Highlights
-
•
Proteomic profiling of urinary exosomes distinguishes between rapidly and slowly progressing ADPKD.
-
•
Proteomic profiling of urinary exosomes distinguishes between good and poor responders to Tolvaptan therapy.
-
•
ADPKD exosomes show stage-dependent changes in donor cell secretion, recipient cell uptake and endosomal vesicular trafficking.
-
•
Urinary exosome profiling offers potential for personalised ADPKD patient management.
Key words: ADPKD progression, Urinary biomarkers, Exosomes, Proteomics, Tolvaptan effects, Vesicular trafficking
Abstract
ADPKD is the most common genetic disease of the kidney leading to end-stage renal disease necessitating renal replacement therapy at any time between the 1st and 8th decades of life due to widely variable rates of disease progression. This presents significant patient anxiety and a significant prognostic and therapeutic challenge. Tolvaptan is the only approved drug licensed to slow ADPKD progression by reducing renal cystic expansion but side-effects can limit its efficacy.
To address the need to identify new biomarkers to monitor progression of ADPKD and to evaluate the therapeutic effects of Tolvaptan, proteomic analysis was conducted on defined (40-100nm) urinary exosomes isolated from ADPKD patients phenotyped and clinically monitored over a 10-year period. Comparative Gene Ontology analysis of Tandem Mass Tag labelled mass spectrometry-derived protein profiles from urinary exosomes from ADPKD patients with rapid (>10ml/min/5 years decline in estimated glomerular filtration rate) versus slow progression showed distinctive patterns of pathway up-regulation. Clear discrimination between rapid and slowly-progressive profiles were seen in all stages functional decline in ADPKD patients whether with mild (>70ml/min), moderate (50-69ml/min) or severe (<49ml/min) disease at onset. Discriminatory pathways and proteins included Notch-, integrin- and growth factor-signalling; microtubular kinase, vesicular proteins and epidermal growth factor substrates.
Confocal microscopy of fluorescently-labelled normal versus ADPKD epithelial cell-derived exosomes in vitro also identified ADPKD-dependent abnormalities in intracellular vesicular trafficking and implicated changes in ADPKD-dependent exosome secretion and target cell uptake as factors underlying urinary exosome excretion biomarker properties.
Comparative proteomic analysis of urinary exosomal proteins in individual patients before and after treatment with Tolvaptan for 4 years also identified distinct patterns of pathway modification dependent on the degree of effectiveness of the therapeutic response. Up-regulation of Wnt-pathway and vesicular proteins were characteristic of urinary exosomes from ADPKD patients with good responses to Tolvaptan while upregulation of angiogenesis pathways and additional molecular forms of vasopressin receptor AVPR2 were characteristic in urinary exosomes of ADPKD patients with poor responses.
Taken together, these studies conclude that proteomic profiling of urinary exosome biomarkers provides a specific, sensitive and practical non-invasive method to identify and monitor the rate of disease progression and the effects of Tolvaptan therapy in individual ADPKD patients. This provides a means to identify those patients most likely to benefit maximally from therapy and to progress towards a personalization of ADPKD prognosis and management.
Graphical abstract
Introduction
Autosomal Dominant Polycystic Kidney Disease (ADPKD) is a common mono-genetic kidney disease leading to renal failure caused by germline mutation in PKD1 in ~85% or PKD2 in ~15% patients with an incidence of ~1:600 -1:1,000 live births [1,2]. ADPKD affects an estimated ~12 million individuals worldwideand accounts for ~10% of the dialysis population[3] presenting a significant burden for patients and healthcare systems. Advances in genotyping and clinical phenotyping typically result in early diagnosis of ADPKD and longitudinal management of symptom development associated with bilateral increases in renal size and progressive loss of renal function. ADPKD culminates in end-stage renal disease (ESRD) at an average of ~53 years in ADPKD patients with PKD1 mutations [1,4,5]. However, there is extreme variability in the rate of disease progression and onset of ESRD varies from 1st to 8th decade [1,6,7]. This presents an urgent need to develop specific, reproducible, longitudinally applicable and universally accessible methods to monitor and predict individual rates of progression and thereby likely age of onset of renal replacement therapy in ADPKD patients. A urinary biomarker assay would fulfil these requirements and provide the additional advantages of being non-invasive and practical. However, due to wide variability in contents of whole urine, a more specific and reproducible approach is essential to provide the basis for assessing the precise status and susceptibility of ADPKD patients to rapid disease progression and of the effectiveness of drug therapy. At present, the vasopressin receptor antagonist, Tolvaptan is the only approved drug therapy aimed at slowing ADPKD disease progression, although others are in development [8]. A universal urinary exosome-specific profiling would not only provide an important monitoring method but has the potential to increase understanding of the underlying biology of ADPKD progression.
ADPKD is characterized by bilateral progressive enlargement of multiple renal tubule-derived epithelial cysts, concomitant loss of functioning nephrons, excessive cyst-lining epithelial cell proliferation and reversed polarity of fluid secretion [1,7,9,10]. Underlying cell biological alterations have been identified in epidermal growth factor (EGF) receptor and cAMP-mediated mitogenic signaling, ion and fluid transporters, and in adhesive cell-cell and cell-matrix interactions [1,7,11,12]. Interstitial fibrosis, ischemia, PKD1 truncation mutations, modifier genes and epigenetic factors have been proposed as progression-promoting candidates in ADPKD [1,8,[13], [14], [15]].
The vasopressin receptor-2 (AVPR-2)-antagonist, Tolvaptan (Otsuka) which targets cAMP/protein kinase A (PKA) pathways is currently the only drug to slow ADPKD progression approved for use in the UK and Europe (since 2015) and USA (since 2018)[16], [17], [18]. Although it has been shown to slow rates of decline in estimated glomerular filtration rate (eGFR) and increases in total kidney volume (TKV) variability in degrees of efficacy as well as adverse side-effects of polyuria and liver toxicity can be limiting [16,18].
Non-invasive methods to predict the risk of rapid progression and efficacy of drug therapies would be highly beneficial for ADPKD patients. Current approaches include sophisticated Mayo imaging classification using MRI-measurements of height-adjusted (h)TKV and detailed genomic analysis combined with clinical phenotyping [19,20]. A urinary biomarker test would provide a simpler, minimally invasive, accessible and globally applicable approach. Urinary and blood biomarkers are increasingly being used to aid diagnosis, prognosis, and therapeutic monitoring of disease. In the kidney, changes in kidney injury molecule-1 (KIM-1), neutrophil gelatinase-associated lipocalin (NGAL), Dickkopf WNT signaling pathway Inhibitor-3 (DKK3), liver-type fatty acid binding protein (L-FABP), tissue inhibitor of metalloproteinases-2 (TIMP-2), insulin growth factor binding protein-7 (IGFBP-7), monocyte chemoattractant protein-1 (MCP-1), matrix metalloproteinase-1 (MMP-1) and Cystatin C have been detected in acute kidney injury (AKI), chronic kidney disease (CKD) and diabetes [21], [22], [23], [24]. However, lack of disease-specificity and severity-sensitivity limits their utility. In ADPKD, clear-cut disease-specific profiles of urinary proteins and miRNA biomarkers have been identified and in both ADPKD and ARPKD reflect increases in cell proliferation and matrix remodeling [25], [26], [27], [28].
Membrane-bound extracellular vesicles (ECVs) in urine and blood range in size from 40nm to 1,000nm and 2 subsets have been defined by size and mode of biogenesis. Exosomes are small (40-100nm) nanoparticles generated by budding invagination of intracellular endosomes into multi-vesicular bodies that are targeted for secretion. By contrast, the larger (125-1,000nm) micro-vesicles (MVs) that are derived by direct exocytic budding of cellular plasma membranes [29,30]. Exosomes are important effectors of cell-cell communication, mediated by transfer of proteins and RNA to specific recipient cells to modify cell function. In the kidney, information transfer can occur locally within a single nephron segment to more distant downstream distal segments [31,32]. Endocytic uptake of exosomes is facilitated by recipient cell clathrin- or dynamin-containing membrane invaginations and protein interactions mediated by exosomal integrin- and tetraspanin-receptors [33,34]. Increases in urinary exosome excretion have been reported in many proliferative disease states including cancer [35], [36], [37], [38]and are associated with reprogramming of differentiation in renal development [39]. Exosomes have also been shown to play important roles in adhesion, matrix modulation and angiogenesis [40].
Since abnormal cell-cell interactions are of central mechanistic importance underlying cystic expansion in ADPKD[1], the current studies were designed to determine whether urinary exosome protein composition could provide a specific indicator profile of ADPKD progression. Specifically, the urinary exosomal proteome was analyzed in detail to evaluate its potential to discriminate between ADPKD patients with rapid compared to slow rates of disease progression at different initial stages of disease severity. Parallel in vitro studies of cell-derived exosomes were designed to gain mechanistic insight into ADPKD stage-dependent changes in recipient cell interactions. The effects of progression and degree of efficacy of Tolvaptan therapy were also evaluated in long-term Tolvaptan-treated ADPKD patients to evaluate the further potential of urinary exosomal proteomics to identify those patients most at risk of rapid progression and/or poor therapeutic response.
