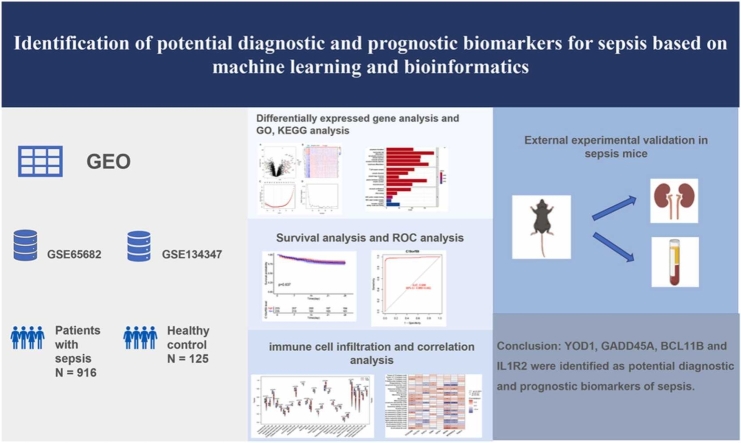
Abstract
Background
To identify potential diagnostic and prognostic biomarkers of the early stage of sepsis.
Methods
The differentially expressed genes (DEGs) between sepsis and control transcriptomes were screened from GSE65682 and GSE134347 datasets. The candidate biomarkers were identified by the least absolute shrinkage and selection operator (LASSO) regression and support vector machine recursive feature elimination (SVM-RFE) analyses. The diagnostic and prognostic abilities of the markers were evaluated by plotting receiver operating characteristic (ROC) curves and Kaplan–Meier survival curves. Gene Set Enrichment Analysis (GSEA) and single-sample GSEA (ssGSEA) were performed to further elucidate the molecular mechanisms and immune-related processes. Finally, the potential biomarkers were validated in a septic mouse model by qRT-PCR and western blotting.
Results
Eleven DEGs were identified between the sepsis and control samples, including YOD1, GADD45A, BCL11B, IL1R2, UGCG, TLR5, S100A12, ITK, HP, CCR7 and C19orf59 (all AUC>0.9). Furthermore, the survival analysis identified YOD1, GADD45A, BCL11B and IL1R2 as the prognostic biomarkers of sepsis. According to GSEA, four DEGs were significantly associated with immune-related processes. In addition, ssGSEA demonstrated a significant difference in the enriched immune cell populations between the sepsis and control groups (all P < 0.05). Moreover, YOD1, GADD45A and IL1R2 were upregulated, and BCL11B was downregulated in the heart, liver, lungs, and kidneys of the septic mice model.
Conclusions
We identified four potential immune-releated diagnostic and prognostic gene markers for sepsis that offer new insights into its underlying mechanisms.
Keywords: Sepsis, Machine learning, Diagnosis, Prognosis, Biomarker
Graphical Abstract
1. Introduction
Sepsis is a state of severe organ dysfunction caused by a disproportionate immune and inflammatory response to infection [1]. The recent Global Burden of Diseases Report states that nearly 50 million sepsis cases have occurred globally, and resulted in 10 million deaths [2]. It is also the most common cause of death in intensive care units (ICU) [3]. The pathophysiology of sepsis is complicated and involves numerous mechanisms, including pathogen invasion, cytokine release, microcirculation dysfunction, and immune imbalance [4]. Recently, sepsis has been defined as sequential organ failure assessment (SOFA) score of 2 or more in the setting of suspected infection [1]. However, the diagnosis and treatment of sepsis can be delayed due to various limitations, such as etiologic diagnosis or laboratory examination [5].
More than one hundred biomarkers have been investigated for sepsis so far [6], including acute-phase proteins, cytokines, chemokines, damage-associated molecular patterns (DAMPs), endothelial cell markers, leukocyte surface markers, non-coding RNAs, miRNA and soluble receptors, as well as metabolites and alterations in gene expression [7]. Several of these biomarkers can be used to detect sepsis onset and prognosis in the early stage. In the clinical setting, procalcitonin (PCT) and C-reactive protein (CRP) are the most common diagnostic biomarkers. Both increase transiently during sepsis but remain in circulation for sufficiently long duration for detection, and can reflect the real-time response. Nevertheless, PCT and CRP are not definitive tests for diagnosing sepsis since their levels also increase in other conditions [8], and they may be more helpful in excluding the possibility of sepsis [9], [10], [11]. Moreover, these biomarkers lack specificity and sensitivity, which limit their clinical use [12]. Therefore, it is crucial to identify novel diagnostic biomarkers that are easy to detect, inexpensive, and can be used to monitor the disease course and treatment response with high sensitivity and specificity.
In recent years, machine learning method has been used to detect and predict biomarkers [13]. A previous study showed that machine learning can accurately predict the onset of sepsis in an ICU patient 4–12 h prior to clinical recognition based on medical data [14]. Similarly, other studies have utilized several machine learning approaches to predict outcomes in patients with sepsis [15], [16], [17]. The aim of this study was to identify novel diagnostic and prognostic biomarkers, as well as potential therapeutic targets for sepsis using machine learning.
2. Materials and methods
2.1. Data acquisition and identification of differentially expressed genes
The GSE65682 [18] and GSE134347 [19] datasets including a total of 916 sepsis patients and 125 healthy controls were downloaded from the GEO database. ‘‘Limma’’ package in the R was used to identify the differentially expressed genes (DEGs) between the sepsis and control samples, with adjusted P value< 0.05 and |log2-fold change (FC)|>2 as the selection criteria. The volcano maps and heatmaps of DEGs were drawn using the ‘‘ggplot2′’ package and the ‘‘pheatmap’’ package in R respectively. Next, two machine learning algorithms were used to identify significant predictors. LASSO is executed using the "glmnet" package in R, which assists in eliminating irrelevant parameters, thus helping in the concentration of selection and regularizing the models. Regularization resolves the overfitting problem, affecting the model’s accuracy level. Lambda (λ) denotes the amount of shrinkage in the lasso regression equation. The best model is selected to minimize the binomial deviance loss. SVM-RFE algorithm was a widely used supervised machine learning protocol for classification and regression and was performed using the "e1071" package. The SVM-RFE algorithm was used to identify genes with high recognition power. The lower the root mean square error (RMSE), the higher the accuracy. A Venn diagram was drawn using the “venn” package to explore the overlapping genes between LASSO regression and SVM-RFE algorithm. Furthermore, feature importance methods, including mean decrease impurity (MDI) method and permutation importance method, were used to assess the contribution of DEGs identified by Lasso regression using Sklearn. SHapley Additive exPlanation (SHAP) method was used to assess the contribution of DEGs identified by the SVM-RFE algorithm using Sklearn.
2.2. GO, KEGG and DO enrichment analysis
The “clusterProfiler”, “enrichplot” and “ggplot2″ packages in R were used to perform GO (including Biological Process, Molecular Function, and Cellular Component), KEGG (including key related pathways) and DO analyses. P < 0.05 and adjusted P < 0.05 were set as the thresholds for GO analysis. P < 0.05 was set as the threshold for KEGG and DO analyses.
2.3. Validation of DEGs and ROC analysis
The DEGs were evaluated by the student’s t-test and P < 0.05 was considered statistically significant. ROC analysis was performed using the "pROC" package in R. And the specificity at 0.85 sensitivity, positive predictive value (PPV), and negative predictive value (NPV) of DEGs was calculated. Area under the curve (AUC) was calculated, and any value greater than 0.7 was indicative of good diagnostic performance.
2.4. Survival analysis
Based on the expression levels of the DEGs, the sepsis patients were divided into the respective high- and low-expression groups, and Kaplan-Meier survival analysis was performed using the “survival” package in R. P < 0.05 was considered statistically significant.
2.5. Construction and verification of nomogram
A nomogram was designed to predict the probability of sepsis based on the expression levels of the DEGs. A calibration curve was constructed to present the association between the predicted probabilities and the observed outcome frequencies. The predictive ability of the nomogram was determined based on the distance between the standard curve (slope of 1) and the prediction curve.
2.6. GSEA and ssGSEA
Gene functional enrichment was performed by Gene Set Enrichment Analysis (GSEA) using data from http://www.gsea-msigdb.org/gsea/index.jsp. NOM P < 0.05 was set as the threshold for significant enrichment. The immune cells associated with the candidate genes were further explored by single-sample GSEA (ssGSEA) using the " GSEABase " and " GSVA " packages. Wilcoxon rank sum test was used to compare the differences in the types of immune cells between sepsis and healthy control groups. Spearman’s analysis was used to evaluate the correlations between the DEGs and immune cells. P < 0.05 was considered statistically significant.
