ABSTRACT
For over a century, physicians have witnessed a common enrichment of bifidobacteria in the feces of breast-fed infants that was readily associated with infant health status. Recent advances in bacterial genomics, metagenomics, and glycomics have helped explain the nature of this unique enrichment and enabled the tailored use of probiotic supplementation to restore missing bifidobacterial functions in at-risk infants. This review documents a 20-year span of discoveries that set the stage for the current use of human milk oligosaccharide-consuming bifidobacteria to beneficially colonize, modulate, and protect the intestines of at-risk, human milk-fed, neonates. This review also presents a model for probiotic applications wherein bifidobacterial functions, in the form of colonization and HMO-related catabolic activity in situ, represent measurable metabolic outcomes by which probiotic efficacy can be scored toward improving infant health.
KEYWORDS: Gut microbiota, human milk oligosaccharides, Bifidobacterium, probiotics, premature infants, neonatal intensive care unit
Consider the fragility of a typical human neonate. To say the infant is born naïve is a stark understatement. Humans are born with immature senses, nascent cognitive abilities, little muscle coordination, an undeveloped immune system, and limited ability to garner food more than reflexive behaviors (sucking, rooting, etc.). Consequently, early postpartum life for humans necessitates a bidirectional interaction within the mother-infant dyad to ensure a stable life trajectory for the infant. Given the overwhelming needs of the neonate ex utero, this stage truly represents a “fourth trimester,” a term coined by anthropologist Sheila Kitzinger1.
There is no question that this bidirectional interaction between mother and infant results in a bewildering number of new experiences for the infant; chief among them is the paradigm shift from predominantly intravenous nutrition facilitated by the placenta to enteral nutrition, produced by the mother in the form of breast milk, a fluid shaped by millions of years of mammalian evolution. While significant emphasis has been placed upon the role of milk in nourishment, evidence from various perspectives argues that acute protection of the infant from infection and inflammation is an equally compelling role for milk. The in utero environment is both supportive and protective, especially from microbial pathogens. Breaches in the microbial barrier to the infant’s environment are typically devastating to a pregnancy. At a relatively early stage of their development, human infants are born, dropped, figuratively and occasionally literally, into the mud. Their world is transformed, from a sterile environment to an environment teeming with microbes. It is this potentially catastrophic environmental transition that has placed a profound selective pressure on the genetics of lactation and the composition and functions of milk throughout mammalian evolution. The most recent investigations of milk support the hypothesis that milk is indeed far more than simple nourishment of the infant, it is a living, active, dynamic biological system sculpted by the evolutionary process to protect the infant from infection and guide the development of both the infant’s innate and adaptive immune systems and the developing intestinal microbiome. It has taken scientists considerable time and resources to discover that human milk is so broadly protective with the parallel emergence of two very different strategies. First, milk delivers antimicrobial proteins, peptides, lipids and enzymes as an awesome array of tactical weapons to protect the infant from pathogenic organisms. Second, milk components shape a symbiotic protective microbiome that influences and interacts with the developing immune system. What mothers have done is truly astonishing: recruit another kingdom of biology to protect their infants. This strategy poses a daunting tactical challenge for mothers: delivering in parallel antimicrobial components to the infant to ensure acute protection and milk components that support specific beneficial microbes within the infant. These components nourish not the infant but a unique bacterial population that provides metabolites and suppresses more inflammatory microbes, all with the selective advantage of protecting the infant!
Early research on a breast milk-gut microbiome association
Since the early invention of the microscope by von Leeuwenhoek, scientists contemplated the utility of the bacteria they witnessed in feces. Early speculations by Frerich in 1846 declared “the bacteria neither aid nor interfere with the digestive processes” a view held by many at that time2. Between 1890 and 1930 three separate lines of research emerged that suggested there was something unusual about the gut microbiome of breast-fed infants. The first was microscopic observations by Henry Tissier3 of fecal smears from healthy breast-fed infants, revealing a near monoculture of “bifid” or Y-shaped bacteria, then termed Bacteria bifidus, an observation repeated frequently throughout the early 1900s2,4–6. This “microbiome” observation differed dramatically from infants who consumed bovine milk, or fermented bovine milks, whose feces contained an assortment of bacterial shapes2,5–7. The second observation was that breast-fed infant feces were consistently more acidic (pH ~ 5) by comparison to that from bovine milk-fed infants which was also more variable (pH ~ 6–8)8–12. The third observation was an early correlation drawn between infants that lack Bacteria bifidus-dominated feces and the increased occurrence of infant gut-borne pathogens3. This led to speculation that human milk, unlike bovine milk, contains a specific component that encouraged growth of Bacteria bifidus and that its metabolism was protective of the infant intestinal environment7. In one sense, this was an early observation of a “microbiome” deficiency among a “dysbiotic” cohort when compared to that in a healthy population – a common strategy employed in comparative gut microbiome studies today.
If Bacteria bifidus were frequently observed in breastfed infant feces in the early 1900s, what components of breast milk were they consuming that were lacking in bovine milk? Solving this mystery took additional decades of research recently elaborated in an excellent historical review by Clemens Kunz13. In short, dissecting of both human and bovine milk revealed specific, non-protein components, responsible for the growth of this key bacterium, which, at that time, was termed Lactobacillus bifidus 9,14 (see Box 1). Gorgy and coworkers used growth of L. bifidus on different fractions of human milk as a guide to reveal a “bifidus” factor that was separable from larger glycoconjugates (like mucins) and from smaller sugars like lactose15. This fraction contained fucose, galactose, and N-acetyl-glucosamine constituents, components that were known associated with blood group glycans, an aligned and emerging scientific area at the time. Subsequent research in the years between 1950 and 2000 documented the remarkable abundance of milk oligosaccharides in human milk by comparison to other milks, notably bovine milk16, as well as isolation and characterization of individual human milk oligosaccharide (HMO) species such as fucosyllactose, lacto-N-tetraose, lacto-N-fucopentaose, and difucosyllactose, among others13.
Box 1.
A taxonomic evolution from Bacteria bifidus to Bifidobacterium longum subsp. infan-tis.
Microbial taxonomy is ever-changing. While Tissier3 identified the predominant bifid-shaped bacteria in the feces of breastfed infants, it took the trained eye of the famous microbiologist Sigurd Orla-Jensen in 1924180 to give a name to these unusual Y-shaped bacteria to the genus, Bifidobacterium. Adoption of the new genus name was not universal, however, and seminal work by Gyorgy14 defined similar infant isolates as Lactobacillus bifidus (later determined to be Bifidobacterium bifidum). Bifidobacterium infantis was first defined as a species in 1963181, with the species name becoming official in 197361. This designation held until 2008, when Mattarelli and colleagues62 revised the species by combining then B. longum, B. infantis, and B. suis into three B. longum subspecies – subsp. longum, subsp. infantis and subsp. suis. This designation still stands today confirmed by numerous pangenomic analysis of the clade57,58. Although the shortened term “B. infantis” is still routinely used in the scientific literature to refer to the subspecies, the official name is Bifidobacterium longum subsp. infantis. Despite the advances in molecular taxonomy, this subspecies has been incorrectly designated for various commercial probiotic products170.
As analytical methods to characterize and purify HMOs advanced from 1950 to 2000, little cognate advance in the understanding of the intestinal bifidobacteria that grow on them took place. During that same time, the subject of probiotics experienced a renaissance with the growth of research on lactic acid bacteria associated with food fermentations and gut health, however research on the interaction between HMOs and infant-borne bifidobacteria was lacking. In the mid 2000s Katayama and colleagues characterized extracellular fucosidase17 and lacto-n-biosidase18 from Bifidobacterium bifidum and Bifidobacterium longum subsp. longum (herein termed BL. longum). If HMOs were truly the “bifidus factor” responsible for enrichment of bifidobacteria in infants, did all bifidobacterial species grow equally? Early efforts by Ward19,20 and LoCascio21,22 clearly showed that only select bifidobacterial strains grow well (i.e. to a high cell density) on HMO as a sole carbon source. Growth rates differed among bifidobacterial species, however individual isolates of Bifidobacterium longum subsp. infantis (herein termed BL. infantis) emerged as the most consistently robust HMO consumers, suggesting it was a characteristic of the subspecies22.
Key to elucidating the relationship between HMOs and bifidobacteria were innovations in profiling of discrete HMO species driven by a series of advances in analytical chemistry techniques. The analytical challenge here is not readily appreciated. More than 300 HMO structures may be possible, although only about 100 are detectible from any individual mother’s milk. Unlike many other metabolites, HMOs contain many isomers, compounds with the same composition but different structures possessing identical masses (e.g. lacto-n-neotetraose vs. lacto-n-tetraose). Differentiating these isomers was the subject of years of challenging analytical chemistry, particularly as separating the similar structures requires new chromatographic techniques. Additionally, the large amount of lactose in milk confounded clinical studies as it swamped any carbohydrate-specific methods of detection. Furthermore, the presence of peptides and proteins diminished the effectiveness of sensitive analytical techniques such as mass spectrometry. Thus, rigorous isolation of HMO pools was necessary to eliminate contamination from lactose as well as from peptides and proteins. Enrichment of the HMOs was accomplished using porous graphitized carbon (PGC), which released lactose in the void volume and retained proteins in the matrix23,24. This method was highly scalable allowing further enrichment of HMOs in gram amounts for biological assays. Although this enrichment procedure is today taken for granted, it was key to determining the biological activity of HMOs as it rigorously removed lactose, which was a contaminant in many early experiments on the biological functions of HMOs.
Initially, matrix-assisted laser desorption/ionization (MALDI) Fourier transform ion cyclotron resonance mass spectrometry (FT ICR MS) was used for compositional profiling of HMOs25,26. This high-performance method for mass detection yielded compositions including the number of hexose, deoxyhexose (fucose), and sialic acid, but no stereochemical information. Coincidentally, the development of nanoflow methods elsewhere coincided with earlier HMO studies. Development of a microfluidic (Lab-in-a-chip) device that included a PGC enrichment column and PGC separation column allowing injection and separation of HMOs produced the most comprehensive method for separating and identifying HMO components27,28. Using this nanoLC chip with a PGC as stationary phase for chromatographic separation and quadrupole time-of-flight (QTOF) analyzers for mass detection, a comprehensive analysis of each individual component could be routinely performed of HMOs in various settings including mother’s milk29–31, urine32, feces,32 and even blood33.
Using purified HMO pools, these methods also allowed precise profiling of which specific HMO structures were metabolized by bacteria34. Strains of BL. infantis appeared to possess a generally similar HMO consumption, although some differences among strains were noted22,35. However, other bifidobacterial species isolates commonly found in the infant gut lacked this more robust phenotype. With the exception of B. bifidum 36, vigorous HMO consumption appears to be an episodic trait among isolates of BL. longum 37, B. breve 38, B. pseudocatenulatum, 39,40 and B. kashiwanohense 41 where most strains of those species do not grow on major components of HMOs, notably the fucosylated and sialylated HMOs. Among the BL. longum, B. breve, B. kashiwnohense, and B. pseudocatenulatum isolates that do robustly grow on HMOs, many consume only a portion of the HMO pool by comparison to BL. infantis 42,43 (Figure 1 depicts the consumption profile of BL. infantis). In general, this results in consumption of some fucosylated HMOs and non-fucosylated/non-sialyated HMOs but generally not the sialylated HMOs.
Figure 1.

Bubble plot representation of simultaneous glycoprofiling of 78 different HMO structures differentially consumed by BL. infantis. The size of the bubble depicts the percent consumption of that specific HMO. Reproduced with permission from Strum et al.42 Copyright (2012) American Chemical Society.
Early work on B. bifidum isolates showed consistently strong growth of HMOs, however it was noted that this species left degradation products of consumption, namely fucose and sialic acid, in the growth media suggesting extracellular hydrolysis of at least some HMOs17,19. Quite different consumption mechanisms elaborated by BL. infantis and B. bifdum illustrate the external (B. bifidum) versus transport and internal (BL. infantis) consumption mechanisms among bifidobacteria (Figure 2). Additional studies clearly showed the external degradation mechanism by B. bifidum enables cross feeding of HMO components of other bifidobacteria in vitro44. Given numerous genera present in the infant gut microbiome have been shown to externally consume HMOs45,46 like B. bifidum, it remains unclear what contribution these other clades might provide to HMO cross-feeding networks. Notably, others have demonstrated how mucin47,48 or HMO49 degradation can release sugar monomers that promote growth of enteric pathogens, suggesting a potential advantage to the host of transport and internal degradation of HMOs by BL. infantis.
