Highlights
-
•
New principles of receptor and catalytic sites’ formation in MIPs were described.
-
•
Computational modeling provides synthesis receptors with desirable selectivity.
-
•
MIP membranes can serve as a basis for biosensors for “point-of-care” applications.
-
•
Approaches to the improvement of working parameters of biosensors were described.
Keywords: Molecular recognition, Biomimetic polymers, Artificial receptors, Molecularly imprinted polymer membranes, Biosensors, Solid-phase extraction
Abstract
The paper is a self-review of works on development of new approaches to formation of mimics of receptor and catalytic sites of biological macromolecules in the structure of highly cross-linked polymer membranes and thin films. The general strategy for formation of the binding sites in molecularly imprinted polymer (MIP) membranes and thin films was described. A selective recognition of a number of food toxins, endocrine disruptors and metabolites is based on the results of computational modeling data for the prediction and optimization of their structure. A strategy proposed for the design of the artificial binding sites in MIP membranes was supported by the research performed by the authors on development of a number of the MIP membrane-based affinity and catalytic biosensors for selective and sensitive measurement (detection limits 0.3–100 nM) of the target analytes. Novel versatile approaches aimed at improving sensitivity of the developed biosensor systems were discussed.
Graphical abstract
1. Introduction
The capability of both eukaryotes and prokaryotes to react on various external influences, as well as to ensure interactions between the cells, is possible due to phenomenon of molecular recognition. Molecular recognition provides signal transduction between living cells and ensures proper functioning of all signaling pathways, including interactions of receptors with ligands, antibodies with complementary antigens, and enzymes with specific substrates. These special features of antibodies, enzymes, and receptors determine their wide application in modern molecular biology, immunology, medicine, and biotechnology areas. Natural biomolecules are being also widely used for the development of biosensors, which are recognized to be the most effective and versatile methods of analytical biotechnology due to their high selectivity, sensitivity, mobility and low cost [1,2]. Thousands of biosensors for the effective detection of a number of analytes in a real time have been developed by the research groups all over the world. However, there are relatively few examples of their successful commercialization. Largely, this is associated with instability of biomolecules in the external environment.
Therefore, the development of highly stable synthetic biomimetic receptors, which would simulate the active sites of biological macromolecules is of a great interest. The method of molecular imprinting [3,4] is one of the most effective approaches to synthesis of the highly-cross-linked biomimetic polymers with “molecular memory”. That is provided by use of target analytes (so-called template molecules) in the synthetic procedure. They are further removed from the biomimetic polymer using standard extraction protocols in an appropriate solvent, resulting in formation of polymers with “molecular memory” determined by formation of “molecular imprints” of the templates. The special feature of these materials is their ability to recognition of the target analytes (Fig. 1) [3,4].
Fig. 1.
The scheme of MIP synthetic procedure (T- template; 1. Formation of molecular complex between the molecule of interest (“template”) and functional monomers; 2. Polymerization in the presence of cross-linker; 3. Removal of the template; 4. “Imprints” contain functional groups, which are complementary to those of template).
Similarly to antibodies, molecularly imprinted polymers (MIPs) can be obtained against a number of substances of different chemical nature [5], [6], [7], while artificial binding sites in biomimetic polymers are very stable [8] and selective [9]. Biomimics are of great interest for the researchers working in both fundamental and applied science. They are widely used as highly selective stationary phase for separation [10,11] as well as man-made enzymes and antibodies in biochemistry and analytical biotechnology [12], [13].
Even though the method on molecular imprinting was proposed in 1970s, the number of publications on MIP synthesis and application is rapidly growing, reaching more than 5500 papers annually. Most of the currently published works are devoted to synthesis of MIPs in the form of polymer particles for different applications [14,15]. However, in this case, the polymer processing procedure is quite labor-intensive and often inefficient. At the same time, a number of significant technological difficulties arise during their application in biosensor technology, immunoassays, and separation.
Synthesis of MIPs in the form of porous polymer membranes and thin films would allow one to avoid problems associated with the integration of MIPs in biosensor device and their immobilization on the surface of ELISA plates, as well as conducting experiments at extremely high pressures in the case of their application as a stationary phase in separation. On the other hand, even though during the last decades the number of publications in MIP area is increasing, most of them are empirical. In most cases, components for the MIP synthesis are chosen without thorough analysis of the ability of various monomers to form receptor sites simulating active sites of biomacromolecules, as well as their ability to form biomimics where most of the formed selective sites are available for interaction with the target analytes.
The present paper is a self-review devoted to development of versatile approaches to the rational design and synthesis of MIP membranes and thin films using computational modeling methods, which is relevant for both understanding molecular recognition processes and for MIP application in modern molecular biology and analytical biotechnology.
2. Design and synthesis of artificial receptors
2.1. Target analytes
The technique of molecular imprinting is a versatile approach since highly selective artificial receptors can be synthesized for a number of low molecular weight substances with different chemical structure [16], [17], [18], [19], [20], [21], [22]. Quite a few works were also published on MIP development for selective recognition of peptides and proteins [23,24] as well as DNA fragments [25,26]. Our attention was mainly focused on several groups of toxic substances – food toxins produced as secondary metabolites by Aspergillus and Fusarium mold species, endocrine disrupting compounds (triazine herbicides, phenols, pharmaceuticals, etc.), and some metabolites (creatinine) (Fig. 2).
Fig. 2.
Chemical structure of some endocrine disruptors, food toxins and metabolites used as template molecules for the synthesis of artificial binding sites in MIP membranes.
In the case of highly toxic templates, application of a “dummy-template approach” was demonstrated to be much more effective as compared to the natural toxins (aflatoxin B1, zearalenone, etc.) [27,28]. The approach assumes application of nontoxic analogues of the natural toxins that provide formation of the selective receptor sites and, at the same time, decreases the cost of the resulting MIPs significantly [27,28]. Application of dummy templates is also very effective in the case of fluorescent templates since the high background fluorescent signals that are normally observed for aflatoxin B1- and zearalenone-imprinted polymers can be avoided [27,28]. The structures of the dummy-templates used for the synthesis of aflatoxin B1- and zearalenone-selective MIP membranes (ethyl-2-oxoceclopentanecarboxylate and cyclododecyl-2,4-dihydroxybenzoate (CDHB)) as compared to the structure of their natural counterparts are shown in Fig. 3.
Fig. 3.
Chemical structure of dummy templates used for the synthesis of mycotoxin-selective receptor sites in MIP membranes as compared to the structures of aflatoxin B1 and zearalenone: a - ethyl-2-oxocyclopentanecarboxylate, b - cyclododecyl-2,4-dihydroxybenzoate (CDHB); c - aflatoxin В1; d – zearalenone. Structural similarity between ethyl-2-oxocyclopentanecarboxylate and aflatoxin B1 is shown by green color, structural similarity between CDHB and zearalenone is shown by red color.
2.2. Development of methods of MIP synthesis in the form of free-standing membranes and thin films on inert surfaces
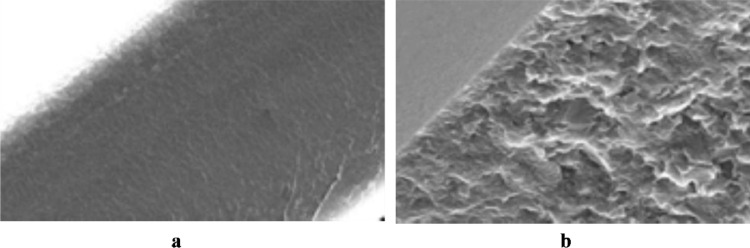
The method of in situ MIP synthesis today remains the most widely used one for obtaining biomimics with different types of selectivity [29]. First of all, this is associated with extremely high degrees of cross-linking of such materials, which ensure that no changes in the structure of artificial binding sites takes place for a long time. Highly-cross-linked MIPs can hardly be obtained in a membrane format due to their high fragility (Fig. 4, a). At the same time, synthesis of MIP membranes combining high degrees of cross-linking with high mechanical stability and capability of generation either optical or electrochemical sensor responses is of great interest for both biosensor technology and other analytical applications.
Fig. 4.
A molecularly imprinted polymer synthesized by the traditional in-situ polymerization method (a), a MIP membrane synthesized in situ using oligourethaneacrylate as a modifier-elastifier (b) and a thin MIP film immobilized on a glass surface (c).
