Abstract
In human embryos, naive pluripotent cells of the inner cell mass (ICM) generate epiblast, primitive endoderm and trophectoderm (TE) lineages, whence trophoblast cells derive. In vitro, naive pluripotent stem cells (PSCs) retain this potential and efficiently generate trophoblast stem cells (TSCs), while conventional PSCs form TSCs at low efficiency. Transient histone deacetylase and MEK inhibition combined with LIF stimulation is used to chemically reset conventional to naive PSCs. Here, we report that chemical resetting induces the expression of both naive and TSC markers and of placental imprinted genes. A modified chemical resetting protocol allows for the fast and efficient conversion of conventional PSCs into TSCs, entailing shutdown of pluripotency genes and full activation of the trophoblast master regulators, without induction of amnion markers. Chemical resetting generates a plastic intermediate state, characterised by co‐expression of naive and TSC markers, after which cells steer towards one of the two fates in response to the signalling environment. The efficiency and rapidity of our system will be useful to study cell fate transitions and to generate models of placental disorders.
Keywords: chemical resetting, naive pluripotency, placenta, pluripotent stem cells, trophoblast stem cells
Subject Categories: Methods & Resources, Stem Cells & Regenerative Medicine
Chemical resetting allows the fast and rapid conversion of conventional pluripotent stem cells (PCGs) into bona fide trophoblast stem cells (TSCs) through a plastic intermediate state, characterized by the co‐expression of naive and TSC markers.
Introduction
In the early blastocyst stage embryo, two cell populations are generated as the result of the first cell fate decision, the inner cell mass (ICM) and the trophectoderm (TE). The TE will generate cytotrophoblasts (CTBs) after implantation (Lee et al, 2016; Okae et al, 2018; Io et al, 2021), while the ICM gives rise to all somatic cells. The ICM of human embryos is more plastic than its murine counterpart, as it is able to generate TE (Guo et al, 2021), as suggested by single‐cell RNA‐sequencing (RNA‐seq) analyses of early embryos (Petropoulos et al, 2016) and recently confirmed by in vitro differentiation of explanted ICM (Guo et al, 2021).
The placenta is composed of cells of both maternal and embryonic origin, the latter known as trophoblast. CTBs are highly proliferative cells that, in turn, give rise to syncytiotrophoblasts (STBs) and extravillous trophoblasts (EVTs; Bischof & Irminger‐Finger, 2005; James et al, 2012). CTBs have been recently captured in vitro as trophoblast stem cells (TSCs; Okae et al, 2018) from human embryos and first‐trimester placental tissues, using a medium (TSC medium) containing epidermal growth factor (EGF), a GSK3 inhibitor (CHIR99021), two inhibitors of transforming growth factor beta (A83‐01 and SB431542), an inhibitor of histone deacetylase (valproic acid, VPA) and an inhibitor of Rho‐associated kinase (Y27632; Okae et al, 2018).
Human naive pluripotent stem cells (PSCs) have been derived either directly from pluripotent cells of the preimplantation embryo (Theunissen et al, 2014; Guo et al, 2016), from resetting of conventional PSCs by transgene expression (Takashima et al, 2014; Theunissen et al, 2014) or using a combination of inhibitors and cytokines (Theunissen et al, 2014; Guo et al, 2017; Bayerl et al, 2021) or by reprogramming from human fibroblasts (Liu et al, 2017; Kilens et al, 2018; Wang et al, 2018; Giulitti et al, 2019). Human naive PSCs retain in vitro the plasticity of the ICM and can readily differentiate towards TE (Guo et al, 2021; Io et al, 2021) and TSCs (Castel et al, 2020; Cinkornpumin et al, 2020; Dong et al, 2020; Bayerl et al, 2021). Interestingly, human naive PSCs express some extraembryonic markers (Dong et al, 2020), potentially explaining their competence for extraembryonic differentiation. Several different combinations and cytokines have been used for the expansion of naive PSCs (Takashima et al, 2014; Theunissen et al, 2014; Szczerbinska et al, 2019; Bayerl et al, 2021; Khan et al, 2021). Among them, we chose the PXGL medium (Guo et al, 2017), containing the cytokine LIF, inhibitors of the MEK and PKC kinases and of tankyrase.
Conventional PSCs are in a pluripotent state primed for differentiation, more akin to the postimplantation epiblast (Hackett & Azim Surani, 2014; Martello & Smith, 2014; Rossant, 2015). It is debated whether conventional PSCs have the competence to form TSCs. When directly exposed to TSC medium, conventional PSCs fail to form TSCs and acquire a neuroectodermal fate (Dong et al, 2020) or stop proliferating and experience elevated cell death (Castel et al, 2020). However, a recent study (Wei et al, 2021) reported the appearance of rare colonies that only after picking and expansion could be stabilised as TSCs.
Several studies reported the formation of trophoblast‐like cells from conventional PSCs after transient BMP stimulation, but those could not be expanded over multiple passages (Xu et al, 2002; Bernardo et al, 2011; Amita et al, 2013; Roberts et al, 2014; Lee et al, 2016; Yabe et al, 2016). The identity of those trophoblast‐like cells has been put into question (Bernardo et al, 2011), as they do not fulfil established criteria for trophoblast identity (Lee et al, 2016). Some authors suggested that those trophoblast‐like cells correspond to amnion (Guo et al, 2021; Io et al, 2021). Indeed, BMP is not involved in trophectoderm specification in the human blastocyst (De Paepe et al, 2019) and conventional PSCs have recently been reported to differentiate into amnion‐like cells in response to BMP (Zheng et al, 2019).
However, over the last year, three studies reported that a transient BMP stimulation followed by culture in the TSC medium converts conventional PSCs to TSCs (Soncin et al, 2018; Wei et al, 2021; Jang et al, 2022). Therefore, it is still debated whether BMP treatment in conventional PSCs induces only trophoblast or amnion, or a combination of both cell types. An interesting possibility is that an amnion‐like intermediate is needed to form TSCs.
Distinguishing between amnion and trophoblast identity is a nontrivial task, as several genes (e.g. TFAP2A and KRT7) are shared between the two cell types. Amnion forms from two waves of epiblast differentiation, called “early” and “late amniogenesis” (Rostovskaya et al, 2022). Rostovskaya and colleagues reported that early amniogenesis occurs via a TE‐like route, as several genes are shared between the two lineages. Conversely, late amniogenesis is governed by a transcriptional programme distinct from TE; thus, late amnion markers might be useful to unambiguously distinguish amnion from TE/trophoblast.
Multiple trajectories might be followed during the conversion of conventional PSCs into TSCs, as demonstrated in the context of reprogramming. Indeed, single‐cell analyses of cells acquiring naive pluripotency revealed the existence of multiple trajectories, depending on the experimental conditions used. In the case of resetting murine primed PSCs, cells either follow a direct trajectory or go via developmental states slightly more advanced (mesoderm) or more primitive (ICM), eventually all converging to the naive pluripotent state (Stuart et al, 2019). In the case of somatic cell reprogramming achieved by a combination of chemicals or using a transcription factor cocktail (Gata3, Eomes, Tfap2c, Myc and Esrrb), cells first transiently acquire an extraembryonic fate (i.e. primitive endoderm) before reaching and stabilising in the naive pluripotent state (Zhao et al, 2015; Benchetrit et al, 2019). Furthermore, different cell populations might be generated, as in the case of trophoblast‐like cells generated along with pluripotent cells, during the reprogramming of human somatic cells (Liu et al, 2020). These trophoblast‐like cells were directly captured as TSCs, without the need for prior stabilisation or expansion in a naive pluripotent state. Overall, these studies indicate that reprogramming protocols confer competence for the generation of both pluripotent and extraembryonic cells.
In this study, we observed that chemical resetting of human conventional PSCs generated a mixed cell population, containing both pluripotent and nonpluripotent cells. Based on the activation of placental imprinted transcripts and markers of TE and TSCs, we hypothesised that the mixed cell population could contain trophoblast‐like cells, which we could capture and expand as TSCs. We were indeed able to convert conventional PSCs to TSCs in an efficient and rapid manner, with no need for prolonged expansion under conditions supporting naive pluripotency.
Results
Activation of TE and trophoblast markers upon chemical resetting of conventional PSCs
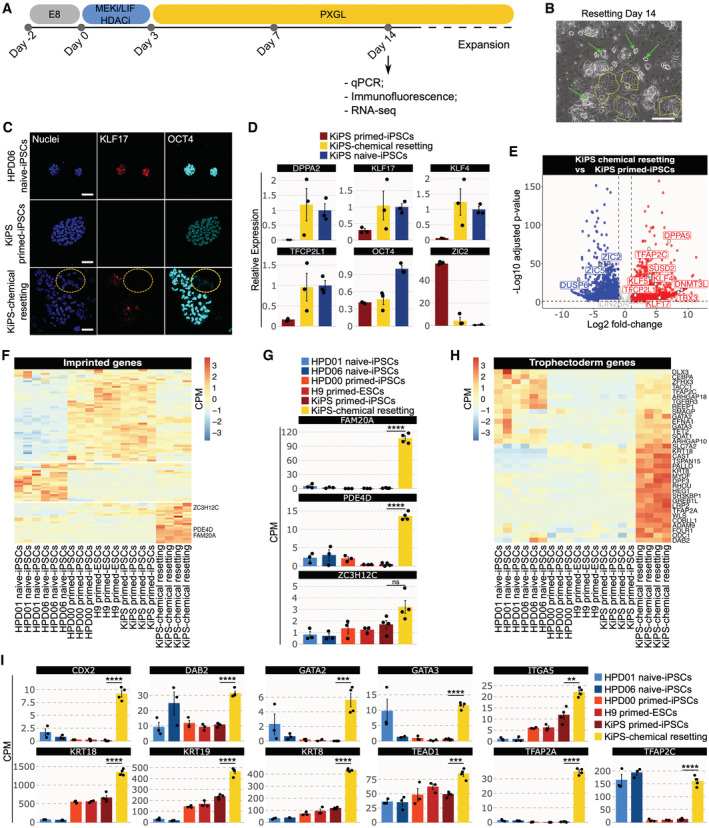
Human conventional induced pluripotent stem cells (iPSCs) obtained from keratinocytes (KiPS, Takashima et al, 2014) were reset to a naive state by transient histone deacetylase (HDAC) inhibition, MEK inhibition and LIF stimulation (Guo et al, 2017; Fig 1A), a process described as chemical resetting. After 7 days, several dome‐shaped, compact colonies started to appear. Several large naive‐like colonies were present at day 14, which we could readily expand for multiple passages in PXGL medium (Bredenkamp et al, 2019; Fig 1B, green arrows). These colonies were composed of cells expressing the naive pluripotency marker KLF17 and the general pluripotency factor POU5F1/OCT4 (Fig 1C).
Figure 1. Activation of TE and trophoblast markers upon chemical resetting of conventional PSCs.
-
AExperimental strategy for chemical resetting of human primed/conventional PSCs to naive pluripotency.
-
BMorphology of KiPS‐chemical resetting after 14 days. Scale bar: 200 μm.
-
CImmunostaining for the naive pluripotency marker KLF17 and the shared pluripotency marker OCT4. Scale bar: 50 μm.
-
DGene expression analysis by qPCR of KiPS primed‐iPSCs, KiPS‐chemical resetting at day 14 and KiPS naive‐iPSCs.
-
ETranscriptome analysis of KiPS‐chemical resetting compared with KiPS primed‐iPSCs. Known primed pluripotency and naive pluripotency markers are highlighted.
-
FHeatmap of 93 imprinted genes detected in pluripotent cells. Three placental imprinted genes are highlighted.
-
GBarplots showing the absolute expression (CPM) of three placental imprinted genes in the reported conditions highlighted in different colours.
-
HHeatmap of trophoblast‐specific genes upregulated in KiPS‐chemical resetting vs. KiPS primed‐iPSCs comparison.
-
IBarplots showing the absolute expression (CPM) of 12 trophoblast genes found to be significantly upregulated in KiPS‐chemical resetting vs KiPS primed‐iPSCs comparison.
Data information: (B and C) Representative images of n = 3 biological replicates are shown. (D, G, I) Bars indicate the mean ± SEM of n = 3 biological replicates shown as dots. (D) Expression was normalised to the mean of HPD06 naive‐iPSCs samples. (G–I) ***P < 0.001, ****P < 0.0001, ns no significance (Student's t‐test).
Source data are available online for this figure.
