Highlights
-
•
Two PIP2 lipids are required to stabilize each Voltage Sensor Domain in the IKS channel.
-
•
The ancillary KCNE1 β-subunits to generate the IKS current interacts with both PIP2 binding sites.
-
•
The key KCNE1 basic residue involved in theses interactions are essential for modulation of the IKS PIP2 sensitivity.
-
•
The key KCNE1 basic residue involved in theses interactions are highly conserved across all members of the KCNE family.
Abstract
The phosphatidyl-inositol-4,5-bisphosphate (PIP2) lipid has been shown to be crucial for the coupling between the voltage sensor and the pore of the potassium voltage-gated KV7 channel family, especially the KV7.1 channel. Expressed in the myocardium membrane, KV7.1 forms a complex with KCNE1 auxiliary subunits to generate the IKS current. Here we present molecular models of the transmembrane region of this complex in its three known states, namely the Resting/Closed (RC), the Intermediate/Closed (IC), and the Activated/Open (AO), robustness of which is assessed by agreement with a range of biophysical data. Molecular Dynamics (MD) simulations of these models embedded in a lipid bilayer including phosphatidyl-inositol-4,5-bisphosphate (PIP2) lipids show that in presence of KCNE1, two PIP2 lipids are necessary to stabilize each state. The simulations also show that KCNE1 interacts with both PIP2 binding sites, forming a tourniquet around the pore and preventing its opening. The present investigation provides therefore key molecular elements that govern the role of PIP2 in KCNE1 modulation of IKS channels, possibly a common mechanism by which auxiliary KCNE subunits might modulate a variety of other ion channels.
Graphical abstract
Introduction
The IKS current is diffused through the plasma membrane of cardiomyocytes, during the last (fourth) phase of the cardiac action potential [1,46,67]. This repolarizing current is conducted by a protein complex derived from the co-expression of four KV7.1 α-subunits and KCNE1 ancillary subunits from the KCNQ1 and KCNE1 genes, respectively. The KV7.1 tetramer forms a voltage-gated potassium (KV) channel, a transmembrane protein that upon modification of membrane potential, opens and carries selectively potassium ions to the extracellular medium, while KCNE1 is a transmembrane peptide which acts as an ancillary β-subunit for several KV channels, including KV7.1 [42]. Together, α and β subunits are forming the IKS channel complex. The transmembrane region of KV7.1 channels is formed by homo-tetramers of 6 transmembrane helical segments. The first four ones (S1 to S4) are forming the voltage-sensor domain (VSD), and the tetrameric organization of the last two (S5 and S6) is forming the pore domain (PD) of the channel. As for most KV channels, the VSD and PD are swapped in KV7.1. The segment S4 in the VSD carries four arginines. Called gating charges, these positively charged residues move across the membrane upon depolarization, interacting sequentially with negatively charged side chains from S2 [70]. The KV7.1 subunit also contains cytoplasmic NTERM and CTERM regions. Specifically, the CTERM region is composed of four cytosolic helices, the first two being connected to the sixth transmembrane segment, the last two being located deeper in the cytosol, forming the tetramerization domain [69]. KCNE1 is a transmembrane polypeptide of 129 aminoacids divided into one extracellular NTERM domain, a helical transmembrane (TMD) domain, and a cytosolic CTERM domain [64].
Numerous mutations of both KCNQ1 and KCNE1 genes are associated with long QT syndromes (LQTS) [28,45,59]. The latter are characterized by an extended cardiac action potential that corresponds to the time interval between Q and T waves in electrocardiograms. The LQT phenomenon reflects the inability of the protein complex to generate its IKS outward current and therefore to return cardiomyocytes membranes toward their resting potential. This delay in repolarization disturbs the propagation of the cardiac action potential within the myocardium tissue, and therefore leads to heart rhythm abnormalities, also known as cardiac arrhythmias. Hence, the IKS channel is a therapeutic target for the treatment of LQTS, whose function must be studied and understood to be able to develop any potential effective drug. Over the last twenty years, this protein complex has been extensively studied using various methods [37].
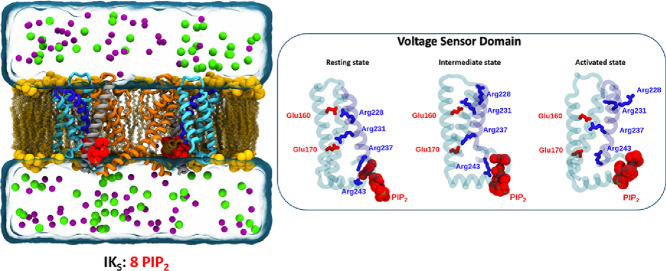
The gating of KV7.1 and IKS channels is triggered by membrane depolarization and involves three stable states of the VSD: Resting, Intermediate, and Activated. The conformations of these states are known to transition from one to another through the motion of three S4 gating charges, R228 (R1), R231 (R2) and R237 (R4) with respect to two binding sites (Table S1). As in VSDs of most voltage-gated ion channels, that of KV7.1 contains two strongly conserved binding sites (residues forming salt bridges with the S4 gating charges): the first one, located in the solvent accessible surface of the VSD, is an acidic residue from S2, E160 (E1). The second one, located deeper in the membrane, is sheltered from the solvent by an aromatic residue from S2, F167, also known as the charge transfer center (CTC), and is composed of two acidic residues, E170 (E2) and D202 (D3) from S2 and S3 segments, respectively [63].
These conformational changes also involve a translation of these gating charges through an aromatic residue from S2, F167, which was suggested to constitute the interface between the solvent accessible surface of the VSD, and its occluded site. This residue is conserved in homologous KV channels such as KV1.2 or KV2.1 [31].
The pore opening is elicited by another mechanism called VSD-PD coupling. The latter occurs in most KV channels and is governed by protein-protein interactions between VSD and PD of distinct α-subunits that couple the activation state of the VSD to the conformation (open or closed) of the PD [55]. The exact mechanism for this “electromechanical” process is not completely determined, yet a functional study revealed that the VSD-PD coupling mechanism of the KV7.1 tetramer occurs in both the Activated and the Intermediate states of the VSD, but not in its Resting state [20]. In our recent integrative study, we unveiled the molecular determinants of the distinct intersubunit coupling interfaces that underlie the VSD-PD coupling in the Intermediate and the Activated states of the KV7.1 channel [21]. In presence of KCNE1, this mechanism appears to be hindered when the VSD is in its Intermediate state and enhanced in its Activated state. Hence, the three functional states known for the IKS channel are described by the “activation state” of the VSD (R, I or A) and by the conformation (O or C) of the pore, and are referred to as RC, IC and AO states.
The phosphatidyl-inositol-4,5-bisphosphate (PIP2) is a membrane phospholipid that participates in the function of many membrane transporters [19], including voltage-gated ion channels. PIP2 is present at a 1% rate in the inner leaflet of the lipid bilayers forming cell membranes, and experimental studies have shown that it interacts with numerous ion channels including KV channels to modulate their activation. This lipid has been shown to be crucial for the coupling between the VSD and the PD for the KV7 channel family [35,74], especially in the KV7.1 channel [[75], [76]].
Hence for instance, the structure of Xenopus Laevis KV7.1 monomer (KCNQ1EM), resolved by cryo-electron microscopy (CryoEM), was locked in a non-physiological conformation of the PD, namely the uncoupled Activated/Closed (AC) state [61]. According to the authors, this might be explained by the absence of PIP2, known to be essential for the coupling of the VSD activated state with PD open state in the KV7 channels family [35].
Since beside KCNQ1EM [61], and the very recently published Cryo-EM structures of the human KV7.1 AO channel in the presence of PIP2 [62], no high-resolution structure of the IKS channel is available, we and others turn to molecular modeling. Quite surprisingly, among the molecular constructs of the IKS channel that have been published the last decade [16,27,53,73], very few were modeled in presence of PIP2. Moreover, those which were modeled with the lipid aimed at validating experiments were limited to the identification of the PIP2 binding sites [14], without providing any molecular insight about the way the lipid interacts with KV7.1 or with its KCNE1 subunits. Recently, a homology model [24] of the IKS complex was subjected to Molecular Dynamics (MD) simulations in a POPC:PIP2 membrane at a 10:1 ratio [25]. Unfortunately, the IKS-PIP2 interactions were not extensively investigated and the study did not provide information about how PIP2 affects the VSD-PD coupling mechanism of the channel. Moreover, despite the fact that several studies have shown that IKS complexes can be expressed in cardiomyocytes with a 4:4 (KV7.1:KCNE1) stoichiometry [43,44] most of the IKS models reported so far [24,27,73] have not been built with this ratio.
