Summary
Environmental stiffness is a crucial determinant of cell function. There is a long-standing quest for reproducible and (human matrix) bio-mimicking biomaterials with controllable mechanical properties to unravel the relationship between stiffness and cell behavior. Here, we evaluate methacrylated human recombinant collagen peptide (RCPhC1-MA) hydrogels as a matrix to control 3D microenvironmental stiffness and monitor cardiac cell response. We show that RCPhC1-MA can form hydrogels with reproducible stiffness in the range of human developmental and adult myocardium. Cardiomyocytes (hPSC-CMs) and cardiac fibroblasts (cFBs) remain viable for up to 14 days inside RCPhC1-MA hydrogels while the effect of hydrogel stiffness on extracellular matrix production and hPSC-CM contractility can be monitored in real-time. Interestingly, whereas the beating behavior of the hPSC-CM monocultures is affected by environmental stiffness, this effect ceases when cFBs are present. Together, we demonstrate RCPhC1-MA to be a promising candidate to mimic and control the 3D biomechanical environment of cardiac cells.
Subject areas: Biomaterials, Cell biology, Materials in biotechnology, Stem cells research
Graphical abstract
Highlights
-
•
We present human RCPhC1-MA with tunable stiffness for 3D in vitro modeling
-
•
HPSC-cardiomyocytes and fibroblasts remain functional for 14 days in this matrix
-
•
The effect of stiffness on ECM production and contractility can be monitored live
-
•
The presence of fibroblasts modulates stiffness-dependent hPSC-CM contractility
Biomaterials; Cell biology; Materials in biotechnology; Stem cells research
Introduction
Physical cues from the cardiac microenvironment, such as extracellular matrix (ECM) stiffness and architecture, strongly determine cardiac cell behavior. Over the last century, animal models have been used abundantly to study cardiac development and (patho)physiology in vivo. However, the cardiovascular system differs between species, often resulting in poor clinical translation.1,2 Moreover, apart from ethical issues, the lack of microenvironmental control and manipulation in vivo makes animal models less suited to provide mechanistic insights at the cellular level.1,2 This prohibits the establishment of cause-effect relationships between environmental cues and cardiac cell behavior in a direct manner, necessitating the development of novel model systems.3,4
In vitro modeling of cardiac (patho)physiology is an emerging method to assess how physical (e.g., ECM stiffness and architecture) and chemical (e.g., composition and ligand density) features of the cardiac microenvironment dictate cell function.5,6,7,8 Especially human cardiac in vitro models can greatly advance our understanding of cardiac (patho)physiology and overcome some of the challenges present in the clinical translation of animal models.3 Two dimensional (2D) in vitro models of monocultures of human cardiac cells demonstrated that myocardial stiffness, progressing from values as low as 0.2-2 kPa9 during development toward a stiffness of 10-20 kPa9,10 in the healthy adult heart, is a crucial determinant of cardiac cell function.11,12,13 Cardiomyocytes (CMs), the force generating cells of the myocardium, detect ECM stiffness via their costameres and focal adhesion complexes at the cell membrane which, in turn, connect to the intracellular cytoskeleton.14 For human embryonic CMs, optimal cell contractility was found on substrates with a stiffness of the developing myocardium.9,15 In turn, the contractility of human CMs was decreased on matrices of pathological ECM stiffness,16 as found in the fibrotic myocardium (20-50 kPa).10
Three-dimensional (3D) models are evolving as well, but show significant variation in type, mechanical behavior, tunability, and reproducibility of matrix-mimicking materials.17,18,19 Moreover, both 2D and 3D models are often limited to contain only one cell type.20 In vivo, however, multiple cardiac cell types reside in a dynamic 3D environment where events such as development and injury alter the myocardial microenvironment, and with that, cell behavior over time.21 A controllable and reproducible 3D environment containing multiple cell types is therefore critical to advance our understanding of how developmental and (patho)physiological stiffness determine cell behavior in vivo.
Biopolymers derived from the ECM have great potential as biomaterials to assess the effect of environmental stiffness on cell behavior because of their intrinsic biocompatibility, along with their inherent biochemical, mechanical, and structural functions.19,22 Hydrogel systems made from these biopolymers are relevant in this realm because of their capacity to mimic the native ECM environment, thanks to their high water content and cytocompatibility, as well as tunable characteristics, including their mechanical properties.23,24,25 Collagen is the most abundant protein in the myocardial ECM, providing structural support and mediating multiple cellular functions,26,27,28 and is thus a promising candidate to mimic the cardiac microenvironment in vitro. To date, hydrogels based on collagen and gelatin, a collagen derivative, have been proposed to serve as a biomimetic microenvironment.29,30,31,32,33,34 However, it remains challenging to systematically tailor the properties of natural collagen hydrogels since these are highly dependent on various fabrication parameters, such as gelation pH and batch-to-batch variation.35 Moreover, these hydrogels are often based on animal-derived collagen, which is associated with immunogenicity, pathogen transmission, and suboptimal clinical translation.36,37
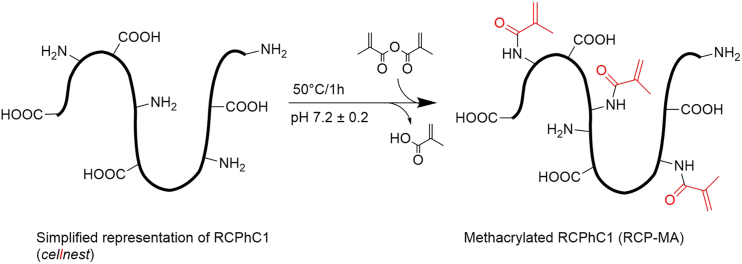
To address these challenges, in this study we introduce and evaluate a new hydrogel biomaterial, based on methacrylated human recombinant collagen peptide (RCPhC1-MA), as a 3D in vitro environment for human pluripotent stem cell-derived CM (hPSC-CMs) and cardiac fibroblast (cFB) (co-)culture. RCPhC1-MA is a human collagen 1 mimicking peptide that has been enriched with arginine-glycine-aspartic acid (RGD) sequences to promote cell adhesion and modified to contain methacrylate groups that enable chemical crosslinking to systematically tailor its mechanical properties (Figure 1). The production method and source material of RCPhC1-MA eliminate the issues of reproducibility and controllable tunability in 3D and increase the potential of clinical translation. Moreover, RCPhC1-MA is not recognized by collagen antibodies enabling the investigation of collagen production solely from the embedded cells. Our data demonstrate that RCPhC1-MA allows i) control of 3D hydrogel stiffness with values in the range of the developing and adult myocardium using photocrosslinking protocols and ii) real-time monitoring of cFB collagen production and hPSC-CM contractility in these hydrogels in response to altering hydrogel stiffness. Using these features, we show that increasing RCPhC1-MA stiffness to ∼10 kPa decreased the contractility of hPSC-CMs in monoculture after eight days of culture, but that contractility was preserved when hPSC-CMs were co-cultured with human fetal cFBs, demonstrating the importance of cardiac cellular crosstalk.
Figure 1.
Reaction scheme for RCPhC1 modification
Results
Characterization of RCPhC1-MA hydrogels
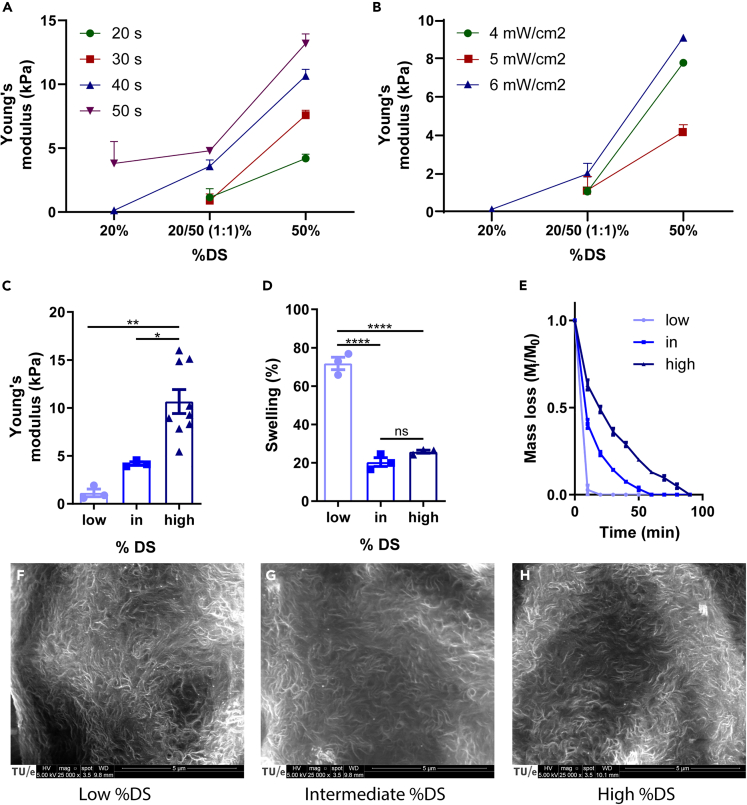
To verify that RCPhC1-MA can be used to obtain hydrogels with the human developmental (0.2-2 kPa9) and physiological (10-20 kPa9,10) cardiac stiffness found in vivo, stiffness characterization was performed on gels obtained via different %DS and photocrosslinking protocols (Figures 2A and 2B). It became evident that the photogelation of the 20% DS RCPhC1-MA required longer exposure times with higher exposure intensities for hydrogel formation. Three cell-free hydrogels were chosen containing: 1) a 1:1 mixture of 20% DS and 50% DS (“low %DS”), 2) 50% DS (“intermediate %DS”), and 3) 100% DS (“high %DS”), resulting in a Young’s modulus of 1.14 ± 0.69 kPa (n = 3), 4.20 ± 0.33 kPa (n = 3), and 10.66 ± 3.74 kPa (n = 9), respectively (Figure 2C). To assess hydrogel stability, the mass swelling percentage and proteolytic degradation rate were determined in hydrogels without cells. The hydrogels showed significantly higher swelling for the low %DS, which is suggested to facilitate nutrient diffusion,38 as compared to the intermediate and high %DS hydrogels (n = 3) (Figure 2D). In addition, the in vitro degradation assay demonstrated an increasing degradation rate with lower %DS (n = 3) (Figure 2E), suggesting that a looser hydrogel network of RCPhC1-MA is more easily degraded by cells as opposed to denser networks, a phenomenon also known for other hydrogels.38 Pore sizes of the RCPhC1-MA hydrogels were theoretically estimated using the rubber elasticity theory,39 showing a decreasing trend with increasing %DS (32.2 nm for low %DS, 25.9 nm for intermediate %DS, and 22.1 nm for high %DS). Moreover, the hydrogel nanostructure was visualized using scanning electron microscopy (SEM), demonstrating a highly nanoporous structure in all hydrogels (Figures 2F-2H).
