Abstract
The purpose of this study is to develop a text clustering–based analysis of COVID-19 research articles. Owing to the proliferation of published COVID-19 research articles, researchers need a method for reducing the number of articles they have to search through to find material relevant to their expertise. The study analyzes 83,264 abstracts from research articles related to COVID-19. The textual data are analysed using singular value decomposition (SVD) and the expectation–maximisation (EM) algorithm. Results suggest that text clustering can both reveal hidden research themes in the published literature related to COVID-19, and reduce the number of articles that researchers need to search through to find material relevant to their field of interest.
Keywords: Abstract clustering, COVID-19 Open Research Data set, expectation–maximisation algorithm, singular value decomposition, text clustering
1. Introduction
In recent years, big data science has emerged as a powerful tool for collecting, storing, managing and analysing data on a large scale. Big data can be characterised as data that are too large or too unconventional to be stored in a traditional row and column database [1]. Textual data are unstructured data and are a form of big data. Text mining uses artificial intelligence and natural language processing (NLP) techniques that turn free text into data that can be analysed using statistical machine learning techniques. Text mining allows researchers to filter through large groups of texts that would take far too long for humans to analyse.
COVID-19, caused by the novel coronavirus SARS-CoV-2, was declared a public health emergency by the World Health Organization (WHO) on 30 January 2020 [2]. COVID-19 is a newly emerging infectious disease, and has motivated a plethora of authors to write about it. By 12 March 2020, it is estimated that over 900 papers, reprints and preliminary reports had been published about COVID-19 [3]. At the time of writing this article, a search for ‘coronavirus’ in the search engine Web of Science generates 7801 peer-reviewed journal articles.
Organisations such as the Center for Disease Control and Prevention, the major academic publishing companies, and research universities have created websites for authors, public health officials and researchers to access the most relevant, up-to-date, articles related to COVID-19. However, even with the assistance of journal lists, it is impossible for a public health official or a researcher to sift through the entirety of this information and find the information that is needed to make decisions and advance research into vaccines, therapeutics and mitigation efforts that could help to slow the spread of the disease. From January 2020 to mid-May 2020, there were an estimated 23,000 papers published related to COVID-19, and the rate of publication was then doubling every 20 days, making it one of the most rapid expansions of the scientific literature [4].
A sound body of literature is an essential prerequisite for scientific research at any level. This is especially true during an outbreak of a novel virus. Many different people will need to access the literature to answer a wide range of questions. For example, a state epidemiologist would need efficient ways to sift through journal articles to find the best ways of managing the surge capacity of local hospitals in order to determine if there are effective measures that need to be put in place to protect vulnerable populations or to learn about those risk factors that have been identified from past epidemiological studies. A virologist would be interested in accessing the literature to conduct a scientific review of how similar SARS-CoV-2 is to other viruses, so as to understand how the virus is transmitted [5]. A clinician would be interested in emerging potential treatment options, as well as nonpharmaceutical interventions, and needs to monitor the most up-to-date information about the treatment of patients [6].
The aim of this article is to use NLP techniques and to conduct a cluster analysis on a set of abstracts of articles related to COVID-19, SARS-CoV-2 and other related coronaviruses. Clustering similar research article abstracts simplifies the search for related articles. Abstract clusters can be thought of as topics of research. Given the large number of publications related to COVID-19, it is difficult for the medical community and researchers to keep up with new information related to the disease. The NLP clustering techniques proposed in this study can assist in reducing the amount of information that a researcher has to sift through in order to find relevant articles and research. There is a growing urgency for these types of text mining techniques to be applied and implemented, so that the researchers and the medical community can more rapidly find the articles most relevant to their research into COVID-19.
The main contributions of this article are as follows:
Apply a clustering technique that applies the expectation–maximisation (EM) algorithm in order to cluster a large corpus related to the COVID-19 corpus.
Individual researchers and research bodies face the genuine problem of sifting through mountains of papers related – to some degree – to their expertise. The techniques used here are aimed at eliminating from researchers’ purview the papers that are less relevant, and so shortening their sifting time.
By enabling researchers to give more attention to their field of expertise, the techniques used here, and the results therefrom, will be making a useful contribution to COVID-19 research.
