Summary
Synaptic maturation is reportedly limited in human induced pluripotent stem cell (iPSC)-derived neurons. Notably, their ability to reach postnatal-like stages and form dendritic spines has been difficult to demonstrate unless using long-term cultured organoids. Recent transcription factor (TF)-based induction methods allow the accelerated generation of differentiated neurons, which offers an unprecedented opportunity to address further progression into late developmental stages. Herein, we report on a comprehensive time-course study of TF-induced iPSC neurons cultured in vitro through an intrinsic maturation program following neurogenesis. Moreover, we determined the transcriptional and morphological sequences of key developmental events associated with spinogenesis, including the conversion of drebrin to its brain-specific isoform A and the N-methyl-D-aspartate (NMDA) receptor subunit switch. TF-induced iPSC neurons successfully acquired structural and functional synaptic maturity, which will critically expand their utility in modeling higher brain functions and disorders.
Subject areas: Cell biology, Neuroscience, Stem cells research
Graphical abstract
Highlights
-
•
Synaptic maturation and spinogenesis of cultured human iPSC-derived neurons
-
•
TF-induced iPSC neurons reproduce postnatal brain development features
-
•
Robust dendritic spine formation is shown by drebrin A expression and localization
-
•
TF-induced iPSC neurons acquire mature functions underlying synaptic plasticity
Cell biology; Neuroscience; Stem cells research
Introduction
Human induced pluripotent stem cell (iPSC)-derived neurons are promising tools for advancing in vitro neurological disease modeling, with potential applications in drug discovery and preemptive medicine.1 Human neurons available on demand offer a substantial opportunity to elucidate cellular responses and pharmacological mechanisms related to higher brain function, which have been previously studied only by animal experimentation or in rodent cell cultures. Synaptically functional neurons are particularly useful for studying neurodegenerative diseases and neurodevelopmental diseases associated with synaptic dysfunction.2,3 However, iPSC-derived neurons generally display limited synaptic maturation, with a considerably slower development than that of rat primary neurons,4,5 incomplete postsynaptic structure formation,6 or undetectable postsynaptic N-methyl-D-aspartate (NMDA) receptor activity.7,8 Particularly, it is unclear if iPSC-derived neurons consistently form dendritic spines.3 Despite a few reports on the morphological observations of spine-like protrusions in long-term cultures9 or brain organoids,10 researchers have not yet demonstrated the spinal functionality in iPSC-derived neurons.
Dendritic spines are postsynaptic structures essential for excitatory synapse transmission and plasticity. They contain high concentrations of the actin cytoskeleton,11 which dynamically controls their motility during synaptic plasticity12 and stabilization during postnatal maturation.13 Spine actin dynamics are governed by drebrin, the actin-binding protein, during neuronal development.14,15 In rat hippocampal neurons, dendritic filopodia form transient excitatory synapses; subsequently, drebrin accumulates into clusters at the synaptic sites to initiate spine morphogenesis and recruit postsynaptic density components, such as postsynaptic density protein 95 (PSD-95).14 Drebrin is highly conserved between rats and humans16; therefore, drebrin cluster formation is expected to be a marker for mature dendritic spines in iPSC-derived neurons.6,17 Furthermore, drebrin distribution changes upon the activation of NMDA receptors,18 which has been proposed as a crucial mechanism in mature synapse functions, such as long-term potentiation and toxicity-induced synaptic loss (19 for review). Therefore, the induction of changes in the drebrin cluster density by glutamate stimulation is used as a functional assay indicative of postsynaptic NMDA receptor activity.
In recent years, researchers have made substantial progress in the development of efficient differentiation methods for iPSC-derived neurons. Traditional approaches involve extrinsic growth factors and morphogens but require complex and variable culture procedures (20 for review). Alternatively, strategies for inducing differentiation by expressing transcription factors (TFs) are becoming popular by enabling rapid and better-controlled neurogenesis (21 for review). The induced expression of a single TF, such as NGN28 or ASL1,22 as well as the introduction of synthetic mRNAs combining multiple neurogenic TFs,23 effectively generates pure neuronal populations. Particularly, mRNA-based approaches offer several technical advantages. mRNAs are rapidly translated without integration into the genome; thus, they facilitate the footprint-free delivery of TFs with high expression. mRNA production and transfection can be easily scaled to any type of iPSC culture, thereby facilitating their application in an industrial context, particularly in the prospect of using various patient-derived iPSC lines. Eventually, multiple TFs can be simultaneously combined, thus potentially increasing the efficiency of cell-type specification.23 However, demonstrating functional maturation in these “TF-induced iPSC neurons” remains challenging21 and is critical for validating their relevance as neuronal models.
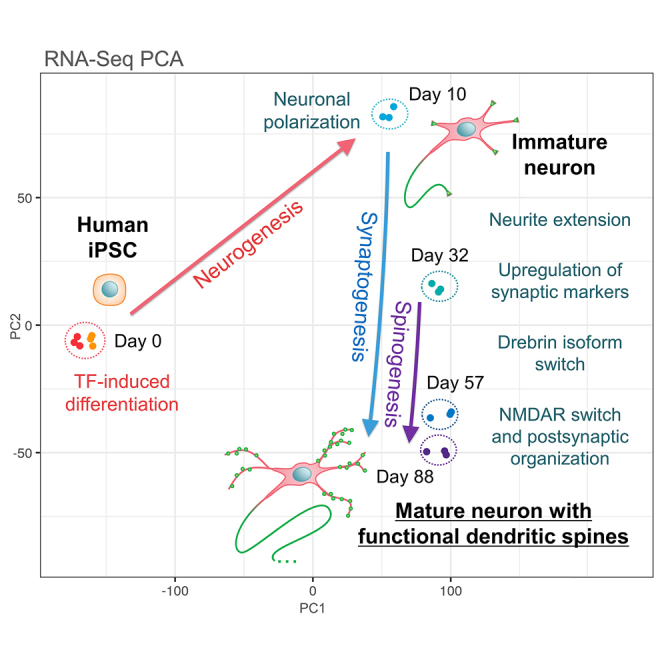
In this study, we report on a comprehensive time-course characterization of TF-induced iPSC neurons in long-term cultures for up to 3 months. RNA-sequencing (RNA-Seq) and immunocytochemistry (ICC) revealed transcriptional and morphological progression from an immature to a completely mature neuronal development stage.
Results
Culture and initial characterization of transcription factor-induced pluripotent stem cell neurons
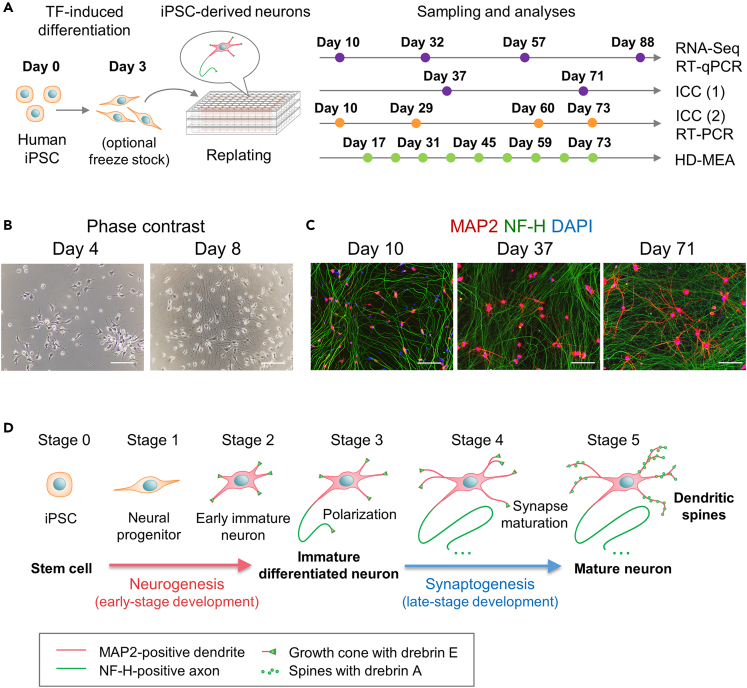
To evaluate neuronal differentiation and maturation, we cultured human iPSC-derived neurons generated by the transient delivery of TF mRNAs.23 The manufacturer froze these TF-induced iPSC neurons on day 3 following the induction of differentiation for long-term storage (Elixirgen Scientific, Baltimore, USA). Optimized culture conditions allowed the maintenance of healthy neurons for 3 months after thawing and replating (Figure 1A). As expected from previous studies,23 we observed the rapid elongation of neurites within few days of culture (Figure 1B). TF-induced iPSC neurons achieved neuronal polarization at day 10, as depicted by the expression and segregated the localization of the dendrite-specific microtubule-associated protein (MAP2) and axon-specific neurofilament heavy chain (NF-H) (Figure 1C). MAP2-positive dendrites remained short in the first month of culture, with most of the cells displaying a single, longer dendrite that presumably established axonal polarity. Interestingly, dendritic growth appeared to be inhibited for a while by possible negative feedback signals during axonal specification (24 for review), then the elongation and branching of MAP2-positive dendrites markedly increased after a dense axonal net has formed in long-term cultures (Figure 1C). We hypothesized that TF-induced iPSC neurons undergo sequential stages of neuronal development, similar to the canonical 5-stage classification established for cultured rat hippocampal neurons25,26 (Figure 1D). TF-induced iPSC neurons rapidly reached stage 3 by day 10 and further progressed into the subsequent stages of dendritic maturation and synaptogenesis. Therefore, we investigated if these neurons could reach stage 5 of development with the complete maturation of dendritic spines.
Figure 1.
The time-course characterization of neuronal development in TF-induced iPSC neurons
(A) An overview of the cell culture and sample collection schedule. The induction of differentiation has been initiated on day 0 by the overexpression of neurogenic TFs in human iPSC. TF-induced iPSC neurons have been cultured and collected for analysis, as indicated by the colored dots. Dots with different colors indicate independent time-course series, using different sets of iPSC cultures from day 0.
(B) Phase-contrast microscopy images of TF-induced iPSC neurons displaying the rapid development of neurites in the early days of culture following the induction of differentiation.
(C) Immunofluorescence images displaying the expression of the axon-specific marker NF-H (green) and the dendritic marker MAP2 (red) on days 10, 32, and 71. Nuclei are stained with DAPI. White scale bar: 100 μm.
