Abstract
Interferon‐induced transmembrane protein 3 (IFITM3) is an antiviral protein that alters cell membranes to block fusion of viruses. Conflicting reports identified opposing effects of IFITM3 on SARS‐CoV‐2 infection of cells, and its impact on viral pathogenesis in vivo remains unclear. Here, we show that IFITM3 knockout (KO) mice infected with SARS‐CoV‐2 experience extreme weight loss and lethality compared to mild infection in wild‐type (WT) mice. KO mice have higher lung viral titers and increases in inflammatory cytokine levels, immune cell infiltration, and histopathology. Mechanistically, we observe disseminated viral antigen staining throughout the lung and pulmonary vasculature in KO mice, as well as increased heart infection, indicating that IFITM3 constrains dissemination of SARS‐CoV‐2. Global transcriptomic analysis of infected lungs shows upregulation of gene signatures associated with interferons, inflammation, and angiogenesis in KO versus WT animals, highlighting changes in lung gene expression programs that precede severe lung pathology and fatality. Our results establish IFITM3 KO mice as a new animal model for studying severe SARS‐CoV‐2 infection and overall demonstrate that IFITM3 is protective in SARS‐CoV‐2 infections in vivo.
Keywords: COVID‐19, IFITM3, IFITM, interferon, SARS‐CoV‐2
Subject Categories: Immunology; Membranes & Trafficking; Microbiology, Virology & Host Pathogen Interaction
IFITM3 limits severity of SARS‐CoV‐2 infections by restricting viral replication and spread, thereby reducing lung inflammation, immune cell infiltration, angiogenesis, and pathology. IFITM3 KO mice are a new model for studying severe SARS‐CoV‐2 infections.

Introduction
IFITM3 is an antiviral restriction factor that inhibits virus fusion with host cell membranes (Li et al, 2013; Desai et al, 2014; Chesarino et al, 2017). IFITM3 potently blocks infection by numerous enveloped viruses of human health concern, including influenza, dengue, and Zika viruses (Majdoul & Compton, 2021). This set of viruses primarily utilizes endocytic pathways for entry into cells, and fuses with endosome membranes where IFITM3 is abundantly localized and poised to restrict infection (Chesarino et al, 2014; Jia et al, 2014). In contrast, viruses that fuse primarily at the plasma membrane, such as Sendai virus, are minimally affected by IFITM3 (Hach et al, 2013; Chesarino et al, 2015). Viruses that are able to fuse at both the plasma membrane or within endosomes, such as human metapneumovirus, can be partially restricted by IFITM3, with the portion of virus that fuses at the plasma membrane largely evading restriction (McMichael et al, 2018). These patterns of restriction hold true for the vast majority of the dozens of viruses that have been tested for IFITM3 inhibition, with the notable exception of the common cold coronavirus OC43, which utilizes dual cell entry pathways similarly to metapneumovirus, but showed enhanced, rather than reduced, infection when IFITM3 was expressed in target cells (Zhao et al, 2014). This unusual finding suggests that IFITM3 may have unanticipated effects on some coronaviruses, leading us and several other groups to examine the impact of IFITM3 and related proteins on SARS‐CoV‐2, which remains a critical threat to worldwide human health.
SARS‐CoV‐2 uses cell surface and endosomal fusion strategies for infection (Hoffmann et al, 2020; Shang et al, 2020). Experiments studying effects of IFITM3 on SARS‐CoV‐2 identified opposing activities in which IFITM3 inhibits virus entry at endosomes but enhances virus entry at the plasma membrane (Shi et al, 2021). The overall effects of IFITM3 remain controversial, with some reports indicating that IFITM3 primarily restricts cellular SARS‐CoV‐2 infection (Peacock et al, 2021; Shi et al, 2021, 2022; Winstone et al, 2021; Xu et al, 2022), while others conclude that IFITM3 is a net enhancer of infection, particularly in lung cells (Prelli Bozzo et al, 2021; Lista et al, 2022; Nchioua et al, 2022). Furthermore, IFITM3 inhibits fusion of SARS‐CoV‐2 infected cells with neighboring cells (syncytia formation), which may be an important mechanism of viral spread within tissues (Buchrieser et al, 2020; Rajah et al, 2021). The overall balance and relevance of the effects of IFITM3 in SARS‐CoV‐2 infection of cells and pathogenesis in vivo remain unclear.
We previously generated IFITM3 knockout (KO) mice to investigate in vivo effects of this antiviral restriction factor and confirmed a profound susceptibility of these mice to severe influenza virus infections (Kenney et al, 2019). These results were consistent with studies linking deleterious single‐nucleotide polymorphisms in the human IFITM3 gene to severe influenza (Everitt et al, 2012; Allen et al, 2017; Zani & Yount, 2018). Although some studies have suggested that IFITM3 gene polymorphisms are risk factors for severe SARS‐CoV‐2 infection (Nikoloudis et al, 2020; Zhang et al, 2020b; Alghamdi et al, 2021; Cuesta‐Llavona et al, 2021; Gomez et al, 2021; Xu et al, 2022), other studies did not identify IFITM3 variants as top candidates (Ellinghaus et al, 2020; Pairo‐Castineira et al, 2021; Pathak et al, 2021). In addition to possible direct effects of IFITM3 gene polymorphisms, defects in interferon responses that may result in decreased IFITM3 levels during infection have been linked to roughly 20% of severe COVID‐19 cases, providing additional circumstances where IFITM3 insufficiency may be impacting disease (Bastard et al, 2020; Zhang et al, 2020a). Given the conflicting reports of IFITM3 activity on SARS‐CoV‐2 in vitro, coupled with the potential, but uncertain, role of IFITM3 in modulating SARS‐CoV‐2 disease severity, we investigated roles of IFITM3 in SARS‐CoV‐2 infection in the mouse model.
Results & Discussion
IFITM3 limits severity of SARS‐CoV‐2 MA10 infection
The exact role of IFITM3 in cellular infections by SARS‐CoV‐2 is a contentious topic, with reports of both restriction and enhancement of infection in vitro (Peacock et al, 2021; Prelli Bozzo et al, 2021; Shi et al, 2021, 2022; Winstone et al, 2021; Lista et al, 2022; Nchioua et al, 2022; Xu et al, 2022). In our laboratory, we have observed that IFITM3 is capable of both restriction and enhancement of infection, a phenomenon that is dependent upon the subcellular localization of the protein (Shi et al, 2021). Moreover, conflicting results in this field are likely confounded by the distinct cells, virus strains, and readouts utilized in each individual report, in addition to key differences in virus stocks that may have arisen through divergent propagation techniques. Thus, direct comparisons of individual studies are difficult, highlighting the need to assess the role of IFITM3 during SARS‐CoV‐2 infection in vivo.
