Abstract
This study aimed to characterize the antimicrobial resistance and virulence of Enterococcus from dogs and cats in Northeast China and evaluate its zoonotic risk based on a total of 469 enterococci strains from 610 samples, including 238 strains of E. faecium and 128 strains of E. faecalis. The isolation rate from police dog samples was 93.79%, pet dog samples was 69.90% and pet cat samples was 76.67%. The differences in the prevalence of E. faecalis among different hosts were statistically significant (P<0.05). The assays showed that most of the virulence genes detected were existed in E. faecalis and police dogs carried the least number of virulence genes. The correlation between enterococcal surface protein (esp) and aggregation substance (asa1) was determined. Enterococci are most resistant to tetracycline and erythromycin, 68.92% of the isolates were classified as multiple drug resistant. Significant differences (P<0.01) were found between E. faecium and E. faecalis in the resistance rates of nine antimicrobials. Four positive and four negative correlations were found between virulence genes and antimicrobial resistance. The results show that Enterococcus colonization and excretion in dogs and cats were related to animal species and living environments. Some correlation between virulence factors and antimicrobial resistance was obtained. This study confirmed the presence of strains carrying multiple virulence factors and antimicrobial resistance at the same time, suggesting a public health risk for dogs and cats as reservoirs of enterococci.
Keywords: antimicrobial resistance, cats, dogs, Enterococcus, virulence factor
The Enterococcus spp. are Gram-positive opportunistic anaerobic bacteria that can be found in a variety of natural environments, including soil and water [4]. Enterococcal infections can cause several human illnesses, including urinary tract infections, sepsis and endocarditis. Enterococci have emerged as hospital-acquired pathogens because of their high resistance to antimicrobials [28], including cephalosporins, aminoglycosides and streptogramins [22]. The potential for enterococci to acquire antimicrobial resistance through plasmid and transposon transfer, chromosomal exchange or mutation poses a substantial barrier to therapeutic approaches [24] and considering the ability of enterococci to acquire and transfer resistance genes, animal hosts are likely donor sources [21].
In addition to antimicrobial resistance, enterococcal virulence contributes to illness development and different virulence factors can cause different pathogenesis. For example, gelatinase (gelE) and exoenzyme (SprE) play specific roles in mediating E. faecalis autolysis and biofilm formation [44], but enterococcal surface protein (esp) promotes upstream urinary tract infection [17]. The most frequently studied are E. faecalis and E. faecium where most putative virulence factors are usually present in E. faecalis, while only a few are present in E. faecium [47].
Apart from human infections, enterococci can cause many livestock infections [9, 27, 46], so there is concern about the possibility of enterococcal transmission between humans and animals. One study concluded that most of the enterococcal strains that infected patients were different from those in livestock, which suggested that those strains from livestock had limited sharing of resistance genes [19]. However, this limitation does not mean zero probability and contrary evidence is found in other studies. Enterococcal strains from humans have been isolated from dogs and pigs [8, 16] and another study confirmed that the distribution of enterococci isolated from humans and dogs was similar [50].
Companion animals, particularly dogs and cats, have evolved into close family members [38], so there is a risk that they can serve as a repository of bacteria and transmit them to humans because of their close intimate physical contact [5]. To investigate the potential risk of companion animals in the transmission of enterococci, this study identified antimicrobial resistance phenotypes and virulence factors of enterococci collected in northeastern China and compared the differences between hosts. The correlation between virulence factors and antimicrobials from these animals was also evaluated.
MATERIALS AND METHODS
Sample collection
Sampling procedures were approved by the Experimental Animal Welfare and Ethics Committee of Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Feces or anal swabs were collected by qualified professionals using sterile swab sticks, between August and November 2021. The samples included 145 police dog samples from a base in Harbin, 345 pet dog samples and 120 pet cat samples from Changchun Pet Hospital. The swabs were placed in physiological saline containing 20% glycerol and stored in liquid nitrogen for a brief period before being transported to the laboratory. All police dogs are healthy. Some pet dogs and cats had concurrent disease not caused by enterococcus infection.
Strain isolation
Each sample was resuspended in saline and an appropriate amount of bacterial suspension was inoculated onto enterococcal agar (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). After incubating at 37°C for 16 to 18 hr, suspicious Enterococcus spp. colonies were selected for continued purification culture.
Species identification
The small black Enterococcus spp. colonies were treated with gram staining, gram-positive spherical bacteria and underwent species and biochemical identification using a BD Phoenix™-100 automated identification system (Becton, Dickinson and Co., Franklin Lakes, NJ, USA) [57].
Polymerase chain reaction (PCR) was used for identification with analysis by enterococcal genus 16S ribosomal RNA gene sequence specific primers listed in Table 1, with reference to the conditions earlier [26] and E. faecium ATCC35667 was used as a positive control. The PCR reaction system was 25 µL in total consisting of 2 × 12.5 µL Taq Master Mix (Com Win Biotech Co., Ltd., Beijing, China), 0.5 µL each of 10 pmol/µL upstream and downstream primers, one µL DNA sample and finally supplemented with ultrapure water to 25 µL. The reaction conditions were pre-denaturation at 94°C for 2 min, denaturation at 94°C for 1 min, annealing at 56°C for 1 min, extension at 72°C for 1 min, all for 30 cycles then a final extension at 72°C for 5 min. The PCR was conducted in a Veriti™ 96-well Thermal Cycler (Applied Biosystem, Carlsbad, CA, USA).
