Highlights
-
•
Dicer complex-dependent siRNAs can activate genes upon dot-1.1 loss.
-
•
Genes upregulated by dot-1.1 (H3K79me) loss are enriched for proteolytic processes.
-
•
Elevated proteolysis likely leads to dot-1.1 lethality.
-
•
27% of genes expressed lower in Argonaute alg-5(-) are elevated by dot-1.1 loss.
Keywords: H3K79 methylation; DOT1L, C. elegans; RNAi; Small RNA; CED-3
Abstract
Methylation of histone H3 at lysine 79 (H3K79) is conserved from yeast to humans and is accomplished by Dot1 (disruptor of telomeric silencing-1) methyltransferases. The C. elegans enzyme DOT-1.1 and its interacting partners are similar to the mammalian DOT1L (Dot1-like) complex. The C. elegans DOT-1.1 complex has been functionally connected to RNA interference. Specifically, we have previously shown that embryonic and larval lethality of dot-1.1 mutant worms deficient in H3K79 methylation was suppressed by mutations in the RNAi pathway genes responsible for generation (rde-4) and function (rde-1) of primary small interfering RNAs (siRNAs). This suggests that dot-1.1 mutant lethality is dependent on the enhanced production of some siRNAs. We have also found that this lethality is suppressed by a loss-of-function of CED-3, a conserved apoptotic protease. Here, we describe a comparison of gene expression and primary siRNA production changes between control and dot-1.1 deletion mutant embryos. We found that elevated antisense siRNA production occurred more often at upregulated than downregulated genes. Importantly, gene expression changes were dependent on RDE-4 in both instances. Moreover, the upregulated group, which is potentially activated by ectopic siRNAs, was enriched in protease-coding genes. Our findings are consistent with a model where in the absence of H3K79 methylation there is a small RNA-dependent activation of protease genes, which leads to embryonic and larval lethality. DOT1 enzymes’ conservation suggests that the interplay between H3K79 methylation and small RNA pathways may exist in higher organisms.
Graphical abstract
Abbreviations
- C. elegans
Caenorhabditis elegans;
- H3K79
histone H3 lysine 79;
- Dot1
disruptor of telomeric silencing-1;
- DOT1L
Dot1(yeast)-like;
- dsRNA
double-stranded RNA;
- RNAi
RNA interference;
- exo-RNAi
RNAi induced by exogenous dsRNA;
- rde-1/4
RNAi-deficient;
- siRNA
small interfering RNA;
- zfp-1
zinc finger protein-1;
- alg-3/4/5
Argonaute (plant)-like gene;
- RdRP
RNA-dependent RNA polymerase;
- sago-2
synthetic secondary siRNA-deficient ArGOnaute mutant.
1. Introduction
C. elegans H3K79 methyltransferase DOT-1.1 and its interacting partner zinc finger protein 1 (ZFP-1) are analogous to the human DOT1L and AF10 (acute lymphoblastic leukemia 1-fused gene from chromosome 10) [1]. Both DOT-1.1 [2] and DOT1L [3,4] are essential for development and have been implicated in enhancer regulation through maintaining open chromatin structure [5,6]. DOT1L plays important roles in erythropoiesis [7], craniofacial development [8], bone morphogenesis [9], neurodevelopment [10,11], cardiac health [12], brown fat generation [13], and immune system function [14], [15], [16]. Inhibition of DOT1L leads to the higher efficiency of iPSC (induced pluripotent stem cell) production [17]. DOT1L is also an oncoprotein initially described in connection to leukemias driven by MLL fusion proteins [18] and later implicated in numerous other cancers (reviewed in [19]), most notably triple-negative breast cancer [20,21]. Notably, DOT1L deficiency allowed increased influenza virus replication in cell culture [22].
In C. elegans, the function of the zfp-1 gene, together with a group of other genes coding for chromatin-interacting proteins, has been connected to modulating double-stranded RNA (dsRNA)-induced gene silencing (i.e. RNA interference or RNAi) [23]. We have recently described the elevation of antisense transcription upon zfp-1 reduction-of-function, which suggested a potential for the generation of ectopic dsRNA and activation of the dsRNA-responsive RNAi pathway [1,2].
Mutations in the rde-1 and rde-4 genes were among the first RNAi-resistant mutants identified in C. elegans [24]. RDE-4 binds dsRNA [25,26] and is critical for Dicer-mediated dsRNA cleavage generating primary siRNAs [25,27], whereas RDE-1 is an Argonaute protein binding primary siRNAs and facilitating their function [28]. In addition to primary siRNAs, more abundant secondary siRNAs are produced by RNA-dependent RNA polymerases (RdRP) and act downstream of primary siRNAs in gene silencing (reviewed in [29]). Whereas RDE-1 and RDE-4 are essential for the exo-RNAi pathway initiated experimentally [24] and important in antiviral defense [30], their role in the numerous known endogenous siRNA pathways that both silence and activate genes is limited (reviewed in [29]).
The idea of the potential ectopic activation of the dsRNA-responsive RNAi pathway when the function of the DOT-1.1 complex is reduced prompted us to test whether lethality of the dot-1.1 deletion mutant could be suppressed by rde-1 or rde-4 null mutants, and, indeed, it was suppressed [2]. This finding strongly suggests that primary siRNAs produced by RDE-4 and bound by RDE-1 cause dot-1.1 mutant lethality. When generating a dot-1.1 deletion via CRISPR-Cas9 we were only able to get heterozygous, and, eventually, homozygous dot-1.1 mutants deficient in H3K79 methylation in the ced-3 mutant background [2]. CED-3 is a conserved cysteine protease acting at the execution phase of apoptosis (reviewed in [31]). Thus, we conclude that dot-1.1(-) lethality involves ectopic siRNAs that eventually cause elevated apoptosis.
Here, we describe molecular analyses of gene expression and primary siRNA abundance changes caused by dot-1.1(knu339) deletion in viable ced-3(n1286) mutant background [32]. We identified groups of genes with significant changes in primary siRNAs accompanied by changes in gene expression. Surprisingly, increased siRNA levels are primarily associated with elevated gene expression in dot-1.1 mutants suggesting positive regulation by RNAi. Indeed, the upregulation, as well as downregulation, of siRNA target genes in dot-1.1(knu339) was dependent on RDE-4. Moreover, we made a connection between siRNA target genes upregulated in dot-1.1(knu339) and genes with reduced expression in the alg-5 Argonaute mutant [33], which suggests positive gene regulation by ALG-5. Importantly, CED-3 expression is positively regulated by alg-5 [33], and we found an enrichment in proteases among siRNA target genes showing elevated expression in dot-1.1(knu339). Thus, based on the previously published and new data, we conclude that increased protease activity driven by RDE-4-dependent siRNAs is the most likely cause of dot-1.1 lethality.
2. Materials and methods
2.1. C. elegans strains and manipulations
N2, Wild-type
MT3002 ced-3(n1286) IV
WM49 rde-4(ne301) III
COP1304 dot-1.1 [knu339 - (pNU1092 - KO loxP::hygR::loxP)] I; ced-3(n1286) IV
AGK782 dot-1.1 [knu339 - (pNU1092 - KO loxP::hygR::loxP)] I; rde-4(ne301) III
The dot-1.1 deletion mutant above is abbreviated to dot-1.1(knu339). Strains were maintained on standard NGM solid agar plates at 20 °C feeding on a spot of E. coli OP50 from an overnight culture.
2.2. RNA extraction from C. elegans
An overnight OP50 culture in LB broth was concentrated 23x. Starved L1 worms were transferred to 150 mm plates seeded with 1.3 ml of the 23x concentrated bacteria. After the worms were grown into adults and started laying eggs, they were collected with M9, bleached with an alkaline hypochlorite solution, and washed 4 times with M9. The surviving eggs were left to hatch overnight in M9. The following day, 1700 – 2500 age-synchronized L1s were added to a plate. Once they grew to egg-laying adults, they were bleached a second time, washed 3 times with M9, and the resulting eggs were suspended in 1 ml TRIzol™ (Thermo Fisher Scientific), and placed at −80 °C. Egg preparations underwent 3 cycles of freezing and thawing to facilitate lysis.
RNA for mRNA- and small RNA-sequencing was purified using the miRNeasy kit (Qiagen) following standard procedures for total RNA extraction, including the optional wash with RWT buffer. RNA for qPCRs was purified using the Direct-zol™ kit (Zymo Research).
2.3. RNA sequencing and data analyses
Three independent biological replicates of worm populations were used for RNA sequencing. Aliquots of the same RNA samples were sent for sequencing of mRNAs* and small RNAs. Illumina NovaSeq 6000 was used by Novogene Corporation. Quality control, mapping, and read-length analyses were performed by Novogene. For small RNA sequencing analyses related to mapping and reads’ length and for mRNA sequencing, genes were mapped to the C. elegans genome with Ensembl (WBcel235, bioproject PRJNA13758, release WS269, Ensembl accession GCA_000002985.3). HISAT2 [34] was used to map mRNA reads and Bowtie was used to map small RNA reads to the genome [35]. miRNA mapping was done separately with miRBase (release 22) as a reference. Analysis of mRNA sequencing was performed by Novogene using StringTie for quantification of reads [36], FPKM normalization, and edgeR for differential expression [37]. Analysis of miRNAs was performed by Novogene using DESEQ2 for differential expression [38] and miREvo [39] and miRDeep2 [40] to predict novel miRNAs.
*Sequenced and analyzed RNAs from the “mRNA sequencing” also include long noncoding RNAs.
Analysis of differentially expressed siRNAs from the small RNA sequencing data was performed with a custom script using QuasR [41]. Briefly, small RNAs were mapped to the C. elegans genome (WBcel235, bioproject PRJNA13758, release WS283, NCBI Refseq accession: GCF_000002985.6, an annotation which lacks piRNAs, which were not considered for siRNA analysis). Alignment was done using the Bowtie option (maxHits = 50) and counting was done using parameter orientation = “same” for sense siRNAs and “opposite” for antisense siRNAs. DESEQ2 [38] was used for the analysis of differential expression.
