Highlights
-
•
MicroRNAs that target human ELOVL6 were predicted using the targetscan database.
-
•
Five miRNAs were identified as the regulators of ELOVL6 by binding to the 3′-UTR.
-
•
miR-135b-5p and miR-135a-5p significantly reduced ELOVL6 activity in GBM cells.
-
•
miR-135a-5p and miR-135b-5p significantly suppressed tumorigenesis of GBM cells.
-
•
These inhibitory effects were reversed by overexpression of ELOVL6 in GBM cells.
Keywords: Cell proliferation, Fatty acid, Glioblastoma, MicroRNA, Migration
Abstract
ELOVL fatty acid elongase 6 (ELOVL6) controls cellular fatty acid (FA) composition by catalyzing the elongation of palmitate (C16:0) to stearate (C18:0) and palmitoleate (C16:1n-7) to vaccinate (C18:1n-7). Although the transcriptional regulation of ELOVL6 has been well studied, the post-transcriptional regulation of ELOVL6 is not fully understood. Therefore, this study aims to evaluate the role of microRNAs (miRNAs) in regulating human ELOVL6. Bioinformatic analysis identified five putative miRNAs: miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p, which potentially bind ELOVL6 3′-untranslated region (UTR). Results from dual-luciferase assays revealed that these miRNAs downregulate ELOVL6 by directly interacting with the 3′-UTR of ELOVL6 mRNA. Moreover, miR-135b-5p and miR-135a-5p suppress cell proliferation and migration in glioblastoma multiforme cells by inhibiting ELOVL6 at the mRNA and protein levels. Taken together, our results provide novel regulatory mechanisms for ELOVL6 at the post-transcriptional level and identify potential candidates for the treatment of patients with glioblastoma multiforme.
1. Introduction
The biosynthesis of fatty acids (FAs) is an important aspect of lipogenesis because FAs are energy stores and components of the membrane structure [1]. Recent studies have indicated that both the quantity and the quality of FAs are important for understanding and preventing metabolic diseases. FAs are categorized according to their chain lengths and the number and position of double bonds [2]. Proper elongation and desaturation play critical roles in regulating the length and degree of unsaturation of FAs; therefore their function and metabolic fate contribute to maintaining lipid homeostasis [2]. FAs are taken through the diet or synthesized de novo by the fatty acid synthase complex and elongated into very long-chain FAs by elongases [2,3]. ELOVL fatty acid elongase 6 (ELOVL6) is a member of mammalian FA elongases and is responsible for the elongation of palmitate (C16:0) to stearate (C18:0) and palmitoleate (C16:1n-7) to vaccinate (C18:1n-7) [4,5]. Because saturated FAs and monounsaturated FAs, with 16–18 carbon atoms, form the bulk of total FAs in cells, ELOVL6 is recognized as a crucial gene in determining overall cellular FA balance. We have shown previously that the disruption of Elovl6 prevents the development of obesity-induced insulin resistance, type 2 diabetes, nonalcoholic steatohepatitis, and atherosclerosis [6], [7], [8], [9], [10]. ELOVL6 also plays important role in hepatocellular carcinoma [11], [12], [13], acute myeloid leukemia [14], lung squamous cell carcinoma [15], and glioblastoma multiforme (GBM) [16,17]. Therefore, the inhibition of ELOVL6 could be a new therapeutic approach for the treatment of metabolic disorders and cancers.
Sterol regulatory element-binding proteins (SREBPs) are transcription factors that regulate the expression of enzymes required for cholesterol, triglyceride, and FA synthesis [18]. Transcriptome analysis of the livers of SREBP-1a transgenic mice identified ELOVL6 as a target of SREBP-1 [19]. The ELOVL6 promoter is directly and primarily activated by nuclear SREBPs. There are two sterol response elements (SREs), namely, proximal SRE-1 and distal SRE-2, and one E-box. Moreover, ELOVL6 expression is under direct transcriptional control of carbohydrate-response element-binding protein (ChREBP). The synergistic effect of SREBP-1c and ChREBP is essential for the maximal induction of Elovl6 expression in the liver [20]. Over the past several years, significant progress has been made in our understanding of the transcriptional regulation of ELOVL6. However, much less is known about the post-transcriptional regulation of ELOVL6 by conserved regulators, such as microRNAs (miRNAs). miRNAs are endogenous small non-coding RNAs that are 21−25 nucleotide in length and direct post-transcriptional regulation of mRNA targets in eukaryotic lineages [21,22]. miRNAs are characterized by short hairpin precursor miRNAs (pre-miRNAs) that mediate gene regulation by forming miRNA-induced silencing complex (miRISC). miRNAs bind to a specific 3′-untranslated region (UTR) of the target mRNA and then downregulate its expression by destabilizing the mRNA and inhibiting translation [23,24]. miRNAs can bind to hundreds of mRNAs, resulting in the regulation of more than one-half of the protein-coding genes in humans [25]. miRNA dysregulation is linked to various human diseases [26]. Bioinformatics-based assessment of complementarity between a miRNA and the seed in the target gene and the experimental evaluation of individual miRNAs are crucial in the discovery and validation of physiologically significant targets for miRNAs. Because ELOVL6 is a key gene involved in various physiologies and pathologies by controlling cellular FA composition, identifying miRNAs that target human ELOVL6 is important for the manipulation of this enzyme. In this study, we identified five miRNAs that are post-transcriptional regulators of human ELOVL6. Interestingly, we observed an interaction between these miRNAs and ELOVL6 in suppressing the growth and migration of GBM cells.
2. Materials and methods
2.1. Cell lines and cultures
Human embryonic kidney 293 (HEK-293), U87-MG, and U118-MG cells were obtained from the American Type Culture Collection. HEK-293 and U118-MG cells were cultured in Dulbecco's modified Eagle's medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. U87-MG cells were cultured in Eagle's minimum essential medium (Fujifilm Wako, Osaka, Japan), supplemented with 10% FBS and 1% penicillin–streptomycin. Cells were incubated at 37 °C in 5% CO2 in a humidified chamber.
2.2. Plasmid construction and transfection
Candidate miRNA precursors were synthesized by GENEWIZ (Tokyo, Japan). The miRNA precursors were cloned into the pmR-ZsGreen1 vector (TaKaRa Bio, Shiga, Japan). To generate the high specificity dual-luciferase reporter vector (pmirGLO-ELOVL6 3′-UTR-WT), human ELOVL6 3′-UTR was divided into three constitutive regions and cloned into the pmir-GLO reporter vector (Promega, Tokyo, Japan). pmirGLO-ELOVL6–3′-UTR-Mut was generated using site-directed mutagenesis. To evaluate ELOVL6 levels, cDNA encoding C-terminal HA-tagged ELOVL6 and 3′-UTR were cloned into pcDNA 3.1(+). To overexpress ELOVL6, a cDNA encoding C-terminal FLAG-tagged ELOVL6 was cloned into pcDNA3.1(+). Cloning results were confirmed by DNA sequencing. Cells were cultured for 24 h and transfected using the X-tremeGENE 9 DNA Transfection Reagent (Merck KGaA, Darmstadt, Germany) or Lipofectamine 2000 (Life Technologies, NY, USA), according to the manufacturer's instructions.
2.3. miRNA mimic
Human mirVANA miRNA mimics hsa-miR-135b-5p (assay ID: MC13044) and hsa-miR-135a-5p (assay ID: MC11126) and the negative control (4,464,058) were obtained from Life Technologies. Cells were transfected with the miRNA mimic using the Lipofectamine RNAiMAX transfection reagent (Life Technologies, NY, USA), according to the manufacturer's instructions.
2.4. Quantitative RT-PCR analysis for miRNA and mRNA expression
Total RNA was isolated using Sepasol-RNA I Super G (Nacalai Tesque, Kyoto, Japan). The expression level of each miRNA was evaluated using the Mir-X miRNA First-Strand Synthesis and SYBR Advantage qPCR Premix (TaKaRa Bio, Shiga, Japan), according to the manufacturer's instructions. U6 snRNA expression level was used for normalization. mRNA expression levels were measured relative to the experimental control with the ΔΔCT method using TB Green Premix Ex Taq II Tli RNaseH Plus (TaKaRa Bio, Shiga, Japan), according to the manufacturer's instructions. mRNA levels were normalized to that of cyclophilin mRNA. The products were analyzed using Applied Biosystems 7300 Real-Time PCR System. Gene-specific primers are listed in Supplementary Table 1.