As previously reported (Guo et al, 2017), the cell population obtained after chemical resetting was heterogeneous (Fig 1B) and also contained flat polygonal cells (Fig 1B, dashed yellow lines) in addition to compact, naive‐like colonies (Fig 1B, green arrows). These cells also did not express pluripotency markers (Fig 1C, yellow dashed circle); thus, we performed molecular analyses for markers of different embryonic cell types with the aim of identifying those cells.
We performed quantitative polymerase chain reaction (qPCR) on the mixed culture obtained and observed activation of the naive‐specific markers DPPA5, KLF17, KLF4 and TFCP2L1 (Fig 1D). ZIC2, a marker specifically expressed by conventional PSCs, was greatly reduced after chemical resetting (Fig 1D). As a control, after resetting, we sorted SUSD2‐positive/CD24‐negative cells (Collier et al, 2017; Bredenkamp et al, 2019) and expanded them in PXGL. This homogenous population of KiPS naive‐iPSCs displays a similar upregulation of naive markers and repression of ZIC2 (blue bars in Fig 1D).
We performed RNA‐seq and confirmed the activation of naive‐specific markers (DPPA5, KLF4/5/17, TFCP2L1 and SUSD2) and the downregulation of markers of conventional PSCs (ZIC2/5, DUSP6 and LIN28A—Fig 1E). We concluded that the chemical resetting generates a mixed culture of naive PSCs and additional undefined cell types, which are unlikely to be conventional PSCs, as they do not express OCT4 (Fig 1C).
Imprinted genes are expressed in a stage‐specific and tissue‐specific manner (Perrera & Martello, 2019). We interrogated RNA‐seq data and observed that naive and conventional PSCs express very distinct sets of imprinted genes (Fig 1F). Strikingly, we observed a group of imprinted genes highly expressed only after chemical resetting. Among them, we found placental transcripts, such as FAM20A, PDE4D and ZC3H12C (Okae et al, 2014; Fig 1G). Placental imprinted genes are highly expressed in the trophoblast cells of the placenta (Hanna, 2020; Martini et al, 2022), so we asked whether other markers of the TE lineage were induced after chemical resetting. We observed that 35 out of 107 expressed genes (P‐value = 4.27 × 10−6, Fisher's exact test) specifically expressed by TE cells in the human embryo (Xiang et al, 2020) were highly induced after chemical resetting (Fig 1H), including KRT19, DAB2, LRP2 and TFAP2A (Fig 1H). We also analysed a wide range of markers expressed by TSCs in vitro and observed strong upregulation after resetting (Fig 1I). Of note, human naive PSCs display spontaneous and low expression levels of some trophoblast markers, consistently with their propensity to generate TSCs (Dong et al, 2020). However, the levels observed after chemical resetting were significantly higher than those of both primed and naive‐iPSCs (Fig 1I). Interestingly, TFAP2C, a gene required for self‐renewal of both naive PSCs and TSCs (Pastor et al, 2018; Guo et al, 2021), was fully activated after chemical resetting.
Our data indicate that chemical resetting leads to the activation of TE and TSC genes.
TSCs are generated via chemical resetting of conventional PSCs
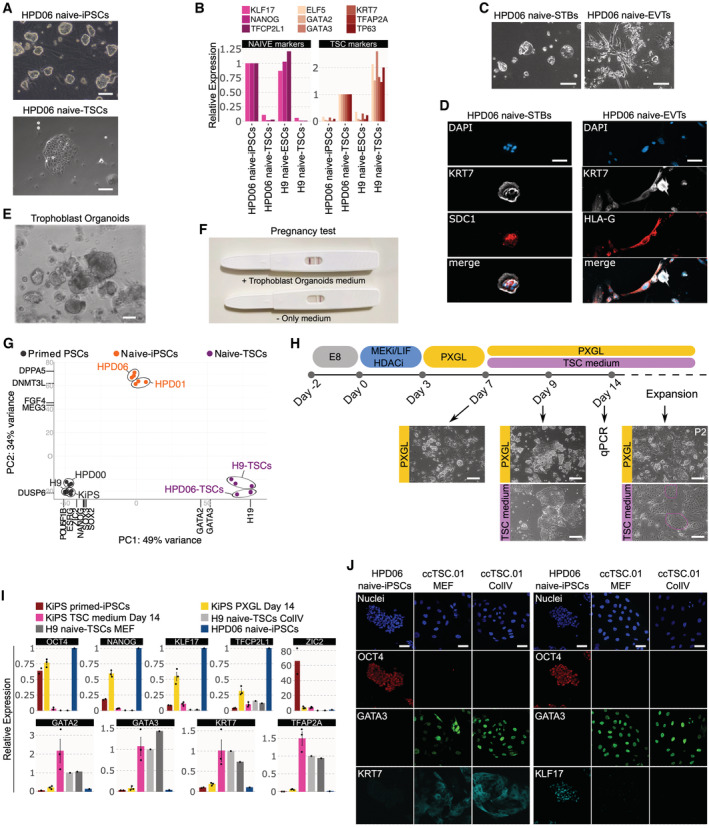
TSCs are stably expandable cell lines recently obtained from early human embryos (Okae et al, 2018) or via differentiation of human naive PSCs (Cinkornpumin et al, 2020; Dong et al, 2020; Guo et al, 2021). We could readily obtain TSCs from naive‐iPSCs, from both human embryonic stem cells (ESCs) and iPSCs (Takashima et al, 2014; Giulitti et al, 2019), and validated their correct identity by the expression of TSC markers GATA2/3, KRT7, TFAP2A, TP63 and ELF5 (Figs 2A and B and EV1A) and by their capacity to differentiate towards STBs and EVTs (Fig 2C and D) and to form trophoblast organoids (Turco et al, 2018) actively producing human chorionic gonadotropin (Fig 2E and F). RNA‐seq data further confirmed a distinct global expression profile of these naive‐iPSCs‐derived TSC lines compared with naive‐iPSCs and conventional PSCs (Fig 2G, see also Appendix Table S1). These TSC lines, named H9 naive‐TSCs and HPD06 naive‐TSCs, served as positive controls in the following experiments.
Figure 2. TSCs are generated via chemical resetting of conventional PSCs.
-
APhase‐contrast images of HPD06 naive‐iPSCs and TSCs derived from HPD06 naive‐iPSCs. Scale bars: 100 μm.
-
BGene expression analysis by RT–qPCR for naive (KLF17, TFCP2L1 and NANOG) and trophoblast (GATA3, GATA2, KRT7, ELF5, TP63 and TFAP2A) marker genes in naive‐iPSCs and TSC cells derived from two different naive‐iPSC lines (HPD06 and H9).
-
CPhase‐contrast images of syncytium trophoblast cells (STBs) derived from HPD06 naive‐TSCs (HPD06 naive‐STBs) and extravillous trophoblast cells (EVTs) derived from HPD06 naive‐TSCs (HPD06 naive‐EVTs). Scale bars: 50 μm.
-
DLeft: Immunostaining for KRT7 and the syncytium trophoblast marker SDC1 of HPD06 naive‐STBs. Scale bars: 50 μm. Right: Immunostaining for KRT7 and the extravillous trophoblast marker HLA‐G of HPD06 naive‐EVTs. Scale bars: 25 μm.
-
EPhase‐contrast images of trophoblast organoids derived from HPD06 naive‐TSCs. Scale bar: 100 μm.
-
FPregnancy test detecting human chorionic gonadotropin in trophoblast organoid medium.
-
GPrincipal component analysis of primed‐iPSCs (HPD00, H9 and KiPS), naive‐iPSCs (HPD06 and HPD01), and TSC cells derived from naive‐iPSCs (HPD06 naive‐TSCs and H9 naive‐TSCs) performed on the top 5,000 most variable genes identified through RNA‐seq.
-
HExperimental scheme of the conversion of conventional PSCs into naive PSCs or TSCs. Morphologies of cells at day 7, day 9 and after 2 passages are shown. Scale bars: 200 μm.
-
IGene expression analysis by qPCR of KiPS primed‐iPSCs, KiPS in PXGL medium at day 14, KiPS in TSC medium at day 14, H9 naive‐TSCs on ColIV, H9 naive‐TSCs on MEF and HPD06 naive‐iPSCs.
-
JImmunostaining for the pluripotency markers OCT4 and KLF17, and the TSC markers GATA3 and KRT7 of HPD06 naive‐iPSC and ccTSC.01 cultured on MEF and on collagen IV (ColIV) after 3 passages. Scale bars: 30 μm.
Data information: (C, D, H, J) Representative images of n = 2 biological replicates are shown. (I) Dots indicate biological replicates, and bars indicate the mean ± SEM of n = 3 biological replicates. Top: naive, primed and general pluripotency markers, expression was normalised to HPD06 naive‐iPSCs; bottom: trophoblast markers, expression was normalised to H9 naive‐TSCs.
Source data are available online for this figure.
Figure EV1. Characterisation of naive‐TSCs and conversion of conventional PSCs.
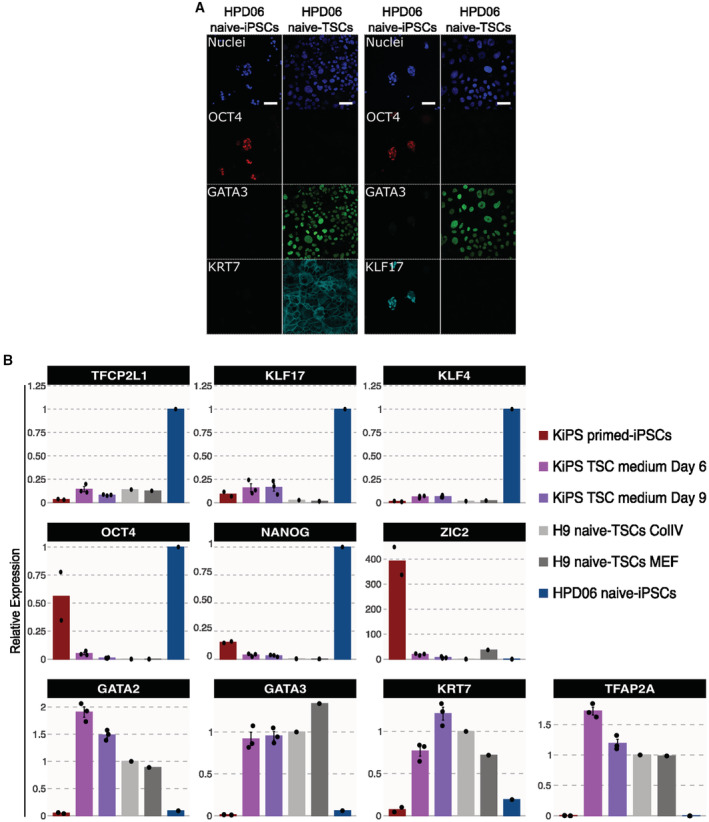
-
AImmunostaining for the pluripotency markers OCT4, KLF17 and the TSC markers GATA3 and KRT7 of HPD06 naive‐iPSC and TSC cells derived from naive‐iPSCs (HPD06 naive‐TSCs). Scale bars: 30 μm.
-
BGene expression analysis by RT–qPCR of KiPS primed‐iPSCs, KiPS in TSC medium at day 6 and 9, H9 naive‐TSCs cultured on ColIV or MEF, and HPD06 naive iPSCs.
Data information: (A) Representative images of n = 3 biological replicates are shown. (B) Dots indicate biological replicates, and bars indicate the mean ± SEM of n = 3 biological replicates. Top and middle: naive, primed and general pluripotency markers, expression was normalised to HPD06 naive‐iPSCs; bottom: trophoblast markers, expression was normalised to H9 naive‐TSCs.
Based on the strong activation of TSC genes (Fig 1) upon chemical resetting, we hypothesised that chemical resetting might endow conventional PSCs with the competence to generate TSCs. To test this hypothesis, we modified the chemical resetting protocol and, after 7 days of resetting, cells were exposed to either naive PSC‐ or TSC‐supporting conditions (Fig 2H). As expected, several dome‐shaped colonies emerged in PXGL (Fig 2H). Transcriptional analysis confirmed the upregulation of naive‐specific and general pluripotency markers (Fig 2I).