Our previous MD study [29] of KV7.1 models in open and closed states, allowed one to localize the PIP2 binding site in the KV7.1 subunit (PIP2 intra), and to characterize the key elements of the KV7.1 modulation by PIP2 in absence of KCNE1. The lipid was shown to participate in VSD/PD coupling of the KV7.1 channel through state-dependent interactions, preventing repulsive forces between basic residues from the helical S2-S3LOOP and S4 in resting state, and between basic residues from the S2-S3LOOP and S6 in open states, which was confirmed in the structure of the human KV7.1 channel in the presence of PIP2 [62]. Our previous MD simulations of KV7.1 also suggested that PIP2 may constitute a third binding site for S4 gating charges. Indeed, the lipid was found to form salt-bridges with R237 (R4) and R243 (R6) in the RC model of KV7.1, and not in the AO model. Moreover, the dependence of this lipid for the function of KV7.1 was also proved to be increased in presence of KCNE1 due to its additional positive charges in its CTERM domain [36]. This study indicates that IKS may carry an additional PIP2 binding site (PIP2 inter) with respect to KV7.1. Therefore, the molecular determinants describing a second PIP2 binding site in KV7.1 subunits in presence of KCNE1 are yet to be investigated. To address these questions, computational chemistry methods are the most insightful ones to unravel the elements of protein function at a molecular level of precision.
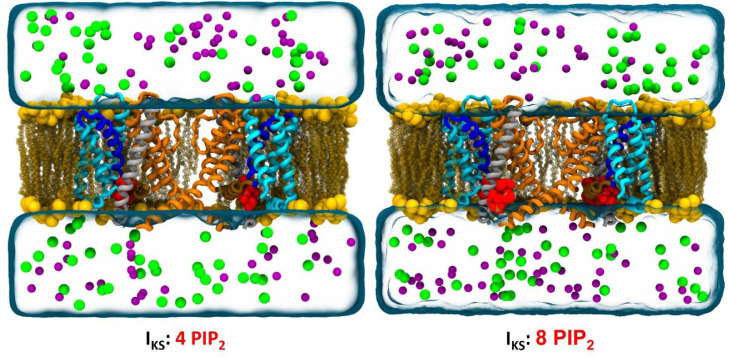
Here we propose three models of the KV7.1 tetramer in distinct metastable states, in presence of KCNE1 subunits corresponding to the RC, IC and AO states of the IKS channel. Each model has been studied either with one or two PIP2 lipids per KV7.1 subunit. A total of 6 systems (Fig. 1) were submitted each to ∼500 ns MD simulations to fully relax the initial constructs within the lipid bilayer. All simulations have been carried out at 0 mV, assuming, as in other works [9,[10], [76]] that the conformations of the channels are stable as well within the simulations time scale. Analyses of these trajectories were then performed to assess the validity of the models by comparing them to the literature.
Fig. 1.
IKS MD systems.
Representation of IKS models in absence (left) and presence (right) of KCNE1’s (gray ribbons) PIP2 binding site, each embedded in a POPC membrane (in yellow spheres) with PIP2 lipids (in red spheres). Two KV7.1 subunits are shown for clarity. The transmembrane segments are represented in ribbons. The VSD segments are colored in cyan, S4 in blue, S4-S5LINKER in brown and PD segments in orange. The membrane-embedded models are surrounded by water (in transparent blue surface) as well as K+ (purple spheres) and Cl− (green spheres) ions.
Results
Validation of the Iks models: 2 PIP2 lipids per subunit are in better agreement with experiments
Molecular modeling methods are based upon the study of representations of chemical and biochemical entities. The main goal of using these computational methods, in the frame of this study, is to gain an atomistic insight on phenomena that were unraveled by experimental studies. Hence, to predict the molecular mechanisms associated to these phenomena, we need to maximize as much as possible the compliance of our models with respect to structural biology data. To achieve this goal, we built our 3D models using structural constraints that were directly drawn from experimental data, and then we monitored the stability of these constraints over the collection of conformations generated by MD simulations, i.e. the trajectories.
To gain a finer insight in molecular determinants allowing for the stabilization of each state of KV7.1 and IKS channels, MD simulations were performed to equilibrate each molecular model within a POPC membrane and PIP2 lipids. In various KV channels, the activation mechanism of the VSD is mostly characterized by state-dependent salt-bridges between basic residues of S4 and acidic residues spread in S2 and S3 segments (Supplementary Fig. S1). This succession of salt-bridges is describing an upward translation of S4, as well as a clockwise rotation during VSD activation. To validate the state dependent structures of our models VSDs, we monitored the distance between each pair of charged groups that are supposed to interact according to the literature. The state-dependent salt-bridges found in the VSD of our IKS models are reported in Table S1.
In 8 PIP2 systems, the interactions between E1 and R228 (R1), and between E2 and R231 (R2), which are specific of the RC state, are both present specifically in our RC model in all subunits (Supplementary Fig. S1). In RC model with 4PIP2 system, the interaction between R1 and E1 is present, but the interaction between R2 and E2 is absent, as R2 interacts with E1 in only two subunits out of four. This first result indicates that the presence of two PIP2 binding sites in IKS models may be crucial for the stabilization of the VSDs.
In the IC state, S4 translates upwards and rotates clockwise, which leads to E1 interacting with R231 (R2), while E2 interacts with R237 (R4). These interactions have been specifically found in IC model of 8PIP2 system, in all subunits for E1-R2 interaction, and in 3 subunits out of 4 for E2-R4 interaction. In the 4PIP2 system, E1-R2 interaction is present as expected, but E2-R4 interaction is not absent. Interestingly, R4 also interacts with D3 in all subunits of both systems. The latter forms the charge transfer center along with F167 from S2 and E2. Therefore, R4-D3 interaction may indicate that R4 is fully anchored in the CTC in the 8PIP2 system and remain partially anchored in the CTC in the 4PIP2 system.
In the AO state, a second upward translation and a clockwise rotation of S4 lead to E1-R4 interaction. This interaction is present only in our AO models of both PIP2 systems. In these models, R6 appeared to be anchored in the CTC in all subunits, interacting with both E2 and D3 (data not shown). Noteworthy, the pairs of salt-bridges determined in our IKS models are also present in their respective states of KV7.1 models. These MD results highlight the different conformations of the VSD which correspond to interaction patterns that remain very stable over time. However, in the 4PIP2 systems, state dependent salt-bridge pairs are satisfying experimental data for AO model, but not for IC and RC models.
Overall, these results suggest that IKS models of 8PIP2 system are in better agreement with experimental results, as their state dependent expected VSD interactions are more satisfied, and more stable throughout the corresponding MD trajectories. Note that distinct patterns of gating charge salt-bridges determined in our molecular models support the sliding helix VSD activation mechanism proposed for the KV channels [12], as well as for the voltage-gated sodium channels [5].
Besides electrostatic interactions, site-directed mutagenesis can provide important information about the relative position of the KCNE1 helices with respect to the KV7.1 subunits. Experiments such as cysteine cross-linking mutagenesis allow one to spot the residues which are close enough to form disulfide bonds. In the framework of such experiments, a disulfide bond can be formed if the Cβ atoms of the corresponding amino acids are within ∼ 13 Å [4]. Thanks to such experimental studies, a significant amount of neighbor residue pairs within KV7.1 channel and IKS complex have been reported. Accordingly, for each neighbor residue pair, we computed the distance between their respective Cβ atoms over our MD trajectories and assumed neighbor residue pairs present in the model if the latter was below 13 Å. Among the seven neighbor residue pairs from KV7.1 (Table S2), the intersubunit pairs assigned to IC state, as well as the intrasubunit ones assigned to the RC state are present in all IKS models (Supplementary Fig. S2, A), regardless of their respective state and regardless of the amount of PIP2 lipids. For the three pairs assigned to the AO state (Supplementary Fig. S2, B), one of those is present in all models, whereas the other two are predominant in most subunits of the AO and RC models, but absent in the IC models. In our KV7.1 models, all these pairs are also present in all subunits.