The application of text mining techniques to a very large number of COVID-19 published papers is novel, as would be the use of the results to sharpen the focus of researchers and, one would hope, therefore accelerate researchers’ finding solutions to the problems caused by the pandemic.
The remaining section of this article is organised as follows: section 2 reviews some of the existing works related to text mining methods for tracking infectious diseases, section 3 provides the detailed methodology used to cluster the abstracts, section 4 presents the results from the text clustering algorithm and section 5 concludes with future works that could be investigated.
2. Literature review
Several research studies have focused on the processing of textual information related to monitoring and tracking infectious diseases. A brief overview of such studies that highlight the significance of text mining is presented here.
In the past several years, much progress has been made in using NLP to advance the understanding of infectious diseases. Jahanbin et al. [7] mined Twitter data to collect social media posts related to influenza, HIV/AIDS, malaria, measles, poliomyelitis, tuberculosis, plague, Ebola and cholera. The authors created a real-time monitoring system to detect where the majority of Tweets related to each of these infectious diseases were occurring around the world. Monitoring the occurrence of an infectious disease is one way in which public health officials can track outbreaks. Ascertaining whether there are many social media feeds discussing an infectious disease has the potential to be a more effective way of detecting outbreaks than traditional public health methods [7].
Currently, researchers are using several different text mining methodologies to understand different aspects of COVID-19. Han et al. [8] used social media posts from China during the early outbreak of COVID-19 to gauge public opinion regarding the disease. The authors utilised a latent Dirichlet allocation model to categorise the posts into seven topics. Then, they examined the relationship between the topics of social media posts and the intensity of the outbreak in the east-central parts of China. The authors found that timely release of information from the Chinese government was helpful in calming public opinion in the early stages of COVID-19. Public opinion can be easily swayed on social media platforms owing to ‘fake news’ and ‘troll opinions’. The type of text mining study that Han et al. [8] conducted can assist the government in understanding public opinion and sentiment towards COVID-19, thus can better support and direct emergency assistance to the public during the pandemic.
In another work, Nguyen [9] provides a recent review of text mining and NLP in the context of COVID-19. The review provides several examples of how researchers use text mining applications on Twitter data to develop infection rate forecasting, and to better understand changing government policies and responses to COVID-19.
There has been success outside the medical field in the text clustering of abstracts in assisting researchers to focus on their field of expertise. Noble and Gamit [10] provide a study in which they cluster the abstract research proposals submitted to the National Science Foundation (NSF) for the years 1985 to 2019. The purpose of clustering the abstracts was to identify similar NSF funding proposals based on the abstracts. Grouping the abstracts into clusters allows awardees to find potential collaborators as well as similar research projects with less manual searching [10]. Carnerud [11] clustered the abstracts of the academic journal International Journal of Quality & Reliability Management, which focuses on the concept of manufacturing improvements and consists of more than 240 volumes from 1984. The focus of the study was to identify published research trends over the years. The research findings of this study allow researchers who are interested in publishing in this particular journal to quickly understand its historical research trends when deciding whether the journal is a good fit for the researcher’s manuscript [11].
3. Methodology
3.1. Data
This study uses the COVID-19 Open Research Data set (CORD-19) [12]. On 16 March 2020, the US President Trump, in collaboration with tech companies and research institutions, issued a call to action for global artificial intelligence researchers to develop novel text and data mining methodologies to assist COVID-19 researchers. The Allen Institute for Artificial Intelligence partnered with the White House, Semantic Scholar and leading research institutes to compile a corpus of COVID-19 and past coronavirus-related articles and publications. The corpus primarily comprises articles published by the WHO, PubMed Central, bioRxiv and medRxiv. The original release consisted of approximately 28,000 papers, and the corpus is updated periodically [12]. The Semantic Scholar team at the Allen Institute for Artificial Intelligence harmonises the articles’ metadata, and converts the pdf articles into JSON files that are machine readable by text mining algorithms. This study utilises 83,264 abstracts from the research articles that were available on the website at the time of this analysis. Figure 1 displays the frequencies of word counts for each of the abstracts in the corpus. The average word count of the abstracts was 75, with the maximum number of words being 137.
