Abstract
The role of the gut-brain axis in maintaining the brain's and gut's homeostasis has been gradually recognized in recent years. The connection between the gut and the brain takes center stage. In this scenario, the nucleotide-binding oligomerization domain leucine-rich repeat and pyrin domain-containing protein 3 (NLRP3) inflammasome promotes inflammatory cell recruitment. It plays a crucial role in coordinating host physiology and immunity. Recent evidence shows how vital the gut-brain axis is for maintaining brain and gut homeostasis. However, more research is needed to determine the precise causal link between changed gut microbiota structure and NLRP3 activation in pathogenic circumstances. This review examines the connection between gut microbiota and the NLRP3 inflammasome. We describe how both dynamically vary in clinical cases and the external factors affecting both. Finally, we suggest that the crosstalk between the gut microbiota and NLRP3 is involved in signaling in the gut-brain axis, which may be a potential pathological mechanism for CNS diseases and intestinal disorders.
Keywords: Gut microbiota, NLRP3 inflammasome, Inflammation, LPS, Gut-brain axis, Neurological disorders
Graphical Abstract
1. Introduction
When the body is exposed to harmful stimuli, the immune system kicks off inflammatory reactions and begins self-defense. Inflammasomes are considered innate immune system receptors and sensors in this process, controlling the development and release of inflammatory factors [1]. However, chronic excess of inflammatory factors in the body sharply raises health risks. According to the latest research, inflammasomes play a role in the onset or progression of a wide range of chronic disorders, including metabolic diseases (e.g., obesity [2], type 2 diabetes [3], [4], atherosclerosis [5], liver disease [6]), cardiovascular disease [7], [8], inflammatory bowel disease (IBD) [9] and neurological disorders (e.g., Parkinson's disease, Alzheimer's disease, and depression) [10], [11]. The nucleotide-binding oligomerization domain leucine-rich repeat and pyrin domain-containing protein 3 (NLRP3), typical of the inflammasome, is a potent driver of inflammatory cell recruitment and a regulator of immune responses in several organs [12]. In addition, the gut microbiome has significant implications for the gastrointestinal tract and immune system, and alterations in its homeostasis are associated with many inflammatory conditions [13]. It is vital to determine if there is an interaction between the gut microbiome and NLRP3 in IBD and neurological illnesses closely associated with the gut microbiome. Groundbreaking evidence suggests that the interaction between the gut microbiome and NLRP3 is dynamically altered in intestinal and peripheral inflammatory diseases [14], [15]. The possible current mechanism of action is that changes in the structure of the gut microbiota alter the intestinal mucosal barrier. This can trigger intestinal and peripheral inflammatory responses, which in turn regulate the physiological functions of other organs [16].
The gut microbiota is involved in many functions of the central nervous system (CNS), such as regulating the permeability of the blood-brain barrier, reducing astrocyte pathogenesis, activating microglia, and expressing myelin-forming genes [17]. The accumulation of central inflammatory factors due to microbial imbalance is essential to the central nervous system and psychiatric disorders. Intestinal bacteria influence brain homeostasis by regulating peripheral inflammation via inflammasome signaling [18], [19]. Most current reviews summarize the independent roles of the gut microbiome or NLRP3 inflammasome activation in regulating gut or central disease physiology. As more evidence of gut microbiome and NLRP3 interactions is discovered, it is necessary to elucidate the specific mechanisms of microbial-immune interactions further. Therefore, this article addresses the interactions between microbes and NLRP3 inflammasome in the gut and CNS for review. In particular, how the dynamic balance of microorganisms affects disease onset and progression through NLRP3 inflammasome should be explored in depth. We aimed to investigate whether the interaction between gut microbes and inflammasomes based on different clinical symptoms is co-occurring. In addition, we hope to find potential directions for the clinical treatment of gut and CNS disorders by analyzing the bidirectional signaling between gut microbiota and NLRP3 inflammasome.
2. Intestinal microbiota is involved in the inflammatory response
More than trillions of microorganisms are present in the intestine, including bacteria, viruses, fungi, and phages. These microorganisms are collectively called the intestinal microbiome, with the major bacterial species including Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. The Firmicutes and Bacteroidetes account for 90% of the intestinal microbiota [20], [21]. The gut microbiome plays an essential physiological role in several aspects, including food digestion, metabolism, maintenance of the intestinal barrier, and regulation of the immune system [22], [23], [24], [25]. Recent studies have shown intestinal microbes increasingly associated with inflammatory responses in host physiological and pathological processes [26], [27]. The inflammatory response related to gut microbes is not only found in gastrointestinal disorders such as IBD, irritable bowel syndrome (IBS), and colorectal cancer (CRC) but also impacts cardiovascular, metabolic, reproductive, autoimmune, and neurodegenerative diseases [26], [28], [29], [30], [31], [32].
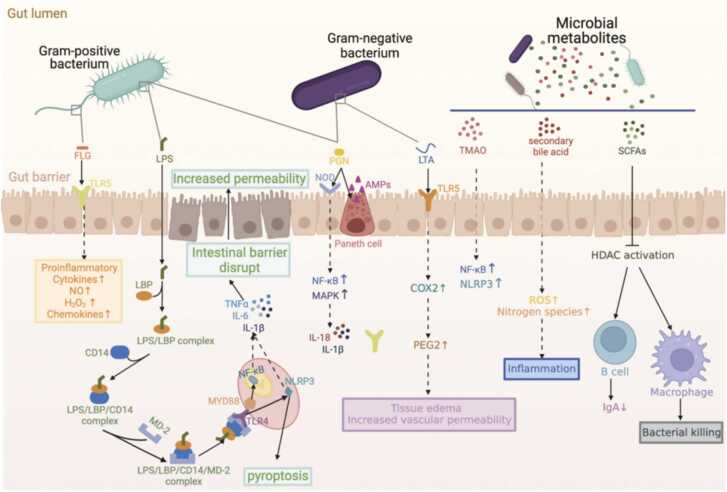
The intestinal mucosal barrier (IEB), as the first line of defense of the organism against pathogenic invasion, is directly influenced by the gut microbiome. The intestinal microbiome maintains the integrity of the IEB by regulating the growth and differentiation of the intestinal epithelial cells (IECs), the expression of tight junction proteins, and mucosal permeability [33]. Alterations in the gut microbiota composition can impair IEB, disrupting intercellular junction function and leading to increased intestinal permeability, increasing inflammatory mediator transport [30], [34]. The gut lamina propria triggers an immune/inflammatory response upon receiving signals from bacterial and metabolic components, thereby linking the gut microbiome to chronic inflammation of the organism [30]. Thus, the inflammatory response induced by intestinal microorganisms through IEB is essential in developing many diseases. The interaction between bacteria and the immune system is crucial in maintaining host homeostasis and regulating physiological and pathological processes [35]. The innate immune system, which responds directly to the gut microbiota, is present in the intestine and consists of Paneth cells, dendritic cells, macrophages, neutrophils, natural killer cells, and mast cells [35], [36]. These cells and epithelial cells possess pattern recognition receptors (PRRs), such as TLR, NLR, C-type lectin receptor (CLR), retinoic acid-inducible gene (RIG)-I-like receptor (RLR), and the deletion 2 (AIM2)-like receptor (ALR) [27]. These receptor families recognize pathogen-associated molecular patterns (PAMPs) or microbial metabolites associated with the gut microbiota. PAMPs are conserved structural components in microorganisms, such as lipopolysaccharide (LPS), peptidoglycan (PGN), Lipoteichoic acid (LTA), and flagellin (FLG), that activate the innate immune system in the intestine by binding to specific receptors. This process regulates microbial-host interactions and immune tolerance (Fig. 1) [37], [38].
Fig. 1.
The intestinal microbiota is involved in regulating intestinal homeostasis and the immune system. Gut microbiota components or metabolites regulate microbial-host interactions and immune tolerance. TLR4 in epithelial and immune cells recognizes LPS and activates NF-κB and NLRP3 inflammasome via MyD88 to produce pro-inflammatory cytokines, ultimately leading to pyroptosis. TLR5 expressed by the basolateral epithelium of the intestine detects bacterial translocation and causes chronic inflammation by recognizing the FLG of Gram-negative bacteria. TRL2 recognizes LTA in the cell wall of Gram-positive bacteria and activates mast cells via PGE2, causing edema and increased vascular permeability. PGN activates defensin synthesis in Paneth cells and increases transcription of pro-inflammatory genes by promoting NF-κB and MAPK activation via NOD family proteins. Microbial metabolites modulate immune responses through different pathways and coordinate host gut health and immune function.
LPS is a significant component of the cell wall of gram-negative bacteria and protects the bacteria from the environment, such as antibiotics or immune cells [39]. Endotoxemia caused by LPS is due to microbiota-associated inflammation in vivo that disrupts the intestinal barrier and increases intestinal permeability, leading to abnormal plasma LPS levels and exacerbating inflammation levels [40], [41]. In immune or epithelial cells, LPS activates toll-like receptor 4 (TLR4) and signaling after TLR4 activation involves activating MyD88 or NLRP3 inflammasome, producing pro-inflammatory cytokine [42], [43]. In addition, LPS activates NLRP3 inflammasome activation via classical and non-classical pathways (without TLR4), causing pyroptosis and the release of inflammatory cytokines [44], [45]. Other PAMPs are recognized by NLR or TLR-like receptor families expressed by epithelial or immune cells in the gut [27]. TLR5 expressed on the basolateral side of the intestinal epithelium detects bacterial translocation by recognizing FLG in Gram-negative bacteria [46]. Activation of TLR5 by FLG induces the synthesis of chemokines, nitric oxide (NO), hydrogen peroxide (H2O2), and pro-inflammatory cytokines [47]. Excessive activation of TLR5 may lead to impaired intestinal barrier integrity and chronic inflammation [48]. TLR2 recognizes LTA in the cell wall of Gram-positive bacteria, causing immune system activation and the development of adaptive immunity [46]. TLR2 recognition of LTA increases cyclooxygenase expression and promotes prostaglandin E2 (PGE2) synthesis [49]. PGE2 activates mast cells, causing typical symptoms of inflammation such as edema and increased vascular permeability in the host. In macrophages, LTA is recognized and stimulates the release of IL-1, IL-6, and TNF-α [49]. Peptidoglycan (PGN) is a cellular structure essential for bacterial growth and has an environmental protection role for bacteria [50]. PGN in the intestinal lumen activates the synthesis of defensins by Paneth cells, which regulate the microbiota and protect against pathogens. Recognition of PGN by NOD family proteins (NOD1, NOD2, and cryopyrin) leads to activation of NF-κB and mitogen-activated protein kinase (MAPK), increases transcription of pro-inflammatory genes, and promotes maturation and secretion of IL-1β and IL-18 [51], [52]. These studies confirm intestinal bacteria influence body homeostasis by modulating the immune system.
