Abstract
Ferroptosis, an iron-dependent lipid peroxidation-driven programmed cell death, is closely related to cancer therapy. The development of druggable ferroptosis inducers and their rational application in cancer therapy are critical. Here, we identified Tubastatin A, an HDAC6 inhibitor as a novel druggable ferroptosis inducer through large-scale drug screening. Tubastatin A directly bonded to GPX4 and inhibited GPX4 enzymatic activity through biotin-linked Tubastatin A putdown and LC/MS analysis, which is independent of its inhibition of HDAC6. In addition, our results showed that radiotherapy not only activated Nrf2-mediated GPX4 transcription but also inhibited lysosome-mediated GPX4 degradation, subsequently inducing ferroptosis tolerance and radioresistance in cancer cells. Tubastatin A overcame ferroptosis resistance and radioresistance of cancer cells by inhibiting GPX4 enzymatic activity. More importantly, Tubastatin A has excellent bioavailability, as demonstrated by its ability to significantly promote radiotherapy-induced lipid peroxidation and tumour suppression in a mouse xenograft model. Our findings identify a novel druggable ferroptosis inducer, Tubastatin A, which enhances radiotherapy-mediated antitumor effects. This work provides a compelling rationale for the clinical evaluation of Tubastatin A, especially in combination with radiotherapy.
1. Introduction
Ferroptosis, a nonapoptotic form of regulated cell death, is driven by excessive accumulation of lipid peroxidation [[1], [2], [3]]. Multiple pathways affect the ferroptotic process by regulating the generation and clearance of lipid peroxidation. Polyunsaturated fatty acid-containing phospholipids, especially arachidonic acid (C20:4)- and adrenic acid (C22:4)-containing phospholipids, undergo peroxidation in the presence of lipoxygenase Lox, cytochrome P450 reductase POR and ferrous iron [2,[4], [5], [6]]. There are four main signalling pathways that mediate lipid peroxidative scavenging, including GPX4-GSH, FSP1-CoQH2, GCH1-BH4 and DHODH-CoQH2. GPX4 is the only enzyme in the GPX family that specifically removes lipid peroxidation. Its activity depends on the level of GSH. SLC7A11 participates in the GPX4 signalling pathway by promoting the transport of cystine and subsequently the synthesis of GSH [7]. The commonly used ferroptosis inducers RSL3 and erastin are direct inhibitors of GPX4 and SLC7A11, respectively. FSP1 scavenges lipid peroxides by trapping lipid peroxyl radicals [8,9]. GCH1 scavenges lipid peroxidation by synthesizing the free radical scavenger BH4 [10]. Mitochondrial DHODH inhibits ferroptosis by reducing CoQ to CoQH2 [11]. Inactivation of these pathways by ferroptosis inducer compounds leads to a rapid accumulation of lipid peroxides and triggers ferroptotic cell death in many cancer cell lines and in vivo tumour models [2,3].
Ferroptosis is associated with many pathological processes, such as ischemia-reperfusion injury, neurodegenerative diseases, and cancer [4,12,13]. Cellular ferroptosis has a certain cancer species selectivity, and breast cancer is one of the sensitive species [4,14,15]. Therefore, induction of ferroptosis may be one of the strategies for the treatment of breast cancer. More importantly, ferroptosis is also involved in cancer immunotherapy and radiotherapy [4,[16], [17], [18]]. Radiotherapy is an important strategy of cancer treatment. Nevertheless, many tumours are refractory to radiotherapy, highlighting the pressing need to develop novel and effective combination therapy. Studies have shown that radiotherapy induced ferroptosis by promoting the generation of ROS and the expression of ACSL4 in cancer cells [[18], [19], [20]]. Inhibition of ferroptosis significantly impairs the efficacy of radiotherapy in vitro and in vivo. These results implied that targeting ferroptosis might enhance the efficacy of radiotherapy in cancer cells. However, limited druggable ferroptosis inducers are currently available for animal experiments and preclinical trials, which greatly restricts the application of ferroptosis inducer in radiotherapy. Tubastatin A is a HDAC6 selective inhibitor. It has been reported that Tubastatin A can affect the oxidative stress of cells by inhibiting HDAC6 [21]. However, whether Tubastatin A regulates ferroptosis remains unclear.
Here, we have identified Tubastatin A (Tub) as a novel GPX4 inhibitor that induced ferroptosis through large-scale drug screening. We showed that IR-mediated GPX4 expression restrained ferroptosis to drive radioresistance in breast cancer. Notably, Tub overcame ferroptosis resistance and radioresistance of cancer cells by inhibiting GPX4 enzymatic activity in vitro and in vivo.
2. Results
2.1. Identification of Tubastatin A as a novel ferroptosis inducer
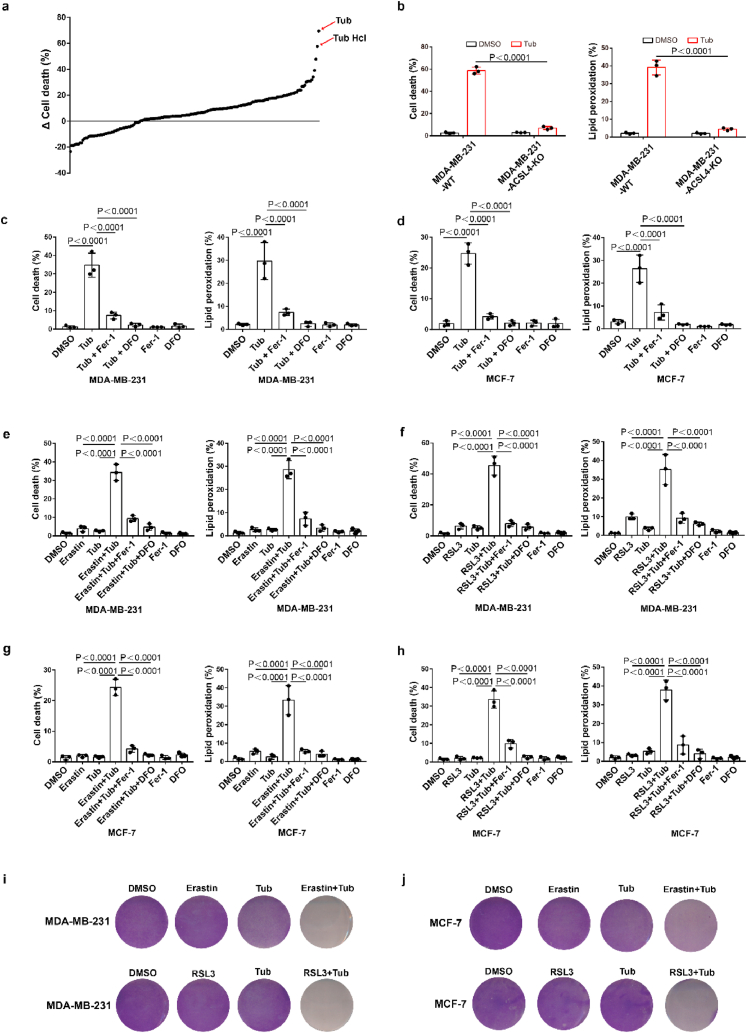
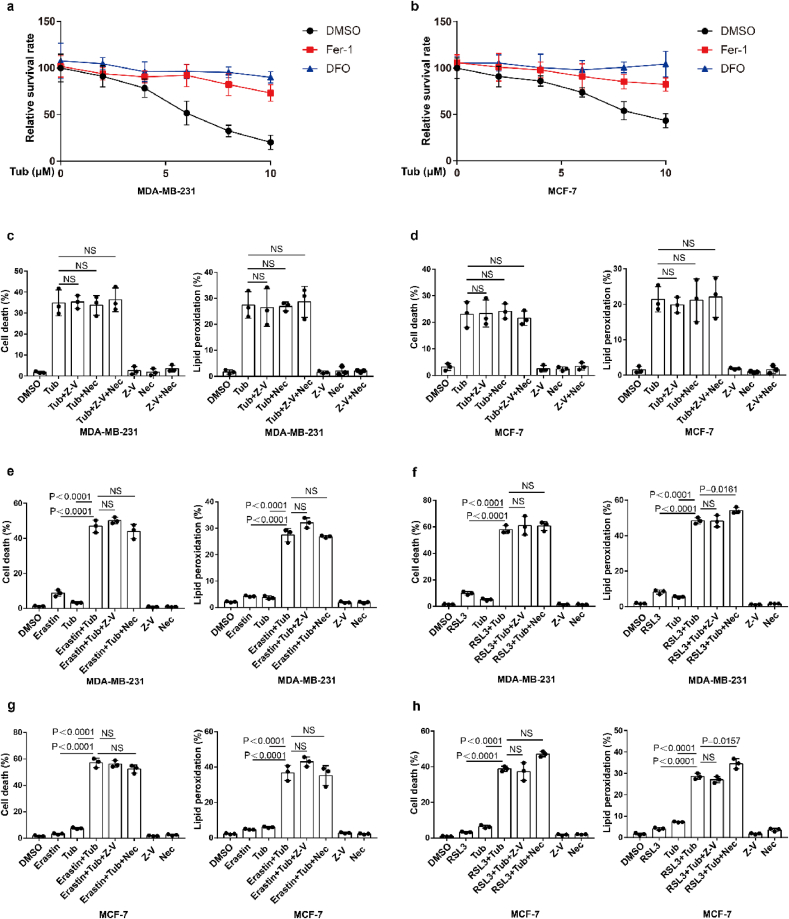
To identify novel small molecule compounds that can induce ferroptosis in cancer cells, we constructed ferroptosis-resistant cell line which ferroptosis driving gene ACSL4 was knocked out in MDA-MB-231 and named MDA-MB-231-ACSL4-KO, and MDA-MB-231-ACSL4-WT was used as a ferroptosis-sensitive cell line. We screened an epigenetic small molecule drug library of 181 compounds for activity in an assay based on cell death in MDA-MB-231-ACSL4-WT and MDA-MB-231-ACSL4-KO cells. Our results showed that Tubastatin A significantly induced cell death in ferroptosis-sensitive cell lines without affecting cell viability of ferroptosis-resistant cell line (Fig. 1a and b). We further used two breast cancer cell lines, MDA-MB-231 and MCF-7, to confirm the results. We found that Tubastatin A could induce ferroptosis and lipid peroxidation in MDA-MB-231 and MCF-7 cells, and Tubastatin A -induced cell death and lipid peroxidation could be fully rescued by the ferroptosis inhibitor Fer-1 (Lipid peroxidation scavenger) or DFO (iron chelator) but not by the apoptosis inhibitor Z-VAD-FMK or the necroptosis inhibitor necrostatin-1s (Fig. 1c and d; Extended Data Fig. 1a–d). Next, we further explored whether Tubastatin A enhanced erastin- or RSL3-induced ferroptosis and lipid peroxidation in cancer cells. Our results showed that treatment with these compounds at low concentrations alone did not effectively induce cell death in MDA-MB-231 and MCF-7 cells. The combination of Tubastatin A and erastin or RSL3 significantly induced cell death and lipid peroxidation in cancer cells (Fig. 1e–j), whereas the increased cell death and lipid peroxidation were substantially restored by treatment with the ferroptosis inhibitor Fer-1 or DFO, rather than the apoptosis inhibitor Z-VAD-FMK or the necroptosis inhibitor necrostatin-1s (Extended Data Fig. 1e–h). These results showed that Tubastatin A and erastin or RSL3 synergistically promoted ferroptosis in cancer cells. Taken together, these results suggest that Tubastatin A is a direct ferroptosis inducer that induces ferroptosis in cancer cells.
Fig. 1.