Figure 2.
RCPhC1-MA material characterization
(A and B) Young’s modulus varies with different illumination time, dosage, and the degree of methacrylation (%DS, n = 3).
(C) Young’s modulus of RCPhC1-MA hydrogels upon varying %DS (n = 3-9).
(D) Mass swelling percentage differs with %DS (n = 3).
(E) Degradation of RCPhC1-MA hydrogels with various %DS in the presence of collagenase (n = 3).
(F-H) SEM of surface relief of RCPhC1-MA gels of varying %DS. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗∗ = p < 0.0001. Scale bar = 5 μm. All data are presented as mean ± SEM. For multiple group comparison, a one-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used.
RCPhC1-MA hydrogels with varying %DS are cytocompatible for human pluripotent stem cell-cardiomyocytes and cardiac fibroblasts
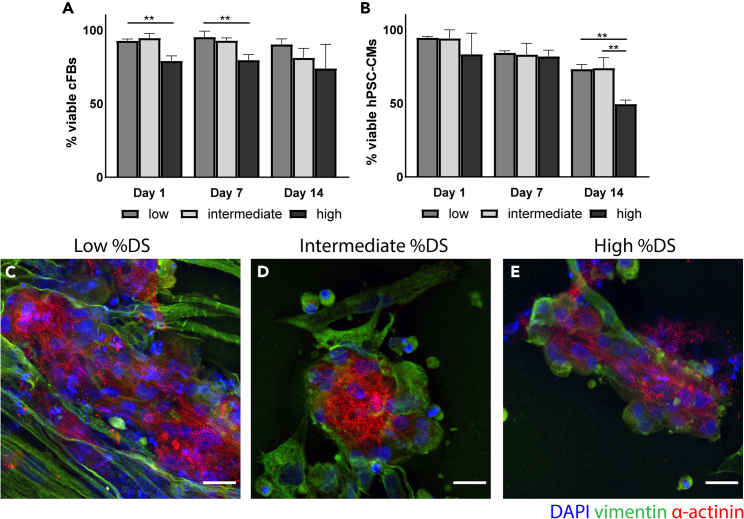
To investigate the cytocompatibility of the RCPhC1-MA constructs, we assessed the viability and morphology of the encapsulated hPSC-CMs and cFBs. To assess the viability of the individual cell types, cell viability assays were performed in monocultures (Figures 3A and 3B). cFB viability was significantly different between the low %DS and the high %DS on day 1 and day 7, showing a lower cell viability in the high %DS hydrogels. After 14 days of culture, cell viability remained high for all %DS hydrogels (90.3 ± 3.7% for low %DS; 81.1 ± 6.5% for intermediate %DS; 73.9 ± 16.45% for high %DS), although slightly lower in the hydrogels with higher %DS (n = 3). For the hPSC-CMs, a decreasing trend in cell viability was found over time and significantly lower viability was found for the high %DS on day 14 (49.50 ± 2.67%), as opposed to the intermediate %DS (73.95 ± 7.08%) and low %DS (73.29 ± 3.04%) (n = 3).
Figure 3.
Cytocompatibility of RCPhC1-MA
(A and B) Quantification of cFB (A) and hPSC-CM (B) viability in 3D culture inside RCPhC1-MA hydrogels with varying %DS over 1, 7, and 14 days (n = 3).
(C-E) hPSC-CMs and cFBs (70:30) cultured in low %DS and intermediate %DS exhibited a clustered morphology, as opposed to the more spread-out morphology in the high %DS. Notable organized sarcomeric α-actinin is found in all constructs after 7 days of encapsulation. ∗∗ = p < 0.01. Scale bar = 20 μm. All data are presented as mean ± SEM. For multiple group comparison, a one-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used.
For the identification of hPSC-CMs, we evaluated the sarcomeric reporter protein α-actinin-mRubyII and for cFB identification, we used the intermediate filament protein vimentin (Figures 3C-3E). After seven days of encapsulation, the hPSC-CMs in co-culture with cFBs in a 70:30 ratio (since CMs occupy ∼70% of the volume of healthy adult myocardium40,41) demonstrated organized sarcomeric structures in all %DS. In the low %DS and intermediate %DS, the hPSC-CMs are clustered and cell elongation, which is typical for adult CMs,42 was not observed. In the high %DS, elongated individual hPSC-CMs were observed. In all %DS RCPhC1-MA constructs, the cFBs were observed in close contact with the hPSC-CMs.
Together, the overall morphology and high cell viability of the cFBs and hPSC-CMs up to 14 days inside the RCPhC1-MA hydrogels demonstrate it provides a beneficial in vitro environment to culture cardiac cells and investigate their response to stiffness change.
Cardiac fibroblasts produce collagen in RCPhC1-MA of varying stiffness
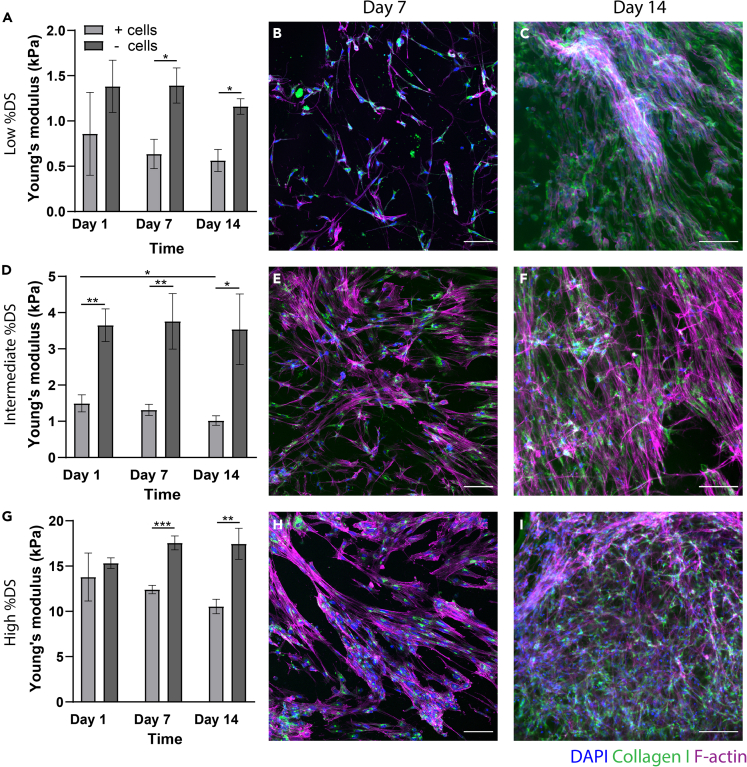
To gain deeper insight into the ECM production of cFBs inside RCPhC1-MA hydrogels of different stiffnesses, overall hydrogel stiffness together with ECM production was determined over time. Cell-free hydrogels were used as controls. The stiffness of the cell-free hydrogels remained constant with time (Figures 4A, 4D, and 4G). Interestingly, however, the stiffness of the cell-laden hydrogels was significantly lower than their cell-free counterparts as of day 1, suggesting that cell encapsulation may have altered the initial network formation of the gel. This is also observed in explanted tissues, where whole tissues (composites of cells and ECM) are reported as softer than decellularized ECM.43
Figure 4.