Figure 2.
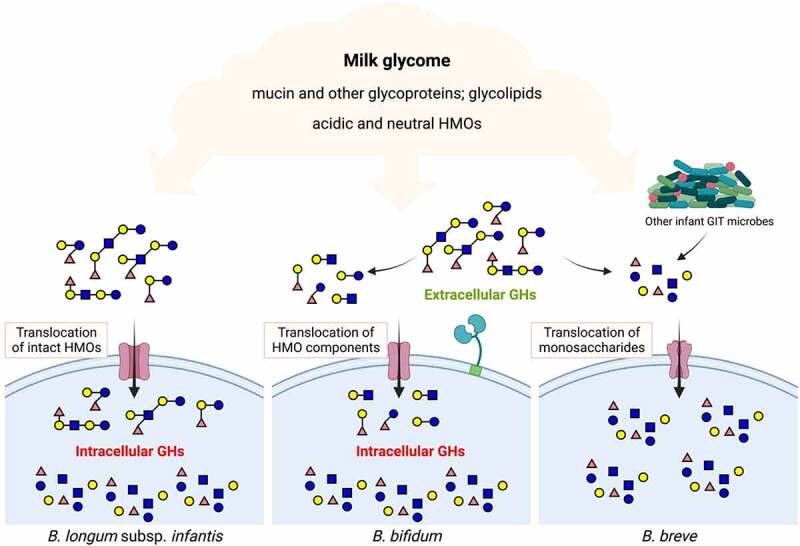
Prototypical strategies for HMO consumption by infant-borne bifidobacteria. GH, glycosyl hydrolase.
An early mechanistic explanation for the robust growth of BL. infantis on HMO emerged from the genome sequence first generated in 200850. At the time, relatively few genes associated with specific HMO catabolism had been identified in any bifidobacterial strain. Within BL. infantis the bulk of the HMO consumption genes were co-localized within a single 43 Kb locus. This HMO cluster contained all glycosyl hydrolases needed to cleave the various HMO linkages, namely intracellular fucosidase51, sialidase52, β-galactosidase53, and N-acetyl-β-hexosaminidase54 activities along with a number of associated ABC transporters55,56. This suite of genes linked to HMO catabolism was confirmed in other BL. infantis isolates35 and subsequent pan-genomic analyses aligned with this view that the subspecies diverged from other B. longum subspecies with HMO consumption as a common phenotype57–60. The genomic work also explained the lack of arabinose utilization by BL. infantis, a long-known phenotype that separates it from other B. longum subspecies61,62. Interestingly, the remnants of arabinose utilization genes are present in BL. infantis however they are disrupted by fucosidase gene and associated permease, suggesting a unique specialization of this subspecies toward the breast milk niche and away from utilization of plant glycans50. A recent analysis of over 30,000 metagenome assembled genomes (MAGs) from the infant gut microbiome clearly shows the unique presence of genes within the 43 Kb HMO cluster among several hundred MAGs of BL. infantis, validating these original observations59 (Figure 3). These researchers also noted a higher relative abundance of BL. infantis among all BL. longum genomes in infant metagenomes worldwide, suggesting that large HMO cluster provides a competitive advantage for colonization of breast-fed infants. For more information on the different mechanistic pathways of HMO consumption by infant-borne bifidobacteria, readers are referred to an excellent review by Katayama and colleagues63.
Figure 3.
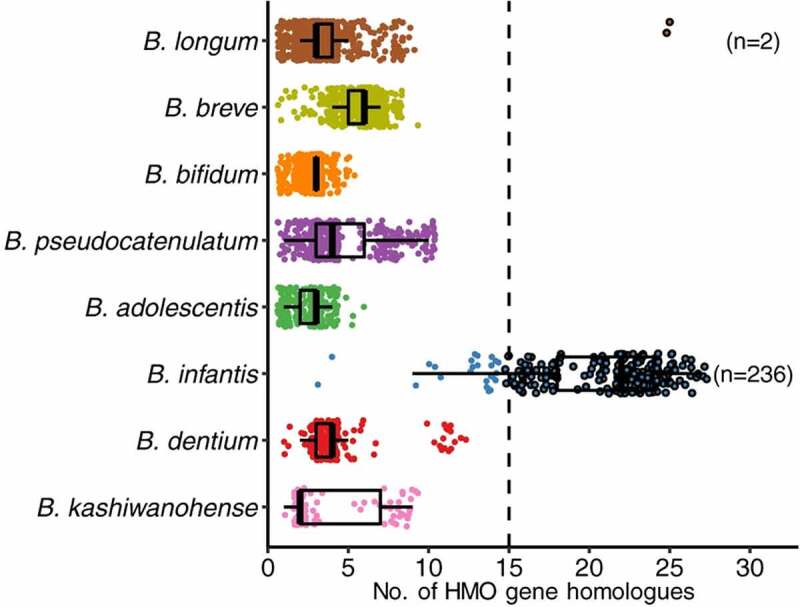
Pan-genomic demonstration of the abundance of HMO related genes in BL. infantis by comparison to other infant-borne bifidobacterial species. (Reproduced with permission from Zeng et al.59).
Are bifidobacteria a common presence in the breast-fed neonate gut or not? What’s old is new again
As insight emerged from early HMO-bifidobacterial work in the mid 2000s, a new technology came along that disrupted the field and put into question the importance of bifidobacteria in infants. Prior to the 2000s, the consensus view was that bifidobacteria routinely represent a predominant sector of the breast-fed infant gut microbiome, a view derived from generations of scientists employing microscopic, culture-based observations and first-generation non-culture-based methods64. Typically, this bifidobacterial predominance was observed after a short window early in lactation, wherein more aerobic taxa (i.e. enterococci, streptococci, staphylococci, Escherichia/Shigella) initially dominate65,66. However, in the mid 2000s, next-generation DNA sequencing provided a breathtaking new capacity to profile the assemblies of uncultured bacteria in the gut, thus providing unique insights into this ecosystem. In 2007, a seminal study by Pat Brown, David Relman, and colleagues at Stanford employed both 16S rRNA gene microarrays and sequencing of 16S rRNA gene amplicons to profile the microbiome of 14 Californian infants over a period of 1 year67. This was the first in-depth look at the “uncultured microbiome” in the developing infant. However, despite probing this dataset in various ways, they did not witness predominance, or even much of presence, of bifidobacteria. At the time, the authors noted:
“Although it is conceivable that there are geographical or demographic differences in the prevalence of Bifidobacteria, we suspect that the emphasis on Bifidobacteria in studies and reviews of the infant GI microbiota may be out of proportion to its prevalence, abundance, and relevance to health”.
As it turned out, numerous studies have since shown geographic differences in bifidobacterial populations among infant gut microbiomes around the world, and their presence is increasingly linked to infant health status65,68–76. In a sense, this early confusion by the first “omics-driven” survey of the infant gut microbiome is a cautionary tale for scientists using new approaches to describe any well-studied system. With this new technology comes the potential for new insights driving an appropriate and compelling desire to re-interpret existing dogma. However, insight through that new window might still be hazy, and in this case, the dogma bit back.
Prior to that seminal study, geographic differences among BL. infantis presence in infants were already a focus of attention. BL. infantis observed in Ghana cohorts was noted to be missing in New Zealand and United Kingdom cohorts77, and differential immunological responses to these bifidobacterial strains were correlated to incidence of atopy64. Once the robust HMO consumption capacity of BL. infantis was established via phenotypic and genotypic means, more researchers examined the geographic distribution of the subspecies among various infant cohorts75,78–80. Notably, BL. infantis appears less prominent in USA and European cohorts70,81,82 in comparison to Bangladeshi83, rural Malawi84, rural Venezuela,84 and Indonesian populations,80 suggesting a possible relationship to the level of industrialization experienced by a population. This geographic disparity was amplified in a recent survey by Sonnenburg and coworkers85 who compared infant metagenomes from the Hadza, a group of modern hunter-gatherers in sub-Saharan Africa, to aged-matched cohorts from Europe and the USA. These researchers also noted a high abundance of BL. infantis, and their HMO-utilization genes, among Hadza infants compared to infant cohorts from Europe or the USA. Fascinatingly, Jarvinen and coworkers recently identified a high population of BL. infantis within infants from rural Mennonite communities by comparison to a neighboring community in New York,74 indicating regional pockets of high BL. infantis exist within the United States.
Why might the “champion” HMO-consuming bifidobacterial subspecies86 be so differentially represented around the world? Sonnenburg and colleagues85 argued that BL. infantis overrepresentation among more rural regions is a reflection of its loss due to a myriad of factors associated with modern lifestyles in more industrialized regions. Indeed, the loss of intestinal taxa associated with industrialization is now the focus of efforts to preserve our “ancestral” microbes with the eventual goal of employing these strains in intestinal health applications87,88. It is easy to compile the various ways modern lifestyles might influence the early infant microbiome, such as use of antibiotics89, birth delivery mode,90 and introduction of formula or complementary feeding91 among others. Such changes may even be magnified across generations, i.e. the grandmother born by cesarean section and formula fed in the 1960s may not have received an optimum colonization from her mother, thus was unable to pass the microbes selected by evolution to her daughter and granddaughter even though each was born vaginally and breast-fed. Recently, Taft and colleagues70 postulated that the level of fecal bifidobacteria, and BL. infantis in particular, is associated with the regional history of breast-feeding practice wherein longer durations of breastfeeding by a population aligned with a higher level of BL. infantis.
Innovation – the translation of functional probiotics into the neonatal intensive care unit
Examined in various surveys, the gut microbiome of premature infants stands in stark difference to that observed in healthy breastfed infants92. This is, perhaps, not surprising as the infant microbiome is shaped by early environmental exposures to microbes, which can be dramatically different in premature infants where mode of delivery, diet (i.e. breast milk or formula), use of antibiotics, respiratory support and use of proton pump inhibitors, among many other interventions, are factors that influence the early life gut microbiome93. In addition, essentially every aspect of the innate and adaptive immune systems of the preterm is immature, poorly regulated and dysfunctional including intestinal motility, acid production, apoptosis, tight junction composition, mucus production, secretion of antimicrobial peptides and mechanisms of regulation of inflammation altering not only the gut microbiome but the host responses thereto. In general, various surveys of premature infant gut microbiomes, both before and after the advent of next-generation sequencing microbiome analyses, have identified a lower overall diversity, increased relative abundance of Enterobacteriaceae and Enterococceae and decreased bifidobacterial populations as hallmarks of premature infant microbiome94,95. By comparison to a healthy bifidobacterial-dominant, term breastfed infant gut microbiota, the “dysbiotic” premature infant microbiota is likely to trigger higher levels of endotoxin, a weakened barrier function, bacterial translocation, and inflammation96. These changes appear to be central to the pathogenesis of necrotizing enterocolitis (NEC) and late-onset sepsis in preterm infants.
