Abstract
Background
SARS-CoV-2 variant surveillance informs vaccine composition and decisions to de-authorize antibody therapies. Though detailed genetic characterization requires whole-genome sequencing, targeted mutation analysis may complement pandemic surveillance efforts.
Methods
This study investigated the qualitative performance of a multiplex oligonucleotide ligation assay targeting 19 spike mutations using 192 whole genome sequenced upper respiratory samples representing SARS-CoV-2 variants of concern.
Results
Initial valid results were obtained from 95.8% [95% confidence interval (CI): 92.0 – 98.2; 184/192] of samples. All eight invalid samples were valid on repeat testing. When comparing SARS-CoV-2 oligonucleotide ligase assay SARS-CoV-2 variant calls with whole genome sequencing, overall positive percent agreement was 100% (95% CI: 98.1 – 100.0; 192/192), as was the positive and negative percent agreement for each of the tested variants; Gamma, Delta, Omicron BA.1, BA.2, and BA.4/BA.5.
Conclusions
This multiplexed oligonucleotide ligation assays demonstrated accurate SARS-CoV-2 variant typing compared to whole genome sequencing. Such an approach has the potential to provide improved turnaround compared to sequencing and more detailed mutation coverage than RT-qPCR.
1. Background
SARS-CoV-2 variants of concern have evolved mutations in the spike gene, many of which encode spike proteins that render variant viruses less susceptible to antibody neutralization and reduce vaccine effectiveness [1,2]. Monitoring the emergence and prevalence of these variants is therefore an important component of the COVID-19 pandemic response. In particular, variant surveillance has informed vaccine composition as well as decisions to revoke the emergency use authorization of monoclonal antibody therapies [3,4].
SARS-CoV-2 variant surveillance is commonly performed using amplicon- or capture-based whole genome sequencing (WGS) on a next-generation sequencing platform [5,6]. Though WGS provides the most detailed information of available methods, turnaround time remains suboptimal, with surveillance data typically delayed by two to four weeks. As a complementary, more rapid approach, TaqPath spike gene target failure and mutation-specific RT-qPCR assays have also been utilized to identify known variants and help prioritize samples for WGS [7], [8], [9]. There is limited literature, however, on intermediate methods that have the potential to provide more comprehensive mutation coverage than RT-qPCR and more timely results than WGS.
In this study, we evaluate the performance of a multiplexed oligonucleotide ligation assay [Meso Scale Discovery (MSD)] using a set of whole genome sequenced respiratory specimens containing variants of concern.
2. Materials and methods
Ethics Statement. This study was conducted with Stanford Institutional Review Board approval (protocol 68234). Individual consent was waived.
Clinical Specimens and Whole Genome Sequencing. This study included 192 whole genome sequenced upper respiratory specimens representing SARS-CoV-2 variants of concern; Gamma (n = 8), Delta (n = 46), Omicron BA.1 (n = 12), BA.2 (n = 34), BA.4 (n = 44), and BA.5 (n = 48). Sequencing was performed as previously described using a laboratory-developed protocol consisting of long-range PCR, followed by fragmentation, then single-end 150-cycle sequencing using MiSeq reagent kit V3 (Illumina, San Diego, CA) [10]. PANGO lineage assignment was performed in December 2022 using https://pangolin.cog-uk.io/ running pangolin version 4.1.3. WGS data was deposited in GISAID (Supplemental Table 1).
MSD® Oligonucleotide Ligation Assay (OLA). Nucleic acid extraction and RT-PCR for the amplification of the complete spike gene are described in the supplemental methods. The OLA format is represented schematically in Fig. 1 . RT-PCR products were diluted 1:50 in nuclease free water prior to use. This study evaluated four multiplexed OLA reactions (Supplemental Table 2). Each OLA reaction contained: 10 µL diluted spike gene RT-PCR product, 0.05 µL Ligase, 2.5 µL DNA Ligase Buffer, 0.5 µL Upstream Probe Mix, and 0.5 µL Downstream Probe Mix in a total volume of 20 µL. Each Upstream OLA probe contains a barcode sequence that is used to specifically identify its OLA product during the detection step (as described below). The thermal cycling reaction conditions were as follows: hold at 95 °C for 2 min, then 30 cycles at 95 °C for 30 s and 58 °C for 2 min. Each OLA run included Lineage A and Variant controls; the control RT-PCR products were diluted 1:250 in nuclease free H2O prior to use. Each run also included two blank OLA reactions performed without DNA ligase. These blank reactions contained 10 µL of the diluted PCR products from RT-PCR wells A1 and A2, respectively, combined with 10 µL of master mix without DNA ligase.