Gene differences considered significant are those with p<0.05. Upregulated genes have log2(fold change) > 0. Downregulated genes have log2(fold change) < 0.
2.4. Quantitative RT-PCR (RT-qPCR)
Three independent biological replicates of worm populations were used. 500 μg of RNA was used for cDNA synthesis using Maxima Reverse Transcriptase (Thermo Scientific), random hexamer primer, and following manufacturer standard procedures. When selecting genes to test dependence on the RNAi pathway for gene expression with qPCR, genes with near-zero average read counts in control or dot-1.1(knu339) from the RNA sequencing data were excluded. Primers were designed to span exon-exon junctions and were confirmed to amplify in wild-type RNA treated with RT (i.e. cDNA) and to not amplify without RT. The act-3 gene was used as a normalization control. Luna® Universal qPCR Master Mix (New England BioLabs) was used for quantitative PCR according to manufacturer specifications. For statistical analysis, a Student's t-test (2 sample, unpaired) was used to compare act-3-normalized cycle threshold (Ct) values between the control and mutant.
2.5. Additional data analyses
2.5.1. Gene overlaps
Representation factor (RF) calculation:
RF = (n1,2)/[(n1 x n2)/N], with n1,2 number of genes common to sets 1 and 2, n1 number of genes in set 1, n2 number of genes in set 2, N total number of genes considered (N = 20,000 genes in C. elegans genome)
(n1,2): observed overlap
[(n1 x n2)/N: expected overlap
The P value corresponds to the normal approximation of the hypergeometric distribution. Representation factor and p-value script by Jim Lund: http://nemates.org/MA/progs/overlap_stats.html
Overlaps were constructed using a common name for each gene, “Public Name”, which was extracted by inputting gene names from each list to WormBase's SimpleMine tool (release WS283) and selecting “Public Name”. To properly correspond and overlap our mRNA sequencing data of dot-1.1; ced-3 vs. ced-3 mutants to the Argonaute mutant small RNA dataset by Seroussi et al. [42], piRNAs and miRNAs were filtered out from the latter dataset before overlapping.
2.5.2. Graphs
Graphs showing small RNA reads were generated with UCSC Genome Browser [43] using WIG files as input, which were generated through QuasR's qExportWig function (binsize=50 L, scaling = TRUE, strand = “+” for sense and “-“ for antisense reads). The displayed read numbers in each bar are normalized to account for the total number of reads in that sample and in the other samples used for comparison. Normalization: the number of alignments per bin (n) for a sample (i) is linearly scaled to the mean total number of alignments over all samples used for comparison (mean(N), which includes ced-3 replicates 1, 2, and 3 and dot-1.1; ced-3 replicates 1, 2, and 3) according to:
n_s = n / N[i] * mean(N) where n_s is the scaled number of alignments in the bin (displayed as a bar) and N[i] is the total number of alignments for the given sample i.
Venn diagrams were generated with the R package eulerr by Johann Larsson. qPCR graphs and statistics were computed with Graphpad Prism 9. The summary model was created with BioRender.com.
2.5.3. Gene ontology enrichment analysis
The clusterProfiler software was used by Novogene for the analyses [44]; the data are available in Supplementary Files 5 and 6. Gene Ontology (GO) is a bioinformatics classification system. It includes three main category branches: cellular component (CC), molecular function (MF), and biological process (BP). GO terms with p-value adjusted (padj) < 0.05 are considered most significantly enriched. p-value adjusted controls for false discovery rate and represents p-values, adjusted for multiple testing with the Benjamini-Hochberg procedure.
3. Results
3.1. Analysis of small RNAs in dot-1.1; ced-3 mutant embryos compared to ced-3(n1286)
We reported that embryonic and larval lethality in the dot-1.1 deletion mutant is suppressed by rde-4 and rde-1 mutations [2]. Therefore, we used embryo preparations to determine changes in small RNA populations between viable dot-1.1; ced-3 double mutant, and ced-3 single mutant strains. RDE-4 binds dsRNA and facilitates dsRNA processing to siRNAs by Dicer [25], [26], [27]. This prompted us to specifically focus on the C. elegans Dicer product (or primary siRNA) populations bearing 5′-monophosphate. We note that Dicer-dependent miRNAs and Dicer-independent piRNAs, also called 21U-RNAs, that contain 5′-monophosphate (reviewed in [29]), would also be detected with our cloning protocol.
3.1.1. dot-1.1 deletion does not cause large changes in 5′-monophosphate containing small RNAs
Overall, there were no dramatic changes in small RNA populations in dot-1.1 mutant worms. Sequencing yielded comparable percentages (∼99%) of mapped small RNA (sRNA) reads (Table S1) and similar small RNA length distribution with the majority of reads in the range of 21–24 nt (Fig. S1), which would correspond to C. elegans piRNAs and miRNAs. There was also a small peak of reads at 26 nt likely corresponding to endogenous 26G RNA produced by Dicer [45]. Similar numbers of reads mapped to mature and precursor miRNAs (Table S2) and repeat sequences (Fig. S2), and a similar percentage of reads corresponded to different classes of non-coding RNAs, including tRNA, rRNA, snRNA, and snoRNA (Table S3).
3.1.2. miRNAs affected by dot-1.1(knu339) predominantly bind to RDE-1
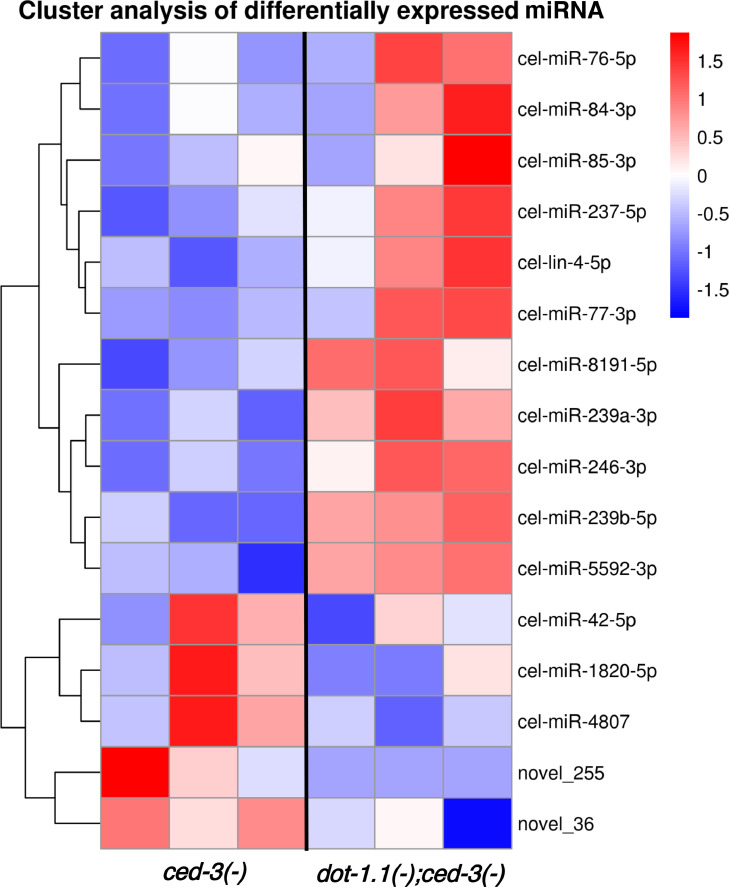
Differential expression analysis revealed 16 significantly misregulated miRNAs: 11 upregulated and 5 downregulated (Fig. 1). The Argonautes ALG-1 and ALG-2 are predominant co-factors of miRNAs that are required for miRNA function (reviewed in [29]). miRNAs have also been found in complexes with RDE-1 [42,46]. Notably, 9 out of 14 annotated miRNAs that change expression in dot-1.1(knu339) were reported to immunoprecipitate (IP) with the RDE-1 Argonaute [42], including three, miR-76–5p, miR-1820–5p, and miR-84–3p found exclusively enriched in RDE-1 IP and not in the other 18 tested Argonaute IPs. Notably, miR-5592–3p was enriched in RDE-1 IP 305-fold compared to input [42] (Table 1). These findings are consistent with the functional connection between DOT-1.1 and RDE-1 that we have reported [2].
Fig. 1.
Hierarchical cluster analysis of differentially expressed miRNAs indot-1.1(knu339).
11 miRNAs are upregulated and 5 are downregulated in dot-1.1(knu339); ced-3(n1286) relative to ced-3(n1286) mutants. The color from blue to red represents the log10(TPM+1) value in ascending order. TPM: transcript per million.
