Abstract
CRISPR–Cas9 genome editing technologies have enabled complex genetic manipulations in situ, including large-scale, pooled screening approaches to probe and uncover mechanistic insights across various biological processes. The RNA-programmable nature of CRISPR–Cas9 greatly empowers tiling mutagenesis approaches to elucidate molecular details of protein function, in particular the interrogation of resistance mechanisms to small molecule drugs, an approach termed CRISPR-suppressor scanning. In a typical CRISPR-suppressor scanning experiment, a pooled library of single-guide RNAs is designed to target across the coding sequence(s) of one or more genes, enabling the Cas9 nuclease to systematically mutate the targeted proteins and generate large numbers of diverse protein variants in situ. This cellular pool of protein variants is then be challenged with drug treatment to identify mutations conferring a fitness advantage. Drug resistance mutations identified with this approach not only can elucidate drug mechanism of action, but also can reveal deeper mechanistic insights into protein structure-function relationships. In this protocol, we outline the framework for a standard CRISPR-scanning experiment. Specifically, we provide instructions for the design and construction of a pooled sgRNA library, execution of a CRISPR-scanning screen, and basic computational analysis of the resulting data.
Basic Protocol 1:
Design and generation of a pooled sgRNA library.
Support Protocol 1:
sgRNA library design using command-line CRISPOR.
Support Protocol 2:
Pooled sgRNA library lentivirus production and titering.
Basic Protocol 2:
Execution and analysis of a CRISPR-suppressor scanning experiment.
Keywords: CRISPR, functional genomics, genetic engineering, tiling mutagenesis, CRISPR-suppressor scanning
INTRODUCTION:
Drug resistance mutations pose key challenges in the clinic and have been instrumental in advancing understanding of biological systems (Freedy and Liau, 2021). Because of their ability to suppress or even reverse the effects of therapeutics, resistance mutations are of considerable concern in drug discovery and development efforts. Nevertheless, identifying such mutations can also provide important insight. Mutations in a target gene conferring resistance to a drug serve as gold-standard evidence of on-target engagement (Schenone et al., 2013). When the protein target is known, resistance mutations can further inform understanding of their structure-function, such as mechanisms of allosteric regulation (Kapoor and Miller, 2017; Freedy and Liau, 2021). In a seminal example, resistance mutations to the ABL kinase inhibitor imatinib were identified outside of the drug-binding pocket, ultimately revealing an allosteric autoinhibitory mechanism controlling ABL activity (Azam et al., 2003; Freedy and Liau, 2021). Crucially, analysis of these distal resistance mutations provided insight into this mechanism prior to the availability of full structural information.
Given the utility of drug resistance mutations, methods to systematically identify them will greatly bolster both basic and translational biology. Starting from a given drug molecule, a key question is which mutations in the target protein (or in interacting proteins for complex members) confer resistance. This can in turn reveal the binding site of the molecule or allosteric regions of the protein crucial to drug function. In such cases, it is inefficient to search genome-wide for naturally occurring mutations that arise spontaneously in response to drug treatment. A more fitting approach would be a targeted mutagenesis strategy that can comprehensively generate mutations in the predefined drug target and preferably in situ.
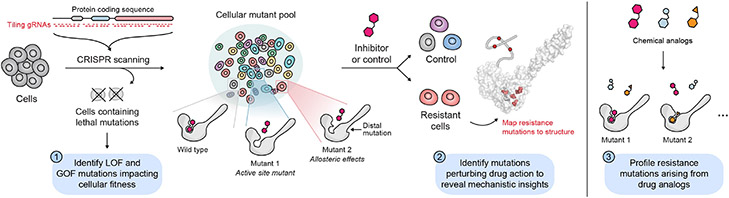
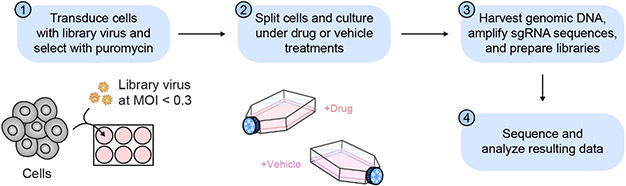
In this context, CRISPR–Cas9 genome editing enables facile RNA-programmable mutagenesis of desired genomic loci, resulting in the systematic generation of diverse mutations. Despite the widespread adoption of CRISPR technologies throughout biology, a relatively underappreciated application is the dissection of protein function through CRISPR–Cas9 tiling mutagenesis, termed CRISPR-scanning (Shi et al., 2015). Here, a library of single-guide RNAs (sgRNAs) tiling across the coding sequence (CDS) of one or more genes is introduced in pooled fashion into cells (Fig. 1). Cas9 cleavage produces double-stranded breaks whose cellular repair in turn generates a spectrum of mutations at each targeted site. The majority of these are insertion/deletion (indel) mutations, including in-frame mutations (Donovan et al., 2017; Ipsaro et al., 2017). Thus, the target protein is systematically mutagenized in situ to create a diverse cellular pool of protein variants, which can be exploited in a variety of ways to study protein sequence-function. For instance, monitoring changes in sgRNA representation as cells are cultured over time can reveal mutations conferring loss-of-function (LOF) or gain-of-function (GOF) effects (Shi et al., 2015; Shen et al., 2015).
Figure 1. Overview of CRISPR-suppressor scanning.
Schematic of a typical CRISPR-suppressor scanning experiment highlighting key insights that can be gained: (1) analysis of sgRNA dropout (e.g., in control vehicle-treated cells) can yield information on how mutations impact cellular fitness, (2) analysis of sgRNA enrichment in drug-treated versus vehicle-treated cells can reveal mutations perturbing drug action, (3) comparing resistance profiles of structurally related drugs can illuminate structure-function relationships.
Importantly, these mutations can also give rise to drug resistance (Ipsaro et al., 2017; Donovan et al., 2017). Thus, by challenging this pool of mutated cells with drug treatment, it is possible to systematically identify mutations conferring resistance. This approach, termed CRISPR-suppressor scanning, has been demonstrated to reveal mechanistic insight into drug mechanism of action as well as target protein function (Ipsaro et al., 2017; Vinyard et al., 2019; Gosavi et al., 2022). Notably, resistance mutations nominated by CRISPR-suppressor scanning can indeed occur far away from the drug binding pocket, even in interacting proteins that do not directly bind the drug (Kwok et al., 2022; Ngan et al., 2022). Drug treatment can additionally be multiplexed in order to reveal drug-specific resistance profiles (Vinyard et al., 2019), opening the door to deep exploration of protein-ligand structure-function.
Here, we outline the basic framework for performing a pooled CRISPR–Cas9 tiling mutagenesis screen with drug selection to identify drug resistance mutations, which we refer to as CRISPR-suppressor scanning. In the first protocol, we describe the design and generation of a pooled sgRNA library. We provide a support protocol to aid automated design of more complex sgRNA libraries. In the second protocol, we describe the execution of a pooled CRISPR-suppressor scanning experiment and analysis of the resulting dataset for follow-up and validation experiments. We also provide a support protocol for production and functional titering of sgRNA library lentivirus.
STRATEGIC PLANNING:
Overview.
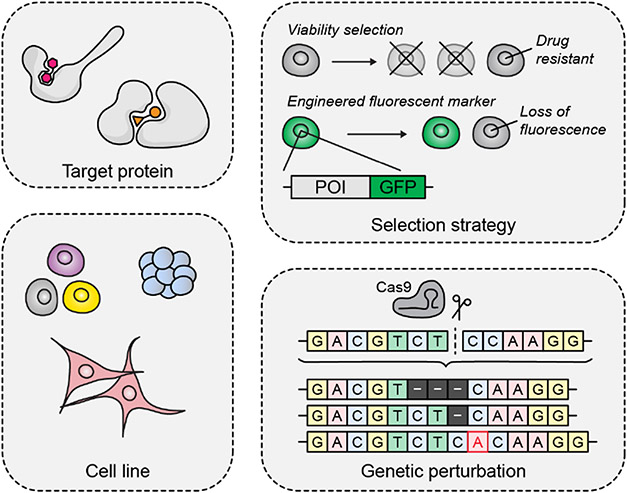
The duration of a CRISPR-suppressor scanning experiment can often take up to 8 weeks depending on the timescale required for drug selection. An additional 2 weeks should be allocated for upstream validation and downstream data analysis. Consequently, the parameters of a CRISPR-suppressor scanning experiment should be carefully evaluated to maximize the chances of a successful screen. In this section, we outline some critical considerations to consider before starting a CRISPR-suppressor scanning experiment (Fig. 2).
Figure 2. Key considerations in planning a CRISPR-suppressor scanning experiment.
Before conducting a CRISPR-suppressor scanning experiment, the target protein and associated selection strategy should be chosen carefully, as well as the cell line model system and form of genetic perturbation employed.
Target and Selection Strategy Considerations.
In designing a CRISPR-suppressor scanning experiment, it is critical to carefully choose the target protein(s) and the selection strategy used to screen for mutations of interest. The choice of selection strategy is dictated by the biological question posed by the study. Most commonly, we screen protein targets using selective inhibitors (or other chemical modulators) that cause defects in cell proliferation, or even death, because resistance then rescues growth in the presence of drug. Thus, enrichment of resistant cells occurs naturally over time without additional manipulation. However, CRISPR-suppressor scanning can also be applied to situations where target inhibition does not significantly affect fitness. In this case, phenotypic readouts such as antibody staining and cell sorting can be used. Additionally, it is possible to engineer a cell line to couple drug resistance to a selection marker, such as a fluorescent tag or antibiotic resistance gene. In such cases, the readout should be designed to minimize false-positive signals due to nonspecific effects and rigorously validated to ensure that it faithfully reports on the desired phenotype.
A related consideration is the stringency of selection, which in this context refers to the drug concentration used for selection. Stringent selections typically enrich for a small set of sgRNAs that confer a significant fitness advantage in the presence of drug, such as complete growth rescue. Such selections may be appropriate for experiments seeking to identify the drug binding site of a known target. However, stringent selections may bottleneck the population, leading to greater variance across replicates and making it more challenging to screen cell lines that are sensitive to cell density. In such cases, a lower dosage of drug can be used to achieve a weaker selection. This can also be advantageous in selecting for more subtle partial resistance phenotypes that would otherwise be outcompeted under more stringent selections, with the caveat that an abundance of hits may require triaging to limit the number of sgRNAs selected for follow-up validation to a reasonable number. Weak selections can also lead to poor signal-to-noise ratios if the enrichment of true positive sgRNAs is not sufficient to identify them with high confidence. In such cases, extending the duration of the screen can enable the expansion of the desired phenotypic subpopulations. Alternatively, a gradual dose escalation can also promote enrichment of more fit variants in a gentler manner by slowly shifting the fitness landscape of the population over time. It is generally prudent to characterize the dose–response relationship of the cell line of interest with respect to the desired drug. While the stringency of selection is experiment-specific and must be determined empirically, concentrations of drug leading to >90% growth inhibition (GI90) would be considered highly stringent. Weaker selections may start at GI50 or even GI30 concentrations.
Cell Line Considerations.
Before beginning a screen, it is important to characterize the response of the chosen cell line to drug treatment. For standard resistance screens, it is important to understand what concentration of drug will inhibit growth of wild-type cells (see above). This can be accomplished through a cell viability assay under varying drug concentrations. If possible, a positive control in the form of a known resistant mutant should also be confirmed. It is also critical to evaluate the properties of the chosen cell line, including seeding density, cell density at confluency, and doubling time, since the growth characteristics of the cell line have significant practical and logistical implications on the resources required to execute the screen. For example, cell lines that are highly sensitive to seeding density or culture conditions may not tolerate more stringent drug selections. The minimum number of cells required to achieve the desired coverage of the sgRNA library should also be calculated beforehand to estimate the resources, including drug stocks, growth medium, and consumables, needed to perform the screen.
Genetic Perturbation Considerations.
CRISPR-suppressor scanning leverages Cas9 nuclease to generate the mutational diversity upon which selection is carried out. Cas9 predominately generates indel mutations, and thus is often sufficient for experiments where the exact type and size of the induced mutation is not a significant concern. Such approaches using Cas9 mutagenesis have been highly successful at identifying in-frame indel resistant alleles (Shi et al., 2015; Ipsaro et al., 2017; Donovan et al., 2017; Neggers et al., 2018; Vinyard et al., 2019; Gosavi et al., 2022). However, there may be specific applications in which it is advantageous to use other CRISPR-based technologies. For example, studies aimed at profiling clinically observed point mutations may benefit from using base editors (Komor et al., 2016). This protocol can be adapted to use alternate CRISPR technologies, with changes to library design and analysis as has been discussed elsewhere (Hanna et al., 2021). However, these tools may introduce greater degrees of complexity that require more extensive validation and testing before performing a screen. Additionally, the chosen perturbation should be validated to ensure that it is compatible with the cell line of interest. For example, some cell lines may be difficult to transduce, thereby restricting the use of certain perturbations or limiting the scope of the screen.
We typically use lentiviral transduction to deliver sgRNAs alongside Cas9 into cells for the screen. It is highly recommended to optimize transduction conditions in the chosen cell line before starting the screen. Because the efficiency of lentiviral delivery and downstream genetic perturbation can vary greatly between cell lines, the desired perturbations should be validated and optimized in the chosen cell line before starting the screen. A simple validation workflow for Cas9 nuclease activity generally involves transducing the cell line of interest with an individual sgRNA and then performing next-generation sequencing (NGS) of the targeted locus to evaluate cutting efficiency at the DNA level and Western blot or flow cytometry analysis of the perturbation at the protein level. We commonly transduce cells with a vector containing GFP and a GFP-targeting sgRNA to monitor the perturbation over time with flow cytometry.
