Highlights
-
•
Combined spin-label EPR and MAS NMR to characterize thylakoid membrane dynamics.
-
•
Assessment of dynamics of bulk, annular and structural lipids and proteins.
-
•
Thylakoid unstacking increases lipid ps-ns dynamics at different membrane depths.
-
•
Thylakoid unstacking increases ms-ms dynamics of protein helices.
Keywords: Photosynthesis, Spin-label EPR, Dynamic spectral-editing NMR, Chlamydomonas reinhardtii, Thylakoid plasticity, Membrane fluidity
Abstract
Chloroplast thylakoid membranes in plants and green algae form 3D architectures of stacked granal membranes interconnected by unstacked stroma lamellae. They undergo dynamic structural changes as a response to changing light conditions that involve grana unstacking and lateral supramolecular reorganization of the integral membrane protein complexes. We assessed the dynamics of thylakoid membrane components and addressed how they are affected by thylakoid unstacking, which has consequences for protein mobility and the diffusion of small electron carriers. By a combined nuclear and electron paramagnetic-resonance approach the dynamics of thylakoid lipids was assessed in stacked and cation-depletion induced unstacked thylakoids of Chlamydomonas (C.) reinhardtii. We could distinguish between structural, bulk and annular lipids and determine membrane fluidity at two membrane depths: close to the lipid headgroups and in the lipid bilayer center. Thylakoid unstacking significantly increased the dynamics of bulk and annular lipids in both areas and increased the dynamics of protein helices. The unstacking process was associated with membrane reorganization and loss of long-range ordered Photosystem II- Light-Harvesting Complex II (PSII-LHCII) complexes. The fluorescence lifetime characteristics associated with membrane unstacking are similar to those associated with state transitions in intact C. reinhardtii cells. Our findings could be relevant for understanding the structural and functional implications of thylakoid unstacking that is suggested to take place during several light-induced processes, such as state transitions, photoacclimation, photoinhibition and PSII repair.
Introduction
The efficiency of photosynthesis is tightly controlled by the specific composition and arrangement of photosynthetic protein complexes in a dynamic thylakoid membrane [1]. Thylakoid organization in algae and plants is a complex and dynamic system [2,3]. The chloroplast thylakoids of plants and green algae form a complicated 3D structure consisting of stacked regions (grana) interconnected by unstacked regions (stromal lamellae) [4]. The stacked region is densely packed with Photosystem II (PSII) and light-harvesting complex II (LHCII), whereas the unstacked region is enriched in Photosystem I (PSI) and ATP synthase. The thylakoid macrostructure is controlled by protein modification, involving phosphorylation or acetylation of LHC polypeptides, by the composition of LHCII, PSI and PSII and membrane-curvature inducing proteins, and by the concentration of mono- and divalent ions [5]. Phosphorylation of LHC and photosystem complexes also plays a role in molecular recognition for stabilizing PS-LHCII interactions [6]. Membrane remodeling has been shown to be crucial for regulatory processes including adaptation to high light [4,7], state transitions to balance the excitation energy between the two photosystems [8,9], non-photochemical quenching (NPQ) for thermal dissipation of excess excitation energy [10], the balance between linear and cyclic electron transfer pathways [11] and the repair cycle of PS II [12]. Several regulatory processes are associated with phosphorylation of PSII and LHCII and partial (un)stacking of thylakoids [6,8,12].
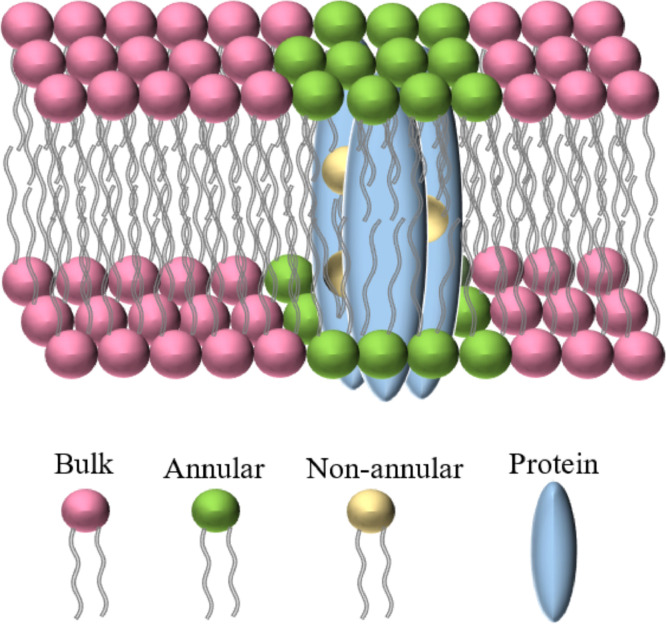
In this study, we address how membrane stacking conditions affect the intrinsic dynamics of the thylakoid proteins and lipids. Biological membrane lipids can be structurally categorized into three groups: (i) the bulk lipids, which do not have specific interaction with proteins; (ii) the annular lipids, which bind to the surface of membrane proteins and form a shell around them and (iii) the non-annular (integral) lipids, which are buried within protein helices or at subunit interfaces within a protein complex [13], [14], [15], see Fig. 1. All three lipid classes play multiple structural and functional roles, which are vital for the function of the photosynthetic machinery [16], [17], [18]. Bulk lipids constitute the lipophilic matrix for photosynthetic proteins, keep the integrity of the membrane and enable the presence of highly curved regions [18]. Annular lipids not only solvate integral membrane proteins but can also affect the function of membrane-embedded proteins [19]. Non-annular lipids play crucial roles in the assembly and functional modulation of the reaction center complexes, e.g. in stabilizing the oligomerization interfaces of PSII dimers and LHCII trimers [16]. Because of the long residence times of non-annular lipids within the binding sites of proteins they are resolved in X-ray and cryo-electron microscopy (EM) structures. The stoichiometry and chemical structure of the non-annular lipids bound to many photosynthetic proteins is known from the crystallographic and cryo-EM data [16,17,20]. However, the information available regarding their dynamics is very limited [14]. Coarse-grain molecular dynamic (CG-MD) simulations of the photosystem II complex embedded in a thylakoid membrane show that non-annular lipids have limited mobility and none of the interfacial lipids leaves the dimer interface [15].
Fig. 1.
Arrangement of bulk lipids (pink), annular lipids (green) around a protein complex (blue) and non-annular lipids (yellow) within a multi-subunit protein complex.
Solid-state NMR spectroscopy is a non-invasive technique that can detect the dynamics of lipids and proteins simultaneously at biologically relevant temperatures. Through NMR dynamic spectral-editing, we have been able to separate the dynamics of integral, non-annular lipids and proteins in thylakoid membranes from those of the annular and bulk lipids [21].
Non-annular, structural lipids that have low dynamics are detected in cross-polarization (CP) based experiments, while annular and bulk lipids with high mobility and low segmental order are detected in insensitive-nuclei enhanced by polarization transfer (INEPT) experiments [22,23]. NMR spectroscopy however does not discriminate between annular and bulk lipids that exchange on an NMR time scale. Similarly, fluorescence-based techniques, which are commonly used to probe membrane fluidity using an external fluorophore, do not easily differentiate those two classes of lipids [24], [25], [26]. In this context, spin-label electron paramagnetic resonance (SL EPR) spectroscopy is the technique of choice to distinguish and characterize the bulk and annular lipids in their native environment [27], [28], [29]. The timescale of the rotational motions of bulk and annular lipids falls nicely into the time window (10−7 to 10−11 s) of EPR detection [17,28,30]. Spin-labeled lipids with stable nitroxide radicals at different positions of their chain can probe the dynamics at different membrane depths. An EPR spectrum of a spin-labeled lipid in a biological membrane generally displays two spectral components, of which one corresponds to the fraction of annular lipids with restricted mobility and the other to the fraction of bulk lipids [31,32]. The fraction of non-annular lipids does not appear in an EPR spectrum as these lipids do not easily exchange with exogenous spin-labeled lipids [17,28]. SL EPR has significantly contributed to the understanding of interactions of annular lipids with proteins in thylakoid membranes [31], [32], [33], [34], [35], [36]. The dynamical properties of plant thylakoid membrane during greening, leaf senescence and oxidative stress were studied using stearic acid spin-label (SASL) EPR [36], [37], [38]. The SL EPR using spin-labeled analogues of the thylakoid membrane lipid components, monogalactosyldiacylglycerol (MGDG), phosphatidylglycerol (PG) and phosphatidylcholine (PC), have been used to investigate the chilling sensitivity of different plants [39] and influence of phosphorylation, illumination and unstacking of thylakoid membranes [40].