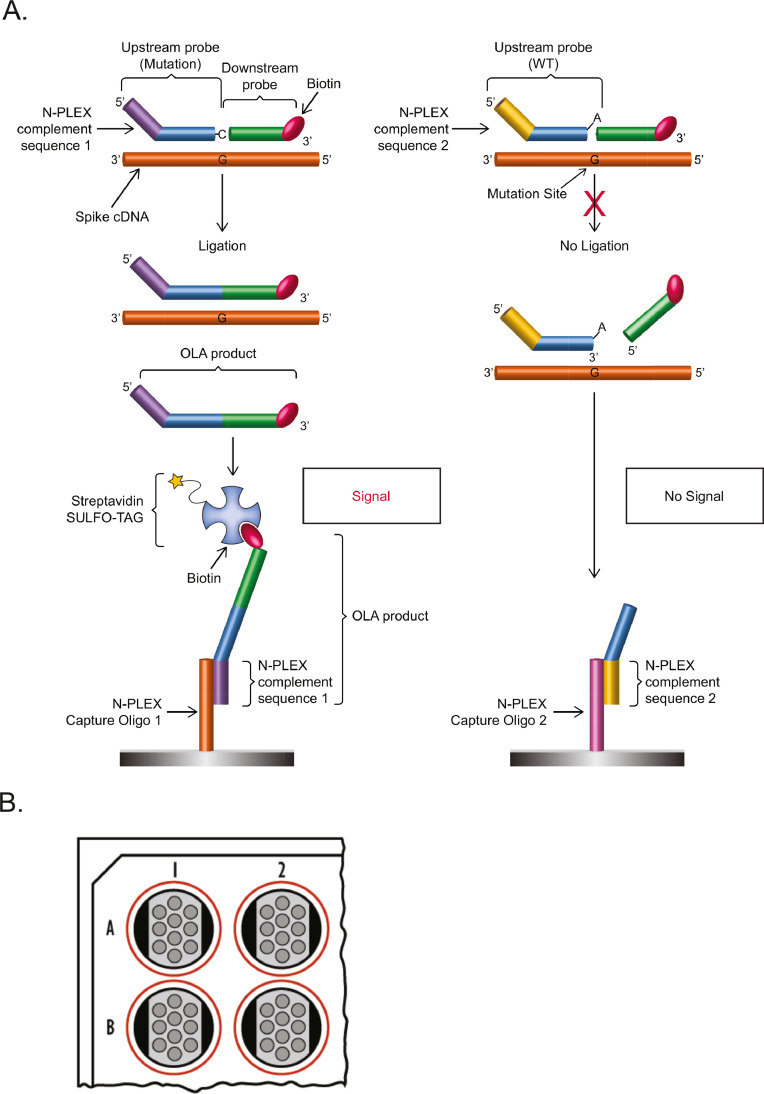
Fig. 1.
(A) SARS-CoV-2 mutation detection via Oligonucleotide Ligation Assay (OLA). For each mutation:wild-type (WT) pair, there is a common downstream oligonucleotide probe targeting spike gene complementary DNA (cDNA) sequence that is labeled with biotin. In addition, there are adjacent upstream probes, one each for the mutant and the WT sequences, that differ by the base at the 3′ end of the oligonucleotide. The upstream probes also comprise different barcode sequences on their 5′ ends that target the probes to specific locations in capture oligonucleotide arrays immobilized within wells of the detection plate. If there is a complete match to the spike gene, the ligation reaction joins the upstream and downstream probes to generate the OLA product. This ligation product is captured on the detection plate and streptavidin-labeled detection reagent (SULFO-TAG™) is added to allow the electrochemiluminescent reaction. If there is not a complete match, ligation does not occur, and capture of the unligated upstream probe does not generate signal. (B) Representation of the wells of an N-PLEX plate. Each well provides an array of 10 unique barcodes.
The product of each OLA reaction was treated with MSD blocking oligonucleotides diluted 1:40 in Hybridization Buffer 2 to prevent bridging of unligated products. Each Blocking Reaction contained 2 µL of the OLA product and 73 µL of blocking mix in a total volume of 75 µL. The Blocking Reaction was incubated at 95 °C for 5 min in a thermalcycler, and then cooled to 4 °C.
MSD OLA Product Detection using Hybridization and Electrochemiluminescence (ECL). The OLA reaction products were detected after hybridizing them, through their barcodes, to arrays of complementary oligonucleotides immobilized on electrodes in the wells of 96-well plates designed for ECL measurements (N-PLEX® plates, MSD). This approach allows N-PLEX plates and reagents to be used for universal detection of different multiplexed OLA reactions, with each well providing an array of 10 unique barcodes (Fig. 1).
