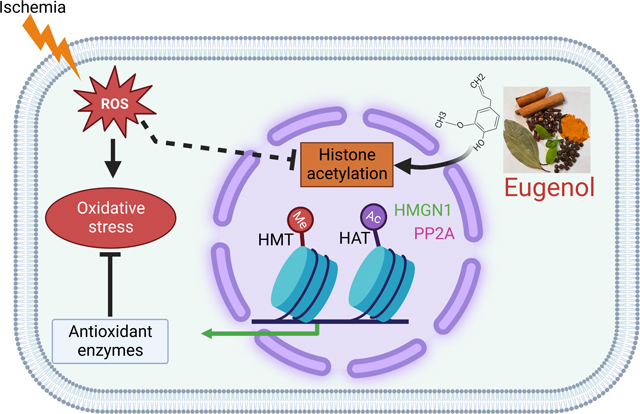
Abstract
Despite clinical advances, ischemia-induced cardiac diseases remain an underlying cause of death worldwide. Epigenetic modifications, especially alterations in the acetylation of histone proteins play a pivotal role in counteracting stressful conditions, including ischemia. In our study, we found that histone active mark H3K27ac was significantly reduced and histone repressive mark H3K27me3 was significantly upregulated in the cardiomyocytes exposed to the ischemic condition. Then, we performed a high throughput drug screening assay using rat ventricular cardiomyocytes during the ischemic condition and screened an antioxidant compound library comprising of 84 drugs for H3K27ac by fluorescence microscopy. Our data revealed that most of the phenolic compounds like eugenol, apigenin, resveratrol, bis-demethoxy curcumin, D-gamma-tocopherol, ambroxol, and non-phenolic compounds like L-Ergothioneine, ciclopirox ethanolamine, and Tanshinone IIA have a crucial role in maintaining the cellular H3K27ac histone marks during the ischemic condition. Further, we tested the role of eugenol on cellular protection during ischemia. Our study shows that ischemia significantly reduces cellular viability and increases total reactive oxygen species (ROS), and mitochondrial ROS in the cells. Interestingly, eugenol treatment significantly restores the cellular acetylation at H3K27, decreases cellular ROS, and improves cellular viability. To explore the mechanism of eugenol-medicated inhibition of deacetylation, we performed a RNAseq experiment. Analysis of transcriptome data using IPA indicated that eugenol regulates several cellular functions associated with cardiovascular diseases, and metabolic processes. Further, we found that eugenol regulates the expression of HMGN1, CD151 and Ppp2ca genes during ischemia. Furthermore, we found that eugenol might protect the cells from ischemia through modulation of HMGN1 protein expression, which plays an active role in regulation of histone acetylation and cellular protection during stress. Thus, our study indicated that eugenol could be exploited as an agent to protect the ischemic cells and could be used to develop a novel drug for treating cardiac disease.
Keywords: Ischemia, antioxidant, eugenol, HMGN1, PP2A, H3K27ac, cardiovascular
Graphical Abstract
1. Introduction:
In the past decade, we have witnessed an unprecedented increase in the number of patients suffering from cardiovascular disorders, including myocardial infarction, as it has become one of the leading causes of mortality and morbidity across the globe [1, 2]. Ischemia-induced myocardial infarction is a syndrome characterized by the reduced blood supply to the tissues resulting in reversible or irreversible tissue damage [3]. Although various pharmacological drugs (nitrates, angiotensin-converting enzyme/receptor inhibitors, beta-blockers, calcium channel blockers, potassium channel openers, free fatty acid oxidation inhibitors) and even revascularization techniques have been adopted to combat myocardial infarction and its associated complications, yet it remains a burden to the health care system [4]. In fact, percutaneous coronary interventions, which are supposed to profoundly reduce the chances of myocardial infarction may predispose individuals to subsequent reperfusion injury [4, 5].
Apparently, the intricate interaction between the oxidant and antioxidant defense system plays a pivotal role in maintaining homeostasis in the cardiomyocytes during normal as well as stressful conditions [6]. It is manifested that ischemic conditions produce metabolic perturbations and dysregulate cardiovascular homeostasis, followed by an exaggerated generation of reactive oxygen/nitrogen species [7]. The reactive oxygen/nitrogen species are considered as the key culprits of ischemic injury [7]. However, various in-vitro and in-vivo studies have revealed that increased antioxidant activity can shield the heart against oxidative damage [8] [9]. In fact, clinical studies have unveiled that boosting the antioxidant defense system may reduce the chances of precipitation of heart attack among individuals [10] [11]. Furthermore, it has also been reported that supplementation of an antioxidant drug along with a pharmacological drug reduced the infarct size in the patients [12].
Intriguingly, besides altering the cellular metabolism, the ischemic injury may also induce epigenetic alterations in the heart. Epigenetic alterations are defined as changes in the gene function without alterations in the DNA sequence [13]. These include post-translational modifications of histone proteins, including acetylation, methylation, ubiquitinylation, phosphorylation, sumoylation, carbonylation, glycosylation, and even DNA methylation [14]. In the chromatin structure, the DNA wraps around the histone proteins, which strongly adhere to each other owing to a strong electrostatic interaction between them because of the opposite charges. However, the acetylation of lysine residues of histone proteins destabilizes the DNA-protein interaction and allows the transformation of condensed chromatin to a loose structure, and facilitates transcriptional activation [15] [14]. On the other hand, methylation of histone proteins usually results in condensation of the chromatin but can be associated with either activation or repression of gene transcription based on which amino acid residues are being methylated.
An array of enzymes called histone acetyltransferases (HATs) and histone deacetylases (HDACs) control acetylation as well as de-acetylation of histone proteins [14], whereas methylation is carried out by DNA methyltransferases (DNMTs). A very recent study has indicated that ischemic condition possibly induces alteration in the expression pattern of aforementioned enzymes [16]. Interestingly, previous preclinical studies have suggested that an increase in histone acetylation exerts a protective effect against ischemic injury [17] [18] [19]. Therefore, the present study has been envisaged to identify the antioxidant compounds which can enhance the H3K27 acetylation during ischemia. Our study showed that the polyphenolic compound eugenol enhances the H3K27 acetylation and improves the viability of ischemic cardiomyocytes.
2. Material and Methods
2.1. Isolation of cells and drug treatment
The animal experiments were approved by the IACUC. Time pregnant Harlan Sprague-Dawley rats were procured from Jackson’s laboratory (Bar Harbor, ME) for this study. The rats had free access to a standard laboratory diet and water. Neonatal rat ventricular cardiomyocytes (NRVCs, primary cardiomyocytes) were isolated from 1–2 days old rat pups as described before [20]. In brief, the left ventricles were dissected from the rat neonates and were subjected to overnight trypsin (0.05%) treatment at 4°C and this was followed by collagenase treatment for 40 min at 37 °C. Cardiomyocytes were separated from other cells by replating them in sterile petri dishes. Later, the cell suspension was plated in gelatin-coated 10 cm plates for isolating protein, in two-chamber slides for carrying out immunostaining, and 96 well plates for drug screening. The cells were cultured in MEM (Grand Island, NY) with 10% FBS (Sigma, St. Louis, MO) and 1X anti-anti (Gibco) for 24h at a cell seeding density of 1.5×106 for 10 cm plate, 1×105 cells for the two-chambered slide and 20,000 cells per well for 96 well plates. Further, MEM media was replaced with DMEM with 2% FBS, and the cells were allowed to grow for another 24h before conducting the experiments. Moreover, total RNA was extracted from the cells which were grown in 6 wells plates at a density of 1×105. For drug screening, cardiomyocytes were treated with antioxidant compounds for 24h at a concentration of 10 μM [21].
