Abstract
In recent years, the treatment of breast cancer has advanced dramatically and neoadjuvant chemotherapy (NAC) has become a common treatment method, especially for locally advanced breast cancer. However, other than the subtype of breast cancer, no clear factor indicating sensitivity to NAC has been identified. In this study, we attempted to use artificial intelligence (AI) to predict the effect of preoperative chemotherapy from hematoxylin and eosin images of pathological tissue obtained from needle biopsies prior to chemotherapy. Application of AI to pathological images typically uses a single machine‐learning model such as support vector machines (SVMs) or deep convolutional neural networks (CNNs). However, cancer tissues are extremely diverse and learning with a realistic number of cases limits the prediction accuracy of a single model. In this study, we propose a novel pipeline system that uses three independent models each focusing on different characteristics of cancer atypia. Our system uses a CNN model to learn structural atypia from image patches and SVM and random forest models to learn nuclear atypia from fine‐grained nuclear features extracted by image analysis methods. It was able to predict the NAC response with 95.15% accuracy on a test set of 103 unseen cases. We believe that this AI pipeline system will contribute to the adoption of personalized medicine in NAC therapy for breast cancer.
Keywords: artificial intelligence, breast cancer, digital pathology, neoadjuvant chemotherapy
Introduction
Neoadjuvant chemotherapy (NAC) is used for locally advanced breast cancer, breast cancer with poor prognosis (triple negative and HER2 positive, lymph node metastasis, or high proliferation rate), or early breast cancer that is amenable to systemic therapy. The greatest benefit of NAC is that the reduction in tumor size allows for improved breast conservation and lower staging, thereby increasing the number of patients who are eligible for surgery. In addition, by suppressing micrometastasis, recurrence and metastasis can be prevented. Although NAC has many advantages, it results in unnecessary chemotherapy for the patients if the drug is ineffective. Therefore, it would be of great significance if we can predict the efficacy of NAC before it is given, and thereby guide treatment selection. There have been several studies on the prediction of response to NAC using MRI and ultrasound [1, 2, 3, 4], and on disease‐free survival and overall survival based on the effect of NAC [5, 6]. However, at present, there is no reliable system to provide decision support for treatment choice.
In the field of digital pathology, artificial intelligence (AI) techniques such as support vector machine (SVM) and convolutional neural network (CNN) have been used to assist in the diagnosis of various tumor pathologies [7, 8, 9, 10, 11]. We have also applied AI to the pathological diagnosis of gastric cancer and breast cancer [12, 13]. Another major advantage of AI is that it can extract information from images beyond what pathologists can determine in the support of pathological diagnosis. To date, attempts have been made to predict the immunophenotype [14] and molecular phenotype [15] of cells from hematoxylin and eosin (H&E)‐stained tissue images. We have also successfully developed a prognostic prediction system for liver cancer and bladder cancer using SVM [16, 17]. However, it has been difficult to predict the actual prognosis with greater than 90% accuracy with those methods. A major reason for this can be attributed to the complex and multifaceted characteristics of cancer atypia. In order to overcome this problem, solutions such as simply increasing the number of AI training cases or training more features have been attempted with only limited success. Instead, we propose an AI‐based prognosis prediction system that uses a novel multi‐model pipeline analysis of H&E‐stained sections of breast cancer extracted prior to NAC treatment. From our experience in AI pathology [16, 18] we have found that CNN can efficiently learn the patterns of structural atypia, while SVM and random forest (RF) combined with fine‐grained nuclear feature extraction are better at learning the patterns of nuclear atypia. Therefore, we constructed an independent AI model for each heteromorphic feature. With this novel pipeline system of multiple AI models, we succeeded in predicting NAC with 95.15% accuracy in a study of 103 unseen test cases. In the future, we believe that such pipeline diagnostic systems that combine AI models to learn the patterns of cancer atypia from various views of a histological sample will become mainstream in digital pathology.
Materials and methods
NAC response criteria
Numerous methods of histopathological determination of the therapeutic effect of preoperative chemotherapy have been reported [19, 20]. The common format of these methods consists of two categories, no or little effect and complete response, with subcategories in between [21]. In addition, treatment efficacy can be determined in four cases: invasive region only; invasive and noninvasive regions; invasive region and lymph nodes; and invasive and noninvasive regions and lymph nodes. In this study, we decided not to collect information on lymph node metastasis because the target specimens were needle biopsy tissue samples prior to NAC. In addition, we did not consider it necessary to limit the therapeutic effect of NAC to the invasive region. Based on the above conditions, we used the response evaluation criteria of the Japanese Breast Cancer Society (JBCS) 2007 [22], which have been reported to be able to stratify prognosis according to the degree of treatment effect [23, 24]. These criteria define four response grades: RG0/1/2/3 ranging from no response to full response (see supplementary material, Table S1).
Clinical information
In this study, a total of 310 female breast cancer patients treated with NAC at Yamaguchi University (26 cases), Tokyo Medical University (136 cases), and Weill Cornell Medicine (148 cases) were included. All cases used were invasive breast cancer, and NAC treatment was chosen as the first‐line treatment. To train the AI models, 207 of the 310 cases (about 70%) were randomly selected from each response grade (RG). The remaining 103 cases were used for evaluation. Details of tumor size, patient age, and cancer subtypes are summarized in Table 1. Sixty percent of the Japanese cases (Yamaguchi University and Tokyo Medical University) occurred in patients aged 40–60 years and 45% of the American cases (Weill Cornel Medicine) occurred in patients in the same age range. Sixteen percent of the American cases occurred in patients aged 30–39 years; therefore, a total of 61% of American cases occurred in patients aged 30–60 years. This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the ethics committees of Yamaguchi University, Tokyo Medical University (SH4140), and Weill Cornell Medicine (1404014987).