Tubastatin A is a novel ferroptosis inducer. (a) MDA-MB-231-ACSL4-WT and MDA-MB-231-ACSL4-KO cells were seeded in 96-well plates overnight. Following treatment with different inhibitor, cell death was detected by MTT assay. Δ Cell death means the death rate of MDA-MB-231-ACSL4-WT cells minus the death rate of MDA-MB-231-ACSL4-KO cells. (b) Cell death and lipid peroxidation measurement in the indicated cells treated with DMSO or Tubastatin A (Tub) for 20 h. Tub, 8 μM Tubastatin. (c–d) Cell death and lipid peroxidation measurement in the indicated cells treated with the indicated compounds for 20 h (c) or 28 h (d). MDA-MB-231, Tub, 8 μM; DFO, the ferroptosis inhibitor Deferoxamine mesylate, 10 μM; Fer-1, the ferroptosis inhibitor Ferrostatin-1, 10 μM. MCF-7, Tub, 10 μM; DFO, 10 μM; Fer-1, 10 μM. (e, f) Cell death and lipid peroxidation measurement in MDA-MB-231 cells treated with the indicated compounds for 20 h. Tub, 4 μM; Erastin, 2.5 μM; RLS3, 2.5 μM; DFO, 10 μM; Fer-1, 10 μM. (g, h) Cell death and lipid peroxidation measurement in MCF-7 cells treated with the indicated compounds for 28 h. Tub, 5 μM; Erastin, 5 μM; RLS3, 2.5 μM; DFO, 10 μM; Fer-1, 10 μM. (i, j) Cell viability measurement in MDA-MB-231 or MCF-7 cells treated with the indicated compounds for 24 h or 32 h, respectively. For MDA-MB-231, Tub, 4 μM; Erastin, 2.5 μM; RLS3, 2.5 μM. For MCF-7, Tub, 5 μM; Erastin, 5 μM; RLS3, 2.5 μM. b-h, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test.
2.2. Tubastatin A induces ferroptosis independent of HDAC6
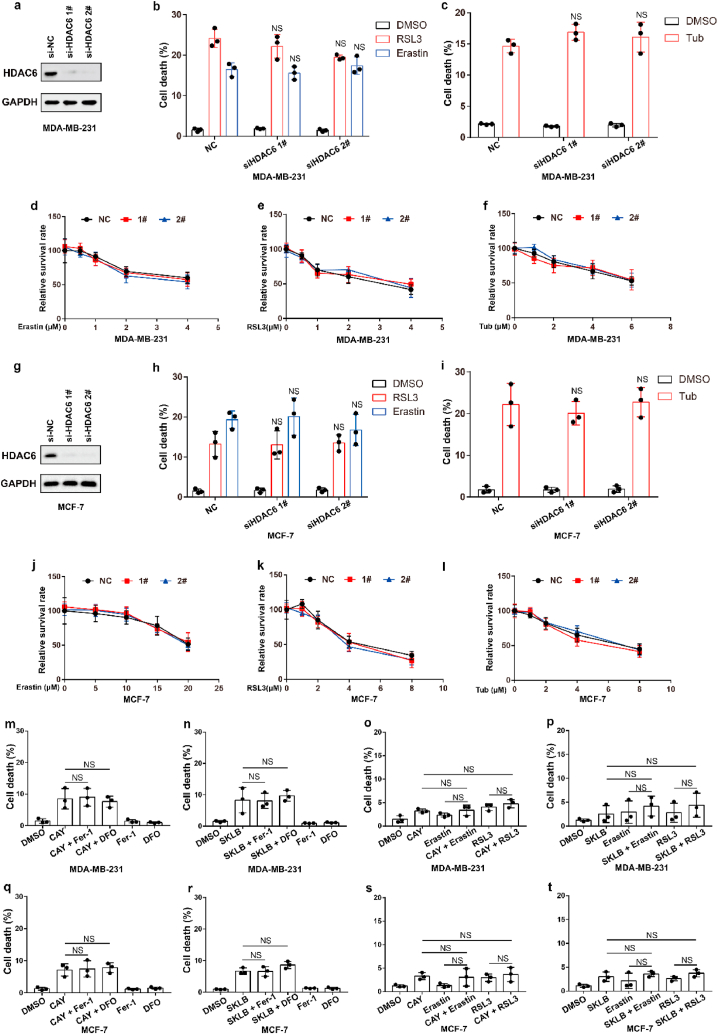
We next sought to study the underlying mechanism by which Tubastatin A induced ferroptosis in cancer cells. Tubastatin A is a specific HDAC6 inhibitor. Therefore, we speculated that Tubastatin A might induce ferroptosis by inhibiting HDAC6. To assess this possibility, HDAC6 was knocked down in MDA-MB-231 or MCF-7 cells. We found that HDAC6 knockdown had no effect on erastin- or RSL3-induced ferroptosis (Fig. 2a, b, d, e, g, h, j, k), suggesting that HDAC6 inhibition could not induce ferroptosis in cancer cells. Our data also showed that knockdown of HDAC6 did not affect Tubastatin A -induced ferroptosis in cancer cells (Fig. 2c, f, i, l). In addition, we treated cancer cells with two other HDAC6 inhibitors (CAY10603 and SKLB-23bb) that differ from the chemical structure of Tubastatin A, and the results showed that CAY10603 or SKLB-23bb neither induced ferroptosis nor promoted erastin- or RSL3-induced ferroptosis in cancer cells (Fig. 2m-t). These data indicate that Tubastatin A induces ferroptosis independent of HDAC6.
Fig. 2.
Tubastatin A induces ferroptosis independent of HDAC6. (a, g) An immunoblot showing the expression of HDAC6 in MDA-MB-231 (a) and MCF-7 (g) cells subjected to the indicated treatments. Data are representative of n = 3 biologically independent experiments. (b) Cell death measurement in the indicated MDA-MB-231 cells treated with DMSO or 6 μM Erastin or 5 μM RSL3 for 20 h. (c) Cell death measurement in the indicated MDA-MB-231 cells treated with DMSO or 8 μM Tub for 20 h. (d–f) Cell viability measurements in MDA-MB-231-NC or HDAC6 knocked-down MDA-MB-231 cells subjected to the indicated treatments for 36 h. (h) Cell death measurement in the indicated MCF-7 cells treated with DMSO or 25 μM Erastin or 8 μM RSL3 for 20 h. (i) Cell death measurement in the indicated MCF-7 cells treated with DMSO or 10 μM Tub for 20 h. (j–l) Cell viability measurements in MCF-7-NC or HDAC6 knocked-down MCF-7 cells subjected to the indicated treatments for 36 h. (m, n) Cell death measurement in MDA-MB-231 cells treated with the indicated compounds for 20 h. CAY, 8 μM CAY10603; SKLB, 8 μM SKLB-23bb. (o, p) Cell death measurement in MDA-MB-231 cells treated with the indicated compounds for 24 h. CAY, 4 μM CAY10603; Erastin, 2.5 μM; RLS3, 2.5 μM; SKLB, 4 μM SKLB-23bb. (q, r) Cell death measurement in MCF-7 cells treated with the indicated compounds for 28 h. CAY, 10 μM CAY10603; SKLB, 10 μM SKLB-23bb. (s, t) Cell death measurement in MCF-7 cells treated with the indicated compounds for 32 h. CAY, 5 μM CAY10603; Erastin, 5 μM; RLS3, 2.5 μM; SKLB, 5 μM SKLB-23bb. b, c, h, i, m-t, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. d-f, j-l, Error bars are mean ± s.d., n = 3 independent repeats. Statistical analysis was performed using a two-way ANOVA analysis.
2.3. Tubastatin A induces ferroptosis by directly inhibiting the enzymatic activity of GPX4
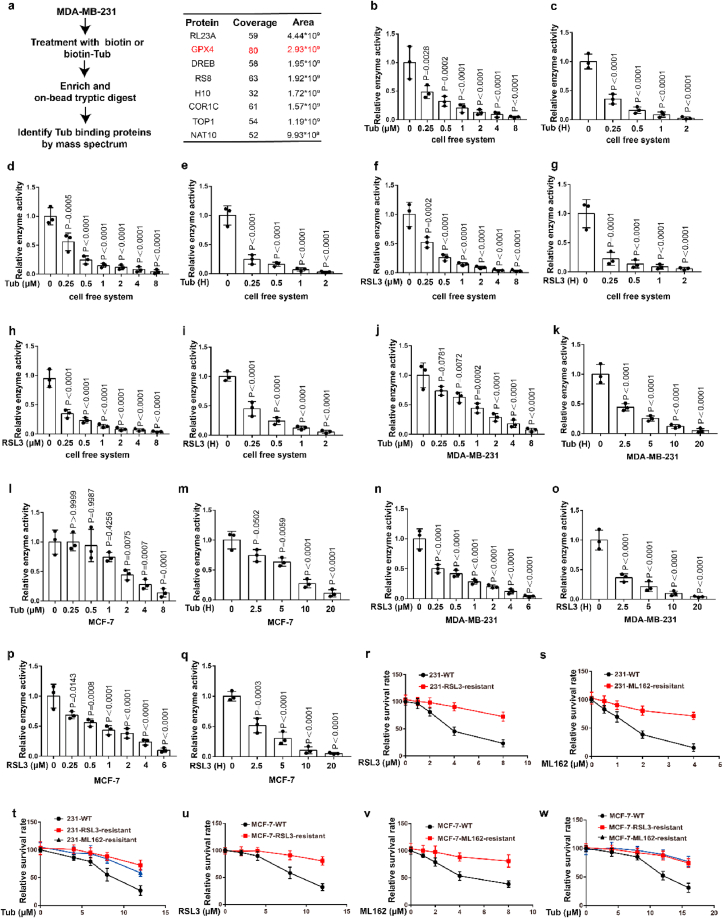
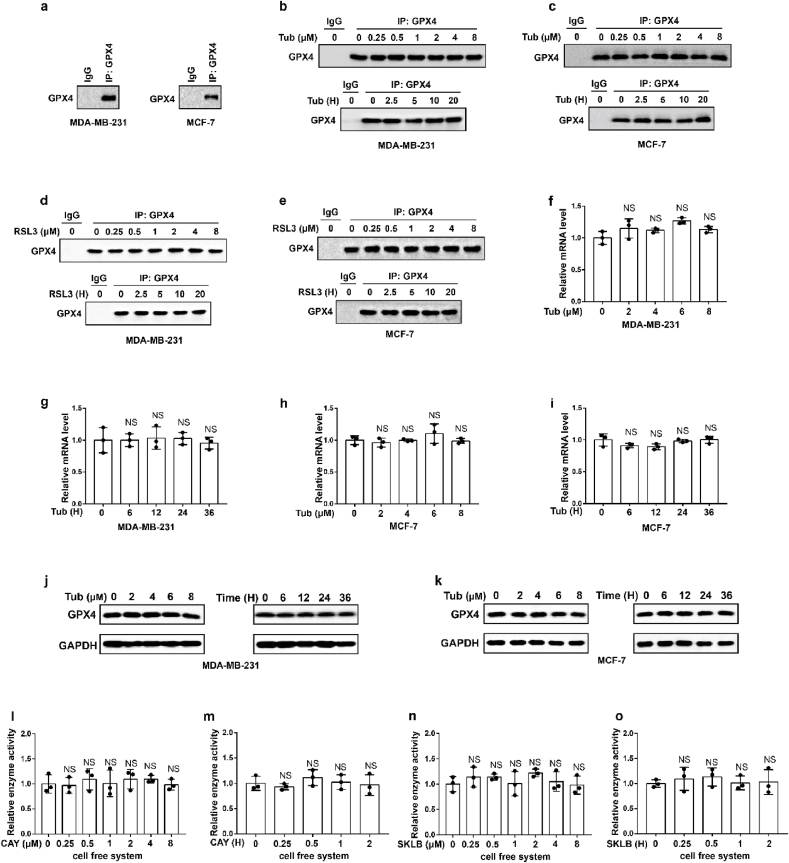
To further explore the molecular mechanism of Tubastatin A-induced ferroptosis in cancer cells, a biotin-streptavidin pulldown was performed with Tubastatin A. Briefly, MDA-MB-231 cells were treated with control or biotin-linked Tubastatin A, and Tubastatin A-binding proteins were enriched on streptavidin beads and subjected to an on-bead trypsin digestion and subsequent LC/LC–MS/MS analysis (Fig. 3a, left). Protein identities were determined by database searches using the SEQUEST algorithm. Surprisingly, GPX4 was identified as a target of Tubastatin A (Fig. 3a, right). GPX4 is an important enzyme that removes lipid peroxidation and subsequently inhibits ferroptosis. Therefore, we further explored whether Tubastatin A regulated ferroptosis by affecting the activity of GPX4. First, we determined whether Tubastatin A was an immediate inhibitor of GPX4 in vitro. We used co-immunoprecipitation to isolate endogenous GPX4 from MDA-MB-231 or MCF-7 cells (Extended Data Fig. 2a). Then, Tubastatin A was added to the test tube containing GPX4, and GPX4 enzyme activity was detected using the glutathione peroxidase assay kit. The results showed that, similar to RSL3, Tubastatin A inhibited GPX4 enzyme activity in a concentration- and time-dependent manner in vitro, even in the nanomolar range (Fig. 3b–i). To further confirm the results, we treated MDA-MB-231 and MCF-7 cells with Tubastatin A or the GPX4 inhibitor RSL3 at the indicated concentrations and times. The cells were collected and co-immunoprecipitation assays were used to isolate endogenous GPX4 from the cells (Extended Data Fig. 2b–e). Then, the GPX4 enzymatic activity of each group was analysed with a glutathione peroxidase assay kit. Our results show that, similar to RSL3, Tubastatin A inhibited GPX4 enzyme activity in a concentration- and time-dependent manner in MDA-MB-231 and MCF-7 cells (Fig. 3j-q). In addition, Tubastatin A did not affect the mRNA and protein levels of GPX4 (Extended Data Fig. 2f–k), and we also found that CAY10603 and SKLB-23bb did not affect the enzymatic activity of GPX4 (Extended Data Fig. 2l-o). To further verify the above results, we successfully constructed MDA-MB-231 cell lines that are resistant to the GPX4 inhibitor RSL3 or ML-162 and named them MDA-MB-231-RSL3-resistant or MDA-MB-231-ML-162-resistant, respectively. During the construction of RSL3 or ML162-resistant cancer cell lines, cancer cells gradually adapt to the inhibited state of GPX4 and survive. These cells may not rely on GPX4 to inhibit ferroptosis, so other drugs that inhibit GPX4 enzyme activity or lower GPX4 may not kill these cancer cells. As expected, MDA-MB-231-RSL3-resistant or MDA-MB-231-ML-162-resistant cell lines also showed obvious resistance to Tubastatin A-mediated ferroptosis (Fig. 3r-w). These results indicated that Tubastatin A induced ferroptosis by targeting GPX4. Taken together, these results suggest that Tubastatin A induces ferroptosis by directly inhibiting GPX4 enzymatic activity.