2.7. Establishment of sepsis model
Animal experiments were approved by Wuhan University and performed in ABSL-3 Lab (animal biosafety level-3 laboratory) of Wuhan University. Sepsis was induced by cecal ligation and puncture (CLP) surgery (n = 6). Briefly, C57BL/6 mice (male, 8 weeks) were anesthetized with pentobarbital sodium, and the cecum was ligated with a 5–0 silk suture at the juncture of the colon and cecum. The cecum was punched twice with a 22-gauge needle and the fecal material was partly squeezed out. The sham-operated mice (n = 6) only underwent laparotomy without cecal ligation and puncture. Finally, the abdominal cavity was closed and fluid resuscitation was performed by subcutaneous injection of preheated saline (37 °C, 50 μl/g body weight). The heart, liver, lungs, and kidneys were harvested 24 h after the operation and stored at −80 ℃ for further analysis. Blood was collected via intracardiac puncture. EDTA-treated blood was centrifuged at 4 ℃ in 1300 g/min to separate the serum and then wad stored at −80 °C for further measurements. In addition, IL-6 was measured to evaluate the CLP sepsis model using ELISA kit following the instructions.
2.8. Western blotting
Western blotting was performed as per standard protocols using primary antibodies for YOD1 (25370-1-AP, Protein Tech, Wuhan, China), GADD45A (A11768, ABclonal, Wuhan, China), BCL11B (A19804, ABclonal, Wuhan, China), IL1R2 (60262-1-Ig, ProteinTech, Wuhan, China) and GAPDH (60004-1-Ig, ProteinTech, Wuhan, China), and an electrochemiluminescence detection system (Tanon, China).
2.9. RT-PCR
RNA was extracted from the tissues using the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China) and reverse transcribed using the ReverTra Ace Kit (Toyobo, Osaka, Japan). The SYBR reaction mix was used for quantitative RT-PCR on the LightCycler96 Real-Time PCR Detection System (Hoffmann-La Roche Ltd., Shanghai, China). All experiments were repeated three times. The primer sequences are listed in Table S1.
2.10. Proteomics
The sera of sham and CLP groups were analyzed by proteomics using the data-independent acquisition (DIA) method. According to the mass-charge ratio (m/z), the scanning range of the mass spectrum was divided into several windows, and the parent ions in each window were broken up and detected. The fragment ion information of all mother ions was collected for qualitative and quantitative analysis. Protein extraction, protease dissociation, database establishment, mass spectrometry, DIA, quality control and protein identification were performed as per routine procedures. DIA raw data files were analyzed by DIA-NN software (V 1.8). SignalP (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) was used to predict secretory proteins.
2.11. Statistical analysis
Data were presented as mean± standard deviation (S.D) or median with ranges based on the normal distribution of the data. Student’s t-test and Mann-Whitney U-test were used to compare the mean or median values of the CLP and sham-operated groups, as appropriate. GraphPad Prism version 8.0, R software version 4.2.0 and Python version 3.11.2 were used for all statistical analyses and graph plotting. P value < 0.05 was considered statistically significant for all the analyses.
3. Results
3.1. Identification of DEGs in sepsis
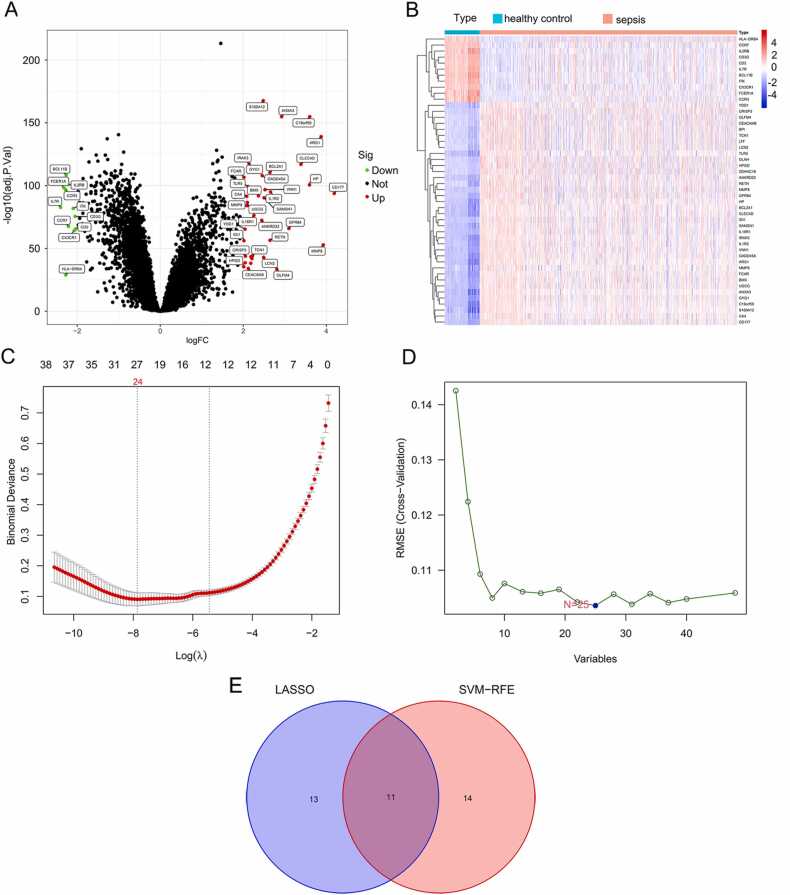
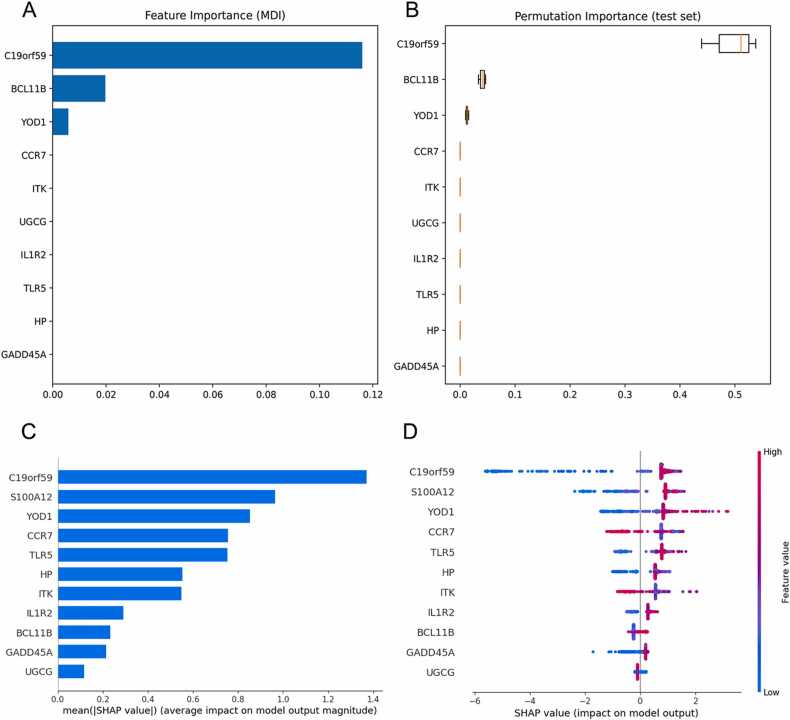
The GSE65682 and GSE134347 datasets were downloaded from the GEO database, and the DEGs between sepsis and normal samples were screened based on |log FC= ≥2 and P < 0.05. As shown in the volcano plot in Fig. 1A, there were 48 DEGs between the two groups, of which 37 genes were upregulated and 11 were downregulated in sepsis. The corresponding heatmaps are shown in Fig. 1B. As Fig. 1C showed, the LASSO algorithm identified 24 DEGs, with penalty parameter tuning conducted by 10–fold cross‐validation. As Fig. 1D showed, the SVM-RFE algorithms identified 25 genes when the RMSE was minimal. The DEGs obtained from the LASSO and SVM‐RFE models were intersected, as shown in the Venn diagram in Fig. 1E, and included YOD1, GADD45A, BCL11B, IL1R2, UGCG, TLR5, S100A12, ITK, HP, CCR7 and C19orf5. The official names of these 11 DEGs are listed in Table 1. In addition, feature importance ranked using the MDI method and permutation importance method showed the importance of these DEGs screened by the Lasso algorithm contributing to sepsis ranked from highest to lowest were C19orf59, BCL11B, YOD1, CCR7, ITK, UGCG, IL1R2, TLR5, HP, and GADD45A (Fig. 2A–B). The SHAP summary plot was depicted to identify the DEGs screened by the SVM-RFE algorithm (Fig. 2C). This plot depicts how high and low genes’ values were in relation to SHAP values. The higher the SHAP value of a gene, the more likely sepsis becomes. And the mean SHAP values ranked from highest to lowest were C19orf59, S100A12, YOD1, CCR7, TLR5, HP, ITK, IL1R2, BCL11B, GADD45A, and UGCG (Fig. 2D).
Fig. 1.