Table 1. Sequences of specific primers and virulence identification primers.
| Gene(s) | Primer sequence (5′ to 3′) | Size (bp) | Reference |
|---|---|---|---|
| ent | ent-F:AGCGCAGGCGGTTTCTTAA | 678 | [26] |
| ent-R:CTCGTTGTACTTCCCATTGT | |||
| cylA | cyl-F:ACTCGGGGATTGATAGGC | 688 | [55] |
| cyl-R:GCTGCTAAAGCTGCGCTT | |||
| esp | esp-F:AGATTTCATCTTTGATTCTTGG | 510 | |
| esp-R:AATTGATTCTTTAGCATCTGG | |||
| asa1 | asaI-F:GCACGCTATTACGAACTATGA | 375 | |
| asaI-R:TAAGAAAGAACATCACCACGA | |||
| gelE | gelE-F:TATGACAATGCTTTTTGGGAT | 213 | |
| gelE-R:AGATGCACCCGAAATAATATA | |||
| hyl | hyl-F:TATGGGTAATGCTGGTCG | 220 | This study |
| hyl-R:GTCCCTTGCTTCGTGTTT | |||
| efaAfs | efaAfs-F:GACAGACCCTCACGAATA | 705 | [30] |
| efaAfs-R:AGTTCATCATGCTGTAGTA | |||
| efaAfm | efaAfm-F:AACAGATCCGCATGAATA | 735 | |
| efaAfm-R:CATTTCATCATCTGATAGTA | |||
| ace | ace-F:CAACCGAATGTGATAGAAA | 411 | This study |
| ace-R:GTAACGGACGATAAAGGA |
Detection of virulence genes
The presence of enterococcal virulence genes, including cytolysin (cylA), esp, aggregation substance (asa1), gelE, hyaluronidase (hyl), endocardial antigens in E. faecalis (efaAfs) and E. faecium (efaAfm) and adhesin in collagen (ace) was investigated by PCR, as shown in Table 1. The primers of hyl (GenBank accession number: WP_002399773) and ace (GenBank accession number: WP_010714416) were designed using Primer 3.0 (https://bioinfo.ut.ee/primer3-0.4.0), with the reaction conditions as described previously [30, 55]. Three different PCR reactions were standardized with one quadruple as cylA-esp-asa1-gelE and two duplex as hyl-efaAfs and efaAfm-ace.
The PCR reaction system was 25 µL as 2 × 12.5 µL Multiplex PCR Buffer, 0.125 µL Multiplex PCR enzyme mix (Takara Bio Inc., Kusatsu, Japan), 0.5 µL upstream and downstream 10 pmol/µL primers, 2 µL template and finally supplemented with ultrapure water to 25 µL. All reaction conditions were consistent as pre-denaturation at 95°C for 15 min, denaturation at 94°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 1 min, all for 30 cycles and final extension at 72°C for 10 min.
Antimicrobial susceptibility
Drug susceptibility testing of enterococcal isolates was performed using the BD Phoenix™-100 automated identification and susceptibility testing system. The panel included 11 classes of a total of 13 antimicrobials with amikacin, ampicillin, ciprofloxacin, erythromycin, gentamicin, linezolid, mupirocin, nitrofurantoin, quinupristin-dalfopristin, rifampin, teicoplanin, tetracycline and vancomycin all present. High-level gentamicin resistance enterococci (HLGR) and vancomycin-resistant enterococci (VRE) strains were screened simultaneously. Susceptibility results were interpreted using Clinical and Laboratory Standards Institute criteria (CLSI, 2020).
Statistical analyzes
Bar graphs were plotted using GraphPad prism version 8.0 software. The SPSS version 20.0 software was used for data statistics. Chi-square tests were used to evaluate the statistical significance of gene prevalence which was detected from different sites and animal species. Binary logistic regression analysis (P<0.05) was applied to assess the relationship between virulence genes detected and drug resistance in the presence or absence of genes, coded as 1 or 0, respectively.
RESULTS
Isolation and identification of Enterococci
In this study, 469 strains were confirmed as enterococci by biochemical identification as shown in Supplementary Table 1. The distribution was 238 E. faecium (50.75%), 128 E. faecalis (16.94%), 91 E. hirae (19.40%), 11 E. gallinarum (2.35%) and one E. avium (0.21%). The isolation rate for police dog samples was 93.79%, 69.90% for pet dog samples and 76.67% for pet cat samples.
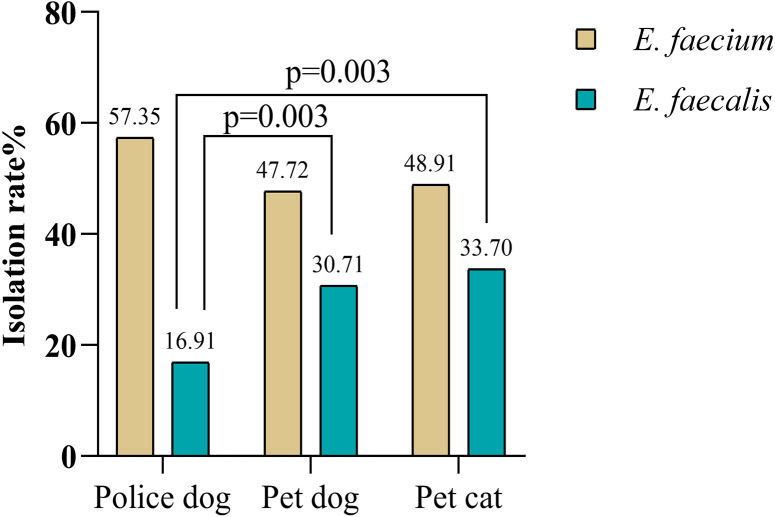
Prevalent isolation rates of E. faecalis from different hosts were statistically significant, with the P values between police dogs and pet dogs, or police dogs and pet cats respectively less than 0.05 (P=0.003), while the differences were not significant for E. faecium (P>0.05) as seen in Fig. 1.