Table 1.
miRNAs changing expression indot-1.1(knu339). Based on small RNA sequencing data. R1–3: replicate 1–3. *Data from Seroussi et al. [42].
| miRNA |
ced-3(n1286) expression levels (TPM) |
dot-1(knu339); ced-3(n1286) expression levels (TPM) |
p-value | Argonaute IP enrichment* | ||||
|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R1 | R2 | R3 | |||
| miR-77 | 2.03 | 1.62 | 2.87 | 3.25 | 26.72 | 31.39 | 1.16E-6 | ALG-1, ALG-2 |
| miR-239a | 9.52 | 17.60 | 8.50 | 34.37 | 74.07 | 39.40 | 1.66E-6 | RDE-1 22x, ERGO-1 |
| miR-237 | 1.68 | 2.79 | 5.17 | 5.90 | 14.21 | 23.79 | 8.65E-5 | ALG-1, RDE-1 2x |
| lin-4 | 1.33 | 0.41 | 1.15 | 1.92 | 4.53 | 7.07 | 0.0011 | ALG-1, RDE-1 3x |
| miR-246 | 20.86 | 29.47 | 21.76 | 36.44 | 62.57 | 59.03 | 0.0015 | ALG-2 |
| miR-85 | 0.35 | 1.12 | 2.41 | 0.81 | 2.84 | 15.70 | 0.0101 | ALG-1, RDE-1 4x |
| miR-76 | 0.07 | 0.81 | 0.23 | 0.37 | 2.71 | 2.12 | 0.013 | RDE-1 12x |
| miR-1820 | 89.33 | 235.98 | 136.37 | 72.80 | 70.82 | 122.34 | 0.014 | RDE-1 9x |
| miR-5592 | 1.40 | 1.32 | 0.69 | 2.51 | 2.77 | 3.01 | 0.026 | ERGO-1, RDE-1 305x |
| miR-8191 | 0.00 | 0.15 | 0.29 | 0.81 | 0.88 | 0.44 | 0.029 | |
| miR-239b | 0.14 | 0.00 | 0.00 | 0.37 | 0.41 | 0.49 | 0.032 | ALG-1, ALG-2, ALG-5, ERGO-1 |
| miR-42 | 15.40 | 71.63 | 39.39 | 10.70 | 32.88 | 23.12 | 0.035 | ERGO-1, ALG-5, RDE-1 |
| miR-84 | 0.28 | 1.01 | 0.57 | 0.52 | 1.83 | 3.23 | 0.043 | RDE-1 174x |
| miR-4807 | 76.73 | 139.85 | 105.08 | 78.26 | 62.91 | 77.86 | 0.046 | ALG-2, ALG-3, ALG-5, ERGO-1 |
3.1.3. siRNA changes caused by dot-1.1 deletion
Next, we analyzed changes in small RNAs corresponding to annotated protein-coding and non-coding genes and selected groups significantly changed in the mutant (File S1, S2). Due to the expectation that primary siRNAs are double-stranded Dicer products, we considered both sense (File S1) and antisense siRNA reads (File S2) separately. The abundance of primary siRNAs is very low, therefore, we largely detected significant changes in either sense or antisense siRNAs corresponding to a particular gene. We are aware that some sense siRNAs could represent mRNA degradation products.
We found 339 genes with significantly upregulated antisense siRNAs and 175 genes with significantly downregulated antisense siRNAs. In the case of sense siRNAs, there were 693 genes with siRNA upregulation and 231 genes with downregulation. Because a lack of RDE-4 and RDE-1 activity suppresses dot-1.1(-) lethality [2], we anticipate that elevated expression of some RDE-4-dependent siRNAs bound by RDE-1 causes changes in gene expression leading to lethality. Therefore, we focus on genes with upregulated siRNAs in the dot-1.1 mutant in further analysis.
3.2. Correlation of gene expression and siRNA changes in dot-1.1 mutant suggest positive gene regulation by RNAi in the embryos
Analysis of significant changes in the mRNA of protein-coding genes in dot-1.1(knu339) revealed 688 downregulated genes and 1291 upregulated ones. Next, we asked whether genes with significantly changed siRNAs were overrepresented among the gene groups changing expression (Table 2). Among the 688 genes with decreased expression in the dot-1.1 mutant, we found 7 genes with elevated antisense siRNAs and 8 genes with elevated sense siRNA. These overlaps are smaller (RF < 1) than expected by chance and indicate a significant depletion of siRNA-changed genes among the downregulated group. Among the 1291 genes with elevated expression in the dot-1.1 mutant, there were 39 genes with upregulated antisense siRNAs and 89 genes with elevated sense siRNAs; both overlaps showed significant overrepresentation above expected by chance (RF > 1). Therefore, we conclude that increased siRNA abundance and elevated expression of corresponding genes in dot-1.1(knu339) show a significant correlation.
Table 2.
Relationship between altered mRNA levels and upregulated primary siRNAs indot-1.1(knu339).
| mRNA change* | siRNA change# | overlap | Rep. factor (RF) | p-value |
|---|---|---|---|---|
| DOWN: 688 | Antisense UP: 399 | 7 | 0.5 | p < 0.033 |
| DOWN: 688 | Sense UP: 693 | 8 | 0.3 | p < 1.14e-04 |
| DOWN: 688 | Both UP: 1085 | 15 | 0.4 | p < 1.54e-05 |
| UP: 1291 | Antisense UP: 399 | 39 | 1.5 | p < 0.006 |
| UP: 1291 | Sense UP: 693 | 89 | 2 | p < 3.29e-10 |
| UP: 1291 | Both UP: 1085 | 126 | 1.8 | p < 5.95e-11 |
Based on mRNA and small RNA sequencing data of dot-1(knu339); ced-3(n1286) and ced-3(n1286) worms. There is significant under-representation (RF < 1 and p < 0.05) of genes with both downregulated mRNA and upregulated siRNA and significant over-representation (RF > 1 and p < 0.05) of genes with both upregulated mRNA and siRNA in dot-1(knu339) mutants. DOWN: number of genes with RNA downregulated in dot-1(knu339) mutants. UP: number of genes with RNA upregulated in dot-1(knu339) mutants. The P-value corresponds to the normal approximation of the hypergeometric distribution (see the Methods Section 2.5.1).
*See Supplementary Files 1 and 2.
#See Supplementary Files 3 and 4.
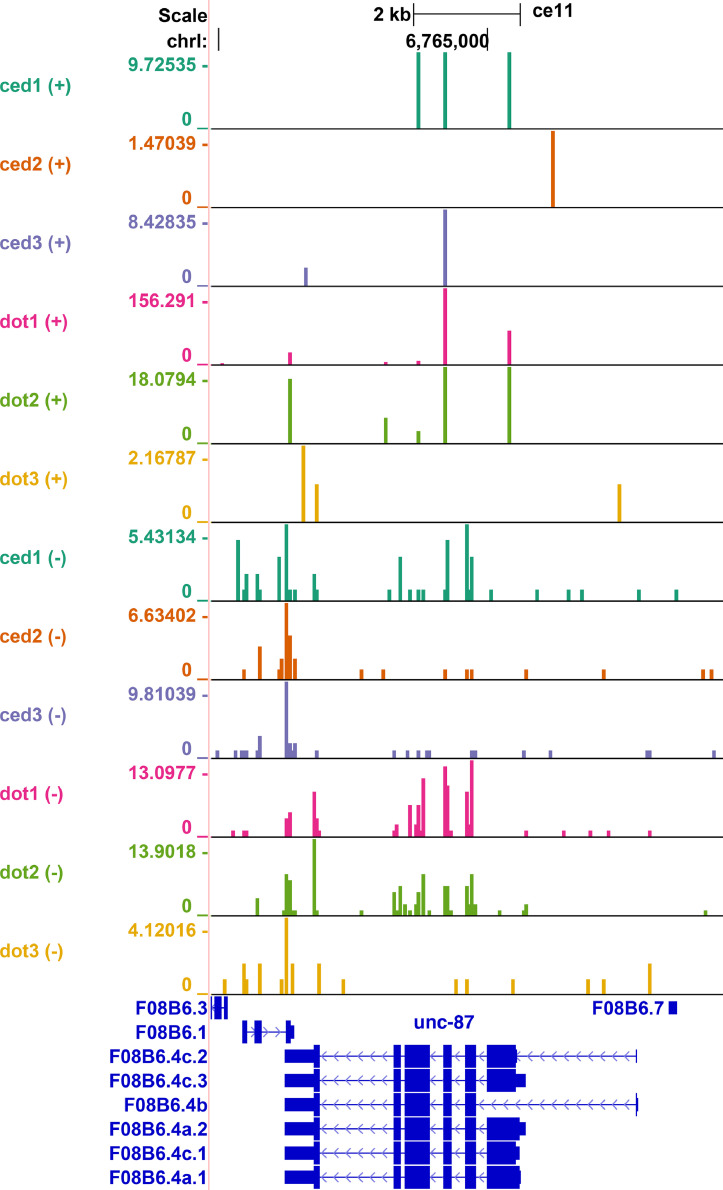
In many cases, changes in both sense and antisense siRNAs were detected but low read counts and variability among the samples did not allow to count them as significant in both directions. Nonetheless, a few examples with significantly elevated sense and antisense siRNAs were identified (Fig. 2).
Fig. 2.
Increase in siRNA production at the unc-87 locus in dot-1.1(knu339). UCSC Genome Browser (http://genome.ucsc.edu) was used for data visualization. The top six tracks (ced1, ced2, ced3, dot1, dot2, and dot3) represent normalized plus strand read counts for three control (ced-3(n1286)) and three dot-1.1(knu339); ced-3(n1286) replicates and the bottom six tracks represent minus strand reads for each sample. The displayed numbers in each sample are normalized to account for the total number of reads in that sample and in the other samples used for comparison, see the Methods Section 2.5.2. for more information. Identical samples are color-coded and data ranges are indicated for each track on the Y-axis. Note the tail/tail overlap between unc-87 coded on the minus DNA strand and the left neighboring gene coded on the sense strand.