The regulatory role of microbial metabolites on the host immune system should also be considered. Common microbial metabolites include short-chain fatty acids (SCFAs), tryptophan catabolites, purines, secondary bile acids, and polyamines[53]. SCFAs are essential to waste products for balancing redox equivalent production in the anaerobic intestinal lumen. Most SCFAs are taken up by intestinal cells to regulate host physiology as energy substrates, regulators of gene expression, and signaling molecules for specific receptors [54], [55], [56], [57]. Butyrate regulates innate immunity by inhibiting histone deacetylase (HDAC) [58]. SCFAs can also modulate adaptive immune responses and promote T cell differentiation to an anti-inflammatory phenotype. In addition, G protein-coupled receptors (GPCRs) in IECs and immune cells regulate inflammatory reactions in response to the activation of SCFAs. In the colonic mucosa, GPCR41 and GPCR43 act as communication vehicles between SCFAs and mast cells and function as immune surveillance [54]. Butyrate acts as a reduction in LPS-induced NF-κB activation via GPR109A [59]. Acetate binding to GPR43 activates NLRP3 inflammasome activation in the colon [60]. Furthermore, high levels of secondary bile acids in the intestine damage the intestinal barrier, accumulating reactive oxygen and nitrogen species and promoting inflammatory diseases [61], [62]. Researchers have recently found that the bacterial-dependent product trimethylamine N-oxide (TMAO) influences the development of multiple diseases by modulating the inflammatory response [63], [64], [65]. Trimethylamine lyase, widely expressed in the gut microbiota, metabolizes dietary nutrients such as choline, betaine, and L-carnitine to form trimethylamine (TMA). The oxidation product of TMA (TMAO) has been identified to induce activation of NLRP3 and NF-κB in the cardiovascular [62], kidney [66], and nervous system [67] related diseases in promoting the development of inflammation. In conclusion, microbial metabolites in the gut cannot be ignored in coordinating host gut health and immune function.
Taken together, this evidence reveals that the gut microbiota and immune system interact to play a critical role in host physiological function. Understanding the relationship between gut microbiome structure and pathogenicity is vital for preventing and treating digestive and systemic diseases. Additionally, knowledge of the specific molecular and cellular mechanisms that activate immune/inflammatory cells is crucial for achieving these goals.
3. NLRP3 inflammasome
The inflammasomes are multi-protein complexes assembled by intracellular PRRs, which play an innate immune defense function in cells [68]. Most inflammasomes consist of receptor protein, connector protein ASC and downstream Caspase family. Receptor proteins are divided into the NLR family and non-NLR proteins. The NLR family includes NLRP1, NLRP2, NLRP3, NLRP6, NLRC4, and NLRP12, all share common features, such as nucleotide-binding domains (NBD) and leucine-rich repeat sequences (LRR). Some NLRs also contain the pyrin domain (PYD) or caspase activation and recruitment domain (CARD) or both [5], [69]. The non-NLR proteins include the PYHIN family and Pyrin. The PYHIN family features a PYD and HIN domain. Its members, such as absent in melanoma 2 (AIM2) and interferon-γ (IFNγ)-inducible protein 16 (IFI16), have emerged as significant recognition factors for viral and bacterial DNA [70], [71]. Pyrin comprises a PYD, two B-boxes, and a coiled-coil structural domain. Pyrin interacts directly with actin to degrade the inflammasome signaling regulators NLRP1, NLRP3, and caspase-1 via autophagy [72].
Recognition of inflammatory receptors leads to sensor activation, oligomerization, and recruitment of junctional proteins called ASC that activate caspase-1, followed by protein hydrolysis to activate IL-1 family cytokines that initiate downstream responses affecting cell fate. Different PRRs activate inflammasome assembly in response to specific PAMPs or endogenous hazards, signaling in the cytoplasmic lysis of host cells through classical and non-classical signaling pathways [73]. For example, NLRP1 recognizes Bacillus anthracis [74], [75]; NLRP3 is activated in response to multiple PAMPs and damage-associated molecular patterns (DAMPs) [76]; NLRC4 is activated by Salmonella flagellin [77]; and the non-NLRs AIM2 and pyrin are triggered by murine cytomegalovirus, bovine pox toxins, and bacterial toxins (e.g., Clostridium difficile toxin B and Clostridium botulinum C3 toxin) [78], [79].
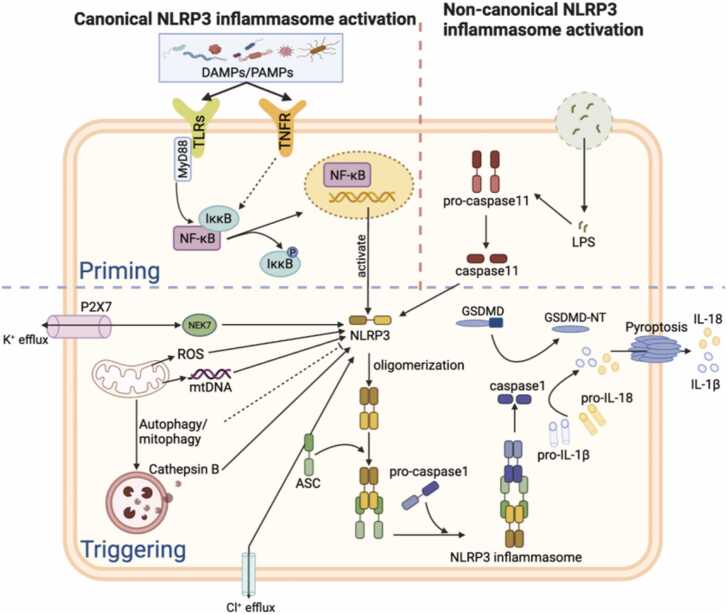
NLRP3 inflammasome has attracted wide attention due to its ability to respond to diverse stimuli and activate inflammation-related factors (Fig. 2). The NLRP3 inflammasome is a multimeric complex comprising NLRP3, ASC, NEK7, and pro-caspase-1. These complexes are widely distributed throughout the immune system, including the thymus, spleen, monocytes/macrophages, B cells, T cells, and neutrophils. They are also in the non-immune system, including the brain, liver, pancreas, dendritic, and epithelial cells [80]. In response to stimulation by environmental stimuli, endogenous danger signals, pathogens, and different PAMPs (e.g., extracellular ATP, viruses, LPS), inflammatory signals such as TLR, NLR, and TNF-α are activated. These signals are considered essential mediators of inflammasome initiation and can trigger expression and post-translational modifications of NLRP3 proteins [81]. In addition, NLRP3 assembly is regulated by phosphorylation and ubiquitination events [82], [83]. Under the stimulation of inflammatory mediators, NLRP3 oligomerizes and binds with the PYD of ASC and recruits pro-caspase-1 via CARD to assemble and activate the inflammasome complex. The activated inflammasome complex prompts ASC to cleave pro-caspase-1 into its active form, caspase-1, which then cleaves pro-IL-1β and pro-IL-18 into IL-1β and IL-18, respectively. IL-1β and IL-18 inflammatory factors increase and induce immune cell recruitment and activate additional pro-inflammatory signaling pathways leading to cellular injury [14]. Moreover, NLRP3 can activate caspase-1 to cause gasdermin D (GSDMD) cleavage, creating membrane pores in the plasma membrane and causing pyroptosis [84], [85]. Currently, NLRP3 inflammasome-induced apoptosis or pyroptosis has been shown to occur in many gastrointestinal and neurological diseases under stress and inflammation [86], [87], [88].
Fig. 2.
The activation mechanism of NLRP3 inflammasome. NLRP3 responds to a variety of PAMPs and DAMPs. Under different stimuli, signals such as TLR and TNFs promote NLRP3 expression. Oligomerized NLRP3 binds to ASC and recruits pro-caspase-1 to complete the assembly and activation of the inflammasome complex, which in turn cleaves pro-caspase-1, pro-IL-1β, and pro-IL-18. Mature IL-1β and IL-18 induce immune cell recruitment and activate pro-inflammatory signaling pathways, leading to cellular injury and inflammatory cell death. In addition, NLRP3 can activate caspase-1 through classical or non-classical pathways causing GSDMD cleavage, generating membrane pores in the plasma membrane, and causing pyroptosis.
4. Correlation between Intestinal microbiota and NLRP3 in inflammation
Studies have found a correlation between the gut microbiome and NLRP3 in the pathological manifestations of many diseases [89], [90], [91]. However, the causal relationship between the bacterial composition and metabolites of the gut microbiome and NLRP3 inflammasome activation has yet to be adequately studied. A recent study demonstrated that the intestinal probiotic Lactobacillus rhamnosus GR-1 can reduce E. coli-induced expression of NLRP3 inflammasome and caspase-1, improve cell morphology and ultrastructure, and limit harmful inflammatory responses [92]. Bifidobacterium longum subsp. infantis and Bacteroides fragilis activate human and murine macrophage NLRP3 inflammasome expression in a phagocytosis-independent manner via K+ efflux. Inhibition of the enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC) effector protein NleA in human myeloid leukemia mononuclear cells (THP-1) impedes the deubiquitination of NLRP3, a process required for inflammasome activation, and reduces caspase-1 activation and IL-1β secretion, which in turn effectively reduces inflammasome lesion formation of the inflammasome [93]. In colitis and colorectal cancer, Lactobacillus casei, Lactobacillus fragilis, and Lactobacillus lactis could negatively regulate NLRP3 and inhibit the assembly of NLRP3 inflammasome [92], [94]. These studies demonstrate that the activation of the NLRP3 inflammasome is an essential mediator of the microbial inflammatory response and subsequent cellular damage in the body.
Under normal physiological conditions, the body's immunity is immune-tolerant to non-pathogenic intestinal microbes and provides effective defense against pathogenic pathogens, a balance essential for maintaining homeostasis [35]. Immune cells actively participate in the microbiome-inflammasome interactions. IgG immune complexes bound to commensal bacteria in the gut stimulate mouse bone marrow-derived macrophages (BMDM) and THP-1 to upregulate the expression of inflammasome-associated genes, including NLRP3 and IL-1β [95]. A recent study showed that activation of the Rho GTPases-activating toxin, CNF1, from E. coli triggers activation of NLRP3 in BMDM and further leads to caspase-1 cleavage and maturation of IL-1β [96]. E. coli isolated from patients with inflammatory bowel disease activates NLRP3 inflammasome, exacerbating the inflammatory response [97]. Overnight stimulation of BMDM by non-hemolytic enterotoxin (NHE) from Bacillus cereus is similarly able to induce activation of NLRP3 inflammasomes and cleavage of GSDMD, ultimately causing cell death [98]. Likewise, Gram-negative bacteria such as Francisella novicida, Salmonella typhimurium, Citrobacter, and E. coli-stimulated BMDM produce IL-1β and IL-18 in an NLRP3-dependent manner [99]. Aspergillus chimaera stimulates newly recruited monocytes to induce NLRP3-dependent IL-1β release, exacerbating intestinal inflammatory injury [100]. Akkermansia muciniphila, in an NLRP3-dependent manner, fosters the enrichment of M1-like macrophages and thus suppresses colorectal cancer tumors[101]. In conclusion, these findings suggest that gut microbes and specific proteins derived from these microbes can activate inflammasomes in immune cells in an NLRP3-dependent manner.