BASIC PROTOCOL 1:
Basic protocol title:
Design and generation of a pooled sgRNA library.
Introductory paragraph:
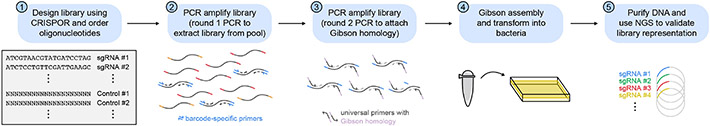
This protocol describes the steps for designing and generating a pooled sgRNA library for use in a CRISPR-suppressor scanning experiment (Fig. 3), and is adapted from previous CRISPR screening protocols (Canver et al., 2018; Joung et al., 2017). More specifically, this protocol outlines how to design and select sgRNAs using the CRISPOR web tool (Concordet and Haeussler, 2018), clone the pooled sgRNA library into a lentiviral vector using Gibson assembly, and validate the sgRNA representation in the library by next-generation sequencing (NGS). This protocol assumes the use of wild-type Streptococcus pyogenes Cas9 (SpCas9) as the Cas nuclease to induce double-stranded breaks and a 1-plasmid system in which both SpCas9 and the sgRNA are expressed from the same lentiviral vector (lentiCRISPRv2, Addgene #52961). However, this protocol can be adapted for the use of alternative Cas nucleases (e.g., S. aureus Cas9, engineered Cas9 variants), genetic perturbations (e.g., base editing), and vector designs (e.g., 2-plasmid system with Cas9 and sgRNA expressed separately). See Strategic Planning for additional discussion. For complex libraries involving multiple genes or genes with many exons, users may wish to automate the sgRNA library design process using command-line CRISPOR (see Support Protocol 1).
Figure 3. Construction of an sgRNA library.
Schematic of Basic Protocol 1, in which an sgRNA library is designed using CRISPOR, amplified through two rounds of PCR, assembled into lentiCRISPRv2 through Gibson assembly, and transformed into bacteria. The plasmid library DNA is then purified and validated by deep sequencing to confirm library representation.
Materials:
Molecular Biology Grade Water (Corning, cat. no. 46-000-CV)
NEBNext Ultra II Q5 Master Mix (New England Biolabs, cat. no. M0544)
Agarose RA (VWR, cat. no. N605-500G)
TAE buffer (50×) (Boston BioProducts, cat. no. BM-250)
100 bp DNA Ladder (New England Biolabs, cat. no. N3231)
Zymoclean Gel DNA Recovery kit (Zymo, cat. no. D4007)
lentiCRISPRv2 (Addgene, cat. no. 52961)
FastDigest Esp3I (Thermo Fisher Scientific, cat. no. FD0454)
FastAP Thermosensitive Alkaline Phosphatase (Thermo Fisher Scientific, cat. no. EF0654)
Dithiothreitol (Thermo Fisher Scientific, cat. no. R0861)
1 kb DNA Ladder (New England Biolabs, cat. no. N3232)
NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, cat. no. E2621)
Isopropanol (VWR, cat. no. BDH1133-1LP)
GlycoBlue Coprecipitant (Invitrogen, cat. no. AM9515)
5 M NaCl (Research Products International, cat. no. S24600-500.0)
Absolute Ethanol (Decon Labs, cat. no. V1016, CAS no. 64-17-5)
TE buffer (Boston BioProducts, cat. no. BM-304A)
LB Agar (Invitrogen, cat. no. 22700-025)
Ampicillin (VWR, cat. no. 102587-686)
245-mm Square BioAssay Dish (Corning, cat. no. 431272)
94-mm Petri Dish (VWR, cat. no. 82050-586)
Endura DUO Electrocompetent Cells (Biosearch Technologies, cat. no. 60242)
Recovery Medium for Endura Cells (Biosearch Technologies, cat. no. 80026-1)
LB Medium (Research Products International, cat. no. L24060)
ZymoPURE II Plasmid Maxiprep Kit (Zymo, cat. no. D4202)
Qubit dsDNA High Sensitivity Assay Kit (Invitrogen, cat. no. Q32851)
Microcentrifuge Tubes (VWR, cat. no. 20170-038)
PCR 8-well Strip Tubes with Domed Caps (VWR, cat. no. 53509-304)
ProFlex Thermocycler (Applied Biosystems, cat. no. 4484073)
Owl EasyCast B1A Mini Gel Electrophoresis System (Thermo Fisher Scientific, cat. no. B1A)
Razor (VWR, cat. no. 55411-050)
NanoDrop OneC UV-Vis Spectrophotometer (Thermo Fisher Scientific, cat. no. ND-ONEC-W)
Vortex-Genie 2 (Scientific Industries, cat. no. SI-0236)
Microcentrifuge (Eppendorf 5424R)
Electroporation Cuvettes (BTX, cat. no. 45-0124)
Electroporator (Eppendorf Eporator)
14-ml Culture Tubes (Corning, cat. no. 352057)
Shaking Incubator (New Brunswick Scientific Excella E24, Eppendorf, cat. no. M1352)
Cell Spreader (VWR, cat. no. 76207-748)
50 ml Conical Tube (VWR, cat. no. 89039-656)
Benchtop Centrifuge (Eppendorf 5810R)
Qubit 3 Fluorometer (Invitrogen, cat. no. Q33216)
Protocol steps with step annotations:
Design of a custom pooled sgRNA library with CRISPOR.
Open the CRISPOR webtool (http://crispor.tefor.net/). For each exon of the gene(s) to be targeted, perform steps 2–4 below.
- Submit a CRISPOR job to design sgRNAs against the exon:
- Under “Step 1,” enter the exon sequence with 20 bp flanking intronic sequences on each side either as a pasted sequence or as genomic coordinates.
- Under “Step 2,” select the genome assembly appropriate to the cell line used. For example, ‘GRCh38/hg38’ should be chosen for human cell lines.
- Under “Step 3,” select the ‘20bp-NGG’ protospacer-adjacent motif, which corresponds to SpCas9.
- Finally, submit the job to design sgRNAs.Ensure that the genomic sequence of the target gene (and not the coding sequence) is used, as libraries designed against the latter may contain sgRNAs spanning splicing junctions, which therefore will not target the genomic sequence. Beyond exonic sequences, 20 bp flanking intronic sequences should be included so that sgRNAs with protospacers extending into the intron but predicted to cut within the exon are not excluded. Each discontinuous sequence (i.e., exon) should be processed separately and the CRISPOR outputs merged later (see Step 5 below).
- After generating each list of sgRNAs, click the ‘Saturating mutagenesis assistant’ link at the top of the table, which will bring you to a second page to set optional parameters and export the CRISPOR output files. At this stage, the most important parameters to consider are:
- Subpool barcodes: The subpool barcode are universal priming sequences appended to either side of the designed sgRNAs for amplification from the synthesized oligonucleotide pool. Pooled oligonucleotide synthesis often requires a minimum number of oligonucleotides and is generally cheaper at scale, so it is common to order multiple pooled sgRNA libraries in the same oligonucleotide pool. The subpool barcode thus enables selective amplification of a specific subset of sgRNAs from the pool.All sgRNAs across multiple exons that will be merged into the same library should be tagged with the same subpool barcode. Ensure that separate sgRNA libraries (e.g., libraries targeting different genes) are marked with distinct barcodes.
- Filters: The CRISPOR sgRNA design tool allows filtering of sgRNAs that do not meet minimum threshold scores for specificity and cutting efficiency. The specificity score is a predicted measure of sgRNA off-target activity, where greater scores signify lower probabilities of off-target cleavage elsewhere in the genome. The efficiency score is a predicted measure of on-target sgRNA cutting activity, where greater scores signify greater probabilities of on-target cleavage at the predicted cut site.Filtering sgRNAs based on scoring criteria is optional but can be prudent depending on the experimental context. We often exclude sgRNAs with specificity scores <20.
- Under the ‘File format’ parameter, select ‘Default: Excel for A and text for B/C/D’ and then click ‘Get A – oligo list’ to download the list of sgRNA sequences and their oligonucleotide sequences for synthesis.You may also download the other files which contain supplementary information and sequences, such as primers for cleavage validation, but these are out of the scope of this protocol.
Merge the output sgRNA/oligonucleotide lists corresponding to each exon of the target gene(s) into a single master spreadsheet. Ensure that there are no duplicated sgRNAs and that all sgRNAs cut within the CDS of the target gene(s).
- Confirm that the full-length oligonucleotide sequences (96–99 bp) under the ‘Oligonucleotide’ column of the exported sgRNA oligo list contain:
- The correct flanking subpool barcode(s) for selective amplification of the desired sgRNA library from the synthesized oligonucleotide pool.All oligonucleotides corresponding to sgRNAs in a given library should contain the same subpool barcode. If multiple libraries are merged into one pooled oligonucleotide order, ensure each subset of sgRNAs are marked with the appropriate distinct barcodes. The full-length oligonucleotide sequences are designed in the following format: 5’–[5’ barcode]–[5’ homology arm]–[sgRNA protospacer]–[3’ homology arm]–[3’ barcode]–3’
- The correct homology arms for Gibson Assembly cloning into the destination vector.The default homology arms appended by CRISPOR are designed for insertion of the sgRNA protospacer between the human U6 promoter (5’) and SpCas9 sgRNA scaffold (3’) sequences corresponding to the commonly used lentiCRISPRv2 (Addgene #52961) and lentiGuide-Puro (Addgene #52963) sgRNA lentiviral vectors. The homology arm sequences can be replaced if a different vector is used.
- Add positive and negative control sgRNAs to the list of sgRNA library sequences and generate the full-length sgRNA oligonucleotide sequences by appending the same flanking subpool barcodes and homology arms as in Step 6.
- Positive controls: sgRNAs targeting essential genes can be used as positive controls for on-target Cas9 activity. Such sgRNAs should be highly depleted in both drug- and vehicle-treated conditions and thus serve as a quality control check for proper Cas9 nuclease behavior and sgRNA depletion. We typically include 3 sgRNAs targeting EEF2, a known pan-essential gene (Wang et al., 2015). Positive controls with regard to the selection strategy can also be used if available, but are generally experiment-specific and chosen based on prior knowledge. For example, a positive control could consist of sgRNAs targeting a separate gene whose knockout causes resistance to the drug through downstream pathways. However, inclusion of this latter type of positive controls should be carefully considered, as controls that confer a significant fitness advantage may preclude enrichment of other sgRNAs in the screen, especially when other sgRNAs of interest only lead to a modest selective advantage.
- Negative controls: Negative control sgRNAs are ones expected to cause neutral effects in the screen, and serve as a normalization reference during data analysis and as a quality control check for identifying true-positive hits. Therefore, their inclusion in each library is critical. The most commonly used negative control sgRNAs are non-targeting controls, which are sgRNAs with no perfect matches in the genome and with minimal off-target cleavage sites. Alternatively, sgRNAs targeting functionally neutral regions of the genome such as safe harbor loci (e.g., AAVS1), intergenic regions, or genes with no effect on fitness can also serve as negative controls. DNA damage due to Cas9-mediated cleavage has a modest impact on fitness independent of gene function, which may lead to slightly different outcomes between non-targeting and functionally neutral negative controls. Therefore, we typically include both types in our libraries. Approximately 1–5% of each final library should correspond to negative control sgRNAs.A list of example control sgRNAs is provided in Table 1. Additional or library-specific controls can be designed manually, but we recommend using sgRNA sequences from publicly available pooled CRISPR libraries (e.g., Brunello, GeCKO) if possible.
Order the full-length sgRNA oligonucleotide sequences as a pool from an oligonucleotide synthesis service such as Twist Biosciences.
Table 1. Control sgRNA sequences.
| Control | sgRNA sequence | Notes |
|---|---|---|
| Positive-1 | TG GAAATATACATCAT AAG A | EEF2 |
| Positive-2 | TGCCAACCTCCGACAAAGGT | EEF2 |
| Positive-3 | GTTGTGCGCGTGCTCGAAGG | EEF2 |
| Negative-1 | CTGACGTGTCTGAAATGAGT | Non-targeting |
| Negative-2 | ATTTCCCTACGGAGATATCC | Non-targeting |
| Negative-3 | ATCAAGTCAGGTTATGCGGG | Non-targeting |
| Negative-4 | GGATACCTGGGCCGACTTTC | Non-targeting |
| Negative-5 | CTCCGTTATGTG G CATGAGA | Non-targeting |
| Negative-6 | CATTGTTATG GCCTCCTCCG | Functionally neutral |
| Negative-7 | GAAAACCTAAGCTAAGTGGT | Functionally neutral |
| Negative-8 | CACGAGACCATATCTTTCAG | Functionally neutral |
| Negative-9 | GAAACTGGGTCAAACGAAGG | Functionally neutral |
| Negative-10 | AT AAGCCAGAT ACAAGACCG | Functionally neutral |
Amplification and cloning of the sgRNA library into the destination vector.
-
9.Resuspend or adjust the concentration of the oligonucleotide pool to 1 ng/μl with molecular biology grade water.The resuspended oligonucleotide pool can be stored short-term (<1 year) at −20 °C or long-term (years) at −80 °C.
