Abstract
Purpose:
The goal of Electronic Medical Records and Genomics (eMERGE) Phase III Network was to return actionable sequence variants to 25,084 consenting participants from 10 different health care institutions across the United States. The purpose of this study was to evaluate system-based issues relating to the return of results (RoR) disclosure process for clinical grade research genomic tests to eMERGE3 participants.
Methods:
RoR processes were developed and approved by each eMERGE institution’s internal review board. Investigators at each eMERGE3 site were surveyed for RoR processes related to the participant’s disclosure of pathogenic or likely pathogenic variants and engagement with genetic counseling. Standard statistical analysis was performed.
Results:
Of the 25,084 eMERGE participants, 1444 had a pathogenic or likely pathogenic variant identified on the eMERGEseq panel of 67 genes and 14 single nucleotide variants. Of these, 1077 (74.6%) participants had results disclosed, with 562 (38.9%) participants provided with variant-specific genetic counseling. Site-specific processes that either offered or required genetic counseling in their RoR process had an effect on whether a participant ultimately engaged with genetic counseling (P = .0052).
Conclusion:
The real-life experience of the multiarm eMERGE3 RoR study for returning actionable genomic results to consented research participants showed the impact of consent, method of disclosure, and genetic counseling on RoR.
Keywords: Consent, eMERGE, Genetic counseling, Genomic medicine, Return of results
Introduction
Increased accessibility to genetic testing provides challenges to medical professionals caring for the growing number of individuals found to have a genetic predisposition to a specific condition or disease. The Electronic Medical Records and Genomics (eMERGE) Network1 is one of several National Institutes of Health–sponsored networks, which include Clinical Sequencing Evidence-Generating Research,2 Clinical Genomic Resource,3 Implementing Genomics in Practice,4 and All of Us5 that are evaluating the use of DNA sequence results in clinical care.
At the time the study was conducted, the American College of Genetics and Genomics (ACMG) had designated 59 genes as highly actionable owing to availability of effective medical management guidelines for individuals with detected disease-causing variants in one of these genes. When an individual elects to have clinical genetic testing, the ACMG recommends that laboratories interrogate and return pathogenic (P) or likely pathogenic (LP) variants in these actionable genes and report them as secondary findings to the individual.6 Although this guideline is based on professional consensus, there is no empirical evidence to direct best practices for returning secondary findings to participants in clinical care or research. For those who receive unanticipated results associated with clinical testing, it has been within the context of clinical care with consent obtained before sample submission. Individuals who undergo genomic sequencing through other avenues, such as research studies or direct-to-consumer (DTC) testing, typically have no primary indication for testing. Thus, all findings in these situations are unexpected because testing was performed outside the clinical context with potentially less access to genetic counseling and anticipatory guidance for gene-specific management. Establishing best practices and guidelines for the return of disease-causing variants, identified through either screening studies or as secondary findings, would support the utility of personalized genomic medicine for a population.7
In addition, the need for best practices for the return of findings is heightened by the expansion of genomic medicine in clinical care, particularly evident in cancer genetics. Specific genetic variants can drive therapeutic options, such as the use of PARP inhibitors for patients with a pathogenic variant in BRCA1 or BRCA2.8 The Office of Public Health Genomics in the Centers for Disease Control and Prevention is considering population screening for highly penetrant genetic disorders and has nominated hereditary breast and ovarian cancer, Lynch syndrome, and familial hyperlipidemia as Tier 1 conditions for initial implementation.9 If this occurs, well-defined processes will be required for consent, testing, and return of genomic results. Moreover, the combination of the clinical utility associated with identifying disease-causing genetic variants and a limited genetic workforce will likely encourage the development of a testing first model before genetic counseling. This highlights the necessity for understanding the potential impact of such a model to guide well-defined processes for genomic testing and return of results (RoR).10
In the eMERGE3 study, approximately 25,000 participants were enrolled from 10 different health care systems and underwent sequencing of about 100 actionable genes, including 58 genes from the ACMG incidental finding list.6 P/LP variants identified in these genes were returned to consented participants. One of the specific aims of eMERGE3 was the evaluation of the RoR process, including consent, methods for RoR, disclosure of results to participants, informing of health care providers, and uploading of results to the electronic health record (EHR). However, there was no predetermined, prescribed process for RoR across the clinical sites, permitting each site to develop their own RoR process. Wiesner et al11 described the wide heterogeneity between the eMERGE site-specific processes, including site-specific source of participants (biorepositories, clinic, or community), consent protocols, method of RoR disclosure to participants, informing of health care providers, and participant engagement in genetic counseling. With the conclusion of eMERGE3, we report on how a clinical site’s RoR process had an effect on results disclosed to a participant and whether genetic counseling was provided. Despite the variation between eMERGE3 sites for RoR, common themes were found in the observed results, illustrating the real-life challenges of returning unsolicited genomic results to participants. Our experience of RoR across the eMERGE3 Network will inform and guide future genomic research projects and implementation efforts.