Figure 1.
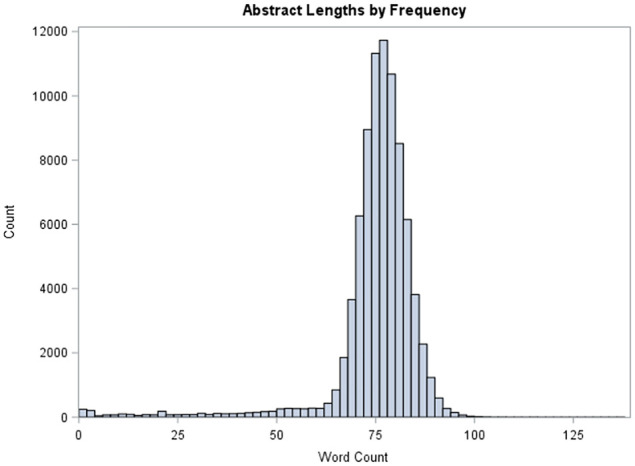
Abstract lengths by frequency.
3.2. Data pre-processing
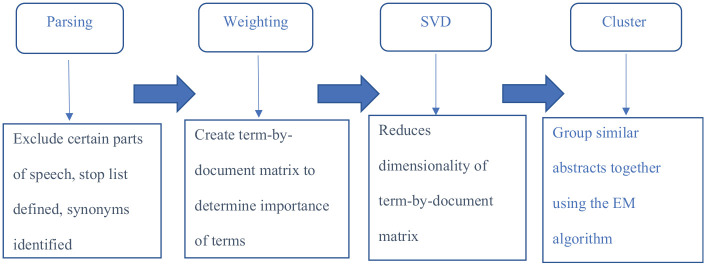
Figure 2 displays each step used in the methodology of clustering the corpus into groups. This analysis uses the SAS Text Miner 14.1 to perform the analysis. One of the first steps is to parse the documents in the corpus into terms that would be used in the clustering algorithm. A term is defined as a character string that has a specific meaning in a given language. Part of the parsing process involves excluding parts of speech that are common, such as conjunctions and prepositions. In addition, parsing requires a stop list to be specified for terms that do not provide any meaning for the analysis. Other than the common stop words that the SAS Text Miner uses, such as ‘the’, ‘at’, ‘can’ and ‘other’, Table 1 shows the additional stop words that are specified. These stop words were compiled by analysing word count frequencies in the corpus.
Figure 2.
Text mining methodology.
Table 1.
Additional stop words specified for the COVID-19 Open Research Data set.
| Stop words |
|---|
| author, preprint, copyright, peer, reviewed, org, https, et al., figure, table, rights, reserved, introduction, permission, biorxiv, medrxiv, licence, Elsevier, PMC, bibref, abstract, volume, issue, cite, ref, ref id, ref spans, issn, result, discussion, study, back matter |
Parsing also requires specifying synonyms so that different terms are processed equivalently. SAS Text Miner has a list of standard synonyms that the software employs. However, for this research, defining synonyms for newly emerging diseases can be difficult. For example, in the early stages of the pandemic, COVID-19 was sometimes referred to as ‘Wuhan seafood market pneumonia’ [8]. A synonym list created specifically for SARS-CoV-2 and COVID-19 terms [13] is imported into SAS Text Miner. Table 2 provides some of the synonyms used. It is not a comprehensive list: in the analysis, 12,915 specialty synonyms are used.
Table 2.
An example of synonyms for the COVID-19 Open Research Data set.
| Synonyms |
|---|
| Covid19, covid 19, coronavirus 2019, coronavirus disease 2019, 2019 novel coronavirus respiratory syndrome, severe acute respiratory syndrome 2019, seafood market pneumonia, 2019 seafood market virus severe disorder, covid19 virus infection, wuhan illness, hcov19 illness, 2019 novel sars cov2 illness, 2019 new sars cov2 illness, 2019 new wuhan coronavirus illness |
The next step is to transpose the parsed terms from the articles into a structured data object via a term-by-document frequency matrix. Each column of the matrix represents the parsed terms that are in each of the articles in the corpus, while the rows represent the articles themselves. Table 3 provides a schematic of what a term-by-document matrix would look like for this analysis. Since the goal of this analysis is to develop categories, the values in the term-by-document frequency matrix are the number of times the term appears in each document. Numerical measures known as term weights (i.e. global weights) can be used to determine the importance of the terms. Because the lengths of the documents in this corpus vary greatly, term weights are used to adjust the frequencies across the documents. This analysis uses entropy as term weights. The entropy in SAS Text Miner is defined as
Table 3.