The first works on synthesis of molecularly imprinted polymers membranes were published by Piletsky et al. in 1990s [30,31]. Membranes based on traditional acrylate polymers were obtained by polymerization of a monomer mixture containing a functional monomer (methacrylic acid) and a cross-linker (ethylene glycol dimethacrylate) in the pores of glass filters. In this case, glass filters were used as an inert substrate to be modified by the MIP and ensure its mechanical stability. It was shown that membranes obtained in the presence of adenosine monophosphate were capable of its further selective recognition [30]. Application of diethyl aminoethyl methacrylate as a functional monomer in the synthetic procedure provided selective recognition of l-phenylalanine with the MIP membranes as compared to d-phenylalanine and l- or d-tyrosine [31].
Sergeyeva et al. developed the method of synthesis of highly cross-linked MIPs in a form of flexible membranes with stable physico-chemical properties [32,33]. In these works, difunctional methacrylate - triethylene glycol dimethacrylate (TEGDM) was selected as a cross-linker and the main component of the polymer network (Fig. 5). As compared to ethylene glycol dimethacrylate traditionally used in molecular imprinting, TEGDM provides formation of MIP membranes with sufficient mechanical stability. Further progress in synthesis of the free-standing membranes (Fig. 4, b) as well as thin MIP films immobilized on inert glass surfaces (Fig. 4, c) was achieved by introducing flexible oligomeric fragments (i.e. oligourethaneacrylate - OUA) into the polymer composition (Fig. 5) [32], [33], [34]. Linear oligomer OUA was selected as a modifier-elastifier because of flexibility of its oligomeric chain, relatively simple synthetic procedure, and a possibility to be easily incorporated in the MIP structure through either thermo-initiated or UV-initiated radical co-polymerization. Application of flexible oligomeric fragments as modifiers-elastifiers was proposed by our group for the first time [32,33].
Fig. 5.
Chemical structure of the main components of the MIP membranes obtained by in situ polymerization: cross-linker – triethylene glycol dimethacrylate (a) and modifier-elastifier - oligourethaneacrylate, n = 35 (b).
Accessibility of the artificial binding sites formed in the MIP structure for the further interaction with target analytes is one of the main issues in successful molecular imprinting. Traditionally that is achieved by addition of organic solvents of different polarity to the monomer mixture (chloroform, acetonitrile, etc.). Synthesis of porous OUA-containing MIP membranes was described for the first time by our group using an additional polymeric porogen and synthesis of molecularly imprinted semi-interpenetrating polymer networks (semi-IPNs) [35]. The MIP membranes were formed in the presence of an inert linear polymer, while its molecular weight can vary from 10,000 to 50,000. Significantly more porous structure (Fig. 6) was formed due to thermodynamical incompatibility between the highly cross-linked and linear polymers in the porous MIP membrane. The water fluxes also increased significantly from 0 to ~ 10,000 l m−2 h−1 [30]. Polyethylene glycol (PEG) MW 20,000 and linear polyurethane MW 40,000 were demonstrated to be the most effective polymeric porogens for this purpose [35]. The molecular modeling was used as a tool, which allows to minimize the number of compositions of the highly-cross-linked component (MIP network) in the semi-IPN-based membranes, while the inert linear semi-IPN component plays a role of a polymeric porogen, which doesn't influence the structure of the artificial binding sites. At the same time, addition of the inert linear polymers significantly improved porosity and permeability of the MIP membranes, providing formation of the systems where the increased number of the artificial binding sites are available for the template binding [30,[35], [36], [37]]. Both, rational design and knowledge of how to make the highly-specific IPN are important tools in making the highly-specific and affinity membrane materials.
Fig. 6.
SEM images of cross-section of the MIP membranes synthesized in situ with dimethylformamide (a) and PEG (MW 20,000) (b) as porogens.
An alternative method of porous MIP membranes' synthesis was developed through grafting modification of the surface of industrial microporous membranes by ~ 10 nm-thick MIP films, combining high selectivity of MIPs with high fluxes of the microporous supports [38], [39], [40], [41], [42], [43]. It was shown that the grafted MIP membranes can generate both electrochemical and optical sensor responses, e.g. effective electrochemical detection of triazine herbicides and optical detection of creatinine was demonstrated [39−45]. For instance, a capacitive sensor for desmetryn detection was constructed using grafting modification of Au electrodes by the thin MIP film [44]. In this case, the method of grafting polymerization from aqueous solutions developed for the modification of inert surfaces was used [38,[40], [41], [42], [43], [44]]. Significant changes in the value of the sensor response were observed after addition of the template (desmetryn) in the range 50–850 nM in the electrochemical cell, while negligible sensor responses were caused by addition of structurally similar substances. An effective colorimetric sensor system based on grafted MIP membranes was also developed for creatinine detection in aqueous solutions. The colorimetric sensor response was generated on the MIP membrane surface after creatinine binding in a further reaction with Prussian blue and potassium ferricyanide, which formed brown-colored complexes in alkaline media. Intensity of staining of the MIP membrane was proportional to creatinine concentration in the sample [39].
2.3. Design of artificial receptor sites
The idea of creation of artificial receptor binding sites in biomimetic polymers is based on assumption that molecular complexes between target analytes and functional monomers are formed before the initiation of the synthetic procedure [3,4]. These complexes are then included in the structure of the acrylate/methacrylate polymer network, which ensures fixation of the active groups of the functional monomer in a certain spatial position for a long time. However, it is recognized that the structure of the formed complexes can be changed during the exothermic radical polymerization, while affinity of the resulting binding sites will be mainly determined by the binding energy between target analytes and functional monomers [45], [46], [47]. Traditionally selection of functional monomers is based on literature analysis as well as general considerations as for the ability of different monomers to form complexes with the analyte followed by extensive synthetic work. Combinatorial approach, which assumes synthesis of large numbers of polymers with further screening of their affinity and selectivity was also proposed [45].
An effective computational method for screening potential functional monomers as for their interaction with target analytes was pioneered by Piletsky et al. [46]. The method assumed fast screening of the library consisting of functional monomers traditionally used in molecular imprinting (Fig. 7) with further selection of potential candidates providing the highest binding energies with the target molecule through non-covalent interactions using LEAPFROG algorithm [46,47].
Fig. 7.
Functional monomers traditionally used in non-covalent molecular imprinting capable of interaction with small organic molecules: 1. – acrylonitrile, 2. – ethylene glycol dimethacrylate, 3. – N,N'-methylenebisacrylamide, 4. – 2-vinylpyridine, 5. – styrene, 6. – 2-(trifluoromethyl)-acrylic acid, 7. – methacrylic acid, 8. – m-divinylbenzene, 9. – acrolein, 10. – acrylamide, 11. – acrylic acid, 12. – diethyl aminoethyl methacrylate, 13. – 4-vinylpyridine, 14. – urocanic acid, 15. – p-divinylbenzene, 16. – - hydroxyethyl methacrylate, 17. – 2-acrylamido-2-methyl-1-propanesulfonic acid, 18. – vinylimidazole, 19. – urocanic acid ethyl ester, 20. – allylamine, 21. – itaconic acid.
The modeling of the MIPs is an effective tool, which allows to select the most suitable monomers and minimize the number of compositions before the optimal is selected. It is possible to do modeling of a molecular complex between the functional monomer and template, or even predict a complete monomeric mixture [44,48].
The modeling of the molecular complexes demonstrated in this review was made using a Linux operating system - based computer equipped with Sybyl 7.3 software (Tripos Inc., Missouri, USA). During modeling, the template minimized to 0.01 kcal mol−1 is screened against the functional monomers using a Leapfrog algorithm. The library consists of 30 monomers and cross-linkers that are commercially available and most commonly used by molecular imprinting community. The polymerizable monomers contain various functional groups (neutral, positively and negatively charged, aromatic and hydrophobic) and are able to interact with a template through ionic and hydrogen bonds, van der Waals’, and dipole-dipole interactions. As a result of the Leapfrog simulation, a table of the monomers sorted by the energy of their complexes with template is produced [47]. Typically, the top monomers showing the lowest energy upon interaction with template, are good candidates for imprinting. The molar ratio between the functional monomers and template could also be optimized using a Molecular Dynamic simulation, formerly known as Simulated annealing, when a virtual box is filled with a template and selected monomers packed in a large quantity. The box is then heated and cooled between 700 K and 300 K. As a result, it is possible to see how many molecules of the particular monomer or monomers could bind to one template molecule, allowing to select the optimal composition for the Molecularly Imprinted Polymer synthesis [49].