Each well of the detection plate was first blocked using 50 µL N-PLEX Blocker solution (MSD). The plate was sealed with an adhesive plate seal and incubated at 37 °C with shaking at 700 rpm for 30 min (Biosan Thermoshaker, PST-60HL). The plate was then washed 3 times with 300 µL 1X Dulbecco's Phosphate Buffered Saline (DPBS), per well. Following these washes, 20 µL of Hybridization Buffer 1 was added to each well, followed by 30 µL of the OLA reaction. The plate was sealed with an adhesive plate seal and incubated at 37 °C with shaking at 700 rpm for 60 min. The plate was cooled to room temperature for 5 min and then washed 3 times with 300 µL 1X DPBS, per well. After these washes, 50 µL of a 1:500 dilution of SULFO-TAG streptavidin in Diluent 54 (MSD) was added to each well. The plate was sealed with an adhesive plate seal, incubated at room temperature with shaking at 700 rpm for 30 min, and then washed 3 times with 300 µL 1X DPBS, per well. After washing, 150 µL of MSD GOLD™ Read Buffer B was added to each well, and the plate was read using the MESO® QuickPlex SQ 120 MM (MSD).
ECL signals were normalized by subtracting the mean background signal from the two blank wells prepared for each reaction set. The normalized ECL signal for the wild-type (Lineage A SARS-CoV-2 virus) or mutation sequence was required to exceed 2500 ECL units to be interpreted as detected. Samples in which more than two wild-type/mutant positions on each plate did not meet these conditions were considered invalid and were repeated. The ratio of ECL signal was then analyzed to determine the presence or absence of the mutant sequence (Supplemental Methods). Variant Lineage was determined based on the mutation patterns described in Table 1 . Starting from the primary sample, the test requires ∼12 h to complete.
Table 1.
SARS-CoV-2 spike mutation patterns used for variant typing by multiplex oligonucleotude ligation assay.
| Variant | 69–70del | N501Y | K417N | K417T | T478K | D215G | L452R | E484K | E484Q | T19R | T95I |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpha | 69–70del | N501Y | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| Beta | WT | N501Y | K417N | NDET | WT | D215G | WT | E484K | NDET | WT | WT |
| Gamma | WT | N501Y | NDET | K417T | WT | WT | WT | E484K | NDET | WT | WT |
| Delta | WT | WT | WT | WT | T478K | WT | L452R | WT | WT | T19R | T95I or WT |
| B.1.1.529/BA.1 | 69–70del | N501Y | K417N | NDET | T478K | NDET | WT | NDET (A) | NDET (A) | WT | T95I |
| BA.2 | WT | N501Y | K417N | NDET | T478K | WT | WT | NDET (A) | NDET (A) | NDET (I) | WT |
| BA.4 | 69–70del | N501Y | K417N | NDET | T478K | WT | L452R | NDET (A) | NDET (A) | NDET (I) | WT |
| BA.5 | 69–70del | N501Y | K417N | NDET | T478K | WT | L452R | NDET (A) | NDET (A) | NDET (I) | WT |
WT, wild-type Wuhan reference sequence; NDET, not detected. BA.1.1.529/BA.1 contains a 3 amino acid insertion (EPE) at spike amino acid 214 such that both D215G and the WT sequences are not detected. In BA.1.1.529/BA.1, BA.2, BA.4, and BA.5, spike amino acid 484 is mutated to Alanine (A) so E484K, E484Q, and the WT sequences are not detected. In BA.2, BA.4, and BA.5, spike amino acid 19 is mutated to Isoleucine (I) so neither T19R nor WT sequences are detected.
3. Results
Upon initial testing with the MSD assay protocol, 95.8% [95% confidence interval (CI): 92.0 – 98.2; 184/192] of samples gave valid results. All eight invalid samples were valid on repeat testing. These repeat samples included 3 BA.2 and 5 Delta variants (Supplemental Table 1). When comparing SARS-CoV-2 OLA variant calls with WGS, overall positive percent agreement (PPA) was 100% (95% CI: 98.1 – 100.0; 192/192), as was the PPA and negative percent agreement for the tested variants (Table 2 ).
Table 2.
