of Toxoplasma Gondii in cats in mainland China. Parasit Vectors. (2017) 10:27. 10.1186/s13071-017-1970-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Chen Y, Zhang W, Chen S, Wen X, Ran X, et al. Prevalence of subclinical mastitis among dairy cattle and associated risks factors in China during 2012-2021: a systematic review and meta-analysis. Res Vet Sci. (2022) 148:65–73. 10.1016/j.rvsc.2022.04.007 [DOI] [PubMed] [Google Scholar]
- 22.Gong QL, Chen Y, Tian T, Wen X, Li D, Song YH, et al. Prevalence of bovine tuberculosis in dairy cattle in China during 2010-2019: a systematic review and meta-analysis. PLoS Negl Trop Dis. (2021) 15:e0009502. 10.1371/journal.pntd.0009502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ZD, Liu Q, Liu HH Li S, Zhang L, Zhao YK, et al. Prevalence of Cryptosporidium, microsporidia and Isospora infection in HIV-infected people: a global systematic review and meta-analysis. Parasit Vectors. (2018) 11:28. 10.1186/s13071-017-2558-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins J, Thomas J, Chandler J, M. C, Li T, Page M, et al. Cochrane Handbook for Systematic Reviews of Interventions (Version 6.3, 2022). Available online at: https://training.cochrane.org/handbook/current (accessed February, 2022).
- 25.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. (2000) 56:455–63. 10.1111/j.0006-341X.2000.00455.x [DOI] [PubMed] [Google Scholar]
- 26.Gong QL, Ge GY, Wang Q, Tian T, Liu F, Diao NC, et al. Meta- analysis of the prevalence of Echinococcus in dogs in China from 2010 to 2019. PLoS Negl Trop Dis. (2021) 15:e0009268. 10.1371/journal.pntd.0009268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Houwelingen HC, Arends LR, Stijnen T. Advanced methods in meta-analysis: multivariate approach and meta-regression. Stat Med. (2002) 21:589–624. 10.1002/sim.1040 [DOI] [PubMed] [Google Scholar]
- 28.Kamphuis C, Mollenhorst H, Hogeveen H. Sensor measurements revealed: Predicting the Gram-status of clinical mastitis causal pathogens. Comput Electron Agric. (2011) 77:86–94. 10.1016/j.compag.2011.03.012 [DOI] [Google Scholar]
- 29.Griffioen K, Cornelissen J, Heuvelink A, Adusei D, Mevius D. Jan van der Wal F. Development and evaluation of 4 loop-mediated isothermal amplification assays to detect mastitis-causing bacteria in bovine milk samples. J Dairy Sci. (2020) 103:8407–20. 10.3168/jds.2019-18035 [DOI] [PubMed] [Google Scholar]
- 30.Cannas da Silva J, Noordhuizen JP, Vagneur M, Bexiga R, Gelfert CC, Baumgartner W. Veterinary dairy herd health management in Europe: constraints and perspectives. Vet Q. (2006) 28:23–32. 10.1080/01652176.2006.9695203 [DOI] [PubMed] [Google Scholar]
- 31.Verbist B, Piessens V, Van Nuffel A, De Vuyst L, Heyndrickx M, Herman L, et al. Sources other than unused sawdust can introduce Klebsiella pneumoniae into dairy herds. J Dairy Sci. (2011) 94:2832–9. 10.3168/jds.2010-3700 [DOI] [PubMed] [Google Scholar]
- 32.Klaas IC, Zadoks RN. An update on environmental mastitis: challenging perceptions. Transbound Emerg Dis. (2018) 65 Suppl 1:166–85. 10.1111/tbed.12704 [DOI] [PubMed] [Google Scholar]
- 33.Hogan J, Smith KL. Managing environmental mastitis. Vet Clin North Am Food Anim Pract. (2012) 28:217–24. 10.1016/j.cvfa.2012.03.009 [DOI] [PubMed] [Google Scholar]