Methods
Clinical samples
Urine samples have been routinely collected as fresh voids from consented diagnosed PKD1-ADPKD patients (Table 1) at 6- to 12-monthly follow-up specialist clinic visits at Royal Free Hospital NHS Foundation Trust since 2011 according to RaDaR guidelines and ethical approval 20772. Informed consent was obtained for experimentation with human subjects. All urine samples were kept on ice and processed within 3 hours of collection. On receipt of samples SigmaFast 10X protease inhibitor cocktail (AEBSF 0.2mM, Aprotinin 0.03µM, Bestatin 0.13µM, E-64 1.4µM, EDTA 0.1mM, Leupeptin 0.1µM, Sigma, Haverhill) was added (final concentration 1X), samples incubated for 10 minutes at room temperature and then centrifuged at 300xg for 15 minutes at 4°C to pellet cell debris. Multiple 5ml aliquots were flash frozen in liquid nitrogen prior to transfer and storage at -80°C in the PKD-Charity-sponsored BioResource Bank. Longitudinal ADPKD sample collections and linked clinical observational, eGFR and renal imaging data recorded in the UCL/Royal Free PKD database (Vital Data) allowed stratification of >250 patients by severity stage (NICE: CKD classification) CKD-1 (20% of patients); CKD-2 (25%); CKD-3 (40%); CKD-4 (15%) as well as by rates of disease progression (loss of eGFR ml/min/year). Some patients have received Tolvaptan therapy for >5 years (Otsuka, TEMPO 3/4, 4/4 and Reprise Trials and NHS England commissioned standard of care). Eleven flash frozen cyst fluid samples collected immediately after nephrectomy according to Institutional Review Board and NIH-approved consented protocols, archived in the PKD Charity-sponsored BioResource Bank were used: 2 from kidneys with simple cysts (non-PKD-related); 3 cysts from 3 patients with early-stage ADPKD-CKD2 and 6 cysts from 6 patients with late-stage ADPKD-CKD4.
Table 1.
Patient data including age, gender, genetics and progression.
| Patient Group | Number | Age (Mean +/- SEM) | Gender | Genetics | Progressionml/min/yr |
|---|---|---|---|---|---|
| Normal | 8 | 42 +/-3 | 4M, 4F | N/A | - |
| CKD ½ | 9 | 41 +/-4 | 4M, 5F | PKD1 | * |
| CKD ¾ | 9 | 55 +/-3 | 5M, 4F | PKD1 | * |
| RapidlyProgressing | 30 | 45 +/-2 | 15M, 15F | PKD1 ter, del,dup, subst,missense | > 2-6 |
| Slowly Progressing | 30 | 52 +/-3 | 15M, 15F | PKD1 ter, dupsubst | < 2 |
| TolvaptanGood | 1 | 59 | F | PKD1 del | < 5 |
| Tolvaptan Good | 1 | 39 | F | PKD1 subst | < 5 |
| TolvaptanPoor | 1 | 54 | F | PKD1 subst | > 5 |
| TolvaptanPoor | 1 | 34 | M | PKD1 subst | > 5 |
| TolvaptanUncertain | 1 | 61 | F | PKD1 del | > 5 |
| TolvaptanUncertain | 1 | 46 | M | PKD1 subst | < 5 |
* 5 patients with rapid progression (>2ml/mi/yr); 4 patients with slow progression.
ter: termination; del: deletion; subst: substitution; dup: duplication.
Urinary exosome isolation and purification
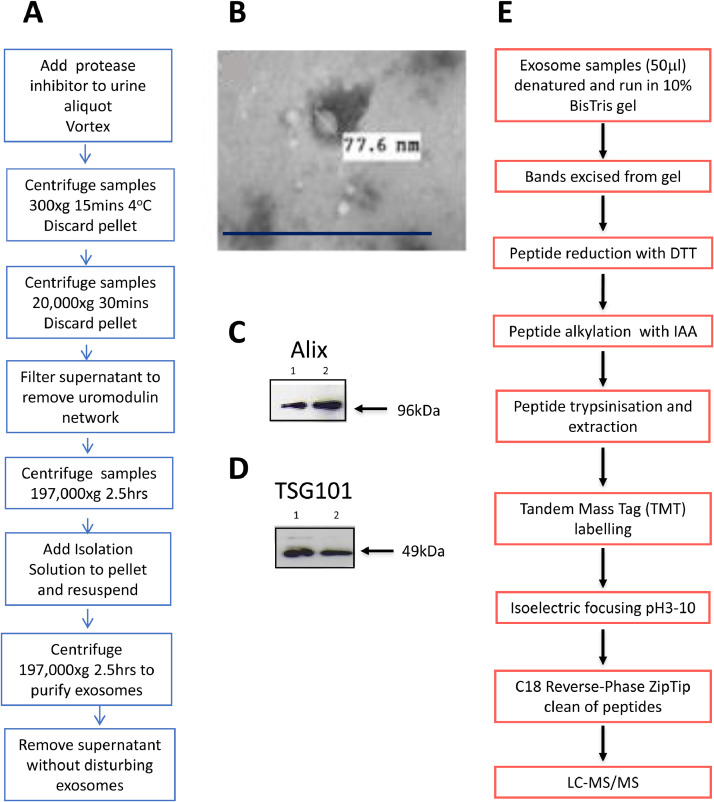
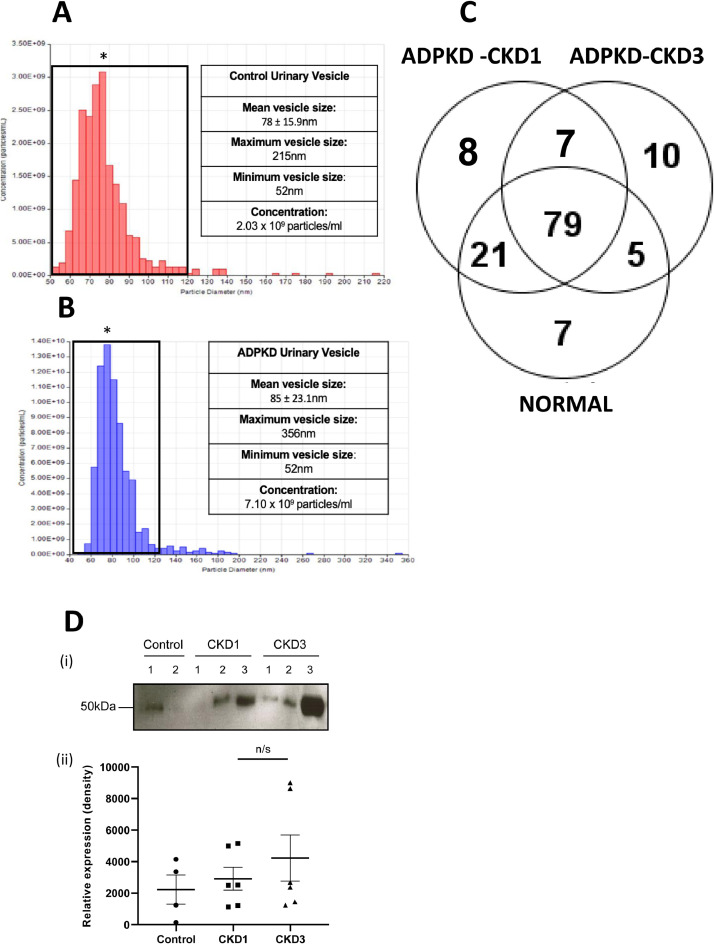
Urinary exosomes were isolated from 74 age-, gender-, stage- and therapy-matched ADPKD patients with PKD1 mutations (34 to 70 years; 50% male, 50% female, CKD stages 1 to 4) in groups with rapidly progressive (≥2-6 ml/min/year) and slowly progressive (<2 ml/min/year) disease; before and after treatment with Tolvaptan as well as from normal subjects. The choice of >2ml ml/min/year as progression rate cut-off was designed to include detection of potential changes in outcome early in the disease process. Stored urine, cyst fluid and conditioned media samples were thawed at room temperature and vortexed for 30 seconds every 2 minutes until completely defrosted. Exosomes were isolated in 10mM Triethanolamine / 250mM sucrose using an optimized differential centrifugation and filtration protocol (Fig. 1). Purity was assessed by size and marker analysis using transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA, NanoSight LM10, Malvern); tunable resistive pulse sensing (TRPS) analysis and Western immunoblotting of the exosomal component proteins Alix and TSG101 (Figs. 1 and 2). Protein concentrations were determined using the Bicinchoninic (BCA) assay (Pierce, ThermoFisher, Dartford) and creatinine measured at 520nm after addition of picric acid (Beckman Coulter, High Wycombe).
Fig. 1.