Positive and negative percent agreement between the SARS-CoV-2 multiplexed oligonucleotide ligation assay and whole genome sequencing.
| Detected WGS* | Not detected WGS | Positive Percent Agreement (95%CI) | Negative Percent Agreement (95%CI) | ||
|---|---|---|---|---|---|
| B.1.1.529/BA.1 | Detected OLA | 12 | 0 | 100.0% (73.5 – 100.0) | 100.0% (98.0 – 100.0) |
| Not Detected OLA | 0 | 180 | |||
| BA.2 | Detected OLA | 34 | 0 | 100.0% (89.7 – 100.0) | 100.0% (97.7 – 100.0) |
| Not Detected OLA | 0 | 158 | |||
| BA.4/BA.5 | Detected OLA | 92 | 0 | 100.0% (95.2 – 100.0) | 100.0% (96.4 – 100.0) |
| Not Detected OLA | 0 | 100 | |||
| Delta | Detected OLA | 46 | 0 | 100.0% (92.3 – 100.0) | 100.0% (97.5 – 100.0) |
| Not Detected OLA | 0 | 146 | |||
| Gamma | Detected OLA | 8 | 0 | 100.0% (63.1 – 100.0) | 100.0% (98.0 – 100.0) |
| Not Detected OLA | 0 | 184 |
Three BA.2 and five Delta variants required repeat testing for detection. OLA, Oligonucleotide Ligation Assay; WGS, whole genome sequencing.
4. Discussion
In this study we determined the performance of multiplexed OLA reactions employing ECL detection in a plate-based format. Though this was the first generation of this research use only test, limited invalid tests were observed, all of which were subsequently valid on repeat testing. Because all repeat tests were performed from the primary sample, further experiments will be required to identify and optimize the step or steps of the testing process (extraction, RT-PCR, OLA) that contributed to these invalid results. Overall, however, the OLA method provided accurate SARS-CoV-2 variant typing compared to WGS.
Strengths of this study include the number of WGS-typed samples tested and the range of variants assessed. As with all mutation-specific typing approaches, this method is limited by the mutations included in the panels. Though the current iteration of this method evaluated 19 mutations, of which 11 contributed to distinguishing between variants, the omicron sublineages BA.4 and BA.5 could not be differentiated from one another. However, the flexibility of the multiplexed OLA, coupled with universal N-PLEX detection plates and reagents, allows rapid development and implementation of new targets for the identification of emerging variants. For example, future versions of this method might include other mutations important for resistance to monoclonal antibodies, as well consolidation of existing targets into fewer reactions.
In summary, this multiplexed OLA method offers a complementary option for SARS-CoV-2 variant typing that has the potential to provide improved turnaround compared to WGS and more detailed mutation coverage than RT-qPCR. Future workflow studies with updated panels will be required to determine the clinical impact and epidemiological utility of such an approach.
Declaration of Competing Interest
FK, TJB, SBH, JNW, and GBS are MSD employees. Testing was performed at Stanford using reagents and instruments provided by MSD to BAP.
Acknowledgements
The authors thank Norm Cyr for graphic arts support.
References
- 1.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barouch D.H. Covid-19 vaccines - immunity, variants, boosters. N. Engl. J. Med. 2022;387:1011–1020. doi: 10.1056/NEJMra2206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Food and Drug Administration. FDA updates sotrovimab emergency use authorization. 2022.
- 4.Food and Drug Administration. FDA announces bebtelovimab is not currently authorized in any US region. 2022.
- 5.Lam C., Gray K., Gall M., Sadsad R., Arnott A., Johnson-Mackinnon J., et al. SARS-CoV-2 genome sequencing methods differ in their abilities to detect variants from low-viral-load samples. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.01046-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber Z., Daviaud C., Delafoy D., Sandron F., Alidjinou E.K., Mercier J., et al. A comparison of high-throughput SARS-CoV-2 sequencing methods from nasopharyngeal samples. Sci. Rep. 2022;12:12561. doi: 10.1038/s41598-022-16549-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMillen T., Jani K., Robilotti E.V., Kamboj M., Babady N.E. The spike gene target failure (SGTF) genomic signature is highly accurate for the identification of Alpha and Omicron SARS-CoV-2 variants. Sci. Rep. 2022;12:18968. doi: 10.1038/s41598-022-21564-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Jean S., Eltringham R., Madison J., Snyder P., Tu H., et al. Mutation-specific SARS-CoV-2 PCR screen: rapid and accurate detection of variants of concern and the identification of a newly emerging variant with spike L452R mutation. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00926-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung P.S., Wang H., Sibai M., Solis D., Yamamoto F., Iwai N., et al. Evaluation of a rapid and accessible reverse transcription-quantitative PCR approach for SARS-CoV-2 variant of concern identification. J. Clin. Microbiol. 2022;60 doi: 10.1128/jcm.00178-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Miller J.A., Verghese M., Sibai M., Solis D., Mfuh K.O., et al. Multiplex SARS-CoV-2 genotyping reverse transcriptase PCR for population-level variant screening and epidemiologic surveillance. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00859-21. [DOI] [PMC free article] [PubMed] [Google Scholar]