Collagen production and hydrogel stiffness with time over 14-day culture period
(A, D, and G) Micro-indentation data depicting hydrogel stiffness over time in cell-free (- cells) and cell-laden (+cells) hydrogel constructs (n = 3 per condition). ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001. All data are presented as mean ± SEM. For multiple group comparison, a one-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used. (B, C, E, F, H, I) Representative fluorescent images demonstrating collagen production (green, CNA-35-OG) and F-actin (magenta, phalloidin) in the RCPhC1-MA constructs with time. Scale bar = 100 μm.
Whereas no collagen was observed in the hydrogels one day after encapsulation (data not shown), the cFBs in the low %DS (0.64 ± 0.11 kPa), intermediate %DS (1.32 ± 0.15 kPa), and high %DS (12.4 ± 0.47 kPa) showed intracellular collagen production after seven and fourteen days of culture (Figures 4B, 4E, and 4H). While no actin stress fibers were observed in the cFBs in the low %DS, these were present in both intermediate and high %DS. After 14 days of culture inside the hydrogels, an increase in cFB cell number was observed and networks of cFBs were formed (Figures 4C, 4F, and 4I). Collagen was merely detected within the cell and no extracellular collagen was observed.
Together, these data suggest that cFBs can synthesize collagen in RCPhC1-MA. Moreover, we demonstrate that the newly produced collagen can be visualized live over time in RCPhC1-MA using the CNA-35-OG probe.
Cell contractility can be monitored live over time in RCPhC1-MA hydrogels and is dependent on culture time and hydrogel stiffness
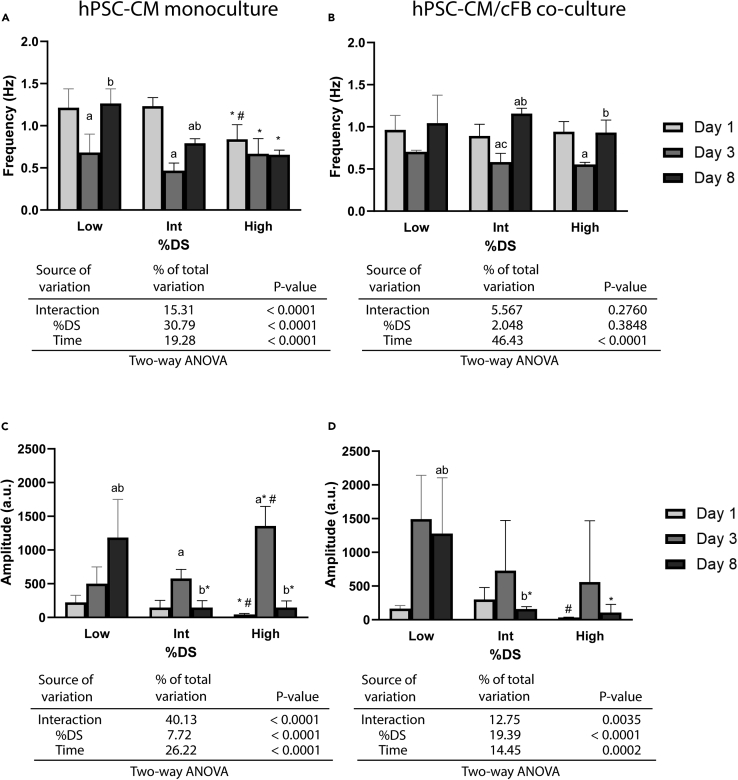
Next, to prove that the RCPhC1-MA hydrogels can be employed for functional studies with hPSC-CMs, contractility measurements were performed using the image-based software tool MUSCLEMOTION.44 Whereas conventional electrophysiological measurement techniques often require customized hardware and advanced informatics, leading to reproducibility issues between laboratories and culture models, MUSCLEMOTION enables reproducible quantitative analysis of cardiac contraction, and was validated against the current golden standards such as patch-clamp analyses.44,45 In assessing the effect of %DS on hPSC-CM contractility, we found a significant decrease in beating frequency with increasing %DS for hPSC-CM monocultures after eight days of culture inside the RCPhC1-MA hydrogels as opposed to one day of culture (Figure 5A). A similar effect was observed for the contraction amplitude (Figure 5C) after eight days of culture. Interestingly, both contraction frequency and amplitude after three days of encapsulation deviated significantly from the values obtained after one and eight days of encapsulation, and contraction frequency and amplitude seem to show opposing trends. It is hypothesized that varying cell-ECM interactions and differences in hPSC-CM maturity differ between one, three, and eight days of encapsulation, possibly contributing to the deviating contraction frequency and amplitude after three days of encapsulation. However, future studies are necessary to explore this further. Videos of isolated beating hPSC-CM clusters after eight days of encapsulations in the different %DS RCPhC1-MA hydrogels can be found in the Videos S1, S2, and S3.
Figure 5.
CM contractility is affected by both culture time and hydrogel stiffness
(A and B) Bar graphs indicating the effect of culture time and %DS on hPSC-CM beating frequency in monocultures (A) and hPSC-CM/cFB co-cultures (B), accompanied by a summary of the two-way Analysis of Variance (ANOVA) results to analyze interaction effects.
(C and D) Bar graphs indicating the effect of culture time and %DS on hPSC-CM contraction amplitude (a.u.) in monocultures (C) and hPSC-CM/cFB co-cultures (D), accompanied by a summary of the two-way ANOVA results to analyze interaction effects. a, significance vs. day 1 group; b, significance vs. day 3 group; c, significance vs. day 8 group; ∗, significance vs. low %DS within each time period; #, significance vs. intermediate %DS within each time period. Significant differences between timepoints and between %DS were determined using one-way ANOVA (n = 3-15 cell clusters analyzed per condition). p < 0.05. All data are presented as mean ± SEM. Moreover, two-way ANOVA was performed to assess the interaction effects of culture time and %DS on hPSC-CM contraction frequency and amplitude. See also Video S1: Encapsulated hPSC-CMs in the low %DS hydrogels after eight days of culture. Related to Figure 5, Video S2: Encapsulated hPSC-CMs in the intermediate %DS hydrogels after eight days of culture. Related to Figure 5, Video S3: Encapsulated hPSC-CMs in the high %DS hydrogels after eight days of culture. Related to Figure 5.
Since hPSC-CMs in vivo co-exist with non-myocytes, such as cFBs, we next co-cultured the hPSC-CMs with cFBs in a 70:30 ratio and assessed how the presence of cFBs altered the contractility of hPSC-CMs in response to varying environmental stiffness. Whereas a strong %DS-dependent effect was found in the monocultures, no %DS-dependent effect on beating frequency was observed in the co-cultures (Figure 5B). Interestingly, significantly higher contraction frequencies were found in the co-culture after eight days of culture inside the intermediate %DS (p < 0.0001) and high% DS (p < 0.0027) RCPhC1-MA hydrogels as opposed to the monoculture (Figures 5A and 5B). Similar to the monocultures, a decreasing trend was observed with increasing %DS for contraction amplitude, although less pronounced and merely significant for the high % DS (Figure 5D). Moreover, while for both contraction frequency and amplitude a similar time-dependent effect was observed in the co-cultures as opposed to the hPSC-CMs monocultures, demonstrating a significant decrease in contract frequency and increase in contraction amplitude after three days of encapsulation, this effect seemed less pronounced (Figures 5B and 5D). Together, these results suggest that the decrease in beating frequency with increasing %DS, found for the hPSC-CM monocultures, was not reflected in the encapsulated co-cultures. This can also be deduced from the two-way ANOVA tables below Figures 5A-5D, indicating a significant %DS-dependent effect for hPSC-CM monocultures on beating frequency, which is not the case for the co-cultures.
Discussion
Physical and mechanical properties of the cellular environment provide key cues to shape cell fate and behavior. In the search for a controllable 3D environment for cardiac cells, we have investigated the suitability of RCPhC1-MA, a highly reproducible human-derived collagen ECM mimic, as an alternative to animal-derived or insufficiently characterized and low reproducibility biomaterials for 3D in vitro modeling. We demonstrated that RCPh1C-MA can serve as a valuable tool providing a human biomimetic cardiac cell environment that can be systematically altered in stiffness. Moreover, we showed the applicability of RCPhC1-MA to monitor cellular contractility and ECM production in real-time.
RCPh1C was modified with MA groups, allowing hydrogel fabrication and control over the mechanical properties, such as swelling, degradation, and Young’s modulus. Our results indicated that RCPh1C-MA with varying degrees of modification can be successfully crosslinked under UV irradiation and form stable 3D hydrogels with mechanical properties in the range of human myocardial tissue from developmental to physiological adult stage (E = 0.2-15 kPa).9,15,46,47 Because of the addition of the crosslinkable MA moieties, RCPhC1-MA hydrogels can reach stiffnesses that are difficult to achieve with conventional collagen gels.48,49 Specifically, a 100% DS RCPhC1-MA hydrogel under optimal conditions (i.e., shielded from oxygen during UV-exposure) yields a Young’s modulus of >100 kPa (Figure S3). Although this stiffness data is obtained via rheology measurements and is not directly comparable to the indentation results presented in the present study, these data suggest that RCPhC1-MA can be an innovative and mechanically more tunable 3D hydrogel material alternative to conventional collagen gels, which have low mechanical properties.35 Moreover, by altering hydrogel stiffness solely through varying the %DS, RCPhC1-MA enables the possibility to alter hydrogel stiffness independently of cell adhesive ligand concentration. This possibility of uncoupling of stiffness and ligand density is highly desirable for mechanobiological studies50,51 and can greatly benefit our understanding of the effect of either stiffness alteration or ligand density on cell behavior.