2.2. Screening of an antioxidant compound library
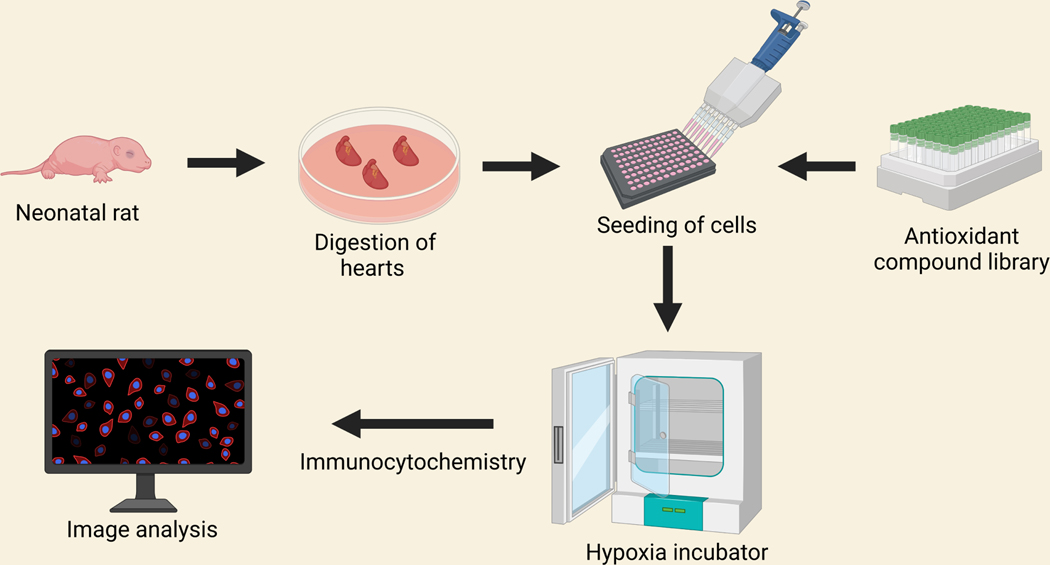
A library of 84 antioxidant compounds was purchased from Enzo Life Sciences (Farmingdale, NY) and was screened during the ischemic condition. For drug screening, NRVCs were plated in 96 well black clear-bottomed plates (Thermo Fisher Scientific) coated with 0.1% gelatin at 20,000 cells/well density. The cells were allowed to settle for 24h and were grown in 2% DMEM media for another 24h. For inducing normoxia condition, the cells were incubated in the DMEM with 2% FBS at 37°C with 5 % CO2. For inducing ischemia, the cells were incubated in DMEM having no glucose, reduced serum media (0.5% FBS, DMEM) with 1X Penicillin-streptomycin [22] using a hypoxia incubator (Eppendorf, Enfield, CT) with a setting of 1% O2, 5% CO2 at 37° C for 24h [21, 23, 24]. For drug screening, in under ischemic conditions, cardiomyocytes were treated with antioxidant compounds for 24h at a concentration of 10 μm for each drug in triplicate. For comparison, three wells of each plate were incubated with DMSO, which served as control. After 24h of incubation, the cells were washed with 1X PBS and then fixed with the 4% PFA for immunocytochemistry as described before [25]. The cells were stained with actinin (Abcam), and H3K27ac (Abcam) antibodies, and the nucleus was stained with DAPI. The plates were stored at 4°C until scanning was done under the fluorescence microscope (Keyence). Automated image capturing was performed using image cytometry, and the captured images were analyzed using Hybrid cell count (Keyence, Osaka, Japan). Apparently, 10 images were analyzed for each well [21] (Figure 2).
Figure 2:
Schematic workflow of antioxidant compound library screening using a 96 well plate during ischemia. Cells were isolated from 1–2 days old neonatal rat heart and seeded onto 96 well plates. Cells were incubated with antioxidant compound library and then incubated in hypoxia incubator for 24 hours. Cells were fixed and immunocytochemistry were performed with H3K27ac and actinin antibodies. Images were captured under fluorescence microscope.
2.3. Protein extraction and western blot analysis
Total cellular protein was extracted from the cells as described before [25, 26]. In brief, the culture cells were washed twice with 1X PBS and lysed using a combination of RIPA buffer (50mM Tris-HCl pH-8.0, 150 mM NaCl, 1% IGEPAL, 12 mM sodium deoxycholate, 1% SDS) and mammalian protease inhibitor (1X, Sigma). Subsequently, the cells were sonicated for 10 seconds with 2 s on and 1 second off setting at 20% amplitude (QSonica, Newtown, CT). To remove the cell debris and unbroken cells, the samples were centrifuged at 10,000g for 10 minutes at 4°C. The supernatant was collected for western blotting, and the protein concentration was determined using a BCA assay kit (Thermo Fisher Scientific, Waltham, MA). In order to perform a western blot, the protein samples were prepared in 1X Laemmli buffer and resolved in SDS-PAGE (Bio-Rad, Hercules, California). The resolved proteins were then transferred to the PVDF membrane using a trans-blot turbo transfer system (Bio-Rad). Next, membranes were incubated with Li-Cor blocking buffer (Li-COR, Lincoln, Nebraska) for an hour at room temperature to block the membranes and to avoid non-specific antibody binding. Afterward, membranes were incubated overnight with the primary antibody in the blocking buffer (Li-COR) at 4 °C. This was followed by washing the membrane with 1X PBST (twice) and with 1X PBS (once) on a shaker for 10 min for each wash. Membranes were then incubated with the secondary antibody tagged with IRDye®680, or 800 (Li-COR) in Li-COR antibody dilution buffer and incubated for 2h at room temperature. The membranes were washed twice with 1X PBST and once with 1X PBS before scanning on the Li-COR scanner (Odyssey). The following antibodies were used for western blotting:H3K27ac, H3K9ac, H3K27me3, and H3K9me3 (Abcam, Waltham, MA), GAPDH (Proteintech, Rosemont, IL), and histone 3, HMGN1 (Cell Signaling, Danvers, MA) [25].
2.4. Immunocytochemistry
For immunocytochemistry, the NRVCs grown in the chamber slides were washed with 1X PBS for two times, and then the cells were fixed by incubating with 4% paraformaldehyde (PFA) at room temperature for 10 min. Next, cells were washed twice with 1X PBS and permeabilized by incubating them with 0.5% Triton X-100 for 10 min at room temperature. The cells were masked with 0.1 M glycine at room temperature for 30 min, followed by washing with 1X PBS. Afterward, the cells were blocked in the blocking buffer (1% BSA, 0.1% Tween 20 in 1X PBS) for 1 h at room temperature. This was followed by overnight incubation of the cells with primary antibody diluted in the blocking buffer at 4°C. The cells were then washed with 1X PBS for twice and were incubated with a secondary antibody tagged with Alexa Fluor (Thermo Fisher Scientific) for 1 h at room temperature. Before counterstaining with a second primary antibody, the cells were again subjected to a blocking buffer for additional 30 min at room temperature and was followed by staining with the primary antibody. The cells were mounted using Vectashield HardSet antifade mounting medium with DAPI (Vector Laboratories, Burlingame, CA), and the images were captured under BZ-X800 Keyence microscope (Keyence, Osaka, Japan) [20] [25].
2.5. Viability assay
For viability assay, NRVCs were seeded in white opaque, flat bottom, transparent white 96 well plate (Thermo Fisher Scientific) at a density of 20,000 cells per well. Cellular viability was detected using the CellTiter-Glo (Promega, Madison, WI) viability assay kit according to manufacturing instructions. In brief, the cells were treated with eugenol (10 μM), C646 (1um), eugenol+C646 (1um) and were kept under ischemic condition (1% O2 and 5% CO2) for 24 hours at 37° C [27]. The CellTiter-Glo reagent was added into the cell plates after 24 hours of incubation in the presence of anti-oxidant drugs under ischemic conditions, and thereafter luminescence was acquired using a microplate reader (Tecan, Switzerland) [27].
2.6. Determination of cellular reactive oxygen species
Cellular ROS level was detected by staining the cells with dihydroethidium (Thermo Fisher Scientific). For ROS detection, NRVCs were plated in a 35 mm culture disk at a density of 2.5×105 cells per well and were allowed to adhere to the plate for at least 24 hours. The cells were treated with antioxidant compounds as well as histone acetyltransferase (HAT) inhibitor for 24 hours and were thereafter subjected to ischemic condition (1% O2 and 5% CO2) for 24 hours at 37° C. After 24 hours period, the media was aspirated, and the cells were incubated with 5 μM dihydroethidium for 5 minutes. Then, the dihydroethidium media was aspirated and fresh media was added, and the cells were observed under a Keyence BZ-X800 fluorescence microscope (Keyence, Osaka, Japan) [28].
2.7. Determination of Mitochondrial Reactive Oxygen Species
Cellular total mitochondrial ROS was detected by staining with MitoSOX red (Thermo Fisher Scientific). NRVCs were plated in 35 mm plates at a density of 2.5×105 cells and were allowed to adhere to the plate for at least 24 h. The cells were treated with antioxidant compounds and were kept under ischemic condition (1% O2 and 5% CO2) for 24 h at 37° C. After 24 h period, the media was aspirated, and the cells were incubated with 1 μM MitoSOX (Thermo Fisher) dye for 30 min in the culture media. The MitoSOX media was aspirated and fresh media was added, and the cells were observed under the fluorescence microscope (Keyence, Osaka, Japan) [29].
2.8. RNA sequence analysis
RNA was isolated from the cardiomyocytes which were subjected to normoxia, eugenol in the presence of normoxia, ischemia and eugenol in the presence of ischemia using the RNeasy Mini kit (Qiagen, Germatown, MD). The quantification of RNA was carried out using Nanodrop 8000 (Thermo Fisher Scientific, Waltham, MA). Also, the quality and integrity of RNA was assessed with Agilent Bioanalyzer (Agilent), and RNA samples with integrity ˃7 were sent for RNA sequencing (Novogene, Sacramento, CA). RANseq data were submitted to Gene Ontology database (accession # GSE203313).
2.9. Functional analysis of differentially expressed genes
GO and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was structured by clusterProfiler R package. The differentially expressed genes after being subjected to ischemic condition and eugenol in the presence of ischemic condition were structured using clusterProfiler R package. GO terms with padj < 0.05 were considered statistically significant. Functional annotation and enrichment analysis, including the functioning of biological processes and their correlation with the development of cardiotoxicity were carried out using Ingenuity pathway analysis (Qiagen).
