Highlights
-
•
Nucleic acid/amyloid proteins interactions often promote protein aggregation.
-
•
Whether the DNA promotes the formation of self-assembly due to direct interactions or indirectly is unknown.
-
•
This question has been addressed for the bacterial amyloid Hfq.
-
•
Possible functional consequence to answer to cellular stresses are discussed.
Keywords: Bacterial amyloid, Functional amyloid, DNA:protein fibrils, DNA induced protein fibrillation, Bacterial adaptation
Abstract
Nucleic acid amyloid proteins interactions have been observed in the past few years. These interactions often promote protein aggregation. Nevertheless, molecular basis and physiological consequences of these interactions are still poorly understood. Additionally, it is unknown whether the nucleic acid promotes the formation of self-assembly due to direct interactions or indirectly via sequences surrounding the amyloid region. Here we focus our attention on a bacterial amyloid, Hfq. This protein is a pleiotropic bacterial regulator that mediates many aspects of nucleic acids metabolism. The protein notably mediates mRNA stability and translation efficiency by using stress-related small non coding regulatory RNA. In addition, Hfq, thanks to its amyloid C-terminal region, binds and compacts DNA. A combination of experimental methodologies, including synchrotron radiation circular dichroism (SRCD), gel shift assay and infrared (FTIR) spectroscopy have been used to probe the interaction of Hfq C-terminal region with DNA. We clearly identify important amino acids in this region involved in DNA binding and polymerization properties. This allows to understand better how this bacterial amyloid interacts with DNA. Possible functional consequence to answer to stresses are discussed.
1. Introduction
Nucleic acids, DNA and RNA, have been described as cofactors that bind to amyloidogenic proteins and facilitate their self-assembly [1, 2]. Even if the implications of this interaction are still unclear, they may have physiological (and possibly pathological) consequences [3]. As an example, the amyloid β peptide (Aβ) involved in Alzheimer's disease interacts with DNA [2], regulates gene transcription [4, 5] and may affect DNA repair process [6]. Thus, a close link between bacterial amyloid self-assembly and gene expression regulation may exist.
Here, we focus our attention on a protein involved in stress response in bacteria, Hfq [7]. We particularly focus our attention on Hfq's amyloid region, involved in DNA binding and condensing properties. In bacteria, the chromosomic DNA is about 2 nm in diameter and more than 1 mm in length. It thus needs to be packed to fit in a length of ∼ 1 μm and an intracellular volume of ∼ 1 μm3. This can be achieved with the help of some proteins, the Nucleoid-Associated Proteins (NAP). The assembly of NAPs and chromosomic DNA forms a condensed structure called nucleoid [8], that occupies about ∼ 0.2 μm3 [9]. More than 10 proteins are essential to achieve this function in E. coli, which play crucial roles in the regulation of several biological processes [10, 11]. For instance, condensed and uncondensed DNA states drastically influence the efficiency of replication and transcription processes [12]. Additionally, when DNA damages such as strand breaks occur, there is a need in the apposition of homologous sequences to repair DNA by recombination and DNA compaction helps this process. This is particularly critical in stress conditions and proteins involved in stress response could have an important role in such a process [13].
Hfq is particularly important for stress response in bacteria [7]. Hfq is an abundant protein that binds to nucleic acids in general, DNA and RNA [14, 15]. Functionally, Hfq controls a large number of bacterial functions [16]. Among these functions, most are related to its RNA-binding properties. Indeed, Hfq facilitates the pairing of small regulatory non-coding RNA (sRNA) with different mRNA target(s) [17]. As sRNA annealing to mRNA often takes place around the Shine-Dalgarno sequence, this usually induces a negative regulation at the post-transcriptional level [18], [19], [20]. Hfq also regulates genetic expression by changing RNA stability [21, 22].
In addition, Hfq can also bind DNA [11, 15], even if its affinity for DNA is lower than for RNA (depending on the sequences KD ranging from nM to µM vs pM to nM, respectively [23, 24]). Hfq binds both linear and circular DNA [25] and in vivo a significant amount of the protein is found in the nucleoid fractions (up to 20%) [26]. Hfq binding to the major groove of DNA B-form leads to a slight reduction in the double stranded DNA helical pitch (∼ 10%) [27]. But Hfq binding also results in the condensation of DNA through protein-protein interactions [24, 28]. This is referred to as “bridging” and observed for other NAPs such as H—NS [29], [30], [31]. Some of the phenotypic effects due to the lack of Hfq, initially associated to sRNA-based regulations, may also be linked to defects in DNA-related processes [28, 32].
Structurally, the N-terminal region of Hfq (NTR, ∼ 65 amino acid residues) shares homologies with the Sm family of protein [33, 34]. Precisely, Hfq-NTR comprises five β-strands that form a strongly bent antiparallel β-sheet, capped by a α-helix. Like Sm proteins, Hfq assembles into a cyclic oligomer (an hexamer in the case of Hfq vs an heptamer for Sm proteins) to form the biologically active unit, a torus. This oligomeric torus binds RNA on both faces [35], [36], [37]. Although the mechanism by which Hfq binds to DNA is not completely clear, it is now established that this NTR domain of the protein binds DNA in a sequence-independent manner [27, 38]. The DNA molecule binds to the proximal face of the Hfq hexamer, i.e. the surface exposing the N-terminal α-helix [27].
In addition to the well-characterized Sm domain, the protein also presents a C-terminal region (CTR), that comprised ∼ 40 amino acid (aa) residues. Hfq-CTR is located at the periphery of the torus [39]. Although no atomic 3D structure is known for this CTR, it has been shown that it self-assembles into an amyloid-like structure in vitro and in vivo [40, 41]. Recently, this CTR region has been shown to play a major role in DNA bridging and compaction [28, 38, 42, 43], but also that it interacts with the membrane [44].
The work reported here further explores the DNA binding properties of the amyloid region of Hfq and its ability to bind and change the structure of DNA. A minimal region of eleven aa has been identified as the nucleation site for the amyloid formation (underlined in the CTR sequence: SRPVSHHSNNAGGGTSSNYHHGSSAQNTSAQQDSEETE) [44]. This minimal sequence is unable to bind DNA, while the CTR does. We suspect that other aa residues are involved in the global assembly of that amyloid structure outside the minimal region of 11 aa, and that some residues may also be involved in DNA binding properties. To answer this question, a combination of experimental methodologies, including synchrotron radiation circular dichroism (SRCD) and infrared (FTIR) spectroscopies, have been used to probe the interaction of Hfq with DNA and to analyze the formation of the amyloid self-assembly.
2. Materials and methods
2.1. Chemicals
All chemicals were purchased from Sigma-Aldrich and Thermofisher scientific.