For pairs involving cross linking between KV7.1 and KCNE1 residues (Table S3), most of those which have been assigned to the AO state of IKS channel are present in both 4PIP2 and 8PIP2 AO models (Supplementary Fig. S3). Surprisingly, the only pair of residues assigned to AO state and not present in our AO model (Q147-K41) is present in the RC models only. Among the six residue pairs assigned to the RC state, five of those are present in the 8PIP2 system, against four of them in the 4PIP2 one. The pairs of residues that are not interacting in RC models are not present in any other model. Finally, for the nine remaining pairs, which were not assigned to any state of the channel, all of them are present in at least one model of each system. Among the three residue pairs that include KV7.1 S6 residues and KCNE1 CTERM residues, only one pair is present in both systems. The second pair is present in the 4PIP2 system only, and the last pair is absent in both systems.
Overall, KV7.1-KV7.1 and KV7.1-KCNE1 residue pairs are mostly present in all IKS models, regardless of the number of PIP2 lipids in the system. Although the presence and the stability of the interactions between each residue pair in our models is testifying for their robustness, these results alone cannot allow to select the best IKS models among their distinct systems.
-
•
Neighbor residue pairs: protein-lipid interactions
Besides the protein-protein interactions, several mutagenesis studies conducted on KV7.1 in absence and presence of KCNE1 also have highlighted ten basic residues from KV7.1 and three others from KCNE1 which can possibly interact with PIP2 (Table S4). To characterize these interactions in our IKS models, we calculated average distances between charged groups of each basic residue and PIP2. Among the thirteen KV7.1 and KCNE1 basic residues which have been shown to be involved in electrostatic interactions with the lipid, ten residues interact with PIP2 in at least one MD trajectory of IKS model in 8PIP2 system.
Results obtained for these models showed that PIP2 inter, located in the inner membrane near S6 and KCNE1 subunits, and PIP2 intra, located in the inner membrane near S2-S3LOOP, are specifically anchored to the VSD and the PD, respectively, in a state independent manner (Supplementary Fig. S4). In each model, residues R190 from the S2-S3LOOP and R249 from the S4-S5LINKER interact with PIP2 intra, while residues K362 and R366 from S6 interact with PIP2 inter. In addition, some basic residues are also interacting with PIP2 in a state dependent manner, such as R192 and R195 from S2-S3LOOP, as well as K358 from S6, whose interactions with PIP2 are favored in AO model, as well as in IC model for R195. State independent interactions involving VSD residues are related to a motion of PIP2, which appears to progressively anchor S2-S3LOOP, while remaining bound to S4-S5LINKER residue R249 during VSD activation. State dependent interactions involving PD residues are related to the movement of S6 upon pore opening. PIP2 inter progressively binds basic residues of S6 cytoplasmic helices as they spread away from the pore axis towards inner membrane surface where PIP2 is localized. Oppositely, the interaction between residue R243 from S4 and the lipid is favored in RC model, which suggests that R243 loses its interaction with PIP2 as S4 translates upward during the VSD activation. In IKS models of the 4PIP2 system, state-independent interactions between VSD residues and PIP2 intra are present, but those between S6 residues and PIP2 are merely present. Contrary to KV7.1 models in which PIP2 intra can bind S6 in AO model, K354 is the only S6 residue that is able to bind PIP2 in both IC and AO models of IKS complex. Residues K358, K362 and R366 are located too far away to reach PIP2 intra in any of these models.
Four residues are not interacting with PIP2 in any IKS system. Residues R181 and K196 from S2-S3LOOP remain too far from PIP2 intra throughout MD simulations. The lack of PIP2 salt-bridges with R181 is not in agreement with the recent structure of KV7.1 and PIP2 [62], whose relative position in regards to KV7.1 might be impacted by the presence of KCNE3 ancillary subunit. Nevertheless, the MD trajectory of IKS AO states showed that R181 rather binds POPC's phosphate groups (data not shown). This might highlight the importance of this residue being anchored in the inner membrane, which was confirmed by recent charge reversal mutatgenesis studies [77]. Indeed, the voltage-dependence of IKS R181E mutant turned out to be shifted towards more depolarized voltages, showing a loss-of-function effect on the channel. Similarly, residue R360 from S6, whose sidechains remain tangent to the conduction pathway, was unable to interact with PIP2 inter. R259 guanidium group, despite being close to PIP2 lipids, cannot get close to the phosphoryl groups of PIP2 because of the steric hindrance induced by presence of KCNE1 subunits.
Surprisingly, the KCNE1 residues R67, K69 and K70 have their sidechains oriented in two opposite directions, allowing for the ancillary subunit to interact with both PIP2 binding sites in a state-dependent fashion in IKS models of 8PIP2 system. In RC model, residue K69 binds PIP2 intra in two opposite subunits, while R67 and K70 are both binding PIP2 inter in all subunits. In IC model, the interaction with K70 is absent, while those with both R67 and K69 are still present. In AO model, K69 is the only KCNE1 residue interacting with PIP2. These subsequent interactions over all our stable state models of IKS of 8PIP2 system suggest that KCNE1 undergoes two clockwise rotations during VSD activation, each occurring during the RC-IC and the IC-AO transitions, respectively. In the IC and RC models, KCNE1 residues are binding both inter and intra PIP2, forming a circle around the cytoplasmic region of S6 helices. This PIP2-KCNE1 circle was not observed in the AO model, as KCNE1 no longer binds PIP2 intra. Thus, the cytoplasmic region of S6 can spread away from the pore axis towards PIP2 inter. Quite interestingly, in the MD trajectories of IKS 4PIP2 system, a likewise PIP2-KCNE1 circle cannot be formed due to the absence of PIP2 inter. Indeed, interactions involving R67 and K70 are impaired, while those involving K69 are conserved. However, in the AO model, the interaction between K69 and PIP2 is impaired, and residues R67, K69 and K70 are facing those of S6 that strongly interact with PIP2 inter in 8PIP2 systems, which might generate electrostatic repulsion energies, leading to the collapse of S6 segments and pore closure.
In summary, our results highlighted an additional PIP2 binding site in IKS channel, with which KCNE1 interacts in a state dependent manner. This additional PIP2 binding site allow for the shaping of a PIP2-KCNE1 circle in the uncoupled RC and IC states model of IKS channel. This may allow for the control of S6 helices conformations by the KCNE1 subunits.
To verify this hypothesis, results of pore radii calculations in our IKS models turn out to be useful. Indeed, results obtained for the IKS models will allow to figure out if the presence of PIP2 inter in the 8PIP2 systems is inducing a change in the width of the conduction pathway.
-
•
Pore radii
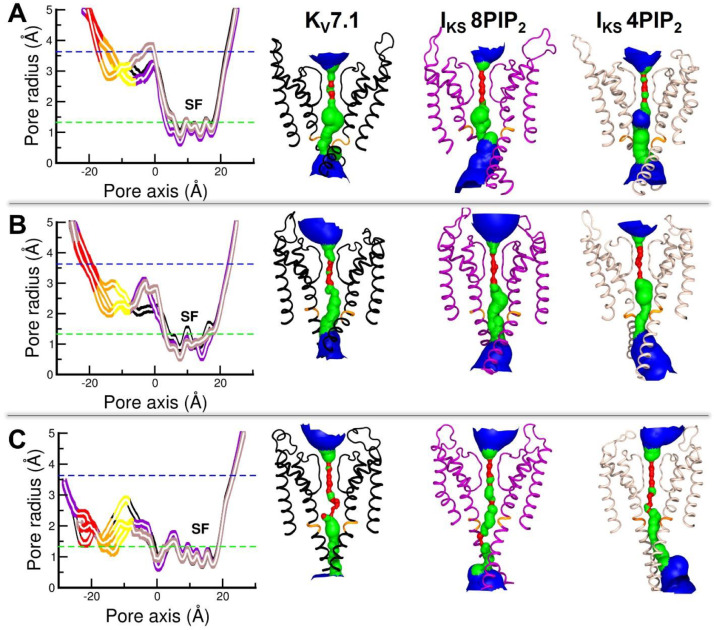
Pore radii calculations have been conducted to select the best IKS model among the systems with 4 PIP2 lipids and the ones with 8PIP2 lipids. Based on the correlation identified between the pore size of a KV channel below the selectivity filter and the free energy associated to the conduction of a potassium ion [66], the analysis of the pore size of our models can allow to assess the reliability of our IKS models with respect to those of KV7.1 model, and also with respect to experimental data. Integrative studies conducted on the Shaker channel [11] highlighted the fact that the cytoplasmic region of KV channel conduction pathway need to be able to accommodate a solvated potassium ion, whose minimal radius is ∼3.6 Å, to be open. Pore radius measurement of the conduction pathway of KCNQ1EM structure [61] in its Activated/Closed state reveals three S6 regions of constriction in the cytoplasmic side of the conduction pathway: the upper constriction is formed by the backbones of G345; the middle constriction is formed by hydrogen bonds between S349 hydroxyl groups, and the lower constriction is formed by the L353 side chains that are oriented towards the center of the pore and binding each other to form a hydrophobic seal. These three regions of constriction present pore radii of ∼2 Å for G345, ∼0.8 Å for S349, and ∼1.15 Å for L353, which may prevent a K+ ion to go through this pathway, as its ionic radius is ∼1.33 Å. This pore radius profile provides a hint on the constriction zones of KV7.1 pore when the channel is decoupled due to the absence of PIP2 lipids. Hence, we compared first the pore radii profiles of IKS MD trajectories in its distinct systems, and then we compared the most robust model of IKS in its three states with those of KV7.1 models.