-
10.Perform library amplification round 1 PCR. The round 1 PCR amplifies the desired sgRNA library from the synthesized oligonucleotide pool using subpool barcode-specific PCR primers. Prepare the round 1 PCR master mix using the following parameters and then divide it into 4 × 50 μl reactions.If the oligonucleotide pool could not be adjusted to 1 ng/μl due to low yield, increase the volume of template DNA or reduce the input amount of template DNA (and adjust the number of cycles) for the PCR master mix. Primer sequences are provided in Table 2.
Component Volume (μl) sgRNA oligonucleotide pool template (1 ng/μl 4 Subpool barcode-specific R1 PCR forward primer (10 μM) 10 Subpool barcode-specific R1 PCR reverse primer (10 μM) 10 2× NEBNext Ultra II Q5 Master Mix 100 Molecular biology grade water 76 -
11.Amplify the sgRNA library using the thermocycling conditions below. Vary the number of cycles used in Step 2 of the thermocycling conditions for each of the 4 replicate reactions. We recommend starting with n = 10, 12, 15, and 20 cycles.The number of PCR cycles used to amplify the sgRNA library should be minimized in order to limit potential PCR biases due to overamplification. Due to the variable nature of custom sgRNA libraries, the optimal number of cycles for library amplification must be empirically determined for each library. The round 1 PCR reaction can be stored short-term (<1 week) at 4 °C or long-term (months to years) at −20 °C.
Step Description Time Temperature 1 Initial denaturation 30 s 98 °C 2 (n cycles) Denaturation 10 s 98 °C Annealing 30 s 63 °C Extension 30 s 72 °C 3 Final extension 2 min 72 °C 4 Hold ∞ 4 °C -
12.
Run 5 μl of each round 1 PCR reaction on a 2% agarose gel in TAE buffer along with 100 bp DNA ladder. Confirm that the PCR products are present at the expected size of ~100 bp, identify the reaction with the minimum number of cycles that produces a visible band, and use that reaction for the subsequent steps.
-
13.
Dilute the selected round 1 PCR reaction 1/10 with molecular biology grade water.
-
14.Perform library amplification round 2 PCR. The round 2 PCR uses universal primers to remove the subpool barcodes and adds more flanking homologous sequence to prepare the sgRNA library for Gibson assembly into the destination vector. Prepare the round 2 PCR master mix using the following parameters and then divide it into 4 × 50 μl reactions.Primer sequences are provided in Table 3.
Component Volume (μl) Round 1 PCR reaction from Steps 10–12 (1/10 diluted) 4 Universal R2 PCR forward primer (10 μM) 10 Universal R2 PCR reverse primer (10 μM) 10 2× NEBNext Ultra II Q5 Master Mix 100 Molecular biology grade water 76 -
15.Perform the round 2 PCR using the same thermocycling conditions as in Step 10. Again, vary the number of cycles used in Step 2 of the thermocycling conditions for each of the 4 replicate reactions. We recommend starting with 10, 12, 15, and 20 cycles.The total number of cycles across both rounds of PCR should be limited to 35–40 cycles to avoid biases from overamplification.
-
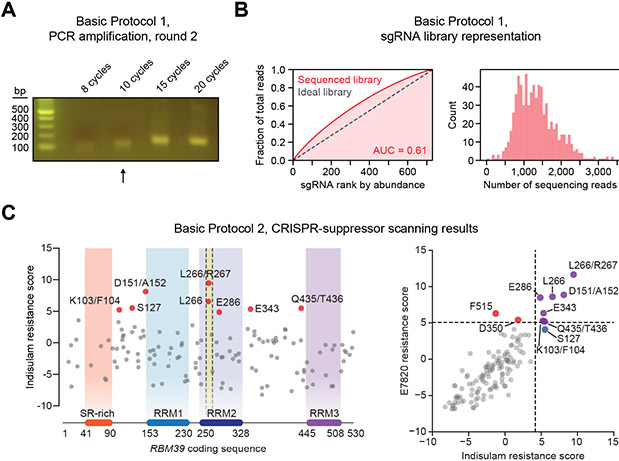
16.Run the round 2 PCR reactions on a 2% agarose gel in TAE buffer along with 100 bp DNA ladder. Confirm that the PCR products are present at the expected size of ~140 bp, identify the reaction with the minimum number of cycles that produces a visible band sufficient for gel extraction, and use this reaction for the subsequent steps.A sample gel is shown in Fig. 5A.
-
17.
Cut out the ~140 bp band selected in Step 16 with a clean razor and perform gel extraction using the Zymoclean Gel DNA Recovery Kit according to the manufacturer’s instructions.
-
18.
Quantify the concentration of the gel-purified round 2 PCR product using a NanoDrop UV-Vis spectrophotometer.
-
19.Perform restriction digest of the vector backbone. To prepare the desired sgRNA library vector backbone for cloning, the plasmid must first be linearized and dephosphorylated. Prepare a master mix for the restriction digest and dephosphorylation of lentiCRISPRv2 (Addgene #52961) using the following parameters and then divide it into 3 × 20 μl reactions.We recommend performing the restriction digest and dephosphorylation with >3 μg of plasmid (1 μg per reaction) as the cut vector can be stored long-term (years) at −20 °C for future use. However, the reaction can be scaled down accordingly if needed. Note that because lentiviral vectors contain repetitive sequences, they cannot be amplified by PCR. Therefore, they must be linearized by restriction digest and not by PCR amplification.
Component Volume (μl) Notes lentiCRISPRv2, diluted to 1 μg/μl 3 1 μg of input DNA per reaction FastDigest Esp3I 3 Esp3I is an isoschizomer of BsmBI FastAP Thermosensitive Alkaline Phosphatase 3 Dithiothreitol (20 mM) 3 10× FastDigest buffer 6 Molecular biology grade water 42 -
20.
Incubate the reactions in a thermocycler at 37 °C for 3 h to perform the restriction digest and dephosphorylation.
-
21.Run the reactions on a 1% agarose gel in TAE buffer along with 1 kb DNA ladder. Confirm the presence of the expected bands at ~13 kb and ~1.9 kb.lentiCRISPRv2 contains an 1,880 bp filler sequence between the Esp3I restriction sites that should be visible on the gel after digestion.
-
22.Cut out the ~13 kb band corresponding to the linearized vector with a clean razor and perform gel extraction using the Zymoclean Gel DNA Recovery Kit according to the manufacturer’s instructions.We recommend cutting out and purifying the three reactions separately and pooling the purified DNA after elution to avoid overloading the column capacity of the gel extraction kit.
-
23.Quantify the concentration of the gel-purified vector backbone using a NanoDrop UV-Vis spectrophotometer.The linearized and dephosphorylated vector backbone can be stored long-term (years) at −20 °C.
-
24.Perform Gibson assembly of the sgRNA library. Perform Gibson assembly to insert the PCR-amplified sgRNAs into the digested lentiCRISPRv2 backbone. Prepare the Gibson assembly master mix using the following parameters and then divide it into 3 × 20 μl reactions.
Component Volume (μl) Notes Gel-purified round 2 PCR product (Step 18) 3 30 ng input (10 ng per reaction) Gel-purified lentiCRISPRv2 backbone (Step 23) 3 120 ng input (40 ng per reaction) 2× NEBuilder HiFi DNA Assembly Master Mix 30 Molecular biology grade water 24 -
25.Incubate the Gibson assembly reactions in a thermocycler at 50 °C for 1 h. After incubation, pool the completed Gibson assembly reactions.Completed Gibson assembly reactions can be stored at −20 °C for up to 1 week.
-
26.Perform isopropanol precipitation. Perform isopropanol precipitation on the pooled Gibson assembly reactions using the following parameters.Although Gibson assembly reactions can be used directly for electroporation, isopropanol precipitation concentrates the assembled library into a smaller volume and removes salts and buffer components that can interfere with electroporation, both of which increase electroporation efficiency.
Component Volume (μl) Pooled Gibson assembly reaction (Step 25) 60 Isopropanol 60 GlycoBlue Coprecipitant 0.6 5 M NaCl solution 1.2 -
27.
Vortex the mixture briefly and incubate at room temperature for 15 min. During the incubation, prepare 2 ml of 80% (v/v) ethanol in water and store at −20 °C to keep ice-cold.
-
28.Centrifuge the mixture at >15,000g at room temperature for 15 min to pellet the precipitated plasmid DNA. The precipitant should be visible as a light blue pellet at the bottom of the microcentrifuge tube.The pellet may be very small and hard to see. Marking the side of the tube on the outer edge of the centrifuge during the spin can help locate the pellet.
-
29.Carefully pipet off the supernatant and wash the pellet twice with 1 ml of ice-cold 80% ethanol per wash (prepared in Step 27).Be careful not to disturb the pellet, as it can be easily dislodged.
-
30.After the second wash, carefully pipet off any residual liquid from the tube. Leave the tube uncapped at room temperature to air-dry for 1–2 min.Allow the pellet to air-dry until all visible liquid has evaporated, but do not allow it to over-dry as it may become difficult to resuspend back into solution.
-
31.Resuspend the pellet in 15 μl of TE buffer (5 μl per Gibson assembly reaction) and quantify the purified sgRNA library using a NanoDrop UV-Vis spectrophotometer or equivalent.To help solubilize the pellet, the TE buffer can be pre-heated to 55 °C or the resuspended pellet can be incubated for 10 min at 55 °C. Purified sgRNA libraries can be stored at −20 °C for months to years.
-
32.Amplify the pooled sgRNA library. The purified sgRNA library must now be transformed to amplify the library and harvested for lentivirus production. We recommend performing 1 electroporation per 5,000–10,000 sgRNAs in the library. Calculate the number of electroporations required.To ensure sufficient sgRNA representation, aim for a total number of colonies at least 50–100× the number of total sgRNAs in the library. Thus, the number of electroporation reactions is dependent on the number of sgRNAs in the library and the transformation efficiency. If adequate coverage is not achieved, scale up the number of electroporations.
-
33.For each electroporation, pre-warm 1 large (245-mm) LB agar plate with ampicillin. In addition, pre-warm 2 standard-sized (94-mm) LB agar plates with ampicillin.To prepare LB agar plates, dissolve LB agar in distilled water and autoclave the mixture to sterilize according to manufacturer’s instructions. Allow the sterilized mixture to cool to 55 °C and add ampicillin to 100 μg/ml final concentration (1:1,000 dilution of a 100 mg/ml sterile-filtered stock solution). Swirl to mix and dispense under aseptic conditions into the large and small dishes. The 2 standard-sized LB agar plates will be used to calculate transformation efficiency and does not need to be scaled per electroporation reaction. If amplifying multiple libraries, you will need 2 standard-sized LB agar plates per library.
-
34.For each electroporation reaction, thaw 25 μl of Endura DUO Electrocompetent cells on ice (10–20 min). Simultaneously, thaw the Lucigen Recovery Medium at room temperature or at 37 °C.Endura DUO Electrocompetent cells come as 50 μl aliquots, each of which can be split into 2 × 25 μl electroporations.
-
35.
Pre-chill a sufficient number of electroporation cuvettes (0.1-cm gap width) for the total number of electroporations.
-
36.
Mix the thawed electrocompetent cells by gently flicking the side of the tube.
-
37.Add 1 μl of the purified sgRNA library for every 25 μl of cells (corresponding to a single electroporation reaction) and mix briefly by stirring with the pipet tip.Avoid pipetting up and down to mix, as this will introduce bubbles and warm air.
-
38.
For each electroporation reaction, carefully transfer 25 μl of the DNA and cell mixture from Step 36 into a pre-chilled electroporation cuvette without introducing bubbles into the gap. Quickly flick the cuvette in a downward motion to deposit the cells at the bottom of the gap evenly.
-
39.Perform the electroporation according to the manufacturer’s protocol. For Lucigen Endura Electrocompetent cells, use the following settings: 10 μF, 600 Ω, 1,800 V.Typical time constants should run between ~4.0–5.0 ms. If the time constant is not within the expected range, the settings can be adjusted to 25 μF, 200 Ω, 1,500 V. If performing electroporation using the raw Gibson assembly reaction mixture rather than isopropanol-purified DNA, use the 1,500 V setting.
-
40.
As quickly as possible after the electroporation is complete, add 1 ml Lucigen recovery medium to the cuvette and resuspend the cells.
-
41.Transfer the resuspended cells to a 14-ml culture tube. Use an additional 1 ml of recovery medium to rinse the cuvette and pool with the rest of the cells in the culture tube.If you are performing multiple electroporations of the same library, you may pool up to 3 electroporation reactions (6 ml total volume) into a single 14-ml culture tube.
-
42.
Recover the cells at 37 °C for 1 h with shaking at 250 rpm.
-
43.Make a 100-fold dilution by adding 10 μl of the bacterial culture to 990 μl of LB medium. Pipet mix thoroughly.If there are multiple cultures of the same library, pool them first before making the dilution.
-
44.
Make a 1,000-fold dilution by adding 100 μl of the 100-fold dilution to 900 μl of LB medium. Pipet mix thoroughly.
-
45.Plate 100 μl each of the 100- and 1,000-fold dilutions on the pre-warmed 94-mm LB agar plates from Step 33. These will serve as 1,000- and 10,000-fold dilutions of the original culture, respectively.These dilutions will be used to estimate transformation efficiency of the library to ensure sufficient coverage and representation of the sgRNAs.