Strikingly, under TSC conditions we observed complete repression of pluripotency markers and rapid activation of TSC markers to levels observed in established TSCs (Fig 2I). The cells obtained, which we named ccTSCs (chemically converted TSCs, ccTSC.01) expanded robustly for several passages, with a stable and very homogeneous morphology. Immunostaining confirmed the presence of TSC markers GATA3 and KRT7, and the complete absence of pluripotency markers OCT4 and KLF17 (Fig 2J). TSCs have been expanded both on a feeder‐layer of mouse embryonic fibroblasts (MEFs) or on Collagen IV (ColIV) coated wells (Okae et al, 2018; Castel et al, 2020; Guo et al, 2021; Io et al, 2021). Once established, we expanded ccTSCs for multiple passages under both conditions with a homogenous expression of TSC markers and similar expansion kinetics (Fig 2J). We conclude that a modified chemical resetting protocol allowed the efficient conversion of conventional PSCs to TSCs.
Rapid and highly efficient generation of ccTSCs
We obtained ccTSCs after 3 days of chemical resetting, followed by 4 days in PXGL, a medium supporting human naive PSCs (Fig 2H–J). We asked whether exposing cells directly to TSC medium right after 3 days of chemical resetting would also lead to ccTSCs generation, skipping an intermediate step in PXGL (Fig 3A).
Figure 3. Rapid and highly efficient generation of ccTSCs.
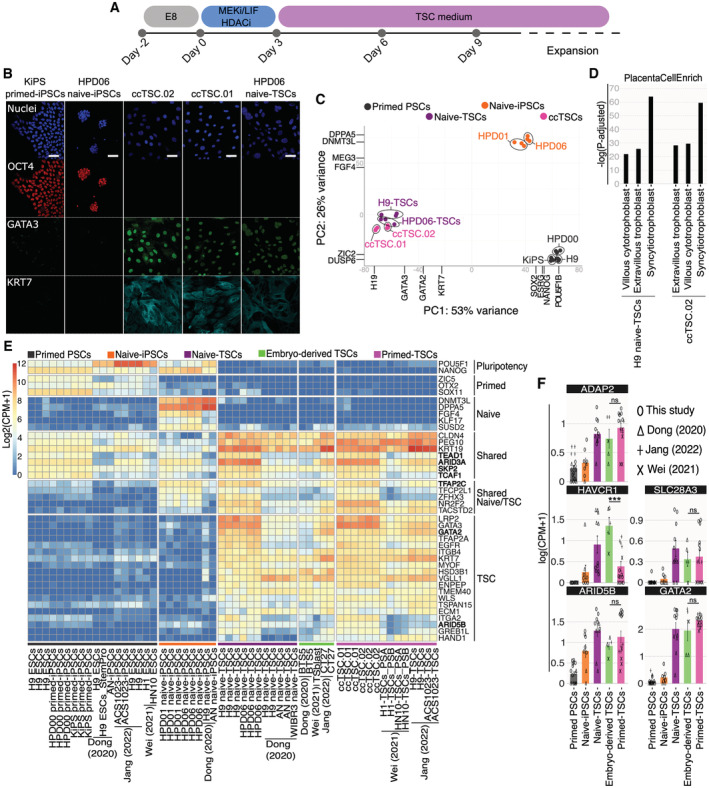
-
AExperimental scheme of the conversion of primed PSCs towards ccTSCs.
-
BImmunostaining for the pluripotency markers OCT4, and the TSC markers GATA3 and KRT7 of KiPS primed‐iPSCs, HPD06 naive‐iPSCs, ccTSC.02 at passage 4, ccTSC.01 at passage 4 and HPD06 naive‐TSCs. Scale bars: 30 μm.
-
CPrincipal component analysis of ccTSCs (ccTSC.01 and ccTSC.02) with primed PSCs (HPD00, H9, and KiPS), naive‐iPSCs (HPD06 and HPD01) and TSCs derived from naive‐iPSCs (HPD06 naive‐TSCs and H9 naive‐TSCs) performed on the top 5,000 most variable genes identified through RNA‐seq.
-
DBarplot showing placenta cell‐specific expression patterns in ccTSC.02 and TSCs derived from naive‐iPSCs (H9 naive‐TSCs).
-
EHeatmap of general, primed, naive, shared, shared between naive and TSCs, and trophoblast‐specific genes in primed‐PSCs, naive‐iPSCs, naive‐TSCs, embryo‐derived TSCs and TSCs derived from primed PSCs (primed‐TSCs) in our study or from published data.
-
FBarplots showing the absolute expression as log(CPM + 1) of ADAP2, HAVCR1, SLC28A3, ARID5B and GATA2 in the reported conditions.
Data information: (B) Representative images of n = 3 biological replicates are shown. (F) Bars indicate the mean ± SEM of at least n = 5 biological replicates from at least two independent studies shown with different symbols. ***P < 0.001, ns no significance (Student's t‐test).
Source data are available online for this figure.
Much to our surprise, after only 3 days in the TSC medium, we detected full activation of TSC markers and repression of markers of both naive and conventional pluripotency (Fig EV1B). Accordingly, the entire cell population (ccTSC.02; Fig EV2A, left panel) displayed a morphology indistinguishable from TSCs obtained from naive‐iPSCs (Fig 2A). The ccTSCs could be readily expanded for > 20 passages and displayed robust and homogenous expression of TSC markers and the absence of pluripotency markers (Figs 3B and EV2B). Moreover, comparing global transcriptomes, both via PCA and correlation analysis revealed that both ccTSC lines clustered with H9 naive‐TSCs and HPD06 naive‐TSCs (Figs 3C and EV2C).
Figure EV2. Transcriptional analysis of BAP, ccTSCs and naive‐TSCs.
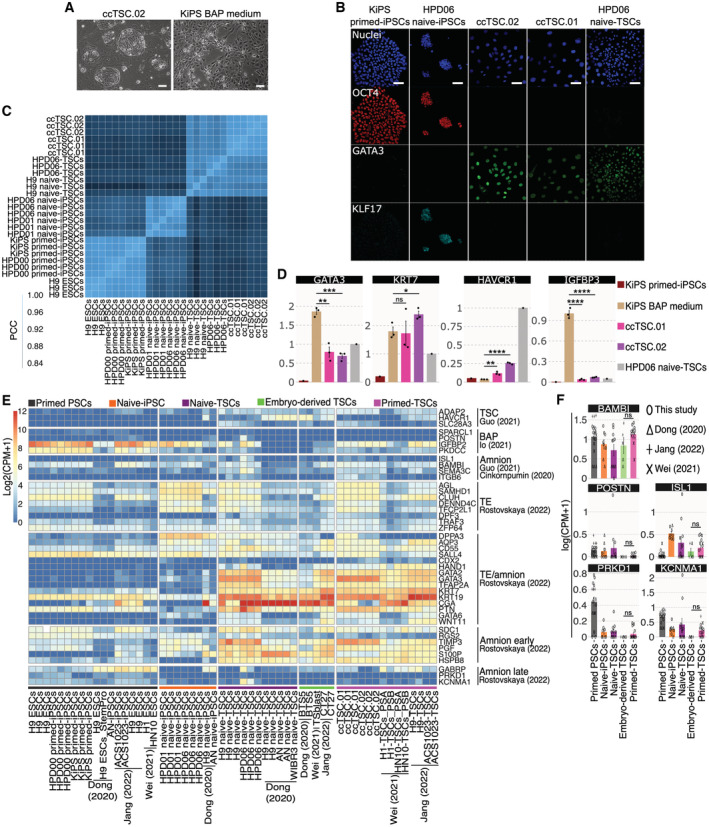
-
APhase‐contrast images of ccTSC.02 at passage 5 and of BAP‐treated KiPS after 3 days of treatment. Scale bar: 200 μm.
-
BImmunostaining for the pluripotency markers OCT4, KLF17 and the TSC marker GATA3 of KiPS primed‐iPSCs, HPD06 naive‐iPSCs, ccTSC.02 at passage 4, ccTSC.01 at passage 4 and HPD06 naive‐TSCs. Scale bars: 30 μm.
-
CCorrelation plot of primed PSCs (H9, HPD00 and KiPS), naive‐iPSCs (HPD01 and HPD06), and TSC cells derived from naive‐iPSCs (H9 naive‐TSCs and HPD06 naive‐TSCs) and ccTSCs (ccTSC.01 and ccTSC.02).
-
DGene expression analysis by qPCR of KiPS primed‐iPSCs, KiPS in BAP medium, KiPS ccTSC.01, KiPS ccTSC.02 and HPD06 naive‐TSCs.
-
EHeatmap of TSC, BAP, amnion, TE, TE/amnion, amnion early, amnion late specific genes in primed PSCs, naive‐iPSC, naive‐TSCs, Embryo‐derived TSCs and primed‐TSCs from our study or from published data.
-
FBarplots showing the absolute expression as log(CPM + 1) of amnion markers in the reported conditions.
Data information: (A and B) Representative images of n = 3 biological replicates are shown. (D) Dots indicate biological replicates and bars indicate the mean ± SEM of n = 3 biological replicates. For trophoblast markers, the expression was normalised to HPD06 naive‐TSCs, for amnion markers the expression was normalised to the mean of KiPS in BAP medium samples. (F) Bars indicate the mean ± SEM of at least n = 5 biological replicates from at least two independent studies shown with different symbols. (D–F) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns no significance (Student's t‐test).
To further characterise the transcriptome of ccTSCs, we used the PlacentaCellEnrich programme (Jain & Tuteja, 2021), which identifies transcriptional programmes of cell types found in first‐trimester human placenta (Vento‐Tormo et al, 2018). The genes upregulated in ccTSC.02, compared with isogenic primed KiPS cells, were highly enriched for different trophoblast types (Fig 3D). A similarly strong enrichment was found comparing H9 naive‐TSCs against their isogenic primed PSCs (H9 ESCs).
Recent studies identified, via CRISPR/Cas9 genetic inactivation, several genes required for TSC identity, such as ARID3A, ARID5B, TCAF1, SKP2, GATA2, TEAD1 and TFAP2C (Guo et al, 2021; Dong et al, 2022). All these functional regulators, together with several TSC markers, were highly expressed in ccTSCs (Fig 3E, bold) to levels comparable, or higher than, TSCs obtained from early human embryos or via differentiation of human naive PSCs (Data ref: Theunissen et al, 2020; Data ref: Jang & Kim, 2021; Data ref: Shan & Wei, 2021).
We conclude that transient inhibition of HDAC and MEK with LIF stimulation is sufficient to allow efficient and rapid conversion of conventional PSCs to ccTSCs, without the need for an intermediate step in a naive PSCs‐supporting medium. The resulting ccTSCs are transcriptionally comparable to both embryo‐derived TSCs and naive‐TSCs.
The ccTSCs transcriptome is compatible with TSCs and not with amnion identity
Transient exposure of conventional PSCs to bone morphogenetic proteins (BMPs), either alone or in combinations with the ALK4/5/7 inhibitor A83‐01, and the FGFR inhibitor PD173074 (BAP medium) has been used to generate cells with some molecular features of trophoblast cells (Amita et al, 2013; Yang et al, 2015). However, several groups reported that BAP cells differ from primary trophoblast cells (Bernardo et al, 2011) and from bona fide TSCs (Dong et al, 2020; Guo et al, 2021; Io et al, 2021), as BAP cells do not self‐renew robustly and collapse in culture after some passages (Jang et al, 2022). It is still debated whether BAP cells have a trophoblast or amnion identity (Bernardo et al, 2011; Lee et al, 2016; Seetharam et al, 2022). Although we have not used BMP ligands in our protocol, we decided to investigate whether our ccTSCs were distinct from BAP cells. We exposed conventional KiPS cells to BAP medium and observed a cell morphology consistent with previous reports (Yang et al, 2015; Io et al, 2021) and completely distinct from our ccTSCs (Fig EV2A). Gene expression analysis confirmed activation of not only KRT7 and GATA3 but also late amnion marker IGFBP3 (Guo et al, 2021; Io et al, 2021; Rostovskaya et al, 2022), while HAVCR1, a marker of bona fide TSCs (Guo et al, 2021; Io et al, 2021; Jang et al, 2022), was not induced in BAP cells (Fig EV2D). In stark contrast, ccTSCs showed robust induction of several TSC markers and negligible expression of IGFBP3, further indicating their bona fide TSC identity (Figs 3E and F and EV2D).
While transient BMP/BAP stimulation by itself does not generate bona fide TSCs, recent studies reported that BMP followed by expansion in the TSC medium converts conventional PSCs to TSCs (Soncin et al, 2018; Wei et al, 2021; Jang et al, 2022). We analysed transcriptomes of TSCs obtained via transient BMP stimulation and found repression of pluripotency markers and activation of TSCs markers (Fig 3E), although some lines failed to activate HAVCR1 and SLC28A3 (Fig 3F, crosses).