- 34.Koovapra S, Bandyopadhyay S, Das G, Bhattacharyya D, Banerjee J, Mahanti A, et al. Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect Genet Evol. (2016) 44:395–402. 10.1016/j.meegid.2016.07.032 [DOI] [PubMed] [Google Scholar]
- 35.Enferad E, Mahdavi S. Antibiotic resistance pattern and frequency of some beta lactamase genes in Klebsiella pneumoniae isolated from raw milk samples in Iran. J Hellenic Vet Med Soc. (2021) 71:2455. 10.12681/jhvms.25925 [DOI] [Google Scholar]
- 36.Salauddin M, Akter MR, Hossain MK, Rahman MM. Isolation of multi-drug resistant Klebsiella sp. from bovine mastitis samples in Rangpur, Bangladesh. J Adv Vet Anim Res. (2019) 6:362–5. 10.5455/javar.2019.f355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Audarya SD, Chhabra D, Sharda R, Gangil R, Sikrodia R, Jogi J, et al. Epidemiology of bovine mastitis and its diagnosis, prevention, and control. In:Oudessa KD, editor. Mastitis in Dairy Cattle, Sheep and Goats. London, FL: IntechOpen Press. (2021) p. Ch. 1. 10.5772/intechopen.100582 [DOI] [Google Scholar]
- 38.Munoz MA, Ahlström C, Rauch BJ, Zadoks RN. Fecal shedding of Klebsiella pneumoniae by dairy cows. J Dairy Sci. (2006) 89:3425–30. 10.3168/jds.S0022-0302(06)72379-7 [DOI] [PubMed] [Google Scholar]
- 39.de Graaf M, Beck R, Caccio SM, Duim B, Fraaij P, Le Guyader FS, et al. Sustained fecal-oral human-to-human transmission following a zoonotic event. Curr Opin Virol. (2017) 22:1–6. 10.1016/j.coviro.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Garch F, Youala M, Simjee S, Moyaert H, Klee R, Truszkowska B, et al. Antimicrobial susceptibility of nine udder pathogens recovered from bovine clinical mastitis milk in Europe 2015-2016: VetPath results. Vet Microbiol. (2020) 245:108644. 10.1016/j.vetmic.2020.108644 [DOI] [PubMed] [Google Scholar]
- 41.Bucław M. The use of inulin in poultry feeding: a review. J Anim Physiol Anim Nutr. (2016) 100:1015–22. 10.1111/jpn.12484 [DOI] [PubMed] [Google Scholar]
- 42.China, MARAPRC . Ministry of Agriculture and Rural Affairs of the People's Republic of China Announcement No. 194. July 9, 2019 [in Chinese]. Available online at: http://www.moa.gov.cn/gk/tzgg_1/gg/201907/t20190710_6320678.htm (accessed July 9, 2019).
- 43.Sudarwanto M, Akineden O, Odenthal S, Gross M, Usleber E. Extended- spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in bulk tank milk from dairy farms in Indonesia. Foodborne Pathog Dis. (2015) 12:585–90. 10.1089/fpd.2014.1895 [DOI] [PubMed] [Google Scholar]
- 44.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. J Antimicrob Chemother. (2009) 53:5046–54. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He T, Wang Y, Sun L, Pang M, Zhang L, Wang R. Occurrence and characterization of bla[[sb]]NDM-5[[/S]]-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu, China. J Antimicrob Chemother. (2017) 72:90–4. 10.1093/jac/dkw357 [DOI] [PubMed] [Google Scholar]
- 46.Evans BA, Amyes SG, OXA. β-lactamases. Clin Microbiol Rev. (2014) 27:241–63. 10.1128/CMR.00117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. (2004) 48:15–22. 10.1128/AAC.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed AM, Shimamoto T. Molecular characterization of antimicrobial resistance in Gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol Immunol. (2011) 55:318–27. 10.1111/j.1348-0421.2011.00323.x [DOI] [PubMed] [Google Scholar]
