Highlights
-
•
Histone modifier treatment afforded protection against diabetes in female NOD mice.
-
•
Infiltration of Ly-6C+, CD4+ and CD8+ cells was reduced in the pancreas of cured mice.
-
•
Protection was accompanied by the regulation of select genes in the spleen and pancreas.
-
•
Epigenetic drug treatment abolished diabetes-causing ability of T lymphocytes.
-
•
Ly-6C+ monocytes of cured mice reduced T-cell mediated diabetes induction.
Keywords: Epigenetics, Genes, Histone deacetylases, Lymphocytes, Macrophages, Trichostatin A, Type 1 diabetes
Abstract
We have previously demonstrated that weekly treatment of female prediabetic NOD mice with a low dose of the histone deacetylase inhibitor Trichostatin A (TSA) bestowed long-lasting, irreversible protection against autoimmune diabetes. Herein we show that drug treatment diminished the infiltration of the pancreas with CD4+, CD8+ T cells, and Ly-6C+ monocytes. Significantly, TSA administration selectively repressed the expression of a set of genes exaggerated during diabetes and constitutively expressed primarily in the spleen and rarely in the pancreas. These genes encode lymphokines, macrophage-associated determinants, and transcription factors. Although the copy numbers of many histone deacetylases increased during diabetes in the spleen and pancreas, only those upregulated in the spleen were rendered sensitive to repression by TSA treatment. Mitogen-activated T lymphocytes derived from drug-treated donors displayed diminished diabetogenic potential following transfer into immunodeficient NOD.scid mice. In the immunocompromised recipients, diabetes caused by the transfer of activated T lymphocytes from untreated diabetic mice was hampered by the co-transfer of highly purified splenic CD11b+Ly-6C+ macrophages from drug-treated mice. However, the transfer of CD11b+Ly-6C+ macrophages from drug-treated mice failed to block ongoing diabetes in wild-type NOD mice. These data demonstrate that the modified gene expression and functional alteration of T lymphocytes and macrophages collectively contribute to diabetes protection afforded by the histone modifier in female NOD mice.
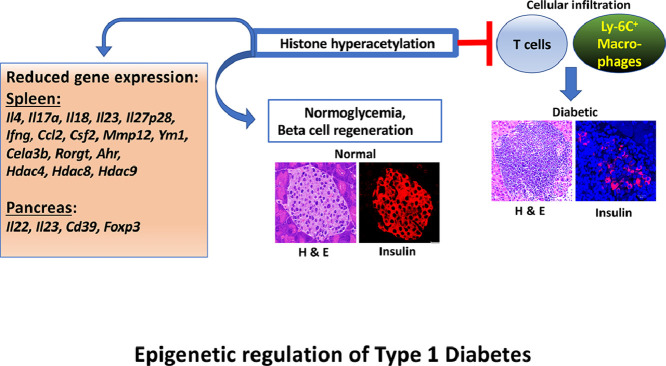
Graphical abstract
Epigenetic regulation of Type 1 Diabetes
1. Introduction
Type 1 diabetes (T1D), a T cell-mediated autoimmune disease, has occurred at an alarming rate in genetically susceptible individuals in recent years [1], [2], [3]. Although genome-wide association studies implicated more than 40 different genetic loci, the causal genes of T1D remain unknown [4]. Lack of familial history and robust concordance among identical twins in developing T1D implied the involvement of environmental factors. Epigenetics provides a mechanism by which external factors can produce a variety of phenotypic variations with identical genotypes. Epigenetic mechanisms include DNA methylation, post-translational modifications of histones, and microRNA-mediated gene regulation [5]. Methylation of cytosine residues in gene promoters results in transcriptional silencing of some [6] but not all genes [7]. Reversible acetylation of the e-amino group of lysine in the histone tails by histone acetyltransferases and deacetylation by histone deacetylases (HDACs) are the best-characterized post-translational modifications of histones [8]. Transcriptional permissiveness correlates with histone acetylation in contrast to deacetylation resulting in gene repression. Non-selective, small molecule HDAC inhibitors can induce chromatin modifications and thereby affect gene expression. Trichostatin A (TSA) derived from Streptomyces platensis is the most potent inhibitor of class I, class IIa, class IIb, and class IV HDACs, which increases histone acetylation resulting in up- and down-regulation of genes in vitro [9] and in vivo [10], [11], [12], [13], [14], [15], [16].
Previously, we have demonstrated the utility of TSA to mitigate autoimmune diabetes [10], [11], [12], [13] and experimental autoimmune encephalomyelitis (EAE) [14], [15], [16] in female NOD mice. Weekly injections of the prediabetic mice between 18 and 24-wks of age with a low dose (500 µg/Kg body weight) of the histone modifier provided long-lasting and irreversible protection against T1D. Disease protection was accompanied by reduced inflammation of the pancreatic islets and regeneration of insulin-producing β-cells without altering the numbers of T regulatory cells [10]. Significantly, TSA treatment abrogated the ability of splenocytes to adoptively transfer T1D into immunodeficient NOD.scid mice, indicating the effect of the epigenetic drug on diabetogenic T lymphocytes selectively [11]. Transcriptome analysis of uninduced splenocytes unraveled the exaggerated expression of inflammatory genes, including the macrophage selective Cela3b (elastase 3) in diabetic mice, which were repressed by TSA treatment. These results are consistent with the possibility that chromatin remodeling by TSA can retard the induction and/or progression of T1D by altering the transcriptional programs of T lymphocytes and macrophages. These results prompted us to investigate further the effects of the epigenetic drug on T lymphocytes and macrophages, respectively implicated in the induction and the manifestation of T1D [17], [18], [19], using quantitative reverse transcriptase-mediated polymerase chain reaction (qRT-PCR) and an adoptive transfer model. Our study unraveled that a select set of exaggerated and constitutively expressed genes encoding cytokines, cell surface determinants, transcription factors, and HDACs was modulated by the histone modifier predominantly in the peripheral lymphoid tissues. Our data also demonstrate distinct effects of TSA on the function of T-cells and macrophages. These data lend novel insights into the mechanisms of T1D and their possible manipulation by epigenetic reprogramming.
2. Materials and materials
2.1. Animals and treatment
The Office of Animal Care and Institutional Biosafety of the University of Illinois at Chicago approved the animal protocol. Experiments were conducted following the NIH guide for the care and use of laboratory animals.
Six to eight-wks old female NOD/ShiLtj (H-2g7) mice purchased from The Jackson Laboratory (Bar Harbor, ME) were maintained under specific pathogen-free conditions and provided with autoclaved animal chow and neutral pH water ad libitum as described [[10], [11], [12], [13], [14], [15], [16], 20]. Mice were injected s.c. with 500 µg/Kg body weight of TSA or DMSO at weekly intervals between 16 and 24-wks of age [[10], [11], [14], [15], [16]]. Non-fasting blood glucose level exceeding 250 mg/dL on two consecutive weekly determinations was considered as diabetic [[10], [11], 20]. Numbers of mice tested are given in individual figure legends.
2.2. Confocal microscopy
The following antibodies were used for staining pancreatic sections: guinea pig anti-insulin Ab (Zymed Laboratories, South San Francisco, CA), tetramethylrhodamine iso-thiocyanate–rabbit antisera raised against guinea pig Ig (Sigma Aldrich, St. Louis, MO), anti-Ly-6C-FITC (clone HK1.4, eBiosciences, San Diego, CA), anti-Ly-6G-FITC (clone 1AB, BD Pharmingen, San Diego, CA), rat monoclonal anti-CD4 (clone RM4-5, eBiosciences-ThermoFisher Scientific, Waltham, MA), DyLight™ 488 goat anti-rat IgG (catalog # 405409, BioLegend, San Diego, CA) and anti-CD8-FITC (clone 53-6.7, eBiosciences). Confocal images were acquired using a Zeiss LSM510 laser scanning microscope and processed by the Zeiss LSM Image browser (4.0 version; Zeiss, Oberkochen, Germany) and Adobe Photoshop Elements version 9.0, as described [20].