Isolation and characterization of urinary exosomes A. Optimized protocol for preparation of urinary exosome from 5ml urine samples: isolation buffer 10mM triethanolamine / 250mM sucrose [64]. B. Transmission electron microscopy of urinary exosomes isolated from normal urine showing characteristic size and shape. Exosomes were fixed in 2%paraformaldehyde/2.5% glutaraldehyde in PBS for 16hours. Drops of samples were pipetted onto formvar-coated copper grids, negatively stained with uranyl acetate and viewed under a JEOL 1200 electron microscope. White insert indicates exosome diameter. Scale bar 0.5μm. C and D. Representative Western immunoblots of exosomal marker proteins in urinary exosomes from normal subjects: C. Alix (96kDa), D. TSG101 (49kDa) (n=2 subjects). E. Workflow for preparation of exosome proteins for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. DTT: dithiothreitol, IAA: iodoacetamide.
Fig. 2.
Comparison of urinary exosomes isolated from ADPKD patients and age-matched normal subjects. Tunable resistive pulse sizing (TRPS) analysis of size distribution and concentration of urinary vesicles isolated from A. normal subjects. B. ADPKD patients. * denotes mean vesicle size in each group (NTA/TRPS n=2, 3 technical repeats, p<0.05). C. Venn diagram showing the number of common, shared and unique proteins in urinary exosomes isolated from normal subjects (n=6). ADPKD patients at CKD1 (n=3) and ADPKD patients at CKD3 (n=3). Proteins were included in each group according to spectral count criteria >2 in 4 out of 6 normal samples and 2 out of 3 of 3 ADPKD samples D. Fetuin-A (AHSG, 50kD) expression in in urinary exosome isolates from normal subjects (n=2), ADPKD-CKD1 (n=3) and ADPKD-CKD3 (n=3) patients: (i) Representative Western immunoblot of n=2 repeat experiments. Exosome proteins were normalized to urine creatinine. (ii) densitometric analysis of Western blots showing means +/- SEM, n=2 experiments. n/s: no significant difference.
Liquid chromatography dual Mass spectrometry (LC-MS/MS)
Tandem Mass Tag (TMT) labelled mass spectrometry was carried out on 50μl urinary exosome samples loaded onto 10% BisTris gels (NuPage, ThermoFisher), and excised bands subjected to peptide reduction by 10mM dithiothreitol (Sigma) and alkylation by 55mM iodoacetic acid, prior to extraction by trypsinization. After TMT labelling high resolution isoelectric focusing was carried out (Agilent 3100, Cheadle) prior to Zip-tip clean-up, chromatographic separation (EASY NanoLC 1200) and tandem Mass Spectrometry (LTQ-Orbitrap Mass Spectrometer, ThermoFisher) at the King’s College London Proteomics Facility.
Proteomic data analysis
MS data were processed using Proteome Discoverer (ThermoFisher) against the Uniprot human database and peptides were identified using the Mascot database. Each of the 12 pooled fraction raw data files were processed together as one TMT10plex experiment and searched as a ‘Mudpit’ using the Mascot search algorithm. Peptides were identified by matching with unique peptides for each protein using the Mascot database with carbamidomethylation (C) as the fixed modification and methionine oxidation as the variable modification with dynamic modifications of TMT10plex (K), TMT10plex (N-terminal). Filters were applied to the data for protein identification for a minimum of 3 peptides and an identification threshold of 95% probability of the confidence interval. To extract and quantify the relative amounts of proteins in the urinary exosome samples, Proteome Discoverer (v1.4) was used to extract the TMT reporter ions for every labelled peptide. To perform a comparison between groups, every peptide needed to have been identified in the database search, had a reporter ion value and a TMT database assigned label. Peptides that were missing any of these parameters were removed prior to quantitative data processing. For protein quantitation, reporter ion intensities of all peptides assigned to a specific protein were summed to give a protein value and compared between samples. In addition, Gene Ontology (GO) pathway analysis was carried out using the Panther Classification system (http://www.pantherdb.org). p-values were calculated using a 2-tailed, equal sample variance t-test; p<0.05 was considered significant. Adjusted p-values were calculated using Bonferroni correction for multiple testing.
Proteomic data normalization
For samples run on two separate occasions (rapid vs slow samples), a total sum scaling method using the reporter ion values was applied across all samples. Briefly, a sum of all the reporter ion values for the entire column of each reporter ion was identified, giving 20 reporter ion values, one value per sample. A median value of the 20 summed reporter ion values was determined and the median value was then divided by the sum value of the reporter ions to give the correction factor for the specific reporter ion. Then each peptide reporter ion value was multiplied by the correction value for the specific reporter ion column. To normalize data to account for potential differences in protein concentration, creatinine values for the samples were used. Sample creatinine absorbance values were divided by the creatinine absorbance value of the lowest sample to obtain a correction factor. The correction factor was applied to each sample and protein values were used to calculate fold-change between rapid and slow samples.
Western Immunoblotting
Exosome samples were solubilized and denatured at 95°C in Laemmli buffer (Biorad, Watford), separated by SDS-polyacrylamide electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Protran 0.45μm pore size, GE Healthcare, Amersham) or polyvinylidene fluoride (PVDF, Immobilon-P 0.45μm, Millipore, Watford). After blocking for 2 hours at room temperature in 50mM Tris-buffered saline containing 0.1% Tween-20 (TBST) and 5% dried milk, membranes were incubated overnight at 4°C with one of the following primary antibodies diluted in blocking solution: anti-Alix (1:500; Millipore), anti-TSG101 (1:500; Abcam, Cambridge), anti-AVPR2 (V2R, 1:1000; Sigma), anti-Dynactin (1:1000; Millipore), anti-vesicular integral membrane protein (VIP)-36 (1:500; Abcam), anti-heat shock proteins (HSP)-90 (1:1000; Abcam), anti-sorting nexin (SNX)-18 (1:1000; GeneTex, Irvine, CA, USA) and anti-Fetuin-A (1:1000, Santa-Cruz, CA, USA) overnight. After washing in TBST and incubation for 1 hour at room temperature with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2000; GE Healthcare; ThermoFisher), protein bands were visualized on X-ray film (high performance chemiluminescence film, Amersham, Oxford) after incubation in chemiluminescence substrate (LumiGLO; Cell Signalling, London).
Immunohistochemistry of human kidney sections
Immunohistochemical analysis was carried out on 4% paraformaldehyde-fixed, paraffin-embedded human kidney sections from 10 age-matched normal, 10 ADPKD-CKD-2 and 10 ADPKD-CKD-4 nephrectomies (PKD Charity BioResource Bank). After graded ethanol de-paraffinization, 3 PBS washes and 5 minutes microwave antigen retrieval in sodium citrate buffer pH6.0, sections were subjected to serum-free protein block (Agilent/Dako) and then incubated for 45 minutes at room temperature in anti-human AVPR2 antibody (1:200 in PBS, Sigma). Avidin/biotin amplification of staining (Vector Laboratories, Peterborough) was carried out followed by incubation in diaminobenzidine as chromogen. Sections were mounted in water soluble mounting media and viewed under a Zeiss microscope using bright-field illumination.
Primary culture of human normal and ADPKD epithelia
Primary normal human collecting tubule cell (NHCT), early-stage (E) ADPKD-CKD-2 and late-stage ADPKD-CKD4 cyst-lining epithelial cells, obtained as cryogenic stocks from the Polycystic Kidney Disease (PKD) Charity BioResource Bank were plated on collagen-coated multi-well plates (Corning, High Wycombe) and grown to confluence in Click/RPMI medium (Sigma/Thermo-Fisher) supplemented with 5µg/ml human transferrin (Sigma), 1x penicillin/streptomycin (Sigma), 2mM glutamine (GlutaMax, ThermoFisher), 5 × 10−8M dexamethasone (Sigma) and 3% exosome-replete fetal bovine serum (FBS, Sera Lab International, Haywards Heath). PKD epithelial cell culture media were also supplemented with 5ug/ml insulin (Sigma) and 5 × 10−12M tri-iodothyronine (Sigma) for optimal growth [41,42].
Cellular exosomes secreted from confluent monolayers of NHCT, early stage ADPKD-CKD2 and late stage ADPKD-CKD4 epithelia were collected in serum-free conditioned media over a 48-hour period, quantified and fluorescently labelled by suspension and incubation for 4 minutes at room temperature, protected from light with the PKH26 lipophilic membrane dye (Sigma). The labelling reaction was stopped with 2ml 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS), samples were ultra-centrifuged at 100,000xg at 4°C for 70 minutes, the supernatant was removed, the pellet washed with PBS by ultracentrifugation and the final labelled exosome pellets resuspended in PBS.
Treatment of Normal Human Collecting Tubule (NHCT) and ADPKD cells with exosomes
5,000 cells/well were plated on collagen-coated 96-well plates (Corning) and cultured in cell type-specific medium containing 3% exosome-replete FBS until ~70% confluent. One day before treatment, cells were washed with PBS and cultured in serum-free medium for 24 hours. Exosomes (1 × 109 particles/ml or an equivalent volume of PBS vehicle were added to 100μl cell-specific, serum-free medium and used to treat cells for 6, 12 or 24 hours.