Table 1.
The details of sample information
| Training | Test | Total | |
|---|---|---|---|
| Number of cases | 207 | 103 | 310 (100%) |
| By subtype | |||
| Triple negative | 35 | 21 | 56 (18%) |
| Hormone receptor + | 96 | 47 | 143 (46%) |
| HER2+ | 27 | 13 | 40 (13%) |
| Hormone receptor/HER2+ | 49 | 22 | 71 (23%) |
| By NAC response grade | |||
| 0: No response | 30 | 10 | 40 (13%) |
| 1: Slight response | 69 | 44 | 113 (36%) |
| 2: Marked response | 49 | 21 | 70 (23%) |
| 3: Complete response | 59 | 28 | 87 (28%) |
| Age | |||
| Average | 53 | 53 | 53 |
| Max | 83 | 91 | 91 |
| Min | 26 | 27 | 26 |
| Size before NAC (cm) | |||
| Average | 4.1 | 3.7 | 3.9 |
| Max | 20 | 13 | 20 |
| Min | 1.3 | 1.3 | 1.3 |
NAC drug regimens
The patients included in this study were treated with an anthracycline (epirubicin or doxorubicin) plus cyclophosphamide‐based (EC or AC respectively) therapy or EC plus 5‐fluorouracil (FEC) and followed by taxane (paclitaxel/docetaxel/Abraxane). For the HER2 positive patients, treatment also included HER2 target drugs, trastuzumab/pertuzumab. For patients treated at Weill Cornell Medicine, eight cases were treated with CBDCA (carboplatin) followed by taxane and six cases also received preoperative endocrine/hormonal therapy (letrozole/anastrozole/exemestane). Patients who were treated with hormonal therapy only were not included in this study.
CNN analysis method
H&E‐stained slide specimens were scanned using a whole slide image (WSI) scanner (NanoZoomer; Hamamatsu Photonics, Hamamatsu, Japan) at ×20 image magnification. For CNN analysis, regions of interest (ROIs) of 256 × 256 pixels were automatically extracted at ×20 magnification to cover the entire needle biopsy specimen with 1 pixel corresponding to 0.46 μm. The ROI images were first color‐normalized using the Macenko method [25]. The image preparation process for CNN analysis is shown in Figure 1B. As the CNN model, we used ResNeXt [26], an improved version of ResNet‐50, with a fully connected head and five class outputs (RG0, RG1, RG2, RG3, and cancer/noncancer). Although the final output of the CNN for this study is only concerned with the response grade (RG0…3), we added an extra output (cancer/noncancer) to help the model learn more relevant features from the data. It has been shown in many studies that this type of multi‐task learning [27, 28] can significantly improve the ability of a deep model to learn by focusing it on more relevant features of the image and avoid over‐learning irrelevant features. The determination of whether an ROI was cancerous or noncancerous was made by a pathologist (MK). For training the CNN model, we used the Pytorch [29] toolset with the stochastic gradient descent optimizer and a learning rate of 1e−3, momentum = 0.9, weight_decay = 1e−3, and cross entropy as the loss function.
Figure 1.
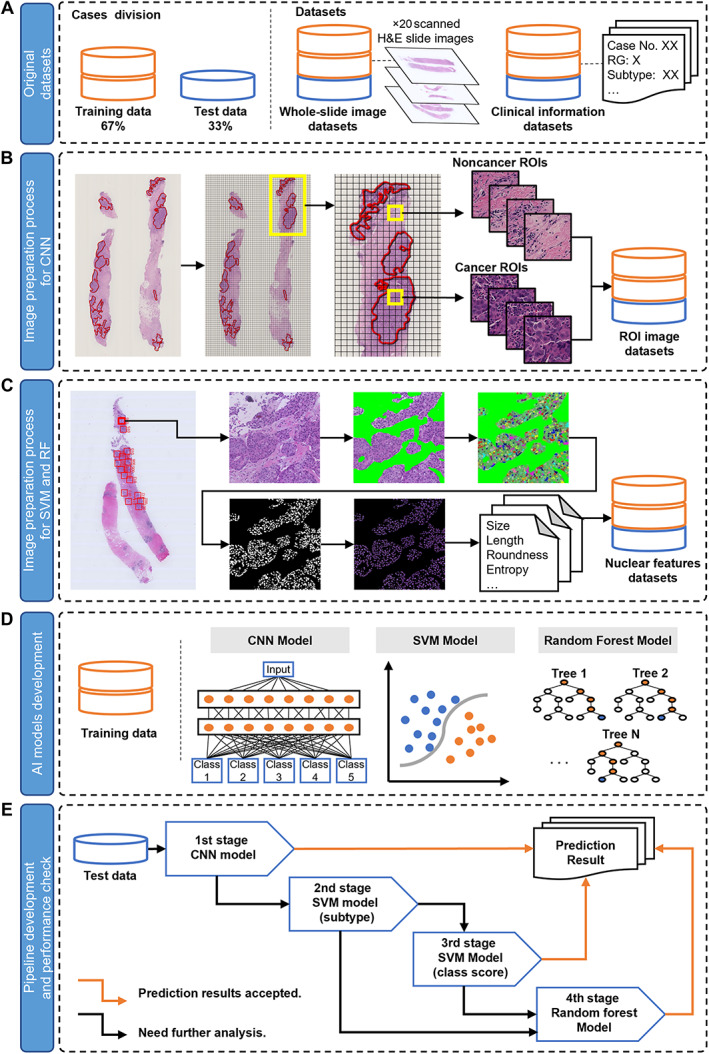
An overview of the dataset preparation and prediction model construction process. (A) Original datasets. (B) Image preparation process for CNN analysis. (C) Image preparation process for SVM and RF analysis. (D) AI models development process. (E) Pipeline development and performance check.