(D) The schematic representation of the hypothetical stages of development in cultured TF-induced iPSC neurons. The stages have been adapted from previously defined stages in cultured rat primary neurons25,26 and modified to include the changes in drebrin subcellular localization during development.19 We have classified the following stages: stage 1, cells at a progenitor state; stage 2, cells beginning neurite elongation; stage 3, differentiated neurons with the establishment of neuronal polarity; stage 4, the late development of MAP2-positive dendrites; and stage 5, the maturation of dendritic components, including postsynaptic structures and drebrin A clusters at the dendritic spines.
TF: transcription factor; iPSC: induced pluripotent stem cell; NF-H: neurofilament heavy chain; MAP2: microtubule-associated protein 2; and DAPI: 4′,6-diamidino-2-phenylindole.
Distinct developmental phases revealed by the RNA-seq analysis of transcription factor-induced pluripotent stem cell neurons
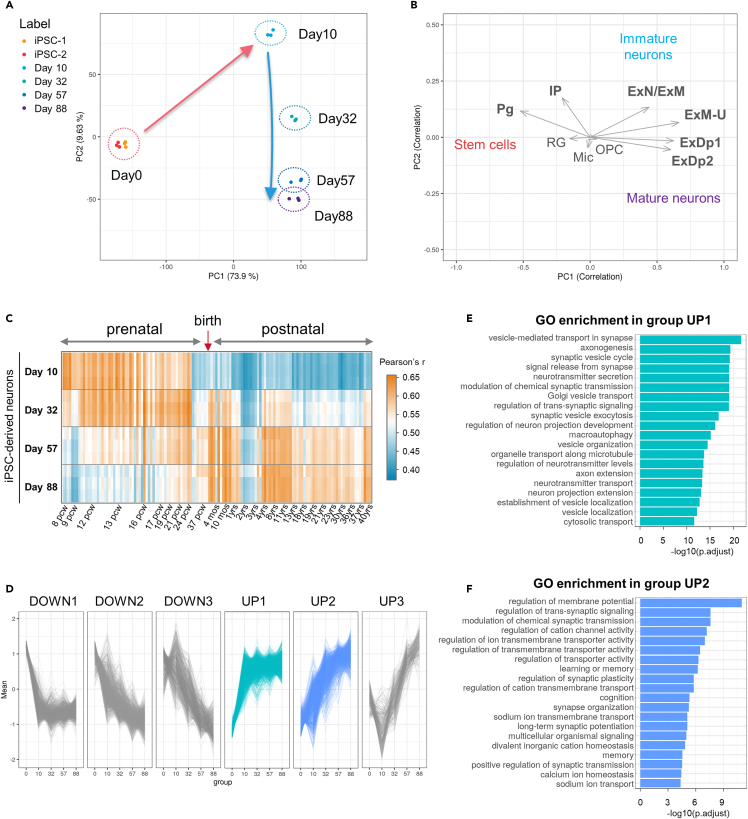
We performed a time-course transcriptome analysis (Figure 2), using bulk RNA-seq of cell samples selected in triplicate on day 10, characterized by complete neuronal differentiation,23 and subsequently on days 32, 57, and 88 (i.e., at approximately 1 month, 2 months, and 3 months). The cells were cultured as spheroids to facilitate the handling of a high number of samples (please refer to STAR Methods); although cellular density might affect neuronal development, we confirmed afterward that there was no significant difference in our conditions since spheroid and monolayer cultures reached similar expression profiles at day 88 (Figure S1). Moreover, we selected two independent cultures of undifferentiated iPSCs as the “day 0” control samples.
Figure 2.
Global transcriptome analysis of cultured TF-induced iPSC neurons by RNA-Seq
(A) Principal component analysis (PCA) of the cell samples at day 0 (iPSC-1 and iPSC-2), days 10, 32, 57, and 88 of culture following the induction of differentiation.
(B) Correlation with the factor loadings of various brain cell types of the human neocortex. The direction most correlated with a specific cell type is represented as the average factor loading vector of the cell type-specific marker genes. The cell types have been selected and annotated according to the definitions from the CoDEx dataset.27
(C) A correlation heatmap of the gene expression in TF-induced iPSC neurons, compared with the developing human brain of the Brainspan dataset.28 The analysis includes data from the prefrontal cortex in the prenatal phase (post-conception week 8-26) and the postnatal/adult phase (4 months-40 years following birth).
(D) Gene expression profiles categorized into six patterns by hierarchical clustering. Each line is plotted from the mean expression levels of a gene with a significant variation across the time-course.
(E and F) The leading 20 significantly enriched Gene Ontology (GO) terms for biological processes in groups UP1 and UP2. The GO enrichment analyses for other groups are reported in Figure S4.
Pg: Cycling progenitors; IP: Intermediate progenitors; ExN: Migrating excitatory neurons; ExM: Maturing excitatory neurons; ExM-U: Maturing excitatory upper enriched; ExDp1: Excitatory deep layer 1; ExDp2: Excitatory deep layer 2; RG: Radial Glia; Mic: Microglia; and OPC: Oligodendrocyte precursor cells.
A principal component analysis (PCA) revealed that triplicate samples from a particular time point clustered together, thus reflecting the high reproducibility of their expression profiles (Figure 2A). The PCA revealed two distinct trajectories as follows: (i) a trajectory with the chief variation along PC1 from day 0 to day 10, reflecting the rapid initial differentiation from iPSCs and (ii) a trajectory along PC2 from days 10-88, thus suggesting a gradual transition in long-term cultures. To estimate the composition of our cell populations at each time point, we compared our data with the brain cell type-specific marker genes defined in the Cortical Development Expression (CoDEx) dataset of the human neocortex27 (Figure 2B). We established the direction of correlation for each cell type by plotting the average vector of the cell-type marker factor loadings in the variable PCA space. Figure S3 summarizes the factor-loading plots for each cell type. The cells at day 0 were expectedly adjacent to the direction of “cycling progenitors” (Pg). This is because undifferentiated iPSC expressed stem cell and proliferation-related genes. On day 10, the cells shifted toward the “Excitatory migrating/maturing neurons” (ExN/ExM) during neurogenesis, followed by cortical “Excitatory upper enriched” (ExM-U) after day 32 and eventually to “Excitatory deep layer” (ExDp) neurons on days 57-88. Thus, the cells progressed to a more mature state resembling the cortical layer neurons following the completion of the cell cycle exit and neurogenesis on day 10. Notably, the glial cells were not correlated in any direction owing to the absence of the majority of markers for radial glia (RG), microglia (Mic), or oligodendrocytes (OPC) from the cultures. We anticipated this finding because we used an optimized feeder cell-free method of neuronal culture to avoid the generation of non-neuronal cells.
In addition, we compared our expression data with the Brainspan dataset,28 which provides a reference for differentially expressed genes during human brain development (Figure 2C). We established Pearson’s correlations between each sample of TF-induced iPSC neurons and the brain prefrontal cortex samples. A heatmap analysis revealed that the profiles at day 10 were closest to the prenatal phase at approximately 8-13 post-conception weeks (PCW) and were poorly correlated with the postnatal phase. Day 32 samples were correlated with the late prenatal phase from 12 PCW to 26 PCW and lost correlations with the early phase before 9 PCW. We observed a substantial transition to the postnatal phase, beginning from day 57. Eventually, day 88 samples were strongly correlated with the postnatal/adult phase and lost the majority of the prenatal correlations. Thus, TF-induced iPSC neurons appeared to reproduce a time-dependent transition to a postnatal state that paralleled in vivo development.
Furthermore, we grouped the genes by the hierarchical clustering of their expression profiles (Figure 2D). A Gene Ontology (GO) enrichment analysis revealed that the genes immediately upregulated and reached a plateau on day 10 (group UP1), and were typically related to axogenesis, neuronal projection, and the formation and trafficking of presynaptic vesicles, thus suggesting active neurite development upon differentiation (Figure 2E). In contrast, the genes that were upregulated after day 32 (group UP2) were involved in the regulation of transmembrane ion transport and mature synaptic functions (learning or memory, cognition, synaptic plasticity, and long-term potentiation) (Figure 2F). This finding suggested an increase in synaptogenesis and neurotransmission activities at future time points, which was consistent with the acquisition of a postnatal profile. Figure S4 describes the other groups in detail. Group UP3 was enriched in purine metabolism, oxidative phosphorylation, and respiratory functions, presumably to meet the high demand for ATP synthesis during neurogenesis29,30 and neurotransmission.31 The downregulated groups were frequently enriched in RNA processing and splicing functions, which reflected the drastic translational changes required to achieve such complex cellular differentiation. Moreover, group DOWN2 contained genes related to nuclear division and chromosome/chromatid segregation, probably upon the cell cycle exit of differentiated neurons.
Collectively, the RNA-seq results indicated that TF-induced iPSC neurons possessed an intrinsic ability to enter the maturation and synaptogenesis phases following the initial rapid neurogenesis phase. Furthermore, the late transition to a postnatal-like profile suggested that the cells followed a progressive sequence of development to reach a completely mature stage.