2.5. Dual-luciferase assay
Cell lysates were collected 48 h after transfection. Cells were harvested to evaluate the firefly and Renilla luminescence activities assay using the Dual-Glo Luciferase assay system (Promega, Tokyo, Japan), according to the manufacturer's instructions. Reporter activity was measured using the multi-mode reader SYNERGY-HTX (Agilent Technologies, CA, USA). Renilla luciferase was used as an internal control.
2.6. Western blotting
Western blotting was performed as described [7]. Aliquots of whole cell lysates (50 μg) were loaded onto SDS-PAGE gels, separated, and transferred to PDVF membranes. Membranes were incubated with primary antibodies against FLAG-tag (Sigma-Aldrich, Cat# F3165, RRID: AB_259,529), HA-tag (Cell Signaling Technology, Cat# 3724, RRID: AB_1,549,585), or GAPDH (Santa Cruz Biotechnology, Cat# sc-32,233, RRID: AB_627,679) overnight at 4 °C. Then, membranes were incubated with the anti-rabbit (Cell Signaling Technology, Cat# 7074, RRID: AB_2,099,233) or anti-mouse (Cell Signaling Technology, Cat# 7076, RRID: AB_330,924) IgG HRP-linked antibody for 1 h at 25 °C. Signal intensity was detected using the Clarity Western ECL Substrate (Bio-Rad Laboratories, Tokyo, Japan) and the Molecular Imager ChemiDoc XRS+ system (Bio-Rad Laboratories, Tokyo, Japan).
2.7. Elovl6 enzyme activity assay
Elovl6 enzyme activity was determined in the microsomes by evaluating the incorporation of [2–14C]malonyl-CoA into exogenous palmitoyl-CoA, as described [5] [7]. Briefly, 48 h after transfection, U87-MG and U118-MG cells were washed with PBS and scraped in ice-cold 0.25 M sucrose and 0.02 M HEPES (pH = 7.5). The cells were washed and resuspended in ice-cold sucrose/HEPES and homogenized. The homogenate was centrifuged at 600 × g for 10 min at 4 °C. The pellet was resuspended in sucrose–HEPES and dounce-homogenized. The suspension was centrifuged at 600 × g for 10 min at 4 °C. The supernatants were combined and centrifuged at 100,000 × g for 60 min at 4 °C. The resultant pellets were resuspended in 100 μL of 0.1 M Tris–HCl (pH = 7.4) and used for the C16:0-CoA elongation assay.
2.8. Fatty acid composition analysis
The fatty acid composition was measured as described previously [27] [7,28] with some modifications. U87-MG and U118-MG cells were transfected with the miRNA mimic using the Lipofectamine RNAiMAX transfection reagent. Forty-eight hours after the transfection, cells were washed with PBS and scraped with 300 μl of methanol containing 1% acetic acid and sonicated on ice, followed by the addition of 2 ml of a 1:2:2 mixture of chloroform:methanol:ethanol and 100 μl of a 2:1:1:1 mixture of chloroform:methanol:ethanol:acetic acid. The mixture was vortexed at room temperature and centrifuged for 10 min at 4 °C; the organic layer was collected and evaporated under reduced pressure. For FA analysis, extracted lipids were methylated with 2.5% H2SO4 in methanol. The FA methyl esters were then extracted with n-hexane and quantified by gas chromatography-mass spectrometry (GC/MS) using a Clarus SQ 8 GC/MS (PerkinElmer Japan, Kanagawa, Japan).
2.9. Cell proliferation assay
Cell proliferation was detected using the Click-iT EdU Imaging Kits (Life Technologies, NY, USA). U87-MG and U118-MG cells were treated with EdU-labeling reagent (final concentration: 5 μM) for 6 h. Then, cells were immediately fixed in 4% paraformaldehyde and processed according to the manufacturer's instructions. Nuclei were counterstained with DAPI. The percentage of EdU-positive cells was counted under a fluorescence microscope.
2.10. Migration assay
Cell migration was performed using Ibidi culture inserts (Nippon Genetics, Tokyo, Japan). The insert consists of two chambers that give a defined 500-µm cell-free gap in the 35-mm tissue culture dishes. The migration of U87-MG and U118-MG cells was evaluated under a phase-contrast microscope and the percentage of wound closure was quantified at 0, 12, and 24 h.
2.11. Cell death assay
Cell death was detected using propidium iodide (PI) and Hoechst 33,342 (Life Technologies, NY, USA) staining. U87-MG and U118-MG cells were stained with PI (1 mg/mL) and counterstained with Hoechst 33,342 (5 μg/mL). The samples were maintained in the dark for 15 min. The percentage of PI-positive cells was counted under a fluorescence microscope.
2.12. Statistical analyses
Values are expressed as means ± SEM. One or two-way ANOVA was used for multiple group comparisons. All analyses were performed using GraphPad Prism version 9 (GraphPad Software Inc., CA, USA), and P <0.05 was considered statistically significant.
3. Results
3.1. Potential miRNAs targeting the 3′-UTR of human ELOVL6
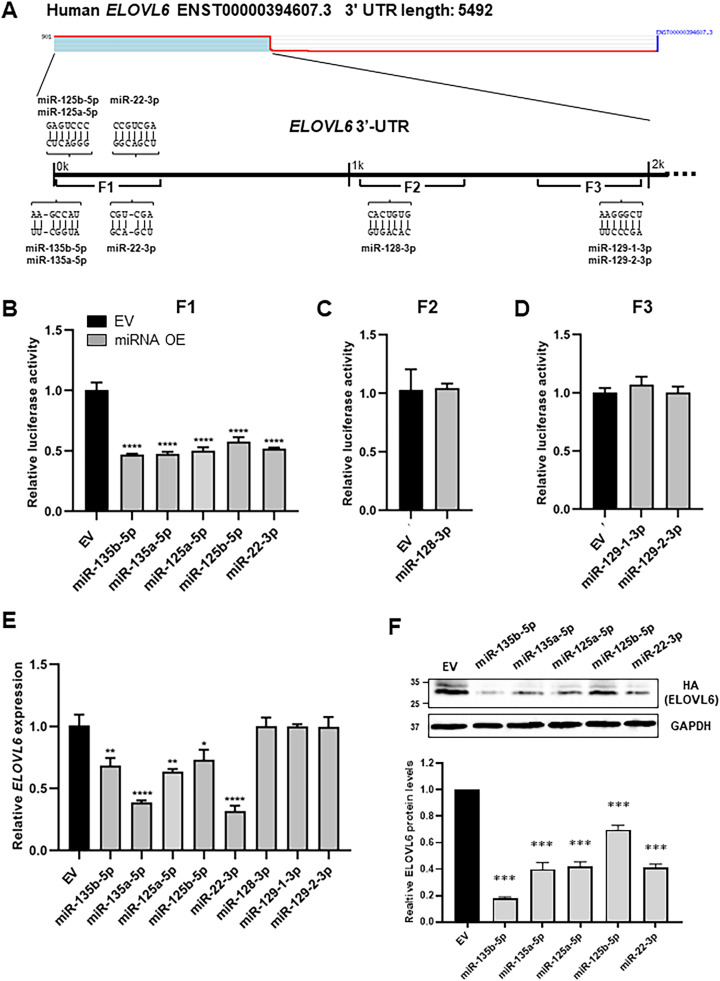
Humans have two lengths of ELOVL6 mRNA, approximately 6.5 kb and 2.7 kb in size, with different lengths of the 3′-UTR [5]. The putative miRNAs that target the common region of the long and short 3′-UTR of ELOVL6 mRNA were predicted using the TargetScanHuman database. Cumulative weighted context++ score (CWCS) ranks based on predicted repression and PCT score of the longest 3′-UTR isoform based upon the confidence that the target site is evolutionarily conserved [29]. Eight microRNAs: miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, miR-22–3p, miR-128–3p, miR-129–1–3p, and miR 129–2–3p were selected to target the 3′-UTR of ELOVL6 mRNA (Table 1). The seed match alignment of candidate miRNAs indicates that they are conserved from rodents to primates. These data suggest that these miRNAs are promising candidates for ELOVL6 regulation.