We stopped at 10 epochs when the loss on the validation set failed to improve.
SVM, RF, and t‐SNE analysis
For SVM and RF analysis, we extract ROI images of 2,048 × 2,048 pixels from WSI images at ×40 magnification. Noncancerous areas (stroma, fibrocystic change areas, and lymphocytes) and cancerous areas were first manually segmented by a pathologist (MK) from the selected ROI images. Then, using commercially available segmentation software, ilastik (version 1.1.8) [30], we performed an automatic nuclear segmentation in the cancerous areas. The image preparation process for SVM and RF analysis is shown in Figure 1C. As a result, a total of 1,563,586 and 952,770 nuclei were segmented from the set of training and test cases, respectively (supplementary material, Table S2). The features related to nuclear atypia were measured with another commercially available specialized cell analysis software: CellProfiler (version 2.1.1) [31]. We focused on identifying features of nuclear atypia in cancer cells that would be helpful in predicting the effect of NAC. In total, we obtained 82 statistical features from the segmented cell nuclei in each ROI. These features included: (1) morphological information related to the shape of the nucleus, such as size, contour length, major axis length, roundness, robustness, and eccentricity, and (2) nuclear texture‐related features, such as second angular momentum, uniformity, and entropy. Finally, we expanded these 82 nuclear features into a total of 960 features using a gray‐level pixel matrix co‐occurrence method named cell feature level co‐occurrence matrix (CFLCM [18]).
For t‐distributed stochastic neighbor embedding (t‐SNE) analysis, a tool for visualizing high‐dimensional datasets to confirm SVM results, we used the R package tsne embedded in R software (version 3.6.1, R Project for Statistical Computing; https://www.r-project.org). The parameter settings for this analysis were set to 1 for perplexity and 300 for step.
Results
CNN model that predicts NAC response included structural atypia
Morphological atypia of cancer can be broadly classified into two categories, structural atypia and nuclear atypia. Structural atypia manifests itself in the way the cancer cells arrange themselves into structures such as glands. Breast cancer is also a cancer that exhibits a variety of glandular structures, and therefore structural atypia is unique in each case. Therefore, we first trained a CNN model to learn the structural variations in breast cancer tissue that predict the effect of NAC. The cases used in the training and evaluation of the CNN were obtained from cases spanning four NAC therapy response grades (RG0/1/2/3, Supplementary Table S1), as well as four cancer subtypes. The number of cases in each class and set are listed in Table 1. We trained the CNN model with 530,173 ROIs from the cancer areas and 76,585 ROIs from the noncancer areas for a total of 606,758 ROIs obtained from the training set of 207 cases (supplementary material, Table S2). The ratio of training set to validation set is 8:2. Then we evaluated the performance of the trained model on the test set of 103 cases (169,533 ROIs in cancer areas and 82,572 ROIs in noncancer areas, 252,105 ROIs in total). The ROI‐level accuracy was 88.8% with a strong‐to‐perfect kappa agreement of 0.85 (the 4‐class confusion matrix is shown in supplementary material, Table S3B). Then, the ROI results were aggregated into case‐level results by averaging the class scores of the ROIs in a case. The results, by case, still showed a strong‐to‐perfect kappa agreement of 0.81 (Table 2A). A visual rendering of the ROI classification (RG0…3 and noncancer) by the CNN model of a representative case is shown in Figure 2. To further explore the performance of the trained model, we used IBM SPSS Statistics for windows (version 28.0) to perform the receiver operating characteristic (ROC) analysis (Figure 3), and calculated the values for area under the curve (AUC) of ROC. This showed that the AUC values of the CNN model on each RG were higher than 0.9 (Figure 3A).
Table 2.
CNN, SVM, and RF model analyses summarized into case‐based results
| (A) | ||||||
|---|---|---|---|---|---|---|
| Prediction | ||||||
| RG0 | RG1 | RG2 | RG3 | Total | ||
| Truth | RG0 | 9 | 0 | 0 | 1 | 10 |
| RG1 | 0 | 34 | 1 | 9 | 44 | |
| RG2 | 1 | 0 | 19 | 1 | 21 | |
| RG3 | 1 | 0 | 0 | 27 | 28 | |
| Total | 11 | 34 | 20 | 38 | 103 | |
| (B) | ||||||
|---|---|---|---|---|---|---|
| Prediction | ||||||
| RG0 | RG1 | RG2 | RG3 | Total | ||
| Truth | RG0 | 5 | 3 | 1 | 1 | 10 |
| RG1 | 0 | 41 | 0 | 3 | 44 | |
| RG2 | 1 | 5 | 13 | 2 | 21 | |
| RG3 | 0 | 2 | 0 | 26 | 28 | |
| Total | 6 | 51 | 14 | 32 | 103 | |
| (C) | ||||||
|---|---|---|---|---|---|---|
| Prediction | ||||||
| RG0 | RG1 | RG2 | RG3 | Total | ||
| Truth | RG0 | 6 | 0 | 2 | 2 | 10 |
| RG1 | 0 | 43 | 0 | 1 | 44 | |
| RG2 | 0 | 4 | 14 | 3 | 21 | |
| RG3 | 0 | 3 | 0 | 25 | 28 | |
| Total | 6 | 50 | 16 | 31 | 103 | |
| (D) | ||||||
|---|---|---|---|---|---|---|
| Prediction | ||||||
| RG0 | RG1 | RG2 | RG3 | Total | ||
| Truth | RG0 | 9 | 1 | 0 | 0 | 10 |
| RG1 | 0 | 42 | 2 | 0 | 44 | |
| RG2 | 0 | 0 | 21 | 0 | 21 | |
| RG3 | 0 | 0 | 0 | 28 | 28 | |
| Total | 9 | 43 | 23 | 28 | 103 | |
(A) CNN model analysis result of test cases (accuracy: 86.4%; 95% CI: 78.3–92.4%; κ = 0.81). (B) SVM model analysis (only RG information was used) result of test cases (accuracy: 82.5%; 95% CI: 73.8–89.3%; κ = 0.74). (C) SVM model analysis (RG and subtype information was used) result of test cases (accuracy: 85.4%; 95% CI: 77.1–91.6%; κ = 0.78). (D) RF model analysis (RG and subtype information was used) result of test cases (accuracy: 97.1%; 95% CI: 91.7–99.4%; κ = 0.96).