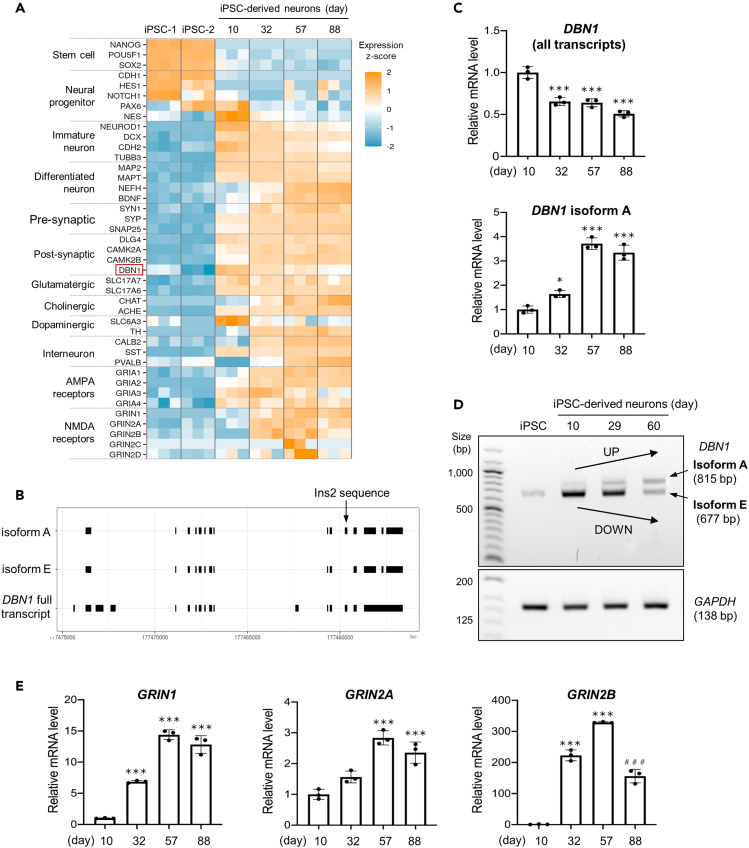
Differential expression of synapse maturation markers in late-stage development
To support the late-stage development of TF-induced iPSC neurons, we examined the time-dependent regulation of major neuron-specific genes (Figure 3A). The stem cell and proliferation markers were expectedly downregulated on day 10. Day 10 neurons expressed a few markers for neural progenitors (PAX6 and NES) and several neuron-specific genes (NEUROD1, DCX, CDH2, TUBB3, MAP2, and MAPT), which confirmed their intermediate progenitor to immature neuron profile. Following day 32, the expression of immature neuron markers decreased, whereas several mature neuron markers (NEFH and BDNF) and presynaptic (SYN1, SYP, and SNAP25) or postsynaptic components (DLG4, CAMK2A and CAMK2B) increased. Moreover, we observed the upregulation of glutamate vesicle transporters (SLC17A6 and SLC17A7) and AMPA receptors (GRIA1-4), followed by NMDA receptors (GRIN1, GRIN2A, and GRIN2B), which indicated the presence of glutamatergic neurons capable of excitatory transmission. In addition, we identified few markers for inhibitory interneurons, cholinergic neurons, and dopaminergic neurons. We also confirmed in an independent experiment by immunocytochemistry that the majority (over 95%) of the cells in our cultures expressed the mature neuron marker NeuN (RBFOX3) and the glutamatergic marker VGlut1 (SLC17A7) at the protein level (Figure S5). Over cell type markers such as the GABAergic neuron marker GAD67 (GAD1) or the dopaminergic neuron marker tyrosine hydroxylase (TH) were rarely detected (less than 5%) by day 84 (Figure S5), suggesting that our cultures contained mostly glutamatergic neurons.
Figure 3.
The differential expression of neuronal and synaptic markers in cultured TF-induced iPSC neurons
(A) Heatmap of the expression profiles of representative stem cells or neuron-specific genes detected by RNA-Seq. The expression levels reported as transcripts per million (TPM) +1 have been log2-transformed and normalized across all the cell samples.
(B) The sequence representation of the two drebrin isoforms identified from the RNA-Seq data by mapping to the DBN1 gene locus of the human reference genome (gene ID: ENSG00000113758.13). The transcript that includes the additional exon 11 has been assumed as the mature brain-specific isoform A and the shorter transcript as the embryonic isoform E.
(C) Relative expression levels of DBN1 (all transcripts) and DBN1 isoform A determined by RT-qPCR. Data are presented as mean ± SD. Statistical analysis by ANOVA with the post hoc Tukey’s test method (∗∗∗p < 0.001, ∗p < 0.05 relative to day 10).
(D) Semi-quantitative RT-PCR of the DBN1 isoform E and isoform A transcripts. The maturation process has been independently reproduced from the RNA-Seq and RT-qPCR experiments by using a different iPSC line. The intensity of the shorter band corresponding to isoform E has increased after the induction of differentiation and decreased in subsequent stages, whereas the larger band corresponding to isoform A has gradually increased. GAPDH has been used as an internal control. (E) Relative expression levels of the NMDA receptor subunits GRIN1, GRIN2A, and GRIN2B by RT-qPCR. Statistical analysis by ANOVA with the post hoc Tukey’s test method (∗∗∗p < 0.001 (relative to day 10), ###p < 0.001 (day 88 relative to day 57 for GRIN2B)). All three subunits have been upregulated in subsequent stages following days 32 or 57; however, GRIN2B appears to be significantly downregulated in future cultures at day 88, thus suggesting the induction of the GluN2B to GluN2A developmental switch.
To further demonstrate the initiation of events leading to synaptic maturation, we examined the following two major transcriptional features indicative of functional maturity: the conversion of drebrin to a mature brain-specific isoform and the developmental switch between NMDA receptor subunits.
Changes in drebrin isoforms occur via alternative splicing during neuronal development. The embryonic isoform drebrin E gets replaced by the brain-specific isoform drebrin A,15 which regulates the formation of dendritic spines by stabilizing postsynaptic actin filaments.32 By mapping the sequence reads to a reference human genome, we confirmed the presence of two different isoforms in our samples (Figure 3B), and could distinguish drebrin A from drebrin E by inserting the Ins2 sequence, as previously described in the human brain.16 RNA-Seq data demonstrated that drebrin (DBN1) was strongly expressed day 10 onwards (Figure 3A). To better distinguish the expression of the two isoforms, we performed reverse transcription-quantitative PCR (RT-qPCR) on aliquots of culture samples, similar to those in the RNA-seq experiment (Figure 3C). In addition, we performed the semi-quantitative electrophoresis of RT-PCR products in an independent time-course culture using primers that allowed the separate detection of the two isoforms (Figure 3D). Both experiments confirmed that drebrin A was predominantly absent at early time points and was upregulated following days 57-60, whereas drebrin E was concomitantly downregulated.
The shift from the NMDA receptor subunit GluN2B (GRIN2B) to GluN2A (GRIN2A) was another important developmental switch, which was closely associated with synaptic plasticity (33,34 for review). To date, NMDA reporter expression is reportedly limited in iPSC-derived neurons until the selection of the most differentiated neurons with a postnatal reporter gene,35 and the subunit switch has been described only in long-term cultured organoids.36 RT-qPCR demonstrated significantly upregulated expression of GRIN1 and GRIN2B on day 32, followed by GRIN2A on day 57 (Figure 3E). GRIN2B was downregulated on day 88. In addition, we repeated this experiment and obtained similar results using an independent time-course culture (Figure S2). Therefore, the switch between the NMDA receptor subunits appeared to occur between days 57 and 88, with a specific repression of GRIN2B, similar to transcriptional transitions previously observed in rat hippocampal postnatal development37 and iPSC-derived organoids.36 Overall, our results were consistent with the suggestion that TF-induced iPSC neurons initiated synapse maturation during the late developmental phase, following day 57, by reproducing hallmark transcriptional features previously characterized in cultured rodent neurons or brain tissues.
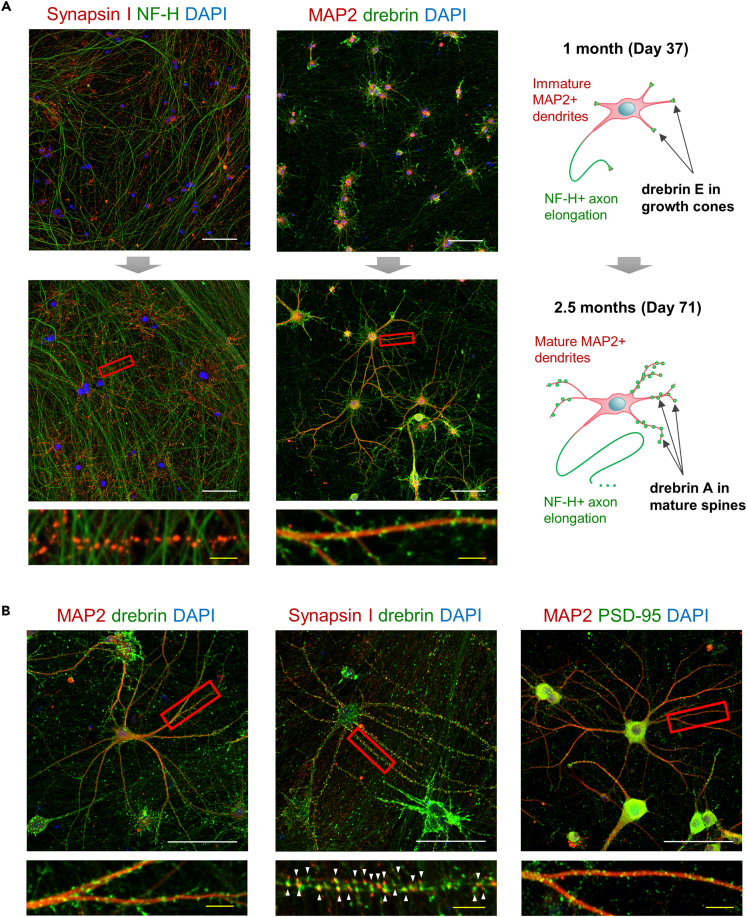
The formation of dendritic spines and synaptic structures in the long-term culture of transcription factor-induced pluripotent stem cell neurons
Consequently, we performed immunocytochemistry to visualize dendritic growth and the formation of synaptic structures. TF-induced iPSC neurons developed short MAP2-positive dendrites and rapidly elongated the neurofilament heavy chain (NF-H)-positive axons in the first month of culture (Figure 1C). Subsequently, MAP2-positive dendrites continued growing and gradually branched (Figure 4A). Concomitantly, the expression of the presynaptic vesicle marker synapsin I increased. Synapsin I was diffused in axons and around cell bodies in early cultures, then aggregated along the mature dendrites in late cultures at day 71, which suggested its concentration at synaptic sites with the progress of neuronal development through synaptogenesis. We further evaluated the subcellular localization of the actin-binding protein, drebrin. On day 37, we detected drebrin around the cell bodies and in the growth cones at the end of neurite development, similar to previous reports of early cultures of rat neurons and immature iPS-derived neurons.4,38 On day 71, drebrin clusters were distributed along the developed MAP2-positive dendrites, thereby indicating a transition to the mature isoform drebrin A, which accumulated into stable dendritic spines.32 The formation of drebrin and synapsin I clusters was reproduced in an independent experiment on day 73 of culture (Figure 4B). Co-labeling synapsin I and drebrin allowed the visualization of closely adjacent presynaptic and postsynaptic clusters, thus suggesting the establishment of synaptic connections at the dendritic spines. Furthermore, the postsynaptic density of PSD-95 was recruited simultaneously and formed visible puncta along mature dendrites (Figure 4B). Thus, both transcriptional and structural evidence demonstrated that TF-induced iPSC neurons reached the final developmental stage 5, with drebrin recruiting the postsynaptic proteins and promoting the formation of dendritic spines.
Figure 4.
Protein expression and the subcellular localization of dendritic and synaptic markers during the maturation phase
(A) Immunofluorescence images (20× objective) and schematic representation of cultured TF-induced iPSC neurons displaying the expression of the axon marker NF-H (left and right, green), dendritic marker MAP2 (middle and right, red), presynaptic vesicle marker synapsin I (left, red), and postsynaptic spine marker drebrin (middle and right, green). Drebrin accumulates around the cell bodies and in growth cones at the end of short dendrites in immature neurons at approximately 1 month of culture. Subsequently, drebrin forms clusters along the mature dendrites following 2.5 months. The bottom row images display magnified views of the red squares, each one including a dendrite surrounded by synapsin I (red), or drebrin (green) clusters.