When functional monomers and a template molecule form multiple point interactions, they generate an affinity-binding site but cross-linker is essential for its stabilization and preservation. A well-selected cross-linker could also minimize a non-specific binding [50].
Molecular dynamics simulation allows visualization of a possible structure of the template-functional monomer complex as well as prediction the number of functional monomers involved in the complex formation and approximate template: functional monomer ratio in the initial mixture of monomers [51].
The effectiveness of this approach was confirmed by numerous works on synthesis of selective MIP adsorbents for microcystin-LR [52], tylosin [53], abacavir [54], biotin [55], ephedrine [47], simazine [56], etc.
All the monomers included in the virtual library of functional monomers, in addition to their ability to form complexes with small organic molecules due to non-covalent interactions, also had unsaturated double bonds necessary for their covalent incorporation into the polymer network (Fig. 7) [52], [53], [54], [55], [56].
The method was proven to be effective for optimization of the MIP membranes’ and thin films’ composition and was comprehensively highlighted in the publications [27,28,36,37,39,41,44,[57], [58], [59]]. It was shown that the effectiveness of the functional monomer for the formation of high affinity sites in MIP membranes and thin films is associated with both binding energy with the analyte as well as the number of its molecules/functional groups participating in the complex formation [27,28,36,37,39,41,44,[57], [58], [59]].
Typical results as for the selection of functional monomer obtained by this method are presented in Fig. 8. It was demonstrated that highly selective artificial binding sites capable of effective recognition of target analytes are formed in the case of participation of at least two molecules/functional groups of a functional monomer in the interaction with the template. On the contrary, participation of only one functional group normally results in formation of either weakly selective or non-selective binding sites in MIP membranes and thin films, which was confirmed by numerous studies [27,28,36,37,39,41,44,[57], [58], [59]].
Fig. 8.
Typical results of computational modeling used for the formation of highly selective artificial receptors for triazine herbicides atrazine (a-c) and desmetryn (d-f) with functional monomers giving the highest binding energy: methacrylic acid (a, e), itaconic acid (b, d), acrylamide (c), 2-acrylamido-2-methyl-1-propansulfonic acid (f).
For example, two molecules of the best functional monomers for atrazine and desmetryn (methacrylic and 2-acrylamido-2-methyl-1-propansulfonic acid, respectively) form complexes with the target analytes (Fig. 8). As a result, the most selective affinity sites in the MIP membranes are formed under their participation, which was confirmed by numerous experimental data [37,44]. Similar situation was observed for the substances, which significantly differ in chemical structure, i.e. creatinine [39], phenol [57] sulfamethoxazole [60], etc.
3. Electrochemical MIP membrane-based sensors
3.1. Electrochemical sensors based on affinity membranes
The selected functional monomers were applied for the synthesis of MIP membranes to be used as a receptor part of the biosensor device. To estimate the effectiveness of the molecular imprinting procedure, all the sensor responses were compared with the responses generated by blank (non-imprinted) polymers obtained from the same mixture of monomers where no template molecules were added and, as a result, no imprinted binding sites were formed [32,33].
Affinity and selectivity of the synthesized MIP membranes towards the target analyte were estimated through evaluation of their electrical conductivity [32,33]. For this purpose, specially constructed electrochemical cell was used and electrical conductivity of the membranes was measured as a function of the analyte concentration. The MIP membranes demonstrated significant changes of their electrical conductivity in response to addition of the target analyte (e.g., triazine herbicide atrazine) in contrast to non-imprinted membranes. The sensor demonstrated fairly good working parameters: detection limit and linear dynamic range achieved for atrazine comprised 5 nmol L−1 and 5–50 nmol L−1, respectively [32,33]. Ability of MIP membranes to bind the template effectively was determined by the type of the functional monomer used in the synthetic procedure, while effectiveness of different functional monomers in formation of highly selective artificial binding sites was in agreement with computational modeling data [37]. The influence of the MIP membrane composition on the ability of the receptor sites to bind the target analyte and generate sensor response was thoroughly investigated. Influence of degree of cross-linking, type and amount of a porogenic solvent were optimized [32,33]. It was shown that polarity of the organic solvent had strong influence on the ability of the MIP membranes to generate conductometric responses. Except for its direct influence on the complex formation in the pre-polymerization mixture, the swelling degrees of the resulting membranes during the analysis should be taken into consideration [32,33]. At the same time, the MIP membranes demonstrated impressive selectivity in binding of atrazine as compared to the other triazine herbicides that was similar to the selectivity of atrazine-selective antibodies. The developed biosensors provided “a real time” analysis within 6–10 min, while their storage stability was estimated as 18 months at room temperature [32,33].
An alternative approach to the development of the MIP-based electrochemical biosensors assumes modification of physical transducer surface with a thin layer of highly-cross-linked MIP by grafting polymerization. The method of modification of inert surfaces from aqueous solutions that is applicable to water-soluble templates [44,61] can be also adapted for modification of gold electrodes. The initiator of photopolymerization is immobilized directly on the surface of the electrode, which is modified with a hydrophobic layer of hexadecanethiol. UV irradiation leads to the transition of the initiator into a triplet state capable of distraction of hydrogen from the substrate surface. This leads to the formation of a radical on the surface of the substrate as well as free radicals, providing synthesis of highly cross-linked polymer chains grafted to the surface of the electrode as it takes place in the case of the inert microfiltration membranes [38,[40], [41], [42]]. The formation of selective receptor sites on the surface of the gold electrode occurs after the extraction of the template as in the case of free-standing MIP membranes. Effective registration of the MIP-analyte binding event was achieved through the development of the capacitive sensor [44]. It was shown that the presence of triazine herbicide desmetryn used as a template in this study caused significant changes of the capacity, proportional to its concentration in the sample [44].
It was confirmed that the type of functional monomer used in the MIP synthesis has a crucial effect on the selectivity of the analyte binding. A pronounced imprinting effect was observed only for polymers synthesized with 2-acrylamido-2-methyl-1-propanesulfonic acid as a functional monomer, which provides high binding energy −90,64 kCal mol−1 with desmetryn along with formation of the bi-functional binding site. The sensor demonstrated low detection limit for desmetryn (100 nM), fast sensor response (10 min), and reasonably high storage stability (12 months) at room temperature [44].
3.2. Towards development of catalytic sensors based on MIPs and MIP membranes
Wide application of MIPs in biosensor technology is restrained by significant difficulties in registration of the MIP-analyte binding event since no electroactive substances are generated. Development of catalytic MIPs capable of highly selective cleavage of target analytes producing electrochemically active substances would provide the possibility of their effective electrochemical detection. Synthesis of catalytic MIPs and MIP membranes possessing tyrosinase activity for the selective recognition of phenols was demonstrated by our group [62,63].
It is widely known that the polyphenol oxidase active center catalyzes oxidation of o-hydroxyphenols to o-quinones, which is accompanied by O2 consumption from the solution (Fig. 9). Two Cu2+ ions in the active center of the enzyme are coordinated by two oxygens of o-hydroxyphenol at a distance ~ 4 Å (Fig. 10) [64].
Fig. 9.
Oxidation of o-hydroxyphenol catalyzed by tyrosinase.
Fig. 10.
Crystal structure of mushroom tyrosinase active center according to Decker et al., 2007 [64].