Materials and Methods
eMERGE3 clinical sites
Briefly, the 10 health care institutions that participated in eMERGE3 were Cincinnati Children’s Hospital and Medical Center (CCHMC), Children’s Hospital of Philadelphia (CHOP), Columbia University (CU), Geisinger (GE), Kaiser Permanente of Washington (KPWA; formerly Group Health Cooperative)/University of Washington (UW), Mayo Clinic (MC), Meharry Medical College (MMC), Northwestern University (NU), Partners HealthCare (PHC) (now Massachusetts General/Brigham), and Vanderbilt University Medical Center (VUMC), with several sites having more than 1 cohort (CCHMC, CU, and MC). The details of the specific cohorts, population demographics, enhanced recruitment based on phenotype or ethnicity, and planned RoR process for eMERGE3 have been previously described.11 Of the 10 clinical sites, 2 sites (CCHMC and CHOP) exclusively enrolled pediatric participants, whereas the other 8 clinical sites only enrolled adults. Depending on the site, participants were recruited from biorepository samples, prospectively from clinical sites, and through community health care systems or a combination of these sources. Methods of disclosure include US mail, phone, clinic appointment, EHR portal, or email. Genetic counseling was offered by all sites, but some sites specifically required/embedded genetic counseling in their RoR protocols (Table 1). The RoR process for each site was approved by each individual site’s institutional internal review board and human subjects committee.
Table 1.
eMERGE3 sites, population type, source of recruitment, whether RoR was required at the time of enrollment, primary planned method for RoR, and whether genetic counseling was embedded or offered in the RoR process
| Group | Institution | Age Group | Source of Participants | RoR Required for Enrollment | Primary Planned Method for RoR | Genetic Counseling with RoRa |
|---|---|---|---|---|---|---|
| Group A | GE | Adult | Biorepository | Yes | Letter | Offered |
| NU | Adult | Clinic, PGx biorepository | Yes | Phone | Offered | |
| VUMC | Adult | Clinic, PGx biorepository | Yes | Letter | Offered | |
| Group B | CU | Adult | Community clinic, biorepository | No | Clinic, portal, letter, email | Embedded |
| KPWA/UW | Adult | Biorepository | No | Clinic | Embedded | |
| MC | Adult | Biorepository | No | Clinic | Embedded | |
| MMC | Adult | Clinic | No | Clinic | Embedded | |
| PHC | Adult | Biorepository | No | Clinic | Embedded | |
| CCHMC | Pediatric | Biorepository, clinic, community | No | Clinic, portal | Embedded | |
| CHOP | Pediatric | Biorepository | No | Clinic | Embedded |
The 3 institutions with adult populations (GE, NU, and VUMC) and genetic counseling offered in the RoR are listed first (Group A), followed by the 5 other adult institutions (CU, KPWA/UW, MC, MMC, PHC) and the 2 pediatric institutions (CCHMC and CHOP) (group B).
CCHMC, Cincinnati Children’s Hospital and Medical Center; CHOP, Children’s Hospital of Philadelphia; CU, Columbia University; eMERGE3, Electronic Medical Records and Genomics Phase III; GE, Geisinger; KPWA, Kaiser Permanente of Washington; MC, Mayo Clinic; MMC, Meharry Medical College; NU, Northwestern University; PGx, pharmacogenomics; PHC, Partners HealthCare; RoR, return of results; UW, University of Washington; VUMC, Vanderbilt University Medical Center.
CU, MC and CCHMC had multiple subpopulations.