Figure 2.
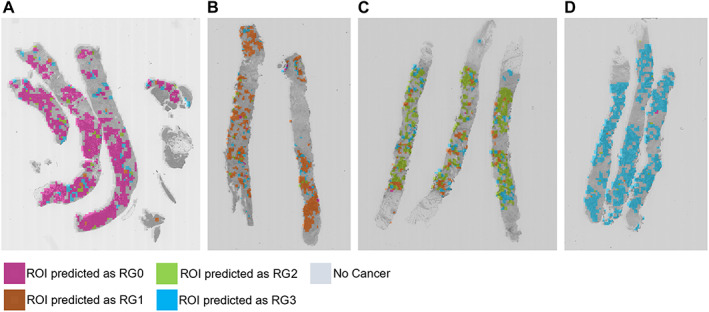
Results of CNN prediction on representative cases. (A) Case #62 is predicted as RG0 by CNN model; 2,646 ROIs are predicted as RG0 among all 3,056 ROIs in the cancer areas, and their average likelihood score is 16.921, which is higher than other RG groups. (B) Case #12 is predicted as RG1 by CNN model; 1,860 ROIs are predicted as RG1 among all 2,317 ROIs in the cancer area, and their average likelihood score is 15.655, which is higher than other RG groups. (C) Case #10 is predicted as RG2 by CNN model; 1,511 ROIs are predicted as RG2 among all 2,208 ROIs in the cancer areas, and their average likelihood score is 17.123, which is higher than other RG groups. (D) Case #15 is predicted as RG3 by CNN model; 2,468 ROIs are predicted as RG3 among all 2,523 ROIs in cancer areas, and their average likelihood score is 24.058, which is higher than other RG groups. The details of the CNN analysis results of these cases are shown in supplementary material, Table S9.
Figure 3.
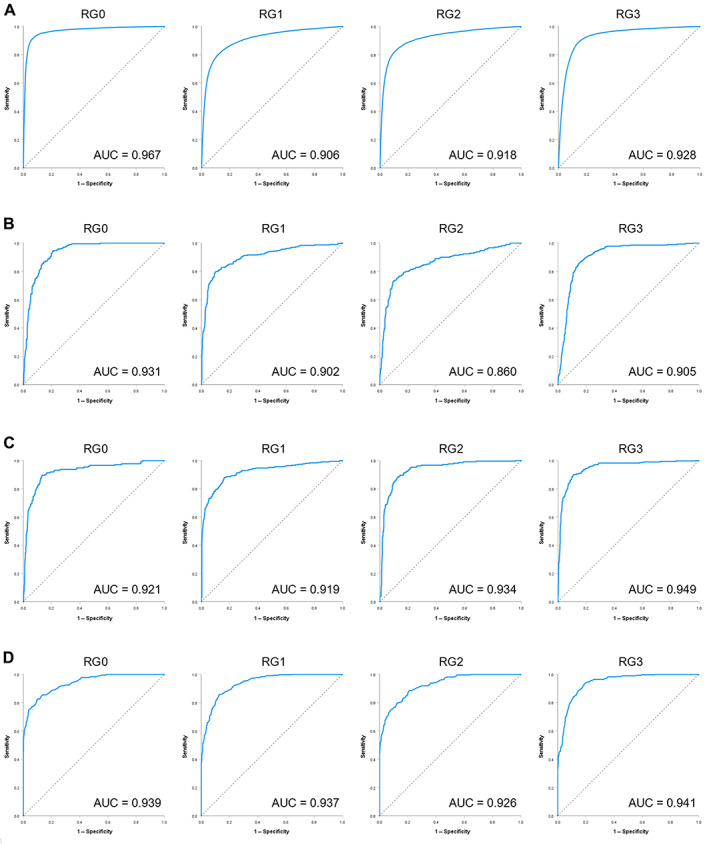
Performance check using ROC analysis on each RG. (A) The ROC curves of CNN prediction model. (B) The ROC curves of SVM prediction model used RG information only. (C) The ROC curves of SVM prediction model used RG and subtype information. (D) The ROC curves of RF prediction model.