Fig. 3.
Tubastatin A induces ferroptosis by directly inhibiting the enzymatic activity of GPX4. (a) (left) Schematic of biotin-streptavidin pulldown method: MDA-MB-231 cells were treated with biotin or biotin-linked Tub, and Tub-binding proteins were enriched on streptavidin beads, subjected to an on-bead trypsin digestion and subsequent LC/LC–MS/MS analysis. (right) Enrichment of proteins based on coverage and area. GPX4 was a top target candidate. (b–i) Relative GPX4 enzyme activity measurement in the indicated test tube treated with Tub or RSL3 for the indicated concentration and time in cell free system. b, c, f, g, we used co-immunoprecipitation to isolate endogenous GPX4 from MDA-MB-231 cells. d, e, h, i, we used co-immunoprecipitation to isolate endogenous GPX4 from MCF-7 cells. b, time: 2 h c, concentration: 8 μM. d, time: 2 h e, concentration: 8 μM. f, time: 2 h g, concentration: 8 μM. h, time: 2 h i, concentration: 8 μM. (j–q) Relative GPX4 enzyme activity measurement in the indicated cell treated with Tub or RSL3 for the indicated concentration and time. j, time: 20 h k, concentration: 8 μM. l, time: 20 h m, concentration: 8 μM. n, time: 20 h o, concentration: 6 μM. p, time: 20 h q, concentration: 6 μM. (r–w) Cell viability measurements in the indicated cells subjected to the indicated treatments for 28 h (r–t) or 36 h (u–w). b-q, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. r-w, Error bars are mean ± s.d., n = 3 independent repeats. Statistical analysis was performed using a two-way ANOVA analysis.
2.4. Cancer cells acquire radioresistance by increasing transcription and protein stability of GPX4
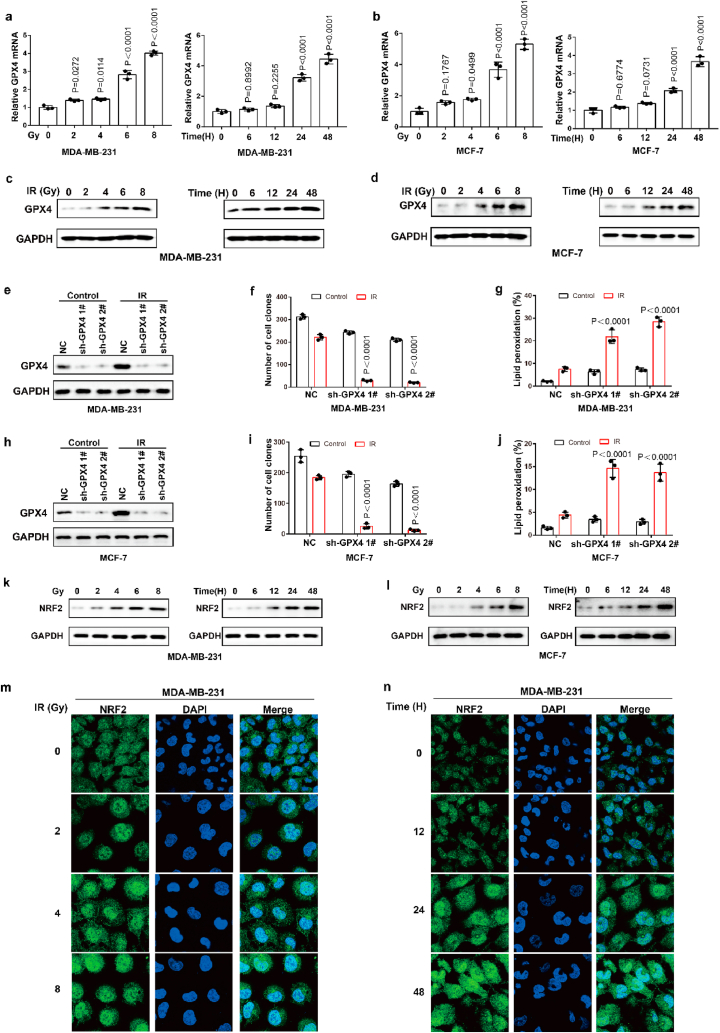
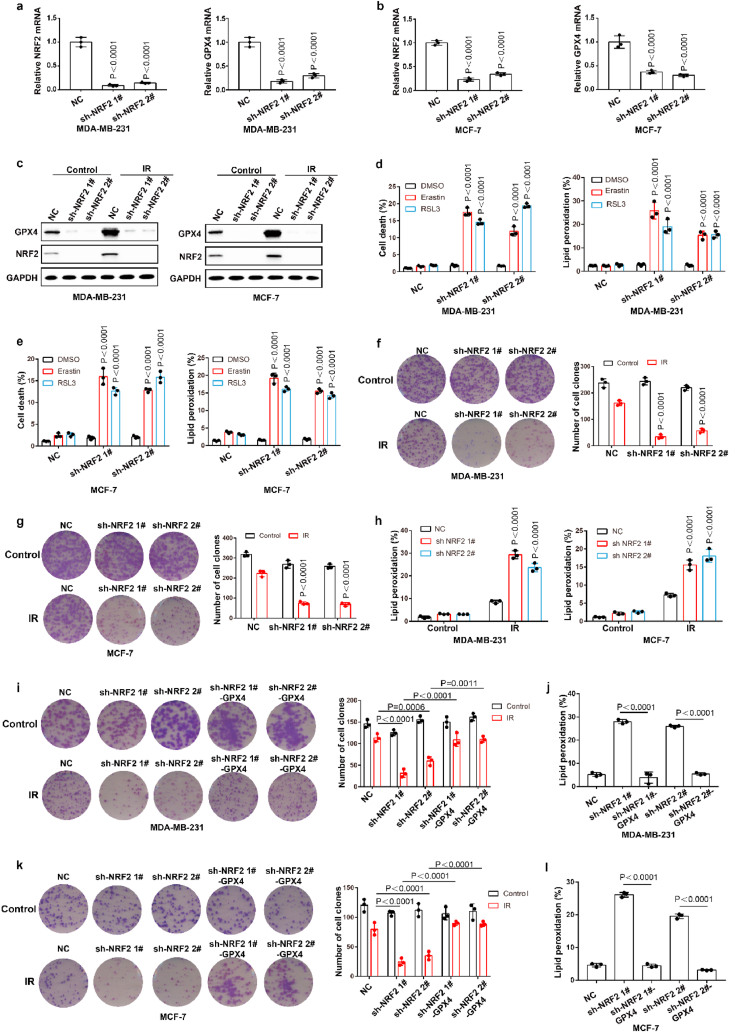
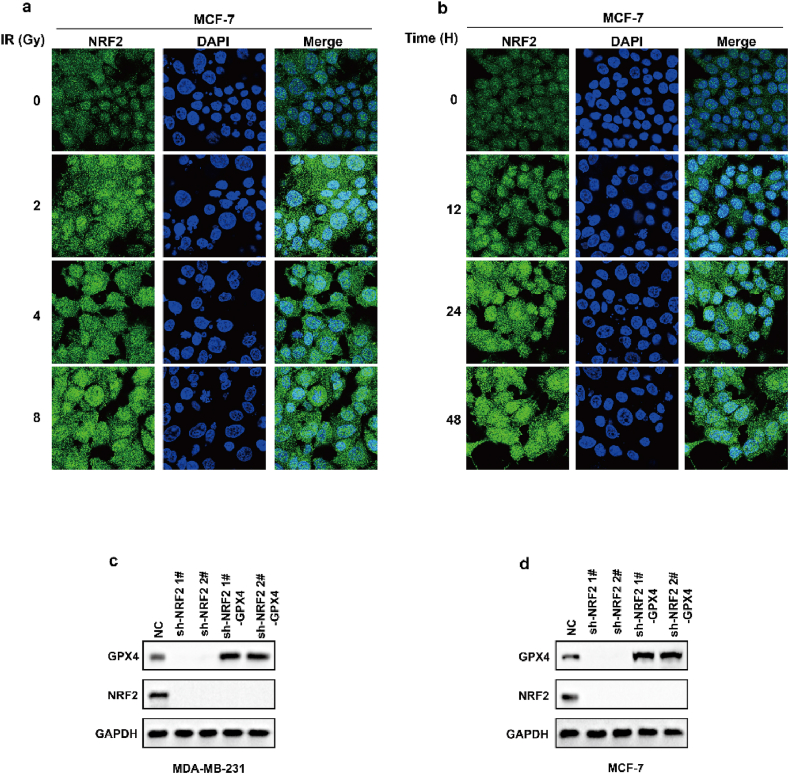
Ionizing radiation (IR) is an important ferroptosis-related pathological process. IR can induce ferroptosis in cancer cells, and blocking ferroptosis reduce the effect of IR, suggesting that ferroptosis contributes to IR-mediated antitumor activity. However, here we found that IR increased the mRNA and protein levels of GPX4 in an intensity-and time-dependent manner in MDA-MB-231 and MCF-7 cells (Fig. 4a–d). As one of the most important ferroptosis inhibitory factors, the elevated protein levels of GPX4 might promote IR tolerance by inhibiting ferroptosis. To assess this possibility, we knocked down GPX4 and detected the cell viability of cancer cells treated with IR. Our data indicated that GPX4 knockdown significantly enhanced the sensitivity of cancer cells to radiotherapy, as shown by reduced cell clones and increased ferroptosis-associated lipid peroxidation (Fig. 4e–j). These results indicated that IR-mediated GPX4 expression promotes ferroptosis resistance and radioresistance in cancer cells. Next, we explored the molecular mechanism of the high expression of GPX4 during IR. Our results showed that IR increased the protein level and nuclear localization of Nrf2 in an intensity- and time-dependent manner in MDA-MB-231 and MCF-7 cells (Fig. 4k-n; Extended Data Fig. 3a and b). Nrf2 is a key transcription factor that can regulate oxidative stress. We hypothesized that IR-mediated Nrf2 activation promoted the expression of GPX4, and subsequently induced radioresistance. To evaluate whether Nrf2 regulated the expression of GPX4, we knocked down Nrf2 and detected the expression of GPX4 in cancer cells treated with or without IR. Nrf2 knockdown significantly inhibited basal and IR-mediated GPX4 mRNA and protein levels in MDA-MB-231 or MCF-7 cells (Fig. 5a–c). In addition, Nrf2 knockdown significantly enhanced erastin- or RSL3-or IR-induced ferroptosis and lipid peroxidation in cancer cells (Fig. 5d–h). Next, we restored GPX4 in cancer cells in which Nrf2 was knocked down (Extended Data Fig. 3c and d). The results showed that GPX4 restoration could reverse the effect of Nrf2 knockdown on IR-induced ferroptosis in cancer cells, as shown by decreased lipid peroxidation and increased colony formation in cancer cells (Fig. 5i-l). Taken together, these results indicated that Nrf2 contributes to enhanced GPX4 expression and subsequent ferroptosis resistance and radioresistance in cancer cells.