Interventions targeted at the gut microbiome are crucial for investigating its specific functions. Common interventions currently include antibiotic treatment, sterile mouse experiments, and flora transplantation. Research and clinical evidence suggest that antibiotic use is associated with ecological and immune dysregulation [102], [103]. In the DSS-induced colitis model, antibiotic-administered NLRP3-/- mice had a higher survival rate, suggesting that the overgrowth of the colonic microbiota significantly contributed to the pathological severity of DSS-induced NLRP3-/- mice [104]. However, it has also been shown that both pancreatic injury and systemic inflammation are improved in antibiotic-treated and GF mice models of acute pancreatitis. Reduced levels of colonic NLRP3 and IL-1β and increased expression of tight junction proteins indicate an enhanced physical barrier of the intestine [90]. In addition, stimulation of BMDMs by fecal contents in specific pathogen-free (SPF) mice resulted in IL-1β production, whereas GF mice did not. In contrast, the inspiration of BMDMs derived from NLRP3-/- mice with fecal contents from SPF mice failed to induce IL-1β production, suggesting that NLRP3 is a crucial mediator of the gut microbiome-induced IL-1β response [100]. Broad-spectrum antibiotic mixtures cause dysregulation of gut microbial ecology in mice and lead to increased expression of NLRP3 inflammasome-associated proteins in serum, small intestine, and cerebral cortex [102], [105]. Fecal microbial transplantation (FMT) is also an effective technique to explore the role of the microbiome. Using the FMT rat model, it was found that susceptibility to atrial fibrillation (AF) in older rats can be transmitted to younger hosts and that circulating LPS and glucose levels are significantly increased, which in turn leads to upregulation of NLRP3 inflammasome expression, enhances atrial fibrosis, and promotes the development [106]. In a mouse Mdr2 knockout (Mdr2-/-) model similar to human primary sclerosing cholangitis (PSC), dysbiosis of the intestinal flora in mice leads to intestinal barrier dysfunction and increased bacterial translocation, which amplifies the hepatic NLRP3-mediated innate immune response. Transfer of the Mdr2-/- microbiota into healthy WT control mice resulted in significant liver injury in recipient mice, highlighting the causal role of gut flora dysbiosis on disease progression [107]. In conclusion, these practical research tools demonstrate that the presence of the gut microbiome is closely associated with the activation of the NLRP3 inflammasome.
The activation of the NLRP3 inflammasome by the gut microbiota has been linked to various disease processes. It has also been found that NLRP3 regulates gut microbial homeostasis and delays disease progression in some diseases [89], [106], [108]. For instance, Cryopyrin-associated periodic syndrome (CAPS) model mice, highly resistant to experimental colitis and colorectal cancer, have a natural enhancement of IL-1β in lamina propria mononuclear phagocytes due to the NLRP3R258W mutation, which leads to excessive NLRP3 activation. The increased IL-1β induced local antimicrobial peptides to promote microbiota remodeling. The remodeled intestinal microbiota neutralizes inflammation due to the increased induction of regulatory T cells (T regs) to maintain intestinal homeostasis. NLRP3 inflammasome in the gut recognizes microbial antigens or metabolites, leading to an innate immune response that produces antimicrobial peptides (AMPs). This helps to reduce pathogen colonization and regulate intestinal microecology [109]. Studies have also shown that NLRP3 is critical to preventing the overgrowth of commensal bacteria in the intestine and reducing bacteremia, which could otherwise exacerbate colitis and damage the intestinal epithelium. For instance, DSS-fed NLRP3-/- and caspase-1-/- mice exhibited increased leukocyte infiltration and elevated chemokine levels, resulting in overall structural damage to the intestinal epithelium [104]. Notably, the relative abundance of the Firmicutes, Aspergillus, and Aspergillus phylum in the intestine of NLRP3 knockout mice was significantly altered compared to WT mice. Transplantation of NLRP3-/- mouse fecal contents into WT mice improved their depression-like behavior, likely by inhibiting the expression of the cyclic RNA HIPK2 and thereby ameliorating depression. This highlights how gut microbes improve astrocyte dysfunction and alleviate depression [110]. These findings underscored the critical role of NLRP3 inflammasome in regulating gut microbiome homeostasis and confirmed the interaction between the gut microbiome and NLRP3 inflammasome.
The environment and diet also influence the gut microbes and the inflammasome. Temperature is an essential aspect of the environment. Temperature stress has been reported to alter the gut-brain axis, including the gut microbiota and neuroinflammation. Different temperatures induce changes in the composition of the gut microbiome and levels of the metabolite SCFA, as well as activation of the central NLRP3 inflammasome [111]. Dietary or age-related ecological dysregulation commonly leads to a loss of SCFA-producing bacteria and SCFA levels. These may trigger intestinal barrier dysfunction and subsequent inflammatory events leading to systemic inflammation and neuroinflammation and neurodegeneration [112], [113]. The nutritional composition of Western, Mediterranean, and ketogenic diets affects the gut microbiome in distinct ways by regulating the NLRP3 inflammasome, which, in turn, influences the function of the gut and blood-brain barrier [114], [115], [116], [117]. Dietary supplements such as dietary fiber, bioflavonoids, or ω3 fatty acids may benefit the organism by modulating the gut microbiome and the activation of the NLRP3 inflammasome [118], [119], [120]. Therefore, targeted modulation of the gut microbiota and inflammasome through environmental improvements or dietary supplementation could represent a promising intervention strategy for diseases related to these factors. Although numerous studies have established a link between the gut microbiome and NLRP3, the precise mechanisms of this interaction require further exploration. Conflicting findings across studies may be attributed to various factors, including differences in animal models or disease contexts. Accordingly, it is crucial to gain a deeper understanding of the specific mechanisms that underlie the interplay between the gut microbiome and NLRP3.
5. Intestinal microbiota and NLRP3 inflammasome regulate intestinal inflammation
In recent years, there has been increasing evidence that NLPR3 functions in intestinal diseases through classical or non-classical pathways and is closely associated with the gut microbiome [89]. Several studies have revealed that in IBD, including Crohn's disease and ulcerative colitis, the expression of NLPR3 inflammasome and its assembly components and downstream inflammatory factors such as IL-1β and IL-18 are involved in protective and pathogenic mechanisms [121], [122]. IBD can develop due to various factors, including changes in antigen levels and abnormal immune responses triggered by the accumulation of commensal bacteria in the intestinal lumen. These factors can lead to damage of the epithelial cells, increased intestinal permeability, and the classic symptoms of IBD, such as chronic inflammation, pain, bleeding, diarrhea, and malnutrition [123], [124], [125], [126]. Significantly, the presence of single-nucleotide polymorphisms (SNPs) in the regulatory elements of NLRP3 has been linked to heightened susceptibility to Crohn's disease in humans, leading to decreased NLRP3 expression and reduced production of IL-1β [127]. The complex role of NLRP3 in chronic intestinal disease is further confirmed by the increased expression of NLRP3 and IL-18 in colon biopsies from a mouse model of colitis and patients with Crohn's disease [127], [128]. These data provide a theoretical basis for assessing the role of the gut microbiome and NLRP3 in intestinal inflammation.
Dextran sulfate sodium (DSS) is a commonly used chemical to induce experimental colitis in animals, which has been shown to activate inflammasomes [100], [129]. NLRP3 inflammasomes were identified as a critical mechanism of intestinal inflammation in a model of DSS colitis [130]. Allen et al. reported that NLRP3 inflammasome-mediated IL-1β protects intestinal epithelial cells [131]. In DSS-induced colitis, NLRP3-/- mice showed decreased expression of the anti-inflammatory cytokine IL-1β, exhibited more severe colitis, a worse prognosis, and an increased risk of inflammation-associated colorectal cancer [15]. However, colonic lamina propria cells isolated from GF mice did not produce IL-1β, highlighting the critical role of the gut microbiota and NLRP3 inflammasome in colitis [100]. Furthermore, NLRP3-/- mice have impaired epithelial proliferation and recovery, increased intestinal permeability, and increased susceptibility of commensal bacteria to translocate to the colonic mucosa and mesenteric lymph nodes after DSS feeding compared to WT mice [132]. In contrast, it has been noted that the deletion of inflammasome-associated component proteins such as NLRP3, ASC, and caspase-1 reduced histopathological signs and sensitivity to DSS in colitis mice compared to controls [130], [133]. Conflicting findings may be attributed to variations in animal feeding environments, which can lead to distinct structural differences in the gut microbiome. Consequently, the modulatory effects of different microbes on NLRP3 can have either positive or negative outcomes. Therefore, clarifying the molecular mechanism and environmental dependence of the interaction between the intestinal microbiome and NLRP3 inflammasome will help explore future therapeutic directions and targets for IBD.
Targeted treatment of chronic intestinal diseases using the interaction between NLRP3 inflammasome and intestinal microecology has recently garnered interest among researchers [15], [134]. Modifying the structure of the intestinal microbiota using anti-inflammatory substances, probiotics, microbiota metabolites, or diet can improve intestinal inflammation [135], [136], [137]. Using a combination of the anti-inflammatory drug Rosuvastatin and Lactobacillus has significantly reduced colonic inflammation induced by DSS/HFD. This combination also corrected flora dysbiosis by inhibiting NF-κB activation and NLRP3 inflammatory inhibition of body assembly. These findings provide a safe and effective treatment strategy for ulcerative colitis [135]. The currently recognized intestinal probiotic Akkermansia muciniphila significantly induces upregulation of NLRP3 in mouse macrophages Raw264.7 cells and BMDM cells and is protective against acute colitis in mice [136]. Dietary supplementation of sanguinarine (SAN), a natural anti-inflammatory component, can significantly alleviate the colonic pathological injury induced by DSS and improve intestinal microbiota imbalance in mice by blocking NLRP3-(Caspase-1)/IL-1β pathway [137]. Dietary fiber is a significant source of short-chain fatty acids, a gut microbial metabolite. When fed a high-fiber diet during DSS-induced colitis, mice lacking NLRP3 but not NLRP6 had an exacerbation compared to their wild-type (WT) counterparts suggesting that the protective effect of dietary fiber is mediated through the NLRP3 inflammasome. GF mice that received FMT from mice fed a high-fiber diet, rather than a zero-fiber diet, showed elevated serum IL-18 levels and reduced symptoms of DSS-induced colitis. This demonstrates that dietary intervention can alleviate colitis by controlling the expansion of pathogenic microbiota and reducing harmful immune responses [60]. However, conflicting findings remain specific to the effects of different fiber diets on colitis [138], [139]. In addition, the immunomodulatory effects of plant-derived extracts such as hydroxytyrosol(HT), phloretin, Saikosaponin-d(SSd), and milk-derived extracellular vesicles (MEV) on the gut via the microbiome and NLRP3 have been successively identified [140], [141], [142].
IBS is a chronic functional gastrointestinal disorder characterized by gut microbiota dysbiosis and chronic low-grade inflammation. Research has shown that the inflammasome inhibitor BAY 11–7082 has therapeutic potential in IBS rats. It can significantly reduce tissue damage caused by edema, neutrophil infiltration, and loss of colonic structure by inhibiting the activation of NLRP3 inflammasome [143]. Bifidobacterium longum has also been found to alleviate visceral hypersensitivity in post-infectious irritable bowel syndrome (PI-IBS) by downregulating IL-18 and IL-1β expression by inhibiting NLRP3 inflammasomes [144]. However, the specific mechanism of action of the gut microbiota and NLRP3 in IBS requires further investigation. The NLRP3 inflammatory microsome-gut microbiota network is crucial in maintaining intestinal homeostasis and coordinating host physiology.