2.10. Statistical analysis
All the experiments were repeated more than three times to get the statistical significance. Statistical analysis was performed using statistical analysis software GraphPad 9.0. Results were presented as a mean ± standard deviation. For statistical significance analysis between the groups, unpaired student t-test or one-way ANOVA was performed (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
3. Results
3.1. Reduction in histone acetylation level in the cardiomyocytes during ischemia
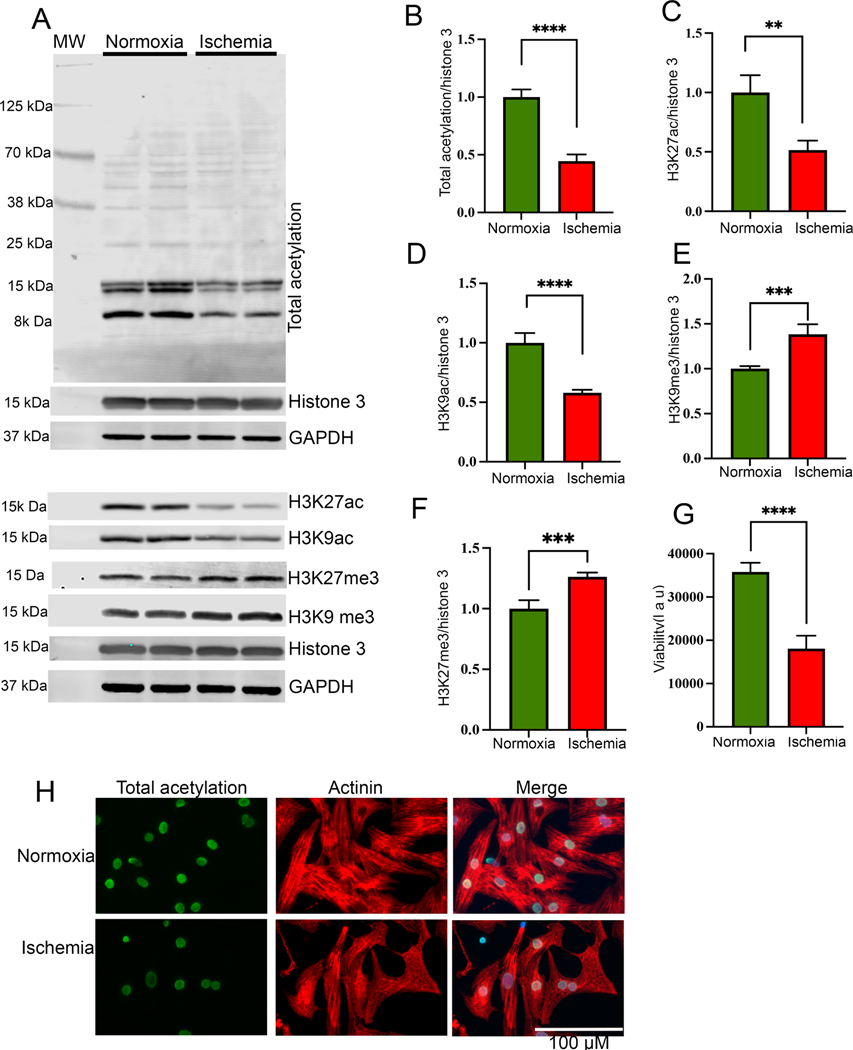
To investigate the role of epigenetic modifications of histone proteins during ischemia, we subjected the rat primary cardiomyocytes to the normoxic condition (control) and to the ischemic condition for 24 h. Total protein was extracted from the cells, and the level of cellular acetylation was detected by western blotting. Our western blot data show that ischemic cells have significantly reduced total acetylation (Fig 1A, B). Additionally, we found that the level of active histone marks H3K27ac and H3K9ac decreased significantly after exposure to the ischemic condition for 24 h (Fig 1A, C, D). Interestingly, we found that the level of suppressive histone mark H3K9me3 and H3K27me3 was significantly increased in the ischemic cells compared to the normoxic cells (Fig 1A, E, F). Furthermore, we found that cellular viability (Fig 1G) was decreased during ischemia, and ROS level was significantly upregulated in the cells (Supplementary Fig 1 A-B).
Figure 1:
Histone acetylation significantly reduced during ischemia. (A-F) Western blots show the cellular acetylation and methylation during normoxia and ischemic condition. NRVCs were incubated in normoxia and ischemic condition for 24 h and western blots were performed using total protein lysate and blots were probed with H3K27ac, H3K9ac, total acetylation, H3K27me3 H3K9me3, GAPDH and histone antibodies. Graphs show the quantification data of the western blot (**, p value<0.05; ***, p value<0.05; ****, p value<0.0001). (G) The graph shows viability of NRVCs after exposing them to normoxic and ischemic conditions (****, p value<0.0001). (H) Representative images show acetylation of NRVCs exposed to normoxia and ischemic condition for 24 h. Cells were fixed with 4% PFA and immunocytochemistry were performed with total acetylation (green) and actinin (red) antibodies and nucleus were stained with DAPI (blue). Images of fixed cells were captured by Keyence microscope and analyzed by BZ-X800 analyzer (n=50 cells in each condition).
3.2. High-throughput drug screening of antioxidant compounds exposed to ischemia
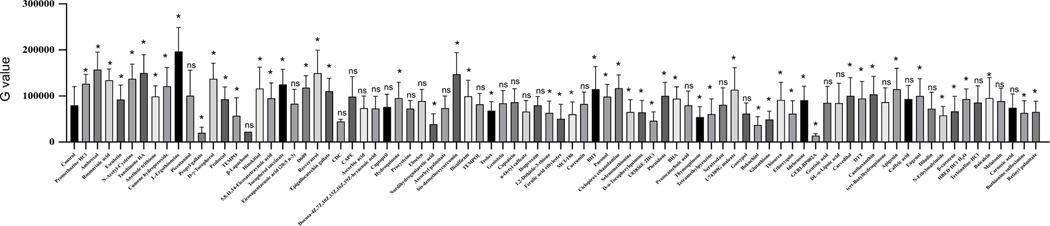
Many antioxidant compounds are known to protect the heart during ischemia-induced cardiac injury [30]. However, the epigenetic regulation during ischemia and the role of antioxidant compounds in modulating the epigenetics to produce cardioprotection through regulation of histone acetylation remains understudied. In this study, we screened an antioxidant compound library against changes in the cellular level of histone acetylation during ischemia (Fig 2). Our drug screening data analysis revealed that most of the phenolic compounds displayed a significant improvement in the H3K27 acetylation intensity (Fig 3). Out of the positive regulator compounds eugenol, apigenin, resveratrol, bis-demethoxy curcumin, D-gamma-tocopherol, ambroxol belong to the phenolic compounds, and L-Ergothioneine, ciclopirox ethanolamine, tanshinone IIA belong to non-phenolic compounds. Also, our cell viability data analysis showed that most of the phenolic compounds and some non-phenolic compounds improved the cellular viability compared to control cells treated with only DMSO (Supplementary Fig 2).
Figure 3: Graph shows quantification of histone acetylation after antioxidant compound library screening using NRVCs during ischemia.
Images of stained cells were analyzed by BZ-X800 analyzer fluorescence intensity were represented by G value.