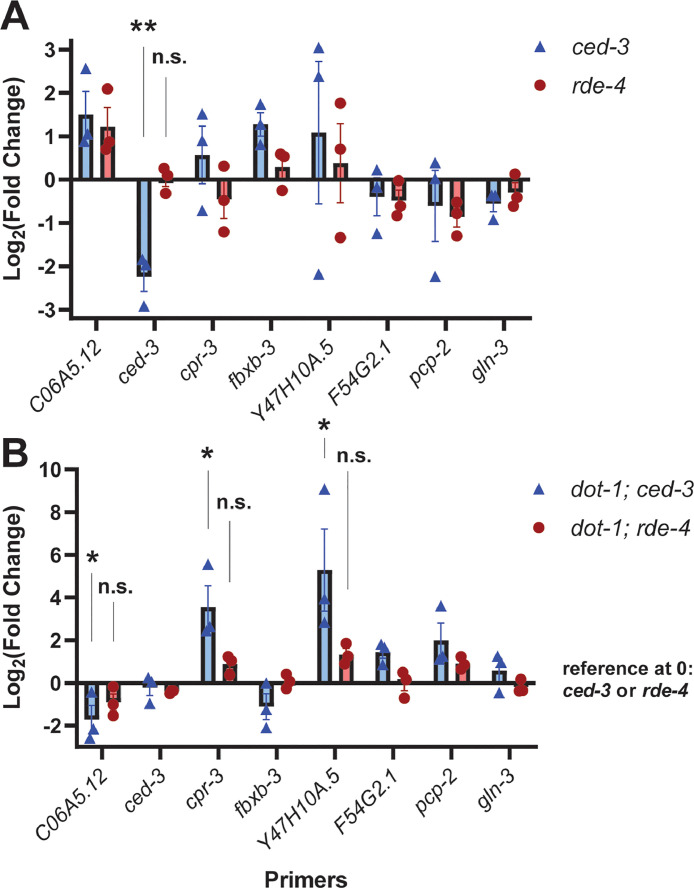
To determine whether siRNA elevation caused changes in gene expression in dot-1.1(knu339) we used the dot-1.1(knu339); rde-4(ne301) double mutant strain. Since RDE-4 is required for primary siRNA production [25], [26], [27] and rde-4(ne301) suppressed the lethality of dot-1.1(knu339), the dependence of gene expression changes on RDE-4 activity would support siRNA-dependent regulation. We evaluated the effect of dot-1.1(knu339) on siRNA target gene expression by comparing dot-1.1(knu339); ced-3(n1286) to ced-3(n1286) background and dot-1.1(knu339); rde-4(ne301) to rde-4(ne301). We chose genes with decreased (C06A5.12, fbxb-3, gln-3) or increased (cpr-3, Y47H10A.5, pcp-2, F54G2.1) expression in dot-1.1(knu339); ced-3(n1286) compared to ced-3(n1286) according to RNA-seq accompanied with significant changes in sense siRNA levels. The cases with sense siRNA changes were chosen to test their prediction value in identifying RNAi-regulated genes. First, we determined how the expression of the chosen siRNA target genes changed in ced-3(n1286) and rde-4(ne301) (Fig. 3A). In both backgrounds we observed variable changes in gene expression compared to wild-type that did not reach statistical significance. Consistent with our sequencing data, the addition of dot-1.1(knu339) to ced-3(n1286) led to expected alterations (increase or decrease) in gene expression in six out of seven cases (except for gln-3); expression changes in three genes (C06A5.12, cpr-3, and Y47H10A.5) reached statistical significance (Fig. 3B). However, the effect of dot-1.1(knu339) on siRNA target gene expression was not observed in the rde-4(ne301) background, indicating the dependence of changes on RDE-4 activity and thus siRNA production (Fig. 3B). Notably, the Y47H10A.5 gene is known to be negatively regulated by the RDE-1/RDE-4 pathway in adult worms [46].
Fig. 3.
Deletion ofdot-1.1results in RDE-4-dependent regulation of genes.
(A) Measurement of gene expression by RT-qPCR in ced-3 and rde-4 mutants, relative to wild-type. ced-3 mRNA expression has been detected by the Northern blotting in ced-3(n1286)[32] and we, therefore, included ced-3 in the panel of tested genes to interrogate its possible regulation by RDE-4 and DOT-1.1.
(B) Measurement of gene expression by RT-qPCR in dot-1.1; ced-3 mutants relative to ced-3 mutants and in dot-1.1; rde-4 mutants relative to rde-4 mutants.
The Student's t-test (2 sample, unpaired) was used to compare act-3-normalized cycle threshold (Ct) values in mutants and their control (see Fig. S3).
**: p<0.01. *: p<0.05. n.s.: not significant. Comparisons without annotation are not significant.
These results suggest that: 1) RDE-4-dependent siRNAs can activate genes, and 2) dot-1.1(knu339) lethality could be due to elevated expression of some genes. We have not anticipated these results because the dsRNA-dependent RNAi pathway is only known to associate with gene silencing (reviewed in [29]).
3.2.1. Genes misregulated by dot-1.1 deletion are enriched in RDE-1-dependent and ALG-4-dependent siRNA targets
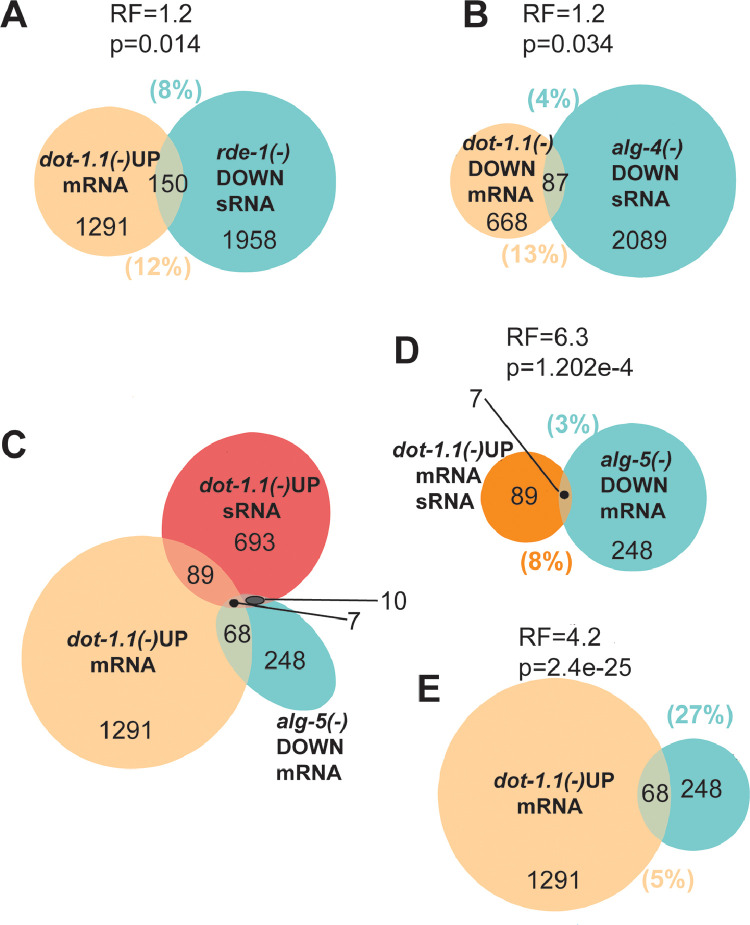
To further understand which downstream component of RNAi could be mediating the increase in gene expression in dot-1.1(knu339) embryos, we utilized comprehensive datasets generated by Claycomb and colleagues that describe siRNA depletion in 19 Argonaute mutants [42]. We compared genes upregulated or downregulated in dot-1.1(knu339) with the lists of siRNA-depleted target genes in the 19 mutant backgrounds [42] (Tables S4, S5). We found only two Argonautes (RDE-1 and SAGO-2) whose target genes were significantly enriched among the genes upregulated in dot-1.1(-) (Fig. 4A, Tables S4, S5). This suggests that RDE-1 and SAGO-2 may act sequentially in the RNAi pathway activated by dot-1.1(knu339), similar to their function in the exo-RNAi pathway induced by dsRNA [28]. On the contrary, there was only one Argonaute (ALG-4) whose target genes were enriched in dot-1.1(-)-downregulated dataset (Fig. 4B). Consistently, genes targeted by alg-4-dependent siRNAs were significantly depleted from the dot-1.1(-)-upregulated dataset (Table S4). We conclude that RDE-1 may activate genes, together with RDE-4, in the dot-1.1(knu339) background, and that ALG-4 may be responsible for the reduced expression of some genes in the dot-1.1 mutant. The latter conclusion is consistent with the increased abundance of the chromatin silencing mark H3K9me2 at ALG-3/4-target genes in dot-1.1(knu339) [6]. Many groups of the Argonaute-dependent siRNA target genes were significantly underrepresented from the groups of genes misregulated by dot-1.1(knu339), including CSR-1, ALG-1, ALG-2, ALG-3, ERGO-1, HRDE-1, NRDE-3, PPW-1, PPW-2, PRG-1, SAGO-1, WAGO-1, and WAGO-4 targets (Tables S4, S5). Therefore, these Argonautes are unlikely to cause aberrant changes in gene expression seen in dot-1.1(knu339).
Fig. 4.
Genes misregulated by deletion ofdot-1.1are enriched in Argonaute-dependent siRNA targets.
There is a significant overlap (RF > 1, p < 0.05) between genes with mRNAs upregulated in dot-1.1 mutants and small RNAs downregulated in rde-1 mutants (A) and mRNAs downregulated in dot-1.1 mutants and small RNAs downregulated in alg-4 mutants (B). Triple overlap of genes with upregulated mRNAs and small RNAs in dot-1.1 mutants and downregulated mRNAs in alg-5 mutants (C). Two of the resulting overlaps are emphasized in (D) and (E). The P-value corresponds to the normal approximation of the hypergeometric distribution (see Methods Section 2.5.1). up: upregulated in the mutant. down: downregulated in the mutant. RF: representation factor. sRNA: small RNA.
3.2.2. Genes upregulated by dot-1.1 deletion include 27% of those found downregulated in alg-5(ram2)
The Argonaute protein ALG-5 has not yet been functionally connected to any specific RNAi-related pathway, although gene expression changes in the alg-5(ram2) null mutant have been reported [33]. Notably, ∼2.6 times as many genes showed reduced expression in the alg-5 mutant (248) compared to the upregulated ones (96) [33]. Remarkably, 27% of these downregulated genes were present in the group of genes with elevated expression in dot-1.1(knu339) (Fig. 4C, E). The downregulated genes in alg-5(ram2) and the set of genes with increases in both sense siRNA and mRNA in dot-1.1(knu339) also significantly overlapped (Fig. 4C, D). Importantly, this overlap includes the genes F54G2.1 and Y47H10A.5, which showed an RDE-4-dependent increase in expression in the dot-1.1(knu339) mutants (Fig. 3B). These findings suggest that ALG-5 plays a role in siRNA-dependent gene activation in the absence of H3K79 methylation.