-
46.Plate the remaining cells on the pre-warmed 245-mm LB agar plate and ensure that the culture is evenly spread across the dish until it is mostly absorbed by the agar and will not drip upon inversion.If the culture is greater than 2 ml (due to multiple electroporations), use 2 ml of culture per large plate.
-
47.Incubate all LB agar plates overnight at 37 °C for 12–14 h.If recombination is an issue, the plates can also be incubated at 30–32 °C for 14-16 h. In either case, it is important to limit the incubation time to avoid bacterial overgrowth, which can introduce potential biases in the sgRNA distribution due to competition or growth rate differences.
-
48.Calculate the transformation efficiency. Count the number of colonies on the 1,000- and 10,000-fold dilution plates and multiply by the respective dilution factor to calculate the number of transformants per ml of culture. Multiply these values by 2 to account for the 2 ml volume of the original culture and then multiply by the number of electroporation reactions to estimate the total number of transformants.For example, if 12 colonies are observed on the 10,000-fold dilution plate and 3 electroporations were performed, the total number of transformants would be 12 × 10,000 × 2 × 3 = 720,000 transformants.
-
49.Divide the total number of transformants by the total number of sgRNAs in the library. Proceed if there are at least 50–100 transformants per sgRNA. Otherwise, use the estimated transformation efficiency to calculate the number of electroporations required to achieve >50–100× colonies per sgRNA and repeat the process.To ensure sufficient sgRNA representation and limit sgRNA dropout, the total number of transformants should be at least 50–100× the number of total sgRNAs in the library.
-
50.
Harvest the colonies from the large LB agar plates by pipetting 10 ml of LB medium and gently scraping off the colonies using a cell spreader. Transfer the liquid to a pre-weighed 50-ml Falcon tube.
-
51.
Repeat Step 50 twice, rinsing the plate with an additional 2 × 10 ml of LB medium and transferring the liquid to the tube for each wash (3 rinses total). This helps to maximize bacterial yield.
-
52.
Pellet the harvested bacteria by centrifugation at 3,000g for 5–10 min and carefully aspirate off the supernatant.
-
53.Weigh the tube to calculate the pellet wet weight and then calculate the number of maxipreps required to process the harvested bacteria.Approximately 0.4–0.5 g of bacteria can be processed per maxiprep.
-
54.
Purify the sgRNA library plasmid DNA by performing a sufficient number of maxipreps using the ZymoPURE II Plasmid Maxiprep Kit according to the manufacturer’s instructions.
-
55.Pool and quantify the concentration of the purified sgRNA library plasmid DNA using a NanoDrop UV-Vis spectrophotometer.The purified sgRNA libraries can be aliquoted and stored long-term (years) at −20 °C.
Table 2. CRISPOR subpool barcodes and round 1 PCR primers for library amplification.
CRISPOR barcodes and associated primers were previously reported in (Canver et al., 2018).
| # | Forward barcode (5’) |
Reverse barcode (3’) |
Forward primer | Reverse primer |
|---|---|---|---|---|
| 1 | CGGGTTCCGT | GCTTAGAATAGAA | CGGGTTCCGTGGAAAGG | TTCTATTCTAAGCGCCTTATTTTAACTTGC |
| 2 | GTTTATCGGGC | ACTTACTGTACC | GTTTATCGGGCGGAAAGG | GGTACAGTAAGTGCCTTATTTTAACTTGC |
| 3 | ACCGATGTTGAC | CTCGTAATAGC | ACCGATGTTGACGGAAAGG | GCTATTACGAGGCCTTATTTTAACTTGC |
| 4 | GAGGTCTTTCATGC | CACAACATA | GAGGTCTTTCATGCGGAAAGG | TATGTTGTGGCCTTATTTTAACTTGC |
| 5 | TATCCCGTGAAGCT | TTCGGTTAA | TATCCCGTGAAGCTGGAAAGG | TTAACCGAAGCCTTATTTTAACTTGC |
| 6 | TAGTAGTTCAGACGC | ATGTACCC | TAGTAGTTCAGACGCGGAAAGG | GGGTACATGCCTTATTTTAACTTGC |
| 7 | GGATGCATGATCTAG | CATCAAGC | GGATGCATGATCTAGGGAAAGG | GCTTGATGGCCTTATTTTAACTTGC |
| 8 | ATGAGGACGAATCT | CACCTAAAG | ATGAGGACGAATCTGGAAAGG | CTTTAGGTGGCCTTATTTTAACTTGC |
| 9 | GGTAGGCACG | TAAACTTAGAACC | GGTAGGCACGGGAAAGG | GGTTCTAAGTTTAGCCTTATTTTAACTTGC |
| 10 | AGTCATGATTCAG | GTTGCAAGTCTAG | AGTCATGATTCAGGGAAAGG | CTAGACTTGCAACGCCTTATTTTAACTTGC |
Table 3. Primer sequences.
Round 2 PCR primers were previously reported in (Canver et al., 2018).
| Primer Name | Notes | Sequence |
|---|---|---|
| Universal R2 PCR forward primer | Round 2 PCR primer for library amplification. | TAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG |
| Universal R2 PCR reverse primer | Round 2 PCR primer for library amplification. | ACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC |
| NGS_P5_0 | P5 primer for NGS library preparation, 0 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTTGTGGAAAGGACGAAACACCG |
| NGS_P5_1 | P5 primer for NGS library preparation, 1 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTTGTGGAAAGGACGAAACACCG |
| NGS_P5_2 | P5 primer for NGS library preparation, 2 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGCTTGTGGAAAGGACGAAACACCG |
| NGS_P5_3 | P5 primer for NGS library preparation, 3 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAGCTTGTGGAAAGGACGAAACACCG |
| NGS_P5_4 | P5 primer for NGS library preparation, 4 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCAACTTGTGGAAAGGACGAAACACCG |
| NGS_P5_6 | P5 primer for NGS library preparation, 6 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCACCTTGTGGAAAGGACGAAACACCG |
| NGS_P5_7 | P5 primer for NGS library preparation, 7 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTACGCAACTTGTGGAAAGGACGAAACACCG |
| NGS_P5_8 | P5 primer for NGS library preparation, 8 nt stagger. | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGAAGACCCTTGTGGAAAGGACGAAACACCG |
| NGS_P7 | Barcoded P7 primer for NGS library preparation. Barcode is indicated by [NNNNNNNN]. | CAAGCAGAAGACGGCATACGAGAT[NNNNNNNN]GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCAATTCCCACTCCTTTCAAGACCT |
Figure 5. Example results for Basic Protocols 1 and 2.
Example results. (A) Agarose gel showing the effect of different cycle numbers on PCR product yield for the round 2 PCR in Basic Protocol 1. Based on these results, 10 cycles (marked with arrow) was chosen for excision and gel purification. (B) Plots showing sgRNA distribution in the sequenced plasmid library obtained in Basic Protocol 1. AUC, area under curve. (C) CRISPR-suppressor scanning results obtained in Basic Protocol 2. Data are from a screen of sgRNAs targeting the RBM39 coding sequence using the drugs indisulam and E7820 (Gosavi et al., 2022). Left, scatterplot showing each sgRNA’s indisulam resistance score plotted against its targeted residue in the coding sequence of RBM39. sgRNAs >2 standard deviations above the mean of negative controls are shown in red. The highlighted yellow region of the RRM2 domain corresponds to the structural degron of RBM39. Right, scatterplot showing E7820 resistance scores compared to indisulam resistance scores for each sgRNA. Dotted lines indicate 2 standard deviations above the mean of negative controls.
Validation of the sgRNA representation in the library by next-generation sequencing.
-
56.Perform library PCR for NGS analysis. Samples are prepared for NGS analysis using a single round of PCR to append the appropriate flanking sequence and barcodes for sample deconvolution. Prepare the library PCR master mix using the following parameters and then divide it into 2 × 25 μl reactions.The P5 primer mix is an equimolar mixture of 8 staggered primers (provided in Table 3). It is important to use a mixture of primers to maintain library diversity on the flow cell and avoid low quality sequencing reads.
Component Volume (μl) sgRNA library plasmid template DNA (20 ng/μl 1 P5 primer mix (10 μM) 1.25 Barcoded P7 primer (10 μM) 1.25 2× NEBNext Ultra II Q5 Master Mix 25 Molecular biology grade water 21.5 -
57.Amplify the sgRNA library using the following thermocycling conditions.
Step Description Time Temperature 1 Initial denaturation 3 min 98 °C 2 (22 cycles) Denaturation 10 s 98 °C Annealing 30 s 63 °C Extension 30 s 72 °C 3 Final extension 2 min 72 °C 4 Hold ∞ 4 °C -
58.
Run the PCR reactions on a 2% agarose gel in TAE buffer along with 100 bp DNA ladder. Confirm the presence of the expected PCR product at ~300 bp.
-
59.
Cut out the ~300 bp band and perform gel extraction using the Zymoclean Gel DNA Recovery Kit according to the manufacturer’s instructions.
-
60.Quantify the DNA concentration of the gel-purified PCR product using the Qubit dsDNA High Sensitivity assay kit according to the manufacturer’s instructions.Do not quantify final libraries using UV-Vis spectrophotometers such as the NanoDrop because this is not as accurate.
-
61.Submit the sample for NGS, ensuring that there are at least 60 cycles for Read 1 and 8 cycles for Index 1.Aim to achieve sequencing coverage of >100 reads per sgRNA in the library.
-
62.Analyze the resulting NGS data using the ‘count_reads’ function in the provided code (https://github.com/liaulab/CRISPR-suppressor_scanning). This function requires two inputs: the FASTQ file with the NGS sequencing reads and a CSV input reference file containing the library sgRNA sequences.This function is adapted from count_spacers.py (Joung et al., 2017). First, the function filters reads by searching for part of the hU6 promoter sequence directly upstream of the sgRNA protospacer sequence. Reads that do not include this sequence are discarded from analysis. Then, it counts ho w many filtered reads perfectly match each given library sgRNA included in the input reference file. Other publicly available packages can also be used, such as CRISPRessoCount in the CRISPResso2 package (Clement et al., 2019) and MAGeCK Count in the MAGeCK package (Li et al., 2014). Refer to the appropriate documentation for detailed instructions for installation and execution of each package.
-
63.Visualize the distribution of library sgRNAs using the ‘qc_plot_dist’ function, which produces both a cumulative distribution plot and a histogram of sgRNA counts.Example plots are shown in Fig. 5B.
-
64.Calculate the following quality control metrics using the tabulated sgRNA read counts and confirm that the library passes benchmarks as follows. Generally, a good quality sgRNA library preparation should have >70% perfect sgRNA matches (percentage filtered reads matching an sgRNA from the input reference file of library sgRNA sequences), <0.5% undetected sgRNAs (percentage sgRNAs in the input reference file with no assigned reads), and a ‘skew ratio’ less than 10 (ratio of filtered reads allocated to top versus bottom 10th percentiles of sgRNAs) (see also (Joung et al., 2017)). If your sgRNA library does not pass these criteria, it is recommended to restart the process from the library amplification round 1 PCR step.Poor statistics can suggest poor quality sequencing reads, cross-contamination by other sgRNA plasmids, issues with library cloning, or an incorrect input reference file. Example statistics are shown in Table 4. See also Troubleshooting.
-
65.
Proceed to Support Protocol 2 to generate lentivirus for the sgRNA library and titering.
Table 4. Validation statistics for a typical sgRNA library.
| Metric | Value |
|---|---|
| Total filtered reads | 1,081,037 |
| Total filtered reads perfectly matching a library sgRNA | 975,982 |
| Total filtered reads without a perfect match | 105,055 |
| Percentage perfect matches | 90.3% |
| Number of undetected library sgRNAs | 0 |
| Percentage of undetected sgRNAs | 0% |
| Skew ratio | 2.74 |
SUPPORT PROTOCOL 1:
Support protocol title:
sgRNA library design using command-line CRISPOR.
Introductory paragraph:
This protocol describes the usage of the UCSC table browser to download FASTA files corresponding to genes of interest, followed by automated sgRNA library design using command-line CRISPOR (Concordet and Haeussler, 2018). This protocol is intended to support users wishing to automate sgRNA generation for complex libraries involving multiple genes or genes with many exons.
Materials:
No materials.
Protocol steps with step annotations:
Generation of FASTA files for each gene of interest.
Open the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables).
- Select the appropriate genome assembly.For screening in human cell lines, users should select ‘Human’ as the genome and ‘GRCh38/hg38’ as the assembly.
Select ‘NCBI RefSeq’ as the track.
- Click ‘paste list’ under the identifiers section. In the following page, paste the RefSeq identifier(s) corresponding to the gene(s) of interest and click ‘Submit.’It is also possible to use alternate identifiers, such as ‘GENCODE V20 (Ensembl 76).’ In this case, ensure that the appropriate alternate track is selected in Step 3.
Select ‘sequence’ under output format.
Click ‘get output’ at the bottom of the page. On the next window, select ‘genomic’ and click ‘submit.’
- Select ‘CDS Exons’ and ‘One FASTA record per region (exon, intron, etc.) with 20 extra bases upstream (5’) and 20 extra downstream (3’)’ in the sequence retrieval region options section.These default settings ensure that the resulting fasta file will contain one record for each CDS exon, with each record consisting of the exon sequence with 20 bp of flanking intronic sequences on either side. The intronic sequences are necessary to avoid excluding sgRNAs whose protospacers extend into the intron but that still cut within the exon. The parameters can be adjusted if desired, for example to include promoter sequences or UTR exons.