2.3. Gene expression analysis
Spleens were harvested from 20-24-wks old diabetic mice and 26-28-wks old mice treated with TSA from 16 to 24-wks of age. Total RNA was extracted from the spleen and pancreas of five individual mice per group/experiment, treated with Turbo DNase, and converted to cDNA using High-capacity cDNA Reverse Transcription kit (Applied Biosystems), as described [[10], [11], [14], [15], [16], 20]. Real-time qRT-PCR was performed in triplicate on Applied Biosystems ViiA7 Real-time PCR system using 1 µl of cDNA equivalent to 100 ng of total RNA and 2X SYBR Green master mix. The primers used in this study were designed and validated previously [[10], [11], [14], [15], [16], 20] and synthesized at the Integrated DNA Technologies (Coralville, IA). The level of specific gene expression in each sample was ascertained using Gapdh as the normalizer and the 2−∆∆CT method.
2.4. Adoptive transfer of diabetes
Spleens were harvested from 20-24-wks old diabetic mice and 26-28-wks old mice treated with TSA between 16 to 24-wks of age. Splenocytes (10×106/ml) were cultured with 5 µg of Concanavalin A (Sigma Aldrich) for two days as described [19]. More than 95% of these cells were CD3+, as determined by flow cytometry. Splenocytes were also stained with PE-Cyanine 5 conjugated anti-CD11b (clone M1/70, eBiosciences) and anti-Ly-6C-FITC antibodies and sorted on a MoFlo Astrios cell sorter, which yielded 2.27 % of the input with > 98% CD11b+Ly-6C+ cells. Both male and female NOD.scid mice were injected i.v. with mitogen-activated 2×106 T lymphocytes with or without sorted 2×105 CD11b+Ly-6C+ cells or purified monocytes alone. Splenocytes were also stained with anti-Ly-6C-FITC and incubated with anti-FITC antibody and purified on a magnetic column (Miltenyi Biotec Inc., Auburn, CA, yielding 2 % of the input with >95% Ly-6C+ cells, as determined by flow cytometry. Female wild-type NOD mice received two i.v. injections of purified 2×105 Ly-6C+ cells at 13 and 21-wksof age.
2.5. Statistics
The difference in diabetes incidence between controls (untreated/DMSO-treated mice) and those treated with TSA was analyzed for statistical significance using the Wilcoxon Signed Rank Test. Gene expression data were analyzed for statistical significance between indicated groups by two-way ANOVA or two-tailed unpaired t-test using the GraphPad Prism (6.0) software and indicated in individual figure legends. A P-value of <0.05 was considered significant.
3. Results
3.1. Protection from T1D is associated with the constrained influx of inflammatory cells into the islets
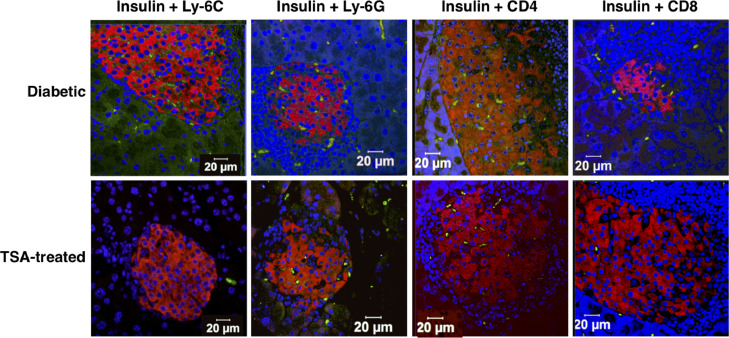
We have consistently observed that 80-100% of 6-8-wks old female NOD mice procured from The Jackson Laboratory and housed in our animal facility developed T1D by 18-wks of age [[10], [11], [12], [13], 20]. Weekly treatment with TSA starting at 16-wks and until 24-wks of age substantially attenuated T1D (Supplementary Fig. 1). Injection of the vehicle DMSO failed to alter the course or intensity of the disease [10], [11], [12], [13]. We have also documented that DMSO treatment did not change gene expression compared to untreated diabetic mice. Since DMSO-treated mice did not display noticeable changes in disease course or gene expression compared to untreated diabetic mice, we compared the litter mates of sex- and age-matched, untreated diabetic mice with TSA-treated mice in all experiments. Treatment with TSA reduced the invasive cellular infiltration of the islets and reversed moderate to severe loss of insulin-producing beta cells [10]. However, we did not characterize the nature of cellular infiltration previously. To this end, pancreatic sections were analyzed by confocal microscopy, which indicated the distribution of Ly-6C+ macrophages, Ly-6G+ neutrophils, CD4+, and CD8+ T-cells across the entire area of the diabetic pancreas (Fig. 1). Treatment with TSA significantly reduced the infiltration of primarily CD4+ cells and, to some extent, CD8+ T-cells implicated in T1D [17]. The influx of Ly-6C+ macrophages but not neutrophils was also reduced by TSA treatment, consistent with a role for macrophages in diabetes [18], [19]. Flow cytometric analysis confirmed that the numbers of CD11b+Ly-6C+ monocytes but not CD11b+F4/80+ macrophages or CD11c+ dendritic cells were also diminished in the spleen after TSA treatment (Supplementary Fig. 2). These data indicated that reduced influx of CD4+ and CD8+ T cell subsets and Ly-6C+ macrophages into the islets is a mechanism of TSA-mediated amelioration of autoimmune diabetes.
Fig. 1.
Cellular infiltration in the pancreas was retarded by TSA treatment.
Diabetic mice and those treated with TSA during 16 and 24-wks of age were sacrificed when they were 26-28-wks old. Sections of the pancreas were stained with guinea pig antisera against insulin followed by TRITC labeled rabbit anti-guinea pig antibody (red), along with FITC-conjugated anti-Ly-6C, anti-Ly-6G, anti-CD4, or anti-CD8 antibodies (green). After counterstaining with Hoechst (blue), the sections were analyzed on a confocal microscope. Representative images from multiple experiments are shown.