Example term-by-document matrix.
| Reaction mixture | Inflammatory disease | Hospitalised patient | . . . . . . . . . . . . . | Respiratorysample | |
|---|---|---|---|---|---|
| Article (document) 1 | 253 | 257 | 215 | . . . . . . . . . . . . . | 209 |
| Article (document) 2 | 154 | 87 | 114 | . . . . . . . . . . . . . | 212 |
| . . . |
. . . . . . . . . . . . . | . . . . . . . . . . . . . | . . . . . . . . . . . . . | . . . . . . . . . . . . . | . . . |
| Article (document) 70,473 | 302 | 132 | 64 | . . . . . . . . . . . | 115 |
where wi is the weight (or entropy) of term i, f is the frequency of terms to be weighted, is the number of times term i appears in the corpus, and n is the total number of documents in the corpus; thus, is the ijth frequency in the unweighted term-by-document frequency matrix. Entropy is an appropriate term for weight because the lengths of the abstracts in this corpus vary greatly. If a term occurs uniquely in a single document, it has a high entropy, but, if that term is spread equally across documents, little information is conveyed and entropy is low [14].
Entropy is the default term weighting scheme in SAS Text Miner. There are other weighting terms that are appropriate for other text mining situations that are described as follows:
Inverse Document Frequency (IDF): this is an appropriate weighting technique if there are a few terms of interest that occur in a few documents across the corpus [15].
Mutual Information: this weighting scheme should be used when there is a categorical response variable and the purpose of the analysis is to classify texts [16].
The term-by-document matrix is extremely large and sparse, and becomes difficult to analyse from a computational standpoint. Singular value decomposition (SVD) is therefore used to reduce the dimensionality of the term-by-document matrix by simplifying the matrix into a product of three components. The term-by-document matrix is denoted by A, where A has m terms and n abstracts, and r denotes the rank of A. A is decomposed into
where U is the term eigenvectors and is an m × r matrix satisfying the orthogonality condition
where Ir × r is an r × r identity matrix, V is the abstract eigenvectors and is an r × n matrix satisfying the orthogonality condition VTV = Ir × r, and is an r × r diagonal matrix of r positive singular values σ1 ≥ σ2 ≥ … ≥ σr [17]. The singular values σi can be thought of as a measure of importance used to decide how many dimensions to keep [17].
SVD prepares the term-by-document matrix for the clustering algorithm. SVD decomposes the data into linearly independent components. These components are the semantic structures of the data [18]. Most SVD components usually have small values and can be ignored. Determining the proper number of dimensions/singular values to use is a crucial part of text mining [19]. A choice of too few dimensions will fail to reveal the latent relationships in the textual data, but too many dimensions may introduce noise [20].
This study utilises cluster analysis to cluster the abstracts of the CORD-19 data set. The cluster analysis approach generates data-driven groupings of abstracts, so that each abstract is assigned to a group (cluster) and abstracts from the same cluster are more similar to each other than abstracts from different groups. SAS Text Miner uses an EM algorithm for clustering [21]. Verma and Maiti [22] provided the mathematical details of the EM algorithm used in SAS Text Miner. The EM algorithm is the default clustering algorithm in SAS Text Miner. The EM algorithm was used to determine the final number of clusters.
In SAS Text Miner, the SVD resolution can be adjusted. Low SVD resolution is the default setting in SAS Text Miner. With a low resolution, SAS Text Miner 14.1 automatically selects a minimum number of SVD components (up to a maximum of 100) in the clustering process. This choice of low resolution for the SVD components is consistent with commonly adopted dimension choices in similar situations [20].