SVM predicts NAC response via nuclear atypia
We next analyzed the nuclear atypia of the cancers. Nuclear atypia is reflected by changes to the shape of the cell's nucleus (enlarged size, irregular contour, etc) and to its texture (condensed chromatin and other variations). Therefore, we first sampled 3,366 ROIs from cancer regions of cases segmented by pathologists and trained a linear SVM model. The model was then evaluated on 1,545 ROIs sampled from the cancer regions of test set cases. The ROI‐level accuracy was found to be 72.6% with a moderate‐to‐strong kappa agreement of 0.6 (see the confusion matrix in supplementary material, Table S4B). Aggregating the results at the case level shows an accuracy of 82.5% and a strong kappa agreement of 0.74 (Table 2B). A visual rendering in Figure 4 shows the SVM model‐predicted RG grade for each selected ROI of a representative case.
Figure 4.
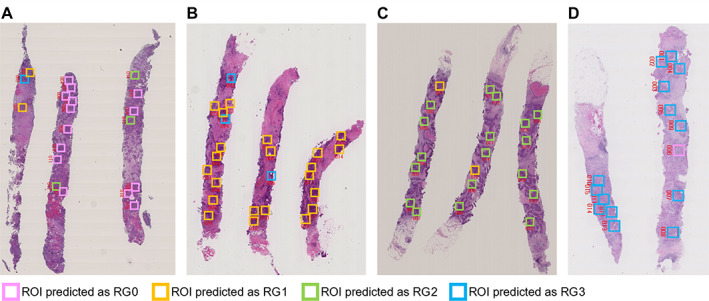
Results of SVM prediction on representative cases. (A) Case #41 is predicted as RG0 by SVM model. Fourteen ROIs are predicted as RG0 among all 15 ROIs selected in the cancer areas, and their average likelihood score is 0.48, which is higher than other RG groups. (B) Case #83 is predicted as RG1 by SVM model. Twenty ROIs are predicted as RG1 among all 26 ROIs selected in the cancer areas, and their average likelihood score is 0.72, which is higher than other RG groups. (C) Case #10 is predicted as RG2 by SVM model. Eighteen ROIs are predicted as RG2 among all 21 ROIs selected in the cancer areas, and their average likelihood score is 0.82, which is higher than other RG groups. (D) Case #65 is predicted as RG3 by SVM model. Fourteen ROIs are predicted as RG3 among all 18 ROIs selected in the cancer areas, and their average likelihood score is 0.78, which is higher than other RG groups. The details of the SVM analysis result of these cases are shown in supplementary material, Table S10. The red numbers near the ROIs indicate the location of the ROIs.
Additional subtype information increased accuracy of SVM and RF model
The test results showed that the prediction accuracy of the SVM was lower than that of the CNN. To visualize the robustness of the classification results, we performed a t‐SNE analysis using the training data. As a result, it was found that several groups of RGs were mixed in one cluster, both in terms of ROI results and cases, as shown in Figure 5A,B. This result suggests that the SVM prediction model is not able to sufficiently separate features in the training phase. To improve the accuracy of the SVM prediction model, we added class information based on the four subtypes of breast cancer, (i.e. triple negative [TN], hormone receptor positive [H+], HER2 positive [HER2+], and hormone receptor positive plus HER2 positive [H+ HER2+]), to the initial RG class information. Since there were no HER2+ cancers in RG0, we obtained a total of 15 classes. Retraining the SVM model using these 15 classes, the test set results improved. We obtained a ROI‐based classification accuracy of 63.7% with a moderate‐to‐strong kappa agreement of 0.58 (supplementary material, Table S5B) and a case‐based classification accuracy of 85.4% with a strong kappa agreement of 0.78 (Table 2C). The AUC values of the SVM model using additional subtype information (AUC = 0.919–0.949) (Figure 3B) were higher than the SVM model using RG information only (AUC = 0.860–0.931) (Figure 3C). To visualize the improved ability of the features to classify the data, we performed the t‐SNE analysis using the expanded classes and the same cases from the training set. While the separation of classes was not perfect when using 4 classes (ROI level Figure 5A and case level Figure 5B), when using 15 classes, all classes are well separated (ROI level Figure 5C and case level Figure 5D), showing that adding the subtype helps classification. Subsequently, we obtained the subtype prediction results of the retrained SVM model. As the subtype information of breast cancer is reflected mostly by the characteristics of the cell nucleus [14, 15], we did not use it for the CNN analysis of structural atypia.
Figure 5.
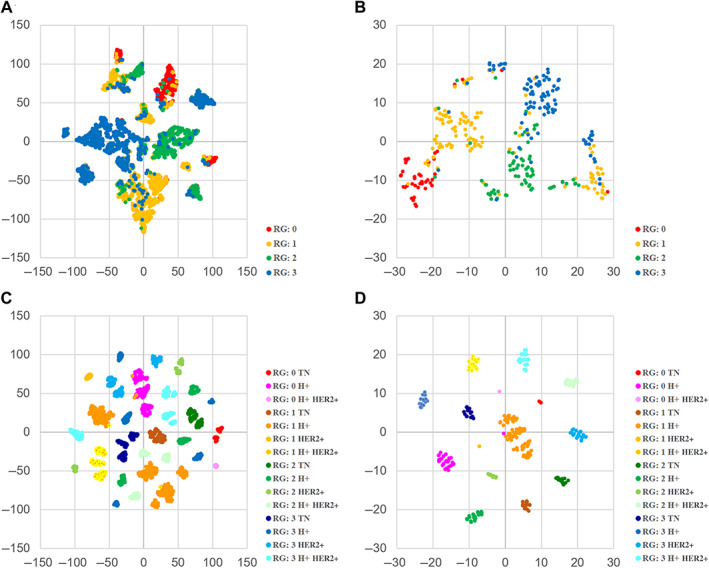
t‐SNE analysis of SVM likelihood score. Two‐dimensional map visualization by t‐SNE to confirm the prediction accuracy of the classification results of training cases. (A) The visualization is based on the prediction result of all cancer region ROIs. SVM prediction model used RG information only. (B) The visualization is based on the prediction result of each case. SVM prediction model used RG information only. (C) The visualization is based on the prediction result of all cancer region ROIs. SVM prediction model used RG and subtype information. (D) The visualization is based on the prediction result of each case. SVM prediction model used RG information and subtype information.