3.3. Functional analysis of gene expression changes occurring in dot-1.1(knu339) embryos
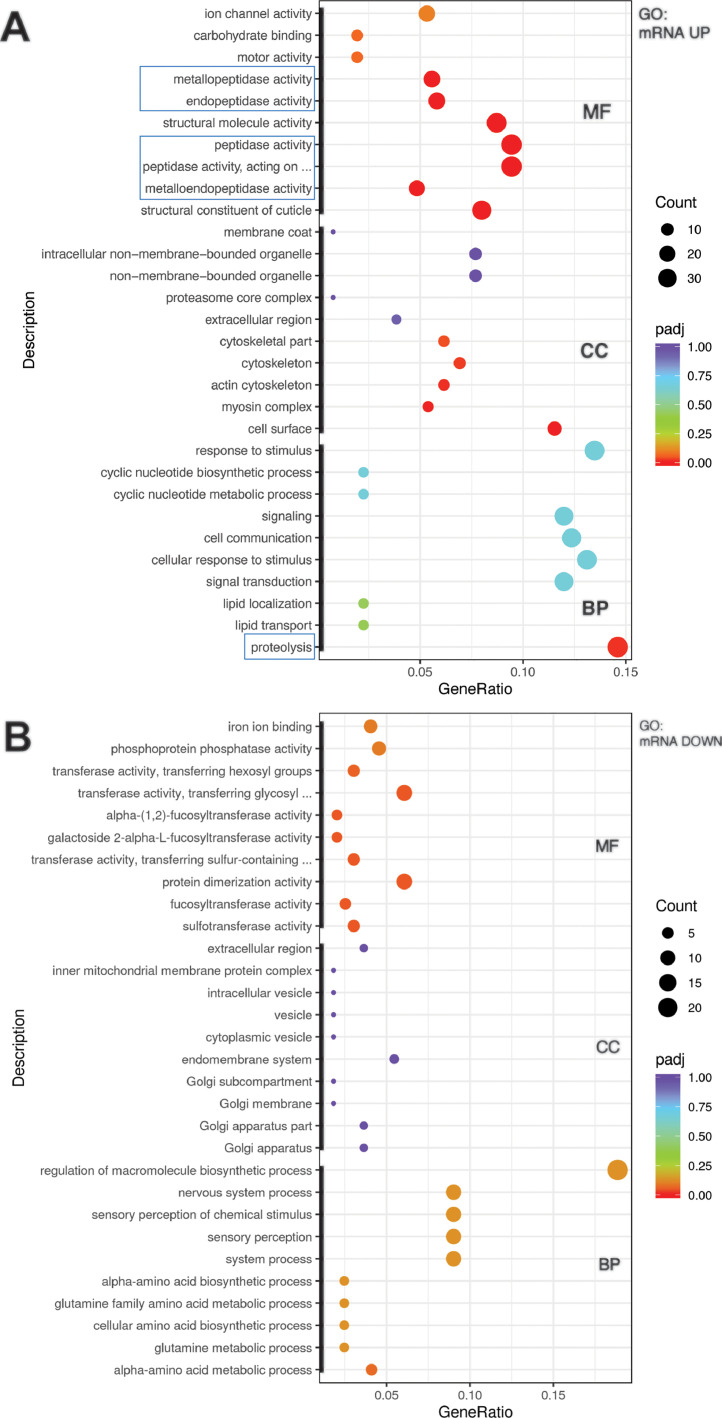
We used Gene Ontology (GO) database for the functional annotation of dot-1.1(knu339) upregulated and downregulated genes. The upregulated group was strongly enriched in proteolysis-related processes, including peptidase activities (Fig. 5A). The cell surface and cuticle structure categories, as well as cytoskeleton and motor activity functions, were also enriched (Fig. 5A). The downregulated group of genes was enriched in metabolic transferase activities, amino acid metabolism, protein dimerization, macromolecule biosynthesis, and nervous system function (Fig. 5B). Notably, we have reported DOT-1.1 complex-mediated control of enhancers regulating neural and sensory-response pathway genes earlier [2].
Fig. 5.
Deletion of dot-1.1 results in the upregulation of proteolytic processes and the downregulation of metabolic and nervous system pathways. Gene ontology (GO) analyses of genes upregulated (A) or downregulated (B) in dot-1.1(-); ced-3(-) compared to ced-3(-). Top ten most significantly affected GO Terms (thick black lines on the Y-axis) in each of the three category branches: cellular component (CC), molecular function (MF), and biological process (BP) are shown. The abscissa is the ratio of the number of differential genes linked with the GO Term to the total number of differential genes, and the ordinate is a GO Term. The size of a point represents the number of genes annotated to a specific GO Term, and the color from red to purple represents the significance level of the enrichment expressed as p-value adjusted (padj). Only red and orange circles represent significant enrichment, the red color signifies enrichment corresponding to (padj) < 0.05. Proteolysis-related terms are highlighted by blue boxes. See Methods Section 2.5.3. and Supplementary Files S5 and S6 for more information.
3.3.1. Soma-enriched genes similar to Y47H10A.5 are upregulated in dot-1.1(knu339) embryos
The Y47H10A.5 gene shows predominant expression in somatic tissues of the worm and is a known RNAi target repressed by an RDE-1-bound miRNA and secondary siRNAs produced by RNA-dependent RNA polymerases (RdRP) [46]. Surprisingly, we found RDE-4-dependent upregulation of Y47H10A.5 in dot-1.1(knu339) (Fig. 3), whereas it is expressed lower in alg-5(ram2) [33]; both findings suggest positive regulation by RNAi. Y47H10A.5 belongs to a group of genes that show a steep increase in mRNA expression from early embryo to late embryo and from embryo to L1 larval stage [47]. A corresponding protein signature has been identified by mass spectrometry (15 proteins) [48] and includes proteins related to muscle function, such as actin and DIM-1 (Disorganized Muscle), as well as galectins (LEC-1–3) and aspartic protease ASP-6. These functional categories are overrepresented among the dot-1.1(knu339) upregulated genes (Fig. 5A). Remarkably, the asp-6 and lec-2 (Fig. 6) genes show a significant increase in both mRNA and siRNA levels in dot-1.1 mutants, and asp-6 is targeted by RDE-1-regulated siRNAs [42]. These observations point to a positive role of endogenous RNAi in supporting an increase in the expression of soma-enriched genes at the late embryogenesis and early larval stages. At the same time, DOT-1.1 negatively modulates the activity of the RNAi pathway(s).
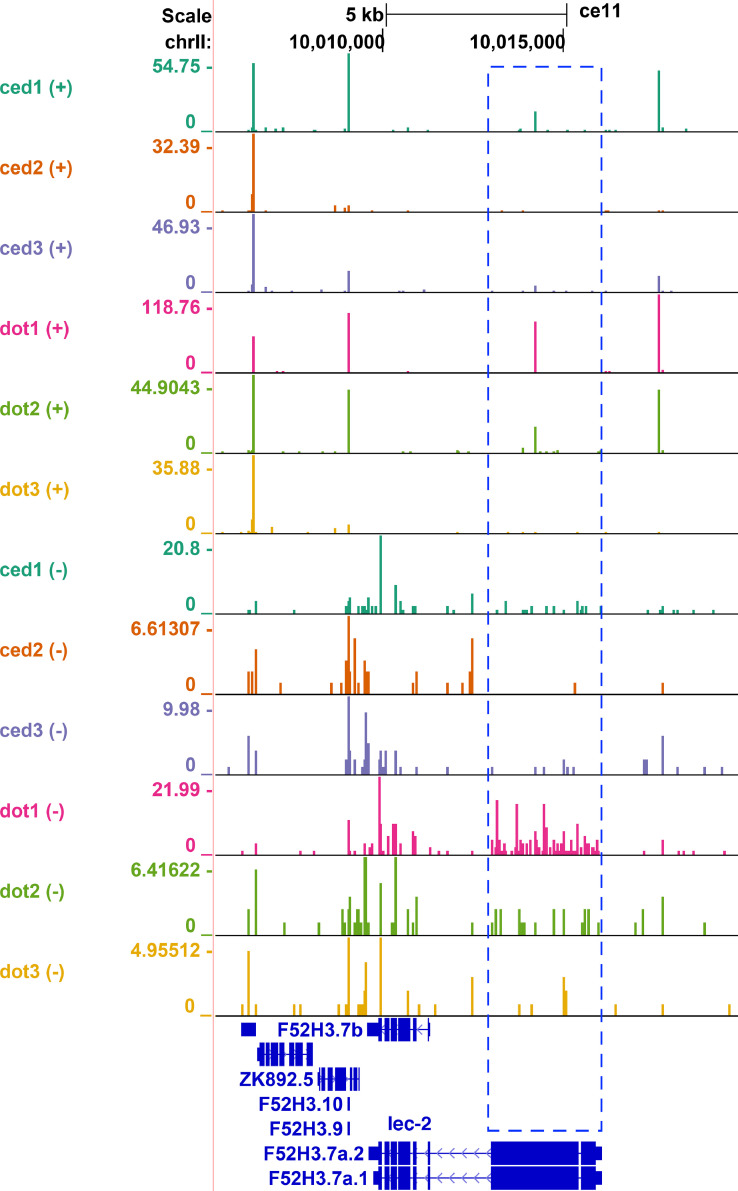
Fig. 6.