6. Intestinal microbiota and NLRP3 inflammasome crosstalk with the gut-brain axis
The gut microbiome regulates the intestinal microenvironment and acts on other organs throughout the body through the immune system [145], [146]. Many studies have proved that the microbiome affects brain homeostasis through the gut-brain axis. Alterations in brain physiological functions are caused by bacterial translocation in the gut, which can be stimulated by various factors such as disease, environment, or diet [111], [147], [148]. The gut-brain axis includes the central nervous system, the central endocrine system, and the central immune system. It consists of the hypothalamic-pituitary-adrenal axis (HPA axis), the sympathetic nervous system, the parasympathetic nervous system (vagus), and the enteric nervous system of the autonomic nervous system, as well as the microorganisms in the gut. Additionally, the gut microbiota plays a significant role in gut-brain communication, forming the microbiota-gut-brain axis (MGB). This network relies heavily on the interaction between gut bacteria and the intestinal epithelial barrier (IEB), the immune system, and neural pathways [145]. The bacteria-immunity interplay facilitates communication between the gut-brain axis, as gut microbiota directly stimulates immune cells in the gut and circulation while regulating the central nervous system and the brain's physiological functions. Moreover, certain microorganisms in the gut diffuse with the blood to the brain and influence inflammation in the CNS [149]. Neurotransmitters regulated by the gut microbiome also play an essential role in transmitting information between the gut and the brain [150], [151]. Therefore, an in-depth exploration of the molecular mechanisms of gut microbiota in gut-brain communication is of great significance for neurological diseases.
Gut microbiota-NLRP3 interactions are involved in the regulation of brain function. Using GF mice, researchers demonstrated that gut bacteria affect the biological processes of the central nervous system in several ways, including development, neurogenesis, neurotransmission, immune activity, and maintenance of the blood-brain barrier [152], [153]. The upregulation of mRNA expression of caspase-1, ASC, IL-1β, and IL-18 in the intestinal and brain tissues of mice treated with antibiotics suggests that the intestinal microbiota modulates the expression of inflammasome components [105]. The impact of the NLRP3 inflammasome on the gut-brain axis has also been demonstrated in animal experiments. Knockdown of NLRP3 in mice led to changes in gut microbiota composition and altered behavior and motility, highlighting the importance of the NLRP3 inflammasome in regulating mood-related activity. Additionally, caspase-1-/- mice displayed a distinct gut microbiota composition and behavioral patterns compared to WT mice, further supporting the role of NLRP3 inflammasome in gut-brain axis interactions [154]. These findings illustrate the regulatory role of the gut microbiota and NLRP3 on the brain. Further exploration and discussion are still needed to clarify the specific cellular mechanisms and factors of gut microbiota and NLRP3 inflammasome in regulating gut-brain communication under physiological and pathological conditions.
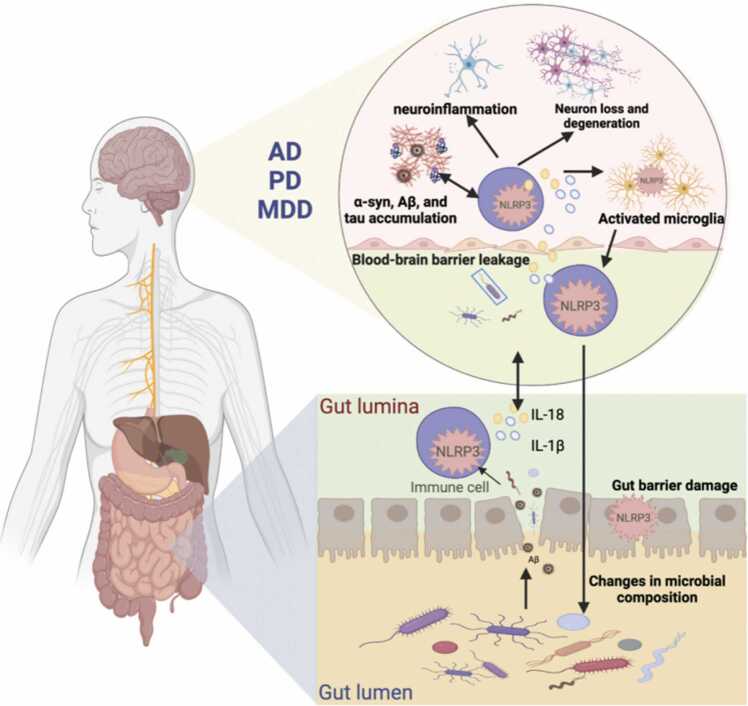
Currently, there is great interest in studying disorders related to the gut-brain axis, particularly neurodegenerative diseases such as Parkinson's (PD) and Alzheimer's (AD), as well as psychiatric disorders including autism spectrum disorders (ASD) and major depressive disorders (MDD). The pathogenesis of these disorders involves oxidative stress, mitochondrial dysfunction, apoptosis, and immune inflammation [155], [156], [157], [158], [159]. Inflammation plays a vital role in the mechanism of action of neurodegenerative diseases, and the activation of neuroglia, especially microglia, is the main hallmark of brain inflammation [160], [161]. Notably, alterations in the gut microbiota composition directly affect the gut mucosal barrier, leading to gastrointestinal dysfunction and triggering a neurogenic/inflammatory response in the gut and periphery [145], [146]. At the same time, the permeability of the BBB is significantly enhanced, and potential neurotoxic factors produced by the microbiome permeate the CNS, leading to neuroinflammation and neurodegeneration [91]. Moreover, the role of NLRP3 inflammasome in activating and releasing IL-1β and IL-18, and thus coordinating host physiology and regulating CNS disorders, is gradually being recognized [162], [163]. Studies have demonstrated that gut microbiota and NLRP3 inflammasome interactions have a regulatory role in the center, and gut bacteria regulate peripheral inflammatory pathways through inflammasome signaling, which affects brain homeostasis [164], [165], [166]. There is evidence to suggest that various stimuli related to inflammation and neurodegenerative processes, such as increased extracellular ATP, β-amyloid (Aβ) fibers, α-synuclein (α-syn) aggregates, reactive oxygen species (ROS), and deubiquitination, may activate the NLRP3 inflammasome, which in turn could affect the composition of gut microbiota [15], [167], [168] (Fig. 3).
Fig. 3.
Interaction between gut microbiota and NLRP3 inflammasome in the gut-brain axis. In patients with AD, PD, and MDD, gut microbes promote activation of NLRP3 in the periphery and the center, activating an inflammatory response that damages both the gut and the blood-brain barrier. This inflammation contributes to central neuroinflammation and neurodegeneration. Additionally, the accumulation of certain proteins such as Aβ, α-syn, and Tau in the brain can stimulate the formation of NLRP3 inflammasome in microglia, further accelerating neurodegeneration. This process may also affect the structure of the gut microbiota.
A typical pathological feature is the massive activation of microglia in the central nervous system for patients with AD and PD. Stimulation by Aβ and α-syn in the brain accelerates the formation of NLRP3 inflammasome in microglia. It induces apoptosis or pyroptosis through the expression of IL-1β, caspase-1, etc., leading to the development of neurodegeneration [164], [169]. At the same time, epidemiology revealed alterations in the intestinal microbiota structure in patients with AD and PD, accompanied by the development of intestinal inflammation [170], [171]. Research has shown a direct effect of microbiota on Aβ amyloidosis in the CNS [172]. In addition, the gut microbiota of AD patients has been shown to produce amyloid peptides that escape from the gastrointestinal tract and accumulate in the brain via the gut-brain upstream pathway, possibly playing a role in the production of pro-inflammatory cytokines, presumably via NLRP3 activation [173]. Thus, NLRP3 inflammasome activation may play a pivotal role in the gut and CNS interaction. APP/PS1 double transgenic mice are an ideal model to study the pathogenesis of AD, and transplantation of intestinal flora from AD patients into APP/PS1 double transgenic mice revealed increased expression of NLRP3 in the intestine and increased expression of inflammatory factors in peripheral blood expression levels of inflammatory factors were also increased. Mice receiving gut microbiota from AD patients had increased microglia activation and expression of neuroinflammatory factors in their brains, leading to more severe cognitive impairment [174]. Additionally, the study investigated the intestinal traits of 5xFAD mice, a transgenic mouse model of AD carrying five familial AD-associated mutations. The results revealed significant alterations in the intestinal microbiota composition of 5xFAD mice compared to WT mice, along with impaired intestinal barrier function and elevated levels of NLRP3 and IL-1β in the gut. Furthermore, the brains of 5xFAD mice exhibited increased expression of NLRP3 inflammasome and IL-1β, as well as an enhanced proliferation of astrocytes and activation of microglia [175]. This evidence suggests that elevated expression of microbial-associated inflammatory components in the gut may be an essential trigger for subsequent inflammatory activation. The intestinal origin of NLRP3 may promote NLRP3 inflammasome-mediated neuroinflammation. Thus, modulating the gut microbiota may be a potential strategy for treating neurologic diseases related to AD in genetically susceptible hosts. Probiotics such as Lactobacillus plantarum MA2 (MA2) have been shown to improve cognitive deficits and anxiety-like behaviors in AD rats by remodeling the structure and composition of the gut microbiota, attenuating intestinal mucosal damage and impeding microglia activation and neuroinflammation via the TLR4/MYD88/NLRP3 signaling pathway [176]. Intestinal microbial metabolites may be another effective treatment for AD. Microbiota-derived indole upregulates the production of aromatic hydrocarbon receptor (AhR) through tryptophan metabolism, inhibits the activation of the NF-κB signaling pathway, and the formation of the NLRP3 inflammasome reduces the inflammatory response and then prevents Aβ accumulation and Tau hyperphosphorylation [67]. In contrast, the gut microbiota-derived metabolite TMAO has been found to stimulate the activation of NLRP3 inflammasome in both the peripheral and the CNS, and its level increases with age-related cognitive dysfunction. Moreover, TMAO has been shown to cause mitochondrial dysfunction, oxidative stress, neuronal aging, and synaptic damage in the brain [67]. Therefore, further understanding the interrelationship between the gut microbiota and NLRP3 and identifying relevant foods or probiotics that regulate its homeostasis could provide new therapeutic directions for neurological diseases.
The relevance of PD, another central disease closely related to neuroinflammation, to the gut microbiome and NLRP3 inflammasome activation is increasingly being understood [177]. A study by Sampson et al. in 2016 showed that transgenic animals that overexpress human αSyn exhibit decreased microglia activation, αSyn inclusions, and motor deficits under GF conditions or when treated with antibiotics to deplete bacteria, as compared to animals with a complex microbiota. Additionally, treatment with SCFAs, produced by gut microbiota, can restore all major disease features in GF mice, confirming the regulatory function of the gut microbiome in PD [178]. Specifically, alterations in the gut microbiota of PD patients were characterized by decreases in Verrucomicrobia, Mucispirillum schaedleri, Porphyromonas, Lactobacillus, and Parabacteroides, as well as increased abundance of Prevotella, associated with increased levels of circulating pro-inflammatory cytokines, including IL-1β [179]. Consistent with this, there was a decrease in SCFA-producing bacteria, reduced colonic IL-1β levels, and impaired IEB in PD patients [180]. When administered to mice with PD, butyrate significantly worsened their motor dysfunction, reduced dopamine and 5-hydroxytryptamine (5-HT) levels, accelerated the loss of dopaminergic neurons, and intensified neuroinflammation mediated by glial cells by increasing the number of microglia and activating astrocytes [181]. Of note, in vitro studies have shown that butyrate significantly increases the expression of pro-inflammatory cytokines(IL-1β, IL-18) in LPS-stimulated BV2 cells and upregulates the expression of NLRP3, indicating a possible association between intestinal microbiota and NLRP3 in the pathogenesis of PD [181]. Nevertheless, additional clinical evidence and experimental investigation are required to elucidate the specific mechanisms by which the gut microbiota and NLRP3 influence the onset and progression of PD.