3.2. TSA treatment selectively repressed the expression of Th1 and Th17 lymphokines, primarily in the spleen
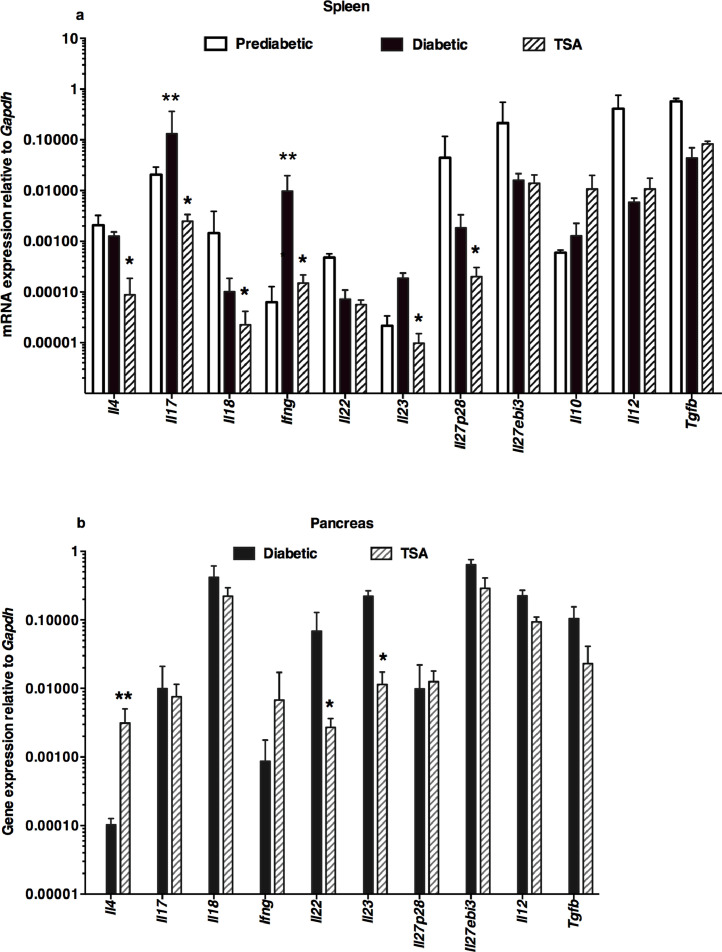
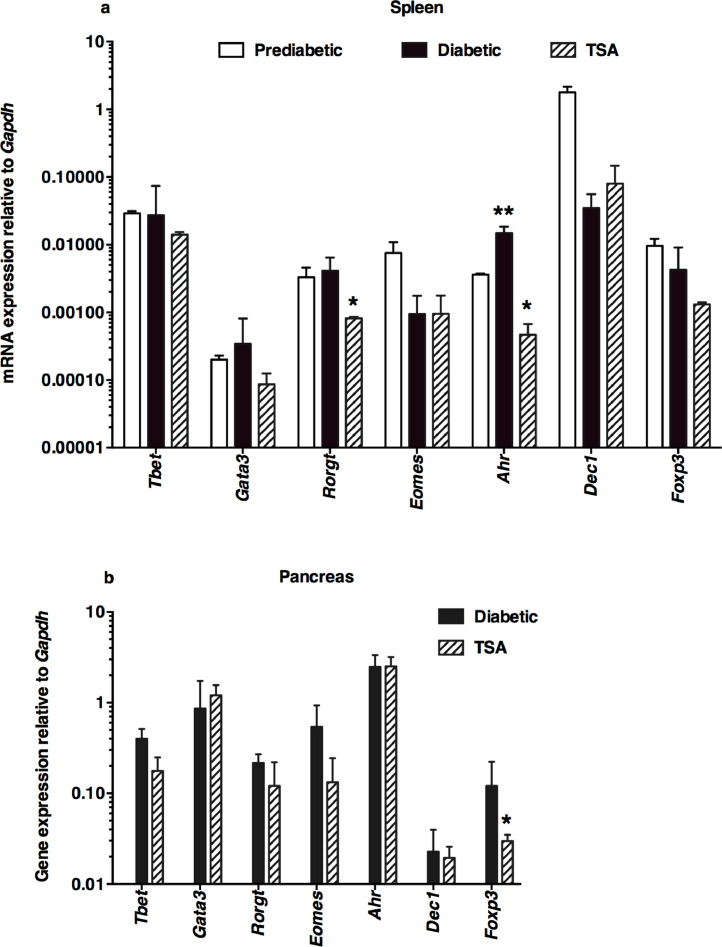
Our previous transcriptome analysis of uninduced splenocytes indicated the downregulation of a few inflammatory genes, including Cela3b (elastase) by TSA treatment [11]. In the current investigation, we analyzed a total of 43 genes selected for their putative roles in T1D. These included genes encoding 11 lymphokines, 13 accessory cell-associated determinants, 7 transcription factors, a chemokine, and 11 genes encoding HDACs. Total RNA was extracted from diabetic and TSA-treated animals, converted to cDNA, and used as the template in the qRT-PCR assay, as described before [10], [11], [12], [13], [14], [15], [16]. The genes critical for Th17 cell development, namely Il17a and Il23 [21], [22], and the Th1-specific Ifng [23], were significantly increased in the spleen of diabetic mice compared to the prediabetic mice displaying normal levels of blood glucose (Fig. 2a). Significantly, TSA treatment diminished the expression levels of these genes exaggerated during diabetes. Although Il4, Il18, and Il27p28 were not upregulated during diabetes, they were nevertheless repressed by the histone modifier treatment. However, other genes such as Il22, Il27ebi3, Il10, Il12, and Tgfb were neither overexpressed during diabetes nor modulated by the histone modifier, indicating that these genes are less likely to participate in diabetogenesis in this model.
Fig. 2.
Differential modulation of lymphokine genes by the histone modifier
Spleen was harvested from 12-wks old prediabetic mice and 25-26-wks old diabetic mice. The pancreas was collected from 25-26-wks old diabetic mice. Mice treated with TSA between 16 and 24-wks of age were sacrificed when they were 25-26-wks old and the spleen and pancreas were harvested. Five mice per group were tested in three different experiments. Total RNA was extracted from the spleen (a) and pancreas (b) of five individual mice, converted to cDNA, and used as the template in qRT-PCR. Data indicate the mean +/- SD. Representative data from three independent experiments are depicted. Asterisks indicate statistical significance in the spleen samples from prediabetic, diabetic, and TSA-treated mice, as assessed by two-way ANOVA (a) and by two-tailed unpaired t-test between diabetic and TSA-treated pancreas (b) (P<0.05). Double asterisks indicated increased values between prediabetic and diabetic mice, whereas a single asterisk denoted the decrease in TSA-treated mice compared to diabetic mice.
Preliminary data indicated that the expression of the genes analyzed herein remained comparable in the pancreas of prediabetic and overtly diabetic mice. Therefore, the gene expression levels in the pancreas of diabetic and TSA-treated mice were compared. Unlike in the spleen, TSA treatment increased the level of Il4 but repressed the transcription of Il22 and Il23 in the pancreas (Fig. 2b). Collectively, these results suggest that TSA treatment that provided protection against diabetes differentially regulated the expression of both exaggerated and constitutively expressed immune response-related genes in the spleen and pancreas.
3.3. Histone modifier regulates the steady-state levels of macrophage-associated cytokines primarily in the spleen
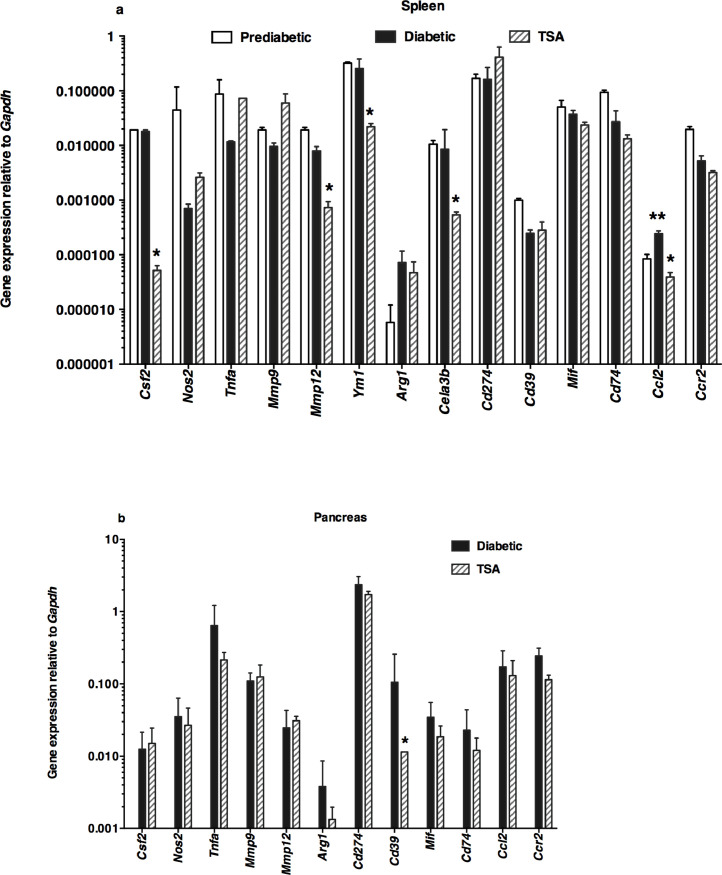
The chemokine CCL2, implicated in the mobilization of myeloid cells from the bone marrow causing insulitis [24], was upregulated in the spleen of diabetic mice, consistent with a role in diabetes (Fig. 3a). The transcription of CCL2 was repressed by TSA treatment. Although the macrophage-selective genes such as Csf2, Mmp12, and Ym1 were not exaggerated during diabetes, they were nevertheless reduced by the histone modifier. Significantly, Cela3b expression was lower in TSA-treated spleens, as shown earlier [11]. Only the steady-state level of Cd39 was subject to regulation by the HDAC inhibitor in the pancreas (Fig. 3b). These data suggest that TSA treatment predominantly diminished the constitutive expression of more macrophage-selective genes in the spleen than the pancreas.
Fig. 3.