3.3. Eugenol positively regulates the histone 3 acetylation during the ischemic condition
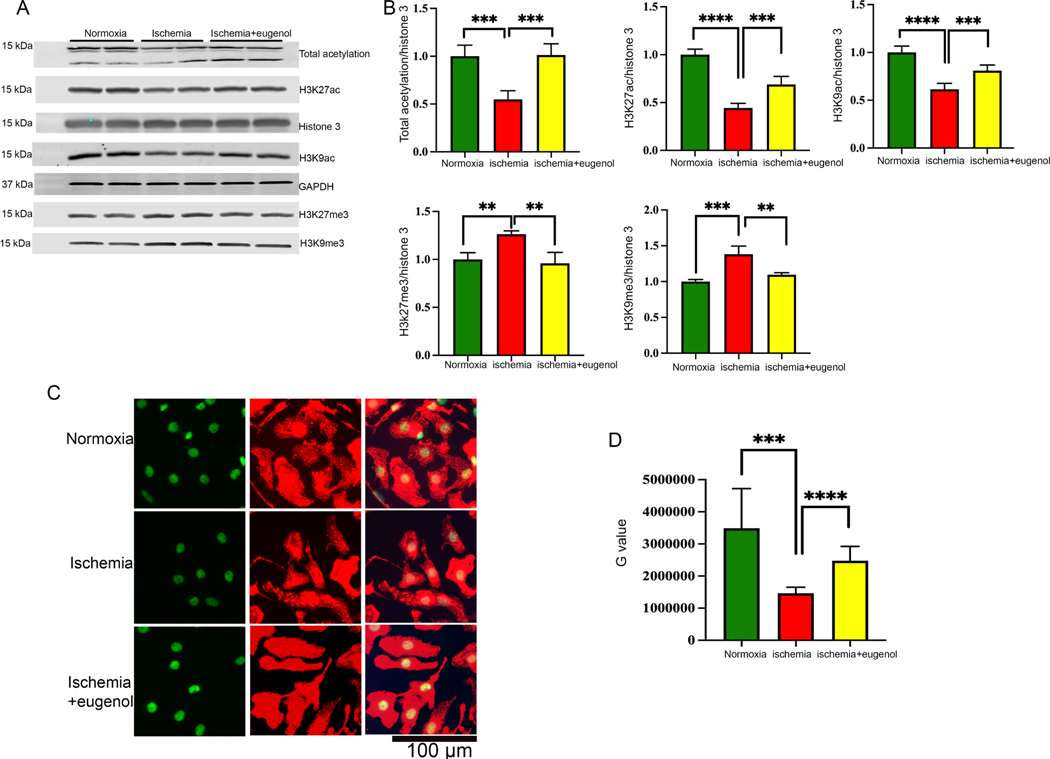
In this study, we found that sustained ischemia for 24 h significantly reduced the total acetylation in NRVCs compared to the cells exposed to normal oxygen condition (Fig 1A, B). Further, we found that during ischemia, both active and repressive markers of gene transcription at H3K27 significantly changed with the overall repression of gene transcription. In order to validate our screening data, we determined the role of eugenol in the regulation of post-translation modification of histone 3 using western blots. NRVCs were treated with eugenol (10 μm) and subjected to ischemic conditions for 24 h. Our western blot analysis showed that eugenol can significantly improve the total cellular acetylation and acetylation of histone 3 at K27, and K9 position (Fig 4A, B). Conversely, we found that the repressive marker of gene transcription H3K27me3 and H3K9me3 level was significantly reduced in the eugenol-treated cells (Fig 4A, B). Overall, our western blot data indicate that eugenol treatment during ischemic stress promotes level of active histone marks in the cardiomyocytes.
Figure 4: Effect of eugenol in cellular acetylation during ischemia.
(A-B) The western blot shows the changes in cellular acetylation (total acetylation, H3K27ac and H3K9ac) and methylation (H3K27me3, H3K9me3) after subjecting the NRVCs to normoxia, ischemia and eugenol during ischemic conditions for a period of 24 hours. The western blot was carried out using total protein lysate and the graph displays the quantification data of the western blot (**, p value<0.05; ***, p value<0.05; ****, p value<0.0001). (C-D) Representative microscopic images show cellular acetylation during ischemia. Cells were incubated with the ischemic condition for 24 hours and fixed with 4% PFA. Immunofluorescence staining of fixed NRVCs were performed with H3K27ac (green) and actinin antibody (red). Also, the nucleus was stained with DAPI (blue). The images were observed and analyzed using the fluorescent microscope. The graph shows the quantification data of the stained NRVCs under normoxic, ischemic conditions and in the presence of eugenol under ischemic conditions (***, p value<0.05; ****, p value<0.0001).
To further investigate the role of eugenol in the regulation of cellular acetylation, we exposed the cardiomyocytes in ischemic condition and performed immunocytochemistry with acetylated antibodies. Images were captured using a fluorescence microscope. Interestingly, in concordance with the western blot data, we found that there was lesser H3K27ac staining in the NRVCs exposed to ischemic conditions in comparison to the normoxia (Fig 4C, D). Intriguingly, eugenol administration under ischemic conditions significantly increased H3K27ac in the NRVCs, along with total acetylation (Fig 4C, D).
3.4. Eugenol improves the cellular viability through reduction of ROS generation
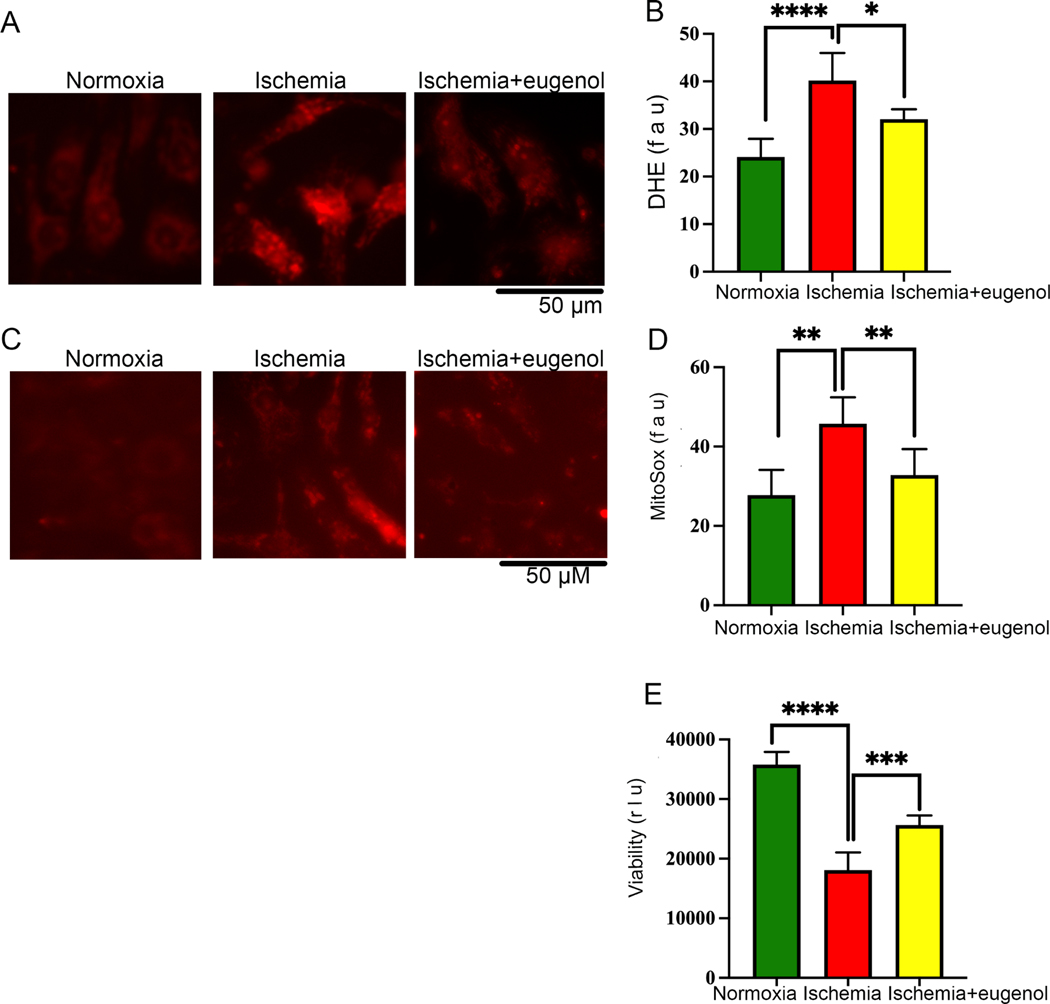
In this study, we also found that prolonged ischemia for 24 hours significantly decreased the cellular viability (Fig 1G) and increased free radical generation in the NRVCs compared to the cells exposed to normoxia. Further, we tested the role of eugenol on cellular ROS production during ischemia. Interestingly, we found that eugenol treatment significantly reduced free radical generation and improved cellular viability under ischemic conditions (Fig 5A, B, E). Moreover, we found that sustained ischemia for 24 hours significantly augmented mitochondrial ROS production in NRVCs compared to the cells exposed to normoxia (Fig 5C, D). However, eugenol administration significantly improved the cellular viability and reduced mitochondrial ROS production in the ischemic cells (Fig 5C, D, E).
Figure 5: Eugenol treatment reduces the free radical generation in cardiomyocytes during ischemia.
NRVCs were exposed to normoxia and ischemic condition for 24 h and then stained with the DHE stain and MitoSox stain and live cell imaging was done with the Keyence florescence microscope. (A-B) Representative microscopic images show NRVCs stained with DHE and the graph shows quantification of DHE staining. (C-D) Representative microscopic images show NRVC stained with MitoSox and the graph shows quantification of MitoSox staining. Quantification was done by ImageJ software. (*, p value<0.05; ****, p value<0.0001). (E) The graph displays the effect of normoxia, ischemia and eugenol in the presence of ischemia on cell viability (***, p value<0.05; ****, p value<0.0001). Cellular viability of NRVCs were determined in presence of eugenol using CellTiter-glow reagent.