Given that BMP has been reported to differentiate conventional PSCs into amnion‐like cells (Zheng et al, 2019), we hypothesised that TSCs obtained via BMP stimulation (Data ref: Jang & Kim, 2021; Data ref: Shan & Wei, 2021) might still express amnion markers. We analysed a large set of amnion markers identified by Rostovskaya and colleagues. As expected, markers shared among TE and amnion were highly expressed by all TSCs, regardless of their origin (Fig EV2E). Also, early amnion markers (e.g. PGF, TIMP3 and S100P) were highly expressed in all TSCs analysed (Fig EV2E), in keeping with early amniogenesis occurring via a TE‐like route. By contrast, the late amnion markers (PRKD1, KCNMA1 and GABRP) were barely detectable in all TSCs (Fig EV2E and F).
Additional amnion markers identified in other studies (Cinkornpumin et al, 2020; Guo et al, 2021), such as BAMBI and ISL1, were expressed at low levels in all TSCs analysed and in PSCs (Fig EV2E and F).
We conclude that TSCs generated from embryos, from naive‐iPSCs, and from conventional PSCs by BMP stimulation or HDAC inhibition, MEK inhibition, and LIF stimulation display a similarly low expression of amnion markers, which does not indicate the acquisition of amnion identity.
We then focussed on the differences between ccTSCs and naive‐TSCs. None of the TSC markers were found differentially expressed between ccTSCs and TSCs obtained from naive‐iPSCs (Fig EV3A, Cluster C). We performed Gene Ontology and KEGG pathways enrichment analyses on the 279 genes (Cluster B) distinguishing ccTSCs from HPD06 naive‐TSCs and failed to find any significant term. However, manual inspection revealed aberrant expression of 6 imprinted genes in TSCs derived from naive‐iPSCs.
Figure EV3. Analysis of amnion and 8CLC markers during conversion.
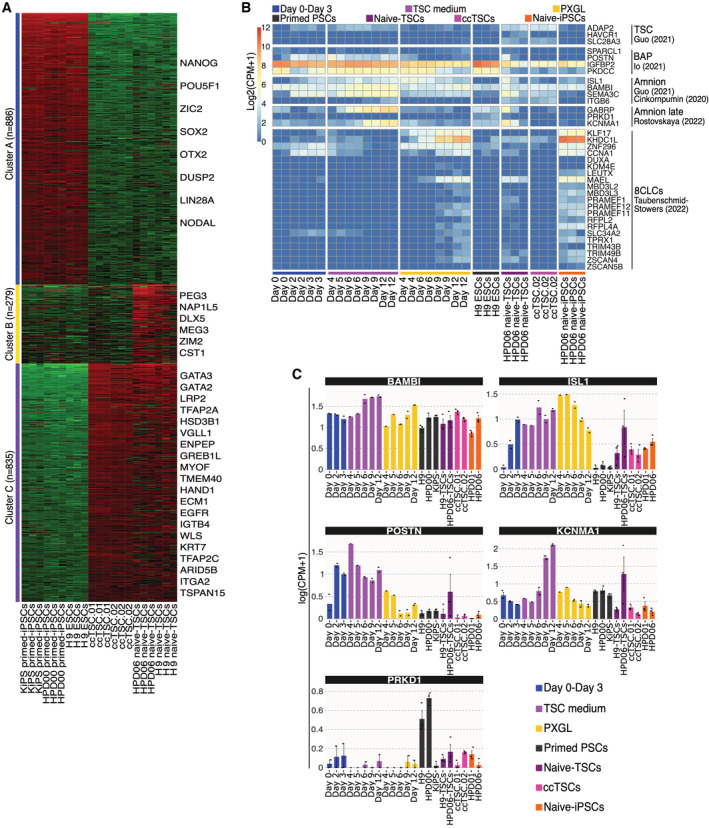
-
AK‐means clustering of the 2000 most variable genes in primed‐iPSCs (KiPS primed‐iPSCs, HPD00 primed‐iPSCs and H9 ESCs), ccTSCs (ccTSC.01 and ccTSC.02), and naive‐TSCs (HPD06 naive‐TSC and H9 naive‐TSCs). Red and green indicate high and low expression, respectively. Representative genes of each cluster are shown.
-
BHeatmap of TSC, BAP, amnion, amnion late and 8CLCs specific genes during the conversion of conventional PSCs into TSCs or naive‐iPSCs, with primed‐ESCs (H9), TSCs derived from naive‐iPSCs (HPD06 naive‐TSCs), ccTSC.02 and naive‐iPSCs (HPD06).
-
CBarplots showing the absolute expression as log(CPM + 1) of amnion markers (BAMBI, ISL1, POSTN, KCNMAI and PRKD1) in the reported conditions highlighted in different colours.
Data information: (C) Dots indicate biological replicates, and bars indicate the mean ± SEM of n = 3 biological replicates.
ccTSCs have the potential to differentiate into STBs and EVTs
To further assess potential differences between ccTSCs and naive‐TSCs, we evaluated the differentiation potential towards STBs and EVTs, a key functional feature of TSCs. Specifically, we tested the differentiation capacity of ccTSC.02, obtained after 3 days in MEKi/LIF/HDACi and directly switched to TSC medium (Fig 3). HPD06 naive‐TSCs, which formed efficiently STBs, EVTs, and Trophoblast organoids, were used as positive controls. STB differentiation of both TSC lines led to upregulation of the STB markers CGA, CGB2, SDC1, PSG3/5 and INHA1 (Io et al, 2021) mRNAs, relative to parental TSCs and isogenic PSCs (Fig 4A, left). SDC1 protein expression was detected only in STBs (Fig 4B, top panel). EVT differentiation also resulted in specific upregulation of the EVT markers VGLL3, NOTUM, SNAI1 and ITGA5 and of HLA‐G protein (Dong et al, 2020; Io et al, 2021; Fig 4A, right and Fig 4B, bottom panel). We conclude that ccTSC.02 has the capacity to give rise to STBs and EVTs, further indicating the correct acquisition of TSC identity.
Figure 4. Differentiation potential of ccTSC.02.
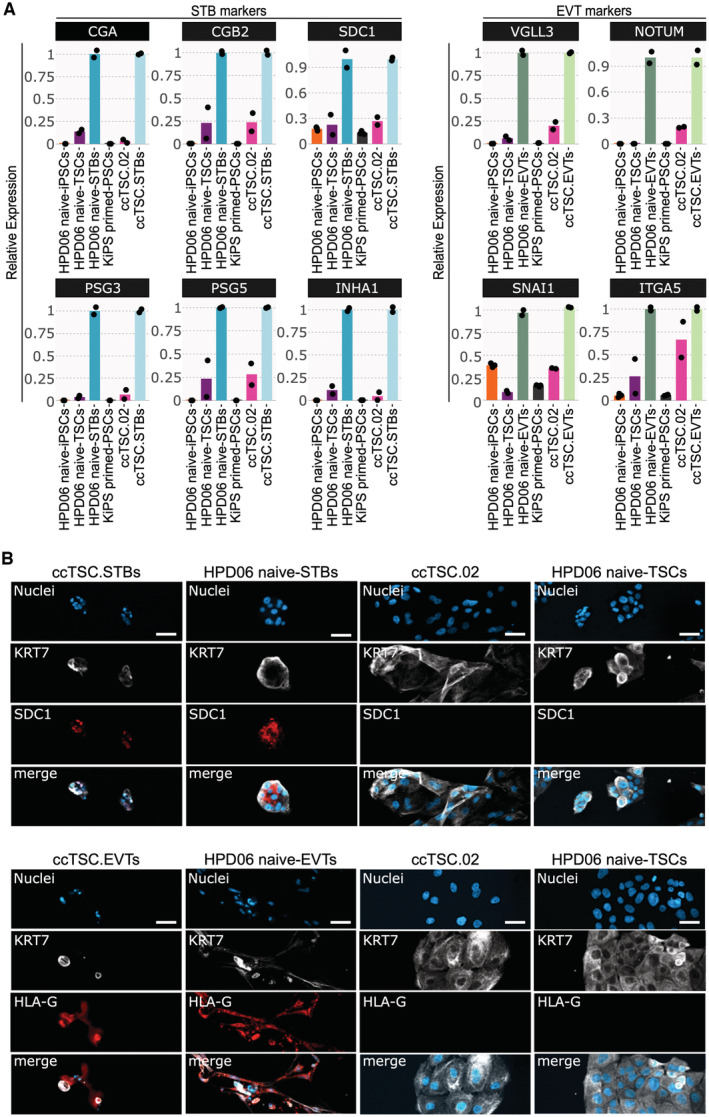
-
AGene expression analysis by qPCR of syncytiotrophoblast (left) and extravillous (right) markers.
-
BTop: Immunostaining for the TSC marker KRT7 and the STB marker SDC1 of ccTSC.02‐STBs, HPD06 naive‐STBs, ccTSC.02 and HPD06 naive‐TSCs. Scale bars: 50 μm. Bottom: Immunostaining for the TSC marker KRT7 and the EVT marker HLA‐G of ccTSC.02‐EVTs, HPD06 naive‐EVTs, ccTSC.02 and HPD06 naive‐TSCs. Scale bars: 25 μm.
Data information: (A) Dots indicate biological replicates. Left: expression was normalised to HPD06 naive‐STBs and ccTSC.02‐STBs; Right: expression was normalised to HPD06 naive‐EVTs and ccTSC.02‐EVTs. (B) Representative images of n = 2 biological replicates are shown.
Source data are available online for this figure.
Conventional PSCs convert to ccTSCs through an intermediate naive‐like pluripotent state
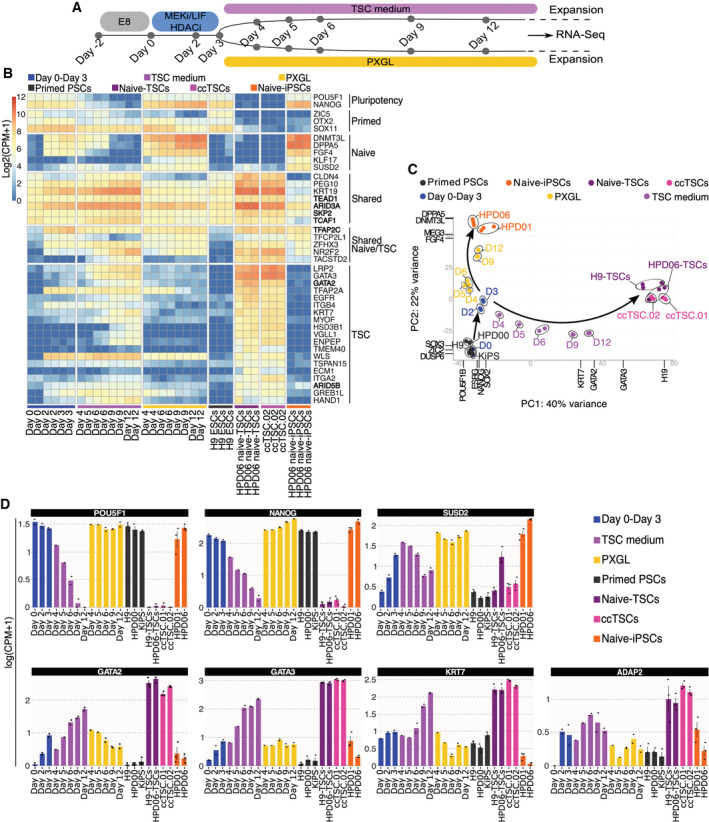
Transient HDAC inhibition, MEK inhibition and LIF stimulation followed by exposure to PXGL are applied for the routine generation of human naive‐iPSCs from conventional PSCs (Guo et al, 2017). Using the same procedure, we could rapidly obtain TSCs through the exposure of conventional PSCs to TSC medium right after 3 days of chemical resetting (Fig 3). We, therefore, wondered whether the KiPS primed‐iPSCs to TSCs conversion proceeds through an intermediate naive‐like pluripotent state with transient naive markers activation or directly to TSCs. We performed RNA‐seq analysis at different timepoints during the HDAC inhibition, MEK inhibition and LIF stimulation step and subsequent stabilisation in PXGL or TSC medium (Fig 5A). During the initial 3 days of chemical resetting, we observed the upregulation of markers of naive pluripotency and of TSCs (Fig 5B). At day 4, the transcriptomes diverged in response to PXGL or TSC media and gradually followed different trajectories towards naive‐iPSCs and stabilised TSC samples (Fig 5C).