Carolina Pellegrini et al. provide a preliminary overview of the interaction between the gut microbiome and NLRP3 inflammasome with psychiatric disorders. Existing evidence suggests a connection between gut microbiota, NLRP3 inflammasome activation, and brain pathology symptoms in psychiatric disorders [167]. Zhang et al. employed NLRP3-/- mice to validate the connection between gut microbiota, NLRP3 inflammasome, and depression. They found that microbiota transplantation from NLRP3-/- mice alleviated depression-like behavior induced by CUS. The underlying mechanism could be attributed to the regulation of circHIPK2 expression, which improved astrocyte dysfunction in mice [110]. A recent groundbreaking study has revealed that CD36, a scavenger receptor, plays a crucial role in maintaining the ecological balance of gut microbiota and regulating NLRP3 in MDD [182]. The study found that CD36 expression was elevated in depressed mice and patients. Interestingly, CD36 deficiency in mice led to decreased hippocampal NLRP3 inflammasome signaling pathway and increased cecum bacterial alpha diversity. Specifically, CD36 knockout mice had a higher abundance of Bacteroides, Rikenella, and Alloprevotella but lower levels of Allobaculum, suggesting that CD36 plays a vital role in shaping the gut microbiota composition, which in turn affects depressive behavior via the inflammasome pathway [182]. Treatment with fecal microbiota transplants derived from normal rats also improved CUS-induced depression-like behavior, with 5-HT reduced in the CUS rat model but significantly increased after fecal microbiota transplantation [165]. Of interest, the application of probiotics for the treatment of mood anxiety and depression is being demonstrated. Mycobacterium bovis reduced basal levels of genes (NLRP3 and NF-κB) involved in microglia priming, thus preventing a stress-induced increase in anxiety-like behavior [183]. Probiotics such as Lactobacillus abrogate the hyperactivation of microglia and reduced expression of dopamine transporter proteins in brain tissue, which are closely associated with depression, by improving ecological dysregulation and lowering levels of TLR4, NLRP3, and IL-1β in intestinal and brain tissue [184], [185]. In conclusion, modulating the gut microbiome and NLRP3 inflammasome to regulate the body's ecological balance and inflammatory homeostasis, as well as utilizing probiotics and their metabolites to regulate gut microbiota structure, could offer novel therapeutic approaches for addressing psychiatric disorders.
7. Prospect
The essential roles of the gut microbiome and inflammasomes in various inflammatory, metabolic, neurological, and cardiovascular diseases are gradually being revealed. At the same time, current evidence supports the role of the gut microbiome and inflammasome interactions in disease regulation. This paper mainly summarizes that in the gut and central system, the gut microbiome and its metabolites interact with NLRP3 inflammasome to regulate the physiological functions or pathological alterations of the body by promoting or inhibiting the cleavage of caspase-1 and the expression of inflammatory factors such as IL-1β and IL-18, and to influence central nervous system diseases through the gut-brain axis. The environment, diet, drugs, and the organism's internal environment also affect this action. Thus, the gut microbiota and NLRP3 inflammasome may be potential targets for treating various pathologies. Depletion through manipulation or targeting of the gut microbiota and genetic deletion or pharmacological modulation of inflammasome signaling may be a new direction to explore in clinical therapy. Therefore, an in-depth understanding of the balance between probiotic and harmful bacteria in the microbiota and between microbiota and inflammasome activation is critical. In addition, whether other inflammasomes, including NLRP6, NLRP1, AIM2, and NLRC4, exist in different diseases in crosstalk with the gut microbiome and NLRP3 must be investigated in depth.
In conclusion, the specific mechanisms of gut microbiome-inflammasome interactions still require further basic and clinical studies. There are still plenty of future research prospects for targeting the gut microbiome and NLRP3 inflammasome to develop therapeutic strategies and drug development for preventing and treating inflammation-associated diseases.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
CRediT authorship contribution statement
Ding Yang: Investigation, Software, Writing – original draft. Zixu Wang: Resources, Writing – review & editing. Yaoxing Chen: Resources, Writing – review & editing. Qingyun Guo: Investigation, Writing – review & editing. Yulan Dong: Supervision, Investigation, Writing – original draft, Writing – review & editing, Funding acquisition. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (grant nos. 31972633, 31572476, 31272483), the Natural Science Foundation of Beijing, China (grant no. 6172022, 6194031), and Bud Program of Beijing Academy of Science and Technology (grant no. BGS202108).
References
- 1.Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 2.Hersoug L.-G., Møller P., Loft S. Role of microbiota-derived lipopolysaccharide in adipose tissue inflammation, adipocyte size and pyroptosis during obesity. Nutr Res Rev. 2018;31:153–163. doi: 10.1017/S0954422417000269. [DOI] [PubMed] [Google Scholar]
- 3.He F., Huang Y., Song Z., Zhou H.J., Zhang H., Perry R.J., et al. Mitophagy-mediated adipose inflammation contributes to type 2 diabetes with hepatic insulin resistance. J Exp Med. 2021;218 doi: 10.1084/jem.20201416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jourdan T., Godlewski G., Cinar R., Bertola A., Szanda G., Liu J., et al. Activation of the Nlrp3 inflammasome in infiltrating macrophages by endocannabinoids mediates beta cell loss in type 2 diabetes. Nat Med. 2013;19:1132–1140. doi: 10.1038/nm.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duewell P., Kono H., Rayner K.J., Sirois C.M., Vladimer G., Bauernfeind F.G., et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Y., Liu Q., Zhang W., Wan Y., Huang C., Zhu X. Ursolic acid reverses liver fibrosis by inhibiting NOX4/NLRP3 inflammasome pathways and bacterial dysbiosis. Gut Microbes. 2021;13:1972746. doi: 10.1080/19490976.2021.1972746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Qazazi R., Lima P.D.A., Prisco S.Z., Potus F., Dasgupta A., Chen K.-H., et al. Macrophage–NLRP3 activation promotes right ventricle failure in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2022;206:608–624. doi: 10.1164/rccm.202110-2274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schunk S.J., Kleber M.E., März W., Pang S., Zewinger S., Triem S., et al. Genetically determined NLRP3 inflammasome activation associates with systemic inflammation and cardiovascular mortality. Eur Heart J. 2021;42:1742–1756. doi: 10.1093/eurheartj/ehab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen S., Prame Kumar K., Stanley D., Moore R.J., Van T.T.H., Wen S.W., et al. Invariant natural killer T cells shape the gut microbiota and regulate neutrophil recruitment and function during intestinal inflammation. Front Immunol. 2018;9:999. doi: 10.3389/fimmu.2018.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinan T.G., Cryan J.F. Microbes, immunity, and behavior: psychoneuroimmunology meets the microbiome. Neuropsychopharmacol. 2017;42:178–192. doi: 10.1038/npp.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inserra A., Rogers G.B., Licinio J., Wong M.-L. The microbiota-inflammasome hypothesis of major depression. BioEssays. 2018;40:1800027. doi: 10.1002/bies.201800027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haneklaus M., O’Neill L.A.J. NLRP3 at the interface of metabolism and inflammation. Immunol Rev. 2015;265:53–62. doi: 10.1111/imr.12285. [DOI] [PubMed] [Google Scholar]
- 13.Brown E.M., Kenny D.J., Xavier R.J. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol. 2019;37:599–624. doi: 10.1146/annurev-immunol-042718-041841. [DOI] [PubMed] [Google Scholar]
- 14.Mangan M.S.J., Olhava E.J., Roush W.R., Seidel H.M., Glick G.D., Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Disco. 2018;17:588–606. doi: 10.1038/nrd.2018.97. [DOI] [PubMed] [Google Scholar]
- 15.Hirota S.A., Ng J., Lueng A., Khajah M., Parhar K., Li Y., et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogala A.R., Oka A., Sartor R.B. Strategies to dissect host-microbial immune interactions that determine mucosal homeostasis vs. intestinal inflammation in gnotobiotic mice. Front Immunol. 2020;11:214. doi: 10.3389/fimmu.2020.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiyama K., Naito Y., Takagi T. Intestinal microbiome as a novel therapeutic target for local and systemic inflammation. Pharmacol Ther. 2019;199:164–172. doi: 10.1016/j.pharmthera.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Mou Y., Du Y., Zhou L., Yue J., Hu X., Liu Y., et al. Gut microbiota interact with the brain through systemic chronic inflammation: implications on neuroinflammation, neurodegeneration, and aging. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.796288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundman M.H., Chen N., Subbian V., Chou Y. The bidirectional gut-brain-microbiota axis as a potential nexus between traumatic brain injury, inflammation, and disease. Brain Behav Immun. 2017;66:31–44. doi: 10.1016/j.bbi.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G., Gasbarrini A., et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7:14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasmi A., Mujawdiya P.K., Pivina L., Doşa A., Semenova Y., Benahmed A.G., et al. Relationship between gut microbiota, gut hyperpermeability and obesity. CMC. 2021;28:827–839. doi: 10.2174/0929867327666200721160313. [DOI] [PubMed] [Google Scholar]
- 23.Guinane C.M., Cotter P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther Adv Gastroenterol. 2013;6:295–308. doi: 10.1177/1756283X13482996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khosravi A., Mazmanian S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr Opin Microbiol. 2013;16:221–227. doi: 10.1016/j.mib.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheithauer T.P.M., Rampanelli E., Nieuwdorp M., Vallance B.A., Verchere C.B., van Raalte D.H., et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.571731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho I., Blaser M.J. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potrykus M., Czaja-Stolc S., Stankiewicz M., Kaska Ł., Małgorzewicz S. Intestinal microbiota as a contributor to chronic inflammation and its potential modifications. Nutrients. 2021;13:3839. doi: 10.3390/nu13113839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anto L., Blesso C.N. Interplay between diet, the gut microbiome, and atherosclerosis: Role of dysbiosis and microbial metabolites on inflammation and disordered lipid metabolism. J Nutr Biochem. 2022;105 doi: 10.1016/j.jnutbio.2022.108991. [DOI] [PubMed] [Google Scholar]