Fig. 4.
IR enhances GPX4 expression by increasing the transcription of GPX4. (a–b) The relative GPX4 mRNA measurement in MDA-MB-231 cells (a) or MCF-7 (b) treated with IR at the indicated intensities and times. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. (c–d) An immunoblot showing the expression of GPX4 in MDA-MB-231 cells (c) or MCF-7 (d) treated with IR at the indicated intensities and times. Data are representative of n = 3 biologically independent experiments. (e, h) An immunoblot showing the expression of GPX4 in MDA-MB-231 (e) and MCF-7 (h) cells subjected to the indicated treatments. Data are representative of n = 3 biologically independent experiments. (f, g) Cell clone measurement (f) or lipid peroxidation measurement (g) in control or GPX4 knocked-down MDA-MB-231 cells subjected to the indicated treatments. For cell clone, 2 Gy IR; for lipid peroxidation measurement, 6 Gy IR. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. (f, g) Cell clone measurement (f) or lipid peroxidation measurement (g) in control or GPX4 knocked-down MDA-MB-231 cells subjected to the indicated treatments. For cell clone, 2 Gy IR; for lipid peroxidation measurement, 6 Gy IR. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. (i, j) Cell clone measurement (i) or lipid peroxidation measurement (j) in control or GPX4 knocked-down MCF-7 cells subjected to the indicated treatments. For cell clone, 2 Gy IR; for lipid peroxidation measurement, 6 Gy IR. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. (k, l) An immunoblot showing the expression of NRF2 in MDA-MB-231 cells (k) or MCF-7 (l) treated with IR at the indicated intensities and times. Data are representative of n = 3 biologically independent experiments. (m, n) MDA-MB-231 cells treated with IR at the indicated intensities and times, then confocal microscopy of the cells was performed after staining with Nrf2 (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Nrf2 contributed to enhanced GPX4 expression and subsequently ferroptosis resistance and radioresistance in cancer cells. (a–b) The relative Nrf2 and GPX4 mRNA measurement in MDA-MB-231 cells (a) or MCF-7 cells (b) stable transfected with negative control (NC) or Nrf2 shRNA. (c) An immunoblot showing the expression of GPX4 and Nrf2 in MDA-MB-231 cells (left) or MCF-7 (right) stable transfected with negative control (NC) or Nrf2 shRNA. Data are representative of n = 3 biologically independent experiments. (d–e) Cell death and lipid peroxidation measurement in the indicated MDA-MB-231 cells (d) or MCF-7 cells (e) treated with the indicated compounds for 16 h (d) or 24 h (e). (d) Erastin, 8 μM; RLS3, 5 μM. (e) Erastin, 10 μM; RLS3, 7.5 μM. (f, g) Cell clone measurement in the indicated MDA-MB-231 cells (f) or MCF-7 cells (g) treated with the 2 Gy IR. (h) Lipid peroxidation measurement in the indicated MDA-MB-231 cells (left) or MCF-7 cells (right) treated with the 6 Gy IR. (i, k) Cell clone measurement in the indicated MDA-MB-231 cells (i) or MCF-7 cells (k) treated with the 2 Gy IR. (j, l) Lipid peroxidation measurement in the indicated MDA-MB-231 cells (j) or MCF-7 cells (l) treated with the 6 Gy IR. a-b, d-l, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test.
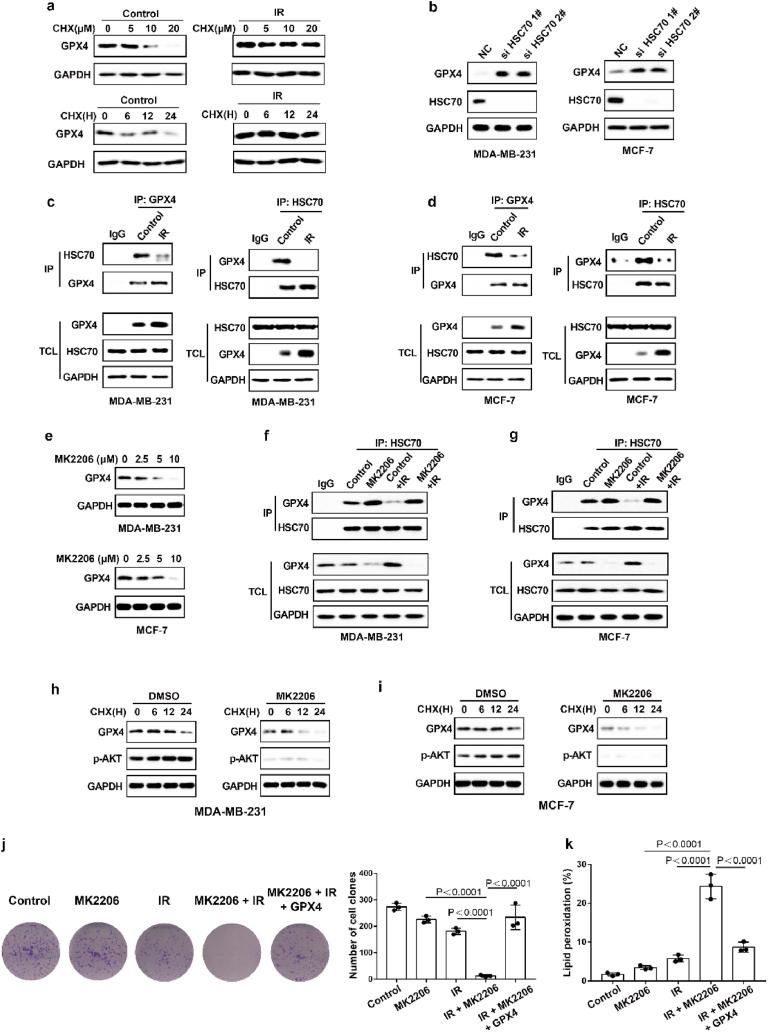
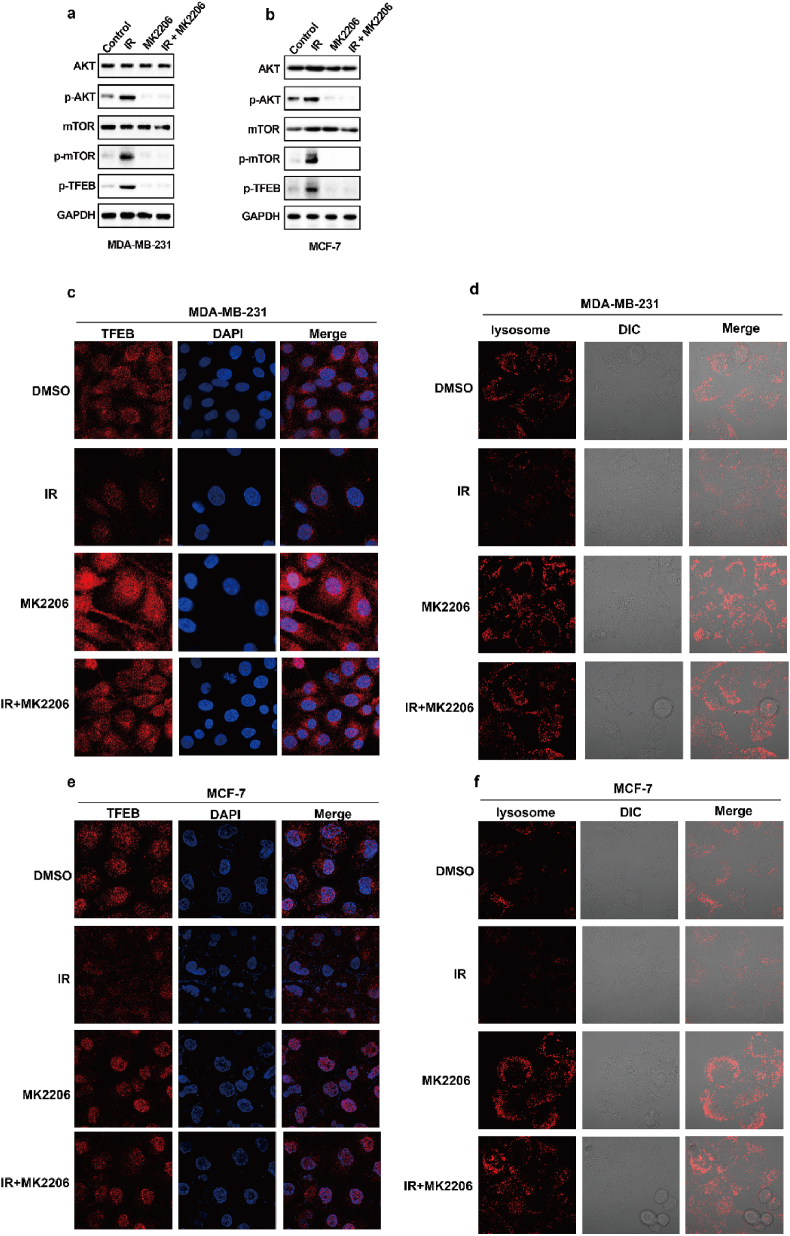
As shown in Fig. 4a–d, the degree of IR-mediated increase in GPX4 mRNA levels does not exactly match the extent of IR-mediated elevation in GPX4 protein levels, and we speculated that IR might also enhance GPX4 expression by promoting GPX4 stability. To determine whether IR enhanced the protein stability of GPX4, we performed GPX4 degradation experiments under treatment with cycloheximide (CHX), a protein synthesis inhibitor. The results show that IR could significantly inhibit the degradation of GPX4 (Fig. 6a). Wu Z et al. found that GPX4 was degraded by chaperone-mediated autophagy. We knocked down HSC70 in MDA-MB-231 and MCF-7 cells, and found that HSC70 knockdown increased the levels of GPX4 protein (Fig. 6b). More importantly, our data showed that IR attenuated the autophagic degradation of GPX4, as shown by decreased binding of GPX4 and HSC70 (Fig. 6c and d). Next, to identify kinases that regulate GPX4 degradation, we screened a kinase inhibitor library for activity in an assay based on the levels of GPX4 protein. Our data showed that the AKT inhibitor MK2206 significantly inhibited GPX4 protein levels in the treatment of cancer cells with IR (Fig. 6e). Moreover, MK2206 enhanced the binding of GPX4 to HSC70 in cancer cells treated with control or IR (Fig. 6f and g). These results suggest that AKT might inhibit the autophagic degradation of GPX4 and then promote the stability of GPX4. To assess this possibility, the cancer cells were treated with IR plus an AKT inhibitor, and MK2206 significantly enhanced the degradation of GPX4 (Fig. 6h and i). Next, we explored the molecular mechanism by which AKT regulates the autophagic degradation of GPX4. We found that IR significantly activated the AKT-mTOR-TFEB signalling pathway and subsequently strongly inhibited the nuclear localization of TFEB (Extended Data Fig. 4a, b, c, e). TFEB is an important transcription factor that mediates lysosomal biosynthesis. These results suggest that IR-mediated AKT activation might inhibit the autophagic degradation of GPX4 by inhibiting lysosomal biosynthesis. Immunofluorescence revealed that the number of lysosomes was significantly reduced in IR-treated cancer cells compared with control cancer cells, whereas the reduced lysosomes were substantially restored by treatment with the AKT inhibitor MK2206 (Extended Data Fig. 4d, f). These results suggest that activation of the AKT-mTOR-TFEB pathway leads to a reduction in lysosomal function, which coincides with reduced binding of HSC70 and GPX4 and subsequently inhibits the autophagic degradation of GPX4.
Fig. 6.