Fig. 1.
Chi-square test and probability values for Enterococcus faecium and E. faecalis.
Virulence gene prevalence
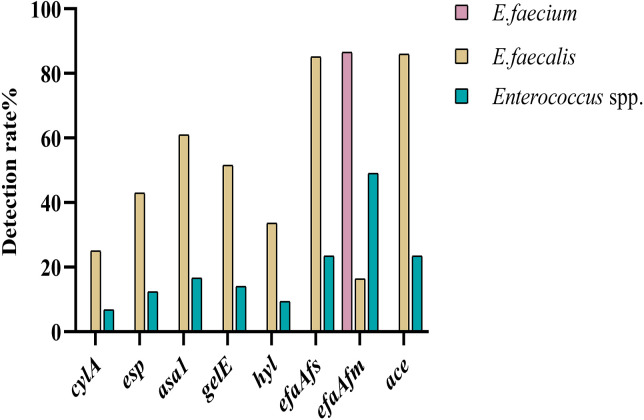
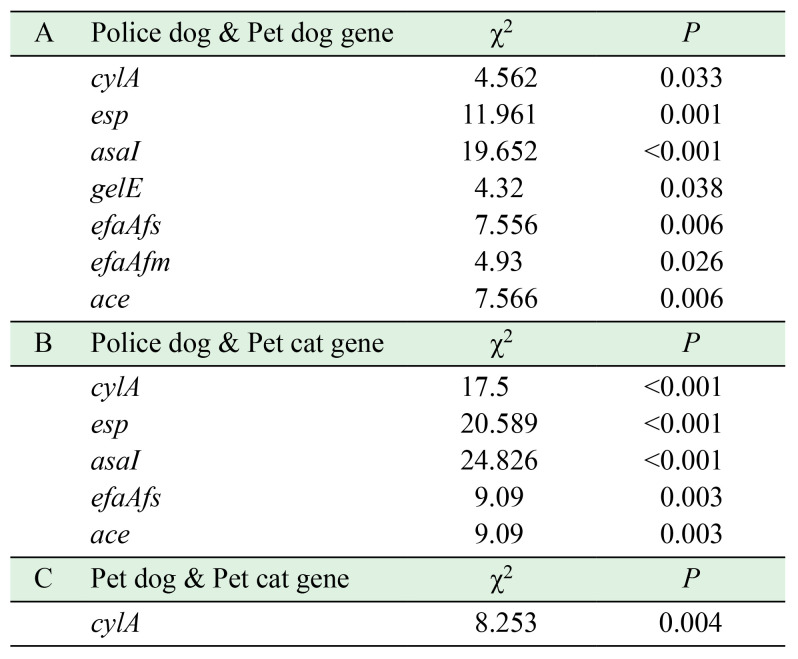
The detection rate of virulence genes is presented in Fig. 2, where the virulence profile differed between E. faecium and E. faecalis. In E. faecium, only the efaAfm gene is present. The detection rates of E. faecalis virulence genes were 32 cylA (25.00%), 55 esp (42.97%), 78 asa1 (60.94%), 66 gelE (51.56%), 43 hyl (33.59%), 109 efaAfs (85.16%), 21 efaAfm (16.41%) and 110 ace (85.94%). The specific virulence profiles statistics are displayed in Supplementary Table 2 and E. faecalis occupied 22 of these species. Virulence genes detected from different hosts were expressed at various levels, with the least virulence detected in police dogs and statistically significant results are shown in Table 2.
Fig. 2.
Detection rate of enterococcal virulence genes. Enterococcus faecium (n=238), E. faecalis (n=128), Enterococcus spp. (n=469).
Table 2. Chi-square tests and probability values of virulence genes of different hosts.
Antimicrobial susceptibility
Table 3 showed that the frequencies of resistance to tetracycline and erythromycin were higher, at 82.61% and 62.61% respectively. The resistance rate of eight species were between 10% and 50% with rifampin 48.26%, quinupristin-dalfopristin 38.04%, amikacin 35.22%, nitrofurantoin 34.13%, ciprofloxacin 32.39%, gentamicin 31.30%, ampicillin 15.00% and linezolid 13.04%. The low resistance rates included mupirocin at 6.96%, teicoplanin 3.26% and vancomycin 0.43%. There were significant differences (P<0.01) in the resistance rates of nine kinds of antimicrobials between E. faecalis and E. faecium (Table 3), including amikacin, ampicillin, ciprofloxacin, gentamicin, linezolid, mupirocin, nitrofurantoin, quinupristin-dalfopristinc and rifampin. Among isolates, 145 HLGR strains (31.52%) and four VRE strains (0.87%) were detected.
Table 3. Resistance rate of Enterococcus faecium and E. faecalis in different species.