For each cluster developed, the root-mean-square standard deviation (RMSSD) is computed. RMSSD can be defined as the measure of goodness of fit, or the average distance between the observations in clusters. A small RMSSD value indicates that clusters are well defined, and that documents within the clusters are very similar to each other. There is no established criterion for choosing a cut-off value for RMSSD, so choosing a cut-off value is a subjective decision [22]. RMSSD was not used in determining the number of clusters for this analysis since it is a subjective decision. RMSSD is reported for each of the clusters developed from the EM algorithm.
4. Results
Table 4 shows the Topic/Cluster of each cluster, the descriptive terms associated with each cluster, the number of abstracts per cluster (frequency), the number of abstracts in each cluster as a percentage of the total number of abstracts and the RMSSD values for each cluster. The results comprise 25 clusters. The themes in the Topic/Cluster column were derived from a close examination of descriptive terms and abstracts in each cluster.
Table 4.
Text clusters for abstract clustering.
| Cluster number | Topic/Cluster (frequency) (percentage) (RMSSD) | Descriptive terms |
|---|---|---|
| 1 | Risk factors (9589) (12%) (0.123) | disease, covid-19, clinical, case, risk, treatment, infection, associate, outcome, present, factor, increase, severe, mortality, diagnosis |
| 2 | Novel inquiries for research (6717) (8%) (0.105) | model, system base, approach, research, propose, process, new, provide, present, information, network, problem, different |
| 3 | Problems in China (6022) (7%) (0.112) | covid-19 coronavirus, case, disease, spread, pandemic, China health, country, outbreak, sars-cov2, infection, cause, world, model |
| 4 | Immune response (5346) (6%) (0.113) | cell, response immune, infection, role, virus, protein, mechanism, expression, system, play, human, viral |
| 5 | Impacts of COVID-19 related to brain issues (5181) (6%) (0.1155) | effect investigate, blood, activity, drug, increase, compare, measure, change, tissue, pressure, different, cerebral, monitor |
| 6 | Testing (4462) (5%) (0.118) | test, virus, antibody, sample, sars-cov-2, assay, coronavirus, infection, detection, viral, serum, pcr, sequence, reaction |
| 7 | Animal to human transmission (4252) (5%) (0.114) | infectious disease, virus, pathogen, human species, emerge, population, control, transmission, outbreak |
| 8 | Genomic structure of virus (4141) (5%) (0.107) | protein, virus, sequence, viral, cell, gene, genome, membrane, replication, host, process, contain, encode, identify |
| 9 | Impact of COVID-19 related to surgical procedures (4002) (5%) (0.093) | laparoscopic, surgery, technique, perform, undergo, procedure, outcome, approach, compare, invasive, resection, minimally, experience |
| 10 | Public and mental health concerns (3985) (5%) (0.100) | health, covid-19, public, pandemic, care, social impact, mental, system, global, challenge, population, service, people, response |
| 11 | Healthcare workforce hazards (3747) (5%) (0.108) | covid-19, healthcare, care, coronavirus, medical health, risk, worker, hospital, practice, personal, survey, challenge, impact |