Identification of DEGs in sepsis. (A) Volcano map of all DEGs in sepsis and healthy control groups. Red plots represent up-regulated mRNAs with P < 0.05 and log2FC > 2. Green plots represent down-regulated genes with P < 0.05 and log2FC < −2. Black plots represent normally expressed mRNAs. (B) Heatmap of all DEGs. The horizontal axis represents the sample, and the vertical axis represents different genes; the red color indicates increased gene expression, and the blue indicates decreased gene expression. (C) λ 1 standard error (λ.1 SE) usually optimizes regularization so that the error and minimum error remain within a standard deviation error. Optimal λ value identified by using 10-fold cross-validation via minimum and 1-SE criteria in the LASSO regression analysis. Two marked dashed lines indicate two special lambda values λ. min and λ.1SE, and the λ between the two values is considered appropriate. λ. 1SE builds the simplest model by using fewer genes. λ. min was more accurate with using a larger number of genes. (D) SVM-RFE algorithm. The horizontal axis represents the number of DEG variables. The vertical axis represents cross-validation RMSEs. The marked plot is the number of DEGs required to obtain the optimal value. (E) Venn diagram of overlapping genes selected by Lasso and SVM-RFE algorithms.
Table 1.
Differentially expressed genes.
| Gene symbol | Official full name |
|---|---|
| YOD1 | YOD1 deubiquitinase |
| GADD45A | Growth arrest and DNA damage-inducible alpha |
| BCL11B | BAF chromatin remodeling complex subunit BCL11B |
| IL1R2 | Interleukin 1 receptor type 2 |
| UGCG | UDP-Glucose ceramide glucosyltransferase |
| TLR5 | Toll-like receptor 5 |
| S100A12 | S100 calcium-binding protein A12 |
| ITK | IL2 inducible T cell kinase |
| HP | Haptoglobin |
| CCR7 | C-C motif chemokine receptor 7 |
| C19orf59 | Microtubule-associated protein 1S |
Fig. 2.
The model's interpretation. (A) Feature importance ranked using mean decrease impurity (MDI) method. (B) Feature importance ranked using permutation importance. (C) The importance ranking of the DEGs according to the mean (|SHapley Additive exPlanations (SHAP) value|). (D) The importance ranking of the DEGs based on SHAP values. The higher SHAP value of a feature is given, the higher risk of death the patient would have. The red part in feature value represents higher value. and blue indicates that the value of a feature is low.
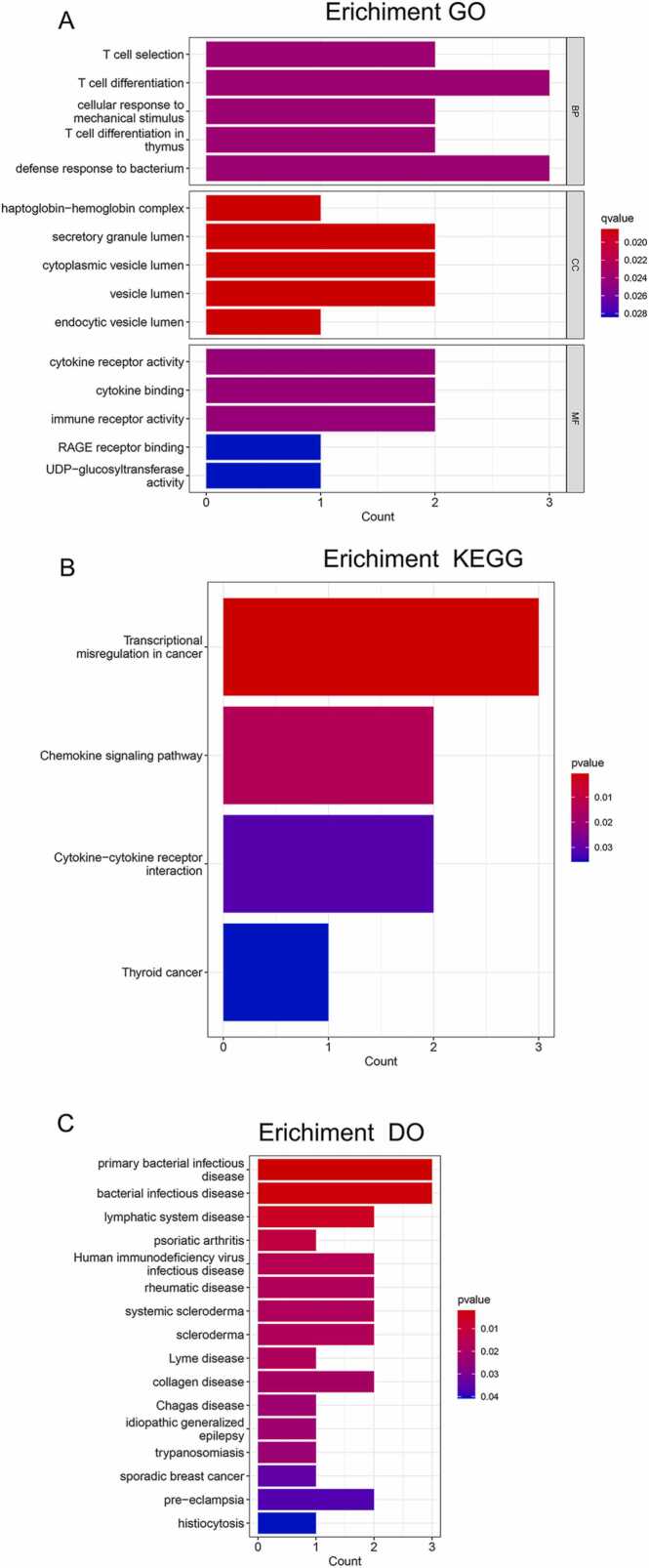
3.2. GO, KEGG and DO enrichment analysis
The 11 DEGs were functionally annotated by GO, KEGG and DO enrichment analyses. The GO enrichment analysis showed that the DEGs were mainly enriched in immune-related functions, including T cell differentiation in the subset of biological process (BP), vesicle lumen in the subset of cell compartment (CC), and immune receptor activity in the subset of molecular function (MF) (Fig. 3A, Table S2). In addition, the significantly enriched KEGG pathways included transcriptional misregulation in cancer, chemokine signaling pathway, cytokine-cytokine receptor interaction, and thyroid cancer (Fig. 3B, Table S3). Finally, the DEGs were closely related to various diseases as per the DO analysis, including dermatitis, skin disease and bacterial infectious disease (Fig. 3C, Table S4).
Fig. 3.
GO, KEGG and DO enrichment analysis. (A) GO enrichment analysis of the 11 DEGs. (B) KEGG enrichment analysis of the 11 DEGs. (C) DO enrichment analysis of the 11 DEGs.
3.3. Expression analysis of DEGs
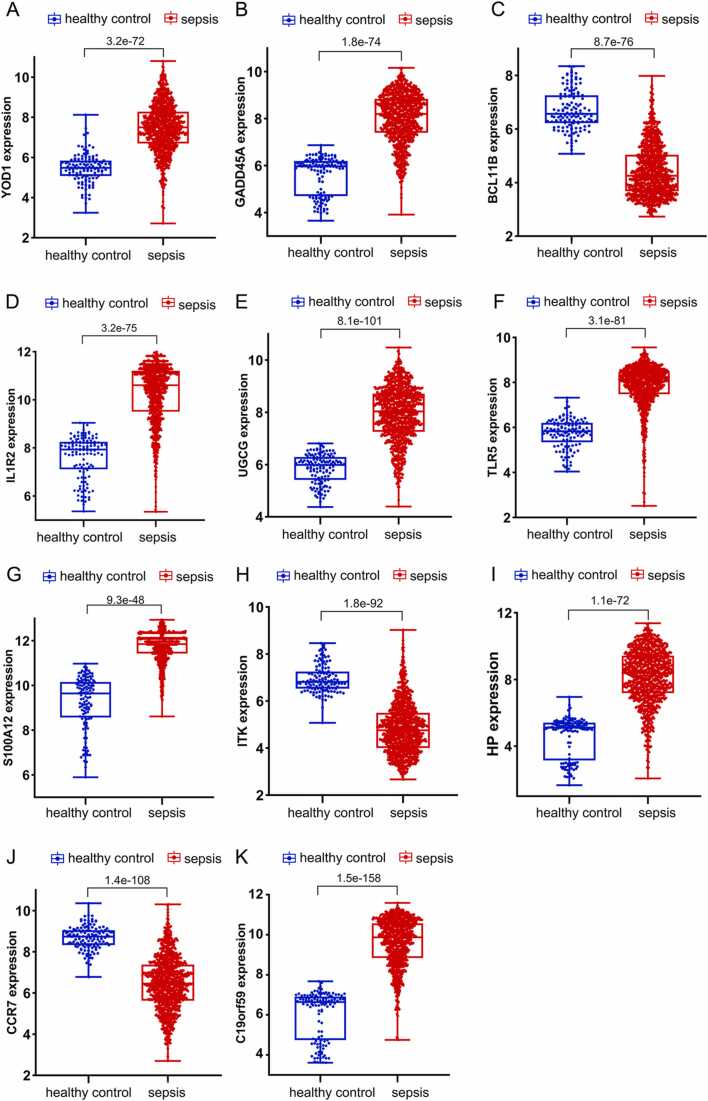
We analyzed the expression levels of the 11 candidate genes in the GSE65682 and GSE134347 datasets. As shown in Fig. 4, YOD1, GADD45A, IL1R2, UGCG, TLR5, S100A12, HP and C19orf59 were significantly upregulated in the sepsis group, whereas BCL11B, ITK and CCR7 were downregulated in the sepsis group compared to the healthy control group (P < 0.05).