We here used a combination of biosynthetic isotope labeling and solid-state NMR and spin-label EPR spectroscopy to investigate the dynamics of C. reinhardtii thylakoid membrane lipids and the consequences of thylakoid unstacking. The rotational diffusion of thylakoid lipids was assessed for stacked and for cation-depletion induced, unstacked thylakoid membranes isolated from the green algae C. reinhardtii. Unstacking of thylakoid membranes is an important process that occurs during state transitions as a response to changes in light quality. The unstacking process was suggested to be associated with the phosphorylation of LHCII and migration of LHCII from PSII to PSI to balance the excitation energy between two complexes [5,7]. Repulsion between the negatively charged phosphoryl groups destabilizes the thylakoid membrane stacks. The repulsive negative charges are neutralized by cations, i.e. Mg2+ in the chloroplast medium [3,41]. Studies on isolated thylakoids clearly demonstrated that the presence of Mg2+ in the medium also regulates thylakoid stacking and unstacking in vitro [41,42] in the absence of phosphorylation changes. Low-temperature and time-resolved fluorescence spectroscopy were applied to measure unstacking-induced changes in functional PSII-LHCII and PSI-LHCI rearrangements. In addition, circular dichroism (CD) spectroscopy was applied to detect the presence of long-range ordered PSII-LHCII stacks that occur in stacked thylakoid membranes in vivo.
Materials and Methods
Cell culturing and thylakoid isolation
Wild-type Chlamydomonas reinhardtii cells (strain cc124) were grown mixotrophically in tris-acetate-phosphate (TAP) medium at pH 7 in a home- built set up, under continuous illumination with white LEDs (∼50 μmol/m2 s) and constant temperature of 25°C. For 13C isotope-label incorporation for NMR experiments, the acetic acid in the TAP medium was replaced by 13C acetic acid (Cambridge Isotopes, Massachusetts, USA).
The isolation of thylakoids was performed according to Chua and Bennoun [43] with some modifications. Cells were harvested in the exponential growth phase and resuspended in buffer A (0.1 M 4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid (HEPES)/KOH, pH 7.5) containing 1 mM MgCl2 and 0.3 M sucrose. Cells were ruptured by sonication and the disrupted cells were centrifuged and resuspended in buffer A with 1 mM MgCl2 and 1.8 M sucrose. The solution was overlaid with 1.3 M sucrose/10 mM Ethylenediaminetetraacetic acid (EDTA) and then 0.5 M sucrose, both in buffer A. The sucrose gradients were ultra-centrifuged in a SW41 swing rotor (Beckmann, California, USA) at 100.000×g. The thylakoid band was extracted using a long needle and kept at 4°C for further use.
The unstacked thylakoid was prepared by twice 10 volume washing of the extracted membrane with 25 mM HEPES/KOH, pH 7.5 containing 5 mM EDTA, then resuspended in the same buffer without EDTA for further experiments.
CD spectroscopy
CD spectra were recorded using a J-815 CD spectrometer (Jasco, Pfungstadt, Germany) equipped with a temperature control unit between 400 and 780 nm at 20°C with a bandwidth of 2 nm and a scanning speed of 100 nm/min in a in a quartz cuvette with 2 mm optical pathlength. The chorophyll concentration of samples was 50 μg/ml.
Fluorescence spectroscopy
Low temperature (77 K) steady-state fluorescence emission spectra were recorded by a FluoroMax spectrofluorometer (HORIBA Jobin-Yvon). The excitation wavelength was set to 400 nm, and emission was detected in the 650–780-nm range. Low temperature time-resolved fluorescence measurements were performed with a (sub)-picoseconds streak-camera setup, which combines a femtosecond laser source (Coherent Vitesse Duo+ a regenerative amplifier Coherent RegA 9000+ an optical parametric amplifier Coherent OPA 9000) with a streak camera detecting system (C5680). The repetition rate was set to 250 kHz. Excitation wavelength was set to 400 nm, and the laser power was adjusted to 80 μW with a laser beam of a ∼1 mm diameter . The fluorescence signal was collected at right angle to the excitation light by a spectrograph (Chromex 250IS, 50 groves/mm ruling, blaze wavelength 600 nm, spectral width 260 nm), with central wavelength set at 720 nm. Scattered excitation light was removed with an optical long-pass filter (550 nm). A 2-ns time window was used, and a high signal-to-noise ratio was achieved by averaging 200 single images, each obtained with exposure time of 10 s. Images were corrected for background and shading and then sliced into traces of ∼2.6 nm width. Samples were directly measured after being frozen in Pasteur pipettes of ∼1 mm diameter for both steady state and time-resolved fluorescence.
Time-resolved fluorescence data obtained with streak camera were analyzed using global analysis with the Glotaran and TIMP software packages. All the decay traces were globally fitted as a sum of exponential decay components convolved with an instrument response function, resulting in decay associated spectra (DASs). See details of global analysis in [44].
SL-EPR spectroscopy
The spin-labeled lipids, 5- and 16-doxyl stearic acid (5- and 16-SASL) were purchased from Sigma-Aldrich, Missouri, USA. For spin-labeling, thylakoid membranes were resuspended in 25mM HEPES buffer, pH 7.5 containing 5 mM MgCl2 for the stacked sample, at 1 mg Chl/ml, and added to a 2 ml glass bottle containing a film of 5-SASL or 16-SASL. The spin label film was prepared on the bottom of the glass bottle by drying 20 µl of a 5 mg/ml n-SASL solution in chloroform. The samples were gently stirred for 40 min in the dark at room temperature. The spin-labelled sample was centrifuged at 14000g for 10 min to get rid of free spin labels. The pellet was transferred into a glass EPR capillary. The EPR measurement of excess supernatant showed that the incubation time was suffiecient to incorporate all the spin labelled lipids into the membrane. As an example, the spectra of 16-SASL in unstacked thylakoid membranes and the excess supernatant from single step centrifugation after incubation of membrane with 16-SASL for 40 minutes are overlaid in Fig. A1.
Continuous-wave (CW) EPR spectra were recorded at 9.88 GHz using an EMX spectrometer (Bruker BioSpin GmbH). The sample capillary was accommodated within a standard 3 mm quartz EPR tube. The measurements were performed at 19 ˚C, modulation amplitude/frequency of 0.15 mT / 100 kHz and microwave power of 0.16 mW.
The EPR spectra were simulated using the EasySpin package [45] based on the Chili function and an isotropic rotation correlation time. The following tensors of g and hyperfine interaction (A, MHz) were used for the simulation: g16-SASL = [2.0092 2.0056 2.0026], A16-SASL = [16 16 91], g5-SASL = [2.0088 2.0061 2.0026] and A5-SASL = [16 15 92.6]. All spectra were simulated using two components. The fraction of each component and its corresponding rotation correlation time were manually adjusted to get the best agreement with the experimental spectra. The minimum difference in the double integral of experimental and simulated spectra was used as a criterion to choose best simulated spectrum. To determine the error margins, the fraction of each component and its corresponding rotation correlation time were changed manually until the difference between the double integral of experimental and simulated spectra was larger than 20% of that of experimental one or a new peak appeared in the simulated spectrum, which was not observed experimentally.
Solid-state NMR spectroscopy
NMR spectra were recorded on a Bruker Avance-I 750 MHz wide bore NMR spectrometer. NMR samples were prepared by centrifugation of fresh thylakoid suspensions into a 4 mm NMR rotor. Magic Angle Spinning was performed with a spinning frequency of 13 kHz. Direct polarization (DP) experiments were performed with 5 s recycle delay and 22 ms acquisition time. For cross- polarization (CP) a contact time of 2 ms, recycle delay of 5 s and acquisition time of 43 ms, ω1C/2π of 40.3 kHz and 1H nutation frequency linearly ramped from 80 to 100 kHz were used. Insensitive nucleus- enhanced polarization-transfer (INEPT) experiments were performed with two delays of 1.25 ms and an acquisition time of 80 ms. NMR data were processed and analyzed in Top-Spin3.5 and MestReNova 12.0.1.