Table 1.
miRNA candidates targeting the 3′-UTR ofELOVL6predicted using TargetScanHuman.
| miRNA | Position in the UTR | Seed match | Context++ score | Context++ score percentile | Weighted context++ score | Conserved branch length | Pct | Mature miRNA sequence | Evolutionary conserved |
|---|---|---|---|---|---|---|---|---|---|
| miR-135b-5p | 18–25 | 8mer | −0.39 | 98 | −0.39 | 4.132 | 0.82 | tatggcttttcattcctatgtga | H, C, R, S, Rb |
| miR-135a-5p | 18–25 | 8mer | −0.39 | 98 | −0.39 | 4.132 | 0.82 | tatggctttttattcctatgtga | H, C, R, S, Rb |
| miR-125a-5p | 27–33 | 7mer-m8 | −0.38 | 97 | −0.38 | 8.549 | 0.91 | tccctgagaccctttaacctgtga | H, C, R, S, M, Rt, Rb |
| miR-125b-5p | 27–33 | 7mer-m8 | −0.38 | 97 | −0.38 | 8.549 | 0.91 | tccctgagaccctaacttgtga | H, C, R, S, M, Rt, Rb |
| miR-22–3p | 87–93, 129–135 | 7 mer-A1, 7mer-m8 |
−0.4 | 98 | −0.4 | 6.851 | 0.82 | aagctgccagttgaagaactgt | H, C, R, S, M, Rt, Rb |
| miR-128–3p | 1143–1149 | 7mer-m8 | −0.21 | 94 | −0.21 | 7.767 | 0.87 | tcacagtgaaccggtctcttt | H, C, R, S, M, Rt, Rb |
| miR-129–1–3p | 1887–1893 | 7mer-m8 | −0.29 | 96 | −0.29 | 6.7 | 0.8 | aagcccttaccccaaaaagtat | H, C, R, S, Rb |
| miR-129–2–3p | 1887–1893 | 7mer-m8 | −0.29 | 96 | −0.29 | 6.7 | 0.8 | aagcccttaccccaaaaagcat | H, C, R, S, Rb |
H: human; C: chimpanzee; R: rhesus monkey; S: squirrel; M: mouse; Rt: rat; Rb: rabbit.
3.2. miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p downregulate mRNA expression and protein abundance of ELOVL6 by directly targeting its 3′-UTR
To verify the interaction of the eight miRNAs with the 3′-UTR of ELOVL6, a reporter construct, which contained firefly and Renilla luciferase genes fused to the ELOVL6 3′-UTR, was cotransfected with the candidate miRNA expressing plasmid in HEK-293 cells. The 3′-UTR of human ELOVL6 is divided into three constitutive fragments (F1, F2, and F3) that contain the predicted miRNA binding sites (Fig. 1A). To overexpress each miRNA candidate, the miR precursor was cloned into the pmR-ZsGreen1 vector and the expression levels of each miRNA were measured. The results from qRT-PCR revealed that all miR candidates were overexpressed compared with the control group (Supplementary Fig. 1). Results from the dual-luciferase assay revealed that the luciferase activity of ELOVL6 3′-UTR was significantly decreased by cotransfection with miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p (Fig. 1B), whereas miR-128–3p, miR-129–1–3p, and miR 129–2–3p did not influence luciferase activity (Figs. 1C and D). Consistently, overexpression of miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, or miR-22–3p significantly reduced ELOVL6 expression in HEK-293 cells (Fig. 1E). To further examine the effect of these miRNAs on ELOVL6 abundance, the expression vector containing HA-tagged ELOVL6 cDNAs fused to the 3′-UTR of ELOVL6 was cotransfected with miRNA expressing plasmids in HEK-293 cells, because the available anti-ELOVL6 antibodies are not specific to ELOVL6. The results from western blotting confirmed that the overexpression of miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, or miR-22–3p could reduce levels of ELOVL6 (Fig. 1F). These results indicate that miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p post-transcriptionally regulate ELOVL6 by directly binding its 3′-UTR.
Fig. 1.
Identification ofELOVL6-targeting miRNAs. A: Schematic representation of the ELOVL6 3′-UTR and the conserved miRNA target sites predicted by TargetScan. F1: Fragment 1, F2: Fragment 2, F3: Fragment 3. B–D: Luciferase activity of the dual-luciferase reporter vector with F1 (B), F2 (C), or F3 (D) of ELOVL6 3′-UTR cotransfected with empty vector (EV) or the predicted miRNA expressing vector in HEK293 cells. E: qRT-PCR analysis of ELOVL6 mRNA in HEK293 cells transfected with EV or the predicted miRNA expressing vector. F: HA-tagged ELOVL6 with the F1 of ELOVL6 3′-UTR was cotransfected with EV, miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, or miR-22–3p into HEK293 cells, and protein levels of HA-ELOVL6 were analyzed by western blotting. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 compared with the EV control. 3′-UTR: 3′-untranslated region.
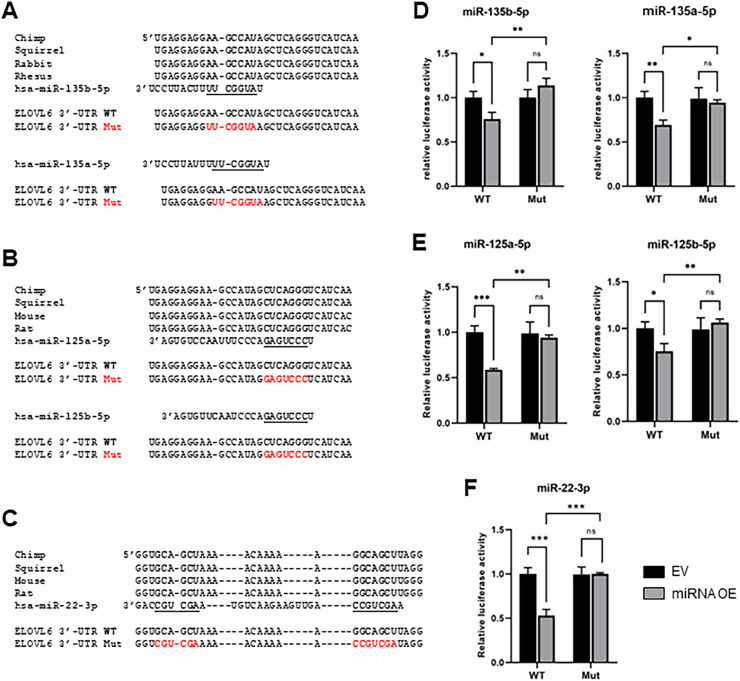
3.3. Specificity of target recognition depends on the seed region of the putative miRNA
To further confirm the binding of miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p to their predicted target regions in the ELOVL6 3′-UTR, mutations were introduced into ELOVL6 3′-UTR, which are complementary in sequence to the recognition site of each miRNA (Figs. 2A–C). Wild-type (WT) reporter, mutant (mut) reporter, and miRNA expression vector were cotransfected into HEK-293 cells, and the dual-luciferase assay was performed. The repressive ability of miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p to the luciferase activity of ELOVL6 3′-UTR was suppressed when the putative 3′-UTR-binding sites were disrupted (Figs. 2D–F). These data demonstrate that miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p directly bind to the putative binding site in ELOVL6 3′-UTR to mediate post-transcriptional repression, and the specificity of target recognition depends on the miRNA seed region.
Fig. 2.