- 29.Arnoriaga-Rodríguez M., Fernández-Real J.M. Microbiota impacts on chronic inflammation and metabolic syndrome - related cognitive dysfunction. Rev Endocr Metab Disord. 2019;20:473–480. doi: 10.1007/s11154-019-09537-5. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh S., Whitley C.S., Haribabu B., Jala V.R. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. 2021;11:1463–1482. doi: 10.1016/j.jcmgh.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liébana-García R., Olivares M., Bullich-Vilarrubias C., López-Almela I., Romaní-Pérez M., Sanz Y. The gut microbiota as a versatile immunomodulator in obesity and associated metabolic disorders. Best Pract Res Clin Endocrinol Metab. 2021;35 doi: 10.1016/j.beem.2021.101542. [DOI] [PubMed] [Google Scholar]
- 32.Salliss M.E., Farland L.V., Mahnert N.D., Herbst-Kralovetz M.M. The role of gut and genital microbiota and the estrobolome in endometriosis, infertility and chronic pelvic pain. Hum Reprod Update. 2021;28:92–131. doi: 10.1093/humupd/dmab035. [DOI] [PubMed] [Google Scholar]
- 33.Wells J.M., Rossi O., Meijerink M., van Baarlen P. Epithelial crosstalk at the microbiota–mucosal interface. Proc Natl Acad Sci USA. 2011;108:4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caron T.J. Tight junction disruption: Helicobacter pylori and dysregulation of the gastric mucosal barrier. WJG. 2015;21:11411. doi: 10.3748/wjg.v21.i40.11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81 doi: 10.1128/MMBR.00036-17. e00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar H., Kawai T., Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 38.Rajaee A., Barnett R., Cheadle W.G. Pathogen- and danger-associated molecular patterns and the cytokine response in sepsis. Surg Infect. 2018;19:107–116. doi: 10.1089/sur.2017.264. [DOI] [PubMed] [Google Scholar]
- 39.Rathinam V.A.K., Zhao Y., Shao F. Innate immunity to intracellular LPS. Nat Immunol. 2019;20:527–533. doi: 10.1038/s41590-019-0368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M., et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 41.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 42.Everts B., Amiel E., Huang S.C.-C., Smith A.M., Chang C.-H., Lam W.Y., et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu J.-K., Kim S.J., Rah S.-H., Kang J.I., Jung H.E., Lee D., et al. Reconstruction of LPS transfer cascade reveals structural determinants within LBP, CD14, and TLR4-MD2 for efficient LPS recognition and transfer. Immunity. 2017;46:38–50. doi: 10.1016/j.immuni.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Barker J.H., Weiss J.P. Detecting lipopolysaccharide in the cytosol of mammalian cells: Lessons from MD-2/TLR4. J Leukoc Biol. 2019;106:127–132. doi: 10.1002/JLB.3MIR1118-434R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi Y.-S. Caspase-11 non-canonical inflammasome: a critical sensor of intracellular lipopolysaccharide in macrophage-mediated inflammatory responses. Immunology. 2017;152:207–217. doi: 10.1111/imm.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Yan H. TLR5: beyond the recognition of flagellin. Cell Mol Immunol. 2017;14:1017–1019. doi: 10.1038/cmi.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hajam I.A., Dar P.A., Shahnawaz I., Jaume J.C., Lee J.H. Bacterial flagellin—a potent immunomodulatory agent. Exp Mol Med. 2017;49 doi: 10.1038/emm.2017.172. e373–e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan K., Pang R., Zhao C., Liu X., Gao W., Zhang J., et al. IL-17-triggered downregulation of miR-497 results in high HIF-1α expression and consequent IL-1β and IL-6 production by astrocytes in EAE mice. Cell Mol Immunol. 2017;14:909–923. doi: 10.1038/cmi.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tominari T., Sanada A., Ichimaru R., Matsumoto C., Hirata M., Itoh Y., et al. Gram-positive bacteria cell wall-derived lipoteichoic acid induces inflammatory alveolar bone loss through prostaglandin E production in osteoblasts. Sci Rep. 2021;11:13353. doi: 10.1038/s41598-021-92744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf A.J., Underhill D.M. Peptidoglycan recognition by the innate immune system. Nat Rev Immunol. 2018;18:243–254. doi: 10.1038/nri.2017.136. [DOI] [PubMed] [Google Scholar]
- 51.Huang Z., Wang J., Xu X., Wang H., Qiao Y., Chu W.C., et al. Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat Microbiol. 2019;4:766–773. doi: 10.1038/s41564-019-0381-1. [DOI] [PubMed] [Google Scholar]
- 52.McDonald C., Inohara N., Nuñez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280:20177–20180. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- 53.Zheng X., Cai X., Hao H. Emerging targetome and signalome landscape of gut microbial metabolites. Cell Metab. 2022;34:35–58. doi: 10.1016/j.cmet.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 54.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.-J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 56.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 57.Morrison D.J., Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shakespear M.R., Halili M.A., Irvine K.M., Fairlie D.P., Sweet M.J. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 59.Chen G., Ran X., Li B., Li Y., He D., Huang B., et al. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine. 2018;30:317–325. doi: 10.1016/j.ebiom.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macia L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S., et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 61.Jia B., Jeon C.O. Promotion and induction of liver cancer by gut microbiome-mediated modulation of bile acids. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X., Geng J., Zhao J., Ni Q., Zhao C., Zheng Y., et al. Trimethylamine N-oxide exacerbates cardiac fibrosis via activating the NLRP3 inflammasome. Front Physiol. 2019;10:866. doi: 10.3389/fphys.2019.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu K., Yuan Y., Yu H., Dai X., Wang S., Sun Z., et al. The gut microbial metabolite trimethylamine N-oxide aggravates GVHD by inducing M1 macrophage polarization in mice. Blood. 2020;136:501–515. doi: 10.1182/blood.2019003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S., Li X., Yang F., Zhao R., Pan X., Liang J., et al. Gut microbiota-dependent marker TMAO in promoting cardiovascular disease: inflammation mechanism, clinical prognostic, and potential as a therapeutic target. Front Pharm. 2019;10:1360. doi: 10.3389/fphar.2019.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X., Li Y., Yang P., Liu X., Lu L., Chen Y., et al. Trimethylamine-N-oxide promotes vascular calcification through activation of NLRP3 (Nucleotide-Binding Domain, Leucine-Rich-Containing Family, Pyrin Domain-Containing-3) inflammasome and NF-κB (Nuclear Factor κB) signals. Arterioscler Thromb Vasc Biol. 2020;40:751–765. doi: 10.1161/ATVBAHA.119.313414. [DOI] [PubMed] [Google Scholar]
- 66.Kapetanaki S., Kumawat A.K., Persson K., Demirel I. The fibrotic effects of TMAO on human renal fibroblasts is mediated by NLRP3, caspase-1 and the PERK/Akt/mTOR pathway. IJMS. 2021;22:11864. doi: 10.3390/ijms222111864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Praveenraj S.S., Sonali S., Anand N., Tousif H.A., Vichitra C., Kalyan M., et al. The role of a gut microbial-derived metabolite, trimethylamine N-oxide (TMAO), in neurological disorders. Mol Neurobiol. 2022 doi: 10.1007/s12035-022-02990-5. [DOI] [PubMed] [Google Scholar]
- 68.Martinon F., Burns K., Tschopp J. The inflammasome. Mol Cell. 2002;10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 69.Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 70.Fernandes-Alnemri T., Yu J.-W., Juliana C., Solorzano L., Kang S., Wu J., et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu H., Yang J., Gao W., Li L., Li P., Zhang L., et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 73.von Moltke J., Ayres J.S., Kofoed E.M., Chavarría-Smith J., Vance R.E. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 74.Boyden E.D., Dietrich W.F. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 75.Sandstrom A., Mitchell P.S., Goers L., Mu E.W., Lesser C.F., Vance R.E. Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science. 2019 doi: 10.1126/science.aau1330. 364:eaau1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Latz E., Xiao T.S., Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 78.Rathinam V.A.K., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sauer J.-D., Witte C.E., Zemansky J., Hanson B., Lauer P., Portnoy D.A. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leemans J.C., Cassel S.L., Sutterwala F.S. Sensing damage by the NLRP3 inflammasome: NLRP3 inflammasome in sterile inflammation. Immunol Rev. 2011;243:152–162. doi: 10.1111/j.1600-065X.2011.01043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGeough M.D., Wree A., Inzaugarat M.E., Haimovich A., Johnson C.D., Peña C.A., et al. TNF regulates transcription of NLRP3 inflammasome components and inflammatory molecules in cryopyrinopathies. J Clin Investig. 2017;127:4488–4497. doi: 10.1172/JCI90699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Groslambert M., Py B. Spotlight on the NLRP3 inflammasome pathway. JIR. 2018;11:359–374. doi: 10.2147/JIR.S141220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Y., Wang H., Kouadir M., Song H., Shi F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019;10:128. doi: 10.1038/s41419-019-1413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coll R.C., Schroder K., Pelegrín P. NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharmacol Sci. 2022;43:653–668. doi: 10.1016/j.tips.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 85.Huang Y., Xu W., Zhou R. NLRP3 inflammasome activation and cell death. Cell Mol Immunol. 2021;18:2114–2127. doi: 10.1038/s41423-021-00740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He X., Yang W., Zeng Z., Wei Y., Gao J., Zhang B., et al. NLRP3-dependent pyroptosis is required for HIV-1 gp120-induced neuropathology. Cell Mol Immunol. 2020;17:283–299. doi: 10.1038/s41423-019-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S., Yuan Y.-H., Chen N.-H., Wang H.-B. The mechanisms of NLRP3 inflammasome/pyroptosis activation and their role in Parkinson’s disease. Int Immunopharmacol. 2019;67:458–464. doi: 10.1016/j.intimp.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 88.Zhen Y., Zhang H. NLRP3 inflammasome and inflammatory bowel disease. Front Immunol. 2019;10:276. doi: 10.3389/fimmu.2019.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larabi A., Barnich N., Nguyen H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16:38–51. doi: 10.1080/15548627.2019.1635384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li X., He C., Li N., Ding L., Chen H., Wan J., et al. The interplay between the gut microbiota and NLRP3 activation affects the severity of acute pancreatitis in mice. Gut Microbes. 2020;11:1774–1789. doi: 10.1080/19490976.2020.1770042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pellegrini C., Antonioli L., Colucci R., Blandizzi C., Fornai M. Interplay among gut microbiota, intestinal mucosal barrier and enteric neuro-immune system: a common path to neurodegenerative diseases. Acta Neuropathol. 2018;136:345–361. doi: 10.1007/s00401-018-1856-5. [DOI] [PubMed] [Google Scholar]