| 12 | Impacts of COVID-19 related to breathing and lung issues (3489) (4%) (0.101) | respiratory, acute, severe, syndrome, disease, sars-cov-2, covid-19, infection, outbreak, pandemic |
| 13 | Child and adult COVID-19 relationship (2864) (3%) (0.104) | child, respiratory, infection, cause, virus, acute, tract, common, clinical, low, associate, illness, adult, human |
| 14 | Clinical trial research efforts (2762) (3%) (0.115) | group, trial, control, randomise, compare, clinical, effect, treatment, design, efficacy, control trial, undergo, outcome, safety, receive |
| 15 | Impacts of COVID-19 related to cancer (2476) (3%) (0.1130) | cancer, ablation, treatment, tumour, purpose, undergo, image, radiofrequency, material, carcinoma, perform, therapy, treat, compare |
| 16 | Literature reviews (2112) (3%) (0.089) | search, systematic, systematic review, database, embase, pubmed, literature, medline, publish, conduct, article, meta-analysis |
| 17 | Critically ill patients (1985) (2%) (0.099) | intensive care unit, hospital, covid-10, admit, critical, mortality, management, practice |
| 18 | Respiratory (1946) (2%) (0.106) | respiratory, lung, ventilation, acute, pulmonary, mechanical, syndrome, distress, pressure, acute respiratory disease syndrome, failure, ards, injury, airway, mechanical ventilation |
| 19 | Related infectious diseases (1837) (2%) (0.098) | influenza, virus, infection, pandemic, h1n1, respiratory, avian, cause, human, outbreak, viral, h5n1 |
| 20 | Heart/blood vessels (1218) (1%) (0.074) | aneurysm, treatment endovascular, artery, treat, intracranial, embolisation, coil, flow, purpose, device, case, cerebral, carotid |
| 21 | Animal coronaviruses (1201) (1%) (0.094) | mouse, virus hepatitis, cell, infection, strain, mhv, coronavirus, murine, infect, model, protein, viral, induce |
| 22 | Stroke/blood vessels (1195) (1%) (0.072) | stroke, acute, ischemic, outcome, endovascular, occlusion, thrombectomy, clinical, therapy, artery, trial, large |
| 23 | Obesity (962) (1%) (0.076) | gastric laparoscopic, gastrectomy, surgery, undergo, bariatric, sleeve, bypass, procedure, weight, loss, perform, complication |
| 24 | Hernia (933) (1%) (0.064) | repair, hernia, mesh, inguinal, abdominal, complication, recurrence, ventral |
| 25 | Cellular (840) (1%) (0.090) | ace2, receptor, enzyme, sars-cov-2, coronavirus, cell, covid-19, angiotensin, respiratory, severe, acute, protein, entry, cause |
| Total | 83,264 |
RMSSD: root-mean-square standard deviation.
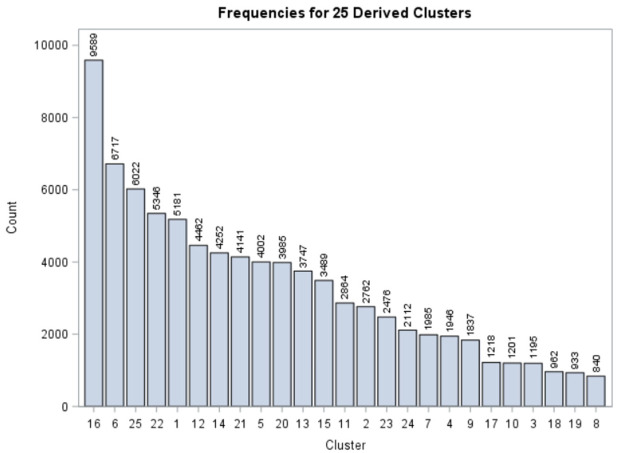
As can be seen in Figure 3, the number of abstracts in the 25 clusters varies greatly, showing the varied levels of interest in COVID-19 topics. The three largest clusters deal with risk factors, new and innovative ways of researching COVID-19 and the problems caused by the virus in China. The three smallest clusters contained articles related to obesity, hernia-related issues and the cellular makeup of the virus. The themes apparent in the clusters illustrate the concerns of the medical community about certain medical conditions. For example, in May 2020, attention was given to the fact that COVID-19 might cause strokes [23], and the cluster number 3 has descriptive terms related to blood vessel issues and stroke.
Figure 3.
Frequencies of abstracts in the 25 clusters.