Increase in siRNA production at the lec-2 locus in dot-1.1(knu339). UCSC Genome Browser (http://genome.ucsc.edu) was used for data visualization and the track order is the same as in Fig. 2. The blue box highlights the lec-2 gene region with a relative (compared to the reads at ZK892.5 and F52H3.7b) increase in sRNA reads in dot-1.1(knu339); ced-3(n1286) samples compared to ced-3(n1286) samples, despite the differences in absolute read counts between the samples.
3.3.2. Functional connection to proteolysis
As we discussed earlier (Table 2), genes with increased primary siRNAs (sense or antisense) were overrepresented among the dot-1.1(knu339) upregulated mRNA group. These include several protease-coding genes, such as asp-6, asp-1, asp-3, asp-12, cpr-4, and nas-30, as well as those corresponding to cuticle components (collagens, galectins) and actin/myosin complex (myo-1, myo-3). Importantly, we have observed large vacuoles in arrested dot-1.1(knu339) larvae [6], which could be due to the elevated activity of the proteases.
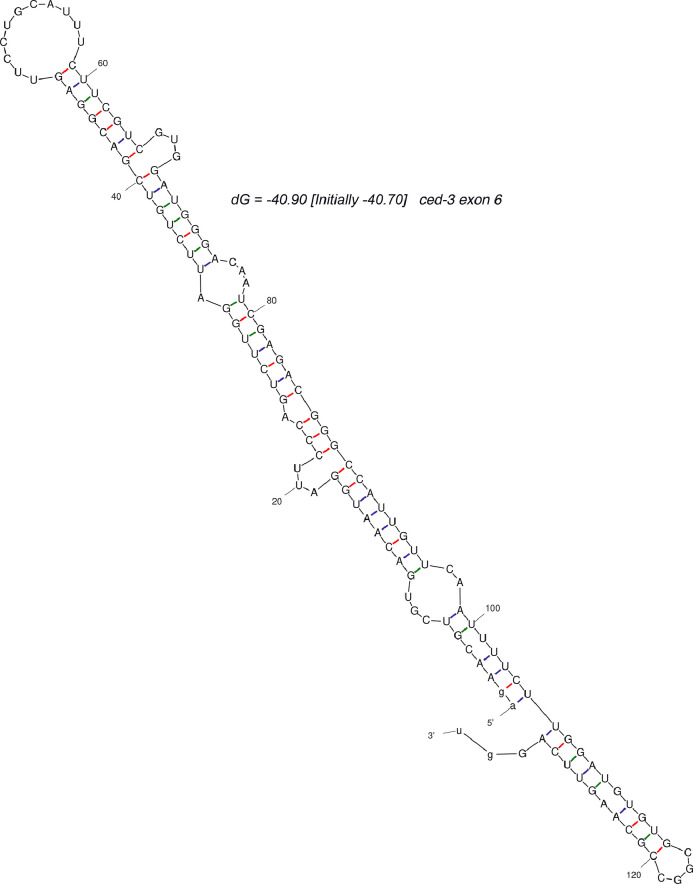
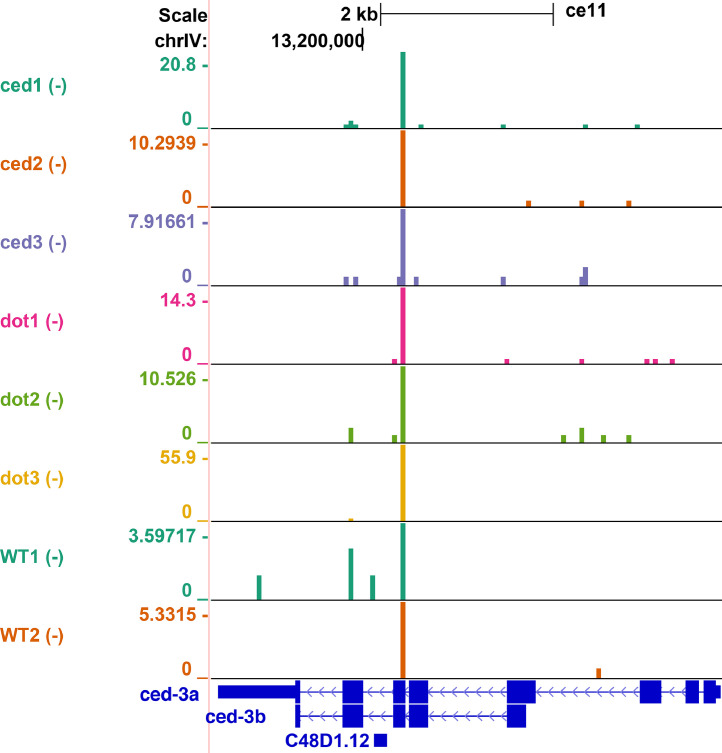
dot-1.1(knu339) lethality is suppressed by ced-3(n1286) loss-of-function [2]. CED-3 is itself an apoptotic protease, and it also promotes the expression of some protease genes, such as pcp-2 [49], which is upregulated in dot-1.1(knu339). We attempted but could not assay regulation of ced-3 by dot-1.1 in our experimental setup using ced-3(n1286) background, which still produces mRNA [32], because of the strongly decreased expression of ced-3 in ced-3(n1286) mutants compared to wild-type (Fig. 3A). However, ced-3 was among the genes expressed lower in alg-5(ram2), and it was also found downregulated in other RNAi-related mutants: rrf-1, rrf-3 (RdRPs) [50] and dcr-1 (Dicer) [51]. Therefore, ced-3 expression may be elevated by a small RNA-mediated process in dot-1.1(knu339) causing mutant lethality. Notably, exon 6 of the ced-3 pre-mRNA folds into a hairpin structure that may be recognized by Dicer (Fig. 7). We detected primary siRNAs corresponding to exon 6 of the longer ced-3 mRNA isoform (Fig. 8), which suggests possible direct regulation by the RNAi pathway(s).
Fig. 7.
Exon 6 of the ced-3 gene folds into a hairpin. The model of the two-dimensional structure of the 128 nt sequence of ced-3 exon 6 flanked by the proximal intronic nucleotides AG and GU was generated by the mfold 4.7 program [52].
Fig. 8.
siRNA production at the ced-3 locus. UCSC Genome Browser (http://genome.ucsc.edu) was used for data visualization, only minus strand reads are shown. The track order is the same as in Fig. 2 with additional two wild-type replicates from a different sequencing experiment shown at the bottom. sRNA reads corresponding to exon 6 are consistently produced in all samples.
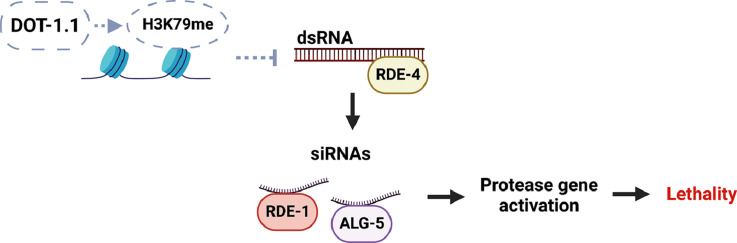
Overall, our analyses of mRNA and siRNA changes observed in dot-1.1(knu339) are consistent with our previously published genetic studies showing suppression of dot-1.1(knu339) lethality by ced-3, rde-4, and rde-1 loss-of-function mutants [2]. Now, we can summarize our findings in a model where the loss of dot-1.1 results in elevated production of RDE-4-dependent siRNAs, likely bound by RDE-1 and ALG-5, and causes elevated expression of protease genes, including ced-3, which leads to embryonic and larval death (Fig. 9). Given that genes expressed lower in dot-1.1(knu339) showed overrepresentation in ALG-4 targets and that our earlier findings detected elevated heterochromatin marks at ALG-3/4 target genes in dot-1.1(knu339) [6], we propose that RDE-4 dependent silencing siRNAs function with ALG-3/4 Argonautes.
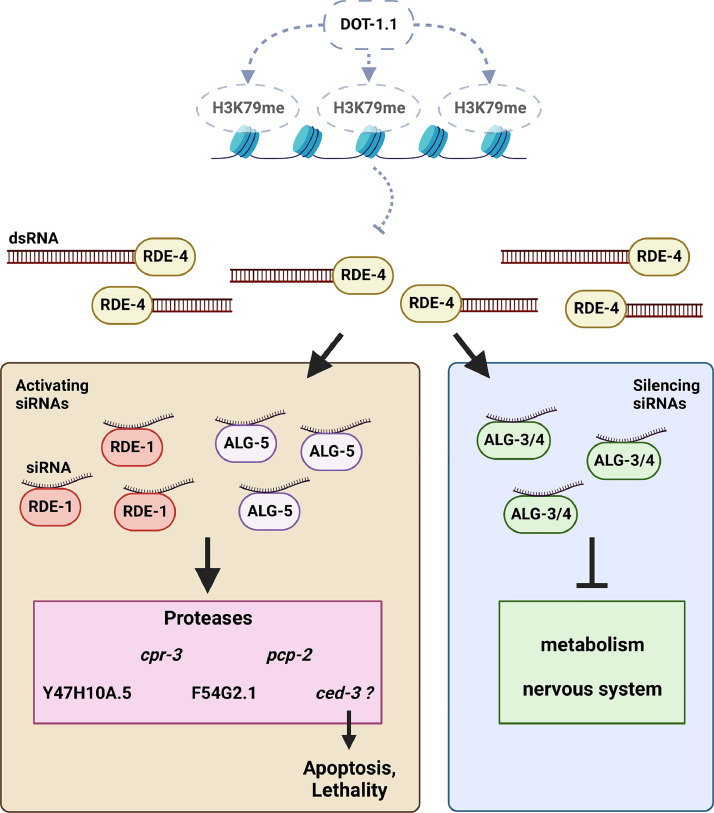
Fig. 9.