IFITM3 is expressed in diverse cell types and has been implicated in protecting the function and survival of respiratory epithelial cells, dendritic cells, and lung‐resident T cells during respiratory virus infections (Bailey et al, 2012; Wakim et al, 2013; Infusini et al, 2015). Likewise, IFITM3 basal expression and interferon induction in lung epithelial cells may be predictive of COVID‐19 disease severity (Hachim et al, 2020; Bridges et al, 2022). Analysis of published single‐cell RNA sequencing data from lungs of control mice and mice infected with a mouse‐adapted SARS‐CoV‐2 strain (Gene Expression Omnibus [GEO] accession No. GSE186360; Vanderheiden et al, 2021) showed Ifitm3 transcripts in epithelial cells and endothelial cells as well as immune cells, with prominent expression in monocytes, dendritic cells, and neutrophils (Fig 1A). Expression of Ifitm3 in these cell populations was enhanced following infection (Fig 1A). For example, the percentage of cells in which Ifitm3 expression was detected in mock vs SARS‐CoV‐2 infected samples increased from 3.8% to 20% in epithelial cells and 12.4% to 22.6% in endothelial cells. To determine the impact of IFITM3 on SARS‐CoV‐2 disease severity, we infected wild‐type (WT) C57BL/6 mice and corresponding IFITM3 KO animals with the mouse‐adapted SARS‐CoV‐2 strain MA10 (Leist et al, 2020). Importantly, our virus stock was generated with a stringent plaque purification and sequencing protocol, allowing us to utilize virus lacking commonly observed tissue culture adaptations that attenuate pathogenicity (Johnson et al, 2021). After infection, we observed a greater than 10% weight loss in WT mice with recovery to full weight by Day 8 postinfection (Fig 1B). In contrast, IFITM3 KO mice lost significantly more weight starting at Day 1 postinfection. This accelerated weight loss continued through Day 5 postinfection, at which point all IFITM3 KOs had either died or met humane endpoint criteria due to severe illness and lack of movement (Fig 1B). To delineate the role of IFITM3 from the broader type I interferon response in protecting against SARS‐CoV‐2 infection, we also infected a cohort of mice lacking the type I interferon receptor (IFNAR). We observed that, similarly to IFITM3 KO mice, IFNAR KO mice exhibited increased weight loss compared with WT mice at Days 3 and 4 postinfection (Fig 1C). However, unlike IFITM3 KO mice, which succumbed to infection, all IFNAR KO mice survived and rapidly recovered weight to baseline values. The less severe outcome in IFNAR KO mice may be due to baseline expression of IFITM3 in IFNAR KOs and/or IFITM3 induction by additional cytokines other than type I IFNs. We conclude that IFITM3 provides essential protection against severe, lethal SARS‐CoV‐2‐induced disease in mice.
Figure 1. IFITM3 limits the severity of SARS‐CoV‐2 MA10 infection.
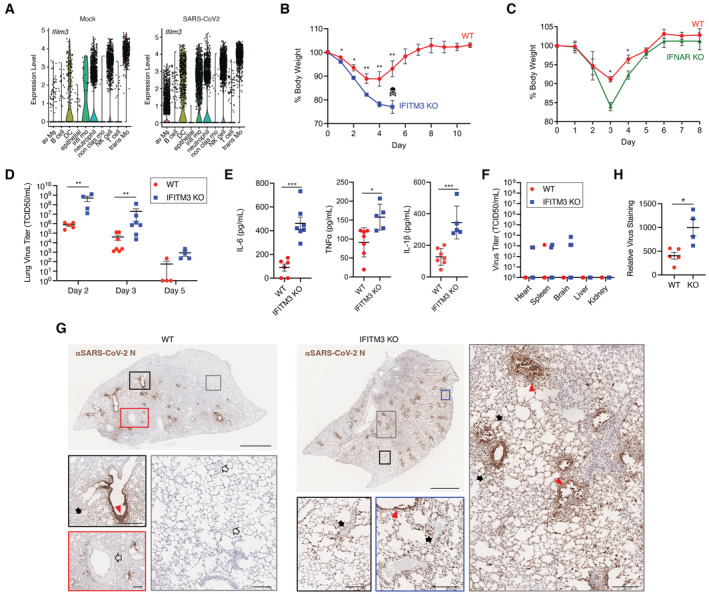
- Analysis of published single‐cell RNA sequencing data from lungs of noninfected mice and mice infected with SARS‐CoV‐2 MA10 (GEO accession No. GSE186360). Av Mϕ, alveolar macrophages; DC, dendritic cells; infl mo, inflammatory monocytes; nonclas mo, nonclassical monocytes; trans mo, transitional/intermediate monocytes.
- WT and IFITM3 KO mice were intranasally infected with 105 TCID50 SARS‐CoV‐2 MA10. Weight loss (data points represent mean and error bars represent SEM, data from three independent experiments [WT n = 11, IFITM3 KO n = 10], skull/crossbones indicates that KO mice did not survive beyond this timepoint, *P < 0.05, **P < 0.01, ANOVA with Bonferoni's multiple comparison's test).
- WT and IFNAR KO mice were intranasally infected with 105 TCID50 SARS‐CoV‐2 MA10. Weight loss (data points represent mean and error bars represent SD, data from one experiment [n = 3 per group], *P < 0.05, ANOVA with Bonferoni's multiple comparison's test).
- Viral titers in lung homogenates at the indicated days postinfection from mice infected as in (B) (bars/error bars represent mean ± SD, data points represent distinct biological samples, data points at the x axis represent samples below the limit of detection, Day 3 data from two independent experiments, Day 2 and 5 data are from single experiments, **P < 0.01, Mann–Whitney test).
- IL‐6 levels from lung homogenates and TNFα and IL‐1β levels from serum on Day 3 postinfection (bars/error bars represent mean ± SD, data points represent distinct biological samples, data from two independent experiments, ***P < 0.001, t‐test).
- Viral titers in extrapulmonary organs on Day 3 postinfection were measured (bars/error bars represent mean ± SD, data points represent distinct biological samples [n = 7 for heart, spleen, brain; n = 5 for kidney and liver], data points at the x axis represent samples below the limit of detection, ns, multiple unpaired t‐tests).
- Representative lung sections from WT or IFITM3 KO animals stained for SARS‐CoV‐2 N protein. Scale bars represent 2 mm for whole lung images. Colored boxes overlaying the whole lung images correspond to the displayed magnified regions and scale bars in these images represent 0.2 mm. Red triangles, infected airways; solid black arrow, noninfected blood vessels; open black arrow, infected blood vessels.