Even before the common use of the term “probiotics”, these differences led to proposals to colonize premature infants using “normal intestinal flora” in hopes of stabilizing their microbiome and preventing the onset of disease97. Of course, this was not a new concept as Tissier7 had discussed such an approach to treat infant disease more than 100 years ago. Early trials on probiotics in premature infants employed commercial strains of Lactobacillus, Bifidobacterium, or even the yeast Saccharomyces98,99. The most recent meta-analyses of prophylactic administration of probiotics in preterm infants include observational cohorts (30 studies, more than 77,000 preterm infants)100 and randomized controlled trials (56 trials, more than 10,000 preterm infants),101 and have demonstrated significant decreases in NEC, late-onset sepsis, and death in this highly vulnerable population. Routine prophylactic administration to preterm infants varies widely from high in Japan, Australia, New Zealand, Germany, and Scandinavia to moderate in Canada and low in the U.K. and U.S.102. Nearly all reviews cited the need for more information on strain specificity and mechanism of action as it is exceedingly difficult to compare probiotic trials of vastly different microbes. For example, it is likely that a mechanism of action of a probiotic yeast Saccharomyces cerevisiae (commonly called Saccharomyces boulardii although “boulardii” is not an accepted species name) is not the same as a probiotic BL. infantis given the microbes evolved within entirely different environments. Others argue that much more consideration on the safety of probiotics in fragile neonates is warranted103. A 2021 statement from the American Academy of Pediatrics echoed these concerns and cautioned against routine use of current commercial probiotics in premature infants citing lack of FDA-regulated, pharmaceutical-grade products and thus the potential for harm104, while the European Society for Pediatric Gastroenterology, Hepatology and Nutrition, the American Gastrointestinal Association and the World Health Organization examined the same data and provided conditional recommendations for routine probiotic use in preterm infants105. Box 2 presents some of the regulatory challenges pertinent to the study and implementation of probiotics in the U.S.
Box 2.
Oversight of U.S. probiotic clinical trials and future clinical directions
In 2004, Underwood and colleagues sought direction from the U.S. Food and Drug Administration (FDA) regarding clinical trials of probiotics. At that time, the view of FDA was that a clinical trial could be performed without the oversight of the investigational new drug (IND) process if the primary outcome was not to prevent, treat, or mitigate a disease, but rather to alter the composition of the intestinal microbiota. The Institutional Review Board (IRB) at UC Davis agreed and approved the early trials116,189–192; the National Institutes of Health also agreed with this approach and provided funding. Subsequently, the FDA changed their guidance, requiring IND oversight for clinical trials of probiotics in preterm infants regardless of the measured outcome. Given the hesitation of probiotic manufacturers to participate in the IND process, it has become more challenging to perform studies of probiotics in U.S. NICUs, though analysis of samples from NICUs outside the U.S.193 and retrospective cohort studies117 are still possible. A large multi-center clinical trial of a Lactobacillus probiotic with IND oversight is nearing completion (Clincaltrials.gov NCT03978000), however to date there are currently no U.S. clinical trials with IND oversight of an HMO-consuming probiotic.
Recent network meta-analyses have summarized efficacy in the prevention of NEC, death, and sepsis of several different probiotic products93,97,187. Unfortunately, most of the clinical trials included in these meta-analyses were relatively small studies comparing a single probiotic product to placebo in different locations with differing baseline incidences of the primary outcomes. To answer the question of which probiotic and dose are most likely to decrease the risk of NEC, death, or sepsis, the ideal studies would be large multicenter cluster-randomized crossover trials adequately powered to look at NEC as an outcome (e.g. multiple NICUs randomized to probiotic A vs. B for 1–2 years then crossed over to the other probiotic). Smaller trials could still be helpful if they included one or more functional outcomes as proposed herein for example, a clinical trial including 100–200 preterm infants randomized to probiotic A or B would not be adequately powered for NEC as a primary outcome but could determine which probiotic lowered fecal pH or increased fecal lactate, acetate, or indole lactic acid. In the absence of such trials, the clinicians must consider whether routine probiotic prophylaxis for very preterm infants is justified and if so which of many probiotics is most likely to improve clinical outcomes. To address the first question, we have advocated for including the parents in this important discussion188 and considering the baseline rates of NEC and late-onset sepsis in a given NICU and the risks associated with probiotic administration (including probiotic sepsis and the potential for contamination of commercial probiotics). To address the second question, key considerations include evidence of clinical benefit (has the product been shown to be efficacious?), evidence of good-manufacturing practices in all aspects of probiotic production and distribution (is the manufacturer able to demonstrate reliable purity and viability of the probiotic strain(s)?), capacity of the local microbiology lab to detect the administered probiotic in infant blood cultures, and insights into the most effective antibiotic treatment options for the rare cases of invasive probiotic sepsis (most bifidobacteria are sensitive to penicillin, ampicillin, vancomycin, and clindamycin but resistant to aminoglycosides and metronidazole)194,195. Development and validation of biomarkers of microbiome and host gut function as advocated in this review would be a significant step forward in identifying infants at highest risk and probiotics with the highest potential benefit. The NEC society is a nonprofit organization and has prepared a probiotic toolkit for clinicians interested in considering probiotic prophylaxis (NECSociety.org).
In addition to regulatory and safety concerns, key questions regarding prophylactic administration of probiotics to preterm infants include optimal strain or combination of strains, optimal dosing and timing of administration, and equally important, optimal performance criteria. What function does a probiotic need to perform to reliably benefit the neonate? Of equal importance, what diagnostics demonstrate that probiotics are performing their function(s)? To consider this we need to understand the ability of a microbe, any microbe, to reliably colonize the infant gut ecosystem. At birth, the infant gut ecosystem is mostly naïve to microbes with open food niches ripe for capture by environmental microbes that enter into the system106. Thus, the early colonizers of the gut microbiome, in concert with the food they have access to, determine the microbial constituents that initially persist. This concept is perhaps best exemplified in the seminal study by Jeff Gordon and colleagues who colonized germ-free mice with input “alien” microbial assemblies derived from human, zebrafish, termite, soil, and estuarine environments107. All alien microbial assemblies colonized the mouse's gastrointestinal tract with their populations differentially shaped over time by the obvious constraints of that particular host and its diet. Given that such diverse microbial assemblies colonized a mouse gut, it is not hard to image how an infant gut becomes colonized by microbes in its environment. In short, a premature infant gut will be colonized with whatever microbes are available in the neonatal intensive care unit environment and that population will become constrained or amplified by diet, antibiotics, and host effects linked to that particular child108. One wonders if this “pioneer” effect is, in part, responsible for the generally beneficial effect seen in meta-analysis of probiotic trials in infants using quite different microbial species (or kingdoms!)109. Perhaps, these probiotics simply arrive early via supplementation into the premature infant gut, are shaped by the constraints of that setting, and even though they are “alien” to that environment, they delay arrival and/or establishment of other potentially problematic clades, such as Enterobacteriaceae, Enterococceaceae and Staphylococcaceae.
Two potential desired functions for a probiotic used in premature infants stem from the two observations made by Tissier3 more than century ago; dominant growth of bifidobacteria in situ is linked to (a) breastfeeding and (b) cognate lowering of the colonic pH. As a mechanistic understanding of the growth of bifidobacteria on HMOs advanced in the last 20 years, an enhanced view of the beneficial role of gut microbiome-generated organic acids, short chain fatty acids (acetate, propionate, and butyrate) and lactic acid produced by the gut microbiome, also emerged110–113. In a landmark study, researchers in Japan demonstrated that production of acetate in situ by metabolically active bifidobacteria was protective against an otherwise lethal Escherichia coli O157:H7 infection114. A key discovery was that only strains with the capacity to ferment fructose, the sugar present in the mouse chow, were protective. Indeed, such strains also persisted longer in the mouse intestine, and the authors postulated that increased persistence drove cognate increases in acetate and thus improved protection. For those studying the differential capacity of bifidobacteria to grow on HMOs, the connection to protection of neonates via a similar mechanism was obvious – robust bifidobacterial colonization driven by robust HMO fermentation should promote robust protection. But was this hypothesis true in humans? Underwood and colleagues first demonstrated HMO-consuming BL. infantis readily colonized premature infants fed breast milk in contrast to Bifidobacterium animalis subsp. lactis, a strain that lacked HMO consumption capacity115. Notably, the BL. infantis strain was even detected in infants who were provided the B. animalis strain implying cross contamination and/or horizontal transfer by BL. infantis into the other infants although a similar transfer did not appear in the reverse direction by B. animalis. Subsequent work by the same group showed high-level colonization of healthy breast-fed term infants with supplementation of BL. infantis (EvivoTM, USA) in comparison to no-probiotic controls116. Within days of supplementation, increases in total bifidobacteria in the feces rose from ~20% relative abundance to roughly 80% an increase driven exclusively by the supplemented BL. infantis. Once probiotic supplementation ceased on day 28, this high level of BL. infantis persisted in the infant microbiome throughout the study suggesting that breastfeeding and its delivery of HMOs was promoting continued, stable, colonization. The fecal biochemistry of the BL. infantis-supplemented infants also changed dramatically. Fecal HMOs plummeted with a corresponding rise in fecal organic acids, acetate, and lactate, the end products of BL. infantis fermentation. Notably, the average fecal pH of the unsupplemented infants in this trial was 5.97 while that for the supplemented infants was 5.15, a level that mirrors those witnessed in the early 1900s in bifidobacterial-dominated babies of the day10. A follow-up study on the same cohort demonstrated the BL. infantis strain persisted at a high level in infants with continued breastfeeding for up to 1 year but the strain was less prevalent in subsets of the cohort who received some formula or antibiotic treatments115. A recent study demonstrated that premature infants supplemented with BL. infantis showed reductions in fecal HMO and increases in fecal organic acids in comparison to a similar cohort supplemented with a non-HMO consuming Lactobacillus reuteri strain117.
Efforts by Hall and colleagues followed a similar strategy supplementing premature infants with a probiotic cocktail (Infloran, Italy) containing Lactobacillus acidophilus and B. bifidum118. Lactobacillus acidophilus is a poor HMO consumer119 and as discussed above, B. bifidum readily consumes HMOs via an external degradation scheme similar to that employed by Bacteroides species45,120. Human milk-fed premature infants supplemented in that cohort exhibited higher levels of B. bifidum and lower amounts of specific HMO species, namely two fucosyl-lactose isomers, in the feces. In addition, fecal acetate and lactate were higher, and the fecal pH was lower, in the supplemented infants suggesting HMO fermentation by B. bifidum colonization. A study by Watkins and colleagues121 also employed Infloran and clearly showed colonization by the Bifidobacterium genus but did not differentiate species, so it is unclear if an HMO-consuming B. bifidum in the Infloran cocktail was indeed a colonizing strain. A recent study employing a different bifidobacterial cocktail (FloraBABY, Renew Life, USA) containing BL. infantis, B. bifidum, BL. longum, and B. breve strains showed engraftment of all strains in premature infants initially for several weeks after supplementation ceased, however loss of BL. infantis occurred by 6 months of age115,122. This diversity in responses likely reflects the use of formula in these children (over 50% had some introduction of formula by then) wherein the more breast milk-glycan focused BL. infantis was more readily lost as formula is introduced115. Surprisingly, even though fecal metabolite analyses were run and differences between the probiotic treated and untreated premature infants were noted, the known fermentation products lactic acid, acetic acid, and 1,2 propanediol (a fermentation product of fucose41 that would be expected from bifidobacterial fermentation of HMOs in these children) were not discussed. Recently, Beck and colleagues123 employed metagenomic sequencing of a large cohort of preterm infants separately receiving either Infloran (B. bifidum and L. acidophilus) or Labinic (the U.K.; contains B. bifidum, BL. infantis and L. acidophilus) and clearly demonstrated that probiotic provision was associated with dramatic changes in the premature infant gut microbiome. Similar to the work by Alcon-Giner118 described above, this analysis showed higher persistence of bifidobacteria than L. acidophilus. However, only a single unknown metabolite was differentiated from an untargeted metabolomic analysis of stool from probiotic treated and untreated premature infants. Thus, it is unclear if robust HMO fermentation and production of acetic and lactic acids by these input bifidobacterial probiotics was a factor.
While these studies in both term and premature infants demonstrated a high bifidobacterial colonization associated with the supplementation of an HMO-consuming bifidobacterial strain into neonates fed human milk, other factors could have influenced this outcome. Certainly, the pioneer effect, discussed above, likely influenced these outcomes. In addition, human milk contains many antimicrobial compounds including lysozyme124, immunoglobins125, lactoferrin126, antimicrobial peptides127–129, glycerol monolaurate,130 and lactoperoxidase131 among others. Even HMOs can function as anti-infectives by binding to pathogens and deflecting their interaction with the host132. Moreover, some milk protein-derived peptides have been shown to directly stimulate bifidobacteria133,134. This constellation of anti- and pro-microbial activities likely play a role in shaping the development of infant gut microbiome and facilitating enrichment of bifidobacteria. So how important is HMO-fermentation by key bifidobacteria in the commonly witnessed enrichment of breastfed infants?