Genetic model of the relationship between DOT-1.1, RNAi pathway components, and siRNA target genes. In the absence of the dot-1.1 function (dotted gray lines), there is reduced H3K79 methylation (H3K79me), resulting in an increase in antisense transcription. The sense and antisense transcripts form dsRNA, which are recognized and bound by RDE-4. The dsRNA is processed by the Dicer complex, which includes RDE-4, and forms pools of 1) activating siRNAs bound by RDE-1 or ALG-5 which upregulate protease gene expression, resulting in apoptosis and lethality dependent on ced-3, as well as 2) silencing siRNAs bound by ALG-3 or ALG-4, resulting in inhibition of genes involved in metabolism and the nervous system.
Created with BioRender.com.
4. Discussion
Genomic analyses described here were motivated by our discovery that the dsRNA-initiated RNAi pathway is responsible for dot-1.1(knu339) lethality since it was suppressed by the rde-4 and rde-1 loss-of-function mutants [2]. In mammals, reduced H3K79 methylation upon downregulation of DOT1L leads to ectopic deposition of silenced chromatin marks, such as H3K9 and H3K27 methylation (reviewed in [4]). Both H3K9me and H3K27me can be induced by the dsRNA-dependent RNAi pathway in C. elegans [[53], [54], [55]]. Thus, in our earlier report, we tested the hypothesis that H3K79me depletion by dot-1.1(knu339) would result in global RNAi-dependent elevation in H3K9 methylation [6]. Although H3K9me was indeed globally elevated at the enhances occupied by DOT-1.1 in dot-1.1(knu339), this increase was not dependent on rde-1 or rde-4. However, we found an increase in H3K9me levels at the genes targeted by siRNAs dependent on ALG-3 and ALG-4 Argonautes [56] and some of them showed potential regulation by rde-1 and rde-4 [6]. Therefore, we concluded that dot-1.1(knu339) lethality was likely due to ectopic RNAi-dependent silencing of some specific genes rather than global perturbations in siRNA populations or chromatin landscape.
Here, consistently with earlier work, we found an enrichment of alg-4-dependent siRNA targets among the genes downregulated in dot-1.1(knu339). However, the connection between the genes upregulated in dot-1.1(knu339) and small RNA-mediated gene regulation was much stronger. First, we found a significant overlap between the upregulated genes and the upregulated siRNAs matching them. Second, we determined that the elevation of mRNA expression driven by dot-1.1(knu339) is not observed in rde-4(-). Third, rde-1-dependent siRNA targets were enriched among the genes upregulated in dot-1.1(knu339), and, finally, these upregulated genes were overrepresented among the group of genes with reduced expression in alg-5(-) [33]. Thus, unexpectedly, we uncovered positive regulation of gene expression by the dsRNA-dependent RNAi pathway in dot-1.1(knu339). We note that the dot-1.1(-)-upregulated genes (e.g. cpr-3, Y47H10A.5, pcp-2) did not show significantly reduced expression in rde-4(-) compared to the wild type. However, they were found downregulated by alg-5(-). ALG-5 is among the least studied C. elegans Argonautes. It is expressed in the germline [33,42] and has been implicated in the regulation of the developmental timing of germline development [33] and specifically in promoting entry into meiosis [57]. Our data suggest that ALG-5 may be involved in siRNA-dependent gene activation and function together with RDE-4 and RDE-1, at least in dot-1.1(knu339) background. Notably, ALG-5 belongs to the sub-family of C. elegans Argonautes which are closely related to the mammalian AGO proteins. There are reports of the gene-activating potential of AGO in mammals (reviewed in [58]). Therefore, it would be interesting to compare the mechanism of action of ALG-5 to those described for AGO in other systems.
The enrichment of proteases among the genes upregulated in dot-1.1(knu339), including rde-4-dependent elevation of cpr-3 (cysteine protease) and pcp-2 (prolyl carboxy peptidase), targeting of asp-6 (aspartyl protease) by rde-1-dependent siRNAs [42], and the requirement of several RNAi genes for proper ced-3 expression [50,51], strongly suggests that the overactive proteolysis background of dot-1.1(knu339) causes its RNAi-dependent lethality. Furthermore, we have been using a ced-3 mutant background for maintaining dot-1.1(knu339), and the newly found RNAi-dependent activation of ced-3 itself is the most likely direct cause of dot-1.1 mutant lethality.
What is the mechanism of elevated RDE-4-dependent siRNA production in dot-1.1(knu339)? Since RDE-4 binds dsRNA [25,26], and since nascent transcription, including antisense transcription, is elevated when DOT-1.1 recruitment to chromatin is compromised by zfp-1 reduction-of-function [1,2], it is logical to assume the answer is elevated dsRNA accumulation at these siRNA-producing loci. We observed primary siRNA production at loci with annotated transcription in both directions, such as tail-tail overlaps and transcription units embedded in large introns. In the case of the ced-3 gene activated by RNAi, the siRNAs are produced from the exon forming a hairpin. Our discovery of RNAi-dependent gene activation in dot-1.1(knu339) opens new directions for mechanistic studies of this process, which can occur at transcriptional and/or post-transcriptional levels.
Our experiments were performed using embryo preparations, the developmental stage when dot-1.1(knu339) mutant generally dies, and we now have molecular readouts of RNAi-dependent gene activation in the embryo, such as cpr-3, and F54G2.1. At the same time, we uncovered an overlap between genes upregulated in dot-1.1(knu339) embryos and downregulated in alg-5(-) adults [33]. Since ALG-5 is only expressed in the germline, there is a possibility that primary siRNAs leading to embryo gene activation are produced in the germline and inherited by the zygote. This scenario has recently been described for maternally produced primary siRNAs inducing zygotic silencing detected via a reporter [59].
Ultimately, activation of RDE-4-dependent RNAi in dot-1.1(knu339) is a result of transcription misregulation in this mutant background. In C. elegans, using Global Run-On (GRO) sequencing (GRO-seq), we found a negative feedback mechanism where the ZFP-1/DOT-1.1 complex negatively modulates transcription of highly expressed genes enriched for DOT-1.1 binding at the promoters [1]. This type of regulation likely occurs genome-wide, since H3K79me deposition follows transcription and therefore low levels of H3K79me are present at all transcriptionally active regions. This notion was corroborated by our subsequent analyses of the original GRO-seq data [2]. Importantly, in differentiated mammalian cells, DOT1L was shown to inhibit a transition from transcription initiation to elongation by RNA Polymerase II, and embryonic stem cells that naturally exclude DOT1L from the nucleus exhibit elevated nascent transcription [60]. Therefore, the elimination of DOT-1.1/DOT1L is expected to be associated with a more pluripotent cell state. Indeed, the dot-1.1 mutation was isolated from an unbiased screen for factors restricting the plasticity of epidermis cells in C. elegans [61].
Moreover, a comprehensive evaluation of Gene Expression Plasticity (GEP), a measure of the capacity of genes to change their expression in different conditions, through the analyses of numerous datasets from four organisms (human, mouse, Drosophila, and C. elegans) identified H3K79 methylation as a factor restricting GEP across these species [62]. It was determined that GEP was poorly correlated with the level or the broadness of gene expression, but genes high in GEP were enriched among immune response categories as well as disease susceptibility gene groups [62]. Considering our finding of the activation of the RDE-4-dependent RNAi pathway upon H3K79me loss, we propose that RNAi activation correlates with GEP increase. The heritability of siRNA changes in C. elegans has recently been evaluated in experimental evolution experiments; it lasted for 2–3 generations [63]. It would be interesting to test the possibility that in the dot-1.1; ced-3 mutant background increased GEP would allow for both a higher frequency of siRNA epimutations and their longer persistence.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM135199. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Gian Paolo Sepulveda for providing valuable discussion during investigation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2023.100080.