- Quantification of lung section staining for multiple mice as depicted in (G) (bars/error bars represent mean ± SD, data points represent distinct biological samples, *P < 0.05, t‐test).
We next measured viral titers at Days 2, 3, and 5 postinfection and found that IFITM3 KO lung titers were significantly higher than WT titers on Days 2 and 3 (Fig 1D). Elevated levels of the inflammatory cytokines IL‐6, TNFα, and IL‐1β were also detected in KO versus WT lungs at Day 3 postinfection (Fig 1E), likely due to higher viral burden in the lung. Alternatively, these results are consistent with feedback inhibition of type I interferon induction and inflammation by IFITM3, as has been previously suggested (Jiang et al, 2017; Stacey et al, 2017; Clement et al, 2022). Viral titers were decreased in both groups between Days 2 and 5 (Fig 1D). However, complete virus clearance was delayed in IFITM3 KOs, with all KO mouse lungs remaining positive for virus at Day 5, while only one of four WT lungs were positive for live virus at this time point (Fig 1D). At Day 3 postinfection, we also detected a low level of live virus in the heart in a subset of KO animals (Fig 1F), suggesting that IFITM3 may have cardioprotective properties similar to what we have previously reported for influenza virus (Kenney et al, 2019, 2022). Indeed, cardiac complications during COVID‐19 infection rapidly emerged as a common and long‐term extrapulmonary concern even in mild cases of the disease (Gupta et al, 2020; Zheng et al, 2021; Puntmann et al, 2022; Xie et al, 2022). Likewise, individuals with prevalent IFITM3 SNPs, which have been linked to increased COVID‐19 severity, may be at heightened risk of these cardiac complications (Nikoloudis et al, 2020; Zhang et al, 2020b; Alghamdi et al, 2021; Cuesta‐Llavona et al, 2021; Gomez et al, 2021; Xu et al, 2022). Infection of the spleen and brain was also enhanced in IFITM3 KO mice, while no virus was detected in liver or kidney tissue in WT or KO animals (Fig 1F). Apart from one mouse that had virus in both the heart and the brain, virus‐positive extrapulmonary samples were from separate animals. Live virus was not detected in any of these extrapulmonary organs in WT or KO animals at Day 5 postinfection. Overall, we observed that the enhanced illness severity in IFITM3 KO mice was accompanied by increased lung viral titers and delayed clearance, as well as more prevalent virus dissemination. Future studies centered on extrapulmonary manifestations, including cardiac pathology, of SARS‐CoV‐2 and the role of IFITM3 in limiting these pathologies are warranted.
Given that IFITM3 generally blocks infection and spread of certain viruses, we investigated the effects of IFITM3 on viral infection patterns in the lungs by staining for viral antigen (N protein) in WT and IFITM3 KO lung sections at Day 2 post‐SARS‐CoV‐2 MA10 infection, representing an early time point at which viral titers are high (Leist et al, 2020). We detected robust staining of the airways in both groups, but observed more disseminated staining throughout IFITM3 KO lungs (Fig 1G). Indeed, large portions of WT lungs showed no staining for viral antigen, while virus was detected throughout KO lungs (Fig 1G). Furthermore, accumulation of viral antigen that likely represents shedding of necrotic, highly infected cells into the bronchioles was observed in KO lungs, even at this early time point (Fig 1G). Infected cells associated with blood vessels could be seen in WT lungs primarily when adjacent to highly infected airways (Fig 1G), while blood vessels associated with infected cells were readily apparent throughout IFITM3 KO lungs (Fig 1G). Quantification of viral staining in lung sections from multiple mice confirmed the significant increase in viral antigen staining in IFITM3 KO versus WT mice (Fig 1H). Overall, viral antigen imaging confirmed increased viral titers (Fig 1D), and also revealed diffuse infection of cells, including the vasculature, in IFITM3 KO lungs.
Loss of IFITM3 in the K18‐hACE2 model of severe SARS‐CoV‐2 infection enhances viral dissemination and accelerates morbidity
Mouse‐adapted SARS‐CoV‐2 differs from the parental human isolate by only seven amino acids (Leist et al, 2020) and utilizes endogenously expressed murine ACE2 as the virus receptor for infection, making it a highly relevant model for studying viral tropism and pathogenesis in vivo. Nonetheless, we also sought to examine the in vivo role of IFITM3 in infection with WA1, the nonadapted SARS‐CoV‐2 parental strain for MA10 that was isolated from humans, which we also stringently propagated and sequenced. For this, we utilized mice expressing human (h)ACE2 under control of a keratin (K18) promoter (K18‐hACE2 mice) crossed with IFITM3 KOs. We observed that K18‐hACE2/IFITM3 KO mice lost significantly more weight than K18‐hACE2 control mice and that all K18‐hACE2/IFITM3 KO mice succumbed to infection by Day 5 (Fig 2A), at which point we ended our experiments. High virus titers at Day 3 postinfection were detected in both control and KO mice, with one of two experiments showing significantly higher titers in IFITM3 KOs and a second experiment showing a trend toward higher titers in the KOs (Fig 2B). Cardiac dissemination of virus was detected in eight of eight K18‐hACE2/IFITM3 KOs compared to one of eight control mice (Fig 2C), confirming that IFITM3 restricts cardiac dissemination of SARS‐CoV‐2. We also observed diffuse viral antigen staining throughout the lungs of both control mice and IFITM3 KOs (Fig 2D). Notably, the pattern of viral antigen staining in K18‐hACE2 mice infected with WA1 was distinct and more widespread throughout the lung tissue compared with staining of WT mouse lungs infected with MA10, which was largely confined to the bronchioles and closely surrounding tissue, even though mice were infected with a 10‐fold higher dose of MA10 (Fig 2D). Indeed, viral burden in the lungs of K18‐hACE2 mice infected with WA1 was as much as four orders of magnitude higher than WT mice infected with MA10 (Figs 1D and 2B). These data suggest that the K18 promoter‐driven expression of hACE2 does not mirror endogenous production of ACE2 in murine lungs and thus allows widespread infection throughout the lung. The extreme virus replication in the K18‐hACE2 model may also explain why the overall effect of IFITM3 on lung viral burden was blunted in this model. We conclude that while the K18‐hACE2 model has proven valuable for investigating effectiveness of vaccines or antivirals, examination of WT mice with MA10 infection may be superior for studying pathogenesis and immune responses since the natural expression of murine ACE2 dictates viral tropism. Nonetheless, we have identified a protective effect of IFITM3 in SARS‐CoV‐2 infections in vivo using both mouse‐adapted and human isolates.