3.5. Eugenol regulates cellular acetylation through modulation of HAT activity
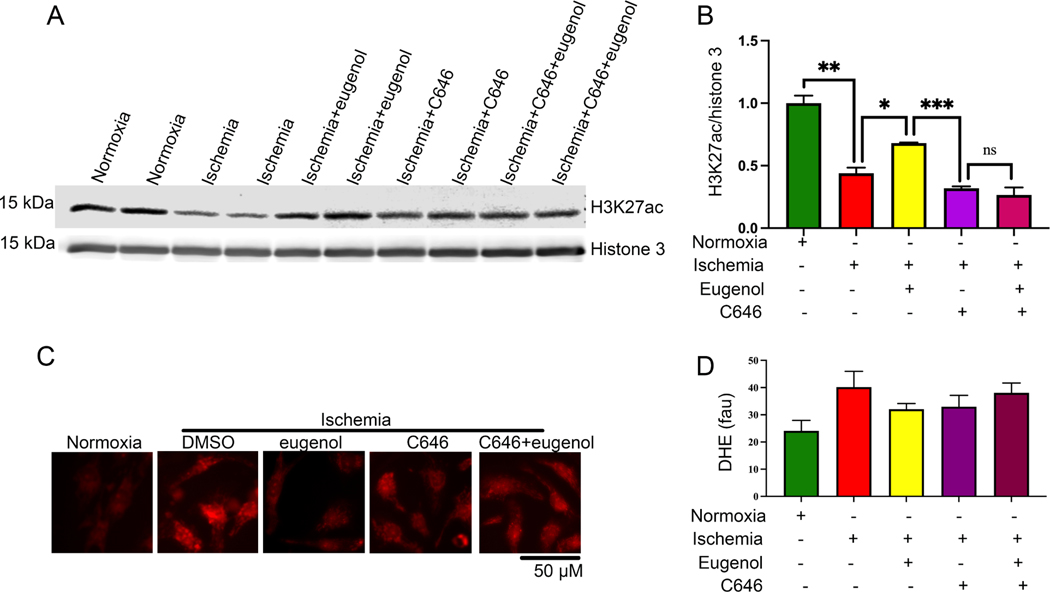
Since, our study suggests that eugenol can preserve histone acetylation, we wanted to explore the effect of eugenol in modulating the function of the cellular histone acetyltransferase (HAT) enzymes. For this, the cells were treated with eugenol alone or with a HAT inhibitor C646 (1μm) and incubated under ischemic conditions for 24 h. The level of histone acetylation was detected by western blot. The western blot data revealed that a eugenol-dependent increase in H3K27ac was abolished in the presence of C646. Our data suggest that eugenol-dependent cytoprotective effects are produced via the activation of HAT enzyme and subsequent increase in H3K27ac (Fig 6A, B). Further, we tested the effect of eugenol on cellular ROS production during ischemia in the presence of HAT inhibitor C646. Interestingly, we found that eugenol-dependent decrease in free radical production was abolished in the presence of HAT inhibitors viz. C646 (1um) (Fig 6C, D). This indicated that eugenol counteracts the deleterious effects of ROS during ischemia by activating the HAT enzyme.
Figure 6: (A-B) HAT blocker inhibits the eugenol mediated acetylation of histone during ischemia.
(A-B) The western blot shows the changes in H3K27ac during normoxia, ischemia, in the presence of eugenol under ischemic conditions, C646 (1μm) (HAT blocker), Eugenol+C646 (1μm) under ischemic conditions. The graph shows the quantification data (*, p value<0.05; **, p value<0.05; ****, p value<0.0001). (C-D) Representative microscopic images show cardiomyocytes stained with DHE. NRVCs were subjected to normoxia, ischemia, eugenol, C646 (1μm) under ischemic conditions, Eugenol+C646 (1μm) under ischemic conditions and thereafter stained with DHE. The graph displays the quantification data of free radical generation during normoxia, ischemia, eugenol in the presence of ischemia, C646 (1μm), Eugenol+C646 (1μm) under ischemic conditions, (*, p value<0.05; ****, p value<0.0001)
3.6. Eugenol treatment regulates cellular antioxidant capacity through modulating the expression of antioxidant transcription factors
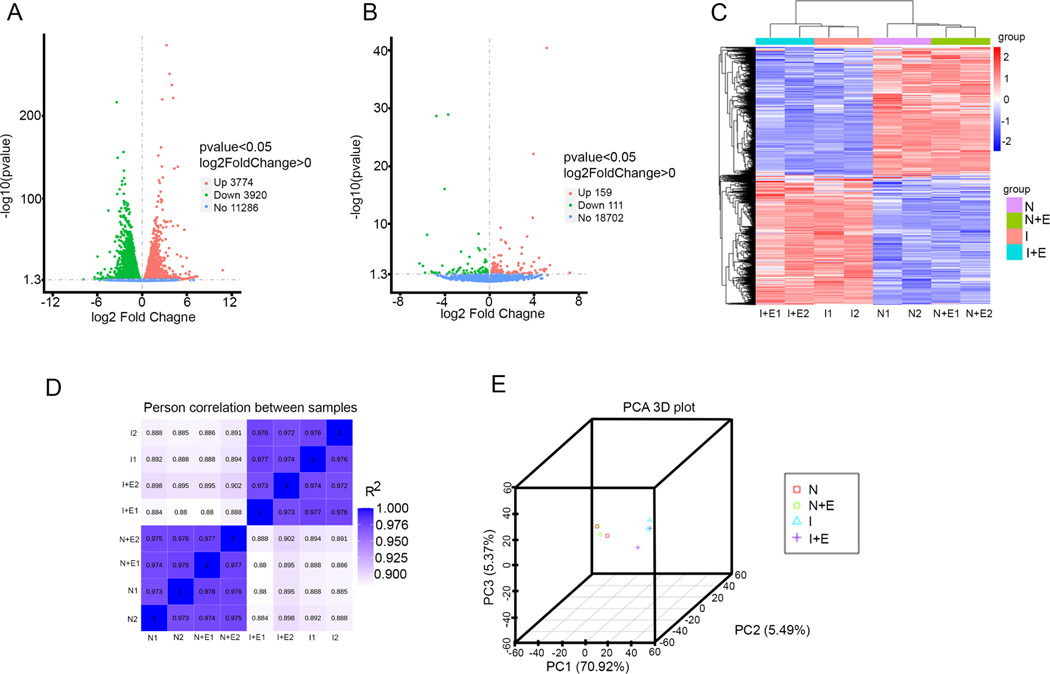
The RNA sequencing and gene expression profiling data revealed that the ischemic condition differentially regulated 7,694 genes in the NRVCs compared to the normoxic condition. To be precise, 3,774 genes were up-regulated, and 3,920 genes were down-regulated (Fig 7A). Also, the administration of eugenol in the presence of the ischemic condition up-regulated 159 genes and down-regulated 111 genes (Fig 7B). The hierarchical clustering heat map indicated the differential expression of genes in the replicate samples subjected to normoxic, eugenol in normoxic condition, ischemic, eugenol in the presence of ischemic condition (Fig 7C). Pearson correlation analysis among the samples shows a distinct cluster of normoxia, eugenol in the presence of normoxia, ischemia, eugenol in the presence of ischemia (Fig 7D). Also, the principal component analysis 3D plot shows the similarity between the samples subjected to different conditions (Fig 7E). To identify the biological processes modulated during ischemic condition, the differentially regulated genes were analyzed using clusterProfiler v.2.3 [31] [32]. Biological processes with -log 10 (padj) ˃1.2 were considered to have statistical significance in the various treatment groups.
Figure 7:
Differential expression analysis of RNA -seq data from NRVCs treated with eugenol and exposed to ischemic condition. (A) Volcano plot shows the differentially expressed genes in the NRVCs exposed to ischemic condition. (B) Volcano plot shows the differentially expressed genes in the eugenol treated NRVCs during ischemic condition. (C) A heat map shows comparison of differentially expressed genes in between NRVCs exposed to normoxia, normoxia with eugenol, ischemia, and ischemia with eugenol condition for 24 hours. (D) The figure indicates the Pearson correlation between the samples. (E) The figure shows principal component analysis (PCA) of RNA-seq data.