Results
Excitonic CD spectroscopy
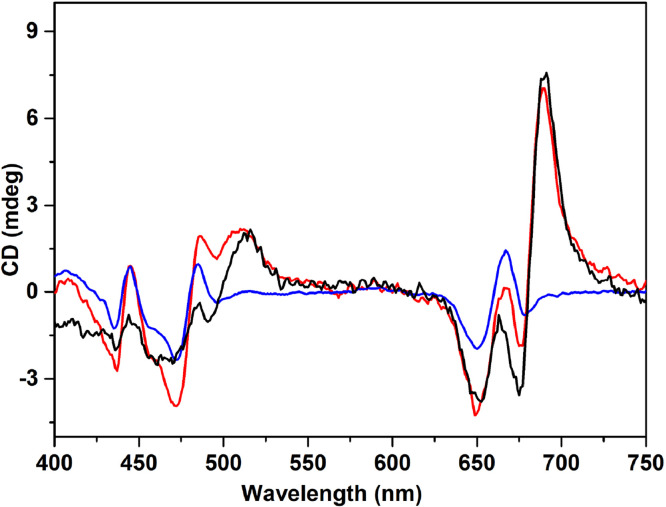
The CD spectra in the visible range of the isolated thylakoid membranes show the typical Psi-type bands (for a full decription of the Psi-type bands see ref. [46]) at around 690 nm and 506 nm, which are assigned to the long-range ordered protein assemblies in the grana, associated with stacked membranes. Both the (+) 690 and (+) 506 bands characteristic of stacking diminish in the EDTA-washed, unstacked sample, see Fig. 2. The CD features observed for both stacked and unstacked samples are compatible with what has been reported in the literature for isolated stacked and unstacked plant thylakoid membranes [47,48]. The CD spectra confirm that cation-depletion-induced unstacking results in a loss of long-range ordered supramolecular structures in the membranes.
Fig. 2.
CD spectra of C. reinhardtii cells (black), stacked (red) and unstacked (blue) thylakoid membrane at 20°C. All samples are in 25 mM Hepes buffer, pH 7.5 plus 5 mM Mg2+ for the stacked membrane and 10% Ficoll for the cells.
Fluorescence spectroscopy and global data analysis
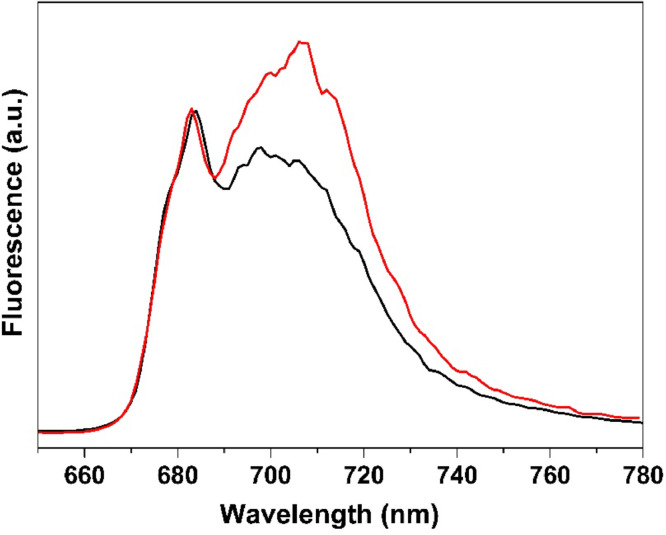
Fig. 3 displays the low temperature steady-state chlorophyll fluorescence spectra of stacked and unstacked thylakoid membranes. Two broad bands with emission at 683 nm and 710 nm were observed, which can be respectively assigned to PSII (together with detached LHCs, if exist) and PSI. Upon thylakoid membrane unstacking, PSI emission was relatively enhanced, in agreement with previous studies [41,42], however, it remains unclear whether PSI emission is enhanced or PSII fluorescence is quenched.
Fig. 3.
77 K steady-state fluorescence spectra of stacked (black) and unstacked (red) thylakoid membranes upon 400 nm excitation. The spectra are normalized at 683 nm.
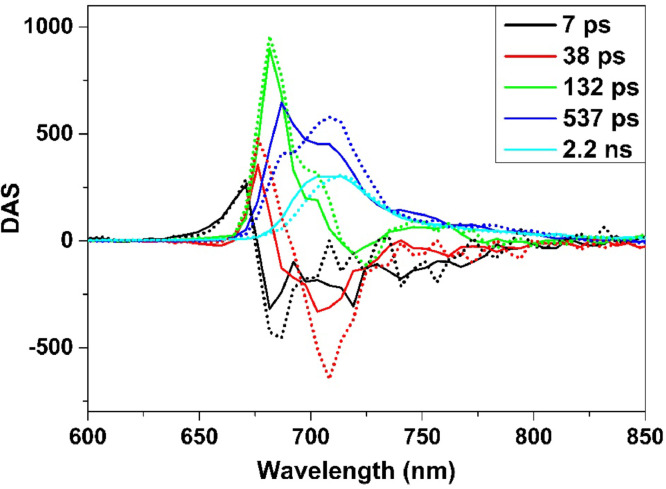
To further address this question, we measured the time-resolved fluorescence with a pico-second streak-camera system. Five components with lifetime ranging from 7 ps to ∼2 ns were estimated by globally fitting all the decay traces of both stacked and unstacked samples. Their amplitudes were plotted over wavelength (Fig. 4), providing so-called Decay Associated Spectra (DASs). Notably, a positive DAS corresponds to a decay of fluorescence, while a negative DAS corresponds to a rise of fluorescence. DAS with a positive amplitude at the higher energy side and a negative amplitude at the lower energy side usually represents a down-hill excitation-energy transfer (EET) process. Hence, the 7 ps component reflects an energy transfer processes as its DAS has a typical EET feature, and most likely this EET takes place within the LHC antenna, which contains two groups of chlorophylls those that emit at ∼670 nm (the positive peak) and populate in succession at ∼681 nm (the negative peak) [49]. Upon unstacking, the negative amplitude increased, suggesting an enhanced EET. The 38 ps component reflects an EET process from bulk chlorophylls to the red-shifted Chls of PSI [49]. This component is significantly affected by unstacking, indicating a more pronounced energy transfer process to PSI. The DAS of the 132 ps component is largely positive, originating from two processes overlapping in time: a LHCs decay (with a peak at 681 nm) and an EET process to/within PSI (with a dip at 720 nm) [49]. Quenched LHCs has often been observed with 77 K time-resolved fluorescence [50, 51], where the 700 nm peak suggests the presence of LHCII aggregates [56]. The amplitude of the 700 nm shoulder increases upon unstacking, suggesting that the amount of quenched LHCs is also increased while the EET process is slightly upregulated. The DAS of the two longest lifetimes reflect the emission of red-shifted chlorophylls from PSII (687∼697 nm) and PSI (708∼713 nm) [49]. Following unstacking, the amplitudes of PSI red-emission in both components (note that in the 2 ns component the increase at 720 nm is counter-balanced by a drop in the vibronic bands of PSII) increase as more energy is funneled either from LHCs to PSI (the so-called shuttle model [52]) or from PSII to PSI (spill-over model [53]). Therefore, it is clear that the PSI fluorescence in the unstacked thylakoid membrane is enhanced because of more energy feed-in.
Fig. 4.
Time-resolved spectroscopy. DAS of stacked (solid) and unstacked thylakoid membranes (dot) upon 400 nm excitation. To simplify the analysis, lifetimes of two samples were forced to be identical during fitting and indicated in the Fig.. DAS were normalized to their time zero emission for direct comparison between two samples.
Solid-state NMR spectroscopy with dynamic spectral editing
We applied 13C MAS NMR-based direct polarization (DP), CP and INEPT for dynamic-based spectral editing to separate dynamical and rigid lipid membrane components. In the DP experiment, a single 90o pulse is applied on 13C and all carbon constituents are detected, irrespective of their dynamics. In the applied CP and INEPT experiments, magnetization is transferred from 1H to 13C for signal enhancement and the detected 13C signal intensities depend on the heteronuclear magnetization transfer efficiencies [21, 54]. In CP, magnetization is transferred via dipolar couplings, which are the direct interaction of nuclear spins through space. CP transfers become ineffective for molecules with significant rapid isotropic motions and CP therefore is selective for detection of rigid molecules. In INEPT, magnetization is transferred via J couplings (indirect interaction between nuclear spins mediated by the bonding electrons) using spin-echo pulses. Fast relaxation in the T2 plane of rigid molecules causes loss of magnetization during the INEPT spin-echo periods, making this experiment sensitive for detection of molecules with large-amplitude, sub-nanosecond motions [21, 54]. T2 is the spin-spin relaxation, which describes the rate of the decay of the magnetization within the xy plane, perpedicular to the applied magnetic field.
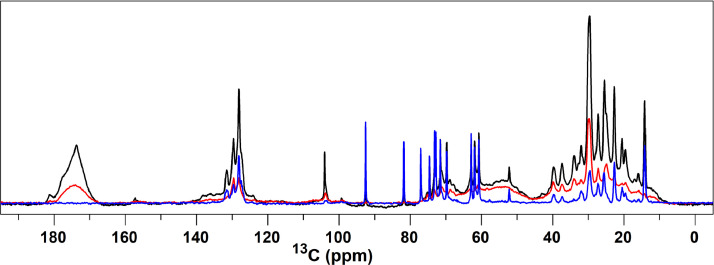
In Fig. 5, the DP, CP and INEPT spectra of stacked thylakoid membranes are overlaid. The DP spectrum contains the signals of all carbon constituents in the sample. In the region between 20-40 ppm protein side-chain 13C signals accumulate as a broad band on which lipid acyl-chain carbon signals are superimposed as sharp peaks [54 - 56]. Protein backbone signals accumulate in the aliphatic 13-carbon region between 50-70 ppm (13Cα) and between 170-180 ppm (13CO) [54, 55]. Signals between 70-110 ppm originate from carbohydrate contents. Those include the galactosyl lipid heads of MGDG (see Fig. A2), digalactosyldiacylglycerol (DGDG) and sulfoquinovosyldiacylglycerols (SQDG) [56, 57] and the natural-abundance 13C signals of the added sucrose that was present in the thylakoid suspensions. The double-bonded 13C signals of the unsaturated lipid chains are visible as sharp peaks in the region between 125-135 ppm and are superimposed on the signals of protein aromatic side-chain carbons [54, 55].