| Antibiotics |
E. faecium
|
E. faecalis
|
The othera | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Police dog | Pet dog | Pet cat | Total | Police dog | Pet dog | Pet cat | |||
| n=232b | n=78 | n=109b | n=45 | n=128 | n=23 | n=74 | n=31 | n=100 | (n=460b) | |
| Amikacinc | 73 (31.47) | 22 (28.21) | 19 (17.43) | 32 (71.11) | 79 (61.72) | 19 (82.61) | 32 (43.24) | 28 (90.32) | 10 (10.00) | 162 (35.22) |
| Ampicillinc | 66 (28.45) | 15 (19.23) | 23 (21.10) | 28 (62.22) | 3 (2.34) | 0 (0.00) | 3 (4.05) | 0 (0.00) | 0 (0.00) | 69 (15.00) |
| Ciprofloxacinc | 82 (35.34) | 20 (25.64) | 36 (33.03) | 26 (57.78) | 56 (43.75) | 10 (43.48) | 29 (39.19) | 17 (54.84) | 11 (11.00) | 149 (32.39) |
| Erythromycin | 159 (68.53) | 50 (64.10) | 69 (63.30) | 40 (88.89) | 85 (66.41) | 10 (43.48) | 50 (67.57) | 25 (80.65) | 44 (44.00) | 288 (62.61) |
| Gentamicinc | 69 (29.74) | 17 (21.79) | 20 (18.35) | 32 (71.11) | 66 (51.56) | 7 (30.43) | 36 (48.65) | 23 (74.19) | 9 (9.00) | 144 (31.30) |
| Linezolidc | 12 (5.17) | 3 (3.85) | 8 (7.34) | 1 (2.22) | 31 (24.22) | 4 (17.39) | 22 (29.73) | 5 (16.13) | 17 (17.00) | 60 (13.04) |
| Mupirocinc | 1 (0.43) | 0 (0.00) | 1 (0.92) | 0 (0.00) | 20 (15.63) | 3 (13.04) | 11 (14.86) | 6 (19.35) | 11 (11.00) | 32 (6.96) |
| Nitrofurantoinc | 121 (52.16) | 29 (37.18) | 63 (57.80) | 29 (64.44) | 11 (8.59) | 2 (8.70) | 9 (12.16) | 0 (0.00) | 25 (25.00) | 157 (34.13) |
| Quinupristin-dalfopristinc | 79 (34.05) | 18 (23.08) | 50 (45.87) | 11 (24.44) | 82 (64.06) | 19 (82.61) | 34 (45.95) | 29 (93.55) | 14 (14.00) | 175 (38.04) |
| Rifampinc | 118 (50.86) | 21 (26.92) | 75 (68.81) | 22 (48.89) | 96 (75.00) | 17 (73.91) | 55 (74.32) | 24 (77.42) | 8 (8.00) | 222 (48.26) |
| Teicoplanin | 3 (1.29) | 0 (0.00) | 3 (2.75) | 0 (0.00) | 4 (3.13) | 0 (0.00) | 4 (5.41) | 0 (0.00) | 8 (8.00) | 15 (3.26) |
| Tetracyclin | 193 (83.19) | 77 (98.72) | 83 (76.15) | 33 (73.33) | 101 (78.91) | 16 (69.57) | 60 (81.08) | 25 (80.65) | 86 (86.00) | 380 (82.61) |
| Vancomycin | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.78) | 0 (0.00) | 1 (1.35) | 0 (0.00) | 1 (1.00) | 2 (0.43) |
Because of the small number of other Enterococcus spp. strains, only E. faecium and E. faecalis were shown in this table. a: Total enterococci other than E. faecalis and E. faecalis. b: Some strains could not be recognized by the BD system, so they were removed from the statistics. c: 9 kinds of antimicrobials with the significant difference in E. faecium and E. faecalis.
The antimicrobial resistance profile statistics are summarized in Supplementary Table 3. In total, 156 resistance profiles were detected against 13 antimicrobials. According to the current criteria for determining multidrug resistance (MDR) in bacteria [34], 68.92% of the isolates were classified as MDR and 98.70% were resistant to at least one class of antimicrobials.
Correlation between virulence gene distribution and drug resistance
Table 4 showed the relationship between the distribution of virulence genes and antimicrobials in E. faecalis. Eight distribution pairings were statistically significant (P<0.05). E. faecalis strains carrying the asa1 gene were detected to be more resistant to gentamicin and erythromycin. The same case for esp and hyl to rifampin. Conversely, E. faecalis strains carrying the gelE gene were detected to be more sensitive to gentamicin and rifampin, hyl to erythromycin, esp to linezolid in the same situation.
Table 4. The logistic regression analysis of antimicrobials and virulence genes in Enterococcus faecalis.
| Antibiotics | Genes | β | P | OR | OR95% (CI) |
|---|---|---|---|---|---|
| Gentamicin | asa1 | 2.144 | 0.001 | 8.532 | 2.439~29.844 |
| gelE | –1.673 | 0.011 | 0.188 | 0.052~0.684 | |
| Erythromycin | asa1 | 2.385 | <0.001 | 10.856 | 3.018~39.049 |
| hyl | –1.742 | 0.013 | 0.175 | 0.045~0.690 | |
| Linezolid | esp | –1.93 | 0.022 | 0.145 | 0.028~0.759 |
| Rifampin | esp | 2.656 | 0.002 | 14.244 | 2.585~78.471 |
| gelE | –1.791 | 0.025 | 0.167 | 0.035~0.802 | |
| hyl | 1.629 | 0.034 | 5.1 | 1.135~22.919 |
OR, odds ratio; CI, confidence interval.
DISCUSSION
The bacterium Enterococcus was once considered a harmless commensal with probiotic properties which enhanced the immune system [41], but it has now become one of the most common pathogens of nosocomial infection [15]. Clinical enterococcal infections in companion animals are rare [52], but studies have proved that such pets can be reservoirs of enterococci [24,25,26]. A community-acquired case of multidrug resistant E. faecalis corneal ulcers caused by a pet cats scratch has been previously reported [39], so enterococci isolated from dogs and cats may cause cross-transfer of pathogenic bacteria [40]. A study demonstrates that enterococcal clones were found in pets in multiple body sites, their human cohabitants, and shared domestic objects [32], in which case regular monitoring of bacterial virulence genes and drug resistance can provide scientific guidance and timely interruption of the source of transmission.