The assessment of functional readouts is essential to understand cell and tissue behavior in vitro. Our data showed that the hPSC-CMs and cFBs survived the encapsulation process and showed viability up to 14 days in culture. Comparable viability of hPSC-CMs and human fetal cFBs was found in methacrylated gelatin (GelMA) hydrogels52,53 and poly(N-isopropylacrylamide) PNIPAAm-gelatin based hydrogels,54 indicating a similar viability of cardiac cells for RCPhC1-MA and gelatin derived hydrogels based on animal material.
Since RCPhC1-MA lacks the triple helical structure found in collagen fibers, it is not recognized by collagen antibodies used in immunohistochemistry or by live collagen probes, such as CNA, allowing the visualization of merely cell-produced collagen in collagenous environments in real-time without the use of cell fixation and biochemical assays. Making use of this advantage, we were able to track collagen production from human cells live in a human collagen mimicking in vitro environment. We visualized cFB-produced collagen on days 7 and 14 after encapsulation, demonstrating the ability of the cFBs to interact with the RCPhC1-MA environment and alter its composition. Whereas collagen gene upregulation in response to increased environmental stiffness is well documented, much less is known about its synthesis and deposition.55,56 Here, we showed an increase in collagen synthesis with time, although no significant upregulation in collagen synthesis with increasing hydrogel stiffness was observed. Since collagen production is increased in myofibroblasts, the lack of an upregulation in collagen synthesis might be explained by the fact that the hydrogel stiffnesses used in this study do not include the stiffness found in fibrotic myocardium (30-70 kPa), which is known to activate cFBs into matrix-producing myofibroblasts.57 Moreover, the lack of extracellular collagen is likely caused by the absence of ascorbic acid in the culture medium, which is necessary for collagen formation.58 Lastly, the cell-laden hydrogels tended to decrease in stiffness over time and it is hypothesized that this is the result of hydrogel remodeling via cell-produced proteinases, more so since RCPhC1-MA is susceptible to proteolytic degradation while newly cFB-produced collagen is expected to be softer than RCPhC1-MA.49
As a marker of cardiac functionality, we quantified contractility by recording phase contrast time-lapse images of monocultures of hPSC-CMs and in co-culture with cFBs. We demonstrated that hPSC-CMs maintained their contractile phenotype up to eight days in culture and adapted their contractility in response to varying RCPhC1-MA stiffness. Furthermore, our results suggested that the presence of cFBs compensates for the decrease in hPSC-CM contractility with increasing environmental stiffness, implying that they are essential to maintain hPSC-CM contractility. These results are in line with in vivo data, where a strong crosstalk between cFBs and CMs is essential to maintain the electrical stability of the cardiac muscle.59 Together, we demonstrated that RCPhC1-MA is suitable for functional studies with hPSC-CMs and cFBs using image analysis-based techniques.
Overall, we have shown that the RCPh1C-MA provides a favorable in vitro environment for culture and controlled manipulation of human cardiac cells. It provides a versatile matrix for cardiac tissue engineering applications and allows for the fabrication of 3D models of (patho)physiological processes in the human myocardium.
Limitations of the study
The presented RCPhC1-MA material and study design also have limitations. Whereas natural collagen gels contain fibers, allowing the compaction of the material, RCPhC1-MA does not form the fibrous structures necessary for material compaction. Therefore, it is challenging to use RCPhC1-MA to build engineered heart tissues (EHTs), the formation of which relies on tissue contraction around (micro)pillars.60,61,62 The lack of material compaction also precludes alignment and conditioning of the cardiac cells, which would better mimic the in vivo environment. Another consequence is that the beating of the hPSC-CMs is not synchronized across the entire gel but is localized in individual clusters. Whereas the applicability of MUSCLEMOTION is extensively validated by comparing its results to golden standard electrophysiological measuring methods, it would still be wise to add electrophysiological measurements instead of solely relying on image analysis-based approaches. Furthermore, whereas the present study is mainly focused on cardiac mechanobiological aspects, future studies should include full cell biological analysis to verify the applicability of the RCPhC1-MA platform to study more cell biological-related characteristics (e.g., growth profiles of the hPSC-CMs and cFBs and their electrophysiological coupling via gap junctions).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Vimentin | Abcam | Ab20346; RRID:AB_445527 |
| Goat-a-mouse IgM AF488 | Molecular Probes | A21042; RRID:AB_2535711 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant collagen polypeptide (RCPhC1) | Fujifilm Manufacturing Europe BV | Irvine Scientific, Catalog ID: 1063967 |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate | Sigma-Aldrich | 900889 |
| Collagenase P | Roche | 11249002001 |
| Propidium Iodide | Invitrogen | P1304MP |
| SYTOX Deep Red Nucleic Acid Stain | Invitrogen | S34859 |
| CNA-35-OG | Krahn, K.N., Bouten, C.V.C., Van Tuijl, S., Van Zandvoort, M.A.M.J., and Merkx, M. (2006). Fluorescently labeled collagen binding proteins allow specific visualization of collagen in tissues and live cell culture. Anal. Biochem. 350, 177–185. 10.1016/J.AB.2006.01.01363 | N.A. |
| Methacrylic anhydride | Sigma-Aldrich | 276685 |
| 2,4,6-Trinitrobenzenesulfonic acid solution, TNBS | Sigma-Aldrich | P2297 |
| Deposited data | ||
| Mechanical characterization | Mendeley data | https://doi.org/10.17632/nvkd84dzdf.2 |
| Cell viability data | Mendeley data | https://doi.org/10.17632/nvkd84dzdf.2 |
| Contractility data | Mendeley data | https://doi.org/10.17632/nvkd84dzdf.2 |
| Experimental models: Cell lines | ||
| DRRAGN human pluripotent stem cell line 3F4 | Ribeiro M.C., Slaats R.H., Schwach V., Rivera-Arbelaez J.M., Tertoolen L.G.J., van Meer B.J., Molenaar R., Mummery C.L., Claessens M.M.A.E., Passier R (2020). A cardiomyocyte show of force: A fluorescent alpha-actinin reporter line sheds light on human cardiomyocyte contractility versus substrate stiffness, J. Mol. Cell. Cardiol. 141, 54–64. 10.1016/j.yjmcc.2020.03.00811 | N.A. |
| Human epicardial-derived fetal cardiac fibroblasts | Dronkers E., Moerkamp A.T., van Herwaarden T., Goumans M.J., Smits A.M. (2018). The Isolation and Culture of Primary Epicardial Cells Derived from Human Adult and Fetal Heart Specimens. J Vis Exp. 2018, 57370. https://doi.org/10.3791/57370.64 | N.A. |
| Software and algorithms | ||
| MUSCLEMOTION | Sala, L., Van Meer, B.J., Tertoolen, L.G.J., Bakkers, J., Bellin, M., Davis, R.P., Denning, C., Dieben, M.A.E., Eschenhagen, T., Giacomelli, E., et al. (2018). Musclemotion: A versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. 122, e5–e16. 10.1161/CIRCRESAHA.117.312067.44 | N.A. |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, prof. dr. Carlijn V.C. Bouten (c.v.c.bouten@tue.nl).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cardiac fibroblasts
Human epicardial-derived cardiac fibroblasts (cFBs) were isolated from human fetal cardiac tissue collected with informed consent and anonymously from elective abortion material of fetuses with a gestational age between 10 and 20 weeks according to the official guidelines of the Leiden University Medical Center, as approved by the local Medical Ethics Committee (number P08.087).64 cFBs were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Invitrogen, Breda, the Netherlands) supplemented with 10% fetal bovine serum (Greiner Bio-one) and 1% Penicillin-Streptomycin (fibroblast culture medium). The cFBs were cultured in flasks coated with 0.1% gelatin from porcine skin in PBS (Sigma-Aldrich) and passaged when 80% confluency was reached. The passage number of the cFBs during experimental procedures was between 7 and 12.
Human pluripotent stem cell-derived cardiomyocytes
The DRRAGN hPSC (derived from human embryonic stem cell line HES3) line 3F4, modified with a double reporter of GFP-NKX2.5 and mRubyIIACTN2 was cultured on vitronectin-coated culture plates and maintained by refreshing Essential 8 (E8) medium (Thermo Fisher, Waltham, Massachusetts, United States) daily and passaging the cells at around 15k cells/cm2 twice a week. The hPSCs were differentiated into cardiomyocytes (hPSC-CM) according to protocol described before.65 On day 13 of differentiation, the cells were dissociated and re-plated on glucose-free matrigel coatings and cultured in glucose-free medium (TDI-Lactate, Table S1) for four days for purification of the hPSC-CM culture. On day 16 of differentiation, the medium was replaced with TDI medium supplemented with growth factors (T3, Dexamethasone & IGF-1, Table S1) for hPSC-CM maturation. On day 20 the hPSC-CMs were used for experiments or cryopreserved for later use.