The possibility of synthesis of both MIP particles and MIP membranes with tyrosinase activity were demonstrated and described in detail in [62,63] by our group. Taking into account the data published by Decker et al. [64], the o-hydroxyphenol-selective sites in both biomimetic catalytic polymers and polymeric membranes were formed using complex of o-hydroxyphenol with two Cu2+ ions as a template, while urocanic acid ethyl ester was selected as a functional monomer due to the presence of imidazole moieties in its structure similarly to imidazole rings present in the natural catalytic center [62], [63], [64]. The free-standing catalytic MIP membranes were synthesized in situ, while the mixture of TEGDM and OUA was used as a cross-linked component of the PEG 20,000-containing semi-IPN. Their catalytic properties were estimated by measuring changes in O2 concentration in an electrochemical cell using portable laboratory O2-meter and O2-sensitive electrode as a transducer. No catalytic sites were formed in blank membranes used in control experiments. MIP membranes synthesized from a monomer mixture where o-hydroxyphenol was replaced by m-hydroxyphenol were used as another control [62,63]. The developed biomimetic sensor was able to detect o-hydroxyphenols in aqueous solutions within the range 0.063–2.5 мМ [63]. The selectivity of the detection was reasonably high: significantly lower catalytic activity was observed for the sensors based on blank membranes. That was also typical for the MIP membranes synthesized using m-hydroxyphenol as a template molecule. Therefore, changes in the structure of the biomimetic catalytic site lead to a significant decrease in the catalytic activity of biomimetic polymers [62,63]. The developed catalytic tyrosinase-mimicking active sites formed in the MIP structure were able to discriminate between o-hydroxyphenols and the other phenols [62,63]. In contract to biosensors based on natural tyrosinase, storage stability of MIP-based sensors at room temperature was estimated as 12 months. This value exceeds storage stability of the natural enzyme-based devices approximately by 25 times.
4. Optical MIP membrane-based and smartphone-based sensors
The special feature of the MIP membranes synthesized according to the methods developed by our group is their ability to generate not only electrochemical, but also optical sensor responses, which can be recorded either by standard laboratory spectrophotometers/spectrofluorimeters or by using new digital approaches including software for digital image analysis and smartphones for the registration and analysis of the optical signals [27,28,34,39,57,58,60,65].
All the developed optical sensors function as follows. At the first stage, the target analytes are to be recognized by the receptor sites in MIP membranes. We demonstrated an effective detection of the bound molecules after UV-irradiation of the MIP membranes, which initiates native fluorescence of the target analytes further registered and quantified using smartphone [27,28,34,58,66,67]. Development of colorimetric sensor systems was also described for the detection of target analytes capable of color complexes' formation [39,57,60,65].
A colorimetric sensor system for phenol detection within the wide range from 50 nmol L−1 to 10 mmol L−1 was described earlier [57,65]. The colorimetric sensor system was constructed using ability of phenol to form pink complexes with 4-aminoantipyrine, while higher analyte concentrations provided higher colorimetric signals. (Fig. 11) [57,65]. Preferential binding of phenol as compared to its structural analogues by the MIP membranes was confirmed in the selectivity studies [57,65].
Fig. 11.
Formation of colored complexes “phenol-4-aminoantipyrine" on the surface of the computationally-designed MIP membranes (a) and MIP membrane-based sensor system for phenol detection (b).
The effectiveness of computational modeling approach used for the selection of functional monomer was also confirmed: the highest colorimetric responses were observed for the itaconic acid-containing biomimics, which provides the highest binding energies of the complex with the target analyte as compared to other functional monomers [57]. The same colored reaction with 4-aminoantipyrine can be also applied for the detection of bisphenol A [68].
Similar principle was used also for the development of the colorimetric sensor system for sulfamethoxazole and creatinine detection based on computationally-designed MIP membranes. Both types of the developed affinity membranes were used in these studies: microfiltration membranes modified with a thin MIP layer as well as MIP membranes synthesized in situ. Ability of sulfamethoxazole to form brown complexes with Prussian blue and potassium ferricyanide was used for its detection [60], while creatinine could be revealed using Jaffe reaction on the surface of the grafted MIP membrane [39]. In all the cases, the analyte-binding sites formed under participation of at least two molecules of the functional monomer demonstrated the best selectivity, virtually no binding of potential interferents was observed [39,60].
Very sensitive fluorescent sensor systems can be designed on the basis of MIP membranes for a number of fluorescent analytes. An extra advantage of this approach is that no additional reaction is needed for revealing MIP-analyte binding event [27,28,58]. The analytes are detected on the MIP membrane surface after its UV-irradiation, while intensity of fluorescence can be measured using both the spectrofluorimeters (including portable devices) and smartphone cameras as detectors of the sensor responses.
The effective method of aflatoxin B1 detection was proposed recently [27]. Aflatoxin B1 attracted our attention as the most toxic secondary metabolite of Apergillus spp. affiliated to the group of aflatoxins. [27]. Analysis of molecular dynamics data allowed us to form aflatoxin B1-binding sites with impressive selectivity. According to computational modeling data [27], acrylamide was selected as a functional monomer for the formation of the aflatoxin B1-selective binding sites in free-standing MIP membranes synthesized in situ. On the one hand, acrylamide provides sufficient binding energy with aflatoxin B1 (−19.06 kCal mol−1). On the other hand, it ensures formation of the imprinted binding sites highly selective for aflatoxin B1, as it interacts with various parts of the molecules of the target analyte and its structural analogues. Therefore, the imprinted sites formed under participation of the dummy template, which mimics the ketolactone part of aflatoxin B1 would not bind difurane part of the other representatives of the group (aflatoxin G2, etc.). At the same time, analysis of computational modeling data allows one to form artificial binding sites with desirable selectivity: it is possible to get materials either with high selectivity towards the target analyte or with group selectivity, capable of recognition of a group of chemically-related substances [27,59]. For instance, effective group-selective recognition of aflatoxins was demonstrated for N, N’-methylenebisacrylamide-containing MIP membranes synthesized in-situ [59]. The developed optical sensors demonstrated reasonably low (14 nmol L−1) detection limit for aflatoxin B1.
The use of smartphones for the recording and further processing of both electrochemical and colorimetric sensor signals is one of the latest trends in bioanalytical chemistry. This significantly simplifies the analytical procedure and, as a result, can provide point-of-care analysis (either at home or in the field) in real time (within minutes). Smartphone-based biosensors are currently being used for both food [69] and water quality control [70] and in medicine (in particular, for early diagnostics and the prescription of adequate treatment) [71]. Smartphone-based electrochemical glucose-sensors have been already reported [72] and more sophisticated smartphone-based affinity SPR-sensors have been also developed [73].
Due to versatility of MIP membranes regarding generation colorimetric and fluorescent responses, they can be effectively combined with a smartphone equipped with high-resolution camera for recording the optical signals. The commercially available smartphone applications for Android 6+ also provide fast and effective data analysis [66,67].
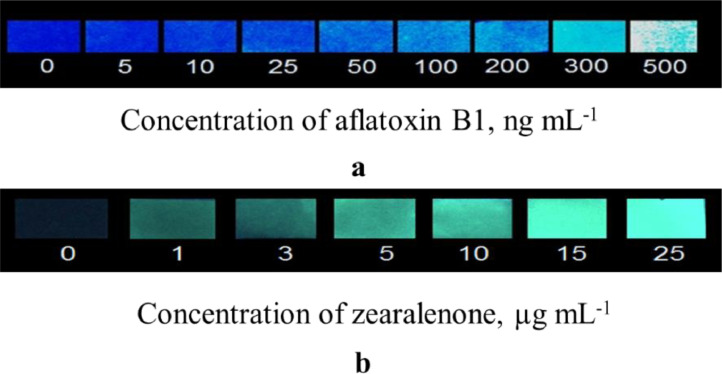
Successful combination of MIP membranes with smartphones was demonstrated by our group recently for the fluorescent detection of widespread food toxins - aflatoxin B1 and zearalenone [64,65]. Both the smartphone-based sensors were based on registration of native fluorescence of aflatoxin B1 and zearalenone (Fig. 12) [66,67].
Fig. 12.
Photos of the aflatoxin B1- (a) and zearalenone-selective (b) MIP membranes’ surfaces after toxins’ binding by the artificial receptor sites from aqueous samples and UV-irradiation.
The novel sensors were also validated and proven to be useful for the mycotoxins’ detection in “real” cereal samples [66,67].