Confocal microscopy of labelled exosome uptake and trafficking in NHCT and ADPKD cells
Cells incubated with labelled exosomes were fixed for 20 minutes with 4% paraformaldehyde (TAAB, Aldermaston) in PBS, washed 3 timed with PBS, and were incubated for 30 minutes protected from light, at room temperature in PBS containing 100µl per well 4’6-diamindion-2-phenylindole (DAPI 1:100, Abcam) to stain nuclear DNA. They were then washed 3 times in PBS and incubated 3 times for 30 minutes protected from light at room temperature in 100µl Wheat Germ Agglutinin (WGA)- Alexa Fluor 488 (1:200 in HBBS, ThermoFisher). Cells were washed 3 times with Hanks’ balance salt solution (HBSS), mounted in FluoroSave mounting media (Calbiochem, Watford) topped by a coverslip for imaging on a fully-motorised Leica SP8 laser-scanning confocal microscope equipped with hybrid detectors, hardware-based autofocus and super-resolution lightning module was used. Microscope control and image acquisition was performed using Leica Application Suite X (LASX, version 3.5.2.18963). Scans were taken at 16-bit and then converted to 8-bit for analysis. ImagePro 10 advance 3D software (ImagePro 10.0.4 build 6912) was used for 3D rendering and 3D segmentation analysis.
Results
Proteomic profiling was carried out on patients from the UCL/Royal Free Hospital ADPKD specialist clinic divided into two separate age – and gender-matched cohorts based on (1) CKD classification (CKD 1 / 2 versus CKD 3 / 4); or (2) rate of disease progression (rapid versus slow) [see Table 1].
Urinary exosomes from ADPKD patients show differential severity stage-dependent characteristics
Urinary exosomes were isolated using an optimized robust and reproducible protocol adapted for small volume (5ml) samples and characterized by electron microscopy, Alix and TSG101 biomarker-content (Fig. 1). NTA/TRPS analyses showed that ADPKD patients excreted higher concentrations of urinary particles/ml (7.10 × 109) than normal subjects (2.03 × 109) and suggested that ADPKD nanoparticles were on average larger (85 ± 23.1nm) and distributed over a wider size-range (52-356nm) than those from normal individuals (size 78±15.9nm; range 52-215nm) (Figs. 2A and B). Using TRPS to exclude non-exosomal (MV) particles of >120nm, comparative proteomic analysis identified differences, not only between ADPKD and normal urinary exosome (40-100nm) protein expression but also between urinary exosomes from ADPKD at different stages of severity (ADPKD-CKD1 compared to ADPKD-CKD3 (Fig. 2C, Table 2). The numbers of >2-fold changed proteins increased with increasing disease severity (5 up- and 19 down-regulated in ADPKD-CKD1 compared to normal exosomes; 15 up- and 31 down-regulated in ADPKD-CKD3 compared to ADPKD-CKD1). Several proteins were up-regulated in both stages of ADPKD compared to normal urinary exosomes, including those of the coagulation and immunomodulatory pathways; while the degree of up-regulation of some proteins, including fetuin-A (α-2-HS-glycoprotein) correlated with increasing stages of disease severity (Fig. 2D). Predominant down-regulated proteins in ADPKD urinary exosomes included γ−glutamyl transpeptidase (GGT), aminopeptidase-N (AMP-N) and megalin consistent with a loss of differentiated membrane and brush border proteins.
Table 2.
Comparisons of numbers of >2-fold up- regulated and >2-fold down-regulated proteins expressed in urinary exosomes from normal subjects; ADPKD patients at different stages of severity CKD1 and CKD3; and ADPKD patients with rapid (>10ml/min decline in eGFR over 5 years) versus slow disease progression and with different levels of renal function at clinical presentation.
| Group comparisons | Numbers of proteins up-regulated >2-fold | Numbers of proteins Down-regulated > 2-fold |
|---|---|---|
| ADPKD-CKD1 v Normal | 5 | 19 |
| ADPKD-CKD1 v ADPKD-CKD3 | 15 | 31 |
| All rapid v slow progression | 59 | 26 |
| Initial eGFR >70ml/min | 83 | 4 |
| Initial eGFR 50-69ml/min | 126 | 13 |
| Initial eGFR <49ml/min | 93 | 61 |
Urinary exosomal proteomics can differentiate between ADPKD patients with rapid versus slow progression
Comparative proteomic expression profiling of urinary exosomes from all 30 ADPKD patients with rapidly or slowly progressing decline in eGFR (> or <10ml/min) over 5 years showed significant differences in numbers and categories of proteins with >2-fold changes in expression levels (Fig. 3A). Larger numbers of urinary exosomal proteins were >2-fold up-regulated (59) or down-regulated (26) in patients with rapid compared to slow progression (Table 2; Supplemental Table 1).GO pathway analysis identified up-regulation of expression proteins from many pathways associated with rapid progression including coagulation, cell division, cytoskeletal organization and matrix-adhesion (Fig. 3B). The most highly up-regulated proteins in the rapidly progressing group were microtubule-associated serine/threonine kinase (MAST)-4 (24x), cytokinesis-associated kinesin-like (KIF) 20B (13x), and dynein heavy chain (8x). The actin-binding proteins A-kinase-anchoring protein (AKAP)-13 and radixin, calcium-dependent annexins-1 and-2 and extracellular matrix proteins fibronectin and tenascin were also >2-fold up-regulated. Fewer pathways were >2-fold down-regulated in rapid progression (Fig. 3C).
Fig. 3.
Proteomic analysis of urinary exosomes isolated from ADPKD patients with rapidly progressive versus slowly-progressing disease ADPKD patients who had a decline in eGFR of >10ml/min over the 5 year period of analysis were designated as rapid progressors while those with relatively stable eGFR (< 10ml/min decline in eGFR) over the same 5 years were designated as slow progessors A. Volcano plot of fold-change in urinary exosomal proteins isolated from samples collected at presentation whose disease subsequently progressed rapidly versus slowly. Log2-fold change (FC) and -Log10 p-values for all proteins identified in exosomes from rapid and slow progressors. Dashed lines: cut-off for significance: Log2FC >1 and -Log10 p-value >1.2 are considered significant. Proteins with statistically significantly different levels of protein expression are shown in red. B. Panther pathway analysis showing up-regulation (proteins >2-fold up-regulated) in urinary exosomes from rapid progressors compared to slow progressors. C. Pathways showing down-regulation (proteins >2-fold down-regulated) in urinary exosomes from rapid compared to slowly progressing ADPKD. D. Key to colour-coding of pathways depicted in pie charts
Differential urinary exosome proteomic profiles from rapidly- and slowly-progressing ADPKD patients with different initial degrees of disease severity
To determine whether the stage of ADPKD disease severity at onset of influenced the capacity to discriminate between rapidly and slowly progressive disease, differential expression proteomics was carried out in 3 groups of ADPKD patients (10/group) stratified by eGFR for degree renal insufficiency. Comparisons between urinary exosomes from rapidly and slowly-progressive patients with mild (eGFRs >70ml/min), moderate (eGFR 50-69ml/min) and severe (eGFR<49ml/min) renal impairment showed clear stage-dependent protein profiles (Fig. 4). These eGFR groupings differed from the cutoffs in the CKD (G1-5) classification scheme to subdivide those patients in G2 and G3a ranges who present to specialist ADPKD tertiary care centres with relatively mild, moderate and severe levels of renal impairment. Volcano plots demonstrated the numbers of proteins with >2-fold significant change in expression in patients with rapid compared to slow progression increased with increasing renal impairment (Fig. 4A-C). At each stage of ADPKD disease severity, larger numbers of proteins were >2-fold up-regulated than were >2-fold down-regulated in rapid compared to slow progression: 83, 126, 93 versus 4, 13, 61, respectively (Table 2). Interestingly, urinary exosomes from rapidly progressing ADPKD patients with an initial eGFR of 50-69ml/min showed the highest numbers of up-regulated proteins (126) while those in the eGFR<49ml/min group showed the highest numbers of down-regulated proteins (61) (Table 2).
Fig. 4.
Proteomic analysis of urinary exosomes isolated from rapidly versus slowly progressing ADPKD in patients with different starting levels of disease severity (eGFR).A, D, G, J, eGFR at presentation of >70ml/min; B, E, H, K eGFR of 50-69ml/min; C, F, I, L eGFR of <49ml/min. A, B, C. Volcano plots of fold-change of proteins in rapid versus slow progressors. Log2 fold-change and -Log10 p-values of all proteins identified in rapid and slow progressors. Dashed lines: cut-off significance: >1 and >1.2, respectively. Proteins with statistically significant different levels of expression are shown in red. D, E, F. Pathway analysis using Panther software of >2-fold up-regulated proteins in urinary exosomes from rapid compared to slowly progressing ADPKD. Key to colour-coding of pathways as in Fig. 3D. G, H, I. Heatmaps of pathways that contained >2 up-regulated proteins showing differences in levels of expression between urinary exosomes from rapidly progressing (PG) compared to slowly-progressing (NPG) ADPKD patients. Key: yellow high expression; blue low expression. J, K, L. Validation of urinary exosome proteomics. (i) Representative Western immunoblots; (ii) densitometric analysis, mean+/-SEM * p<0.05, n/s not statistically significant. HSP90 (Ji and ii) was characteristic of urinary exosomes from patients with starting eGFR of >70ml/min; SNX18 (Ki and ii) was characteristic of urinary exosomes from patients with starting eGFR 50-69ml/min; and VIP36 (Li and ii) was characteristic of urinary exosomes from patients with starting eGFR <49ml/ min.