(B) Detailed representative fluorescence images (40× objective) displaying the accumulation of pre- and postsynaptic markers along the developed dendrites at day 73 of an independently reproduced cell culture. The bottom row images display magnified views of the red squares, each one including a dendrite surrounded by synapsin I (red), drebrin (green), or PSD-95 (green) clusters. In the middle image, co-labeling the presynaptic synapsin I and the postsynaptic drebrin displays green clusters adjacent to red clusters, as indicated by white arrows, which suggest the formation of synaptic connectivity. Nuclei have been stained with DAPI. White scale bar: 100 μm. Yellow scale bar in magnified dendrite images: 10 μm.
The functional activity of excitatory postsynaptic proteins
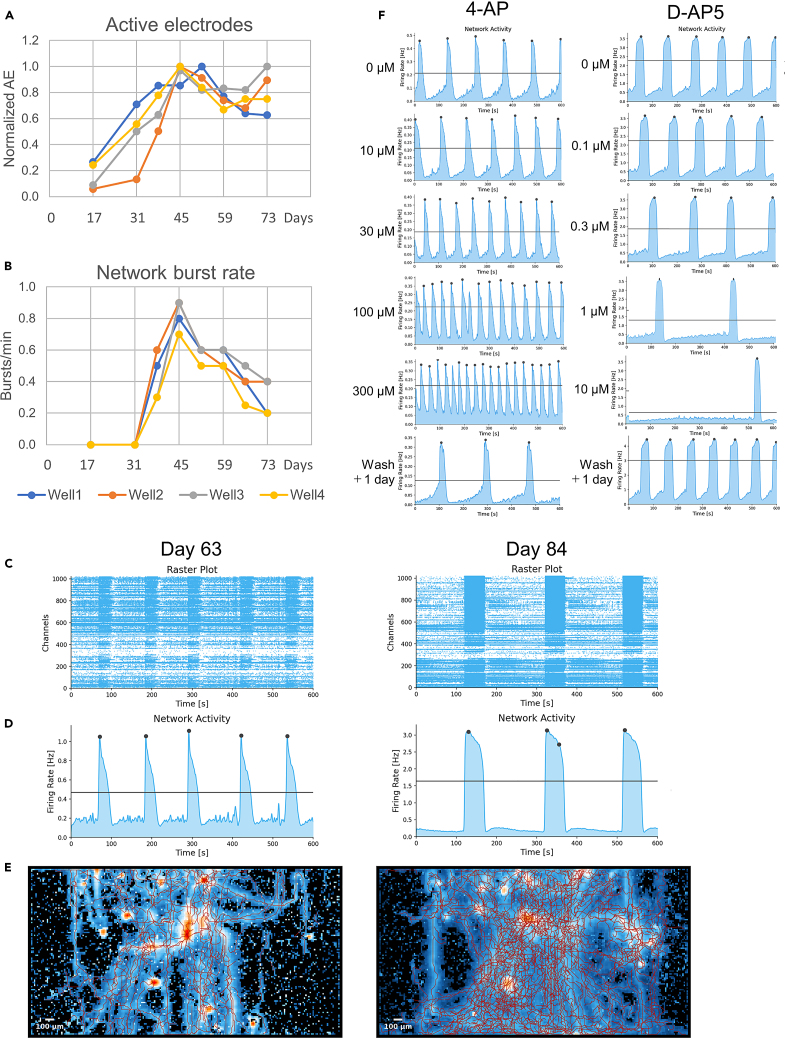
We characterized the electrophysiological responses of TF-induced iPSC neurons using a high-density microelectrode array (HD-MEA) system. Spontaneous firing increased after approximately 1 month of culture (days 31-36), as depicted by the increase in the number of active electrodes recording extracellular spike activity (Figure 5A). Network bursts appeared on day 36 and reached a maximal rate on day 45, concomitantly with the increase in synaptic gene expression (Figure 5B). Synchronized network bursts are generally considered indicators of the establishment of neuronal connections, and are used to characterize iPSC-derived neuronal cultures.5,39 The network bursts were stably measured for up to 2 months (day 59), following which the burst rate decreased at future time points. The population spike rate histograms at a late time point (day 84) showed a strong increase in burst duration, spike per network burst, and interburst interval (Figures 5C and 5D). Accordingly, Axon Tracking analysis showed that axonal propagation and network complexity significantly increased over the culture area during that period (Figure 5E). Thus, the burst rate decrease was not attributed to a loss of neuronal activity but might be rather related to the maturation of network connectivity and increased stability of oscillations. More investigation would be required to understand the molecular basis of the change in electrophysiological activities associated with neuronal maturation.
Figure 5.
The functional characterization of synaptic activities in TF-induced iPSC neurons
(A) The detection of the electrophysiological response of TF-induced iPSC neurons cultured on HD-MEA. The firing activity has been determined as the proportion of active electrodes (AE) recording extracellular spikes (relatively normalized to the maximal number of AE per well). The local field potential has been recorded from four independent wells of cultured TF-induced iPSC neurons.
(B) The variation of network activity defined as the rate of spontaneous synchronous bursts.
(C) Representative raster plot of firing spikes simultaneously recorded by 1,024 electrodes in one well for 600 s at day 63 and day 84 of culture.
(D) Representative population spike rate histograms corresponding to the above raster plots and showing the network burst activity. The network burst rate decreased between day 63 and day 84, concomitantly to an increase in burst duration, mean firing rate per burst, and interburst interval.
(E) Axon Tracking map showing the spatial distribution of action potential amplitude and the reconstructed propagation paths for neurons resolved over the 4 mm × 2 mm sensor area containing 26,400 electrodes.
(F) Pharmacological response assay with representative population spike rate histograms simultaneously recorded by 1,024 electrodes in each well at week 8 of culture. The cultures have been treated with increasing concentrations of neuroactive compounds, with 1-min stabilization and 10-min recording between each treatment. The cultures have been washed with three medium changes and left for 1 day to assess the declining effect of the compounds.
We also assessed the cellular responses to neuroactive compounds on day 59, during which the network bursts were the most stable and reproducible (Figure 5F). Exposure to the seizure-inducing potassium channel blocker 4-aminopyridine (4-AP) resulted in a concentration-dependent increase in the burst frequency. Exposure to the NMDA receptor antagonist D-2-amino-5-phosphonopentanoic acid (D-AP5) elicited a concentration-dependent reduction in the burst frequency and an increase in the interburst intervals. Therefore, TF-induced iPSC neurons develop proper excitatory synaptic functions and expectedly respond to the addition of drugs affecting neurotransmission.
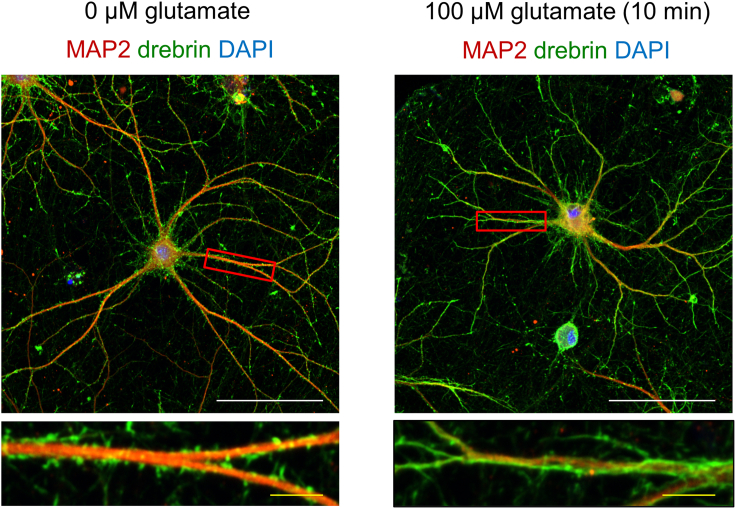
Network burst activity may be indicative of only immature synapses that randomly connect and synchronize neighboring cells; therefore, we used drebrin as a marker of the postsynaptic glutamate-dependent NMDA receptor activity to further ensure the detection of mature synaptic functions in dendritic spines. Drebrin exits dendritic spines upon glutamate exposure and migrates into dendritic shafts via a mechanism that requires postsynaptic NMDA receptor-dependent calcium ion influx.18 This molecular event, termed drebrin exodus, plays an essential role in synaptic plasticity,18,40 and the initiation of synaptic damage by excitotoxicity or neurodegeneration.41 Using a protocol similar to that established for rodent cells,17 we demonstrated that the application of 100 μM glutamate for 10 min was sufficient to induce a decrease in the drebrin cluster density without damaging the dendrites (Figure 6). Drebrin expression concomitantly increased in the dendritic shafts, which appeared to be homogeneously stained by the anti-drebrin antibody. This study replicated the drebrin exodus mechanism in human stem cell-derived neurons. Taken together, mature TF-induced iPSC neurons consisted of functional dendritic spines, including postsynaptic NMDA receptor-dependent activity.
Figure 6.
The effect of glutamate treatment on mature TF-induced iPSC neurons
The cells have been treated with 100 μM glutamate for 10 min before immunostaining. The images in the bottom row display magnified views of the dendrites in the red squares. The 0 μM glutamate control neurons display drebrin clusters along the sides of the dendrite. Glutamate treatment induces the drebrin exodus from the spines, such that the dendritic shafts appear homogeneously stained in green. Cell cultures similar to those of the RNA-Seq analysis and the first ICC at day 73 have been used. White scale bar: 100 μm. Yellow scale bar in magnified dendrite images: 10 μm.
Discussion
In this study, we have characterized the transcriptional and morphological changes associated with the late-stage development of TF-induced iPSC. We have successfully induced the maturation of dendritic spines and provided evidence that the TF-induced iPSC neurons acquire features resembling a postnatal stage. iPSC-derived neuronal models were frequently reported to retain immature characteristics.3,6,35 Although several recent transcriptomic studies identified subsets of cells with the ability to acquire postnatal-like signatures35,42,43), the correspondence between a global postnatal transition and the period of dendritic spine formation and activation of developmental switches in vitro was previously demonstrated only in long-term cultured organoids.10,36 Herein, we have shown that pure cultures of TF-induced iPSC neurons can reproducibly undergo a postnatal-like transition at one-third of the time required in previous organoid studies (60-70 days, compared with 180-250 days in organoids). TF-induced iPSC neurons rapidly differentiate and acquire neuronal features.8,23 This accelerated neurogenesis offers two major advantages. First, it considerably reduces the time to generate differentiated neurons equivalent to stage 3 of neuronal development, which allows the allocation of greater culture time for subsequent stages. Second, homogeneous neuronal cultures allow robust and synchronized entry into the subsequent maturation phase, such that we could establish the transition from stage 3 to completely developed stage 5 neurons with unprecedented temporal resolution.