Figure 5. Transcriptional analysis of ccTSCs generation.
-
AExperimental scheme of the conversion of conventional PSCs into naive‐iPSCs or TSCs.
-
BHeatmap of primed, naive and trophoblast‐specific genes during the conversion of conventional PSCs into TSCs or naive‐iPSCs, with primed‐ESCs (H9), TSCs derived from naive‐iPSCs (HPD06 naive‐TSCs), ccTSC.02 and naive‐iPSCs (HPD06).
-
CPrincipal component analysis of the time point conversion of conventional PSCs into naive‐iPSCs or TSCs, with primed‐PSC (HPD00, H9 and KiPS), naive‐iPSCs (HPD06 and HPD01), TSCs derived from naive‐iPSCs (HPD06 naive‐TSCs and H9 naive‐TSCs) and ccTSCs (ccTSC.01 and ccTSC.02) and performed on the top 5,000 most variable genes identified through RNA‐seq.
-
DBarplots showing the absolute expression as log(CPM + 1) of general pluripotency (OCT4 and NANOG), naive (SUSD2) and TSC‐specific genes (GATA2, GATA3, KRT7 and ADAP2) in the reported conditions highlighted in different colours.
Data information: (D) Dots indicate biological replicates and bars indicate the mean ± SEM of n = 3 biological replicates.
Source data are available online for this figure.
Expression of naive‐specific markers (e.g. SUSD2 and TFCP2L1) was further consolidated in PXGL or gradually switched off in TSC medium (Fig 5B–D). Similarly, the expression of TSC markers, like TFAP2A, ADAP2, GATA2/3 and KRT7, was detected early during the chemical resetting and gradually decreased in PXGL or further increased in TSC medium (Fig 5B–D). Our data indicate that divergent trajectories towards naive‐iPSCs and stabilised TSCs proceed through a shared intermediate state, which can be reached in 3 days of chemical resetting and in which the cell population is highly responsive to differentiation stimuli.
We then investigated the intermediate state reached at day 3 of conversion, asking whether the ability to form naive‐iPSCs and trophoblast arises from an earlier developmental state. Two recent studies described the in vitro stabilisation of the human 8‐cell (8C) embryo state. The resulting 8C‐like cells (8CLCs) are endowed with embryonic and extraembryonic potential (Mazid et al, 2022; Taubenschmid‐Stowers et al, 2022). We analysed a set of 8CLCs markers during conversion and in established TSCs and PSCs and failed to detect any significant upregulation, indicating that cells at day 3 do not resemble 8CLCs (Fig EV3B).
The conversion might occur via an amnion‐like intermediate or might generate amnion together with TSCs. We measured the expression of unambiguous amnion markers (Fig EV2E) and detected rapid upregulation of only ISL1 and POSTN during the first 3 days. We conclude that cells do not acquire a transient amnion identity during conversion (Fig EV3B and C).
The late amnion markers KCNMA1 and GABRP were upregulated (Fig EV3B and C) at day 9 in TSC medium. In established ccTSCs, all these amnion markers were expressed at negligible levels. These results might suggest that either a fraction of amnion is generated during the conversion, but these cells do not expand in TSC conditions or, alternatively, that some amnion markers are erroneously activated by transcription factors shared among TE and amnion identity (e.g. TFAP2A).
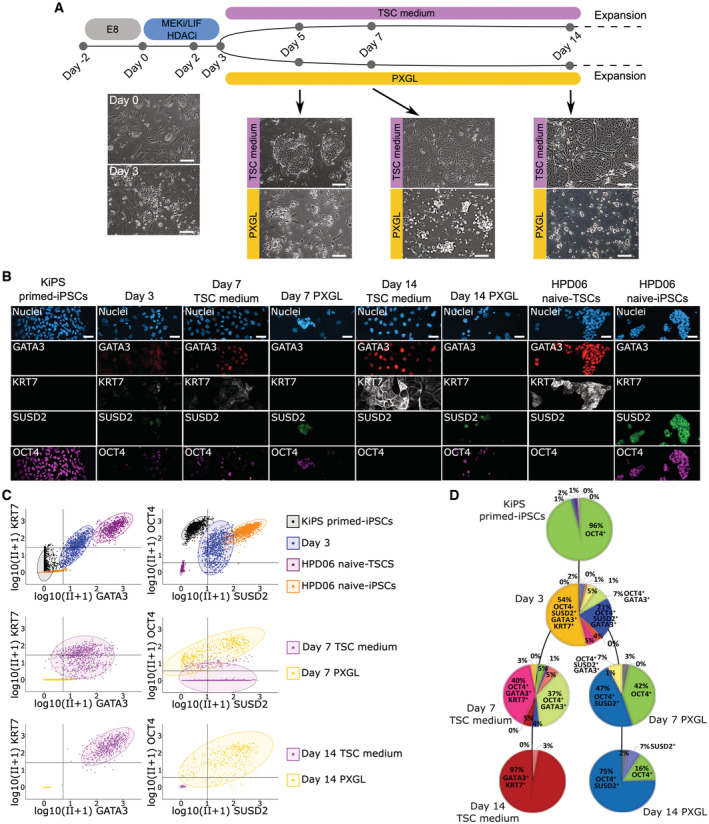
Our bulk transcriptome analyses indicate rapid upregulation of a large number of TSC and naive pluripotency markers during the first 3 days of conversion (Fig 5). We then asked whether the same cells expressed markers of both lineages or if we had a mixed population of TSCs and naive‐iPSCs already at day 3 during our modified resetting protocol (Fig 6A). To distinguish between the two hypotheses, we measured the protein levels of SUSD2, OCT4, GATA3 and KRT7. SUSD2 is commonly used to isolate naive PSCs (Bredenkamp et al, 2019); OCT4 is a general pluripotency regulator (Hay et al, 2004; Matin et al, 2004; Zaehres et al, 2005; Wang et al, 2012); GATA3 is functionally required for TSCs maintenance (Dong et al, 2022) and together with KRT7 has been used to confirm the generation of TSCs from conventional PSCs (Soncin et al, 2018; Wei et al, 2021; Jang et al, 2022).
Figure 6. Single‐cell analysis of ccTSCs and naive‐iPSC generation.
-
AExperimental scheme of the conversion of conventional PSCs into naive‐iPSCs or TSCs. Morphologies of cells at day 0, day 3, day 5, day 7 and day 14 in TSC medium or in PXGL are shown. Scale bars: 200 μm.
-
BImmunostaining for the TSC markers GATA3 and KRT7, the naive pluripotency marker SUSD2 and general pluripotency markers OCT4 of KiPS primed‐iPSCs, KiPS at day 3 of the conversion, KiPS at day 7 in TSC medium or PXGL, KiPS at day 14 in TSC medium or PXGL, HPD06 naive‐TSCs and HPD06 naive‐iPSCs. Scale bars: 25 μm.
-
CSingle‐cell scatter plots showing the log(CPM + 1) integrated intensity of TSC markers GATA3 and KRT7, naive pluripotency marker SUSD2 and general pluripotency marker OCT4 of KiPS primed‐iPSCs, KiPS at day 3 of the conversion, KiPS at day 7 in TSC medium or PXGL, KiPS at day 14 in TSCs medium or PXGL, HPD06 naive‐TSCs and HPD06 naive‐iPSCs. Black lines represent the threshold used to define positivity to a given marker.
-
DPie charts representing patterns of activation of TSC markers GATA3 and KRT7, naive pluripotency marker SUSD2 and general pluripotency marker OCT4 in KiPS primed‐iPSCs, KiPS at day 3 of the conversion, KiPS at day 7 in TSC medium or PXGL, and KiPS at day 14 in TSC medium or PXGL.
Data information: (A) Representative images of n = 4 biological replicates are shown. (B) Representative images of n = 2 biological replicates are shown. (C) Fluorescence intensity was measured by immunofluorescence in 5–12 independent fields from 2 independent experiments. (D) The number of cells exhibiting each specific activation pattern is reported as % of total cells.
Source data are available online for this figure.
As expected, conventional PSCs (KiPS at day 0) expressed only OCT4 (96%) (Fig 6B–D). On day 3, we observed co‐expression of all 4 markers in 54% of cells, of OCT4, SUSD2 and GATA3 in 21% of cells, and of OCT4 and GATA3 in 7%. The fractions of cells expressing only pluripotency markers (i.e. OCT4 and SUSD2), or only TSC markers (i.e. GATA3 and KRT7) were negligible (1% and 0%, respectively). These results indicate that at day 3 most of the cells co‐express pluripotency and TSC markers, rather than being a mixed population of TSCs and naive‐iPSCs. Indeed, the morphology of cells at day 3 does not resemble neither naive PSCs nor TSCs (Fig 6A).
Next, we monitored the response to the two different signalling environments (PXGL and TSC media). At day 7 in TSC medium, we observed downregulation of SUSD2 and perduring expression of OCT4 together with TSC markers (Fig 6B–D). A similar co‐expression of pluripotency and TSC markers has been reported during the conversion of naive‐iPSCs to TSCs (Zijlmans et al, 2022). At day 14 in TSC medium 97% of cells were GATA3 and KRT7 double‐positive, indicating efficient conversion (Fig 6B–D).
At day 7 in PXGL, we observed downregulation of TSC markers, although 7% of cells retained GATA3 expression. Most of the cells expressed OCT4, either alone (42%) or in combination with SUSD2 (47%). At day 14 in PXGL, the fraction of OCT4 and SUSD2 expressing cells increased to 75%, while the remaining 25% expressed either OCT4, SUSD2, or none of the two (Fig 6D). This heterogeneity is consistent with the mixed morphology (Figs 2H and 6A) and the expression of naive and TE markers (Fig 1) we observed during resetting in PXGL.
In sum, these results indicate that at day 3 of conversion, the majority of cell co‐express markers of naive PSCs and TSCs and the subsequent exposure to different signalling environments steer the cells towards the two alternative cell fates.
Discussion
We reported the rapid and efficient conversion of conventional PSCs to TSCs without the need for genetic modifications or prolonged expansion under conditions supporting naive pluripotency.
Previous studies reported resetting of conventional PSCs to naive‐iPSCs, followed by differentiation towards TE and to TSCs (Castel et al, 2020; Dong et al, 2020; Guo et al, 2021; Io et al, 2021). These multistep protocols require the isolation of the cell type of interest by fluorescence‐activated cell sorting and long‐term expansion, possibly to enrich some rare populations. In particular, a stable naive phenotype is obtained only after isolation and expansion for some passages in naive pluripotency‐sustaining conditions. We were thus rather surprised by the rapidity with which we obtained a homogeneous TSCs population with a negligible expression of amnion markers from conventional PSCs through HDAC inhibition, MEK inhibition and LIF stimulation. Such conversion might occur either directly or via a transient naive pluripotent state. Interestingly, in the case of reprogramming of somatic cells, a trophoblast‐like identity was transiently acquired before naive pluripotency establishment (Liu et al, 2020), raising the possibility that also during chemical resetting cells acquire a transient TE or trophoblast identity on their path towards the naive state. Conversely, a recent study also proposed the co‐expression of embryonic and extraembryonic genes as a functional attribute of naive pluripotency, suggesting that the trophoblast identity can be acquired via a transient naive‐like state (Dong et al, 2020). In agreement with this idea, we detected co‐expression of naive‐iPSC and TSC markers at the single‐cell levels after 3 days of chemical conversion, indicating an intermediate cellular state responsive to different external stimuli.
The conversion occurs in the presence of HDAC inhibitor, LIF and MEK inhibitor. The HDAC inhibitor and LIF might affect the epigenome of conventional PSCs, by increasing histone acetylation and reducing DNA methylation (Betto et al, 2021). It should also be noted that HDAC inhibitors are used as anticancer drugs as they induce cell cycle arrest and apoptosis in cancer cell lines. HDAC inhibitors are also used to enhance reprogramming or cell differentiation, but rarely for cell expansion. Conversely, the HDAC inhibitor VPA allows for the robust expansion of TSCs (Okae et al, 2018), suggesting that high levels of histone acetylation might be an intrinsic and peculiar feature of human TSCs. In this view, our findings also imply that epigenetic modifications and extensive methylation could be a limiting factor in the competence of conventional PSCs (Hackett & Azim Surani, 2014; Martello & Smith, 2014; Rossant, 2015) to directly form bona fide TSCs.