2.2. Hfq CTR peptides
Hfq-related peptides were chemically synthetized (Proteogenix, France) and prepared as described previously in Fortas et al. [40]. These peptides correspond to the amyloid CTR domain of Hfq (residues 64 to 102) and are referred to as Hfq-CTR throughout the manuscript. The sequences of the various CTR mutants are:
| WT: | SRPVSHHSNNAGGGTSSNYHHGSSAQNTSAQQDSEETE |
| R66A: | SAPVSHHSNNAGGGTSSNYHHGSSAQNTSAQQDSEETE |
| S65A,S69A,S72A: | ARPVAHHANNAGGGTSSNYHHGSSAQNTSAQQDSEETE |
| S80A,S81A: | SRPVSHHSNNAGGGTAANYHHGSSAQNTSAQQDSEETE |
| S87A,S88A: | SRPVSHHSNNAGGGTSSNYHHGAAAQNTSAQQDSEETE |
| S93A,S98A: | SRPVSHHSNNAGGGTSSNYHHGSSAQNTAAQQDAEETE |
| H70A,H71A,H84A,H85A: | SRPVSAASNNAGGGTSSNYAAGSSAQNTSAQQDSEETE |
| H70A,H71A: | SRPVSAASNNAGGGTSSNYHHGSSAQNTSAQQDSEETE |
| H84A,H85A: | SRPVSHHSNNAGGGTSSNYAAGSSAQNTSAQQDSEETE |
| Y83A | SRPVSHHSNNAGGGTSSNAHHGSSAQNTSAQQDSEETE |
| E99A,E100A,E102A: | SRPVSHHSNNAGGGTSSNYHHGSSAQNTSAQQDSAATA |
| D97A,E99A,E100A,E102A: | SRPVSHHSNNAGGGTSSNYHHGSSAQNTSAQQASAATA |
| G76A,G77A,G78A: | SRPVSHHSNNAAAATSSNYHHGSSAQNTSAQQDSEETE |
Hfq-CTR peptides were reconstituted in water at 20 mg/mL. We determined that the pH used in our condition (∼ 5) was the most appropriate to form the complex with DNA. Indeed, the net charge of the WT peptide was calculated as +1 at pH 5, 0 at pH 6, −2 at pH 7 and −3 at pH 8. The positive charge of the peptide at pH 5 allows its interaction with DNA, while increasing pH abolishes this interaction. At this concentration, the peptide shows self-buffering properties [45]. The positive charge of the peptide could be more pronounced at pH 4, but we estimated that this condition was too far of the E. coli physiological pH [46]. Note that in vivo the CTR of the protein is not found isolated but bound to the Hfq-NTR part that may change the microenvironment of CTR aminoacids and could drastically change their pKa and thus peptide net charge at physiological pH [47, 48].
2.3. Preparation of the complexes for SRCD and FTIR analysis
Complexes between (dA:dT)59 DNA (Eurogentec) and Hfq-CTR peptides were prepared as described previously [28, 43]. Briefly, complexes between Hfq-CTR DNA were prepared at pH 5 and used at a final concentration of 1.8 mM and 7.3 mM, respectively. The stoichiometry was 1 Hfq-CTR per 4 base pair. The choice for this 59 base-pair homopolymeric DNA sequence was made because (i) Hfq CTR has the higher affinity for AT-rich sequences; (ii) Hfq-CTR has a low affinity for short DNAs, that in turn cannot be analysed using SRCD as they don't have significant signal; (iii) using a random sequence will not give any relevant KD as a random sequence is heterogeneous and therefore the KD changes all along the sequence according to the nucleotide sequence to which Hfq-CTR is bound; (iv) more important the structure of DNA may drastically change the SRCD and even more the FTIR spectra and cannot be subtracted accurately for our spectral analysis. As salts don't change drastically the affinity of the CTR for DNA (Sup Fig. S1), we preferred to omit additional salts in our preparations in order to get a broader spectral band (170–320 nm) [42, 49]. Furthermore, this procedure corresponds to the one used previously for the accumulation of reference dataset (https://pcddb.cryst.bbk.ac.uk/). Samples were analyzed after at least 2 weeks minimum to allow peptide self-assembly on DNA that is not instantaneous [28] (while DNA:Hfq-CTR simple interaction is, see 2.4 and 2.5).
2.4. Electrophoretic mobility shift assay (EMSA)
Binding of Hfq–CTRs to dsDNA was investigated with an EMSA. The DNA fragment was incubated with Hfq–CTR WT or mutants for 20 min at room temperature in water at pH5. We observed that this short time is sufficient to allow the interaction of the CTR with DNA (while amyloidogenesis takes more time, see 2.3.). EMSA was carried out using a non-denaturing gradient 4–12% polyacrylamide gel (Bio-rad). The gel was run for 2 h at room temperature with TAE buffer (40 mM Tris-Acetate, 1 mM ethylenediaminetetraacetic acid, pH 8.0), then stained with GelRed nucleic acid stain (Biotium) and imaged with a G-BOX system (Syngene, Cambridge, UK). The concentration of Hfq-CTR peptides and DNA fragment were 1 µM and 100 nM, respectively.
2.5. Fluorescence anisotropy
Equilibrium constant dissociation (KD) were determined using fluorescence anisotropy [24]. Measurements were collected with an Eclipse fluorimeter (Agilent) equipped with polarizers. The extrinsic fluorophore was attached to one DNA strand. Here we used 5′-fluoresceinated dA20 and non labelled dT20 oligonucleotides (Eurogentec); the duplex of oligonucleotides were formed after stoichiometric addition of the two oligonucleotides in the same tube, heating at 90 °C for 3 min and then slowly cooled down at 20 °C. Note that for anisotropy measurements we used low molecular weight (shorter) oligonucleotides than for EMSA, as fluorescence anisotropy can be used only if the molecular size of the protein–DNA complex is sufficiently different from the free fluorescing DNA. 1 nM of fluoresceinated dsDNA was added to the cuvette (1 ml) and titrated by Hfq-CTR (concentration ranging from 0 to few µM, see Fig. 2). Samples were incubated 60 s prior to each measurement, ensuring equilibrium binding (DNA binding is almost instantaneous). The measurement was performed at 298 K and pH 5, samples were excited at 490 nm and emission was measured at 520 nm. The normalization of fluorescence anisotropy was carried out after determination of the Amax value, which was obtained at saturating Hfq-CTR concentrations. A/Amax ratios were plotted versus Hfq-CTR concentrations. The curves were fitted by the nonlinear least-squares regression method, assuming a bimolecular model, with Hfq-CTR (monomeric) concentration in excess compare to dA20:dT20. Binding affinities were confirmed with at least two different experiments.
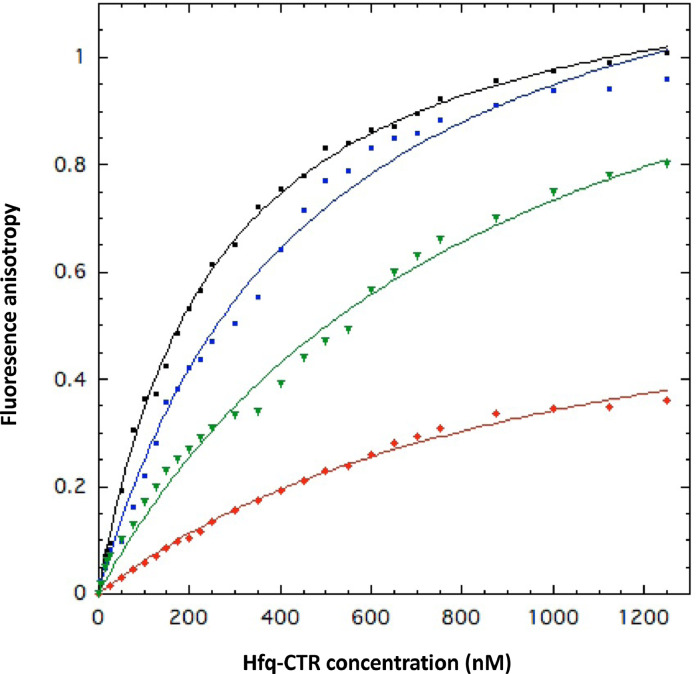
Fig. 2.