Mutations in miRNA binding sites in the 3′-UTR ofELOVL6disturb the effects ofELOVL6-targeting miRNAs (A–C) Sequence alignments of binding sites of miR-135b-5p and miR-135a-5p (A), miR-125a-5p and miR 125b-5p (B), and miR-22–3p (C) in ELOVL6 3′-UTR from different species and mutations that were introduced into specific binding sites. The seed regions are highlighted. (D–F) Validation of specific target recognition using the dual-luciferase assay in miR-135b-5p and miR-135a-5p (D), miR-125a-5p and miR 125b-5p (E), and miR-22–3p (F). WT: Wild-type, Mut: Mutant. ns: not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
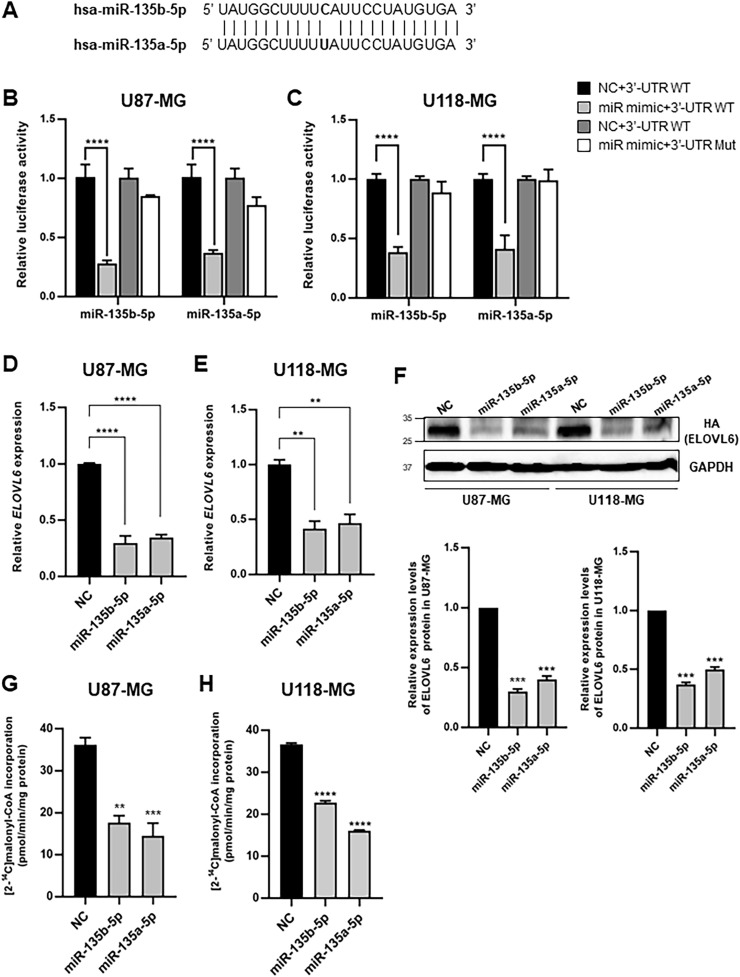
3.4. ELOVL6 is the direct target of miR-135b-5p and miR-135a-5p in GBM cell lines
ELOVL6 expression is high in the brain. ELOVL6 is overexpressed in a subset of patients with GBM and is associated with a poor prognosis [16]. miR-135b expression was downregulated in glioblastoma cell lines and stem-like cells [30]. Glia-enriched miR-135a was reportedly downregulated in malignant glioma and correlates inversely with pathological grade [31]. Therefore, we hypothesized that the downregulation of miR-135 would increase ELOVL6 expression and involve glioma formation. The mature sequences of miR-135b-5p and miR-135a-5p differ by only one nucleotide (Fig. 3A). To validate the interaction between miR-135 and ELOVL6 in GBM, we performed the dual-luciferase reporter assay. Transfection of U87-MG or U118-MG GBM cells with miR-135b-5p or miR-135a-5p mimics significantly reduced the luciferase activity of the wild-type ELOVL6 3′-UTR (WT-ELOVL6) compared with that of the nontarget miRNA mimic transfected control (Figs. 3B and C). By contrast, mutations in the binding site of miR-135b-5p or miR-135a-5p in the ELOVL6 3′-UTR (mut-ELOVL6) canceled the effect of miR-135b-5p or miR-135a-5p mimics in GBM cell lines. Transfection with the miR-135b-5p or miR-135a-5p mimics significantly repressed the expression of endogenous ELOVL6 mRNA in both U87-MG and U118-MG cells (Figs. 3D and E), and the degree of downregulation was greater than that of tumor necrosis factor receptor-associated factor 5 (TRAF5), a target of miR-135a-5p in glioma cells [32] (Supplementary Fig. 2). Consistent with the endogenous expression pattern, the level of C-terminal HA-tagged ELOVL6, expressed from a vector encoding the HA-ELOVL6 cDNA fused to the 3′-UTR of ELOVL6, was significantly reduced in the presence of miR-135b-5p and miR-135a-5p mimics (Fig. 3F). Furthermore, the palmitoyl-CoA (C16:0) elongation activity was significantly reduced by miR-135b-5p and miR-135a-5p mimics in GBM cells (Figs. 3G and H). Taken together, these results indicate that miR-135b-5p and miR-135a-5p are post-transcriptional regulators of ELOVL6 in GBM cell lines.
Fig. 3.
miR-135b-5p and miR-135a-5p decrease the abundance of ELOVL6 at both mRNA and protein levels in GBM cells (A) Alignment between mature miR-135b-5p and miR-135a-5p indicating one nucleotide difference. (B, C) Luciferase activity of the dual-luciferase reporter vector with the F1 of ELOVL6 3′-UTR cotransfected with miRNA mimics or negative control (NC) in U87-MG (B) and U118-MG cells (C). 3′-UTR WT containing wild-type ELOVL6 3′-UTR; 3′-UTR Mut containing mutations in specific miRNA target sites of ELOVL6 3′-UTR indicated in Fig. 2A; EV or predicted miRNA expressing vector in HEK293 cells. (D, E) Expression levels of ELOVL6 in U87-MG (D) and U118-MG cells (E) transfected with the miR-135b-5p mimic, miR-135a-5p mimic, or NC were measured by qRT-PCR. (F) HA-tagged ELOVL6 and F1 of ELOVL6 3′-UTR were cotransfected with the miR-135b-5p mimic, miR-135a-5p mimic, or NC into U87-MG and U118-MG cells, and levels of HA-ELOVL6 were analyzed by western blotting. (G, H) The palmitoyl-CoA elongation activity assay in U87-MG (G) and U118-MG GBM (H) cells transfected with the miR-135b-5p mimic, miR-135a-5p mimic, or NC. **P < 0.01; ***P < 0.001, ****P < 0.0001. GBM: glioblastoma multiforme.
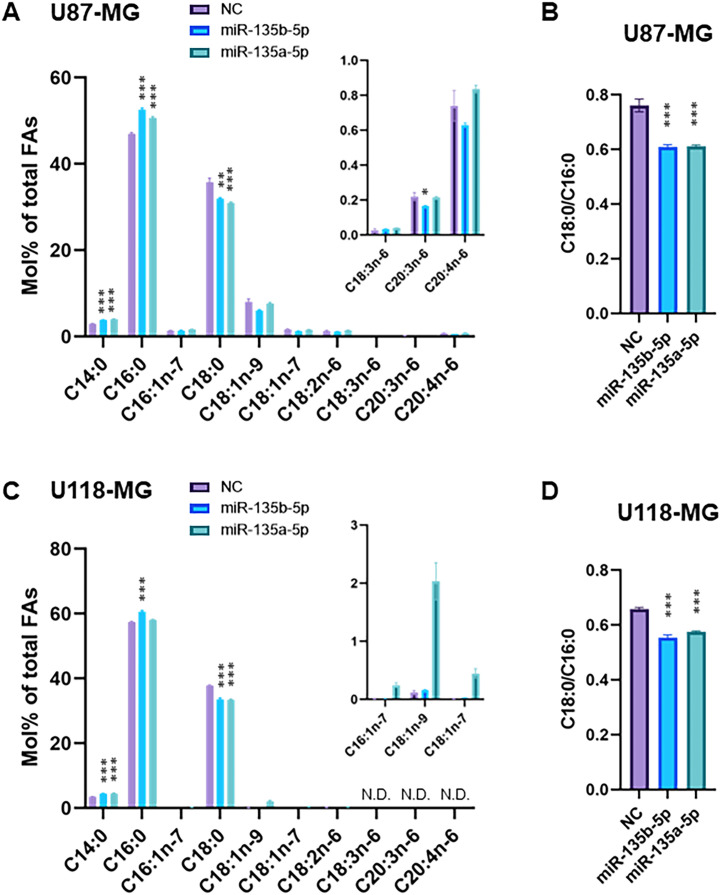
3.5. miR-135b-5p and miR-135a-5p alters fatty acid composition in GBM cells
To further investigate the role of miR-135b-5p and miR-135a-5p in FA metabolism, we analyzed the FA composition of U87-MG and U118-MG cells transfected with miRNA mimics or negative control by GC/MS. Consistent with reduced ELOVL6 activity, the FA composition in U87-MG cells transfected with the miR-135b-5p or miR-135a-5p mimic displayed a significant decrease in C18:0 and an increase in C16:0 and C14:0 FA composition compared with that in U87-MG cells transfected with the negative control mimic (Fig. 4A). The ratio for Elovl6 action (18:0/16:0) was consistently reduced by the miR-135b-5p and miR-135a-5p mimic in U87-MG cells (Fig. 4B). The same trend was also seen in U118-MG cells (Fig. 4C and D). These results, thus, suggest that miR-135b-5p and miR-135a-5p plays a role in regulating FA composition by modulating the abundance of ELOVL6 at both mRNA and protein level and its function in GBM cells.