Appendix. Supplementary materials
Data availability
Raw sequencing data have been submitted to the NCBI GEO database with accession number GSE223865
References
- 1.Cecere G., Hoersch S., Jensen M.B., Dixit S., Grishok A. The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol. Cell. 2013:50. doi: 10.1016/j.molcel.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esse R., Gushchanskaia E.S., Lord A., Grishok A. DOT1L complex suppresses transcription from enhancer elements and ectopic RNAi in caenorhabditis elegans. RNA. 2019:25. doi: 10.1261/rna.070292.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G.A., Kadam S., Zhai H., Valdez R., et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLos Genet. 2008;4 doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wille C.K., Sridharan R. Connecting the DOTs on cell identity. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.906713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godfrey L., Crump N.T., Thorne R., Lau I.J., Repapi E., Dimou D., Smith A.L., Harman J.R., Telenius J.M., Oudelaar A.M., et al. DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nat. Commun. 2019:10. doi: 10.1038/s41467-019-10844-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esse R., Grishok A. Caenorhabditis elegans deficient in DOT-1.1 exhibit increases in h3k9me2 at enhancer and certain RNAi-regulated regions. Cells. 2020:9. doi: 10.3390/cells9081846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Y., Yang Y., Ortega M.M., Copeland J.N., Zhang M., Jacob J.B., Fields T.A., Vivian J.L., Fields P.E. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogoh H., Yamagata K., Nakao T., Sandell L.L., Yamamoto A., Yamashita A., Tanga N., Suzuki M., Abe T., Kitabayashi I., et al. Mllt10 knockout mouse model reveals critical role of Af10-dependent H3K79 methylation in midfacial development. Sci. Rep. 2017;7:11922. doi: 10.1038/s41598-017-11745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutter P.A., Karki S., Crawley I., Singh V., Bernt K.M., Rowe D.W., Crocker S.J., Bayarsaihan D., Guzzo R.M. Mesenchyme-specific loss of Dot1L histone methyltransferase leads to skeletal dysplasia phenotype in mice. Bone. 2021;142 doi: 10.1016/j.bone.2020.115677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franz H., Villarreal A., Heidrich S., Videm P., Kilpert F., Mestres I., Calegari F., Backofen R., Manke T., Vogel T. DOT1L promotes progenitor proliferation and primes neuronal layer identity in the developing cerebral cortex. Nucleic. Acids. Res. 2019;47:168–183. doi: 10.1093/nar/gky953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrari F., Arrigoni L., Franz H., Izzo A., Butenko L., Trompouki E., Vogel T., Manke T. DOT1L-mediated murine neuronal differentiation associates with H3K79me2 accumulation and preserves SOX2-enhancer accessibility. Nat. Commun. 2020;11:5200. doi: 10.1038/s41467-020-19001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen A.T., Xiao B., Neppl R.L., Kallin E.M., Li J., Chen T., Wang D.-.Z., Xiao X., Zhang Y. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev. 2011;25:263–274. doi: 10.1101/gad.2018511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi D., Nguyen H.P., Dinh J., Viscarra J.A., Xie Y., Lin F., Zhu M., Dempersmier J.M., Wang Y., Sul H.S. Dot1L interacts with Zc3h10 to activate Ucp1 and other thermogenic genes. Elife. 2020;9:1–48. doi: 10.7554/eLife.59990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwesi-Maliepaard E.M., Aslam M.A., Alemdehy M.F., van den Brand T., McLean C., Vlaming H., van Welsem T., Korthout T., Lancini C., Hendriks S., et al. The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:20706–20716. doi: 10.1073/pnas.1920372117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aslam M.A., Alemdehy M.F., Kwesi-Maliepaard E.M., Muhaimin F.I., Caganova M., Pardieck I.N., van den Brand T., van Welsem T., de Rink I., Song J.-.Y., et al. Histone methyltransferase DOT1L controls state-specific identity during B cell differentiation. EMBO Rep. 2021;22:e51184. doi: 10.15252/embr.202051184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kealy L., di Pietro A., Hailes L., Scheer S., Dalit L., Groom J.R., Zaph C., Good-Jacobson K.L. The histone methyltransferase DOT1L is essential for humoral immune responses. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108504. [DOI] [PubMed] [Google Scholar]
- 17.Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B., et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold O., Barbosa K., Deshpande A.J., Zhu N. The role of DOT1L in normal and malignant hematopoiesis. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.917125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrova E., Salvati A., Pecoraro G., Lamberti J., Melone V., Sellitto A., Rizzo F., Giurato G., Tarallo R., Nassa G., et al. Histone methyltransferase DOT1L as a promising epigenetic target for treatment of solid tumors. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.864612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregoire J.-.M., Fleury L., Salazar-Cardozo C., Alby F., Masson V., Arimondo P.B., Ausseil F. Identification of epigenetic factors regulating the mesenchyme to epithelium transition by RNA interference screening in breast cancer cells. BMC Cancer. 2016;16:700. doi: 10.1186/s12885-016-2683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurani H., Razavipour S.F., Harikumar K.B., Dunworth M., Ewald A.J., Nasir A., Pearson G., van Booven D., Zhou Z., Azzam D., et al. DOT1L is a novel cancer stem cell target for triple-negative breast cancer. Clin. Cancer Res. 2022;28:1948–1965. doi: 10.1158/1078-0432.CCR-21-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcos-Villar L., Díaz-Colunga J., Sandoval J., Zamarreño N., Landeras-Bueno S., Esteller M., Falcón A., Nieto A. Epigenetic control of influenza virus: role of H3K79 methylation in interferon-induced antiviral response. Sci. Rep. 2018;8:1230. doi: 10.1038/s41598-018-19370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui M., Kim E.B., Han M. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. Elegans. PLos Genet. 2006;2:719–732. doi: 10.1371/journal.pgen.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The Rde-1 gene, RNA interference, and transposon silencing in C. Elegans. Cell. 1999:99. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 25.Parker G.S., Eckert D.M., Bass B.L. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of DsRNA to SiRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabara H., Yigit E., Siomi H., Mello C.C. The DsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 27.Parrish S., Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 28.Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.-C.G., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C. Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. CellCell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 29.Grishok A. Biology and mechanisms of short RNAs in caenorhabditis elegans. Adv. Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- 30.Félix M.-.A., Ashe A., Piffaretti J., Wu G., Nuez I., Bélicard T., Jiang Y., Zhao G., Franz C.J., Goldstein L.D., et al. Natural and experimental infection of caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen G.M. Caspases: the executioners of apoptosis. Biochem. J. 1997;326(Pt 1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J., Shaham S., Ledoux S., Ellis H.M., Horvitz H.R., The C. Elegans cell death gene Ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 33.Brown K.C., Svendsen J.M., Tucci R.M., Montgomery B.E., Montgomery T.A. ALG-5 is a MiRNA-associated argonaute required for proper developmental timing in the caenorhabditis elegans germline. Nucleic. Acids. Res. 2017;45:9093–9107. doi: 10.1093/nar/gkx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson M.D., McCarthy D.J., Smyth G.K. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen M., Shen Y., Shi S., Tang T.MiREvo. An integrative MicroRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics. 2012;13:140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel MicroRNA genes in seven animal clades. Nucleic. Acids. Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaidatzis D., Lerch A., Hahne F., Stadler M.B.QuasR. Quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seroussi, U.; Lugowski, A.; Wadi, L.; Lao, R.X.; Willis, A.R.; Zhao, W.; Sundby, A.E.; Charlesworth, A.G.; Reinke, A.W.; Claycomb, J.M. A comprehensive survey of C. elegans argonaute proteins reveals organism-wide gene regulatory networks and functions. bioRxiv 2022, 10.1101/2022.08.08.502013. [DOI] [PMC free article] [PubMed]
- 43.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu G., Wang L.-.G., Han Y., He Q.-.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han T., Manoharan A.P., Harkins T.T., Bouffard P., Fitzpatrick C., Chu D.S., Thierry-Mieg D., Thierry-Mieg J., Kim J.K. 26G Endo-SiRNAs regulate spermatogenic and zygotic gene expression in caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corrêa R.L., Steiner F.A., Berezikov E., Ketting R.F. MicroRNA-directed SiRNA biogenesis in caenorhabditis elegans. PLos Genet. 2010;6 doi: 10.1371/journal.pgen.1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerstein M.B., Lu Z.J., van Nostrand E.L., Cheng C., Arshinoff B.I., Liu T., Yip K.Y., Robilotto R., Rechtsteiner A., Ikegami K., et al. Integrative analysis of the caenorhabditis elegans genome by the ModENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabuse Y., Nabetani T., Tsugita A. Proteomic analysis of protein expression profiles during caenorhabditis elegans development using two-dimensional difference gel electrophoresis. Proteomics. 2005;5:2876–2891. doi: 10.1002/pmic.200401154. [DOI] [PubMed] [Google Scholar]
- 49.Rao W., Isaac R.E., Keen J.N. An analysis of the caenorhabditis elegans lipid raft proteome using GeLC-MS/MS. J. Proteomics. 2011;74:242–253. doi: 10.1016/j.jprot.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Lee R.C., Hammell C.M., Ambros V. Interacting endogenous and exogenous RNAi pathways in caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welker N.C., Habig J.W., Bass B.L. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007:13. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic. Acids. Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu S.G., Pak J., Guang S., Maniar J.M., Kennedy S., Fire A. Amplification of SiRNA in caenorhabditis elegans generates a transgenerational sequence-targeted Histone H3 Lysine 9 methylation footprint. Nat. Genet. 2012:44. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao H., Zhu C., Zong D., Weng C., Yang X., Huang H., Liu D., Feng X., Guang S. The nrde pathway mediates small-RNA-directed histone H3 Lysine 27 trimethylation in caenorhabditis elegans. Curr. Biol. 2015;25:2398–2403. doi: 10.1016/j.cub.2015.07.051. [DOI] [PubMed] [Google Scholar]
- 55.Burkhart K.B., Guang S., Buckley B.A., Wong L., Bochner A.F., Kennedy S. A Pre-MRNA-associating factor links endogenous SiRNAs to chromatin regulation. PLos Genet. 2011;7 doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conine C.C., Batista P.J., Gu W., Claycomb J.M., Chaves D.A., Shirayama M., Mello C.C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner J.L., Jyo E.M., Mohammad A., Fox P., Jones V., Mardis E., Schedl T., Maine E.M. TRIM-NHL protein, NHL-2, modulates cell fate choices in the C. elegans germ line. Dev. Biol. 2022;491:43–55. doi: 10.1016/j.ydbio.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalantari R., Chiang C.-.M., Corey D.R. Regulation of mammalian transcription and splicing by nuclear RNAi. Nucleic. Acids. Res. 2016;44:524–537. doi: 10.1093/nar/gkv1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almeida M.V., de Jesus Domingues A.M., Ketting R.F. Maternal and zygotic gene regulatory effects of endogenous RNAi pathways. PLos Genet. 2019;15 doi: 10.1371/journal.pgen.1007784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wille C.K., Zhang X., Haws S.A., Denu J.M., Sridharan R. Antagonistic H3K79me-H3K9ac crosstalk determines elongation at housekeeping genes to promote pluripotency. bioRxiv. 2022 doi: 10.1101/2022.09.26.509534. [DOI] [Google Scholar]
- 61.Rahe D.P., Hobert O. Restriction of cellular plasticity of differentiated cells mediated by chromatin modifiers, transcription factors and protein kinases. G3: genes, genomes. Genetics. 2019;9:2287–2302. doi: 10.1534/g3.119.400328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao L., Zhao Z., He F., Du Z. Multivariable regulation of gene expression plasticity in metazoans. Open Biol. 2019;9 doi: 10.1098/rsob.190150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beltran T., Shahrezaei V., Katju V., Sarkies P. Epimutations driven by small RNAs arise frequently but most have limited duration in caenorhabditis elegans. Nat. Ecol. Evol. 2020;4:1539–1548. doi: 10.1038/s41559-020-01293-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data have been submitted to the NCBI GEO database with accession number GSE223865