KD measurements of WT and mutated Hfq-CTR:dsDNA complexes using fluorescence anisotropy. In this case a dA:dT20 dsDNA was used. WT Hfq-CTR (black) has an equilibrium dissociation constant KD = 260 ± 10 nM; mutant Hfq-CTR S87A,S88A (blue) has a KD = 460 ± 30 nM. The mutants Hfq-CTR Y83A (red) and Hfq-CTR E99A,E100A,E102A (green) have lower affinities with KD = 1000 ± 75 nM and 890 ± 80 nM, respectively. For other mutants, namely Hfq-CTR S80A,S81A; Hfq-CTR R66A; Hfq-CTR S65A,S69A,S72A; Hfq-CTR S93A,S98A; Hfq-CTR H70A,H71A,H84A,H85A and Hfq-CTR G76A, G77A, G78A no complex is formed, titration curve was flat and not shown, in agreement with EMSA result.
2.6. Synchrotron radiation circular dichroism (SRCD)
SRCD measurements were carried out on DISCO beamline at SOLEIL Synchrotron as described in Malabirade et al. [28] (proposals 20,171,061 and 20,200,007). Samples (∼ 4 µl) were loaded into a CaF2 circular cell of 33 µm pathlength [50]. Spectral acquisitions of 1 nm steps at 1.2 s integration time were recorded in triplicate between 320 and 180 nm. (+)-camphor-10-sulfonic acid (CSA) was used to calibrate amplitudes and wavelength positions of the experiment. Data analyses (averaging, baseline subtraction, smoothing, scaling and spectral summations) were carried out with CDtoolX [51]. Spectra are presented in units of mdeg versus nm maintaining the same molar ratios for all presented samples. Due to the origin of absorption, spectra of mixed samples (polynucleotides + peptides) could not be standardized to ∆e.
2.6. Infrared spectroscopy (FTIR)
Infrared (FTIR) spectroscopy analysis was performed as described in Partouche et al. [41]. Briefly, we acquired IR transmission spectra by depositing peptides on a CaF2 surface. The deposits were then dried at room temperature. For each deposit, spectra were acquired at different positions using a Thermo Scientific IN10 infrared microscope (Villebon sur Yvette, France). Infrared absorption measurements were recorded between 4000 and 400 cm−1 with 128 scans, but only the region between 1750 and 1550 cm−1 was analyzed and compared for the different peptides.
3. Results
3.1. Binding of Hfq-CTR mutants to DNA
We first analyzed the interaction of the Hfq-CTR mutant peptides with DNA using EMSA. Note we didn't evaluate the effect of individual amino acids properties, but focused on the effect of short regions of the protein that include specific aa of interest.
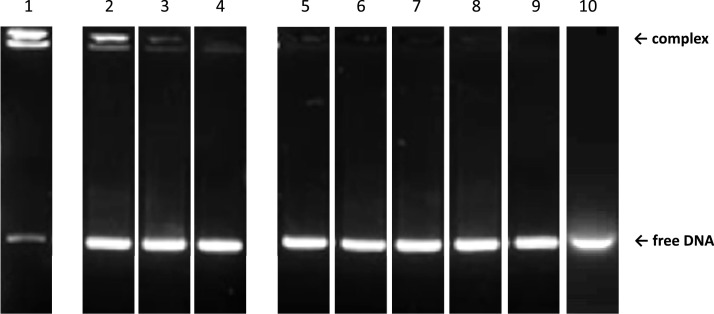
The EMSA results are shown on Fig. 1. A clear band shift is observed in the case of the Wild Type (WT) Hfq-CTR, confirming a significant binding. Nevertheless, EMSA is not adapted to measure KD in solution and migration in the gel matrix often influences the stability of peptide-DNA complex [52]. Furthermore, we also noticed that increasing CTR concentration to titrate DNA results in very high molecular weight complexes (possibly (CTRn:AT59)n) that stay in the well of the gel and cannot be quantified accurately (Sup Fig S2). We thus used anisotropy fluorescence measurements to determine accurately equilibrium dissociation constants KD of the various complexes (Fig. 2). Anisotropy fluorescence measurements gave a KD value of 260 ± 10 nM for CTR, a value in agreement with previous reports [24, 43]). We tested the effect of salt addition on the stability of this complex (100 mM and 300 mM NaCl) and observed only a slightly better affinity at 100 mM (KD = 240 ± 11 nM vs 260 ± 10 nM), while increasing NaCl concentration to 300 mM reduces the stability of the complex, in agreement with previous reports [53, 54] (Sup. Fig. S1, KD = 455 ± 22 nM). Addition of 1 mM MgCl2 does not change KD (259 ± 9 nM, Sup. Fig. S1). This is however not surprising as DNA and peptide solution used to form the complex contain traces of Mg2+ (no EDTA has been added to our samples). The mutant Hfq-CTR S87A,S88A binds dsDNA with an affinity slightly lower than WT Hfq-CTR (KD = 460 ± 30 nM, Fig. 2). The mutants Hfq-CTR Y83A and Hfq-CTR E99A,E100A,E102A bind DNA but to with a significantly lower affinity than WT Hfq-CTR (KD = 1000 ± 75 nM and 890 ± 80 nM, respectively, Fig. 2). Note that mutating D97A in addition to E99A,E100A,E102A has the same effect (sup. Fig. S3). Finally, no band shift was observed upon addition of Hfq-CTR S80A,S81A, Hfq-CTR S65A,S69A,S72A, Hfq-CTR S93A,S98A, Hfq-CTR H70A,H71A,H84A,H85A, Hfq-CTR R66A mutants and Hfq-CTR G76A,G77A,G78A, indicating that these mutants do not bind DNA, at least to a degree which can be detected with the EMSA and anisotropy assays. The same result was obtained for the double mutants Hfq-CTR H70A,H71A and Hfq-CTR H84A,H85A than with Hfq-CTR H70A,H71A,H84A,H85A (Sup. Fig. S3). This indicates that both histidine repetitions are important for DNA binding.
Fig. 1.
EMSA analysis of Hfq-CTR wild type (WT) and mutants in the presence of DNA. Lane 1: Control, Hfq-CTR WT (this control was reprinted with permission from Biomacromolecules 2020, 21, 3668–3677. Copyright 2020 American Chemical Society); lane 2: Hfq-CTR S87A,S88A; lane 3: Hfq-CTR E99A,E100A,E102A; lane 4: Hfq-CTR Y83A. In these case a complex is formed with the peptide. Note that the complex, when it forms, migrates on the top of the gel (but not in the well that is not visible here), indicating that numerous peptides are bound to DNA. Sometimes 2 complexes of different size are present, probably corresponding to different numbers of CTRs bound to DNA [43]. Taking into account the bridging properties of the CTR [38], we suspect these two bands could be (CTRn:AT59) and (CTRn:AT59)2, where 2 (CTRn:AT59) are bridged. Lane 5: Hfq-CTR S80A,S81A; Lane 6: Hfq-CTR R66A; Lane 7: Hfq-CTR S65A,S69A,S72A; Lane 8: Hfq-CTR S93A,S98A; Lane 9: Hfq-CTR H70A,H71A,H84A,H85A; Lane 10: Hfq-CTR G76A, G77A, G78A. In these cases no complex is formed.