LIF might promote the conversion by boosting oxidative phosphorylation (Carbognin et al, 2016) or by inducing and maintaining naive pluripotency, as reported in murine cells (Yang et al, 2010; Martello et al, 2013; Carbognin et al, 2016), although it is still not clear whether LIF is strictly required to sustain naive pluripotency in human cells (Theunissen et al, 2014; Bayerl et al, 2021; Khan et al, 2021).
MEK inhibition is commonly used to stabilise naive pluripotency in human (Gafni et al, 2013; Takashima et al, 2014; Theunissen et al, 2014; Guo et al, 2017; Szczerbinska et al, 2019; Khan et al, 2021) and murine cells (Dunn et al, 2014; Martello & Smith, 2014). MEK inhibition has also been shown to promote the transition of naive PSCs towards TE (Guo et al, 2021; Io et al, 2021) in the absence of other stimuli supporting human naive pluripotency, such as TGF‐beta signalling, PKC inhibition and SRC inhibition (Takashima et al, 2014; Theunissen et al, 2014; Guo et al, 2017; Szczerbinska et al, 2019; Bayerl et al, 2021; Khan et al, 2021; Osnato et al, 2021). Thus, MEK inhibition seems to have a dual role, as stabiliser of human naive pluripotency and inducer of TE identity. This could explain the enhanced plasticity of human naive PSCs.
While we were finalising our manuscript, a study from the Hanna laboratory reported the generation of TSCs at high efficiency from conventional PSCs using a combination of TGF‐beta receptor inhibitor and Vitamin C in N2B27 medium (Viukov et al, 2022). Interestingly, no induction of naive genes was detected, indicating the existence of multiple trajectories followed by conventional PSCs towards a TSC state. It is also interesting to note that TGF‐beta inhibition in conventional PSCs in E8 or mTeSR medium results in neuroectoderm differentiation (Muñoz‐Sanjuán & Brivanlou, 2002; Vallier et al, 2004; Zorzan et al, 2020). Thus, some parameters like the basal medium composition, the duration of the treatment or the inhibitor concentration might affect the outcome of TGF‐beta inhibition. For example, TGF‐beta inhibition, combined with MEK inhibition, in N2B27 medium results in trophoblast differentiation of naive PSCs (Guo et al, 2021). It will be interesting to test which signalling pathways, epigenetic modifications and metabolic pathways are needed for the generation of naive‐iPSCs or TSCs from conventional PSCs.
Expression of imprinted genes can be a bona fide marker of good quality TSC identity, as widespread placental‐specific imprinting has been observed in humans (Hanna, 2020). However, prolonged expansion of human naive PSCs might lead to an impaired expression of imprinted genes (Giulitti et al, 2019; Perrera & Martello, 2019), which might be inherited in naive‐TSCs. In line with this, we observed the expression of placental imprinted genes during chemical resetting and the aberrant expression of 6 imprinted genes in TSCs derived from naive‐iPSCs. During the revision of our manuscript, a study from Arima laboratory reported that activation of the imprinted microRNA cluster C19MC occurs in naive‐TSCs but not in TSCs converted from conventional PSCs through BMP stimulation, and it confers differentiation potential into trophoblast lineages (Kobayashi et al, 2022). It will be interesting in future to systematically compare the epigenome and the expression of imprinted genes of TSCs obtained through different protocols, like differentiation of naive PSCs (Dong et al, 2020; Guo et al, 2021; Io et al, 2021), conversion of conventional PSCs with (Soncin et al, 2018; Wei et al, 2021; Jang et al, 2022; Kobayashi et al, 2022) or without BMP stimulation (this study and Viukov et al, 2022) or somatic cell reprogramming (Liu et al, 2020), and to functionally test the consequences of altered imprinted gene expression.
Our results have practical applications, given that conventional iPSC lines have been generated from patients with placental disorders (Hanna, 2020). It will be extremely easy to convert such iPSCs to TSCs, from which placental organoids or specific placental cell types of interest could be obtained.
Materials and Methods
Culture of hPSCs
Human primed PSCs Keratinocytes induced Pluripotent Stem Cells (KiPS, Takashima et al, 2014) and HPD00 (Giulitti et al, 2019), and hESCs (H9) were cultured in feeder‐free on precoated plates with 0.5% growth factor‐reduced Matrigel (CORNING 356231; vol/vol in PBS with MgCl2/CaCl2, Sigma‐Aldrich D8662) in E8 medium (made in‐house according to Chen et al (2011)) at 37°C, 5% CO2, 5% O2. Cells were passaged every 3–4 days at a split ratio of 1:8 following dissociation with 0.5 mM EDTA (Invitrogen AM99260G) in PBS without MgCl2/CaCl2 (Sigma‐Aldrich D8662), pH8. KiPS line was derived by reprogramming of human keratinocytes (Takashima et al, 2014) (Invitrogen) with Sendai viruses encoding for OSKM and kindly provided by Austin Smith's laboratory.
Human naive‐ESCs (reset H9, were provided by Austin Smith's laboratory and were generated via transient expression of NK2 and previously described in Takashima et al, 2014) and human naive‐iPSCs (HPD06 and HPD01 were previously generated by direct reprogramming from somatic cells and described in Giulitti et al, 2019) were cultured on mitotically inactivated mouse embryonic fibroblasts (MEFs; DR4 ATCC) in PXGL medium (Bredenkamp et al, 2019) or in RSeT medium (Stem Cell Technologies 05969). The PXGL medium was prepared as follows: N2B27 (DMEM/F12 [Gibco 11320‐074], and Neurobasal in 1:1 ratio [Gibco 21103‐049], with 1:200 N2 Supplement [Gibco 17502‐048], and 1:100 B27 Supplement [Gibco 17504‐044], 2 mM L‐glutamine [Gibco 25030‐024], 0.1 mM 2‐mercaptoethanol [Sigma‐Aldrich M3148]) supplemented with 1 μM PD0325901 (Axon Medchem), 2 μM XAV939 (Axon Medchem), 2 μM Gö6983 (Axon Medchem) and 10 ng/ml human LIF (produced in‐house). Human naive‐iPSCs were passaged as single cells every 4 days at a split ratio of 1:3 or 1:4 following dissociation with TrypLE (Gibco 12563‐029) for 10 min (min) at room temperature (RT). The ROCK inhibitor (Y27632, Axon Medchem 1683) was added in the naive medium only for 24 h after passaging. None of the cell lines used in this study are listed in the ICLAC register of misidentified cell lines. All cell lines were mycoplasma‐negative (Mycoalert, Lonza).
Culture of hTSCs
hTSCs were cultured as described previously (Okae et al, 2018). Briefly, a 12‐well plate was coated with 5 μg/ml Collagen IV (Corning, 354233) at 37°C overnight or on mitotically inactivated mouse embryonic fibroblasts. Cells were cultured in 0.8 ml TS medium (DMEM/F12 supplemented with 0.1 mM 2‐mercaptoethanol, 0.2% FBS, 0.5% Penicillin–Streptomycin, 0.3%, BSA [Gibco 15260‐037], 1% ITS‐X [Gibco, 51500], 1.5 μg/ml L‐ascorbic acid [Sigma A4544], 50 ng/ml EGF [peprotech AF‐100‐15], 2 μM CHIR99021 [Axon Medchem cat. nos 1386 and 1408], 0.5 μM A83‐01 [Axon Medchem 1421], 1 μM SB431542 [Axon Medchem 1661], 0.8 mM VPA [HDACi, Sigma, P4543], and 5 μM Y‐27632) and in 5% CO2 and 5% O2. Media were changed every 2 days, and cells were passed using TrypLE Express every 3–4 days at a ratio of 1:8. Unless otherwise specified, hTSCs between passages 5 and 20 were used for experiments. All cell lines were mycoplasma‐negative (Mycoalert, Lonza).
Culture of BAP‐treated primed pluripotent stem cells
KiPS were dissociated into single cells with TrypLE. The cells were cultured on precoated plates with 0.5% growth factor‐reduced Matrigel at a density of 2 × 104 cells/cm2 with MEF‐conditioned medium supplemented with 10 ng/ml BMP4 (Peprotech, 120‐05ET), 1 μM A83‐01, 0.1 μM PD173074 (Axon, 1673), and 10 μM Y‐27632, as described previously (Io et al, 2021). DMEM/Ham's F‐12 medium containing 0.1 mM 2ME, 1% ITS‐X supplement, 1% NEAA, 2 mM L‐glutamine, and 20% KSR was cultured on MEF for 24 h, and the supernatant was collected and used as MEF‐conditioned medium. The medium was changed daily.
Chemical resetting
For the chemical resetting from primed to naive‐iPSC, KiPS were seeded at 10,000 cells/cm2 on mitotically inactivated MEFs in E8 medium with 10 μM ROCKi (added only for 24 h). Two days after plating (day 0), the medium was changed to MEKi/LIF/HDACi (N2B27 with 1 μM PD0325901 [MEKi], 10 ng/ml human LIF and 1 mM VPA [HDACi]). Following 3 days in PD03/LIF/HDACi, the medium was changed to PXGL for a further 10–11 days before passing in PXGL medium on MEFs (see Fig 1A). For the chemical resetting from primed to ccTSCs, KiPS were seeded at 10,000 cells/cm2 on mitotically inactivated MEFs in E8 medium. Two days later (day 0) medium was changed to MEKi/LIF/HDACi. Following 3 days in MEKi/LIF/HDACi, the medium was changed to PXGL for a further 4 days and then switched with TSC medium in the case of ccTSC.01 (see Fig 2H) or directly to TSC medium in the case of ccTSC.02 (see Fig 5A). After resetting, ccTSCs were passaged as TSCs (see above).
Sorting of naive cells
KiPS chemical resetting to naive at passage 6–7 were dissociated into single cells using TrypLE and passed through 50‐μm cell strainers (VWR). Conjugated antibodies and Fixable Viability Dye‐eF780 (eBioscience) were mixed with 50 μl of Brilliant stain buffer (BD Biosciences, 566349) and applied to 50 μl of cells (500,000 cells per reaction). Cells were incubated for 30 min at 4°C in the dark and washed with 2% foetal bovine serum (FBS) in PBS and centrifuged at 300 g for 3 min. Cells were resuspended in 2% FBS in PBS and analysed with a BD FACSAria Fusion for cell sorting. The following fluorescent conjugated antibodies and flow cytometer laser and filter settings were used: CD24‐BUV395 (BD Bioscience, 563818, 1.25 μl per test; detected using the 355‐nm laser with 379/28 filters), CD90.2‐APC‐Cy7 (BD Bioscience, 561641, 2.5 μl per test; 640‐nm laser with 780/60 filters), Fixable Viability Dye‐eF780 (ThermoFisher, 65‐0865‐14, 0.6 μl per test; 640‐nm laser with 780/60 filters) and SUSD2‐FITC (MACS Miltenyi Biotec, 130‐106‐327, 0.25 μl per test; 488‐nm laser with 530/30 filters).
Conversion of human naive‐iPSCs into TSCs
Naive‐iPSCs did a pretreatment on MEF with TS medium for 24 h, whereupon naive‐iPSCs were single cell dissociated by TrypLE Express, and 0.25–0.5 × 106 cells were seeded on a 12‐well plate precoated with 5 μg/ml Collagen IV or on mitotically inactivated mouse embryonic fibroblasts and cultured in 0.8 ml TS medium. Cells were cultured in 5% CO2 and 5% O2; media were changed every 2 days and passaged upon 80–100% confluency at a ratio of 1:2 to 1:4. During derivation from naive‐iPSCs, the cells grew slowly during the initial few passages. Between passages 5 and 10, highly proliferative TSCs emerged.
Trophoblast organoids
To obtain trophoblast organoids, we used the protocol previously described in Turco et al (2018). TSCs were single cell dissociated by TrypLE Express, and approximately 50 K cells were resuspended in Matrigel (Corning 356231) on ice. Drops (25 μl) were plated per well into a 24‐well culture plate, set at 37°C for 10 min, and overlaid with 400 μl trophoblast organoid medium (TOM, previously described in Turco et al (2018)). Cultures were maintained in 5% CO2 and 5% O2 in a humidified incubator at 37°C. The medium was replaced every 2–3 days. Small organoid clusters became visible by around day 7 and were passaged when at least 50% had reached a diameter of 200–300 μm (usually between days 7 and 10). Mechanical disruption was achieved by pipetting the drops of Matrigel with TrypLE Express to obtain a single‐cell suspension, after that spin dawn at 4°C and resuspend the pellet directly in Matrigel at the correct proportion. Frozen stocks of organoids were made in 70% TOM, 20% FBS and 10% DMSO freezing medium and stored in liquid nitrogen.