eMERGE3 sequencing
The Clinical Laboratory Improvement Amendments/College of American Pathologists–certified sequencing laboratories for eMERGE3, the Partners HealthCare Laboratory for Molecular Medicine and the Baylor College of Medicine Human Genome Sequencing Center Clinical Laboratory, used the eMERGEseq platform with 109 genes, including 58 actionable genes from the ACMG list,6 and 1555 single nucleotide variants (SNVs) in additional genes. The consensus panel returned by nearly all sites included 67 genes and 14 SNVs that were deemed actionable by the eMERGE Network;1 additional genes returned were determined by the site’s protocol or existing biobank protocols established before eMERGE3. Genes not returned by a given clinical site were not included on participant reports for that site. There was also variability in the types of results disclosed. For example, 1 site disclosed heterozygous status for common autosomal recessive conditions, 4 sites disclosed pharmacogenomic information, and 6 sites disclosed null result information.12 Variants of uncertain significance were disclosed for colorectal cancer at the KPWA/UW site in conjunction with enhanced recruitment at that site for participants with colorectal cancer and colon polyps. Sequencing laboratories began releasing results to clinical sites in November 2017, and for the purposes of this report, were considered complete as of April 2020 (see eMERGE Clinical Annotation Working Group13 for details on distribution of returned findings). For the purpose of this study, we only considered participants who received P/LP results for the eMERGEseq consensus 67-gene panel and 14 clinically relevant SNVs.13
There were specific considerations for RoR at the pediatric clinical sites. CCHMC enrolled 2 cohorts, a biobank cohort and a prospective adolescent cohort. Because the biobank cohort represented all age ranges and was not prospectively given a choice about the type of results to return, CCHMC requested that only the genes for medically actionable conditions during childhood be analyzed by the sequencing laboratory, fewer than the 67 consensus gene panel. All genes for medically actionable conditions, including those for adult-onset conditions, were analyzed for CCHMC’s prospective adolescent cohort, but returned results were restricted to those that matched adolescent/parent dyad choices.14,15 A genetic counselor returned P/LP results to both the adolescent and parent by phone. Adolescents in the CCHMC prospective cohort were not given the option to change which results they wanted to learn once they turned 18. For the CCHMC and CHOP biobank participants, children/adolescent results were returned to parents. Although genes associated with adult-onset conditions were analyzed for CHOPs biobank participants, P/LP variants for these conditions were withheld until the participant turned 18, consented as an adult to the biobank, and then recontacted for results disclosure.16
Data collection
Data used in this manuscript were obtained from a detailed questionnaire that was developed by a subgroup of authors (K.A.L, G.L.W., and A.K.R.) and distributed during the second year of RoR to investigators at each of the clinical sites. The goal of the questionnaire was to gather detailed information regarding the RoR process and the challenges and barriers encountered. The questionnaire also assessed sites’ use of genetic counseling in RoR, whether genetic counseling was offered or specifically embedded in the site-specific RoR process. The eMERGE3 Coordinating Center at VUMC tracked and recorded RoR data for each clinical site. Variables that were examined for impact on RoR included differences between adult and pediatric populations, how participants were consented into eMERGE3, RoR disclosure process, and whether participants engaged in genetic counseling. For the purposes of analysis, sites were grouped by consent process, whether participant engagement was needed for results disclosure, and whether genetic counseling was offered (group A) or embedded/required (group B) in the RoR processes (Table 1). Group A (GE, NU, and VUMC) required participant consent to RoR at the time of enrollment, used disclosure methods that did not require participant engagement, and offered genetic counseling with RoR. Group B (CCHMC, CHOP, CU, KPWA/UW, MC, Meharry Medical College, and PHC) allowed participants to decline RoR after samples were submitted for sequencing, required participant engagement for RoR, and embedded genetic counseling in their RoR process. Chi-square analysis was performed to evaluate the ability of group A and group B to disclose genomic results to participants and to evaluate participant engagement in genetic counseling between group A and group B.
Results
Of the 25,084 participants in eMERGE3, 1444 had at least 1 P/LP variant eligible for RoR; of these 1444 participants, 35 had P/LP variants in 2 different genes. The overall frequency of P/LP variants among eMERGE participants was 5.7%, ranging from 2.8% to 10.3% at an individual site depending on initial sampling method and site-specific criteria for RoR. P/LP variants were disclosed to 1077 of the 1444 eMERGE3 participants (74.6%) (Table 2). Comparing adult and pediatric populations, overall, 82.5% of adults had results disclosure ranging from 24.7% to 97.5% across adult cohorts and clinical sites, whereas 24.2% of pediatric participants had RoR ranging between 20.9% and 83.3% across cohorts and clinical sites.
Table 2.
eMERGE3 sites listed with total number of participants, number of participants with P/LP variants, and number of participants with disclosed P/LP variants
| Institution | Total Number of Participants | Number of Participants with P/LP Variants | Number of Participants with Returned P/LP |
|---|---|---|---|
| CCHMC prospective adolescent | 160 | 6 (3.8%) | 5 (83.3%) |
| CCHMC biobank | 2840 | 91 (3.2%) | 19 (20.9%) |
| CHOP | 2990 | 101 (3.4%) | 24 (23.8%) |
| Columbia IMAgene | 341 | 30 (8.8%) | 28 (93.3%) |
| Columbia- prospective | 1120 | 65 (5.8%) | 51 (78.5%) |
| Columbia -retrospective | 1135 | 73 (6.4%) | 18 (24.7%) |
| Geisinger | 2500 | 263 (10.5%) | 244 (92.8%) |
| KPWA/UW | 2500 | 96 (3.8%) | 58 (60.4%) |
| Mayo - Rochester | 2535 | 121 (4.8%) | 118 (97.5%) |
| Mayo - Arizona | 500 | 10 (2.0%) | 9 (90.0%) |
| Meharry | 500 | 19 (3.8%) | 14 (73.7%) |
| Northwestern | 3000 | 279 (9.3%) | 255 (88.5%) |
| Partners Healthcare | 2500 | 65 (2.6%) | 25 (35.5%) |
| VUMC | 2454 | 225(9.1%) | 209 (92.9%) |
| Total | 25,084 | 1444 (5.7%) | 1077 (74.6%) |
CCHMC, CU and MC have multiple listings representing different cohorts.