Moreover, this comprehensive time-course study allowed us to assemble the sequence of events leading to synapse maturation in TF-induced iPSC neurons. Neuronal polarization occurred first, with the rapid upregulation of axonal extension and transport-related genes, including drebrin isoform E, which is involved in growth cone dynamics. Consequently, the dendrites began growing and branching at a slower pace, along with the upregulation of synaptic genes and the increase in network burst activity, as measured by MEA. This eventually led to the acquisition of a postnatal-like profile, including the expression of the dendritic spine marker drebrin isoform A and NMDA receptor subunits. Spine formation occurred subsequently, and the postsynaptic density PSD-95 began to be recruited with a progress in spine stabilization. This final period also coincided with the transcriptional switch between NMDA receptor subunits, thus suggesting the increased incorporation of GluN2A-containing receptors at postsynaptic sites contributed to the acquisition of synaptic plasticity. In this regard, we successfully demonstrated postsynaptic NMDA receptor activity using a functional assay that reproduced the actin dynamics underlying the initiation of structure-based synapse plasticity.
Overall, the integration of transcriptome analysis with ICC and functional assays allowed us to depict a coherent picture of undescribed synapse maturation process in iPSC-induced neurons. We also demonstrated the novel finding that drebrin subcellular localization changes with the stages of development and upon glutamate stimulation in iPSC-derived neurons. Thus, we successfully demonstrated the conservation of developmental events previously described in rodent neurons. Our results highlighted the advantages of using TF-induced iPSC neurons to model late-stage synaptic development, as demonstrated by the formation of functional dendritic spines.
Limitations of the study
We aimed in this study at establishing a simple and reproducible synaptic maturation process in iPSC-derived neuronal monocultures in vitro. Therefore, our system does not take into account any interaction with glial cells. Interestingly, TF-induced iPSC neurons appeared to possess the endogenous ability to progress through the late-stage maturation program and acquire postnatal-like features in vitro. iPSC-derived neurons are often co-cultured with astrocytes to enhance maturation5,7,8,42; however, our cultures did not require the addition of astrocytes. They also did not generate any detectable glial cells because of the use of an antiproliferative agent (CultureOne, Thermo Fisher44) to avoid contamination by non-neuronal cells. Feeder cell-free cultures allow the straightforward analysis of neuron-specific data, which facilitates the characterization of transcriptional features and mechanisms of action attributable to neurons alone. Nevertheless, it is worth mentioning that co-culture studies with glial cells would be interesting for future in-depth comparisons with neuronal functions in vivo.
Obtaining complete maturity is still time-consuming, compared with the rapid neurogenesis phase. This warrants further investigations to identify the determinants of the slower maturation program, which would allow better control over the entire process. One limitation of the TF-induced iPSC neurons could be the generation of heterogeneous cell populations, as previously reported in studies using NGN2 induction.35,45 Indeed, our cultures may not be entirely homogeneous, considering the expressed neuronal subtype markers, besides the most obvious glutamatergic neuron-related markers. Our RNA-seq and electrophysiological studies were limited to global population analysis of whole cultures. Single-cell analyses may facilitate the complete characterization and possible control of the neuronal subtype variations in the future. Still, in the present study, the consistency between the transcriptional, morphological, and functional results across several independent time-course cultures in different formats indicate that our neurons reached a globally mature spiny neuron profile in a robust manner. Therefore, we confidently propose that the TF-induced iPSC neurons reached synapse maturation through a sequence of key developmental events, comparable to that in rodent neurons or brain tissues, and our data may serve as the basis for additional improvements in neuronal specification.
Conclusions
We demonstrated that TF-induced iPSC neurons could replicate the transcriptional and structural features of neuronal development. Our results enhanced the relevance of TF-induced iPSC neurons as a developmental model, and the data may serve as a reference for future studies on late-stage maturation. Moreover, we demonstrated that TF-induced iPSC neurons could reach dendritic spine maturation, which is essential for postsynaptic efficacy and plasticity. Modeling spinogenesis in human cells and clarifying the conservation of mechanisms previously described in rodents would be particularly useful for validating non-animal in vitro assays, such as the evaluation of synaptic loss by quantifying drebrin clusters.17,46 The availability of reliable tools to study human synaptic functions would substantially contribute to the future development of disease models and assays that are physiologically relevant to higher brain function and cognitive disorders.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-MAP-2 | Millipore | Cat# AB5622-I; RRID: AB_2800501 |
| Rabbit polyclonal anti-synapsin I | Sigma-Aldrich | Cat# S193; RRID: AB_261457 |
| Rabbit polyclonal anti-Tyrosine Hydroxylase, N1C1 | GeneTex | Cat# GTX102424; RRID: AB_2038197 |
| Rabbit polyclonal anti-VGLUT1 | Proteintech | Cat# 55491-1-AP; RRID: AB_2881348 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat# ab177487; RRID: AB_2532109 |
| Mouse monoclonal anti-drebrin, M2F6 | MBL | Cat# D029-3; RRID: AB_591275 |
| Mouse monoclonal anti-PSD-95, 6G6-1C9 | Thermo Fisher | Cat# MA1-045; RRID: AB_325399 |
| Mouse monoclonal anti-NF-H | BioLegend | Cat# 801601; RRID: AB_2564641 |
| Mouse monoclonal anti-beta III tubulin, 2G10 | Abcam | Cat# ab78078; RRID: AB_2256751 |
| Mouse monoclonal anti-GAD67, 1G10.2 | Millipore | Cat# MAB5406; RRID: AB_2278725 |
| Alexa Fluor 594 conjugated goat anti-rabbit | Thermo Fisher | Cat# A32740; RRID: AB_2762824 |
| Alexa Fluor 488 conjugated goat anti-mouse | Thermo Fisher | Cat# A32723; RRID: AB_2633275 |
| Biological samples | ||
| Quick-Neuron Excitatory - Human iPSC-derived Neurons (F, 74 yr donor) - Healthy Control | Elixirgen Scientific | Cat# EX-SeV-CW50065 |
| Total RNA from the CIRM iPS cell line CW50065 | Elixirgen Scientific | N/A (provided as a gift) |
| Chemicals, peptides, and recombinant proteins | ||
| Poly(ethyleneimine) solution | Sigma-Aldrich | Cat# 181978 |
| Pierce 20X Borate Buffer | Thermo Fisher | Cat# 28341 |
| Laminin Mouse Protein, Natural | Thermo Fisher | Cat# 23017015 |
| DPBS, no calcium, no magnesium | Thermo Fisher | Cat# 14190144 |
| Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher | Cat# A1413202 |
| Quick-Neuron Excitatory - Maintenance Medium | Elixirgen Scientific | Cat# 2EX-MM |
| Y-27632 2HCl | Selleckchem | Cat# S1049 |
| Neurobasal Plus Medium | Thermo Fisher | Cat# A3582901 |
| B-27 Plus Supplement (50x) | Thermo Fisher | Cat# A3582801 |
| CultureOne Supplement (100X) | Thermo Fisher | Cat# A3320201 |
| Neuron Culture Medium | Wako Chemicals | Cat# 148-09671 |
| L(+)-Ascorbic Acid | Wako Chemicals | Cat# 012-04802 |
| GlutaMAX Supplement | Thermo Fisher | Cat# 35050-061 |
| Penicillin-Streptomycin | Thermo Fisher | Cat# 15140122 |
| RNaseZap RNase decontamination solution | Ambion | Cat# AM9780 |
| Nuclease-free water | Invitrogen | Cat# 10977023 |
| β-Mercaptoethanol, 14.3 M | Sigma | Cat# M6250-100mL |
| Tris buffer, pH 8.0, 1 M | Ambion | Cat# 9010 |
| Tween 20 | Sigma | Cat# P9416-100ML |
| Magnesium chloride, 1 M | Ambion | Cat# 9010 |
| RNase inhibitor, cloned 40 U μl-1 | Ambion | Cat# AM2682 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787-100ML |
| dNTP mix 10 mM each | Thermo Fisher | Cat# 18427-088 |
| Superscript II reverse transcriptase | Thermo Fisher | Cat# 18064-014 |
| Betaine for Molecular Biology | Wako Chemicals | Cat# 028-16294 |
| KAPA HiFi HotStart ReadyMix 2× | KAPA Biosciences | Cat# KK26010 |
| Ethanol, molecular biology grade | Wako Chemicals | Cat# 054-07225 |
| Agencourt AMPure XP beads | Beckman Coulter | Cat# A63881 |
| ERCC spike-in mix 1 | Ambion | Cat# 4456740 |
| 16%-Paraformaldehyde Aqueous Solution | Nacalai Tesque | Cat# 15710 |
| 1M Phosphate Buffer Solution | Muto Pure Chemicals | Cat# 11281 |
| Triton X-100 | Nacalai Tesque | Cat# 12967-32 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat# A9647 |
| DAPI solution (1 mg/mL) | Thermo Fisher | Cat# 62248 |
| Sodium Azide | Nacalai Tesque | Cat# 31233-71 |