We mapped the average pore radii to see if these residues are forming a constriction in our IKS models. The MD trajectories of our models suggest that the backbone of G345 is oriented toward the center of the pore, while the side chains of both S349 and L353 remain tangent to the pore surface in all models. Nevertheless, the average pore radii estimated on these regions (Fig. 2) shed light on state dependent opening / closure of the conduction pathways. The models indicate, in the AO models (Fig. 2A), values are between 2.7 Å and 5 Å. In the Intermediate models (Fig. 2B), these values are between 2 Å and 4.5 Å. In the resting models (Fig. 2C), the average pore radii at the level of constricted regions assume values between 1 Å and 3 Å for the three models.
Fig. 2.
Validation of IKS models through pore radii calculations
The graphs report the average pore radii (left) along the conduction pathway of KV7.1 (black curves) IKS 8PIP2 (purple curves) and IKS 4PIP2 (beige curves) models in A. AO state B. Intermediate state and C. RC state. Averages radii at the levels of residues G345, S349 and L353 are depicted in yellow, orange and red curves, respectively. The right panels show cartoon representations of KV7.1 (black) IKS 8PIP2 (purple) and IKS 4PIP2 (beige) models. Potassium ionic (1.33 Å) and hydrodynamic radii (3.6 Å) are represented by green and blue dashed lines in the graphs, respectively. Pore solvent accessible surfaces are colored as follows. Pore radii values inferior to K+ ionic radius, are colored in red. Pore radii values ranging between K+ ionic radius and K+ hydrodynamic radius, are colored in green. Pore radii values superior to K+ hydrodynamic radius, are colored in blue.
To discriminate one IKS model from another, we compared for each state, the average pore radii values obtained at the level of the constriction zones formed by G345, S349 and L353 in IKS models with those of KCNQ1EM structure. In the AO state, KV7.1 ionic current is increased in presence of KCNE1, thus IKS AO models should present wider pore radii than KCNQ1EM structure at the level of constriction regions. Among our AO models (Fig. 2A), the IKS 8PIP2 system have the least constrictions. Pore radii values of ∼ 3 Å, 3.9 Å and 4.5 Å in average at the levels of G345, S349 and L353, were respectively found. The IKS 4PIP2 AO model in contrast appears to be more constricted at the level of S349, as its average pore radii is 0.9 Å lower than those of other AO models. A decrease of 1 Å in the radius value of a nanopore can lead to a significant rise in the free energy cost necessary for conduction [11,66]. This result, in addition to the number of protein-lipid interactions we found in IKS AO models, suggest that the IKS AO model is in better agreement with experimental data when embedded in 8PIP2 system than in 4PIP2 system.
In the Intermediate state, the ionic current is abolished in presence of KCNE1, so IKS models should present a smaller pore radius than in KV7.1. In the intermediate models (Fig. 2B), the pore radii of the constriction zone of L353 are ∼3.6 Å in all models. Therefore, we only considered G345 and S349 constriction zones. The KV7.1 model present the highest average pore radii values, (∼ 2.7 Å and 2.9 Å at the levels of G345 and S349, respectively) compared to the IKS models (both ∼ 2 Å). Hence, in a nutshell, the results obtained for intermediate models suggest that KV7.1 conduction pathway is ∼0.8 Å narrower in presence of KCNE1, regardless of the number of PIP2 lipids in the system which agrees with experiments.
In summary, constriction zones of RC models (Fig. 2C), present pore radii values below the minimal hydrodynamic radius of K+ ions of 3.6 Å, which suggest that in RC models, the pore is closed regardless of the presence of KCNE1 or the number of PIP2 lipids.
The differences observed between the average pore radii calculated over the MD trajectories obtained for KV7.1 and IKS models indicate that KCNE1 may increase the tightening of the inner pore in the RC and IC states, while inducing a closer proximity between S6c region and the inner membrane surface in the AO state, which agrees with the experimental studies. The comparison of the average pore radii obtained for the IKS models of both 4PIP2 and 8PIP2 systems with those obtained for the KV7.1 models indicate that the MD trajectories of the IKS models of 8PIP2 system fit better the ionic conductance measures of the IKS channel [71] compared to the KV7.1 channel.
Discussion and conclusion
The present work provides tridimensional models of three states of the KV7.1 channel in presence of KCNE1, whose features are mostly satisfying the structural constraints drawn from experimental studies. Our previous computational study of KV7.1 models [29] highlighted two components of VSD-PD coupling: a protein-protein component and a protein-lipid component. Protein-protein components of this mechanism were mostly characterized by electrostatic interactions between residues from the S4-S5LINKER and residues from S6, while the protein-lipid components were characterized by state- dependent interactions between PIP2 and the KV7.1 subunits.
In our IKS models, considering a second PIP2 binding site per subunit was required in order to optimize the agreement with experimental results. This work shows that in contrast to the case of KV7.1, in IKS models, PIP2 lipids engage in state-independent interactions with KV7.1 subunits: PIP2 intra, which is present in both KV7.1 and IKs models, predominantly binds the lower VSD basic residues, while PIP2 inter, presumably most required in IKS channels, binds the basic residues from S6 and KCNE1. In each subunit, PIP2 inter is located between CTERM regions of both the KCNE1 helix and the S6 segment (Supplementary Fig. S4). In 8PIP2 IKS models (Supplementary Fig. S4, A, upper panel), PIP2 inter remain bound to the S6 residues K362 and R366 in a state independent manner, as well as to the KCNE1 residues R67 K69 and K70 in a state dependent manner. In the 4PIP2 IKS AO system, which is lacking the PIP2 inter (Supplementary Fig. S4, B, upper panel) the basic residues from S6 and from KCNE1 might repel each other and prevent S6 Cter from reaching PIP2 intra located in the inner membrane surface in AO state, as observed.
The molecular determinants of this second PIP2 binding site we identified in our IKS models are supported by a previous functional study [36] which shed light on several residues of KCNE1 subunit participating in KCNE1-PIP2 interactions in IKS channels. Our results are also in agreement with an integrative study including both experimental and computational approaches [14], which aimed at identifying KV7.1 interactions with PIP2 in IKS that highlighted the existence of two PIP2 binding sites for this complex, one for VSD residues and a second one for PD residues.