In addition to the linear SVM, we also trained a RF model, which is a nonlinear classification method. For RF experiments, we used the same setting as for the SVM and predicted 15 classes. On the test set, we obtained a ROI‐level accuracy of 72.3% with a strong kappa agreement of 0.66 (supplementary material, Table S6B) and a case‐level accuracy of 97.1% with a perfect kappa agreement of 0.96 (Table 2D). To confirm the potential accuracy of the RF model, the out‐of‐bag (OOB) error was calculated. The results showed that the OOB error rate using the training data was 17.76% (supplementary material, Table S7D), indicating a potential accuracy of the RF model of 82.24%, which is in line with our empirical findings.
Development of integrated pipeline system
As shown above, we established three independent models for predicting the effects of NAC based on the characteristics of morphological variants of cancer. To further improve the prediction accuracy, we combined these models into a pipeline system consisting of four stages (Figure 6).
Figure 6.
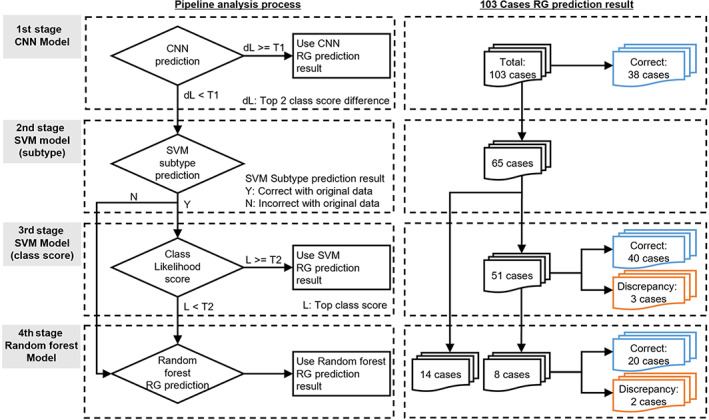
Prognosis prediction pipeline system using three AI models. The pipeline consists of a CNN model as the first step, a distributor as the second step, an SVM model as the third step, and an RF model as the fourth step. Of the 103 cases, 38 cases were determined in the first pipeline and 65 cases for which determination was withheld in the first pipeline were moved to the second pipeline. Of the 65 cases, the second pipeline distributed 51 cases to the third pipeline and 14 cases to the fourth pipeline.
For the first stage of the pipeline, we decided to use the CNN model because it had the highest potential accuracy among the three models, as supported by the OOB error. If the CNN can reliably classify a sample, the pipeline system returns the CNN result. On the other hand, when the difference in class output score is small between the top two classes, it means that the model hesitates between two classes and therefore cannot reliably classify the sample. In that case, the pipeline continues into the second stage. The threshold for the score difference is obtained empirically by using a value that allows all training examples correctly classified by the CNN to be classified by the first stage of the pipeline.
In the second stage of the pipeline, the SVM model trained with 15 classes is applied. The subtype prediction results (TN, H+, HER2+, H+ HER2+) of the SVM model are used to compare with the original results to assign to subsequent stages. If the SVM model predicts the wrong cancer subtype we decide to use the RF model instead and go to stage 4. If the SVM predicts the correct cancer subtype, we move to stage 3, where the class score is tested for its confidence level. If the confidence is high (score ≥ 0.5) the pipeline system returns the SVM result.
If the fourth stage of the pipeline is reached, the RF model is applied, and its result is returned by the pipeline.
Prognosis prediction using the pipeline system
We used the test data of 103 cases to verify the accuracy of the pipeline system (Figure 6). The results of the first stage analysis showed that for 38 of 103 patients the CNN model was able to make a final decision. There were no prediction discrepancies in those cases. The remaining 65 cases were then analyzed in the second stage, and the cases were assigned to subsequent stages. As a result, 51 cases were distributed to the third stage, and 14 cases were distributed to the fourth stage. In the third stage, 43 of 51 cases were confirmed by the SVM model, and 8 cases below the threshold of 0.5 were left to be determined in the fourth stage by the RF model. Of the 43 cases with final results in the third stage, there were 3 cases in which the prediction did not match the results. Of these three cases, two were RG0 H+ cases, and the third stage incorrectly predicted RG2. One case was an RG1 H+ case and the third stage incorrectly predicted RG2. Finally, using the RF model in the fourth stage, we analyzed 14 cases distributed from the second stage and 8 cases judged to be pending in the third stage. As a result, among those 22 cases, 2 cases did not match the results. Of these two cases, one was an RG0 H+ case, which the fourth stage incorrectly predicted as RG1. The other case was RG2 HER2+, and the fourth stage incorrectly predicted RG3. Overall, the fusion pipeline predicted 98 of 103 cases correctly, with an accuracy of 95.15% (95% confidence interval [CI]: 89.03–98.41%). This fusion pipeline was able to improve the accuracy by approximately 10% over each single model. Detailed results of the integrated pipeline system are shown in supplementary material, Table S8.