Fig. 5.
Overlaid DP (black), CP (red) and INEPT (blue) 13C NMR spectra of stacked thylakoid membranes recorded at 10°C.
The CP spectrum (Fig. 5, in red) is selective for rigid carbon components with motions on micro- to millisecond timescales. Those include the majority of membrane protein segments and the integral, non-annular lipids that are protein-bound. For a perfect solid, optimal CP-based 1H-13C transfer efficiencies would theoretically give a 4-fold enhancement of the integrated CP signal intensities compared to DP, based on the 1H and 13C respective gyromagnetic ratios. As shown in Fig. 5, the 13 carbon signal intensities in the CP spectrum (red) however are equal to or lower than the DP intensities (black). This indicates that the integral membrane proteins and non-annular lipids possess significant dynamics in the (sub)microsecond range.
The INEPT spectrum (Fig. 5, in blue) is selective for carbon components that undergo rapid, isotropic motions on (sub)nanosecond time scales. For biological membranes, those components typically are the annular and mobile bulk lipids and highly dynamic protein segments. The INEPT spectrum contains sharp peaks that are characteristic of lipid moieties and lacks typical protein signals. Large signal intensities are observed of the methyl carbons that resonate at 15 ppm and of the double-bond carbons resonating at 128 ppm and have been assigned to the lipid 13C10 carbons of the unsaturated bonds that are close to the lipid end tails [54]. Those signals show that the unsaturated lipids undergo (sub)nanosecond dynamics and have highly mobile tails.
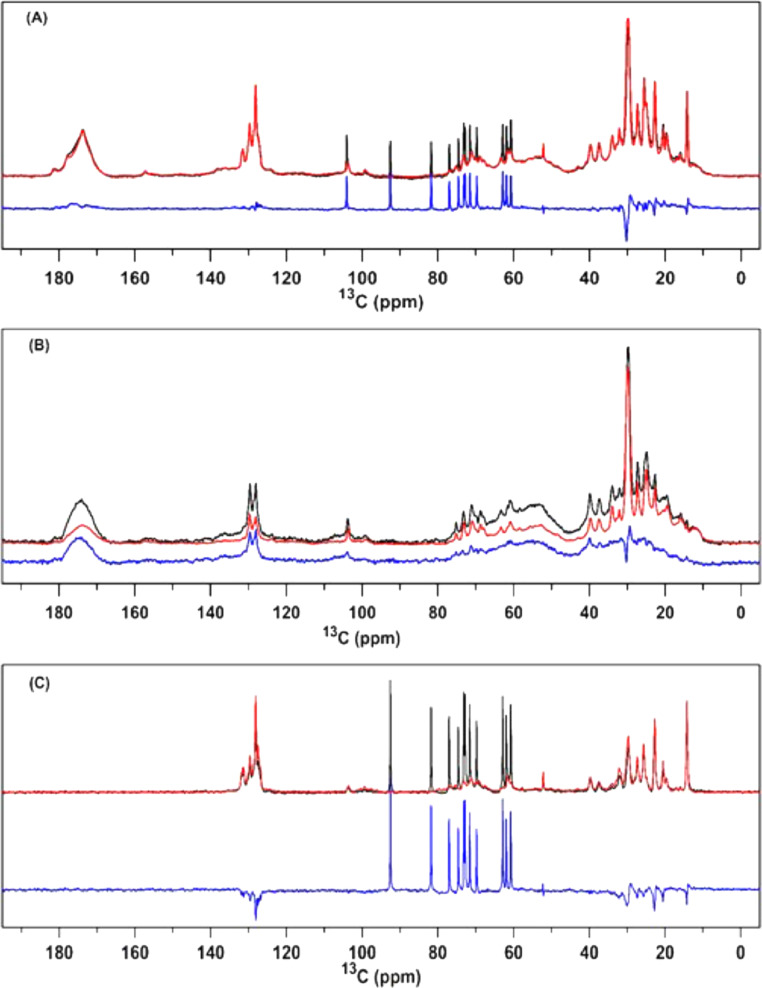
Fig. 6 compares the NMR spectra of stacked and unstacked thylakoid membranes. The DP spectra for the stacked and unstacked samples are normalized to the 13C carbonyl peak of protein at 175 ppm. Accordingly, the CP and INEPT spectra are both scaled using the scaling factor that was used to normalize the DP spectra. The two absolute DP spectra and the difference spectrum of stacked minus unstacked are presented in Fig. 6A. Note that a washing step for removal of cations was used for unstacking, washing out the added sucrose, as shown by the disappearance of the sucrose signals in the unstacked spectrum. Apart from the sucrose peaks, the DP difference spectrum contains weak positive signals in the carbonyl region, a negative peak at 30 ppm and two small band-shift signals at 22 ppm and 15 ppm.
Fig. 6.
Overlaid DP, CP, INEPT 13C NMR spectra of stacked (black) and unstacked (red) thylakoid membranes recorded at 10°C. The blue spectra show the difference spectrum of stacked minus unstacked. (A) DP, (B) CP and (C) INEPT.
The DP spectral intensities should be proportional to the number of spins and not depend on the dynamics of molecules. However, under the chosen experimental conditions, components with very slow T1 relaxation times that exceed the applied recycle delays between the series of scans will not be acquired with the same efficiency. T1 is the spin-lattice relaxation, which describes the rate of the decay of the magnetization in the z-direction, parallel to the applied magnetic field. Lipid acyl chains in particular can have long T1s, which could explain the negative peak at 30 ppm in the DP difference spectrum. Further, the small band shifts may be a side effect of the spectral processing phasing procedures and are not further interpreted. The overall flat shape of the difference spectrum confirms that the cation-depletion unstacking procedure and washing with EDTA did not result in losses of specific thylakoid protein or lipid components.
Fig. 6B shows the CP spectra of stacked and unstacked thylakoid membranes and the CP difference spectrum of stacked minus unstacked. In contrast to the DP spectrum, the intensity of the CP spectrum is drastically reduced in the unstacked state. The CP stacked minus unstacked difference spectrum shows a typical protein spectrum, hence, revealing that there is a notable loss of protein rigidity in the unstacked state. The carbonyl peak maximum of the stacked-minus-unstacked difference spectrum is shifted downfield compared to the maximum of the carbonyl band in the absolute CP spectra. In 13C protein NMR spectra, contributions of protein loop and strand carbonyls typically accumulate in the right wing of the broad carbonyl band between 170-180 ppm [58]. We therefore take a closer look at the maximum position of the carbonyl band in CP difference spectrum. The carbonyl peak maximum of the stacked-minus-unstacked difference spectrum is shifted downfield compared to the maximum of the carbonyl band in the absolute CP spectra. This indicates that there is a significant loss of signal predominantly in the left wing of the carbonyl band where the signals from helical contributions accumulate [59, 60]. We therefore conclude that unstacking enlarges the micro- to millisecond dynamics of the protein helical segments.
The INEPT difference spectrum of stacked minus unstacked in Fig. 6C contains negative peaks for single-bond (10–40 ppm) and for double-bond (120–140 ppm) 13-carbons, indicating increased INEPT efficiency for the unstacked membranes. The difference peaks can be attributed to the accumulated signals of lipid 13-carbons located at different positions along the lipid tails. Because non-annular lipids are not visible in the INEPT spectrum, those are signals from bulk or annular lipids that have fast dynamics at (sub)nanosecond timescales, indicating an increase of annular and bulk lipid dynamics in the unstacked state.