Differential regulation of accessory cell-associated determinants following TSA treatment
The expression levels of genes associated with accessory cells were analyzed in (a) the spleen and (b) the pancreas. Gene expression was analyzed by qRT-PCR using the same cDNA preparations as shown in Fig. 2. Data indicate arithmetic mean +/- SD of three determinations. Representative data from three independent experiments are depicted (n=5 mice per group/experiment). Asterisks indicate statistical significance in the spleen samples from prediabetic, diabetic, and TSA-treated mice, as assessed by two-way ANOVA (a) and by two-tailed unpaired t-test between diabetic and TSA-treated pancreas (b) (P<0.05). Increased values between prediabetic and diabetic mice were indicated by double asterisks, whereas the decrease in values in TSA-treated mice compared to diabetic mice was denoted by a single asterisk.
3.4. Epigenetic drug impacts the expression of transcription factors sparingly in the spleen
Notably, the transcription factor Ahr involved in nitric oxide and arginine production, and alteration of M1/M2 macrophage polarization [25] were transcriptionally increased in the spleen of diabetic mice and reduced following TSA treatment (Fig. 4a). Only the constitutive expression of Rorgt, crucial for the transcription of IL-17A [26], was repressed by TSA treatment. Surprisingly, Tbet and Gata3, respectively involved in the transcription of IFN-γ and IL-4, and other transcription factors such as Eomes, Dec1, and Foxp3 examined were neither transcriptionally upregulated during diabetogenesis nor subject to epigenetic regulation. The level of the T regulatory cell-selective transcript Foxp3 remained unaltered in the spleen of TSA-treated mice (Fig. 3a), consistent with the lack of influence on FoxP3+ T regulatory cells observed earlier [10]. Interestingly, TSA treatment reduced the level of Foxp3 transcript in the pancreas (Fig. 4b). These data suggest that TSA-mediated protection against diabetes is associated with selective repression of the transcription factors Ahr and Rorgt in the lymphoid organ without altering the transcription of the T regulatory-selective Foxp3 or other transcription factors examined.
Fig. 4.
Selective regulation of the transcription factor genes by TSA
The expression levels of genes encoding various transcription factors were analyzed in (a) the spleen and (b) pancreas by qRT-PCR using the same cDNA preparations assayed in Fig. 2 and 3. Data indicate the mean +/- SD of three determinations. Representative data from three independent experiments are depicted (n=5 mice per group/experiment). Double asterisks indicate a statistically significant increase between prediabetic and diabetic mice (*P<0.05), whereas a single asterisk denotes a decrease in the value between diabetic and TSA-treated groups (**P<0.01). Statistical significance was assessed respectively by two-way ANOVA (a) and two-tailed unpaired t-test between diabetic and TSA-treated pancreas (b) as recommended by the GraphPad Prism software.
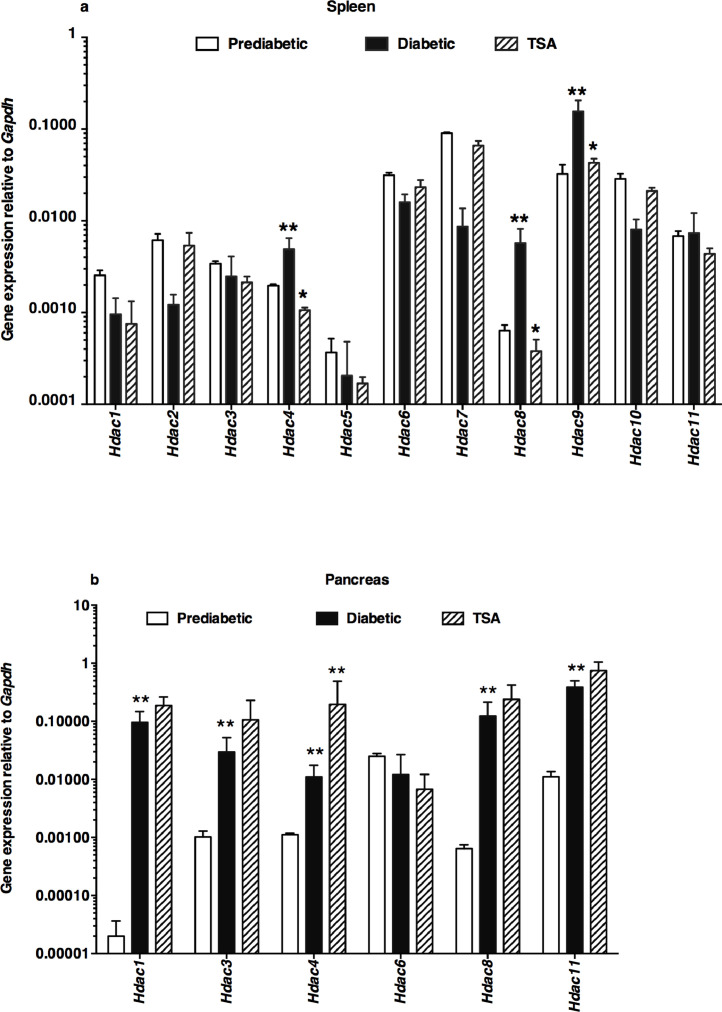
3.5. Epigenetic modifier differentially modulated HDACs in the spleen and pancreas
Treatment of NOD mice with TSA induced the hyperacetylation of histone H3 in the spleen and pancreas of NOD mice [10]. However, it is unclear whether it can lead to selective expression of class I (Hdac1, Hdac2, Hdac3, and Hdac8), class IIa (Hdac4, Hdac5, Hdac7, and Hdac9), class IIb (Hdac6 and Hdac10), or class IV (Hdac11) HDAC genes in the lymphoid system and the target organ pancreas. In the spleen of prediabetic, normoglycemic, 12-wks old female NOD mice, the basal levels of the Hdac5 and Hdac8 were minimal (Fig. 5a). During diabetes manifestation, the transcription of Hdac4, Hdac8, and Hdac9 genes was increased, which was repressed by TSA treatment. Interestingly, Hdac1, Hdac3, Hdac4, Hdac8, and Hdac11 were upregulated in the pancreas of diabetic mice (Fig. 5b). Whereas most of these genes were not repressed by the histone modifier, Hdac4 was upregulated in drug-treated mice. Notably, Hdac2, Hdac5, Hdac7, Hdac9, and Hdac10 genes could not be reliably detected in the pancreas of prediabetic, overtly diabetic, or TSA-treated mice by qRT-PCR in repeated experiments (n=25-30 mice). Thus, TSA-mediated protection against T1D was accompanied by a reduction of the exaggerated expression of Hdac4, Hdac8, and Hdac9 genes in the spleen, while Hdac4 was upregulated in the pancreas.
Fig. 5.
Differential modulation of Hdac genes by the epigenetic drug
The expression levels of Hdac genes in the spleen (a) and pancreas (b) were determined in cDNA samples illustrated in Fig. 2-4. Data indicate the mean +/- SD of three determinations. Representative data from three independent experiments are depicted (n=5 mice per group/experiment). Double asterisks indicated an increase in the values between prediabetic and diabetic mice. The decrease in values in TSA-treated mice compared to diabetic mice was denoted by a single asterisk. Statistical significance was assessed using two-way ANOVA (a) and by a two-tailed unpaired t-test between diabetic and TSA-treated pancreas (b). *P<0.05 and **P<0.01.