Fig. 4.
Expression analysis of the 11 candidate DEGs in the GSE65682 and GSE134347 datasets between sepsis and healthy control groups. The relative expression levels of (A) YOD1, (B) GADD45A, (C) BCL11B, (D) IL1R2, (E) UGCG, (F) TLR5, (G) S100A12, (H) ITK, (I) HP, (J) CCR7 and (K) C19orf59 mRNAs are shown.
3.4. ROC analysis
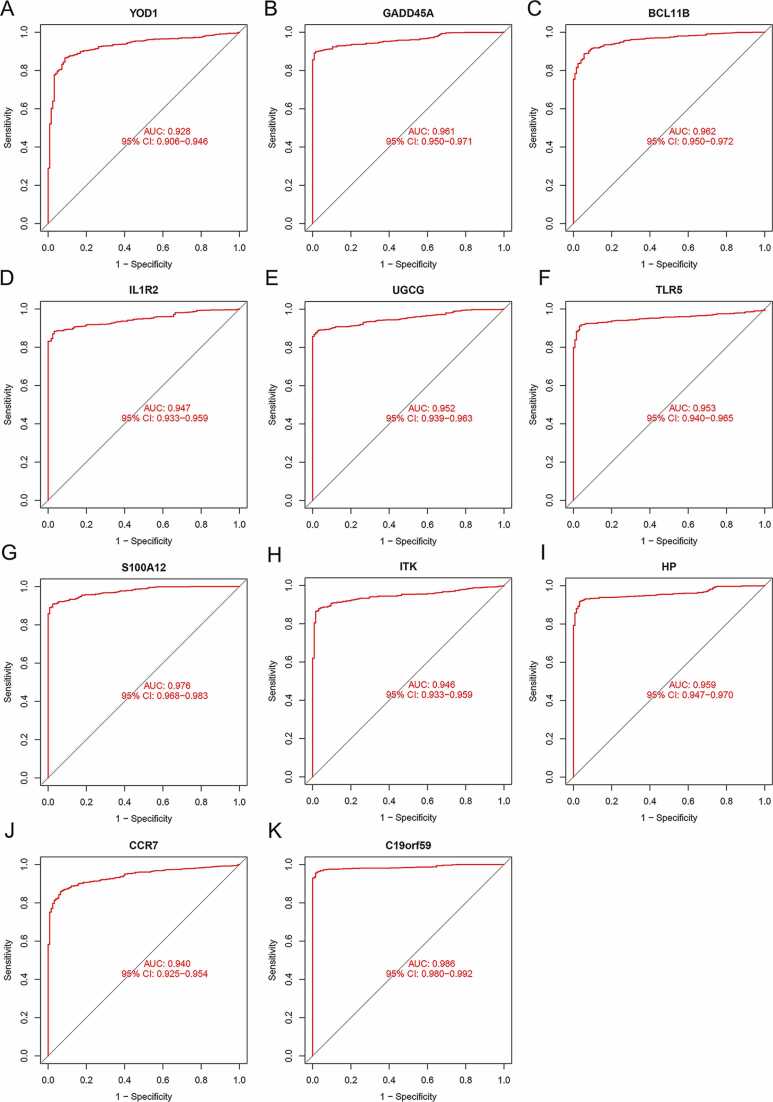
The diagnostic values of the candidate genes were analyzed by plotting ROC curves. As shown in Table 2, when the sensitivity was at 0.85, the specificity, PPV, and NPV of YOD1 in detecting sepsis were 0.912, 0.986, and 0.452, respectively. The specificity, PPV, and NPV of GADD45A were 1, 1, and 0.475, respectively. The specificity, PPV, and NPV of BCL11B were 0.960, 0.994, and 0.467, respectively. The specificity, PPV, and NPV of IL1R2 were 0.976, 0.996, and 0.469, respectively. The specificity, PPV, and NPV of UGCG were 1, 1, and 0.477, respectively. The specificity, PPV, and NPV of TLR5 were 0.984, 0.997, and 0.471, respectively. The specificity, PPV, and NPV of S100A12 were 1, 1, and 0.477, respectively. The specificity, PPV, and NPV of ITK were 0.984, 0.997, and 0.471, respectively. The specificity, PPV, and NPV of HP were 0.992, 0.999, and 0.473, respectively. The specificity, PPV, and NPV of CCR7 were 0.936, 0.990, and 0.459, respectively. The specificity, PPV, and NPV of C19orf59 were 1, 1, and 0.475, respectively. Furthermore, the AUC values of YOD1 (0.928), GADD45A (0.961), BCL11B (0.962), IL1R2 (0.947), UGCG (0.952), TLR5 (0.953), S100A12 (0.976), ITK (0.946), HP (0.959), CCR7 (0.940) and C19orf59 (0.986) were all above 0.9 (Fig. 5A-K). Taken together, these 11 DEGs had excellent diagnostic power for sepsis.
Table 2.
Specificity at 0.85 sensitivity, PPV, and NPV of these 11 DEGs.
| gene | specificity | PPV | NPV |
|---|---|---|---|
| YOD1 | 0.912 | 0.986 | 0.452 |
| GADD45A | 1.000 | 1.000 | 0.475 |
| BCL11B | 0.960 | 0.994 | 0.467 |
| IL1R2 | 0.976 | 0.996 | 0.469 |
| UGCG | 1.000 | 1.000 | 0.477 |
| TLR5 | 0.984 | 0.997 | 0.471 |
| S100A12 | 1.000 | 1.000 | 0.477 |
| ITK | 0.984 | 0.997 | 0.471 |
| HP | 0.992 | 0.999 | 0.473 |
| CCR7 | 0.936 | 0.990 | 0.459 |
| C19orf59 | 1.000 | 1.000 | 0.475 |
Fig. 5.
Diagnostic value of DEGs for sepsis in the GSE65682 and GSE134347 datasets. The ROC curves of (A) YOD1, (B) GADD45A, (C) BCL11B, (D) IL1R2, (E) UGCG, (F) TLR5, (G) S100A12, (H) ITK, (I) HP, (J) CCR7 and (K) C19orf59 are shown.
3.5. Survival analysis
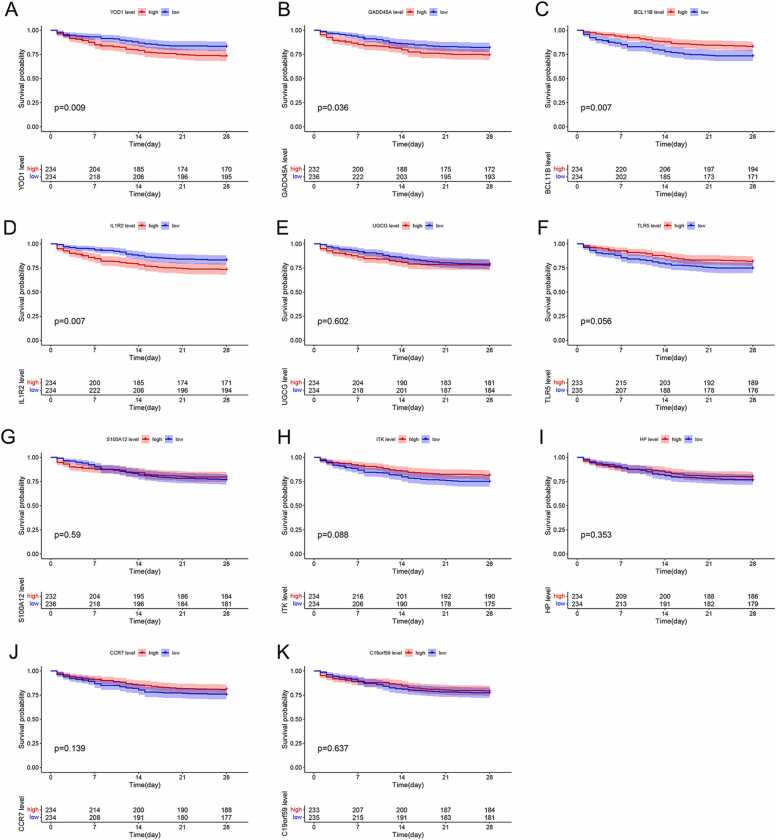
To further verify the prognostic value of the 11 genes, we demarcated 468 sepsis patients in the GSE65682 dataset into the high- and low-expression groups of the respective genes. High expression levels of YOD1 (P = 0.009), GADD45A (P = 0.036) and IL1R2 (P = 0.007) were associated with poor prognosis (Fig. 6A, B and D), whereas higher BCL11B levels was correlated to favorable prognosis (P = 0.007; Fig. 6C). Thus, YOD1, GADD45A, BCL11B and IL1R2 were identified as potential prognostic biomarkers for sepsis.