IR enhances GPX4 expression by increasing the protein stability of GPX4. (a) An immunoblot showing the expression of GPX4 in MDA-MB-231 cells treatment with control or IR with or without CHX. Data are representative of n = 3 biologically independent experiments. (b) An immunoblot showing the expression of GPX4 and Hsc70 in MDA-MB-231 cells (left) or MCF-7 (right) transfected with negative control (NC) or Hsc70 siRNA. Data are representative of n = 3 biologically independent experiments. (c) Endogenous GPX4 (left) or Hsc70 (right) was immunoprecipitated from MDA-MB-231 cells subjected to the indicated treatments for 16 h, followed by immunoblotting using an antibody to GPX4 and Hsc70. IR, 4 Gy. Data are representative of n = 3 biologically independent experiments. (d) Endogenous GPX4 (left) or Hsc70 (right) was immunoprecipitated from MCF-7 cells subjected to the indicated treatments for 16 h, followed by immunoblotting using an antibody to GPX4 and Hsc70. IR, 6 Gy. Data are representative of n = 3 biologically independent experiments. (e) An immunoblot showing the expression of GPX4 in IR-treated MDA-MB-231 cells (top) or MCF-7 cells (bottom) treatment with MK2206 at the indicated concentration and time. Data are representative of n = 3 biologically independent experiments. (f, g) Endogenous Hsc70 was immunoprecipitated from MDA-MB-231 cells (f) or MCF-7 cells (g) subjected to the indicated treatments for 12 h, followed by immunoblotting using an antibody to GPX4 and Hsc70. (f) IR, 4 Gy; MK2206 2.5 μM. (g) IR, 6 Gy; MK2206 2.5 μM. Data are representative of n = 3 biologically independent experiments. (h, i) An immunoblot showing the expression of GPX4 in MDA-MB-231 cells (h) or MCF-7 cells (i) treatment with DMSO or MK2206 with or without CHX. Data are representative of n = 3 biologically independent experiments. (j) Cell clone measurement in the indicated MDA-MB-231 cells treated with the 2 Gy IR with or without 2.5 μM MK2206. (k) Lipid peroxidation measurement in the indicated MDA-MB-231 cells treated with the 6 Gy IR with or without 2.5 μM MK2206. j, k, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test.
In addition, MK2206 significantly enhanced IR-induced ferroptosis and lipid peroxidation in cancer cells (Fig. 6j and k). Next, we restored GPX4 in cancer cells treated with MK2206. The results show that GPX4 restoration can reverse the effect of MK2206 on IR-induced ferroptosis in cancer cells, as shown by decreased lipid peroxidation and increased colony formation in cancer cells (Fig. 6j and k). Taken together, these results indicate that AKT-mediated inhibition of GPX4 degradation contributes to enhanced GPX4 expression and subsequent ferroptosis resistance and radioresistance in cancer cells.
2.5. Tubastatin A overcomes radioresistance of cancer cells in vitro and in vivo by inhibiting GPX4 and boosting ferroptosis
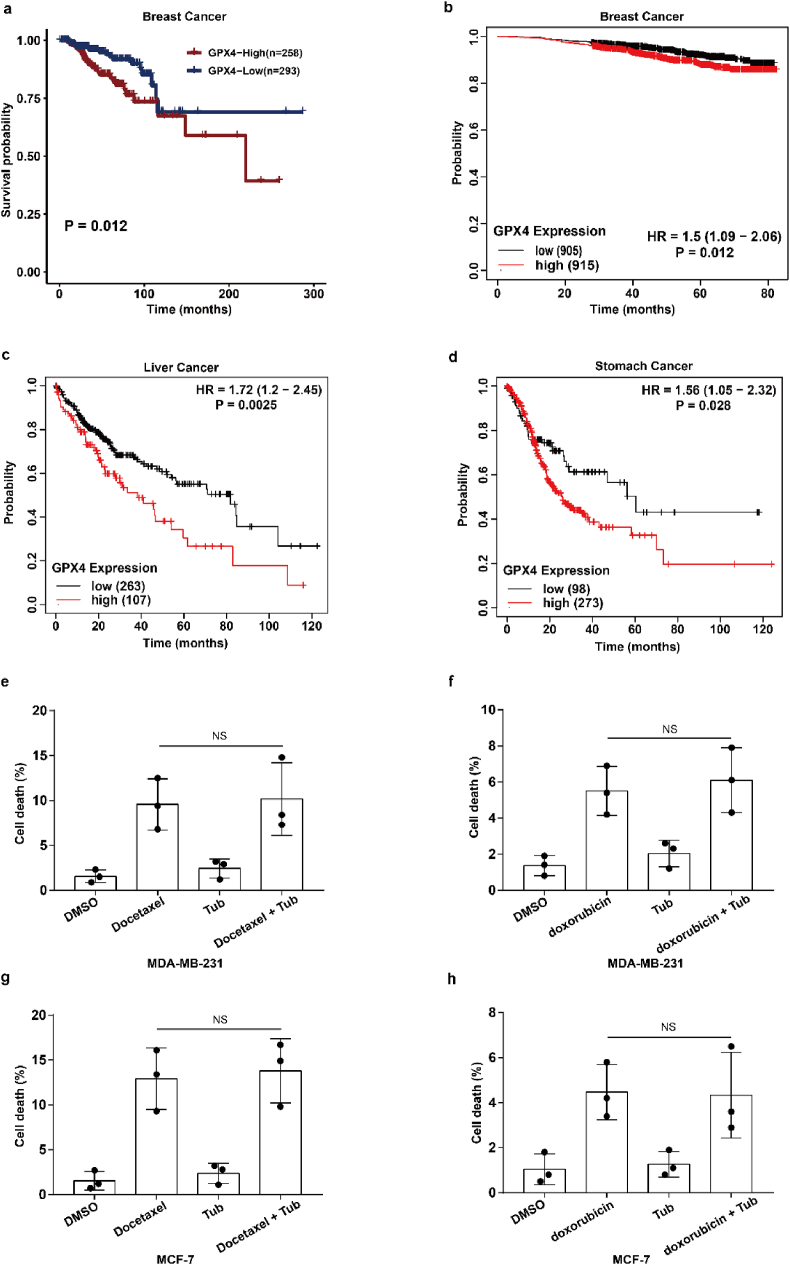
To assess the clinical significance of IR-mediated GPX4 expression, we explored the relationship between GPX4 expression and radiotherapy response in cancer patients. Our results show that patients with high expression of GPX4 had a shorter survival period after radiotherapy, which indicates that high expression of GPX4 might mediate radioresistance and weaken the efficacy of radiotherapy in clinical practice (Extended Data Fig. 5a). In addition, Kaplan-Meier survival analysis showed that breast cancer, liver cancer and gastric cancer patients with high GPX4 expression had worse overall survival, suggesting that GPX4 should be a potential tumour prognostic indicator (Extended Data Fig. 5b–d). Taken together, these results suggest that cancer cells acquire radioresistance by increasing the expression of GPX4, and GPX4 could be used for clinical application as a predictor of the response to cancer radiotherapy.
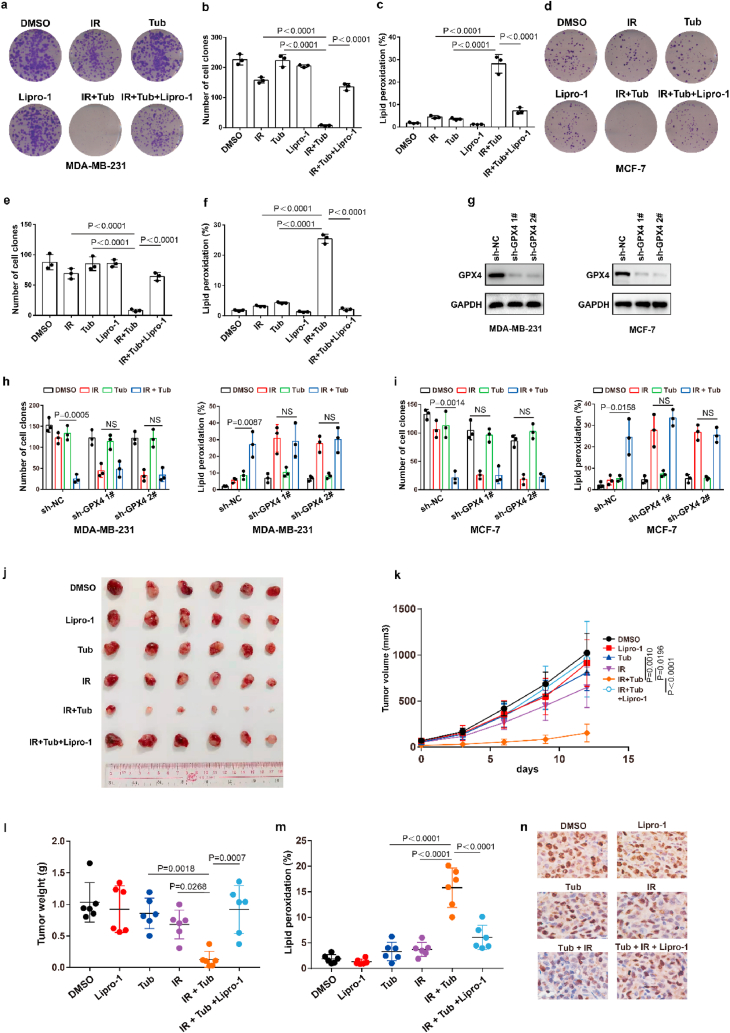
Our above results showed that Tubastatin A inhibited GPX4 enzymatic activity by directly targeting GPX4. Thus, we speculated that Tubastatin A might overcome the radioresistance of cancer cells by inhibiting GPX4. To assess this possibility, MDA-MB-231 and MCF-7 cells were treated with IR with DMSO or Tubastatin A or Lipro-1. The results showed that Tubastatin A and IR synergistically reduced the colonies and increased lipid peroxidation in MDA-MB-231 and MCF-7 cells, whereas the reduced cell colonies and increased lipid peroxidation were substantially restored by treatment with the ferroptosis inhibitor Lipro-1 (Fig. 7a–f). In addition, we found that Tubastatin A combined with docetaxel or doxorubicin could not enhance the tumour suppressive effect of docetaxel or doxorubicin in MDA-MB-231 and MCF-7 cells (Extended Data Fig. 5e–h). These results suggest that Tubastatin A can specifically sensitize radiotherapy by enhancing ferroptosis. Then, we explored whether Tubastatin A enhanced the radiosensitivity of cancer cells in a GPX4-dependent manner. We knocked down GPX4 and detected cell death and lipid peroxidation in MDA-MB-231 and MCF-7 cells treated with IR with Tub or DMSO. Our results showed that GPX4 knockdown increased IR-mediated ferroptosis in MDA-MB-231 and MCF-7 cells. More importantly, Tubastatin A significantly enhanced IR-induced ferroptosis in wild-type cancer cells, but not in GPX4 knockdown cancer cells (Fig. 7g–i). These results indicate that GPX4 is the target of Tubastatin A, and Tubastatin A enhances the sensitivity of cancer cells to IR in a GPX4-dependent manner.
Extended Data Fig. 1.
Tubastatin A promotes ferroptosis, rather than apoptosis or necrosis. (a, b) Cell viability measurements in MDA-MB-231 (a) or MCF-7 (b) cells subjected to the indicated treatments for 32 h or 36 h. Error bars are mean ± s.d., n = 3 independent repeats. Statistical analysis was performed using a two-way ANOVA analysis. (c–d) Cell death and lipid peroxidation measurement in the indicated cells treated with the indicated compounds for 20 h (c) or 28 h (d). MDA-MB-231, Tub, 8 μM; Z-V, 10 μM Z-VAD-FMK; and Nec, 2 μM necrostatin-1s. MCF-7, Tub, 10 μM; Z-V, 10 μM; and Nec, 2 μM. (e–h) Cell-death and lipid-peroxidation measurements in MDA-MB-231 or MCF-7 subjected to the indicated treatments for 20 h or 28 h. MDA-MB-231; Tub, 4 μM; Erastin, 2.5 μM; RSL3, 2.5 μM; Z-V, 10 μM Z-VAD-FMK; and Nec, 2 μM necrostatin-1s. MCF-7; Tub, 5 μM; Erastin, 5 μM; RSL3, 2.5 μM; Z-V, 10 μM Z-VAD-FMK; and Nec, 2 μM necrostatin-1s. c-h, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test.
Extended Data Fig. 2.
Tubastatin A did not affect the mRNA and protein level of GPX4.