Figure 2. IFITM3 limits the severity of SARS‐CoV‐2 WA1 infection in K18‐hACE2 mice.
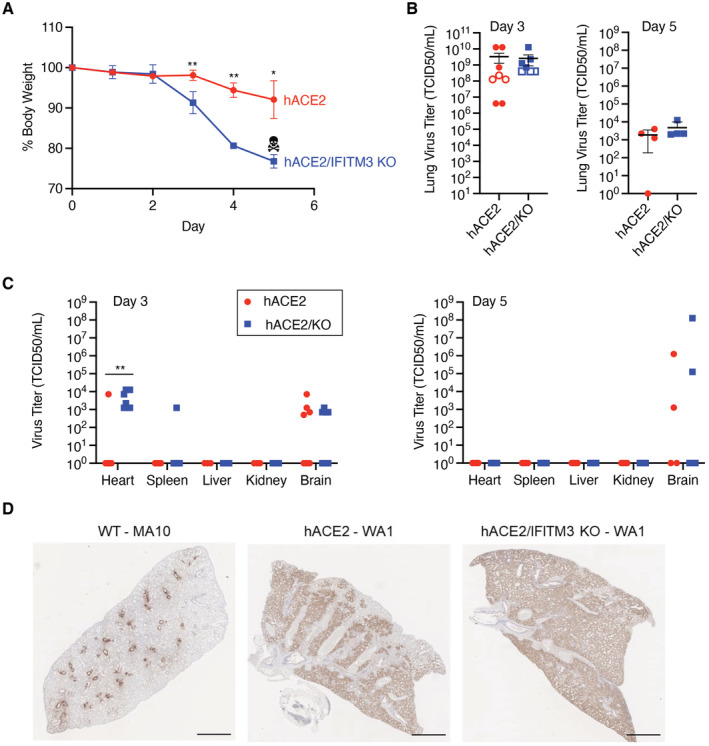
- Weight loss (data points represent mean and error bars represent SEM, data from two independent experiments [hACE2 n = 7, hACE2/IFITM3 KO n = 8], skull/crossbones indicates that KO mice did not survive beyond this timepoint, *P < 0.05, **P < 0.01, ANOVA with Bonferoni's multiple comparison's test).
- Viral titers in lung homogenates at the indicated days postinfection (bars/error bars represent mean ± SD, data points represent distinct biological samples, data for Day 3 are from two independent experiments, ns, t‐test).
- Viral titers in extrapulmonary organs on the indicated days postinfection were measured (bars/error bars represent mean ± SD, data points represent distinct biological samples, Day 3 data for heart, spleen, and brain are from two independent experiments, Day 3 data for liver and kidney and Day 5 data are from single experiments, data points at the x axis represent samples below the limit of detection, **P < 0.01, Mann–Whitney test).
- Representative lung sections from WT mice infected with SARS‐CoV‐2 MA10 (105 TCID50) or K18‐hACE2 or K18‐hACE2/IFITM3 KO mice infected with SARS‐CoV‐2 WA1 (104 TCID50) stained for SARS‐CoV‐2 N protein at Day 5 postinfection. Scale bars represent 2 mm.
Lung histopathology and immune cell infiltration are exacerbated in the absence of IFITM3
To mechanistically characterize roles of IFITM3 in limiting SARS‐CoV‐2 lung pathogenesis, we performed hematoxylin and eosin staining on lung sections from WT and IFITM3 KO animals after infection with SARS‐CoV‐2 MA10. All lungs from infected animals showed areas of consolidated tissue, cellular infiltration, and inflammation (Fig 3A). We observed that larger portions of the lungs were afflicted in IFITM3 KO samples, and thus quantified inflamed and consolidated areas versus open airspace in sections from individual infected WT and IFITM3 KO mouse lungs. This analysis confirmed increased pathology in infected IFITM3 KO versus WT lungs at Day 5 postinfection (Fig 3B), correlating with the increased illness and viral titers that we observed in these animals (Fig 1B and D). This conclusion was supported further by immunohistochemical staining and quantification of CD45‐positive immune cell infiltration into the lung, which was elevated in IFITM3 KO versus WT mice at Day 5 (Fig 4A and B). Thus, lung pathology resulting from SARS‐CoV‐2 infection was exacerbated in IFITM3 KO mice.
Figure 3. SARS‐CoV‐2‐induced lung histopathology is exacerbated in the absence of IFITM3.
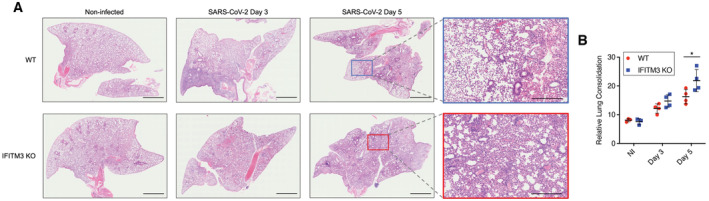
- Representative lung images from mice at the indicated times postinfection were stained with hematoxylin and eosin. Scale bars represent 2 mm for whole lung images and 500 μm for zoomed images.
- Staining quantifications from multiple mice are shown (bars/error bars represent mean ± SD, data points represent distinct biological samples, *P < 0.05, ANOVA with Bonferoni's multiple comparison's test).
Figure 4. Elevated immune cell infiltration in response to SARS‐CoV‐2 infection in IFITM3 KO lungs.
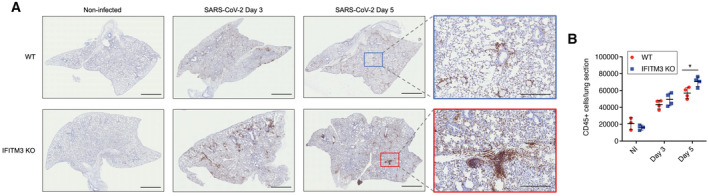
- Representative lung images from mice at the indicated times postinfection were stained with anti‐CD45. Scale bars represent 2 mm for whole lung images and 500 μm for zoomed images.
- Staining quantifications from multiple mice are shown (bars/error bars represent mean ± SD, data points represent distinct biological samples, *P < 0.05, ANOVA with Bonferoni's multiple comparison's test).