We used EMSA to confirm that the monomeric form of the peptide interacts with DNA and then polymerize on it (Fig. 1), but we also confirm that this interaction occurs with pre-polymerized peptide (Sup. Fig. S4). Due to the aggregation of the peptide, it was not possible in this case to measure KD using fluorescence anisotropy. The EMSA result suggests that the amyloid form interacts with DNA with an affinity probably similar to that of the monomeric peptide binding (Sup. Fig. S4). As explained previously, the KD cannot de measured accurately using EMSA. Nevertheless, we conclude that although the amyloid-DNA interaction promotes amyloid formation, the cross-β structure does not seem to significantly enhance DNA binding in the case of Hfq-CTR.
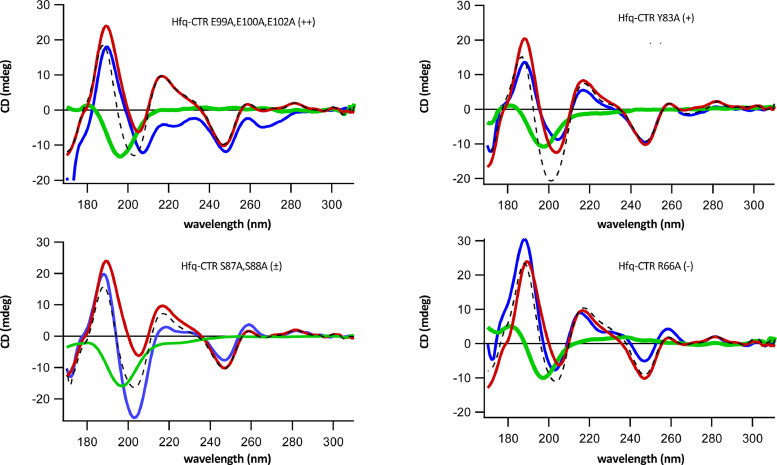
Then the SRCD spectra pertaining to WT-CTR and mutants have been investigated. Fig. 3 shows how the different CTR influence DNA structure (see also Sup Fig. S5). Spectra of the individual components are measured with equivalent DNA and CTR concentrations. Significant spectral changes in the 220 nm region and 260–280 nm spectral band correlate with amyloid formation or base-tilting and base-pairing of AT sequences, respectively [28, 42]. In contrast, when the addition of CTR mutant to DNA induces no significant difference in the SRCD spectrum, this means the interaction between Hfq-CTR and DNA may be inexistent or that the interaction occurs but does not result in a structural change. In these cases, the experimentally observed spectrum is close to the recomposed spectrum obtained from the combination of the individual components. Four main behaviors are observed: (i) such as for Hfq-CTR E99A,E100A,E102A that binds DNA and significantly influences DNA structure (similarly to Hfq-CTR WT as shown in Malabirade et al. [28]); (ii) such as for Hfq-CTR Y83A that influences DNA structure but to a lesser extent. (iii) such as Hfq-CTR S87A,S88A which bind DNA but does not drastically influence its structure; and finally (iv) such as Hfq-CTR R66A, Hfq-CTR S65A,S69A,S72A, Hfq-CTR S80A,S81A, Hfq-CTR S93A,S98A, Hfq-CTR G76A, G77A, G78A and H70A,H71A,H84A,H85A that do not bind dsDNA and thus do not influence its structure. An example of each behavior is shown below.
Fig. 3.
SRCD spectra of the CTR/DNA complex (blue), DNA (red) and CTR (green). Spectra of the individual components are measured with equivalent DNA and CTR concentrations. The dotted spectrum represents the relevant combination of the spectra pertaining to the individual components. (++), (+), (±) and (-) indicate the four main behaviors observed. An example of each behavior is presented. For instance for Hfq-CTR E99A,E100A,E102A/DNA spectra (top left corner, (++), a clear different spectrum is measured (blue) compared to the theoretical addition of the peptide and DNA spectra (dotted line). The same analysis for all other Hfq-CTR mutants with (-) behaviors are presented in Sup. Fig. S5. The qualitative comparison considering amplitude and peak position shifts between theoretical and measured spectra always correlates with EMSA and FTIR results.
3.2. Self-assembly of Hfq-CTR mutants
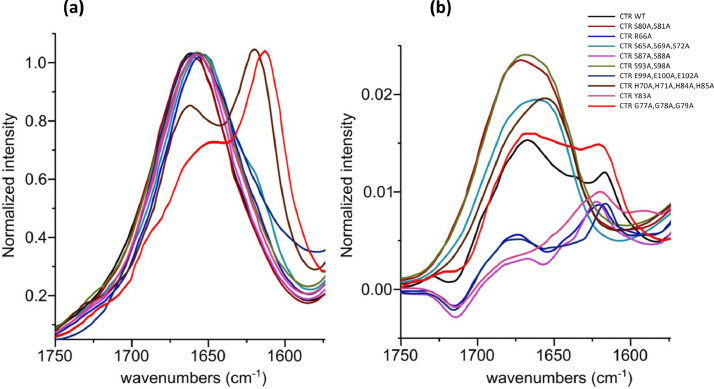
We finally analyzed the amyloid assembly of the CTRs using Fourier Transform InfraRed (FTIR) spectroscopy (Fig. 4). A peak around 1630–1640 cm−1 is indicative of intramolecular β-sheets, while intermolecular hydrogen-bonding in a cross-β structure in amyloids induces a shift to lower wavenumbers and an absorption band around 1620 cm−1 [55, 56]. In Fig. 4, we clearly observe this band at ∼ 1620 cm−1 for some mutants. The analysis was performed in the presence or absence of DNA. We can conclude that in the absence of DNA, the peptides that self-assemble spontaneously are Hfq-CTR R66A, Hfq-CTR S65A,S69A,S72A and Hfq-CTR H70A,H71A,H84A,H85A, and Hfq-CTR G76A,G77A,G78A (Fig 4a). The spontaneous self-assembly of Hfq-CTR H70A,H71A,H84A,H85A at a higher concentration than that used in Sup. Fig. S5 was confirmed by SRCD analysis, with a strong signal at 220 nm (not shown). Note also for the mutant Hfq-CTR G76A,G77A,G78A the shift of the peak at ∼1620 cm−1 to a slightly lower wavenumber, indicating a strengthening of the amyloid structure [55]. No clear spontaneous self-assembly was observed for CTR-WT, as expected [28]. This indicates that the mutations of the hydrophilic aa R66, S65, S69, S72, H70, H71, H84, H85 and even more for G76, G77, G78 into more hydrophobic alanine stabilize the amyloid assembly [57].
Fig. 4.
FTIR spectra of Hfq-CTR in the presence or absence of DNA. (a) peptides alone. (b) difference spectrum obtained by subtracting the (dA:dT)59 contribution from the complex spectrum. We clearly observe in the Amide I band a contribution at ∼ 1620 cm−1, indicative of the formation of the amyloid structure in the presence of DNA.
In the presence of DNA, the peptides that self-assemble are Hfq-CTR WT, Hfq-CTR S87A,S88A, Hfq-CTR R66A, Hfq-CTR Y83A, Hfq-CTR G76A,G77A,G78A and Hfq-CTR E99A,E100A,E102A (Fig 4b). Curiously, Hfq-CTR S65A,S69A,S72A and Hfq-CTR H70A,H71A,H84A,H85A do not seem to self-assemble on DNA while they self-assemble spontaneously. The concentration used to test self-assembly in the absence of DNA was however higher (∼ 3x).