Differentiation of STBs and EVTs
Differentiation of TSCs into terminal cell types was performed as described previously (Okae et al, 2018), with minor modifications. Prior to differentiation, TSCs were grown to about 80% confluency, and then single cell dissociated using TrypLE Express.
For STB differentiation, 6‐well plates were coated with 2.5 μg/ml Collagen IV overnight. 2 × 105 TSCs were seeded per well in 2 ml STB medium (DMEM/F12 supplemented with 0.1 mM β‐mercaptoethanol, 0.5% penicillin–streptomycin, 0.3% BSA, 1% ITS‐X, 2.5 μM Y‐27632, 2 μM Forskolin [Sigma‐Aldrich, F3917], and 4% KSR). The media were changed at day 3, and on day 6, the cells were ready for downstream analysis. For EVT differentiation, 6‐well plates were coated with 1 μg/ml Collagen IV overnight. 1.2 × 105 TSCs were seeded per well in 2 ml EVT medium [DMEM/F12 supplemented with 0.1 mM β‐mercaptoethanol, 0.5% penicillin–streptomycin, 0.3% BSA, 1% ITS‐X, 2.5 μM Y‐27632, 7.5 mM A83‐01, 100 ng/ml NRG1 (Cell Signalling, 5218SC), 4% KSR]. Matrigel was added to the final concentration of 2% after resuspending the cells in the medium. On day 3, the medium was replaced with EVT medium without NRG1 and with Matrigel at 0.5% final concentration. On day 6, the medium was replaced with EVT medium without NRG1 and KSR, and with Matrigel at 0.5% final concentration. The cells were ready for downstream analysis on day 8.
Immunofluorescence
Immunofluorescence analysis was performed on 1% Matrigel‐coated glass coverslip in wells. Cells were fixed in 4% Formaldehyde (Sigma‐Aldrich 78775) in PBS for 10 min at RT, washed in PBS, permeabilised for 1 h in PBS + 0.3% Triton X‐100 (PBST) and in the case of TSCs we permeabilised only for 10 min in PBST at RT in order to avoid cell detachment, and blocked in PBST +5% of Horse serum (ThermoFisher 16050‐122) for 5 h at RT. Cells were incubated overnight at 4°C with primary antibodies (Appendix Table S2) in PBST+3% of Horse serum. After washing with PBS, cells were incubated with secondary antibodies (Alexa, Life Technologies) (Appendix Table S2) for 45 min at RT. Nuclei were stained with DAPI (4′,6‐diamidino‐2‐phenylindole, Sigma‐Aldrich F6057).
Images were acquired with a Zeiss LSN700, a Leica SP5, or a Leica SP8 confocal microscope using ZEN 2012 or LeicaLeica TCS SP5 LAS AF (v2.7.3.9723) software, respectively.
Images analysis
Fluorescence intensity was quantified using Cell Profiler software (v4.2.1). DAPI staining was used to identify individual nuclei of cells and the cytoplasm surrounding each nucleus. MEFs were recognised based on the nucleus size and negativity for all markers. Between 5 and 12 independent fields from 3 replicates were quantified for each sample, for a total of 600–900 cells per sample. Single cell measured integrated intensity values were considered for downstream data analysis and plotted as log(Integrated Intensity+1). Protein‐specific presence corresponding to active and inactive protein states in the cell population was identified setting the integrated fluorescence intensity threshold to 99% of the cumulative density function in samples where the protein is expressed (GATA3 and KRT7 in KiPS primed‐iPSCs and HPD06 naive‐iPSCs, SUSD2 in KiPS primed‐iPSCs and HPD06 naive‐TSCs, OCT4 in HPD06 naive‐TSCs for OCT4). Identified activation thresholds were as follows: thrGATA3 = 4.27, thrOCT4 = 2.68, thrKRT7 = 27.01 and thrSUSD2 = 9.25. Patterns of protein expression (16 total patterns, considering each protein as present/absent) were identified by counting the number of cells exhibiting each specific activation pattern and reported as % of total cells.
Quantitative PCR
Total RNA was isolated using Total RNA Purification Kit (Norgen Biotek 37500), and complementary DNA (cDNA) was made from 500 ng using M‐MLV Reverse Transcriptase (Invitrogen 28025‐013) and dN6 primers. For real‐time PCR SYBR Green Master mix (Bioline BIO‐94020) was used. Primers are detailed in Appendix Table S3. Three technical replicates were carried out for all quantitative PCR. GAPDH was used as an endogenous control to normalise expression. qPCR data were acquired with QuantStudio™ 6&7 Flex Software 1.0.
RNA sequencing and analysis
Quant Seq 3′ mRNA‐seq Library Prep kit (Lexogen) was used for library construction. Library quantification was performed by fluorometer (Qubit) and bioanalyser (Agilent). Sequencing was performed on NextSeq500 ILLUMINA instruments to produce 5 million reads (75 bp SE) for the sample.
For the analysis of imprinted genes and TSC markers, the reads were trimmed using BBDuk (BBMap v. 37.87), with parameters indicated in the Lexogen data analysis protocol. After trimming, reads were aligned to the Homo sapiens genome (GRCm38.p13) using STAR (v. 2.7.6a). The gene expression levels were quantified using featureCounts (v. 2.0.1). Genes were sorted removing those that had a total number of counts below 10 in at least 3 samples. All RNA‐seq analyses were carried out in the R environment (v. 4.0.0) with Bioconductor (v. 3.7). We computed differential expression analysis using the DESeq2 R package (v. 1.28.1) (Love et al, 2014). Transcripts with an absolute value of log2 Fold Change (log2FC) > 1 and an adjusted P‐value < 0.05 (Benjamini–Hochberg adjustment) were considered significant and defined as differentially expressed for the comparison in the analysis. Heatmaps were made using counts‐per‐million (CPM) values with the pheatmap function from the pheatmap R package (v.1.0.12) on differentially expressed genes or selected markers. Volcano plots were computed with log2FC and − log10 adjusted P‐value from DESeq2 differential expression analysis output using the ggscatter function from the ggpubr R package (v. 0.4.0). Barplots were made using CPM values with the ggbarplot function from the ggpubr R package.
For the time point analysis during resetting and the following stabilisation in PXGL or TSC medium, transcript quantification was performed from raw reads using Salmon (v1.6.0) (Patro et al, 2017) on transcripts defined in Ensembl 105. Gene expression levels were estimated with tximport R package (v1.20.0; Soneson et al, 2016), and differential expression analysis was computed using the DESeq2 R package (v. 1.28.1; Love et al, 2014). Transcripts with an absolute value of log2FC ≥ 3 and an adjusted P‐value < 0.05 (Benjamini–Hochberg adjustment) were considered significant. Principal component analysis was performed on variance stabilised data (vst function from DESeq2 R package v 1.32.0; Love et al, 2014) using prcomp function on the top 5,000 most variable genes. Heatmaps were performed using the pheatmap function (pheatmap R package v1.0.12) on log2 CPM data of selected markers. All analyses except salmon were performed in R version 4.1.1. Pairwise correlations were computed with “cor” function from “stats” R package (v 4.1.3) using Pearson as correlation coefficient. For the PlacentaCellEnrich programme analysis (Jain & Tuteja, 2021), we first identified the DEGs (log2FC > 1 or < −1, adjusted P‐value < 0.05) between ccTSC.02 and isogenic conventional KiPS, or between H9 naive‐TSCs and conventional H9. The two lists of DEGs were uploaded on the web tool at https://placentacellenrich.gdcb.iastate.edu, using the Vento‐Tormo datasets and default parameters.
Statistics and reproducibility
No statistical method was used to predetermine sample size, but our sample sizes are similar to those commonly used in our field of research. No data were excluded from the analyses. For experiments with cell lines, we randomly allocated a fraction of each cell population to different biological replicates. For the analysis of immunostaining, we analysed random fields. Other kinds of experiments were not randomised. Data collection and analysis were not performed blind to the conditions of the experiments, but data analyses have been performed with identical parameters and software. Data representation and statistical analyses were performed using R software (v4.0.0) unless stated otherwise. All error bars indicate the standard error of the mean (± SEM). Results were analysed by unpaired two‐tailed Student's t‐test, as indicated in figure legends. Data distribution was assumed to be normal, but this was not formally tested. For each dataset, sample size n refers to the number of biological replicates as stated in the figure legends. All qPCR experiments were performed with three technical replicates.
Author contributions
Irene Zorzan: Conceptualization; formal analysis; supervision; validation; investigation; visualization; methodology; project administration; writing—review and editing. Riccardo, Massimiliano Betto: Formal analysis; validation; investigation; visualization; methodology. Giada Rossignoli: Formal analysis; validation; investigation; visualization; writing—review and editing. Mattia Arboit: Data curation; formal analysis; visualization. Andrea Drusin: Formal analysis; validation; investigation; visualization; methodology. Clelia Corridori: Formal analysis; visualization. Paolo Martini: Data curation; formal analysis; visualization. Graziano Martello: Conceptualization; resources; supervision; funding acquisition; writing—original draft; project administration; writing—review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank all the members of the Martello Laboratory for discussions, suggestions and technical support, particularly L. Diamante. We also thank S. Thompson at the Babraham Institute Flow Core and P. Rugg‐Gunn for technical support. G.M.'s Laboratory is supported by grants from the Giovanni Armenise–Harvard Foundation, the Telethon Foundation (GJC21157) and an ERC Starting Grant (MetEpiStem). Open Access Funding provided by Universita degli Studi di Padova within the CRUI‐CARE Agreement.
EMBO reports (2023) 24: e55235
Contributor Information
Irene Zorzan, Email: irene.zorzan@babraham.ac.uk.
Graziano Martello, Email: graziano.martello@unipd.it.
Data availability
RNA‐seq data for this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE184562. We also included available RNA‐seq data for primed PSCs, naive‐iPSCs, TSCs derived from naive‐iPSCs, TSCs derived from primed PSCs and embryo‐derived TSCs (GSE138688, GSE178162 and GSE135695) from (Data ref: Theunissen et al, 2020; Data ref: Jang & Kim, 2021; Data ref: Shan & Wei, 2021).