3.6. TSA treatment differentially affected the function of T lymphocytes and macrophages
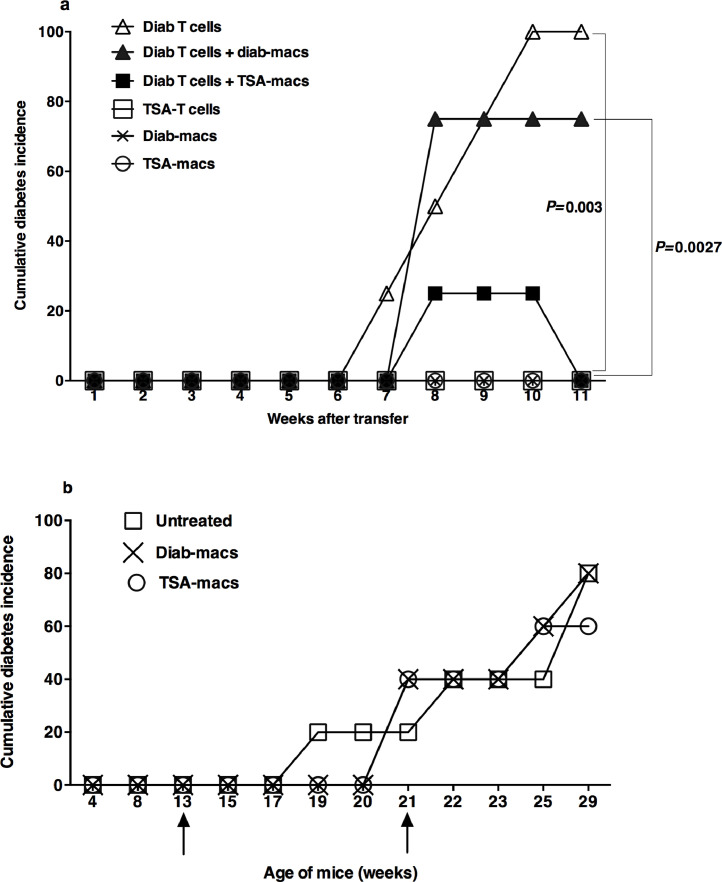
We next examined whether epigenetically modulated genes could impact the diabetogenic potential of T lymphocytes and the presumptive effector function of macrophages in diabetes [18], [19]. To this end, we transferred activated T-cells and sorted macrophages from diabetic mice and those treated with TSA and cured of diabetes, individually or in combination, into the immunodeficient NOD.scid mice. The data shown in Fig. 6a indicate that the transfer of Concanavalin A-activated T lymphocytes from diabetic mice induced diabetes in 100% of the immunodeficient NOD.scid recipients by 9-wks, similar to an earlier report [19]. This observation is consistent with the possibility that activated T-cells can cause diabetes independent of endogenous macrophages since NOD.scid mice lack T-cells, B-cells, and functionally competent macrophages [27]. However, similarly activated T lymphocytes derived from TSA-treated mice failed to induce the disease in NOD.scid recipients, consistent with the data obtained using whole splenocytes [11]. These results indicate that epigenetic modulation of the genome abolished the diabetogenic potential of T lymphocytes.
Fig. 6.
Epigenetic regulation of T cells and macrophages
(a) Diabetes was monitored in NOD.scid mice (10-12-wks old) injected i.v. with Concanavalin A-activated 2×106 T lymphocytes from diabetic or TSA-treated NOD mice (26-28-wks of age) (P=0.0027). Mice were also co-transferred with mitogen-activated diabetic T-cells and 2×105 sorted CD11b+Ly-6C+ macrophages from the spleens of diabetic mice or those treated with TSA and diabetes monitored (P=0.003). Some mice received only macrophages isolated from diabetic mice or TSA-treated mice. Four mice per group were tested. Representative data from two independent experiments are shown. Statistical significance between specified groups was assessed using a two-tailed, unpaired t-test. (b) Wild-type female NOD mice were injected with purified 2×105 Ly6-C+ macrophages from diabetic mice or those treated with TSA at 13 and 21-wks of age, as indicated by the arrows. Diabetes was monitored weekly. Five mice per group were tested. The data shown are from two independent experiments.
Interestingly, the adoptive transfer of sorted CD11b+Ly-6C+ macrophages from diabetic or TSA-treated mice alone failed to cause diabetes in the immunodeficient mice, indicating that the macrophages are not the effectors of T1D. This observation is at variance with the proposal that macrophages are the effectors of diabetes since cytotoxicity of macrophages was observed against the beta cells in vitro [19]. Surprisingly, the co-transfer of CD11b+Ly-6C+ macrophages purified from the TSA-treated but not diabetic mice reduced the frequency of T-cell mediated diabetes in NOD.scid recipients. These data suggest that the epigenetically modulated macrophages could block the execution of diabetes by T lymphocytes in an immunodeficient environment. However, the transfer of purified splenic Ly6-C+ macrophages from TSA-treated mice during the prediabetic stage and shortly after the onset of diabetes failed to impede the disease progression in wild-type NOD mice (Fig. 6b). This is in stark contrast to diabetes protection afforded by the neonatal transfer with M2-type macrophages polarized from the bone marrow in vitro [28]. These data suggest that although the TSA-treated macrophages can directly suppress the diabetogenic potential of T lymphocytes in an immunodeficient environment, they fail to retard the progression of diabetes in unmanipulated, immunocompetent wild-type NOD mice. Thus, the transfer of TSA-treated macrophages is insufficient to quell the overwhelming effects of diabetogenic T lymphocytes in immunocompetent mice.
4. Discussion
Although many intervention strategies have been reported to delay the onset of diabetes in female NOD mice, the success of translating these studies into T1D treatment remains grim [29]. We have reported a novel strategy in which weekly administration of a low dose of the potent epigenetic drug TSA between 18 and 24-wks of age bestowed irreversible diabetes protection in female NOD mice [10], [11], [12], [13]. The immunosuppressive regimens currently used for treating T1D patients affect the T-cells and inflammatory cells indiscriminately. The data presented herein indicate that the action of the histone modifier is selective. This is evidenced by diminished trafficking patterns of CD4+ and CD8+ T-cells into the pancreas (Fig. 1), without decreasing their numbers [10]. Interestingly, the drug treatment reduced the numbers of CD11b+Ly-6C+ macrophages and their infiltration into the pancreas while sparing the CD11b+F4/80+ macrophages and CD11c+ dendritic cells (Supplementary Fig. 2). Moreover, the histone modifier increased the numbers of the CD4+ + CD62L+ but not the CD4+ + FoxP3+ T regulatory cells [10]. Interestingly, TSA treatment endowed the ability of CD11+Ly-6C+ macrophages to block the diabetogenic potential of activated T lymphocytes co-transferred in an immunodeficient environment (Fig. 6a). Importantly, epigenetic programming deprived the ability of splenocytes [11], activated T lymphocytes (Fig. 6a) [30], and polarized Th1 and Th17 cells [30] to adoptively transfer diabetes into immunodeficient NOD.scid mice. These data attest to the selective nature of the small molecule inhibitor on immune cells leading to robust T1D control.
Another critical point unraveled is that the histone modifier influenced the expression of only a small [61] number of genes out of 43 examined, regardless of whether they were induced or remained unaltered in the peripheral lymphoid tissues and pancreas during diabetogenesis (Fig. 2-5). Earlier, we showed that protection from diabetes was associated with H3 hyperacetylation in both the spleen and pancreas, resulting in altered gene expression [10]. Notably, we observed that the activation of drug-treated splenic T lymphocytes resulted in exaggerated IFN-γ transcription [10], indicating a protective in T1D. In contrast, unseparated and uninduced splenocytes revealed lower levels of Ifng expression following TSA treatment (Fig. 2a), suggesting a pathogenic role instead. The discrepancy between these results could be because the steady-state level of IFN-γ in uninduced whole splenocytes reflects Ifng expression not only by CD4+ cells but also by CD8+ and NK-T cells. These data are in line with the contradictory roles of IFN-γ in T1D unraveled by previous studies. Although IFN-γ-producing Th1 cells were shown to mediate diabetes in neonatal NOD mice [31], the genetic absence of IFN-γ delayed but did not prevent diabetes [32]. Paradoxically, a protective role of IFN-γ in diabetes was also reported [33], [34]. Collectively, these data reiterate that most of the expression levels of genes observed during diabetes could be due to the effect rather than the cause of the disease.