4. Discussion
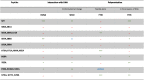
The properties of the various Hfq-CTR peptides analyzed are summarized in table 1. We confirm that the self-assembly and DNA binding properties of Hfq involves the same region of the protein, outside of the Sm ring, and show that some aa residues are involved in both DNA binding and amyloid assembly properties. Thus, a crosstalk between amyloid aggregation and DNA binding probably exists in Hfq, which explains that DNA promotes amyloid assembly [28]. Although the amyloid-DNA interaction promotes amyloid formation, the cross-β structure does not significantly enhance DNA binding in the case of Hfq-CTR. This differs from other amyloids [58].
Table 1.
Summary of mutant properties in the CTR amyloid region of Hfq.
 |
Note that if amyloidogenesis requires time in vitro, in contrast the complex formation is almost instantaneous. The difference in time scales for division and amyloidogenesis could appear surprising, but it must be stressed that in vitro conditions are different to the conditions found in vivo. Precisely (i) the CTR here is not bound to the NTR and this influences the process of self-assembly; (ii) possible cofactors that may accelerate amyloidogenesis such as other proteins are absent, and (iii) more important the conditions for crowding and confinement in vitro have not been taken into account yet. Thus, we strongly believe the amyloidogenesis goes faster in vivo and that it is compatible with cell division time.
4.1. Aminoacids involved in peptide:nucleic acid interactions
Protein interactions with nucleic acids are based on hydrogen bonding, electrostatic contacts, and hydrophobic interactions. In our previous work, we suspected that hydrogen-bonding with the relatively abundant histidine and serine residues of Hfq-CTR could play a role in the binding of the CTR to DNA [38]. With this work we confirm that the histidines repetitions in the CTR of Hfq are critical for both DNA binding, amyloid self-assembly, and consequently for the conformational change in the DNA structure.
We also identify that the positively charged arginine 66 plays a role in DNA binding. Nucleic acids usually promote amyloid formation from short basic sequences [58]. This could also apply to Hfq-CTR, but only one arginine is present and charged positively at pH ∼ 5 (Hfq-CTR pI = 5.6, thus at pH 5 the CTR exhibits 4 negatively charged residues vs one positively charged residue; charges on histidines should be only partial). Note that positively charged residues are more prevalent in pathological amyloids than in functional amyloids and this seems to apply to Hfq-CTR [59].
This CTR region is particularly rich in serines, and we identify that the serine repetition S80,81 is involved in DNA binding, but not the repetition S87,88. Serines-rich regions including serine 65,69 72, 93 and 98 also play a role in DNA binding.
Finally, we conclude that mutations of Y83 and S87,88 may not be involved directly in DNA binding, but that they abolish partially the structural change of DNA induced by the CTR. We suspect that the tyrosine could play a role in base stacking, as already observed for other nucleic-acid binding proteins [60], while the S87,88 repetion may change DNA conformation due to the formation of H-bonds with the nucleic acid. The possibility that interactions between peptides and DNA could lead to the formation of amyloid:DNA fibres whose properties are distinct from peptidic-only fibers are currently investigated.
4.2. Aminoacids involved in self-assembly
We already identified that a 11 aa sequence is the minimal region necessary to form the amyloid structure (underlined):
SRPVSHHSNNAGGGTSSNYHHGSSAQNTSAQQDSEETE [28].
We hypothetize this region of 11 aa forms a “steric zipper”, i.e. pairs of self-complementary β-sheets formed by short sequences found in amyloids [61]:
Nter-SAQNTSAQQDS-Cter
Cter-SDQQASTNQAS-Nter
In these “zippers”, pairs of asparagines and glutamines can form hydrogen bonds along the fibrils. Interestingly, these steric zippers are thermostable and this applies to Hfq-CTR (Tm = 62.0 °C for the 11 aa region and ∼80 °C for 38 aa Hfq-CTR [62, 63]).
Here we observe that the replacement of the hydrophilic R66, S65, S69, S72 and even more for the H70, H71, H84 and H85 into the hydrophobic alanine [57] results in a stronger self-assembly. Indeed, a high prevalence of aliphatic residues (valine, alanine, leucine and isoleucine) was reported for pathological and bacterial amyloids [59]. Note that these aa (indicated in bold above) are not very close to the 11 aa “zipper”. But it is likely these R66, S65, S69, S72, H70, H71, H84 and H85 are close to “zipper” in the tri-dimensional structure of the CTR when it folds and that changing them into an hydrophobic aa results in a more stable structure.
Interestingly, Hfq-CTR presents a sequence that is characteristic of functional amyloids [59]. Indeed, a prevalence of glycines (∼ 18%), serines (∼ 10%) and asparagine (∼ 7%) is observed for functional amyloids [59]. This prevalence is also observed for Hfq-CTR with ∼ 8% of glutamine, ∼ 10% of glycines and ∼ 24% of serines. This prevalence of glycines and serines was proposed to be involved in the formation of hydrophilic and flexible interfaces necessary to have reversible fibrils. This contrasts with irreversible aggregation of pathological amyloids mediated by hydrophobic residues [59]. Note also that functional amyloids in bacteria contain a particularly high prevalence of alanine, asparagine and threonine. This also applies to Hfq-CTR with ∼ 10% of asparagines, ∼ 8% of alanines and ∼ 8% of threonines. As shown with its GRAVY index (Fig. 4), this applies to Hfq-CTR that contains ∼ 13% of hydrophobic uncharged residues (blue, F, I, L, M, V, W, A, P), ∼ 13% of hydrophilic basic residues (red, R, K, H), ∼ 11% of hydrophilic acidic residues (purple, D, E) and ∼ 63% of hydrophilic uncharged residues (green G, S, T, C, N, Q, Y).
In the presence of DNA, we identified that the mutation of tyrosine 83, serines 87,88 and glutamate 99,110 and 102 into alanine does not abolish the self-assembly property of the CTR on DNA, while the mutations of serines 80,81, and 93,98 abolish it, regardless of the presence or absence of DNA.
The two mutated Hfq-CTR that give the strongest amyloid signals in IR for the protein alone (Hfq-CTR S65A,S69A,S72A and Hfq-CTR H70A,H71A,H84A,H85A) no longer form amyloid in contact with DNA. Nevertheless, these mutants do not interact with DNA and the concentration used to test their self-assembly alone was higher than that used in the presence of DNA.
4.3. Crosstalk between amyloid aggregation and dna binding
In the sequence below, we indicate which amino acid residues are presumably involved in DNA binding (in red), in amyloid assembly (highligthed in yellow) and in DNA structural change (highligthed in blue).
![]()
As shown, some aa are involved in both DNA binding and in the formation of the amyloid structure (red highlighted in yellow). Our results thus indicate that Hfq-CTR binds to DNA mainly with serine and histidine tracks which may form H-bonds with DNA, as expected [28]. The interaction with DNA also occurs thanks to the positive arginine 66, well conserved among Hfq orthologs [64] (Fig. 5). This arginine probably interacts with the negative charge of DNA phosphate. On the other hand, the hydrophobic tyrosine 83, even if it induces a conformational change of DNA when bound to the protein, is not involved directly in DNA binding and only partially conserved among various Hfqs. Note that residues 83–86 have been shown to be motionally restricted, and proposed to serve as binding sites for incoming RNAs [65].