The enterococcal microbiota of the intestinal tract of dogs and cats are predominantly E. faecalis and E. faecium [53] as shown in this study, where they were the main isolates. The isolation rates of E. faecalis in pet dogs and cats were significantly (P<0.05) different from those of police dogs. This meant that the prevalence of E. faecalis was statistically significant across hosts, while E. faecium was not. In a previous study, the effect of dog breeds on gut microbiomes was noted [33], which may explain the differences in isolation rates. In China, a police dog is a large breed working dog, while pet breeding is more varied according to the owner’s preference. Analysis of the virulence genes showed that police dogs carried the lowest number and pet dogs and cats carried comparable levels. This result may reflect the influence of environment or exposure in the route of transmission, as police dogs live in a simple environment with a single population of contacts, but pets are frequently exposed to a variety of external environments and have closer contact with humans.
Virulence factors are determinants of infection-causing strains [54]. The efaA gene was the most frequently detected adhesin gene. In this study, efaAfm was found in 86.55% of E. faecium strains and efaAfs in 85.16% of E. faecalis strains. However, the presence of only efaAfm gene seems to have no value as a risk indicator in E. faecium strains [29]. The E. faecium and E. faecalis strains showed significantly different patterns in the incidence of virulence determinants and multiple virulence determinants were found in E. faecalis [11]. More attention has focused on E. faecalis, as most of the virulence factors in this study were found in these strains. The ace gene encoded a collagen-binding protein involved in the pathogenesis of endocarditis and its incidence rate was second only to efaA. The esp gene for enterococcal surface protein and the asa1 gene for aggregation substance are associated with biofilm formation [13]. In the study, the detection rate of esp in E. faecalis was 42.97%, of which 94.55% also carried the asa1 gene, demonstrating a strong association between the two genes.
The hyaluronidase encoded by the chromosomal hyl gene, is a degradative enzyme associated with tissue damage [35]. It has been shown that most hyl positive strains were also positive for esp in E. faecium [43]. The results suggested that the same conclusion does not exist for E. faecalis, with only 20.93% of strains matching this profile. The gelE gene encodes a zinc metalloprotease, with hydrolytic capacity. The results of this study indicated that 51.56% of enterococcal isolates were gelatinase producers. The cylA gene is a determinant of lysin production and enables the bacteria to evade the host immune response by destroying cells such as macrophages and neutrophils [56]. The detection rate of cylA was the lowest in this study, which is consistent with the results of a study related to human clinical infection [18].
This study found that 82.03% (105/128) of strains carried three and more virulence genes in E. faecalis. Based on the idea that the pathogenic potential may be due to virulence being multifactorial and associated with different genes, focusing on the role of individual virulence genes is no longer sufficient and the superposition of multiple virulence genes may pose new challenges for disease treatment [5].
Another factor affecting treatment after enterococcal infection is resistance to antimicrobials. A study has mentioned that tetracyclines and erythromycin belong to the class of antimicrobials commonly used in small animal veterinary medicine [20]. In China, there are no exact statistics on the use of antimicrobials in companion animals, but we found that tetracyclines and macrolides were among the top sales by the China Veterinary Drug Association (http://www.cvda.org.cn/index.html). We speculate that the usage amount may be responsible for the high rate of tetracycline and erythromycin resistance in this study. In this study, 67.28% of the tetracycline resistant phenotypic strains were also resistant to erythromycin, which is consistent with the idea that enterococci tend to be resistant to both erythromycin and tetracycline antimicrobials [7].
Enterococci also have an acquired resistance to aminoglycosides and β-lactams. The resistance rate of E. faecium isolates was significantly higher for aminoglycosides, while ampicillin resistance was higher for E. faecalis isolates [36]. Compared with these results, it must be pointed out that although the rate of drug resistant strains was different, the results both confirmed the severity of drug resistance. This study showed that enterococcal strains had low levels of resistance to glycopeptide resistant drugs. Strains resistant to vancomycin and teicoplanin were assigned to vanA phenotype, while those susceptible to teicoplanin but resistant to vancomycin were considered as the vanB phenotype [49], but in this study, only one isolate was classified as vanA. In this study, nine kinds of antimicrobial susceptibilities differed between E. faecium and E. faecalis. The E. faecalis is inherently resistant to quinupristin-dalfopristin, which may explain the high rate of resistance to this antibiotic E. faecalis. For other antimicrobial agents in this experiment, until more conclusive evidence is available, the high resistance can only be attributed to the irregular use of antimicrobial drugs.
Evaluating risk factors that the presence of multi-drug resistant (MDR) Enterococcus strains in animals is crucial for public and environmental health [3, 6, 31]. In one study, 67.23% of E. faecium and 93.75% of E. faecalis isolates were found to be MDR strains [14], but contrary to the findings of Shahraki [48], this study found a higher rate of MDR in E. faecalis. The high rate of MDR results was not only present in companion animals but similar results were obtained in isolates from food, animals and the environment [10, 12, 14]. The current concern is that the acquired antimicrobial resistance among enterococci makes treatment very difficult [12], so it is crucial to monitor the resistance characteristics of isolates and regulate the use of antimicrobials in veterinary medicine.
The current study identified two types of resistant strains, where HLGR strains were found to exist in four species of enterococci. The HLGR not only affects the synergistic use of antimicrobials but has spread in hospitals [45], so its presence needs attention. The VRE strain which can lead to a more severe risk of infection, was found in only four strains and all of them were E. faecalis. Vancomycin is considered the last line of defense for enterococcal therapy and the advent of VRE presents an even greater challenge. The use of avoparcin was discontinued as a precautionary measure [37] and the study of linezolid and quinupristin-dalfopristin as alternative molecules for VRE infection [51] all aim to avoid the development of drug resistance, but they do not achieve complete blockage.