Loss of IFITM3 leads to aberrant inflammatory and angiogenic gene expression programs
We next examined the global impact of loss of IFITM3 on transcriptional programs at Day 2 postinfection with SARS‐CoV‐2 MA10 by performing lung RNA sequencing. Dimensionality reduction approaches showed divergence of gene signatures in infected WT versus KO lungs (Fig 5A), corresponding to 1,865 differentially expressed genes between the groups (≥ |2| fold change, P‐value < 0.05 (Fig 5B); a list of differentially expressed genes, expression values, fold changes, and P‐values are shown in Dataset EV1). GO term enrichment analysis of genes upregulated in infected IFITM3 KO versus WT lungs revealed significant increases in antiviral defense responses, angiogenesis, and inflammation (Fig 5C; Dataset EV2). Among the antiviral and inflammatory genes increased in KO versus WT animals were multiple type I interferon genes, including Ifnb1 and Ifna4, and canonical interferon‐stimulated genes, such as Oas1b, Mx1, and Ifit2 (Fig 5D). Interestingly, type I interferons provide a double‐edged sword in COVID‐19, limiting virus replication early in infection while also contributing to pathological inflammation (Israelow et al, 2020; Sposito et al, 2021). Expression of chemokine genes Ccl5 and Cxcl9 was also potentiated in KO lungs relative to WT (Fig 5D), consistent with the increased recruitment of CD45‐positive immune cells that we observed (Fig 3C). Classic angiogenesis‐driving genes with significantly elevated expression in KO versus WT animals included Vegfa, Hif3a, Wnt7a, and Pdgfa (Fig 5E), consistent with previous sequencing data of SARS‐related disease (Gralinski et al, 2013). Furthermore, angiogenesis and thrombosis rapidly emerged as hallmarks of SARS‐CoV‐2‐associated disease (Ackermann et al, 2020). The observed upregulation of angiogenic gene programs in infected IFITM3 KOs may result from direct infection of blood vessel cells or from infected cells mediating bystander signaling. We also noted that several coagulation‐associated genes had enhanced upregulation in the KO animals, such as Vwf, F2r, F2rl3, Plat, Fgg, F3, and Thbd (Dataset EV1), consistent with exacerbated endothelial cell dysfunction occurring in the IFITM3 KO animals.
Figure 5. SARS‐CoV‐2 infection induces increased inflammatory and angiogenic gene expression programs in IFITM3 KO lungs.
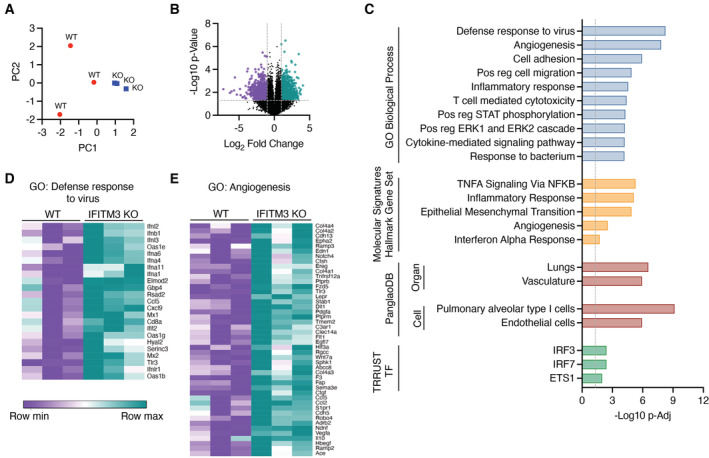
- Principal components analysis comparing WT and KO RNA samples.
- Volcano plot of differential gene expression comparing IFITM3 KO versus WT. Purple, 958 genes downregulated in KO versus WT; Green, 907 genes upregulated in KO versus WT.
- Upregulated genes in KO versus WT were subjected to pathway and ontology analysis. The top 10 most significant GO Biological Processes are shown (−log10 P‐Elim pruning value is graphed), the top five most significant Hallmark gene sets, the significant associations from PanglaoDB analysis, and the significant associations from TRRUST transcription factor analysis are shown.
- Relative gene expression heat maps for genes from the top two most significant GO Biological Processes as shown in (C).
These statistically significant increases in activation of inflammatory antiviral and angiogenesis pathways in KO lungs were further confirmed by Molecular Signature Database Hallmark gene set enrichment analysis (Fig 5C). Likewise, analysis with the PanglaoDB database of single‐cell RNA sequencing experiments identified significant associations with gene signatures specific to pneumocytes and endothelial cells within genes upregulated in IFITM3 KOs (Fig 5C). Furthermore, TRRUST Transcriptional Regulatory Network database analysis identified interferon regulatory factors, IRF3 and IRF7, as well as the pro‐angiogenic protein ETS1, as transcriptional factors likely involved in upregulation of genes in infected IFITM3 KO versus WT lungs (Fig 5C). Thus, multiple independent analyses of our data converged on inflammatory interferon and angiogenic/endothelial nodes as key pathways that precede the uniquely severe manifestations of SARS‐CoV‐2 infections occurring in the absence of IFITM3. Analysis of the downregulated genes in IFITM3 KO versus WT lungs indicated association with muscle gene signatures, possibly suggesting infection and elimination of lung smooth muscle cells by SARS‐CoV‐2 infection or disruption of regenerative responses (Dataset EV2). These results are consistent with our findings that IFITM3 restrains SARS‐CoV‐2 replication and spread through the lungs and cardiovascular system, thus limiting induction of pathological interferon and angiogenesis gene expression programs.
In sum, although IFITM3 may be capable of promoting virus replication in certain cell culture conditions, we conclude that it has a net beneficial effect in limiting SARS‐CoV‐2 disease severity in the mouse model. IFITM3 KO mice represent an important tool for investigating pathological mechanisms active in severe SARS‐CoV‐2 infections, including inflammation, cardiac pathologies, and angiogenic and pro‐thrombotic signals. In addition to uncovering the utility of IFITM3 KO mice for understanding SARS‐CoV‐2 pathogenesis mechanisms, the results of our study demonstrate that IFITM3 restrains virus replication, limits activation of inflammatory and angiogenic hallmarks of severe disease, and overall limits pathology in SARS‐CoV‐2 infections in vivo.
Materials and Methods
Biosafety
All experiments with SARS‐CoV‐2 were performed in the Ohio State University (OSU) Biosafety Level 3 (BSL3) facility. Experimental procedures were approved by the OSU Institutional Biosafety Committee and OSU BSL3 Advisory Group. Research samples analyzed outside the BSL3 containment were decontaminated according to experimentally validated and institutionally approved protocols.