Select ‘Exons in upper case, everything else in lower case’ in the sequence formatting options section.
Click ‘get sequence.’
Generation of libraries using command-line CRISPOR.
-
10.Clone the CRISPOR GitHub repository by using the following commands:
git clone https://github.com/maximilianh/crisporWebsite.git git remote set-url origin https://github.com/maximilianh/crisporWebsite.git git pull origin master
-
11.Install the CRISPOR dependencies according to the documentation at the GitHub repository (https://github.com/maximilianh/crisporWebsite).Note that CRISPOR uses python2.7.
-
12.Create a directory to hold the relevant genome assembly (this should correspond to that used in Step 2), and download the genome from http://crispor.tefor.net/genomes/ to this directory by following the instructions on the GitHub repository.Many commonly used genome assemblies, such as hg38 and mm39, are available in this fashion from the CRISPOR website.
-
13.Run CRISPOR from the command line using the command below, substituting the appropriate filenames (including paths, if the files are not in your current directory). ‘crispor.py’ refers to the CRISPOR executable file, ‘genome_directory’ refers to the directory to which the genome assembly was downloaded in Step 12, and ‘input.fasta’ refers to the FASTA file downloaded in Step 9. CRISPOR will produce a TSV file with sgRNA sequences and metrics, which will be saved to the ‘output.tsv’ filename.
python crispor.py genome_directory input.fasta output.tsv
Ensure that python2.7 is installed to run this command.
SUPPORT PROTOCOL 2:
Support protocol title:
Pooled sgRNA library lentivirus production and titering.
Introductory paragraph:
In order to introduce the pooled sgRNA library into the cell line of interest for screening, lentivirus must first be produced from the purified sgRNA plasmid library. Subsequently, the lentiviral titer must be assessed in order to determine critical parameters for performing the cellular transduction for the CRISPR-suppressor scanning experiment. This Support Protocol describes a basic procedure for lentivirus production in HEK293T cells and determination of lentiviral titer in a commonly used screening cell line, K562, using puromycin for selection.
CAUTION: Lentivirus is considered a Biosafety Level 2+ (BSL-2+) pathogen. Follow all appropriate guidelines and regulations for the use and handling of lentivirus and lentiviral waste.
Materials:
HEK293T Cells (ATCC, cat. no. CRL-3216, RRID:CVCL_0063)
D10 Medium (see Reagents and Solutions)
Opti-MEM (Gibco, cat. no. 31985062)
pMD2.G (Addgene #12259)
psPAX2 (Addgene #12260)
FuGENE HD transfection reagent (Promega, cat. no. E2311)
K562 cells (ATCC, cat. no. CCL-243, RRID:CVCL_0004)
R10 Medium (see Reagents and Solutions)
Polybrene (Santa Cruz Biotechnology, cat. no. sc-134220)
Puromycin (Gibco, cat. no. A1113803)
Biosafety Cabinet (Thermo Fisher Scientific, cat. no. 13-261-304)
15-cm TC-treated Cell Culture Dish (Corning, cat. no. 353025)
Sterile Microcentrifuge Tubes (Axygen, cat. no. MCT-175-C-S)
Humidified CO2 Incubator (Thermo Fisher Scientific, cat. no. 51033558)
0.45-μm, 50-ml Tube Vacuum Filter System (Corning, cat. no. 430314)
Cryotubes (Sigma-Aldrich, cat. no. Z760951-400EA)
Benchtop Centrifuge (Eppendorf 5810R)
12-well Tissue Culture Plate (Corning, cat. no. 353043)
15-ml Conical Tube (Corning, cat. no. 352097)
Protocol steps with step annotations:
Lentivirus production in HEK293T cells.
- 20–24 h before transfection, seed a 15-cm dish with 15–18 × 106 HEK293T cells in a total volume of 25–30 ml of D10 medium. This should correspond to approximately 40–50% confluency, so that the cells reach 80–90% confluency the following day.All cell culture should be conducted under aseptic conditions in a biosafety cabinet. HEK293T cells should be maintained and regularly passaged according to the manufacturer’s recommendations. Do not let the cells exceed 70–80% confluency prior to seeding for lentiviral production. For optimal lentivirus production, use healthy HEK293T cells that are low-passage number (passage <10). The amount of lentivirus (i.e., the number of plates to seed) required for a screen is dependent on the lentiviral titer, the transduction efficiency in the cell line of interest, and the scale of your sgRNA library. One 15-cm dish is generally sufficient for a small library (<3,000 sgRNAs) if the screen is performed in a cell line that is amenable to transduction (e.g., K562). If your cell line is not easily transduced, it may be prudent to scale up the lentivirus production.
Examine the cells the next day. If the cells have reached 80–90% confluency, proceed with the lentivirus plasmid transfection.
- Lentivirus plasmid transfection. Prepare the lentivirus plasmid transfection mixture in a sterile microcentrifuge tube using the following parameters. Ensure that the FuGENE HD transfection reagent is added last and pipetted directly into the solution and not along the walls of the tube.Lentivirus production requires the co-transfection of accessory helper plasmids that encode the necessary genes for packaging and generating lentiviral particles. We use pMD2.G (VSV-G envelope plasmid, Addgene #12259) and psPAX2 (gag/pol packaging plasmid Addgene #12260). The amounts in the table below can be scaled up if multiple 15-cm dishes of HEK293T cells are to be transfected.
Component Amount Opti-MEM (no supplements) 1,200 μl pMD2.G (Addgene #12259) 3 μg psPAX2 (Addgene #12260) 6 μg Purified sgRNA library plasmid 9 μg FuGENE HD transfection reagent 60 μl Mix thoroughly, either by gentle pipet mixing or flicking the tube, but take care not to introduce bubbles into the mixture.
- Incubate the transfection mixture at room temperature for 10–15 min.Do not allow the transfection mixture to incubate for longer than 15 min, as this may lead to lower transfection efficiency.
- Pipet the entire transfection mixture dropwise onto the confluent 15-cm dish and then gently rock the plate to disperse the mixture evenly.Take care not to dislodge the cells from the plate by pipetting or shaking too vigorously.
Incubate the cells at 37 °C for 6–7 h.
- After incubation, carefully aspirate off the medium and replace with 30 ml of fresh pre-warmed D10 medium. Return the cells to the incubator.When replacing with fresh D10 medium, care should be taken to dispense the medium slowly down the side of the dish in order to avoid dislodging the cells from the plate.
- Harvesting the lentivirus. The fresh D10 medium from Step 8 will begin accumulating lentiviral particles over the next several days (hereafter referred to as ‘lentiviral supernatant’). 48 h after medium exchange, collect the lentiviral supernatant from the cells and clarify it by filtration through a 0.45-μm, 50-ml tube filter system to remove cellular debris.Lentivirus particles are delicate and should be treated gently to maintain good titer. When pipetting, avoid introducing air into the sample. Additionally, do not subject lentivirus to vortexing, multiple freeze-thaw cycles, or storage at room temperature for extended periods of time. Ensure that a 0.45-μm filter, and not a 0.22-μm filter, is used, as the small pore size of the latter may decrease lentiviral titer.
- Aliquot the filtered lentiviral supernatant into cryotubes and snap-freeze on dry ice before storing the aliquots at −80 °C.The frozen lentiviral supernatant can be stored for up to 1 year at −80 °C with minimal decreases in viral titer. Lentivirus aliquots should be made in volumes suitable for single-use applications, as multiple freeze-thaw cycles will significantly decrease titer. We usually freeze lentivirus in 0.5–1 ml aliquots. Flash freezing in liquid nitrogen can also be performed.
Determining the functional lentiviral titer in K562 cells.
-
11.Before starting, determine the antibiotic concentration for selection in your cell line of interest. If not known, perform an antibiotic kill curve to identify the optimal concentration.The antibiotic is used to select for cells that have been successfully transduced with the sgRNA library. In this protocol, we will use 2 μg/ml of puromycin to select for K562 cells transduced with lentiCRISPRv2. However, the optimal concentration of antibiotic should be determined empirically for each new cell line and antibiotic.
-
12.Thaw an aliquot of lentivirus on ice.For best results, lentivirus should be thawed slowly on ice. However, rapid thawing in a 37 °C water bath is also possible. In this case, take care to remove the aliquot from the water bath as soon as it appears completely thawed (no visible ice) to avoid heating up the lentivirus.
-
13.Centrifuge 9 × 106 K562 cells at 300g for 5 min to pellet them. Then, carefully aspirate off the supernatant and resuspend the cells in fresh pre-warmed R10 medium containing 16 μg/ml of polybrene to a concentration of 1.5 × 106 cells/ml.K562 cells should be maintained and regularly passaged according to the manufacturer’s (e.g., ATCC) recommendations. The polybrene concentration in this mixture is 2× the final concentration. Polybrene can cause cytotoxicity at high concentrations, so avoid leaving the cells too long at this step. The optimal polybrene concentration for lentiviral transduction is cell line specific. For K562 cells, the final polybrene concentration should be 8 μg/ml.
-
14.
Pre-warm the centrifuge to 37 °C.
-
15.
Seed 6 wells of a 12-well plate with 1 mL each of the cell/polybrene/R10 mixture from Step 12, for a total of 1.5 × 106 cells/well.
-
16.Prepare a titration curve by diluting various volumes of lentiviral supernatant in R10 medium to a total volume of 1 ml. We recommend starting with 400 μl, 200 μl, 100 μl, 50 μl, 25 μl, and 0 μl (no-infection control) of lentiviral supernatant.The transduction efficiency is highly dependent on the cell line, and the volumes tested here can be adjusted as necessary. The 0 μl (no lentivirus) negative control condition is essential for functional titering. This condition cannot be omitted. If more conditions are desired, the number of wells can be scaled as needed.
-
17.Add the 1 ml lentivirus/R10 mixtures from Step 16 to the seeded wells from Step 15 and pipet mix thoroughly, taking care not to introduce air bubbles.The final transduction conditions are as follows: 0.75 × 106 K562 cells/ml (1.5 × 106 cells/well), 8 μg/ml polybrene, 2 ml total volume.
-
18.
Transduce the K562 cells by centrifugation at 1,800g for 90 min at 37 °C. Add 1 ml of R10 medium to each seeded well and return the cells to the incubator.
-
19.48 h after transduction, resuspend and transfer the transduced cells to sterile 15-ml tubes and centrifuge them at 300g for 5 min to pellet them. Aspirate the supernatant and resuspend the cells in R10 medium to a concentration of 4 × 105 cells/ml.Due to the spinfection, the K562 cells may be slightly adherent to the bottom of the wells. Pipet up and down thoroughly to dislodge them from the plate.
-
20.For each transduction condition, seed 2 wells each of a 24-well plate with 500 μl of the resuspended cells (2 × 105 cells). Then, add 500 μl of R10 medium to 1 well and 500 μl of R10 medium with 4 μg/ml puromycin to the other well.The final seeding conditions should be 2 × 105 cells/ml in 1 ml of R10 medium with 0 or 2 μg/ml puromycin.
-
21.Regularly assess the cell viability of the plated cells daily by light microscopy or flow cytometry. Passage cells as necessary, recording viable cell counts before and after passaging. After 3–5 d of antibiotic selection, when the no-infection control (0 μl lentivirus) is 80–90% confluent in the no puromycin treatment condition and there are <5–10% viable cells in the 2 μg/ml puromycin treatment condition, proceed to the next step.If these conditions are not met within 7 d of antibiotic selection, you may need to re-optimize the antibiotic selection conditions and repeat.
-
22.
Quantify the viable cell counts for all wells.
-
23.For each lentiviral transduction condition, calculate the transduction efficiency as the viable cell count in the puromycin treatment condition divided by the cell viability in the no treatment condition (adjusted accordingly if the cells were passaged using the counts pre- and post-passage). The curve should ideally have at least several conditions which fall below <30% transduction efficiency. If all tested conditions have >50% transduction efficiency, repeat the titering process with smaller lentiviral supernatant volumes.At transduction rates <50%, the transduction efficiency should be linearly correlated to the volume of lentiviral supernatant used. At higher transduction rates, the transduction efficiency will begin to plateau with increasing volumes of lentiviral supernatant. Example titering results are shown in Table 5. Note that results may vary considerably depending on the quality of lentivirus and the cell line used.
-
24.Select the lentiviral transduction condition with the desired transduction efficiency and proceed to Basic Protocol 2 to begin the CRISPR-suppressor scanning experiment. We normally perform transductions for CRISPR-suppressor scanning experiments at a multiplicity of infection (MOI) < 0.3, which corresponds to a transduction efficiency of ~26%, but transduction rates <50% are reasonable.The transduction efficiency is used to estimate the MOI. At higher MOIs (i.e., higher transduction efficiency), there is a higher probability that a cell will be infected more than once, which is undesirable as the presence of multiple sgRNAs makes it difficult to determine which perturbation is responsible for the observed phenotype. This can lead to false positives and a lower signal-to-noise ratio. The MOI is calculated assuming a Poisson distribution of infection events. Expected transduction outcomes for various MOIs are shown in Table 6. At an MOI of 0.3, ~26% of cells are infected and ~86% of infected cells have a single integrant. At an MOI of 0.5, ~39% of cells are infected and ~77% of infected cells have a single integrant. Thus, the rate of multiple integrations is less likely at lower MOIs, but at the expense of lower transduction efficiency, which will require scaling up the number of cells in the initial transduction in order to achieve sufficient coverage of the sgRNA library. Selecting an optimal MOI will therefore depend on the transduction efficiency in your desired cell line, the number of sgRNAs in the library, and other logistical factors (e.g., cell densities at seeding and at maximal confluency).