Insight into this question has recently emerged. Applications of bovine milk oligosaccharides (that are somewhat similar to HMOs135) and a B. animalis strain exhibited a synbiotic enrichment in vitro, in fermentations of infant feces136. Heiss and colleagues137 demonstrated that provision of a single HMO (2’-fucosyllactose) along with a cognate 2’-fucosyllactose-consuming Bifidobacterium pseudocatenulatum strain39 enabled a dramatic five-log increase in B. pseudocatenulatum in a wild-type mouse, reaching up to 60% relative abundance in the gut, an increase not witnessed with a different B. pseudocatenulatum strain that could not grow on 2’-fucosyllactose. Several aspects of this work are noteworthy. The dramatic enrichment of the B. pseudocatenulatum strain both persisted and increased after supplementation of the probiotic ceased as long as the 2’-fucosyllactose was provided. Once provision ceased, the supplemented B. pseudocatenulatum population rapidly declined. In addition, unlike the naïve infant gut, the adult wild-type mice employed in this study possessed a fully developed microbiome harboring normal colonization resistance138,139. Thus, the synbiotic application of HMO plus HMO-consuming bifidobacteria was able to override colonization resistance, indicating that HMO alone can provide an exclusive metabolic niche to the cognate bifidobacterial strain similar to that previously witnessed between porphyrin polysaccharide and select Bacteroides strains140,141. Another paper recently reinforced this HMO-bifidobacterial synbiotic function, demonstrating that pools of purified HMOs combined with BL. infantis promoted engraftment of the strain in both human subjects and gnotobiotic mice containing humanized gut microbiome142. Notably, a prior attempt by different authors but with a similar synbiotic combination (pooled HMOs and BL. infantis) did not show significant engraftment in mice143. While more research is clearly needed in this area, it appears that under some conditions, HMOs alone are capable of enriching cognate HMO-consuming strains in the absence of priority effects or other microbiota modifying factors inherent to breast milk. Importantly, these findings suggest a route to engraft HMO-fermenting bifidobacteria in infants not receiving human milk. The increasing use of pasteurized donor human milk in preterm infants in the NICU is also relevant to this discussion as standard pasteurization techniques utilized by milk banks denature bioactive proteins and peptides, while HMOs are heat-resistant. It is likely that diminished activity of the previously noted human milk components (lactoferrin, immunoglobulins, lactoferrin, antimicrobial peptides, and lactoperoxidase) in donor milk may be partially offset by the abundance of HMOs in donor milk (a pooled product including HMOs from a variety of women). Additional research assessing the impact of individual HMOs, groups of HMOs (e.g. fucosylated vs. sialylated) and single donor vs. pooled donor milk on the gut microbiome and functional outcomes is needed.
Impact of bifidobacteria on infant health: data from preclinical models and infant biomarker studies
A century ago researchers linked the acid produced in the breast-fed infant colon to preservation of health status and, when absent, its association with disease. As stated by Marriott and Davidson in 192312
“In a number of instances, it was possible to study the same infants, first while normal, and subsequently when suffering from an infection. There was regularly a marked decrease in gastric acidity in the presence of infection”.
As discussed above, we now know much more of the mechanism by which bifidobacteria ferment HMOs, producing organic acids, and lowering colonic pH. But does this action improve infant health? Henrick and colleagues144 compiled data on the fecal pH of breast-fed infants in studies performed over the last century, noting a clear increase in pH over time and hypothesized different connections to inflammation and immune disorders. Lower fecal pH in infants has been directly linked to reduced stunting145 and, recently, morbidity and mortality146 in cohorts from less developed regions.
While this reduction in pH is known to come from the production of acetic and lactic acids, other metabolites produced by infant-borne bifidobacteria have been shown to beneficially modulate intestinal function. Indole lactic acid (ILA), a catabolite of tryptophan produced by many intestinal bacteria147, including most bifidobacteria148, is known to interact with the host aryl hydrocarbon receptors, a key factor in intestinal homeostasis acting on barrier function, epithelial renewal, and various immune cell types149. ILA is present at a higher level in feces dominated by bifidobacteria (particularly BL. infantis) and is directly anti-inflammatory toward intestinal cells150–153. Henrick and colleagues154 showed that breastfed infants supplemented with BL. infantis had suppressed Th2 and Th17 responses and increased interferon β response compared to no-probiotic controls. Fecal water from the supplemented infants contained high levels of ILA, which in turn upregulated immunostimulatory galectin-1 in Th2 and Th17 cells during polarization. This work provides a nascent mechanistic rationale for why HMO-driven bifidobacterial enrichments in early life may be negatively associated with immune-related diseases78,155. More recently, Laursen and colleagues151 discovered a larger range of aromatic lactic acids correlated with infant-borne bifidobacterial populations, all linked to a specific aromatic lactic dehydrogenase gene uniquely present in specific infant-borne species—BL. longum, B. breve, B. bifidum, and BL. infantis.
Direct demonstrations of the potential health benefits from active, HMO-consuming bifidobacterial probiotics have also emerged in both animal models and human trials. Jena and colleagues156 showed that synbiotic applications of BL. infantis and bovine milk oligosaccharides reversed nonalcoholic steatohepatitis in a mouse model. Heiss and colleagues137 demonstrated high-level colonization by B. pseudocatenulatum, driven by simultaneous 2’-fucosyllactose provision, dramatically improved colitis within a mouse model. In both healthy and premature infant cohorts, early colonization by supplemented BL. infantis exhibited lowered fecal calprotectin and proinflammatory cytokines157 as well as reduced the incidence of antimicrobial resistance genes117,157–159.
Impact of bifidobacteria on infant health: data from clinical studies
Reports on the clinical outcomes of the use of validated, HMO-consuming, bifidobacterial probiotics in premature infants are relatively rare precisely because most probiotic strains employed in the NICU are not assessed for HMO-growth capacity. Indeed, some have speculated that the lack of efficacy in reducing NEC in large clinical trial employing a B. breve strain160 was due, in part, to the lack of HMO consumption and colonization by the provided strain161,162. A number of studies using the probiotic product Infloran containing L. acidophilus and either BL. infantis or B. bifidum (bifidobacterial species that consume HMO) have been shown to reduce the risk of NEC163–169. While these studies looked at clinical outcomes, they did not score for bifidobacterial colonization or cognate changes in fecal biochemistries so no direct links to HMO-fermentation can be ascribed. Moreover, analysis of these studies can be confusing as they differentially record the same Infloran product as containing either species BL. infantis or B. bifidum. Around the same time, our group demonstrated that many probiotic products containing bifidobacterial species (particularly BL. infantis) actually had different species in the product than the label indicated170. Contamination of NICU-focused commercial probiotics with alternative bifidobacterial species was recently demonstrated by Beck et al.123 where a B. animalis species, not described on the product label, was observed both directly in the probiotic cocktail and in the supplemented infants. Clearly, a needed component of future NICU probiotic trials is precise validation of input probiotic species.
Recent observational studies employing probiotics in the NICU provide more direct links that the HMO-bifidobacterial axis may be a factor to infant health outcomes. In a companion paper to the study by Alcon-Giner et al.118 that demonstrated functional evidence of HMO-fermentation of the input B. bifidum, Robertson and colleagues162 followed a larger cohort of 982 premature infants over a 10-year period, the latter 5 years of which the infants received a probiotic which changed from a cocktail of B. bifidum and L. acidophilus (Infloran) to one containing B. bifidum, BL. infantis, and L. acidophilus (Labinic). As seen in the studies described above, NEC and late-onset sepsis rates were reduced with use of HMO-fermenting bifidobacterial probiotics, however reductions in all-cause mortality were not statistically significant. In a separate study employing a BL. infantis probiotic (EvivoTM) Tobias and colleagues171 demonstrated statistically significant reductions in NEC incidence and NEC-associated mortality in very low birthweight infants (N = 483 in cohort), including an extremely low birth weight subgroup (Figure 4). Robust colonization and evidence of HMO fermentation in situ for this specific probiotic have been previously validated115,116,159. Why might these two recent studies show differences in mortality? First, the overall incidence of NEC was higher in the NICU examined in the Tobias et al.171 study, which facilitated a lower overall cohort number needed to reach statistical significance per NEC outcomes. However, additional factors such as notable differences in probiotic species, format (cocktail vs. single strain), formulation (HMO-activated strains vs. not) and dosage may have influenced these outcomes.
Figure 4.
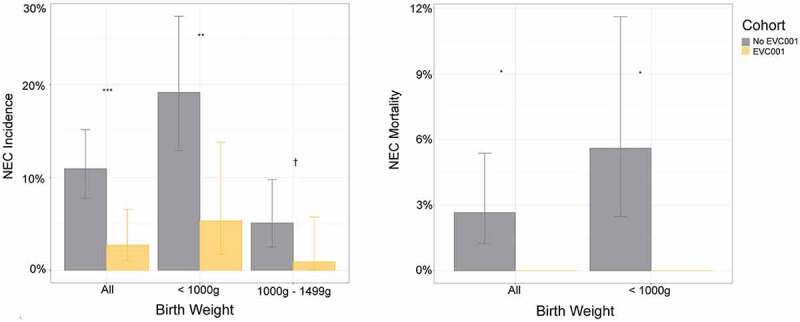
NEC incidence and mortality in the observational study employing BL. infantis EVC001 (EvivoTM) or no probiotic controls. Left panel, NEC incidence by birth weight and cohort. Error bars show 95% CIs around the estimates. ***P < .001; **P < .01;†P < .1, Fisher exact test. Right panel, NEC-related mortality rates by birth weight and cohort. Error bars show 95% CIs around the estimates. *P < .05, Fisher exact test. Reproduced with permission from Tobias et al.171.
Finally, a recent study focused on infants with severe acute malnutrition (SAM) illustrates how a focus on the HMO-fermenting bifidobacteria can help repair growth functions in these at-risk populations as well. Barrett and colleagues161 demonstrated that supplementation of BL. infantis (EvivoTM) in breastfed infants with SAM resulted in significant colonization by the probiotic strain which correlated with improved weight gain and reduced inflammation.
Summary
Research over the last 20 years has provided key insight into the molecular mechanism of consumption of HMOs by infant-borne bifidobacteria as well as applications of these HMO-consuming strains in the NICU. This new mechanistic understanding of a very old observation – the enrichment of bifidobacteria in breastfed infants – now drives new questions that challenge the concept of a probiotic “function”. While numerous reviews detail the litany of potential mechanisms associated with probiotic functions,172,173 little research has scored those precise functions for specific host responses and beneficial clinical outcomes in actual human clinical trials employing these probiotics. Indeed, since most probiotics do not robustly colonize the host, it is perhaps not surprising that efficacy in their use across a range of clinical targets is questioned174 (see Box 3). We posit that breastfed neonate presents an unusual case where an overt mechanistic link has emerged between select supplemented probiotics and improvement in infant health. The assembled research described herein argues select HMO-consuming bifidobacteria readily colonize the breast-fed infant gut and ferment HMOs to produce organic acids and other key metabolites (such as ILA) which, in turn, beneficially condition the gastrointestinal environment and restrict entry by other, often problematic, microbial taxa. However, numerous questions, research gaps, and debates remain. Several such questions are elaborated below.
Box 3.
The problem with probiotics.
Since the early proposal as a general concept by Metchnikov182 to the development of the first commercial product by Shirota183, probiotics have been simultaneously popular and controversial174,184–186. The current definition of a probiotic as “Live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” originated in 2001 by FAO/WHO178. Many current best-selling probiotics were developed decades ago wherein appropriate commercial concerns (oxygen tolerance, fermentation scale up, strain stability, shelf life) were important considerations for their commercialization in order to deliver a live microbe in “adequate amounts”. Efficacy toward a specific health target was not the sole criteria early on, but rather a more general effort to mitigate “dysbiosis”, typically in the form of increased presence of the probiotic and thus lowering of other pathobiont clades. In general, commercial probiotics have been shown not to persist, much less increase, in the host beyond supplementation and various probiotic trials have not shown significant alterations in the host gut microbiome187. This failure of probiotics to contribute quantitatively to the host microbiome is not surprising. Colonization resistance inherent to established gut microbiomes provides protection from exogenous bacterial interlopers, due in part to the established and resilient food networks that form within a health gut ecosystem. Suez and coworkers174 proposed that the lack of persistence, level of colonization and associated mucosal interaction is a likely factor impacting efficacy in probiotic trials.