- 49.Diab M, Hamze M, Bonnet R, Saras E, Madec JY, Haenni M. OXA-48 and CTX-M-15 extended-spectrum beta-lactamases in raw milk in Lebanon: epidemic spread of dominant Klebsiella pneumoniae clones. J Med Microbiol. (2017) 66:1688–91. 10.1099/jmm.0.000620 [DOI] [PubMed] [Google Scholar]
- 50.Munoz MA, Welcome FL, Schukken YH, Zadoks RN. Molecular epidemiology of two Klebsiella pneumoniae mastitis outbreaks on a dairy farm in New York State. J Clin Microbiol. (2007) 45:3964–71. 10.1128/JCM.00795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X. Etiology investigation and drug sensitive test of the pathogenic bacteria of cow mastitis in Baotou region (in Chinese) (MS thesis: ). Northwest A&F University, Xianyang, China (2008). [Google Scholar]
- 52.Botrel MA, Haenni M, Morignat E, Sulpice P, Madec JY, Calavas D. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog. Dis. (2010) 7:479–87. 10.1089/fpd.2009.0425 [DOI] [PubMed] [Google Scholar]
- 53.Mekibib B, Furgasa M, Abunna F, Megersa B, Regassa A. Bovine mastitis: Prevalence, risk factors and major pathogens in dairy farms of Holeta Town, Central Ethiopia. Vet World. (2010) 3:397–403. 10.5455/vetworld.2010.397-403 [DOI] [Google Scholar]
- 54.Zanella G, Mikcha J, Bando E, Siqueira V, Machinski M, Jr. Occurrence and antibiotic resistance of coliform bacteria and antimicrobial residues in pasteurized cow's milk from Brazil. J Food Prot. (2010) 73:1684–7. 10.4315/0362-028x-73.9.1684 [DOI] [PubMed] [Google Scholar]
- 55.Guo CY. Studies on the cow mastitis and environment pathogenic bacteria of an intensive cow farm (in Chinese) (MS thesis: ). Inner Mongolia Agricultural University, Hohhot, China (2011). [Google Scholar]
- 56.Kalmus P, Aasmäe B, Kärssin A, Orro T, Kask K. Udder pathogens and their resistance to antimicrobial agents in dairy cows in Estonia. Acta Vet Scand. (2011) 53:1–7. 10.1186/1751-0147-53-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abera M, Habte T, Aragaw K, Asmare K. Sheferaw. D. Major causes of mastitis and associated risk factors in smallholder dairy farms in and around Hawassa, Southern Ethiopia. Trop Anim Health Prod. (2012) 44:1175–9. 10.1007/s11250-011-0055-3 [DOI] [PubMed] [Google Scholar]
- 58.Ba KM, Li XY. Isolation and identification of mastitis pathogenic bacteria and their pathogenicity and drug sensitivity tests in dairy cows in the Hohhot area (in Chinese). Mod Anim Husb. (2012) 2012:15–8. 10.14070/j.cnki.15-1150.2012.12.013 [DOI] [Google Scholar]
- 59.Haftu R, Taddele H, Gugsa G, Kalayou S. Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large-scale dairy farms of Northern Ethiopia. Trop. Anim Health Prod. (2012) 44:1765–71. 10.1007/s11250-012-0135-z [DOI] [PubMed] [Google Scholar]
- 60.Liu MC. The investigation and treatment with Chinese herbal medicine of subclinical mastitis of dairy cattle in Liuzhou (in Chinese) (MS thesis: ). Nanjing Agricultural Univesity, Nanjing, China (2012). [Google Scholar]
- 61.Wang XY, Li HS, Li JX, Wang XH, Meng JR, Yang F, et al. Identification of bacteria isolated from dairy cows with clinical type mastitis and analysis of drug resistance (in Chinese). China Anim Husb Vet Med. (2012) 39:195–8. [Google Scholar]