GO pathway analysis demonstrated stage-dependence of differential up-regulation of urinary exosomal proteins associated with rapid progression (Fig. 4D to F). Notch- and integrin-mediated pathway up-regulation were characteristic of mild impairment (eGFR>70ml/min); apoptosis pathways were characteristic of moderate impairment (eGFR 50-69ml/min); while cell migration and EGFR signaling pathways were characteristic of severe impairment (eGFR <40ml/min). Glycolysis pathways first showed as up-regulated in the moderate group and increased further in the severe group while coagulation pathways were equally prevalent in all groups. Heat map analysis demonstrated clear discrimination between pathways up-regulated in urinary exosomes from patients with rapid progression in the 3 severity groups (Fig. 4G to I). In addition to confirmation of differential expression of Notch, integrin, apoptosis and EGFR pathways, additional roles for Toll receptor signaling (in the 50-69ml/min group), cell migration, Fas and Ras GTPase signaling in <49ml/min group were identified.
Analysis of individual proteins (Supplemental Table 2) identified plakoglobin as the most highly up-regulated protein in urinary exosomes from patients with rapid progression with a starting eGFR of >70ml/min as well as MAST-4, KIF2A, dynein, dynactin and the ATP-dependent and HS-90 chaperone proteins (Fig. 4J). Several actin-binding integrin-related proteins were also up-regulated including α-actinin-4, AKAPs-9 and -13, radixin, and tetraspanin-1 (CD-9). In the moderate severity group with a starting eGFR of 50-69ml/min the most prevalent differentially up-regulated protein was the endosomal trafficking protein SNX-18 (Fig. 4K). Differential up-regulation of many other vesicle-trafficking proteins was associated with rapid progression in this group, including multi-vesicular body (MVB) proteins 1, 2a, 2b and the vacuolar sorting proteins 4A, B,13A and AP. The cell-cell adhesion-related desmosomal protein, desmoplakin was also up-regulated as were α−actinin-4, AKAP-9, tetraspanin-1, fibronectin and tenascin. Interestingly, the vesicle-mediated transporter proteins glucose transporter-1 (GLUT1) and aquaporin-2 (AQP2), ATPBP and endoplasmic reticulum (ER)-ATPaseswere uniquely >2-fold up-regulated in urinary exosomes rapidly progressive patients from this group of.In the severely affected group (with starting eGFR<49ml/min) the most highly up-regulated protein was matrix-adhesion-related vitronectin. Pro-EGF ligand, EGFR substrates 8 and 8L and the endocytic VIP-36 (Fig. 4L) were also uniquely >2-fold up-regulated in this group. Differential upregulation of tetraspanin-1, prominins-1 and -2, ezrin and ras were also characteristic of urinary exosomes from severely affected ADPKD patients with rapid progression.
Urinary exosomes from ADPKD patients with good therapeutic responses to Tolvaptan show different proteomic profiles from patients with poor responses
Immunohistochemistry showed AVPR2 localization in basal cell membranes of normal human medullary collecting tubules as well as apical membranes and luminal vesicles of ADPKD-CKD2 and ADPKD-CKD4 cyst-lining epithelia (Fig. 5A to D). Immunoblot analysis confirmed higher levels of expression of 40kDa AVPR2 in urinary exosomes from ADPKD patients compared to normal subjects (Fig. 5E (i) and (ii)). Intriguingly, additional 70kDa, 35kDa and 25kDa molecular weight forms of AVPR2 were identified in urinary exosomes from patients with rapid but not slow progression (Fig. 5E(i) right panel). Higher levels of 40kDa AVPR2 were also detected in cyst fluids from early-stage ADPKD-CKD2 and late-stage ADPKD-CKD4 patients compared to non-PKD simple cysts (Fig. 5E(iii)). Interestingly, additional ~70kDa, 35kDa and 25kDa molecular weight forms of AVPR2 were also seen in 2 of the 4 cyst fluid samples (Fig. 5E(iii) lanes 2 and 4).
Fig. 5.
Vasopressin receptor-2 (AVPR2) expression and responses to its inhibitor, Tolvaptan.A-D. Immuno-histochemical localization of AVPR2 in human kidneys. A. IgG control. B. Positive staining (brown reaction product) is localized to medullary collecting ducts of normal human kidneys. C. AVPR2 is highly expressed in cyst-lining epithelial apical cell membranes and associated with particles in cystic lumens of ADPKD-CKD2 kidneys and D. in cystic cell membranes lining large and small epithelial cysts in ADPKD-CKD4 kidneys. Original magnifications x 20. E. Representative Western immunoblots and densitometric analysis of AVPR2 expression in exosomes isolated from urine (i) and (ii) and cyst fluid samples (iii). Densitometric analysis: mean +/-SEM *p<0.05 showed significantly increased levels of AVPR2 expression in urinary exosomes isolated from ADPKD patents compared to normal subjects (ii). Low levels of 40kD AVPR2 were seen in exosomes isolated from non-ADPKD simple cysts (SC) compared to ADPKD-CKD2 (early-stage) or ADPKD-CKD4 (late-stage) cyst fluid exosomes (iii). Additional 70, 35, 25 and 15kD bands of AVPR2 were highly expressed in urinary exosomes from patients with rapidly progressive (PG) ADPKD (i) and in 2 out of 4 ADPKD cyst fluid exosome samples (iii). F-K. Proteomic pathway (Panther) analysis of proteins >2-fold up-regulated in urinary exosomes isolated from ADPKD patients immediately before and after 4 years of Tolvaptan therapy showed distinctly different patterns in patients who responded well (F, G) compared to those who responded poorly to Tolvaptan therapy (H, I). Patients in whom the response could not be categorized displayed both patterns (J, K). Key to pie-chart colour-coding of pathways as in Fig. 3D.
Urinary exosomes isolated from 6 ADPKD patients immediately before and 4 years after onset of Tolvaptan therapy were subjected to comparative proteomic profiling. A good response was defined as a substantial reduction in the rate of decline in eGFR after therapy (>8ml/min over 4 years; >2ml/min/year). Poor responders showed little or no change in disease progression after therapy. Pre- versus post-Tolvaptan therapy urinary exosome proteomic expression profiling showed that many more urinary exosome proteins were >2-fold up-regulated (464 and 14) and far fewer were down-regulated (107 and 5, respectively) following a good response compared with poor responders (Table 3). Up-regulation of Wnt/β-catenin, platelet-derived growth factor (PDGF) and migration pathways, JAK/STAT, MAPK signaling, cytoskeletal and vesicular trafficking proteins were characteristic of a good response while up-regulation of angiogenesis, vascular endothelial growth factor (VEGF)-signaling, cadherin-13 adhesion, ciliary zinc finger DZIP1 and molecular chaperone HSP-70 proteins were associated with a poor response (Fig. 5F-I).
Table 3.
Comparisons of numbers of >2-fold up- regulated and >2-fold down-regulated proteins expressed in urinary exosomes from ADPKD patients treated with Tolvaptan for 5 years: effects of disease progression and therapeutic efficacy.
| eGFR at onset of treatment | Progression rate/year | Response to Tolvaptan | Up-regulated Proteins | Down-regulated Proteins |
|---|---|---|---|---|
| 58ml/min | 3.3ml/min | Good | 464 | 14 |
| 32ml/min | 5ml/min | Good | 107 | 5 |
| 52ml/min | 5ml/min | Poor | 58 | 2175 |
| 30ml/min | 7.5ml/min | Poor | 49 | 346 |
| 75ml/min | 5ml/min | Uncertain | 38 | 336 |
| 38ml/min | 6.25ml/min | Uncertain | 4 | 767 |
In this index cohort, although the stage of severity at initiation of treatment (eGFR 75-30ml/min) was not correlated with the efficacy of Tolvaptan response, higher rates of progression over the 5 years prior to onset of treatment (>5ml/min/year) appeared to be associated with poor responses. Interestingly, in 2 patients with uncertain/intermediate responses to Tolvaptan, one, with a rapid rate of progression (6.25ml/min/year) prior to therapy showed a urinary exosome proteomic profile resembling that of a poor responder while the other with a slower pre-treatment progression rate showed a proteomic expression profile resembling good responders (Figs. 5 J and K, Table 3).
Uptake and vesicular trafficking of cell-derived exosomes by renal epithelia in vitro depends on ADPKD stage of severity
Immunoblot analysis of the marker protein Alix (96kD) showed that confluent monolayers of renal epithelia in vitro secreted exosomes into their serum-free conditioned media (Fig. 6A, NHCT). NTA of exosomal isolates confirmed cell-type and ADPKD disease stage-dependence of cell-derived exosomal isolates with highest levels secreted by early stage ADPKD-CKD2 cells (17 × 1011/ml) > NHCT (3.6 × 1011 /ml) > late-stage ADPKD-CKD4 cells (2.4 × 1011/ml). Confocal tracking of cell uptake of fluorescently (PKH26)-labelled exosomes showed initial attachment to the outer surface of the recipient cell followed by internalization via invaginations of the cell plasma membrane (Fig. 6B, ADPKD). Super-resolution 3D image analysis showed time-dependent incorporation into intracellular vesicles (Fig. 6C). Z-axis profile analysis of WGA-labelled recipient NHCT cells demonstrated that intracellular vesicular trafficking of PKH26-labelled endosomal vesicles accumulated predominantly in the apical cortical areas of the cytoplasm after 24 hours of incubation (Fig. 6D).