Question – Is a greater focus on probiotic function in neonates needed? In terms of the breast-fed infant gut, we argue that microbiota function and its impact on the host should be the chief focus in clinical cohorts assessing the use of probiotics in neonates. Each year brings ever more elaborate microbiome analyses of infant cohorts. Early work on severe acute malnutrition by Jeff Gordon and coworkers employed the use of microbial composition as a measure of “gut microbiota age”, determining a persistent “immaturity” in the gut microbiota of afflicted infants175. This concept of using microbial taxa alone to score the chronological age or maturity of the gut microbiome infers a cognate functional component to that microbiome. However, in the absence of a scored microbiota function (pH, organic acids, etc.), how informative is this measurement in neonates? The recent comprehensive metagenomic analysis of probiotic use in the NICU by Beck and colleagues123 deemed the probiotic-treated neonates to have experienced an “accelerated maturation” due to the presence of a supplemented bifidobacterial population. However, that same cohort did not witness significant differences in fecal organic acids. Is the mere presence of the supplemented probiotic in the naïve infant gut all that is needed for proper maturation? If so, would continuous delivery of nonviable (i.e. dead) bifidobacterial probiotics have performed the same function? Perhaps, the unusual abundance of HMOs in human breast milk can now be explained by the resulting metabolic output of a robust fermenter-like BL. infantis and the functions of those metabolites within the intestine of neonates.
A crude, but useful, analogy emerges from the use of starter cultures in food and beverage fermentations. For centuries, wine and dairy fermentations were carried out wherein the indigenous microbiota (yeast and lactic acid bacteria, respectively) performed the main fermentations. However, advances in fermentation science brought the use of select starter cultures that, upon supplementation, far more reproducibly drove those fermentations. Further sophistication in starter culture science drove novel strain selection as well as strain inoculation and management practices, all for the purpose of maintaining a robust fermentation, a function that was easily measured (e.g., alcohol or acid production or sugar loss). In these industries, a real-time assessment of the fermentation is the norm and any fermentation that lacks proper functional outputs is considered at risk and remediation would start immediately. Now that we better understand the factors behind HMO-driven bifidobacterial colonization of infants, should the NICU adopt a similar approach with premature infants? We argue that there is enough evidence to suggest a path for the NICU to routinely track such fecal fermentation products (e.g. decreased fecal HMOs, increased fecal organic acids, decreased fecal pH) as a means to provide clinicians with critical real-time insight into the nature of the gut fermentation occurring in premature infants. Such routine measurements will similarly provide data the scientific community needs to accurately and quantitatively compare and contrast probiotic trials and ultimately provide an evidential basis to enhance effective treatments.
Question – What version of probiotic(s) should be used in the NICU? As described above, there are various probiotic formats containing HMO-consuming strains. Two main themes emerge: (a) use of single strains versus a cocktail of strains and (b) employing strains that catabolize HMO externally (i.e. B. bifidum type) vs. internally (i.e. BL. infantis type) (as discussed previously, see Figure 2). Strain cocktails are a common format for probiotics. The general rationale for cocktails is obvious – they provide more functions and/or duplications of functions in the face of different host environments. In a sense, this mirrors the concept that multiplicity and diversity of function providing a robust resilience to the adult human gut microbiome to meet a diversity of diets and ecological interfaces (e.g. bacteriocins and bacteriophage), among other interactions. However, the naïve infant gut is a unique environment provided with a unique food, breast milk, which itself is selective for HMO-consuming bacteria, primarily bifidobacteria. Recent work from Katayama and colleagues provides a nascent view of how HMOs might drive different bifidobacterial communities to form176. The ability to supplement HMO-consuming bifidobacteria early in life enables neonatologists to direct a bifidobacterial community structure of the early breast-fed infant gut123. Choices of internal vs. external HMO degradation formats (Figure 2) will also likely influence community structure as external degradation would be expected to prompt cross-feeding interactions among other gut microbes, be they good40,44 or bad49. As argued above, perhaps a better rationale for the use of any probiotic is one of function. Which format more reproducibly delivers the most robust HMO-fermentation in situ? As this is readily measured, more research on such functional outputs of NICU probiotic use will help decipher this question. Finally, the use of non-HMO consuming species, or postbiotics177, in breastfed infants and approaches to optimizing the microbiota of formula-fed infants need more research on possible mechanistic rationales for their efficacy.
Conclusion
In the last two decades the field of microbiology has experienced an incredible transformation. The advent of “omics” tools, particularly next-generation sequencing to determine uncultured microbial communities has provided a wealth of insight into our gut microbiome and its function. At the same time, general knowledge of probiotics and their use by both physicians and the public has increased dramatically,178 yet significant challenges to their clinical efficacy remain174. The use of HMO-consuming bifidobacterial probiotics in neonates represents a novel, tailored, use where the direct function of the implanted probiotic is readily observed and can be related to clinical efficacy. We propose that similarly “functional” synbiotic applications179 may drive new discoveries, inventions and translations (see Box 4) to advance human health in a range of settings.
Box 4.
The route from discovery to invention to translation.
“To him who devotes his life to science nothing can give more happiness than increasing the number of discoveries, but his cup of joy is full when the results of his studies immediately find practical applications.” Louis Pasteur 188
The central goal of research in the life sciences is discovery; how does the world work? Breakthroughs in our understanding of how biology functions from atomic-level mechanisms to ecological behavior of entire communities do not, however, immediately identify how such discoveries can be translated into practice. Invention is the cognitive process of recognizing a utility that can be provided by explicitly acting upon new understanding. Scientists, innovators, and regulators alike are faced with major challenges in building utility from the discoveries of microbiome research. Indeed, for the average microbiologist or neonatologist embracing a new discovery, application is not always so easily recognizable. This was the case with the UC Davis Milk Bioactives group (initiated by authors German, Lebrilla and Mills), a multidisciplinary team that examined the HMO-bifidobacterial axis. Early discovery of specific bifidobacteria, most notably BL. infantis, consume the bulk of HMOs in mother’s milk21 suggested a unique partnership between these abundant oligosaccharides and specific bifidobacterial taxa. The initial excitement of this discovery was the normal academic joy of recognizing a clear and compelling research path to identify the mechanism underlying this phenotype. However, finding a utility for this phenotype was not obvious at the time. It was with the connection to neonatologist (and coauthor) Mark Underwood and witnessing premature infants in his NICU suffering from NEC and infections that the “real world” intersected with our otherwise lofty academic pursuits. The Milk Bioactives group immediately recognized that, BL. infantis, the “champion” HMO consumer, should be able to populate the premature infant gut, reduce suffering, and improve outcomes. As a consequence, the company Evolve Biosciences (now Infinant Health) was launched jointly by the University of California, Davis, and the scientists responsible for the discovery research. To date, over 50,000 infants have been supplemented with BL. infantis EVC001.
What were the key elements that facilitated this translation? First and foremost, it was the interdisciplinary nature of the UCD Milk Bioactives group. The historic academic model of siloed research can be an anathema to the kind of integrated thinking that advances understanding of complex biology, but it also mutes innovative thinking. A second component was the increasing recognition that translation in the form of entrepreneurial activity is a required (and very much desired) component of a University’s public service. This has not always been the case, particularly at land grant universities where translation was/is typically done in the form of publishing and extension. Thankfully, the fruits of this new thinking are being realized and the field of microbiome science has no shortage of university-fostered startup companies addressing a range of applications.
Acknowledgments
The authors acknowledge the tremendous contributions from the many students, postdoctoral researchers, research staff, and faculty who have participated in the UC Davis Milk Bioactives Group, past and present. DAM thanks Dr. You-Tae Kim for help with Figure 2.
Funding Statement
Research performed by the UC Davis Milk Bioactives group has been supported by Peter J Shields Endowed Chair in Dairy Food Science (DAM), the UC Davis RISE Program, the California Dairy Research Foundation, USDA grant 2008-35200-18776 and NIH grants HD059127, R21AT006180, R01AT007079, R01AT008759, R01HD061923, R01HD065122, and U01CA179582 and NIH Training grants F32HD093185, F32AT006642, and F32AT008533.
Author contributions
The authors jointly wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Disclosure statement
DAM, JBG, and CBL are co-founders of Infinant Health, a company focused on probiotic-based manipulation of the infant gut microbiota; BCD Biosciences, a company advancing novel bioactive glycans; and Matrubials Inc., a company advancing milk-based antimicrobial peptides. None of these companies had any role in the conceptualization, design, analysis, or preparation of this manuscript.