Our analysis indicated the possible contribution of IL-17A to diabetes as reported [35], but the Th17 cells were required to convert to Th1 cells to cause diabetes in immunodeficient NOD.scid mice [36], [37]. Yet others have shown that T1D depends on both IL-17 and IFN-γ signaling [38]. Our data showing the repression of both Il17a and Ifng in TSA-treated mice (Fig. 2a) [30] that attained normoglycemia supports this contention. A closer analysis unraveled that the TSA treatment prevented the upregulation of Il23, required for the generation of Th17 cells [22] (Fig. 2a). In addition, TSA treatment also diminished the steady-state levels of several genes implicated in T1D such as Ccl2 [24], Il4 [39], Il18 [40], and Il27p28 [41] in the spleen (Fig. 2a, Fig. 3a). A novel finding is that the gene encoding the transcription factor Ahr [25] was accentuated in the spleen during diabetes and repressed by the histone modifier (Fig. 4a). In addition, the basal level expression of the transcription factor Rorgt, crucial for IL-17A transcription [26], was suppressed in the spleen of drug-treated mice (Fig. 4a). Interestingly, Il22, Il23 (Fig. 2b), and Cd39 (Fig. 3b) implicated in diabetes [42] were similarly repressed in the pancreas of mice cured of diabetes. Interestingly, Foxp3, expressed by the T regulatory cells crucial for immunoregulation [43], was decreased in the pancreas after TSA treatment (Fig. 4b), indicating that the regulation of T1D is independent of CD4+FoxP3+ cells, as we reported earlier [10]. This is similar to a model of T1D delay mediated by the administration of the migration inhibitory factor antagonist that repressed FoxP3 expression [44]. Overall, our data provide supporting evidence for the contribution of previously known genes such as Il17a and Ccl2, and importantly Il23 and Ahr in diabetes manifestation, more so in lymphoid tissues than in the pancreas. These data reiterate the notion that genes upregulated during diabetogenesis may be the effect rather than the cause of diabetes. Further work is necessary to parse the causative role of these genes in autoimmune diabetes.
For the first time, we show herein that diabetogenesis involves exaggerated expression of Hdac4, Hdac8, and Hdac9 genes in the secondary lymphoid organ, which was repressed by the histone modifier (Fig. 5a). Although many Hdac genes were also exaggerated in the pancreas of diabetic mice, surprisingly, they were not sensitive to TSA treatment (Fig. 5b). This was not due to the lack of access of the drug to the pancreas as TSA treatment increased Hdac4 expression. Thus, the Hdac genes are differentially regulated by histone hyperacetylation in the lymphoid tissue vs. the target organ, consistent with the active role of T-cells in diabetes induction. Interestingly, we noted that the Hdac3 gene was not repressed either in the spleen or pancreas by low doses of TSA (Fig. 5a-b), which is at odds with the finding that administering large amounts of the HDAC3 selective inhibitor BRD3308 conferred marginal protection against diabetes in NOD mice [45]. However, BRD3308 reduced cytokine-induced apoptosis of dispersed primary islets in vitro and improved glycemia and insulin secretion in obese diabetic rats [46], [47], [48]. Whether the BRD3308 inhibitor can exert immunosuppressive effects as TSA [10], [11], [12], [13], [14], [15], [16] (Fig. 1-6) remains undetermined. Interestingly, TSA blocked the inflammatory cytokine-mediated beta-cell toxicity in vitro more effectively than the BRD3308 inhibitor [48]. Since TSA is endowed with both immunomodulatory [[10], [11], [12], [13], [14], [15], [16], Fig. 1-6] and beta-cell protective effects [48], it seems to be an ideal choice of drug for treating autoimmune diabetes with an extremely low dose regimen compared to a large amount of the HDAC3-selective BRD3308 inhibitor required to exert marginal protection [45].
The diminished diabetogenic potential of whole splenocytes [10] and the enriched T lymphocytes (Fig. 6a) [30] by epigenetic reprogramming reiterates the importance of T-cells in diabetes. In addition to T lymphocytes, macrophages had been proposed to play a crucial role during the effector phase of T1D [18], [19]. Both antigen presentation to cytotoxic CD8+ T cells [18] and direct cytotoxicity on dispersed islet cells in vitro [19] have been attributed to macrophages. However, the actual participation of macrophages in diabetogenesis in vivo has been challenging to prove. We observed that the CD11b+Ly-6C+ macrophages from diabetic mice failed to transfer diabetes to immunodeficient mice, indicating that the macrophages do not serve as effectors of T1D (Fig. 6a), unlike previously proposed [19]. Unexpectedly, we found that TSA treatment endowed the CD11b+Ly-6C+ macrophages with the ability to suppress diabetes upon co-transfer with diabetogenic T cells into NOD.scid recipients (Fig. 6a). However, the transfer of Ly-6C+ macrophages derived from TSA-treated mice failed to suppress ongoing diabetes in wild-type NOD mice (Fig. 6b). This paradoxical finding appears similar to the failure of polarized Th2 [39] and Th17 cells [36] derived from the BDC2.5 TCR transgenic mice to transfer diabetes into neonatal wild-type NOD mice while readily transferring the disease to immunodeficient NOD.scid mice. Similarly, we observed that the transfer of macrophages from TSA-treated mice could only suppress diabetes when co-transferred with activated T-cells into NOD.scid mice and not into wild-type NOD mice (Fig.6a, b). These intriguing findings could be attributed to the lack of endogenous immunoregulatory mechanisms in immunodeficient mice facilitating the action of transferred cells without opposition, unlike the immunocompetent wild-type NOD mice. Our data are also different from diabetes suppression in NOD mice mediated by the injection of M2-type macrophages polarized in vitro from bone-marrow-derived monocytes using a cocktail of IL-4/IL-10/TGF-β [28]. Apparently, TSA treatment could not convert the macrophages into M2-type macrophages, as described [28]. Therefore, this strategy, transfer of macrophages from TSA-treated mice, may not be helpful for T1D treatment. Although histone hyperacetylation has been proposed to impact monocytes and macrophages central to tissue homeostasis and immune responses [49], [50], little evidence exists about the functional modulation of macrophages under diabetic conditions. Previously, we observed that NOD mice treated with TSA and cured of T1D displayed a lower level of the pro-inflammatory gene Cela3b (elastase 3) [11], which was validated herein (Fig. 3a). Moreover, TSA treatment downregulated macrophage-specific genes such as Csf2, Mmp12, and Ym1. Further studies are required to determine whether these changes could contribute to the suppressive function of macrophages observed following transfer into the lymphopenic NOD.scid mouse.
Conclusions
The most potent HDAC inhibitor, TSA, alters the ‘histone code’ within 2-4 hr of drug treatment in vitro, which results in up-and down-regulation of a small set of inducible genes (2%) without affecting the house-keeping gene, GAPDH, leading to a variable transcriptional outcome in human cancer cells [51], [52], [53]. The selectivity of TSA treatment was further indicated by the selective repression of genes encoding CD25, CD154, and CD28 in activated primary human CD4+ cells [54], [55]. Therapeutic benefits of TSA were indicated by the reduction of overexpressed CD154 and IL-10 and restoration of IFN-γ levels in T helper cells of systemic lupus erythematosus patients in vitro [56]. Previous investigations indicated selective action on CD4+ T cells in animal models of lupus [57], colitis [58], multiple sclerosis [14], [15], [16], and asthma [59]. In addition, neutrophils [15] and antibody-producing B-cells [60] were respectively diminished in EAE and lupus mice by TSA treatment. Collectively, these data are consistent with the possibility that abnormally expressed genes in tumors and autoimmune diseases are primarily targeted by the HDAC inhibitor TSA. The data presented herein and elsewhere [[10], [11], [12], [13], 30] indicate that the histone modifier mediated protection against T1D involves the modified expression of a select set of immune response-related genes such as Il4, Il17a, Il18, Il23, Il27p28, Ifng, Ccl2, Rorgt, and Ahr in the spleen, and Il22, Il23, Cd39, and Foxp3 in the pancreas. Notably, the macrophage-specific Csf2, Mmp12, Ym1, Cela3b, and Ccl2 were downregulated in the spleen while Cd39 was suppressed in the pancreas. Interestingly, Hdac4, Hdac8, and Hdac9 were subject to regulation by the histone modifier in the spleen, and none of the 11 Hdac genes were regulated in the pancreas of TSA-treated mice. Compared to the spleen, the relatively small proportion of the genes regulated by TSA treatment in the pancreas is consistent with a more active role of lymphoid cells in the induction and manifestation of autoimmune diabetes than the target organ pancreas. This is at variance with the view that the pancreas has an active role in manifestion of autoimmune diabetes [61]. Although the small number of genes regulated by TSA in the peripheral lymphoid tissues and the pancreas could serve as biomarkers of T1D [11], [12], [13], the peripheral blood is easily accessible to glean insights into diabetes pathogenicity. Further research is required to parse the genes that cause autoimmune diabetes and those act as bystanders.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Arathi Jayaraman: Methodology, Writing – review & editing. Maria Arianas: Methodology, Writing – review & editing. Sundararajan Jayaraman: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Mark Holterman is acknowledged for the support of this work. The expert help of the Imaging Core and Flow Cytometry Facility at the University of Illinois at Chicago in acquiring confocal images and high-speed sorting of macrophages, respectively, is acknowledged.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2021.100031.