| L-Glutamic acid monosodium salt hydrate | Sigma-Aldrich | Cat# G5889 |
| Critical commercial assays | ||
| Quick-Neuron Excitatory - SeV Kit | Elixirgen Scientific | Cat# EX-SeV |
| RNeasy Micro Kit (50) | Qiagen | Cat# 74004 |
| Qubit RNA HS Assay | Thermo Fisher | Cat# Q32852 |
| Qubit dsDNA HS Assay | Thermo Fisher | Cat# Q32851 |
| Nextera XT DNA library preparation kit, 96 samples | Illumina | Cat# FC-131-1096 |
| Nextera XT Index Kit v2 Set A (96 indexes, 384 samples) | Illumina | Cat# FC-131-2001 |
| NextSeq 500/550 Mid Output Kit v2.5 (150 Cycles, 75PE) | Illumina | Cat# 20024904 |
| D5000 ScreenTape | Agilent Technologies | Cat# 5067-4626 |
| D5000 Reagents | Agilent Technologies | Cat# 5067-5593 |
| ReverTra Ace qPCR RT Master Mix | Toyobo | Cat# FSQ-201 |
| KOD SYBR qPCR Mix | Toyobo | Cat# QKD-201 |
| PrimeScript RT-PCR Kit | Takara Bio | Cat# RR014A |
| Deposited data | ||
| Raw and analyzed data: scRNA-seq | This Paper | GEO: GSE224751 |
| Broad Institute's human reference genome GRCh38 | TOPMed RNA-seq pipeline | https://github.com/broadinstitute/gtex-pipeline/blob/master/TOPMed_RNAseq_pipeline.md |
| GENCODE 34 annotations | TOPMed RNA-seq pipeline | https://github.com/broadinstitute/gtex-pipeline/blob/master/TOPMed_RNAseq_pipeline.md |
| CoDex (Cortical Development Expression) dataset | Polioudakis et al., 201927 | http://solo.bmap.ucla.edu/shiny/webapp/ |
| BrainSpan developmental transcriptome dataset RNA-Seq Gencode v10 summarized to genes |
Li et al., 201828 | https://www.brainspan.org/ |
| Experimental models: Cell lines | ||
| CIRM iPS cell line CONTROL: no cognitive decline | FCDI | CW50065 |
| Human iPS cell line derived from healthy individual | RIKEN BRC | HPS0381 : 1231A3 |
| Oligonucleotides | ||
| TSO AGCAGTGGTATCAACGCAGAGTACATrGrG+G |
IDT Picelli et al., 201447 |
N/A |
| Oligo-dT30VN AAGCAGTGGTATCAACGCAGAGTACT30VN |
IDT Picelli et al., 201447 |
N/A |
| ISPCR oligo AAGCAGTGGTATCAACGCAGAGT |
IDT Picelli et al., 201447 |
N/A |
|
TBP (PrimeTime qPCR Primers) Forward: CAG CAA CTT CCT CAA TTC CTT G Reverse: GCT GTT TAA CTT CGC TTC CG |
IDT | Hs.PT.58v.39858774 |
|
DBN1 all isoforms Forward: CAC GGA GAT CCA CGA TGC AG Reverse: TGC CAG GCC ATA GGT CAA TGA G |
IDT Togo et al., 20216 |
N/A |
|
DBN1 isoform A Forward: TTC ATA AAG GCA TCG GAC AGT GG Reverse: ATG GGA GGG AGG AAG AGA GGT TTG G |
IDT Togo et al., 20216 |
N/A |
|
GAPDH Forward: CCA CTT TGT CAA GCT CAT TTC CT Reverse: TCT CTT CCT CTT GTG CTC TTG CT |
IDT Togo et al., 20216 |
N/A |
|
GRIN2B (PrimeTime qPCR Primers) Forward: CTC TCT GTG CTG CCG TTG Reverse: CTT AGC TGC CTT CAT GAT CCA |
IDT | Hs.PT.58.26188572 |
|
GRIN2B (PrimeTime qPCR Primers) Forward: CCA TGG ATG CAG CTG TAG ATA C Reverse: CAG TTC CGA CAT TGC TTT ATG G |
IDT | Hs.PT.58.20804834 |
|
GRIN2A (PrimeTime qPCR Primers) Forward: ACG ATG CTG AGA TGG TTG TC Reverse: CTG GTG GTG ATT GTG CTG AA |
IDT | Hs.PT.58.2281813 |
|
GRIN1 (PrimeTime qPCR Primers) Forward: GTG GAT GGC TAA CTA GGA TGG Reverse: CTC CTG GAA GAT TCA GCT CAA |
IDT | Hs.PT.58.39141804 |
|
GRIN2A (PrimeTime qPCR Primers) Forward: GCA GAA ACA ATG AGC AGC ATC Reverse: CAA GAA GTA ATG GCA CCG TCT |
IDT | Hs.PT.58.26949410 |
|
GRIN2B (PrimeTime qPCR Primers) Forward: CAT CAC AAA CAT CAT CAC CCA TAC Reverse: CTT CAT AGA GAC AGG CAT CAG T |
IDT | Hs.PT.58.40419546 |
|
DBN1 (for semi-quantitative RT-PCR) Forward: ATT GCC CAG CGG CCT GAC AAC CCA AGG GAG Reverse: TCT GCC AGG GTG ACC TCA GTA CCC GAG GGT G |
Hiroyuki Yamazaki, Gunma University spikar@gunma-u.ac.jp | N/A (provided as a gift) |
| Software and algorithms | ||
| PRINSEQ++ (v1.2) | Ho and Patrizi, 202148 | https://github.com/Adrian-Cantu/PRINSEQ-plus-plus |
| HISAT2 (v2.1.0) | Kim et al., 201949 | https://github.com/DaehwanKimLab/hisat2 |
| StringTie (v2.0.6) | Kovaka et al., 201950 | https://github.com/gpertea/stringtie |
| R version 4.1.0 (2021-05-18) | The R Project for Statistical Computing | https://www.r-project.org/; RRID: SCR_001905 |
| ClusterProfiler | Yu et al., 201251 | https://guangchuangyu.github.io/software/clusterProfiler/ |
| Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| CellPathfinder (for the CQ1 confocal system) | Yokogawa Electric Corporation | https://www.yokogawa.com/solutions/products-platforms/life-science/high-content-analysis/analysis-software/cellpathfinder/ |
| MaxLab Live Software (for the MaxTwo Multiwell HD-MEA platform) | Maxwell Biosystems | https://www.mxwbio.com/products/maxone-mea-system-microelectrode-array/maxlab-live-software/ |
| Other | ||
| NextSeq 500 System | Illumina | Cat# SY-415-1001 |
| Agilent 4200 TapeStation | Agilent Technologies | Cat# G2991BA |
| Qubit 3.0 Fluorometer | Thermo Fisher | Cat# Q33216 |
| BZ-X800 Microscope | Keyence | https://www.keyence.com/products/microscope/fluorescence-microscope/bz-x700/ |
| CQ1 Confocal System | Yokogawa Electric Corporation | https://www.yokogawa.com/solutions/products-platforms/life-science/high-content-analysis/cellvoyager-cq1/ |
| QuantStudio 12 K Flex System | Thermo Fisher | https://www.thermofisher.com/jp/ja/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/quantstudio-systems/models/quantstudio-12-flex.html |
| MaxTwo Multiwell HD-MEA platform | Maxwell Biosystems | https://www.mxwbio.com/products/maxtwo-multiwell-microelectrode-array/ |
| E-Gel EX Agarose Gels, 1% | Thermo Fisher | Cat# G401001 |
| 96-well Cell Culture Plates | Eppendorf | Cat# 0030 730.135 |
| 96-well Low attachment U-bottom plates | Greiner | Cat# 650970 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Waka Lin (waka.lin@jp.ricoh.com).
Materials availability
This study did not generate new unique reagents. All material sources are listed in the key resources table.
Experimental model and subject details
Cell cultures
We used cryopreserved human iPSC-derived neurons (Quick-Neuron Excitatory - Human iPSC-derived Neurons, Elixirgen Scientific, Cat# EX-SeV-CW50065) for the experiments, as indicated by the green and purple dots in Figure 1A. According to the manufacturer, these neurons were derived from the iPSC line CW50065 of the California Institute for Regenerative Medicine repository and frozen on day 3 following differentiation by the introduction of a proprietary cocktail of TFs, similar to a published method.23 The TF-induced iPSC neurons were thawed and plated according to the manufacturer’s instructions, with few modifications. We prepared the plating medium using the recommended Maintenance Medium (Elixirgen Scientific, Cat# EX-MM), with the addition of 10 μM ROCK inhibitor Y27632. For monolayer cultures, 96-well microplates were coated with 0.05% of polyethyleneimine solution (PEI) diluted in 1X borate buffer and 20 mM of laminin diluted in phosphate-buffered saline. The cells were plated at a density of 5×104 cells/cm2 in 160 μL of the plating medium per well. Six-well HD-MEA plates (Maxwell Biosciences) were coated with 0.05% of PEI diluted in borate buffer, following which 1/100 Geltrex was diluted in the plating medium. We plated 1×105 cells in a 10 μL droplet over the electrodes and incubated for 30 min until cell attachment, followed by the addition of 1.5 mL of the plating medium per well. For spheroid formation, 1×104 cells in 100 μL plating medium were dispensed into each well of the low-attachment U-bottom 96 well plates. The cells were maintained in culture at 37°C in a CO2 incubator. Half of the media was changed on the first day following plating using the Maintenance Medium (Elixirgen Scientific), and subsequently every 3 days or 4 days (twice a week) until day 10. Following 10 days, the medium was changed to the Neurobasal Plus Medium (Thermo Fisher Scientific) supplemented with 2% of B-27 Plus, 1% of GlutaMAX, 1% of CultureOne, 10% of Neuron Culture Medium (Wako Chemicals), 2 mM of ascorbic acid, and 1% of penicillin-streptomycin. For the experiments presented as orange dots in Figure 1A, we used a TF-based differentiation kit from the similar manufacturer (Quick-Neuron Excitatory-SeV Kit, Elixirgen Scientific, Cat# EX-SeV) to generate neurons in-house from the iPSC line HPS0381:1231A3 provided by the RIKEN BRC Cell Bank. We performed neuronal differentiation following the manufacturer’s instructions, and long-term cultures were maintained from day 10 under similar conditions as above.
Method details
RNA-seq library construction
Neuronal cell spheroids were collected, washed once in PBS by centrifugation, and immediately frozen and stored in a -150°C freezer. 3 spheroids were pooled in 1 microtube to constitute 1 biological sample, and N = 3 sample replicates were taken at each time point. We extracted total RNA using the RNeasy Micro Kit (Qiagen) following the manufacturer’s user manuals. We measured the concentration of the purified total RNA using the Qubit RNA HS Assay (Thermo Fisher), then we used 1 ng of RNA per sample for cDNA library preparation according to the Smart-seq 2 method.47 The libraries were sequenced as paired-end 75 bp reads on the NextSeq 550 system (Illumina).