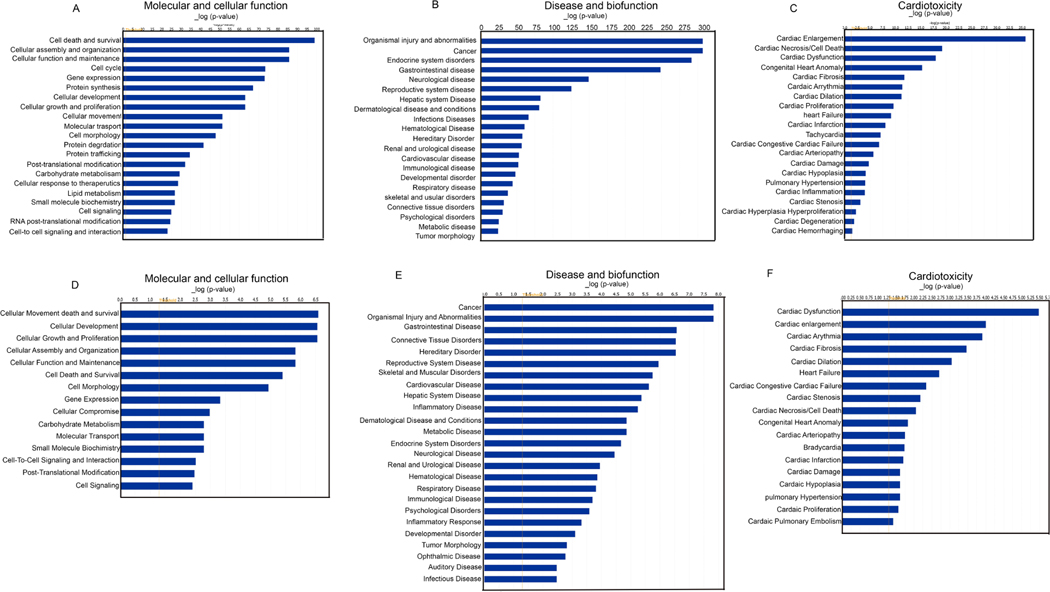
Our analysis revealed that during ischemic condition, both the upregulated and down-regulated genes controlled post-translational modification of histone proteins. Also, the upregulated genes monitored cell death and survival, cellular assembly and organization, cellular function and maintenance, cell cycle, protein synthesis, transport, etc. The down-regulated genes controlled cell signaling, cell-to-cell signaling and interaction, small molecule biochemistry, etc (Fig 8A). Apparently, it was found that the administration of eugenol under ischemic condition up-regulated intracellular transduction signaling and down-regulated negative regulation of metabolic processes, negative regulation of nucleic acid templated transcription, negative regulation of biosynthetic processes etc. To explore the association of differentially expressed genes with biological function, disease, and cardiotoxicity; the differentially expressed genes were evaluated using Ingenuity pathway analysis (IPA). Intriguingly, IPA analysis of differentially expressed genes unveiled that during ischemic condition various biological processes including cell death and survival, post-translational modification, cellular assembly and organization, cellular movement, cellular functioning and maintenance were considerably modulated (Fig 8A). Also, it was associated with cardiovascular disorder among the top listed disease and bio-function (Fig 8B) and the cardiotoxic effects exhibited included cardiac necrosis (Fig 8C). Our analysis also shows that cardiac cell death, cardiac dysfunction, cardiac infarction etc were the most significantly enriched cardiotoxicities associated with differentially expressed genes. However, in the presence of eugenol the cellular functions that were most affected included cellular movement, cellular development, cellular assembly and organization, post-translation modification etc (Fig 8D). Disease and biofunction analysis revealed that the differentially expressed genes were associated with cardiovascular disease as the most common disorders (Fig 8E) and there was modulation in the cardiotoxicity (Fig 8F). Further our IPA analysis also revealed that eugenol treatment restores the expression of several gene expressions associated with cellular viability (Supplementary Fig 3 A). To validate the transcriptional regulators, we performed a western blot to assay the expression of HMGN1 in ischemic cells. Interestingly we found that the expression of HMGN1 level was significantly reduced in ischemic cells, and eugenol treatment restored the expression of HMGN1 (Supplementary Fig 3 B-C).
Figure 8: IPA analysis shows major pathways regulated during ischemia and effect of eugenol.
(A-C) Figures show that regulation of cellular pathways during ischemia. (D-F) Figures indicate effect of eugenol treatment in cellular function during ischemia. RNA-seq data were analyzed by IPA software.
4. Discussion:
Earlier studies suggest an inextricably interlinked connection between the change in oxygen concentration and induction of epigenetic alterations in the cardiomyocytes [14, 33]. Epigenetic alterations, especially acetylation, and methylation of histone proteins, play a pivotal role in modulating cell homeostasis during ischemic conditions [14, 33, 34]. Our study found that the ischemic condition significantly reduces the total acetylation in the NRVCs compared to normoxic conditions. Also, we found that ischemia remarkably decreases H3K27ac and H3K9ac in the NRVCs compared to the normoxic group. Additionally, we found that oxygen deprivation considerably increases H3K27me3 and H3K9me3 in the NRVCs exposed to ischemic conditions compared to the normoxic group. Previous studies have also indicated that ischemic conditions significantly modulate histone acetylation and methylation [14, 33, 34], which can be a potential contributor to mediating ischemic injury [33, 35]
A plethora of studies have indicated that an imbalance between oxidants and antioxidants elevates the cells’ oxidative stress level, which tends to be a potential contributor in the pathogenesis of myocardial infarction [2, 36, 37]. However, it is manifested that the magnitude of the imbalance between oxidants and antioxidants can be counteracted via exogenous administration of antioxidants [36–38]. Very recently, a study indicated that the administration of antioxidant compounds in patients suffering from myocardial infarction may protect against the precipitation of myocardial infarction [39]. Based on the previous studies, we hypothesized that certain antioxidant compounds may possess the ability to restore the reduced acetylation during the ischemic condition. Using the H3K27ac staining, we screened a 84 polyphenolic and non-polyphenolic antioxidant compounds library. Amazingly, we found that among the 84 antioxidant compounds, most of the phenolic compounds like eugenol, apigenin, resveratrol, bis-demethoxy curcumin, D-gamma-tocopherol, ambroxol, and non-phenolic compounds L-Ergothioneine, ciclopirox ethanolamine, Tanshinone IIA facilitated the maintenance of the cellular H3K27 histone marks during the ischemic condition and improved the cellular viability. Earlier study shows that eugenol have protective role during cellular stress. However, epigenetic mechansiam of eugenol mediated cellular protection is not clear yet. Using the western blot, we further tested the role of eugenol in the post-translational modification of histone, and we found that it significantly improves H3K27ac in the NRVCs exposed to the ischemic conditions. Also, we found that eugenol administration under ischemic conditions led to the development of intense immunofluorescence stains in the NRVCs in comparison to the ischemic group. Apart from improving acetylation, we found that eugenol administration remarkably enhanced the cell viability under ischemic conditions. This is further corroborated by previous studies which indicated that eugenol administration improved cell viability [21] and reduced ischemia-reperfusion injury in the intestine and brain [21, 40].
Further, we found that the ischemic condition remarkably increases the level of ROS and mitochondrial ROS in the NRVCs. However, eugenol administration significantly reduces the generation of total ROS and mitochondrial ROS in the NRVCs under the ischemic condition. This is substantiated by previous studies indicating that eugenol administration significantly reduces oxidative stress-induced damage and improves the antioxidant defense response in the cells [41] [40] [42]. In corroboration with our study, Kumar et al reported that eugenol acts as a protective antioxidant agent during cadmium-induced oxidative stress in rat liver tissues through positive regulation of cellular antioxidant enzymes, including superoxide dismutase (SOD), catalase (CAT), glutathione-S-transferase (GSH)[43]. Additionally, the authors showed that eugenol protects the cells from cadmium induced cellular stress through reduction of inflammatory cytokines i. e. TNF-α, IL-6. Similarly, another study by Bai and co-workers have reported that eugenol treatment (1.5%) enhanced CAT activity and facilitated the decomposition of H2O2 to limit H2O2-induced oxidative damage in fresh‐cut yam subjected to four-day storage. Further this study found that eugenol can reduce the oxidative stress in the cell through inhibition of lipid membrane peroxidation. Interestingly, the authors found that eugenol administration potentiated the scavenging capacity of DPPH (1,1-diphenyl-2-picrylhydrazyl (DPPH)-2,2-diphenyl-1picrylhydrazyl), ABTS (2,2’-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid), and FRAP (iron-reducing antioxidant capacity). Altogether, the study indicated that eugenol enhances antioxidant capacity of the cells via abrogating ROS-induced membrane lipid metabolism [44].
Intriguingly, in our study we found that eugenol-dependent increase in H3K27ac was abolished in the presence of HAT blocker C646. This possibly indicated that eugenol-dependent protective effects against prolonged ischemia were mediated via HAT-dependent increase in H3K27ac. Apart from this, we found that eugenol-dependent decline in ROS production was considerably abolished in the presence of C646. This indicates that eugenol exhibits protective effect against sustained ischemic injury via limiting the production of ROS.
Our RNA sequencing data revealed that ischemic condition significantly down-regulated 3920 genes and up-regulated 3774 genes in comparison to normoxia. However, the administration of eugenol under ischemic conditions upregulated 159 genes and downregulated 111 genes in comparison to ischemia. Using the IPA software, we found that various biological processes involved in biosynthesis and metabolism were down-regulated during ischemia. Among all the processes, cell death and survival pathways were considerably altered. Interestingly, we found that administration of eugenol under ischemic conditions remarkably upregulated the expression of certain down-regulated genes under ischemic conditions. Based on IPA data analysis, we found that the high mobility group nucleosome binding domain (HMGN1) is down-regulated during the ischemic condition compared to the normoxic condition. However, eugenol administration under ischemic conditions restored the expression of HMGN1. HMGN1 is a protein-coding gene which upon binding to the nucleosomal DNA facilitates the formation of transcriptionally active chromatin [45, 46].