Fig. 6.
Exosome-recipient cell interactions: secretion, uptake and intracellular vesicular trafficking. A. Representative Western immunoblot of marker protein Alix(96kDa, arrow) in exosomes isolated from serum-free conditioned media (CM) of confluent monolayers of normal human collecting tubule (NHCT) epithelia from 3 separate donors. B. Confocal microscopy imaging after 12 hours of uptake of PKH26 fluorescent dye (red)-labelled exosomes from ADPKD-CKD4 CM into ADPKD-CKD4 cystic epithelial cells whose membranes were labelled with wheat germ agglutinin (WGA, green) using a fully motorized Leica SP8 laser scanning confocal microscope. XY analysis showed exosome attachment to the external surface of the cell and incorporation into invaginated plasma membrane pits (high power, right panel). C. 3D analysis of stacked images from apical to basal membranes (Y axis) and from front to back (X axis) of NHCT cells incubated for 12 hours with NHCT-derived exosomes showed intake into the cytoplasm via a vesicular mode of intracellular trafficking. D. Z-stack analysis comparing the relative overlap of PKH26-labelled exosomes (red) with apical and basal cell membranes (green) demonstrated intracellular accumulation of exosomes in the apical, cortical third of the epithelial cell. E. NHCT cells incubated for 6, 12 and 24-hours with 109/ml PKH26 (red) pre-labelled exosomes derived from NHCT, ADPKD-CKD2 or ADPKD-CKD4 tubule or cystic epithelial monolayers in vitro showed different patterns of time-dependent intracellular vesicular trafficking and accumulation. WGA labelled cell membranes green; DAPI labelled nuclei blue. Control cells were treated for 24 hours with PKH dye only.
Characteristics of uptake of equal numbers of cell-derived exosomes into recipient NHCT cells were shown to be donor cell-type dependent (Table 4). After 6 hours of incubation 1.7-fold and 5-fold higher levels of uptake of ADPKD-CKD2 and ADPKD-CKD4-derived exosomes, respectively, were identified by comparison to NHCT-derived exosomes. While exosomal uptake of ADPKD-CKD2 exosomes increased with time up to 24 hours, uptake of ADPKD-CKD4 exosomes decreased after 12 hours of incubation (Table 4). These results suggested that exosomes secreted from the more highly proliferative, differentiated and metabolically active ADPKD cystic epithelia in early CKD2-stage kidneys were able to interact more productively with the endocytic machinery of normal NHCT cells than those exosomes derived from more de-differentiated later-stage CKD4 kidneys. Time- and ADPKD stage-dependent differences in intracellular vesicular trafficking and accumulation were confirmed by parallel confocal image analysis (Fig. 6E). Normal NHCT cell-derived exosomes incorporated into NHCT cells (homotypic controls) were localized in intracellular endocytic vesicles after 6 hours of incubation and accumulated in larger groups of perinuclear vesicles by 24 hours. Similar patterns of vesicular trafficking of ADPKD-CKD2 cell-derived exosomes were seen after 6 and 12 hours although more diffuse cytoplasmic accumulation after 24 hours of incubation. By contrast, strikingly different patterns of ADPKD-CKD4 exosome accumulation were seen characterized by the marked accumulation of large multi-vesicular aggregates of PKH-labelled exosomes by 24 hours of incubation.
Table 4.
Uptake of exosomes by NHCT cells incubated with PKH26-labelled exosomes isolated from NHCT, ADPKD-CKD2 and ADPKD-CKD4 cystic epithelial cells. Cells were fixed after 6, 12 and 24 hours of incubation and stacked images of XY, XZ and YZ planes imaged on a fully-motorized Leica SP8 laser-scanning confocal microscope. Segmentation analysis was carried out to determine concentrations of intracellular exosomes, cell volumes and cell numbers.
| Exosome origin | Numbers taken up after 6h incubation | Numbers taken up after 12h incubation | Numbers taken up after 24h incubation |
|---|---|---|---|
| NHCT | 104 | 107 | 159 |
| ADPKD-CKD2 | 173 | 596 | 921 |
| ADPKD-CKD4 | 542 | 476 | 112 |
Discussion
The results show that the optimized protocol for isolation of size-selected 40-100nm exosomes from 5ml urine samples is practical, scalable and reproducible providing high sensitivity for proteomic definition of this specific subset of endosome-derived urinary ECV particles. The exclusion of the larger, 125-1,000nm cell-membrane-derived MV subset from the exosomes preparations was confirmed by the absence of larger proteins and fragments such as fibrocystin that were previously detected in studies of mixed urinary ECVs [26].Storage of urinary exosomes at -80°C after addition of protease inhibitors prior to freezing and vortexing during defrosting has been established previously and it has been shown that there is no significant loss in the exosomal yield compared to the use of fresh urine samples []26-28]. Isolation of exosomes has also been described after long-term storage at -80°C of urine samples ranging from 7 months to 20 years [28], [29], [30], [31], [32].
Quantitative analysis in vivo and in vitro suggested that the increased numbers of urinary exosomes excreted by ADPKD patients were due to stage-dependent biological abnormalities in exosomes secreted by cystic cells. In normal kidneys, the majority of tubule epithelial cell-derived exosomes are taken up by downstream nephron segments and a small proportion are excreted in urine. By contrast, in ADPKD kidneys, although only ~60-70% of cysts may retain contact with their nephron of origin [6, 65] total numbers of excreted urinary exosomes increased due increasing abnormalities in the properties of ADPKD cell-derived exosomes and associated loss of tubule cell uptake. In addition, ADPKD patients are polyuric due to progressive loss of urinary concentrating ability which would further contribute to increased urinary volumes.
As first described by Hogan et al al [19] urinary exosome proteomic profiling clearly distinguished between normal subjects and ADPKD patients in this study. It was noted, however, that expression levels of some proteins, including polycystin-1 fragments and fibrocystin-like transmembrane proteins were less prevalent in our size-excluded (NTA/TRPS) small (40-100nm) exosome (EV) preparations than reported in larger exosome-like vesicle (ELV) (≥100-1,000nm) or microvesicle (MV) preparations. This finding is consistent with the distinctive biogenesis of EVs from endocytic pathways and of MVs from plasma-membrane vesicular pathways.
Differential proteomic expression analysis of our small size (40-100nm)-defined urinary exosome preparations also distinguished between ADPKD patients at different stages of severity (CKD-1 to -4), which were associated with losses of differentiated tubule brush border proteins and increases in coagulation and immunomodulation proteins. Significant differences were also seen between ADPKD patients with rapid (eGFR decline >10m/min over 5 years; >2 - 6ml/min/year) compared to slower rates of progression. Further subdivision into groups with mild, moderate or severe renal impairment in the CKD2 to 3b range at onset of progression showed interesting patterns of differentially up-regulated proteins and pathways. These included maximal up-regulation of Notch-pathway and MAST-4 proteins in rapidly progressing ADPKD patients with mild disease (>70ml/min eGFR); of apoptosis and sorting nexin vesicular proteins in rapidly progressing ADPKD patients with moderate disease (50-69ml/min eGFR); and of migration pathways and EGFR substrate proteins in rapidly progressing ADPKD patients with severe disease (<49ml/min eGFR). These pathways and proteins have previously been linked with ADPKD cystic expansion [1,7,[43], [44], [45], [46], [47], [48]]. Changes in cell-cell adhesion proteins plakoglobin and desmoplakin in rapidly progressing ADPKD patients were consistent with the previously described pathogenic role of cell-cell adhesion disruption cell-cell adherens and desmosomal junctions in ADPKD cysts [49], [50], [51].Cell-matrix abnormalities in ADPKD were reflected by significant changes in urinary exosome fibronectin and vitronectin while switches in prevalence of up-regulation of actin-plasma membrane crosslinker proteins radixin and ezrin in urinary exosomes of rapidly progressing ADPKD patients were consistent with cytoskeletal involvement in cystic pathogenesis [1,7,8,44,[51], [52], [53], [54], [55], [56]].
Increased cell proliferation is a key feature of ADPKD cyst expansion, particularly in early stage disease [1,7]. Not surprisingly, urinary exosomes from rapidly progressing ADPKD patients showed significant up-regulation of cytokinesis-related MAST-4 and EGFR kinase substrates 8 and 8L proteins, which normally increase proliferative responses to EGF [57]. Integral membrane proteins prominins 1 and 2 are also significantly up-regulated consistent with disruptions of cell shape, spreading and migration that characterize ADPKD cystic epithelia in vitro [54,58].