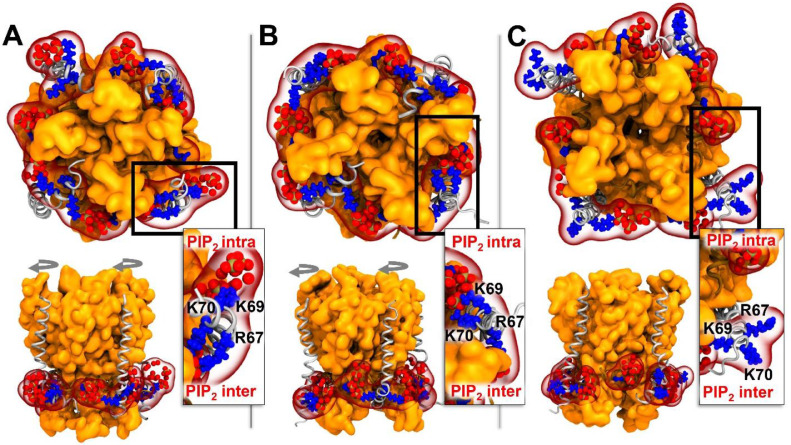
Noteworthy, KCNE1 residues bind both PIP2 inter and PIP2 intra in the RC and IC states, which may induce a tighter packing of PD segments. Indeed, these KCNE1-PIP2 interactions form a circle of electrostatic interactions around the cytoplasmic region of S6 (Fig. 4A, B). This circle may act as a tourniquet (Fig. 4A, B, top panels), inducing a shrinkage of the conduction pathway, leading to the lower pore radii values we obtained for IKS RC and IC models of 8PIP2 with respect to IKS RC and IC models of 4PIP2, respectively. In the AO models of IKS, KCNE1 loses its interactions with PIP2 intra and binds only PIP2 inter (Fig. 4C). Through this interaction, one can predict that KCNE1 is pulling S6 helices towards the inner leaflet of the membrane, as PIP2 inter remains bound to S6 basic residues.
Fig. 4.
Structural mapping of KCNE1 basic residues and PIP2 lipids in the pore domain of IKS models in 8PIP2 systems
Intracellular view (top panels) and side view (bottom panels) of KV7.1 pore domain segments (in orange surface) along with S4-S5LINKER (in brown ribbons), KCNE1 subunits (in gray ribbons) and PIP2 phosphate groups (in red spheres) in A. RC model, B. IO model and C. AO model. KCNE1 basic residues R67, K69 and K70 are shown in blue spheres (See Tab S4 for their detailed interactions). S4-S5LINKER residue R249 that interact with PIP2 intra in a state independent fashion is shown in blue sticks. The successive rotational movements of KCNE1 subunits predicted to occur during RC-IC and IC-AO transitions are shown in gray circled arrows. For each state, a detailed intracellular view of KCNE1 basic residues interactions (framed in black) are depicted on the right side of the side view panels.
In the RC and IC models, the KCNE1 basic residues form electrostatic interactions with both PIP2 binding sites. This network of interactions forms a circle around S6 CTERM helices which seems to prevent those helices from moving away from the pore axis and therefore preventing the conduction pathway of the IKS channel from expending. Nevertheless, these pore radii differences between the studied IKS models estimated from the MD trajectories did not allow us to determine if these models are genuinely able to conduct potassium ions. Further studies allowing for the calculation of the free energies associated to ion translocation, beyond the scope of this work, would be required to address this issue.
As a matter of fact, a recent computational study, which consisted in the use of machine learning methods to generate a conformational space of IKS channels, have yielded to the design of a structure-based predictor of IKS channel experimental properties including its subconductance and gating current [54]. The two sequential translations and rotations of S4 and the rotations of KCNE1 leading to VSD activation we predicted from our models are supported by the results obtained with this structure-based predictor.
The results reported by Li et al. [36] have shown that KCNE1 increases PIP2 sensitivity 100-fold over channels formed by the pore-forming KV7.1 α-subunits alone. In this study the authors identified four residues (R67, K69, K70, and H73) in proximal C-terminus of KCNE1 as key determinants of PIP2 sensitivity. Mutations of these key residues in KCNE1 (R67C, R67H, K70M, and K70N) are associated with long QT syndrome [17,28,32]. They reduce IKS currents and PIP2 sensitivity. Application of exogenous PIP2 to these mutants restores wild-type channel activity. The results reported in the study of Li et al. [36] reveal the vital role of PIP2 for KCNE1 modulation of IKS channels, confirming the previous studies that highlighted the inhibitory effects of PIP2 membrane depletion on IKS channel function, by inducing PIP2 depletion through the co-transfection of a IKS channel construct [8,68] and a PIP2-phosphatase in Human Embryonic Kidney (HEK-293) cells [56,60]. Furthermore, other studies reported that PIP2 acts as a second messenger [38] of various ion channels. In the case of IKS channel, PIP2 participates in the transduction of sympathetic signaling pathways induced by stress [40] or exercise [13], leading to a left-shift on IKS voltage-dependence of activation [47] and to a 2-fold increase of IKS current amplitude [39], respectively.
The increase in PIP2 membrane levels appeared to restore the function of several loss-of-function mutations (R174C, R243C and R336Q) of KCNQ1 gene [49] that are related to impaired sympathetic stimulation pathways of IKS channel [41]. Specifically, this observation suggests that both sympathetic pathways mutants activate the IKS channel by strengthening its interactions with VSD residues R174 and R243 as well as with PD residue R366. The present investigation, confirming that two of these residues are involved in specific interactions with PIP2, provides the key molecular elements that govern such a role. Indeed, our models suggest that residues R243 and R366 bind PIP2 intra and PIP2 inter in a state-dependent and a state-independent fashion, respectively. This observation agrees with the sympathetic stimulations of the VSD mutants and PD mutants cited above which yielded to different sensitivities to PIP2 membrane levels. Altogether, these results indicate a possible difference of binding affinity between both PIP2 binding sites in IKS channel.
Moreover, the family of ion channel β-subunits (KCNE1-5) contains several members that have been reported to modulate the activity of a variety of channel α-subunits in ion channel complexes. Many of these channel α-subunits or channel complexes are also modulated by PIP2. The KCNE1 basic residues listed above that are essential for modulation of the IKS PIP2 sensitivity are highly conserved across all members of the KCNE family of peptides (Fig. 3), suggesting that modulation of PIP2 sensitivity may be a common mechanism of current modulation by the KCNE β-subunits. As we built models of the transmembrane regions of both KV7.1 and KCNE1 subunits, the recently reported PIP2 binding sites, located in the distal cytoplasmic region of KV7.1 [65] and KV7.3 subunits [7], could not have been addressed in the frame of this study.
Fig. 3.
Conserved basic residues in the of KCNE ancillary subunits.
The picture shows a sequence alignment of the transmembrane domains (TMD) of KCNE subunits, highlighted in cyan. The conserved basic residues, located at the end of the TMD, near their CTERM domains, are highlighted in blue.
However, it is important to note that the KV7.3 residues identified by Choveau et al. [7] functional studies include three KV7 conserved basic residues R242, K358 and K366, (which correspond to R243, K354 and K362, respectively, in KV7.1) that turned out to be crucial for KV7.3 pore-opening. For similar reasons, the modulation effects of Calmodulin on KV7.1 gating kinetics, which has been reported by several experimental studies [[65], [78],7] could not be properly addressed in the frame of this study, as a recent integrative study recently reported that Calmodulin is crucial for the transition of KV7.1 IO-AO transition, in the absence of KCNE1 [77].
The present analysis demonstrated the coherence of the simulation of our IKS models in a system that contain eight PIP2 molecules instead of four, each located in the respective binding sites of KV7.1 and KCNE1 subunits, in order to fit the experimental data as much as possible. Hence, the robustness of the resulting MD trajectories will be used for ongoing work that features the investigation of the molecular determinants of the VSD-PD coupling and pore opening mechanisms of the KV7.1 channel, in both the absence [21] and the presence of the KCNE1 ancillary subunit.
Material and methods
To investigate the structural determinants of PIP2 binding sites of the IKS channel, we first needed to build molecular models which had to be as trustworthy as possible with respect to experimental data available. Since no high-resolution structure of the KV7.1 channel was available yet, we used homology modeling to build several models of the channel embedded within a lipid bilayer [48]. For KV7.1, the activation mechanism involves three stable states, i.e., RC, IC, and AO states. Accordingly, each state was modeled to obtain a larger spectrum of possible α-subunits conformations, allowing for the prediction of possible transition mechanisms for IKS channel. For these models, the NTERM and CTERM cytoplasmic regions of KV7.1 were ignored. Only residues 122 to 366 from KCNQ1 human sequence, corresponding to the transmembrane region of the channel, were considered. To fully characterize the modulation of KV7.1 by the KCNE1 subunits, we built each state model along with the TMD (residues 39 to 76) of the human KCNE1 NMR structure [64] using a KV7.1:KCNE1 subunit ratio of 4:4.