Table 5. Example sgRNA library titering results.
| Volume of virus (μl) | Percentage of cells infected |
|---|---|
| 100 | 65.67 |
| 50 | 60.42 |
| 25 | 39.95 |
| 12.5 | 33.49 |
| 6.25 | 15.75 |
| 0 | 1.68 |
Table 6. Theoretical rates of transduction events with different MOIs.
| MOI | % All Cells Successfully Transduced |
% Transduced Cells with 1 Lentiviral Integration |
% Transduced Cells with >1 Lentiviral Integration |
|---|---|---|---|
| 1 | 63.2 | 58.2 | 41.8 |
| 0.5 | 39.3 | 77.1 | 22.9 |
| 0.3 | 25.9 | 85.7 | 14.3 |
| 0.2 | 18.1 | 90.3 | 9.7 |
| 0.1 | 9.5 | 95.1 | 4.9 |
| 0.05 | 4.9 | 97.5 | 2.5 |
| 0.01 | 1.0 | 99.5 | 0.5 |
BASIC PROTOCOL 2:
Basic protocol title:
Execution and analysis of a CRISPR-suppressor scanning experiment.
Introductory paragraph:
This Basic Protocol describes the steps for performing a CRISPR-suppressor scanning experiment and basic analysis of the resulting NGS dataset (Fig. 4). The design and parameters for a CRISPR-suppressor scanning experiment is usually highly context-specific and tailored for the biological question at hand. Important considerations include the selection of sgRNAs for inclusion in the library, the type and stringency (e.g., drug concentration) of selection pressure, and the cell line of interest and desired MOI (see Strategic Planning for an in-depth discussion). This protocol assumes the use of lentiCRISPRv2 (Addgene #52961), which encodes SpCas9 and the sgRNA in a 1-plasmid system, and K562 as the cell line of interest for a prototypical CRISPR-suppressor scanning experiment using a small-molecule drug selection.
Figure 4. CRISPR-suppressor scanning execution and analysis.
Schematic of Basic Protocol 2, in which the sgRNA library is introduced into cells by lentiviral transduction, followed by treatment of cells with drug or vehicle to perform CRISPR-suppressor scanning. Genomic DNA is purified and the sgRNA abundances are quantified by deep sequencing.
Materials:
K562 cells (ATCC, cat. no. CCL-243, RRID:CVCL_0004)
R10 Medium (see Reagents and Solutions)
Polybrene (Santa Cruz Biotechnology, cat. no. sc-134220)
Puromycin (Gibco, cat. no. A1113803)
Dimethylsulfoxide (DMSO) (Sigma-Aldrich, cat. no. D8418-100ML)
QIAamp DNA Blood Mini Kit (Qiagen, cat. no. 51104)
RNase A (Qiagen, cat. no. 19101)
NEBNext Ultra II Q5 Master Mix (New England Biolabs, cat. no. M0544)
Molecular Biology Grade Water (Corning, cat. no. 46-000-CV)
Agarose RA (VWR, cat. no. N605-500G)
TAE buffer (50×) (Boston BioProducts, cat. no. BM-250)
100 bp DNA Ladder (New England Biolabs, cat. no. N3231)
Zymoclean Gel DNA Recovery kit (Zymo, cat. no. D4007)
Qubit dsDNA High Sensitivity Assay Kit (Invitrogen, cat. no. Q32851)
Biosafety Cabinet (Thermo Fisher Scientific, cat. no. 13-261-304)
12-well Tissue Culture Plate (Corning, cat. no. 353043)
Humidified CO2 Incubator (Thermo Fisher Scientific, cat. no. 51033558)
15-ml Conical Tube (Corning, cat. no. 352097)
Benchtop Centrifuge (Eppendorf 5810R)
24-well Tissue Culture Plate (Corning, cat. no. 353047)
T-25 Tissue Culture Flasks (Corning, cat. no. 430639)
Sterile Microcentrifuge tubes (Axygen, cat. no. MCT-175-C-S)
NanoDrop OneC UV-Vis Spectrophotometer (Thermo Fisher Scientific, cat. no. ND-ONEC-W)
PCR 8-well Strip Tubes with Domed Caps (VWR, cat. no. 53509-304)
ProFlex Thermocycler (Applied Biosystems, cat. no. 4484073)
Owl EasyCast B1A Mini Gel Electrophoresis System (Thermo Fisher Scientific, cat. no. B1A)
Razor (VWR, cat. no. 55411-050)
Qubit 3 Fluorometer (Invitrogen, cat. no. Q33216)
Protocol steps with step annotations:
Initial transduction of K562 cells with the pooled sgRNA library.
- Calculate the number of transduced cells required to achieve the desired level of sgRNA coverage by multiplying the desired number of cells per sgRNA by the total number of sgRNAs in the library. For example, to achieve 1,000× coverage of a library of 1,000 sgRNAs would require 1 × 106 transduced cells (1,000 cells per sgRNA × 1,000 sgRNAs).We recommend aiming for >500–1,000× coverage of the library in order to ensure sufficient representation of all sgRNAs.
- Calculate the number of cells required for the initial transduction using the minimum number of transduced cells required (from Step 1) and the transduction efficiency of the selected transduction conditions (determined from Support Protocol 1). For example, if the selected conditions have a transduction efficiency rate of 25%, then a minimum of 4 × 106 cells would be required for the initial transduction, as only 1 × 106 will be transduced.It is highly recommended to perform the transduction using an excess of cells (1.2–1.5 × the number calculated here), as any variability in the transduction rate, sample loss due to attrition, and other factors can cause the number of transduced cells to dip below the desired coverage.
- Calculate the number of wells of a 12-well plate required to transduce the desired number of K562 cells and perform the transduction using identical conditions as performed previously during the lentiviral titering (see Support Protocol 1). In addition to these transductions, include a no-infection control well where R10 medium is added instead of lentiviral supernatant.All cell culture should be conducted under aseptic conditions in a biosafety cabinet. It is critical that the transduction conditions (e.g., plate type, cells per well, volume of lentiviral supernatant, polybrene concentration, spinfection parameters) are kept consistent between the titering and the transduction for the screen in order to ensure reproducibility. The lentiviral transduction efficiency can be affected by all of these parameters, so using identical conditions is essential to maintain consistent transduction rates. If the transduction needs to be scaled up from the titering in order to accommodate a sufficient number of cells, do not increase the cell density in each well. Rather, scale up the number of wells for transduction, while keeping the conditions constant across each well.
- 48 h after transduction, resuspend and pool the transduced cells in sterile centrifuge tubes and centrifuge them at 300g for 5 min to pellet them. Then, aspirate the supernatant and resuspend the cells in R10 medium to a concentration 4 × 105 cells/ml.At this point, you may pool together all the cells from the wells transduced with the sgRNA library but remember to process the no-infection control cells separately.
For the transduced and no-infection control conditions, seed 2 wells of a 24-well plate with 2 × 105 cells in a total volume of 1 ml of R10 medium, with 1 well containing no puromycin and the other well containing 2 μg/ml puromycin. There should be 4 wells total: (i) transduced cells + 0 μg/ml puromycin, (ii) transduced cells + 2 μg/ml puromycin, (iii) no-infection control + 0 μg/ml puromycin, (iv) no-infection control + 2 μg/ml puromycin.
- Seed the remaining transduced K562 cells in a sufficient number of T-25 tissue culture flasks at a density of 2 × 105 cells/ml in R10 medium with a final concentration of 2 μg/ml puromycin.Depending on the volume needed, larger flasks can be used.
Allow the antibiotic selection to continue for the same duration of time as during the titering (3–5 d). Then, quantify the cell viability of the cells in the small-scale antibiotic selection (i.e., the seeded wells of the 24-well plate) using the same method as performed during the titering.
- Calculate the transduction efficiency for the transduced condition as the cell viability (e.g., number of cells) in the puromycin treatment condition divided by the cell viability in the no treatment condition. The small-scale selection plate can be discarded at this point. Then, assess whether the results of the screen transduction are consistent with the results from the previous transduction for titering:
- The no-infection control with no puromycin treatment is ~80–90% confluent.
- The no-infection control with puromycin treatment has <5–10% viable cells.
- The calculated transduction efficiency for the transduced condition is roughly comparable to the results from the titering. In particular, ensure that an MOI < 0.3 was achieved and that the number of cells successfully transduced achieves adequate coverage of the library.It is common to observe some small variability between independent transductions. If the transduction efficiency is lower than expected, re-calculate the number of transduced cells using the new transduction rate to ensure that the desired coverage of the library is met. If the transduction efficiency is higher than expected, you may proceed if the new MOI is acceptable.
If the transduction rate of the screen is acceptable, resuspend and transfer the transduced cells to sterile centrifuge tubes and pellet the cells by centrifugation at 300g for 5 min. Aspirate the supernatant and resuspend the cells in R10 medium.
Pool the antibiotic-selected cells together and count the number of cells.
- Seed 6 × T-25 flasks with the antibiotic-selected cells, making sure that the total number of cells in each flask exceeds the minimum number of cells to maintain coverage of the library. The 6 flasks will serve as 3 replicates each for the drug and vehicle treatment conditions.For example, to maintain 1,000× coverage of a 1,000 sgRNA library, a minimum of 1 × 106 cells is required per flask. We recommend exceeding this number by 1.2–1.5× to avoid dipping below this threshold due to sample attrition or other factors. Depending on the culture volume required (which depends on the number of cells needed to maintain coverage and will vary with library size), larger flasks can also be used. In general, do not exceed the volume of medium per flask recommended by the manufacturer, as doing so could negatively impact cell growth.
Add R10 medium with drug to 3 flasks to reach a seeding density of 2–4 × 105 cells/ml and the desired final drug concentration for selection.
Add R10 medium with vehicle (usually DMSO) to the other 3 flasks to reach an identical seeding density and vehicle concentration as the drug-treated conditions.
- Maintain the cells in culture, regularly passaging the cells and refreshing the medium with drug or vehicle as needed. When passaging the cells, record the cell counts and ensure that the minimum number of cells required to maintain sufficient coverage of the library is met. If this is not possible (i.e., due to drug treatment), try to retain the maximal number of cells possible during each passage to avoid introducing bottlenecks.The cells should be passaged and maintained according to the recommended guidelines (e.g., ATCC guidelines). It is likely that the vehicle-treated conditions will grow more quickly than the drug-treated conditions, so cell counts should be systematically monitored to prevent cells from reaching maximal confluency. The cells under drug selection will show varying growth defects depending on the stringency of selection. It is common for cell populations under high selection pressure (e.g., IC90) to fall far below the minimal coverage guidelines due to low viability. In such cases, the cells should be given enough time for resistant cells to expand and recover from treatment before passaging or taking cell pellets. If this is not intentional, restart the screen and lower the selection pressure to reduce the impact on cell viability. Over time, it is common for the drug-treated populations to become less sensitive to drug and exhibit smaller growth defects. This is often a positive indication that the screen is working as intended, as the drug-resistant populations become more dominant in the pool.
- After the desired duration of time has passed, end the screen by harvesting the cells for each condition and pelleting them by centrifugation at 300g for 5 min. The cell pellets can be divided into multiple aliquots but ensure that each pellet contains more than the minimum number of cells required to maintain coverage of the library. After removing supernatant, snap-freeze the cell pellets on dry ice.The length of time required is highly context dependent. If desired, cells can be harvested at intermediate timepoints before the end of the screen (e.g., during regular passages) in order to provide insight on the sgRNA population over time. The frozen cell pellets can be stored at −80 °C for up to a year.
Isolation of genomic DNA (gDNA) from the cell pellets and preparation of NGS samples.
-
16.Thaw the cell pellets and extract the gDNA from the pellets using the QIAamp DNA Blood Mini Kit according to the manufacturer’s instructions.Perform the optional RNase treatment step to ensure high-quality gDNA extractions.
-
17.
Quantify the purified gDNA using a NanoDrop UV-Vis spectrophotometer or equivalent.
-
18.PCR of the sgRNA cassette from gDNA for NGS analysis. Samples are prepared for NGS analysis using an analogous workflow to the sgRNA library plasmid validation workflow (see Basic Protocol 1). For each sample, prepare PCR reactions using the following parameters. Scale up the number of reactions for each sample such that the gDNA equivalent of the minimum number of cells to maintain coverage is used or all the extracted gDNA is amplified. Each 50 μl reaction can accommodate up to 2.5 μg of gDNA template. All reactions corresponding to the same biological sample (e.g., replicate 1 of drug-treated condition) should use the same barcoded P7 primer. Different biological samples (e.g., two replicates of drug treatment) should be prepared with different barcoded primers so that they can be sequenced together and demultiplexed.A rough estimate for the gDNA mass in a single human diploid cell is 6.6 pg. Therefore, 1 × 106 cells contain approximately 6.6 μg DNA. For K562 cells, which are near-triploid, this is equivalent to 9.9 μg. Due to loss during the gDNA extraction process, it is common to have less gDNA than the expected yield, in which case all of the gDNA should be amplified. Regardless of how much of the isolated gDNA is amplified, we recommend dividing the gDNA across ≥3 reactions to minimize any technical artifacts arising from PCR bias. The P5 primer mix is an equimolar mixture of 8 staggered primers (provided in Table 3). It is important to use a mixture of primers to maintain library diversity on the flow cell and avoid low quality sequencing reads.