- 62.Kateete DP, Kabugo U, Baluku H, Nyakarahuka L, Kyobe S, Okee M, et al. Prevalence and antimicrobial susceptibility patterns of bacteria from milkmen and cows with clinical mastitis in and around Kampala, Uganda. PLoS ONE. (2013) 8:e63413. 10.1371/journal.pone.0063413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nam HM, Lim SK, Jang GC, Joung DY, Kim HJ, Lee CS, et al. Culture results from quarter milk samples submitted to veterinary diagnostic laboratories during January November 2012 in Korea. Prev Vet Med. (2013) 37:111–9. 10.13041/jpvm.2013.37.3.111 [DOI] [Google Scholar]
- 64.Thompson-Crispi KA, Miglior F, Mallard BA. Incidence rates of clinical mastitis among Canadian Holsteins classified as high, average, or low immune responders. Clin Vaccine Immunol. (2013) 20:106–12. 10.1128/CVI.00494-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramírez NF, Keefe G, Dohoo I, Sanchez J, Arroyave O, Ceron J, et al. Herd- and cow-level risk factors associated with subclinical mastitis in dairy farms from the high plains of the northern Antioquia, Colombia. J Dairy Sci. (2014) 97:4141–50. 10.3168/jds.2013-6815 [DOI] [PubMed] [Google Scholar]
- 66.Supre K, Lommelen K, De Meulemeester L. Antimicrobial susceptibility and distribution of inhibition zone diameters of bovine mastitis pathogens in Flanders, Belgium. Vet Microbiol. (2014) 171:374–81. 10.1016/j.vetmic.2014.02.045 [DOI] [PubMed] [Google Scholar]
- 67.Timofte D, Maciuca IE, Evans NJ, Williams H, Wattret A, Fick JC, et al. Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 beta-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob Agents Chemother. (2014) 58:789–94. 10.1128/AAC.00752-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuenzalida MJ, Fricke PM, Ruegg PL. The association between occurrence and severity of subclinical and clinical mastitis on pregnancies per artificial insemination at first service of Holstein cows. J Dairy Sci. (2015) 98:3791–805. 10.3168/jds.2014-8997 [DOI] [PubMed] [Google Scholar]
- 69.Langoni H, Guiduce MVS, Nóbrega DB, da Silva RC, Richini-Pereira VB, Salina A, et al. Research of Klebsiella pneumoniae in dairy herds. Pesq Vet Bras. (2015) 35:9–12. 10.1590/S0100-736X2015000100003 [DOI] [Google Scholar]
- 70.Xie LH, Ji W, Liu JH, Gao C, Qi RB, Han CL, et al. Study on the status of endogenous β-lactamase residues in raw cow's milk in Beijing (in Chinese). China Dairy. (2015) 2015:51–6. 10.16172/j.cnki.114768.2015.02.021 [DOI] [Google Scholar]
- 71.Levison LJ, Miller-Cushon EK, Tucker AL, Bergeron R, Leslie KE, Barkema HW, et al. Incidence rate of pathogen-specific clinical mastitis on conventional and organic Canadian dairy farms. J Dairy Sci. (2016) 99:1341–50. 10.3168/jds.2015-9809 [DOI] [PubMed] [Google Scholar]
- 72.Rodríguez Pérez RO. Frequency and antimicrobial susceptibility of mastitis-causing bacteria in cattle from a Conache dairy of the Laredo-Trujillo district 2015 [in Spanish] (MS thesis: ). Trujillo National University, Trujillo, Peru (2016). [Google Scholar]
- 73.Gao J, Barkema HW, Zhang L, Liu G, Deng Z, Cai L, et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. (2017) 100:4797–806. 10.3168/jds.2016-12334 [DOI] [PubMed] [Google Scholar]
- 74.Schabauer A, Pinior B, Gruber CM, Firth CL, Kasbohrer A, Wagner M, et al. The relationship between clinical signs and microbiological species, spa type, and antimicrobial resistance in bovine mastitis cases in Austria. Vet Microbiol. (2018) 227:52–60. 10.1016/j.vetmic.2018.10.024 [DOI] [PubMed] [Google Scholar]