Finally, we compared our pipeline system with a simple majority voting method. Using the majority voting, we were able to successfully predict 97 of 103 test cases with an accuracy of 94.17% (95% CI: 87.75–97.83%) (supplementary material, Table S8). The superior performance of the pipeline system can be explained by a multi‐redundancy safety mechanism that takes advantage of the characteristics of the combined systems, and minimizes the effects of errors in each system, leading to improved prognostic accuracy. On the other hand, the majority voting approach still provides excellent result, is more general and does not require empirical thresholds.
Discussion
In this study, we successfully developed a system to predict the effect of NAC on breast cancer by combining multiple machine learning models into a pipeline system. We used a CNN model in the first stage of the pipeline to predict the RG based on the tissue structure, which follows the diagnostic process of pathologists, where the entire slide is viewed at low magnification. Then, cases that could not be confirmed in the first stage of the pipeline were analyzed using only nuclear information by the SVM and RF models. This approach is similar to the process by which pathologists definitively confirm a diagnosis at high magnification. We measured model confidence from its output scores to decide whether a model's result was to be trusted and used or whether another model would be applied. The linear SVM model was used before the RF model, which is nonlinear and has the possibility of overfitting. In the end, the pipeline system developed in this study achieved approximately 10% higher accuracy than any of the single model. In recent years, ensemble learning, in which multiple independent models are developed and the results of each model are combined by majority vote, has become a popular method for AI learning and we show that this approach also works very well in our setting, reaching a slightly lower performance than the pipeline system. The use of a CNN model greatly contributes to the overall accuracy, being able to learn the many structural variants of breast cancer. We show that combining deep learning CNN models and classic machine learning SVM/RF models closely matches the histological characteristics of cancer by focusing on both structural patterns and precise nuclear features, and leads to improved accuracy.
In SVM and RF models, the prediction accuracy of RG was remarkably improved by adding the information of the four subtypes of breast cancer. We confirmed this finding with a t‐SNE visual analysis that showed an improved and clearer clustering of the classes when adding cancer subtype information. This result shows that there may be common factors that define the subtype of breast cancer and nuclear characteristics. In both SVM and RF models, it is possible to rank the input features in order of importance for classification. We plan to perform such analysis in the future to understand which particular nuclear phenotypes are expressed by each RG or cancer subtype.
Considering future clinical applications, the automation of the entire system is a very important point. While the CNN model was trained and evaluated from ROIs sampled automatically from the training images, the SVM and RF models were trained and evaluated using ROIs that were manually sampled from the annotated cancer regions. Considering a fully automatic system for clinical use, we would need to apply the SVM and RF classifier to all the ROIs classified by the CNN as cancerous but which RG class prediction is deemed unreliable by the pipeline. Based on the promising results of this study, we plan to evaluate such a fully automated system in a future work.
Author contributions statement
BS, AS and MK conceived and designed the study. BS, AS and MK developed the methodology. All authors were involved in investigation, acquisition, analysis and interpretation of data. MK drafted the paper. BS and AS prepared the figures. BS, EC and MK reviewed and edited the paper. AHM, HPG and EC supervised the study. MK acquired funding. All authors had final approval of the paper.
Ethics approval and consent to participate
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the ethics committees of Yamaguchi University, Tokyo Medical University (SH4140), and Weill Cornell Medicine (1404014987).
Supporting information
Table S1. JBCS histopathological criteria for the assessment of therapeutic response in breast cancer (2007 version)
Table S2. Number of ROIs for the CNN, SVM, and RF models
Table S3. The prediction result for each ROI by CNN model
Table S4. The prediction result for each ROI by the SVM model, trained by RG information only
Table S5. The prediction result for each ROI by the SVM model, trained by RG and subtype information
Table S6. The prediction result for each ROI by the RF model, trained by RG and subtype information
Table S7. CNN, SVM, and RF model analysis summarized into case‐based results on training cases
Table S8. Test case results of the pipeline system and majority voting method
Table S9. Detailed analysis results of CNN heat map example cases
Table S10. Detailed analysis results of SVM heat map example cases
Acknowledgements
This work was supported by grants from the JSPS KAKENHI grant numbers (17H04067 and 21H02706 to MK, 18K07027 to AS). This work was also supported in part by the Strategic Research Foundation Grant‐aided Project (S1511011) for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MK).
No conflicts of interest were declared.
Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The self‐developed algorithm CFLCM used for this study is available at the following link: https://github.com/Shen-tokyomed/Breast_AI_CFLCM_tool.