-
•
Homology modeling of KV7.1 in its distinct states
Homology modeling, also known as comparative modeling, aims at building a protein structure from its primary sequence, starting from the premise that two proteins with similar primary sequences will be displaying similar folds (3D structures). To adjust the salt-bridge patterns of the VSD with respect to experimental results [71,70], and thereby obtain distinct activation state models of the VSD, the charged group of E160 (E1) was constrained to be in close proximity with:
-
-R237 (R4) in activated/open (AO) model, using the refined crystallographic structure of KV1.2 [6] as a template;
- -
-
-R228 (R1) in Resting/Closed (RC) model, using the ε conformation of KV1.2 refined structure obtained from the aforementioned study.
-
-
The alignment of the KV7.1 human sequence and KV1.2 rat sequence was first conducted automatically, using ClustalW2 [33]. For the PD, the percentage of sequence identity between KV7.1 and KV1.2 is 36%. For the VSD, the percentage is 19.5%. This lower sequence identity is mainly due to the S2-S3 loop, which is longer in KV7.1 sequence than in the one of KV1.2. To overcome this discrepancy, this alignment was refined manually and locally. Indeed, we specifically aligned KV7.1 important residues (conserved acidic residue of S2, conserved CTC, S4 conserved gating charges) with similar KV1.2 ones. Plus, insertions and deletions were concentrated in the loop regions. Eventually, without S2-S3 loop, the percentage of identity between KV7.1 and KV1.2 was increased to 25% for the VSD.
For each stable state of KV7.1, fifty models were generated using MODELLER [15]. This software performs constrained modeling, a technique which consists in using template coordinates and sequence alignment information as constraints for the building of 3D models. S2-S3 loop has been modeled from a template 3D structure extracted from NMR data [50], which suggests a helical structure for this connecting loop. Since MODELLER allows one to add specific geometric restraints, several ones were applied according to site-mutagenesis results to increase the reliability of these models with respect to experimental data. Other constraints drawn from 3D models of IKS channel stable states [27] were also used to predict the position of KCNE1 subunits with respect to KV7.1.
Among these 50 obtained models, the best 10 were selected according to their potential energy values, calculated using DOPE (Discrete Optimizing Protein Energy) [57] knowledge-based scoring function, implemented in MODELLER. The stereo-chemical quality of these models were evaluated using PROCHECK software [34]. For each stable state, the structure presenting the highest number of Phi and Psi torsion angles in Ramachandran's plot well favored areas (>95%), as well as the lowest number of torsion angles in the disfavored areas (<5%), was chosen to perform molecular dynamics simulations to study our models.
-
•
Molecular Dynamics simulations
To reproduce the behavior of IKS channels in their natural environment, the three models were embedded in lipid bilayers prior to the simulations. To do this, we used a method available in the input generator of CHARMM [26], which consists in adding lipid molecules around the protein structure [72]. Since Phosphatidylcholine (PC) lipids are the most abundant lipids found in cell membranes, Palmitoyl-Oleyl PC (POPC) lipids were selected to build the bilayer in each of our nine systems. To incorporate phospholipids in the correct binding sites of the channels, a PIP2 molecule was added within the inner leaflet of the bilayer, at the bottom of each VSD (PIP2 intra) with respect to experimental studies [14,75] and computational results of MD simulations conducted on KV7.1 subunits along with PIP2 [29,76]. We embedded the RC, IC and AO states models in two distinct environments. In the first system, referred to as 4PIP2, we added only PIP2 intra, corresponding to the binding site of KV7.1 subunits. In the second system, referred to as 8PIP2 we added a second PIP2 molecule (PIP2 inter), at the bottom of KCNE1 subunits, following data from experiments that highlighted a second PIP2 binding site in IKS channels [36] (cf. Fig. 1.)
Simulations were carried out using the NPT ensemble for the equilibration of the systems, at 300 K, and 1 atm. (Noteworthy, POPC has a transition temperature above 300 K, i.e. it is in its liquid crystal phase). Chemical bond lengths between hydrogen and heavy atoms were all constrained at their equilibrium values so that a time-step of 2.0 fs could be used. The systems (lipids+channel) were surrounded by a 150 mM [KCl] solution. We used the CHARMM36 force-field [22], along with CMAP correction [3] and NAMD code [52] to perform all MD calculations.
The MD simulations were conducted in four steps, during which motion constraints were applied on the whole system, and then gradually released. The first step of 200 ps aimed at fully solvating the protein in the membrane, by letting water molecules rearrange themselves around the protein. Accordingly, constraints were set up on all IKS atoms. The second step of 6 ns was run to relax the side chains of the protein, so the constrains were kept only on IKS backbone. During these two steps, the positions of PIP2 phosphorus atoms of in the system were kept constant, to maintain this lipid in its correct binding sites. The third step was conducted to allow PIP2 lipids to rearrange around the protein complex and within the lipid bilayer. Hence the constraints on PIP2 phosphorus atoms were removed but we maintained on the backbone of IKS complex. This step of 70 ns was also conducted to let the density of the system reach a constant value. Finally, the last step corresponds to the so-called production phase. This step, performed without any specific forces on any coordinate of the system, lasts approximately 500 ns. As our main goal is to obtain models of membrane-embedded IKS channels at the equilibrium, only the backbone of the selectivity filter (corresponding to the voltage-gated ion channel conserved 311-TTIGYG-316 sequence) was spatially constrained, to prevent ion conduction during the simulations.
-
•
Validation of IKS models by MD simulation analyses: Strategy
We considered here only the production phase of MD simulations. To assess the reliability of the models with respect to experimental data, we confronted, for each state of the IKS channel, the 2 systems against a set of results obtained from biophysical characterization. To achieve this, we first gathered all the cross-linking studies conducted on IKS channel to identify: (i) The VSD salt-bridge patterns, (ii) The KV7.1 intersubunit and intrasubunit neighbor residues, (iii) The KCNE1/KV7.1 neighbor residues and (iv) The KV7.1 and KCNE1 residues which bind PIP2. Cysteine cross-linking studies assume that mutated cysteine residues can form disulfide bonds with a significant formation rate constant if the distance between their Cβ atoms is ⩽ 13.2 Å [4]. Accordingly, we evaluated the Cβ-Cβ distances of each pair of residues in the MD trajectories we obtained for our IKS models and reported their average values (see tables S2-S3 (Supporting Information)).
A pair was considered as fulfilled in the model if the average distance between the Cβ atoms pair was below ∼ 13 Å for at least three channel subunits out of four. These distances were monitored every 2 ns over the MD trajectories, using a designed program written in TCL language which we executed within the scripting interface of Visual Molecular Dynamics (VMD) software [23]. We investigated the electrostatic interactions and the hydrogen bonds that are expected between the S4 gating charges and the S2 negative ones (Table S1), as well as the ones between basic side chains from the cytoplasmic region of KCNE1 and PIP2 (Table S4). These were monitored every 2 ns and were considered satisfied if the average distance between the respective charge moieties were below 3.5 Å, which is the average distance encountered for ion pairs in protein NMR structures [30].
For these calculations, we considered that the positive charge of arginine side chains is delocalized between three terminal nitrogen atoms of its guanidium group, and the negative charges of glutamate and aspartate side chains are shared by two oxygen atoms from their respective carboxyl group. For PIP2, its five negative charges are delocalized between eight oxygen atoms from its three phosphate groups (Supplementary Fig. S2). Hence, the salt bridges they form with the channel residues were monitored by computing each of the atom pair combinations of terminal nitrogen atoms from Arg guanidium group and terminal oxygen atoms from Glu or Asp carboxyl groups. The distance graphics were designed using R scripting language (Heiner [18]). For each pair of residues, an interaction was considered present if found in at least three subunits out of four.
Starting from the premise that pore radius of open KV channels gates energetically favors ionic conduction [2,51,66], one expects that channel models with larger pore radii are likely to facilitate ion conduction and therefore generate similar ionic currents as those recorded experimentally. Pore radii and pore solvent accessible surfaces of all models were calculated and generated every 20 ns of their respective MD trajectory, both using HOLE program [58]. For each model, we reported the average pore radii values along the pore axis, i.e. the conduction pathway. For each state, we compared the obtained pore radii with those calculated for the KV7.1 models built in the frame of a previous study. Indeed, KCNE1 is known to enhance KV7.1 ionic currents in the AO state, while abolishing this current in both the Intermediate and the RC states. Pore surfaces were rendered with VMD.
For each IKS state, the MD trajectory which presented the highest number of salt bridges in agreement with experimental data, and the most relevant average pore radii with respect to those obtained for the KV7.1 models, was selected as the best model of IKS channel complex.
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2023.100073.