To gain further insight in the specific dynamics changes that occur as a result of membrane unstacking, we compared the stacked-minus-unstacked CP difference spectrum to a difference spectrum of a stacked thylakoid sample that was measured at two different temperatures (5 °C minus 19 °C, see Fig. A3). We observe that CP signal intensity is reduced over the whole spectral range at the higher temperature, consistent with an overall loss of rigidity and increase of thermal motions. In contrast, the changes between the stacked and the unstacked CP spectrum are more specific. The stacked-minus-unstacked difference CP spectrum in Fig. 6B has low intensity in the region between 0 and 30 ppm where the aliphatic and methyl 13C signals occur and lacks the sharp peaks that are typical of lipid acyl chains. Moreover, the 13CO peak maximum of the protein carbonyls is shifted downfield for the stacked-minus-unstacked spectrum compared to the temperature difference spectrum, showing that unstacking predominantly increases the dynamics of helical protein contents. Together, the observations reveal that upon membrane unstacking selective molecular motions are enhanced in the micro- to millisecond range.
EPR spectroscopy
We used two spin-labeled lipids (5- and 16-SASL), which carry the paramagnetic nitroxide group bound to the 5th and 16th carbon atom, respectively, of the stearic acid chain. The position of the nitroxide group determines its depth in the thylakoid membrane. The 5- and 16-SASL are used to probe the dynamics near the head groups of the lipid bilayer and in the center of the membrane, respectively. The mobility of spin labels close to lipid-water phase is known to be lower than that of in the middle of the membrane. This is due to the restricted motion close to the lipid head groups i.e. the lipid-water interface. This restricted motion results in broader EPR line shape [34, 61].
As in other thylakoid membrane systems studied [31,36, 39] the quantitative analysis of the EPR spectra of spin-labeled lipids in stacked and unstacked thylakoid membranes is based on the assumption that the spectra consist of two components: those that derive from SASL molecules in the bulk (i) and those from SASL molecules that are in interaction with the outer surface of proteins, i.e. the annular lipids (ii). However, it might be an oversimplification to assume uniform interaction of annular lipids with proteins [32]. The amount of each component as well as the corresponding rotation correlation time, τ, of the spin-labeled lipid was determined by simulating the EPR line-shape and the parameters obtained are given in Table 1. The τ1 and τ 2 values represent the fluidity of the annular and bulk lipid environment, respectively (the larger the τ value the lower the fluidity). The two components C1 and C2 reflect the fractions of annular and bulk lipid, respectively.
Table 1.
The contribution of the two fit components, immobile (C1) and mobile (C2), which reflects the fractions of annular and bulk lipid, respectively and the corresponding rotation correlation time (τ).
| C1 (%) | τ1 (ns) | C2 (%) | τ2 (ns) | |
|---|---|---|---|---|
| Stacked, 16-SASL |
40 ± 5 | 8.91 ± 0.74 | 60 ± 5 | 2.19 ± 0.15 |
| Unstacked, 16-SASL | 39 ± 7 | 7.10 ± 0.68 | 61 ± 7 | 1.18 ± 0.09 |
| Stacked, 5-SASL |
43 ± 15 | 12.88 ± 1.80 | 57 ± 15 | 7.08 ± 1.02 |
| Unstacked, 5-SASL | 48 ± 12 | 12.31 ± 1.56 | 52 ± 12 | 5.62 ± 0.81 |
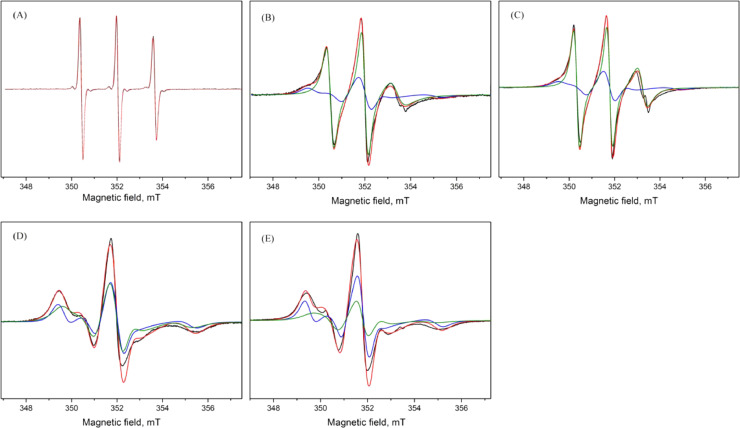
The experimental and simulated EPR spectra of 16-SASL and 5-SASL in stacked and unstacked membranes are shown in Fig. 7. To illustrate the contribution of the two components to the spectra, the simulated spectrum of each component is also presented.
Fig. 7.
Experimental and simulated EPR spectra recorded at 19°C of 16-SASL in solution (A) and in stacked (B) and unstacked (C) membranes, and of 5-SASL in stacked (D) and unstacked (E) membranes. Experimental spectra are shown in black and the simulated spectra are overlaid in red (total spectrum), blue (the immobile component) and green (mobile component).
Comparing the spectra of 16-SASL in stacked and unstacked membranes, the three lines between 350 mT and 354 mT have similar intensities, whereas an additional signal around 349 mT becomes visible in the former (Fig. 7B and 7C). As discussed below, the additional signal belongs to the annular lipid component, for which the τ is longer in the spectrum of stacked membrane than in the unstacked spectrum. The spectral lines of 5-SASL in the membrane are more broadened, ranging from 348 mT to 354 mT, than those of 16-SASL (Fig. 7D and 7E). This is due to the restricted motion close to the lipid head groups i.e. the lipid-water interface. The restricted motion of 5-SASL in bulk lipids results in overlapping of the two components of each spectrum, as the difference in τ1 and τ2 values get smaller compared to that of 16-SASL. Accordingly, the degeneracy in τ values gives rise to a larger uncertainty in the estimated contribution of each component. From the EPR spectra it can be seen that both annular and bulk lipids have shorter τ values, which means higher mobility, in the unstacked membrane.
Discussion
Supramolecular organization and excitation-energy transfer in stacked and unstacked membranes
The CD experiments confirm that cation-depleted unstacking results in the loss of long-range ordered supramolecular LHCII-PSII structures. The fluorescence results indicate the drastic increase of PSI emission as more energy is funneled either from LHCs to PSI, shuttle model or from PSII to PSI, spill-over model, which suggests a more random distribution of LHC, PSI and PSI complexes in the unstacked membrane. Interestingly, similar changes of time-resolved fluorescence kinetics were reported by Wlodarczyk et al. [51] when studying state-transition of intact cells.
Enhanced dynamics of annular and bulk lipids in unstacked membranes
The spin-label EPR experiments report that thylakoid unstacking induces a significant increase in dynamics of both the bulk and annular lipids and increases membrane fluidity at the lipid-water interface as well as in the hydrophobic interior of the membrane. The changes in the rotation correlation time with unstacking of the 16-SALS label positioned at the center of the bilayer are larger compared to that of 5-SALS that is positioned close to the lipid-water interface. Thus, unstacking has a larger effect on lipid fluidity in the hydrophobic interior of the membrane than on the mobility of the lipid headgroups. For both labels, the changes in τ2 are larger than the changes in τ1, indicating more extensive larger changes in mobility of the bulk lipids than of the annular lipids. The increased dynamics is in agreement with NMR INEPT data that report an overall increase in ns-ps motions of the mobile annular and bulk lipids.
The fractions of bulk and annular lipids in stacked membrane are estimated to be 0.60 and 0.40, respectively, for 16-SALS label, and do not appreciably change in the unstacked state. The quantity of each component is within the range of those found previously for the stacked membranes from higher plants [31,37,39,62]. The fraction of annular lipids, determined in this way, depends on the type of spin-labelled lipids and the organism. For instance, in case of the 12- MGDG spin label, it ranges from 0.25 for lettuce to 0.67 for cucumber [39]. Using a 5-SALS spin label, the reported fraction was between 0.38 and 0.74 for different stages of thylakoid development during barley greening [37]. The fractions of annular lipids in pea thylakoid using 12-MGDG and 12-PC spin label were found to be 0.36 and 0.39, respectively, [31] which are comparable with the number found in the present study (0.40). We did not find any EPR study using spin-labelled lipids on the thylakoid membrane from C. reinhardti to compare with our data. The absolute percentage of annular lipids that we report might not represent the actual fraction of annular to bulk in Cr thylakoid membrane, but the focus of our study is the comparison of stacked and unstacked membranes. We observe that the annular to lipid fraction does not change significantly in stacked and unstacked membrane, whereas there are significant changes in lipid dynamics. In their early work, Knowles et al. [40] did not observe changes in dynamics in pea thylakoids during unstacking using a PG spin label. A recent study of Petrova et al. [41] however reports an increase of lipid-phase fluidity in unstacked pea thylakoids, using lipophilic fluorescence probes, in line with our results. The discrepancy could be due to the selectivity of PG for strong interactions with proteins that might be the same for stacked and unstacked membranes.
In plant thylakoid membranes, apart from bilayer lipid phase, non-bilayer phases have been observed [17, 63]. To best of our knowledge, thylakoid non-bilayer lipid phases has not been reported for Chlamydomonas reinhardtii although the lipid composition that for Chlamydomonas also contain high amounts of the non-bilayer lipid MGDG. We did not observe any significant improvements in the simulation of our SL EPR spectra by adding an extra component to represent the third lipid group, i.e. non-bilayer, apart from bulk and annular lipids.