References
- 1.Kitzinger S. The fourth trimester? Midwife Health Visit Community Nurse. 1975;11:118–25. [PubMed] [Google Scholar]
- 2.Gerstley JR. Some factors influencing the fecal flora of infants. Arch Pediatr Adolesc Med. 1932;43(3):555. doi: 10.1001/archpedi.1932.01950030025003. [DOI] [Google Scholar]
- 3.Tissier MH. Recherches sur la flore intestinale normale et pathologique du nourisson [Doctoral dissertation]. 1900. [Google Scholar]
- 4.Brown EW. Studies of infant feeding xvi. Am J Dis Child. 1922;23(3):243. doi: 10.1001/archpedi.1922.01910390062005. [DOI] [Google Scholar]
- 5.Logan WR. The intestinal flora of infants and young children. J Pathol Bacteriol. 1913;18(1):527–551. doi: 10.1002/path.1700180154. [DOI] [Google Scholar]
- 6.Upton MF. The anaerobic intestinal flora of normal breast-fed and artificially fed infants. Arch Pediatr Adolesc Med. 1929;37(6):1221. doi: 10.1001/archpedi.1929.01930060098013. [DOI] [Google Scholar]
- 7.Tissier MH. Repartition des microbes dans l’intestin du nourrisson. Ann Inst Pasteur (Paris). 1905;19:109–123. [Google Scholar]
- 8.Gyorgy P. A hitherto unrecognized biochemical difference between human milk and cow’s milk. Pediatrics. 1953;11(2):98–108. doi: 10.1542/peds.11.2.98. [DOI] [PubMed] [Google Scholar]
- 9.Barbero GJ, Runge G, Fischer D, Crawford MN, Torres FE, Gyorgy P. Investigations on the bacterial flora, pH, and sugar content in the intestinal tract of infants. J Pediatr. 1952;40(2):152–163. doi: 10.1016/S0022-3476(52)80176-3. [DOI] [PubMed] [Google Scholar]
- 10.Norton RC. The hydrogen ion concentration of the stools of new-born infants. Arch Pediatr Adolesc Med. 1926;32(2):183. doi: 10.1001/archpedi.1926.04130080023002. [DOI] [Google Scholar]
- 11.Tisdall FF. Studies on the acidity (hydrogen ion concentration) of infants’ stools. Arch Pediatr Adolesc Med. 1924;27(4):312. doi: 10.1001/archpedi.1924.01920100017003. [DOI] [Google Scholar]
- 12.Marriott WM. The acidity of the gastric contents of infants. Arch Pediatr Adolesc Med. 1923;26(6):542. doi: 10.1001/archpedi.1923.04120180041004. [DOI] [Google Scholar]
- 13.Kunz C. Historical aspects of human milk oligosaccharides. Adv Nutr. 2012;3(3):430S–439S. doi: 10.3945/an.111.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyorgy P, Norris RF, Rose CS. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch Biochem Biophys. 1954;48(1):193–201. doi: 10.1016/0003-9861(54)90323-9. [DOI] [PubMed] [Google Scholar]
- 15.Gyorgy P, Jr H, Kuhn R, Cs R, Bifidus factor . The rate of dialysis. Arch Biochem Biophys. 1954;48(1):209–213. doi: 10.1016/0003-9861(54)90325-2. III. [DOI] [PubMed] [Google Scholar]
- 16.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20(1):699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 17.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-l -fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol. 2004;186(15):4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71(6):3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51(11):1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 20.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72(6):4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55(22):8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 22.Locascio RG, Niñonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol. 2009;2(3):333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruhaak LR, Lebrilla CB. Analysis and role of oligosaccharides in milk. BMB Rep. 2012;45(8):442–451. doi: 10.5483/BMBRep.2012.45.8.161. [DOI] [PubMed] [Google Scholar]
- 24.Wu LD, Ruhaak LR, Lebrilla CB. Analysis of milk oligosaccharides by mass spectrometry. Methods Mol Biol. 2017;1503:121–129. [DOI] [PubMed] [Google Scholar]
- 25.Park Y, Lebrilla CB. Application of Fourier transform ion cyclotron resonance mass spectrometry to oligosaccharides. Mass Spectrom Rev. 2005;24(2):232–264. doi: 10.1002/mas.20010. [DOI] [PubMed] [Google Scholar]
- 26.Finke B, Mank M, Daniel H, Stahl B. Offline coupling of low-pressure anion-exchange chromatography with MALDI-MS to determine the elution order of human milk oligosaccharides. Anal Biochem. 2000;284(2):256–265. doi: 10.1006/abio.2000.4680. [DOI] [PubMed] [Google Scholar]
- 27.Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10(2):856–868. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9(8):4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totten SM, Wu LD, Parker EA, Davis JCC, Hua S, Stroble C, Ruhaak LR, Smilowitz JT, German JB, Lebrilla CB. Rapid-throughput glycomics applied to human milk oligosaccharide profiling for large human studies. Anal Bioanal Chem. 2014;406(30):7925–7935. doi: 10.1007/s00216-014-8261-2. [DOI] [PubMed] [Google Scholar]
- 30.Vinjamuri A, Davis JCC, Totten SM, Wu LD, Klein LD, Martin M, Quinn EA, Scelza B, Breakey A, Gurven M, et al. Human milk oligosaccharide compositions illustrate global variations in early nutrition. J Nutr. 2022;152(5):1239–1253. doi: 10.1093/jn/nxac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niñonuevo MR, Perkins PD, Francis J, Lamotte LM, LoCascio RG, Freeman SL, Mills DA, German JB, Grimm R, Lebrilla CB. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J Agric Food Chem. 2008;56(2):618–626. doi: 10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- 32.De Leoz MLA, Wu S, Strum JS, Niñonuevo MR, Gaerlan SC, Mirmiran M, German JB, Mills DA, Lebrilla CB, Underwood MA. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. Anal Bioanal Chem. 2013;405(12):4089–4105. doi: 10.1007/s00216-013-6817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruhaak LR, Stroble C, Underwood MA, Lebrilla CB. Detection of milk oligosaccharides in plasma of infants. Anal Bioanal Chem. 2014;406(24):5775–5784. doi: 10.1007/s00216-014-8025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Leoz MLA, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, Mills DA, Lebrilla CB. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. 2015;14(1):491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76(22):7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr. 2012;3(3):422S–429S. doi: 10.3945/an.111.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB, Mills DA. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep. 2016;6(1):35045. doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, German JB, Lebrilla CB, Mills DA. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79(19):6040–6049. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shani G, Hoeflinger JL, Heiss BE, Masarweh CF, Larke JA, Jensen NM, Wickramasinghe S, Davis JC, Goonatilleke E, El-Hawiet A, et al. Fucosylated human milk oligosaccharide foraging within the species Bifidobacterium pseudocatenulatum is driven by glycosyl hydrolase content and specificity. Appl Environ Microbiol. 2021;88(2):AEM0170721. doi: 10.1128/AEM.01707-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawson MAE, O’neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, Chalklen L, Hall LJ. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14(2):635–648. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunesova V, Lacroix C, Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16(1):248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strum JS, Kim J, Wu S, De Leoz MLA, Peacock K, Grimm R, German JB, Mills DA, Lebrilla CB. Identification and accurate quantitation of biological oligosaccharide mixtures. Anal Chem. 2012;84(18):7793–7801. doi: 10.1021/ac301128s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. 2016;7(1):11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018;26(4):339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012;18(Suppl 4):12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pichler MJ, Yamada C, Shuoker B, Alvarez-Silva C, Gotoh A, Leth ML, Schoof E, Katoh T, Sakanaka M, Katayama T, et al. Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat Commun. 2020;11(1):3285. doi: 10.1038/s41467-020-17075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engevik MA, Engevik AC, Engevik KA, Auchtung JM, Chang-Graham AL, Ruan W, Luna RA, Hyser JM, Spinler JK, Versalovic J. Mucin-degrading microbes release monosaccharides that chemoattract clostridioides difficile and facilitate colonization of the human intestinal mucus layer. ACS Infect Dis. 2021;7(5):1126–1142. doi: 10.1021/acsinfecdis.0c00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y-L, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun. 2015;6(1):8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105(48):18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom H-J, Joachimiak A, Lebrilla CB, Mills DA. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78(3):795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286(14):11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrido D, Ruiz-Moyano S, Jimenez-Espinoza R, Eom H-J, Block DE, Mills DA. Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol. 2013;33(2):262–270. doi: 10.1016/j.fm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrido D, Ruiz-Moyano S, Mills DA. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe. 2012;18(4):430–435. doi: 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrido D, Kim JH, German JB, Raybould HE, Mills DA . Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE. 2011;6(3):e17315. doi: 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J-H, An HJ, Garrido D, German JB, Lebrilla CB, Mills DA, de Crécy-Lagard V, de Crécy-Lagard V. Proteomic analysis of Bifidobacterium longum subsp. infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLoS ONE. 2013;8(2):e57535. doi: 10.1371/journal.pone.0057535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarracchini C, Milani C, Lugli GA, Mancabelli L, Fontana F, Alessandri G, Longhi G, Anzalone R, Viappiani A, Turroni F, et al. Phylogenomic disentangling of the Bifidobacterium longum subsp. infantis taxon. Microb Genom. 2021;7(7). doi: 10.1099/mgen.0.000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albert K, Rani A, Sela DA. Comparative pangenomics of the mammalian gut commensal Bifidobacterium longum. Microorganisms. 2019;8(1):7. doi: 10.3390/microorganisms8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng S, Patangia D, Almeida A, Zhou Z, Mu D, Paul Ross R, Stanton C, Wang S. A compendium of 32,277 metagenome-assembled genomes and over 80 million genes from the early-life human gut microbiome. Nat Commun. 2022;13(1):5139. doi: 10.1038/s41467-022-32805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonnenburg ED, Sonnenburg JL. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol. 2019;17(6):383–390. doi: 10.1038/s41579-019-0191-8. [DOI] [PubMed] [Google Scholar]
- 61.Reuter G. Designation of type strains for bifidobacterium species. Int J Syst Bacteriol. 1971;21(4):273–275. doi: 10.1099/00207713-21-4-273. [DOI] [Google Scholar]
- 62.Mattarelli P, Bonaparte C, Pot B, Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58(4):767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- 63.Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao J-Z, Kitaoka M, Katayama T. Varied pathways of infant gut-associated bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2019;12(1):71. doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tannock GW. New perceptions of the gut microbiota: implications for future research. Gastroenterol Clin North Am. 2005;3434(3):361–382. doi: 10.1016/j.gtc.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Tannock GW, Ercolini D, Ercolini D. Building robust assemblages of bacteria in the human gut in early life. Appl Environ Microbiol. 2021;87(22):e0144921. doi: 10.1128/AEM.01449-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO, Ruan Y, Ruan Y. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barratt MJ, Ahmed T, Gordon JI. Gut microbiome development and childhood undernutrition. Cell Host Microbe. 2022;30(5):617–626. doi: 10.1016/j.chom.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, Raqib R, Underwood MA, Mills DA, Stephensen CB. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. 2019;143(2). doi: 10.1542/peds.2018-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taft DH, Lewis ZT, Nguyen N, Ho S, Masarweh C, Dunne-Castagna V, Tancredi DJ, Huda MN, Stephensen CB, Hinde K, et al. Bifidobacterium species colonization in infancy: a global cross-sectional comparison by population history of breastfeeding. Nutrients. 2022;14(7):1423. doi: 10.3390/nu14071423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tannock GW, Lee PS, Wong KH, Lawley B. Why don’t all infants have bifidobacteria in their stool? Front Microbiol. 2016;7:834. doi: 10.3389/fmicb.2016.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vatanen T, Plichta DR, Somani J, Münch PC, Arthur TD, Hall AB, Rudolf S, Oakeley EJ, Ke X, Young RA, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol. 2019;4(3):470–479. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA, Suen G, Suen G. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere. 2018;3(5). doi: 10.1128/mSphere.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seppo AE, Bu K, Jumabaeva M, Thakar J, Choudhury RA, Yonemitsu C, Bode L, Martina CA, Allen M, Tamburini S, et al. Infant gut microbiome is enriched with Bifidobacterium longum ssp. infantis in old order Mennonites with traditional farming lifestyle. Allergy. 2021;76(11):3489–3503. doi: 10.1111/all.14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu J, Lawley B, Wong G, Otal A, Chen L, Ying TJ, Lin X, Pang WW, Yap F, Chong Y-S, et al. Ethnic diversity in infant gut microbiota is apparent before the introduction of complementary diets. Gut Microbes. 2020;11(5):1362–1373. doi: 10.1080/19490976.2020.1756150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jordan A, Carding SR, Hall LJ. The early-life gut microbiome and vaccine efficacy. Lancet Microbe. 2022;3(10):e787–94. doi: 10.1016/S2666-5247(22)00185-9. [DOI] [PubMed] [Google Scholar]