A significant progress in sensitivity of the developed fluorescent MIP-membrane-based sensors was demonstrated recently [34] through application of plasmon-enhanced fluorescence phenomenon. It is widely recognized, that metal nanoparticles can modify properties of fluorescent molecules in the case of overlapping fluorescence and local surface plasmon resonance spectra, occasioning plasmon-enhanced fluorescence phenomenon [74]. The versatile method for the in situ synthesis of Ag nanoparticles in close proximity to artificial receptor sites in toxin-imprinted biomimetic membranes and thin films has been proposed in our recent work [34]. For example, in situ formation of 30–70 nm diameter Ag nanoparticles in the aflatoxin B1-selective biomimetics, allowed us to decrease the detection limit for aflatoxin B1 from 10 ng mL−1 to 0.3 ng mL−1. Wide linear dynamic range (0.3–25 ng mL−1) along with superior selectivity was typical for the developed biosensor system [34]. The proposed approach is very versatile and can be adapted to the formation of selective materials towards the other fluorescent molecules, which differ in chemical structure (i.e. fluorescent mycotoxin zearalenone, bisphenol A, etc.)
All the sensors based on the MIP-membranes and thin films demonstrated extremely good storage stability at room temperature, which comprised 12 and 18 months for grafted MIP membranes and MIP membranes synthesized in situ, respectively. At the same time, the data obtained using biosensors based on artificial toxin-selective biomimetic polymers were in accordance with those obtained by standard ELISA and HPLC methods. All the developed sensors were validated as for their effectiveness for detection of target analytes in “real” samples in numerous studies (extracts of food products and feeding stuffs; tape, drinking, natural, and wastewaters; pharmaceutical drugs, etc.) [27,28,34,39,57,58,60,[65], [66], [67]].
5. Versatile approach to the highly-sensitive detection of the analytes. Solid-phase extraction based on porous MIP membranes
Another large area for the application of polymers-biomimics in a form of porous membranes is their use for the selective pre-concentration of diluted samples of target analytes. This can significantly decrease detection limits of existing traditional biochemical methods, biosensors, and instrumental analytical methods. Porous membranes can also compete with traditional SPE adsorbents in selective removal of interfering components from “real” samples, they also can be used for their storage and transportation. Computational MIP membranes can be rapidly custom-made for a variety of chemical substances. At the same time, they have advantages as compared to traditional SPE adsorbents, first of all, surpassing them in selectivity, adsorption capability and productivity [27,[35], [36], [37], [38], [39], [40], [41], [42], [43],49,59,60,63,65,68].
Both types of the MIP membranes synthesized either in situ or by grafting modification of the inert microfiltration membranes with a thin MIP layer were shown to be an effective stationary phase for SPE of small organic molecules [27,[35], [36], [37], [38], [39], [40], [41], [42], [43],49,59,60,63,65,68]. The porous MIP membranes synthesized in situ obtained by bulk radical cross-linking polymerization of methacrylate monomers demonstrated increased porosity and water fluxes, which depended on the type and concentration of the linear polymer in the semi-IPN [35], [36], [37]. Application of polyethylene glycol MW 20,000 and linear polyurethane MW 40,000 provided an increase in the water fluxes up to ~ 10,000 l m−2 h−1 and ~ 15,000 l m−2 h−1, respectively, as compared to 0 l m−2 h−1 for the membranes obtained without “linear porogens”. The values of selective adsorption towards target analytes were increased proportionally to the amount of the linear semi-IPN component, which is apparently associated with better accessibility of their supramolecular binding sites. The adsorption capacity of semi-IPN-based herbicide-imprinted membranes varied from 6.53 to 12.5 mg g−1 of polymer (up to ∼25% of the theoretically calculated value) depending on chemical nature of the linear polymer, which has dramatic influence on both the morphology of the MIP membrane and the structure of its binding sites [35,52,53].
The method proposed by our group for the synthesis of the IPN-based MIP membranes can be easily adapted for various analytes. We demonstrated their effectiveness for the pre-concentration of diluted samples of a number of endocrine disruptors and toxins: triazine herbicides, phenol, bisphenol A, mycotoxins, etc. The type of selectivity of the affinity membranes can be varied by changing type of the functional monomer used for the polymer synthesis. Estimation of binding energy together with molecular dynamics data can provide synthesis of artificial binding sites either with group or individual selectivity for different purposes [27,[35], [36], [37], [38], [39], [40], [41], [42], [43],49,59,60,63,65,68]. As a result, the MIP membranes demonstrated selectivity comparable to the selectivity of natural antibodies and receptors.
The composite MIP membranes obtained by the thin-layer modification of the inert microfiltration supports with MIPs is another promising alternative to the traditional SPE adsorbents. Polypropylene, polyamide, polyvynylidenefluoride, and cellulose microfiltration membranes can be successfully modified with biomimetic polymer layers. Surface modification normally provides the following degrees of modification with MIP layers: 670–1500 µg cm−2 for modification procedure from aqueous solutions and 50–800 µg cm−2 for modification from organic solvents [38,[40], [41], [42]]. Importantly, MIP modification does not influence productivity of the inert microfiltration supports, which comprises ~12,000 l м−2⋅h−1.
It was shown that the most important factors affecting the selectivity of composite MIP membranes are the type and amount of the functional monomer, as well as the cross-linking agent used for the surface modification [38,[40], [41], [42]]. At the same time, in contrast to the IPN-based affinity membranes, the composite membranes can be developed not only for the templates soluble in organic solvents, but also for the water-soluble target analytes. The adsorption capacity of composite MIP membranes modified under optimized conditions comprise 5–6.8 μg cm−2, which corresponds to ∼40% of the theoretically calculated value [38,[40], [41], [42]]. Both types of the porous MIP membranes were shown to be effective in solid-phase extraction and ensured effective preliminary concentration (up to 100 times) of diluted samples of the target analytes.
6. Future outlook
Clear evidence of effectiveness of the computational modeling approach for the design of affinity binding sites in the biomimetic polymers, membranes and thin films were presented. Analysis of molecular dynamics data can provide synthesis of biomimetic polymers with predictable properties (e.g. affinity and selectivity) towards a number of target analytes with different chemical structures. Other obvious benefits of biomimetic polymers are their low cost and relatively easy, time-effective synthetic procedure, as well as significantly higher stability as compared to their natural counterparts. Mainly, the effectiveness of this approach was confirmed for the small organic molecules that are of significant interest for environmental monitoring, evaluation of water and food quality, and in diagnostics. The developed versatile methods of the MIP membranes synthesis assume mild polymerization conditions. Moreover, generic methods of artificial binding sites formation were developed for target analytes with different types of solubility: for water-soluble ones and for those soluble in organic solvents. This might provide in a combination with computer-aided design further prospective for synthesis of the MIP membranes towards a number of much more “sophisticated” biological molecules (proteins, DNA, oligonucleotides, etc.) that are of significant interest for medical diagnostics (e.g. genetic disorders, hereditary and infection diseases, cancer, etc.) as well as development of smart biosensors for their effective point-of-care detection. This might significantly improve treatment of chronic diseases, provide effective early diagnostics, promote the development of personalized medicine, and, as a result, decrease expenditures on medical care. Further investigation of functioning of artificial receptor sites in biomimetic polymers using computational modeling methods would definitely add to our understanding of molecular recognition phenomena.
7. Conclusions
New principles and versatile methods of synthesis of receptor and catalytic sites mimicking those of natural enzymes and receptors in the structure of molecularly imprinted polymer membranes and thin films were proposed. It is possible to highlight that the research objectives have been successfully achieved and highly stable, selective and sensitive biosensors for various applications of Analytical Biotechnology were developed. Combination of molecular imprinting with computational modeling approach provides an effective method for synthesis of artificial receptors with desirable selectivity (either with individual or group selectivity) for a number of target analytes of various chemical structure (herbicides, pharmaceuticals, metabolites, food toxins, etc.). Porous MIP membranes with artificial receptor sites in their structure can be successfully used as selective elements of electrochemical and optical sensor devices for various “point-of-care” applications and as SPE stationary phase for the effective pre-concentration of diluted samples of target analytes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Financial support from National Academy of Sciences of Ukraine is gratefully acknowledged. The authors are grateful to Armed Forces of Ukraine and all the other defenders of Ukraine who provide our safety and made this work possible.
Data availability
Data will be made available on request.