Appendix. Supplementary materials
References
- 1.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. KvLQT1 and IsK (minK) proteins associate to form the IKS cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 2.Beckstein O., Tai K., Sansom M.S.P. Not ions alone: barriers to ion permeation in nanopores and channels. J. Am. Chem. Soc. 2004;126:14694–14695. doi: 10.1021/ja045271e. [DOI] [PubMed] [Google Scholar]
- 3.Buck M., Bouguet-Bonnet S., Pastor R.W., MacKerell A.D. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Careaga C.L., Falke J.J. Thermal motions of surface α-Helices in the D-Galactose chemosensory receptor. J. Mol. Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catterall W.A. Molecular properties of voltage-sensitive sodium channels. New Insights Cell Membr. Transp. Process. 1986:3–20. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Wang Q., Ni F., Ma J. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc. Natl. Acad. Sci. USA. 2010;107:11352–11357. doi: 10.1073/pnas.1000142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choveau F.S., De la Rosa V., Bierbower S.M., Hernandez C.C., Shapiro M.S. Phosphatidylinositol 4,5-bisphosphate (PIP 2) regulates KCNQ3 K + channels by interacting with four cytoplasmic channel domains. J. Biol. Chem. 2018;293:19411–19428. doi: 10.1074/jbc.RA118.005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahimène S., Alcoléa S., Naud P., Jourdon P., Escande D., Brasseur R., Thomas A., Baró I., Mérot J. The N-terminal juxtamembranous domain of KCNQ1 is critical for channel surface expression: implications in the Romano-Ward LQT1 syndrome. Circ. Res. 2006;99:1076–1083. doi: 10.1161/01.RES.0000250262.12219.95. [DOI] [PubMed] [Google Scholar]
- 9.Delemotte L., Tarek M., Klein M.L., Amaral C., Treptow W. Intermediate states of the Kv1.2 Vage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. 2011;108:6109–6114. doi: 10.1073/pnas.1102724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delemotte L., Kasimova M.A., Klein M.L., Tarek M., Carnevale V. Free-energy landscape of ion-channel voltage-sensor–domain activation. Proc. Natl. Acad. Sci. 2015;112:124–129. doi: 10.1073/pnas.1416959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Díaz-Franulic I., Sepúlveda R.V., Navarro-Quezada N., González-Nilo F., Naranjo D. Pore dimensions and the role of occupancy in unitary conductance of Shaker K channels. J. Gen. Physiol. 2015;146:133–146. doi: 10.1085/jgp.201411353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durell S.R., Guy H.R. Atomic scale structure and functional models of voltage-gated potassium channels. Biophys. J. 1992;62:238–250. doi: 10.1016/S0006-3495(92)81809-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvir M., Strulovich R., Sachyani D., Ben-Tal Cohen I., Haitin Y., Dessauer C., Pongs O., Kass R., Hirsch J.A., Attali B., et al. Long QT mutations at the interface between KCNQ1 helix C and KCNE1 disrupt IKS regulation by PKA and PIP2. J. Cell Sci. 2014;127:3943–3955. doi: 10.1242/jcs.147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckey K., Wrobel E., Strutz-Seebohm N., Pott L., Schmitt N., Seebohm G. Novel Kv7.1-phosphatidylinositol 4,5-bisphosphate interaction sites uncovered by charge neutralization scanning. J. Biol. Chem. 2014;289:22749–22758. doi: 10.1074/jbc.M114.589796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.Y., Pieper U., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007;50 doi: 10.1002/0471140864.ps0209s50. 2.9.1-2.9.31. [DOI] [PubMed] [Google Scholar]
- 16.Gofman Y., Shats S., Attali B., Haliloglu T., Ben-Tal N. How does KCNE1 regulate the Kv7.1 potassium channel? Model-structure, mutations, and dynamics of the Kv7.1-KCNE1 complex. Structure. 2012;20:1343–1352. doi: 10.1016/j.str.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Hedley P.L., Jørgensen P., Schlamowitz S., Wangari R., Moolman-Smook J., Brink P.A., Kanters J.K., Corfield V.A., Christiansen M. The genetic basis of long QT and short QT syndromes: a mutation update. Hum. Mutat. 2009;30:1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- 18.Schwarte Heiner, Masarotto Guido, Falcon Seth, Bates Douglas, Chambers John, Dalgaard Peter, Gentleman Robert, Hornik Kurt, Ihaka Ross, Kalibera Tomas, et al. R Foundation for Statistical Computing; Vienna, Austria: 2017. R: A language and Environment For Statistical Computing. [Google Scholar]
- 19.Hilgemann D.W., Feng S., Nasuhoglu C. The complex and intriguing lives of PIP2 with ion channels and transporters. Sci. STKE Signal Transduct. Knowl. Environ. 2001;2001 doi: 10.1126/stke.2001.111.re19. re19–re19. [DOI] [PubMed] [Google Scholar]
- 20.Hou P., Eldstrom J., Shi J., Zhong L., McFarland K., Gao Y., Fedida D., Cui J. Inactivation of KCNQ1 potassium channels reveals dynamic coupling between voltage sensing and pore opening. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou P., Kang P.W., Kongmeneck A.D., Yang N.-.D., Liu Y., Shi J., Xu X., White K.M., Zaydman M.A., Kasimova M.A., et al. Two-stage electro–mechanical coupling of a KV channel in voltage-dependent activation. Nat. Commun. 2020;11:676. doi: 10.1038/s41467-020-14406-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J., Mackerell A.D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Jalily Hasani H., Ahmed M., Barakat K. A comprehensive structural model for the human KCNQ1/KCNE1 ion channel. J. Mol. Graph. Model. 2017;78:26–47. doi: 10.1016/j.jmgm.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Jalily Hasani H., Ganesan A., Ahmed M., Barakat K.H. Effects of protein-protein interactions and ligand binding on the ion permeation in KCNQ1 potassium channel. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 27.Kang C., Tian C., Sönnichsen F.D., Smith J.A., Meiler J., George A.L., Vanoye C.G., Kim H.J., Sanders C.R. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel † ‡. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapplinger J.D., Tester D.J., Salisbury B.A., Carr J.L., Harris-Kerr C., Pollevick G.D., Wilde A.A.M., Ackerman M.J. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION® long QT syndrome genetic test. Hear. Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasimova M.A., Zaydman M.A., Cui J., Tarek M. PIP2-dependent coupling is prominent in Kv7.1 due to weakened interactions between S4-S5 and S6. Sci. Rep. 2015;5:7474. doi: 10.1038/srep07474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S., Nussinov R. Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys. J. 2002;83:1595–1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix J.J., Bezanilla F. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6444–6449. doi: 10.1073/pnas.1103397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai L.-.P., Su Y.-.N., Chiang F.-.T., Juang J.-.M., Liu Y.-.B., Ho Y.-.L., Chen W.-.J., Yeh S.-.J., Wang C.-.C., Ko Y.-.L., et al. Denaturing high-performance liquid chromatography screening of the long QT syndrome-related cardiac sodium and potassium channel genes and identification of novel mutations and single nucleotide polymorphisms. J. Hum. Genet. 2005;50:490–496. doi: 10.1007/s10038-005-0283-3. [DOI] [PubMed] [Google Scholar]
- 33.Larkin M.A.A., Blackshields G., Brown N.P.P., Chenna R., Mcgettigan P.A.A., McWilliam H., Valentin F., Wallace I.M.M., Wilm A., Lopez R., et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Li Y., Gamper N., Hilgemann D.W., Shapiro M.S. Regulation of Kv7 (KCNQ) K+ channel open probability by phosphatidylinositol 4,5-bisphosphate. J. Neurosci. 2005;25:9825–9835. doi: 10.1523/JNEUROSCI.2597-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Zaydman M.A., Wu D., Shi J., Guan M., Virgin-Downey B., Cui J. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc. Natl. Acad. Sci. USA. 2011;108:9095–9100. doi: 10.1073/pnas.1100872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liin S.I., Barro-Soria R., Larsson H.P. The KCNQ1 channel – remarkable flexibility in gating allows for functional versatility. J. Physiol. 2015;593:2605–2615. doi: 10.1113/jphysiol.2014.287607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logothetis D.E., Petrou V.I., Adney S.K., Mahajan R. Channelopathies linked to plasma membrane phosphoinositides. Pflugers Arch. Eur. J. Physiol. 2010;460:321–341. doi: 10.1007/s00424-010-0828-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marx S.O., Kurokawa J., Reiken S., Motoike H., D'Armiento J., Marks A.R., Kass R.S. Requirement of a macromolecular signaling complex for β adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 40.Matavel A., Lopes C.M.B. PKC activation and PIP2 depletion underlie biphasic regulation of IKs by Gq-coupled receptors. J. Mol. Cell. Cardiol. 2009;46:704–712. doi: 10.1016/j.yjmcc.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matavel A., Medei E., Lopes C.M.B. PKA and PKC partially rescue Long QT type 1 phenotype by restoring channel-PIP 2 interactions. Channels. 2010;4:3–11. doi: 10.4161/chan.4.1.10227. [DOI] [PubMed] [Google Scholar]