CCHMC, Cincinnati Children’s Hospital and Medical Center; CHOP, Children’s Hospital of Philadelphia; eMERGE3, Electronic Medical Records and Genomics Phase III; KPWA, Kaiser Permanente of Washington; LP, likely pathogenic; MC, Mayo Clinic; P, pathogenic; UW, University of Washington; VUMC, Vanderbilt University Medical Center.
Clinical sites differ in whether a participant was required to have results disclosed as part of the inclusion criteria at the time of enrollment in eMERGE3 (group A) or whether a participant could decline RoR after they were included in a site’s eMERGE3 cohort (group B). Although consented biorepository samples were used for both group A and B, the requirement for RoR at the time of enrollment ultimately affected the proportion of participants with RoR. Of the 767 participants in group A, 92.3% of participants had P/LP results disclosed. This is compared with the 677 participants across 7 clinical sites in group B, where 54.5% of participants completed results disclosure. When separating the adult and pediatric sites in group B, the 5 eMERGE3 adult sites disclosed results to 66.9% of participants, compared with 24.2% of the participants at the 2 pediatric sites.
Site-specific processes for RoR included an in-person appointment with a medical geneticist and/or genetic counselor, phone call (typically with a genetic counselor or other health care professional), use of a patient portal in the EHR, and use of US postal mail. Some sites included multiple contact methods within their RoR process. CU and PHC allowed participants to choose their RoR disclosure method. GE and VUMC (group A) used US postal mail as the initial contact. However, GE’s initial mail outreach informed the participant that information important to their health was available and that the study team would contact them with specific information,17 whereas VUMC’s letter contained information about the specific gene involved and the identified P/LP variant with instructions to discuss with their primary care. Genetic counselors at NU used an unscheduled phone call to disclose results as the initial outreach. Similar to GE, MC and KPWA/UW (group B) used letters notifying participants that there was a clinically relevant result but required participants to contact the site and meet with a medical geneticist and/or genetic counselor to complete result disclosure. When consented participants could not be reached by the planned RoR process, some sites used other methods, such as certified US postal mail, for disclosure. GE, by using multiple methods to notify a participant that a result was available, including mail, patient portal, and telephone outreach, was able to ultimately disclose 92.8% of results. By using US mail, VUMC disclosed results to 92.9% of their participants. Group A sites were able to disclose results largely without participant engagement to a similar percentage of their participants, ranging from 88.5% to 92.9%. The group B sites, which required a participant initiation to activate a portal or to schedule an appointment with a health care provider, had a broader variability in their ability to disclose results, ranging from 20.9% to 97.5%, with an overall average of 74.6% of participants with results disclosure. Of note, although letters containing eMERGE3 results were sent by US postal mail, it is unknown whether these participants opened their mail and were aware of their results. Ultimately, results for all eMERGE3-consented participants were included in the EHR. However, it was unknown how many eMERGE3 participants learned of their results from their EHR.
Of the 1444 participants with at least 1 P/LP variant, 367 (25.4%) did not have results disclosed (Figure 1). Of these, 116 were no longer eligible for RoR based on institution-specific protocol(s) or because they were deceased. Across all sites, 33 (2.3%) actively declined RoR. These were participants who previously consented to RoR or had consented biorepository samples included in the eMERGE3 cohort but actively declined results when offered disclosure. Another 134 (9.3%) were considered to have passively withdrawn from eMERGE3 because they did not respond to the site-specific RoR process after multiple attempts. A total of 75 participants could not be located for RoR. At the time of the data freeze for this analysis, 9 participants were still in the RoR process.
Figure 1. Outcome of disclosure of P/LP variants to eMERGE3 participants.