References
- 1.Singh K.R., Nayak V., Singh R.P. Advanced Biosensors for Virus Detection. Academic Press; 2022. Future aspects of biosensor-based devices in disease detection; pp. 423–439. [DOI] [Google Scholar]
- 2.Bastidas S.C. Encyclopedia of Sensors and Biosensors (vol. 1, pp. 429-457) Elsevier; 2021. Biosensors: biosensors With Signal Amplification. [DOI] [Google Scholar]
- 3.Wulff G., Sarhan A., Zabrocki K. Enzyme-analogue built polymers and their use for the resolution of racemates. Tetrahedron Lett. 1973;14(44):4329–4332. doi: 10.1016/S0040-4039(01)87213-0. [DOI] [Google Scholar]
- 4.Arshady R., Mosbach K. Synthesis of substrate-selective polymers by host-guest polymerization. Die Makromolekulare Chemie: Macromolecular Chem. Phys. 1981;182(2):687–692. doi: 10.1002/macp.1981.021820240. [DOI] [Google Scholar]
- 5.Speltini A., Scalabrini A., Maraschi F., Sturini M., Profumo A. Newest applications of molecularly imprinted polymers for extraction of contaminants from environmental and food matrices: a review. Anal. Chim. Acta. 2017;974:1–26. doi: 10.1016/j.aca.2017.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Moein M.M. Advancements of chiral molecularly imprinted polymers in separation and sensor fields: a review of the last decade. Talanta. 2021;224 doi: 10.1016/j.talanta.2020.121794. [DOI] [PubMed] [Google Scholar]
- 7.Luliński P. Molecularly imprinted polymers based drug delivery devices: a way to application in modern pharmacotherapy. a review. Mater. Sci. Eng.: C. 2017;76:1344–1353. doi: 10.1016/j.msec.2017.02.138. [DOI] [PubMed] [Google Scholar]
- 8.Kupai J., Razali M., Buyuktiryaki S., Kecili R., Szekely G. Long-term stability and reusability of molecularly imprinted polymers. Polym. Chem. 2017;8(4):666–673. doi: 10.1039/C6PY01853J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cennamo N., Arcadio F., Seggio M., Maniglio D., Zeni L., Bossi A.M. Spoon-shaped polymer waveguides to excite multiple plasmonic phenomena: a multisensor based on antibody and molecularly imprinted nanoparticles to detect albumin concentrations over eight orders of magnitude. Biosens. Bioelectron. 2022;217 doi: 10.1016/j.bios.2022.114707. [DOI] [PubMed] [Google Scholar]
- 10.Kholová A., Lhotská I., Erben J., Chvojka J., Švec F., Solich P., Šatínský D. Comparison study of nanofibers, composite nano/microfiber materials, molecularly imprinted polymers, and core-shell sorbents used for on-line extraction-liquid chromatography of ochratoxins in Tokaj wines. Microchem. J. 2021;170 doi: 10.1016/j.microc.2021.106680. [DOI] [Google Scholar]
- 11.Kubo T., Kuroda K., Tominaga Y., Naito T., Sueyoshi K., Hosoya K., Otsuka K. Effective determination of a pharmaceutical, sulpiride, in river water by online SPE-LC–MS using a molecularly imprinted polymer as a preconcentration medium. J. Pharm. Biomed. Anal. 2014;89:111–117. doi: 10.1016/j.jpba.2013.10.040. [DOI] [PubMed] [Google Scholar]
- 12.Chen T., Wei S., Cheng Z., Liu J. Specific detection of monosaccharide by dual-channel sensing platform based on dual catalytic system constructed by bio-enzyme and bionic enzyme using molecular imprinting polymers. Sens. Actuators B. 2020;320 doi: 10.1016/j.snb.2020.128430. [DOI] [Google Scholar]
- 13.Rajpal S., Mishra P. Next generation biosensors employing molecularly imprinted polymers as sensing elements for in vitro diagnostics. Biosens. Bioelectron X. 2022;11 doi: 10.1016/j.biosx.2022.100201. [DOI] [Google Scholar]
- 14.Kitayama Y., Isomura M. Molecularly imprinted polymer particles with gas-stimuli responsive affinity toward target proteins prepared using switchable functional monomer. Polymer (Guildf) 2020;203 doi: 10.1016/j.polymer.2020.122781. [DOI] [PubMed] [Google Scholar]
- 15.Shahar T., Tal N., Mandler D. Molecularly imprinted polymer particles: formation, characterization and application. Colloids Surf. A. 2016;495:11–19. doi: 10.1016/j.colsurfa.2016.01.027. [DOI] [Google Scholar]
- 16.Ferreira V.R., Azenha M.A., Pereira C.M., Silva A.F. Molecularly imprinted polymers for enhanced impregnation and controlled release of l-tyrosine. React. Funct. Polym. 2018;131:283–292. doi: 10.1016/j.reactfunctpolym.2018.07.017. [DOI] [Google Scholar]
- 17.Nawaz N., Bakar N.K.A., Mahmud H.N.M.E., Jamaludin N.S. Molecularly imprinted polymers-based DNA biosensors. Anal. Biochem. 2021;630 doi: 10.1016/j.ab.2021.114328. [DOI] [PubMed] [Google Scholar]
- 18.Pareek S., Jain U., Balayan S., Chauhan N. Ultra-sensitive nano-molecular imprinting polymer-based electrochemical sensor for Follicle-Stimulating Hormone (FSH) detection. Biochem. Eng. J. 2022;180 doi: 10.1016/j.bej.2021.108329. [DOI] [Google Scholar]
- 19.Khulu S., Ncube S., Nuapia Y., Madikizela L.M., Mavhunga E., Chimuka L. Development and application of a membrane assisted solvent extraction-molecularly imprinted polymer based passive sampler for monitoring of selected pharmaceuticals in surface water. Water Res. 2022 doi: 10.1016/j.watres.2022.119145. [DOI] [PubMed] [Google Scholar]
- 20.Quílez-Alburquerque J., Descalzo A.B., Moreno-Bondi M.C., Orellana G. Luminescent molecularly imprinted polymer nanocomposites for emission intensity and lifetime rapid sensing of tenuazonic acid mycotoxin. Polymer (Guildf) 2021;230 doi: 10.1016/j.polymer.2021.124041. [DOI] [Google Scholar]
- 21.Liu X., Chen L., Gao Y., Li J., Sun J., Gan T. Molecularly imprinted polymer capped Au@ HKUST− 1 nanocapsules-based electrochemical sensing platform for monitoring isoproturon herbicide in water at sub− nanomole level. J. Environ. Chem. Eng. 2022;10(3) doi: 10.1016/j.jece.2022.107661. [DOI] [Google Scholar]
- 22.Zhang J., Qin L., Yang Y., Liu X. Porous carbon nanospheres aerogel based molecularly imprinted polymer for efficient phenol adsorption and removal from wastewater. Sep. Purif. Technol. 2021;274 doi: 10.1016/j.seppur.2021.119029. [DOI] [Google Scholar]
- 23.Lee M.H., Lin C.C., Kutner W., Thomas J.L., Lin C.Y., Iskierko Z., …, Lin H.Y. Peptide-imprinted conductive polymer on continuous monolayer molybdenum disulfide transferred electrodes for electrochemical sensing of Matrix Metalloproteinase-1 in lung cancer culture medium. Biosens. Bioelectron X. 2022 doi: 10.1016/j.biosx.2022.100258. [DOI] [Google Scholar]
- 24.Zhang W., Zhang Y., Wang R., Zhang P., Zhang Y., Randell E., …, Jia Q. A review: development and application of surface molecularly imprinted polymers toward amino acids, peptides, and proteins. Anal. Chim. Acta. 2022;340319 doi: 10.1016/j.aca.2022.340319. [DOI] [PubMed] [Google Scholar]