Virus stocks and titering
SARS‐CoV‐2 strain USA‐WA1/2020 (WA1) and mouse‐adapted SARS‐CoV‐2 strain MA10 (generated by the laboratory of Dr. Ralph Baric, University of North Carolina) were obtained from BEI Resources (NR‐52281 & NR‐55329). Both viruses were plaque purified on Vero‐TMPRSS2 cells (kindly provided by Dr. Shan‐Lu Liu, The Ohio State University), and viral genomes from individual plaques were sequenced to identify virus lacking mutations in the furin cleavage site of the Spike protein. Virus plaques with intact furin cleavage sites were propagated on Vero‐TRMPSS2 cells. Virus‐containing supernatants were aliquoted and flash frozen in liquid nitrogen, and stored at −80°C. Virus stocks were again sequenced to ensure the absence of furin cleavage site mutations. Viral stocks and research samples were titered via TCID50 measurements on Vero E6 cells using 10‐fold serial dilutions in triplicate. The presence of virus replication in each well was assessed by visual examination of cytopathic effect along with antibody staining for N protein antigen (mouse monoclonal antibody 40143‐MM08, Sino Biological) with fluorescent secondary antibody detection. TCID50 titers were calculated using the Reed–Muench method. Cell lines were confirmed mycoplasma‐negative prior to use.
Mouse husbandry and infections
All mice (mus muluscus) used in this study were of the C57BL/6 background. IFITM3 KO mice with a 53 base pair deletion in exon 1 of the Ifitm3 gene were described previously (Kenney et al, 2019). K18‐hACE2 hemizygous mice were purchased from Jackson Laboratories (B6.Cg‐Tg(K18‐ACE2)2Prlmn/J, Strain #:034860, RRID:IMSR_JAX:034860). IFITM3 KO mice were crossed with K18‐hACE2 mice to obtain F1 mice possessing a K18‐hACE2 allele with heterozygous IFITM3 KO. F1 mice were then bred with IFITM3 KO mice to generate mice carrying hACE2 transgene and homozygous KO of IFITM3. All animals were housed in sterile ventilated cages within a specific pathogen‐free environment. Male and female mice between 6 and 12 weeks of age were used in our experiments. Mice were infected intranasally under anesthesia with isoflurane. Mouse organs were collected and homogenized in PBS for ELISAs or titering, homogenized in Trizol for extraction of RNA, or fixed in 10% formalin for histology. All procedures were approved by the OSU IACUC (protocol 2020A00000054) and were performed in accordance with guidelines for the ethical use of animals.
ELISAs and histology
IL‐6 ELISAs were performed on fluid from lung homogenates using an R&D Systems Duoset ELISA kit (catalog # DY406) according to the manufacturer's instructions. Quantification of serum IL‐1β and TNFα was performed using a mouse V‐PLEX Proinflammatory Panel 1 kit (Meso Scale Diagnostics) and was analyzed by the OSU Center for Clinical and Translational Science. For lung histology, lung tissue samples were fixed in formalin for 7 days at 4°C, followed by embedding in paraffin. For H&E and anti‐CD45 staining, lungs were sectioned, stained, and imaged by the OSU Comparative Pathology and Mouse Phenotyping Shared Resource. Quantification of these images was performed using ImageJ software and the color deconvolution method. Staining for SARS‐CoV‐2 antigen was performed and imaged by Histowiz (Histowiz.com, Brooklyn, NY, USA).
RNA sequencing and informatics
RNA was extracted from lung tissue using TRIzol (Invitrogen). RNA library preparation and sequencing was performed by GENEWIZ, LLC./Azenta US, Inc (South Plainfield, NJ, USA) with the following methods as provided by GENEWIZ: “The RNA samples received were quantified using Qubit 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) and RNA integrity was checked using TapeStation (Agilent Technologies, Palo Alto, CA, USA). The RNA sequencing libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina using manufacturer's instructions (New England Biolabs, Ipswich, MA, USA). Briefly, mRNAs were initially enriched with Oligod(T) beads. Enriched mRNAs were fragmented for 15 min at 94°C. First strand and second strand cDNA were subsequently synthesized. cDNA fragments were end repaired and adenylated at 3′ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by PCR with limited cycles. The sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA), and quantified by using Qubit 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA). The sequencing libraries were multiplexed and clustered onto a flowcell. After clustering, the flowcell was loaded onto the Illumina HiSeq instrument according to manufacturer's instructions. The samples were sequenced using a 2 × 150 bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de‐multiplexed using Illumina bcl2fastq 2.17 software. One mis‐match was allowed for index sequence identification.”
Resulting fastq files were analyzed by ROSALIND® (https://rosalind.bio/), with a HyperScale architecture developed by ROSALIND, Inc. (San Diego, CA). Read Distribution percentages, violin plots, identity heatmaps, and sample MDS plots were generated as part of the QC step. Statistical analysis for differential gene expression was performed using the “limma” R library (Ritchie et al, 2015). The principal components analysis and volcano plots were re‐formatted in PRISM graphing software using values downloaded from ROSALIND. Heatmaps were generated using gene expression values downloaded from ROSALIND and Morpheus software (https://software.broadinstitute.org/morpheus). Hypergeometric distribution was used to analyze the enrichment of pathways, gene ontology, and other ontologies. The topGO R library was used to determine local similarities and dependencies between GO terms in order to perform Elim pruning correction. Several database sources were referenced for enrichment analysis, including MSigDB (Subramanian et al, 2005; Liberzon et al, 2011), TRRUST (Han et al, 2018), and PanglaoDB (Franzen et al, 2019). Enrichment was calculated relative to a set of background genes relevant for the experiment.
Single‐cell RNA sequencing data mining
Single‐cell analysis of Ifitm3 expression in murine (C57BL/6J) lungs (mock vs Day 4 postinfection with SARS‐CoV‐2 MA10) was performed on published data (GEO dataset accession No. GSE186360) exactly as previously described (Vanderheiden et al, 2021).
Statistical analysis
Statistical analysis was performed using the appropriate comparisons (t‐test, ANOVA) under the assumptions that (i) the responses for each factor level had a normal population distribution, (ii) these distributions had the same variance, and (iii) the data were independent. Datasets that did not meet the first criterion were subjected to Mann–Whitney tests. Samples were not blinded but were collected and analyzed in an equivalent, unbiased fashion.
Author contributions
Adam D Kenney: Formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Ashley Zani: Formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Jeffrey Kawahara: Investigation. Adrian C Eddy: Investigation. Xiao‐Liang Wang: Resources. KC Mahesh: Resources. Mijia Lu: Resources. Jeronay Thomas: Data curation. Jacob E Kohlmeier: Data curation. Mehul S Suthar: Data curation. Emily A Hemann: Investigation. Jiangrong LI: Resources. Mark E Peeples: Resources. Luanne Hall‐Stoodley: Resources. Adriana Forero: Formal analysis; visualization. Chuanxi Cai: Resources. Jianjie Ma: Resources; funding acquisition. Jacob S Yount: Conceptualization; data curation; formal analysis; supervision; funding acquisition; visualization; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors do not have any conflicts of interest to disclose relating to this work.