Exploring the correlation between resistance to antimicrobial agents and virulence factors of enterococcal strains can be of great significance for appropriate treatment initiatives by veterinary practitioners. One study suggested some correlation between antimicrobial resistance and different virulence determinants [2], which may increase morbidity and mortality [42]. In this study, a negative correlation between rifampicin and gelE was observed, which might be due to linezolid combined with rifampicin having a good therapeutic effect on biofilms [23]. In addition to the known significant association of the esp gene with resistance to ciprofloxacin, erythromycin, and tetracycline [1], it was also found that esp was related to linezolid and rifampicin resistance in this study. A definitive conclusion was not made and further studies are needed to examine the association of pathogenicity with multi-virulence and multi-drug resistance.
Virulence factors and drug resistance associated with plasmids were also considered. This has been described in other studies, as the plasmid addiction system, with the detection of transferable linezolid resistance genes (optrA) in enterococci and the facilitation of plasmid binding and exchange by aggregation substance [29, 41, 53]. Although such findings are still few, these studies deserve to be pursued in-depth as more specific routes of transmission.
In conclusion, this study found that E. faecalis in dogs and cats was more common and contained more virulence factors than E. faecium, with an association between virulence factors and antimicrobial resistance. Comparing isolation rates in enterococcal species, the results suggested that both the hosts and the environment influence the results of enterococcal harboring in dogs and cats. Based on the current results, more comprehensive surveillance of companion animals is recommended, as is the standardization of antimicrobial use in veterinary medicine to interrupt the possible transmission risk of Enterococcus spp. More studies should be conducted to find the cause of enterococcal pathogenicity.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supplementary
Acknowledgments
This work was funded by the National Science and Technology Major Project of China (grant no. 2018ZX10733402).
REFERENCES
- 1.Arabestani MR, Nasaj M, Mousavi SM. 2017. Correlation between infective factors and antibiotic resistance in Enterococci clinical isolates in West of Iran. Chonnam Med J 53: 56–63. doi: 10.4068/cmj.2017.53.1.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbabi L, Boustanshenas M, Rahbar M, Majidpour A, Shayanfar N, Afshar M, Adabi M, Owlia P, Talebi-Taher M. 2016. The correlation between resistance to antimicrobial agents and harboring virulence factors among enterococcus strains isolated from clinical samples. JMBR 6: 35. doi: 10.5539/jmbr.v6n1p35 [DOI] [Google Scholar]
- 3.Bang K, An JU, Kim W, Dong HJ, Kim J, Cho S. 2017. Antibiotic resistance patterns and genetic relatedness of Enterococcus faecalis and Enterococcus faecium isolated from military working dogs in Korea. J Vet Sci 18: 229–236. doi: 10.4142/jvs.2017.18.2.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben BO, Smaoui S. 2019. Enterococci: between emerging pathogens and potential probiotics. International BR 2019: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Said L, Dziri R, Sassi N, Lozano C, Ben Slama K, Ouzari I, Torres C, Klibi N. 2017. Species distribution, antibiotic resistance and virulence traits in canine and feline enterococci in Tunisia. Acta Vet Hung 65: 173–184. doi: 10.1556/004.2017.018 [DOI] [PubMed] [Google Scholar]
- 6.Cinquepalmi V, Monno R, Fumarola L, Ventrella G, Calia C, Greco MF, Vito D, Soleo L. 2012. Environmental contamination by dog’s faeces: a public health problem? Int J Environ Res Public Health 10: 72–84. doi: 10.3390/ijerph10010072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui P, Feng L, Zhang L, He J, An T, Fu X, Li C, Zhao X, Zhai Y, Li H, Yan W, Li H, Luo X, Lei C, Wang H, Yang X. 2020. Antimicrobial resistance, virulence genes, and biofilm formation capacity among Enterococcus species from Yaks in Aba Tibetan autonomous prefecture, China. Front Microbiol 11: 1250. doi: 10.3389/fmicb.2020.01250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damborg P, Top J, Hendrickx APA, Dawson S, Willems RJL, Guardabassi L. 2009. Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections. Appl Environ Microbiol 75: 2360–2365. doi: 10.1128/AEM.02035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolka B, Chrobak-Chmiel D, Czopowicz M, Szeleszczuk P. 2017. Characterization of pathogenic Enterococcus cecorum from different poultry groups: Broiler chickens, layers, turkeys, and waterfowl. PLoS One 12: e0185199. doi: 10.1371/journal.pone.0185199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dos Santos LDR, Furlan JPR, Gallo IFL, Ramos MS, Savazzi EA, Stehling EG. 2021. Occurrence of multidrug-resistant Enterococcus faecium isolated from environmental samples. Lett Appl Microbiol 73: 237–246. doi: 10.1111/lam.13508 [DOI] [PubMed] [Google Scholar]
- 11.Eaton TJ, Gasson MJ. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol 67: 1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Zamkan MA, Mohamed HMA. 2021. Antimicrobial resistance, virulence genes and biofilm formation in Enterococcus species isolated from milk of sheep and goat with subclinical mastitis. PLoS One 16: e0259584. doi: 10.1371/journal.pone.0259584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahmy N, Abdel-Gawad A, Rezk G, Mahmoud E. 2021. Characterization of Enterococci isolated from intensive care unit (ICU); Distribution of virulence markers, virulence genes and antibiotic resistance pattern. Microbes Infect 2: 725–735. [Google Scholar]