Fig. 4.
miR-135b-5p and miR-135a-5p regulate fatty acid composition in GBM cells (A) Fatty acid composition of U87-MG cells transfected with miR-135b-5p mimic, miR-135a-5p mimic or negative control (NC) mimic for 48 h (n = 4 per group). (B) The ratio of stearate (C18:0) to palmitate (C16:0) in U87-MG cells transfected with miR-135b-5p mimic, miR-135a-5p mimic or negative control mimic for 48 h (n = 4 per group). (C) Fatty acid composition of U118-MG cells transfected with miR-135b-5p mimic, miR-135a-5p mimic, or negative control mimic for 48 h (n = 4 per group). (D) The ratio of stearate (C18:0) to palmitate (C16:0) in U118-MG cells transfected with miR-135b-5p mimic, miR-135a-5p mimic, or negative control (NC) mimic for 48 h (n = 4 per group). *P < 0.05; **P < 0.01; and ***P < 0.001 vs. NC. N.D.: not detected.
3.6. miR-135b-5p and miR-135a-5p suppress GBM cell growth and migration, whereas restoration of ELOVL6 attenuates their inhibitory effects
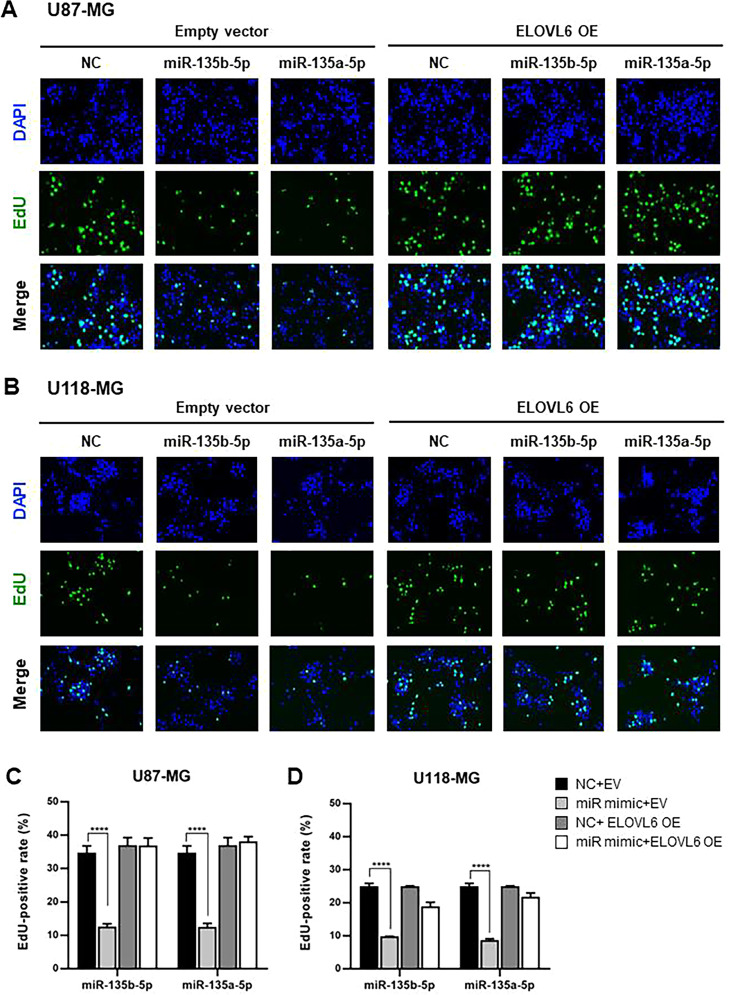
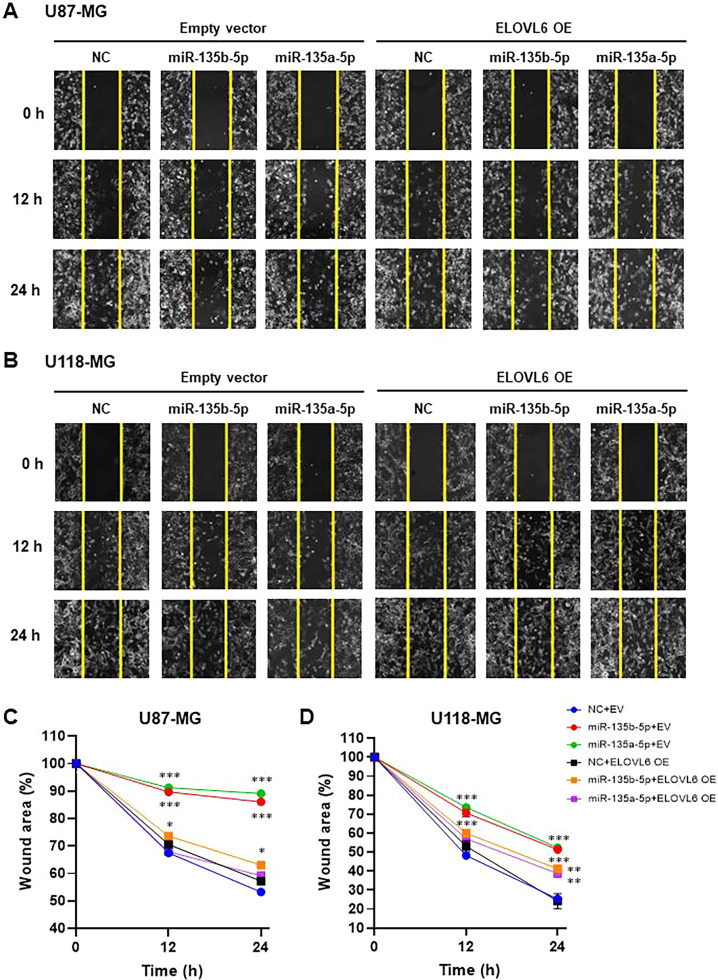
miR-135a and miR-135b have been shown to play important roles in the proliferation and migration of glioblastoma cells [30,32,33] [34]. This prompted us to investigate the contribution of ELOVL6 to miR-135a- and miR-135b-mediated regulation of the proliferation and migration of GBM cells. Consistent with other reports, transfection of miR-135a-5p or miR-135b-5p mimics significantly suppressed ethynyl deoxyuridine (EdU) uptake in U87-MG and U118-MG cells (Fig. 5). By contrast, the overexpression of ELOVL6 (Supplementary Fig. 3) completely canceled the inhibitory effect of miR-135b-5p and miR-135a-5p. Next, we assessed the rate of cell migration and displayed it as a plot of the closure area as a function of time. Consistent with cell proliferation results, transfection of miR-135a-5p or miR-135b-5p mimics significantly inhibited the migration of U87-MG and U118-MG cells compared with the control at 12 and 24 h (Fig. 6). Remarkably, overexpression of ELOVL6 in miR-135a-5p or miR-135b-5p mimic transfected U87-MG and U118-MG cells reversed the inhibition on migration at 12 and 24 h. No significant increase in cell death was detected after treatment with the miR-135b-5p or miR-135a-5p mimics compared with controls (Supplementary Fig. 4). Together, these results indicate that miR-135b-5p and miR-135a-5p regulate the proliferation and migration of U87-MG and U118-MG cells, at least in part, by changing ELOVL6-dependent lipid metabolism.
Fig. 5.
miR-135b-5p and miR-135a-5p suppress cell proliferation in glioblastoma cells by inhibiting ELOVL6 (A, B) Representative fluorescence microscopy images of EdU cell proliferation assay in U87-MG (A) and U118-MG (B) cells transfected with the miR-135b-5p mimic, miR-135a-5p mimic, or NC and ELOVL6 or EV. (C, D) Percentage of EdU-positive cells in U87-MG (C) and U118-MG (D) cells treated as in (A) and (B), respectively. ****P < 0.0001. EdU, 5-ethynyl-2′-deoxyuridine; DAPI, 4′,6-diamidino-2-phenylindole. EV: pcDNA 3.1 (+) empty vector, OE: overexpression.
Fig. 6.
miR-135b-5p and miR-135a-5p suppress cell migration in glioblastoma cells by inhibiting ELOVL6 (A, B) Representative images of the wound healing assay in U87-MG (A) and U118-MG (B) cells transfected with the miR-135b-5p mimic, miR-135a-5p mimic, or NC and ELOVL6 or EV at 0, 12, and 24 h. (C, D) Percentage of the migrated area following the wound healing assay in U87-MG (C) and U118-MG (D) cells treated as in (A) and (B), respectively. **P < 0.01, ***P < 0.001 compared to NC. OE: overexpression, EV: pcDNA 3.1 (+) empty vector.