Component Amount Purified gDNA template Up to 2.5 μg P5 primer mix (10 μM) 1.25 μl Barcoded P7 primer (10 μM) 1.25 μl 2× NEBNext Ultra II Q5 Master Mix 25 μl Molecular biology grade water To 50 μl -
19.Amplify the samples using the following thermocycling conditions.The number of cycles used for amplification can be optimized depending on the input amount of template gDNA. Typically, 20–25 cycles should be sufficient for bands that can be gel extracted. Avoid overamplification of the sgRNA cassette to minimize any PCR biases. We recommend testing several different cycle numbers on an extra sample in advance to empirically determine the right cycle number to use.
Step Description Time Temperature 1 Initial denaturation 3 min 98 °C 2 (22 cycles) Denaturation 10 s 98 °C Annealing 30 s 63 °C Extension 30 s 72 °C 3 Final extension 2 min 72 °C 4 Hold ∞ 4 °C -
20.
Run the completed reactions on a 2% agarose gel in TAE buffer along with 100 bp DNA ladder. Confirm the presence of the expected PCR products at ~300 bp.
-
21.
Cut out the ~300 bp bands and perform gel extraction using the Zymoclean Gel DNA Recovery Kit according to the manufacturer’s instructions.
-
22.Quantify the DNA concentration of the gel-purified PCR product using the Qubit dsDNA High Sensitivity assay kit according to the manufacturer’s instructions.Do not quantify final libraries using UV-Vis spectrophotometers such as the NanoDrop because this is not as accurate.
-
23.Submit the samples for NGS, ensuring that there are at least 60 cycles for Read 1 and 8 cycles for Index 1.The minimum sequencing coverage (reads per sgRNA) should be equivalent to the minimum cell coverage used in the screen. Therefore, if a 1,000 sgRNA library was screened at 1,000× coverage (1 × 106 cells), then aim for >1,000 reads per sgRNA (1 × 106 reads) for each sequenced sample.
Analysis of the CRISPR-suppressor scanning NGS data.
-
24.Extract sgRNA read counts from the NGS data using the ‘batch_count’ function in the provided code (https://github.com/liaulab/CRISPR-suppressor_scanning). This function requires two inputs: a CSV batch sample sheet and a CSV input reference file containing the library sgRNA sequences.This function uses the ‘count_reads’ function from Basic Protocol 1, which is adapted from count_spacers.py (Joung et al., 2017). Other publicly available packages can also be used to analyze screening data, such as CRISPRessoCount in the CRISPResso2 package (Clement et al., 2019) and MAGeCK Count in the MAGeCK package (Li et al., 2014). Refer to the appropriate documentation for detailed instructions for installation and execution of each package.
-
25.Pre-process sgRNA read count data. Analysis of CRISPR-suppressor scanning data first requires several pre-processing steps in order to normalize and calculate relative sgRNA enrichment scores. Perform the following transformations on the tabulated sgRNA read counts for each sample, including the sgRNA library plasmid (sequenced in Basic Protocol 1). This can be carried out manually or using the provided ‘batch_process’ function.
- Calculate the total number of reads for all sgRNAs in the sample.
- For each sgRNA, multiply the read count by a scaling factor of 1 × 106 and divide by the total number of reads (calculate ‘reads per million’).
- Add a pseudocount of 1 to each sgRNA and then log2-transform the resulting scores.
- Normalize each sample to the sgRNA library by subtracting the log2-transformed scores of each sgRNA in the library from their scores in the sample.This transformation first normalizes sgRNA read counts to the sequencing depth of each sample and converts the raw read counts into ‘reads per million.’ The pseudocount of 1 is added to avoid log2-transformation on sgRNAs with zero reads. Normalization of the sample scores to the sgRNA library, which serves as a proxy for initial abundances at the start of the screen, enables the calculation of relative sgRNA enrichment or depletion over time. CRISPR-suppressor scanning experiments can be analyzed without sequencing data from the sgRNA library plasmid, but we strongly recommend including it for downstream analysis.
-
26.
Calculate the mean of the library-normalized sgRNA scores across the set of replicates for each condition (e.g., week 8 drug treatment, replicates 1–3). This is accomplished as part of the ‘batch_process’ function.
-
27.Calculate sgRNA ‘fitness’ and ‘resistance’ scores. The sgRNA ‘fitness’ and ‘resistance’ scores are the two primary measures that we typically use to analyze CRISPR-suppressor scanning data. The fitness score serves as a measure of an sgRNA’s relative impact on cellular fitness in the absence of an external selection pressure and is calculated by comparing sgRNA abundances under vehicle treatment versus the plasmid library. The resistance score serves as a measure of an sgRNA’s relative effect on fitness in the presence and absence of selection (i.e., drug treatment) and is calculated by comparing sgRNA abundances under drug versus vehicle treatment. Perform the following transformations on the library-normalized, replicate-averaged scores from Step 26 using the provided ‘compare_conds’ function to calculate sgRNA fitness and resistance scores (note: this function does not perform the normalization to the mean of negative control sgRNAs, which should be performed manually).
- Fitness scores: To obtain sgRNA fitness scores for a given condition (e.g., week 8 vehicle treatment), calculate the mean score of the negative control sgRNAs and subtract it from all sgRNA scores.
- Resistance scores: To obtain sgRNA resistance scores for a given pair of conditions (e.g., week 8 drug treatment vs. week 8 vehicle treatment), subtract the sgRNA scores in vehicle treatment from the corresponding sgRNA scores in drug treatment. From these vehicle-normalized scores, calculate the mean score of the negative control sgRNAs and subtract it from all sgRNA scores.While the final normalization to the mean of the negative control sgRNAs is not necessary, we typically perform this normalization for analysis and visualization purposes, as it adjusts the values such that 0 represents the mean of the negative control sgRNAs. Thus, positive and negative scores represent sgRNAs that are enriched and depleted, respectively, relative to the negative control sgRNAs.
-
28.Interpreting sgRNA fitness and resistance scores and quality control. After calculating the fitness and resistance scores, the resulting dataset can be analyzed to identify hits in the screen for further downstream validation. Before calling hits, however, we strongly recommend performing quality control checks to evaluate the screen. Examples of quality control criteria are listed below.
- Evaluate the percentage of undetected sgRNAs and the skew ratio for each sample.It is common to observe more undetected sgRNAs and greater skew ratios in the screen samples compared to the plasmid library, as sgRNAs deplete and drop out over the duration of the screen. Typically, the number of undetected sgRNAs and skew ratios are greater in drug- versus vehicle-treated conditions due to the selection pressure. If these values are significantly higher under drug treatment, this can be indicative of ‘jackpotting,’ in which a handful of sgRNAs confer a significant fitness advantage and thus take over the population. Large numbers of undetected sgRNAs and skew ratios observed across both drug and vehicle treatment may arise if the targeted gene is highly essential in the cell line of interest. Additionally, if multiple time points were sequenced, it is typical to observe increasing numbers of undetected sgRNAs and skew ratios over time, with drug-treated samples exhibiting greater values than vehicle-treated samples at matched time points. This is generally indicative that the Cas nuclease and selection pressure are operating as expected.
- Evaluate the log2-transformed sgRNA scores for each replicate (from Step 25).Screen sample replicates should exhibit consistent changes in the sgRNA scores. Replicates should exhibit greater correlation or similarity to each other than across conditions (e.g., drug versus vehicle). It is common to observe greater variance between replicates at longer screening timescales (e.g., at week 8 versus week 2). Greater variance is also expected when there is significant population bottlenecking (e.g., the target gene is highly essential, stringent selection pressure). If multiple time points were sequenced, correlation between drug- and vehicle-treated samples should decrease over time due to selection. Earlier time points should exhibit greater correlations to the plasmid library and greater inter-replicate correlations than later time points.
- Analyzing positive control sgRNAs.Positive control sgRNAs targeting known pan-essential genes (e.g., EEF2, housekeeping genes) are included in the library to validate the presence of Cas9 nuclease activity and evaluate the efficiency of the perturbation in the cell line of interest. These sgRNAs should be highly depleted across both drug- and vehicle-treated samples. If this depletion is not observed, then validation of on-target Cas nuclease activity may be required. Another form of positive control sgRNAs include ones known to cause drug resistance, such as ones targeting the drug-binding site of the target protein or other proteins in the pathway known to be important for drug action. This is experiment-specific and may not be available depending on the model or the biological process under study. If included, however, confirm that these controls lead to the expected behavior.
- Evaluate the negative control sgRNAs.Negative control sgRNAs should not exhibit any significant systematic enrichment or depletion. Modest enrichment of negative control sgRNAs may be observed at later time points, especially in screens targeting highly essential genes, due to a relative fitness advantage of the negative controls. Later time points may also exhibit greater variance in negative control sgRNA fitness scores due to clonal drift and other factors. Neutral gene-targeting controls may exhibit some depletion relative to non-targeting controls due to cytotoxicity associated with Cas nuclease-mediated cleavage. If the protein of interest is known to be essential, sgRNAs targeting the protein CDS should exhibit lower fitness scores relative to the negative control sgRNAs. Additionally, it is also common for sgRNAs targeting known protein domains (e.g., UniProt or Pfam annotated) to exhibit lower fitness scores than those targeting inter-domain regions (e.g., linkers).
-
29.Calling enriched sgRNAs and downstream validation. If the screen exhibits the expected behavior and passes quality control checks, then enriched sgRNAs can be selected for further validation and follow-up. The criteria for calling hits can be adjusted depending on the experiment, but we generally use a minimum enrichment threshold cutoff methodology, which is typically set to the mean resistance score plus 2 standard deviations of the negative control sgRNAs. After identifying enriched sgRNAs, we typically perform functional validation through independent transductions of individual enriched sgRNAs in an arrayed format. Subsequently, we split the transduction into two treatment conditions (i.e., drug and vehicle) and assess whether the sgRNA recapitulates the desired phenotype (i.e., resistance or growth rescue).Since we normally perform normalization to the mean of the negative control sgRNAs, we call ‘enriched’ sgRNAs as those exceeding 2 standard deviations of the negative control sgRNA resistance scores. Example data are provided in Fig. 5C.
REAGENTS AND SOLUTIONS:
D10 Medium
500 ml DMEM, High Glucose, Pyruvate (Gibco, cat. no. 11995073)
50 ml Fetal Bovine Serum (Peak Serum, cat. no. PS-FB2)
5 ml Penicillin/Streptomycin (Gibco, cat. no. 15140163)
Final composition: DMEM with ~10% (v/v) Fetal Bovine Serum and 1% (v/v) Penicillin/Streptomycin
Mix ingredients under aseptic conditions in a biosafety cabinet and store up to 1 month at 4 °C.
R10 Medium
500 ml RPMI-1640 Medium (Gibco, cat. no. 11875119)
50 ml Fetal Bovine Serum (Peak Serum, cat. no. PS-FB2)
5 ml Penicillin/Streptomycin (Gibco, cat. no. 15140163)
Final composition: RPMI-1640 with ~10% (v/v) Fetal Bovine Serum and 1% (v/v) Penicillin/Streptomycin
Mix ingredients under aseptic conditions in a biosafety cabinet and store up to 1 month at 4 °C.
COMMENTARY:
Background Information:
Genome editing is a powerful method for evaluating the effects of genetic perturbations in an endogenous context and has aided the discovery and dissection of molecular mechanisms underlying numerous biological processes. Historically, in situ mutagenesis was performed using agents such as chemical mutagens or ionizing radiation, but these approaches lack specificity and induce mutations that are costly and laborious to identify (Muller, 1927; Grimm, 2004; O’Loughlin and Gilbert, 2018). Although zinc-finger nucleases and transcription-activator like effector nucleases later enabled greater precision and control for in situ mutagenesis, the need to design new nucleases on an ad hoc basis has limited their broad utility (Urnov et al., 2010; Joung and Sander, 2013). With the advent of RNA-programmable CRISPR–Cas9 genome editing technology, it became possible to target Cas9 to arbitrary genomic loci of interest in a facile manner. This technology has empowered complex genetic manipulations at scale in live cells such as genome-wide gene knockout screens (Wang et al., 2015).
The protocols described here present a method applying CRISPR–Cas9 genome editing to in situ tiling mutagenesis with small-molecule inhibitors to interrogate drug resistance pathways and mechanism of action, termed CRISPR-suppressor scanning. Cas9 cleavage forms double-stranded breaks, whose subsequent repair by error-prone non-homologous end-joining and microhomology-mediated end-joining generates mutations upon which drug selection can act. Our lab and others have applied this approach to uncover mechanistic insights across a diverse number of protein targets and cell lines (Ipsaro et al., 2017; Donovan et al., 2017; Neggers et al., 2018; Vinyard et al., 2019; Gosavi et al., 2022), demonstrating the versatility of CRISPR-suppressor scanning as a general approach. Crucially, Cas9 mutagenesis produces a diverse spectrum of mutations, many of which are indel mutations.