- 14.Finisterra L, Duarte B, Peixe L, Novais C, Freitas AR. 2021. Industrial dog food is a vehicle of multidrug-resistant enterococci carrying virulence genes often linked to human infections. Int J Food Microbiol 358: 109284. doi: 10.1016/j.ijfoodmicro.2021.109284 [DOI] [PubMed] [Google Scholar]
- 15.Fiore E, Van Tyne D, Gilmore MS. 2019. Pathogenicity of Enterococci. Microbiol Spectr 7: 10.1128/microbiolspec.GPP3-0053-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freitas AR, Coque TM, Novais C, Hammerum AM, Lester CH, Zervos MJ, Donabedian S, Jensen LB, Francia MV, Baquero F, Peixe L. 2011. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J Clin Microbiol 49: 925–931. doi: 10.1128/JCM.01750-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao W, Howden BP, Stinear TP. 2018. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol 41: 76–82. doi: 10.1016/j.mib.2017.11.030 [DOI] [PubMed] [Google Scholar]
- 18.Gholizadeh P, Aghazadeh M, Ghotaslou R, Ahangarzadeh Rezaee M, Pirzadeh T, Köse Ş, Ganbarov K, Yousefi M, Kafil HS. 2020. CRISPR-cas system in the acquisition of virulence genes in dental-root canal and hospital-acquired isolates of Enterococcus faecalis. Virulence 11: 1257–1267. doi: 10.1080/21505594.2020.1809329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouliouris T, Raven KE, Ludden C, Blane B, Corander J, Horner CS, Hernandez-Garcia J, Wood P, Hadjirin NF, Radakovic M, Holmes MA, de Goffau M, Brown NM, Parkhill J, Peacock SJ. 2018. Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. MBio 9: e01780–e18. doi: 10.1128/mBio.01780-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guardabassi L, Schwarz S, Lloyd DH. 2004. Pet animals as reservoirs of antimicrobial-resistant bacteria Review. J Antimicrob Chemother 54: 321–332. doi: 10.1093/jac/dkh332 [DOI] [PubMed] [Google Scholar]
- 21.Hammerum AM. 2012. Enterococci of animal origin and their significance for public health. Clin Microbiol Infect 18: 619–625. doi: 10.1111/j.1469-0691.2012.03829.x [DOI] [PubMed] [Google Scholar]
- 22.Hollenbeck BL, Rice LB. 2012. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3: 421–433. doi: 10.4161/viru.21282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmberg A, Mörgelin M, Rasmussen M. 2012. Effectiveness of ciprofloxacin or linezolid in combination with rifampicin against Enterococcus faecalis in biofilms. J Antimicrob Chemother 67: 433–439. doi: 10.1093/jac/dkr477 [DOI] [PubMed] [Google Scholar]
- 24.Iweriebor BC, Obi LC, Okoh AI. 2015. Virulence and antimicrobial resistance factors of Enterococcusspp. isolated from fecal samples from piggery farms in Eastern Cape, South Africa. BMC Microbiol 15: 136. doi: 10.1186/s12866-015-0468-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson CR, Fedorka-Cray PJ, Davis JA, Barrett JB, Frye JG. 2009. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J Appl Microbiol 107: 1269–1278. doi: 10.1111/j.1365-2672.2009.04310.x [DOI] [PubMed] [Google Scholar]
- 26.Jahan M, Krause DO, Holley RA. 2013. Antimicrobial resistance of Enterococcus species from meat and fermented meat products isolated by a PCR-based rapid screening method. Int J Food Microbiol 163: 89–95. doi: 10.1016/j.ijfoodmicro.2013.02.017 [DOI] [PubMed] [Google Scholar]
- 27.Jung A, Rautenschlein S. 2014. Comprehensive report of an Enterococcus cecorum infection in a broiler flock in Northern Germany. BMC Vet Res 10: 311. doi: 10.1186/s12917-014-0311-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kajihara T, Nakamura S, Iwanaga N, Oshima K, Takazono T, Miyazaki T, Izumikawa K, Yanagihara K, Kohno N, Kohno S. 2015. Clinical characteristics and risk factors of enterococcal infections in Nagasaki, Japan: a retrospective study. BMC Infect Dis 15: 426. doi: 10.1186/s12879-015-1175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubašová I, Strompfová V, Lauková A. 2017. Safety assessment of commensal enterococci from dogs. Folia Microbiol (Praha) 62: 491–498. doi: 10.1007/s12223-017-0521-z [DOI] [PubMed] [Google Scholar]
- 30.Lauková A, Strompfová V, Ščerbová J, Pogány Simonová M. 2019. Virulence factor genes incidence among Enterococci from sewage sludge in Eastern Slovakia following safety aspect. BioMed Res Int 2019: 2735895. doi: 10.1155/2019/2735895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leite-Martins L, Mahú MI, Costa AL, Bessa LJ, Vaz-Pires P, Loureiro L, Niza-Ribeiro J, de Matos AJF, Martins da Costa P. 2015. Prevalence of antimicrobial resistance in faecal enterococci from vet-visiting pets and assessment of risk factors. Vet Rec 176: 674–674. doi: 10.1136/vr.102888 [DOI] [PubMed] [Google Scholar]
- 32.Leite-Martins L, Meireles D, Bessa LJ, Mendes Â, de Matos AJ, da Costa PM. 2014. Spread of multidrug-resistant Enterococcus faecalis within the household setting. Microb Drug Resist 20: 501–507. doi: 10.1089/mdr.2013.0217 [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Sun Q, Li Y, Guan Z, Wei J, Li B, Liu K, Shao D, Mi R, Liu H, Qiu Y, Ma Z. 2022. Analysis and comparison of gut microbiome in young detection dogs. Front Microbiol 13: 872230. doi: 10.3389/fmicb.2022.872230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 35.Meena B, Anburajan L, Varma KS, Vinithkumar NV, Kirubagaran R, Dharani G. 2020. A multiplex PCR kit for the detection of three major virulent genes in Enterococcus faecalis. J Microbiol Methods 177: 106061. doi: 10.1016/j.mimet.2020.106061 [DOI] [PubMed] [Google Scholar]