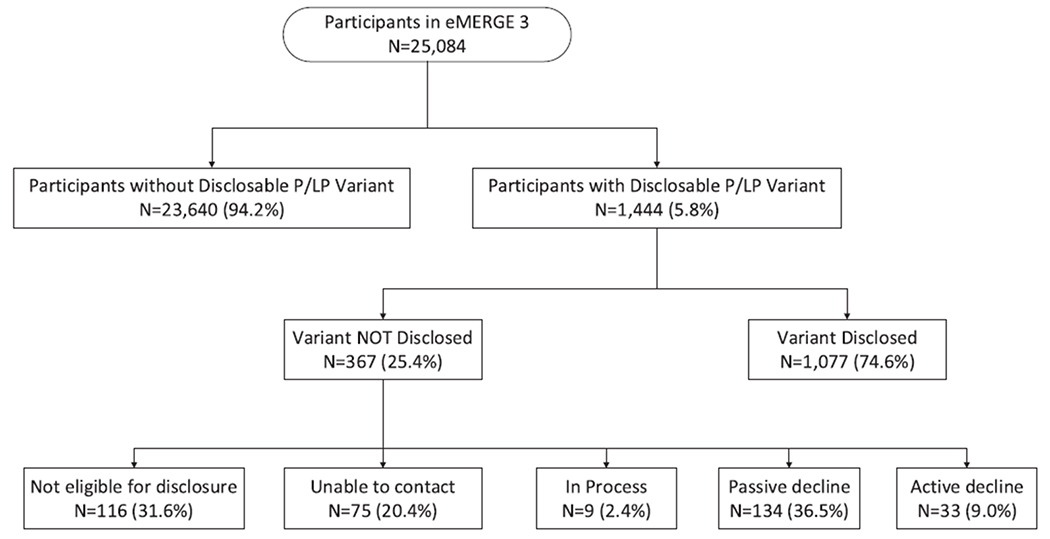
Diagram illustrating the number of enrolled participants in eMERGE3, participants with return of results, and the breakdown of numbers and reasons why participants did not have return of results. eMERGE3, Electronic Medical Records and Genomics Phase III; LP, likely pathogenic; P, pathogenic.
A full genetic counseling session for eMERGE3 participants was considered to include a discussion of health implications of the identified disease-associated variant, management and surveillance recommendations, and family risk information. As part of the site-specific protocol, genetic counselors may disclose results without providing genetic counseling. Overall, 562 (38.9%) of the 1444 eMERGE3 participants engaged in genetic counseling as part of site-specific protocols for RoR. Genetic counseling could occur by phone or clinic visit (Figure 2) and was offered at group A sites as part of RoR processes but was embedded in the RoR process at group B sites. Of the 767 participants at group A sites with P/LP variants, 708 had RoR, with 272 (38.4%) formally engaging in genetic counseling. Of the 677 participants at group B sites with P/LP variants, 369 participants had results disclosed, and 290 (42.8%) of those engaged in genetic counseling. There is a significant difference (P = .0052) between sites that did not require genetic counseling for RoR (group A; 38.4%) and sites where it was embedded for RoR (group B; 42.8%). This difference is more pronounced (P = .00001) when comparing the overall RoR completion between these groups (group A, 92.3% disclosed vs group B, 54.5% disclosed; P = .00001). This likely reflects the combined differences in the site processes of recruitment, consent, and requirements for RoR disclosure.
Figure 2. Genetic counselling and the return process for P/LP results.
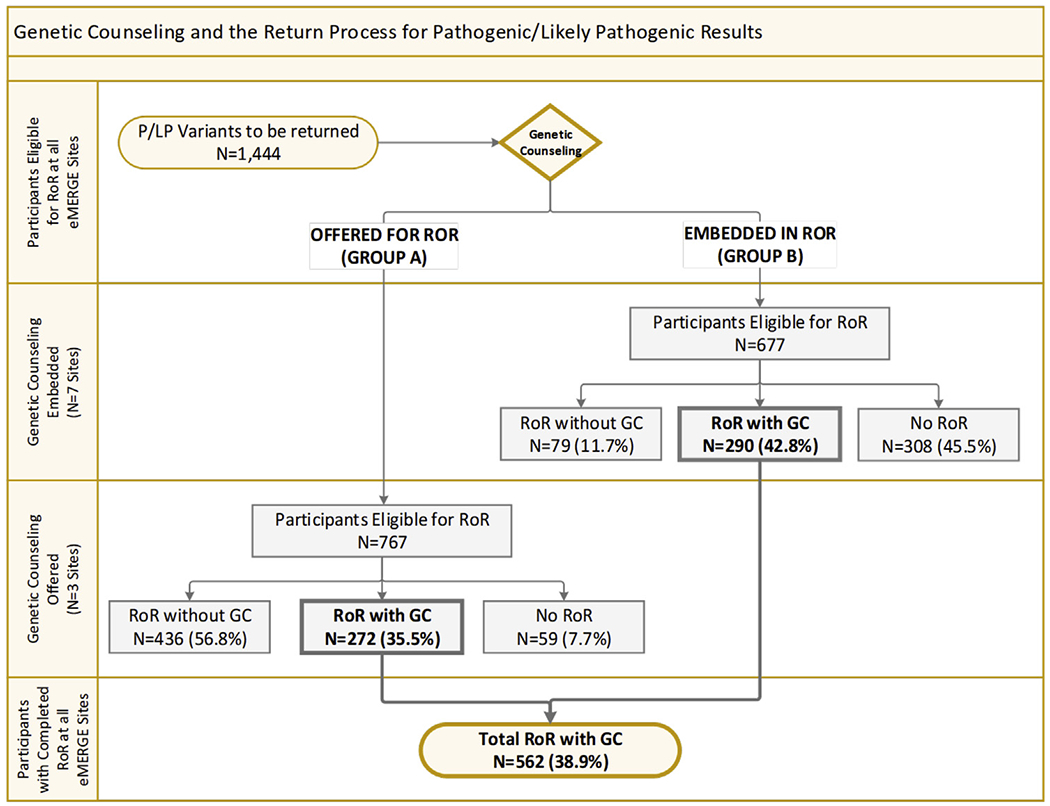
Diagram illustrating RoR and genetic counselling for participants, broken down between 3 sites that did not have genetic counselling embedded in RoR (group A: Geisinger, Northwestern University, Vanderbilt University Medical Center) vs the 7 sites that had genetic counselling embedded in their RoR process (group B: Cincinnati Children’s Hospital and Medical Center, Children’s Hospital of Philadelphia, Columbia, Kaiser Permanente of Washington/University of Washington, Mayo, Meharry, Partners HealthCare). eMERGE3, Electronic Medical Records and Genomics Phase III; LP, likely pathogenic; P, pathogenic; RoR, return of results.
Discussion
One of the great promises of genomic medicine is to provide tailored management and surveillance to individuals with a highly penetrant disease-causing genetic variant with the hope of decreasing or ameliorating associated disease. With actionable genomic variants increasingly identified by the expansion of genetic testing in research, clinical care, and DTC platforms, there is a growing need to identify best practices for returning such results and ultimately, incorporating the information regarding disease-associated genetic variants into an individual’s health management.