- 42.McCrossan Z.A., Abbott G.W. The MinK-related peptides. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Murray C.I., Westhoff M., Eldstrom J., Thompson E., Emes R., Fedida D. Unnatural amino acid photo-crosslinking of the IKs channel complex demonstrates a KCNE1:KCNQ1 stoichiometry of up to 4:4. Elife. 2016;5 doi: 10.7554/eLife.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajo K., Ulbrich M.H., Kubo Y., Isacoff E.Y. Stoichiometry of the KCNQ1 - KCNE1 ion channel complex. Proc. Natl. Acad. Sci. USA. 2010;107:18862–18867. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Napolitano C., Priori S.G., Schwartz P.J., Bloise R., Ronchetti E., Nastoli J., Bottelli G., Cerrone M., Leonardi S. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. J. Am. Med. Assoc. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 46.Nerbonne J.M., Kass R.S. Molecular physiology of cardiac repolarization. Physiol. Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 47.O-Uchi J., Rice J.J., Ruwald M.H., Parks X.X., Ronzier E., Moss A.J., Zareba W., Lopes C.M. Impaired IKs channel activation by Ca2+-dependent PKC shows correlation with emotion/arousal-triggered events in LQT1. J. Mol. Cell. Cardiol. 2015;79:203–211. doi: 10.1016/j.yjmcc.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Page R.C., Li C., Hu J., Gao F.P., Cross T.A. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magn. Reson. Chem. 2007;45:S2–S11. doi: 10.1002/mrc.2077. [DOI] [PubMed] [Google Scholar]
- 49.Park K.-H.H., Piron J., Dahimene S., Mérot J., Baró I., Escande D., Loussouarn G., Mérot J., Baró I., Escande D., et al. Impaired KCNQ1-KCNE1 and phosphatidylinositol-4,5-bisphosphate interaction underlies the long QT syndrome. Circ. Res. 2005;96:730–739. doi: 10.1161/01.RES.0000161451.04649.a8. [DOI] [PubMed] [Google Scholar]
- 50.Peng D., Kim J.-H.H., Kroncke B.M., Law C.L., Xia Y., Droege K.D., Van Horn W.D., Vanoye C.G., Sanders C.R. Purification and structural study of the voltage-sensor domain of the human KCNQ1 potassium ion channel. Biochemistry. 2014;53:2032–2042. doi: 10.1021/bi500102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peter C., Hummer G. Ion transport through membrane-spanning nanopores studied by molecular dynamics simulations and continuum electrostatics calculations. Biophys. J. 2005;89:2222–2234. doi: 10.1529/biophysj.105.065946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramasubramanian S., Rudy Y. The Structural basis of IKs ion channel activation: mechanistic insights from molecular simulations. Biophys. J. 2018;114:2584–2594. doi: 10.1016/j.bpj.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramasubramanian S., Rudy Y. The Structural basis of IKs ion-channel activation: mechanistic insights from molecular simulations. Biophys. J. 2018;114:2584–2594. doi: 10.1016/j.bpj.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roux B. Dissecting the coupling between the voltage sensor and pore domains. Neuron. 2006;52:568–569. doi: 10.1016/j.neuron.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Royal A.A., Tinker A., Harmer S.C. Phosphatidylinositol-4,5-bisphosphate is required for KCNQ1/KCNE1 channel function but not anterograde trafficking. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0186293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen M., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., Sansom M.S.P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 59.Splawski I., Shen J., Timothy K.W., Lehmann M.H., Priori S., Robinson J.L., Moss A.J., Schwartz P.J., Towbin J.A., Vincent G.M., et al. Spectrum of mutations in long-QT syndrome genes. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 60.Suh B.C., Inoue T., Meyer T., Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J., MacKinnon R. Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell. 2017;169:1042–1050. doi: 10.1016/j.cell.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun J., MacKinnon R. Structural basis of human KCNQ1 modulation and gating. Cell. 2020;180:340–347. doi: 10.1016/j.cell.2019.12.003. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao X., Lee A., Limapichat W., Dougherty D.A., MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian C., Vanoye C.G., Kang C., Welch R.C., Kim H.J., George A.L., Sanders C.R., Hak J.K., George A.L., Sanders C.R., et al. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome †. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tobelaim W.S., Dvir M., Lebel G., Cui M., Buki T., Peretz A., Marom M., Haitin Y., Logothetis D.E., Hirsch J.A., et al. Ca 2+ -Calmodulin and PIP2 interactions at the proximal C-terminus of Kv7 channels. Channels. 2017;11:686–695. doi: 10.1080/19336950.2017.1388478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treptow W., Tarek M. Molecular restraints in the permeation pathway of ion channels. Biophys. J. 2006;91:L26–L28. doi: 10.1529/biophysj.106.087437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tristani-Firouzi M., Sanguinetti M.C. Voltage-dependent inactivation of the human K + channel KvLQT1 is eliminated by association with minimal K + channel (minK) subunits. J. Physiol. 1998;510:37–45. doi: 10.1111/j.1469-7793.1998.037bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang W., Xia J., Kass R.S. MinK-KvLQT1 fusion proteins, evidence for multiple stoichiometries of the assembled I(sK) channel. J. Biol. Chem. 1998;273:34069–34074. doi: 10.1074/jbc.273.51.34069. [DOI] [PubMed] [Google Scholar]
- 69.Wiener R., Haitin Y., Shamgar L., Fernández-Alonso M.C., Martos A., Chomsky-Hecht O., Rivas G., Attali B., Hirsch J.A. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. J. Biol. Chem. 2008;283:5815–5830. doi: 10.1074/jbc.M707541200. [DOI] [PubMed] [Google Scholar]
- 70.Wu D., Delaloye K., Zaydman M.A., Nekouzadeh A., Rudy Y., Cui J. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 2010;135:595–606. doi: 10.1085/jgp.201010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu D., Pan H., Delaloye K., Cui J. KCNE1 remodels the voltage sensor of Kv7.1 to modulate channel function. Biophys. J. 2010;99:3599–3608. doi: 10.1016/j.bpj.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., Qi Y., Lee J., Monje-Galvan V., Venable R.M., et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y., Wang Y., Meng X.-Y.Y., Zhang M., Jiang M., Cui M., Tseng G.-N.N. Building KCNQ1/KCNE1 channel models and probing their interactions by molecular-dynamics simulations. Biophys. J. 2013;105:2461–2473. doi: 10.1016/j.bpj.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaydman M.A., Cui J. PIP2 regulation of KCNQ channels: biophysical and molecular mechanisms for lipid modulation of voltage-dependent gating. Front. Physiol. 2014;5:195. doi: 10.3389/fphys.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaydman M.A., Silva J.R., Delaloye K., Li Y., Liang H., Larsson H.P., Shi J., Cui J. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc. Natl. Acad. Sci. 2013;110:13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaydman M.A., Kasimova M.A., McFarland K., Beller Z., Hou P., Kinser H.E., Liang H., Zhang G., Shi J., Tarek M., et al. Domain–domain interactions determine the gating, permeation, pharmacology, and subunit modulation of the IKs ion channel. Elife. 2014;3:e03606. doi: 10.7554/eLife.03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang P.W., Westerlund A.M., Shi J., White K.M.F., Dou A.K., Cui A.H., Silva J.R., Delemotte L., Cui J. Calmodulin acts as a state-dependent switch to control a cardiac potassium channel opening. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shamgar L., Ma L., Schmitt N., Haitin Y., Peretz A., Wiener R., Hirsch J., Pongs O., Attali B. Calmodulin Is Essential for Cardiac I KS Channel Gating and Assembly. Circ. Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.