While these protocols exploit Cas9 nuclease-mediated cleavage and subsequent repair of the double-stranded break to generate a large spectrum of coding mutations, other CRISPR-based technologies, such as base editors, have also successfully been employed to search for drug resistance mutations (Hanna et al., 2021; Cuella-Martin et al., 2021; Kim et al., 2022; Sangree et al., 2022). These observations testify to the versatility of CRISPR-based approaches in studying protein-drug interactions and drug resistance. As CRISPR–Cas9 technologies continue to mature, increasingly sophisticated perturbations such as prime editing (Anzalone et al., 2019) will not only expand the range of accessible mutational space but also enable greater precision and manipulation of the desired perturbation. We anticipate that such improvements, in conjunction with advancements in computational and structural techniques, will further increase the resolution and power of CRISPR-suppressor scanning as an approach for uncovering structure-activity relationships.
Critical Parameters:
Many parameters should be carefully designed before beginning a CRISPR-suppressor scanning experiment. As discussed in the Strategic Planning section, important considerations include the choice of target, selection strategy, model cell line, and method of perturbation (Fig. 2). Other critical parameters include the design and selection of sgRNAs for the library, as well as details related to the selection pressure (e.g., drug concentration or duration of treatment) and phenotypic readout (e.g., cell viability, staining for markers, or fluorescence). In our experience, the most critical parameter for a successful screen is the selection pressure and care should be taken to ensure its compatibility with the other experimental parameters, such as the biological process under study and the chosen cell line. The quality of the data generated from a genetic screen is often directly correlated to the quality of the selection and a poorly crafted selection generally results in poor quality data.
For example, a common consideration for most CRISPR-suppressor scanning experiments is determining the stringency of the selection (i.e., the drug concentration). The stringency of selection can be quantified in terms of the percent growth inhibition of wild-type cells. High concentrations of drug corresponding to the GI90 will produce stringent selections that generally enrich for a handful of sgRNAs conferring a strong resistance phenotype, such as those targeting the drug binding site. For cytotoxic drugs, stringent selections may also bottleneck the cellular population, rendering the surviving cells too sparse to recover or causing significant inter-replicate variability, poor signal-to-noise, or jackpotting of a small subset of sgRNAs. These issues can be exacerbated by the unique design choices of each experiment, such as those involving cell lines that are highly sensitive to culture conditions, and thus warrant deeper consideration. Weaker selection pressures, for example concentrations of drug corresponding to GI50, may be necessary where significant cytotoxicity is a concern or in experiments aiming to identify more subtle phenotypes, such as partial growth rescue. However, for weaker selections, it is critical to ensure that a sufficiently strong selection pressure is applied to enrich true positive sgRNAs at a good signal-to-noise relative to controls. In cases where this is challenging, it may be worthwhile to consider a gradual escalation of drug in the dosing regimen or extending the screen for a longer duration to promote the expansion of the desired phenotypic subpopulations over time.
Troubleshooting:
Table 7.
Troubleshooting CRISPR-suppressor scanning experiments
| Problem | Possible Cause | Solution |
|---|---|---|
| Insufficient number of colonies is observed following library cloning transformation. |
|
|
| Sequenced sgRNA library shows poor statistics (e.g., many undetected sgRNAs, high skew ratio). |
|
|
| Low lentiviral titer is observed. |
|
|
| Number of cells transduced during the screen is unexpectedly high or low. |
|
|
| Low viability of cells is observed following drug selection during CRISPR-suppressor scanning experiment. |
|
|
| Analysis of CRISPR-suppressor scanning results shows high replicate variability or unexpected behavior of controls. |
|
|
Understanding Results:
To ensure reliable results in the CRISPR-suppressor scanning, it is critical to ensure that the starting sgRNA library is of high quality. During library construction, care should be taken to minimize the cumulative rounds of PCR amplification used to amplify sgRNA inserts. Thus, Basic Protocol 1 recommends testing different cycle numbers for these PCR steps. An example of results from this can be found in Fig. 1A, based upon which 10 cycles was empirically chosen for the round 2 PCR. Sequencing of the plasmid library, as detailed in Basic Protocol 1, is also important to verify library composition. The composition of an example library is shown in Fig. 1B and Table 4. Due to the size of these libraries, it is not feasible or realistic to achieve a perfectly uniform distribution of sgRNAs. However, we routinely achieve sgRNA libraries with even coverage for a majority of sgRNAs, as indicated by a skew ratio below 10 (Table 4) and an area under the curve in the cumulative distribution plot close to 0.5 (Fig. 1B). It is also normal for a small percentage of filtered reads to not match perfectly to any library sgRNAs, for instance due to errors in sgRNA oligonucleotide synthesis, PCR errors introduced during cloning or sequencing preparation, or other sequencing artifacts. However, it is reasonable to expect 70-90% of reads to match perfectly to the library sgRNAs (Table 4). With regard to library lentivirus, the efficacy can greatly vary depending on the cell line of interest. An example of typical titering results for high-quality lentivirus is provided in Table 5.
The ultimate output of a CRISPR-suppressor scanning experiment is a table of resistance scores for each sgRNA. These data are most clearly visualized as a scatterplot depicting each sgRNA’s resistance score plotted against its cut site in the CDS of the gene (Fig. 5C, left). This presentation is helpful for understanding how resistant mutations are distributed across the CDS, and whether specific regions or domains are particularly important for drug function. We typically define enriched sgRNAs as those with resistance scores greater than 2 standard deviations above the mean of negative controls—these correspond to sgRNAs whose mutational outcomes confer resistance to the drug used in the screen. The example results shown in Fig. 5C show a collection of hit sgRNAs targeting the RBM39 gene conferring resistance to indisulam, a molecular glue that binds to RBM39 and DCAF15 and induces degradation of RBM39 (Gosavi et al., 2022). A key point of validation for a CRISPR-suppressor screen is the identification of known or expected resistance mechanisms. Here, sgL266 and sgL266/R267, two sgRNAs targeting the structural degron of RBM39 that is necessary for indisulam-mediated degradation, both registered as strongly enriched hits (Fig. 5C). However, CRISPR-suppressor scanning can also nominate hits in unexpected regions of the protein, such as sgD151/A152, which is distal to the structural degron. Upon identifying these hits, users can then perform follow-up sequencing of the edited site(s) to characterize the specific mutations responsible for resistance, which can enable downstream assays to further investigate aspects of protein structure-function.
Users may also be interested in comparing the effects of structurally related chemical modulators in order to gain insight into structure-activity relationships. As an example of this application, our lab has investigated differences between resistance profiles comparing indisulam to a related drug, E7820 (Fig. 5C, right). Testifying to similarities between the two drugs, the results are well-correlated between the two screens and most enriched hits are shared, including the aforementioned sgL266, sgL266/R267, and sgD151/A152 hits. However, this experiment also identified sgRNAs conferring resistance specifically to indisulam (sgS127) or specifically to E7820 (sgF515 and sgD350).
Time Considerations:
Basic Protocol 1: 2 weeks (1 day for library design, 1 week for library cloning, and 1 week for sequencing validation).
Support Protocol 1: 1 day.
Support Protocol 2: 1 week.
Basic Protocol 2: 4–10 weeks (2–8 weeks for transduction and CRISPR-suppressor scanning, depending on the experiment; 2 weeks for library preparation, sequencing, and analysis).
ACKNOWLEDGEMENTS:
K.C.N. and N.Z.L. were supported by National Science Foundation Graduate Research Fellowships (grant no. DGE1745303). C.L. was supported by a Herchel Smith Graduate Fellowship. B.B.L. is a Damon Runyon-Rachleff Innovator supported in part by the Damon Runyon Cancer Research Foundation (grant no. DDR 60S-20). This research was additionally supported by award no. 1DP2GM137494 from the National Institute of General Medical Sciences and startup funds from Harvard University.
Footnotes
CONFLICT OF INTEREST STATEMENT:
B.B.L. is on the scientific advisory board of H3 Biomedicine, holds a sponsored research project with H3 Biomedicine, is a scientific consultant for Imago BioSciences, and is a shareholder and member of the scientific advisory board of Light Horse Therapeutics. The remaining authors declare no competing interests.
INTERNET RESOURCES:
Tool for designing sgRNAs targeting given input sequences (Basic Protocol 1).
https://genome.ucsc.edu/cgi-bin/hgTables
UCSC Table Browser for downloading fasta files inputs for command-line CRISPOR (Support Protocol 1).
https://github.com/liaulab/CRISPR-suppressor_scanning
GitHub repository containing functions and example files associated with this protocol (Basic Protocols 1 and 2).
DATA AVAILABILITY STATEMENT:
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
LITERATURE CITED:
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Latek RR, and Daley GQ 2003. Mechanisms of Autoinhibition and STI-571/Imatinib Resistance Revealed by Mutagenesis of BCR-ABL. Cell 112:831–843. [DOI] [PubMed] [Google Scholar]
- Canver MC, Haeussler M, Bauer DE, Orkin SH, Sanjana NE, Shalem O, Yuan G-C, Zhang F, Concordet J-P, and Pinello L 2018. Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nature Protocols 13:946–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, et al. 2019. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nature Biotechnology 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet J-P, and Haeussler M 2018. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research 46:gky354-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuella-Martin R, Hayward SB, Fan X, Chen X, Huang J-W, Taglialatela A, Leuzzi G, Zhao J, Rabadan R, Lu C, et al. 2021. Functional interrogation of DNA damage response variants with base editing screens. Cell 184:1081–1097.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KF, Hegde M, Sullender M, Vaimberg EW, Johannessen CM, Root DE, and Doench JG 2017. Creation of Novel Protein Variants with CRISPR/Cas9-Mediated Mutagenesis: Turning a Screening By-Product into a Discovery Tool. PLoS ONE 12:e0170445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedy AM, and Liau BB 2021. Discovering new biology with drug-resistance alleles. Nature Chemical Biology 17:1219–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosavi PM, Ngan KC, Yeo MJR, Su C, Li J, Lue NZ, Hoenig SM, and Liau BB 2022. Profiling the Landscape of Drug Resistance Mutations in Neosubstrates to Molecular Glue Degraders. ACS Central Science 8:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S 2004. The art and design of genetic screens: mammalian culture cells. Nature Reviews Genetics 5:179–189. [DOI] [PubMed] [Google Scholar]
- Hanna RE, Hegde M, Fagre CR, DeWeirdt PC, Sangree AK, Szegletes Z, Griffith A, Feeley MN, Sanson KR, Baidi Y, et al. 2021. Massively parallel assessment of human variants with base editor screens. Cell 184:1064–1080.e20. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Shen C, Arai E, Xu Y, Kinney JB, Joshua-Tor L, Vakoc CR, and Shi J 2017. Rapid generation of drug-resistance alleles at endogenous loci using CRISPR-Cas9 indel mutagenesis. PLOS ONE 12:e0172177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, and Sander JD 2013. TALENs: a widely applicable technology for targeted genome editing. Nature Reviews Molecular Cell Biology 14:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, and Zhang F 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nature Protocols 12:828–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, and Miller RM 2017. Leveraging Chemotype-Specific Resistance for Drug Target Identification and Chemical Biology. Trends in Pharmacological Sciences 38:1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lee S, Cho S, Park J, Chae D, Park T, Minna JD, and Kim HH 2022. High-throughput functional evaluation of human cancer-associated mutations using base editors. Nature Biotechnology 40:874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, and Liu DR 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok HS, Freedy AM, Siegenfeld AP, Morriss JW, Waterbury AL, Kissler SM, and Liau BB 2022. Drug addiction mutations unveil a methylation ceiling in EZH2-mutant lymphoma. bioRxiv:2022.04.04.486977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biology 15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ 1927. Artificial Transmutation of the Gene. Science 66:84–87. [DOI] [PubMed] [Google Scholar]
- Neggers JE, Kwanten B, Dierckx T, Noguchi H, Voet A, Bral L, Minner K, Massant B, Kint N, Delforge M, et al. 2018. Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes. Nature Communications 9:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan KC, Hoenig SM, Gosavi PM, Tanner DA, Lue NZ, Garcia EM, Lee C, and Liau BB 2022. Activity-based CRISPR Scanning Uncovers Allostery in DNA Methylation Maintenance Machinery. bioRxiv:2022.05.14.491958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin TA, and Gilbert LA 2018. Functional Genomics for Cancer Research: Applications In Vivo and In Vitro. Annual Review of Cancer Biology 3:1–19. [Google Scholar]
- Sangree AK, Griffith AL, Szegletes ZM, Roy P, DeWeirdt PC, Hegde M, McGee AV, Hanna RE, and Doench JG 2022. Benchmarking of SpCas9 variants enables deeper base editor screens of BRCA1 and BCL2. Nature Communications 13:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenone M, Dančík V, Wagner BK, and Clemons PA 2013. Target identification and mechanism of action in chemical biology and drug discovery. Nature Chemical Biology 9:232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe J-S, Suzuki Y, Pappin DJ, Joshua-Tor L, and Vakoc CR 2015. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Molecular Cell 60:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, and Vakoc CR 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature Biotechnology 33:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, and Gregory PD 2010. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics 11:636–646. [DOI] [PubMed] [Google Scholar]
- Vinyard ME, Su C, Siegenfeld AP, Waterbury AL, Freedy AM, Gosavi PM, Park Y, Kwan EE, Senzer BD, Doench JG, et al. 2019. CRISPR-suppressor scanning reveals a nonenzymatic role of LSD1 in AML. Nature Chemical Biology 15:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM 2015. Identification and characterization of essential genes in the human genome. Science 350:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.