4. Discussion
Elongation and desaturation are central steps in the de novo synthesis of long-chain FAs [2]. ELOVL6 is a microsomal enzyme responsible for the elongation of C16 saturated and monounsaturated FAs into C18 FAs, thereby potentially affecting >50% of the total cellular lipids [5,35]. Thus, a better understanding of the regulation of cellular FAs that mediate ELOVL6 expression and function will help understand the complex system to regulate FA length and maintain lipid homeostasis. miRNAs direct the post-transcriptional silencing of mRNA targets by binding to their complementary sequences in the 3′-UTR, thereby repressing translation and accelerating mRNA degradation [36]. miRNAs, including miR-33, miR-122, miR-27a/b, mi-R378, miR-34a, and miR-21, have emerged as important regulators of lipid metabolism [37]. In this study, nine miRNAs were predicted to be regulators of human ELOVL6 through TargetScanHuman. We have identified that miR-135b-5p, miR-135a-5p, miR-125a-5p, miR-125b-5p, and miR-22–3p post-transcriptionally regulate ELOVL6 expression by directly binding to their recognition element in the 3′-UTR, whereas miR-128–3p, miR-129–1–3p, and miR-129–2–3p do not affect ELOVL6 expression. One of the possible reasons why miR-128–3p, miR-129–1–3p, and miR-129–2–3p could not target ELOVL6 can be attributed to miRNA targeting efficacy in the 3′-UTR, such as site conservation, target site abundance, site type, site context, UTR size, local AU content, and affinity and regulatory impact of RNA-binding proteins (RBPs) [25,[38], [39], [40], [41], [42], [43]]. For instance, in the ELOVL6 3′-UTR, miR-128–3p is located at 1143–1149 bp and miR-129–1–3p and miR-129–2–3p are located at 1887–1893 bp, whereas the identified putative miRNAs are located at the beginning of the 3′-UTR. Grimson et al. reported that miRNA sites in the middle of long UTRs might be less accessible to the miRNA silencing complex because they have the opportunity to generate occlusive contacts with segments from either side [38]. miR-125a was shown to negatively regulate Elovl6 expression during porcine intramuscular preadipocyte differentiation [44] and obesity-associated insulin resistance and dysregulated lipid metabolism in mouse liver [45]. Moreover, miR-125b-5p was linked to the control of hepatic lipid metabolism by targeting Elovl6 in geese [46]. miR-22–3p was found to suppress hepatic Elovl6 expression in chicken [47]. miR-22 is a lipid and folate regulator in breast cancer cells and prevents de novo lipogenesis and FA elongation by suppressing the expression of ATP citrate lyase and ELOVL6 [48]. In this study, we demonstrated that these miRNAs also regulate ELOVL6 expression in human cells. miR-125a-5p, miR-125b-5p, and miR-22–3p are highly conserved among species, including humans, chimpanzees, rhesus monkeys, squirrels, rabbits, rats, and mice, suggesting the important role of these miRNAs in regulating FA metabolism by directly targeting ELOVL6 in mammals. miR-135b-5p is located on chromosome 1q32.1 and is highly expressed in the epididymis, spinal cord, thyroid, brain, and adipocyte [49]. miR-135a-5p is located on chromosome 3p21.2 and is highly expressed in the thyroid, epididymis, brain, and spinal cord [49]. With only a single nucleotide difference in their mature sequences, they target the same gene [50] and have neuroprotective roles in the serotonergic activity, Alzheimer's disease, and oxygen-glucose deprivation and reoxygenation-induced brain injury [50,51]. Studies have reported that the upregulation of miR-135b-5p or miR-135a-5p increases the sensitivity of drug-resistant breast cancer cells and exerts tumor suppressive effects in osteosarcoma, prostate cancer, ovarian cancer, glioma and glioblastoma stem-like cells [[30], [31], [32], [52], [53], [54], [55], [56]]. In this study, we aim to examine the interaction of miR-135b-5p and miR-135a-5p with ELOVL6 in one of the most malignant tumors—GBM. To investigate the molecular mechanism by which miR-135b-5p and miR-135a-5p associate with ELOVL6 during GBM development, we performed a series of in vitro assays initially. Interestingly, for the first time, miR-135b-5p and miR-135a-5p were identified as regulators of ELOVL6 and their functional targets in GBM cells. miR-135b-5p and miR-135a-5p mimic suppressed the ELOVL6 3′-UTR luciferase reporter activity, and this effect was abolished by mutating the miR-135b-5p and miR-135a-5p binding sites. miR-135b-5p and miR-135a-5p mimic also led to a significant reduction in ELOVL6 expression at the mRNA and protein level that induced a significant decrease in the relative proportion of C18:0 in both U87-MG and U118-MG cells. Results from proliferation and migration assays revealed that the overexpression of miR-135b-5p and miR-135a-5p suppressed the proliferation and migration of GBM cells. These results indicate that miR-135b-5p and miR-135a-5p function as tumor suppressors in part by repressing ELOVL6 expression during the development of GBM. Interestingly, miR-125 and miR-22 were also reported to suppress GBM cell proliferation by targeting hexokinase 2 [57] and sirtuin 1 [58], respectively. Consistently, ELOVL6 is overexpressed in patients with GBM and is hypomethylated in GBM [59]. Results from a glial-specific RNAi screen and bioinformatics analysis of patients with glioma revealed that high expression of ELOVL6 significantly correlated with poor survival in patients with high-grade gliomas. Thus, ELOVL6 is important for glial function and a potential biomarker for glioma [17].
In addition, Elovl6 is associated with poor prognosis in patients with hepatocellular carcinoma, and Elovl6 promotes oncogenic activity in cancers of the liver, breast, colon, prostate, and pancreas [13,[60], [61], [62]]. Interestingly, inhibiting Elovl6 reduced tumor growth in liver cancer, lung squamous cell carcinoma, and retinoblastoma-deficient tumor cells [15,56,60]. Thus, cancers expressing high levels of ELOVL6 could be good clinical targets of miR-135, miR-125, and miR-22. Further investigation is required to clarify the mechanisms by which these miRNAs regulate ELOVL6, FA metabolism, and tumorigenesis in GBM and other cancers.
Aberrant lipid metabolism is a hallmark of tumors, including GBM [63,64]. GBM cells enhance de novo FA synthesis to produce energy sources, membrane components, and signaling molecules for intense proliferation, survival, dissemination, and adaptation to the tumor microenvironment. Palmitate and stearate are two common saturated FAs in cancer cells, which are preferentially incorporated into membrane-forming phospholipids and sphingolipids [15,63]. We have provided evidence of the expected decrease in the stearate to palmitate in miR-135b-5p or miR-135a-5p treated GBM cells. Although the underlying mechanism remains to be elucidated, miR-135b-5p and miR-135a-5p might affect membrane fluidity and local membrane structure and influence the localization, topology, and signaling of membrane proteins by modulating ELOVL6-mediated FA chain elongation.
In summary, we identified five conserved miRNAs that regulate the post-transcriptional level of ELOVL6 by binding with the 3′-UTR of human ELOVL6 mRNA. These may therefore play an important role in lipid metabolism. Furthermore, we showed a key role for miR-135b-5p and miR-135a-5p in the regulation of cell proliferation and migration of GBM cells by targeting ELOVL6. These findings will help us understand the endogenous regulation of ELOVL6 mRNA and the complex regulatory mechanisms of lipid metabolism in metabolic disorders and cancers. Moreover, we have identified ELOVL6-targeting miRNAs as potential targets for the development of novel strategies for cancer treatment.
Author contributions
T.M., N.I., and H. Shimano designed the project; N.I., M.Shimizu., and T.M. performed the experiments; K.M., H.O., S.H., R.S., Y. Okajima, T.S., E.M., S.H., Y.M., Y. Osaki, Y.A., H. Suzuki., H.S., Y.T., M.Sekiya., N.Y., and Y.N. analyzed and interpreted the data; and N.I., T.M., and H. Shimano prepared the manuscript. All authors reviewed the manuscript.
Data availability
The majority of the data generated or analyzed during this study are included here and in the supplementary material, but additional datasets generated and/or analyzed during this study are available from the corresponding author upon reasonable request.
Declaration of Competing Interest
The authors declare that there is no conflict of interest associated with this manuscript.
Acknowledgments
This work was supported by research funds from the grants-in-aid for Scientific Research (B) 18H03189 (to T.M.) and Scientific Research (B) 21H03371 (to T.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The authors thank Katsuko Okubo and Chizuko Fukui for technical assistance, and the members of Shimano laboratory for discussion and helpful comments on the manuscript. The authors would like to thank Enago (www.enago.jp) for the English language review. Dr. Hitoshi Shimano is the guarantor of this work, and as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2023.100078.