Worth noting is that the concentrations of the thylakoid preparations used for fluoresence, EPR and NMR measurements are quite different. However, in all three spectroscopic methods, the local dynamics is measured, which results from the interaction of neighbouring lipids and proteins within one membrane. In this case the concentration of thylakoid membrane should not affect the local dynamics as long as the sample is well hydrated, which is routinely checked by monitoring the shape of the 1H water signal when setting up the NMR experiments. In addition, we did not observe any aggregate formation in EPR and NMR samples after dilution of the suspensions.
Enhanced molecular fluctuations of protein helices in unstacked membranes
From the CP difference spectrum, we conclude that unstacking enlarges the micro- to millisecond dynamics of the protein helical segments. The large change in CP signal intensity of the difference spectrum suggests that a substantial fraction of the protein helices is affected by the unstacking. We would expect that the water-exposed protein outer-loop segments of LHCII, which mediate stacking via inter-protein interactions in the aqueous phase at the stromal side, also have enlarged dynamics in the unstacked state. It is possible however that those segments are not detected by dynamic-spectral edited NMR. Because the loop segments are very flexible [64,65], they might be too dynamic for NMR detection via CP, while the dynamics of protein carbonyl atoms falls outside the detection window of INEPT-based NMR. Restrained molecular fluctuations of individual transmembrane helices in the 102-104 Hz range (the detection window of CP-based NMR) have been observed by NMR spectroscopy for the membrane protein bacteriorhodopsin when it forms hexagonal lattice assemblies in purple membranes [59,60]. Stacked thylakoid membranes show a typical Psi-type CD signature of ordered LHCII-PSII arrays that may hinder the molecular fluctuations of individual LHC and PSII transmembrane helices. In thylakoid membranes most of the supercomplexes remain unaffected in the unstacked state and it can be inferred that the lateral order, stabilized by transversal stacking, involves motion-limited lipid-protein rather than inter-helix interactions. Consistent with this thought, we also observe enhanced dynamics of the annular lipids in the unstacked state.
Thylakoid dynamics and functional implications
The combined results demonstrate that cation-depletion-induced unstacking of C. reinhardtii thylakoid membranes leads to reorganization of protein complexes and to an increase of membrane fluidity, similar to the effects that have been reported for plant thylakoid membranes [5,42]. Depletion of Mg2+ fully releases electrostatic, hydrostructural and van der Waals forces that constrain LHCII/PSII in transversal frame [66], [67], [68]. The remaining interactions are lateral protein-protein or protein-lipid interactions. Upon full unstacking, the protein (super)complexes are free to diffuse in the membrane, leading to redistribution of LHCII over PSII and PSI and/or PSI-PSII interactions (spillover). According to the fluorescence analysis, new, functional interactions between the complexes are formed. Bulk lipid dynamics increases according to INEPT NMR and spin-label EPR results, and helix fluctuations are enlarged in the unstacked state. This interestingly suggests that release of structural constraints that involve physical contacts at the membrane exterior in the transversal plane, increases the molecular dynamics of protein and lipid in the membrane interior.
Although CD spectra indicate that there is a loss of pigment-protein super-structures with long-range order, we show that this is not accompanied by substantially enlarged fractions of annular lipids for solvating released proteins. Dynamic thylakoid stacking plays a role in balancing linear and cyclic photosynthetic electron transfer (LET and CET) [11], by regulating diffusion of mobile electron carriers via the diameter of grana and number of membrane stacks per grana. Our results suggest that membrane unstacking, apart from controlling the grana dimensions, may influence the diffusion of electron carriers such as plastoquinone by increasing the fluidity of the lipid bilayer interior. Membrane fluidity is also important for the PSII repair cycle, where damaged D1 proteins have to migrate from the grana to the stroma lamellae [6, 12]. Partial de-stacking of thylakoid membranes has been observed under high-light conditions [6,69] and was proposed to increase PSII mobility. An unexpected result from our study is the increased dynamics of the protein helices in the unstacked state. In combination with the increased lipid phase fluidity this could facilitate lateral diffusion of membrane protein complexes like PSII and facilitate reorganization of proteins in the membrane. Increased protein dynamics may also have implications for the functioning of LHC complexes that are the most abundant protein complexes in thylakoid membranes. Micro- and millisecond conformational fluctuations in individual light-harvesting complexes can induce quenching of light excitations [70] and it has been demonstrated that LHC proteins are able to control exited-state interactions via their conformational dynamics and protein-protein interactions [71]. Our results suggest that molecular motions, which may regulate the function of individual LHCs, are controlled by the membrane stacking conditions and thus by the thylakoid supramolecular architecture.
In this work, we investigated the effects of thylakoid unstacking induced by varying a sole parameter, namely the Mg 2+ ion concentration. Nevertheless, the fluorescence lifetimes of stacked versus unstacked C. reinhardtii thylakoid membranes measured here very much resemble those of C. reinhardtii cells in state I versus state II conditions [51], which suggests that similar functioning membrane states can be induced in different ways.
Conclusion
We demonstrate that the combined nuclear magnetic resonance and electron paramagnetic resonance approach is a unique non-invasive way for the simultaneous assessment of lipid and protein dynamics at physiological temperature and in a complex native membrane. Based on the results, we conclude that the unstacking of thylakoid membrane significantly increases membrane fluidity close to the lipid headgroup and, surprisingly, in the hydrophobic interior of the membrane and strongly enhances the dynamics of membrane bulk and annular lipids. Moreover, we show that thylakoid unstacking enhances protein helix conformational fluctuations, which may regulate the functions of individual LHCs. Our results may serve as a steppingstone toward a better understanding of the structural dynamic consequences of thylakoid organization, which involve vertical grana unstacking.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
FN was financially supported by a Fundamental Research on Matter (FOM) Projectruimte grant of the Netherlands Organization of Scientific Research (NWO) under grant nr. 680.91.15.19. AP and LT were financially supported by a CW-VIDI grant of the Netherlands Organization of Scientific Research (NWO) under grant nr. 723.012.103.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2021.100015.