Fig. 5.
Multiple sequence alignment of various bacterial Hfq CTRs. E. coli Hfq CTR aa presumably involved in DNA binding are indicated in red, in amyloid assembly in yellow and in DNA structural change in cyan (see 4.3). For GRAVY index hydrophobic uncharged residues are in blue, hydrophilic basic in red, hydrophilic acidic in purple and hydrophilic uncharged in dark green. The CTR regions are variable in length among bacteria and only those with a CTR comparable to that of E. coli Hfq are shown. In the alignment, strictly conserved R66 is indicated in hot pink; aa conserved in most Hfqs CTR are indicated in light pink. Other aa are indicated in yellow and green.
Finally, it has been shown that the Alzheimer's Aβ also binds to DNA and that a consensus sequence KGGRKTGGGG is important for this property. This peptide-DNA interaction is sequence specific and mutations in the G-repetition abolishes peptide-DNA interaction [66]. Here we show that Hfq has also a repetition of G that is partially conserved and that play a role in DNA binding. Indeed, mutating this repetition of G abolishes DNA interaction and has also been shown to have a function in bacterial physiology [67]. Nevertheless, according to the type of interaction involved in the formation of the complex (hydrogen-bonding [38]), we suspect a more important role for the histidine and serine residues in DNA interaction. The Glycine repetition may rather give a conformational freedom to this region, allowing interaction with DNA of other amino acid residues that makes H-bonds with DNA.
As for the conserved acidic tail of the CTR (D97SE99E100TE102), that has been described to compete against non-specific RNA and could mimic nucleic acid binding to the NTR ring [64], our experiments here tend to show that it is probably not involved in DNA binding. This was however expected due to the negative charges of these aa.
5. Conclusions
The evidence for a role of amyloids in DNA-related processes is more and more obvious [2]. This is for instance the case of bacterial amyloids such as in curli that may induce immune response when bound to DNA [68] or of the RepA protein that may influence DNA replication [69]. But this also applies to human amyloids responsible of neurological disorders, such as Alzheimer's Aβ. Indeed, these amyloids bind apparently non-specifically to DNA and have been reported inside the nucleus [4]. Amyloid toxicity may thus be also the consequence of aberrant DNA:amyloid interaction, affecting normal genetic expression [70]. Here we analyze in details the interaction of a bacterial amyloid, the Hfq C-terminal region with DNA. We identify important amino acids involved both in DNA binding and polymerization properties, and show that there is a crosstalk between these two processes. In particular we show that histidines, serines and glycines-tracts are particularly important for the amyloid:DNA interaction and self-assembly. This thus shed a new light on residues that may have a functional role in other amyloids, and that my influence their function and/or toxicity.
Furthermore, Hfq is also well known to interact with RNA. Even if the interaction of Hfq-CTR with RNA is still controversial [39, 71, 72], it may occur as in the case of a reported interaction between PrP (the prion protein) and RNA indicating that RNA may trigger PrP assembly [73]. Taking into account the important role of Hfq in RNA metabolism, this property should be analyzed further in the future. Finally, similarly to Alzheimer's Aβ, cross β sheet-induced DNA condensation could be inhibited by divalent ions, such as Cu2+ and Zn2+ [74]. Indeed, due to these histidines repetition, Hfq naturally binds to divalent ion, and for instance Ni2+ used to purify His-tagged proteins [75]. This property may also drastically influence self-assembly, DNA/RNA binding properties and answer to stresses, such as oxidative stress. This however needs to be investigated.
Author contributions
Florian Turbant: Investigation, Formal analysis, Methodology, Software. Omar El Hamoui: Investigation, Formal analysis, Methodology, Software. David Partouche: Investigation, Formal analysis, Methodology, Software. Christophe Sandt: Investigation, Formal analysis, Methodology, Software, Supervision, Writing – review & editing. Florent Busi: Writing – review & editing, Formal analysis, Visualization. Frank Wien: Data curation, Writing – review & editing, Formal analysis, investigation, methodology, Software, Supervision, Validation, Visualization. Véronique Arluison: Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Methodology, Resources, Funding acquisition, Project administration, Validation, Visualization.
Funding
This work was supported by CNRS, CEA, synchrotron SOLEIL. SRCD measurements on DISCO beamline at the SOLEIL Synchrotron were performed under proposals 20171061 and 20200007. This study contributes to the IdEx Université de Paris ANR-18-IDEX-0001. This work is supported by a public grant overseen by the French National research Agency (ANR) as part of the « Investissements d'Avenir » program, through the "ADI 2021″ project funded by the IDEX Paris-Saclay, ANR-11-IDEX-0003–02
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Jehan Waeytens (ULB, Bruxelles) for many fruitful discussions and to Marianne Bombled (LLB, CEA Saclay) for technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2021.100029.
Contributor Information
Frank Wien, Email: frank.wien@synchrotron-soleil.fr.
Véronique Arluison, Email: veronique.arluison@u-paris.fr.
Appendix. Supplementary materials
Supplementary Materials: The following are available online. Sup Figure S1: KD measurements of WT Hfq-CTR:dsDNA complexes using fluorescence anisotropy in the presence of 100 mM NaCl, 300 mM NaCl and 1 mM MgCl2. Sup Figure S2: EMSA analysis of (dA:dT)59 titration by Hfq-CTR. Sup Figure S3: EMSA analysis of additional Hfq-CTR mutants in the presence of DNA. Sup Figure S4: EMSA analysis of pre-polymerized Hfq-CTR in the presence of DNA. Sup Figure S5: SRCD spectra of other CTR/DNA complexes
References
- 1.Nandi P.K., Leclerc E., Nicole J.C., Takahashi M. DNA-induced partial unfolding of prion protein leads to its polymerisation to amyloid. J. Mol. Biol. 2002;322:153–161. doi: 10.1016/s0022-2836(02)00750-7. [DOI] [PubMed] [Google Scholar]
- 2.Stewart K.L., Radford S.E. Amyloid plaques beyond Abeta: a survey of the diverse modulators of amyloid aggregation. Biophys. Rev. 2017;9:405–419. doi: 10.1007/s12551-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordeiro Y., Macedo B., Silva J.L., Gomes M.P.B. Pathological implications of nucleic acid interactions with proteins associated with neurodegenerative diseases. Biophys. Rev. 2014;6:97–110. doi: 10.1007/s12551-013-0132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barucker C., Harmeier A., Weiske J., Fauler B., Albring K.F., Prokop S., Hildebrand P., Lurz R., Heppner F.L., Huber O., Multhaup G. Nuclear translocation uncovers the amyloid peptide Abeta42 as a regulator of gene transcription. J. Biol. Chem. 2014;289:20182–20191. doi: 10.1074/jbc.M114.564690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barucker C., Sommer A., Beckmann G., Eravci M., Harmeier A., Schipke C.G., Brockschnieder D., Dyrks T., Althoff V., Fraser P.E., Hazrati L.N., George-Hyslop P.S., Breitner J.C., Peters O., Multhaup G. Alzheimer amyloid peptide abeta42 regulates gene expression of transcription and growth factors. J. Alzheimers Dis. 2015;44:613–624. doi: 10.3233/JAD-141902. [DOI] [PubMed] [Google Scholar]