Appendix. Supplementary materials
References
- 1.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2007;30(Suppl 1):S42–S47. doi: 10.2337/dc07-S042. [DOI] [PubMed] [Google Scholar]
- 2.Hyttinen V., Kaprio J., Kinnunen L., Koskenvuo M., Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52:1052–1055. doi: 10.2337/diabetes.52.4.1052. [DOI] [PubMed] [Google Scholar]
- 3.Shaw J.E., Sicree R.A., Zimmet PZ P.Z. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res. Clin. Pract. 2010;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Todd J.A. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 6.Bird A. Genetic determinants of the epigenome in development and cancer. Swiss Med. Wkly. 2017;147:w14523. doi: 10.4414/smw.2017.14523. [DOI] [PubMed] [Google Scholar]
- 7.Jayaraman A., Jayaraman S. DNA hypermethylation does not negatively impact the transcription of the TNF-α gene in an acute T-cell leukemia. Epigenomics. 2019;11:1753–1763. doi: 10.2217/epi-2019-0015. [DOI] [PubMed] [Google Scholar]
- 8.Micelli C., Rastelli G. Histone deacetylases: structural determinants of inhibitor selectivity. Drug Discov. Today. 2015;20:718–735. doi: 10.1016/j.drudis.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 9.de Ruijter J., van Gennip A.H., Caron N.H., Kemp S., van Kuilenburg A.B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel T., Patel V., Singh R., Jayaraman S. Chromatin remodeling resets the immune system to protect against autoimmune diabetes in mice. Immunol. Cell Biol. 2011;89:640–649. doi: 10.1038/icb.2010.144. [DOI] [PubMed] [Google Scholar]
- 11.Jayaraman S., Patel A., Jayaraman A., Patel V., Holterman M., Prabhakar B B. Transcriptome analysis of epigenetically modulated genome indicates signature genes in manifestation of type 1 diabetes and its prevention in NOD mice. PLoS One. 2013;8:e55074. doi: 10.1371/journal.pone.0055074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayaraman S. Novel methods of type 1 diabetes treatment. Discov. Med. 2014;17:347–355. PMID: 24979255. [PubMed] [Google Scholar]
- 13.Jayaraman S. In: The epigenetics of autoimmunity. Zhang R., editor. Vol. 5. Elsevier Inc; Amsterdam, The Netherlands: 2018. Type 1 diabetes and epigenetics; pp. 188–205. (Translational Epigenetics Series). [Google Scholar]
- 14.Jayaraman A., Soni A., Prabhakar B.S., Holterman M., Jayaraman S. The epigenetic drug Trichostatin A ameliorates experimental autoimmune encephalomyelitis via T cell tolerance induction and impaired influx of T cells into the spinal cord. Neurobiol. Dis. 2017;108:1–12. doi: 10.1016/j.nbd.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Jayaraman A., Sharma M., Prabhakar B., Holterman M., Jayaraman S. Amelioration of progressive autoimmune encephalomyelitis by epigenetic regulation involves selective repression of mature neutrophils during the preclinical phase. Exp. Neurol. 2018;304:14–20. doi: 10.1016/j.expneurol.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman A., Avgush K., Kulam R., Soni A., Khan A., Kerdjoudj M., Jayaraman S. Treatment of autoimmune encephalomyelitis with a histone deacetylase inhibitor. Analyzing the role of immune-response genes. Free Neuropathology. 2020;1:19. doi: 10.17879/freeneuropathology-2020-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christianson S.W., Shultz L.D., Leiter E.H. Adoptive transfer of diabetes into immunodeficient NOD-scid/scid mice. Relative contributions of CD4+ and CD8+ T-cells from diabetic versus prediabetic NOD.NON-Thy-1a donors. Diabetes. 1993;42:44–55. doi: 10.2337/diab.42.1.44. [DOI] [PubMed] [Google Scholar]
- 18.Jun H.S., C.S Yoon, Zbytnuik L., van Rooijen N., Yoon J.W. The role of macrophages in T cell-mediated autoimmune diabetes in nonobese diabetic mice. J. Exp. Med. 1999;189:347–358. doi: 10.1084/jem.189.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calderon B., Suri A., Unanue E.R. In CD4+ T-cell-induced diabetes, macrophages are the final effector cells that mediate islet beta-cell killing: studies from an acute model. Am. J. Pathol. 2006;169:2137–2147. doi: 10.2353/ajpath.2006.060539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayaraman S., Patel T., Patel V., Ajani S., Garza R., Jayaraman A., Kwon S., Singh R., Rondelli D., Prabhakar B.S., Holterman M. Transfusion of nonobese diabetic mice with allogeneic newborn blood ameliorates autoimmune diabetes and modifies the expression of selected immune response genes. J. Immunol. 2010;184:3008–3015. doi: 10.4049/jimmunol.0903615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaffen S.L. Recent advances in the IL-17 cytokine family. Curr. Opin. Immunol. 2011;23:613–619. doi: 10.1016/j.coi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGeachy M.J., Cua D.J. The link between IL-23 and Th17 cell-mediated immune pathologies. Semin. Immunol. 2007;19:372–376. doi: 10.1016/j.smim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Skapenko A., Schulze-Koops H. Analysis of Th1/Th2 T-cell subsets. Methods Mol. Med. 2007;136:87–96. doi: 10.1007/978-1-59745-402-5_7. [DOI] [PubMed] [Google Scholar]
- 24.Martin A.P., Rankin S., Pitchford S., Charo I.F., Furtado G.C., Lira S.A. Increased expression of CCL2 in insulin-producing cells of transgenic mice promotes mobilization of myeloid cells from the bone marrow, marked insulitis, and diabetes. Diabetes. 2008;57:3025–3033. doi: 10.2337/db08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Climaco-Arvizu S., Domínguez-Acosta O., Cabañas-Cortés M.A., Rodríguez-Sosa M., Gonzalez F.J., Vega L., Elizondo G. Aryl hydrocarbon receptor influences nitric oxide and arginine production and alters M1/M2 macrophage polarization. Life Sci. 2016;155:76–84. doi: 10.1016/j.lfs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Piganelli J.D., Martin T., Haskins K. Splenic macrophages from the NOD mouse are defective in the ability to present antigen. Diabetes. 1998;47:1212–1218. doi: 10.2337/diab.47.8.1212. [DOI] [PubMed] [Google Scholar]
- 28.Parsa R., Andresen P., Gillett A., Mia S., Zhang X.M., Mayans S., Holmberg D., Harris R.A. Adoptive transfer of immunomodulatory M2 macrophages prevents type 1 diabetes in NOD mice. Diabetes. 2012;61:2881–2892. doi: 10.2337/db11-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoda L.K., Young D.L., Ramanujan S., Whiting C.C., Atkinson M.A., Bluestone J.A., Eisenbarth G.S., Mathis D., Rossini A.A., Campbell S.E., Kahn R., Kreuwel H.T. A comprehensive review of inventions in the NOD mouse and implications for translation. Immunity. 2005;23:115–126. doi: 10.1016/j.immuni.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Patel V., Jayaraman A., Jayaraman S. doi. bioRxiv; 2021. (Epigenetic reprogramming ameliorates type 1 diabetes by decreasing the generation of Th1 and Th17 subsets and restoring self-tolerance in CD4+ T cells). .07.24.453657. [DOI] [PubMed] [Google Scholar]
- 31.Katz J.D., Benoist C., Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- 32.Hultgren B., Huang X., Dybdal N., Stewart T.A. Genetic absence of gamma-interferon delays but does not prevent diabetes in NOD mice. Diabetes. 1996;45:812–817. doi: 10.2337/diab.45.6.812. [DOI] [PubMed] [Google Scholar]