Fig. 6.
Prognostic value of DEGs for patients in the GSE65682 dataset. The Kaplan-Meier survival curves of the high- and low-expression groups of (A) YOD1, (B) GADD45A, (C) BCL11B, (D) IL1R2, (E) UGCG, (F) TLR5, (G) S100A12, (H) ITK, (I) HP, (J) CCR7 and (K) C19orf59 are shown.
3.6. Nomogram for the prediction of sepsis
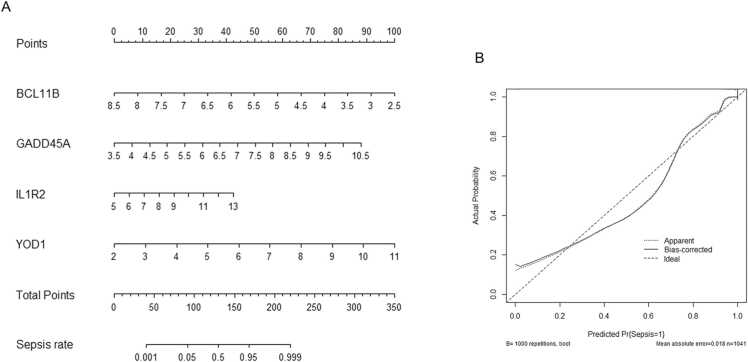
Based on the findings above, we constructed a prognostic nomogram based on the gene expression levels of YOD1, GADD45A, BCL11B and IL1R2 for risk assessment of sepsis patients (Fig. 7A). The scores calculated for each gene predicted the probability of sepsis. The overall calibration was reduced, but the apparent calibration curve and the bias-corrected curve did not deviate too far from the ideal curve. The predicted incidence of sepsis was slightly lower than the actual incidence, when the prediction probability was between 30 % and 70 %. The maximum difference in the predicted and actual incidence was seen at 60 % auxiliary diagnostic probability, and the predicted incidence was about 10 % lower (Fig. 7B).
Fig. 7.
Nomogram prediction model. (A) Nomogram to predict the sepsis rate based on the GSE65682 and GSE134347 datasets. (B) Calibration curve for the predictive ability of the nomogram.
3.7. GSEA
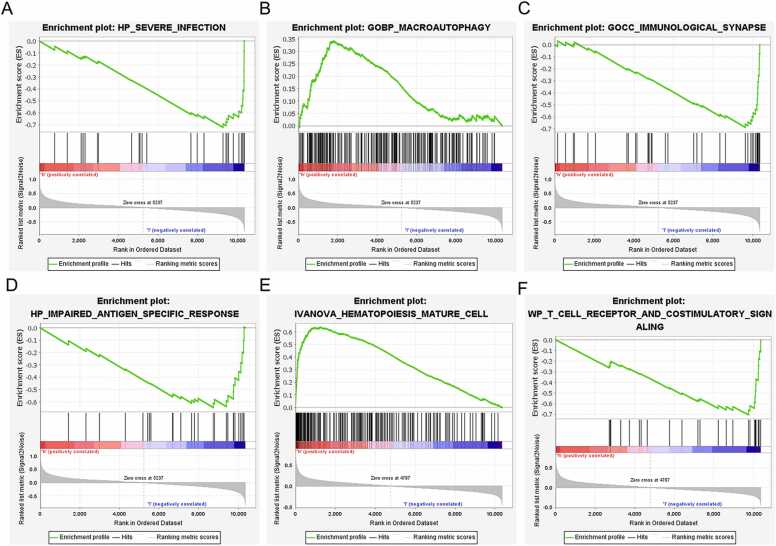
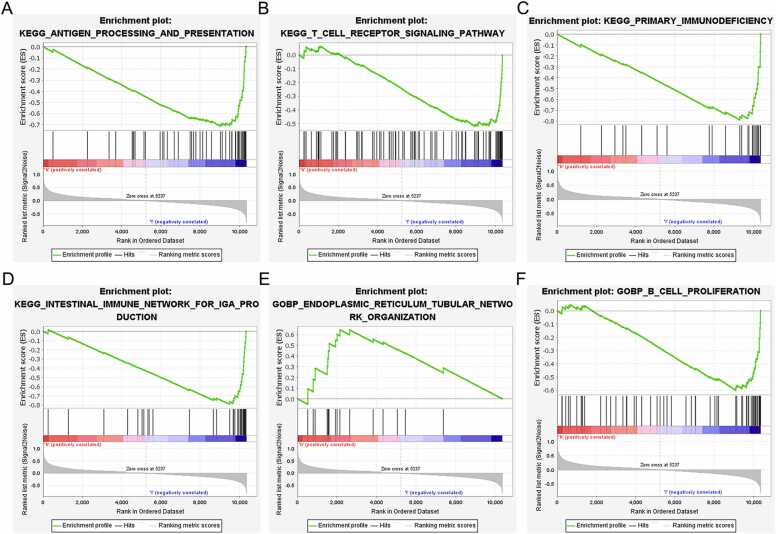
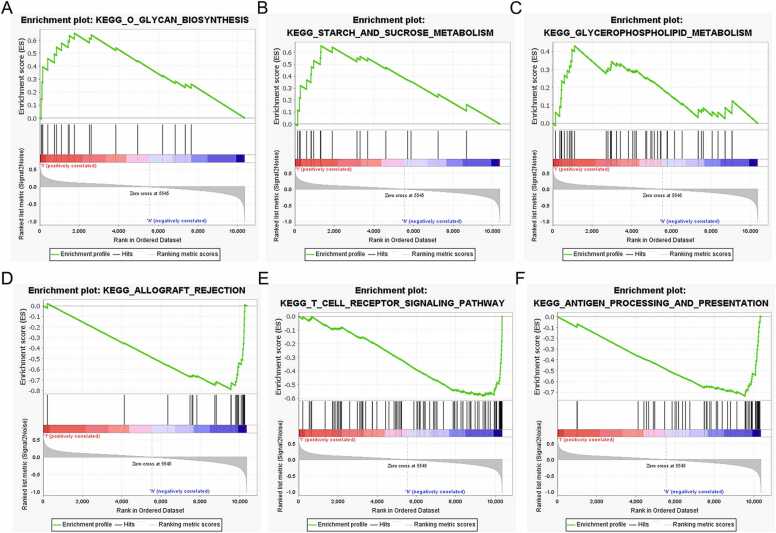
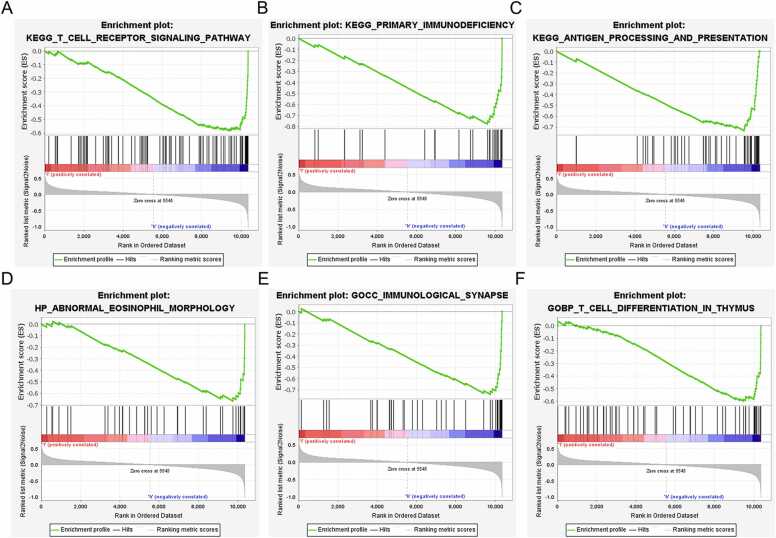
GSEA showed that YOD1 is associated with severe infection, macroautophagy, immunological synapse, impaired antigen specific response, hematopoiesis mature cell and T cell receptor signaling (Fig. 8A–F). GADD45A is primarily involved in mechanisms regarding antigen processing, T cell receptor signaling pathway, primary immunodeficiency, intestinal immune network, endoplasmic reticulum tubular network organization and B cell proliferation (Fig. 9A–F). Furthermore, BCL11B showed significant association with T cell receptor signaling pathway, antigen processing and presentation, T cell morphology, cellular defense response and T cell differentiation (Fig. 10A–F), and IL1R2 was associated with included T cell receptor, primary immunodeficiency, antigen processing and presentation, abnormal eosinophil morphology, immunological synapse and T cell differentiation in thymus (Fig. 11A–F).
Fig. 8.
GSEA results of YOD1. (A) Severe infection. (B) Macroautophagy. (C) Immunological synapse. (D) Impaired antigen specific response. (E) Hematopoiesis mature cell. (F) T cell receptor and costimulatory signaling.
Fig. 9.