Immunocytochemistry
Cultured TF-induced iPSC neurons were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer for 20 min at room temperature and washed 3 times with PBS, permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked with 3% bovine serum albumin in PBS for 1 h. The cells were incubated overnight at 4°C with primary antibody diluted in PBS containing 3% bovine serum album. The following primary antibodies were used: rabbit anti-MAP-2 (1:2,000) (Millipore), anti-synapsin Ⅰ (1:1,000) (Sigma-Aldrich), mouse anti-drebrin (1:1,000) (MBL International), anti-PSD-95 (1:500) (Thermo Fisher), anti-NF-H (1:1,000) (Bio Legend), anti-vGlut1 (1:50) (Proteintech), anti-TH (1:200) (GeneTex), anti-NeuN (Abcam) (1:100) and anti-GAD67 (1:500) (Millipore) antibodies. The cells were washed with PBS and incubated with secondary antibody and DAPI solution (1:1,000) (Thermo Fisher) for 2 hours at room temperature. The following secondary antibodies were used: Alexa Fluor 594 conjugated goat anti-rabbit (1:250) (Thermo Fisher), Alexa Fluor 488 conjugated goat anti-mouse (1:250) (Thermo Fisher) antibodies. The cells were washed and stored in PBS containing 0.1% sodium azide. We acquired phase-contrast images using a CKX53 inverted microscope (Olympus). We acquired fluorescence images using the BZ-X800 Microscope (Keyence) or the CellVoyager CQ1 Confocal system (Yokogawa Electric) through a 20 × or 40 × 0.45 numerical aperture objective lens. For the confocal microscopy, we compiled for each field Z-stack images from 3 slices with 3 μm interval for the 20 × lens or 7 slices with 1 μm interval for the 40 × lens. All the data were collected at 2000 × 2000 resolution at 16 bits/pixel. We processed the images with the CellPathfinder analysis software (Yokogawa Electric) to construct a single 2D image with an increased depth of field by maximum intensity projection.
Glutamate assay
We added 1 mM Glutamate solution to each well for a final concentration of 100 uM, and the cells were incubated at 37°C for 10 min, which is known to be enough to induce a loss of drebrin clusters without affecting cell viability in rat hippocampal neuron cultures.17 We added the same volume of water was added for the “0 μm” control wells. At the end of the 10 min incubation, we fixed the cells immediately with 4% paraformaldehyde in 0.1 M phosphate buffer to proceed with immunocytochemistry.
RT-qPCR
We used the same total RNA samples as those used in the RNA-Seq experiments. We synthesized cDNA from 10 ng RNA using the ReverTra Ace qPCR RT Master Mix (Toyobo) according to the manufacturer’s instructions. SYBR green qPCR reactions were prepared using gene-specific primers for DBN1, DBN1 isoform A, GRIN2A, and GRIN2B as listed in the key resources table and the KOD SYBR qPCR Mix (Toyobo). We run qPCR analyses on the QuantStudio 12 K Flex System (Thermo Fisher). We tested every primers in order to demonstrate a linear amplification efficiency > 90% with R2 > 0.98. We tested several reference genes from a panel previously recommended for use in iPSC-Derived neural stem cells and progenitors,52 and we chose TBP as the most stably expressed in our conditions.
Semi-quantitative RT-PCR
TF-induced iPSC neurons cultured in 96-well plates were collected by scraping and pipetting, then centrifuged at 200 g for 1 min. The cell pellets were immediately frozen and stored in a -150°C freezer. We extracted total RNA using the RNeasy Micro Kit (Qiagen) and measured their concentration using the Qubit RNA HS Assay (Thermo Fisher). We performed RT-PCR using the PrimeScript RT-PCR Kit (Takara Bio) and gene-specific primers for DBN1. We run electrophoresis in a 1% E-Gel EX agarose gel (Thermo Fisher) to visualize the specific PCR products. The primers were designed to include the Ins2 sequence, which is removed by alternative splicing in drebrin isoform E, so that the band corresponding to isoform E appears shorter (677 bp) than isoform A (815 bp).
HD-MEA recordings
We used the MaxTwo Multiwell HD-MEA platform (Maxwell Biosystems) to record the extracellular field potential of the TF-induced iPSC neurons cultured on CMOS-based microelectrodes.53 Spontaneous firing and network burst activities were recorded every week using the “Activity Scan Assay” and “Network Assay” modules of the MaxLab Live software as previously described.39 Recording and visualization of axonal propagation was performed using the “Axon Tracking Assay” module for 30 neurons reconstructed by spike sorting with the software default parameters and mapping with “Number of spikes threshold = 20” and “Footprint completeness threshold = 0.75”.
Quantification and statistical analysis
RNA-seq data processing
To process the RNA-seq reads, we removed low-quality and low-complexity reads using PRINSEQ++ version 1.248 with the parameters “-trim_tail_left 10 -trim_tail_right 10 -trim_qual_left 30 -trim_qual_right 30 -ns_max_n 0 -min_len 30 -threads 8”. We mapped the reads to the human reference genome GRCh38 and the GENCODE 34 annotations, including the ERCC spike-in references using HISAT2 version 2.1.0 with default parameters.49 Finally, we estimated the mRNA expression levels as transcripts per million (TPM) using StringTie version 2.0.6.50
PCA and correlation with the factor loadings of the human neocortex cell types
We selected the genes displaying an expression > 0 TPM in more than 3 samples to perform the principal component analysis (PCA). All TPM values were transformed to log2(TPM+1). We performed the PCA of the transformed TPM values using the R command “prcomp” with the parameter “scale = F”. PCA factor loadings were calculated as the Pearson's correlation coefficient between the PC1 and PC2 components and the TPM of each gene.
We used the factor loadings of cell type-specific marker genes to estimate the direction most correlated with each cell type. The marker gene sets defining each cell type of the human neocortex were obtained from the CoDEx database (http://solo.bmap.ucla.edu/shiny/webapp/).27 For a specific cell type, the average factor loading vector was defined as .
V: Vector of the direction correlated with a specific cell type.
G: PC1/PC2 factor loading vector.
i: marker gene of the specific cell type.
n: total number of the marker genes for the specific cell type.
Correlation with the brainspan dataset
The analysis was performed similarly to a previous method35 with a few modifications. We downloaded the normalized RPKM expression values from the BrainSpan atlas of the developing human brain (http://www.brainspan.org).28 We extracted the data corresponding to the prefrontal cortex (including the medial (MFC), orbital (OFC), dorso-lateral (DFC), and ventrolateral prefrontal cortex (VFC)) from 8 weeks post conception (WPC) to 40 years and we removed the genes that had a standard deviation less than or equal to 1 across all of the samples. We processed similarly the FPKM expression data of the TF-induced iPSC neurons from day 10 to day 88 to remove the genes that had a standard deviation less than or equal to 1 across the time-course. We retained the genes remaining in common between the two datasets for further analysis and we calculated the Pearson’s correlation (r) between the log2(RPKM+1) values from the Brainspan prefrontal cortex regions at different ages and the log2(FPKM+1) values from different time points of the TF-induced iPSC neurons.
Hierarchical clustering and Gene Ontology analysis
To select the genes with a significant variation of expression among the five time-points (day 0, 10, 33, 63 and 88), we performed an analysis of variance (ANOVA) for each gene using the R command “anova”. To adjust the p-values for multiple tests, we set a threshold for the false discovery rate (FDR < 0.05). A total of 2,438 genes had a significantly different expression between time points, and standardized Z-scores were calculated across the samples. We performed a hierarchical clustering of the defined 2,438 genes according to the Z-score, using the R commands “dist” with the parameter “method = "euclidean"”, and “hclust” with the parameter “method = "ward.D2"”. To assign the genes to each cluster, we used the R command “cutree” with the parameter “k = 6”. Finally, we performed a GO enrichment analysis for each cluster using the R command “enrichGO” of the package “clusterProfiler”51 with the parameters “OrgDb = org.Hs.eg.db, keyType = 'SYMBOL', ont = "BP", pAdjustMethod = "BH", qvalueCutoff = 0.05”. Finally, we extracted for each cluster the top 20 significantly enriched GO terms ranked by their adjusted p-value.
RT-qPCR analysis
We collected 3 biological replicate samples at each time point, and we run RT-qPCR in duplicate for each sample. We calculated the relative mRNA expression levels by the comparative threshold cycle (Ct) method. The difference ΔCt was first calculated between the gene of interest and the reference gene TBP. Then the mean fold-change in expression was reported at each time-point relatively to the expression at day 10. For multiple comparisons, we performed an ordinary one-way ANOVA with the post hoc Tukey’s test method using the Prism 7 software, and we considered p-values <0.001 to be statistically significant.
Acknowledgments
We would like to thank Y. Iijima for the RT-qPCR experimental support; K. Aiba, T. Tanaka, and M. Seo (Elixirgen Scientific Inc.) for their advice on cell culture and for providing undifferentiated iPSC total RNA samples; H. Yamazaki (Gunma University) for the advice on drebrin RT-PCR and for providing drebrin isoform-specific primer sequences; and T. Shirao (Alzmed, Inc.) and T. Hosoya (Ricoh Company, Ltd.) for their insightful discussions on this research.
Author contributions
Conceptualization, W.L. and Y.S.; methodology, W.L., Y.K., and H.W.; investigation, W.L., S.S, S.Y., Y.K., and K.M.; formal analysis, S.S., H.W., and Y.E.; visualization, W.L., S.S., and H.W.; writing – original draft, W.L., S.Y., and H.W.; writing – review & editing, W.L. and Y.S.; supervision, W.L and Y.S.
Declaration of interests
Y.S. is a scientific consultant to Ricoh Company, Ltd.
Published: February 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106285.