- 77.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, et al. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol. 2004;11(4):686–690. doi: 10.1128/CDLI.11.4.686-690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen A-M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davis JCC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, Mills DA, Lebrilla CB, Zivkovic AM. Growth and morbidity of Gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. 2017;7(1):40466. doi: 10.1038/srep40466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lawley B, Otal A, Moloney-Geany K, Diana A, Houghton L, Heath AL, Taylor RW, Tannock GW, Elkins CA. Fecal Microbiotas of Indonesian and New Zealand children differ in complexity and bifidobacterial taxa during the first year of life. Appl Environ Microbiol. 2019;85(19). doi: 10.1128/AEM.01105-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casaburi G, Duar RM, Brown H, Mitchell RD, Kazi S, Chew S, Cagney O, Flannery RL, Sylvester KG, Frese SA, et al. Metagenomic insights of the infant microbiome community structure and function across multiple sites in the United States. Sci Rep. 2021;11(1):1472. doi: 10.1038/s41598-020-80583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin Y-S, German JB, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3(1):13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134(2):e362–72. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olm MR, Dahan D, Carter MM, Merrill BD, Yu FB, Jain S, Meng X, Tripathi S, Wastyk H, Neff N, et al. Robust variation in infant gut microbiome assembly across a spectrum of lifestyles. Science. 2022;376(6598):1220–1223. doi: 10.1126/science.abj2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res. 2015;77(1–2):229–235. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bello MGD, Knight R, Gilbert JA, Blaser MJ. Preserving microbial diversity. Science. 2018;362(6410):33–34. doi: 10.1126/science.aau8816. [DOI] [PubMed] [Google Scholar]
- 88.Sonnenburg JL, Sonnenburg ED. Vulnerability of the industrialized microbiota. Science. 2019;366(6464). doi: 10.1126/science.aaw9255. [DOI] [PubMed] [Google Scholar]
- 89.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber DA, Wu F, Gi PP, Chen Y, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davis EC, Castagna VP, Sela DA, Hillard MA, Lindberg S, Mantis NJ, Seppo AE, Järvinen KM. Gut microbiome and breast-feeding: implications for early immune development. J Allergy Clin Immunol. 2022;150(3):523–534. doi: 10.1016/j.jaci.2022.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Healy DB, Ryan CA, Ross RP, Stanton C, Dempsey EM. Clinical implications of preterm infant gut microbiome development. Nat Microbiol. 2022;7(1):22–33. doi: 10.1038/s41564-021-01025-4. [DOI] [PubMed] [Google Scholar]
- 93.Zwittink RD, Renes IB, van Lingen RA, van Zoeren-Grobben D, Konstanti P, Norbruis OF, Martin R, Groot Jebbink LJM, Knol J, Belzer C. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur J Clin Microbiol Infect Dis. 2018;37(3):475–483. doi: 10.1007/s10096-018-3193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henderickx JGE, Zwittink RD, van Lingen RA, Knol J, Belzer C. The preterm gut microbiota: an inconspicuous challenge in nutritional neonatal care. Front Cell Infect Microbiol. 2019;9:85. doi: 10.3389/fcimb.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goldmann DA. The bacterial flora of neonates in intensive care-monitoring and manipulation. J Hosp Infect. 1988;11(Suppl A):340–351. doi: 10.1016/0195-6701(88)90209-5. [DOI] [PubMed] [Google Scholar]
- 96.Casaburi G, Wei J, Kazi S, Liu J, Wang K, Tao G-Z, Lin P-Y, Dunn JCY, Henrick BM, Frese SA, et al. Metabolic model of necrotizing enterocolitis in the premature newborn gut resulting from enteric dysbiosis. Front Pediatr. 2022;10:893059. doi: 10.3389/fped.2022.893059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lawrence G, Bates J, Gaul A. Pathogenesis of neonatal necrotising enterocolitis. Lancet. 1982;1(8264):137–139. doi: 10.1016/S0140-6736(82)90383-X. [DOI] [PubMed] [Google Scholar]
- 98.Deshpande G, Rao S, Patole S. Probiotics for prevention of necrotising enterocolitis in preterm neonates with very low birthweight: a systematic review of randomised controlled trials. Lancet. 2007;369(9573):1614–1620. doi: 10.1016/S0140-6736(07)60748-X. [DOI] [PubMed] [Google Scholar]
- 99.Morgan RL, Preidis GA, Kashyap PC, Weizman AV, Sadeghirad B, Chang Y, Florez ID, Foroutan F, Shahid S, Zeraatkar D, et al. Probiotics reduce mortality and morbidity in preterm, low-birth-weight infants: a systematic review and network meta-analysis of randomized trials. Gastroenterology. 2020;159(2):467–480. doi: 10.1053/j.gastro.2020.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deshmukh M, Patole S. Prophylactic probiotic supplementation for preterm neonates—a systematic review and meta-analysis of nonrandomized studies. Adv Nutr. 2021;12(4):1411–1423. doi: 10.1093/advances/nmaa164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharif S, Meader N, Oddie SJ, Rojas-Reyes MX, McGuire W. Probiotics to prevent necrotising enterocolitis in very preterm or very low birth weight infants. Cochrane Database Syst Rev. 2020;10(10):CD005496. doi: 10.1002/14651858.CD005496.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbian ME, Patel RM. Probiotics for prevention of necrotizing enterocolitis: where do we stand? Semin Perinatol. 2022;47(1):151689. doi: 10.1016/j.semperi.2022.151689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neu J. Routine probiotics for premature infants: let’s be careful! J Pediatr. 2011;158(4):672–674. doi: 10.1016/j.jpeds.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Poindexter B, Cummings J, Hand I, Adams-Chapman I, Aucott SW, Puopolo KM, Goldsmith JP, Kaufman D, Martin C, Mowitz M, et al. Use of probiotics in preterm infants. Pediatrics. 2021;147(6). doi: 10.1542/peds.2021-051485. [DOI] [PubMed] [Google Scholar]
- 105.WHO recommendations for care of the preterm or low-birth-weight infant. Geneva: World Health Organization; 2022. https://www.ncbi.nlm.nih.gov/books/NBK586704/. [PubMed] [Google Scholar]
- 106.Duar RM, Henrick BM, Casaburi G, Frese SA. Integrating the ecosystem services framework to define dysbiosis of the breastfed infant gut: the role of B. infantis and human milk oligosaccharides. Front Nutr. 2020;7:33. doi: 10.3389/fnut.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159(2):253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taft DH, Ambalavanan N, Schibler KR, Yu Z, Newburg DS, Ward DV, Morrow AL. Intestinal microbiota of preterm infants differ over time and between hospitals. Microbiome. 2014;2(1):36. doi: 10.1186/2049-2618-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Deshpande G, Rao S, Patole S, Bulsara M. Updated meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Pediatrics. 2010;125(5):921–930. doi: 10.1542/peds.2009-1301. [DOI] [PubMed] [Google Scholar]
- 110.Bollrath J, Powrie F. Feed your Tregs more fiber. Science. 2013. Immunology;341(6145):463–464. doi: 10.1126/science.1242674. [DOI] [PubMed] [Google Scholar]
- 111.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469(7331):543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 115.O’brien CE, Meier AK, Cernioglo K, Mitchell RD, Casaburi G, Frese SA, Henrick BM, Underwood MA, Smilowitz JT. Early probiotic supplementation with B. infantis in breastfed infants leads to persistent colonization at 1 year. Pediatr Res. 2022;91(3):627–636. doi: 10.1038/s41390-020-01350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, Xu G, Davis JCC, Lebrilla CB, Henrick BM, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2(6). doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Larke JA, Kuhn-Riordon K, Taft DH, Sohn K, Iqbal S, Underwood MA, Mills DA, Slupsky CM. Preterm infant fecal microbiota and metabolite profiles are modulated in a probiotic specific manner. J Pediatr Gastroenterol Nutr. 2022;75(4):535–542. doi: 10.1097/MPG.0000000000003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alcon-Giner C, Dalby MJ, Caim S, Ketskemety J, Shaw A, Sim K, Lawson MAE, Kiu R, Leclaire C, Chalklen L, et al. Microbiota supplementation with Bifidobacterium and Lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study. Cell Rep Med. 2020;1(5):100077. doi: 10.1016/j.xcrm.2020.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58(9):5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Turroni F, Bottacini F, Foroni E, Mulder I, Kim J-H, Zomer A, Sánchez B, Bidossi A, Ferrarini A, Giubellini V, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Watkins C, Murphy K, Dempsey EM, O’shea CA, Murphy BP, O’toole PW, Ross RP, Stanton C, Ryan CA. Dose-interval study of a dual probiotic in preterm infants. Arch Dis Child Fetal Neonatal Ed. 2019;104(2):F159–64. doi: 10.1136/archdischild-2017-313468. [DOI] [PubMed] [Google Scholar]
- 122.Samara J, Moossavi S, Alshaikh B, Ortega VA, Pettersen VK, Ferdous T, Hoops SL, Soraisham A, Vayalumkal J, Dersch-Mills D, et al. Supplementation with a probiotic mixture accelerates gut microbiome maturation and reduces intestinal inflammation in extremely preterm infants. Cell Host Microbe. 2022;30(5):696–711.e5. doi: 10.1016/j.chom.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 123.Beck LC, Masi AC, Young GR, Vatanen T, Lamb CA, Smith R, Coxhead J, Butler A, Marsland BJ, Embleton ND, et al. Strain-specific impacts of probiotics are a significant driver of gut microbiome development in very preterm infants. Nat Microbiol. 2022;7(10):1525–1535. doi: 10.1038/s41564-022-01213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Larsen IS, Jensen BAH, Bonazzi E, Choi BSY, Kristensen NN, Schmidt EGW, Süenderhauf A, Morin L, Olsen PB, Hansen LBS, et al. Fungal lysozyme leverages the gut microbiota to curb DSS-induced colitis. Gut Microbes. 2021;13(1):1988836. doi: 10.1080/19490976.2021.1988836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dunne-Castagna VP, Mills DA, Lönnerdal B. Effects of milk secretory immunoglobulin a on the commensal microbiota. Nestle Nutr Inst Workshop Ser. 2020;94:158–168. [DOI] [PubMed] [Google Scholar]
- 126.Lönnerdal B, Iyer S. Lactoferrin: molecular structure and biological function. Annu Rev Nutr. 1995;15(1):93–110. doi: 10.1146/annurev.nu.15.070195.000521. [DOI] [PubMed] [Google Scholar]
- 127.Liepke C, Zucht H-D, Forssmann W-G, Ständker L. Purification of novel peptide antibiotics from human milk. J Chromatogr B Biomed Sci Appl. 2001;752(2):369–377. doi: 10.1016/S0378-4347(00)00516-8. [DOI] [PubMed] [Google Scholar]
- 128.Jones EM, Smart A, Bloomberg G, Burgess L, Millar MR. Lactoferricin, a new antimicrobial peptide. J Appl Bacteriol. 1994;77(2):208–214. doi: 10.1111/j.1365-2672.1994.tb03065.x. [DOI] [PubMed] [Google Scholar]
- 129.Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12(5):2295–2304. doi: 10.1021/pr400212z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schlievert PM, Kilgore SH, Seo KS, Leung DYM. Glycerol monolaurate contributes to the antimicrobial and anti-inflammatory activity of human milk. Sci Rep. 2019;9(1):14550. doi: 10.1038/s41598-019-51130-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shin K, Tomita M, Lönnerdal B. Identification of lactoperoxidase in mature human milk. J Nutr Biochem. 2000;11(2):94–102. doi: 10.1016/S0955-2863(99)00082-0. [DOI] [PubMed] [Google Scholar]
- 132.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4glcnac), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278(16):14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 133.Liepke C, Adermann K, Raida M, Mägert H-J, Forssmann W-G, Zucht H-D. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur J Biochem. 2002;269(2):712–718. doi: 10.1046/j.0014-2956.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 134.Oda H, Wakabayashi H, Yamauchi K, Sato T, Xiao J-Z, Abe F, Iwatsuki K. Isolation of a bifidogenic peptide from the pepsin hydrolysate of bovine lactoferrin. Appl Environ Microbiol. 2013;79(6):1843–1849. doi: 10.1128/AEM.03343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Robinson RC. Structures and metabolic properties of bovine milk oligosaccharides and their potential in the development of novel therapeutics. Front Nutr. 2019;6:50. doi: 10.3389/fnut.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marsaux B, Van den Abbeele P, Ghyselinck J, Prioult G, Marzorati M, Bogićević B. Synbiotic Effect of Bifidobacterium lactis CNCM I-3446 and bovine milk-derived oligosaccharides on infant gut microbiota. Nutrients. 2020;12(8):2268. doi: 10.3390/nu12082268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heiss BE, Ehrlich AM, Maldonado-Gomez MX, Taft DH, Larke JA, Goodson ML, Slupsky CM, Tancredi DJ, Raybould HE, Mills DA. Bifidobacterium catabolism of human milk oligosaccharides overrides endogenous competitive exclusion driving colonization and protection. Gut Microbes. 2021;13(1):1986666. doi: 10.1080/19490976.2021.1986666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Olsan EE, Byndloss MX, Faber F, Rivera-Chávez F, Tsolis RM, Bäumler AJ. Colonization resistance: the deconvolution of a complex trait. J Biol Chem. 2017;292(21):8577–8581. doi: 10.1074/jbc.R116.752295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Albright MBN, Louca S, Winkler DE, Feeser KL, Haig S-J, Whiteson KL, Emerson JB, Dunbar J. Solutions in microbiome engineering: prioritizing barriers to organism establishment. Isme J. 2022;16(2):331–338. doi: 10.1038/s41396-021-01088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557(7705):434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kearney SM, Gibbons SM, Erdman SE, Alm EJ. Orthogonal dietary niche enables reversible engraftment of a gut bacterial commensal. Cell Rep. 2018;24(7):1842–1851. doi: 10.1016/j.celrep.2018.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Button JE, Autran CA, Reens AL, Cosetta CM, Smriga S, Ericson M, Pierce JV, Cook DN, Lee ML, Sun AK, et al. Dosing a synbiotic of human milk oligosaccharides and B. infantis leads to reversible engraftment in healthy adult microbiomes without antibiotics. Cell Host Microbe. 2022;30(5):712–725.e7. doi: 10.1016/j.chom.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 143.Musilova S, Modrackova N, Hermanova P, Hudcovic T, Svejstil R, Rada V, Tejnecky V, Bunesova V. Assessment of the synbiotic properites of human milk oligosaccharides and Bifidobacterium longum subsp. infantis in vitro and in humanised mice. Benef Microbes. 2017;8(2):281–289. doi: 10.3920/BM2016.0138. [DOI] [PubMed] [Google Scholar]