- 92.Wu Q., Liu M.-C., Yang J., Wang J.-F., Zhu Y.-H. Lactobacillus rhamnosus GR-1 ameliorates escherichia coli-induced inflammation and cell damage via attenuation of ASC-independent NLRP3 inflammasome activation. Appl Environ Microbiol. 2016;82:1173–1182. doi: 10.1128/AEM.03044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yen H., Sugimoto N., Tobe T. Enteropathogenic Escherichia coli uses NleA to inhibit NLRP3 inflammasome activation. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dou X., Qiao L., Chang J., Yan S., Song X., Chen Y., et al. Lactobacillus casei ATCC 393 and it’s metabolites alleviate dextran sulphate sodium-induced ulcerative colitis in mice through the NLRP3-(Caspase-1)/IL-1β pathway. Food Funct. 2021;12:12022–12035. doi: 10.1039/d1fo02405a. [DOI] [PubMed] [Google Scholar]
- 95.Castro-Dopico T., Dennison T.W., Ferdinand J.R., Mathews R.J., Fleming A., Clift D., et al. Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis. Immunity. 2019;50(1099–1114) doi: 10.1016/j.immuni.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dufies O., Doye A., Courjon J., Torre C., Michel G., Loubatier C., et al. Escherichia coli Rho GTPase-activating toxin CNF1 mediates NLRP3 inflammasome activation via p21-activated kinases-1/2 during bacteraemia in mice. Nat Microbiol. 2021;6:401–412. doi: 10.1038/s41564-020-00832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De la Fuente M., Franchi L., Araya D., Díaz-Jiménez D., Olivares M., Álvarez-Lobos M., et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int J Med Microbiol. 2014;304:384–392. doi: 10.1016/j.ijmm.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fox D., Mathur A., Xue Y., Liu Y., Tan W.H., Feng S., et al. Bacillus cereus non-haemolytic enterotoxin activates the NLRP3 inflammasome. Nat Commun. 2020;11:760. doi: 10.1038/s41467-020-14534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Man S.M., Karki R., Sasai M., Place D.E., Kesavardhana S., Temirov J., et al. IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell. 2016;167(382–396) doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seo S.-U., Kamada N., Muñoz-Planillo R., Kim Y.-G., Kim D., Koizumi Y., et al. Distinct commensals induce interleukin-1β via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity. 2015;42:744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fan L., Xu C., Ge Q., Lin Y., Wong C.C., Qi Y., et al. A. Muciniphila suppresses colorectal tumorigenesis by inducing TLR2/NLRP3-mediated M1-like TAMs. Cancer Immunol Res. 2021;9:1111–1124. doi: 10.1158/2326-6066.CIR-20-1019. [DOI] [PubMed] [Google Scholar]
- 102.Feng Y., Huang Y., Wang Y., Wang P., Song H., Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sabui S., Skupsky J., Kapadia R., Cogburn K., Lambrecht N.W., Agrawal A., et al. Tamoxifen-induced, intestinal-specific deletion of Slc5a6 in adult mice leads to spontaneous inflammation: involvement of NF-κB, NLRP3, and gut microbiota. Am J Physiol-Gastrointest Liver Physiol. 2019;317:G518–G530. doi: 10.1152/ajpgi.00172.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zaki MdH Boyd K.L., Vogel P., Kastan M.B., Lamkanfi M., Kanneganti T.-D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lowe P.P., Gyongyosi B., Satishchandran A., Iracheta-Vellve A., Cho Y., Ambade A., et al. Reduced gut microbiome protects from alcohol-induced neuroinflammation and alters intestinal and brain inflammasome expression. J Neuroinflamm. 2018;15:298. doi: 10.1186/s12974-018-1328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Y., Zhang S., Li B., Luo Y., Gong Y., Jin X., et al. Gut microbiota dysbiosis promotes age-related atrial fibrillation by lipopolysaccharide and glucose-induced activation of NLRP3-inflammasome. Cardiovasc Res. 2022;118:785–797. doi: 10.1093/cvr/cvab114. [DOI] [PubMed] [Google Scholar]
- 107.Liao L., Schneider K.M., Galvez E.J.C., Frissen M., Marschall H.-U., Su H., et al. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut. 2019;68:1477–1492. doi: 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 108.Zhang X.-N., Yu Z.-L., Chen J.-Y., Li X.-Y., Wang Z.-P., Wu M., et al. The crosstalk between NLRP3 inflammasome and gut microbiome in atherosclerosis. Pharm Res. 2022;181 doi: 10.1016/j.phrs.2022.106289. [DOI] [PubMed] [Google Scholar]
- 109.Manko-Prykhoda A., Allain T., Motta J.-P., Cotton J.A., Feener T., Oyeyemi A., et al. Giardia spp. promote the production of antimicrobial peptides and attenuate disease severity induced by attaching and effacing enteropathogens via the induction of the NLRP3 inflammasome. Int J Parasitol. 2020;50:263–275. doi: 10.1016/j.ijpara.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 110.Zhang Y., Huang R., Cheng M., Wang L., Chao J., Li J., et al. Gut microbiota from NLRP3-deficient mice ameliorates depressive-like behaviors by regulating astrocyte dysfunction via circHIPK2. Microbiome. 2019;7:116. doi: 10.1186/s40168-019-0733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yi W., Cheng J., Wei Q., Pan R., Song S., He Y., et al. Effect of temperature stress on gut-brain axis in mice: Regulation of intestinal microbiome and central NLRP3 inflammasomes. Sci Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2020.144568. [DOI] [PubMed] [Google Scholar]
- 112.Noble E.E., Hsu T.M., Kanoski S.E. Gut to brain dysbiosis: mechanisms linking western diet consumption, the microbiome, and cognitive impairment. Front Behav Neurosci. 2017:11. doi: 10.3389/fnbeh.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Obrenovich M. Leaky gut, leaky brain? Microorganisms. 2018;6:107. doi: 10.3390/microorganisms6040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mattson M.P., Longo V.D., Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Res Rev. 2017;39:46–58. doi: 10.1016/j.arr.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mischley L.K., Lau R.C., Bennett R.D. Role of diet and nutritional supplements in parkinson’s disease progression. Oxid Med Cell Longev. 2017;2017:1–9. doi: 10.1155/2017/6405278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi A., Shi H., Wang Y., Liu X., Cheng Y., Li H., et al. Activation of Nrf2 pathway and inhibition of NLRP3 inflammasome activation contribute to the protective effect of chlorogenic acid on acute liver injury. Int Immunopharmacol. 2018;54:125–130. doi: 10.1016/j.intimp.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 117.Traba J., Geiger S.S., Kwarteng-Siaw M., Han K., Ra O.H., Siegel R.M., et al. Prolonged fasting suppresses mitochondrial NLRP3 inflammasome assembly and activation via SIRT3-mediated activation of superoxide dismutase 2. J Biol Chem. 2017;292:12153–12164. doi: 10.1074/jbc.M117.791715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin C., Chao H., Li Z., Xu X., Liu Y., Bao Z., et al. Omega-3 fatty acids regulate NLRP3 inflammasome activation and prevent behavior deficits after traumatic brain injury. Exp Neurol. 2017;290:115–122. doi: 10.1016/j.expneurol.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 119.Tőzsér J., Benkő S. Natural compounds as regulators of NLRP3 inflammasome-mediated IL-1 β production. Mediat Inflamm. 2016;2016:1–16. doi: 10.1155/2016/5460302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Youm Y.-H., Nguyen K.Y., Grant R.W., Goldberg E.L., Bodogai M., Kim D., et al. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome–mediated inflammatory disease. Nat Med. 2015;21:263–269. doi: 10.1038/nm.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Q.-L., Yin H.-R., He Q.-Y., Wang Y. Targeting the NLRP3 inflammasome as new therapeutic avenue for inflammatory bowel disease. Biomed Pharmacother. 2021;138 doi: 10.1016/j.biopha.2021.111442. [DOI] [PubMed] [Google Scholar]
- 122.Song Y., Zhao Y., Ma Y., Wang Z., Rong L., Wang B., et al. Biological functions of NLRP3 inflammasome: A therapeutic target in inflammatory bowel disease. Cytokine Growth Factor Rev. 2021;60:61–75. doi: 10.1016/j.cytogfr.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 123.Geremia A., Biancheri P., Allan P., Corazza G.R., Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13:3–10. doi: 10.1016/j.autrev.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 124.Kudelka M.R., Stowell S.R., Cummings R.D., Neish A.S. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol. 2020;17:597–617. doi: 10.1038/s41575-020-0331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neurath M.F. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 126.Ni J., Wu G.D., Albenberg L., Tomov V.T. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Villani A.-C., Lemire M., Fortin G., Louis E., Silverberg M.S., Collette C., et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Monteleone G., Trapasso F., Parrello T., Biancone L., Stella A., Iuliano R., et al. Bioactive IL-18 expression is up-regulated in Crohn’s disease. J Immunol. 1999;163:143–147. [PubMed] [Google Scholar]
- 129.Umiker B., Lee H.-H., Cope J., Ajami N.J., Laine J.-P., Fregeau C., et al. The NLRP3 inflammasome mediates DSS-induced intestinal inflammation in Nod2 knockout mice. Innate Immun. 2019;25:132–143. doi: 10.1177/1753425919826367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bauer C., Duewell P., Mayer C., Lehr H.A., Fitzgerald K.A., Dauer M., et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–1199. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- 131.Allen I.C., TeKippe E.M., Woodford R.-M.T., Uronis J.M., Holl E.K., Rogers A.B., et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Song-Zhao G.X., Srinivasan N., Pott J., Baban D., Frankel G., Maloy K.J. Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. Mucosal Immunol. 2014;7:763–774. doi: 10.1038/mi.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bauer C., Duewell P., Lehr H.-A., Endres S., Schnurr M. Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig Dis. 2012;30:82–90. doi: 10.1159/000341681. [DOI] [PubMed] [Google Scholar]
- 134.Vicentini F.A., Keenan C.M., Wallace L.E., Woods C., Cavin J.-B., Flockton A.R., et al. Intestinal microbiota shapes gut physiology and regulates enteric neurons and glia. Microbiome. 2021;9:210. doi: 10.1186/s40168-021-01165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saber S., Abd El-Fattah E.E., Yahya G., Gobba N.A., Maghmomeh A.O., Khodir A.E., et al. A novel combination therapy using rosuvastatin and lactobacillus combats dextran sodium sulfate-induced colitis in high-fat diet-fed rats by targeting the TXNIP/NLRP3 interaction and influencing gut microbiome composition. Pharmaceuticals. 2021;14:341. doi: 10.3390/ph14040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qu S., Fan L., Qi Y., Xu C., Hu Y., Chen S., et al. Akkermansia muciniphila Alleviates Dextran Sulfate Sodium (DSS)-Induced Acute Colitis by NLRP3 Activation. Microbiol Spectr. 2021;9 doi: 10.1128/Spectrum.00730-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li X., Wu X., Wang Q., Xu W., Zhao Q., Xu N., et al. Sanguinarine ameliorates DSS induced ulcerative colitis by inhibiting NLRP3 inflammasome activation and modulating intestinal microbiota in C57BL/6 mice. Phytomedicine. 2022;104 doi: 10.1016/j.phymed.2022.154321. [DOI] [PubMed] [Google Scholar]
- 138.Llewellyn S.R., Britton G.J., Contijoch E.J., Vennaro O.H., Mortha A., Colombel J.-F., et al. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology. 2018;154(1037–1046) doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Singh V., Yeoh B.S., Walker R.E., Xiao X., Saha P., Golonka R.M., et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut. 2019;68:1801–1812. doi: 10.1136/gutjnl-2018-316250. [DOI] [PubMed] [Google Scholar]