Supporting information
Dataset EV1
Dataset EV2
Acknowledgments
The authors would like to thank Dr. Kara Corps (OSU Comparative Pathology and Mouse Phenotyping Core Facility) for overseeing lung sectioning and staining, and Dr. Eugene Oltz (OSU) for critical reading and editing of the manuscript. Research in the Yount laboratory is supported by NIH Grants AI130110, AI151230, HL157215, HL154001, and CA260582, and an American Lung Association COVID‐19 and Emerging Respiratory Viruses Research Award. ADK is supported by an institutional T32 postdoctoral fellowship (NIH grant AI165391). AZ was supported by an NSF‐GRFP fellowship grant. MEP and KCM are supported by NIH grant U19AI42733. The Graphical Abstract for this article was created using BioRender via a subscription maintained by EAH.
EMBO reports (2023) 24: e56660
Data availability
The datasets produced in this study are available in the following databases: RNA‐seq data: Gene Expression Omnibus GSE194062 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE194062).
References
- Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A et al (2020) Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID‐19. N Engl J Med 383: 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghamdi J, Alaamery M, Barhoumi T, Rashid M, Alajmi H, Aljasser N, Alhendi Y, Alkhalaf H, Alqahtani H, Algablan O et al (2021) Interferon‐induced transmembrane protein‐3 genetic variant rs12252 is associated with COVID‐19 mortality. Genomics 113: 1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EK, Randolph AG, Bhangale T, Dogra P, Ohlson M, Oshansky CM, Zamora AE, Shannon JP, Finkelstein D, Dressen A et al (2017) SNP‐mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med 23: 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, Huang IC, Kam C, Farzan M (2012) Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 8: e1002909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V et al (2020) Autoantibodies against type I IFNs in patients with life‐threatening COVID‐19. Science 370: eabd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges JP, Vladar EK, Huang H, Mason RJ (2022) Respiratory epithelial cell responses to SARS‐CoV‐2 in COVID‐19. Thorax 77: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel‐Benhassine F, Van der Werf S et al (2020) Syncytia formation by SARS‐CoV‐2‐infected cells. EMBO J 40: e106267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, McMichael TM, Hach JC, Yount JS (2014) Phosphorylation of the antiviral protein interferon‐inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J Biol Chem 289: 11986–11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, McMichael TM, Yount JS (2015) E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog 11: e1005095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, Compton AA, McMichael TM, Kenney AD, Zhang L, Soewarna V, Davis M, Schwartz O, Yount JS (2017) IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep 18: 1740–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Forbester JL, Marsden M, Sabberwal P, Sommerville MS, Wellington D, Dimonte S, Clare S, Harcourt K, Yin Z et al (2022) IFITM3 restricts virus‐induced inflammatory cytokine production by limiting Nogo‐B mediated TLR responses. Nat Commun 13: 5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta‐Llavona E, Albaiceta GM, García‐Clemente M, Duarte‐Herrera ID, Amado‐Rodríguez L, Hermida‐Valverde T, Enríquez‐Rodriguez AI, Hernández‐González C, Melón S, Alvarez‐Argüelles ME et al (2021) Association between the interferon‐induced transmembrane protein 3 gene (IFITM3) rs34481144 / rs12252 haplotypes and COVID‐19. Curr Res Virol Sci 2: 100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB (2014) IFITM3 restricts influenza a virus entry by blocking the formation of fusion pores following virus‐endosome hemifusion. PLoS Pathog 10: e1004048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernández J, Prati D, Baselli G, Asselta R et al (2020) Genomewide association study of severe Covid‐19 with respiratory failure. N Engl J Med 383: 1522–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ et al (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen O, Gan LM, Bjorkegren JLM (2019) PanglaoDB: a web server for exploration of mouse and human single‐cell RNA sequencing data. Database 2019: baz046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J, Albaiceta GM, Cuesta‐Llavona E, Garcia‐Clemente M, Lopez‐Larrea C, Amado‐Rodriguez L, Lopez‐Alonso I, Melon S, Alvarez‐Arguelles ME, Gil‐Pena H et al (2021) The interferon‐induced transmembrane protein 3 gene (IFITM3) rs12252 C variant is associated with COVID‐19. Cytokine 137: 155354 [DOI] [PubMed] [Google Scholar]
- Gralinski LE, Bankhead A 3rd, Jeng S, Menachery VD, Proll S, Belisle SE, Matzke M, Webb‐Robertson BJ, Luna ML, Shukla AK et al (2013) Mechanisms of severe acute respiratory syndrome coronavirus‐induced acute lung injury. mBio 4: e00271‐13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY et al (2020) Extrapulmonary manifestations of COVID‐19. Nat Med 26: 1017–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach JC, McMichael T, Chesarino NM, Yount JS (2013) Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti‐influenza a virus activity. J Virol 87: 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim MY, Al Heialy S, Hachim IY, Halwani R, Senok AC, Maghazachi AA, Hamid Q (2020) Interferon‐induced transmembrane protein (IFITM3) is upregulated explicitly in SARS‐CoV‐2 infected lung epithelial cells. Front Immunol 11: 1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E et al (2018) TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 46: D380–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine‐Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infusini G, Smith JM, Yuan H, Pizzolla A, Ng WC, Londrigan SL, Haque A, Reading PC, Villadangos JA, Wakim LM (2015) Respiratory DC use IFITM3 to avoid direct viral infection and safeguard virus‐specific CD8+ T cell priming. PLoS One 10: e0143539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ et al (2020) Mouse model of SARS‐CoV‐2 reveals inflammatory role of type I interferon signaling. J Exp Med 217: e20201241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng YM, Liu SL, Guo F, Geng Y, Qiao W et al (2014) Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol 16: 1080–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L‐Q, Xia T, Hu Y‐H, Sun M‐S, Yan S, Lei C‐Q, Shu H‐B, Guo J‐H, Liu Y (2017) IFITM3 inhibits virus‐triggered induction of type I interferon by mediating autophagosome‐dependent degradation of IRF3. Cell Mol Immunol 15: 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK et al (2021) Loss of furin cleavage site attenuates SARS‐CoV‐2 pathogenesis. Nature 591: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AD, McMichael TM, Imas A, Chesarino NM, Zhang L, Dorn LE, Wu Q, Alfaour O, Amari F, Chen M et al (2019) IFITM3 protects the heart during influenza virus infection. Proc Natl Acad Sci USA 116: 18607–18612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AD, Aron SL, Gilbert C, Kumar N, Chen P, Eddy A, Zhang L, Zani A, Vargas‐Maldonado N, Speaks S et al (2022) Influenza virus replication in cardiomyocytes drives heart dysfunction and fibrosis. Sci Adv 8: eabm5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist SR, Dinnon KH 3rd, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ et al (2020) A mouse‐adapted SARS‐CoV‐2 induces acute lung injury and mortality in standard laboratory mice. Cell 183: 1070–1085.