Contributor Information
Takashi Matsuzaka, Email: t-matsuz@md.tsukuba.ac.jp.
Hitoshi Shimano, Email: hshimano@md.tsukuba.ac.jp.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guillou H., Zadravec D., Martin P.G., Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog. Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Jakobsson A., Westerberg R., Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Moon Y.A., Shah N.A., Mohapatra S., Warrington J.A., Horton J.D. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2001;276:45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzaka T., Shimano H., Yahagi N., Yoshikawa T., Amemiya-Kudo M., Hasty A.H., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Takahashi A., Yato S., Sone H., Ishibashi S., Yamada N. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 2002;43:911–920. [PubMed] [Google Scholar]
- 6.Matsuzaka T., Shimano H., Yahagi N., Kato T., Atsumi A., Yamamoto T., Inoue N., Ishikawa M., Okada S., Ishigaki N., Iwasaki H., Iwasaki Y., Karasawa T., Kumadaki S., Matsui T., Sekiya M., Ohashi K., Hasty A.H., Nakagawa Y., Takahashi A., Suzuki H., Yatoh S., Sone H., Toyoshima H., Osuga J., Yamada N. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007;13:1193–1202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- 7.Matsuzaka T., Kuba M., Koyasu S., Yamamoto Y., Motomura K., Arulmozhiraja S., Ohno H., Sharma R., Shimura T., Okajima Y., Han S.I., Aita Y., Mizunoe Y., Osaki Y., Iwasaki H., Yatoh S., Suzuki H., Sone H., Takeuchi Y., Yahagi N., Miyamoto T., Sekiya M., Nakagawa Y., Ema M., Takahashi S., Tokiwa H., Shimano H. Hepatocyte ELOVL Fatty Acid Elongase 6 Determines Ceramide Acyl-Chain Length and Hepatic Insulin Sensitivity in Mice. Hepatology. 2020;71:1609–1625. doi: 10.1002/hep.30953. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H., Matsuzaka T., Nakano Y., Motomura K., Tang N., Yokoo T., Okajima Y., Han S.I., Takeuchi Y., Aita Y., Iwasaki H., Yatoh S., Suzuki H., Sekiya M., Yahagi N., Nakagawa Y., Sone H., Yamada N., Shimano H. Elovl6 deficiency improves glycemic control in diabetic db/db mice by expanding beta-cell mass and increasing insulin secretory capacity. Diabetes. 2017;66:1833–1846. doi: 10.2337/db16-1277. [DOI] [PubMed] [Google Scholar]
- 9.Matsuzaka T., Atsumi A., Matsumori R., Nie T., Shinozaki H., Suzuki-Kemuriyama N., Kuba M., Nakagawa Y., Ishii K., Shimada M., Kobayashi K., Yatoh S., Takahashi A., Takekoshi K., Sone H., Yahagi N., Suzuki H., Murata S., Nakamuta M., Yamada N., Shimano H. Elovl6 promotes nonalcoholic steatohepatitis. Hepatology. 2012;56:2199–2208. doi: 10.1002/hep.25932. [DOI] [PubMed] [Google Scholar]
- 10.Saito R., Matsuzaka T., Karasawa T., Sekiya M., Okada N., Igarashi M., Matsumori R., Ishii K., Nakagawa Y., Iwasaki H., Kobayashi K., Yatoh S., Takahashi A., Sone H., Suzuki H., Yahagi N., Yamada N., Shimano H. Macrophage Elovl6 deficiency ameliorates foam cell formation and reduces atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2011;31:1973–1979. doi: 10.1161/ATVBAHA.110.221663. [DOI] [PubMed] [Google Scholar]
- 11.Muir K., Hazim A., He Y., Peyressatre M., Kim D.Y., Song X., Beretta L. Proteomic and lipidomic signatures of lipid metabolism in NASH-associated hepatocellular carcinoma. Cancer Res. 2013;73:4722–4731. doi: 10.1158/0008-5472.CAN-12-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibasaki Y., Horikawa M., Ikegami K., Kiuchi R., Takeda M., Hiraide T., Morita Y., Konno H., Takeuchi H., Setou M., Sakaguchi T. Stearate-to-palmitate ratio modulates endoplasmic reticulum stress and cell apoptosis in non-B non-C hepatoma cells. Cancer Sci. 2018;109:1110–1120. doi: 10.1111/cas.13529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Y.C., Feng Y.H., Wu H.T., Huang Y.S., Tung C.L., Wu P., Chang C.J., Shiau A.L., Wu C.L. Elovl6 is a negative clinical predictor for liver cancer and knockdown of Elovl6 reduces murine liver cancer progression. Sci. Rep. 2018;8:6586. doi: 10.1038/s41598-018-24633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anelli L., Zagaria A., Coccaro N., Tota G., Impera L., Minervini C.F., Pastore D., Minervini A., Casieri P., Specchia G., Albano F. A novel t(4;16)(q25;q23.1) associated with EGF and ELOVL6 deregulation in acute myeloid leukemia. Gene. 2013;529:144–147. doi: 10.1016/j.gene.2013.07.105. [DOI] [PubMed] [Google Scholar]
- 15.Marien E., Meister M., Muley T., Gomez Del Pulgar T., Derua R., Spraggins J.M., Van de Plas R., Vanderhoydonc F., Machiels J., Binda M.M., Dehairs J., Willette-Brown J., Hu Y., Dienemann H., Thomas M., Schnabel P.A., Caprioli R.M., Lacal J.C., Waelkens E., Swinnen J.V. Phospholipid profiling identifies acyl chain elongation as a ubiquitous trait and potential target for the treatment of lung squamous cell carcinoma. Oncotarget. 2016;7:12582–12597. doi: 10.18632/oncotarget.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shergalis A., Bankhead A., 3rd, Luesakul U., Muangsin N., Neamati N. Current 2sze3ioblastoma. Pharmacol. Rev. 2018;70:412–445. doi: 10.1124/pr.117.014944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi K.C., Tsai W.C., Wu C.L., Lin T.Y., Hueng D.Y. An adult drosophila glioma model for studying pathometabolic pathways of gliomagenesis. Mol. Neurobiol. 2019;56:4589–4599. doi: 10.1007/s12035-018-1392-2. [DOI] [PubMed] [Google Scholar]
- 18.Shimano H., Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 19.Kumadaki S., Matsuzaka T., Kato T., Yahagi N., Yamamoto T., Okada S., Kobayashi K., Takahashi A., Yatoh S., Suzuki H., Yamada N., Shimano H. Mouse Elovl-6 promoter is an SREBP target. Biochem. Biophys. Res. Commun. 2008;368:261–266. doi: 10.1016/j.bbrc.2008.01.075. [DOI] [PubMed] [Google Scholar]
- 20.Bae J.S., Oh A.R., Lee H.J., Ahn Y.H., Cha J.Y. Hepatic Elovl6 gene expression is regulated by the synergistic action of ChREBP and SREBP-1c. Biochem. Biophys. Res. Commun. 2016;478:1060–1066. doi: 10.1016/j.bbrc.2016.08.061. [DOI] [PubMed] [Google Scholar]
- 21.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 25.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hesse M., Arenz C. MicroRNA maturation and human disease. Methods Mol. Biol. 2014;1095:11–25. doi: 10.1007/978-1-62703-703-7_2. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y., Matsuzaka T., Tahara-Hanaoka S., Shibuya K., Shimano H., Nakahashi-Oda C., Shibuya A. Elovl6 regulates mechanical damage-induced keratinocyte death and skin inflammation. Cell Death. Dis. 2018;9:1181. doi: 10.1038/s41419-018-1226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranaka H., Hayashi A., Minami K., Kitajima S., Kohno S., Nishimoto Y., Nagatani N., Suzuki M., Kulathunga L.A.N., Sasaki N., Okada N., Matsuzaka T., Shimano H., Tada H., Takahashi C. A distinct function of the retinoblastoma protein in the control of lipid composition identified by lipidomic profiling. Oncogenesis. 2017;6:e350. doi: 10.1038/oncsis.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lulli V., Buccarelli M., Martini M., Signore M., Biffoni M., Giannetti S., Morgante L., Marziali G., Ilari R., Pagliuca A., Larocca L.M., De Maria R., Pallini R., Ricci-Vitiani L. miR-135b suppresses tumorigenesis in glioblastoma stem-like cells impairing proliferation, migration and self-renewal. Oncotarget. 2015;6:37241–37256. doi: 10.18632/oncotarget.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S., Lin Y., Xu D., Chen J., Shu M., Zhou Y., Zhu W., Su X., Zhou Y., Qiu P., Yan G. MiR-135a functions as a selective killer of malignant glioma. Oncogene. 2012;31:3866–3874. doi: 10.1038/onc.2011.551. [DOI] [PubMed] [Google Scholar]