Abnormalities in intra-vesicular trafficking associated with polarization of membrane receptors and transporters are characteristic of ADPKD epithelia [1,[59], [60], [61]]. Interestingly, urinary exosomes from rapidly progressing ADPKD patients were characterized by significant levels of up-regulation of vesicular sorting nexins, vacuolar sorting proteins, and vesicle-mediated transporters GLUT-1 and AQP-2 in ADPKD patients with moderate disease. It remains to be determined whether urinary exosome analysis will not only be valuable in detecting and predicting rapid rates of progression of ADPKD but also in leading to additional insights into cellular mechanisms underpinning progression at different levels of disease severity.
In an index cohort of ADPKD patients treated for 4-years with Tolvaptan, prior rapid progression over 5 years as well as urinary exosome expression of additional AVPR-2 molecular forms were associated with poor therapeutic responses. Proteomic profiling of urinary exosomes identified efficacy-related differences not from the primary cAMP/PKA pathway but from previously identified alternative AVPR-2 targets including cell-cell adhesion and actin cytoskeleton remodeling pathways [62,63].Up-regulation of urinary exosomal Wnt/β-catenin and PDGF-signaling proteins was characteristic of a good response while up-regulation of angiogenesis and VEGF were indicative of a poor response. It was of interest that both “rapid” and “poor” profiles were identified in an intermediate group but larger-scale studies will be needed in the future to determine any predictive potential when urine samples from larger numbers of ADPKD patients undergoing long-term Tolvaptan treated become available.
Overall, these studies suggest that proteomic profiling of urinary exosomes offers strong potential for the development of a routinely and universally applicable, non-invasive test with high specificity and reproducibility to identify and monitor those ADPKD patients at the highest risk of rapidly progressive disease and of responding to Tolvaptan therapy. This biomarker approach might also be of value in health care settings where MRI based- renal volume and genetic analyses are not readily available. The ultimate goal for ADPKD patients is to develop a readily accessible reliable and reproducible test to predict outcomes with regard to progression and response to drug therapies. The development of a urinary exosome protein expression “atlas” would facilitate the identification of individual patients who are most in urgent need and most likely to benefit from long-term drug therapy. This provides another step towards the goal of increasingly personalized assessment of ADPKD prognosis, management and susceptibility to effective drug therapies.
Author contributions
PW and JN designed the study
KR, HH and AMB carried out the experiments
KR and HH analyzed the data
PW, KLR, HH and JN drafted and revised the paper
All authors approved the final version of the manuscript
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
This work was supported by the Royal Free Charity (KR PhD fellowship), PKD Charity (KR PhD fellowship; JN Small Research Grant) and Rosetrees/Stoneygate Trust (PW Research Grant). Samples were stored in the PKD Charity-sponsored Bioresource Bank at the Royal Free. We thank Prof. D. Gale, Royal Free, London for identification of ADPKD patient cohorts; Mr. I. Chatworthy and Mrs. A. Carbajal, Royal Free Electron Microscopy Unit for TEM analysis of exosomes; Mr. J. Suthar, UCL School of Pharmacy for help with NTA of exosomes; Mr. S. Lynham, King’s College, London Proteomics Facility for bioinformatics advice; as well as Prof. D. Bockenhauer, UCL/GOSH and Prof. J. Dear, Queen’s Medical Research Institute, Edinburgh for their analytical and insightful discussion.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2021.100013.
Contributor Information
Katie L. Raby, Email: katie.raby.15@alumni.ucl.ac.uk.
Harry Horsely, Email: h.horsely@ucl.ac.uk.
Jill T. Norman, Email: j.norman@ucl.ac.uk.
Patricia D. Wilson, Email: patricia.wilson@ucl.ac.uk.
Appendix. Supplementary materials
References
- 1.Wilson P.D. Molecular and cellular aspects of polycystic kidney disease. New Engl. J. Med. 2004;350:151–164. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- 2.Harris P.C., Torres V.E. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J. Clin. Invest. 2014;124:2315–2324. doi: 10.1172/JCI72272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annual reports . 2010. 2019. UK Renal Registry. [Google Scholar]
- 4.Hateboer N., et al. Comparison of phenotypes of polycystic kidney disease types 1 and 2. European PKD1-PKD2 study group. Lancet. 1999;353:103–107. doi: 10.1016/s0140-6736(98)03495-3. [DOI] [PubMed] [Google Scholar]
- 5.Lanktree M., et al. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J. Am. Soc. Nephrol. 2018;29:2593–2600. doi: 10.1681/ASN.2018050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson P.D., Goilav B. Cystic Disease of the kidney. Annu. Rev. Pathol. Mech. Dis. 2007;2:341–368. doi: 10.1146/annurev.pathol.2.010506.091850. [DOI] [PubMed] [Google Scholar]
- 7.Norman J. Fibrosis and progression of Autosomal Dominant Polycystic Kidney Disease. Biochim. Biophys. Acta. 2011;1812:1327–1336. doi: 10.1016/j.bbadis.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weimbs T, Shillingford J.M., Torres J., Kruger S.L., Bourgeois B.C. Emerging targeted strategies for the treatment of autosomal dominant polycystic kidney disease. CKJ. 2018;11(Suppl 1) doi: 10.1093/ckj/sfy089. i27-i38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terryn S., Ho A., Beauwens R., Devuyst O. Fluid transport and cystogenesis in autosomal dominant polycystic kidney disease BBA MBD. 2011;1812:1314–1321. doi: 10.1016/j.bbadis.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Ong A.C.M., Harris P.C. A polycystin-centric view of cyst formation and disease. Kidney Int. 2015;88:699–710. doi: 10.1038/ki.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polgar K, et al. Disruption of polycystin-1 function interferes with branching morphogenesis of the ureteric bud in developing mouse kidneys. Dev. Biol. 2005;286:16–30. doi: 10.1016/j.ydbio.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 12.Drummond I. Polycystins, focal adhesions and extracellular matrix interactions. Biochim. Biophys. Acta. 2011;1812:1322–1326. doi: 10.1016/j.bbadis.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastos A., Piontek K. Onuchic L, F. Pkd1 haploinsufficiency increases renal damage and induces microcyst formation following ischemia/reperfusion. J. Am. Soc. Nephrol. 2010;21:1062. doi: 10.1681/ASN.2008040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gall Cornec-Le, E. Audrezet, M.P. Chen, J.M Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrol. 2013;24:1006–1013. doi: 10.1681/ASN.2012070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosetti S., Harris P.C. Genotype-phenotype correlations in Autosomal Dominant and Autosomal Recessive polycystic Kidney Disease. J. Am. Soc. Nephrol. 2007;18:1374–1380. doi: 10.1681/ASN.2007010125. [DOI] [PubMed] [Google Scholar]
- 16.Torres V.E. Tolvaptan in patients with Autosomal Dominant Polycystic Kidney Disease. N. Engl. J. Med. 2012;367:2407–2418. doi: 10.1056/NEJMoa1205511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres V.E., et al. REPRISE trial investigators. Tolvaptan in later-stage autosomal dominant polycystic kidney disease. N. Engl. J. Med. 2017;377:1930–1942. doi: 10.1056/NEJMoa1710030. [DOI] [PubMed] [Google Scholar]
- 18.Bennett H., McEwan P., Hamilton K., O’Reilly K. Modelling the long-term benefits of tolvaptan therapy on renal function decline in autosomal dominant polycystic kidney disease: an exploratory analysis using the ADPKD outcomes model. BMC Nephrol. 2019;20:136–144. doi: 10.1186/s12882-019-1290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irazabel M.V., et al. Prognostic enrichment design in clinical trials for autosomal dominant polycystic kidney disease: The TEMPO 3:4 clinical trial. Kidney Int. Rep. 2016;1:3213–3220. doi: 10.1016/j.ekir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornec-Le Gall E., Blais J.D., et al. Can we further enrich autosomal dominant polycystic kidney disease clinical trials for rapidly progressive patients? Application of the PROPKD score in the TEMPO trial. Nephrol. Dial. Transplant. 2018;33:645–652. doi: 10.1093/ndt/gfx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alderson H.V., Ritchie J.P. The associations of blood kidney injury molecule-1 and neutrophil gelatinase-associated lipocalin with progression from CKD to ESRD. Clin. J. Am. Soc. Nephrol. 2016;11:2141–2149. doi: 10.2215/CJN.02670316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadkami G.N., et al. Association of urinary biomarkers of inflammation, injury and fibrosis with renal function decline: the ACCORD trial. Clin. J. Am. Soc. Nephrol. 2016;11:1343–1352. doi: 10.2215/CJN.12051115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zewinger S, et al. Dickkopf-3 (DKK3) in urine identifies patients with short-term risk of eGFR loss. J. Am. Soc. Nephrol. 