(a–d) MDA-MB-231 or MCF-7 cells were treated with Tub or RSL3 at the indicated concentrations and times. The cells were collected and co-immunoprecipitation assays were used to isolate endogenous GPX4 from the cells. (e) Co-immunoprecipitation assays were used to isolate endogenous GPX4 from MDA-MB-231 or MCF-7 cells. (f–i) The relative GPX4 mRNA measurement in MDA-MB-231 cells (f, g) or MCF-7 cells (h, i) treated with Tub at the indicated concentrations and times. (j, k) The relative GPX4 protein measurement in MDA-MB-231 cells (j) or MCF-7 cells (k) treated with Tub at the indicated concentrations and times. f-i, statistical analysis was performed using an unpaired two-tailed Student's t-test. (l–o) Relative GPX4 enzyme activity measurement in the indicated test tube treated with CAY (CAY10603) or SKLB (SKLB-23bb) for the indicated concentration and time in cell free system. We used co-immunoprecipitation to isolate endogenous GPX4 from MDA-MB-231 cells. l, time: 2 h m, concentration: 8 μM. n, time: 2 h o, concentration: 8 μM. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. a-e, j, k, Data are representative of n = 3 biologically independent experiments.
Extended Data Fig. 3.
IR enhances GPX4 expression by increasing the transcription of GPX4. (a, b) MCF-7 cells treated with IR at the indicated intensities and times, then confocal microscopy of the cells were performed after staining with Nrf2 (green). (c, d) An immunoblot showing the expression of GPX4 and NRF2 in MDA-MB-231 (c) or MCF-7 (d) cells stable transfected with negative control (NC) or NRF2 shRNA with or without GPX4 overexpression. Data are representative of n = 3 biologically independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Extended Data Fig. 4.
Radiotherapy-mediated decrease of autophagic degradation by the AKT-mTOR-TFEB-lysosomal synthesis pathway. (a–b) An immunoblot showing the expression of the indicated protein in MDA-MB-231 cells (a) or MCF-7 cells (b) subjected to the indicated treatments for 12 h. (a) IR, 4 Gy; MK2206 2.5 μM. (b) IR, 6 Gy; MK2206 2.5 μM. Data are representative of n = 3 biologically independent experiments. (c) MDA-MB-231 cells subjected to the indicated treatments for 12h, then confocal microscopy of the cells was performed after staining with TFEB (red). IR, 4 Gy; MK2206 2.5 μM. (d) MDA-MB-231 cells subjected to the indicated treatments for 12h, then confocal microscopy of the cells was performed after staining with lysotracker (red). IR, 4 Gy; MK2206 2.5 μM. (e) MCF-7 cells subjected to the indicated treatments for 12h, then confocal microscopy of the cells was performed after staining with TFEB (red). IR, 6 Gy; MK2206 2.5 μM. (f) MCF-7 cells subjected to the indicated treatments for 12h, then confocal microscopy of the cells was performed after staining with lysotracker (red). IR, 6 Gy; MK2206 2.5 μM. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Extended Data Fig. 5.
GPX4 works as a predictor of the response to cancer radiotherapy. (a) Kaplan–Meier analysis of overall survival for breast cancer patients who have undergone radiotherapy. (b–d) Kaplan–Meier analysis of overall survival for breast cancer patients (b), liver cancer patients (c), stomach cancer patients (d). (e–h) Cell-death measurements in MDA-MB-231 or MCF-7 subjected to the indicated treatments for 24 h or 28 h. MDA-MB-231; Tub, 4 μM; docetaxel, 1.25 μM; doxorubicin, 1.25 μM. MCF-7; Tub, 5 μM; docetaxel, 1.25 μM; doxorubicin, 1.25 μM. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test.
Extended Data Fig. 6.
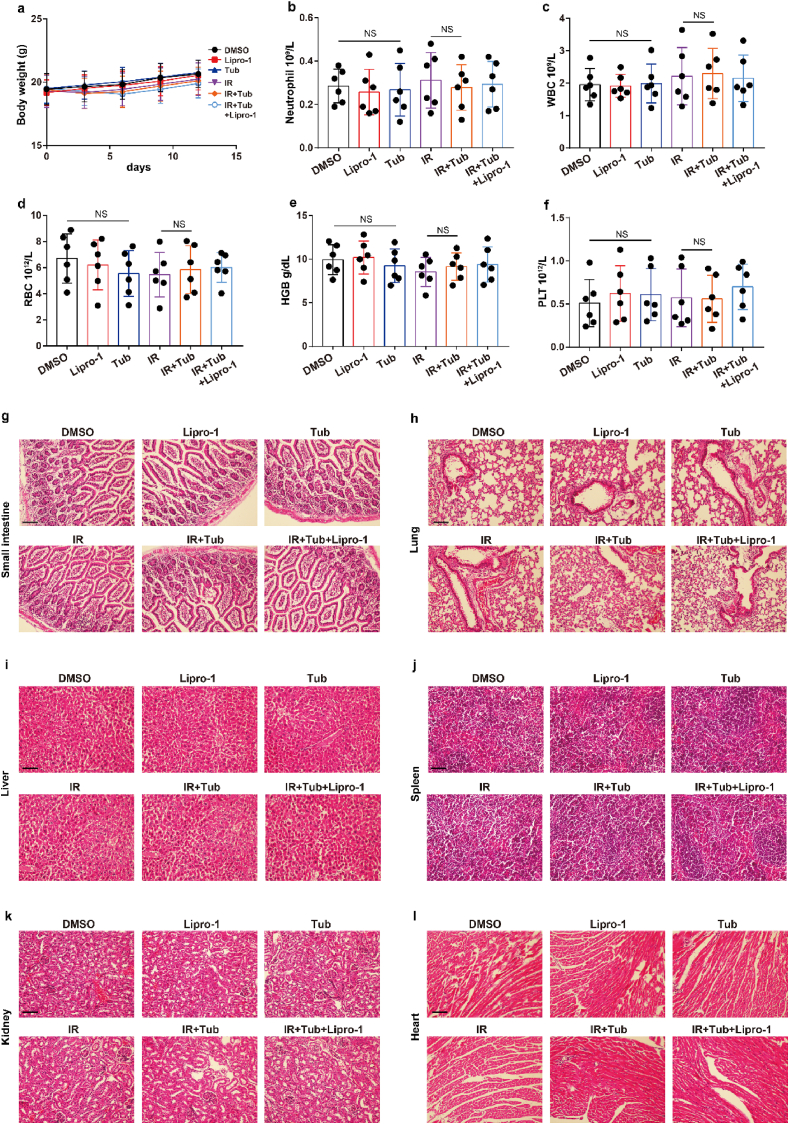
Tubastatin A has no significant toxicity of bone marrow, intestine, liver, kidney, spleen, lung, and heart. (a) Body weight of the mice in experiment Fig. 7J was measured. (b–f) Blood samples were drawn from the mice in experiment Fig. 7J, and blood cell counts were performed with the Sysemx XN-3000 Hematology System (Sysmex America, Inc.) Data represent
mean ± SD. Significance determined by two tailed Student's t-test. WBC, white blood cell; RBC, red blood cell; HGB, hemoglobin; PLT, platelet. (g–l) Small intestine, lung, liver, spleen, kidney, and heart samples were obtained from mice in experiment Fig. 6b. H&E staining was performed on processed, sliced samples. Scale bar: 200 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 7.
Tubastatin A enhances radiosensitivity of cancer cells by boosting ferroptosis. (a–f) Cell clones and lipid peroxidation measurement in MDA-MB-231 cells (a–c) or MCF-7 (d–f) treated with the indicated compounds or IR. (a–c) Cell clones: IR, 2 Gy; lipro-1, 5 μM; Tub, 4 μM. Lipid peroxidation: IR, 6 Gy; lipro-1, 5 μM; Tub, 5 μM. (d–f) Cell clones: IR, 2 Gy; lipro-1, 5 μM; Tub, 5 μM. Lipid peroxidation: IR, 6 Gy; lipro-1, 5 μM; Tub, 7.5 μM. b, c, e, f, Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. (g) An immunoblot showing the expression of GPX4 in MDA-MB-231 (left) or MCF-7 (right) cells stable transfected with negative control (NC) or GPX4 shRNA. Data are representative of n = 3 biologically independent experiments. (h, i) The number of cell clones (h) and lipid peroxidation (i) in the indicated cancer cells were treated with the indicated compounds. MDA-MB-231, cell clones: IR, 2 Gy; Tub, 4 μM. Lipid peroxidation: IR, 6 Gy; Tub, 5 μM. MCF-7, cell clones: IR, 2 Gy; Tub, 5 μM. Lipid peroxidation: IR, 6 Gy; Tub, 7.5 μM. Data are the mean ± s.d.; n = 3 biologically independent experiments. Statistical analysis was performed using an unpaired two-tailed Student's t-test. (j, k) Tumour volumes of MDA-MB-231 xenograft tumours with the indicated treatments after exposure to 10 Gy of IR. Error bars are means ± SD, n = 6 independent repeats. P values were determined using two-way ANOVA. (l) Weights of MDA-MB-231 xenograft tumours with the indicated treatments at different time points (days) after exposure to 10 Gy of IR. Error bars are means ± SD, n = 6 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. (m) Relative lipid peroxidation in tumour cells isolated from the indicated tumours. Error bars are means ± SD, n = 6 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. (n) Representative immunohistochemical images of ki67 in tumours tissues in each group.
To further confirm Tubastatin A-mediated radiosensitization in vivo, MDA-MB-231 cells were subcutaneously inoculated into nude mice. Then, the mice were treated with IR and either Tubastatin A or Lipro-1 or DMSO. As expected, Tubastatin A significantly enhanced IR-induced ferroptosis in vivo, and Lipro-1 treatment substantially restored the reduced tumour growth and increased lipid peroxidation in mice (Fig. 7j-m). We also found that the treatment of Tubastatin A did not affect the body weight of mice, the number of blood cells (including white blood cell, red blood cell, platelet) and the level of hemoglobin (HGB) and the tissue morphology of intestine, liver, kidney, spleen, lung, and heart, indicating that Tubastatin A has no significant toxicity to bone marrow, intestine, liver, kidney, spleen, lung, and heart (Extended Data Fig. 6). In addition, our data revealed that tumour tissues from Tubastatin A and IR treatment mice had weak staining for Ki67 compared with those from other groups of mice (Fig. 7n). Taken together, these results show that increased GPX4 during IR can promote IR resistance and that Tubastatin A sensitizes cancer cells to IR-induced ferroptosis in vitro and in vivo by directly targeting GPX4.
3. Discussion
Ferroptosis, an iron-dependent and lipid peroxidation-driven programmed cell death, is implicated in the occurrence, development and treatment of cancer [[22], [23], [24], [25], [26], [27], [28]]. Ferroptosis has been reported to be involved in radiotherapy- and immunotherapy-mediated cell death [17,19,[29], [30], [31], [32], [33]], which breaks the long-standing model that apoptosis is the theme of cancer therapy, suggesting that ferroptosis may be the next breakthrough in cancer therapy. Here, we performed drug screening in ferroptosis-resistant cancer cell lines, and we identified Tubastatin A as a novel ferroptosis inducer through small molecule drug library screening. Tubastatin A induced ferroptosis by directly interacting with and inhibiting the enzymatic activity of GPX4 in cancer cells, which is independent of HDAC6 inhibition. We found that knockdown of HDAC6 had no effect on ferroptosis induced by the ferroptosis inducers erastin or RSL3 or Tubastatin A in cancer cells. However, one study indicated that knockdown of HDAC6 could weakly inhibit ferroptosis induced by extracellular histone H3 in RAW264.7 macrophages [34]. This different conclusion may be due to different cell models and methods of inducing ferroptosis. We also found that IR activates not only Nrf2-mediated GPX4 transcription but also inhibits lysosome-mediated GPX4 degradation, which subsequently induces ferroptosis tolerance and radioresistance in cancer cells. Tubastatin A overcame ferroptosis resistance and radioresistance of cancer cells by inhibiting GPX4 enzymatic activity. More importantly, Tubastatin A has excellent bioavailability, as demonstrated by its ability to significantly promote radiotherapy-induced lipid peroxidation and tumour suppression in a mouse xenograft model. Considering that Tubastatin A has stronger efficacy in vivo, it can be used as a single drug or in combination with radiotherapy in clinically in the future.