References
- 1. Sutton EJ, Onishi N, Fehr DA, et al. A machine learning model that classifies breast cancer pathologic complete response on MRI post‐neoadjuvant chemotherapy. Breast Cancer Res 2020; 22: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tudorica A, Oh KY, Chui SY, et al. Early prediction and evaluation of breast cancer response to neoadjuvant chemotherapy using quantitative DCE‐MRI. Transl Oncol 2016; 9: 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tran WT, Gangeh MJ, Sannachi L, et al. Predicting breast cancer response to neoadjuvant chemotherapy using pretreatment diffuse optical spectroscopic texture analysis. Br J Cancer 2017; 116: 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tadayyon H, Sannachi L, Gangeh MJ, et al. A priori prediction of neoadjuvant chemotherapy response and survival in breast cancer patients using quantitative ultrasound. Sci Rep 2017; 7: 45733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. von Minckwitz G, Blohmer JU, Costa SD, et al. Response‐guided neoadjuvant chemotherapy for breast cancer. J Clin Oncol 2013; 31: 3623–3630. [DOI] [PubMed] [Google Scholar]
- 6. Yang Y, Im SA, Keam B, et al. Prognostic impact of AJCC response criteria for neoadjuvant chemotherapy in stage II/III breast cancer patients: breast cancer subtype analyses. BMC Cancer 2016; 16: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raciti P, Sue J, Ceballos R, et al. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod Pathol 2020; 33: 2058–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kather JN, Krisam J, Charoentong P, et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med 2019; 16: e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song Z, Zou S, Zhou W, et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun 2020; 11: 4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Veta M, Kornegoor R, Huisman A, et al. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol 2012; 25: 1559–1565. [DOI] [PubMed] [Google Scholar]
- 11. Nawaz S, Heindl A, Koelble K, et al. Beyond immune density: critical role of spatial heterogeneity in estrogen receptor‐negative breast cancer. Mod Pathol 2015; 28: 766–777. [DOI] [PubMed] [Google Scholar]
- 12. Oikawa K, Saito A, Kiyuna T, et al. Pathological diagnosis of gastric cancers with a novel computerized analysis system. J Pathol Inform 2017; 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamamoto Y, Saito A, Tateishi A, et al. Quantitative diagnosis of breast tumors by morphometric classification of microenvironmental myoepithelial cells using a machine learning approach. Sci Rep 2017; 7: 46732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naik N, Madani A, Esteva A, et al. Deep learning‐enabled breast cancer hormonal receptor status determination from base‐level H&E stains. Nat Commun 2020; 11: 5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rawat RR, Ortega I, Roy P, et al. Deep learned tissue “fingerprints” classify breast cancers by ER/PR/Her2 status from H&E images. Sci Rep 2020; 10: 7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saito A, Toyoda H, Kobayashi M, et al. Prediction of early recurrence of hepatocellular carcinoma after resection using digital pathology images assessed by machine learning. Mod Pathol 2021; 34: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tokuyama N, Saito A, Muraoka R, et al. Prediction of non‐muscle invasive bladder cancer recurrence using machine learning of quantitative nuclear features. Mod Pathol 2022; 35: 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saito A, Numata Y, Hamada T, et al. A novel method for morphological pleomorphism and heterogeneity quantitative measurement: named cell feature level co‐occurrence matrix. J Pathol Inform 2016; 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buchbender C, Kuemmel S, Hoffmann O, et al. FDG‐PET/CT for the early prediction of histopathological complete response to neoadjuvant chemotherapy in breast cancer patients: initial results. Acta Radiol 2012; 53: 628–636. [DOI] [PubMed] [Google Scholar]
- 20. Kaise H, Shimizu F, Akazawa K, et al. Prediction of pathological response to neoadjuvant chemotherapy in breast cancer patients by imaging. J Surg Res 2018; 225: 175–180. [DOI] [PubMed] [Google Scholar]
- 21. Horii R, Akiyama F. Histological assessment of therapeutic response in breast cancer. Breast Cancer 2016; 23: 540–545. [DOI] [PubMed] [Google Scholar]
- 22. Kurosumi M, Akashi‐Tanaka S, Akiyama F, et al. Histopathological criteria for assessment of therapeutic response in breast cancer (2007 version). Breast Cancer 2008; 15: 5–7. [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi K, Horii R, Ito Y, et al. Prognostic significance of histological therapeutic effect in preoperative chemotherapy for breast cancer. Pathol Int 2016; 66: 8–14. [DOI] [PubMed] [Google Scholar]
- 24. Mukai H, Arihiro K, Shimizu C, et al. Stratifying the outcome after neoadjuvant treatment using pathological response classification by the Japanese Breast Cancer Society. Breast Cancer 2016; 23: 73–77. [DOI] [PubMed] [Google Scholar]
- 25. Macenko M, Niethammer M, Marron JS , et al. A method for normalizing histology slides for quantitative analysis. In 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE: Boston, 2009; 1107–1110.
- 26. Xie S, Girshick R, Dollár P, et al. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, 2017; 1492–1500.
- 27. Graziani M, Otálora S, Muller H , et al. Guiding CNNs towards relevant concepts by multi‐task and adversarial learning. arXiv preprint arXiv:200801478 2020. [Not peer reviewed].
- 28. Caruana R. Multitask learning. Mach Learn 1997; 28: 41–75. [Google Scholar]
- 29. Paszke A, Gross S, Massa F, et al. Pytorch: an imperative style, high‐performance deep learning library. Adv Neural Inf Process Syst 2019; 32: 8024–8035. [Google Scholar]
- 30. Berg S, Kutra D, Kroeger T, et al. Ilastik: interactive machine learning for (bio)image analysis. Nat Methods 2019; 16: 1226–1232. [DOI] [PubMed] [Google Scholar]
- 31. Kamentsky L, Jones TR, Fraser A, et al. Improved structure, function and compatibility for CellProfiler: modular high‐throughput image analysis software. Bioinformatics 2011; 27: 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. JBCS histopathological criteria for the assessment of therapeutic response in breast cancer (2007 version)
Table S2. Number of ROIs for the CNN, SVM, and RF models
Table S3. The prediction result for each ROI by CNN model
Table S4. The prediction result for each ROI by the SVM model, trained by RG information only
Table S5. The prediction result for each ROI by the SVM model, trained by RG and subtype information
Table S6. The prediction result for each ROI by the RF model, trained by RG and subtype information
Table S7. CNN, SVM, and RF model analysis summarized into case‐based results on training cases
Table S8. Test case results of the pipeline system and majority voting method
Table S9. Detailed analysis results of CNN heat map example cases
Table S10. Detailed analysis results of SVM heat map example cases
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The self‐developed algorithm CFLCM used for this study is available at the following link: https://github.com/Shen-tokyomed/Breast_AI_CFLCM_tool.