- 33.Trembleau S., Penna G., Gregori S., Giarratana N., Adorini L. IL-12 administration accelerates autoimmune diabetes in both wild-type and IFN-gamma-deficient nonobese diabetic mice, revealing pathogenic and protective effects of IL-12-induced IFN-gamma. J. Immunol. 2003;170:5491–5501. doi: 10.4049/jimmunol.170.11.5491. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J., Huang Z., Sun R., Tian Z., Wei H. IFN-γ induced by IL-12 administration prevents diabetes by inhibiting pathogenic IL-17 production in NOD mice. J. Autoimmun. 2012;38:20–28. doi: 10.1016/j.jaut.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Bellemore S.M., Nikoopour E., Krougly O., Lee-Chan E., Fouser L.A., Singh B. Pathogenic T helper type 17 cells contribute to type 1 diabetes independently of interleukin-22. Clin. Exp. Immunol. 2016;183:380–388. doi: 10.1111/cei.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Orozco N., Chung Y., Chang S.H., Wang Y.H., Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur. J. Immunol. 2009;39:216–224. doi: 10.1002/eji.200838475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bending D., De la Peña H., Veldhoen M., Phillips J.M., Uyttenhove C., Stockinger B., Cooke A A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuriya G., Uchida T., Akazawa S., Kobayashi M., Nakamura K., Satoh T., Horie I., Kawasaki E., Yamasaki H., Yu L., Iwakura Y., Sasaki H., Nagayama Y., Kawakami A., Abiru N. Double deficiency in IL-17 and IFN-γ signalling significantly suppresses the development of diabetes in the NOD mouse. Diabetologia. 2013;56:1773–1780. doi: 10.1007/s00125-013-2935-8. [DOI] [PubMed] [Google Scholar]
- 39.Pakala S.V., Kurrer M.O., Katz J.D. T helper 2 (Th2) T cells induce acute pancreatitis and diabetes in immune-compromised nonobese diabetic (NOD) mice. J. Exp. Med. 1997;186:299–306. doi: 10.1084/jem.186.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marleau A.M., Sarvetnick N.E. IL-18 is required for self-reactive T cell expansion in NOD mice. J. Autoimmun. 2011;36:263–277. doi: 10.1016/j.jaut.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciecko A.E., Foda B., Barr J.Y., Ramanathan S., Atkinson M.A., Serreze D.V., Geurts A.M., Lieberman S.M., Chen Y.G. Interleukin-27 is essential for type 1 diabetes development and Sjögren syndrome-like Inflammation. Cell Rep. 2019;29:3073–3086. doi: 10.1016/j.celrep.2019.11.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chia J.S., McRae J.L., Cowan P.J., Dwyer K.M. The CD39-adenosinergic axis in the pathogenesis of immune and nonimmune diabetes. J. Biomed. Biotechnol. 2012;2012 doi: 10.1155/2012/320495.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hori S., Nomura T., Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 44.Korf H., Breser L., Van Hoeck J., Godoy J., Cook D.P., Stijlemans B., De Smidt E., Moyson C., Monteiro Carvalho Mori Cunha J.P., Rivero V., Gysemans C., Mathieu C. MIF inhibition interferes with the inflammatory and T cell-stimulatory capacity of NOD macrophages and delays autoimmune diabetes onset. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dirice E., Ng R.W.S., Martinez R., Hu J., Wagner F.F., Holson E.B., Wagner B.K., Kulkarni R.N. Isoform-selective inhibitor of histone deacetylase 3 (HDAC3) limits pancreatic islet infiltration and protects female nonobese diabetic mice from diabetes. J. Biol. Chem. 2017;292:17598–17608. doi: 10.1074/jbc.M117.804328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lundh M., Christensen D.P., Damgaard Nielsen M., Richardson S.J., Dahllöf M.S., Skovgaard T., Berthelsen J., Dinarello C.A., Stevenazzi A., Mascagni P., Grunnet L.G., Morgan N.G., Mandrup-Poulsen T. Histone deacetylases 1 and 3 but not 2 mediate cytokine-induced beta cell apoptosis in INS-1 cells and dispersed primary islets from rats and are differentially regulated in the islets of type 1 diabetic children. Diabetologia. 2012;55:2421–2431. doi: 10.1007/s00125-012-2615-0. [DOI] [PubMed] [Google Scholar]
- 47.Lundh M., Galbo T., Poulsen S.S., Mandrup-Poulsen T. Histone deacetylase 3 inhibition improves glycaemia and insulin secretion in obese diabetic rats. Diabetes Obes. Metab. 2015;17:703–707. doi: 10.1111/dom.12470. [DOI] [PubMed] [Google Scholar]
- 48.Wagner F.F., Lundh M., Kaya T., McCarren P., Zhang Y.L., Chattopadhyay S., Gale J.P., Galbo T., Fisher S.L., Meier B.C., Vetere A., Richardson S., Morgan N.G., Christensen D.P., Gilbert T.J., Hooker J.M., Leroy M., Walpita D., Mandrup-Poulsen T., Wagner B.K., Holson E.B. An isochemogenic set of inhibitors to define the therapeutic potential of histone deacetylases in β-cell protection. ACS Chem. Biol. 2016;11:363–374. doi: 10.1021/acschembio.5b00640. [DOI] [PubMed] [Google Scholar]
- 49.Das Gupta K, Shakespear M.R., Iyer A., Fairlie D.P., Sweet M.J. Histone deacetylases in monocyte/macrophage development, activation and metabolism: refining HDAC targets for inflammatory and infectious diseases. Clin. Transl. Immunology. 2016;5:e62. doi: 10.1038/cti.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roger T., Lugrin J., Roy D.Le, Goy G., Mombelli M., Koessler T., Ding X.C., Chanson A.L., Reymond M.K., Miconnet I., Schrenzel J., François P., Calandra T. Histone deacetylase inhibitors impair innate immune responses to 43 Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 51.Marks P.A., Richon V.M., Breslow R., Rifkind R.A. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 52.Van Lint C., Emiliani S., Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyeracetylation. Gene Expr. 1996;5:245–253. PMID: 8723390. [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi I., Miyaji H., Yoshida T., Sato S., Mizukami T. Selective inhibition of IL- 2 gene expression by trichostatin A, a potent inhibitor of mammalian histone deacetylase. J. Antibiot. 1996;49:453–457. doi: 10.7164/antibiotics.49.453. [DOI] [PubMed] [Google Scholar]
- 54.Moreira J.M., Scheipers P., Sørensen P. The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses. BMC Cancer. 2003;3:30. doi: 10.1186/1471-2407-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skov S., Rieneck K., Bovin L.F., Skark K., Tomra S., Michelsen B.K., Ødum N. Histone deacetylase inhibitors: a new class of immunosuppressors targeting a novel signal pathway essential for CD154 expression. Blood. 2003;101:1430–1438. doi: 10.1182/blood-2002-07-2073. [DOI] [PubMed] [Google Scholar]
- 56.Mishra N., Brown D.R., Olorenshaw I.M., Kammer G.M. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-gamma gene and protein expression in lupus T cells. Proc. Natl. Acad. Sci. USA. 2001;98:2628–2633. doi: 10.1073/pnas.051507098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mishra N., Reilly C.M., Brown D.R., Ruiz P., Gilkeson GS G.S. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J. Clin. Invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glauben R., Batra A., Fedke I., Zeitz M., Lehr H.A., Leoni F., Mascagni P., Fantuzzi G., Dinarello C.A., Siegmund B. Histone hyperacetylation is associated with amelioration of experimental colitis in mice. J. Immunol. 2006;176:5015–5022. doi: 10.4049/jimmunol.176.8.5015. [DOI] [PubMed] [Google Scholar]
- 59.Choi J-H., Oh S-W., Kang M-S., Kwon H.J., Oh G-T., Kim D-Y. Trichostatin A attenuates airway inflammation in mouse asthma model. Clin. Exp. Allergy. 2005;35:89–96. doi: 10.1111/j.1365-2222.2004.02006.x. [DOI] [PubMed] [Google Scholar]
- 60.Reilly C.M., Regna N., Mishra N. HDAC inhibition in lupus models. Mol. Med. 2011;17:417–425. doi: 10.2119/molmed.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roep BO, Thomaidou S, van Tienhoven R, Zaldumbide A Type 1 diabetes mellitus as a disease of the β-cell (do not blame the immune system?) Nat Rev Endocrinol. 2021;17:150–161. doi: 10.1038/s41574-020-00443-4. 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.