Intriguingly, we found that during ischemic condition, acetylation is significantly reduced and eugenol administration under ischemic condition restores the decline in acetylation possibly by increasing HMGN1 gene expression. Thus, our RNA-seq data suggest that eugenol administration under ischemic condition replenishes HMGN1 gene expression which aids in the maintenance of open chromatin structure and facilitates transcriptional changes via increasing H3K27ac and H3K9ac to elicit cardioprotective effects. Similar to our study, another study shows that eugenol protects HEK‐293 and NIH‐3T3 cells from oxidative stress through regulating the expression of nuclear transcription factor erythroid 2 p45‐related factor 2 (Nrf2) protein. Further through gain in and knockdown approach the authors showed that Nrf2 protects the cells through diminishing the cellular ROS level [47]. Mechanistically, it was found that eugenol administration considerably increased the transcriptional activity of Nrf2 (nuclear transcription factor erythroid 2 p45‐related factor 2) as well the concentration of Nrf2 protein in HEK‐293 and NIH‐3T3 cells [47]. Further, it was demonstrated that Nrf2 modulates the antioxidant capacity of cells through the regulation of histone acetylation and modulation of antioxidant gene expression [48]. Additonally, our study shows that eugenol modulates the expression of Serine/threonine-protein phosphatase 2A catalytic submint alpha (PPP2Ca) gene, which encodes the catalytic subunit of Serine/threonine-protein phosphatase 2A (PP2A). Previous study shows that PP2A regulates the cellular epigenetic modification through the modulation of chromatin remodeling enzymes [49]. Apparently, a study has revealed that the expression of and activity of PP2Ais signfificantly reduced during hypoxic condition [50]. However, overexpression of PP2A induced protective effect during ischemia [51]. In our study, we unfolded the fact that the found that expression of PPP2Ca decreased during ischemia and eugenol restored the expression of PP2Ca.
Apart from this, we also found that the expression of LOC100911730 (CD151) gene was also reduced during ischemia compared to normoxia. Apparently, the gene CD151 encodes a protein which is a member of transmembrane 4 superfamily and mediates signal transduction for cellular growth and development [52–54]. In fact, the delivery of CD151 gene in in-vivo models of ischemia-reperfusion injury promoted neovascularization (angiogenesis and arteriogenesis), mitigated myocardial dysfunction via enhancing blood flow [55, 56], and augmented the expression of vascular endothelial growth factor (VEGF) [57] to improve the left ventricular function. Based on these studies, we have postulated that the expression of CD151 gene plays a robust role in regulating cell adhesion as well as cell survival during stressful conditions, including ischemia. Furthermore, in concordance with above-mentioned studies, our study found that the administration of eugenol under ischemic conditions possibly increases the expression of CD151 gene to induce cytoprotective effects. Owing to the crucial role of CD151, PPP2Ca and HMGN1 gene expression in modulating ischemia these can be exploited as a novel therapeutic target for the treatment of ischemic or other vascular disorders.
Conclusion:
Our study revealed that during stressful conditions, including ischemia there is a significant reduction in the acetylation of histone proteins, especially at H3K27ac and H3K9ac positions. These changes at the transcriptional level modulate the oxidative stress and viability in the cells to a large extent. However, eugenol administration under ischemic conditions significantly restored the decline in the acetylation of histone proteins at the H3K27ac and H3K9ac positions. This was accompanied with improved cellular viability and reduced oxidative stress. Also, the RNA-sequence analysis unveiled that there is modulation in the expression of three genes viz. HMGN1, PPP2Ca, and CD151 under ischemic conditions. But, eugenol administration under ischemic conditions replenished the decline in the expression of the HMGN1, PPP2Ca and CD151 genes. It is manifested that restoring HMGN1 and PPP2Ca gene expression may assist in maintaining open chromatin structure and facilitate transcriptional change via acetylation of histone 3.
Supplementary Material
Supplementary figure 1A Representative images of NRVCs exposed to normoxia, ischemia and thereafter subjected to dihydroethidium staining. Figure 1B The graph displays the quantification data of free radical generation during normoxia and ischemia (****, p value<0.0001)
Supplementary figure 2: The graph shows viability of antioxidant compound treated NRVCs during ischemic condition. NRVCs were incubated with the antioxidant compound and then exposed to ischemic condition for 24 hours. Cellular viability was determined by CellTiter-Glow reagent.
(A) IPA analysis shows that during ischemia several gene expression associated with cellular viability significantly decreased and eugenol treatment protect cells through reversing the ischemic effect. (B) Western blot and graph shows that during ischemia expression of HMGN1 significantly reduced and eugenol treatment improve the expression of HMGN1 (**, p value<0.001).
Highlights.
Ischemic condition considerably reduced active histone mark H3K27ac and significantly up-regulated repressive histone mark H3K27me3 in the cardiomyocytes.
Eugenol administration under ischemic condition induced transcriptional changes and replenished active histone mark H3K27ac but reduced repressive histone mark H3K27me3 in the cardiomyocytes.
Eugenol administration under ischemic condition reduces oxidative stress and improves cell viability through regulation of histone acetyltransferase activity.
Eugenol possibly protects the cells from ischemia through modulation of HMGN1 and CD151 gene expression
Acknowledgement:
The authors are thankful to National Heart, Lung and Blood Institute grant 1R01HL141045-01A1 for funding and supporting us. We also thank Shiridhar Kashyap for his help in the data analysis. Additionally, we thank past and present members of the Gupta laboratory for providing technical help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mensah GA, Roth GA, and Fuster V, The Global Burden of Cardiovascular Diseases and Risk Factors: 2020 and Beyond. J Am Coll Cardiol, 2019. 74(20): p. 2529–2532. [DOI] [PubMed] [Google Scholar]
- 2.Randhawa PK and Gupta MK, Melatonin as a protective agent in cardiac ischemia-reperfusion injury: Vision/Illusion? Eur J Pharmacol, 2020. 885: p. 173506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandoval Y. and Jaffe AS, Type 2 Myocardial Infarction: JACC Review Topic of the Week. J Am Coll Cardiol, 2019. 73(14): p. 1846–1860. [DOI] [PubMed] [Google Scholar]
- 4.Mirza AJ, et al. , Coronary artery perforation complicating percutaneous coronary intervention. Asian Cardiovasc Thorac Ann, 2018. 26(2): p. 101–106. [DOI] [PubMed] [Google Scholar]
- 5.Shimonaga T, et al. , Myocardial Injury after Percutaneous Coronary Intervention for In-Stent Restenosis Versus de novo Stenosis. Intern Med, 2015. 54(18): p. 2299–305. [DOI] [PubMed] [Google Scholar]
- 6.Münzel T CG, Maack C, Bonetti NR, Fuster V, Kovacic JC, Impact of Oxidative Stress on the Heart and Vasculature: Part 2 of a 3-Part Series. J Am Coll Cardiol, 2017. 70(2): p. 212–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Montero J, et al. , Myocardial reperfusion injury and oxidative stress: Therapeutic opportunities. World J Cardiol, 2018. 10(9): p. 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adlam VJ, et al. , Targeting an antioxidant to mitochondria decreases cardiac ischemia-reperfusion injury. FASEB J, 2005. 19(9): p. 1088–95. [DOI] [PubMed] [Google Scholar]
- 9.Lan H, et al. , Melatonin protects circulatory death heart from ischemia/reperfusion injury via the JAK2/STAT3 signalling pathway. Life Sci, 2019. 228: p. 35–46. [DOI] [PubMed] [Google Scholar]
- 10.Hill MF, et al. , Reduction in oxidative stress and modulation of heart failure subsequent to myocardial infarction in rats. Exp Clin Cardiol, 2005. 10(3): p. 146–53. [PMC free article] [PubMed] [Google Scholar]
- 11.Wan S., et al., Total flavonoids from Anchusa italica Retz. Improve cardiac function and attenuate cardiac remodeling post myocardial infarction in mice. J Ethnopharmacol, 2020. 257: p. 112887. [DOI] [PubMed] [Google Scholar]
- 12.Pasupathy S, et al. , Early Use of N-acetylcysteine With Nitrate Therapy in Patients Undergoing Primary Percutaneous Coronary Intervention for ST-Segment-Elevation Myocardial Infarction Reduces Myocardial Infarct Size (the NACIAM Trial [N-acetylcysteine in Acute Myocardial Infarction]). Circulation, 2017. 136(10): p. 894–903. [DOI] [PubMed] [Google Scholar]
- 13.Patsouras MD and Vlachoyiannopoulos PG, Evidence of epigenetic alterations in thrombosis and coagulation: A systematic review. J Autoimmun, 2019. 104: p. 102347. [DOI] [PubMed] [Google Scholar]
- 14.Tang J. and Zhuang S, Histone acetylation and DNA methylation in ischemia/reperfusion injury. Clin Sci (Lond), 2019. 133(4): p. 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verdone L, Caserta M, and Di Mauro E, Role of histone acetylation in the control of gene expression. Biochem Cell Biol, 2005. 83(3): p. 344–53. [DOI] [PubMed] [Google Scholar]