- 75.Suleiman TS, Karimuribo ED, Mdegela RH. Prevalence of bovine subclinical mastitis and antibiotic susceptibility patterns of major mastitis pathogens isolated in Unguja island of Zanzibar, Tanzania. Trop Anim Health Prod. (2018) 50:259–66. 10.1007/s11250-017-1424-3 [DOI] [PubMed] [Google Scholar]
- 76.Tomazi T, Ferreira GC, Orsi AM, Goncalves JL, Ospina PA, Nydam DV, et al. Association of herd-level risk factors and incidence rate of clinical mastitis in 20 Brazilian dairy herds. Prev Vet Med. (2018) 161:9–18. 10.1016/j.prevetmed.2018.10.007 [DOI] [PubMed] [Google Scholar]
- 77.Wang J. Investigation on the causes of clinical mastitis in bovine, isolation, identification and drug sensitivity test (in Chinese) (MS thesis: ). Northwest A&F University, Xianyang, China (2018). [Google Scholar]
- 78.Bach KD, Sipka A, McArt JA. A Case study: evaluating quarter and composite milk sampling for detection of subclinical intramammary infections in dairy cattle. Prev Vet Med. (2019) 163:51–7. 10.1016/j.prevetmed.2018.12.013 [DOI] [PubMed] [Google Scholar]
- 79.Feng XH, Du L, Wang LF, Zhang SF, Song J, Hu ZG. Isolation and identification of Klebsiella and analysis of drug resistance from mastitis dairy cows in Shanghai and Hebei in 2017 (in Chinese). Ani Hus Feed Sci. (2019) 40:108–12. 10.16003/j.cnki.issn1672-5190.2019.01.029 [DOI] [Google Scholar]
- 80.Hozyen HF, Ibrahim ES, Khairy EA, El-Dek SI. Enhanced antibacterial activity of capped zinc oxide nanoparticles: a step towards the control of clinical bovine mastitis. Vet World. (2019) 12:1225–32. 10.14202/vetworld.2019.1225-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou MX, Zuo XX, Xv Y, Bao X, Zhang JQ, Yang ZP, et al. Isolation, identification, drug resistance, and conserved antigen analysis of mastitis pathogenic bacteria from cows in a dairy farm in Jiangsu, China (in Chinese). J Yangzhou Univ. (2019) 40:54–60. 10.16872/j.cnki.1671-4652.2019.06.009 [DOI] [Google Scholar]
- 82.Fuenzalida MJ, Ruegg PL. Molecular epidemiology of nonsevere clinical mastitis caused by Klebsiella pneumoniae occurring in cows on 2 Wisconsin dairy farms. J. Dairy Sci. (2020) 103:3479–92. 10.3168/jds.2019-17464 [DOI] [PubMed] [Google Scholar]
- 83.Jiang LZ, Xv Y, Lin ZP, Ma F, Xing GD, Zhou MX, et al. Isolation, identification, and drug resistance of recalcitrant pathogens of mastitis in a dairy farm in northern Jiangsu province (in Chinese). Acta Agric Jiangxi. (2020) 32:115–9. 10.19386/j.cnki.jxnyxb.2020.02.21 [DOI] [Google Scholar]
- 84.Abboud Z, Galuppo L, Tolone M, Vitale M, Puleio R, Osman M, et al. Molecular characterization of antimicrobial resistance and virulence genes of bacterial pathogens from bovine and caprine mastitis in Northern Lebanon. Microorganisms. (2021) 9:1148. 10.3390/microorganisms9061148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng J, Zhou M, Nobrega DB, Cao Z, Yang J, Zhu C, et al. Virulence profiles of Klebsiella pneumoniae isolated from 2 large dairy farms in China. J Dairy Sci. (2021) 104:9027–36. 10.3168/jds.2020-20042 [DOI] [PubMed] [Google Scholar]
- 86.Chung LK, Sahibzada S, Annandale HC, Robertson ID, Waichigo FW, Tufail MS, et al. Bacterial pathogens associated with clinical and subclinical mastitis in a Mediterranean pasture-based dairy production system of Australia. Res Vet Sci. (2021) 141:103–9. 10.1016/j.rvsc.2021.10.005 [DOI] [PubMed] [Google Scholar]