Method details
Materials
Recombinant collagen polypeptide RCPhC1, commercially available as Cellnest, was a gift from Fujifilm Manufacturing Europe BV (Tilburg, the Netherlands). The protein sequence of human type I collagen alpha 1 chain from 716 to 779 (GenBank P02452) was used to design RCPhC1.37 All chemicals were obtained from Sigma-Aldrich (Zwijndrecht, the Netherlands) unless noted otherwise.
Methacrylation of RCPhC1
RCPhC1 was functionalized with methacrylamide moieties using an optimized protocol, adapted from Sutter et al.66 and Van Den Bulcke et al.67 Karl-Fischer analysis was used to correct “dry” RCPhC1 weights for initial moisture content (mc) to obtain the true dry weight.68,69 For RCPhC1 functionalization, a 10 w/w% solution was prepared by dissolving 20.0 g (g) RCPhC1 freeze-dried sponge (0.666 mmol amines per g dry RCPhC1, including N-terminal amines) in 180 g phosphate buffered saline (PBS) solution. The appropriate amount of methacrylic anhydride was added to the RCPhC1 solution at 50°C and stirred for 1 h. During this time, the solution pH was continuously monitored and maintained between 7 and 7.4 by addition of 1 M sodium hydroxide solution. At reaction end, the solution was diluted to 1 L volume using highly purified water, sterile filtered using a 0.2 μm PES filter membrane and dialyzed for 2 days at 25°C against a 14 kDa MWCO cellulose membrane (Sigma-Aldrich). The product solution was then freeze-dried at −15°C and stored at −20°C until use. An amount of 1.174 mmol methacrylic anhydride (MAnh) was required to completely functionalize 1 mmol amines, corresponding to 117.6 μL MAnh per g dry (moisture-free) RCPhC1. Proportionally reduced amounts of reagent were used to functionalize RCPhC1 with different degrees of substitution (%DS).
NMR spectroscopy
Lyophilized samples were dissolved in deuterated water to a concentration of 15–30 mg/mL and measured at 25°C in a Bruker Avance III HD Nanobay 400 MHz spectrometer. The vinylic protons in the coupled methacrylamide moieties were used to quantify the level of substitution using published random-coil polypeptide chemical shifts.70 The peaks due to the vinylic protons of bound methacrylamide moieties appeared at chemical shift values between 5.4 and 6.5 ppm and were well separated from the other peaks (Figure S1). The known RCPhC1 composition with 571 amino acid residues was then used to derive the %DS. The combined integral for the valine (Val), isoleucine (Ile), and leucine (Leu) methylic protons can be normalized to 288H as internal reference, since all combined methylic protons of Val, Ile, and Leu make up a total of 288 protons per molecule. The quantification of attached methacrylamide moieties is then given by the following equation:
| (Equation 1) |
where Ha and Hb represent the vinylic protons in methacrylamide moieties bound to either lysine or terminal glycine residues. These make a maximum of 68 vinylic protons (33 Lys plus a terminal amine equals 34 methacrylamides with each 2 vinylic protons) per molecule. and represent integrals, calculated by the NMR software. The integral limits are noted as subscripts.
Trinitrobenzene sulphonic acid assay
A trinitrobenzene sulphonic acid assay (TNBS)71 was used as an independent method to evaluate the modification levels of RCPhC1-MA. The residual lysine residues were labeled by dissolving 10 mg dry sample in 2 mL solution containing 1% TNBS and 4% sodium bicarbonate. The mixture was kept at 37°C for 3 h. Subsequently, the samples were acidified with 6 N hydrochloric acid and diluted 40 times to record the extinction at 345 nm using a double-beam CARY100 spectrophotometer (VARIAN) using 10 mm quartz-cuvettes and a value of 1.48 × 104 M−1cm−1 for the ε-TNP-Lys molar extinction coefficient. The DS-level was then determined by considering the RCPhC1-MA DS-dependent molecular weight increase and converting the number of residual amines into the modification level. The sample moisture content was determined to correct weighed quantities of RCPhC1 and RCPhC1-MA.
Gel permeation chromatography
Samples were dissolved at a concentration of 1 mg/mL in highly purified water with 1% sodium dodecyl sulphate added at a pH of 5.3. Measurements were done using a Waters 2695 Alliance instrument with TSK SuperSW3000 and TSK SuperSW2000 columns and a Waters 2489 UV/VIS detector (Figure S2).
RCPhC1-MA hydrogel fabrication
Lyophilized RCPhC1-MA was stored at −30°C until further use. Hydrogels were formed by radical crosslinking of solubilized RCPhC1-MA in the presence of Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, Sigma-Aldrich, 20 mg/mL in PBS) (Figure 1). In short, lyophilized RCPhC1-MA was brought to room temperature (RT) and dissolved in PBS (10 w/v%). The gel mixture was placed on a roller bank for 15 min at RT to allow it to dissolve completely. Right before crosslinking, LAP was added to the gel mixture to achieve a working concentration of 1 mg/mL. The pre-polymer solution was sterilized before hydrogel fabrication. When used for cell encapsulation, pelleted cells were resuspended in the pre-polymer mixture to a final concentration of 1 million cells/mL. The total gel volume was corrected for the volume of the cell pellet. 50 μL of pre-polymer solution was pipetted on hydrophobic polydimethylsiloxane (PDMS, Sylgard 184; Dow Corning, Michigan, United States) substrates. The PDMS substrates were obtained by pouring PDMS with curing agent (10:1) in a square mold (2 × 2 cm) in a Petri dish and curing the constructs for 120 min at 65°C. The RCPhC1-MA pre-polymer solution was crosslinked by exposure to UV-light (wavelength 365 nm) of 5 mW/cm2 for 30 s, as calibrated with an R2000 UV Radiometer. The hydrogel was removed from the PDMS surface, immersed in culture medium in non-adhesive well plates and incubated at 37°C. Cell-free hydrogels were fabricated for physicochemical characterization and as controls.
Hydrogel physicochemical characterization
Micro-indentation
Hydrogel stiffness was probed in PBS at 37°C with using a CellScale microtester (CellScale, ON, Canada). Tungsten beams with a nominal diameter (BD) of 0.20 mm, modulus (BM) of 4.11 × 105 MPa, and length (BL) of 50 mm with a stainless-steel sphere of 500 μm in diameter attached at its end were used for the indentations. The vertical displacement (z) of the beam was controlled in micrometer resolution by means of an electromotor. The microtester is equipped with two cameras to measure the vertical deflection of the beam (d). The force (F) applied to the hydrogel during indentation was computed by:
| (Equation 2) |
where BIcorresponds to the beam area moment of inertia, which for a cylindrical beam is given by:
| (Equation 3) |
The indentation depth (δ) can be calculated by:
| (Equation 4) |
where d0 is the deflection offset and z0 the beam displacement at the contact point between the tip and the sample. F-δ curves recorded were analyzed with the Hertz contact mechanical model for a sphere indenting a semi-infinite half-space:
| (Equation 5) |
whereR is the sphere radius of the tip and ν the Poisson’s ratio of the sample (assumed to be 0.5). The Young’s modulus of the sample (E) was computed for each F-δ curve by non-linear least-squares fitting using a custom-built code (MATLAB, The MathWorks Inc., MA). The fitting was performed for the loading curve. Measurements were performed in acellular and cell-laden hydrogels for different timepoints (24 h, 7 days, and 14 days after fabrication) and UV doses. In each hydrogel, 5 F-δ curves were recorded at 0.025 Hz and a loading rate of ∼75 μm/s.
Swelling and degradation assays
To analyze hydrogel swelling, the cell-free hydrogels were weighed directly after fabrication (M0) and after reaching equilibrium (24 h, MI) at 37°C and 80–85% humidity (cell culture incubator). A swelling percentage (S) could then be calculated using S = (MI – M0)/M0 ∗ 100. To examine the degradation rate, a collagenase-mediated degradation test was performed. Three cell-free hydrogels per %DS were immersed in collagenase P solution (Roche, 0.5 mg/mL) at RT on a shaker plate. Weight of the hydrogels was measured at different time points (0, 10, 20, 30, 40, 50 min, etc.) until complete degradation.
Morphological analysis of hydrogel microstructure using scanning electron microscopy
To determine the hydrogel micro- and nanostructure, cell-free RCPhC1-MA hydrogels were fixed 24 h after fabrication in 2.5% glutaraldehyde solution in PBS at room temperature. The fixed hydrogels washed twice with MilliQ and dehydrated in a graded ethanol series (50, 60, 70, 80, 90 and 95% ethanol in water) for 10 min each, and ethanol in hexamethyldisilane (HMDS) (30, 50, 60, 100% HMDS in ethanol) for 15 min each. The samples were visualized using high vacuum and an electron density of 5 kV with spot 4 at 100x magnification and 25.000× magnification by a scanning electron microscope (SEM, Quanta 600F, GEI, Eindhoven).