- 16.Uzdensky AB and Demyanenko S, Histone acetylation and deacetylation in ischemic stroke. Neural Regen Res, 2021. 16(8): p. 1529–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granger A, et al. , Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J, 2008. 22(10): p. 3549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao TC, et al. , HDAC inhibition elicits myocardial protective effect through modulation of MKK3/Akt-1. PLoS One, 2013. 8(6): p. e65474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, et al. , HDAC inhibition induces autophagy and mitochondrial biogenesis to maintain mitochondrial homeostasis during cardiac ischemia/reperfusion injury. J Mol Cell Cardiol, 2019. 130: p. 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon SH, et al. , Electrically stimulable indium tin oxide plate for long-term in vitro cardiomyocyte culture. Biomater Res, 2020. 24: p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun X, et al. , Eugenol Attenuates Cerebral Ischemia-Reperfusion Injury by Enhancing Autophagy via AMPK-mTOR-P70S6K Pathway. Front Pharmacol, 2020. 11: p. 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDougal AD and Dewey CF Jr., Modeling oxygen requirements in ischemic cardiomyocytes. J Biol Chem, 2017. 292(28): p. 11760–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li HT, et al. , Chronic hypoxia differentially regulates alpha 1-adrenergic receptor subtype mRNAs and inhibits alpha 1-adrenergic receptor-stimulated cardiac hypertrophy and signaling. Circulation, 1995. 92(4): p. 918–25. [DOI] [PubMed] [Google Scholar]
- 24.He F, et al. , Downregulation of tripartite motif protein 11 attenuates cardiomyocyte apoptosis after ischemia/reperfusion injury via DUSP1-JNK1/2. Cell Biol Int, 2022. 46(1): p. 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashyap S, et al. , Antiretroviral Drugs Regulate Epigenetic Modification of Cardiac Cells Through Modulation of H3K9 and H3K27 Acetylation. Front Cardiovasc Med, 2021. 8: p. 634774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, et al. , Knockdown of LRP6 activates Drp1 to inhibit survival of cardiomyocytes during glucose deprivation. Biomed Pharmacother, 2018. 103: p. 1408–1414. [DOI] [PubMed] [Google Scholar]
- 27.Li J, et al. , The Inhibitory Effect of WenxinKeli on H9C2 Cardiomyocytes Hypertrophy Induced by Angiotensin II through Regulating Autophagy Activity. Oxid Med Cell Longev, 2017. 2017: p. 7042872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. , FNDC5 alleviates oxidative stress and cardiomyocyte apoptosis in doxorubicin-induced cardiotoxicity via activating AKT. Cell Death Differ, 2020. 27(2): p. 540–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi S, et al. , Mitochondrial Fission and Mitophagy Coordinately Restrict High Glucose Toxicity in Cardiomyocytes. Front Physiol, 2020. 11: p. 604069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W. and Kang PM, Oxidative Stress and Antioxidant Treatments in Cardiovascular Diseases. Antioxidants (Basel), 2020. 9(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G, et al. , clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 2012. 16(5): p. 284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zha Y., et al., Comprehensive Analysis of Survival-Related lncRNAs, miRNAs, and mRNAs Forming a Competing Endogenous RNA Network in Gastric Cancer. Front Genet, 2021. 12: p. 610501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JW, et al. , Ischemic preconditioning alters the epigenetic profile of the brain from ischemic intolerance to ischemic tolerance. Neurotherapeutics, 2013. 10(4): p. 789–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, et al. , Role of epigenetic regulation in myocardial ischemia/reperfusion injury. Pharmacol Res, 2021. 170: p. 105743. [DOI] [PubMed] [Google Scholar]
- 35.Stanzione R, et al. , Pathogenesis of Ischemic Stroke: Role of Epigenetic Mechanisms. Genes (Basel), 2020. 11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boarescu PM, et al. , Antioxidant and Anti-Inflammatory Effects of Curcumin Nanoparticles on Drug-Induced Acute Myocardial Infarction in Diabetic Rats. Antioxidants (Basel), 2019. 8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasparetto C, et al. , Antioxidant vitamins reduce oxidative stress and ventricular remodeling in patients with acute myocardial infarction. Int J Immunopathol Pharmacol, 2005. 18(3): p. 487–96. [DOI] [PubMed] [Google Scholar]
- 38.Guo Y, et al. , A chitosan-vitamin C based injectable hydrogel improves cell survival under oxidative stress. Int J Biol Macromol, 2022. 202: p. 102–111. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigo R, et al. , Joint Cardioprotective Effect of Vitamin C and Other Antioxidants against Reperfusion Injury in Patients with Acute Myocardial Infarction Undergoing Percutaneous Coronary Intervention. Molecules, 2021. 26(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saleh H. and El-Shorbagy HM, Mechanism underlying methyl eugenol attenuation of intestinal ischemia/reperfusion injury. Appl Physiol Nutr Metab, 2017. 42(10): p. 1097–1105. [DOI] [PubMed] [Google Scholar]
- 41.Said MM, The protective effect of eugenol against gentamicin-induced nephrotoxicity and oxidative damage in rat kidney. Fundam Clin Pharmacol, 2011. 25(6): p. 708–16. [DOI] [PubMed] [Google Scholar]
- 42.Chniguir A, et al. , Eugenol prevents fMLF-induced superoxide anion production in human neutrophils by inhibiting ERK1/2 signaling pathway and p47phox phosphorylation. Sci Rep, 2019. 9(1): p. 18540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A, et al. , Protective effect of eugenol on hepatic inflammation and oxidative stress induced by cadmium in male rats. Biomed Pharmacother, 2021. 139: p. 111588. [DOI] [PubMed] [Google Scholar]
- 44.Bai T, et al. , Scavenging of ROS After Eugenol Treatment as Mechanism of Slowing Down Membrane Lipid Metabolism to Maintain the Surface Color of Fresh-Cut Yam. Food and Bioprocess Technology, 2022. 15(8): p. 1821–1835. [Google Scholar]
- 45.Lim JH, et al. , Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J, 2005. 24(17): p. 3038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, et al. , High-mobility group nucleosome binding domain 1 (HMGN1) functions as a Th1-polarizing alarmin. Semin Immunol, 2018. 38: p. 49–53. [DOI] [PubMed] [Google Scholar]
- 47.Ma L, et al. , Eugenol protects cells against oxidative stress via Nrf2. Exp Ther Med, 2021. 21(2): p. 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alam MM, et al. , Glucocorticoid receptor signaling represses the antioxidant response by inhibiting histone acetylation mediated by the transcriptional activator NRF2. J Biol Chem, 2017. 292(18): p. 7519–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tinsley SL and Allen-Petersen BL, PP2A and cancer epigenetics: a therapeutic opportunity waiting to happen. NAR Cancer, 2022. 4(1): p. zcac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elgenaidi IS and Spiers JP, Hypoxia modulates protein phosphatase 2A through HIF-1alpha dependent and independent mechanisms in human aortic smooth muscle cells and ventricular cardiomyocytes. Br J Pharmacol, 2019. 176(11): p. 1745–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gergs U, et al. , Protein Phosphatase 2A Improves Cardiac Functional Response to Ischemia and Sepsis. Int J Mol Sci, 2022. 23(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Termini CM and Gillette JM, Tetraspanins Function as Regulators of Cellular Signaling. Front Cell Dev Biol, 2017. 5: p. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li GX, et al. , Tetraspanin18 regulates angiogenesis through VEGFR2 and Notch pathways. Biol Open, 2021. 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C, et al. , Endothelial Cell-Specific Inactivation of TSPAN12 (Tetraspanin 12) Reveals Pathological Consequences of Barrier Defects in an Otherwise Intact Vasculature. Arterioscler Thromb Vasc Biol, 2018. 38(11): p. 2691–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuo H, et al. , CD151 gene delivery after myocardial infarction promotes functional neovascularization and activates FAK signaling. Mol Med, 2009. 15(9–10): p. 307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, et al. , CD151 mediates netrin-1-induced angiogenesis through the Src-FAK-Paxillin pathway. J Cell Mol Med, 2017. 21(1): p. 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu H., Tan J, and Yin Q, Effects of recombinant adeno-associated virus-mediated CD151 gene transfer on the expression of rat vascular endothelial growth factor in ischemic myocardium. Exp Ther Med, 2015. 9(1): p. 187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1A Representative images of NRVCs exposed to normoxia, ischemia and thereafter subjected to dihydroethidium staining. Figure 1B The graph displays the quantification data of free radical generation during normoxia and ischemia (****, p value<0.0001)
Supplementary figure 2: The graph shows viability of antioxidant compound treated NRVCs during ischemic condition. NRVCs were incubated with the antioxidant compound and then exposed to ischemic condition for 24 hours. Cellular viability was determined by CellTiter-Glow reagent.
(A) IPA analysis shows that during ischemia several gene expression associated with cellular viability significantly decreased and eugenol treatment protect cells through reversing the ischemic effect. (B) Western blot and graph shows that during ischemia expression of HMGN1 significantly reduced and eugenol treatment improve the expression of HMGN1 (**, p value<0.001).