- 87.Deng B, Wang XX, Liu Y, Sun XH, Han YY, Feng DS. Analysis of virulence genes and drug resistance of Klebsiella pneumoniae in raw milk in Shanghai (in Chinese). China Dairy Cattle. (2021) 2021:35–40. 10.19305/j.cnki.11-3009/s.2021.06.009 [DOI] [Google Scholar]
- 88.Duse A, Persson-Waller K, Pedersen K. Microbial aetiology, antibiotic susceptibility and pathogen-specific risk factors for udder pathogens from clinical mastitis in dairy cows. Animals. (2021) 11:2113. 10.3390/ani11072113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frechette A, Fecteau G, Cote C, Dufour S. Clinical mastitis incidence in dairy cows housed on recycled manure solids bedding: a Canadian cohort study. Front Vet Sci. (2021) 8:742868. 10.3389/fvets.2021.742868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nobrega DB, Calarga AP, Nascimento LC, Chande Vasconcelos CG, de Lima EM, Langoni H, et al. Molecular characterization of antimicrobial resistance in Klebsiella pneumoniae isolated from Brazilian dairy herds. J. Dairy Sci. (2021) 104:7210–24. 10.3168/jds.2020-19569 [DOI] [PubMed] [Google Scholar]
- 91.Tsuka T, Ozaki H, Saito D, Murase T, Okamoto Y, Azuma K, et al. Genetic characterization of CTX-M-2-producing Klebsiella pneumoniae and Klebsiella oxytoca associated with bovine mastitis in Japan. Front Vet Sci. (2021) 8:659222. 10.3389/fvets.2021.659222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Y, Peng Y, Jiang J, Gong Z, Zhu H, Wang K, et al. Isolation and characterization of multidrug-resistant Klebsiella pneumoniae from raw cow milk in Jiangsu and Shandong provinces, China. Transbound Emerg Dis. (2021) 68:1033–9. 10.1111/tbed.13787 [DOI] [PubMed] [Google Scholar]
- 93.Dyson R, Charman N, Hodge A, Rowe SM, Taylor LF. A survey of mastitis pathogens including antimicrobial susceptibility in southeastern Australian dairy herds. J. Dairy Sci. (2022) 105:1504–18. 10.3168/jds.2021-20955 [DOI] [PubMed] [Google Scholar]
- 94.Yang Y, Higgins CH, Rehman I, Galvao KN, Brito IL, Bicalho ML, et al. Genomic diversity, virulence, and antimicrobial resistance of Klebsiella pneumoniae strains from cows and humans. Appl Environ Microbiol. (2019) 85:6. 10.1128/AEM.02654-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. (2015) 112:E3574–81. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. (2010) 192:3144–58. 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio. (2019) 10:2. 10.1128/mBio.00089-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanevsky-Mullarky I, Nedrow AJ, Garst S, Wark W, Dickenson M, Petersson-Wolfe CS, et al. Short communication: comparison of virulence factors in Klebsiella pneumoniae strains associated with multiple or single cases of mastitis. J Dairy Sci. (2014) 97:2213–8. 10.3168/jds.2013-7140 [DOI] [PubMed] [Google Scholar]
- 99.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. (2018) 18:37–46. 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- 100.Steeneveld W, Hogeveen H, Barkema HW, van den Broek J, Huirne RB. The influence of cow factors on the incidence of clinical mastitis in dairy cows. J Dairy Sci. (2008) 91:1391–402. 10.3168/jds.2007-0705 [DOI] [PubMed] [Google Scholar]
- 101.Podder MP, Rogers L, Daley PK, Keefe GP, Whitney HG, Tahlan K. Klebsiella species associated with bovine mastitis in Newfoundland. PLoS ONE. (2014) 9:e106518. 10.1371/journal.pone.0106518 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.