As a first step toward developing general guidance, we report on the impact of heterogeneous RoR processes across 10 clinical sites in the eMERGE3 Network and identify factors impacting RoR. The purpose of comparing rates of RoR between clinical sites was not to measure which process was more or less successful in disclosing results to participants but to illuminate what barriers influence the disclosure of actionable genomic results to research participants, including both children and adults, and whether participants choose to engage in genetic counseling as part of RoR. This analysis highlights issues regarding consent for RoR at the time of enrollment in eMERGE3, methods of disclosing results to participants and whether that method of disclosure required participant engagement, and finally, participant engagement in genetic counseling.
eMERGE3 sites that included only participants who consented to RoR at the time of enrollment (group A) had the highest number of participants with results disclosed. Group B had a lower RoR in sites that used samples from previously consented biorepository participants but not specifically for eMERGE3. Attempts to reconsent these individuals after sequencing resulted in many participants, either actively or passively, withdrawing from the study; therefore, sites allowing participants to opt out of disclosure at different points in the RoR process ultimately disclosed fewer results.
Pediatric sites faced the unique challenge of children transitioning to age 18 years between sample collection and site receipt of the Clinical Laboratory Improvement Amendments report, necessitating reconsent as an adult before results could be returned. In addition, CHOPs internal review board required withholding of results for adult-onset conditions to biobank participants until they turned 18 and signed consent for RoR.16 Of the 116 participants across eMERGE3 that were no longer eligible to receive their results, 65 (56%) were from the 2 pediatric sites.
The method of RoR, and whether result disclosure required participant engagement, affected RoR. Active participant engagement in care is required to schedule an in-person or phone appointment with a healthcare professional or to activate a portal for results disclosure. We suspect that RoR processes that required participant engagement in genetic counseling allowed more opportunity for participants to passively withdraw from or actively decline results disclosure.
Although it is essential to examine the underlying reasons for participants’ acceptance or refusal of an eMERGE3 actionable result, the demographics of these participants and reasons why the 33 participants actively declined or 134 participants passively declined RoR were not possible to measure across the eMERGE3 Network for this project. However, a site-specific analysis by Henrikson et al18 of 1131 eMERGE3 participants at KPWA/UW who did not respond to requests for disclosure (ie, passive refusal) of their eMERGE3 results suggests that passive refusal may represent a true desire not to receive genomic information. On follow-up contact with these 1131 participants, 71% actively refused RoR, citing reasons of insurance concerns, privacy, and not wanting the results.