Radiotherapy is an important treatment for cancer. Many kinds of cancer are resistant to radiotherapy [[35], [36], [37]]. Thus, elucidating the molecular mechanism of cancer cell resistance to radiotherapy and targeting this mechanism to enhance the radiosensitivity of cancer are urgent problems to be solved in the field of cancer therapy. Previous studies have shown that enhanced autophagy, reduced apoptosis, and enhanced DNA damage repair facilitated radioresistance in cancer cells [[38], [39], [40], [41], [42], [43]]. Here, our study shows that cancer cells can acquire radioresistance by enhancing the levels of GPX4 protein. These data suggest that GPX4-mediated ferroptosis resistance is one of the important mechanisms by which cancer cells develop radioresistance. More importantly, we found that Tub can directly inhibit the enzyme activity of GPX4 and subsequently significantly enhance the radiosensitivity of cancer cells. These results suggest that targeting ferroptosis resistance is an important breakthrough in the field of radiosensitization.
Ferroptosis contributes to immunotherapy- and radiotherapy-mediated antitumor activity. Induction of ferroptosis in cancer cells is a new and important strategy to enhance the sensitivity of cancer radiotherapy and immunotherapy. RSL3 has potent effects of inducing ferroptosis in cancer cells in vitro; however, it cannot be used as antitumor drugs due to their low bioavailability in vivo [44,45]. The identification of new druggable GPX4 inhibitor is an urgent problem to be solved. Here, we discovered and identified Tubastatin A as a novel GPX4 inhibitor that induces ferroptosis through large-scale drug screening. Tubastatin A has better bioavailability in vivo and meets one of the prerequisites for clinical trials. Compared with RSL3, Tubastatin A is more promising in future clinical application as a single drug or in combination with radiotherapy or immunotherapy. However, the presence of hydroxamic in Tubastatin A enables it to inhibit HDAC6 at the same time, which may cause certain side effects in future clinical applications. Therefore, the most perfect solution is to modify the Tubastatin A drug structure on the basis of analyzing the crystal structure of the Tubastatin A-GPX4 complex. Ideally, the hydroxamic acid should be removed while maintaining the structure of Tubastatin A that inhibits GPX4. In addition, the analysis of the key molecular skeleton of Tubastatin A inhibiting GPX4 also provides a new direction for the development of new GPX4 inhibitors. This will be a very challenging but meaningful research to be done in the future.
4. Methods
Cancer Cell lines. The human breast cancer cell line MDA-MB-231 was donated by Prof. Mien-Chie Hung's laboratory at The University of Texas MD Anderson Cancer Centre, and MCF-7 cells were obtained from ATCC. Cells were cultured in DMEM with 7% foetal bovine serum (FBS) at 37 °C in an incubator with a humidified atmosphere of 20% O2 and 5% CO2. All cells were cultured in 10-cm plates and subcultured into 6-well plates for cell death and lipid peroxidation measurements. The cells were treated with reagents including the HDAC6 inhibitor Tub (TOPSCIENCE, T6161), the ferroptosis inducers erastin (Selleck, S7242) or RSL3 (Selleck, S8155) and the ferroptosis inhibitors Fer-1 (Selleck, S7243) or DFO (Selleck, S5742).
4.1. Cell lines construction
For stable GPX4 or NRF2 shRNA expression, the retroviral vector (pSUPER. puro) encoding hairpin RNA sequences was constructed. In order to overexpress GPX4 in NRF2 stable knockdown cells, we stably transfected PCDH-neo-GPX4 plasmid into NRF2 knockdown cancer cells. G418 was used to screen cancer cells overexpressing GPX4.
To construct RSL3 or ML162 resistant cell lines, cancer cells were treated with low concentrations of RSL3 or ML162 for one week. Surviving cancer cells were cultured with normal medium for three days. The cancer cells were then treated with higher concentrations of the drug for a week. Follow this strategy for 4 cycles, gradually increasing the drug concentration during the process. Finally, surviving cancer cells were cultured with normal cell culture media. Cell survival assays were used to verify the resistance of cancer cells to RSL3 and ML162.
Cell death assays. Propidium iodide staining followed by flow cytometry analysis was used to detect cell death. MDA-MB-231 and MCF-7 cells were seeded in six-well plates at a density of 1.5 × 105 cells per well, and the cells were pretreated with Fer-1 or DFO. On the second day, the cells were treated with the HDAC6 inhibitor Tub or the ferroptosis inducer erastin or RSL3 with or without other compounds. After a period of time, viable cells and dead cells were collected, stained with 5 μg ml-1 PI, and analysed using flow cytometry. At least 10,000 cells were analysed in each group, and all experiments were repeated at least three times.
Lipid peroxidation assay. MDA-MB-231 or MCF-7 cells were seeded in 6-well plates at a density of 1.5 × 105 per well, and the cells were treated with Fer-1 or DFO. On the second day, the cells were treated with -Tub and erastin or RSL3 with or without other compounds for the appropriate time. Then, the cells were collected and stained with 2.5 μM BODIPY 581/591C11 at 37 °C for 20 min and analysed using flow cytometry. For BODIPY 581/591C11 staining, the signals from both nonoxidized C11 (PE channel) and oxidized C11 (FITC channel) were monitored. The ratio of the MFI of FITC to the MFI of PE was calculated for each sample. Cells undergoing lipid peroxidation were defined as those having a high FITC/PE fluorescence ratio. The boundaries that defined what constituted an increased FITC/PE fluorescence ratio were set based on untreated cancer cells, a condition that represents cells with little or no lipid peroxidation. Lipid peroxidation-positive cells were defined as cells with a FITC/PE fluorescence ratio greater than 98% of the untreated cells. At least 10000 cells were analysed in each group, and all experiments were repeated at least three times.
Immunoblotting and immunoprecipitation. For immunoblotting, the protein concentration of each sample was determined after the cells were lysed in RIPA buffer. Twenty micrograms of protein was separated using SDS‒PAGE and transferred to a PVDF membrane. For immunoprecipitation, Pierce immunoprecipitation lysis buffer containing phosphatase or protease inhibitors was applied to lyse cells. The lysate containing 500 μg protein was incubated with 2 μg primary antibody at 4 °C overnight. On the next day, 30 μl of Pierce protein A/G agarose beads was added to the protein-antibody mixture and then incubated at 4 °C for 2 h. Then, the beads were washed five times with cold Pierce immunoprecipitation lysis buffer, the supernatant was discarded, and the beads were heated to 100 °C for 10 min with 60 μl of 2 × SDS. To denature the protein fully, the protein was removed, and the bottom of the EP tube was gently removed every 2 min. Finally, the target protein was detected using western blotting. Antibodies included Hsc70 (Abcam, ab51052), GPX4 (Abcam, ab125066), normal rabbit IgG (CST, 2729S), and NRF2 (Proteintech, 16396-1-AP).
Immunohistochemical (IHC) staining Tumour tissues in mice were excised and fixed with 4% paraformaldehyde overnight. Tissue embedding was carried out the next day. The sliced tissue was immersed in EDTA citrate buffer and microwaved to complete antigen retrieval. Then, the primary antibodies against GPX4 (Abcam, ab125066) and Ki67 (Epitomics, 2642-1) were incubated overnight at 4 °C. The next day, the sliced tissue was incubated with HRP-conjugated secondary antibody for 30 min. Haematoxylin was applied for nuclear counterstaining. Image acquisition was performed using a Nikon camera and software. The percentage of positively stained cancer cells per image was analysed.
4.2. Irradiation and clonogenic survival assay
To determine the effect of Tub on IR-induced ferroptosis, MDA-MB-231 and MCF-7 cells were seeded in 6-well plates at a density of 1000 cells per well, and the cells were pretreated with Fer-1 or DFO for 12 h. On the second day, the cells were irradiated at doses from 0 to 4 Gy and 250 MU/min and cultured in normal medium. Fresh medium containing different compounds was added to the plates every 2 days. After incubation for two weeks, the cells were stained with 0.5% crystal violet dissolved in 20% methanol. The colonies in each well were counted visually.
Irradiation and cell line-derived xenograft model. This study complied with all relevant ethical regulations. All procedures involving mice and experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Sun Yat-sen University Cancer Centre. Female 5-week-old athymic nude mice were obtained from Sun Yat-sen University. All mice were kept under specific-pathogen free conditions in the animal facility of Sun Yat-sen University Cancer Centre. Cancer cells were suspended and counted in 1 × DMEM, and 2 × 106 MDA-MB-231 cells were injected into mice subcutaneously. When the tumours reached 50–100 mm3, the mice were randomly assigned to different treatment groups. Tumours were irradiated with a JL Shepherd Mark I-68A irradiator at a dose of 10 Gy. Tub was dissolved in solvent containing 1% DMSO, 30% polyethylene glycol, 1% Tween 80 and 68% H2O and then subcutaneously administered to mice at a dose of 2.5 mg/kg once a day. Lipro-1 diluted in PBS was intraperitoneally injected daily at a dose of 10 mg/kg. Tub or Lipro-1 was administered three times before irradiation followed by continued daily administration until the endpoint, as indicated in the corresponding figures. The tumour volume was measured once every three days until the endpoint and calculated according to the equation volume = length × width2 × 1/2.
Biotin-streptavidin pulldown method. MDA-MB-231 cells (2 × 106) were seeded into 10 cm dishes. The next day, the cells were treated with 10 μM biotin or biotin-linked Tub. After 24 h of treatment, cancer cells were lysed, and biotin-streptavidin pulldown was performed. Upon incorporation of biotin, Tub-bound proteins were enriched on streptavidin beads and subjected to on-bead trypsin digestion and subsequent LC/LC–MS/MS analysis. Protein identities were determined with database searches using the SEQUEST algorithm.
GPX4 enzyme activity detection. MDA-MB-231 or MCF-7 cells were seeded in a 10-cm dish at a density of 2 × 106. On the second day, the cancer cells were treated with Tub at the indicated concentrations and times. The cells were collected and co-immunoprecipitation assays were used to isolate endogenous GPX4 from the cells. Then, the GPX4 enzymatic activity of each group was analysed with a glutathione peroxidase assay kit according to the manufacturer’s protocol (Abcam, ab102530). To measure the inhibitory effect of Tub on GPX4 enzymatic activity in vitro, MDA-MB-231 or MCF-7 cells were seeded in a 10-cm dish at a density of 2 × 106. On the second day, the cells were collected and co-immunoprecipitation assays were used to isolate endogenous GPX4 from the cells. Then, Tub was added to the test tube containing GPX4 at the indicated concentrations and times, and the GPX4 enzymatic activity of each group was analysed with a glutathione peroxidase assay kit according to the manufacturer’s protocol (Abcam, ab102530).
Statistics. Statistical analyses were performed with GraphPad Prism 8.0 (GraphPad, La Jolla, CA, USA) and SPSS 20 software. The results were analysed by unpaired Student's t-test, 1-way or 2-way ANOVA followed by either Dunnett’s or Tukey’s test. All statistical tests were two-sided and a P value less than 0.05 was considered significant.
Study approval. The mouse experiments complied with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Sun Yat-sen University Cancer Centre.
Author contributions
XZ, HZ, JH, RD and SL initiated the study, designed the experiments, and wrote the manuscript. JL, ZY, LL, and GF performed the animal experiments. YC and BH performed the immunohistochemical staining. YG and DY performed the enzyme activity detection experiments. ZL and JC performed the biotin-streptavidin pulldown experiments. All co-authors have seen and approved the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the Natural Science Foundation of China (grant nos.82130079, 82272708, 82073917, 81972855, Natural Science Foundation of Guangdong Province (grant no. 2023A1515011525), National Postdoctoral Program for Innovative Talents (grant no. BX2021391).
Contributor Information
Rong Deng, Email: dengrong@sysucc.org.cn.
Jia-Jia Huang, Email: huangjiaj@sysucc.org.cn.
Xiao-Feng Zhu, Email: zhuxfeng@mail.sysu.edu.cn.
Data availability
Data will be made available on request.