GSEA results of GGADD45A. (A) Antigen processing and presentation. (B) T cell receptor signaling pathway. (C) Primary immunodeficiency. (D) Intestinal immune network for IgA production. (E) Endoplasmic reticulum tubular network organization. (F) B cell proliferation.
Fig. 10.
GSEA results of BCL11B. (A) O glycan biosynthesis. (B) Starch and sucrose metabolism. (C) Glycerophospholipid metabolism. (D) Allograft rejection. (E) T cell receptor signaling pathway. (F) Antigen processing and presentation.
Fig. 11.
GSEA results of IL1R2. (A) T cell receptor signaling pathway. (B) Primary immunodeficiency. (C) Antigen processing and presentation. (D) Abnormal eosinophil. (E) Immunological. (F) T cell Differentiation.
3.8. Immune cell enrichment analysis
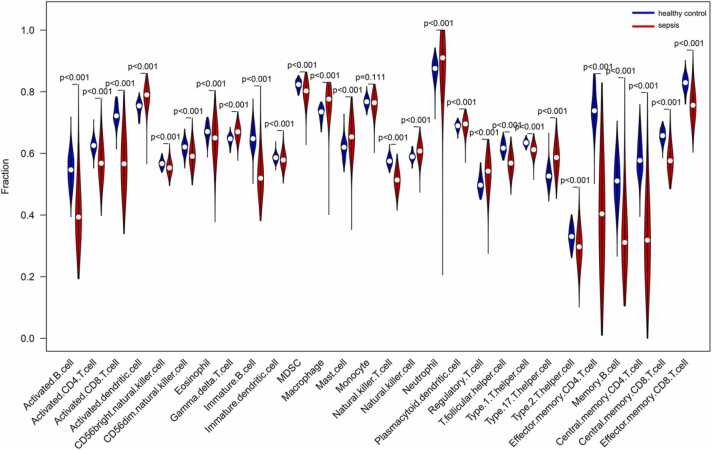
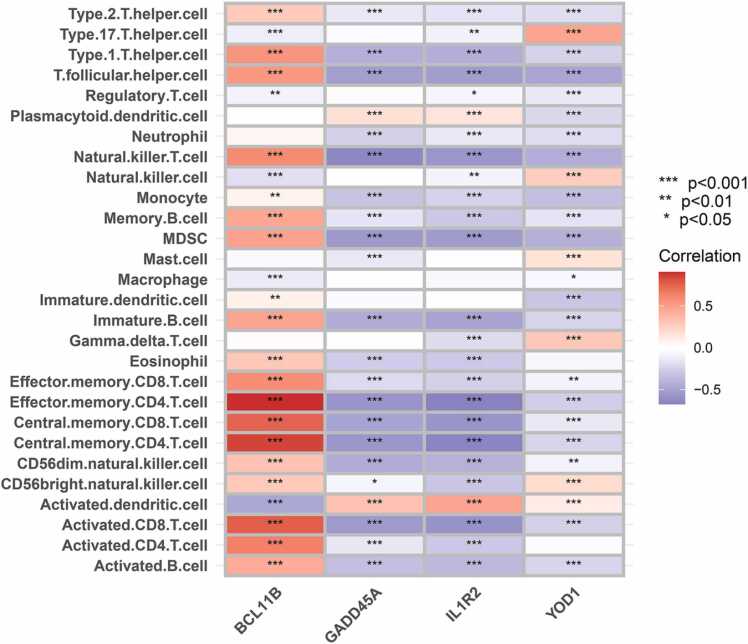
To further explore the immune functions that might involve the four sepsis biomarkers, we performed ssGSEA to analyze the relationship of these four genes with multiple immune cell populations. The sepsis group had a greater abundance of activated dendritic cells (DCs), gamma delta T cells, macrophages, mast cells, natural killer (NK) cells, neutrophils, serum cytoid DCs, regulatory T cells (Tregs) and type 17T helper (Th17) cells compared to the healthy controls. The latter had greater abundance of activated B cells, activated CD4 T cells, activated CD8 T cells, CD56bright NK cells, CD56dim NK cells, eosinophils, immature B cells, immature DCs, MDSCs, NK T cells, T follicular helper cells, type 1T helper (Th1) cells, type 2T helper (Th2) cells, effector memory CD4 T cells, memory B cells, central memory CD4 T cells, central memory CD8 T cells, and effector memory CD8 T cells compared to the sepsis group (all P values<0.001) (Fig. 12). Furthermore, BCL11B showed a significant positive correlation with the abundance of Th1 cells, NK T cells, memory B cells, MDSCs, immature B cells, effector memory CD8 T cells, effector memory CD4 T cells, central memory CD8 T cells, central memory CD4 T cells, activated CD8 T cells, activated CD4 T cells and activated B cells. On the other hand, GADD45A was negatively correlated with the abundance of NK T cells, MDSCs, effector memory CD4 T cells, central memory CD4 T cells and activated CD8 T cells. IL1R2 showed a negative correlation with effector memory CD4 T cells, central memory CD4 T cells and activated CD8 T cells, and YOD1 with Th17 cells (all P values <0.001) (Fig. 13).
Fig. 12.
Comparison of immune cell proportions between healthy control and sepsis groups.
Fig. 13.
Correlation between immune cells and the biomarker genes. Red represents a positive correlation and blue represents a negative correlation.
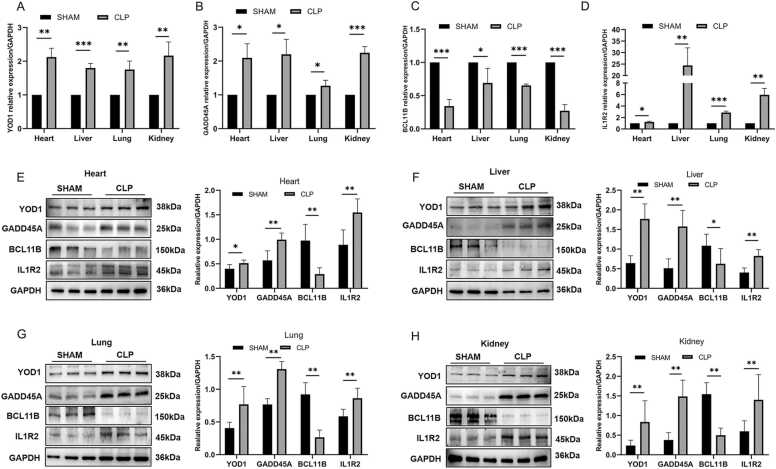
3.9. Levels of YOD1, GADD45A, BCL11B and IL1R2 in septic mice
ELISA results showed that IL-6 was upregulated in sepsis group, indicating that the sepsis model was successfully constructed (P values<0.05) (Fig. S1). In addition, the relative expression levels of YOD1, GADD45A and IL1R2 mRNAs were all significantly higher and that of BCL11B mRNA were down-regulated in the heart, liver, lung and kidney of the CLP group (all P values<0.05) (Fig. 14A-D). Similar results were observed with western blotting (all P values<0.05) (Fig. 14E–H). Furthermore, proteomics analysis showed that IL1R2 was significantly elevated in septic serum (P values<0.05; Fig. S2). However, YOD1, GADD45A and BCL11B were not been detected in the sepsis group, most likely since these proteins are secretory as validated using SignalP [20].
Fig. 14.
Validation of biomarkers in a mouse model of sepsis. (A-D) YOD1, GADD45A, BCL11B and IL1R2 mRNA levels in the heart, liver, lungs and kidneys (n = 3, compared with the Mann-Whitney test); (E-H) YOD1, GADD45A, BCL11B and IL1R2 protein levels in the heart, liver, lung and kidney tissues (n = 6, *p < 0.05, **p < 0.01, ***p < 0.001).
4. Discussion
We identified 11 potential sepsis biomarkers by screening the DEGs between healthy controls and sepsis patients in two GEO databases. While all genes showed high diagnostic sensitivity and specificity in the ROC curve analysis, only YOD1, GADD45A, BCL11B and IL1R2 were associated with patient survival, and were used to construct a predictive nomogram. GSEA showed that YOD1 is mainly involved in severe infection, macroautophagy and immune-related mechanisms, GADD45A plays a key role in endoplasmic reticulum stress (ERS) and immune-related mechanisms, and BCL11B and IL1R2 are mainly involved in immune-related processes. Furthermore, ssGSEA revealed that the four genes are closely associated with immune cells. Finally, YOD1, GADD45A and IL1R2 mRNA and protein were elevated in a mouse model of sepsis, whereas BCL11B was downregulated.