References
- Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T (2013) Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA 110: E1212–E1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayerl J, Ayyash M, Shani T, Manor YS, Gafni O, Massarwa R, Kalma Y, Aguilera‐Castrejon A, Zerbib M, Amir H et al (2021) Principles of signaling pathway modulation for enhancing human naive pluripotency induction. Cell Stem Cell 28: 1549–1565.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchetrit H, Jaber M, Zayat V, Sebban S, Pushett A, Makedonski K, Zakheim Z, Radwan A, Maoz N, Lasry R et al (2019) Direct induction of the three pre‐implantation blastocyst cell types from fibroblasts. Cell Stem Cell 24: 983–994.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC et al (2011) BRACHYURY and CDX2 mediate BMP‐induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9: 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betto RM, Diamante L, Perrera V, Audano M, Rapelli S, Lauria A, Incarnato D, Arboit M, Pedretti S, Rigoni G et al (2021) Metabolic control of DNA methylation in naive pluripotent cells. Nat Genet 53: 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof P, Irminger‐Finger I (2005) The human cytotrophoblastic cell, a mononuclear chameleon. Int J Biochem Cell Biol 37: 1–16 [DOI] [PubMed] [Google Scholar]
- Bredenkamp N, Stirparo GG, Nichols J, Smith A, Guo G (2019) The cell‐surface marker sushi containing domain 2 facilitates establishment of human naive pluripotent stem cells. Stem Cell Rep 12: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbognin E, Betto RM, Soriano ME, Smith AG, Martello G (2016) Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J 35: 618–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel G, Meistermann D, Bretin B, Firmin J, Blin J, Loubersac S, Bruneau A, Chevolleau S, Kilens S, Chariau C et al (2020) Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep 33: 108419 [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga‐Otto K, Howden SE, Diol NR, Propson NE et al (2011) Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8: 424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinkornpumin JK, Kwon SY, Guo Y, Hossain I, Sirois J, Russett CS, Tseng H‐W, Okae H, Arima T, Duchaine TF et al (2020) Naive human embryonic stem cells can give rise to cells with a trophoblast‐like transcriptome and methylome. Stem Cell Rep 15: 198–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, Corcoran AE, Walker R, Douagi I, Lanner F et al (2017) Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell 20: 874–890.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe C, Aberkane A, Dewandre D, Essahib W, Sermon K, Geens M, Verheyen G, Tournaye H, Van de Velde H (2019) BMP4 plays a role in apoptosis during human preimplantation development. Mol Reprod Dev 86: 53–62 [DOI] [PubMed] [Google Scholar]
- Dong C, Beltcheva M, Gontarz P, Zhang B, Popli P, Fischer LA, Khan SA, Park K, Yoon E‐J, Xing X et al (2020) Derivation of trophoblast stem cells from naïve human pluripotent stem cells. Elife 9: e52504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Fu S, Karvas RM, Chew B, Fischer LA, Xing X, Harrison JK, Popli P, Kommagani R, Wang T et al (2022) A genome‐wide CRISPR‐Cas9 knockout screen identifies essential and growth‐restricting genes in human trophoblast stem cells. Nat Commun 13: 2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG (2014) Defining an essential transcription factor program for naïve pluripotency. Science 344: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben‐Yosef D, Kalma Y, Viukov S, Maza I, Zviran A et al (2013) Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286 [DOI] [PubMed] [Google Scholar]
- Giulitti S, Pellegrini M, Zorzan I, Martini P, Gagliano O, Mutarelli M, Ziller MJ, Cacchiarelli D, Romualdi C, Elvassore N et al (2019) Direct generation of human naive induced pluripotent stem cells from somatic cells in microfluidics. Nat Cell Biol 21: 275–286 [DOI] [PubMed] [Google Scholar]
- Guo G, Von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J (2016) Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep 6: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Rostovskaya M, Clarke J, Dietmann S, Baker D, Sahakyan A, Myers S, Bertone P, Reik W et al (2017) Epigenetic resetting of human pluripotency. Development 144: 2748–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Stirparo GG, Strawbridge SE, Spindlow D, Yang J, Clarke J, Dattani A, Yanagida A, Li MA, Myers S et al (2021) Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 28: 1040–1056.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Azim Surani M (2014) Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15: 416–430 [DOI] [PubMed] [Google Scholar]
- Hanna CW (2020) Placental imprinting: emerging mechanisms and functions. PLoS Genet 16: e1008709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Sutherland L, Clark J, Burdon T (2004) Oct‐4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22: 225–235 [DOI] [PubMed] [Google Scholar]
- Io S, Kabata M, Iemura Y, Semi K, Morone N, Minagawa A, Wang B, Okamoto I, Nakamura T, Kojima Y et al (2021) Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 28: 1023–1039.e13 [DOI] [PubMed] [Google Scholar]
- Jain A, Tuteja G (2021) PlacentaCellEnrich: a tool to characterize gene sets using placenta cell‐specific gene enrichment analysis. Placenta 103: 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JL, Carter AM, Chamley LW (2012) Human placentation from nidation to 5 weeks of gestation. Part I: what do we know about formative placental development following implantation? Placenta 33: 327–334 [DOI] [PubMed] [Google Scholar]
- Jang Y, Kim J (2021) Gene Expression Omnibus GSE178162 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178162) [DATASET]
- Jang YJ, Kim M, Lee B‐K, Kim J (2022) Induction of human trophoblast stem‐like cells from primed pluripotent stem cells. Proc Natl Acad Sci USA 119: e2115709119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Park K, Fischer LA, Dong C, Lungjangwa T, Jimenez M, Casalena D, Chew B, Dietmann S, Auld DS et al (2021) Probing the signaling requirements for naive human pluripotency by high‐throughput chemical screening. Cell Rep 35: 109233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilens S, Meistermann D, Moreno D, Chariau C, Gaignerie A, Reignier A, Lelièvre Y, Casanova M, Vallot C, Nedellec S et al (2018) Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat Commun 9: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Okae H, Hiura H, Kubota N, Kobayashi EH, Shibata S, Oike A, Hori T, Kikutake C, Hamada H et al (2022) The microRNA cluster C19MC confers differentiation potential into trophoblast lineages upon human pluripotent stem cells. Nat Commun 13: 3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CQE, Gardner L, Turco M, Zhao N, Murray MJ, Coleman N, Rossant J, Hemberger M, Moffett A (2016) What is trophoblast? A combination of criteria define human first‐trimester trophoblast. Stem Cell Rep 6: 257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Nefzger CM, Rossello FJ, Chen J, Knaupp AS, Firas J, Ford E, Pflueger J, Paynter JM, Chy HS et al (2017) Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat Methods 14: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Liu X, Ouyang JF, Rossello FJ, Tan JP, Davidson KC, Valdes DS, Schröder J, Sun YBY, Chen J, Knaupp AS et al (2020) Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 586: 101–107 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Smith A (2014) The nature of embryonic stem cells. Annu Rev Cell Dev Biol 30: 647–675 [DOI] [PubMed] [Google Scholar]
- Martello G, Bertone P, Smith A (2013) Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J 32: 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini P, Sales G, Diamante L, Perrera V, Colantuono C, Riccardo S, Cacchiarelli D, Romualdi C, Martello G (2022) BrewerIX enables allelic expression analysis of imprinted and X‐linked genes from bulk and single‐cell transcriptomes. Commun Biol 5: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, Moore HD, Andrews PW (2004) Specific knockdown of Oct4 and β2‐microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells 22: 659–668 [DOI] [PubMed] [Google Scholar]
- Mazid MA, Ward C, Luo Z, Liu C, Li Y, Lai Y, Wu L, Li J, Jia W, Jiang Y et al (2022) Rolling back human pluripotent stem cells to an eight‐cell embryo‐like stage. Nature 605: 315–324 [DOI] [PubMed] [Google Scholar]
- Muñoz‐Sanjuán I, Brivanlou AH (2002) Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci 3: 271–280 [DOI] [PubMed] [Google Scholar]
- Okae H, Chiba H, Hiura H, Hamada H, Sato A, Utsunomiya T, Kikuchi H, Yoshida H, Tanaka A, Suyama M et al (2014) Genome‐wide analysis of DNA methylation dynamics during early human development. PLoS Genet 10: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T (2018) Derivation of human trophoblast stem cells. Cell Stem Cell 22: 50–63.e6 [DOI] [PubMed] [Google Scholar]
- Osnato A, Brown S, Krueger C, Andrews S, Collier AJ, Nakanoh S, Quiroga Londoño M, Wesley BT, Muraro D, Brumm AS et al (2021) TGFβ signalling is required to maintain pluripotency of human naïve pluripotent stem cells. Elife 10: e67259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Liu W, Chen D, Ho J, Kim R, Hunt TJ, Lukianchikov A, Liu X, Polo JM, Jacobsen SE et al (2018) TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nat Cell Biol 20: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrera V, Martello G (2019) How does reprogramming to pluripotency affect genomic imprinting? Front Cell Dev Biol 7: 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F (2016) Single‐cell RNA‐seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165: 1012–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Loh KM, Amita M, Bernardo AS, Adachi K, Alexenko AP, Schust DJ, Schulz LC, Telugu BPVL, Ezashi T et al (2014) Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? Reproduction 147: D1–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J (2015) Mouse and human blastocyst‐derived stem cells: vive les differences. Development 142: 9–12 [DOI] [PubMed] [Google Scholar]
- Rostovskaya M, Andrews S, Reik W, Rugg‐Gunn PJ (2022) Amniogenesis occurs in two independent waves in primates. Cell Stem Cell 29: 744–759.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharam AS, Vu HTH, Choi S, Khan T, Sheridan MA, Ezashi T, Roberts RM, Tuteja G (2022) The product of BMP‐directed differentiation protocols for human primed pluripotent stem cells is placental trophoblast and not amnion. Stem Cell Rep 17: 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Wei Y (2021) Gene Expression Omnibus GSE135695 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135695) [DATASET]
- Soncin F, Khater M, To C, Pizzo D, Farah O, Wakeland A, Arul Nambi Rajan K, Nelson KK, Chang C‐W, Moretto‐Zita M et al (2018) Comparative analysis of mouse and human placentae across gestation reveals species‐specific regulators of placental development. Dev Camb Engl 145: dev156273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD (2016) Differential analyses for RNA‐seq: transcript‐level estimates improve gene‐level inferences. F1000Res 4: 1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart HT, Stirparo GG, Lohoff T, Bates LE, Kinoshita M, Lim CY, Sousa EJ, Maskalenka K, Radzisheuskaya A, Malcolm AA et al (2019) Distinct molecular trajectories converge to induce naive pluripotency. Cell Stem Cell 25: 388–406.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerbinska I, Gonzales KAU, Cukuroglu E, Ramli MNB, Lee BPG, Tan CP, Wong CK, Rancati GI, Liang H, Göke J et al (2019) A chemically defined feeder‐free system for the establishment and maintenance of the human naive pluripotent state. Stem Cell Rep 13: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W et al (2014) Resetting transcription factor control circuitry toward ground‐state pluripotency in human. Cell 158: 1254–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenschmid‐Stowers J, Rostovskaya M, Santos F, Ljung S, Argelaguet R, Krueger F, Nichols J, Reik W (2022) 8C‐like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell 29: 449–459.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L et al (2014) Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15: 471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T, Dong C, Gontarz P, Zhang B (2020) Gene Expression Omnibus GSE138688 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138688) [DATASET]
- Turco MY, Gardner L, Kay RG, Hamilton RS, Prater M, Hollinshead MS, McWhinnie A, Esposito L, Fernando R, Skelton H et al (2018) Trophoblast organoids as a model for maternal–fetal interactions during human placentation. Nature 564: 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA (2004) Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol 275: 403–421 [DOI] [PubMed] [Google Scholar]
- Vento‐Tormo R, Efremova M, Botting RA, Turco MY, Vento‐Tormo M, Meyer KB, Park J‐E, Stephenson E, Polański K, Goncalves A et al (2018) Single‐cell reconstruction of the early maternal–fetal interface in humans. Nature 563: 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viukov S, Shani T, Bayerl J, Aguilera‐Castrejon A, Oldak B, Sheban D, Tarazi S, Stelzer Y, Hanna JH, Novershtern N (2022) Human primed and naïve PSCs are both able to differentiate into trophoblast stem cells. Stem Cell Rep 17: 2484–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N (2012) Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10: 440–454 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao C, Hou Z, Yang Y, Bi Y, Wang H, Zhang Y, Gao S (2018) Unique molecular events during reprogramming of human somatic cells to induced pluripotent stem cells (iPSCs) at naïve state. Elife 7: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wang T, Ma L, Zhang Y, Zhao Y, Lye K, Xiao L, Chen C, Wang Z, Ma Y et al (2021) Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Sci Adv 7: eabf4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Yin Y, Zheng Y, Ma Y, Li Y, Zhao Z, Guo J, Ai Z, Niu Y, Duan K et al (2020) A developmental landscape of 3D‐cultured human pre‐gastrulation embryos. Nature 577: 537–542 [DOI] [PubMed] [Google Scholar]
- Xu R‐H, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20: 1261–1264 [DOI] [PubMed] [Google Scholar]
- Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM (2016) Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA 113: E2598–E2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JCR, Smith A (2010) Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 7: 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Adachi K, Sheridan MA, Alexenko AP, Schust DJ, Schulz LC, Ezashi T, Roberts RM (2015) Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci USA 112: E2337–E2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz‐Eldor J, Daley GQ (2005) High‐efficiency RNA interference in human embryonic stem cells. Stem Cells 23: 299–305 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhao T, Guan J, Zhang X, Fu Y, Ye J, Zhu J, Meng G, Ge J, Yang S et al (2015) A XEN‐like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 163: 1678–1691 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL et al (2019) Controlled modelling of human epiblast and amnion development using stem cells. Nature 573: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans DW, Talon I, Verhelst S, Bendall A, Van Nerum K, Javali A, Malcolm AA, van Knippenberg SSFA, Biggins L, To SK et al (2022) Integrated multi‐omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat Cell Biol 24: 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzan I, Pellegrini M, Arboit M, Incarnato D, Maldotti M, Forcato M, Tagliazucchi GM, Carbognin E, Montagner M, Oliviero S et al (2020) The transcriptional regulator ZNF398 mediates pluripotency and epithelial character downstream of TGF‐beta in human PSCs. Nat Commun 11: 2364 [DOI] [PMC free article] [PubMed] [Google Scholar]