- 144.Henrick BM, Hutton AA, Palumbo MC, Casaburi G, Mitchell RD, Underwood MA, Smilowitz JT, Frese SA, Oh J. Elevated fecal ph indicates a profound change in the breastfed infant gut microbiome due to reduction of bifidobacterium over the past century. mSphere. 2018;3(2). doi: 10.1128/mSphere.00041-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hossain MS, Das S, Gazi MA, Alam MA, Haque NMS, Mahfuz M, Ahmed T, Damman CJ. Association of faecal pH with childhood stunting: results from a cross-sectional study. BMJ Peds Open. 2019;3(1):e000549. doi: 10.1136/bmjpo-2019-000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Washabaugh JR. First foods. Intestinal Ecology, and Early Life Health and Growth Outcomes [Doctoral dissertation].Boulder (CO): University of Colorado; 2021. [Google Scholar]
- 147.Liu M, Nieuwdorp M, de Vos WM, Rampanelli E. Microbial tryptophan metabolism tunes host immunity, metabolism, and extraintestinal disorders. Metabolites. 2022;12(9):834. doi: 10.3390/metabo12090834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aragozzini F, Ferrari A, Pacini N, Gualandris R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol. 1979;38(3):544–546. doi: 10.1128/aem.38.3.544-546.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23(6):716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 150.Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, Zhu W, Walker WA. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020;88(2):209–217. doi: 10.1038/s41390-019-0740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, Pekmez CT, Rivollier A, Michaelsen KF, Mølgaard C, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. 2021;6(11):1367–1382. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sakurai T, Odamaki T, Xiao J-Z. Production of indole-3-lactic acid by Bifidobacterium strains isolated from human infants. Microorganisms. 2019;7(9):340. doi: 10.3390/microorganisms7090340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ehrlich AM, Pacheco AR, Henrick BM, Taft D, Xu G, Huda MN, Mishchuk D, Goodson ML, Slupsky C, Barile D, et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020;20(1):357. doi: 10.1186/s12866-020-02023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, Olin A, Wang J, Mikes J, Tan Z, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184(15):3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 155.Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 156.Jena PK, Sheng L, Nagar N, Wu C, Barile D, Mills DA, Wan YJ. Synbiotics Bifidobacterium infantis and milk oligosaccharides are effective in reversing cancer-prone nonalcoholic steatohepatitis using western diet-fed FXR knockout mouse models. J Nutr Biochem. 2018;57:246–254. doi: 10.1016/j.jnutbio.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, Underwood MA, Smilowitz JT. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. 2019;86(6):749–757. doi: 10.1038/s41390-019-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, Smilowitz JT, Underwood MA. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2019;8(1):131. doi: 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nguyen M, Holdbrooks H, Mishra P, Abrantes MA, Eskew S, Garma M, Oca C-G, McGuckin C, Hein CB, Mitchell RD, et al. Impact of probiotic B. infantis EVC001 feeding in premature infants on the gut microbiome, nosocomially acquired antibiotic resistance, and enteric inflammation. Front Pediatr. 2021;9:618009. doi: 10.3389/fped.2021.618009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Costeloe K, Hardy P, Juszczak E, Wilks M, Millar MR, Probiotics in Preterm Infants Study Collaborative Group . Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial. Lancet. 2016;387(10019):649–660. doi: 10.1016/S0140-6736(15)01027-2. [DOI] [PubMed] [Google Scholar]
- 161.Millar M, Seale J, Greenland M, Hardy P, Juszczak E, Wilks M, Panton N, Costeloe K, Wade WG. The microbiome of infants recruited to a randomised placebo-controlled probiotic trial (PiPS Trial). EBioMedicine. 2017;20:255–262. doi: 10.1016/j.ebiom.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Robertson C, Savva GM, Clapuci R, Jones J, Maimouni H, Brown E, Minocha A, Hall LJ, Clarke P. Incidence of necrotising enterocolitis before and after introducing routine prophylactic Lactobacillus and Bifidobacterium probiotics. Arch Dis Child Fetal Neonatal Ed. 2020;105(4):380–386. doi: 10.1136/archdischild-2019-317346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Denkel LA, Schwab F, Garten L, Geffers C, Gastmeier P, Piening B, Kollmann TR. Protective effect of dual-strain probiotics in preterm infants: a multi-center time series analysis. PLoS ONE. 2016;11(6):e0158136. doi: 10.1371/journal.pone.0158136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guthmann F, Kluthe C, Bührer C. Probiotics for prevention of necrotising enterocolitis: an updated meta-analysis. Klinische Pädiatrie. 2010;222(05):284–290. doi: 10.1055/s-0030-1254113. [DOI] [PubMed] [Google Scholar]
- 165.Lin H-C, Hsu C-H, Chen H-L, Chung M-Y, Hsu J-F, Lien R, Tsao L-Y, Chen C-H, B-H S. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008;122(4):693–700. doi: 10.1542/peds.2007-3007. [DOI] [PubMed] [Google Scholar]
- 166.Hoyos AB. Reduced incidence of necrotizing enterocolitis associated with enteral administration of Lactobacillus acidophilus and Bifidobacterium infantis to neonates in an intensive care unit. Int J Infect Dis. 1999;3(4):197–202. doi: 10.1016/S1201-9712(99)90024-3. [DOI] [PubMed] [Google Scholar]
- 167.Guthmann F, Arlettaz Mieth RP, Bucher HU, Bührer C. Short courses of dual-strain probiotics appear to be effective in reducing necrotising enterocolitis. Acta Paediatr. 2016;105(3):255–259. doi: 10.1111/apa.13280. [DOI] [PubMed] [Google Scholar]
- 168.Repa A, Thanhaeuser M, Endress D, Weber M, Kreissl A, Binder C, Berger A, Haiden N. Probiotics (Lactobacillus acidophilus and Bifidobacterium infantis) prevent NEC in VLBW infants fed breast milk but not formula [corrected]. Pediatr Res. 2015;77(2):381–388. doi: 10.1038/pr.2014.192. [DOI] [PubMed] [Google Scholar]
- 169.Härtel C, Pagel J, Rupp J, Bendiks M, Guthmann F, Rieger-Fackeldey E, Heckmann M, Franz A, Schiffmann J-H, Zimmermann B, et al. Prophylactic use of Lactobacillus acidophilus/Bifidobacterium infantis probiotics and outcome in very low birth weight infants. J Pediatr. 2014;165(2):285–289.e1. doi: 10.1016/j.jpeds.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 170.Lewis ZT, Shani G, Masarweh CF, Popovic M, Frese SA, Sela DA, Underwood MA, Mills DA. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr Res. 2016;79(3):445–452. doi: 10.1038/pr.2015.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tobias J, Olyaei A, Laraway B, Jordan BK, Dickinson SL, Golzarri-Arroyo L, Fialkowski E, Owora A, Scottoline B. Bifidobacterium longum subsp. infantis EVC001 administration is associated with a significant reduction in the incidence of necrotizing enterocolitis in very low birth weight infants. J Pediatr. 2022;244:64–71.e2. doi: 10.1016/j.jpeds.2021.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lebeer S, Bron PA, Marco ML, Van Pijkeren J-P, O’connell Motherway M, Hill C, Pot B, Roos S, Klaenhammer T. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol. 2018;49:217–223. doi: 10.1016/j.copbio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 173.Sanders ME, Benson A, Lebeer S, Merenstein DJ, Klaenhammer TR. Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr Opin Biotechnol. 2018;49:207–216. doi: 10.1016/j.copbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 174.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25(5):716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 175.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ojima MN, Jiang L, Arzamasov AA, Yoshida K, Odamaki T, Xiao J, Nakajima A, Kitaoka M, Hirose J, Urashima T, et al. Priority effects shape the structure of infant-type Bifidobacterium communities on human milk oligosaccharides. Isme J. 2022;16:2265–2279. doi: 10.1038/s41396-022-01270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, Sanders ME, Shamir R, Swann JR, Szajewska H, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. 2021;18(9):649–667. doi: 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 179.Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, Verbeke K, Scott KP, Holscher HD, Azad MB, Delzenne NM, et al. The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. 2020;17(11):687–701. doi: 10.1038/s41575-020-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Orla-Jensen . La classification des bactéries lactiques. Le Lait. 1924;4(36):468–474. doi: 10.1051/lait:19243627. [DOI] [Google Scholar]
- 181.Reuter G. Comparative studies on the bifidus flora in the feces of infants and adults. with a contribution to classification and nomenclature of bifidus strains. Zentralbl Bakteriol Orig. 1963;191:486–507. [PubMed] [Google Scholar]
- 182.Metchnikoff E. The prolongation of life. Optimistic Studies. G. P. Putnam’s Sons. 1908. [Google Scholar]
- 183.Ozen M, Dinleyici EC. The history of probiotics: the untold story. Benef Microbes. 2015;6(2):159–165. doi: 10.3920/BM2014.0103. [DOI] [PubMed] [Google Scholar]
- 184.de Kruif P. The microbe hunters. New York: Blue Ribbon Books; 1926. [Google Scholar]
- 185.Atlas RM. Probiotics–snake oil for the new millennium? Environ Microbiol. 1999;1(5):377–382. doi: 10.1046/j.1462-2920.1999.00063.x. [DOI] [PubMed] [Google Scholar]
- 186.Berg RD. Probiotics, prebiotics or “conbiotics”? Trends Microbiol. 1998;6(3):89–92. doi: 10.1016/S0966-842X(98)01224-4. [DOI] [PubMed] [Google Scholar]
- 187.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3(106):106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rene Dubois The dreams of reason: science and utopias xiixii, Vol. 167New York: Columbia University Press, Ann Am Acad Pol Soc Sci. 1962. pp. 199 [Google Scholar]
- 189.Underwood MA, Salzman NH, Bennett SH, Barman M, Mills DA, Marcobal A, Tancredi DJ, Bevins CL, Sherman MP. A randomized placebo-controlled comparison of 2 prebiotic/probiotic combinations in preterm infants: impact on weight gain, intestinal microbiota, and fecal short-chain fatty acids. J Pediatr Gastroenterol Nutr. 2009;48(2):216–225. doi: 10.1097/MPG.0b013e31818de195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Underwood MA, Kalanetra KM, Bokulich NA, Lewis ZT, Mirmiran M, Tancredi DJ, Mills DA. A comparison of two probiotic strains of bifidobacteria in premature infants. J Pediatr. 2013;163(6):1585–1591.e9. doi: 10.1016/j.jpeds.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ellis CL, Bokulich NA, Kalanetra KM, Mirmiran M, Elumalai J, Haapanen L, Schegg T, Rutledge JC, Raff G, Mills DA, et al. Probiotic administration in congenital heart disease: a pilot study. J Perinatol. 2013;33(9):691–697. doi: 10.1038/jp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Powell WT, Borghese RA, Kalanetra KM, Mirmiran M, Mills DA, Underwood MA. Probiotic administration in infants with gastroschisis: a pilot randomized placebo-controlled trial. J Pediatr Gastroenterol Nutr. 2016;62(6):852–857. doi: 10.1097/MPG.0000000000001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Underwood MA, Davis JCC, Kalanetra KM, Gehlot S, Patole S, Tancredi DJ, Mills DA, Lebrilla CB, Simmer K. Digestion of human milk oligosaccharides by Bifidobacterium breve in the premature Infant. J Pediatr Gastroenterol Nutr. 2017;65(4):449–455. doi: 10.1097/MPG.0000000000001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xiao J-Z, Takahashi S, Odamaki T, Yaeshima T, Iwatsuki K. Antibiotic susceptibility of bifidobacterial strains distributed in the Japanese market. Biosci Biotechnol Biochem. 2010;74(2):336–342. doi: 10.1271/bbb.90659. [DOI] [PubMed] [Google Scholar]
- 195.Esaiassen E, Hjerde E, Cavanagh JP, Simonsen GS, Klingenberg C, Ledeboer NA, Norwegian Study Group on Invasive Bifidobacterial Infections . Bifidobacterium bacteremia: clinical characteristics and a genomic approach to assess pathogenicity. J Clin Microbiol. 2017;55(7):2234–2248. doi: 10.1128/JCM.00150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]