There are currently no specific biomarkers for detecting sepsis, and diagnosis largely relies on clinical data. Serum procalcitonin (PCT) and C-reactive protein (CRP) are the common biomarkers for the clinical diagnosis of sepsis [12]. However, the accuracy and validity of these biomarkers need to be further explored. A meta-analysis of 77 individual PCTS for the diagnosis of sepsis showed sensitivity and specificity of 75–85 % and 31–83 % respectively [21], [22]. Two meta-analyses (78 pure adult trials) did not consistently demonstrate the value of a single CRP assay in patients with suspected sepsis, which indicated a range of sensitivity and specificity of 60 %−80 % and 31 %−61 % respectively [21], [23]. In addition, some recent studies also found potential gene biomarkers for sepsis from GEO datasets. Li et al. [24] identified key genes (SLC2A6, C1ORF55, DUSP5 and RHOB) from GSE54514 and GSE25504 datasets by performing LASSO regression and ROC analyses. Gong et al. [25] identified nine genes (LRG1, ELANE, TP53, LCK, TBX21, ZAP70, CD247, ITK and FYN) as potential new biomarkers from three datasets (GSE95233, GSE57065, and GSE28750) by developing ROC analyses, and validated their results with RT-qPCR. Huo et al. [26] identified BCL11B and CEACAM6 as promising biomarkers for sepsis risk assessment from three GEO datasets (GSE36809, GSE37069, GSE74224, and GSE65682) by performing weighted gene co-expression network analysis (WGCNA). In addition, Lang et al. [27] had verified that IL1R2 as a diagnostic marker for sepsis. In our study, we identified YOD1, GADD45A, BCL11B and IL1R2 as biomarkers for sepsis from two datasets (GSE65682 and GSE134347) by performing machine learning methods such as LASSO regression and SVM-RFE algorithm, as well as ROC analysis and survival analysis. The prognostic genes were validated in murine sepsis model. While BCL11B and IL1R2 have also been reported elsewhere in the context of sepsis, the role of YOD1 and GADD45A in sepsis has not been studied so far. Therefore, this is the first time to study the function of YOD1 and GADD45A in sepsis, and demonstrate that the combination of these four genes can effectively diagnose sepsis and predict the prognosis.
YOD1 is a deubiquitinase [28] that binds to p97 or valosin-containing protein (VCP/p97, Cdc48 in yeast), an abundant and conserved type II ATPase [29], to clear damaged lysosomes by autophagy [30]. Furthermore, VCP/p97 and YOD1 also participate in endoplasmic reticulum-associated degradation (ERAD) [31]. However, the specific function of YOD1 in sepsis is still unknown. Our findings indicate that YOD1 expression increases significantly in sepsis and is associated with macroautophagy.
GADD45A gene belongs to the GADD45 family and encodes a ubiquitously expressed protein in normal adult and embryonic tissues. It is often induced by DNA damage and other stress signals, and regulates genes involved in growth arrest and apoptosis [32]. Activation of transcription factor-4 (ATF-4) plays a central role in cellular stress responses, such as arsenic exposure, leucine depletion, proteasome inhibition and ERS, via GADD45A induction [33], [34], [35]. Furthermore, GADD45A promotes apoptosis by upregulating p38 and JNK, and its levels increase significantly during apoptosis. Knocking down GADD45A can reduce DNA damage-induced apoptosis [36], [37]. A recent study showed that ERS mediates apoptosis in the kidneys of septic mice, and that promoting ERS can reduce apoptosis and sepsis-associated acute renal injury [38]. Other studies showed that GADD45A inhibited autophagy in tumor cells by influencing the interaction between BECN1 and PIK3C3, and induced cell death [39]. In this study, GADD45A was significantly elevated in sepsis, and associated with ERS and apoptosis in sepsis. Thus, GADD45A might play an essential role in sepsis by regulating ERS and apoptosis.
BCL11B is a transcription factor belonging to the zinc finger protein family (Cys2-His2), and regulates gene expression by inducing chromatin rearrangement and inhibiting the elongation factor B (P-TEFb) complex [40]. Previous studies have shown that BCL11B inhibited apoptosis, and knocking out the gene activated the apoptotic pathway via loss of mitochondrial membrane potential and elevation of BclxL, Caspase8 and Caspase9 [41]. BCL11B is a critical regulator of T cell differentiation, and aberrant allele-specific deregulation of BCL11B can promote leukemic development of ambiguous lineage. BCL11B is also aberrantly expressed in AIDS, cancer, cardiac hypertrophy and sepsis [42]. One study reported that BCL11B is a promising biomarker for estimating the risk of sepsis, and its high expression is associated with improved survival of sepsis patients regardless of age and endocrine type [26]. Another study showed that BCL11B repression by has-miR-150 affected the pathogenesis of neonatal sepsis [43]. In this study, high BCL11B expression indicated better survival in patients with sepsis prognosis, and functional enrichment analysis showed that BCL11B is related to the T cell immune process in sepsis.
IL1R2 (interleukin 1 receptor 2) is a decoy receptor of the interleukin 1 (IL1) receptor family that can competitively bind to IL1β and prevent its binding to IL1R1, thereby blocking IL-1β signals transduction in inflammatory diseases [44]. IL1R2 expression is restricted under normal conditions but increases rapidly during inflammation. Previous studies have shown that IL1R2 is correlated with the severity of infection in critically ill patients with sepsis, tuberculosis, acute Neisseria meningitides infection, experimental endotoxemia, surgical trauma, acute respiratory distress syndrome and necrotizing enterocolitis in premature infants [27], [45], [46], [47]. In addition, IL1R2 was reported to be a more effective marker of sepsis with Gram+ or Gram- bacterial infection compared to PCT [27]. Our results are consistent with these previous reports.
However, our study has certain limitations that ought to be considered. First, we did not study the molecular mechanism of the DEGs in sepsis. Second, all data presented in this paper are based on public datasets and were only validated in mice. Further verification is required on clinical cohorts.
5. Conclusion
We identified YOD1, GADD45A, BCL11B and IL1R2 as potential diagnostic and prognostic biomarkers of sepsis, which likely regulate sepsis-related immune processes. Our findings provide new insights into the mechanisms of sepsis, as well as potential therapeutic targets that are worth further investigation.
Author contributions
Li Ke: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. Yasu Lu and Han Gao: Methodology, Visualization, Writing – original draft. Chang Hu: Methodology, Writing – review & editing. Jiahao Zhang and Qiuyue Zhao: Validation, Writing – review & editing. Zhongyi Sun and Zhiyong Peng: Conceptualization, Writing – review & editing, Supervision. All authors have read and agreed to the published version of the manuscript. All authors reviewed and approved the final version of the manuscript.
Ethics Statement
Animal experiments in our study were reviewed and approved by the Animal Care and Use Committee of Wuhan University (IACUC: ZN2021182).
Funding
This work was funded by the National Natural Science Foundation of China (grants 81772046 and 81971816 to Dr. Peng), and the Subject Cultivation Project of Zhongnan Hospital of Wuhan University (Zhiyong Peng, No. ZNXKPY2021001).
CRediT authorship contribution statement
Li Ke: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. Yasu Lu and Han Gao: Methodology, Visualization, Writing – original draft. Chang Hu: Methodology, Writing – review & editing. Jiahao Zhang and Qiuyue Zhao: Validation, Writing – review & editing. Zhongyi Sun and Zhiyong Peng: Conceptualization, Writing – review & editing, Supervision. All authors have read and agreed to the published version of the manuscript. All authors reviewed and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors gratefully acknowledge the data provided by patients and researchers participating in GEO.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.03.034.
Contributor Information
Zhongyi Sun, Email: sunzhongyi@whu.edu.cn.
Zhiyong Peng, Email: Pengzy5@hotmail.com.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material. Levels of IL-6 in serum of sham and CLP mice
.
Supplementary material. Levels of IL1R2 in serum of sham and CLP mice
.
Data availability
All data were available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). All the experimental data analyzed and displayed in the present manuscript are available from the corresponding author upon reasonable request.