- 25.You M., Yang S., Jiao F., Yang L.Z., Zhang F., He P.G. Label-free electrochemical multi-sites recognition of G-rich DNA using multi-walled carbon nanotubes–supported molecularly imprinted polymer with guanine sites of DNA. Electrochim. Acta. 2016;199:133–141. [Google Scholar]
- 26.Ogiso M., Minoura N., Shinbo T., Shimizu T. Detection of a specific DNA sequence by electrophoresis through a molecularly imprinted polymer. Biomaterials. 2006;27(22):4177–4182. doi: 10.1016/j.biomaterials.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Sergeyeva T., Yarynka D., Piletska E., Lynnik R., Zaporozhets O., Brovko O., Piletsky S., El'skaya A. Fluorescent sensor systems based on nanostructured polymeric membranes for selective recognition of Aflatoxin B1. Talanta. 2017;175:101–107. doi: 10.1016/j.talanta.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Sergeyeva T., Yarynka D., Dubey L., Dubey I., Piletska E., Linnik R., Antonyuk M., Ternovska T., Brovko O., Piletsky S., El'skaya A. Sensor based on molecularly imprinted polymer membranes and smartphone for detection of Fusarium contamination in cereals. Sensors. 2020;20(15):4304–4324. doi: 10.3390/s20154304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahhoseini F., Azizi A., Bottaro C.S. A critical evaluation of molecularly imprinted polymer (MIP) coatings in solid phase microextraction devices. TrAC Trends Anal. Chem. 2022 doi: 10.1016/j.trac.2022.116695. [DOI] [Google Scholar]
- 30.Piletsky S.А., Dubey I.Ya., Fedoryak D.М., Kukhar V.P. Substrate-selective polymeric membranes. Selective transfer of nucleic acid components. Biopolym. Cell. 1990;6(5):55. doi: 10.7124/bc.00028D. [DOI] [Google Scholar]
- 31.Piletsky S.А., Butovich I.A., Kukhar V.P. Design of molecular sensors based on substrate-selective polymeric membranes. J. Analyt. Chem. Moscow. 1992;47(9):1681–1684. [Google Scholar]
- 32.Sergeyeva T.A., Piletsky S.A., Brovko A.A., Slinchenko E.A., Sergeeva L.M., El'Skaya A.V. Selective recognition of atrazine by molecularly imprinted polymer membranes. Development of conductometric sensor for herbicides detection. Anal. Chim. Acta. 1999;392(2–3):105–111. doi: 10.1016/S0003-2670(99)00225-1. [DOI] [Google Scholar]
- 33.Sergeyeva T.A., Piletsky S.A., Brovko A.A., Slinchenko E.A., Panasyuk T.L., Sergeeva L.M., El'Skaya A.V. Conductimetric sensor for atrazine detection based on molecularly imprinted polymer membranes. Analyst. 1999;124(3):331–334. doi: 10.1039/A808484J. [DOI] [Google Scholar]
- 34.Sergeyeva T., Yarynka D., Lytvyn V., Demydov P., Lopatynskyi A., Stepanenko Y., Brovko O., Pinchuk A., Chegel V. Highly-selective and sensitive plasmon-enhanced fluorescence sensor of aflatoxins. Analyst. 2022;147(6):1135–1143. doi: 10.1039/D1AN02173G. [DOI] [PubMed] [Google Scholar]
- 35.Sergeyeva T.A., Piletsky S.A., Piletska E.V., Brovko O.O., Karabanova L.V., Sergeeva L.M., El'skaya A., Turner A.P. In situ formation of porous molecularly imprinted polymer membranes. Macromolecules. 2003;36(19):7352–7357. doi: 10.1021/ma030105x. [DOI] [Google Scholar]
- 36.Sergeyeva T.A., Brovko O.O., Piletska E.V., Piletsky S.A., Goncharova L.A., Karabanova L.V., Sergeeva L.M., El'skaya A.V. Porous molecularly imprinted polymer membranes and polymeric particles. Anal. Chim. Acta. 2007;582(2):311–319. doi: 10.1016/j.aca.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Sergeyeva T.A., Piletska O.V., Piletsky S.A., Sergeeva L.M., Brovko O.O., El'ska G.V. Data on the structure and recognition properties of the template-selective binding sites in semi-IPN-based molecularly imprinted polymer membranes. Mater. Sci. Eng.: C. 2008;28(8):1472–1479. doi: 10.1016/j.msec.2008.04.006. [DOI] [Google Scholar]
- 38.Sergeyeva T.A., Matuschewski H., Piletsky S.A., Bendig J., Schedler U., Ulbricht M. Molecularly imprinted polymer membranes for substance-selective solid-phase extraction from water by surface photo-grafting polymerization. J. Chromatogr. A. 2001;907(1–2):89–99. doi: 10.1016/S0021-9673(00)01053-0. [DOI] [PubMed] [Google Scholar]
- 39.Sergeyeva T.A., Gorbach L.A., Piletska E.V., Piletsky S.A., Brovko O.O., Honcharova L.A., Lutsyk O.D., Sergeeva L.M., Zinchenko O.A., El'skaya A.V. Colorimetric test-systems for creatinine detection based on composite molecularly imprinted polymer membranes. Anal. Chim. Acta. 2013;770:161–168. doi: 10.1016/j.aca.2013.01.048. [DOI] [PubMed] [Google Scholar]
- 40.Matuschewski H., Sergeyeva T.A., Bendig J., Piletsky S.A., Ulbricht M., Schedler U. Advanced Environmental and Chemical Sensing Technology (Vol. 4205, pp. 65-74) SPIE; 2001. Surface engineering: molecularly imprinted affinity membranes by photograft polymerization. [DOI] [Google Scholar]
- 41.Sergeyeva T.A., Matuschewski H., Pletsky S.A., Schedler U., Ulbricht M. Development of molecularly imprinted polymer membranes with specificity to triazine herbicides, prepared by the surface photografting technique. Biopolym. Cell. 2004;20(4):307. doi: 10.7124/bc.0006B4. [DOI] [Google Scholar]
- 42.Piletsky S.A., Matuschewski H., Schedler U., Wilpert A., Piletska E.V., Thiele T.A., Ulbricht M. Surface functionalization of porous polypropylene membranes with molecularly imprinted polymers by photograft copolymerization in water. Macromolecules. 2000;33(8):3092–3098. doi: 10.1021/ma991087f. [DOI] [Google Scholar]
- 43.Hattori K., Hiwatari M., Iiyama C., Yoshimi Y., Kohori F., Sakai K., Piletsky S.A. Gate effect of theophylline-imprinted polymers grafted to the cellulose by living radical polymerization. J. Memb. Sci. 2004;233(1–2):169–173. doi: 10.1016/j.memsci.2003.12.013. [DOI] [Google Scholar]
- 44.Sergeyeva T.A., Panasyuk-Delaney T.L., El'skaya A.V., Piletska O.V., Piletsky S.A. Capacitive sensor for environmental monitoring based on thin films of molecularly imprinted polymers. Computational modeling for optimization of the polymers-biomimics composition. Ukr Biochem. J. 2006;78(2):121–130. [PubMed] [Google Scholar]
- 45.Morrill P.R., Gupta G., Sproule K., Winzor D., Christensen J., Mollerup I., Lowe C.R. Rational combinatorial chemistry-based selection, synthesis and evaluation of an affinity adsorbent for recombinant human clotting factor VII. J. Chromatogr. B. 2002;774(1):1–15. doi: 10.1016/s1570-0232(02)00103-4. [DOI] [PubMed] [Google Scholar]
- 46.Piletsky S.A., Day R.M., Chen B., Subrahmanyam S., Piletska O., Turner A.P.F. Rational design of MIPs using computational approach. International Patent PCT/GB01/00324; 2001.