Supplemental information
Data and code availability
Raw and normalized RNA-seq data have been deposited in NCBI's Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. The GEO Series accession number is GEO: GSE224751 and is also included in the key resources table. This paper also analyzes existing, publicly available data. References and source URLs for the datasets are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Okano H., Yamanaka S. IPS cell technologies: significance and applications to CNS regeneration and disease. Mol. Brain. 2014;7:1–12. doi: 10.1186/1756-6606-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taoufik E., Kouroupi G., Zygogianni O., Matsas R. Synaptic dysfunction in neurodegenerative and neurodevelopmental diseases: an overview of induced pluripotent stem-cell-based disease models. Open Biol. 2018;8:180138. doi: 10.1098/rsob.180138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson E.S., Newell-Litwa K. Stem cell models of human synapse development and degeneration. Mol. Biol. Cell. 2018;29:2913–2921. doi: 10.1091/mbc.E18-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohara Y., Koganezawa N., Yamazaki H., Roppongi R.T., Sato K., Sekino Y., Shirao T. Early-stage development of human induced pluripotent stem cell-derived neurons. J. Neurosci. Res. 2015;93:1804–1813. doi: 10.1002/jnr.23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odawara A., Katoh H., Matsuda N., Suzuki I. Physiological maturation and drug responses of human induced pluripotent stem cell-derived cortical neuronal networks in long-term culture. Sci. Rep. 2016;6:26181–26214. doi: 10.1038/srep26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Togo K., Fukusumi H., Shofuda T., Ohnishi H., Yamazaki H., Hayashi M.K., Kawasaki N., Takei N., Nakazawa T., Saito Y., et al. Postsynaptic structure formation of human iPS cell-derived neurons takes longer than presynaptic formation during neural differentiation in vitro. Mol. Brain. 2021;14:149. doi: 10.1186/s13041-021-00851-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam R.S., Töpfer F.M., Wood P.G., Busskamp V., Bamberg E. Functional maturation of human stem cell-derived neurons in long-term cultures. PLoS One. 2017;12:e0169506–e0169526. doi: 10.1371/journal.pone.0169506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirwan P., Turner-Bridger B., Peter M., Momoh A., Arambepola D., Robinson H.P.C., Livesey F.J. Development and function of human cerebral cortex neural networks from pluripotent stem cells in vitro. Development. 2015;142:3178–3187. doi: 10.1242/dev.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P., et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matus A., Ackermann M., Pehling G., Byers H.R., Fujiwara K. High actin concentrations in brain dendritic spines and postsynaptic densities. Proc. Natl. Acad. Sci. USA. 1982;79:7590–7594. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer M., Kaech S., Knutti D., Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/S0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 13.Dunaevsky A., Tashiro A., Majewska A., Mason C., Yuste R. Developmental regulation of spine motility in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 1999;96:13438–13443. doi: 10.1073/pnas.96.23.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi H., Sekino Y., Tanaka S., Mizui T., Kishi S., Shirao T. Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J. Neurosci. 2003;23:6586–6595. doi: 10.1523/jneurosci.23-16-06586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki C., Sekino Y., Hanamura K., Fujisawa S., Mahadomrongkul V., Ren Y., Shirao T. Drebrin A is a postsynaptic protein that localizes in vivo to the submembranous surface of dendritic sites forming excitatory synapses. J. Comp. Neurol. 2005;483:383–402. doi: 10.1002/cne.20449. [DOI] [PubMed] [Google Scholar]
- 16.Toda M., Shirao T., Minoshima S., Shimizu N., Toya S., Uyemura K. Molecular cloning of cDNA encoding human drebrin E and chromosomal mapping of its gene. Biochem. Biophys. Res. Commun. 1993;196:468–472. doi: 10.1006/bbrc.1993.2273. [DOI] [PubMed] [Google Scholar]
- 17.Hanamura K., Koganezawa N., Kamiyama K., Tanaka N., Oka T., Yamamura M., Sekino Y., Shirao T. High-content imaging analysis for detecting the loss of drebrin clusters along dendrites in cultured hippocampal neurons. J. Pharmacol. Toxicol. Methods. 2019;99:106607. doi: 10.1016/j.vascn.2019.106607. [DOI] [PubMed] [Google Scholar]
- 18.Sekino Y., Tanaka S., Hanamura K., Yamazaki H., Sasagawa Y., Xue Y., Hayashi K., Shirao T. Activation of N-methyl-d-aspartate receptor induces a shift of drebrin distribution: disappearance from dendritic spines and appearance in dendritic shafts. Mol. Cell. Neurosci. 2006;31:493–504. doi: 10.1016/j.mcn.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Shirao T., Hanamura K., Koganezawa N., Ishizuka Y., Yamazaki H., Sekino Y. The role of drebrin in neurons. J. Neurochem. 2017;141:819–834. doi: 10.1111/jnc.13988. [DOI] [PubMed] [Google Scholar]
- 20.Tao Y., Zhang S.C. Neural subtype specification from human pluripotent stem cells. Cell Stem Cell. 2016;19:573–586. doi: 10.1016/j.stem.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flitsch L.J., Laupman K.E., Brüstle O. Transcription factor-based fate specification and forward programming for neural regeneration. Front. Cell. Neurosci. 2020;14:121. doi: 10.3389/fncel.2020.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chanda S., Ang C.E., Davila J., Pak C., Mall M., Lee Q.Y., Ahlenius H., Jung S.W., Südhof T.C., Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goparaju S.K., Kohda K., Ibata K., Soma A., Nakatake Y., Akiyama T., Wakabayashi S., Matsushita M., Sakota M., Kimura H., et al. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci. Rep. 2017;7:42367–42412. doi: 10.1038/srep42367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takano T., Funahashi Y., Kaibuchi K. Neuronal polarity: positive and negative feedback signals. Front. Cell Dev. Biol. 2019;7:69. doi: 10.3389/fcell.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dotti C.G., Sullivan C.A., Banker G.A. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arimura N., Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 27.Polioudakis D., de la Torre-Ubieta L., Langerman J., Elkins A.G., Shi X., Stein J.L., Vuong C.K., Nichterwitz S., Gevorgian M., Opland C.K., et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron. 2019;103:785–801.e8. doi: 10.1016/j.neuron.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M., Santpere G., Imamura Kawasawa Y., Evgrafov O.V., Gulden F.O., Pochareddy S., Sunkin S.M., Li Z., Shin Y., Zhu Y., et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:139–148. doi: 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fumagalli M., Lecca D., Abbracchio M.P., Ceruti S. Pathophysiological role of purines and pyrimidines in neurodevelopment: unveiling new pharmacological approaches to congenital brain diseases. Front. Pharmacol. 2017;8:941–1018. doi: 10.3389/fphar.2017.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata R., Vanderhaeghen P. Regulatory roles of mitochondria and metabolism in neurogenesis. Curr. Opin. Neurobiol. 2021;69:231–240. doi: 10.1016/j.conb.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangaraju V., Calloway N., Ryan T.A. Activity-driven local ATP synthesis is required for synaptic function. Cell. 2014;156:825–835. doi: 10.1016/j.cell.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanamura K., Kamata Y., Yamazaki H., Kojima N., Shirao T. Isoform-dependent regulation of drebrin dynamics in dendritic spines. Neuroscience. 2018;379:67–76. doi: 10.1016/j.neuroscience.2018.02.038. [DOI] [PubMed] [Google Scholar]
- 33.Paoletti P., Bellone C., Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 34.Vieira M., Yong X.L.H., Roche K.W., Anggono V. Regulation of NMDA glutamate receptor functions by the GluN2 subunits. J. Neurochem. 2020;154:121–143. doi: 10.1111/jnc.14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S., et al. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–2523. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon A., Yoon S.J., Tran S.S., Makinson C.D., Park J.Y., Andersen J., Valencia A.M., Horvath S., Xiao X., Huguenard J.R., et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat. Neurosci. 2021;24:331–342. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodenas-Ruano A., Chávez A.E., Cossio M.J., Castillo P.E., Zukin R.S. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat. Neurosci. 2012;15:1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizui T., Kojima N., Yamazaki H., Katayama M., Hanamura K., Shirao T. Drebrin e is involved in the regulation of axonal growth through actin-myosin interactions. J. Neurochem. 2009;109:611–622. doi: 10.1111/j.1471-4159.2009.05993.x. [DOI] [PubMed] [Google Scholar]
- 39.Ronchi S., Buccino A.P., Prack G., Kumar S.S., Schröter M., Fiscella M., Hierlemann A. Electrophysiological phenotype characterization of human iPSC-derived neuronal cell lines by means of high-density microelectrode arrays. Adv. Biol. 2021;5:e2000223. doi: 10.1002/adbi.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizui T., Sekino Y., Yamazaki H., Ishizuka Y., Takahashi H., Kojima N., Kojima M., Shirao T. Myosin II ATPase activity mediates the long-term potentiation-induced exodus of stable F-actin bound by drebrin a from dendritic spines. PLoS One. 2014;9:e85367. doi: 10.1371/journal.pone.0085367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishizuka Y., Hanamura K. Drebrin in alzheimer’s disease. Adv. Exp. Med. Biol. 2017;1006:203–223. doi: 10.1007/978-4-431-56550-5_12. [DOI] [PubMed] [Google Scholar]
- 42.Burke E.E., Chenoweth J.G., Shin J.H., Collado-Torres L., Kim S.K., Micali N., Wang Y., Colantuoni C., Straub R.E., Hoeppner D.J., et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat. Commun. 2020;11:462–514. doi: 10.1038/s41467-019-14266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandes H.J.R., Patikas N., Foskolou S., Field S.F., Park J.-E., Byrne M.L., Bassett A.R., Metzakopian E. Single-cell transcriptomics of Parkinson’s disease human in vitro models reveals dopamine neuron-specific stress responses. Cell Rep. 2020;33:108263. doi: 10.1016/j.celrep.2020.108263. [DOI] [PubMed] [Google Scholar]
- 44.Ni P., Noh H., Shao Z., Zhu Q., Guan Y., Park J.J., Arif F., Park J.M., Abani C., Beaudreault C., et al. Large-scale generation and characterization of homogeneous populations of migratory cortical interneurons from human pluripotent stem cells. Mol. Ther. Methods Clin. Dev. 2019;13:414–430. doi: 10.1016/j.omtm.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H.C., He Z., Ebert S., Schörnig M., Santel M., Nikolova M.T., Weigert A., Hevers W., Kasri N.N., Taverna E., et al. NGN2 induces diverse neuron types from human pluripotency. Stem Cell Rep. 2021;16:2118–2127. doi: 10.1016/j.stemcr.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaBarbera K.M., Limegrover C., Rehak C., Yurko R., Izzo N.J., Knezovich N., Watto E., Waybright L., Catalano S.M. Modeling the mature CNS: a predictive screening platform for neurodegenerative disease drug discovery. J. Neurosci. Methods. 2021;358:109180. doi: 10.1016/j.jneumeth.2021.109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 48.Ho K.H., Patrizi A. Assessment of common housekeeping genes as reference for gene expression studies using RT-qPCR in mouse choroid plexus. Sci. Rep. 2021;11:3278–3315. doi: 10.1038/s41598-021-82800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019;20:278–313. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Augustyniak J., Lenart J., Lipka G., Stepien P.P., Buzanska L. Reference gene validation via RT–qPCR for human iPSC-derived neural stem cells and neural progenitors. Mol. Neurobiol. 2019;56:6820–6832. doi: 10.1007/s12035-019-1538-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müller J., Ballini M., Livi P., Chen Y., Radivojevic M., Shadmani A., Viswam V., Jones I.L., Fiscella M., Diggelmann R., et al. High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab Chip. 2015;15:2767–2780. doi: 10.1039/c5lc00133a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and normalized RNA-seq data have been deposited in NCBI's Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. The GEO Series accession number is GEO: GSE224751 and is also included in the key resources table. This paper also analyzes existing, publicly available data. References and source URLs for the datasets are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.