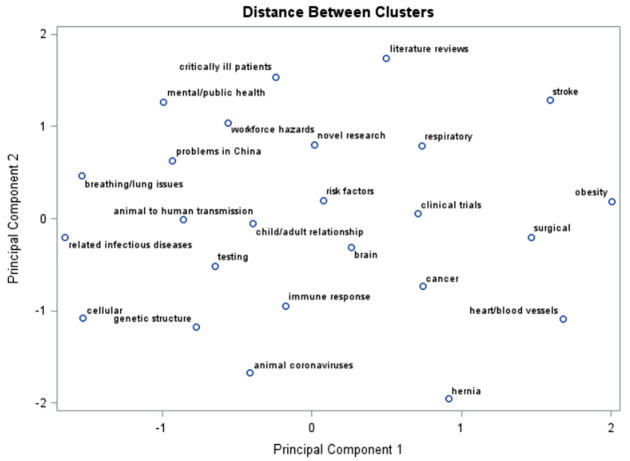
Figure 4 displays a scatter plot showing the distances between each cluster in a Cartesian coordinate system. The topics derived from Table 4 are used as the labels for each cluster. This visual representation allows researchers to gauge how similar a certain cluster of abstracts is to another cluster. The closer the clusters are to each other in the coordinate system, the more similar are the abstracts in each cluster. For example, it is readily seen that abstracts related to cellular and genetic structure of the virus are more similar than those related to obesity or hernia-related issues.
Figure 4.
Scatter plot showing the distances between the clusters.
5. Conclusion
Using a text mining clustering approach, this study was able to assess the differing foci of a large number of COVID-19-related abstracts. This article proposes a text clustering approach to assist researchers in narrowing down the range of research articles they need to digest. The rate of increase in the quantity of research and publications related to COVID-19 shows no signs of falling. With the increasing numbers of preprint journal publications and online journals, and the growth of social media, information related to COVID-19 can be disseminated widely and rapidly. Helping researchers to find articles and information relevant to their research interest in COVID-19 is imperative.
The text clustering analysis results also offer insight into emerging research themes. This analysis shows that researchers are paying close attention to the clinical management and risk factors of the disease, emerging issues in China and the effects of underlying conditions such as obesity and heart disease. Other issues, such as the treatment of the virus as it relates to vulnerable populations, do not appear, so far, to be as well-studied.
A likely practical implication of this work is that it has the potential to reduce the time a researcher needs to spend sifting through articles to find those related to their research. Other disciplines have experienced these benefits. Bebe and Clark [24] experimented with developing a tool that reduced the number of irrelevant search returns for forensic investigators. Their proposed approach assisted investigators in finding relevant text searches that were related to the objective of their investigation more quickly. To realise the reduction of time a researcher has to search for relevant articles, the methodology presented in this article would need to be incorporated into a search engine, and the clustering algorithm would need to be evaluated. In addition, Strikanth and Sakthivel [25] evaluated the speed and accuracy of a text clustering methodology using a multimedia web search engine. Therefore, future work related to this article would include incorporating the text clustering methodology in a search engine and evaluating its performance and speed.
Another area for future work related to this research would use the initial results presented here as the basis of a predictive model that could categorise research articles. The themes that were derived for each cluster could be used as a label (target) for a training data set. Then, using predictive models, new research articles could be scored and assigned their predicted cluster label. Such predictive models would produce a probability of group membership for each article. A researcher could examine the probabilities, and determine whether a particular article is likely to be of interest, based on the predicted probabilities. This would be another step forward in assisting researchers in their search for related publications.
Another future study could compare the text clustering results of this analysis of abstracts with a similar analysis of full articles. It would be interesting to investigate whether clustering the full documents would produce similar groups. If similar groups are indeed formed using the full documents, using only the abstracts would be shown to be equally useful, and incur less processing time and fewer computational resources. In addition, comparing the results of this analysis of abstracts with an analysis using only keywords would also provide useful insight in terms of validating the results shown in this article. Moreover, if similar results could be achieved using keywords, then this would have a significant impact on the time a researcher would take to sift through the body of COVID-19 literature.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship and/or publication of this article.
ORCID iD: Billie S Anderson  https://orcid.org/0000-0002-1327-7004
https://orcid.org/0000-0002-1327-7004
References
- 1.Davenport TH. Big data at work: Dispelling the myths, uncovering the opportunities. Boston, MA: Harvard Business Review Press, 2014. [Google Scholar]
- 2.Xiang Y-T, Li W, Zhang Qet al. Timely research papers about COVID-19 in China. Lancet 2020; 395: 684–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callaway E, Cyranoski D, Mallapaty Set al. The coronavirus pandemic in five powerful charts. Nature: Int Weekly J Sci 2020; 579: 482. [DOI] [PubMed] [Google Scholar]
- 4.Brainard S. Scientists are drowning in COVID-19 papers. Can new tools keep them afloat? Science, https://www.sciencemag.org/news/2020/05/scientists-are-drowning-covid-19-papers-can-new-tools-keep-them-afloat# (2020, accessed 30 August 2020).