RoR processes did not necessarily imply engagement with genetic counseling because site-specific processes may or may not have had genetic counseling embedded in their RoR process. In total, across the eMERGE3 Network, 38.9% of the 1444 participants with P/LP variants had results disclosed and received genetic counseling. The number of participants who actually received genetic counseling is likely higher because some participants had prior knowledge and previous genetic counseling for their disease-associated variant. We are anecdotally aware that genetic counseling was provided to some eMERGE3 participants by physicians or healthcare providers that were not medical geneticists or genetic counselors. However, this information was not tracked as an outcome for eMERGE3 participants. As the knowledge of the impact of genetics and genomics disseminates across all fields of health care, it will be necessary to determine whether individuals who do not formally engage with genetic services receive and understand the necessary information for themselves and family members at risk. Studies have shown that individuals who receive information about a pathogenic variant in the context of genetic counseling are more likely to receive tailored management recommendations and are more likely to have cascade testing of family members.18
The strength of the eMERGE3 Network is the heterogeneity of RoR processes that reflect the multiple differences in health care organization structure and resources in the United States. Despite this complexity, certain patterns become evident when considering factors that impact result disclosure and can guide best practices for genomic medicine. This study highlights that when participants in genomic research are recruited with specific enrollment criteria to receive results and use passive methods for result disclosure, without required genetic counseling, results can be returned to nearly all participants. But is this the best practice? Additional analysis of factors having an effect on the willingness of participants to engage with their genomic test results represents important information for directing both clinical practice and future research studies. Given the low proportion of participants who had genetic counseling observed across eMERGE3, other methods of providing genetic care need to be evaluated. The historic paternalism around genetic diagnosis and the perceived need for genetic counseling as the sole way to disclose results and provide management and information to family members may be creating barriers for identifying individuals with genetic risk.19 Research studies are needed to determine whether more individuals at risk for a highly penetrant genetic condition are best identified by population screening vs attempting cascade screening through an index patient.
The results reported here have some limitations and must be understood in the context of the eMERGE3 Network. First, the percentage of P/LP variants reported here (5.8%) is notably higher than the frequency of secondary findings reported previously (3.02%)13 owing to the heterogeneous nature of the participant populations across the 10 eMERGE sites. Sites varied by enrolling for phenotypic indications or enrichment for P/LP findings.17 Second, this project focused on the systems-based impact and effectiveness of the RoR processes. The impact of processes downstream, such as use of health care services and cascade testing, are beyond the scope of this evaluation. Finally, each individual site RoR processes differed in the required point of contact and level of engagement in RoR, causing some of the observed variability; however, this heterogeneity illuminates the factors important to the clinical utility of genomic screening in the real world that can guide future research investigations and clinical care implementation.
Genetic and genomic testing and research will continue to expand through clinical care, DTC, and research projects such as All of Us. The list of genes considered actionable by the ACMG was 59 genes at the time of eMERGE3 and has recently been expanded to 73 genes.20 There may be routine adult population screening for hereditary breast and ovarian cancer, Lynch syndrome, and familial hyperlipidemia because the Centers for Disease Control and Prevention considers these Tier 1 conditions as having a high impact on public health. Finally, the SARS-CoV-2 pandemic has had a dramatic impact on transitioning health care to virtual care, which will likely continue and increase access to further impact genetic care. To conclude, the eMERGE3 RoR project provides results of an observational, pragmatic study of genomic medicine implementation across different health care systems and populations. Barriers experienced by sites and adaptations to the eMERGE3 RoR processes illuminate issues of consent, contact, engagement, and participation in genetic counseling to be considered before broader implementation of DNA sequence results into health care.
Acknowledgments
The authors thank the Coordinating Center for the Electronic Medical Records and Genomics (eMERGE) Network for assistance in meeting and technical support.
This phase of the eMERGE Network was initiated and funded by the National Human Genome Research Institute through the following grants: U01HG008657 (Group Health Cooperative/University of Washington), U01HG008685 (Brigham and Women’s Hospital), U01HG008672 (Vanderbilt University Medical Center), U01HG008666 (Cincinnati Children’s Hospital Medical Center), U01HG006379 (Mayo Clinic), U01HG008679 (Geisinger Clinic), U01HG008680 (Columbia University Health Sciences), U01HG008684 (Children’s Hospital of Philadelphia), U01HG008673 (Northwestern University), U01HG008701 (Vanderbilt University Medical Center serving as the Coordinating Center), U01HG008676 (Partners Healthcare/Broad Institute), U54MD007593-10 (Meharry Medical College), and U01HG008664 (Baylor College of Medicine). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Ethics Declaration
This study was reviewed and approved by the Institutional Review Board and Human Subjects Committee of each of the 10 institutions involved in the Electronic Medical Records and Genomics Phase III (eMERGE3) Network: Cincinnati Children’s Hospital and Medical Center, Children’s Hospital of Philadelphia, Columbia University, Geisinger, Kaiser Permanente of Washington/University of Washington, Mayo Clinic, Meharry Medical Center, Northwestern University, Partners Healthcare Center, and Vanderbilt University Medical Center.