Appendix. Supplementary materials
References
- 1.Kirchhoff H. Molecular crowding and order in photosynthetic membranes. Trends Plant Sci. 2008;13:201–207. doi: 10.1016/j.tplants.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Garab G. Hierarchical organization and structural flexibility of thylakoid membranes. Biochim. Biophys. Acta - Bioenerg. 2014;1837:481–494. doi: 10.1016/j.bbabio.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Lambrev P.H., Akhtar P. Macroorganisation and flexibility of thylakoid membranes. Biochem. J. 2019;476:2981–3018. doi: 10.1042/BCJ20190080. [DOI] [PubMed] [Google Scholar]
- 4.Johnson M.P., Wientjes E. The relevance of dynamic thylakoid organisation to photosynthetic regulation. Biochim. Biophys. Acta - Bioenerg. 2020;1861 doi: 10.1016/j.bbabio.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Fristedt R., Granath P., Vener A.V. A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwai M., Takahashi Y., Minagawa J. Molecular Remodeling of Photosystem II during State Transitions in Chlamydomonas reinhardtii. Plant Cell Online. 2008;20:2177–2189. doi: 10.1105/tpc.108.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Herbstova, S. Tietz, C. Kinzel, M. V. Turkina, H. Kirchhoff, Architectural switch in plant photosynthetic membranes induced by light stress, Proc. Natl. Acad. Sci. 109 (2012) 20130–20135. https://doi.org/10.1073/pnas.1214265109. [DOI] [PMC free article] [PubMed]
- 8.Ünlü C., Drop B., Croce R., van Amerongen H. State transitions in Chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc. Natl. Acad. Sci. 2014;111:3460–3465. doi: 10.1073/pnas.1319164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nawrocki W.J., Santabarbara S., Mosebach L., Wollman F.A., Rappaport F. State transitions redistribute rather than dissipate energy between the two photosystems in Chlamydomonas. Nat. Plants. 2016;2:1–7. doi: 10.1038/NPLANTS.2016.31. [DOI] [PubMed] [Google Scholar]
- 10.Johnson M.P., Goral T.K., Duffy C.D.P., Brain A.P.R., Mullineaux C.W., Ruban A.V. Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell. 2011;23:1468–1479. doi: 10.1105/tpc.110.081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood W.H.J., Macgregor-chatwin C., Barnett S.F.H., Mayneord G.E., Huang X., Hobbs J.K., Hunter C.N., Johnson M.P. Dynamic thylakoid stacking regulates the balance between linear and cyclic photosynthetic electron transfer. Nat. Plants. 2018:4. doi: 10.1038/s41477-017-0092-7. [DOI] [PubMed] [Google Scholar]
- 12.Yoshioka-Nishimura M. Close Relationships between the PSII Repair Cycle and Thylakoid Membrane Dynamics. Plant Cell Physiol. 2016;57:1115–1122. doi: 10.1093/pcp/pcw050. [DOI] [PubMed] [Google Scholar]
- 13.Hsia C.-Y., Richards M.J., Daniel S. A review of traditional and emerging methods to characterize lipid–protein interactions in biological membranes. Anal. Methods. 2015;7:7076–7094. doi: 10.1039/C5AY00599J. [DOI] [Google Scholar]
- 14.Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. Lipids in photosystem II: Interactions with protein and cofactors. Biochim. Biophys. Acta - Bioenerg. 2007;1767:509–519. doi: 10.1016/j.bbabio.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Francesc-Xabier Contreras B.B., Ernst Andreas Max, Wieland Felix. Vol. 3. 2011. Heidelberg, Specificity of Intramembrane Protein– Lipid Interactions, Cold Spring Harb. (Perspect. Biol.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizusawa N., Wada H. The role of lipids in photosystem II. Biochim. Biophys. Acta - Bioenerg. 2012;1817:194–208. doi: 10.1016/j.bbabio.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Páli T., Garab G., Horváth L.I., Kóta Z. Functional significance of the lipid-protein interface in photosynthetic membranes. Cell. Mol. Life Sci. 2003;60:1591–1606. doi: 10.1007/s00018-003-3173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Boudière, M. Michaud, D. Petroutsos, F. Rébeillé, D. Falconet, O. Bastien, S. Roy, G. Finazzi, N. Rolland, J. Jouhet, M.A. Block, E. Maréchal, Glycerolipids in photosynthesis : Composition, synthesis and trafficking, 1837 (2014) 470–480. https://doi.org/10.1016/j.bbabio.2013.09.007. [DOI] [PubMed]
- 19.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta - Biomembr. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 20.X. Sheng, X. Liu, P. Cao, M. Li, Z. Liu, Structural roles of lipid molecules in the assembly of plant PSII−LHCII supercomplex, Biophys. Reports. 4 (2018) 189–203. https://doi.org/10.1007/s41048-018-0068-9. [DOI] [PMC free article] [PubMed]
- 21.Azadi Chegeni F., Perin G., Sai Sankar Gupta K.B., Simionato D., Morosinotto T., Pandit A. Protein and lipid dynamics in photosynthetic thylakoid membranes investigated by in-situ solid-state NMR. Biochim. Biophys. Acta - Bioenerg. 2016;1857:1849–1859. doi: 10.1016/j.bbabio.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Xiwei Zheng D.S.H., Bi Cong, Brooks Marissa. Cardiolipin interaction with subunit c of ATP synthase: Solidstate NMR characterization. Anal. Chem. 2015;25:368–379. doi: 10.1016/j.cogdev.2010.08.003.Personal. [DOI] [Google Scholar]
- 23.Huster D. Solid-state NMR spectroscopy to study protein-lipid interactions. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 2014;1841:1146–1160. doi: 10.1016/j.bbalip.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 24.H. Mullineaux, Conrad W and Kirchhoff, Role of Lipids in the Dynamics of Thylakoid Membranes Conrad, Lipids Photosynth. Essent. Regul. Funct. (2009) 283–294. https://doi.org/10.1007/978-90-481-2863-1.
- 25.Kirchhoff C.W.M. Using Fluorescence Recovery After Photobleaching to Measure Lipid Diffusion in Membranes. Methods Mol. Biol. 2007;400:267–275. doi: 10.1007/978-1-59745-519-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto Y., Hori H., Kai S., Ishikawa T., Ohnishi A., Tsumura N., Morita N. Quality control of Photosystem II: reversible and irreversible protein aggregation decides the fate of Photosystem II under excessive illumination. Front. Plant Sci. 2013;4:1–9. doi: 10.3389/fpls.2013.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh D. Electron spin resonance in membrane research: Protein-lipid interactions from challenging beginnings to state of the art. Eur. Biophys. J. 2010;39:513–525. doi: 10.1007/s00249-009-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.rg H. Kleinschmidt J. Methods Mol. Biol. 2013. Lipid-Protein Interactions: Methods and Protocols. [DOI] [Google Scholar]
- 29.East J.M., Melville D., Lee A.G. Exchange Rates and Numbers of Annular Lipids for the Calcium and Magnesium Ion Dependent Adenosinetriphosphatase. Biochemistry. 1985;24:2615–2623. doi: 10.1021/bi00332a005. [DOI] [PubMed] [Google Scholar]
- 30.Marsh L.I. Derek and Horváth, Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim. Biophys. Acta - Rev. Biomembr. 1998;1376:267–296. doi: 10.1016/S0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 31.G. Li, P.F. Knowles, D.J. Murphy, I. Nishida, D. Marsh, Spin-Label ESR Studies of Lipid-Protein Interactions in Thylakoid Membranes, Biochemistry. 28 (1989) 7446–7452. https://doi.org/10.1021/bi00444a044. [DOI] [PubMed]
- 32.Ivancich A., Horváth L.I., Droppa M., Horváth G., Farkas T. Spin label EPR study of lipid solvation of supramolecular photosynthetic protein complexes in thylakoids. BBA - Biomembr. 1994;1196:51–56. doi: 10.1016/0005-2736(94)90294-1. [DOI] [PubMed] [Google Scholar]
- 33.Li G., Horváth L.I., Knowles P.F., Murphy D.J., Marsh D. Spin label saturation transfer ESR studies of protein-lipid interactions in Photosystem II-enriched membranes. BBA - Biomembr. 1989;987:187–192. doi: 10.1016/0005-2736(89)90543-9. [DOI] [Google Scholar]
- 34.Ligeza A., Tikhonov A.N., Hyde J.S., Subczynski W.K. Oxygen permeability of thylakoid membranes: electron paramagnetic resonance spin labeling study. Biochim. Biophys. Acta - Bioenerg. 1998;1365:453–463. doi: 10.1016/S0005-2728(98)00098-X. [DOI] [PubMed] [Google Scholar]
- 35.Vishnyakova E.A., Ruuge A.E., Golovina E.A., Hoekstra F.A., Tikhonov A.N. Spin-labeling study of membranes in wheat embryo axes. 1. Partitioning of doxyl stearates into the lipid domains. Biochim. Biophys. Acta - Biomembr. 2000;1467:380–394. doi: 10.1016/S0005-2736(00)00234-0. [DOI] [PubMed] [Google Scholar]
- 36.Calucci L., Navari-Izzo F., Pinzino C., Sgherri C.L.M. Fluidity changes in thylakoid membranes of durum wheat induced by oxidative stress: A spin probe EPR study. J. Phys. Chem. B. 2001;105:3127–3134. doi: 10.1021/jp002963q. [DOI] [Google Scholar]
- 37.Z. Kota, L.I. Horvath, M. Droppa, G. Horvath, T. Farkas, T. Pali, Protein assembly and heat stability in developing thylakoid membranes during greening, Proc. Natl. Acad. Sci. 99 (2002) 12149–12154. https://doi.org/10.1073/pnas.192463899. [DOI] [PMC free article] [PubMed]
- 38.Jajić I., Wiśniewska-Becker A., Sarna T., Jemioła-Rzemińska M., Strzałka K. EPR spin labeling measurements of thylakoid membrane fluidity during barley leaf senescence. J. Plant Physiol. 2014;171:1046–1053. doi: 10.1016/j.jplph.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Li G., Knowles P.F., Murphy D.J., Marsh D. Lipid-Protein Interactions in Thylakoid Membranes of Chilling-Resistant and Chilling-Sensitive Plants Studied by Spin Label Electron-Spin-Resonance Spectroscopy. J. Biol. Chem. 1990;265:16867–16872. [PubMed] [Google Scholar]