References
- 1.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., 3rd, Stockwell B.R. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassannia B., Vandenabeele P., Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H.L., Hu B.X., Li Z.L., Du T., Shan J.L., Ye Z.P., Peng X.D., Li X., Huang Y., Zhu X.Y., Chen Y.H., Feng G.K., Yang D., Deng R., Zhu X.F. PKCbetaII phosphorylates ACSL4 to amplify lipid peroxidation to induce ferroptosis. Nat. Cell Biol. 2022;24:88–98. doi: 10.1038/s41556-021-00818-3. [DOI] [PubMed] [Google Scholar]
- 4.Lei G., Zhuang L., Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer. 2022;22:381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan B., Ai Y., Sun Q., Ma Y., Cao Y., Wang J., Zhang Z., Wang X. Membrane damage during ferroptosis is caused by oxidation of phospholipids catalyzed by the oxidoreductases POR and CYB5R1. Mol. Cell. 2021;81:355–369 e310. doi: 10.1016/j.molcel.2020.11.024. [DOI] [PubMed] [Google Scholar]
- 6.Zou Y., Li H., Graham E.T., Deik A.A., Eaton J.K., Wang W., Sandoval-Gomez G., Clish C.B., Doench J.G., Schreiber S.L. Cytochrome P450 oxidoreductase contributes to phospholipid peroxidation in ferroptosis. Nat. Chem. Biol. 2020;16:302–309. doi: 10.1038/s41589-020-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., Roberts M.A., Tong B., Maimone T.J., Zoncu R., Bassik M.C., Nomura D.K., Dixon S.J., Olzmann J.A. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doll S., Freitas F.P., Shah R., Aldrovandi M., da Silva M.C., Ingold I., Goya Grocin A., Xavier da Silva T.N., Panzilius E., Scheel C.H., Mourao A., Buday K., Sato M., Wanninger J., Vignane T., Mohana V., Rehberg M., Flatley A., Schepers A., Kurz A., White D., Sauer M., Sattler M., Tate E.W., Schmitz W., Schulze A., O'Donnell V., Proneth B., Popowicz G.M., Pratt D.A., Angeli J.P.F., Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 10.Kraft V.A.N., Bezjian C.T., Pfeiffer S., Ringelstetter L., Muller C., Zandkarimi F., Merl-Pham J., Bao X., Anastasov N., Kossl J., Brandner S., Daniels J.D., Schmitt-Kopplin P., Hauck S.M., Stockwell B.R., Hadian K., Schick J.A. GTP cyclohydrolase 1/Tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 2020;6:41–53. doi: 10.1021/acscentsci.9b01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao C., Liu X., Zhang Y., Lei G., Yan Y., Lee H., Koppula P., Wu S., Zhuang L., Fang B., Poyurovsky M.V., Olszewski K., Gan B. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E., Noel K., Jiang X., Linkermann A., Murphy M.E., Overholtzer M., Oyagi A., Pagnussat G.C., Park J., Ran Q., Rosenfeld C.S., Salnikow K., Tang D., Torti F.M., Torti S.V., Toyokuni S., Woerpel K.A., Zhang D.D. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma N., Vinik Y., Saroha A., Nair N.U., Ruppin E., Mills G., Karn T., Dubey V., Khera L., Raj H., Maina F., Lev S. Synthetic lethal combination targeting BET uncovered intrinsic susceptibility of TNBC to ferroptosis. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi J., Zhu J., Wu J., Thompson C.B., Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2020;117:31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao P., Wang W., Wang W., Kryczek I., Li X., Bian Y., Sell A., Wei S., Grove S., Johnson J.K., Kennedy P.D., Gijon M., Shah Y.M., Zou W. CD8(+) T cells and fatty acids orchestrate tumor ferroptosis and immunity via ACSL4. Cancer Cell. 2022;40:365–378 e366. doi: 10.1016/j.ccell.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Green M., Choi J.E., Gijon M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A., Xia H., Zhou J., Li G., Li J., Li W., Wei S., Vatan L., Zhang H., Szeliga W., Gu W., Liu R., Lawrence T.S., Lamb C., Tanno Y., Cieslik M., Stone E., Georgiou G., Chan T.A., Chinnaiyan A., Zou W. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang X., Green M.D., Wang W., Yu J., Choi J.E., Jiang L., Liao P., Zhou J., Zhang Q., Dow A., Saripalli A.L., Kryczek I., Wei S., Szeliga W., Vatan L., Stone E.M., Georgiou G., Cieslik M., Wahl D.R., Morgan M.A., Chinnaiyan A.M., Lawrence T.S., Zou W. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019;9:1673–1685. doi: 10.1158/2159-8290.CD-19-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei G., Zhang Y., Koppula P., Liu X., Zhang J., Lin S.H., Ajani J.A., Xiao Q., Liao Z., Wang H., Gan B. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye L.F., Chaudhary K.R., Zandkarimi F., Harken A.D., Kinslow C.J., Upadhyayula P.S., Dovas A., Higgins D.M., Tan H., Zhang Y., Buonanno M., Wang T.J.C., Hei T.K., Bruce J.N., Canoll P.D., Cheng S.K., Stockwell B.R. Radiation-induced lipid peroxidation triggers ferroptosis and synergizes with ferroptosis inducers. ACS Chem. Biol. 2020;15:469–484. doi: 10.1021/acschembio.9b00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyk J., Daly C., Janssen-Bienhold U., Kennedy B.N., Richter-Landsberg C. HDAC6 inhibition by tubastatin A is protective against oxidative stress in a photoreceptor cell line and restores visual function in a zebrafish model of inherited blindness. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Shi J., Liu X., Feng L., Gong Z., Koppula P., Sirohi K., Li X., Wei Y., Lee H., Zhuang L., Chen G., Xiao Z.D., Hung M.C., Chen J., Huang P., Li W., Gan B. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 2018;20:1181–1192. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubellacker J.M., Tasdogan A., Ramesh V., Shen B., Mitchell E.C., Martin-Sandoval M.S., Gu Z., McCormick M.L., Durham A.B., Spitz D.R., Zhao Z., Mathews T.P., Morrison S.J. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 2020;585:113–118. doi: 10.1038/s41586-020-2623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J., Viswanathan S.R., Chattopadhyay S., Tamayo P., Yang W.S., Rees M.G., Chen S., Boskovic Z.V., Javaid S., Huang C., Wu X., Tseng Y.Y., Roider E.M., Gao D., Cleary J.M., Wolpin B.M., Mesirov J.P., Haber D.A., Engelman J.A., Boehm J.S., Kotz J.D., Hon C.S., Chen Y., Hahn W.C., Levesque M.P., Doench J.G., Berens M.E., Shamji A.F., Clemons P.A., Stockwell B.R., Schreiber S.L. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Z., Liu J., Long F., Kang R., Kroemer G., Tang D., Yang M. The lipid flippase SLC47A1 blocks metabolic vulnerability to ferroptosis. Nat. Commun. 2022;13:7965. doi: 10.1038/s41467-022-35707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller F., Lim J.K.M., Bebber C.M., Seidel E., Tishina S., Dahlhaus A., Stroh J., Beck J., Yapici F.I., Nakayama K., Torres Fernandez L., Bragelmann J., Leprivier G., von Karstedt S. Elevated FSP1 protects KRAS-mutated cells from ferroptosis during tumor initiation. Cell Death Differ. 2023;30:442–456. doi: 10.1038/s41418-022-01096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anandhan A., Dodson M., Shakya A., Chen J., Liu P., Wei Y., Tan H., Wang Q., Jiang Z., Yang K., Garcia J.G., Chambers S.K., Chapman E., Ooi A., Yang-Hartwich Y., Stockwell B.R., Zhang D.D. NRF2 controls iron homeostasis and ferroptosis through HERC2 and VAMP8. Sci. Adv. 2023;9 doi: 10.1126/sciadv.ade9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang K., Yin X., Zhang Q., Yin J., Tang Q., Xu M., Wu L., Shen Y., Zhou Z., Yu H., Yan S. STC2 activates PRMT5 to induce radioresistance through DNA damage repair and ferroptosis pathways in esophageal squamous cell carcinoma. Redox Biol. 2023;60 doi: 10.1016/j.redox.2023.102626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amos A., Jiang N., Zong D., Gu J., Zhou J., Yin L., He X., Xu Y., Wu L. Depletion of SOD2 enhances nasopharyngeal carcinoma cell radiosensitivity via ferroptosis induction modulated by DHODH inhibition. BMC Cancer. 2023;23:117. doi: 10.1186/s12885-022-10465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q., Zheng W., Guan J., Liu H., Dan Y., Zhu L., Song Y., Zhou Y., Zhao X., Zhang Y., Bai Y., Pan Y., Zhang J., Shao C. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ. 2023;30:137–151. doi: 10.1038/s41418-022-01051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen D., Luo J., Chen L., Ma W., Mao X., Zhang Y., Zheng J., Wang Y., Wan J., Wang S., Ouyang J., Yi H., Liu D., Huang W., Zhang W., Liu Z., McLeod H.L., He Y. PARPi treatment enhances radiotherapy-induced ferroptosis and antitumor immune responses via the cGAS signaling pathway in colorectal cancer. Cancer Lett. 2022;550 doi: 10.1016/j.canlet.2022.215919. [DOI] [PubMed] [Google Scholar]
- 33.Cai H., Ren Y., Chen S., Wang Y., Chu L. Ferroptosis and tumor immunotherapy: a promising combination therapy for tumors. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1119369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q., Zhang Q., Cao P., Shi C., Zhang L., Wang L., Gong Z. NOD2-mediated HDAC6/NF-kappab signalling pathway regulates ferroptosis induced by extracellular histone H3 in acute liver failure. J. Cell Mol. Med. 2022;26:5528–5538. doi: 10.1111/jcmm.17582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loibl S., Poortmans P., Morrow M., Denkert C., Curigliano G. Breast cancer. Lancet. 2021;397:1750–1769. doi: 10.1016/S0140-6736(20)32381-3. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Mu X., He H., Zhang X.D. Cancer radiosensitizers. Trends Pharmacol. Sci. 2018;39:24–48. doi: 10.1016/j.tips.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Darvish L., Bahreyni Toossi M.T., Azimian H., Shakeri M., Dolat E., Ahmadizad Firouzjaei A., Rezaie S., Amraee A., Aghaee-Bakhtiari S.H. The role of microRNA-induced apoptosis in diverse radioresistant cancers. Cell. Signal. 2023;104 doi: 10.1016/j.cellsig.2022.110580. [DOI] [PubMed] [Google Scholar]
- 38.Ouellette M.M., Zhou S., Yan Y. Cell signaling pathways that promote radioresistance of cancer cells. Diagnostics. 2022;12 doi: 10.3390/diagnostics12030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor M.A., Das B.C., Ray S.K. Targeting autophagy for combating chemoresistance and radioresistance in glioblastoma. Apoptosis. 2018;23:563–575. doi: 10.1007/s10495-018-1480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaffray D.A. Image-guided radiotherapy: from current concept to future perspectives. Nat. Rev. Clin. Oncol. 2012;9:688–699. doi: 10.1038/nrclinonc.2012.194. [DOI] [PubMed] [Google Scholar]
- 41.Wei J., Zhu K., Yang Z., Zhou Y., Xia Z., Ren J., Zhao Y., Wu G., Liu C. Hypoxia-induced autophagy is involved in radioresistance via HIF1A-associated beclin-1 in glioblastoma multiforme. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng W., Chen Q., Liu H., Zeng L., Zhou Y., Liu X., Bai Y., Zhang J., Pan Y., Shao C. SDC1-dependent TGM2 determines radiosensitivity in glioblastoma by coordinating EPG5-mediated fusion of autophagosomes with lysosomes. Autophagy. 2023;19:839–857. doi: 10.1080/15548627.2022.2105562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J., Song C., Gu J., Li C., Zang W., Shi L., Chen L., Zhu L., Zhou M., Wang T., Li H., Qi S., Lu Y. RBBP4 regulates the expression of the Mre11-Rad50-NBS1 (MRN) complex and promotes DNA double-strand break repair to mediate glioblastoma chemoradiotherapy resistance. Cancer Lett. 2023;557 doi: 10.1016/j.canlet.2023.216078. [DOI] [PubMed] [Google Scholar]
- 44.Badgley M.A., Kremer D.M., Maurer H.C., DelGiorno K.E., Lee H.J., Purohit V., Sagalovskiy I.R., Ma A., Kapilian J., Firl C.E.M., Decker A.R., Sastra S.A., Palermo C.F., Andrade L.R., Sajjakulnukit P., Zhang L., Tolstyka Z.P., Hirschhorn T., Lamb C., Liu T., Gu W., Seeley E.S., Stone E., Georgiou G., Manor U., Iuga A., Wahl G.M., Stockwell B.R., Lyssiotis C.A., Olive K.P. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. 2020;368:85–89. doi: 10.1126/science.aaw9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Tan H., Daniels J.D., Zandkarimi F., Liu H., Brown L.M., Uchida K., O'Connor O.A., Stockwell B.R. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 2019;26:623–633 e629. doi: 10.1016/j.chembiol.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.