- 32.Luo W., Sun C., Zhou J., Wang Q., Yu L., Bian X.W., Zhou X., Hua D., Wang R., Rao C., Jiang Z., Shi C., Yu S. miR-135a-5p functions as a glioma proliferation suppressor by targeting tumor necrosis factor receptor-associated factor 5 and predicts patients' prognosis. Am. J. Pathol. 2019;189:162–176. doi: 10.1016/j.ajpath.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Lin J., Wen X., Zhang X., Sun X., Yunzhi L., Peng R., Zhu M., Wang M., Zhang Y., Luo W., Luo G., Zhang Y. miR-135a-5p and miR-124-3p inhibit malignancy of glioblastoma by downregulation of syndecan binding protein. J. Biomed. Nanotechnol. 2018;14:1317–1329. doi: 10.1166/jbn.2018.2579. [DOI] [PubMed] [Google Scholar]
- 34.Mokgautsi N., Wen Y.T., Lawal B., Khedkar H., Sumitra M.R., Wu A.T.H., Huang H.S. An integrated bioinformatics study of a novel niclosamide derivative, NSC765689, a potential GSK3β/β-Catenin/STAT3/CD44 suppressor with anti-glioblastoma properties. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22052464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan C.Y., Virtue S., Bidault G., Dale M., Hagen R., Griffin J.L., Vidal-Puig A. Brown adipose tissue thermogenic capacity is regulated by Elovl6. Cell Rep. 2015;13:2039–2047. doi: 10.1016/j.celrep.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 37.van Battum E.Y., Verhagen M.G., Vangoor V.R., Fujita Y., Derijck A., O'Duibhir E., Giuliani G., de Gunst T., Adolfs Y., Lelieveld D., Egan D., Schaapveld R.Q.J., Yamashita T., Pasterkamp R.J. An image-based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting kruppel-like factor 4. J. Neurosci. 2018;38:613–630. doi: 10.1523/JNEUROSCI.0662-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., Cappello T., Wang L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B. 2015;5:145–150. doi: 10.1016/j.apsb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 42.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S., Kim S., Chang H.R., Kim D., Park J., Son N., Park J., Yoon M., Chae G., Kim Y.K., Kim V.N., Kim Y.K., Nam J.W., Shin C., Baek D. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 2021;12:5057. doi: 10.1038/s41467-021-25078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du J., Xu Y., Zhang P., Zhao X., Gan M., Li Q., Ma J., Tang G., Jiang Y., Wang J., Li X., Zhang S., Zhu L. MicroRNA-125a-5p affects adipocytes proliferation, differentiation and fatty acid composition of porcine intramuscular fat. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19020501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R., Wang M., Li E., Yang Y., Li J., Chen S., Shen W.J., Azhar S., Guo Z., Hu Z. Dysregulation of microRNA-125a contributes to obesity-associated insulin resistance and dysregulates lipid metabolism in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158640. [DOI] [PubMed] [Google Scholar]
- 47.Chen F., Zhang H., Li J., Tian Y., Xu J., Chen L., Wei J., Zhao N., Yang X., Zhang W., Lu L. Identification of differentially expressed miRNAs in the fatty liver of Landes goose (Anser anser) Sci. Rep. 2017;7:16296. doi: 10.1038/s41598-017-16632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Z., Li H., Zheng H., Jiang K., Yan F., Tian Y., Kang X., Wang Y., Liu X. Hepatic ELOVL6 mRNA is regulated by the gga-miR-22-3p in egg-laying hen. Gene. 2017;623:72–79. doi: 10.1016/j.gene.2017.04.040. [DOI] [PubMed] [Google Scholar]
- 49.Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stahler C., Meese E., Keller A. Distribution of miRNA expression across human tissues. Nucleic. Acids. Res. 2016;44:3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Issler O., Haramati S., Paul E.D., Maeno H., Navon I., Zwang R., Gil S., Mayberg H.S., Dunlop B.W., Menke A., Awatramani R., Binder E.B., Deneris E.S., Lowry C.A., Chen A. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–360. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 51.Duan Q., Sun W., Yuan H., Mu X. MicroRNA-135b-5p prevents oxygen-glucose deprivation and reoxygenation-induced neuronal injury through regulation of the GSK-3beta/Nrf2/ARE signaling pathway. Arch. Med. Sci. 2018;14:735–744. doi: 10.5114/aoms.2017.71076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., Qin Y., Chen P., Luo Q., Shi H., Jiang X. miR‑135b‑5p enhances the sensitivity of HER‑2 positive breast cancer to trastuzumab via binding to cyclin D2. Int. J. Mol. Med. 2020;46:1514–1524. doi: 10.3892/ijmm.2020.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Xia F., Zhang F., Cui Y., Wang Q., Liu H., Wu Y. miR-135b-5p enhances doxorubicin-sensitivity of breast cancer cells through targeting anterior gradient 2. J. Exp. Clin. Cancer Res. 2019;38:26. doi: 10.1186/s13046-019-1024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang N., Tao L., Zhong H., Zhao S., Yu Y., Yu B., Chen X., Gao J., Wang R. miR-135b inhibits tumour metastasis in prostate cancer by targeting STAT6. Oncol. Lett. 2016;11:543–550. doi: 10.3892/ol.2015.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang W., Jiang Y., Mu X., Xu L., Cheng W., Wang X. MiR-135a functions as a tumor suppressor in epithelial ovarian cancer and regulates HOXA10 expression. Cell Signal. 2014;26:1420–1426. doi: 10.1016/j.cellsig.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Ren R., Wu J., Zhou M.Y. MiR-135b-5p affected malignant behaviors of ovarian cancer cells by targeting KDM5B. Eur. Rev. Med. Pharmacol. Sci. 2020;24:11469. doi: 10.26355/eurrev_202011_23761. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J., Chen G., Gao Y., Liang H. HOTAIR/miR-125 axis-mediated Hexokinase 2 expression promotes chemoresistance in human glioblastoma. J. Cell. Mol. Med. 2020;24:5707–5717. doi: 10.1111/jcmm.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H., Lu Q., Fei X., Shen L., Jiang D., Dai D. miR-22 inhibits the proliferation, motility, and invasion of human glioblastoma cells by directly targeting SIRT1. Tumour Biol. 2016;37:6761–6768. doi: 10.1007/s13277-015-4575-8. [DOI] [PubMed] [Google Scholar]
- 59.Vyazunova I., Maklakova V.I., Berman S., De I., Steffen M.D., Hong W., Lincoln H., Morrissy A.S., Taylor M.D., Akagi K., Brennan C.W., Rodriguez F.J., Collier L.S. Sleeping Beauty mouse models identify candidate genes involved in gliomagenesis. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0113489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giovannucci E., Harlan D.M., Archer M.C., Bergenstal R.M., Gapstur S.M., Habel L.A., Pollak M., Regensteiner J.G., Yee D. Diabetes and cancer: a consensus report. CA Cancer J. Clin. 2010;60:207–221. doi: 10.3322/caac.20078. [DOI] [PubMed] [Google Scholar]
- 61.Caan B.J., Coates A.O., Slattery M.L., Potter J.D., Quesenberry C.P., Jr., Edwards S.M. Body size and the risk of colon cancer in a large case-control study. Int. J. Obes. Relat. Metab. Disord. 1998;22:178–184. doi: 10.1038/sj.ijo.0800561. [DOI] [PubMed] [Google Scholar]
- 62.Bianchini F., Kaaks R., Vainio H. Overweight, obesity, and cancer risk. Lancet Oncol. 2002;3:565–574. doi: 10.1016/s1470-2045(02)00849-5. [DOI] [PubMed] [Google Scholar]
- 63.Broadfield L.A., Pane A.A., Talebi A., Swinnen J.V., Fendt S.M. Lipid metabolism in cancer: new perspectives and emerging mechanisms. Dev. Cell. 2021;56:1363–1393. doi: 10.1016/j.devcel.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 64.Lee H., Kim D., Youn B. Targeting oncogenic rewiring of lipid metabolism for glioblastoma treatment. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232213818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of the data generated or analyzed during this study are included here and in the supplementary material, but additional datasets generated and/or analyzed during this study are available from the corresponding author upon reasonable request.
Data will be made available on request.