- 40.Li G., Knowles P.F., Murphy D.J., Marsh D. Lipid-protein interactions in stacked and destacked thylakoid membranes and the influence of phosphorylation and illumination. Spin label ESR studies. BBA - Biomembr. 1990;1024:278–284. doi: 10.1016/0005-2736(90)90355-R. [DOI] [PubMed] [Google Scholar]
- 41.Petrova N., Todinova S., Paunov M., Kovács L., Taneva S., Krumova S. Thylakoid membrane unstacking increases LHCII thermal stability and lipid phase fluidity. J. Bioenerg. Biomembr. 2018;50:425–435. doi: 10.1007/s10863-018-9783-7. [DOI] [PubMed] [Google Scholar]
- 42.Van Der Weij-De Wit C.D., Ihalainen J.A., Van Grondelle R., Dekker J.P. Excitation energy transfer in native and unstacked thylakoid membranes studied by low temperature and ultrafast fluorescence spectroscopy. Photosynth. Res. 2007;93:173–182. doi: 10.1007/s11120-007-9157-1. [DOI] [PubMed] [Google Scholar]
- 43.Chua N., Bennoun P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl. Acad. Sci. 1975;72:2175–2179. doi: 10.1073/pnas.72.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Stokkum I.H.M., Larsen D.S., Van Grondelle R. Global and target analysis of time-resolved spectra. Biochim. Biophys. Acta - Bioenerg. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Stoll S., Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Tóth T.N., Rai N., Solymosi K., Zsiros O., Schröder W.P., Garab G., Van Amerongen H., Horton P., Kovács L. Fingerprinting the macro-organisation of pigment-protein complexes in plant thylakoid membranes in vivo by circular-dichroism spectroscopy. Biochim. Biophys. Acta - Bioenerg. 2016;1857:1479–1489. doi: 10.1016/j.bbabio.2016.04.287. [DOI] [PubMed] [Google Scholar]
- 47.Tóth T.N., Rai N., Solymosi K., Zsiros O., Schröder W.P., Garab G., Van Amerongen H., Horton P., Kovács L. Fingerprinting the macro-organisation of pigment-protein complexes in plant thylakoid membranes in vivo by circular-dichroism spectroscopy. Biochim. Biophys. Acta - Bioenerg. 2016;1857:1479–1489. doi: 10.1016/j.bbabio.2016.04.287. [DOI] [PubMed] [Google Scholar]
- 48.Karlický V., Kurasová I., Ptáčková B., Večeřová K., Urban O., Špunda V. Enhanced thermal stability of the thylakoid membranes from spruce. A comparison with selected angiosperms. Photosynth. Res. 2016;130:357–371. doi: 10.1007/s11120-016-0269-3. [DOI] [PubMed] [Google Scholar]
- 49.Tian L., Nawrocki W.J., Liu X., Polukhina I., van Stokkum I.H.M., Croce R. pH dependence, kinetics and light-harvesting regulation of nonphotochemical quenching in Chlamydomonas. Proc. Natl. Acad. Sci. 2019;116:8320–8325. doi: 10.1073/pnas.1817796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasil'ev S., Irrgang K.D., Schrötter T., Bergmann A., Eichler H.J., Renger G. Quenching of chlorophyll a fluorescence in the aggregates of LHCII: Steady state fluorescence and picosecond relaxation kinetics. Biochemistry. 1997;36:7503–7512. doi: 10.1021/bi9625253. [DOI] [PubMed] [Google Scholar]
- 51.Wlodarczyk L.M., Dinc E., Croce R., Dekker J.P. Excitation energy transfer in Chlamydomonas reinhardtii de fi cient in the PSI core or the PSII core under conditions mimicking state transitions. BBA - Bioenerg. 2016;1857:625–633. doi: 10.1016/j.bbabio.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, M. Iwai, Y. Takahashi, J. Minagawa, Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii, Proc. Natl. Acad. Sci. 103 (2006) 477–482. https://doi.org/10.1073/pnas.0509952103. [DOI] [PMC free article] [PubMed]
- 53.Barber J. An explanation for the relationship between salt-induced thylakoid stacking and the chlorophyll fluorescence changes associated with changes in spillover of energy from photosystem II to photosystem I. FEBS Lett. 1980;118:1–10. doi: 10.1016/0014-5793(80)81207-5. [DOI] [Google Scholar]
- 54.Azadi-Chegeni F., Schiphorst C., Pandit A. In vivo NMR as a tool for probing molecular structure and dynamics in intact Chlamydomonas reinhardtii cells. Photosynth. Res. 2017;135:227–237. doi: 10.1007/s11120-017-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnold A.A., Genard B., Zito F., Tremblay R., Warschawski D.E., Marcotte I. Identification of lipid and saccharide constituents of whole microalgal cells by 13C solid-state NMR. Biochim. Biophys. Acta - Biomembr. 2015;1848:369–377. doi: 10.1016/j.bbamem.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 56.Beal C.M., Webber M.E., Ruoff R.S., Hebner R.E. Lipid analysis of Neochloris oleoabundans by liquid state NMR. Biotechnol. Bioeng. 2010;106:573–583. doi: 10.1002/bit.22701. [DOI] [PubMed] [Google Scholar]
- 57.Coddington J.M., Johns S.R., Leslie D.R., Willing R.I., Bishop D.G. 13C Nuclear magnetic resonance studies of the composition and fluidity of several chloroplast monogalactosyldiacylglycerols. Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 1981;663:653–660. doi: 10.1016/0005-2760(81)90076-X. [DOI] [PubMed] [Google Scholar]
- 58.Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C.F., Tolmie D.E., Wenger R.Kent, Yao H., Markley J.L. BioMagResBank. Nucleic. Acids. Res. 2008;36:402–408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krebs M.P., Isenbarger T.A. Structural determinants of purple membrane assembly. Biochim. Biophys. Acta - Bioenerg. 2000;1460:15–26. doi: 10.1016/S0005-2728(00)00126-2. [DOI] [PubMed] [Google Scholar]
- 60.Tuzi S., Naito A., Saitô H. Local protein structure and dynamics at kinked transmembrane α-helices of [1-13C] Pro-labeled bacteriorhodopsin as revealed by site-directed solid-state 13C NMR. J. Mol. Struct. 2003;654:205–214. doi: 10.1016/S0022-2860(03)00250-3. [DOI] [Google Scholar]
- 61.Marsh D. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. 2001;98:7777–7782. doi: 10.1073/pnas.131023798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuki Nakamura Y. Springer International Publishing Switzerland; 2016. Lipids in Plant and Algae Development. [DOI] [Google Scholar]
- 63.Garab G., Ughy B., de Waard P., Akhtar P., Javornik U., Kotakis C., Šket P., Karlický V., Materová Z., Špunda V., Plavec J., van Amerongen H., Vígh L., Van As H., Lambrev P.H. Lipid polymorphism in chloroplast thylakoid membranes – as revealed by 31P-NMR and time-resolved merocyanine fluorescence spectroscopy. Sci. Rep. 2017;7:13343. doi: 10.1038/s41598-017-13574-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sunku K., De Groot H.J.M., Pandit A. Insights into the photoprotective switch of the major light-harvesting complex II (LHCII): A preserved core of arginine-glutamate interlocked helices complemented by adjustable loops. J. Biol. Chem. 2013;288:19796–19804. doi: 10.1074/jbc.M113.456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dockter C., Muller A.H., Dietz C., Volkov A., Polyhach Y., Jeschke G., Paulsen H. Rigid core and flexible terminus: Structure of solubilized light-harvesting chlorophyll a/b complex (LHCII) measured by EPR. J. Biol. Chem. 2012;287:2915–2925. doi: 10.1074/jbc.M111.307728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puthiyaveetil S., Van Oort B., Kirchhoff H. Surface charge dynamics in photosynthetic membranes and the structural consequences. Nat. Plants. 2017;3:1–9. doi: 10.1038/nplants.2017.20. [DOI] [PubMed] [Google Scholar]
- 67.Albanese P., Tamara S., Saracco G., Scheltema R.A., Pagliano C. How paired PSII–LHCII supercomplexes mediate the stacking of plant thylakoid membranes unveiled by structural mass-spectrometry. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-020-15184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zsiros O., Ünnep R., Nagy G., Almásy L., Patai R., Székely N.K., Kohlbrecher J., Garab G., Dér A., Kovács L. Role of Protein-Water Interface in the Stacking Interactions of Granum Thylakoid Membranes—As Revealed by the Effects of Hofmeister Salts. Front. Plant Sci. 2020;11:1–14. doi: 10.3389/fpls.2020.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khatoon M., Inagawa K., Pospišil P., Yamashita A., Yoshioka M., Lundin B., Horie J., Morita N., Jajoo A., Yamamoto Y., Yamamoto Y. Quality control of photosystem II: Thylakoid unstacking necessary to avoid further damage to the D1 protein and to facilitate D1 degradation under light stress in spinach thylakoids. J. Biol. Chem. 2009;284:25343–25352. doi: 10.1074/jbc.M109.007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kondo T., Gordon J.B., Pinnola A., Dall'Osto L., Bassi R., Schlau-Cohen G.S. Microsecond and millisecond dynamics in the photosynthetic protein LHCSR1 observed by single-molecule correlation spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2019;166:11247–11252. doi: 10.1073/pnas.1821207116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Natali A., Gruber J.M., Dietzel L., Stuart M.C.A., Van Grondelle R., Croce R. Light-harvesting Complexes (LHCs) cluster spontaneously in membrane environment leading to shortening of their excited state lifetimes. J. Biol. Chem. 2016;291:16730–16739. doi: 10.1074/jbc.M116.730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.