References
- 1.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) Jama-J Am Med Assoc. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr F.B., Yende S., Angus D.C. Epidemiology of severe sepsis. Virulence. 2014;5:4–11. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gotts J.E., Matthay M.A. Sepsis: pathophysiology and clinical management. Bmj-Brit Med J. 2016;353:i1585. doi: 10.1136/bmj.i1585. [DOI] [PubMed] [Google Scholar]
- 5.Angus D.C., van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 6.Pierrakos C., Vincent J.L. Sepsis biomarkers: a review. Crit Care. 2010;14:R15. doi: 10.1186/cc8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barichello T., Generoso J.S., Singer M., Dal-Pizzol F. Biomarkers for sepsis: more than just fever and leukocytosis-a narrative review. Crit Care. 2022;26:14. doi: 10.1186/s13054-021-03862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J.H., Kim D.H., Jang H.R., Kim M.J., Jung S.H., Lee J.E., et al. Clinical relevance of procalcitonin and C-reactive protein as infection markers in renal impairment: a cross-sectional study. Crit Care. 2014;18:640. doi: 10.1186/s13054-014-0640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori T., Nishiyama H., Kato H., Ikegami S., Nagayama M., Asami S., et al. Clinical value of procalcitonin for patients with suspected bloodstream infection. Am J Clin Pathol. 2014;141:43–51. doi: 10.1309/AJCP4GV7ZFDTANGC. [DOI] [PubMed] [Google Scholar]
- 10.Chan Y.L., Liao H.C., Tsay P.K., Chang S.S., Chen J.C., Liaw S.J. C-reactive protein as an indicator of bacterial infection of adult patients in the emergency department. Chang Gung Med J. 2002;25:437–445. [PubMed] [Google Scholar]
- 11.Faix J.D. Biomarkers of sepsis. Crit Rev Cl Lab Sci. 2013;50:23–36. doi: 10.3109/10408363.2013.764490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papafilippou L., Claxton A., Dark P., Kostarelos K., Hadjidemetriou M. Nanotools for sepsis diagnosis and treatment. Adv Health Mater. 2021;10 doi: 10.1002/adhm.202001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radakovich N., Nagy M., Nazha A. Machine learning in haematological malignancies. Lancet Haematol. 2020;7:e541–e550. doi: 10.1016/S2352-3026(20)30121-6. [DOI] [PubMed] [Google Scholar]
- 14.Nemati S., Holder A., Razmi F., Stanley M.D., Clifford G.D., Buchman T.G. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Crit Care Med. 2018;46:547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu C., Li L., Li Y., Wang F., Hu B., Peng Z. Explainable machine-learning model for prediction of in-hospital mortality in septic patients requiring intensive care unit readmission. Infect Dis Ther. 2022;11:1695–1713. doi: 10.1007/s40121-022-00671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu C., Li L., Huang W., Wu T., Xu Q., Liu J., et al. Interpretable machine learning for early prediction of prognosis in sepsis: a discovery and validation study. Infect Dis Ther. 2022;11:1117–1132. doi: 10.1007/s40121-022-00628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou N., Li M., He L., Xie B., Wang L., Zhang R., et al. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J Transl Med. 2020;18:462. doi: 10.1186/s12967-020-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters-Sengers H., Butler J.M., Uhel F., Schultz M.J., Bonten M.J., Cremer O.L., et al. Source-specific host response and outcomes in critically ill patients with sepsis: a prospective cohort study. Intens Care Med. 2022;48:92–102. doi: 10.1007/s00134-021-06574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scicluna B.P., Uhel F., van Vught L.A., Wiewel M.A., Hoogendijk A.J., Baessman I., et al. The leukocyte non-coding RNA landscape in critically ill patients with sepsis. Elife. 2020:9. doi: 10.7554/eLife.58597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen H. Predicting secretory proteins with signalP. Methods Mol Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 21.Simon L., Gauvin F., Amre D.K., Saint-Louis P., Lacroix J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis. 2004;39:206–217. doi: 10.1086/421997. [DOI] [PubMed] [Google Scholar]
- 22.Tan M., Lu Y., Jiang H., Zhang L. The diagnostic accuracy of procalcitonin and C-reactive protein for sepsis: a systematic review and meta-analysis. J Cell Biochem. 2019;120:5852–5859. doi: 10.1002/jcb.27870. [DOI] [PubMed] [Google Scholar]
- 23.Kondo Y., Umemura Y., Hayashida K., Hara Y., Aihara M., Yamakawa K. Diagnostic value of procalcitonin and presepsin for sepsis in critically ill adult patients: a systematic review and meta-analysis. J Intensive Care. 2019;7:22. doi: 10.1186/s40560-019-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Huang B., Yi W., Wang F., Wei S., Yan H., et al. Identification of potential early diagnostic biomarkers of sepsis. J Inflamm Res. 2021;14:621–631. doi: 10.2147/JIR.S298604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong F.C., Ji R., Wang Y.M., Yang Z.T., Chen Y., Mao E.Q., et al. Identification of potential biomarkers and immune features of sepsis using bioinformatics analysis. Mediat Inflamm. 2020;2020:3432587. doi: 10.1155/2020/3432587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huo J., Wang L., Tian Y., Sun W., Zhang G., Zhang Y., et al. Gene co-expression analysis identified preserved and survival-related modules in severe blunt trauma, burns, sepsis, and systemic inflammatory response syndrome. Int J Gen Med. 2021;14:7065–7076. doi: 10.2147/IJGM.S336785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang Y., Jiang Y., Gao M., Wang W., Wang N., Wang K., et al. Interleukin-1 receptor 2: a new biomarker for sepsis diagnosis and gram-negative/gram-positive bacterial differentiation. Shock. 2017;47:119–124. doi: 10.1097/SHK.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 28.Ernst R., Mueller B., Ploegh H.L., Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka K., Sasagawa Y., Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta. 2012;1823:130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., et al. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. Embo J. 2017;36:135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasset L., Petris G., Cesaratto F., Burrone O.R. The VCP/p97 and YOD1 proteins have different substrate-dependent activities in endoplasmic reticulum-associated degradation (ERAD) J Biol Chem. 2015;290:28175–28188. doi: 10.1074/jbc.M115.656660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amanullah A., Azam N., Balliet A., Hollander C., Hoffman B., Fornace A., et al. Cell signalling: cell survival and a Gadd45-factor deficiency. Nature. 2003;424(741):742. doi: 10.1038/424741b. [DOI] [PubMed] [Google Scholar]
- 33.Song L., Li J., Zhang D., Liu Z.G., Ye J., Zhan Q., et al. IKKbeta programs to turn on the GADD45alpha-MKK4-JNK apoptotic cascade specifically via p50 NF-kappaB in arsenite response. J Cell Biol. 2006;175:607–617. doi: 10.1083/jcb.200602149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang Q., Bhatia D., Zhang Y., Meighan T., Castranova V., Shi X., et al. Incorporation of an internal ribosome entry site-dependent mechanism in arsenic-induced GADD45 alpha expression. Cancer Res. 2007;67:6146–6154. doi: 10.1158/0008-5472.CAN-07-0867. [DOI] [PubMed] [Google Scholar]
- 35.Salvador J.M., Brown-Clay J.D., Fornace A.J. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- 36.Takekawa M., Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 37.Mita H., Tsutsui J., Takekawa M., Witten E.A., Saito H. Regulation of MTK1/MEKK4 kinase activity by its N-terminal autoinhibitory domain and GADD45 binding. Mol Cell Biol. 2002;22:4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang N., Huang R., Zhang J., Xu D., Li T., Sun Z., et al. TIMP2 mediates endoplasmic reticulum stress contributing to sepsis-induced acute kidney injury. Faseb J. 2022;36 doi: 10.1096/fj.202101555RR. [DOI] [PubMed] [Google Scholar]
- 39.Zhang D., Zhang W., Li D., Fu M., Chen R., Zhan Q. GADD45A inhibits autophagy by regulating the interaction between BECN1 and PIK3C3. Autophagy. 2015;11:2247–2258. doi: 10.1080/15548627.2015.1112484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lennon M.J., Jones S.P., Lovelace M.D., Guillemin G.J., Brew B.J. Bcl11b-A critical neurodevelopmental transcription factor-roles in health and disease. Front Cell Neurosci. 2017;11:89. doi: 10.3389/fncel.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karanam N.K., Grabarczyk P., Hammer E., Scharf C., Venz S., Gesell-Salazar M., et al. Proteome analysis reveals new mechanisms of Bcl11b-loss driven apoptosis. J Proteome Res. 2010;9:3799–3811. doi: 10.1021/pr901096u. [DOI] [PubMed] [Google Scholar]
- 42.Le Douce V., Cherrier T., Riclet R., Rohr O., Schwartz C. CTIP2, a multifunctional protein: cellular physiopathology and therapeutic implications. M S-Med Sci. 2014;30:797–802. doi: 10.1051/medsci/20143008019. [DOI] [PubMed] [Google Scholar]
- 43.Huang L., Qiao L., Zhu H., Jiang L., Yin L. Genomics of neonatal sepsis: has-miR-150 targeting BCL11B functions in disease progression. Ital J Pedia. 2018;44:145. doi: 10.1186/s13052-018-0575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Qiang J., Yang X., Wang D., Rehman A.U., He X., et al. IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv Sci. 2020;7:1901728. doi: 10.1002/advs.201901728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovach M.A., Stringer K.A., Bunting R., Wu X., San M.L., Newstead M.W., et al. Microarray analysis identifies IL-1 receptor type 2 as a novel candidate biomarker in patients with acute respiratory distress syndrome. Resp Res. 2015;16:29. doi: 10.1186/s12931-015-0190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan K.Y., Leung F.W., Lam H.S., Tam Y.H., To K.F., Cheung H.M., et al. Immunoregulatory protein profiles of necrotizing enterocolitis versus spontaneous intestinal perforation in preterm infants. Plos One. 2012;7 doi: 10.1371/journal.pone.0036977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller B., Peri G., Doni A., Perruchoud A.P., Landmann R., Pasqualini F., et al. High circulating levels of the IL-1 type II decoy receptor in critically ill patients with sepsis: association of high decoy receptor levels with glucocorticoid administration. J Leukoc Biol. 2002;72:643–649. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material. Levels of IL-6 in serum of sham and CLP mice
Supplementary material. Levels of IL1R2 in serum of sham and CLP mice
Data Availability Statement
All data were available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). All the experimental data analyzed and displayed in the present manuscript are available from the corresponding author upon reasonable request.