- 140.Li P., Wu M., Xiong W., Li J., An Y., Ren J., et al. Saikosaponin-d ameliorates dextran sulfate sodium-induced colitis by suppressing NF-κB activation and modulating the gut microbiota in mice. Int Immunopharmacol. 2020;81 doi: 10.1016/j.intimp.2020.106288. [DOI] [PubMed] [Google Scholar]
- 141.Tong L., Hao H., Zhang Z., Lv Y., Liang X., Liu Q., et al. Milk-derived extracellular vesicles alleviate ulcerative colitis by regulating the gut immunity and reshaping the gut microbiota. Theranostics. 2021;11:8570–8586. doi: 10.7150/thno.62046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wu M., Li P., An Y., Ren J., Yan D., Cui J., et al. Phloretin ameliorates dextran sulfate sodium-induced ulcerative colitis in mice by regulating the gut microbiota. Pharm Res. 2019;150 doi: 10.1016/j.phrs.2019.104489. [DOI] [PubMed] [Google Scholar]
- 143.Allander E., Lindahl B.I. The Mediterranean Osteoporosis Study (MEDOS): theoretical and practical issues of a major international project on hip fracture epidemiology. Bone. 1993;14(Suppl 1):S37–S43. doi: 10.1016/8756-3282(93)90348-e. [DOI] [PubMed] [Google Scholar]
- 144.Gu Q.-Y., Zhang J., Feng Y.-C. Role of NLRP3 inflammasome in Bifidobacterium longum -regulated visceral hypersensitivity of postinfectious irritable bowel syndrome. Artif Cells, Nanomed, Biotechnol. 2016;44:1933–1937. doi: 10.3109/21691401.2015.1111238. [DOI] [PubMed] [Google Scholar]
- 145.Cryan J.F., O’Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 146.Cryan J.F., O’Riordan K.J., Sandhu K., Peterson V., Dinan T.G. The gut microbiome in neurological disorders. Lancet Neurol. 2020;19:179–194. doi: 10.1016/S1474-4422(19)30356-4. [DOI] [PubMed] [Google Scholar]
- 147.Foster J.A., Rinaman L., Cryan J.F. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol Stress. 2017;7:124–136. doi: 10.1016/j.ynstr.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kaur H., Nookala S., Singh S., Mukundan S., Nagamoto-Combs K., Combs C.K. Sex-dependent effects of intestinal microbiome manipulation in a mouse model of alzheimer’s disease. Cells. 2021;10:2370. doi: 10.3390/cells10092370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao K., Mu C., Farzi A., Zhu W. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr. 2020;11:709–723. doi: 10.1093/advances/nmz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. doi: 10.1016/j.brainres.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Erny D., Hrabě de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Luczynski P., McVey Neufeld K.-A., Oriach C.S., Clarke G., Dinan T.G., Cryan J.F. Growing up in a bubble: using germ-free animals to assess the influence of the gut microbiota on brain and behavior. IJNPPY. 2016;19:pyw020. doi: 10.1093/ijnp/pyw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wong M.-L., Inserra A., Lewis M.D., Mastronardi C.A., Leong L., Choo J., et al. Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol Psychiatry. 2016;21:797–805. doi: 10.1038/mp.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kaufmann F.N., Costa A.P., Ghisleni G., Diaz A.P., Rodrigues A.L.S., Peluffo H., et al. NLRP3 inflammasome-driven pathways in depression: Clinical and preclinical findings. Brain Behav Immun. 2017;64:367–383. doi: 10.1016/j.bbi.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 156.Musgrove R.E., Helwig M., Bae E.-J., Aboutalebi H., Lee S.-J., Ulusoy A., et al. Oxidative stress in vagal neurons promotes parkinsonian pathology and intercellular α-synuclein transfer. J Clin Investig. 2019;129:3738–3753. doi: 10.1172/JCI127330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pei L., Wallace D.C. Mitochondrial etiology of neuropsychiatric disorders. Biol Psychiatry. 2018;83:722–730. doi: 10.1016/j.biopsych.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sabatino J.J., Pröbstel A.-K., Zamvil S.S. B cells in autoimmune and neurodegenerative central nervous system diseases. Nat Rev Neurosci. 2019;20:728–745. doi: 10.1038/s41583-019-0233-2. [DOI] [PubMed] [Google Scholar]
- 159.Süß P., Hoffmann A., Rothe T., Ouyang Z., Baum W., Staszewski O., et al. Chronic peripheral inflammation causes a region-specific myeloid response in the central nervous system. Cell Rep. 2020;30(4082–4095) doi: 10.1016/j.celrep.2020.02.109. [DOI] [PubMed] [Google Scholar]
- 160.Subhramanyam C.S., Wang C., Hu Q., Dheen S.T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin Cell Dev Biol. 2019;94:112–120. doi: 10.1016/j.semcdb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 161.Kwon H.S., Koh S.-H. Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl Neurodegener. 2020;9:42. doi: 10.1186/s40035-020-00221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gordon R., Albornoz E.A., Christie D.C., Langley M.R., Kumar V., Mantovani S., et al. Inflammasome inhibition prevents α-synuclein pathology and dopaminergic neurodegeneration in mice. Sci Transl Med. 2018 doi: 10.1126/scitranslmed.aah4066. 10:eaah4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heneka M.T., McManus R.M., Latz E. Inflammasome signalling in brain function and neurodegenerative disease. Nat Rev Neurosci. 2018;19:610–621. doi: 10.1038/s41583-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 164.Bai H., Zhang Q. Activation of NLRP3 inflammasome and onset of Alzheimer’s disease. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.701282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rao J., Qiao Y., Xie R., Lin L., Jiang J., Wang C., et al. Fecal microbiota transplantation ameliorates stress-induced depression-like behaviors associated with the inhibition of glial and NLRP3 inflammasome in rat brain. J Psychiatr Res. 2021;137:147–157. doi: 10.1016/j.jpsychires.2021.02.057. [DOI] [PubMed] [Google Scholar]
- 166.Rogers G.B., Keating D.J., Young R.L., Wong M.-L., Licinio J., Wesselingh S. From gut dysbiosis to altered brain function and mental illness: mechanisms and pathways. Mol Psychiatry. 2016;21:738–748. doi: 10.1038/mp.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pellegrini C., Antonioli L., Calderone V., Colucci R., Fornai M., Blandizzi C. Microbiota-gut-brain axis in health and disease: Is NLRP3 inflammasome at the crossroads of microbiota-gut-brain communications. Prog Neurobiol. 2020;191 doi: 10.1016/j.pneurobio.2020.101806. [DOI] [PubMed] [Google Scholar]
- 168.Pellegrini C., Antonioli L., Lopez-Castejon G., Blandizzi C., Fornai M. Canonical and non-canonical activation of NLRP3 inflammasome at the crossroad between immune tolerance and intestinal inflammation. Front Immunol. 2017:8. doi: 10.3389/fimmu.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Panicker N., Sarkar S., Harischandra D.S., Neal M., Kam T.-I., Jin H., et al. Fyn kinase regulates misfolded α-synuclein uptake and NLRP3 inflammasome activation in microglia. J Exp Med. 2019;216:1411–1430. doi: 10.1084/jem.20182191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bonfili L., Cecarini V., Berardi S., Scarpona S., Suchodolski J.S., Nasuti C., et al. Microbiota modulation counteracts Alzheimer’s disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci Rep. 2017;7:2426. doi: 10.1038/s41598-017-02587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vogt N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C., et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep. 2017;7:13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brandscheid C., Schuck F., Reinhardt S., Schäfer K.-H., Pietrzik C.U., Grimm M., et al. Altered gut microbiome composition and tryptic activity of the 5xFAD Alzheimer’s mouse model. JAD. 2017;56:775–788. doi: 10.3233/JAD-160926. [DOI] [PubMed] [Google Scholar]
- 173.Pistollato F., Sumalla Cano S., Elio I., Masias Vergara M., Giampieri F., Battino M. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr Rev. 2016;74:624–634. doi: 10.1093/nutrit/nuw023. [DOI] [PubMed] [Google Scholar]
- 174.Shen H., Guan Q., Zhang X., Yuan C., Tan Z., Zhai L., et al. New mechanism of neuroinflammation in Alzheimer’s disease: the activation of NLRP3 inflammasome mediated by gut microbiota. Prog Neuropsychopharmacol Biol Psychiatry. 2020;100 doi: 10.1016/j.pnpbp.2020.109884. [DOI] [PubMed] [Google Scholar]
- 175.Shukla P.K., Delotterie D.F., Xiao J., Pierre J.F., Rao R., McDonald M.P., et al. Alterations in the gut-microbial-inflammasome-brain axis in a mouse model of Alzheimer’s disease. Cells. 2021;10:779. doi: 10.3390/cells10040779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang Y., Wang D., Lv H., Dong Q., Li J., Geng W., et al. Modulation of the gut microbiota and glycometabolism by a probiotic to alleviate amyloid accumulation and cognitive impairments in AD rats. Mol Nutr Food Res. 2022 doi: 10.1002/mnfr.202200265. [DOI] [PubMed] [Google Scholar]
- 177.Jackson A., Forsyth C.B., Shaikh M., Voigt R.M., Engen P.A., Ramirez V., et al. Diet in Parkinson’s disease: critical role for the microbiome. Front Neurol. 2019;10:1245. doi: 10.3389/fneur.2019.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167(1469–1480) doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lin C.-H., Chen C.-C., Chiang H.-L., Liou J.-M., Chang C.-M., Lu T.-P., et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J Neuroinflamm. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Perez-Pardo P., Dodiya H.B., Engen P.A., Forsyth C.B., Huschens A.M., Shaikh M., et al. Role of TLR4 in the gut-brain axis in Parkinson’s disease: a translational study from men to mice. Gut. 2019;68:829–843. doi: 10.1136/gutjnl-2018-316844. [DOI] [PubMed] [Google Scholar]
- 181.Qiao C.-M., Sun M.-F., Jia X.-B., Li Y., Zhang B.-P., Zhao L.-P., et al. Sodium butyrate exacerbates Parkinson’s disease by aggravating neuroinflammation and colonic inflammation in MPTP-induced mice model. Neurochem Res. 2020;45:2128–2142. doi: 10.1007/s11064-020-03074-3. [DOI] [PubMed] [Google Scholar]
- 182.Bai S., Wang W., Wang T., Li J., Zhang S., Chen Z., et al. CD36 deficiency affects depressive-like behaviors possibly by modifying gut microbiota and the inflammasome pathway in mice. Transl Psychiatry. 2021;11:16. doi: 10.1038/s41398-020-01130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Frank M.G., Fonken L.K., Dolzani S.D., Annis J.L., Siebler P.H., Schmidt D., et al. Immunization with Mycobacterium vaccae induces an anti-inflammatory milieu in the CNS: Attenuation of stress-induced microglial priming, alarmins and anxiety-like behavior. Brain Behav Immun. 2018;73:352–363. doi: 10.1016/j.bbi.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Avolio E., Fazzari G., Zizza M., De Lorenzo A., Di Renzo L., Alò R., et al. Probiotics modify body weight together with anxiety states via pro-inflammatory factors in HFD-treated Syrian golden hamster. Behav Brain Res. 2019;356:390–399. doi: 10.1016/j.bbr.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 185.Qiu X., Wu G., Wang L., Tan Y., Song Z. Lactobacillus delbrueckii alleviates depression-like behavior through inhibiting toll-like receptor 4 (TLR4) signaling in mice. Ann Transl Med. 2021;9:366. doi: 10.21037/atm-20-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]