- 6.Colas J., Chessel N., Ouared A., Gruz-Gibelli E., Marin P., Herrmann F.R., Savioz A. Neuroprotection against Amyloid-beta-Induced DNA Double-Strand Breaks Is Mediated by Multiple Retinoic Acid-Dependent Pathways. Neural Plast. 2020;2020 doi: 10.1155/2020/9369815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman S. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019;294:11685–11700. doi: 10.1074/jbc.REV119.005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luijsterburg M.S., Noom M.C., Wuite G.J., Dame R.T. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Skoko D., Yoo D., Bai H., Schnurr B., Yan J., McLeod S.M., Marko J.F., Johnson R.C. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman C.J. Nucleoid-associated proteins and bacterial physiology. Adv. Appl. Microbiol. 2009;67:47–64. doi: 10.1016/S0065-2164(08)01002-2. [DOI] [PubMed] [Google Scholar]
- 11.Azam T.A., Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 12.Cagliero C., Grand R.S., Jones M.B., Jin D.J., O’Sullivan J.M. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic. Acids. Res. 2013;41:6058–6071. doi: 10.1093/nar/gkt325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhawale A., Bindal G., Rath D., Rath A. DNA repair pathways important for the survival of Escherichia coli to hydrogen peroxide mediated killing. Gene. 2021;768 doi: 10.1016/j.gene.2020.145297. [DOI] [PubMed] [Google Scholar]
- 14.Vogel J., Luisi B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cech G.M., Szalewska-Palasz A., Kubiak K., Malabirade A., Grange W., Arluison V., Wegrzyn G. The Escherichia Coli Hfq Protein: an Unattended DNA-Transactions Regulator. Front Mol Biosci. 2016;3:36. doi: 10.3389/fmolb.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobrero P., Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman S., McCullen C.A., Guillier M., Vanderpool C.K., Majdalani N., Benhammou J., Thompson K.M., FitzGerald P.C., Sowa N.A., FitzGerald D.J. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storz G., Opdyke J.A., Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Kavita K., de Mets F., Gottesman S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018;42:53–61. doi: 10.1016/j.mib.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Morita T., Maki K., Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Lay N., Schu D.J., Gottesman S. Bacterial small RNA-based negative regulation: hfq and its accomplices. J. Biol. Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folichon M., Arluison V., Pellegrini O., Huntzinger E., Regnier P., Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic. Acids. Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geinguenaud F., Calandrini V., Teixeira J., Mayer C., Liquier J., Lavelle C., Arluison V. Conformational transition of DNA bound to Hfq probed by infrared spectroscopy. Phys. Chem. Chem. Phys. 2011;13:1222–1229. doi: 10.1039/c0cp01084g. [DOI] [PubMed] [Google Scholar]
- 25.Takada A., Wachi M., Kaidow A., Takamura M., Nagai K. DNA binding properties of the hfq gene product of Escherichia coli. Biochem. Biophys. Res. Com. 1997;236:576–579. doi: 10.1006/bbrc.1997.7013. [DOI] [PubMed] [Google Scholar]
- 26.Diestra E., Cayrol B., Arluison V., Risco C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS One. 2009;4:e8301. doi: 10.1371/journal.pone.0008301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orans J., Kovach A.R., Hoff K.E., Horstmann N.M., Brennan R.G. Crystal structure of an Escherichia coli Hfq Core (residues 2-69)-DNA complex reveals multifunctional nucleic acid binding sites. Nucleic. Acids. Res. 2020;48:3987–3997. doi: 10.1093/nar/gkaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malabirade A., Partouche D., El Hamoui O., Turbant F., Geinguenaud F., Recouvreux P., Bizien T., Busi F., Wien F., Arluison V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018;8:16792. doi: 10.1038/s41598-018-35060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber S. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr. Opin. Microbiol. 2014;22:102–110. doi: 10.1016/j.mib.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Qin L., Erkelens A.M., Ben Bdira F., Dame R.T. The architects of bacterial DNA bridges: a structurally and functionally conserved family of proteins. Open Biol. 2019;9 doi: 10.1098/rsob.190223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiggins P.A., Dame R.T., Noom M.C., Wuite G.J. Protein-mediated molecular bridging: a key mechanism in biopolymer organization. Biophys. J. 2009;97:1997–2003. doi: 10.1016/j.bpj.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parekh V.J., Niccum B.A., Shah R., Rivera M.A., Novak M.J., Geinguenaud F., Wien F., Arluison V., Sinden R.R. Role of Hfq in Genome Evolution: instability of G-Quadruplex Sequences in E. coli. Microorganisms. 2019;8 doi: 10.3390/microorganisms8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilusz C.J., Wilusz J. Lsm proteins and Hfq: life at the 3′ end. RNA Biol. 2013;10:592–601. doi: 10.4161/rna.23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mura C., Randolph P.S., Patterson J., Cozen A.E. Archaeal and eukaryotic homologs of Hfq: a structural and evolutionary perspective on Sm function. RNA Biol. 2013;10:636–651. doi: 10.4161/rna.24538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Link T.M., Valentin-Hansen P., Brennan R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan R.G., Link T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher M.A., Pearson R.F., Moller T., Valentin-Hansen P., Brennan R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malabirade A., Jiang K., Kubiak K., Diaz-Mendoza A., Liu F., van Kan J.A., Berret J.F., Arluison V., van der Maarel J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic. Acids. Res. 2017;45:7299–7308. doi: 10.1093/nar/gkx431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arluison V., Folichon M., Marco S., Derreumaux P., Pellegrini O., Seguin J., Hajnsdorf E., Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 40.Fortas E., Piccirilli F., Malabirade A., Militello V., Trepout S., Marco S., Taghbalout A., Arluison V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015;35 doi: 10.1042/BSR20140128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partouche D., Militello V., Gomesz-Zavaglia A., Wien F., Sandt C., Arluison V. In situ characterization of Hfq bacterial amyloid: a Fourier-transform infrared spectroscopy study. Pathogens. 2019;8 doi: 10.3390/pathogens8010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wien F., Martinez D., Le Brun E., Jones N.C., Vronning Hoffmann S., Waeytens J., Berbon M., Habenstein B., Arluison V. The Bacterial Amyloid-Like Hfq Promotes In Vitro DNA Alignment. Microorganisms. 2019;7 doi: 10.3390/microorganisms7120639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Hamoui O., Yadav I., Radiom M., Wien F., Berret J.-.F., van der Maarel J.R.C., Arluison V. Interactions between DNA and the Hfq Amyloid-like Region Trigger a Viscoelastic Response. Biomacromolecules. 2020;21:3668–3677. doi: 10.1021/acs.biomac.0c00747. [DOI] [PubMed] [Google Scholar]