- 47.Piletsky S.A., Karim K., Piletska E.V., Day C.J., Freebairn K.W., Legge C., Turner A.P.F. Recognition of ephedrine enantiomers by molecularly imprinted polymers designed using a computational approach. Analyst. 2001;126(10):1826–1830. doi: 10.1039/B102426B. [DOI] [Google Scholar]
- 48.Karim K., Cowen T., Guerreiro A., Piletska E., Whitcombe M.J., Piletsky S.A. A protocol for the computational design of high affinity molecularly imprinted polymer synthetic receptors. Glob. J. Biotechnol. Biomater. Sci. 2017;3(1):001–007. doi: 10.17352/gjbbs.000009. [DOI] [Google Scholar]
- 49.Piletska E., Kumire J., Sergeyeva T., Piletsky S. Rational design and development of affinity adsorbents for analytical and biopharmaceutical applications. J. Chin. Adv. Mater. Soc. 2013;1(3):229–244. doi: 10.1080/22243682.2013.839207. [DOI] [Google Scholar]
- 50.Muhammad T., Nur Z., Piletska E.V., Yimit O., Piletsky S.A. Rational design of molecularly imprinted polymer: the choice of cross-linker. Analyst. 2012;137(11):2623–2628. doi: 10.1039/C2AN35228A. [DOI] [PubMed] [Google Scholar]
- 51.Piletska E.V., Romero-Guerra M., Chianella I., Karim K., Turner A.P., Piletsky S.A. Towards the development of multisensor for drugs of abuse based on molecular imprinted polymers. Anal. Chim. Acta. 2005;542(1):111–117. doi: 10.1016/j.aca.2005.03.067. [DOI] [Google Scholar]
- 52.Chianella I., Lotierzo M., Piletsky S.A., Tothill I.E., Chen B., Karim K., Turner A.P. Rational design of a polymer specific for microcystin-LR using a computational approach. Anal. Chem. 2002;74(6):1288–1293. doi: 10.1021/ac010840b. [DOI] [PubMed] [Google Scholar]
- 53.Piletsky S., Piletska E., Karim K., Foster G., Legge C., Turner A. Custom synthesis of molecular imprinted polymers for biotechnological application: preparation of a polymer selective for tylosin. Anal. Chim. Acta. 2004;504(1):123–130. doi: 10.1016/S0003-2670%2803%2900814-6. [DOI] [Google Scholar]
- 54.Chianella I., Karim K., Piletska E.V., Preston C., Piletsky S.A. Computational design and synthesis of molecularly imprinted polymers with high binding capacity for pharmaceutical applications-model case: adsorbent for abacavir. Anal. Chim. Acta. 2006;559(1):73–78. doi: 10.1016/j.aca.2005.11.068. [DOI] [Google Scholar]
- 55.Piletska E., Piletsky S., Karim K., Terpetschnig E., Turner A. Biotin-specific synthetic receptors prepared using molecular imprinting. Anal. Chim. Acta. 2004;504(1):179–183. doi: 10.1016/S0003-2670(03)00813-4. [DOI] [Google Scholar]
- 56.Piletska E.V., Turner N.W., Turner A.P., Piletsky S.A. Controlled release of the herbicide simazine from computationally designed molecularly imprinted polymers. J. Controlled Release. 2005;108(1):132–139. doi: 10.1016/j.jconrel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Sergeyeva Т.А., Gorbach L.A., Slinchenko О.А., Goncharova L.A., Piletska O.V., Brovko О.О., Sergeeva L.M., El'ska G.V. Towards development of colorimetric test-systems for phenols detection based on computationally-designed molecularly imprinted polymer membranes. Mater. Sci. Eng.: C. 2010;30(3):431–436. doi: 10.1016/j.msec.2009.12.012. [DOI] [Google Scholar]
- 58.Sergeyeva T.A., Piletska O.V., Honcharova L.A., Brovko O.O., Piletsky S.A., El'ska H.V. Sensor system based on molecular-imprinted polymer membranes for the selective recognition of aflatoxin B1. Ukr. Biochem. J. 2008;80(3):84–93. [PubMed] [Google Scholar]
- 59.Sergeyeva T.A., Piletska O.V., Brovko O.O., Honcharova L.A., Piletsky S.A., El'ska H.V. Aflatoxin-selective molecularly-imprinted polymer membranes based on acrylate-polyurethane semi-interpenetrating polymer networks. Ukr. Biochem. J. 2007;79(5):109–115. [PubMed] [Google Scholar]
- 60.Sergeyeva T.A., Piletska O.V., Gorbach L.A., Ivanova A.V., Brovko O.O., El'ska H.V. Sensor system for sulfamethoxazole detection based on molecularly imprinted polymer membranes. Scientif. News Natl. Techn. Univ. Ukraine Kyiv Polytechnic Inst. 2015;3:61–67. [Google Scholar]
- 61.Panasyuk T.L., Mirsky V.M., Piletsky S.A., Wolfbeis O.S. Electropolymerized molecularly imprinted polymers as receptor layers in capacitive chemical sensors. Anal. Chem. 1999;71(20):4609–4613. doi: 10.1021/ac9903196. [DOI] [Google Scholar]
- 62.Piletsky S.A., Nicholls I.A., Rozhko M.I., Sergeyeva T.A., Piletska E.V., El'skaya A.V., Karube L. Molecularly imprinted polymer tyrosinase mimics. Ukr. Biochem. J. 2005;77:67–78. [PubMed] [Google Scholar]
- 63.Sergeyeva T.A., Slinchenko O.A., Gorbach L.A., Matyushov V.F., Brovko O.O., Piletsky S.A., Elska G.V. Catalytic molecularly imprinted polymer membranes: development of the biomimetic sensor for phenols detection. Anal. Chim. Acta. 2010;659(1–2):274–279. doi: 10.1016/j.aca.2009.11.065. [DOI] [PubMed] [Google Scholar]
- 64.Decker H., Schweikardt T., Nillius D., Salzbrunn U., Jaenicke E., Tuczek F. Similar enzyme activation and catalysis in hemocyanins and tyrosinases. Gene. 2007;398(1–2):183–191. doi: 10.1016/j.gene.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 65.Sergeyeva T.A., Chelyadina D.S., Gorbach L.A., Brovko O.O., Piletska E.V., Piletsky S.A., Sergeeva L.M., El'skaya A.V. Colorimetric biomimetic sensor systems based on molecularly imprinted polymer membranes for highly-selective detection of phenol in environmental samples. Вiopolym. Cell. 2014;30(3):209–215. doi: 10.7124/bc.000898. [DOI] [Google Scholar]
- 66.Yarynka D., Sergeyeva T., Piletska E., Linnik R., Antonyuk M., Brovko O., Piletsky S.A., El'skaya A. Validation of aflatoxin B1 MIP membrane-based smartphone sensor system for real sample applications. Вiopolym. Cell. 2021;37(5):346–356. doi: 10.7124/bc.000A60. [DOI] [Google Scholar]
- 67.Yarynka D.V., Sergeyeva T.A., Piletska E.V., Stepanenko Y., Brovko O.O., Piletsky S.A., El'skaya A.V. Zearalenone-selective biomimetic-based sensor system and its validation for real samples’ analysis. Biopolym. Cell. 2021;37(6):438–446. doi: 10.7124/bc.000A69. [DOI] [Google Scholar]
- 68.Sergeyeva T.A., Satyr A.V., Piletska E.V., Gorbach L.A., Brovko O.O., El'skaya A.V. Biosensor system for detection of bisphenol A in aqueous solutions. Polym. J. 2016;38(3):261–266. [Google Scholar]
- 69.Yang T., Luo Z., Bewal T., Li L., Xu Y., Jafari S.M., Lin X. When smartphone enters food safety: a review in on-site analysis for foodborne pathogens using smartphone-assisted biosensors. Food Chem. 2022 doi: 10.1016/j.foodchem.2022.133534. [DOI] [PubMed] [Google Scholar]
- 70.Cheng Y., Wang H., Zhuo Y., Song D., Li C., Zhu A., Long F. Reusable smartphone-facilitated mobile fluorescence biosensor for rapid and sensitive on-site quantitative detection of trace pollutants. Biosens. Bioelectron. 2022;199 doi: 10.1016/j.bios.2021.113863. [DOI] [PubMed] [Google Scholar]
- 71.Ma L., Yin L., Li X., Chen S., Peng L., Liu G., Man S. A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jędrzak A., Kuznowicz M., Rębiś T., Jesionowski T. Portable glucose biosensor based on polynorepinephrine@ magnetite nanomaterial integrated with a smartphone analyzer for point-of-care application. Bioelectrochemistry. 2022;145 doi: 10.1016/j.bioelechem.2022.108071. [DOI] [PubMed] [Google Scholar]
- 73.Xiao C., Eriksson J., Suska A., Filippini D., Mak W.C. Print-and-stick unibody microfluidics coupled surface plasmon resonance (SPR) chip for smartphone imaging SPR (Smart-iSRP) Anal. Chim. Acta. 2022;1201 doi: 10.1016/j.aca.2022.339606. [DOI] [PubMed] [Google Scholar]
- 74.Li M., Cushing S.K., Wu N. Plasmon-enhanced optical sensors: a review. Analyst. 2015;140(2):386–406. doi: 10.1039/C4AN01079E. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.