Conflicts of Interest
R.C.G. receives compensation for advising AIA, Applied Therapeutics, Humanity, and Verily and is a cofounder of Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers, and care systems. All other authors declare no conflicts of interest.
Data Availability
All data sets summarized in this article can be requested on the Electronic Medical Records and Genomics website https://emerge-network.org.
References
- 1.eMERGE Consortium. Harmonizing clinical sequencing and interpretation for the eMERGE III Network. Am J Hum Genet. 2019;105(3):588–605. 10.1016/j.ajhg.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendola LM, Berg JS, Horowitz CR, et al. The Clinical Sequencing Evidence-Generating Research Consortium: integrating genomic sequencing in diverse and medically underserved populations. Am J Hum Genet. 2018;103(3):319–327. 10.1016/j.ajhg.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm HL, Berg JS, Brooks LD, et al. ClinGen–the clinical genome resource. N Engl J Med. 2015;372(23):2235–2242. 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weitzel KW, Alexander M, Bernhardt BA, et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics. 2016;9:1. 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.All of Us Research Program Investigators, Denny JC, Rutter JL, et al. The “All of Us” research program. N Engl J Med. 2019;381(7):668–676. 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. Published correction appears in Genet Med 2017;19(4):484 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 7.Pal T, Lee JH, Besharat A, et al. Modes of delivery of genetic testing services and the uptake of cancer risk management strategies in BRCA1 and BRCA2 carriers. Clin Genet. 2014;85(1):49–53. 10.1111/cge.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCann KE, Hurvitz SA. Advances in the use of PARP inhibitor therapy for breast cancer. Drugs Context. 2018;7:212540. 10.7573/dic.212540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grzymski JJ, Elhanan G, Morales Rosado JA, et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat Med. 2020;26(8):1235–1239. 10.1038/s41591-020-0982-5. [DOI] [PubMed] [Google Scholar]
- 10.Stoll K, Kubendran S, Cohen SA. The past, present and future of service delivery in genetic counseling: keeping up in the era of precision medicine. Am J Med Genet C Semin Med Genet. 2018;178(1):24–37. 10.1002/ajmg.c.31602. [DOI] [PubMed] [Google Scholar]
- 11.Wiesner GL, Kulchak Rahm A, Appelbaum P, et al. Returning results in the genomic era: initial experiences of the eMERGE network. J Pers Med. 2020;10(2):30. 10.3390/jpm10020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn KS, Lynch J, Aufox S, et al. Returning negative results from large-scale genomic screening: experiences from the eMERGE III network. Am J Med Gene At. 2021;185(2):508–516. 10.1002/ajmg.a.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.eMERGE Clinical Annotation Working Group. Frequency of genomic secondary findings among 21,915 eMERGE network participants. Genet Med. 2020;22(9):1470–1477. 10.1038/s41436-020-0810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers MF, Martin LJ, Prows CA. Adolescents’ and parents’ genomic testing decisions: associations with age, race, and sex. J Adolesc Health. 2020;66(3):288–295. 10.1016/j.jadohealth.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prows CA, Marsolo K, Myers MF, Nix J, Hall ES. Adapting clinical systems to enable adolescents’ genomic choices. ACI Open. 2020;4(2):e126–e131. 10.1055/s-0040-1718747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoell C, Wynn J, Rasmussen LV, et al. Participant choices for return of genomic results in the eMERGE Network. Genet Med. 2020;22(11):1821–1829. 10.1038/s41436-020-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz MLB, McCormick CZ, Lazzeri AL, et al. A model for genome-first care: returning secondary genomic findings to participants and their healthcare providers in a large research cohort. Am J Hum Genet. 2018;103(3):328–337. 10.1016/j.ajhg.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrikson NB, Scrol A, Leppig KA, Ralston JD, Larson EB, Jarvik GP. Preferences of biobank participants for receiving actionable genomic test results: results of a recontacting study. Genet Med. 2021;23(6):1163–1166. 10.1038/s41436-021-01111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter S, Taylor D, Bates I. Institutionalized paternalism? Stakeholders’ views on public access to genetic testing. J Health Serv Res Policy. 2006;11(3):155–161. 10.1258/135581906777641758. [DOI] [PubMed] [Google Scholar]
- 20.Miller DT, Lee K, Chung WK, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23(8):1381–1390. Published correction appears in Genet Med. 2021;23(8):1582-1584. 10.1038/s41436-021-01172-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data sets summarized in this article can be requested on the Electronic Medical Records and Genomics website https://emerge-network.org.