- 44.Malabirade A., Morgado-Brajones J., Trepout S., Wien F., Marquez I., Seguin J., Marco S., Velez M., Arluison V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017;7:10724. doi: 10.1038/s41598-017-11157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahrenburg S., Karow A.R., Garidel P. Buffer-free therapeutic antibody preparations provide a viable alternative to conventionally buffered solutions: from protein buffer capacity prediction to bioprocess applications. Biotechnol. J. 2015;10:610–622. doi: 10.1002/biot.201400531. [DOI] [PubMed] [Google Scholar]
- 46.Wilks J.C., Slonczewski J.L. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 2007;189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehler E.L., Fuxreiter M., Simon I., Garcia-Moreno E.B. The role of hydrophobic microenvironments in modulating pKa shifts in proteins. Proteins. 2002;48:283–292. doi: 10.1002/prot.10153. [DOI] [PubMed] [Google Scholar]
- 48.Urry D.W., Peng S.Q., Parker T.M., Channe Gowda D., Hearris R. Relative Significance of Electrostatic- and Hydrophobic-Induced pKa Shifts in a Model Protein: the Aspartic Acid Residue. Angewandte Chemie. 1993;32:1440. [Google Scholar]
- 49.Le Brun E., Arluison V., Wien F. Application of Synchrotron Radiation Circular Dichroism for RNA Structural Analysis. Methods in molecular biology. 2020;2113:135–148. doi: 10.1007/978-1-0716-0278-2_11. [DOI] [PubMed] [Google Scholar]
- 50.Wien F., Wallace B.A. Calcium fluoride micro cells for synchrotron radiation circular dichroism spectroscopy. Appl. Spectrosc. 2005;59:1109–1113. doi: 10.1366/0003702055012546. [DOI] [PubMed] [Google Scholar]
- 51.Miles A.J., Wallace B.A. CDtoolX, a downloadable software package for processing and analyses of circular dichroism spectroscopic data. Protein Sci. 2018;27:1717–1722. doi: 10.1002/pro.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fried M.G., Bromberg J.L. Factors that affect the stability of protein-DNA complexes during gel electrophoresis. Electrophoresis. 1997;18:6–11. doi: 10.1002/elps.1150180103. [DOI] [PubMed] [Google Scholar]
- 53.Hwang W., Arluison V., Hohng S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic. Acids. Res. 2011;39:5131–5139. doi: 10.1093/nar/gkr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hopkins J.F., Panja S., McNeil S.A., Woodson S.A. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic. Acids. Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruysschaert J.-.M., Raussens V. ATR-FTIR Analysis of Amyloid Proteins. Methods in molecular biology. 2018;1777:69–81. doi: 10.1007/978-1-4939-7811-3_3. [DOI] [PubMed] [Google Scholar]
- 56.Shivu B., Seshadri S., Li J., Oberg K.A., Uversky V.N., Fink A.L. Distinct beta-sheet structure in protein aggregates determined by ATR-FTIR spectroscopy. Biochemistry. 2013;52:5176–5183. doi: 10.1021/bi400625v. [DOI] [PubMed] [Google Scholar]
- 57.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 58.Braun S., Humphreys C., Fraser E., Brancale A., Bochtler M., Dale T.C. Amyloid-associated nucleic acid hybridisation. PLoS One. 2011;6:e19125. doi: 10.1371/journal.pone.0019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragonis-Bachar P., Landau M. Functional and pathological amyloid structures in the eyes of 2020 cryo-EM. Curr. Opin. Struct. Biol. 2021;68:184–193. doi: 10.1016/j.sbi.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Liebert K., Hermann A., Schlickenrieder M., Jeltsch A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 2004;341:443–454. doi: 10.1016/j.jmb.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 61.Park J., Kahng B., Hwang W. Thermodynamic selection of steric zipper patterns in the amyloid cross-beta spine. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Partouche D., Turbant F., El Hamoui O., Campidelli C., Bombled M., Trepout S., Wien F., Arluison V. Epigallocatechin Gallate Remodelling of Hfq Amyloid-Like Region Affects Escherichia coli Survival. Pathogens. 2018;7 doi: 10.3390/pathogens7040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turbant F., Partouche D., El Hamoui O., Trepout S., Legoubey T., Wien F., Arluison V. Apomorphine Targets the Pleiotropic Bacterial Regulator Hfq. Antibiotics (Basel) 2021;10 doi: 10.3390/antibiotics10030257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santiago-Frangos A., Jeliazkov J.R., Gray J.J., Woodson S.A. Acidic C-terminal domains autoregulate the RNA chaperone Hfq. Elife. 2017;6 doi: 10.7554/eLife.27049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wen B., Wang W., Zhang J., Gong Q., Shi Y., Wu J., Zhang Z. Structural and dynamic properties of the C-terminal region of the Escherichia coli RNA chaperone Hfq: integrative experimental and computational studies. Phys. Chem. Chem. Phys. 2017;19:21152–21164. doi: 10.1039/c7cp01044c. [DOI] [PubMed] [Google Scholar]
- 66.Maloney B., Lahiri D.K. The Alzheimer’s amyloid beta-peptide (Abeta) binds a specific DNA Abeta-interacting domain (AbetaID) in the APP, BACE1, and APOE promoters in a sequence-specific manner: characterizing a new regulatory motif. Gene. 2011;488:1–12. doi: 10.1016/j.gene.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma A., Dubey V., Sharma R., Devnath K., Gupta V.K., Akhter J., Bhando T., Verma A., Ambatipudi K., Sarkar M., Pathania R. The unusual glycine-rich C terminus of the Acinetobacter baumannii RNA chaperone Hfq plays an important role in bacterial physiology. J. Biol. Chem. 2018;293:13377–13388. doi: 10.1074/jbc.RA118.002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tursi S.A., Lee E.Y., Medeiros N.J., Lee M.H., Nicastro L.K., Buttaro B., Gallucci S., Wilson R.P., Wong G.C.L., Tukel C. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giraldo R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17388–17393. doi: 10.1073/pnas.0702006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jimenez J.S. Protein-DNA interaction at the origin of neurological diseases: a hypothesis. J. Alzheimers Dis. 2010;22:375–391. doi: 10.3233/JAD-2010-100189. [DOI] [PubMed] [Google Scholar]
- 71.Olsen A.S., Moller-Jensen J., Brennan R.G., Valentin-Hansen P. C-Terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 2010;404:173–182. doi: 10.1016/j.jmb.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 72.Vecerek B., Rajkowitsch L., Sonnleitner E., Schroeder R., Blasi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic. Acids. Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adler V., Zeiler B., Kryukov V., Kascsak R., Rubenstein R., Grossman A. Small, highly structured RNAs participate in the conversion of human recombinant PrP(Sen) to PrP(Res) in vitro. J. Mol. Biol. 2003;332:47–57. doi: 10.1016/s0022-2836(03)00919-7. [DOI] [PubMed] [Google Scholar]
- 74.Yu H., Ren J., Qu X. Time-dependent DNA condensation induced by amyloid beta-peptide. Biophys. J. 2007;92:185–191. doi: 10.1529/biophysj.106.093559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Milojevic T., Sonnleitner E., Romeo A., Djinovic-Carugo K., Blasi U. False positive RNA binding activities after Ni-affinity purification from Escherichia coli. RNA Biol. 2013;10:1066–1069. doi: 10.4161/rna.25195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials: The following are available online. Sup Figure S1: KD measurements of WT Hfq-CTR:dsDNA complexes using fluorescence anisotropy in the presence of 100 mM NaCl, 300 mM NaCl and 1 mM MgCl2. Sup Figure S2: EMSA analysis of (dA:dT)59 titration by Hfq-CTR. Sup Figure S3: EMSA analysis of additional Hfq-CTR mutants in the presence of DNA. Sup Figure S4: EMSA analysis of pre-polymerized Hfq-CTR in the presence of DNA. Sup Figure S5: SRCD spectra of other CTR/DNA complexes