- 36.Mousavi SH, Peeri-Doghaheh H, Mohammadi-Ghalehbin B, Teimourpour R, Maleki D, Khademi F, Arzanlou M. 2020. High-level resistance to aminoglycosides and ampicillin among clinical isolates of Enterococcus species in an Iranian referral hospital. Iran J Microbiol 12: 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson O. 2012. Vancomycin resistant enterococci in farm animals-occurrence and importance. Infect Ecol Epidemiol 2: 16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overgaauw PAM, Vinke CM, Hagen MAEV, Lipman LJA. 2020. A one health perspective on the human-companion animal relationship with emphasis on zoonotic aspects. Int J Environ Res Public Health 17: 3789. doi: 10.3390/ijerph17113789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng CH, Cheng CK, Chang CK, Chen YL. 2009. Multiresistant enterococci: a rare cause of complicated corneal ulcer and review of the literature. Can J Ophthalmol 44: 214–215. doi: 10.3129/i09-010 [DOI] [PubMed] [Google Scholar]
- 40.Pillay S, Zishiri OT, Adeleke MA. 2018. Prevalence of virulence genes in Enterococcus species isolated from companion animals and livestock. Onderstepoort J Vet Res 85: e1–e8. doi: 10.4102/ojvr.v85i1.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos S, Silva V, Dapkevicius MLE, Igrejas G, Poeta P. 2020. Enterococci, from harmless bacteria to a pathogen. Microorganisms 8: 1118. doi: 10.3390/microorganisms8081118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao C, Dhawan B, Vishnubhatla S, Kapil A, Das B, Sood S. 2020. Emergence of high-risk multidrug-resistant Enterococcus faecalis CC2 (ST181) and CC87 (ST28) causing healthcare-associated infections in India. Infect Genet Evol 85: 104519. doi: 10.1016/j.meegid.2020.104519 [DOI] [PubMed] [Google Scholar]
- 43.Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, Klare I, Nallapareddy SR, Huang W, Murray BE. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis 187: 508–512. doi: 10.1086/367711 [DOI] [PubMed] [Google Scholar]
- 44.Sadaka A, Durand ML, Gilmore MS. 2012. Bacterial endophthalmitis in the age of outpatient intravitreal therapies and cataract surgeries: host-microbe interactions in intraocular infection. Prog Retin Eye Res 31: 316–331. doi: 10.1016/j.preteyeres.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saffari F, Darehkordi H, Ahmadrajabi R. 2021. Clonal dissemination of high-level gentamicin-resistant isolates of Enterococcus faecalis within a university hospital in southeastern Iran. Wien Med Wochenschr 171: 18–23. doi: 10.1007/s10354-019-00716-2 [DOI] [PubMed] [Google Scholar]
- 46.Sanciu G, Marogna G, Paglietti B, Cappuccinelli P, Leori G, Rappelli P. 2013. Outbreak of mastitis in sheep caused by multi-drug resistant Enterococcus faecalis in Sardinia, Italy. Epidemiol Infect 141: 582–584. doi: 10.1017/S0950268812000647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sava IG, Heikens E, Huebner J. 2010. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect 16: 533–540. doi: 10.1111/j.1469-0691.2010.03213.x [DOI] [PubMed] [Google Scholar]
- 48.Shahraki S, Rabi NMM. 2017. Determination of virulence factors in clinical multidrug resistance Enterococci isolates at Southeast of Iran. Jundishapur J Microbiol 10: e45514. doi: 10.5812/jjm.45514 [DOI] [Google Scholar]
- 49.Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Aghazadeh M, Milani M. 2012. Survey of virulence determinants among vancomycin resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens of hospitalized patients of North west of Iran. Open Microbiol J 6: 34–39. doi: 10.2174/1874285801206010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simjee S, White DG, McDermott PF, Wagner DD, Zervos MJ, Donabedian SM, English LL, Hayes JR, Walker RD. 2002. Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: evidence of gene exchange between human and animal enterococci. J Clin Microbiol 40: 4659–4665. doi: 10.1128/JCM.40.12.4659-4665.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smoglica C, Vergara A, Angelucci S, Festino AR, Antonucci A, Marsilio F, Di Francesco CE. 2022. Evidence of linezolid resistance and virulence factors in Enterococcus spp. isolates from wild and domestic ruminants, Italy. Antibiotics (Basel) 11: 223. doi: 10.3390/antibiotics11020223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talaga-Ćwiertnia K, Bulanda M. 2018. Drug resistance in the genus Enterococcus−current problem in humans and animals. Postepy Mikrobiol 57: 244–250. [Google Scholar]
- 53.Torres C, Alonso CA, Ruiz-Ripa L, León-Sampedro R, Del Campo R, Coque TM. 2018. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol Spectr 6: 10.1128/microbiolspec.ARBA-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urshev Z, Yungareva T. 2021. Initial safety evaluation of Enterococcus faecium LBB.E81. Biotechnol Biotec Eq 35: 11–17. doi: 10.1080/13102818.2020.1840438 [DOI] [Google Scholar]
- 55.Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. 2004. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol 42: 4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuksekdag Z, Ahlatcı NS, Hajikhani R, Darilmaz DO, Beyatli Y. 2021. Safety and metabolic characteristics of 17 Enterococcus faecium isolates. Arch Microbiol 203: 5683–5694. doi: 10.1007/s00203-021-02536-8 [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Xie W, Lin X, Baloch AR, Zhang X. 2012. Antimicrobial resistance in enterococci isolates from pet dogs in Xi’an, China. Pak Vet J 32: 462–464. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.