Analysis of the hydrogel macromolecular network using rubber elasticity theory
A hydrogel macromolecular network can be described by rubber elasticity theory to yield the average chain molecular weight between junctions (cross-links) (g/mol) The relation between and Youngs’s modulus E is given by Equation 6, adapted from Žigon-Branc et al.,39 using , where μ is the Poisson number, 0.5 for ideal rubbers.
| (Equation 6) |
in which E is Young’s modulus (Pa, or Nm−2), c is the dry polymer concentration (g·m−3), R is the gas constant (J/(mol∗K)), T the absolute temperature (K), equals the polymer number-average molecular weight before crosslinking (g/mol), and Q is the volumetric swelling ratio. The value of Q is given by , with ρRCPMA 1.35 g/cm3 for the solid RCPhC1-MA density,72 1 g/cm3 for the water density and q being the unitless mass swelling ratio. After solving Equation 6 for , the mesh size ξ is calculated using the Canal-Peppas equation Equation 7 73, using a factor 3 instead of 2 in the numerator of the first term.74
| (Equation 7) |
in which l (1.44 Å) is the arithmetic average of single bond lengths in the polymer backbone, Cn is the Flory characteristic ratio 8.26 for gelatin,74 Mr (89.6 g/mol) is the average RCPhC1-MA residue unit weight, calculated from the amino acid sequence.
Cell viability
To assess the viability of monocultures of cFBs and hPSC-CMs in RCPhC1-MA hydrogels, the hydrogels were carefully washed with PBS and incubated with 1 μg/mL calcein AM and 750 nM propidium iodide (Invitrogen) (cFBs), and with 1 μg/mL calcein AM and 1 μM of SYTOX Deep Red Nucleic Acid Stain (hPSC-CMs), for 20 min at 37°C protected from light. The SYTOX Deep Red Nucleic Acid Stain replaced the propidium iodide used in the cFB viability assay because the hPSC-CMs express the α-actinin reporter in the red channel. After incubation, the hydrogels were washed with PBS and directly imaged with an inverted microscope (Leica DMi8, Leica, Mannheim, Germany) using the 10x/0.32 objective. Images were taken at three random representative spots per condition and hydrogels treated with ethanol were used as negative control. The viability was analyzed on day 1, 7 and 14 after seeding.
Contractility measurements
The automated open-source software package MUSCLEMOTION was used for all contraction analysis.44 The contraction amplitude (in arbitrary unit, a.u) is an optical measure of contraction that is widely used for cardiac in vitro studies44 and correlates with the force of contraction, as has been validated with single adult CMs, iPSC-CMs, cardiac spheroids, and engineered heart tissues.44 The principle underlying this software is based on an algorithm that quantifies absolute changes in pixel intensity between a reference frame and the frame of interest. To ensure consistency between recordings, a brightness and contrast check was performed to standardize the mean gray value across all images, using ImageJ. To avoid calculation artifacts, a field-of-view (FOV) of 260 × 260 μm was used, containing one cell cluster per FOV. Three to five FOVs were quantified per experimental group for three independent experiments (n = 3). Contractions were computed in the cell-laden hydrogels at day 1, 3 and 8 after cell encapsulation.
Immunofluorescence staining
The viable fluorescent probe CNA-35-OG63 was used to visualize cell-produced collagen in the RCPhC1-MA gels at day 1, 7, and 14. In brief, the CNA-35-OG probe was diluted in culture medium (1:100) and added to the hydrogels with cFBs, the ECM supporting cells of the myocardium. After 20 min of incubation at 37°C, the medium was refreshed, and samples were imaged immediately. For immunofluorescence staining, the cell-laden gels were washed with PBS trice before and after fixation, and fixed in 3.7% formaldehyde (Merck, Darmstadt, Germany) for 20 min. The gels were permeabilized with 0.5% Triton X-100 (Merck) in PBS for 10 min and blocked for non-specific antibody binding with 10% horse serum (Sigma-Aldrich) in PBS for 40 min. Next, the cell-laden gels were incubated overnight with the primary antibodies at 4°C. The gels were washed 6 times with PBS for 5 min before incubating with the secondary antibodies in PBS and phalloidin for 1.5 h. All used primary and secondary antibodies and dyes are listed in Table S2. The gels were washed twice with PBS before incubating with DAPI in PBS for 5 min to visualize the cell nuclei. Finally, the cell-laden gels were washed 4 times with PBS for 5 min and stored protected from light at 4°C. Before microscopic imaging, the hydrogels were placed between two microscopic slides to flatten the gel for improved visualization. Maximum intensity projections were created from z-stacks of 7.5–10 μm (5 stacks per projection). All immunofluorescent samples were analyzed with a confocal fluorescence microscope (Leica SP8X, Leica, Mannheim, Germany), using a 20x/0.75 objective.
Quantification and statistical analysis
All data are presented as mean ± SEM and three independent experiments were performed using three replicates per condition, unless indicated otherwise. For two group comparisons, a Student’s t test was used at 95% confidence interval after normality and equal variances were checked with Shapiro-Wilk test and Brown-Forsythe test, respectively. For comparison between multiple groups, a one-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used after normality and equal variances were checked with Shapiro-Wilk test and Brown-Forsythe test, respectively. two-way ANOVA was performed to assess the interaction effects of culture time and %DS on hPSC-CM contraction frequency and amplitude after heteroscedasticity and normality of residuals were checked with Spearman’s test and Anderson-Darling test, respectively. For contraction amplitude, the assumptions of heteroscedasticity and normality of residuals were met when a log-transformation was performed on the original data. For all comparisons, p < 0.05 was considered statistically significant.
Acknowledgments
This research was financially supported by the Gravitation Program “Materials Driven Regeneration,” funded by the Netherlands Organization for Scientific Research (024.003.013). N. A. Kurniawan and C.V.C. Bouten acknowledge financial support from the European Research Council (NAK: 851960; CVCB: 101054726). I. Jorba acknowledges financial support from the Dutch Research Council (OCENW.XS21.4.146). M. Vissers (Multiscale lab, Eindhoven University of Technology) is gratefully acknowledged for his help in performing the SEM imaging. Milan van Wezel from the Institute for Complex Molecular Systems Animation Studio is kindly acknowledged for his help in creating the graphical abstract.
Author contributions
Conceptualization, D.M., I.J., and L.K.; methodology and investigation, D.M., I.J., H.B., and B.G.; writing – original draft, D.M. and I.J.; writing – review and editing, D.M., I.J., L.K., N.A.K., and C.V.C.B.; funding acquisition, I.J., N.A.K., and C.V.C.B.; resources, R.P., H.B., and M.G; supervision, N.A.K. and C.V.C.B.
Declaration of interests
H.A. van Boxtel is an employee of Fujifilm Manufacturing Europe B.V. The results of this study were not influenced by him or any other employee of Fujifilm Manufacturing Europe B.V.
Published: March 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106423.