- 5.Harapan H, Itoh N, Yufika Aet al. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health 2020; 13: 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu H, Tu S, Gao Set al. The epidemiological and clinical features of COVID-19 and lessons from this global infectious public health event. J Infect 2020; 81: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahanbin K, Rahmanian F, Rahmanian Vet al. Application of Twitter and web news mining in infectious disease surveillance systems and prospects for public health. GMS Hyg Infect Control 2019; 14: Doc19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han X, Wang J, Zhang Met al. Using social media to mine and analyze public opinion related to COVID-19 in China. Int J Environ Res Public Health 2020; 17: 2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen TT. Artificial intelligence in the battle against coronavirus (COVID-19): a survey and future research directions, 2020, https://www.researchgate.net/publication/340487417_Artificial_Intelligence_in_the_Battle_against_Coronavirus_COVID-19_A_Survey_and_Future_Research_Directions
- 10.Noble J, Gamit H. Unsupervised contextual clustering of abstracts. In: SAS Conference Proceedings: SAS Global Forum 2020, Cary, NC, 16 June 2020. [Google Scholar]
- 11.Carnerud D. Exploring research quality and reliability management through text mining methodology. Int J Qual Reliab Manag 2017; 34: 975–1014. [Google Scholar]
- 12.Wang LL, Lo K, Chandrasekhar Yet al. CORD-19: the Covid-19 open research dataset, 2020, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7251955/
- 13.Kazemi Rashed S, Frid J, Aits S. English dictionaries, gold and silver standard corpora for biomedical natural language processing related to SARS-CoV-2 and COVID-19, 2020, https://arxiv.org/abs/2003.09865
- 14.Luther S, Berndt D, Finch Det al. Using statistical text mining to supplement the development of an ontology. J Biomed Inform 2011; 44: S86–S93. [DOI] [PubMed] [Google Scholar]
- 15.Reincke U. Profiling and classification of scientific documents with SAS Text Miner. In: Paper presented at Third & Knowledge Discovery Workshop, Karlsruhe, 30 October 2003, pp. 6–8. Cary, NC: SAS Publishing. [Google Scholar]
- 16.Xu Y, Jones GFJ, Li Jet al. A study on mutual information-based in feature selection for text categorization. J Comput Inf Syst 2020; 3: 1007–1012. [Google Scholar]
- 17.SAS Institute. Text analytics using SAS text miner course notes. Cary, NC: SAS Publishing, 2019. [Google Scholar]
- 18.Evangelopoulos N, Zhang X, Prybutok VR. Latent semantic analysis: five methodological recommendations. Eur J Inf Syst 2012; 21: 70–86. [Google Scholar]
- 19.Albright R. Taming text with the SVD. Cary, NC: SAS Publishing, 2004. [Google Scholar]
- 20.Guan J, Levitan A, Goyal S. Text mining using latent semantic analysis: an illustration through examination of 30 years of research at JIS. J Inf Syst 2018; 32: 67–86. [Google Scholar]
- 21.Chakraborty G, Pagolu M, Garla S. Text mining and analysis practical methods, examples, and case studies using SAS. Cary, NC: SAS Publishing, 2013. [Google Scholar]
- 22.Verma A, Maiti J. Text-document clustering-based cause and effect analysis methodology for steel plant incident data. Int J Inj Contr Saf Promot 2018; 25: 416–426. [DOI] [PubMed] [Google Scholar]
- 23.Neal S, Zheng C. Covid-19 through the lens of the peer-reviewed literature. Significance 2020; 17: 10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bebe N, Clark J. Digital forensic text string searching: Improving information retrieval effectiveness by thematically clustering search results. Digit Investig 2007; 4S: 49–54. [Google Scholar]
- 25.Strikanth D, Sakthivel S. Vantage point latent semantic indexing for multimedia web document search. Cluster Comput 2019; 22: 10587–10594. [Google Scholar]