2018;29:2722–2733. doi: 10.1681/ASN.2018040405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malhotra R., et al. Urine markers of kidney tubule cell injury and kidney function decline in SPRINT trial participants with CKD. Clin. J. Am. Soc. Nephrol. 2020;15:349–358. doi: 10.2215/CJN.02780319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Dov IZ., et al. MicroRNA as potential biomarkers in Autosomal Dominant Polycystic Kidney Disease progression: description of miRNA profiles at baseline. PLOS ONE. 2014;9:e86586. doi: 10.1371/journal.pone.0086856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogan M.C., et al. Identification of biomarkers for PKD1 using urinary exosomes. J. Am. Soc. Nephrol. 2015;26:1661–1670. doi: 10.1681/ASN.2014040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salih M., et al. DIPAK consortium. Proteomics of urinary vesicles links plakins and complement to polycystic kidney disease. J. Am. Soc. Nephrol. 2016;27:3079–3092. doi: 10.1681/ASN.2015090994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruschi M., et al. Proteomic analysis of urinary microvesicles and exosomes in medullary sponge kidney disease and Autosomal Dominant Polycystic Kidney Disease. Clin. J. Am. Soc. Nephrol. 2019;14:834–843. doi: 10.2215/CJN.12191018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles and friends. J. Cell Biol. 2013;4:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause M., Samoylenko A., Vainio S.J. Exosomes as renal inductive signals in health and disease, and their application as diagnostic markers and therapeutic agents. Frontiers Cell Dev. Biol. 2015;3:1–13. doi: 10.3389/fcell.2015.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knepper M., Pisitikun T. Exosomes in urine: who would have thought….? Kidney Int. 2007;72:1043–1045. doi: 10.1038/sj.ki.5002510. [DOI] [PubMed] [Google Scholar]
- 32.Gildea J.J., et al. Exosomal transfer from human renal proximal tubule cells to distal tubule and collecting duct cells. Clin. Biochem. 2014;47:89–94. doi: 10.1016/j.clinbiochem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hemler M.E. Tetraspanin proteins mediate cellular penetration, invasion and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 34.Rana S., Yue S, Stadel D., Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012;44:1574–1584. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Stahl A., Johansson K., Mossberg M, Kahn R., Karpman D. Exosomes and microvesicles in normal physiology, pathophysiology and renal diseases. Pediatr. Nephrol. 2019;34:11–30. doi: 10.1007/s00467-017-3816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hood J.L., San R.S., Wickline S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 37.Rak J. Extracellular vesicles-biomarkers and effectors of the cellular interactome in cancer. Front. Pharmacol. 2013;4:1–4. doi: 10.3389/fphar.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erozenci L.A., Bottger F., Bijnsdorp I.V., Jimenez C.R. Urinary exosomal proteins as (pan) cancer biomarkers: insights from the proteome. FEBS Lett.593. 2019:1580–1597. doi: 10.1002/1873-3468.13487. [DOI] [PubMed] [Google Scholar]
- 39.Kwon S.H., Liu K.D., Mostov K.E. Intercellular transfer of GPRC5B via exosomes drives HGF-mediated outward growth. Curr. Biol. 2014;24:199–204. doi: 10.1016/j.cub.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borges F.T., et al. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 2013;24:385–392. doi: 10.1681/ASN.2012101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson P.D., Dillingham M.A., Breckon R, Anderson R.J. Defined human renal tubular epithelia in culture: growth, characterization, and hormonal response. Am. J. Physiol. 1985;248 doi: 10.1152/ajprenal.1985.248.3.F436. F436-F443. [DOI] [PubMed] [Google Scholar]
- 42.Wilson P.D., Schrier R.W., Breckon R.D., Gabow P.A. A new method for studying human polycystic kidney disease epithelia in culture. Kidney Int. 1986;30:371–378. doi: 10.1038/ki.1986.194. [DOI] [PubMed] [Google Scholar]
- 43.Sheldon H., Heilkamp E.Turley. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–2394. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- 44.Wilson P.D., Geng L., Li X., Burrow CR. The PKD1 gene product, “Polycystin-1”, is a tyrosine-phosphorylated protein that co-localizes with α2β1-integrin in focal clusters in adherent renal epithelia. Lab Invest. 1999;79:1311–1323. [PubMed] [Google Scholar]
- 45.Lee K., Boctor S., Barisoni L.M.C., Gusella GL. Inactivation of integrin-β1 prevents the development of polycystic kidney disease after the loss of polycystin-1. J. Am. Soc. Nephrol. 2015;26:888–895. doi: 10.1681/ASN.2013111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goilav B., Satlin L.M., Wilson P.D. Pathways of apoptosis in human autosomal recessive and autosomal dominant polycystic kidney disease. Pediatr. Nephrol. 2008;23:1473–1482. doi: 10.1007/s00467-008-0851-9. [DOI] [PubMed] [Google Scholar]
- 47.Wilson S.J., Amsler K., Hyink D., Burrow C.R., Wilson P.D. Inhibition of HER-2(neu/ErbB2) restores normal function and structure to polycystic kidney disease (PKD) epithelia. Biochem. Biophys. Acta. 2006;1762:647–655. doi: 10.1016/j.bbadis.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Castelli M., et al. Regulation of the microtubular cytoskeleton by Polycystin-1 favors focal adhesions turnover to modulate cell adhesion and migration. BMC Cell Biol. 2015;16:1–16. doi: 10.1186/s12860-015-0059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roitbak T., et al. A polycystin-1 multiprotein complex is disrupted in polycystic kidney diseases cells. Mol. Biol. Cell. 2004;15:1334–1346. doi: 10.1091/mbc.E03-05-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silberberg M., Charron A.J., Wandinger-Ness A. Mispolarization or desmosomal proteins and altered cell adhesion in Autosomal Dominant Polycystic Kidney Disease. Am. J. Physiol. Renal Physiol. 2005;288:F1153–F1163. doi: 10.1152/ajprenal.00008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng L., Burrow C.R., Li H, Wilson P.D. Modification of polycystin-1 multiprotein complexes by calcium and tyrosine phosphorylation. Biochem. Biophys. Acta. 2001;1535:21–35. doi: 10.1016/s0925-4439(00)00079-x. [DOI] [PubMed] [Google Scholar]
- 52.Kuo N., Norman J.T., Wilson P.D. Acidic FGF regulation of hyperproliferation of fibroblasts in human Autosomal Dominant Polycystic Kidney Disease. Biochem. Mol. Med. 1997;61:178–191. doi: 10.1006/bmme.1997.2583. [DOI] [PubMed] [Google Scholar]
- 53.Verma D., et al. Flow induced adherens junction remodeling driven by cystoskeletal forces. Exp. Cell Res. 2017;359:327–336. doi: 10.1016/j.yexcr.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Nigro E.A., et al. Polycystin-1 regulates actomyosin contraction and the cellular response to extracellular stiffness. Sci. Rep. 2019;9:16640. doi: 10.1038/s41598-019-53061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puleo J.I., et al. Mechanosensing during directed cell migration requires dynamic actin polymerization at focal adhesions. J. Cell Biol. 2019;218:4215–4235. doi: 10.1083/jcb.201902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naz F., Anjum F., Islam A., Ahmad F., Hassan I. Microtubule affinity-regulating kinase 4: structure function, and regulation. Cell Biochem. Biophys. 2013;67:485–499. doi: 10.1007/s12013-013-9550-7. [DOI] [PubMed] [Google Scholar]
- 57.Fazioli F., et al. Eps8, a substrate for the EGFR kinase enhances EGF-dependent mitogenic signals. EMBO J. 1993;12:3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joly D., et al. The polycystin1-C terminal fragment stimulates ERK-dependent spreading of renal epithelial cells. J. Biol. Chem. 2006;281:26329–26339. doi: 10.1074/jbc.M601373200. [DOI] [PubMed] [Google Scholar]
- 59.Wilson P.D. Apico-basal polarity in polycystic kidney disease epithelia. Biochim. Biophys. Acta. 2011;1812:1239–1248. doi: 10.1016/j.bbadis.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Devuyst O, Burrow CR, Smith BL, Agre P, Knepper MA, Wilson PD. Expression of aquaporins-1 and -2 in human kidneys during nephrogenesis and in autosomal dominant polycystic kidney disease. Am. J. Physiol. 1996;271 doi: 10.1152/ajprenal.1996.271.1.F169. F169-F183. [DOI] [PubMed] [Google Scholar]
- 61.Zheleznova N.N., Wilson P.D., Staruschenko A. Epidermal growth factor-mediated proliferation and sodium transport in. normal and PKD epithelial cells. Biochim. Biophys. Acta. 2011;1812:1301–1313. doi: 10.1016/j.bbadis.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffert J.D., et al. Dynamics of the G protein-coupled vasopressin V2 receptor signaling network revealed by quantitative phosphor-proteomics. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014613. M111.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oosthuyzen W., et al. Vasopressin regulates extracellular vesicle uptake by kidney collecting duct cells. J. Am. Soc. Nephrol. 2016;27:3345–3355. doi: 10.1681/ASN.2015050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gonzales P., et al. Isolation and purification of exosomes in urine. Methods Mol. Biol. 2010;641:89–99. doi: 10.1007/978-1-60761-711-2_6. [DOI] [PubMed] [Google Scholar]
- 65.Sullivan L, et al. Epithelial transport in polycystic kidney disease. Physiol. Rev. 1998;78:1165–1191. doi: 10.1152/physrev.1998.78.4.1165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.