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC et al (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9: e1003124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics 27: 1739–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista MJ, Winstone H, Wilson HD, Dyer A, Pickering S, Galao RP, De Lorenzo G, Cowton VM, Furnon W, Suarez N et al (2022) The P681H mutation in the spike glycoprotein of the alpha variant of SARS‐CoV‐2 escapes IFITM restriction and is necessary for type I interferon resistance. J Virol 96: e0125022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdoul S, Compton AA (2021) Lessons in self‐defence: inhibition of virus entry by intrinsic immunity. Nat Rev Immunol 22: 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael TM, Zhang Y, Kenney AD, Zhang L, Zani A, Lu M, Chemudupati M, Li J, Yount JS (2018) IFITM3 restricts human Metapneumovirus infection. J Infect Dis 218: 1582–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nchioua R, Schundner A, Kmiec D, Prelli Bozzo C, Zech F, Koepke L, Graf A, Krebs S, Blum H, Frick M et al (2022) SARS‐CoV‐2 variants of concern hijack IFITM2 for efficient replication in human lung cells. J Virol 96: e0059422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloudis D, Kountouras D, Hiona A (2020) The frequency of combined IFITM3 haplotype involving the reference alleles of both rs12252 and rs34481144 is in line with COVID‐19 standardized mortality ratio of ethnic groups in England. PeerJ 8: e10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo‐Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, Walker S, Parkinson N, Fourman MH, Russell CD et al (2021) Genetic mechanisms of critical illness in COVID‐19. Nature 591: 92–98 [DOI] [PubMed] [Google Scholar]
- Pathak GA, Singh K, Miller‐Fleming TW, Wendt FR, Ehsan N, Hou K, Johnson R, Lu Z, Gopalan S, Yengo L et al (2021) Integrative genomic analyses identify susceptibility genes underlying COVID‐19 hospitalization. Nat Commun 12: 4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez‐David RY et al (2021) The furin cleavage site in the SARS‐CoV‐2 spike protein is required for transmission in ferrets. Nat Microbiol 6: 899–909 [DOI] [PubMed] [Google Scholar]
- Prelli Bozzo C, Nchioua R, Volcic M, Koepke L, Kruger J, Schutz D, Heller S, Sturzel CM, Kmiec D, Conzelmann C et al (2021) IFITM proteins promote SARS‐CoV‐2 infection and are targets for virus inhibition in vitro . Nat Commun 12: 4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntmann VO, Martin S, Shchendrygina A, Hoffmann J, Ka MM, Giokoglu E, Vanchin B, Holm N, Karyou A, Laux GS et al (2022) Long‐term cardiac pathology in individuals with mild initial COVID‐19 illness. Nat Med 28: 2117–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A et al (2021) SARS‐CoV‐2 alpha, beta, and delta variants display enhanced spike‐mediated syncytia formation. EMBO J 40: e108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 43: e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F (2020) Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci USA 117: 11727–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Kenney AD, Kudryashova E, Zani A, Zhang L, Lai KK, Hall‐Stoodley L, Robinson RT, Kudryashov DS, Compton AA et al (2021) Opposing activities of IFITM proteins in SARS‐CoV‐2 infection. EMBO J 40: e106501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Chiramel AI, Li T, Lai KK, Kenney AD, Zani A, Eddy AC, Majdoul S, Zhang L, Dempsey T et al (2022) Rapalogs downmodulate intrinsic immunity and promote cell entry of SARS‐CoV‐2. J Clin Invest 132: e160766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposito B, Broggi A, Pandolfi L, Crotta S, Clementi N, Ferrarese R, Sisti S, Criscuolo E, Spreafico R, Long JM et al (2021) The interferon landscape along the respiratory tract impacts the severity of COVID‐19. Cell 184: 4953–4968.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MA, Clare S, Clement M, Marsden M, Abdul‐Karim J, Kane L, Harcourt K, Brandt C, Fielding CA, Smith SE et al (2017) The antiviral restriction factor IFN‐induced transmembrane protein 3 prevents cytokine‐driven CMV pathogenesis. J Clin Invest 127: 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheiden A, Thomas J, Soung AL, Davis‐Gardner ME, Floyd K, Jin F, Cowan DA, Pellegrini K, Shi PY, Grakoui A et al (2021) CCR2 signaling restricts SARS‐CoV‐2 infection. mBio 12: e0274921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim LM, Gupta N, Mintern JD, Villadangos JA (2013) Enhanced survival of lung tissue‐resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol 14: 238–245 [DOI] [PubMed] [Google Scholar]
- Winstone H, Lista MJ, Reid AC, Bouton C, Pickering S, Galao RP, Kerridge C, Doores KJ, Swanson CM, Neil SJD (2021) The polybasic cleavage site in SARS‐CoV‐2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol 95: e02422‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Xu E, Bowe B, Al‐Aly Z (2022) Long‐term cardiovascular outcomes of COVID‐19. Nat Med 28: 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wang G, Zhao F, Huang Y, Fan Z, Mei S, Xie Y, Wei L, Hu Y, Wang C et al (2022) IFITM3 inhibits SARS‐CoV‐2 infection and is associated with COVID‐19 susceptibility. Viruses 14: 2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani A, Yount JS (2018) Antiviral protection by IFITM3 in vivo . Curr Clin Microbiol Rep 5: 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada‐Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C et al (2020a) Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science 370: eabd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qin L, Zhao Y, Zhang P, Xu B, Li K, Liang L, Zhang C, Dai Y, Feng Y et al (2020b) Interferon‐induced transmembrane protein 3 genetic variant rs12252‐C associated with disease severity in coronavirus disease 2019. J Infect Dis 222: 34–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo JT (2014) Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci USA 111: 6756–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng KI, Feng G, Liu WY, Targher G, Byrne CD, Zheng MH (2021) Extrapulmonary complications of COVID‐19: a multisystem disease? J Med Virol 93: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]