Supplemental information
Data and code availability
-
•
All data generated in this study have been deposited at Mendeley data and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Milani-Nejad N., Janssen P.M.L. Small and large animal models in cardiac contraction research: advantages and disadvantages. Pharmacol. Ther. 2014;141:235–249. doi: 10.1016/J.PHARMTHERA.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusunose K., Penn M.S., Zhang Y., Cheng Y., Thomas J.D., Marwick T.H., Popović Z.B. How similar are the mice to men? Between-species comparison of left ventricular mechanics using strain imaging. PLoS One. 2012;7:e40061. doi: 10.1371/JOURNAL.PONE.0040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savoji H., Mohammadi M.H., Rafatian N., Toroghi M.K., Wang E.Y., Zhao Y., Korolj A., Ahadian S., Radisic M. Cardiovascular disease models: a game changing paradigm in drug discovery and screening. Biomaterials. 2019;198:3–26. doi: 10.1016/J.BIOMATERIALS.2018.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuppinger C. 3D cardiac cell culture: a critical Review of current technologies and applications. Front. Cardiovasc. Med. 2019;6:87. doi: 10.3389/fcvm.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaghan N.I., Hadipour-Lakmehsari S., Lee S.H., Gramolini A.O., Simmons C.A. Modeling cardiac complexity: advancements in myocardial models and analyticaltechniques for physiological investigation and therapeutic development invitro. APL Bioeng. 2019;3 doi: 10.1063/1.5055873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorba I., Mostert D., Hermans L.H.L., Van Der Pol A., Kurniawan N.A., Bouten C.V.C. In Vitro methods to model cardiac mechanobiology in health and disease. Tissue Eng. Part C Methods. 2021;27:139–151. doi: 10.1089/ten.tec.2020.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra A., Tabdanov E., Patel H., Janmey P.A., Kresh J.Y. Cardiac myocyte remodeling mediated by N-cadherin-dependent mechanosensing. Am. J. Physiol. Heart Circ. Physiol. 2011;300:H1252–H1266. doi: 10.1152/AJPHEART.00515.2010/SUPPL_FILE/FIGURES. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childers R.C., Lucchesi P.A., Gooch K.J. Decreased substrate stiffness promotes a hypofibrotic phenotype in cardiac fibroblasts. Int. J. Mol. Sci. 2021;22:6231. doi: 10.3390/IJMS22126231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majkut S., Dingal P.C.D., Discher D.E. Stress sensitivity and mechanotransduction during heart development. Curr. Biol. 2014;24:R495–R501. doi: 10.1016/J.CUB.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry M.F., Engler A.J., Woo Y.J., Pirolli T.J., Bish L.T., Jayasankar V., Morine K.J., Gardner T.J., Discher D.E., Sweeney H.L. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol. - Hear. Circ. Physiol. 2006;290:2196–2203. doi: 10.1152/AJPHEART.01017.2005/ASSET/IMAGES/LARGE/ZH40050667170007.JPEG. [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro M.C., Slaats R.H., Schwach V., Rivera-Arbelaez J.M., Tertoolen L.G.J., van Meer B.J., Molenaar R., Mummery C.L., Claessens M.M.A., Passier R. A cardiomyocyte show of force: a fluorescent alpha-actinin reporter line sheds light on human cardiomyocyte contractility versus substrate stiffness. J. Mol. Cell. Cardiol. 2020;141:54–64. doi: 10.1016/j.yjmcc.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Young J.L., Engler A.J. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32:1002–1009. doi: 10.1016/J.BIOMATERIALS.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin I.L., Hool L., Choi Y.S. Interrogating cardiac muscle cell mechanobiology on stiffness gradient hydrogels. Biomater. Sci. 2021;9:6795–6806. doi: 10.1039/D1BM01061A. [DOI] [PubMed] [Google Scholar]
- 14.Samarel A.M. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am. J. Physiol. - Hear. Circ. Physiol. 2005;289 doi: 10.1152/AJPHEART.00749.2005/ASSET/IMAGES/LARGE/ZH40120563640003.JPEG. [DOI] [PubMed] [Google Scholar]
- 15.Majkut S., Idema T., Swift J., Krieger C., Liu A., Discher D.E. Optimal development of matrix elasticity. Curr. Biol. 2013;23:2434–2439. doi: 10.1016/J.CUB.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Münch J., Abdelilah-Seyfried S. Sensing and responding of cardiomyocytes to changes of tissue stiffness in the diseased heart. Front. Cell Dev. Biol. 2021;9:403. doi: 10.3389/FCELL.2021.642840/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majid Q.A., Fricker A.T.R., Gregory D.A., Davidenko N., Hernandez Cruz O., Jabbour R.J., Owen T.J., Basnett P., Lukasiewicz B., Stevens M., et al. Natural biomaterials for cardiac tissue engineering: a highly biocompatible solution. Front. Cardiovasc. Med. 2020;7 doi: 10.3389/fcvm.2020.554597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pushp P., Gupta M.K. Cardiac tissue engineering: a role for natural biomaterials. Adv. Struct. Mater. 2021;140:617–641. doi: 10.1007/978-3-030-54027-2_18/FIGURES/2. [DOI] [Google Scholar]
- 19.Majid Q.A., Fricker A.T.R., Gregory D.A., Davidenko N., Hernandez Cruz O., Jabbour R.J., Owen T.J., Basnett P., Lukasiewicz B., Stevens M., et al. Natural biomaterials for cardiac tissue engineering: a highly biocompatible solution. Front. Cardiovasc. Med. 2020;7:192. doi: 10.3389/FCVM.2020.554597/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippi M., Stadiotti I., Pompilio G., Sommariva E. Human cell modeling for cardiovascular diseases. Int. J. Mol. Sci. 2020;21:6388–6427. doi: 10.3390/IJMS21176388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzahor E., Dimmeler S. A coalition to heal—the impact of the cardiac microenvironment. Science. 2022;80:377. doi: 10.1126/SCIENCE.ABM4443/ASSET/0B42F6B4-7C41-4669-A6AC-0C824B8FF181/ASSETS/IMAGES/LARGE/SCIENCE.ABM4443-FA.JPG. [DOI] [PubMed] [Google Scholar]
- 22.Perea-Gil I., Prat-Vidal C., Bayes-Genis A. In vivo experience with natural scaffolds for myocardial infarction: the times they are a-changin. Stem Cell Res. Ther. 2015;6:248–325. doi: 10.1186/S13287-015-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saludas L., Pascual-Gil S., Prósper F., Garbayo E., Blanco-Prieto M. Hydrogel based approaches for cardiac tissue engineering. Int. J. Pharm. 2017;523:454–475. doi: 10.1016/J.IJPHARM.2016.10.061. [DOI] [PubMed] [Google Scholar]
- 24.Weinberger F., Mannhardt I., Eschenhagen T. Engineering cardiac muscle tissue: a maturating field of research. Circ. Res. 2017;120:1487–1500. doi: 10.1161/CIRCRESAHA.117.310738. [DOI] [PubMed] [Google Scholar]
- 25.Kurniawan N.A., Bouten C.V. Mechanobiology of the cell–matrix interplay: catching a glimpse of complexity via minimalistic models. Extreme Mech. Lett. 2018;20:59–64. doi: 10.1016/J.EML.2018.01.004. [DOI] [Google Scholar]
- 26.Bashey R.I., Martinez-Hernandez A., Jimenez S.A. Isolation, characterization, and localization of cardiac collagen type VI. Associations with other extracellular matrix components. Circ. Res. 1992;70:1006–1017. doi: 10.1161/01.RES.70.5.1006. [DOI] [PubMed] [Google Scholar]
- 27.Horn M.A., Trafford A.W. Aging and the cardiac collagen matrix: novel mediators of fibrotic remodelling. J. Mol. Cell. Cardiol. 2016;93:175–185. doi: 10.1016/j.yjmcc.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fratzl P. Springer Science and Business Media LLC; 2008. Collagen Structure and Mechanics P. Fratzl. [Google Scholar]
- 29.Antoine E.E., Vlachos P.P., Rylander M.N. Review of collagen I hydrogels for bioengineered tissue microenvironments: characterization of mechanics, structure, and transport. Tissue Eng. Part B Rev. 2014;20:683–696. doi: 10.1089/TEN.TEB.2014.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichol J.W., Koshy S.T., Bae H., Hwang C.M., Yamanlar S., Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31:5536–5544. doi: 10.1016/J.BIOMATERIALS.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pepelanova I., Kruppa K., Scheper T., Lavrentieva A. Gelatin-Methacryloyl (GelMA) hydrogels with defined degree of functionalization as a versatile toolkit for 3D cell culture and extrusion bioprinting. Bioengineering. 2018;5 doi: 10.3390/BIOENGINEERING5030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakr M.A., Sakthivel K., Hossain T., Shin S.R., Siddiqua S., Kim J., Kim K. Recent trends in gelatin methacryloyl nanocomposite hydrogels for tissue engineering. J. Biomed. Mater. Res. 2022;110:708–724. doi: 10.1002/JBM.A.37310. [DOI] [PubMed] [Google Scholar]
- 33.Chin I.L., Amos S.E., Jeong J.H., Hool L., Hwang Y., Choi Y.S. Volume adaptation of neonatal cardiomyocyte spheroids in 3D stiffness gradient GelMA. J. Biomed. Mater. Res. 2022 doi: 10.1002/JBM.A.37456. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin I.L., Amos S.E., Jeong J.H., Hool L., Hwang Y., Choi Y.S. Mechanosensation mediates volume adaptation of cardiac cells and spheroids in 3D. Mater. Today. Bio. 2022;16 doi: 10.1016/J.MTBIO.2022.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antoine E.E., Vlachos P.P., Rylander M.N. Review of collagen i hydrogels for bioengineered tissue microenvironments: characterization of mechanics, structure, and transport. Tissue Eng. Part B Rev. 2014;20:683–696. doi: 10.1089/ten.teb.2014.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Browne S., Zeugolis D.I., Pandit A. Collagen: finding a solution for the source. Tissue Eng. 2013;19:1491–1494. doi: 10.1089/TEN.TEA.2012.0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fushimi H., Hiratsuka T., Okamura A., Ono Y., Ogura I., Nishimura I. Recombinant collagen polypeptide as a versatile bone graft biomaterial. Commun. Mater. 2020;1:13–87. doi: 10.1038/s43246-020-00089-9. [DOI] [Google Scholar]
- 38.Drury J.L., Mooney D.J. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24:4337–4351. doi: 10.1016/S0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 39.Žigon-Branc S., Markovic M., Van Hoorick J., Van Vlierberghe S., Dubruel P., Zerobin E., Baudis S., Ovsianikov A. Impact of hydrogel stiffness on differentiation of human adipose-derived stem cell microspheroids. Tissue Eng. 2019;25:1369–1380. doi: 10.1089/TEN.TEA.2018.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou P., Pu W.T. Recounting cardiac cellular composition. Circ. Res. 2016;118:368–370. doi: 10.1161/CIRCRESAHA.116.308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto A.R., Ilinykh A., Ivey M.J., Kuwabara J.T., D’antoni M.L., Debuque R., Chandran A., Wang L., Arora K., Rosenthal N.A., Tallquist M.D. Revisiting cardiac cellular composition. Circ. Res. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veerman C.C., Kosmidis G., Mummery C.L., Casini S., Verkerk A.O., Bellin M. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cell. Dev. 2015;24:1035–1052. doi: 10.1089/SCD.2014.0533/ASSET/IMAGES/LARGE/FIGURE2.JPEG. [DOI] [PubMed] [Google Scholar]
- 43.Alcaraz J., Otero J., Jorba I., Navajas D. Bidirectional mechanobiology between cells and their local extracellular matrix probed by atomic force microscopy. Semin. Cell Dev. Biol. 2018;73:71–81. doi: 10.1016/J.SEMCDB.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 44.Sala L., Van Meer B.J., Tertoolen L.G.J., Bakkers J., Bellin M., Davis R.P., Denning C., Dieben M.A.E., Eschenhagen T., Giacomelli E., et al. Musclemotion: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. 2018;122:e5–e16. doi: 10.1161/CIRCRESAHA.117.312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickenden A.D. Overview of electrophysiological techniques. Curr. Protoc. Pharmacol. 2000;11:11. doi: 10.1002/0471141755.PH1101S64. [DOI] [PubMed] [Google Scholar]
- 46.Villemain O., Correia M., Khraiche D., Podetti I., Meot M., Legendre A., Tanter M., Bonnet D., Pernot M. Myocardial stiffness assessment using shear wave imaging in pediatric hypertrophic cardiomyopathy. JACC. Cardiovasc. Imaging. 2018;11:779–781. doi: 10.1016/J.JCMG.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Carbone R.G., Monselise A., Bottino G., Negrini S., Puppo F. Stem cells therapy in acute myocardial infarction: a new era? Clin. Exp. Med. 2021;21:231–237. doi: 10.1007/s10238-021-00682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mason B.N., Starchenko A., Williams R.M., Bonassar L.J., Reinhart-King C.A. Tuning 3D collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 2013;9:4635–4644. doi: 10.1016/J.ACTBIO.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi J., Mahajan G., Kothapalli C.R. Three-dimensional collagenous niche and azacytidine selectively promote time-dependent cardiomyogenesis from human bone marrow-derived MSC spheroids. Biotechnol. Bioeng. 2018;115:2013–2026. doi: 10.1002/BIT.26714. [DOI] [PubMed] [Google Scholar]
- 50.Berger A.J., Linsmeier K.M., Kreeger P.K., Masters K.S. Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen. Biomaterials. 2017;141:125–135. doi: 10.1016/j.biomaterials.2017.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janmey P.A., Fletcher D.A., Reinhart-King C.A. Stiffness sensing by cells. Physiol. Rev. 2020;100:695–724. doi: 10.1152/physrev.00013.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basara G., Ozcebe S.G., Ellis B.W., Zorlutuna P. Tunable human myocardium derived decellularized extracellular matrix for 3d bioprinting and cardiac tissue engineering. Gels. 2021;7 doi: 10.3390/GELS7020070/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bracco Gartner T.C.L., Deddens J.C., Mol E.A., Magin Ferrer M., van Laake L.W., Bouten C.V.C., Khademhosseini A., Doevendans P.A., Suyker W.J.L., Sluijter J.P.G., Hjortnaes J. Anti-fibrotic effects of cardiac progenitor cells in a 3D-model of human cardiac fibrosis. Front. Cardiovasc. Med. 2019;6:52. doi: 10.3389/fcvm.2019.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navaei A., Truong D., Heffernan J., Cutts J., Brafman D., Sirianni R.W., Vernon B., Nikkhah M. PNIPAAm-based biohybrid injectable hydrogel for cardiac tissue engineering. Acta Biomater. 2016;32:10–23. doi: 10.1016/J.ACTBIO.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 55.Karamichos D., Brown R.A., Mudera V. Collagen stiffness regulates cellular contraction and matrix remodeling gene expression. J. Biomed. Mater. Res. 2007;83:887–894. doi: 10.1002/JBM.A.31423. [DOI] [PubMed] [Google Scholar]
- 56.Herum K.M., Choppe J., Kumar A., Engler A.J., McCulloch A.D. Mechanical regulation of cardiac fibroblast profibrotic phenotypes. Mol. Biol. Cell. 2017;28:1871–1882. doi: 10.1091/MBC.E17-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Putten S., Shafieyan Y., Hinz B. Mechanical control of cardiac myofibroblasts. J. Mol. Cell. Cardiol. 2016;93:133–142. doi: 10.1016/J.YJMCC.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 58.Hakimi O., Poulson R., Thakkar D., Yapp C., Carr A. Ascorbic acid is essential for significant collagen deposition by human tenocytes in vitro. Oxid. Antioxid. Med. Sci. 2014;3:119. doi: 10.5455/OAMS.030514.OR.063. [DOI] [Google Scholar]
- 59.Saucerman J.J., Tan P.M., Buchholz K.S., McCulloch A.D., Omens J.H. Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 2019;16:361–378. doi: 10.1038/S41569-019-0155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Spreeuwel A.C.C., Bax N.A.M., Bastiaens A.J., Foolen J., Loerakker S., Borochin M., Van Der Schaft D.W.J., Chen C.S., Baaijens F.P.T., Bouten C.V.C. The influence of matrix (an)isotropy on cardiomyocyte contraction in engineered cardiac microtissues. Integr. Biol. (United Kingdom) 2014;6:422–429. doi: 10.1039/c3ib40219c. [DOI] [PubMed] [Google Scholar]
- 61.Goldfracht I., Protze S., Shiti A., Setter N., Gruber A., Shaheen N., Nartiss Y., Keller G., Gepstein L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020;11:15–75. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo J., Huebsch N. Modeling the response of heart muscle to mechanical stimulation in vitro. Curr. Tissue Microenviron. Rep. 2020;1:61–72. doi: 10.1007/s43152-020-00007-8. [DOI] [Google Scholar]
- 63.Krahn K.N., Bouten C.V.C., Van Tuijl S., Van Zandvoort M.A.M., Merkx M. Fluorescently labeled collagen binding proteins allow specific visualization of collagen in tissues and live cell culture. Anal. Biochem. 2006;350:177–185. doi: 10.1016/J.AB.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 64.Dronkers E., Moerkamp A.T., van Herwaarden T., Goumans M.J., Smits A.M. The isolation and culture of primary epicardial cells derived from human adult and fetal heart specimens. J. Vis. Exp. 2018;2018 doi: 10.3791/57370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G., et al. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:733–745. doi: 10.1016/J.CELREP.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sutter M., Siepmann J., Hennink W.E., Jiskoot W. Recombinant gelatin hydrogels for the sustained release of proteins. J. Control. Release. 2007;119:301–312. doi: 10.1016/J.JCONREL.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Van Den Bulcke A.I., Bogdanov B., De Rooze N., Schacht E.H., Cornelissen M., Berghmans H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules. 2000;1:31–38. doi: 10.1021/bm990017d. [DOI] [PubMed] [Google Scholar]
- 68.Fischer K. Neues Verfahren zur maßanalytischen Bestimmung des Wassergehaltes von Flüssigkeiten und festen Körpern. Angew. Chem. Int. Ed. Engl. 1935;48:394–396. doi: 10.1002/ANGE.19350482605. [DOI] [Google Scholar]
- 69.Tavar E., Turk E., Kreft S. Simple modification of karl-fischer titration method for determination of water content in colored samples. J. Anal. Methods Chem. 2012;2012:379724. doi: 10.1155/2012/379724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wishart D.S., Bigam C.G., Sykes B.D. H, 13C and random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 71.Bubnis W.A., Ofner C.M. The determination of epsilon-amino groups in soluble and poorly soluble proteinaceous materials by a spectrophotometric method using trinitrobenzenesulfonic acid. Anal. Biochem. 1992;207:129–133. doi: 10.1016/0003-2697(92)90513-7. [DOI] [PubMed] [Google Scholar]
- 72.Rose P.I. Encyclopedia of Polymer Science and Engineering. Wiley & Sons; 1987. Gelatin; pp. 5128–5136. [Google Scholar]
- 73.Canal T., Peppas N.A. Correlation between mesh size and equilibrium degree of swelling of polymeric networks. J. Biomed. Mater. Res. 1989;23:1183–1193. doi: 10.1002/JBM.820231007. [DOI] [PubMed] [Google Scholar]
- 74.Ma S., Natoli M., Liu X., Neubauer M.P., Watt F.M., Fery A., Huck W.T.S. Monodisperse collagen-gelatin beads as potential platforms for 3D cell culturing. J. Mater. Chem. B. 2013;1:5128–5136. doi: 10.1039/c3tb20851f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data generated in this study have been deposited at Mendeley data and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.